Abstract
Drought is a major constraint to rice (Oryza sativa) yield and its stability in rainfed and poorly irrigated environments. Identifying genomic regions influencing the response of yield and its components to water deficits will aid in our understanding of the genetics of drought tolerance and development of more drought tolerant cultivars. Quantitative trait loci (QTL) for grain yield and its components and other agronomic traits were identified using a subset of 154 doubled haploid lines derived from a cross between two rice cultivars, CT9993-510 to 1-M and IR62266-42 to 6-2. Drought stress treatments were managed by use of a line source sprinkler irrigation system, which provided a linearly decreasing level of irrigation coinciding with the sensitive reproductive growth stages. The research was conducted at the Ubon Rice Research Center, Ubon, Thailand. A total of 77 QTL were identified for grain yield and its components under varying levels of water stress. Out of the total of 77 QTL, the number of QTL per trait were: 7-grain yield (GY); 8-biological yield (BY); 6-harvest index (HI); 5-d to flowering after initiation of irrigation gradient (DFAIG); 10-total spikelet number (TSN); 7-percent spikelet sterility (PSS); 23-panicle number (PN); and 11-plant height (PH). The phenotypic variation explained by individual QTL ranged from 7.5% to 55.7%. Under well-watered conditions, we observed a high genetic association for BY, HI, DFAIG, PSS, TSN, PH, and GY. However, only BY and HI were found to be significantly associated with GY under drought treatments. QTL flanked by markers RG104 to RM231, EMP2_2 to RM127, and G2132 to RZ598 on chromosomes 3, 4, and 8 were associated with GY, HI, DFAIG, BY, PSS, and PN under drought treatments. The aggregate effects of these QTL on chromosomes 3, 4, and 8 resulted in higher grain yield. These QTL will be useful for rainfed rice improvement, and will also contribute to our understanding of the genetic control of GY under drought conditions at the sensitive reproductive stage. Close linkage or pleiotropy may be responsible for the coincidence of QTL detected in this experiment. Digenic interactions between QTL main effects for GY, BY, HI, and PSS were observed under irrigation treatments. Most (but not all) DH lines have the same response in measure of productivity when the intensity of water deficit was increased, but no QTL by irrigation treatment interaction was detected. The identification of genomic regions associated with GY and its components under drought stress will be useful for marker-based approaches to improve GY and its stability for farmers in drought-prone rice environments.
In rainfed lowland areas, one of the major abiotic constraints depressing rice (Oryza sativa) production is water stress, including deficit (drought) or excess water (flood; O'Toole and Chang, 1979; Herdt, 1991; Lin and Shen, 1993). Drought stress is a serious limiting factor to rice production and yield stability in rainfed rice areas (Dey and Upadhyaya, 1996). The occurrence of drought can be assessed by following variables such as weather conditions, soil moisture, and crop condition over a particular growing season. In rice, the effect of drought varies with the variety, degree, and duration of stress and its coincidence with different growth stages. Rice's susceptibility to water stress is more pronounced at the reproductive stage and causes the greatest reduction in grain yield when stress coincides with the irreversible reproductive processes (Matsushima, 1966; Cruz and O'Toole, 1984).
Drought tolerance has been considered as a valid breeding target to partially compensate for the loss in yield. Phenotypic traits associated directly with drought tolerance are unclear; however, several investigations noted that deep rooting (Ekanayake et al., 1985; Lilley and Fukai, 1994; Pantuwan et al., 1996; Wade et al., 1996) and osmotic adjustment (Ludlow and Muchow, 1990; Jongdee and Cooper, 1998; Zhang et al., 1999b) are associated with drought tolerance. Until recently, lack of concept, direction, and protocol has remained a significant obstacle to genetic improvement of drought tolerance (Blum et al., 1996). Selecting rice for drought tolerance based on unknown genetic mechanisms will result in inefficient genetic improvement. Standard assays used to measure drought tolerance must be enhanced since they are important to success in genetic improvement. The line source sprinkler system can be used to maintain variable water application rates across plots, thus allowing the assessment of different levels of water supply at specific growth stages (Hanks et al., 1976).
Molecular tools facilitate the identification and genomic locations of genes controlling traits related to drought tolerance using quantitative trait loci (QTL) analysis. This paper aims to improve understanding of genetic responses of agronomically important traits such as grain yield, biological yield, harvest index, number of panicles per hill, total spikelet number per panicle, days to flowering after initiation of the irrigation gradient, percent spikelet sterility, and plant height to drought stress of different intensities coinciding with the flowering stage of rice. Grain yield and its components are important traits for breeders since the ultimate goal in breeding programs is to obtain high and stable yield. After identifying the components contributing to grain yield, the relationship of yield and its components to morphophysiological traits will help to assess and understand the mechanisms of drought tolerance. Thus, morphophysiological traits act as secondary traits in relation to yield and its components. The line source sprinkler irrigation system was employed to conduct drought phenotyping experiments under field conditions to create a linear gradient of drought intensity. The number, genomic locations, and effects of QTL were determined. Epistasis and QTL × environment (Q × E) interactions were also investigated. This information will help us understand the genetic control of yield and its components under various intensities of drought stress. The results obtained may also be directly applicable in improving drought tolerance in rice. Correlating genetic information with physiological and morphological traits related to drought tolerance will allow the development of rice varieties tolerant to drought stress in a particular rice ecosystem.
RESULTS AND DISCUSSION
Manipulation of Flowering Date
GY is generally considered to be the most important trait for rice farmers in rainfed lowland areas where drought developed late in the wet season (Cooper, 1999). Late season drought contributed to yield loss between 13% and 35% in northeast Thailand (Jongdee et al., 1997). Stable and high yields of rainfed lowland rice under drought conditions can be obtained by having appropriate phenology to avoid late season drought and high potential yield under well-watered conditions. These are primary characteristics for food security in rainfed rice farming systems as reviewed by Fukai et al. (1999). Fukai et al. (1999) also emphasized the ability of rice plants to maintain high leaf water potential as another trait relevant to stabilize yield in rainfed rice planting areas. As mentioned by Fukai et al. (1999), any attempt to successfully study drought tolerance, especially late season drought, needs to minimize the effect of the flowering time because it is a major determinant of GY and its components. In this study, we tried to eliminate or at least minimize the effect of flowering time on GY and its components by synchronizing the flowering date of the DH population in the experiment as shown in Figure 1. Most of the DH lines flowered synchronously under W0. There are, however, some lines that experienced water stress before or after flowering stage. Genotypic variance was statistically significant for DFAIG under W0. This may be explained by the fact that open field experiments such as this one encounter environmental factors such as temperature and photoperiod that allow changes in the plant phenology (Fukai, 1999). These factors are also made more complex by water availability, soil fertility, and manner of planting rice in rainfed lowland. The variability of water availability within the toposequence in rainfed lowlands hinders the precise identification of flowering time of rice. Therefore, in order to determine the optimum flowering time of a genotype for a particular region of interest, knowledge of water availability patterns in different seasons will be of great help.
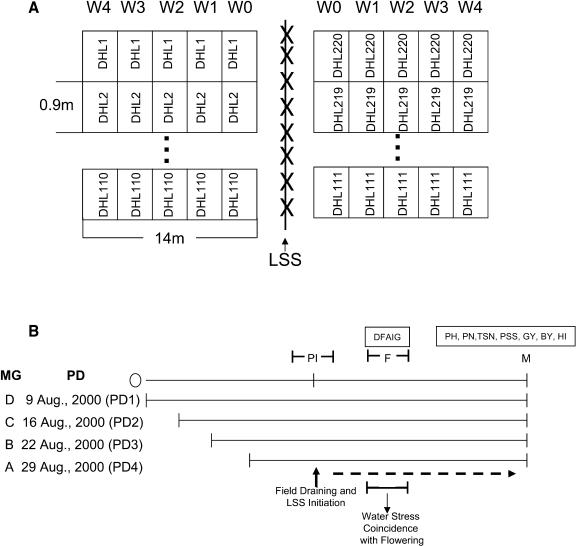
Figure 1.
A, Spatial schematic depicting the experimental design. Orientation of plots; line source sprinkler (LSS) irrigation gradients; resulting irrigation levels (W0–W4); DHL1, DHL2, DHL110, DHL111, DHL219, and DHL220 represent the arrangement of the DH population in the plots. B, Temporal schematic depicting the experimental design. Staggered planting date (PD) of the four maturity groups (MG) resulting in the expected synchrony of reproductive growth stage (flowering, F) coincident with water stress; recording of flowering (DFAIG) stage of each DH line; and measurement of above ground biomass (BY), grain yield (GY), and yield components (PH, PN, TSN, PSS, and HI) at maturity (M) for the 220 DH lines and two parental lines.
Effects of Irrigation Treatments
Standing water was drained out of the experimental field when most of the DH lines reached panicle initiation and then line source sprinkler (LSS) was applied until the plants reached maturity. This established a water gradient resulting in varying levels of drought stress. Although different levels of stress were created by LSS irrigation, wind was a factor making the water application not uniform. Because this problem was paramount to the experimental treatments, sprinkler irrigation was carried out in the early morning hours when wind speed was low. Before the end of water application, the amount of irrigation distributed to the field was known by determining the amount of water collected in catch cans versus the distance of the sprinkler line that showed a slightly bell-shaped relationship and the yield-to-water relationship averaged over all genotypes was remarkably linear (data not shown). ANOVA analysis indicated a statistically significant correlation of irrigation treatments for most of the measured traits using the average values of DH lines (Table III) indicating that the application of water gradients using the LSS method was successfully established.
Table III.
Trait mean values, phenotypic variation, mean squares from experiment-wise analysis of variance of grain yield and its components, and broad-sense heritability of doubled haploid population, CT9993 × IR62266, under line source sprinkler experiment at Ubon Rice Research Center, Thailand, during wet season of 2000
| Trait | Irrigation Treatment | CT9993 | IR62266 | DH Population
|
I Effectf | G Effectg | I × G Effect | H2 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Range | Skewness | ||||||||
| GY | W0 | 1.63a | 3.43b | 1.99a | 0.41–5.02 | 5.52 | ||||
| W1 | 1.56a | 2.76b | 1.80b | 0.18–4.99 | 3.77 | |||||
| W2 | 1.32a | 1.79a | 1.39c | 0.01–3.47 | 3.44 | ** | ** | ns | 0.39 | |
| W3 | 1.23a | 0.73a | 1.09d | 0.03–3.32 | 3.74 | |||||
| W4 | 0.49a | 0.20a | 0.64e | 0.00–2.83 | 8.74 | |||||
| BY | W0 | 4.39a | 8.16b | 5.40a | 1.65–11.64 | 7.43 | ||||
| W1 | 4.28a | 6.95b | 5.07b | 1.97–11.3 | 6.20 | |||||
| W2 | 4.31a | 5.92a | 4.48c | 1.51–9.08 | 3.96 | ** | ** | ns | 0.33 | |
| W3 | 3.81a | 5.40a | 4.31d | 1.18–8.44 | 4.54 | |||||
| W4 | 2.84a | 4.74a | 3.92e | 0.68–8.94 | 4.49 | |||||
| HI | W0 | 0.37a | 0.42a | 0.37a | 0.11–2.19 | 60.97 | ||||
| W1 | 0.36a | 0.40a | 0.35b | 0.04–0.73 | −2.10 | |||||
| W2 | 0.30a | 0.30a | 0.30c | 0.00–0.69 | −1.95 | ** | ** | ns | 0.46 | |
| W3 | 0.33a | 0.12b | 0.24d | 0.01–0.47 | −1.28 | |||||
| W4 | 0.15a | 0.03b | 0.15e | 0.00–0.75 | 5.93 | |||||
| DFAIG | W0 | 28.00a | 30.50a | 24.05a | 0.00–46.00 | 0.24 | ||||
| W1 | 23.00a | 30.00a | 23.14a | 0.00–52.00 | 1.24 | |||||
| W2 | 20.50a | 31.50a | 24.15a | 0.00–63.00 | 9.30 | * | ** | ns | 0.49 | |
| W3 | 25.00a | 43.50a | 27.86b | 4.00–63.00 | 8.06 | |||||
| W4 | 47.00a | 55.50a | 38.58c | 0.00–63.00 | 1.60 | |||||
| TSN | W0 | 127.00a | 94.00a | 105.44a | 45.00–221.00 | 5.46 | ||||
| W1 | 147.50a | 94.00b | 106.01a | 44.00–215.00 | 4.99 | |||||
| W2 | 149.00a | 91.00b | 104.56b | 41.00–189.00 | 3.41 | ns | ** | ns | 0.53 | |
| W3 | 131.00a | 99.50a | 108.46b | 9.00–207.00 | 2.28 | |||||
| W4 | 122.00a | 85.50a | 106.27b | 9.00–215.00 | 1.44 | |||||
| PSS | W0 | 33.45a | 28.30a | 36.59a | 12.00–99.30 | 7.56 | ||||
| W1 | 34.90a | 33.15a | 37.70a | 10.80–95.50 | 5.77 | |||||
| W2 | 35.25a | 41.90a | 42.65b | 5.10–86.50 | 2.86 | ** | ** | ns | 0.45 | |
| W3 | 49.65a | 58.10a | 51.00c | 13.30–99.50 | 3.23 | |||||
| W4 | 53.95a | 69.45a | 58.57d | 13.10–100.00 | 0.34 | |||||
| PN | W0 | 5.35a | 10.65b | 8.57a | 3.00–19.30 | 6.02 | ||||
| W1 | 5.70a | 10.15b | 8.21ab | 4.00–15.30 | 4.45 | |||||
| W2 | 5.95a | 9.55b | 7.93b | 4.00–17.00 | 5.53 | ns | ** | ns | 0.53 | |
| W3 | 5.00a | 9.65b | 7.81c | 3.30–21.50 | 12.39 | |||||
| W4 | 6.00a | 7.85a | 7.70d | 3.30–16.80 | 7.48 | |||||
| PH | W0 | 78.60a | 72.00a | 68.91a | 38.50–113.30 | 2.28 | ||||
| W1 | 80.65a | 67.65a | 67.20b | 39.50–114.30 | 3.29 | |||||
| W2 | 83.15a | 61.10b | 63.72c | 33.80–106.30 | 3.58 | ** | ** | ns | 0.71 | |
| W3 | 84.10a | 55.80b | 62.17d | 34.80–116.30 | 4.96 | |||||
| W4 | 76.65a | 50.80b | 58.00e | 27.30–116.80 | 6.45 | |||||
Trait, (see Table 2). Irrigation Treatments W0–W4, (see Table 1). Skewness, determines the normality of the distribution of traits. Normal distribution of traits must fall within +2 or −2. firrigation effect. ggenotype effect. H2, plot base broad-sense heritability of traits. a to e, indicate significant difference of the trait mean values among irrigation treatments. *, level of significance (** significant at 99%; * significant at 95%). ns, not significant.
Genotype by environment (G × E) interaction was a major contributor to the phenotypic variation for most agronomically important traits in rainfed lowland rice (IRRI, 1993; Cooper and Somrith, 1997; Wade et al., 1999). Different combinations of soil type, temperature, amount of water availability, and other agrohydrology with plant phenology were reported to influence the G × E interaction (Cooper and Somrith, 1997; Fukai et al., 1999). LSS and synchronization successfully minimized the variation contributed by the soil type and plant phenology, although other environmental factors could not be controlled. In this experiment, G × E interaction accounted for a small portion of the total sum of squares compared to the genotype and irrigation treatment, and showed that all traits measured were nonstatistically significant at P < 0.05 − P < 0.01 (Table III). This further verifies that the level and intensity of drought stress were uniformly developed from floral initiation until harvesting and water supply is the major discriminating factor among treatments.
Phenotypic Variation and Broad-Sense Heritability
Table III displays the grand means, ranges, and broad-sense heritabilities of measured traits for CT9993, IR62266, and the DH population. The parents showed statistically significant differences at P < 0.05 and P < 0.01 for the traits GY, BY, HI, TSN, PN, and PH but not for DFAIG and PSS (Table III). The phenotypic distributions in the DH lines for the traits mentioned did not show discrete classes but approximately fitted a normal distribution, indicating that all measured traits were quantitatively inherited in nature. Transgressive segregation in both directions was observed for most traits (Table III) under every drought intensity (W0 to W4), indicating that both parents transmitted favorable alleles for each trait.
Broad-sense heritabilities (H2) computed across five water regimes were relatively moderate (Table III). PN and PH had the highest H2 at 0.61 and 0.73, respectively, while GY and BY had the lowest H2 at 0.32 and 0.31, respectively. This indicates that traits such as GY and BY are more prone and easily affected by drought stress than traits such as PN and PH. It can be noted that the trend of trait heritability increased until W2, and declined at W3 and W4.
Phenotypic Correlation and Factors Contributing to Grain Yield
Correlation between measured traits and GY at different water levels was evaluated at P < 0.05 and P < 0.01 for each irrigation treatment as shown in Table IV. Highly significant positive correlation between GY and BY ranging from 0.73 at W1 to 0.54 at W4 was found, indicating that genetic improvement in the GY would likely be accompanied by improvement of BY. Positive correlation between GY and HI was also highly significant (Table IV). As drought stress increased, the correlation between GY and HI increased dramatically, indicating that HI is also a primary determinant of GY under stress. Therefore, genetic improvement of HI would also improve GY (Fukai et al., 1999; Babu et al., 2003). Negative correlations between GY and DFAIG and between GY and PSS were also highly significant under all water levels. The strength of correlation between GY and DFAIG increased to 0.76 at W4. This result suggests that DH lines flowering late after the onset of drought suffered more yield loss (Fukai et al., 1999). The correlations between GY and PSS were not much different under well watered and drought conditions and it should be noted that a high PSS was found even at W0. Also, PSS can be caused by the genetic incompatibility between the two parents, and may confound the effect of drought in this population. The significant difference of the grand mean for PSS under well-watered and severe drought suggested that drought stress indeed affected PSS in the DH lines (Jongdee et al., 2002; Table III). PN showed a significant correlation with GY only under well-watered and mild drought stresses (at W0 to W2). At severe drought stresses (W3 and W4) the correlation was not significant. In contrast, TSN was positively correlated with GY only under stress at W2 (r = 0.25) and W4 (r = 0.27). The results suggest that genotypes with high spikelet number per panicle produced better grain yield under stress. Correlation between GY and PH was slightly significant (r = 0.21) only under severe stress.
Table IV.
Correlation coefficients (r) of plant production traitsa and phenology traits with grain yield at various water levelsb
| Trait (Irrigation Treatment) | GY (W0) | GY (W1) | GY (W2) | GY (W3) | GY (W4) |
|---|---|---|---|---|---|
| GY (W0) | 1.00 | ||||
| GY (W1) | 0.76** | 1.00 | |||
| GY (W2) | 0.57** | 0.68** | 1.00 | ||
| GY (W3) | 0.44** | 0.53** | 0.71** | 1.00 | |
| GY (W4) | 0.45** | 0.44** | 0.49** | 0.61** | 1.00 |
| BY (W0) | 0.76** | 0.56** | 0.36** | 0.20** | 0.22** |
| BY (W1) | 0.59** | 0.77** | 0.46** | 0.29** | 0.21** |
| BY (W2) | 0.46** | 0.60** | 0.74** | 0.43** | 0.27** |
| BY (W3) | 0.39** | 0.51** | 0.54** | 0.67** | 0.35** |
| BY (W4) | 0.41** | 0.48** | 0.36** | 0.33** | 0.66** |
| HI (W0) | 0.51** | 0.40** | 0.33** | 0.35** | 0.32** |
| HI (W1) | 0.52** | 0.71** | 0.55** | 0.51** | 0.46** |
| HI (W2) | 0.45** | 0.50** | 0.80** | 0.64** | 0.50** |
| HI (W3) | 0.35** | 0.40** | 0.59** | 0.88** | 0.59** |
| HI (W4) | 0.40** | 0.72** | 0.48** | 0.64** | 0.91** |
| DFAIG (W0) | −0.51** | −0.39** | 0.44** | 0.46** | −0.53** |
| DFAIG (W1) | −0.43** | −0.46** | −0.48** | −0.48** | −0.49** |
| DFAIG (W2) | −0.36** | −0.38** | −0.57** | −0.55** | −0.46** |
| DFAIG (W3) | −0.30** | −0.33** | −0.53** | −0.65** | −0.52** |
| DFAIG (W4) | −0.33** | −0.36** | −0.48** | −0.58** | −0.72** |
| TSN (W0) | 0.22** | 0.12* | 0.15** | 0.13* | 0.12* |
| TSN (W1) | 0.25** | 0.26** | 0.18** | 0.18** | 0.11ns |
| TSN (W2) | 0.17** | 0.19** | 0.27** | 0.15* | 0.09ns |
| TSN (W3) | 0.08ns | 0.12* | 0.14** | 0.21** | 0.14* |
| TSN (W4) | 0.25** | 0.22** | 0.23** | 0.26** | 0.29** |
| PSS (W0) | −0.36** | −0.36** | −0.37** | −0.29** | −0.24** |
| PSS (W1) | −0.33** | −0.33** | −0.37** | −0.30** | −0.24** |
| PSS (W2) | −0.29** | −0.28** | −0.43** | −0.37** | −0.26** |
| PSS (W3) | −0.20** | −0.23** | −0.40** | −0.50** | −0.43** |
| PSS (W4) | −0.15** | −0.20** | −0.31** | −0.37** | −0.40** |
| PN (W0) | 0.24** | 0.19** | 0.10ns | 0.05ns | 0.09ns |
| PN (W1) | 0.21** | 0.24** | 0.12* | 0.11* | 0.11* |
| PN (W2) | 0.16** | 0.19** | 0.15** | 0.05ns | 0.14* |
| PN (W3) | 0.10ns | 0.12* | 0.12* | 0.09ns | 0.14* |
| PN (W4) | 0.16** | 0.14* | 0.07ns | 0.07ns | 0.15** |
| PH (W0) | 0.21** | 0.11* | 0.05ns | 0.04ns | −0.01ns |
| PH (W1) | 0.16** | 0.21** | 0.15** | 0.12* | 0.05ns |
| PH (W2) | 0.13* | 0.20** | 0.29** | 0.23** | 0.11* |
| PH (W3) | 0.13* | 0.13* | 0.25** | 0.38** | 0.19** |
| PH (W4) | 0.15** | 0.13* | 0.16* | 0.24** | 0.34** |
In this study, BY, HI, DFAIG, TSN, PSS, and PH were the traits determining GY under well-watered condition (Table V). Path coefficient analysis permits the separation of the correlation coefficient into components of direct and indirect effect on GY. GY under well-watered condition was important in determining GY under water-limiting conditions. Better performance of cultivars with high potential yield under rainfed lowland regions was proposed (Pantuwan et al., 1996; Fukai et al., 1999) and demonstrated by Rajatasereekul et al. (1997). Although yield potential has no direct genetic relationship to drought tolerance, under drought stress it contributed to higher grain yield. On the other hand, GY under very severe stress was mainly related to BY and HI. In a study conducted for pearl millet under drought stress condition, HI and BY were also found to increase yield under drought stress (Yadav et al., 2002).
Table V.
Factors contributing to grain yield at various water treatment levels based on path coefficient analysis
| Traits | GY (W0) | GY (W1) | GY (W2) | GY (W3) | GY (W4) |
|---|---|---|---|---|---|
| BY | 0.3723*a | 0.3364** | 0.3084** | 0.2540** | 0.1737** |
| 0.0200b | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| HI | 2.9000** | 4.7999** | 4.2181** | 4.1542** | 3.9644** |
| <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | |
| DFAIG | −0.0100** | ||||
| <0.0001 | ns | ns | ns | ns | |
| TSN | 0.0014* | ||||
| 0.0138 | ns | ns | ns | ns | |
| PSS | −0.0072** | ||||
| <0.0001 | ns | ns | ns | ns | |
| PN | ns | ns | ns | ns | ns |
| PH | −0.0043** | ||||
| 0.0003 | ns | ns | ns | ns | |
| R2 Value | 90.99 | 97.10 | 96.05 | 96.71 | 94.70 |
QTL Detection
The number, genomic locations, and effects of QTL associated with GY and its components were summarized in Table VI.
Table VI.
QTL detected for grain yield and its components by simple and composite interval mapping via MQTL1 in a doubled haploid population of 154 lines from the cross IR62266 and CT9993 under different irrigation treatments
| Trait (Irrigation treatment) | Chrom | QTL Name | Marker Interval | Peak Marker | LOD | Effect | R2 | PVE |
|---|---|---|---|---|---|---|---|---|
| GY (W0) | 4 | qgy4.1 | ME4_9–RZ565 | RZ565 | 3.04 | 0.42 (I) | 9.8 | 9.8** |
| GY (W1) | (4)a | qgy4.2 | EMP2_2–RZ565 | ME10_11 | 3.10 | 0.36 (I) | 9.4 | |
| 6 | qgy6.1 | RZ682–EM14_9 | RZ682 | 3.26 | 0.41 (I) | 11.3 | 17.3** | |
| GY (W2) | ||||||||
| GY (W3) | (3) | qgy3.1 | EM11_9–RM231 | RM231 | 2.60 | 0.40 (C) | 11.5 | |
| (4) | qgy4.3 | ME10_11–RZ565 | ME4_9 | 2.57 | 0.36 (I) | 7.2 | 21.7** | |
| GY (W4) | (3) | qgy3.2 | EM11_9–RM231 | RM231 | 2.84 | 0.35 (C) | 11.6 | |
| (10) | qgy10.1 | EM14_10–EM11_5 | R1629 | 3.32 | 0.29 (I) | 9.5 | 18.8** | |
| BY (W0) | (4) | qby4.1 | RM273–RM317 | RM317 | 2.79 | 0.69 (I) | 11.6 | 11.6** |
| BY (W1) | (11) | qby11.1 | EM17_10–ME10_16 | ME10_16 | 2.62 | 0.81 (I) | 7.9 | 7.9** |
| BY (W2) | 11 | qby11.2 | ME4_14–ME10_16 | ME4_14 | 3.81 | 0.78 (I) | 13.5 | 13.5** |
| BY (W3) | 9 | qby9.1 | R41–ME9_3 | ME9_3 | 3.37 | 0.62 (I) | 11.3 | |
| 11 | qby11.3 | ME4_14–ME6_2 | ME4_14 | 5.46 | 0.79 (I) | 18.8 | 31.0** | |
| BY (W4) | (8) | qby8.1 | ME2_11–G187 | ME2_11 | 2.95 | 0.61 (I) | 9.4 | |
| 9 | qby9.2 | RM201–RM215 | RG667 | 3.56 | 0.65 (I) | 11.6 | ||
| 10 | qby10.1 | G333–EM18_17 | EM18_17 | 3.37 | 0.56 (I) | 11.1 | 20.6** | |
| HI (W0) | 3 | qhi3.1 | RG104–RM81 | RM231 | 2.86 | 0.07 (C) | 17.2 | 17.2** |
| HI (W1) | 3 | qhi3.2 | RG104–RM81 | RM231 | 5.43 | 0.07 (C) | 24.4 | 24.4** |
| HI (W2) | 3 | qhi3.3 | RG104–RM81 | RM231 | 5.02 | 0.09 (C) | 21.2 | 21.2** |
| HI (W3) | 3 | qhi3.4 | RG104–RM81 | RM231 | 6.27 | 0.11 (C) | 24.8 | 24.8** |
| HI (W4) | (1) | qhi1.1 | CDO345–RZ909 | RZ909 | 2.75 | 0.07 (I) | 7.8 | |
| 3 | qhi3.5 | RG104–RM81 | RM231 | 5.24 | 0.10 (C) | 18.6 | 23.2** | |
| DFAIG (W0) | 3 | qdfaig3.1 | RG104–CDO20 | EM11_9 | 3.87 | 5.86 (C) | 18.1 | 18.1** |
| DFAIG (W1) | 3 | qdfaig3.2 | RG104–CDO20 | RM231 | 3.94 | 5.55 (C) | 18.1 | 18.1** |
| DFAIG (W2) | 3 | qdfaig3.3 | RG104–RM81 | EM11_9 | 3.21 | 6.60 (C) | 10.2 | 10.2** |
| DFAIG (W3) | 3 | qdfaig3.4 | RG104–RM231 | RM231 | 4.73 | 10.45 (C) | 16.5 | 16.5** |
| DFAIG (W4) | 3 | qdfaig3.5 | RG104–RM81 | RM231 | 3.59 | 13.45 (C) | 15.8 | 15.8** |
| TSN (W0) | (4) | qtsn4.1 | RG908–ME6_10 | ME6_10 | 2.95 | 20.47 (C) | 9.9 | |
| (4) | qtsn4.2 | RG939–RG476 | RG476 | 2.86 | 17.86 (C) | 10.6 | ||
| (5) | qtsn5.1 | RG313–ME5_13 | RG313 | 2.90 | 17.00 (C) | 9.1 | ||
| 9 | qtsn9.1 | K936–C313 | K936 | 3.90 | 19.13 (C) | 5.7 | ||
| TSN (W1) | 4 | qtsn4.3 | EMP3_10–RG214 | RG939 | 4.20 | 20.78 (C) | 13.4 | 22.5* |
| 9 | qtsn9.2 | K936–C313 | ME2_17 | 4.73 | 21.04 (C) | 17.4 | 23.2* | |
| TSN (W2) | (4) | qtsn4.4 | RG939–RG476 | RG939 | 2.93 | 18.21 (C) | 10.3 | |
| 9 | qtsn9.3 | K936–C313 | EMP3_2c | 4.60 | 21.47 (C) | 14.4 | 19.6* | |
| TSN (W3) | 3 | qtsn3.1 | EM19_11–C746 | C746 | 3.39 | 17.31 (C) | 9.2 | |
| 4 | qtsn4.5 | EMP3_10–RG214 | RM252 | 4.53 | 16.52 (C) | 12.1 | 17.7* | |
| TSN (W4) | 9 | qtsn9.4 | K936–C313 | K936 | 3.94 | 20.26 (C) | 10.4 | 9.8** |
| PSS (W0) | (4) | qpss4.1 | RG214–RM127 | RG620 | 2.71 | 6.37 (C) | 8.2 | |
| 5 | qpss5.1 | EM15_4–CDO202 | EM15_4 | 3.15 | 8.16 (C) | 10.0 | ||
| 9b | qpss9.1 | RM219–EM14_6 | RM219 | b | 4.89 (C) | 8.3 | 17.0* | |
| PSS (W1) | 5 | qpss5.2 | EM15_4–CDO202 | C1230 | 4.00 | 9.25 (C) | 12.0 | 12.0** |
| PSS (W2) | 5 | qpss5.3 | EM15_4–CDO202 | RG119 | 4.62 | 11.56 (C) | 15.0 | |
| 8 | qpss8.1 | RM256–RM210 | RM210 | 3.39 | 8.08 (C) | 11.2 | 21.6** | |
| PSS (W3) | ||||||||
| PSS (W4) | (8) | qpss8.2 | ME5_4–RM256 | G2132 | 6.12 | 7.53 (C) | 7.8 | 7.8** |
| PN (W0) | 1 | qpn1.1 | RG109–CDO345 | CDO345 | 5.94 | 2.24 (I) | 21.7 | |
| 4 | qpn4.1 | ME6_10–EMP2_2 | EMP2_2 | 5.37 | 2.23 I) | 17.9 | ||
| 4 | qpn4.2 | ME4_9–RG214 | RG476 | 6.38 | 2.10 (I) | 21.3 | ||
| 8 | qpn8.1 | ME2_11–EM18_5 | EM14_1 | 9.06 | 2.77 (I) | 23.1 | 52.6** | |
| PN (W1) | 1 | qpn1.2 | ME4_18–CDO345 | CDO345 | 5.46 | 2.20 (I) | 20.7 | |
| (2) | qpn2.1 | ME2_4–R3128 | ME2_4 | 2.84 | 1.41 (C) | 11.2 | ||
| 4 | qpn4.3 | ME6_10–EMP2_2 | EMP2_2 | 4.40 | 2.00 (I) | 15.0 | ||
| 4 | qpn4.4 | ME4_9–RG214 | RG476 | 4.60 | 1.71 (I) | 16.3 | ||
| 8 | qpn8.2 | ME2_11–EM18_5 | EM14_1 | 6.36 | 2.25 (I) | 18.3 | 47.2* | |
| PN (W2) | 1 | qpn1.3 | RG109–CDO345 | CDO345 | 4.82 | 2.01 (I) | 18.6 | |
| 4 | qpn4.5 | RG939–RG214 | RG476 | 5.48 | 1.74 (I) | 18.9 | ||
| 4 | qpn4.6 | ME6_10–EMP2_2 | EMP2_2 | 4.62 | 1.92 (I) | 15.8 | ||
| 8 | qpn8.3 | RM210–RG598 | EM14_1 | 6.09 | 2.07 (I) | 17.2 | ||
| 10 | qpn10.1 | EM11_5–G333 | G333 | 3.17 | 1.22 (I) | 12.0 | 55.3* | |
| PN (W3) | 1 | qpn1.4 | ME4_18–CDO345 | CDO345 | 5.30 | 2.06 (I) | 20.5 | |
| 4 | qpn4.7 | ME6_10–RZ565 | EMP2_2 | 6.03 | 2.31 (I) | 20.2 | ||
| 4 | qpn4.8 | RG939–RG214 | RG476 | 3.76 | 1.55 (I) | 12.7 | ||
| 7 | qpn7.1 | EAAM17_5–ME4_3 | ME10_6 | 3.54 | 1.58 (I) | 11.3 | ||
| 8 | qpn8.4 | G187–RG598 | EM14_1 | 5.52 | 2.09 (I) | 15.3 | 50.5* | |
| PN (W4) | 1 | qpn1.5 | ME4_18–CDO345 | CDO345 | 5.21 | 2.09 (I) | 20.8 | |
| 4 | qpn4.9 | EM15_3–EMP2_2 | EMP2_2 | 6.40 | 2.29 (I) | 21.3 | ||
| 4 | qpn4.10 | ME4_9–RM317 | RG476 | 4.14 | 1.70 (I) | 16.7 | ||
| 7 | qpn7.2 | EAAM17_5–ME4_5 | ME10_6 | 3.37 | 1.51 (I) | 11.5 | 50.3** | |
| PH (W0) | 1 | qph1.1 | RM102–RZ909 | RG109 | 13.33 | 16.15 (C) | 46.1 | |
| 8 | qph8.1 | ME6_13–EM18_5 | EM14_1 | 7.48 | 15.08 (C) | 20.6 | ||
| 10 | qph10.1 | RM222–RG257 | RG257 | 3.30 | 8.50 (C) | 16.5 | 55.7* | |
| PH (W1) | 1 | qph1.2 | RM102–RZ909 | RG109 | 12.08 | 15.27 (C) | 41.1 | |
| 8 | qph8.2 | ME6_13–EM18_5 | EM14_1 | 7.66 | 15.80 (C) | 21.2 | 48.8** | |
| PH (W2) | 1 | qph1.3 | RM102–CDO345 | RG109 | 11.18 | 13.95 (C) | 36.2 | |
| 8 | qph8.3 | ME2_11–RG598 | EM14_1 | 8.12 | 15.73 (C) | 20.2 | 42.6** | |
| PH (W3) | 1 | qph1.4 | C813–CDO345 | RM104 | 9.79 | 13.08 (C) | 32.6 | |
| 8 | qph8.4 | ME2_11–RG598 | EM14_1 | 8.10 | 15.14 (C) | 19.1 | 39.5** | |
| PH (W4) | 1 | qph1.5 | RM102–CDO345 | RG109 | 6.51 | 10.49 (C) | 23.1 | |
| 8 | qph8.5 | ME2_11–RG598 | RZ997 | 7.61 | 13.49 (C) | 18.6 | 31.3** |
Individual QTL are designated with the italicized abbreviation of the trait and the chromosome number. When more than one QTL affecting a trait is identified on the same chromosome, they are distinguished by decimal numbers
MQTL1, refer to “Materials and Methods”.
Trait, refer to Table 2.
Irrigation treatment W0–W4, refer to Table 1.
Chrom, refers to the chromosome number
LOD, maximum-likelihood LOD score for the QTL. A statistically computed estimate of the likelihood of a QTL being present at the particular location on the genome.
R2, the fitted model explains the variability of the trait at a particular water treatment level.
PVE, the total R2 explained by the variability of the trait for all detected QTL at a particular water level.
C and I, allele contributing positive effect (C, CT9993; I, IR62266).
*, level of significance of total PVE (*** significant at 99%; ** significant at 95%).
chromosome numbers in parentheses are identified QTL with LOD score near the threshold.
significant only in composite interval mapping.
Grain Yield
IR62266 produced higher GY than CT9993 under fully irrigated conditions (W0). The GY of IR62266 decreased dramatically as drought stress became more severe while the GY of CT9993 declined slower than IR62266. The extended effects of yield potential (Blum, 1988) were also observed under mild stress at W2 in which the GY of IR62266 was reduced by approximately 50% but still remained higher than that of CT9993. The GY of CT9993 and DH lines was reduced by approximately 50% under W3. As mentioned by Pantuwan et al. (2002), a drought intensity that causes a 50% yield reduction is considered a critical point for the expression of drought tolerance mechanisms in rice. Therefore, we considered W3 and W4 the most appropriate intensities of drought stress to monitor drought tolerance mechanisms in this experiment. Under W3 and W4, CT9993 produced higher GY than IR62266 and DH lines segregated tremendously for GY, which ranged from 0 to 2.83 tons/ha (Table III). Transgressive segregation for GY in both directions indicated that both parents contributed favorable alleles under severe drought stress.
Seven QTL located on chromosomes 3, 4, 6, and 10 were identified for GY. The IR62266 alleles of QTL on chromosomes 4, 6, and 10 increased GY in all water levels (W0, W1, W3, and W4), while the CT9993 alleles of QTL on chromosome 3 increased GY only under severe drought stress (W3 and W4). The QTL on chromosome 4 designated as qgy4.1, qgy4.2, and qgy4.3 flanked by markers ME10_11 and RZ565 were identified under well-watered condition (W0), mild stress (W1), and severe stress (W3), respectively. The QTL on chromosome 6 was identified only under W1. Considering the coincidence of QTL on chromosome 4 and allelic contribution by IR62266 at both QTL locations, these QTL might contribute to the increased yield potential derived from IR62266. The QTL on chromosome 3 (qgy3.1 and qgy3.2) flanked by markers EM11_9 to RM231 were identified only in the severe drought stress in which, as mentioned earlier, W3 and W4 were the appropriate stress intensities to monitor drought tolerance. CT9993 contributed the allele that increases grain yield for the two QTL (qgy3.1 and qgy3.2) on chromosome 3. These QTL might contribute to the maintenance of GY through the control of complex biochemical and physiological processes associated with drought tolerance. The QTL on chromosome 10 designated as qgy10.1 was detected only under W4. The detected QTL, qgy3.2 and qgy10.1, supported the evidence of transgressive segregation for GY under severe drought stress. These data suggest that unique configurations of multiple alleles may be required for high levels of drought tolerance while maintaining GY. No QTL was detected for W2, although the segregation for GY ranged from 0.01 to 3.47 t/ha in the DH lines. Previous studies by Xing et al. (2002) and Yu et al. (1997) found that epistatic interactions play an important role in determining GY and its components in rice, and most of them do not have main effects at the single-locus level. This finding probably explains why no QTL was detected for W2. Total phenotypic variation explained (PVE) by detected QTL for GY ranged from 9.8% to 21.7%. These QTL collectively accounted for 32% to 68% of estimated genotypic variation. The residual variation that cannot be accounted for by the QTL may be due to epistatic interactions.
Fourteen QTL for GY were reported by Zhang et al. (1999a) using the same mapping population. Out of 14 QTL, eight were from the phenotypic evaluations conducted in Thailand and the other six were from Israel. Only the qgy6.1 detected in this experiment coincided with the QTL reported by Zhang. The rest of the QTL for yield under control conditions were not common between QTL analyses done by Zhang for the phenotypic data from Thailand and Israel in years 1996 and 1997, respectively, and in this study. This striking difference indicates the complex nature of yield and G × E interaction for GY. Interestingly, most of the QTL for GY under well-watered conditions reported by Zhang et al. (1999a) were coincidentally found or linked with QTL for flowering date in the same experiment. The report by Zhang et al. (1999a) clearly indicated that flowering time was a major determinant of GY in this mapping population. In this study, the QTL contributed by IR62266 alleles for GY were identified in chromosomes 4 (qgy4.3) and 10 (qgy10.1) under W3 and W4, respectively. Babu et al. (2003) identified the same QTL in India. As was mentioned, the qgy3.1 and qgy3.2 contributed by CT9993 alleles were identified on chromosome 3 only under severe drought stress. This indicates that the presence of these QTL made it possible to maintain GY even under severe drought.
Biological Yield
IR62266 produced higher BY than CT9993 under all irrigation treatments, but the difference was only significant under W0 and W3. The BY of IR62266 was reduced by approximately 50% under severe drought stress. That of CT9993 was reduced by only 25%, but suggesting IR62266 still out-yielded CT9993. Evidence of transgressive segregation was also observed. This may indicate that both parents contribute alleles to increase BY.
Eight QTL were detected for BY. These QTL were localized in different chromosomal locations for each irrigation treatment except for the QTL on chromosome 11 designated as qby11.1, qby11.2, and qby11.3 that were found in the same region flanked by markers ME4_14 to EM17_16 under W1, W2, and W3, respectively. A single QTL was identified under W0, W1, and W2. Two QTL, qby9.1 and qby11.3, located on chromosomes 9 and 11 were detected under W3. Three QTL located on chromosomes 8, 9, and 10, designated as qby8.1, qby9.2, and qby10.1, were identified under W4. IR62266 alleles contributed to higher BY for all identified QTL. Phenotypic variation explained by QTL ranged from 7.9% to 31%.
The QTL for canopy temperature were also located in the same regions of chromosomes 9 and 10 (data not shown). The radiation load on the leaf canopy affects the leaf temperature and transpiration. This in turn has a relation to the water uptake and nutrient assimilation necessary for grain production. Stem reserves are an important source of carbohydrates and nitrogen for grain filling, especially at times when transient photosynthesis is inhibited by drought and other factors like heat or leaf disease that occurred during grain filling. The efficiency of stem reserves in overcoming the effect of drought during grain filling is also dependent on the amount of reserves in the stem before flowering. It can be seen in the result of this study that GY under W4 is highly correlated with BY under the same water condition.
Harvest Index
HI is the ratio of GY to BY. HI indicates the efficiency of translocation of food assimilates from the vegetative tissue to the reproductive tissue. Thus for breeders, it serves as a means to predict grain growth and yield in many crops. The HI of IR62266 and CT9993 was not significantly different under well-watered to mild stresses. Their values were in a range of 0.30 to 0.42 (Table III). The HI of IR62266, however, decreased dramatically to 0.12 and 0.03, as drought became more severe (W3 and W4) while the HI of CT9993 decreased slowly to 0.33 and 0.15 for the same water stress levels, respectively. Phenotypic correlations between HI and GY were high under all conditions (Table V). The strength of correlation between HI and GY were directly proportional to the severity of the stress, increasing from 0.68 (P < 0.01) in very mild stress (W1) to 0.91 (P < 0.001) in severe stress (W4). The increasing value of the correlation coefficient of HI and GY as drought intensity increase further confirms the importance of HI in determining GY. Better maintenance of HI may have contributed to high GY under drought stress. In pearl millet, QTL that contribute to increased drought tolerance through an ability to maintain HI and BY was also reported (Yadav et al., 2002). Six QTL for HI were detected on chromosomes 1 and 3 in all irrigation treatments. The QTL on chromosome 3 designated as qhi3.1, qhi3.2, qhi3.3, qhi3.4, and qhi3.5 were mapped to the same position at the RG104-RM231 interval. CT9993 alleles of these QTL contributed higher HI. The PVE of these QTL ranged from 17.2% to 24.8% (Table VI). The IR62266 allele contributed higher HI on the QTL detected on chromosome 1 designated as qhi1.1 that was expressed only under severe drought stress (W4).
Interestingly, the QTL specific to better yield performance under drought stress, qgy3.1 and qgy3.2, were clearly located in the same marker interval. This result showed that close linkage or pleiotropy might be responsible for the high correlation of GY with HI and the coincidence of the QTL for both traits. As it was mentioned, the favorable QTL for GY were located in this region was contributed by CT9993. The same region was reported to harbor QTL for days to 50% flowering under control and stress conditions by Zhang et al. (2001) and Babu et al. (2003).
Days to 50% Flowering after Initiation of the Irrigation Gradient
Ontogenetic characters, especially appropriate flowering time, play an important role in drought avoidance of rainfed lowland rice (Fukai et al., 1999). Rajatasereekul et al. (1997) demonstrated the effect of flowering time in determining grain yield and Jongdee et al. (2002) pointed out that phenology is the most important factor. Timing, intensity, and occurrence of water deficit have been associated with the delay of heading or flowering (Fukai et al., 1999). If the stress occurred in the vegetative stage and if the stress is not severe, there might not be much effect on the heading or flowering. However, if the occurrence is at the end of the vegetative stage, there may be a delay in panicle initiation that may affect GY. In this experiment, the average value of DFAIG of DH lines under different irrigation treatments was not significantly different (Table III). CT9993 and IR62266 flowered at approximately the same time in all irrigation treatments. The wide range of flowering time in the DH lines indicated that the synchronizing approach used was partially successful. However, QTL analysis was carried out using the DFAIG values. Five QTL for DFAIG were identified on chromosome 3. These QTL designated as qdfaig3.1, qdfaig3.2, qdfaig3.3, qdfaig3.4, and qdfaig3.5 were mapped in the vicinity of markers RG104 to RM231, where the QTL for GY (qgy3.1 and qgy3.2) and HI (qhi3.1, qhi3.2, qhi3.3, qhi3.4, and qhi3.5) were also mapped. CT9993 alleles of all QTL decreased DFAIG but increased GY and HI. Negative correlations between DFAIG and GY were observed under all conditions (Table IV). The correlation was strongest (r2 = −0.7632) when the drought stress was most severe. This suggests that lines with late flowering exposed to more intense and longer water deficit had grain yield reduction as a consequence.
The QTL for DFAIG detected in this experiment were located in the same position as the QTL for days to heading mapped by Zhang et al. (2001), Babu et al. (2003), and Price et al. (1997). Under water deficit, the ability to maintain GY is mainly due to ability to maintain flowering time and ability to maintain high HI. The coincidence of QTL for GY, DFAIG, and HI under severe drought stress supported this result. Close linkage or pleiotropy may be the genetic basis for the coincidence of these QTL.
Total Spikelet Number
Total spikelet number per panicle was not significantly different between the two parents and was not affected by drought stresses (Table III). It indicates that the onset of drought stress was probably beyond the floral initiation stage. Ten QTL for TSN were identified on chromosomes 3, 4, 5, and 9. The QTL on chromosomes 4 and 9 were consistently identified in different irrigation treatments. The QTL on chromosome 4 were located between EMP3_10 and RG214 markers. Same genomic locations were identified by Hittalmani et al. (2003) for the same trait. The QTL on chromosome 9 were located between K936 and C313 markers. Of all QTL, CT9993 contributed the favorable allele for higher number of spikelet per panicle. Collectively, the total phenotypic variance explained by these QTL ranged from 10.4% to 23.3%. TSN was not a factor contributing to GY under well-watered conditions but it was a factor under drought stress (Table V). The QTL for TSN on chromosome 4 were linked with the QTL for PN, indicating that genotypes with a high PN may have a low TSN.
Percent Spikelet Sterility
Sheoran and Saini (1996) reported that changes in carbohydrate levels and enzyme activities, associated with inhibition of starch accumulation in pollen, are potential causes of spikelet sterility. PSS was also found affected by a slower rate of panicle exertion due to water stress (O'Toole and Namuco, 1983). Initially at W0, IR62266 had lower PSS than CT9993 and as water stress intensified (W3 and W4), PSS was drastically affected especially for IR62266. GY in all irrigation treatments showed negative correlations with PSS indicating that drought stress occurring during the reproductive stage increased the PSS and consequently decreased the GY. This evidence was reported by Boonjung and Fukai (1996), in which the yield reduction of 40% was due to the increment of PSS when drought occurred during grain filling period (Jongdee et al., 2002). Under well watered conditions, the level of PSS of IR62266 and CT9993 was not significantly different. The PSS of IR62266 increased dramatically as drought became more severe, while the PSS of CT9993 increased more slowly (Table III), although the difference was not significant at P < 0.05. The differences of PSS were large in the DH lines under all irrigation treatments and increased as stress became more severe. Three QTL, designated qpss4.1, qpss5.1, and qpss9.1 were identified on chromosomes 4, 5, and 9 under the well-watered treatment. When the water deficit was mild as in W1 and W2, QTL designated as qpss5.2 and qpss5.3 were also identified on chromosome 5. These QTL were located in the marker interval, EM15_4 to CDO20, the same as qpss5.1. As the level of water stress increased to W2 and W4, QTL such as qpss8.1 and qpss8.2 were identified. The qpss8.1 and qpss8.2 both mapped in the interval of ME5_4 to RM256. It can be hypothesized that qpss4.1 and qpss9.1 are not expressed under drought conditions as these QTL were replaced by QTL on chromosome 8 but this needs further verification. CT9993 alleles contributed to higher PSS in all detected QTL.
Zhang et al. (2001) reported QTL for spikelet sterility on chromosome 9 in the same mapping population. However, this QTL was at different position as compared with qpss9.1. Interestingly, the qpss9.1 was in the same position as the QTL for osmotic adjustment (OA), designated as oa9.1, reported by Zhang et al. (2001). The qpss8.1 and qpss8.2, only expressed in response to mild or severe drought stress, was also mapped to the same position as the QTL for OA designated oa8.1 by Zhang et al. (2001) and QTL for OA located at RG1 to RM80 by Robin et al. (2003). IR62266 alleles at both QTL increase OA and also spikelet fertility under drought stress. Lilley and Ludlow (1996) also reported the QTL for OA in the same position as the qpss8.1 and qpss8.2 using a different mapping population. As pointed out by Sheoran and Saini (1996) and Saini and Lalonde (1998), pollen development is very sensitive to drought stress because of an inhibition of accumulation of starch in the pollen. It is possible that drought stress might not only inhibit an accumulation of starch in the pollen but also inhibit the accumulation of solutes in cells. The consequence is high PSS and low OA. Physiobiological mechanisms for maintaining spikelet fertility via OA under drought stress need to be well investigated and care must be taken before any conclusion are drawn.
Panicle Number
IR62266 produced a higher number of panicles than CT9993 under all irrigation treatments except W4, in which both produced the same number (Table III). Average values of PN of the DH lines were not significantly different across the water regimes. This indicates the developmental stage at which water deficit was encountered was beyond panicle initiation. PN was determined before the onset of drought stress. Phenotypic correlation between PN and GY was low (r2 = 0.1707 − 0.3357). The correlation was lower when drought stress was more severe. It is suggested that PN was not a major factor in the loss in GY by drought stress.
A total of 23 QTL for PN were identified on chromosomes 1, 2, 4, 7, 8, and 10 (Table IV), collectively explaining 47.2% to 55.3% of phenotypic variance. Out of the total, the number of QTL per chromosome were: 5-chromosome 1; 1-chromosome 2; 10-chromosome 4; 2-chromosome 7; 4-chromosome 8; and 1-chromosome 10. IR62266 alleles of all QTL contributed high PN except for the QTL on chromosome 2. The QTL on chromosomes 1, 4, and 8 were identified in all irrigation treatments except that the QTL on chromosome 8 was not detected under W4. The QTL on chromosome 1 designated qpn1.1, qpn1.2, qpn1.3, qpn1.4, and qpn1.5 were mapped to the ME4_18-CDO345 interval. The QTL on chromosome 8 designated as qpn8.1, qpn8.2, qpn8.3, and qpn8.4 were mapped to the G187 -EM18_5 interval. The QTL on chromosome 4 were found significantly linked to ME6_10-EMP2_2 and ME4_9-RG214 intervals. Two QTL on chromosome 7 designated as qpn7.1 and qpn7.2 were detected only as water stress became severe (W3 and W4). These QTL were located between EAAM17_5 and ME4_3 markers.
Plant Height
PH was also affected by drought (Table III) but was not correlated with GY in all irrigation treatments (Table IV). The PH of IR62266 and CT9993 was not significantly different (P < 0.05) under well watered (W0) and very mild stress (W1). The difference, however, was observed under mild and severe drought stresses. Drought had little effect on the PH of the CT9993 but did affect IR62266. The reduction in PH was 21.2 cm for IR62266 and 2 cm for CT9993 under W4. Eleven QTL for PH were identified on chromosomes 1, 8, and 10 under different irrigation treatments (Table VI). The QTL on chromosome 1 designated as qph1.1, qph1.2, qph1.3, qph1.4, and qph1.5 were located in the C813-RZ909 interval, where the semi-dwarfing locus, sd-1, was reported (Price et al., 2000). The QTL on chromosome 8 designated qph8.1, qph8.2, qph8.3, qph8.4, and qph8.5 were mapped to the ME6_13 and EM18_5 interval. Under severe stress, the approximate location of QTL for PH was the same as the QTL for BY and PSS. The additional QTL on chromosome 10 designated qph10.1 was linked to the RG257 marker. Greater PH was contributed by the CT9993 allele for all detected PH QTL. Phenotypic variance explained by these QTL was between 31.3% and 55.7%.
QTL × QTL Interactions
The genetic improvement of adaptation to rainfed lowland environments has been focused on improving higher and more stable yields (Mackill et al., 1999). However, the genetic mechanism of grain yield in rice is not well understood. As pointed out by Xing et al. (2002), main effects, epistatic interactions, and environmental interactions of QTL are all-important genetic components of quantitative traits such as grain yield and traits contributing to grain yield. Understanding the genetic components underlying the expression of quantitative traits such as GY and traits contributing to grain yield will contribute to a more efficient breeding program. In this experiment, most of the traits were found controlled by main-effect QTL. We realized that a larger mapping population is required for estimating higher order QTL × QTL interactions. However, we proceeded to test for interactions between significant main-effect QTL that were earlier reported to be involved in epistatic interactions, in the form of additive by additive interactions (Xing et al., 2002).
In most cases, the phenotypic variance explained is largely from the genetic variance (δ2g) or main-effect QTL in this experiment. The δ2g , which cannot be explained by detected QTL, may be attributed to QTL with smaller effects, and/or due to QTL × QTL interactions referred to as epistatic interaction. Epistasis is the phenotypic effect of interaction among alleles at multiple loci (Xing et al., 2002) that play an important role in the genetic basis of quantitative traits (Lark et al., 1995; Li et al., 1997; Yu et al., 1997). We understand that the distances between marker loci and the QTL biased the estimated effects of epistasis. In the experiment, the calculations were, however, based on the closest marker with the main-effect QTL. Epistatic interactions between main-effect QTL were identified for many traits such as GY, HI, BY, and PSS under different irrigation treatments.
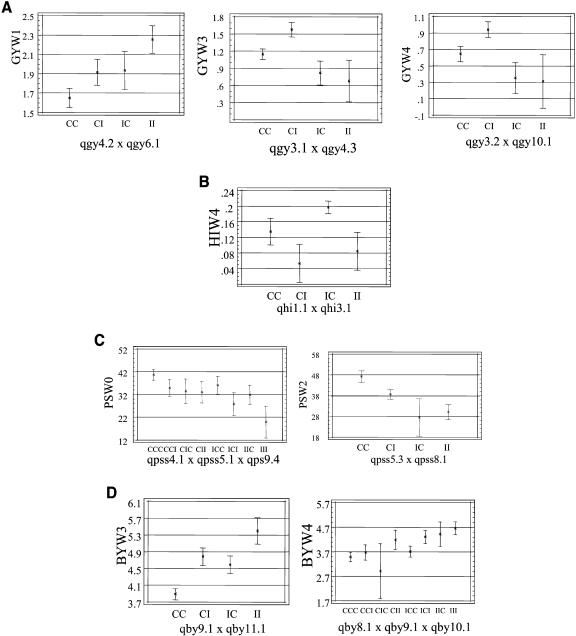
For example, the qgy4.2 and qgy6.1 identified at W1 showed an additive by additive interaction (Fig. 2A) where higher GY was contributed by IR62266 alleles at both loci. DH lines possessing the IR62266 allele at these two loci produced an average GY of 2.25 t/ha, while those possessing the CT9993 allele produced an average yield of 1.65 t/ha under W1. On the other hand, DH lines having one CT9993 and one IR62266 allele each from either of the two loci produced an average GY of 1.91 to 1.93 t/ha. As water stress became more severe at W3 and W4, the additive by additive interaction was also identified between significant main-effect QTL. At W3, the presence of CT9993 allele on the qgy3.1 and IR62266 allele on the qgy4.3 in the population was found to be significant in maintaining yield. The DH lines having this allelic combination produced an average yield of 1.58 t/ha under the W3 (Fig. 2A). The DH lines possessing CT9993 alleles at both loci produced higher yield (1.15 t/ha) than those having IR62266 allele in both loci, which produced an average GY of 0.68 t/ha under W3. The additive by additive interaction between significant main-effect QTL (qgy3.2 × qgy10.1 interaction) was repeatedly identified at W4. The average GY of DH lines with the CT9993 allele at the qgy3.2 and IR62266 allele at the qgy10.1 was 0.95 t/ha. Those with the CT9993 allele at both loci had average GY of 0.64 t/ha and those with the IR62266 allele at both loci had an average GY of 0.31 t/ha.
Figure 2.
Additive by additive interactions between/among main QTL. A, Additive interactions of GY QTL identified in W1, W3, and W4. B, Additive interactions of HI QTL at W4. C, PSS QTL showing additive interactions at W0 and W2. D, QTL for BY identified at W3 and W4 showing additive interaction.
Additive by additive interactions of main effect QTL were also identified for HI at W4. Two QTL (qhi1.1 and qhi3.5) were detected in which qhi3.5 was contributed by CT9993 and qhi1.1 was contributed by IR62266. The presence of the IR62266 allele at the qhi1.1 and CT9993 allele at the qhi3.5 resulted in comparatively high HI of 0.2 at W4. DH lines possessing an opposite allelic profile of these loci have a low HI value of 0.05 at the same stress condition (Fig. 2B). Main-effect QTL for PSS and BY also showed additive by additive interactions. Low PSS (Fig. 2C) was contributed by the IR62266 alleles at all detected QTL loci at W0. IR62266 alleles contributed higher BY that also showed additive by additive interactions (Fig. 2D).
Epistasis, in the form of additive by additive interactions, played a role in controlling the expression of GY and its components in this experiment. Yu et al. (1997), Li et al. (1997), and Xing et al. (2002) reported large numbers of interactions for grain yield and yield components in different genetic backgrounds, and additive by additive interactions were predominant. Digenic interactions between loci, neither of which show main effects at the single locus level, were also reported by Li et al. (1997) and Xing et al. (2002), explaining a portion of genetic variance of grain yield and its components in rice. Two-locus interaction would suggest that the effect of one QTL locus is dependent on another QTL locus. In this experiment, GY under stress was controlled by at least two QTL, which showed epistatic interaction. Therefore, in transferring QTL for GY under stress, both QTL must be considered.
QTL × Environment Interactions
The complexity of rainfed lowland environments and the incidence of large genotype-by-environment (G × E) interactions were documented and led to the implementation of multi-environmental trials to determine the adaptation of rice in rainfed conditions (Cooper, 1999). Fukai et al. (1999) also mentioned that in rainfed lowland rice, the G × E variance component is large relative to the genotypic component. Genotype by environment interaction is an important factor determining the stability of crop production in unfavorable environments. In this experiment, we aimed to gain knowledge of the genetic control of grain yield and its components under various intensities of drought stress in the target environment. G × E interactions occur due to differences in the extent of genetic variance among environments, which is also known as variation for broad adaptation (Cooper, 1999). For most of the traits, the mean trait values were decreased as the intensity of water deficit increased. However, evidence of crossover G × E interaction for all measured traits was not found in this experiment (Table III). The result suggests that DH lines did react to stress evenly even where the mean of measured traits across irrigation treatments did differ. Genotypes and environments themselves contributed most of the variation observed for all traits. This result was contrary to the findings of other groups in which the genotype by environment interaction is a predominant contributor to the genetic component under multi-location trails. This result is, however, an indication of the success of implementing a managed environment by means of the LSS method and synchronization, which reduced the effects of the other environmental factors and flowering time. Experimental errors could also be a factor contributing to the individual phenotypic differences. Broad sense heritability of grain yield and yield components in each water regime was relatively low. This implies that not only gene(s) but also environmental and experimental errors contributed to the individual phenotypic differences.
Different irrigation treatments had a significant effect on GY and its components, except for the traits TSN and PN. There was no QTL by environment interaction detected when all irrigation treatments were combined for QTL analysis (Table III). Comparing the number and locations of QTL detected in different environments will provide genetic information on the possible QTL that may perform differently under different environmental conditions. The comparison could not provide direct estimates for QTL by environment interactions. Direct estimation of QTL by environment interaction cannot be done with QTL information alone but it is possible to infer from the QTL data that crossover interactions might occur at the level of the genotype of a particular line. Different combinations of individual genes may give rise to the behavior of multi-gene genotypes, thus may allow the observation of crossover G × E interaction (Cooper, 1999).
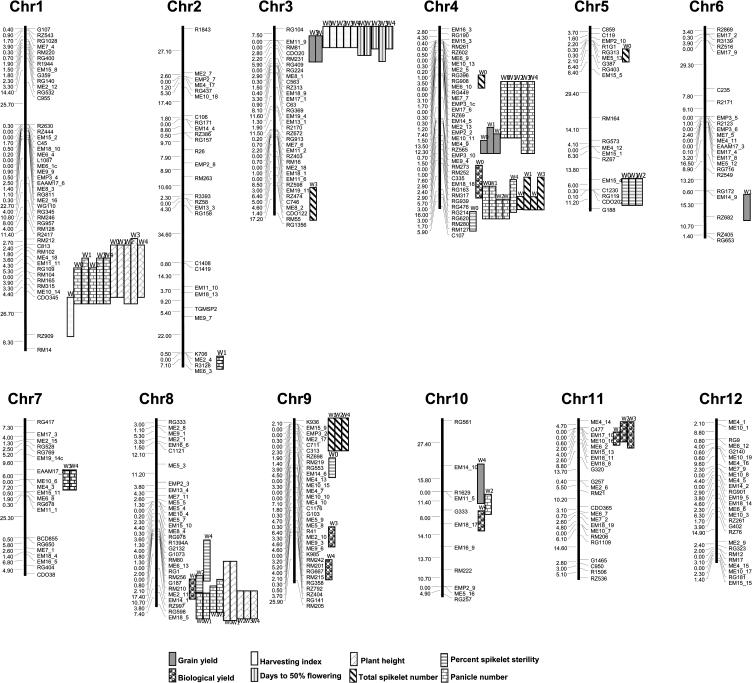
Coincidence of QTL for Grain Yield and Its Components
QTL for GY overlapped or were linked with several QTL for yield components. For example, the region on chromosome 3 flanked by the markers RG104 and RM231 contained the qdfaig3.1, qdfaig3.2, qdfaig3.3, qdfaig3.4, qdfaig3.5, qgy3.1, qgy3.2, qhi3.1, qhi3.2, qhi3.3, qhi3.4, and qhi3.5 (Fig. 3). This result was supported by high correlation between traits. Under severe stress, the correlation coefficient between GY and HI was 0.91, and −0.76 between GY and DFIAG (Table IV). This result implies that molecular mechanisms of drought tolerance to overcome the drastic yield reduction under severe drought stress involved traits such as maintaining high harvest index and less delay in flowering time. The physiological processes involved in this regard are still unknown. The large sink size and efficient transport of assimilates from leaves and stems into developing spikelets were reported to determine grain yield in rice (Cui et al., 2003). This finding by Cui may be tested and evaluated under drought stress to assess the genetic relationship of the traits under stress.
Figure 3.
Chromosomal locations of grain yield and yield components QTL identified in the CT9993 × IR62266 DH population by water or irrigation treatment levels.
The RZ69-RZ565 interval on chromosome 4 is another example of coincidence of QTL locations. This region contained the qgy4.1, qgy4.2, qgy4.3, qpn4.1, qpn4.2, qpn4.3, qpn4.4, qpn4.6, qpn4.7, qpn4.9, and qpn4.10 for GY and PN. The QTL for GY and PN were mapped near the QTL for PSS and TSN (Fig. 3). CT9993 contributed the favorable alleles for all QTL in this region except PSS.
The region on chromosome 1 flanked by the markers C813 and RZ909 markers contained qhi1.1, qpn1.1, qpn1.2, qpn1.3, qpn1.4, and qpn1.5. The QTL for PH was also located in this region. In this experiment, the onset of stress developed after panicle initiation. The colocation of qhi1.1, qpn1.5, and qph1.5 therefore suggested that high PN (IR62266 contributed favorable allele) contributed to a high HI, and consequently high yield under stress. This finding was supported by the significant positive correlation (r2 = 0.1789) between GY and PN at W4. QTL identified for trait-related drought tolerance such as total root dry weight (Zhang et al., 2001), relative water content, leaf rolling, and leaf drying (Babu et al., 2003) were reportedly located in this region. CT9993 contributed all favorable alleles in this region. Interestingly, the favorable alleles were contributed by IR62266 for yield components and CT9993 for trait-related drought tolerance. Therefore it is possible that selection for drought tolerance using this region as a target may incur a penalty on grain yield via decreasing the yield components.
Coincidence of QTL for BY, PSS, PN, and PH was observed on chromosome 8 in the G187-RG997 interval (Fig. 3). Most favorable alleles at QTL were contributed by IR62266, except for PH. QTL for osmotic adjustment (Zhang et al., 2001) and cell-membrane stability (Tripathy et al., 2000) were also contributed by IR62266. CT9993 contributed favorable alleles at QTL for basal root thickness (Zhang et al., 2001) and PH. This region could be a good target for yield improvement under drought stress.
CONCLUSIONS
The drought experiment conducted at Ubon Ratchatani Thailand in the wet season of 2000 allows the identification of QTL for grain yield and its components under various drought intensities. The genetic variation of measured traits is mainly contributed by main effect QTL. However, digenic interactions between main effect QTLs were observed for GY, BY, HI, and PSS. Phenotypic variance explained by QTL ranged from 7.8% to 55%. In this experiment, manipulation of flowering time by staggering planting date and managed drought intensities by using LSS minimized the variance contributed by the G × E interaction. Thus, G × E interaction for all measured traits was not observed in this experiment. Coincidence of QTL for GY and its components were identified in many regions especially on chromosomes 3, 4, and 8 suggesting a tight linkage or pleiotropy. These QTL coincided with QTL for root system and osmotic adjustment detected in the same mapping population. The aggregate effects of these QTL resulted in better management of GY under drought stress. These QTL could therefore be of interest for rice breeders to use as targets to improve GY under stress through the selection of QTL by molecular markers. This information will be useful for rice improvement by marker-assisted selection.
MATERIALS AND METHODS
Plant Materials
A doubled haploid (DH) population was derived from a cross between CT9993-510-1-M (Oryza sativa; abbreviated as CT9993, an upland japonica type) and IR62266-42-6-2 (Oryza sativa; abbreviated as IR62266, an indica type). These breeding lines show variations in potential yield, osmotic adjustment (OA), and root characters such as deep and thick rooting system. This population was developed at Centro Internacional de Agricultura Tropical, Columbia, and the International Rice Research Institute, Philippines. Several collaborating research institutes have used this population for the genetic study of traits associated with drought tolerance (Blum et al., 1999; Zhang et al., 2001). In the current study, the 220 DH lines and parents were used for phenotypic evaluations of the following traits: grain yield (GY); harvest index (HI); biological yield (BY); days to flowering after initiation of irrigation gradient (DFAIG); percent spikelet sterility (PSS); total spikelet number (TSN); panicle number (PN); and plant height (PH) in this study. For the map construction and QTL analyses, 154 DH lines were randomly selected and (Zhang et al., 2001) were used in this study.
Experimental Design and Growth Conditions
The field experiment was conducted under a rainfed lowland environment at Ubon Ratchathani Rice Research Center (latitude 15° 19′ 52.35″ N, longitude 104° 40′ 55.15″ E, altitude 110 m), located in northeast Thailand, during the 2000 wet season. The 220 DH lines, and the parents CT9993 and IR62266, were first evaluated under irrigated conditions in the 1996 wet season. The DH lines showed a wide range of flowering dates. In order to synchronize the flowering date in succeeding experiments, the flowering dates of 220 DH lines were used to group the DH lines into four maturity groups. Four seeding dates were then staggered at 6 to 7 d-intervals i.e. the latest flowering lines were seeded early and the earliest flowering lines were seeded last (Fig. 1).
Seeds were sown by hand on a slightly acidic, infertile, sandy loam soil with low organic matter and total nitrogen, at a rate of 4 to 6 seeds/hill in rows 0.15 m apart and hills within each row spaced 0.20 m apart. Plots were replicated twice and each replication was arranged in randomized complete block design. Plot size was 0.90 × 14.0 m (Fig. 1; 6 rows of 70 hills) and arranged perpendicularly to the line source sprinkler system (LSS; Hanks et al., 1976; Cruz and O'Toole, 1984). A mixed commercial chemical fertilizer was applied at the rate of 19-38-38 kg N-P2O5-K2O ha−1 at 32 d after seeding (DAS) of the first planted maturity group and 19 kg N ha−1 was applied at 60 DAS.
Surface irrigation was applied during the vegetative stage. When the majority of the lines reached the panicle initiation stage, standing water was drained from the field and LSS irrigation was applied thereafter and continued until maturity. LSS irrigation produced gradients of five moisture levels, from high to low. In order to minimize the effect of wind, irrigation by the LSS was applied everyday between 4 am and 8 am. Water treatments were assigned, i.e. W0 was a full irrigation condition (control) and W1 to W4 were the four levels of water deficit (from mild to severe) as described in Table I. Each water treatment was arranged perpendicularly to the LSS. A schematic diagram illustrating LSS with the water gradient creating different water stress levels and the total amount of water applied using catch cans are shown in Figure 1. Areas of W0, W1, W2, W3, and W4 received 9.4 mm, 5.4 mm, 2.9 mm, 0.9 mm, and 0.0 mm of average water applied per day, respectively, and the average pan evaporation was 4.14 ± 0.20 mm per day during the period of water stress treatment.
Table I.
Irrigation treatments and codes
| Treatment Code | Treatment Description |
|---|---|
| W0 | control or well-watered condition; received 9.4 mma water/d |
| W1 | categorized as very mild stress condition; received 5.4 mma water/d |
| W2 | categorized as mild stress condition; received 2.9 mma water/d |
| W3 | categorized as severe stress condition; received 0.9 mma water/d |
| W4 | categorized as very severe stress condition; received 0.0 mma water/d |
Average amount of water applied per day determined using catch cans.
Phenotyping
Determination of Yield, Yield Components, and Agronomic Traits
All data obtained were from irrigation treatments specified above as W0, W1, W2, W3, and W4. Abbreviations for the irrigation treatments and traits can be found in Tables I and II.
Table II.
Trait codes, full names, and units
| Trait Code | Trait Full Name and Unit |
|---|---|
| GY | Grain yield (t/ha) |
| BY | Biological yield (t/ha) |
| HI | Harvest index (ratio grain weight to biological yield) |
| DFAIG | Days to flowering after initiation of irrigation gradient (d) |
| TSN | Total spikelet number (per 0.96m2) |
| PSS | Percent spikelet sterility (%) |
| PN | Panicle number (hill−1) |
| PH | Plant height (cm) |
DFAIG was determined starting from water drainage until 50% of the total panicles in each plot were fully exerted. When each line reached maturity, PN per hill and PH were determined from five randomly sampled plants per plot. At harvest, filled and unfilled spikelets were counted to determine TSN and PSS. PSS was calculated from the filled and unfilled grain numbers per panicle. The above-ground plant parts (panicles, stems, and leaves) of the center four rows in each plot (0.96 m2) were harvested. Samples were sun dried for 3 weeks and then weighed to determine BY. Grains were threshed from the samples, dried in a hot air oven at 70°C for 5 d, and then weighed to determine GY. HI was calculated as a ratio of grain yield to the total above ground biological yield.
Statistical Analysis
ANOVA appropriate for the specified experimental and treatment design was performed on each measured trait listed in Table II using the multi-factor ANOVA procedure in the STATGRAPHICS 3.0 (Manugistics, 1997). Irrigation treatments were treated as fixed effects, replications and genotypes as random effects. Broad-sense heritability of the traits was calculated across five irrigation treatments. Broad-sense heritability (H2) based on DH lines was estimated according to the following equation: 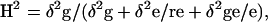 where r is the number of replications, e is the number of irrigation treatments, δ2g is the genetic variance, δ2ge is the variance of genotype × irrigation treatment interaction, and δ2e is the residual variance. Phenotypic correlations of the traits obtained from the experiment were computed using the genotypic means. Path-coefficient analysis using multiple regression was used to determine the factors contributing to GY at different irrigation levels.
where r is the number of replications, e is the number of irrigation treatments, δ2g is the genetic variance, δ2ge is the variance of genotype × irrigation treatment interaction, and δ2e is the residual variance. Phenotypic correlations of the traits obtained from the experiment were computed using the genotypic means. Path-coefficient analysis using multiple regression was used to determine the factors contributing to GY at different irrigation levels.
QTL Analysis
QTL analysis was conducted based on the subset of 154 DH lines. The 154 DHL were originally used for the linkage map construction at Texas Tech University (Zhang et al., 2001). This map contained 315 markers consisting of 145 restriction fragment length polymorphism (RFLP), 17 simple sequence repeat (SSR), and 153 amplified fragment length polymorphism (AFLP). Twenty SSR markers were screened and combined into the existing linkage map. The new linkage map was then reconstructed with 335 markers using MAPMAKER EXP.3.0 software (Lincoln et al., 1992). Three SSR markers were found unlinked; therefore, the final map was constructed with 332 markers. The average distance between markers was 4.4 cm. The Kosambi centimorgan function was used to express the distance between linked markers. The assignment of linkage groups to their corresponding chromosomes was according to published maps of Kurata et al. (1994) and Causse et al. (1994).
Simple interval mapping and simplified composite interval mapping procedures were performed to determine the single-locus QTL using the computer program MQTL (Tinker and Mather, 1995). The analysis for each data set was computed with 1,000 permutations, a 5 cM walking speed, and a Type I error rate of 5%. Log of the odds (LOD) score threshold for each trait was determined based on permutations. The lowest and highest determined LOD thresholds were 2.4 and 3.2. QTL with LOD score near the threshold were included and placed in parentheses. Nomenclature for QTL was modified from that described by McCouch et al. (1997). Associations of markers with traits and number of detected QTL were reconfirmed using simple and multiple regression analysis in the computer program STATGRAPHICS 3.0 (Manugistics, 1997). ANOVA analysis was also performed to determine digenic interactions between QTL main effect using the same software.
Acknowledgments
The authors would like to thank Dr. Apichart Vanavichit and Dr. Somvong Tragoonrung for kindly providing the facilities of DNA analysis at Rice Gene Discovery Unit and DNA Technology Laboratory and also Dr. Peerasak Srinives for his comments. Thanks to Dr. J.C. O'Toole for helpful comments on the manuscript.
This work was supported by grants (2000FS057 and 2000FS058) from the Rockefeller Foundation of New York.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.035527.
References
- Babu CR, Nguyen BD, Chamarerk V, Shanmugasundaram P, Chezhian P, Juyaprakash P, Ganesh SK, Palchamy A, Sadasivam S, Sarkarung S, et al. (2003) Genetic analysis of drought resistance in rice by molecular markers: association between secondary traits and field performance. Crop Sci 43: 1457–1469 [Google Scholar]
- Blum A (1988) Drought Resistance: Plant Breeding for Stress Environments. CRC Press, Boca Raton, FL, pp 43–77
- Blum A, Munns R, Passioura JB, Turner NC, Sharp RE, Boyer JS, Nguyen HT, Hsiao TC (1996) Letters to the editors: genetically engineered plants resistant to soil drying and salt stress: how to interpret osmotic relations. Plant Physiol 110: 1051–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum A, Mayer J, Golan G, Sinmena B (1999) Drought tolerance of a doubled-haploid line population if rice in the field. In O Ito, J O'Toole, B Hardy, eds, Genetic Improvement of Rice for Water-Limited Environments. International Rice Research Institute, Los Banos, Philippines, pp 319–330
- Boonjung H, Fukai S (1996) Effects of soil water deficit at different growth stages on rice growth and yield under upland conditions. 2. Phenology, biomass production and yield. Field Crops Res 48: 47–55 [Google Scholar]
- Causse MA, Fulton TM, Cho YG, Ahn SN, Chunwongse J, Wu K, Xiao J, Yu Z, Ronald PC, Harrington SE, et al. (1994) Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138: 1251–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M (1999) Concepts and strategies for plant adaptation research in rainfed lowland rice. Field Crops Res 64: 13–34 [Google Scholar]
- Cooper M, Somrith B (1997) Implications of genotype-by-environment interactions for yield adaptation of rainfed lowland rice: influence of flowering date on yield variation. In S Fukai, M Cooper, J Salisbury, eds, Proceedings of Workshop, Plant Breeding Strategies for Rainfed Lowland Rice in Drought Prone Environments. ACIAR, Canberra, Australia, pp 104–114
- Cruz RT, O'Toole JC (1984) Dry land rice response to an irrigation gradient at flowering stage. Agron J 76: 178–183 [Google Scholar]
- Cui HK, Peng SB, Xing YZ, Yu SB, Xu CG, Zhang Q (2003) Molecular dissection of the genetic relationships of source, sink and transport tissue with yield traits in rice. Theor Appl Genet 106: 649–658 [DOI] [PubMed] [Google Scholar]
- Dey MM, Upadhyaya HK (1996) Yield loss due to drought, cold and submergence in Asia. In RE Evenson, RW Herdt, M Hossain, eds, Rice Research in Asia: Progress and Priorities. CAB International, Wallingford, UK, 291–303
- Ekanayake IJ, Garrity DP, Masajo TM, O'Toole JC (1985) Root pulling resistance in rice: inheritance and association with drought tolerance. Euphytica 34: 905–913 [Google Scholar]
- Fukai S (1999) Phenology in rainfed lowland rice. Field Crops Res 64: 51–60 [Google Scholar]
- Fukai S, Pantuwan G, Jongdee B, Cooper M (1999) Screening for drought resistance in rainfed lowland rice. Field Crops Res 64: 61–74 [Google Scholar]
- Hanks RJ, Keller J, Rasmussen VP, Wilson GD (1976) Line source sprinkler for continuous variable irrigation-crop production studies. Soil Sci Soc Am J 40: 426–429 [Google Scholar]
- Herdt RW (1991) Research priorities for rice biotechnology. In GS Khush, GH Toenniessen, eds, Rice Biotechnology. CAB International, Wallingford, UK, 19–54
- Hittalmani S, Huang N, Courtois B, Venuprasad R, Shashidhar HE, Zhuang JY, Zheng KL, Liu GF, Wang GC, Sidhu JS, et al. (2003) Identification of QTL for growth and grain yield-related traits in rice across nine locations in Asia. Theor Appl Genet 107: 679–690 [DOI] [PubMed] [Google Scholar]
- International Rice Research Institute (1993) 1993–1995 IRRI Rice Almanac. IRRI, Los Banos, Philippines
- Jongdee B, Mitchell JH, Fukai S (1997) Modeling approach for estimation of rice yield reduction due to drought in Thailand. In S Fukai, M Cooper, J Salisbury, eds, Breeding Strategies for Rainfed Lowland Rice in Drought-Prone Environments, Proceedings of an International Workshop, Ubon Ratchathani, Thailand, November 5–8, 1996. ACIAR Proceedings No. 77, pp 65–73
- Jongdee B, Cooper M (1998) Genetic variation for grain yield of rice under water deficit condition. In Proceedings of the 9th Australian Agronomy Conference, Wagga Wagga, Australia
- Jongdee B, Fukai S, Cooper M (2002) Leaf water potential and osmotic adjustment as physiological traits to improve drought tolerance in rice. Field Crops Res 76: 153–163 [Google Scholar]
- Kurata N, Nagamura Y, Yamamoto K, Harushima Y, Sue N, Wu J, Antonio BA, Shomura A, Shimizu T, Lin SY, et al. (1994) A 300 kilobase interval genetic map of rice including 833 expressed sequences. Nat Genet 8: 365–372 [DOI] [PubMed] [Google Scholar]
- Lark KG, Chase K, Adler FR, Mansur LM, Orf JJ (1995) Interactions between quantitative trait loci in soybean in which trait variation at one locus is conditional upon a specific allele at another. Proc Natl Acad Sci USA 92: 4656–4660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZK, Picson SRM, Park WS, Paterson AH, Stansel JW (1997) Epistasis for three grain yield components in rice Oryza sativa L. Genetics 145: 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley JM, Fukai S (1994) Effects of timing and severity of water deficit on four diverse rice cultivars. III. Phenological development, crop growth and grain yield. Field Crops Res 37: 225–234 [Google Scholar]
- Lilley JM, Ludlow MM (1996) Expression of osmotic adjustment and dehydration tolerance in diverse rice lines. Field Crops Res 48: 185–197 [Google Scholar]
- Lin JY, Shen M (1993) Rice production constraints in China. In RE Everson, ed, Rice Production in Asia: Progress and Priorities. CAB International, Wallingford, UK, pp 161–178
- Lincoln SM, Daly, Lander E (1992) Constructing Genetic Maps with MAPMAKER/EXP. 3.0. Whitehead Institute Technical Report, Ed 3. Whitehouse Institute, Cambridge, MA
- Ludlow MM, Muchow RC (1990) A critical evaluation of traits for improving crop yields in water-limited environments. Adv Agron 43: 107–153 [Google Scholar]
- Mackill DJ, Nguyen HT, Zhang J (1999) Use of molecular markers in plant improvement programs for rainfed lowland rice. Field Crops Res 64: 177–185 [Google Scholar]
- Manugistics (1997) Manugistics, Rockville, MD
- Matsushima S (1966). Crop Science in Rice: Theory of Yield Determination and Its Application. Fuji Publishing, Tokyo, pp 365
- McCouch SR, Cho YG, Yano M, Paul E, Blinstrub M, Morishima H, Kinoshita T (1997) Report on QTL nomenclature. Rice Genet Newsl 14: 11–13 [Google Scholar]
- O'Toole JC, Chang TT (1979). Drought resistance in cereals-rice: a case study. In H Mussell, RC Staples, eds, Physiology of Crop Plants. John Wiley & Sons, New York, pp 374–405
- O'Toole JC, Namuco OS (1983) Role of panicle exertion in water stress induced sterility. Crop Sci 23: 1093–1097 [Google Scholar]
- Pantuwan G, Ingram KT, Sharma PK (1996) Rice root system development under rainfed conditions. Proceedings of the Thematic Conference on Stress Physiology, Rainfed Lowland Rice Research Consortium, Lucklow, India, February 28–March 5, 1994. International Rice Research Centre, Manila, Philippines, pp 198–206
- Pantuwan G, Fukai S, Cooper M, Rajatasereejul S, O'Toole JC (2002) Yield response of rice (Oryza sativa L.) genotypes to different types of drought under rainfed lowlands, Part 1. Grain yield and yield components. Field Crops Res 73: 153–168 [Google Scholar]
- Price AH, Young EM, Tomos AD (1997) Quantitative trait loci associated with stomatal conductance, leaf rolling and heading date mapped in upland rice (O. sativa). New Phytol 137: 83–91 [Google Scholar]
- Price AH, Steele KA, Moore BJ, Barraclough PB, Clark LJ (2000) A combined RFLP and AFLP linkage map of upland rice (Oryza sativa L.) used to identify QTLs for root penetration ability. Theor Appl Genet 100: 49–56 [Google Scholar]
- Rajatasereekul S, Sriwisut S, Porn-uraisanit P, Ruangsook S, Mitchell JH, Fukai S (1997) Phenology requirement for rainfed lowland rice in Thailand and Lao PDR. In Breeding Strategies for Rainfed Lowland Rice in Drought-Prone Environments, Proceedings of an International Workshop, Ubon Ratchatani, Thailand, November 5–8, 1996. ACIAR, Canberra, Australia, pp 97–103
- Robin S, Pathan MS, Courtois B, Lafitte R, Carandang C, Lanceras S, Amante M, Nguyen HT, Li Z (2003) Mapping osmotic adjustment in an advanced back-cross inbred population of rice. Theor Appl Genet 107: 1288–1296 [DOI] [PubMed] [Google Scholar]
- Saini HS, Lalonde S (1998) Injuries to reproductive development under water stress, and their consequences for crop productivity. J Crop Prod 1: 223–248 [Google Scholar]
- Sheoran IS, Saini HS (1996) Drought-induced male sterility in rice: changes in carbohydrate levels and enzyme activity associated with the inhibition of starch accumulation in pollen. Sex Plant Reprod 9: 161–169 [Google Scholar]
- Tinker NA, Mather DE (1995) MQTL: Software for Simplified Composite Interval Mapping of QTL in Multiple Environments, JQTL
- Tripathy JN, Zhang J, Robin S, Nguyen ThT, Nguyen HY (2000) QTLs for cell-membrane stability mapped in rice (Oryza sativa L.) under drought stress. Theor Appl Genet 100: 1197–1202 [Google Scholar]
- Wade LJ, McLaren CG, Samson BK, Regmi KR, Sarkarung S (1996) The importance of site characterization for understanding genotype by environment interactions. In M Cooper, GL Hammer, eds, Plant Adaptation and Crop Improvement. CAB International, Wallingford, UK, pp 549–562
- Wade LJ, McLaren CG, Quintana L, Harnpichitvitaya D, Rajatasereekul S, Sarawgi AK, Kumar A, Ahmed HU, Sarwoto, Singh AK, et al. (1999) Genotype by environment interactions in diverse rainfed lowland rice environment. Field Crops Res 64: 35–50 [Google Scholar]
- Xing YZ, Tan YF, Hua JP, Sun XL, Xu CG, Zhang Q (2002) Characterization of the main effects, epistatic effects and their environmental interactions of QTLs on the genetic basis of yield traits in rice. Theor Appl Genet 105: 248–257 [DOI] [PubMed] [Google Scholar]
- Yadav RS, Hash CT, Bidinger FR, Cavan GP, Howarth CJ (2002) Quantitative trait loci associated with traits determining grain and stover yield in pearl millet under terminal drought-stress conditions. Theor Appl Genet 104: 67–83 [DOI] [PubMed] [Google Scholar]
- Yu SB, Li JX, Xu CG, Tan YF, Gao YJ, Li XH, Zhang Q, Saghau Maroof MA (1997) Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci USA 94: 9226–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zheng HG, Ali ML, Tripathy JN, Aarti A, Pathan MS, Sarial AK, Robin S, Nguyen TT, Babu RC, et al. (1999. a) Molecular dissection of drought tolerance in rice: from physio-morphological traits to field performance. In JC O'Toole, O Ito, B Hardy, eds, Genetic Improvement of Rice for Water-Limited Environments. Proceedings of the Workshop on Genetic Improvement of Rice for Water-Limited Environments, December 1–3, 1998. International Rice Research Institute, Los Banos, Philippines
- Zhang J, Nguyen HT, Blum A (1999. b) Genetic analysis of osmotic adjustment in crop plants. J Exp Bot 50: 291–302 [Google Scholar]
- Zhang J, Zheng HG, Aarti A, Pantuwan G, Nguyen TT, Tripathy JN, Sarial AK, Robin S, Babu RC, Nguyen BD, et al. (2001) Locating genomic regions associated with components of drought resistance in rice: comparative mapping within and across species. Theor Appl Genet 103: 19–29 [Google Scholar]