Abstract
Background
Mutation in the tyrosine kinase domain of epidermal growth factor receptor (EGFR) is a common feature observed in lung adenocarcinoma. A fusion gene between echinoderm microtubule-associated protein-like 4 (EML4) and the intracellular domain of anaplastic lymphoma kinase (ALK), named EML4-ALK, has been identified in a subset of non-small-cell lung cancer (NSCLC) tumors. The objective of this study was to determine the prevalence of EGFR mutations and EML4-ALK fusions in Indian patients with NSCLC (adenocarcinoma) as well as evaluate their clinical characteristics.
Patients and methods
Patients with NSCLC, adenocarcinoma histology, whose tumors had been tested for EGFR mutational status, were considered for this study. ALK gene rearrangement was detected by fluorescence in situ hybridization using the Vysis ALK Break Apart Rearrangement Probe Kit. ALK mutation was tested in samples that were negative for EGFR mutation.
Results
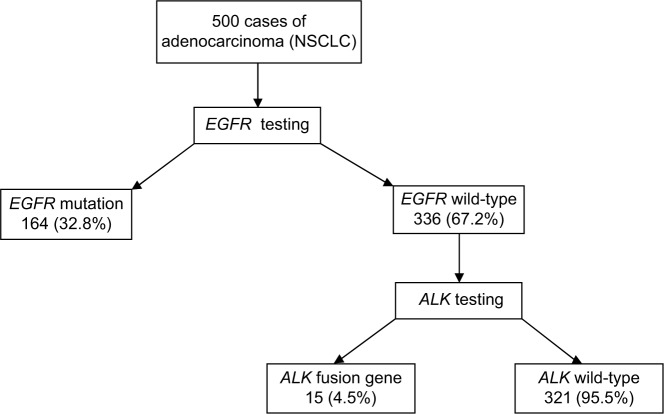
A total of 500 NSCLC adenocarcinoma patients were enrolled across six centers. There were 337 (67.4%) men and 163 (32.6%) women with a median age of 58 years. One hundred and sixty-four (32.8%) blocks were positive for EGFR mutations, whereas 336 (67.2%) were EGFR wild-type. Of the 336 EGFR-negative blocks, EML4-ALK fusion gene was present in 15 (4.5%) patients, whereas 321 (95.5%) tumors were EML4-ALK negative. The overall incidence of EML4-ALK fusion gene was 3% (15/500).
Conclusion
The incidence of EGFR mutations (33%) in this Indian population is close to the reported incidence in Asian patients. EML4-ALK gene fusions are present in lung adenocarcinomas from Indian patients, and the 3% incidence of EML4-ALK gene fusion in EGFR mutation-negative cases is similar to what has been observed in other Western and Asian populations. The mutual exclusivity of EML4-ALK and EGFR mutations suggests implementation of biomarker testing for tumors harboring ALK rearrangements in order to identify patients that can benefit from newer targeted therapies.
Keywords: NSCLC, Crizotinib, Vysis ALK Break Apart Rearrangement Probe Kit
Introduction
Lung cancer is one of the leading causes of cancer-related death in many parts of the world, including India. The overall 5-year survival rate remains at 15% despite significant improvements in the detection and treatment of lung cancer.1 Treatment outcomes using various chemotherapy (eg, taxane, platinum, gemcitabine, vinorelbine, pemetrexed) remain poor for advanced non-small-cell lung cancer (NSCLC).2 Adenocarcinoma is the most commonly occurring form of NSCLC and usually presents at an advanced stage with limited treatment options.
The introduction of targeted therapies into the treatment of NSCLC has improved survival of patients. However, it needs an accurate histological classification as well as testing of tumors for biomarkers that are predictive of response. Activating mutations in exons 18−21 of the tyrosine kinase domain of the gene for epidermal growth factor receptor (EGFR) are known to correlate with a high likelihood of response to EGFR tyrosine kinase inhibitors (TKIs), and these findings have paved the way for a new era of personalized treatment for NSCLC.3,4
Recently, a fusion gene between echinoderm microtubule-associated protein-like 4 (EML4) and the intracellular domain of anaplastic lymphoma kinase (ALK), named EML4-ALK, has been identified in some NSCLC tumors.5,6 EML4 ALK fusion protein, when inhibited by ALK and c met inhibitors in a selected subset of patients, has shown response rates of more than 60%.7
In an East Asian population, the EML4-ALK fusion gene positivity was found to be 5%, mostly in young patients who were never-smokers.8 However, a higher incidence of ALK mutations was reported in Chinese NSCLC patients, particularly in those with adenocarcinoma lacking EGFR/K-RAS mutation. EML4-ALK and EGFR mutations were mutually exclusive, suggesting that ALK mutations may be an important oncogenic factor and a potential therapeutic target in EGFR wild-type lung cancer.6
An oral ATP-competitive TKI of ALK and c-MET, crizotinib (PF-02341066), has shown impressive clinical activity in advanced NSCLCs, especially in tumors harboring ALK rearrangements. An impressive response rate of 57% and 72% progression-free survival were seen in an expanded cohort of 86 patients treated with crizotinib 250 mg twice daily. All these patients were negative for EGFR mutation and amplification of MET, another target for crizotinib, which suggests the therapeutic effect through inhibition of ALK.9 Treatment with crizotinib has been associated with higher 1- and 2-year overall survival, of 77% and 64%, respectively, in patients with advanced NSCLC.10 Crizotinib has been reported to be safe, with preliminary evidence of improved symptoms and clinically meaningful antitumor activity in patients with pretreated ALK-rearranged NSCLC.11 In a Phase III, open-label trial comparing crizotinib with chemotherapy in locally advanced or metastatic ALK-positive lung cancer patients, crizotinib demonstrated a median progression-free survival of 7.7 months, as compared to 3 months in the chemotherapy group (P<0.001). The response rates were 65% with crizotinib compared to 20% in the chemotherapy group (P<0.001).12
Various studies of NSCLC patients of Indian ethnicity have reported the presence of EGFR mutations in the range of 23%–44%.13–15 However, there is only one published report from India on EML 4 ALK mutations, with a limited number of patients.16 Although the presence of EML 4 ALK mutation seems to be mutually exclusive of the presence of EGFR or KRAS mutations in NSCLC, there are a few reports of concurrent existence of both the mutations.17–19 The present study was carried out to detect the prevalence of EGFR mutations and EML4-ALK fusions in EGFR mutation-negative cases of NSCLC (adenocarcinoma) as well as to evaluate the clinical characteristics of EGFR mutations and EML4-ALK fusions in EGFR mutation-negative cases of NSCLC.
Materials and methods
Patients with NSCLC, adenocarcinoma histology, whose tumors had been tested for EGFR mutational status by the amplification-refractory mutation system without enrichment of tumor samples during the period 2010 to 2014, were considered for this study. Stored tissue blocks from six centers were used for this study. Permission was obtained from the ethics committee at each center before the start of the study. Clinical characteristics and treatment details were collected from the patient’s medical records in a de-identified manner. Formalin-fixed, paraffin-embedded tissue blocks were used for the mutation analysis. ALK mutation was tested in samples that were negative for EGFR mutation. ALK gene rearrangement was detected by fluorescence in situ hybridization (FISH) using the Vysis ALK Break Apart Rearrangement Probe Kit (Abbott Molecular, Abbott Park, IL, USA), which identifies all rearrangements of ALK irrespective of other fusion partners.
The Break Apart Rearrangement Probe contained two differently labeled probes on opposite sides of the breakpoint of the ALK gene. A SpectrumOrange (red) labeled 250 kb probe at the 3′ end of ALK and a SpectrumGreen (green) labeled 300 kb probe at the 5′ end of ALK. Sections of 4–6 μm thickness of formalin-fixed, paraffin-embedded tissue were cut and placed onto slides precoated with poly-L-lysine. The slides were deparaffinized, dehydrated, and then put into 10 mM HCl. After drying the slides, they were rinsed with 2.5% sodium thiocyanate solution and steamed in a microwave for 2 minutes. Next, the slides were put into 10 mM sodium citrate buffer and boiled in the microwave for 4 minutes. The slides were immersed in a pepsin–HCl solution at 37°C for 25 minutes and then dehydrated. The probe mixture was added to the slide, the cover slip applied, the edges sealed, and the slide placed in a hybridizer at 80°C for 5 minutes to denature the probe. The hybridizer was sealed and the slides were incubated inside it at 37°C for 16–20 hours to allow hybridization. Post–hybridization washes with saline sodium citrate were given and 4,6-diamidino-2-phenylindole (DAPI) counterstain was applied. The signals for each probe were evaluated under a fluorescence microscope using an oil immersion objective.
There were two patterns of ALK rearrangement. One was break-apart, where the red and green signals were split, and the distance between these two signals was ≥2 signal diameters.16 In the other pattern, the green signal was lost, and only isolated or single red signals (3′ ALK) were observed, and these denote deletion in the 5′ ALK region in association with 2p inversion.16 A minimum of 50 tumor cells were counted, and cases were considered positive for ALK rearrangement when >15% of the tumor cells showed split signals or single red signals; the rest of the cases were classified as ALK FISH negative.20–22 While scoring, normal lung tissue and lymphocyte nuclei were not counted. If no signals were detected, the test was repeated (if enough tissue was present). When the repeat test also showed no signals, the result of “test uninterpretable” was issued and the cause of uninterpretability reported. The FISH results were evaluated and the reasons for uninterpretable results investigated.
Statistical analysis was performed using Pearson’s chi-squared or Fisher’s exact test – whichever was appropriate for categorical variables. Logistic regression was performed to compare the study groups. A two-sided P<0.05 was considered significant. Binary logistic regression with a single independent variable was performed; therefore, statistical correction has not been applied to the P-values reported in this paper. Statistical analysis was performed using SAS software (v 9.3; SAS Institute Inc., Cary, NC, USA).
Results
We included 500 NSCLC adenocarcinoma patients in this study across six centers. There were 337 (67.4%) men and 163 (32.6%) women with a median age of 58 years (range 21 to 90 years). Of the 500 patients, 250 (50%) were never-smokers and 357 (71.4%) had stage IV disease at the time of initial diagnosis. Baseline patient characteristics are presented in Table 1.
Table 1.
Summary of patient demographics and tumor characteristics (N=500)
| Characteristics | N (%) |
|---|---|
| Median age, years (range) | 58 (21–90) |
| Sex | |
| Male | 337 (67.4%) |
| Female | 163 (32.6%) |
| Smoking | |
| Smokers | 164 (32.8%) |
| Never-smokers | 250 (50%) |
| Unknown | 86 (17.2%) |
| Basis of diagnosis | |
| Histology | 500 (100%) |
| Grade | |
| Well-differentiated | 18 (3.6%) |
| Moderately differentiated | 53 (10.6%) |
| Poorly differentiated | 91 (18.2%) |
| Unknown | 338 (67.6%) |
| Stage | |
| I | 4 (0.8%) |
| II | 12 (2.4%) |
| III | 58 (11.6%) |
| IV | 357 (71.4%) |
| Unknown | 69 (13.8%) |
The results of molecular testing are shown in Table 2 and Figure 1. One hundred and sixty-four (32.8%) blocks were positive for EGFR mutations, whereas 336 (67.2%) were EGFR wild-type. Of the 336 EGFR-negative blocks, EML4-ALK fusion gene was present in 15 (4.5%) patients, whereas 321 (95.5%) tumors were EML4-ALK negative. The overall incidence of EML4-ALK fusion gene was 3% (15/500). Chemotherapy alone or in combination with radiotherapy and oral targeted therapy was the foremost treatment modality adopted to treat these patients.
Table 2.
Results of molecular testing
| Characteristics | N (%) |
|---|---|
| EGFR mutations | |
| Positive | 164 (32.8%) |
| Wild-type | 336 (67.2%) |
| EML4-ALK fusion gene | |
| Positive | 15 (4.5%) |
| Wild-type | 321 (95.5%) |
Figure 1.
Molecular testing results.
Abbreviation: NSCLC, non-small-cell lung cancer.
The association of each of the individual factors with regard to EGFR and EML4-ALK fusion gene mutation is shown in Table 3. While sex and cigarette smoking were significantly associated with EGFR mutation (P<0.05) the EML4-ALK fusion gene mutation showed a significant association with sex and age (P<0.05). There was a borderline association of cigarette smoking with EML4-ALK fusion gene mutation (P=0.053).
Table 3.
Association of each of the individual factors vis-à-vis EGFR and EML4-ALK fusion gene mutations
| Serial number | Variable | EGFR mutations
|
EML4-ALK fusion gene mutations
|
||
|---|---|---|---|---|---|
| χ2 | P-value | χ2 | P-value | ||
| 1 | Sex | 8.7257 | 0.0031 | 7.2785 | 0.007 |
| 2 | Age | 1.1984 | 0.5493 | 23.6355 | <0.0001 |
| 3 | Cigarette smoking | 29.1236 | <0.0001 | 3.2547 | 0.0533 |
| 4 | Tobacco chewing | 0.104 | 0.7471 | 3.3185 | 0.2344 |
| 5 | Alcohol intake | 1.2887 | 0.2563 | 3.4131 | 0.2295 |
| 6 | Grade | 1.5262 | 0.4662 | 2.401 | 0.428 |
| 7 | Stage of disease | 11.4349 | 0.0221 | 1.1379 | 0.1769 |
| 8 | Performance status | 8.4289 | 0.1887 | 6.6411 | 0.2288 |
Table 4 presents the distribution of EGFR and EML4-ALK gene mutations across the study groups.
Table 4.
Distribution of EGFR and EML4-ALK gene mutations
| Variable | EGFR
|
EML4-ALK fusion gene
|
||||
|---|---|---|---|---|---|---|
| Wild-type n (%) | Mutated n (%) | P-value | Wild-type n (%) | Mutated n (%) | P-value | |
| Sex | 0.003 | 0.009 | ||||
| Female | 95 (58.3) | 68 (41.7) | 86 (90.5) | 9 (9.5) | ||
| Male | 241 (71.5) | 96 (28.5) | 235 (97.5) | 6 (2.5) | ||
| Age, years | 0.549 | 0.0006 | ||||
| 20–40 | 25 (67.6) | 12 (32.4) | 19 (76) | 6 (24) | ||
| 40–60 | 153 (64.8) | 83 (35.2) | 146 (96.1) | 6 (3.9) | ||
| >60 | 158 (69.6) | 69 (30.4) | 156 (98.1) | 3 (1.9) | ||
| Smoking history | <0.001 | 0.151 | ||||
| Never-smokers | 142 (56.8) | 108 (43.2) | 134 (94.4) | 8 (5.6) | ||
| Smokers | 135 (82.3) | 29 (17.7) | 132 (97.1) | 4 (2.9) | ||
| Unknown | 59 (68.6) | 27 (31.4) | 53 (91.4) | 5 (8.6) | ||
| Stage | 0.346 | 0.644 | ||||
| I | 4 (80) | 1 (20) | 4 (100) | 0 | ||
| II | 6 (54.5) | 5 (45.5) | 6 (100) | 0 | ||
| III | 60 (81.1) | 14 (18.9) | 59 (98.3) | 1 (1.7) | ||
| IV | 246 (63.6) | 141 (36.4) | 236 (95.9) | 10 (4.1) | ||
| Unknown | 20 (87) | 3 (13) | 16 (80) | 4 (20) | ||
Discussion
The last decade has been an exciting period for pulmonary oncology research, wherein targeted therapies have improved median overall survival for patients with metastatic NSCLC to more than 1 year.3 Certain clinical subgroups, namely the never-smoking and minimal former-smoking patients, have seen even greater survival advantage. The uncontested benefit of EGFR TKIs for patients with EGFR activating mutations has clearly demonstrated the feasibility and power of tumor molecular profiling as a guide for clinical treatment decisions and has paved the way for a new era of personalized treatment for NSCLC.
Studies across different ethnicities have reported the incidence of EGFR mutation in NSCLC at the rate of 10%–15% in North Americans and Europeans,23–25 10% in African-Americans,26 24% in Koreans,27 50.5% in Taiwanese,28 26.3% in Japanese,29 38.1% in Chinese,30 and 23.2% in Indian14 populations. Two other studies from India reported EGFR mutation frequencies of 25.9% and 51.8%, respectively, along with female dominance.13,15 Regional differences have also been reported, with a higher incidence, at 65%, in the southern Indian population as compared to 33% in northern Indian population.14,15,31–33 The incidence of EGFR mutations (32.8%) in our study is close to the reported incidence in Asian patients (47%) and much higher than that of Caucasian patients (13%).34 Our study also confirms a higher incidence of EGFR mutation in the southern Indian population (47%) as compared to the northern Indian population (27%). The incidences of EGFR mutations in eastern and western Indian populations were 33% and 26%, respectively. In the present study, a higher EGFR mutation rate was observed in females than in males (41.7% versus 28.5%; P=0.003), and the results were found to be statistically significant. This observation is consistent with previously published studies that have shown female sex dominance in cases of EGFR mutation.14,35,36 Although we included only those patients whose tumors had been tested for EGFR mutational status, the data for NSCLC in the never-smoker population (50%) was in range of previously published reports from India (29.4% to 54.7%).14,15,37,38 The results of our study demonstrate a positive correlation of EGFR mutation in the never-smoker group as compared to smokers (43.2% versus 17.7%; P<0.001). Overall, the results of our study are in line with the findings of other Indian studies, with the advantage of having data from all the four geographical regions (east, west, north, and south) of the country.
EML4 ALK fusion protein represents another molecular type, and, when inhibited by ALK and c met inhibitors in a selected subset of patients, has shown response rates of more than 60%.4 The initial reports of EML4 ALK fusion protein date back to 2007, when a few studies reported NSCLC harboring a fusion gene between EML4 and the intracellular domain of ALK.5,39 Crizotinib is an inhibitor of receptor tyrosine kinases including ALK, hepatocyte growth factor receptor (HGFR), c-Met, and Recepteur d’Origine Nantais (RON).9 Unlike with EGFR, there are very few reports on EML4 ALK mutation across different ethnicities. The overall incidence of ALK gene rearrangements in NSCLC is estimated to be 3%, with an incidence of up to 13% in East Asian populations.5,8 Among 226 NSCLC tumor samples available in an East Asian population, the EML4-ALK fusion gene was found in about 5% of cases, mostly in young patients who were never-smokers.8 Another study, by Zhang et al reported a 13% incidence of ALK mutations in Chinese NSCLC patients, particularly in those with adenocarcinoma lacking EGFR/K-RAS mutation.6 In this study, EML4-ALK and EGFR mutations were found to be mutually exclusive, suggesting ALK mutations as a potential therapeutic target in EGFR wild-type lung cancer. A perspective review on the potential of crizotinib in NSCLC by Bang reports the prevalence of ALK fusion gene in unselected and adenocarcinoma-enriched patient populations across 14 studies as 3.4% and 4.5%, respectively.40 The total numbers of patients across 14 studies included in this review were 2,864 and 1,820 for the unselected and adenocarcinoma-enriched patient populations, respectively.
To date, there is only one published report from India on EML4 ALK mutations demonstrating a positivity of 2.7%.16 Our study confirms that EML4-ALK gene fusions are present in lung adenocarcinomas from Indian patients, and the 3% incidence of EML4-ALK gene fusion in EGFR mutation-negative cases is similar to what has been observed in other Western and Asian populations.40–42 In the present study, a higher EML4-ALK fusion gene mutation rate was observed in females than in males (9.5% versus 2.5%; P=0.009), and the results were found to be statistically significant. Age was categorized in three groups in this study, and a higher EML4-ALK fusion gene mutation rate (24%) was observed in the youngest age group, of 20–40 years (P=0.006), as compared to 3.9% and 1.9% in the age groups of 40–60 years and >60 years, respectively. This finding is consistent with previously published reports on EML4 ALK fusion gene mutations.
Conclusion
Our study reports the first multicenter data on EML4 ALK fusion gene mutations in EGFR mutation-negative Indian NSCLC patients, with the limitation of being a retrospective study that lacks survival data. However, it establishes the importance of EML4 ALK fusion gene testing in Indian NSCLC population to detect tumors harboring ALK rearrangements which can benefit from new targeted therapies. Prospective studies to investigate the survival benefit are warranted.
Acknowledgments
This study was supported by Pfizer’s Investigator Initiated Research Grant.
Footnotes
Disclosure
The authors report no conflicts of interest in this work. No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Reck M, Heigener DF, Mok T, Soria JC, Rabe KF. Management of non-small-cell lung cancer: recent developments. Lancet. 2013;382:709–719. doi: 10.1016/S0140-6736(13)61502-0. [DOI] [PubMed] [Google Scholar]
- 3.Okamoto I, Mitsudomi T, Nakagawa K, Fukuoka M. The emerging role of epidermal growth factor receptor (EGFR) inhibitors in first-line treatment for patients with advanced non-small cell lung cancer positive for EGFR mutation. Ther Adv Med Oncol. 2010;2:301–307. doi: 10.1177/1758834010370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsao AS, Papadimitrakopoulou VA. The future of NSCLC: molecular profiles guiding treatment decisions. Oncology (Williston Park) 2011;25:607–614. [PubMed] [Google Scholar]
- 5.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 6.Zhang X, Zhang S, Yang X, et al. Fusion of EML4 and ALK is associated with development of lung adenocarcinomas lacking EGFR and KRAS mutations and is correlated with ALK expression. Mol Cancer. 2010;9:188. doi: 10.1186/1476-4598-9-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D, Ahn M, Yang P, et al. Updated results of a global phase II study with crizotinin in advanced ALK-positive non-small cell lung cancer (NSCLC) Ann Oncol. 2012;23 Abstr 1230 PD. [Google Scholar]
- 8.Wong DW, Leung EL, So KK, et al. University of Hong Kong Lung Cancer Study Group The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009;115:1723–1733. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]
- 9.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw AT, Yeap BY, Solomon BJ, et al. Impact of crizotinib on survival in patients with advanced, ALK-positive NSCLC compared with historical controls. J Clin Oncol. 2011;29(suppl) abstr 7507. [Google Scholar]
- 11.Crino L, Kim D, Riely GJ, et al. Initial Phase II results with crizotinib in advanced ALK-positive non-small cell lung cancer (NSCLC): PROFILE 1005. J Clin Oncol. 2011;29(suppl) abstr 7514. [Google Scholar]
- 12.Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368(25):2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 13.Sahoo R, Harini VV, Babu VC, et al. Screening for EGFR mutations in lung cancer, a report from India. Lung Cancer. 2011;73(3):316–319. doi: 10.1016/j.lungcan.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Chougule A, Prabhash K, Noronha V, et al. Frequency of EGFR mutations in 907 lung adenocarcinoma patients of Indian ethnicity. PLoS One. 2013;8(10):e76164. doi: 10.1371/journal.pone.0076164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doval DC, Azam S, Batra U, et al. Epidermal growth factor receptor mutation in lung adenocarcinoma in India: a single center study. J Carcinog. 2013;12:12. doi: 10.4103/1477-3163.114970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desai SS, Shah AS, Prabhash K, Jambhekar NA. A year of anaplastic large cell kinase testing for lung carcinoma: pathological and technical perspectives. Indian J Cancer. 2013;50(2):80–86. doi: 10.4103/0019-509X.117007. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka H, Hayashi A, Morimoto T, et al. A case of lung adenocarcinoma harboring EGFR mutation and EML4-ALK fusion gene. BMC Cancer. 2012;12:558. doi: 10.1186/1471-2407-12-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JK, Kim TM, Koh Y, et al. Differential sensitivities to tyrosine kinase inhibitors in NSCLC harboring EGFR mutation and ALK trans-location. Lung Cancer. 2012;77(2):460–463. doi: 10.1016/j.lungcan.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 19.Popat S, Vieira de Araújo A, Min T, et al. Lung adenocarcinoma with concurrent exon 19 EGFR mutation and ALK rearrangement responding to erlotinib. J Thorac Oncol. 2011;6(11):1962–1963. doi: 10.1097/JTO.0b013e31822eec5e. [DOI] [PubMed] [Google Scholar]
- 20.Varella-Garcia M, Cho Y, Lu X, et al. ALK gene rearrangements in unselected caucasians with non-small cell lung carcinoma (NSCLC) J Clin Oncol. 2010;28:18s. (suppl; abstr 10533) [Google Scholar]
- 21.Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodig SJ, Mino-Kenudson M, Dacic S, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res. 2009;15:5216–5223. doi: 10.1158/1078-0432.CCR-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 24.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 25.Dogan S, Shen R, Ang DC, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18:6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cortes-Funes H, Gomez C, Rosell R, et al. Epidermal growth factor receptor activating mutations in Spanish gefitinib-treated non-small-cell lung cancer patients. Ann Oncol. 2005;16:1081–1086. doi: 10.1093/annonc/mdi221. [DOI] [PubMed] [Google Scholar]
- 27.Jang TW, Oak CH, Chang HK, Suo SJ, Jung MH. EGFR and KRAS mutations in patients with adenocarcinoma of the lung. Korean J Intern Med. 2009;24:48–54. doi: 10.3904/kjim.2009.24.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu SG, Gow CH, Yu CJ, et al. Frequent epidermal growth factor receptor gene mutations in malignant pleural effusion of lung adenocarcinoma. Eur Respir J. 2008;32:924–930. doi: 10.1183/09031936.00167407. [DOI] [PubMed] [Google Scholar]
- 29.Usui K, Ushijima T, Tanaka Y, et al. The frequency of epidermal growth factor receptor mutation of nonsmall cell lung cancer according to the underlying pulmonary diseases. Pulm Med. 2011;2011:290132. doi: 10.1155/2011/290132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang YS, Yang JJ, Zhang XC, et al. Impact of smoking status and pathologic type on epidermal growth factor receptor mutations in lung cancer. Chin Med J (Engl) 2011;124:2457–2460. [PubMed] [Google Scholar]
- 31.Aggarwal S, Patil S, Minhans S, Pungliya M, Soumitra N. A study of EGFR mutation in nonsmoker NSCLC: striking disparity between north and south India patients. J Clin Oncol. 2012;30(suppl) abstr e18041. [Google Scholar]
- 32.Pungliya M, Sachin M, Soumittra N, Shekar P, Shyam A. A study of incidence of EGFR mutations in non-smoker adenocarcinoma of the lung: disparity between north and south indian patients. J Cancer Ther Res. 2014;3:4. [Google Scholar]
- 33.Mehta J. Molecular epidemiology of epidermal growth factor receptor mutations in lung cancers in Indian population. Indian J Cancer. 2013;50(2):102–106. doi: 10.4103/0019-509X.117019. [DOI] [PubMed] [Google Scholar]
- 34.Sekine I, Yamamoto N, Nishio K, Saijo N. Emerging ethnic differences in lung cancer therapy. Br J Cancer. 2008;99:1757–1762. doi: 10.1038/sj.bjc.6604721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smits AJ, Kummer JA, Hinrichs JW, et al. EGFR and KRAS mutations in lung carcinomas in the Dutch population: increased EGFR mutation frequency in malignant pleural effusion of lung adenocarcinoma. Cell Oncol (Dordr) 2012;35:189–196. doi: 10.1007/s13402-012-0078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka T, Matsuoka M, Sutani A, et al. Frequency of and variables associated with the EGFR mutation and its subtypes. Int J Cancer. 2010;126:651–655. doi: 10.1002/ijc.24746. [DOI] [PubMed] [Google Scholar]
- 37.Krishnamurthy A, Vijayalakshmi R, Gadigi V, Ranganathan R, Sagar TG. The relevance of “Nonsmoking-associated lung cancer” in India: a single-centre experience. Indian J Cancer. 2012;49:82–88. doi: 10.4103/0019-509X.98928. [DOI] [PubMed] [Google Scholar]
- 38.Malik PS, Sharma MC, Mohanti BK, et al. Clinico-pathological profile of lung cancer at AIIMS: a changing paradigm in India. Asian Pac J Cancer Prev. 2013;14:489–494. doi: 10.7314/apjcp.2013.14.1.489. [DOI] [PubMed] [Google Scholar]
- 39.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 40.Bang YJ. The potential for crizotinib in non-small cell lung cancer: a perspective review. Ther Adv Med Oncol. 2011;3(6):279–291. doi: 10.1177/1758834011419002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukui T, Yatabe Y, Kobayashi Y, et al. Clinicoradiologic characteristics of patients with lung adenocarcinoma harboring EML4-ALK fusion oncogene. Lung Cancer. 2012;77:319–325. doi: 10.1016/j.lungcan.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 42.Paik JH, Choi CM, Kim H, et al. Clinicopathologic implication of ALK rearrangement in surgically resected lung cancer: a proposal of diagnostic algorithm for ALK-rearranged adenocarcinoma. Lung Cancer. 2012;76:403–409. doi: 10.1016/j.lungcan.2011.11.008. [DOI] [PubMed] [Google Scholar]