Abstract
Arabidopsis natural variation was used to analyze the genetics of plant growth rate. Screening of 22 accessions revealed a large variation for seed weight, plant dry weight and relative growth rate but not for water content. A positive correlation was observed between seed weight and plant area 10 d after planting, suggesting that seed weight affects plant growth during early phases of development. During later stages of plant growth this correlation was not significant, indicating that other factors determine growth rate during this phase. Quantitative trait locus (QTL) analysis, using 114 (F9 generation) recombinant inbred lines derived from the cross between Landsberg erecta (Ler, from Poland) and Shakdara (Sha, from Tadjikistan), revealed QTLs for seed weight, plant area, dry weight, relative growth rate, chlorophyll fluorescence, flowering time, and flowering-related traits. Growth traits (plant area, dry weight, and relative growth rate) colocated at five genomic regions. At the bottom of chromosome 5, colocation was found of QTLs for leaf area, leaf initiation speed, specific leaf area, and chlorophyll fluorescence but not for dry weight, indicating that this locus might be involved in leaf development. No consistent relation between growth traits and flowering time was observed despite some colocations. Some of the QTLs detected for flowering time overlapped with loci detected in other recombinant inbred line populations, but also new loci were identified. This study shows that Arabidopsis can successfully be used to study the genetic basis of complex traits like plant growth rate.
Analysis of plant growth is an essential step in the understanding of plant performance and productivity (Leister et al., 1999) and may reveal different strategies of plants to survive under limiting conditions.
Growth rate and, more specifically, relative growth rate (RGR) are comprehensive traits of plants, which characterize to a large extent plant performance and are also important components of fitness (McGraw and Garbutt, 1990). These parameters integrate morphological and physiological traits of plants. RGR is an inherent quantitative trait that may vary among plant species, occurring in a wide range of habitats. Plants in favorable environments often have an inherently high RGR, whereas those from less favorable habitats have an inherently low RGR, even when grown in the same favorable conditions (Grime and Hunt, 1975; Poorter and Remkes, 1990). In addition, plant growth rate is also affected by developmental changes such as the onset of flowering or the formation of storage organs.
Various parameters have been used to evaluate growth rate, including measurement of fresh or dry weight, root to shoot ratio, shoot number, or shoot length (Li et al., 1998; Leister et al., 1999). The measurement of fresh or dry weight is destructive and hence large numbers of plants are required to analyze growth in time. Although the analysis of growth by measuring the area covered by a plant instead of measuring its weight has been applied successfully (Smith and Spomer, 1987; Smith et al., 1989; Motooka et al., 1991), its use is hampered by complicated experimental designs. A nondestructive approach would be preferable, e.g. using image analysis. For Arabidopsis, which in its vegetative phase grows as a flat rosette with limited leaf overlap, Leister et al. (1999) showed that the use of digital video and image analysis was very effective in the determination of plant growth (rate) nondestructively, even during early developmental stages.
Growth rate can be seen as the integration of a wide range of processes, and thus genetic variation for such a complex trait may depend on many genes. Since also within species heritable differences in growth and morphology can be found (Maloof, 2003) these traits are amenable for genetic analysis. Complex polygenic traits can be studied genetically by quantitative trait loci (QTL) analysis. QTL analysis allows the dissection of quantitative genetic variation to the contribution of different loci. When mechanistically related traits map to similar map positions, this might suggest that variation for these traits at this locus is controlled by the same gene and in genetic terminology is pleiotropic. The extensive natural variation that occurs in Arabidopsis is being exploited increasingly as a source of genetic variation for the analysis of important adaptive traits, e.g. flowering time, plant and seed size, seed dormancy, pathogen resistance, and tolerance to abiotic stresses (for review, see Alonso-Blanco and Koornneef, 2000; Koornneef et al., 2004). Recombinant inbred lines (RILs) provide an immortal population, as each individual is practically homozygous, and large numbers of genetically identical individuals can be obtained, allowing repeated measurements of various traits in different conditions (Alonso-Blanco and Koornneef, 2000; Doerge, 2002).
We have used Arabidopsis natural variation to analyze growth rate by image analysis of plant leaf area, and by measuring a series of related parameters. From a greenhouse experiment involving approximately 130 Arabidopsis accessions, from a wide range of habitats, 22 accessions (Table I) were selected based on obvious differences in growth characteristics, carbohydrate content, and/or because they were used in generating Arabidopsis mapping populations (www.natural-eu.org). These accessions were studied to get insight in differences in various growth-related traits, which, when present, can be genetically analyzed further in segregating populations such as recombinant inbred lines (RILs). To investigate the genetic basis of differences in growth and growth-related traits and to see if relationships between traits in the selection of accessions might be due to a common genetic basis, we analyzed growth-related traits by QTL mapping. For this we used a newly developed RIL population derived from the cross between the laboratory accession Landsberg erecta (Ler), originating from northern Europe (Rédei, 1992), and the accession Shakdara (Sha), originating from high altitudes in Tadjikistan (Khurmatov, 1982). These parental accessions were not the extremes in the accession screen, but they showed a considerable variation for various growth traits as was also observed in their progeny.
Table I.
Names, stock numbers, origin, fresh and dry weight, water content and seed weight for 22 Arabidopsis accessions
| Name | Stock No | Country | Longitude | Latitude | Fresh Wt | Dry Wt | %WC | Seed Wt |
|---|---|---|---|---|---|---|---|---|
| (gm) | (gm) | (mg) | ||||||
| Amel-1 | CS 22526 | Netherlands | E 5.6 | N 53.4 | 1.43 | 0.19 | 87.0 | 0.026 |
| Nes-1 | CS 10041 | Netherlands | E 5.8 | N 53.3 | 0.87 | 0.14 | 83.9 | 0.030 |
| Nes-3 | CS 10042 | Netherlands | E 5.8 | N 53.2 | 0.83 | 0.11 | 86.4 | 0.028 |
| Oerd-2 | CS 10299 | Netherlands | E 5.9 | N 53.2 | 1.47 | 0.20 | 86.4 | 0.031 |
| Oerd-4 | CS 10040 | Netherlands | E 5.9 | N 53.2 | 1.03 | 0.12 | 88.4 | 0.030 |
| Cerv-1 | CS 22523 | Italy | E 12.5 | N 41.9 | 1.70 | 0.23 | 86.3 | 0.020 |
| Rome-1 | CS 22524 | Italy | E 12.5 | N 41.9 | 1.10 | 0.16 | 85.5 | 0.017 |
| Ler | N 20 | Poland | E 15.2 | N 52.7 | 0.97 | 0.13 | 86.2 | 0.020 |
| Col-2 | CS 907 | Poland | E 15.7 | N 52.7 | 0.65 | 0.08 | 88.5 | 0.022 |
| Cvi | N 8580 | Cape Verde Islands | W 24.4 | N 14.9 | 1.35 | 0.15 | 88.9 | 0.033 |
| An-1 | N 944 | Belgium | E 4.4 | N 51.2 | 0.37 | 0.06 | 84.5 | 0.021 |
| Bla-10 | JA 10185 | Spain | E 2.8 | N 41.7 | 0.93 | 0.12 | 86.8 | 0.022 |
| Kond | CS 6175 | Tadjikistan | E 68.5 | N 38.5 | 1.50 | 0.20 | 86.9 | 0.019 |
| Ely-1a | CS 6088 | UK | W 0.3 | N 52.4 | 0.53 | 0.07 | 87.5 | 0.020 |
| Eri | CS 22548 | Sweden | E 15 | N 56.4 | 0.97 | 0.12 | 87.2 | 0.019 |
| Hog | CS 6179 | Tadjikistan | E 68.5 | N 38.5 | 1.35 | 0.16 | 88.5 | 0.018 |
| Kas-2 | N 1264 | India | E 71.8 | N 34.3 | 0.17 | 0.03 | 84.0 | 0.028 |
| Sid-1 | CS 6077 | UK | E 15.4 | N 51.4 | 0.53 | 0.06 | 88.8 | 0.016 |
| Sha | CS 929 | Tadjikistan | E 71.3 | N 37.3 | 0.90 | 0.10 | 88.9 | 0.019 |
| Pak-3 | JW 10214 | Pakistan | E 73.4 | N 33.9 | 0.15 | 0.01 | 93.3 | 0.032 |
| Ik | JW 10223 | Japan | E 135.1 | N 35.5 | 1.30 | 0.19 | 85.4 | 0.030 |
| Kyo-1 | JW 10231 | Japan | E 135.8 | N 35.0 | 0.70 | 0.15 | 79.3 | 0.021 |
Since RGR depends on the gain of biomass via photosynthesis and on the starting mass of the plant, i.e. ultimately the seed from which it grows, we determined the seed weight and chlorophyll fluorescence as a nondestructive parameter for photosynthetic capacity. Allocation of biomass within the plant is expected to change upon flower induction and hence flowering time and related parameters were also analyzed in this study.
RESULTS
Variation among the Accessions
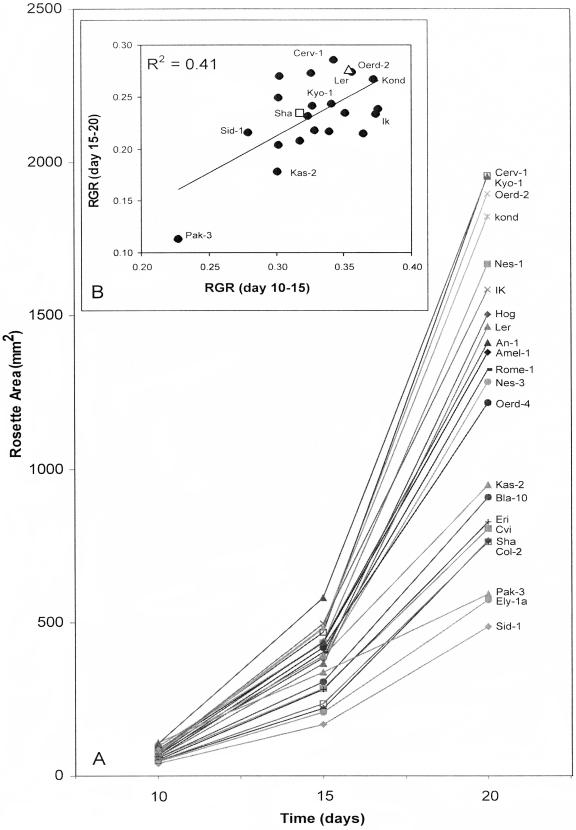
Screening the 22 accessions revealed a large variation for seed weight, growth rate, and plant fresh and dry weight but less for water content (Table I, Fig. 1). The seed weight of these accessions was not correlated with the latitude at which the accessions had originally been collected as suggested before by Li et al. (1998). Seed weight showed a positive correlation with plant area at 10 d after planting, suggesting that seed weight affected plant growth during early phases of development (Fig. 1A). During later stages of plant growth this correlation was not significant, indicating that other factors determined plant growth at that phase (Fig. 1B). Final plant dry weight correlated with RGR (based on area) especially during the last period (Fig. 1, C and D). A positive correlation was observed between area at day 20 and the plant dry weight at day 35 (R2 = 0.56, data not shown). We found a large variation between accessions for their growth rate as well as for their relative growth rate (on the basis of plant area) with Pak-3 having the lowest and Cerveteri-1, Kyoto-1, Oerd-2, and Kond the highest rates (Fig. 2, A and B). Ten days after planting, Pak-3 showed the largest area, but gradually its growth rate decreased due to early senescence, which was observed also for Kas-2 (Fig. 2A).
Figure 1.
Correlation between seed weight and plant area at day 10 (A) and day 20 (B) for 22 Arabidopsis accessions. Correlation between dry weight at 35 d and relative growth rate (RGR) day 10–15 (C) and day 15–20 (D) on the basis of plant area. Δ and □ correspond to Ler and Sha mean values, respectively.
Figure 2.
Growth rate curves for 22 Arabidopsis accessions determined by plant area (A). The correlation between relative growth rate (RGR) (day 10–15) and RGR (day 15–20) on the basis of plant area (B).
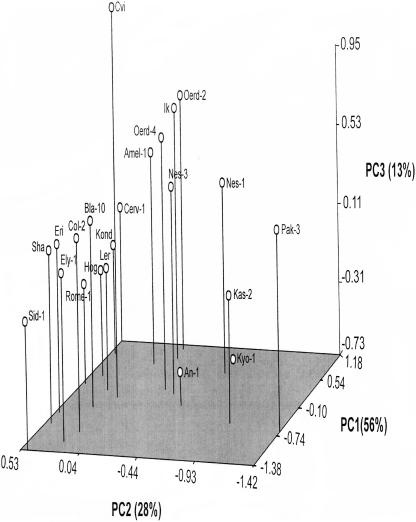
A principle component analysis indicated that the first three principle components (PCs) explained 97% of the variation for the six traits: fresh weight (FW), dry weight (DW), seed weight (SW), total leaf area 1 (TLA1), total leaf area 2 (TLA2), and total leaf area 3 (TLA3). PC1 showed a large variation between accessions and is mainly determined by growth related parameters (TLA3, TLA2, and DW). On the second function (PC2), TLA1, SW, and FW were the most important traits. On the third function (PC3), SW was the main variable discriminating between the accessions. Accessions with a large initial area (TLA1) and high seed weight (SW) were situated on the left side of the graph. The related accessions Oerd2, Oerd4, Nes1 and Nes3, all collected in the dunes of the island Ameland in the north of The Netherlands, as well as IK, had high SW and moderate TLA1, were grouped in the middle (Fig. 3). PC1 discriminated between the smallest accession in final plant size (Sid-1) and the largest one (Cerveteri-1), while PC3, which was determined mainly by seed weight, gave the largest seeded accession Cvi a separate position that contrasted most with the low seed weight accession An-1.
Figure 3.
Principle component analysis of variables: fresh and dry weight, seed weight, total leaf area1, total leaf area2 and total leaf area3 for the 22 accessions. The first principle component (PC1) is determined by growth-related parameters; total leaf area3, total leaf area2, and dry weight. The second principle component (PC2) is determined mainly by total leaf area1, seed weight, and fresh weight. In the third principle component (PC3), seed weight was the most important variable discriminating between the accessions.
Genetic Variation among the Ler × Sha RILs
For all traits analyzed significant variation was observed between RILs as indicated by the broad sense heritabilities ranging from 0.86 to 0.33 for flowering time traits and number of side branches, respectively (Table II and Fig. 4). Transgression beyond the parental values was observed for all traits including those for which parental values hardly differed, such as chlorophyll fluorescence. This amount of genetic variation indicated that QTL mapping was likely to reveal QTLs for most of the traits.
Table II.
Parental values, ranges and heritabilities in the Ler × Sha RILs of all measured traits
| Trait | Ler value | Sha value | Range | Mean | Heritability |
|---|---|---|---|---|---|
| Seed weight (mg) | 0.018 | 0.017 | 0.014–0.023 | 0.018 | nd |
| Total leaf area1 (mm2) | 130 | 153 | 48–365 | 161 | 0.65 |
| Total leaf area2 (mm2) | 436 | 551 | 155–1319 | 555 | 0.71 |
| Total leaf area3 (mm2) | 2191 | 2556 | 576–7418 | 2703 | 0.78 |
| Relative growth rate 2-1(area) | 0.3 | 0.32 | 0.22–0.36 | 0.31 | nd |
| Relative growth rate 3-2(area) | 0.27 | 0.26 | 0.08–0.32 | 0.26 | nd |
| Relative growth rate 3-1(area) | 0.28 | 0.28 | 0.13–0.33 | 0.28 | nd |
| Dry weight young plant (mg) | 2.5 | 3.5 | 1.4–8 | 3.5 | 0.68 |
| Dry weight old plant (mg) | 37 | 46 | 9–74 | 33 | 0.74 |
| Relative growth rate (weight) | 0.34 | 0.32 | 0.15–0.35 | 0.28 | nd |
| Water content (%) | 89 | 89.4 | 85.1–92.1 | 89.5 | 0.52 |
| Specific leaf area (mm2.mg−1) | 175 | 160 | 90–350 | 162 | nd |
| Speed of leaf initiation | 13 | 14 | 8–19 | 13.7 | nd |
| Chlorophyll fluorescence | 0.72 | 0.71 | 0.65–0.75 | 0.71 | 0.53 |
| Flowering time SD (days) | 33.2 | 32 | 21–48.7 | 35.3 | 0.86 |
| Flowering time LD (days) | 25.2 | 28.8 | 21.3–46.3 | 29.3 | 0.83 |
| Total leaf number | 7.9 | 9.5 | 6.6–31.1 | 10.6 | 0.86 |
| Rosette leaf number | 6 | 7.7 | 4.8–24.2 | 8.4 | 0.86 |
| Cauline leaf number | 1.9 | 1.8 | 1.3–6.9 | 2.3 | 0.59 |
| Total plant length (cm) | 17.5 | 37.4 | 11–50.4 | 26.7 | 0.85 |
| Plant length till 1st silique (cm) | 7.4 | 8.7 | 4–21.1 | 9.5 | 0.74 |
| Inflorescence length (cm) | 10.1 | 28.7 | 7–29.3 | 17.2 | 0.80 |
| Number of side branches | 1.9 | 2.4 | 0.4–3.9 | 1.83 | 0.33 |
nd, not determined because only one replica per line was analyzed.
Figure 4.
Frequency distribution of nonnormalized data of some traits in the Ler × Sha RIL population. Growth traits: Seed weight (A), Plant area at 10 d (B), Relative growth rate calculated on the basis of plant area (C), Dry weight 1 at 15 d (D), Dry weight 2 at 25 d (E), Relative growth rate calculated on the basis of plant dry weight (F). Flowering traits: Flowering time scored in long day (LD) conditions (G), Total leaf number counted in LD (H), Total plant length measured in LD (I), Flowering time scored in Short Day (SD) conditions (J), Leaf number counted at 24 d in SD referring to speed of leaf initiation (K), Chlorophyll fluorescence measured in SD (L). The average parental value is indicated with an arrow for both parents, L for Ler and S for Sha, and the horizontal bars represent the se for these parental values.
QTL Mapping
Seed Weight
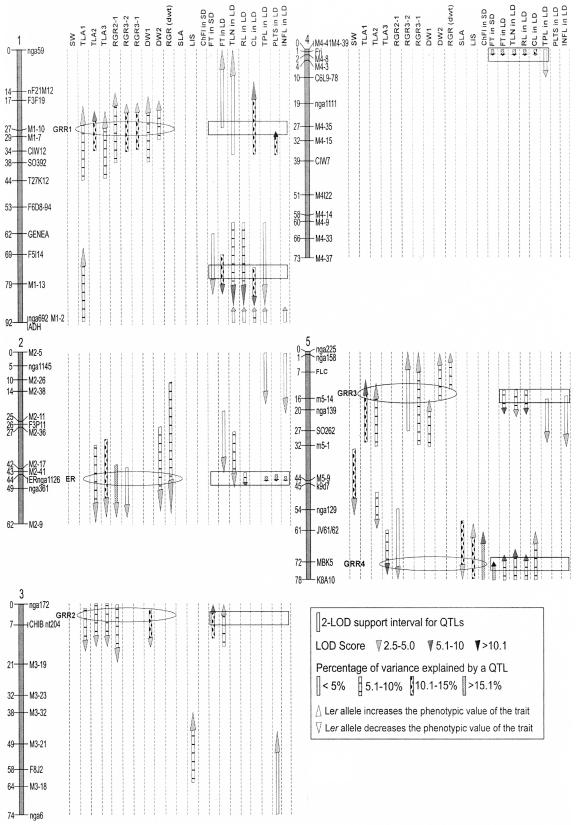
Although the difference in seed weight between the Ler and Sha parents was small (Fig. 4A and Table II), QTL mapping revealed one QTL on chromosome 5 (Fig. 5). This QTL, for which the Sha allele increased seed weight, explained 14.3% of the variance.
Figure 5.
The Ler × Sha linkage map showing the locations of QTLs for the traits analyzed. The lengths of the arrows indicate the 2-LOD support intervals. The direction of the arrow's head indicates the allelic effect: upward, Ler increasing and Sha decreasing; downward, Sha increasing and Ler decreasing. The gray scale of the arrow's head indicates the LOD score value. The filling pattern of the arrow's stem refers to the percentage of variance explained by each QTL. The ovals indicate QTLs of growth regions, GRR1 until GRR4 and the ER region, while the rectangles indicate the flowering time related QTLs.
Plant Total Leaf Area and RGR
Figure 5 and Table III summarize the QTLs found in Ler × Sha RILs for total leaf area (TLA) (four, five, and four QTLs for TLA1, TLA2, and TLA3, respectively). The detected QTLs showed a total explained phenotypic variance of 34%, 43%, and 37.5% for TLA1, TLA2, and TLA3, respectively (Table III). For TLA1, the Ler alleles increased plant area at three QTLs (at msat1-10, nga692, and msat5-14), whereas at the CHIB locus the Sha allele increased the area. The Sha alleles at ER, CHIB, and nga129 increased the TLA2, whereas the Ler alleles did so at msat1-10 and SO262. For TLA3, the Sha alleles at ER, CHIB, and MBK5 loci, and the Ler allele at msat1-10 increased the area.
Table III.
Characteristics of the detected QTLs explaining growth traits, flowering time and flowering related traits in Ler × Sha RIL population
| Trait | QTL | Map position | LOD score | % of variance | Additive allele effect |
|---|---|---|---|---|---|
| Seed weight (mg) | |||||
| K9D7 | 5–45 | 3.6 | 14.3 | −0.12 | |
| Toal leaf area1 (mm2) | 34 | ||||
| M1-10 | 1–26.7 | 3.08 | 6 | 13 | |
| nga692 | 1–91.9 | 3.18 | 6.1 | 14 | |
| CHIB | 3–6.8 | 4.55 | 9 | −17 | |
| m5-14 | 5–16.3 | 6.13 | 12.9 | 19 | |
| Total leaf area2 (mm2) | 43 | ||||
| M1-10 | 1–26.7 | 5.53 | 11.1 | 68 | |
| ER | 2–43.7 | 3.66 | 7.1 | −55 | |
| CHIB | 3–6.8 | 4.7 | 9.3 | −64 | |
| SO262 | 5–27.2 | 4.16 | 8.1 | 62 | |
| nga 129 | 5–53.9 | 3.83 | 7.4 | −65 | |
| Total leaf area3 (mm2) | 37.5 | ||||
| M1-10 | 1–26.7 | 3.57 | 8.1 | 346 | |
| ER | 2–43.7 | 4.37 | 10.1 | −379 | |
| CHIB | 3–6.8 | 3 | 6.7 | −331 | |
| MBK5 | 5–72.4 | 5.36 | 12.6 | −441 | |
| Relative growth rate 2-1 (area) | 34.3 | ||||
| M1-10 | 1–26.7 | 2.89 | 7.4 | 0.07 | |
| ER | 2–43.7 | 3.51 | 17.4 | −0.09 | |
| CHIB | 3–6.8 | 2.99 | 5.2 | −0.06 | |
| MBK5 | 5–72.4 | 2.75 | 4.3 | −0.04 | |
| Relative growth rate 3-2 (area) | 18.3 | ||||
| M1-10 | 1–26.7 | 3.25 | 10.2 | 0.11 | |
| nga361 | 2–48.6 | 2.58 | 3.5 | −0.09 | |
| nga225 | 5–0 | 2.88 | 4.6 | 0.10 | |
| Relative growth rate 3-1 (area) | 16.7 | ||||
| M1-10 | 1–26.7 | 3.45 | 10.4 | 0.12 | |
| nga225 | 5–0 | 2.69 | 6.3 | 0.09 | |
| Dry weight 1 (mg) | 25.8 | ||||
| M1-10 | 1–26.7 | 3.24 | 8.3 | 0.39 | |
| CHIB | 3–6.8 | 4.23 | 11.1 | −0.44 | |
| SO262 | 5–27.2 | 2.52 | 6.4 | 0.30 | |
| Dry weight 2 (mg) | 24.1 | ||||
| M1-10 | 1–26.7 | 2.65 | 6.8 | 3.73 | |
| ER | 2–43.7 | 3.8 | 10 | −4.58 | |
| nga225 | 5–0 | 2.83 | 7.3 | 3.68 | |
| Relative growth rate (weight) | 20.4 | ||||
| ER | 2–43.7 | 2.75 | 6.7 | −0.11 | |
| nga225 | 5–0 | 3.33 | 8.2 | 0.12 | |
| M1-10 × CHIB | 5.5 | ||||
| Specific leaf area (mm2.mg−1) | |||||
| MBK5 | 5–72.4 | 3.41 | 13.1 | −16 | |
| Leaf initiation speed | 20.6 | ||||
| F8J2 | 3–58 | 2.56 | 7.8 | 0.59 | |
| MBK5 | 5–72.4 | 4.06 | 12.8 | 0.78 | |
| Chlorophyll fluorescence (SD) | 27.6 | ||||
| MBK5 | 5–72.4 | 6.28 | 21.4 | 0.008 | |
| m4-14 × MBK5 | 6.2 | ||||
| Flowering time SD (days) | 52.4 | ||||
| M1-13 | 1–78.7 | 3.77 | 4.8 | −1.19 | |
| CHIB | 3–6.8 | 7.44 | 11 | 1.78 | |
| FRI | 4–0.8 | 11.48 | 18.7 | −2.33 | |
| K8A10 | 5–77.7 | 11.08 | 17.9 | 2.31 | |
| Flowering time LD (days) | 66.6 | ||||
| nga59 | 1–0 | 3.38 | 3.6 | 1.05 | |
| M1-13 | 1–78.7 | 8.92 | 11 | −2.13 | |
| M2-36 | 2–26.6 | 3.69 | 4 | −1.05 | |
| CHIB | 3–6.8 | 4.83 | 5.5 | 1.15 | |
| FRI | 4–0.8 | 17.14 | 25.5 | −2.52 | |
| m5-14 | 5–16.3 | 5.84 | 7 | −1.32 | |
| K8A10 | 5–77.7 | 8.06 | 10 | 1.63 | |
| Total leaf number | 63.4 | ||||
| nga59 | 1–0 | 2.51 | 3 | 0.71 | |
| M1-13 | 1–78.7 | 7.24 | 9.7 | −1.54 | |
| ADH | 1–92.3 | 3.63 | 4.7 | 1.07 | |
| ER | 2–43.7 | 4.84 | 6.1 | −1.00 | |
| FRI | 4–0.8 | 15.47 | 25.2 | −1.81 | |
| m5-14 | 5–16.3 | 4.88 | 6.4 | −0.98 | |
| K8A10 | 5–77.7 | 6.13 | 8.3 | 1.07 | |
| Rosette leaf number | 63.8 | ||||
| M1-13 | 1–78.7 | 6.69 | 8 | −1.24 | |
| ADH | 1–92.3 | 3.79 | 4.5 | 0.91 | |
| ER | 2–43.7 | 7.45 | 9 | −1.01 | |
| FRI | 4–0.8 | 17 | 25.9 | −1.58 | |
| m5-14 | 5–16.3 | 6.13 | 7.5 | −0.88 | |
| K8A10 | 5–77.7 | 7.07 | 8.9 | 0.93 | |
| Cauline leaf number | 48.1 | ||||
| F3F19 | 1–17.2 | 5.61 | 11.4 | 0.22 | |
| M1-13 | 1–78.7 | 6.69 | 13.8 | −0.31 | |
| ADH | 1–92.3 | 2.73 | 5.2 | 0.20 | |
| FRI | 4–0.8 | 5.86 | 12 | −0.24 | |
| K8A10 | 5–77.7 | 2.97 | 5.7 | 0.17 | |
| Total plant length (cm) | 81.7 | ||||
| F5I14 | 1–69.1 | 2.7 | 1.5 | −1.51 | |
| ADH | 1–92.3 | 4.99 | 2.9 | 1.70 | |
| M2-26 | 2–10 | 3.34 | 1.8 | −1.49 | |
| ER | 2–43.7 | 46.26 | 70.1 | −9.24 | |
| FRI | 4–0.8 | 4.47 | 2.5 | −1.68 | |
| SO262 | 5–27.2 | 4.98 | 2.9 | −1.74 | |
| Plant length till 1st silique (cm) | 60.6 | ||||
| CIW12 | 1–34.5 | 10.93 | 14.5 | 1.20 | |
| ER | 2–43.7 | 23.95 | 42.6 | −2.10 | |
| F8J2 | 3–58 | 3.14 | 3.5 | 0.65 | |
| Inflorescence length (cm) | 79.9 | ||||
| nga692 | 1–91.9 | 4.14 | 2.7 | 1.52 | |
| nga1145 | 2–5.3 | 2.61 | 1.7 | −1.09 | |
| ER | 2–43.7 | 43.6 | 72.4 | −7.19 | |
| SO262 | 5–27.2 | 4.59 | 3.1 | −1.49 |
Four, three, and two QTLs were found for RGR2-1, RGR3-2, and RGR3-1, respectively. The detected QTLs showed a total explained phenotypic variance of 34.3%, 18.3%, and 16.7% for RGR2-1, RGR3-2, and RGR3-1, respectively (Table III). For RGR2-1, the Sha alleles at ER, CHIB, and MBK5 and the Ler allele at msat1-10 increased plant growth rate. At the nga361, the Sha allele increased the RGR3-2 values, whereas the Ler alleles did so at the msat1-10 and nga225 loci. For RGR3-1, at two QTLs (msat1-10 and nga225), the Ler alleles increased growth rates.
At the top of chromosome 1 (msat1-10), colocation was found of the loci for TLA1, TLA2, TLA3 and for all three RGR parameters, as well as with flowering-related traits, i.e. flowering time (FT), total leaf number (TLN), cauline leaf number (CL), and plant length until first silique (PLTS). Colocation of QTLs for these different traits could also be observed at the bottom of chromosome 2 at the ER locus, at the CHIB marker near the top of chromosome 3 and at the top and bottom of chromosome 5 (Fig. 5). Colocation of these QTLs, at the top of chromosome 3, with a QTL for speed of germination (Clerkx et al., 2004) was observed, the Sha allele increasing the speed of germination.
The two detected QTLs for relative growth rate as based on dry weight (RGRdw) colocated with the QTLs for RGR calculated on the basis of plant area.
Plant Dry Weight and Relative Growth Rate
Despite the small differences in plant dry weight (DW) between Ler and Sha, large variation was found between the RILs for dry weight of young (DW1) as well as older plants (DW2; Fig. 4, D and E). Three QTLs were detected for each parameter, explaining together 25.8% and 24.1% of the total variance, respectively (Fig. 5, Table III). With the exception of msat1-10, QTLs for DW1 and DW2 were at different positions. For CHIB and SO262 only DW1 QTLs were detected, whereas ER and nga225 revealed QTLs for DW2, indicating that growth in different phases of development may be controlled by different genes but also by the same genes. In agreement with the observed transgression QTLs in which either Ler alleles increased growth (msat1-10, SO262, and nga225) and those for which the Sha alleles (CHIB and ER) lead to higher DW were found. Based on DW1 and DW2, the RGR for DW could be calculated, which revealed QTLs at ER and nga225, which are the DW2 specific QTLs. In contrast the DW2 QTL at msat1-10 is due to the growth in the first phase, which is continued.
For RGWdw a significant interaction between msat1-10 and CHIB, which represented the DW1 QTL, was found explaining 5.5% of the phenotypic variance.
As might be expected plant total area and plant dry weight, were strongly correlated (R2 = 0.61) at day 15.
Specific Leaf Area, Leaf Initiation Speed, and Chlorophyll Fluorescence
For specific leaf area (SLA) only one QTL, located near MBK5 and explaining 13.1% of the phenotypic variance, was detected (Fig. 5). This QTL, for which the Sha allele has a higher value, indicating thinner leaves, colocated with QTLs for leaf initiation speed (LIS), TLA, RGR, and FT traits.
Two QTLs controlling the speed of leaf initiation were found at the F8J2 and MBK5 (Fig. 5) explaining 7.8% and 12.8% of the phenotypic variance, respectively. For the detected QTLs, the Ler alleles increased the rate of leaf development compared with the Sha alleles. Although the QTL for LIS at MBK5 colocated with one of the flowering time QTLs, this was not the case for the QTL at F8J2. No significant correlation (R2 = 0.15) was observed between leaf number at day 24 and FT in short day (SD) indicating that a higher leaf initiation speed does not account for the major variation in flowering time.
One QTL for chlorophyll fluorescence (ChFl), also located near the MBK5 and explaining 21.4% of the phenotypic variance, was detected, which together with the interaction between MBK5 and msat4-14 (a minor QTL) explained 27.6% of the total variance (Fig. 5). For the detected QTL, the Ler allele increased the photosynthetic capacity of the plant compared with the Sha allele.
Flowering Time and Flowering-Related Traits including Plant Length and Branching
In Arabidopsis, flowering time is often correlated with the number of leaves formed prior to flowering, and one might expect that the mass of the vegetative parts may influence the elongation of the inflorescence and, therefore, may affect total plant height. Furthermore, the number of leaves determines the number of axillary buds that might develop into secondary inflorescence, thus affecting branching. Because of the expected correlation the data obtained for these parameters are discussed together. The expected relationships were indeed observed in the present material as indicated by the strong correlation between flowering time and total leaf number (TLN, R2 = 0.86), rosette leaf number (RL, R2 = 0.86), and cauline leaf number (CL, R2 = 0.61).
Flowering time differences between Ler and Sha were relatively small and the relative order of both genotypes depended on the day-length condition, Sha being slighter earlier in short day (SD), but later in long day (LD) condition (Table II). However, variation between RILs was considerable and had the same magnitude in LD and SD conditions, with a highly significant correlation (R2 = 0.71) (Fig. 4, G and J). Figure 5 and Table III summarize the QTLs found in Ler × Sha RILs for flowering time in LD and SD conditions (seven and four QTLs, respectively). Detected QTLs showed a total explained phenotypic variance of 66.6% and 52.4%, respectively. In SD, at two of the four detected QTLs, viz, at msat1-13 and FRI, Sha alleles delayed flowering, whereas Sha alleles at CHIB and K8A10 accelerated flowering. In LD these four QTLs were also detected and showed the same allelic effects. In addition two QTLs were detected in LD, at msat2-36 and msat5-14, where the Sha alleles delayed flowering and one QTL, at nga 59, where the Sha allele promoted flowering.
Seven QTLs were found for TLN, six of them colocating with FT QTLs in LD condition, with similar contributions and allelic effect (Fig. 5, Table III). At CHIB a minor QTL (at the border of significance) could be detected.
Rosette leaf number and cauline leaf number, being the two components of TLN, showed six and five QTLs, explaining 63.8% and 48.1% of variance, respectively. One QTL, specific for cauline leaf number, colocating with FT (LD) but not with RL, was found on chromosome 1, near marker F3F19. For RL, two QTLs at chromosome 2 and top of chromosome 5 colocated with FT but not with CL QTLs, indicating that although flowering time is intimately linked with number of leaves initiated before the transition to flowering, the number of elongating internodes is under separate genetic control. The remaining QTLs for RL and CL colocated with each other and with QTLs for TLN and FT (Fig. 5).
For total plant length (TPL) and its two length components (length until the first silique and inflorescence length), the total explained variance was relatively high (81.7%, 60.6%, and 79.9%, respectively), which was largely due to the effect of the ER locus, explaining 70.1%, 42.6%, and 72.4% of the observed variation. The remaining five QTLs for TPL contribute little and for all loci, except ADH, the Sha alleles increased plant length (Table III).
For PLTS and inflorescence length (INFL) fewer QTLs were detected per trait (Table III). One locus at marker CIW12 might be specific for PLTS since it did not colocate with any other QTLs for TPL or INFL. For INFL, two loci at the bottom of chromosome 1 and near the middle of chromosome 5, colocated with TPL but not with PLTS (Fig. 5), suggesting that they might only be responsible for the increase in the internode length between the flowers.
No QTLs could be detected for the number of side branches derived either from the axillary buds of the rosette leaves or the cauline leaves, which may be due to the fact that many genes with small effects segregate in this population or due to the low heritability (0.33).
DISCUSSION
Variation specifically for growth of leaves among Arabidopsis accessions as such has not been studied, in contrast to hypocotyl growth (Maloof et al., 2001; Borevitz et al., 2002; Botto and Smith, 2002) and flowering time (for review, see Koornneef et al., 2004). A study by Li et al. (1998) compared growth for a number of accessions and tried to relate growth variation with other parameters such as seed weight and geographical distribution. In addition, growth was studied in genetic analyses of nitrogen-use efficiency (Rauh et al., 2002; Loudet et al., 2003). Ungerer et al. (2002) analyzed leaf size together with developmental traits dealing with flowering and plant architecture, while Pérez-Pérez et al. (2002) used Arabidopsis natural variation for genetic analysis of leaf morphology and leaf area.
Growth-Related Traits
In this article we provide a genetic analysis of traits related to plant growth. A comparison of the more extreme phenotypes among a collection of Arabidopsis accessions showed that large differences for growth rate exist, which may be different between accessions during consecutive phases of development. Differences in biomass may result from differences in seed mass, emergence time, or variation in RGR (Van Andel and Biere, 1990; Poorter and Navas, 2003). Differences in RGR can be explained by differences in leaf area per unit plant mass (LAR; leaf area ratio) or by differences in the rate of increase in plant mass per unit leaf area (ULR; unit leaf rate; Evans, 1972). In this study we found that RGR and final dry weight were not correlated with seed weight as was described by Li et al. (1998). However, early growth, determined as rosette area, was significantly correlated with seed weight. This correlation weakened during subsequent growth, implying that other factors started to dominate growth rate. A 2-fold difference was observed for seed weight; the low latitude accessions from the Cape Verde Islands and Pakistan but also accessions from the Dutch island of Ameland had heavy seeds, disproving the negative correlation between seed size and latitude suggested by Li et al. (1998). In the latter study this correlation was based almost exclusively on the Cvi accession.
The extensive heritable variation present in natural populations is shown in the analysis of a new RIL population derived from the cross Ler × Sha, in which we studied a number of traits directly related to biomass production as well as to flowering. For most traits we detected heritable variation and QTLs could be mapped. The highest percentages of explained variation were obtained for flowering time and related traits, which have a high heritability. Less variation could be attributed to specific loci for growth-related traits and even less for parameters that were derived from two measured parameters, for which the variation of both measurements is added up. The usefulness of nondestructive growth measurements is clearly shown by the higher explained variance of leaf area than for dry weight, which is most likely due to the fact that more plants could be measured per genotype.
QTLs that were found for leaf area, dry weight, and RGR colocated in many cases, which is expected since they all measure different, but related, aspects of overall plant growth. However, in several cases no colocations were found for these growth-related traits. This indicates that some loci may have an overall effect on plant growth, whereas others specifically regulate certain processes that contribute more to some but less to other of the measured parameters, or act during a specific phase of growth. For example the QTLs on top of chromosome 3 were found mainly for the earlier phases, indicating that this QTL has a development-specific effect.
Colocation of QTLs for traits that are less obviously related might suggest pleiotropy. In case developmental changes such as flowering would be influenced by growth or vice versa, this would be reflected by colocation. Similarly, one could predict that larger late flowering plants would have longer stems. When traits have a causal relationship the allelic effects should also be in the same direction and a high overall correlation of these traits in the RIL population should be observed. Since only two out of the four FT QTLs found in SD, where growth analysis was performed, colocate with growth QTLs but have opposite allelic effects for the two traits and because the overall correlation between DW2 and FT was not significant (R2 = 0.03), we conclude that both traits are genetically different. Although a flowering time QTL is found in the ER region, we do not consider this a pleiotropic effect because the line with the ER wild-type allele in Ler background does not show this effect (data not shown).
For plant length the strongest effect is due to the ER locus, where the Sha allele promotes both growth and length. However, at the top part of chromosome 5 the QTLs for growth and total plant length colocate but the alleles act in opposite direction, which indicates that at this locus rosette growth might have a trade-off with total plant length. A weak, but significant, overall correlation was found between FT and length when the lines with mutant and wild-type ER alleles were treated separately. The highest correlation was between FT and length until the first silique, which was R2 = 0.49 for ER plants and 0.19 for er plants. The relationship between both traits is also suggested by colocations at three positions with allelic effects in the same direction (Fig. 5).
For plant growth-related traits we found five regions with QTLs (Fig. 5). The effects of the loci were never more than 2-fold. The characteristic of the QTLs around msat1-10 near the top of chromosome 1, which is called GRR1 (Growth Rate 1), is that it affects all parameters and therefore, growth as such during the vegetative phase of development. This locus might be the same as DM10.1 described by Loudet et al., (2003) in the Bay × Sha population, in which the Sha allele also has a negative effect. An interesting colocation found at this position, but not elsewhere, is between the number of cauline leaves and the length until the first silique. When the two traits are combined, this implies that the Ler allele promotes formation of cauline leaves and stimulates elongation of internodes between leaves.
The second growth-related QTL region (ER) is around the ERECTA locus and very likely the ERECTA gene itself, since the analysis of a near-isogenic line, having the wild-type ER allele in a Ler genetic background, showed similar differences with Ler for the same traits (data not shown). Interestingly, the growth effects of this locus were not detected at the earlier phases of development. As shown before for both Col × Ler and Cvi × Ler RIL populations (Alonso-Blanco et al., 1999; Pérez-Pérez et al., 2002; Ungerer et al., 2002), this locus always makes a major contribution to plant length and leaf size.
Torii et al. (1996), Yokoyama et al. (1998), and Shpak et al. (2003) provided arguments that the ER gene plays a role in coordination of cell growth patterns within the organ primordia initiated from the shoot apical meristem. The gene is predominantly expressed in the shoot apical meristems and in organ primordia. The expression is weak during early plant development but increases with the transition from the vegetative to the reproductive growth phase, in agreement with the absence of effects during the early phase of growth. Douglas et al. (2002) also showed that the ER gene influences multiple processes during Arabidopsis development, including internode and pedicle elongation, leaf and silique morphogenesis, and thickness of stem tissue.
A third locus for growth on top of chromosome 3, named GRR2, mainly affected early growth. When comparing the accessions it was noted that early plant growth correlated positively with seed weight (Fig. 1A). However, in the Ler × Sha RIL population, the GRR2 locus affected early growth, but not seed weight. The finding of a QTL for speed of germination at that position (Clerkx et al., 2004) may suggest the cause of this early growth to be related to seed vigor, giving plants a faster start. Loudet et al. (2003) found a QTL for dry mass, which they named DM3.2 at the same position, for which the Sha allele also increases growth. This effect is rather small and was only observed when plants were grown at low nitrogen (3 mm) conditions (Loudet et al., 2003).
The locus near nga139 on top of chromosome 5 (GRR3) has not been described in other populations. It might actually consist of two loci that did not show up as significant in all analyses. A locus on top of this chromosome was described as DM10.7 by Loudet et al. (2003).
Probably the most interesting new QTL region is at the bottom of chromosome 5 (GRR4), where possibly two QTLs are located. Besides QTLs for growth rate and FT, also loci affecting LIS, SLA, and ChFl were found in this region, the latter two not being found in the other regions. Interestingly a higher rate of leaf initiation due to the Ler allele coincided with smaller leaves and lower growth rate, suggesting that the leaves that are formed are smaller and also thinner as indicated by the reduction of SLA by the Ler allele. The effect on chlorophyll fluorescence suggests that the physiology of these leaves is also different.
Flowering Time
In this study we have analyzed the flowering behavior of two early Arabidopsis accessions. They differed slightly in their flowering phenotype (measured as both FT and TLN) and in their response to photoperiod length. However, variation between segregating RILs derived from crosses between these two accessions showed a large variation as shown also in other crosses, viz, between Ler and Cvi (Alonso-Blanco et al., 1998) and between Bay-0 and Sha (Loudet et al., 2002), and larger than that between Ler and Col (Jansen et al., 1995). The significant correlation between flowering time in SD and in LD conditions (R2 = 0.71) indicates that flowering time in both conditions is predominantly controlled by the same genetic factors in the Ler × Sha RILs. The flowering behavior differences between the Ler × Sha lines in both LD and SD conditions can be mainly attributed to QTLs located at msat1-13, CHIB, FRI, and K8A10. Ler alleles at CHIB and K8A10 result in lateness, while at msat1-13 and FRI Ler alleles are earlier, thus explaining the similar behavior of the parental lines and the transgression in the RILs. Three other loci that were found in LD only are located at nga59, msat2-36, and msat5-14, where Ler alleles at the first locus give lateness while for the other two loci the Ler alleles lead to earliness. Previously it was shown that the FRI and the FLC loci determine flowering time differences between very late, vernalization-responsive accessions and early ones (Johanson et al., 2000; Gazzani et al., 2003; Michaels et al., 2003). Sequence analysis has shown that Sha contains a wild-type FRI gene (Gazzani et al., 2003; Michaels et al., 2003) in contrast to Ler (Johanson et al., 2000) making it most likely that the FRI locus is the gene for this QTL. This is further supported by the flowering time QTL, detected by Loudet et al. (2002) in the Sha × Bay-0 population. We could not find any QTL at the FLC locus, probably because both the Sha and Ler parents carry weak alleles at this locus (Koornneef et al., 1994; Loudet et al., 2002; Gazzani et al., 2003; Michaels et al., 2003). The Sha accession was slightly less sensitive to changes in photoperiod length compared to Ler. Three of the detected QTLs might be specific to day length and, therefore, affect day length sensitivity, viz, at ADH (not significant for FT but detected for TLN) and at msat2-36, in LD condition only, and at CIW7 (minor QTL for FT) in SD condition. The first two QTLs might be the same found to be specific for photoperiod in the Bay-0 × Sha population in SD and LD, respectively (Loudet et al., 2002). Some of the flowering-related QTLs that we found in the Ler × Sha population colocalized with previously published QTLs detected in other populations (Kowalski et al., 1994; Clarke et al., 1995; Jansen et al., 1995; Kuittinen et al., 1997; Alonso-Blanco et al., 1998; Loudet et al., 2002).
Concluding Remarks
Screening a number of Arabidopsis accessions revealed different patterns for growth. In this study we could identify a number of QTLs affecting plant growth. These loci appear to have different physiological functions, as concluded from colocations of QTLs for different traits. Especially the GRR4 locus near marker MBK5 looks very interesting because it affects a plethora of physiological effects including speed of leaf initiation, specific leaf area, and chlorophyll fluorescence. However, it should be emphasized that due to the inaccuracy of QTL mapping in a population of this size, it cannot be excluded that independent but linked genes control these apparent pleiotropic effects. This should further be investigated by fine mapping, which is most effectively done when no other QTLs segregate, i.e. using near-isogenic lines (NILs, see Alonso-Blanco and Koornneef, 2000). In addition the loci GRR1 and 3, which might be related to nitrogen-use efficiency (Loudet et al., 2003) deserve further study. The GRR3 QTL has the intriguing property that it affects early seedling growth. Because of the complexity of comprehensive traits like growth we cannot speculate on candidates for the QTLs, except for the ERECTA locus for which isogenic lines prove the involvement of the ERECTA gene. For flowering time several previously detected QTLs were found as well as a few new ones. Natural allelic variation for FRI and FLC is already studied at the molecular level (Johanson et al., 2000; Le Corre et al., 2002; Gazzani et al., 2003; Michaels et al., 2003). This may indicate the direction for future research, aiming to understand causes and consequences of natural genetic variation.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The seeds from different accessions were sown in petri dishes on water-saturated filter paper, followed by a 4-d cold treatment at 4°C, and then transferred to a climate room at 25°C and 16 h light for 2 d before planting in 7-cm pots with standard soil. In all descriptions of experiments, time is referred to as days after planting. Details of the selected 22 accessions are given in Table I. These accessions (24 plants/accession) were grown under controlled conditions in a growth cabinet, with 70% relative humidity, 22°C, 12-h day length and light intensity 25 Wm−2, for a detailed growth analysis. Plants were placed on carts, and the carts were shuffled daily to avoid minor environmental differences within the growth cabinet.
F9 plants of a new set of 114 RILs, obtained by single-seed descent of F2 plants derived from the cross Ler × Sha, were analyzed for flowering time and growth-related traits in two different experiments. The first one was carried out in an air-conditioned green house supplemented with additional light (model SON-T plus 400W, Philips, Eindhoven, The Netherlands) providing a day length of at least 16 h light which is a long day (LD), and maintained at a temperature between 22°C and 25°C (day) and 18°C (night). The second one was carried out in a growth cabinet under 12 h light, which is a mild short day (SD) treatment for Arabidopsis. In the greenhouse experiment 12 plants/RIL were grown in the same conditions as mentioned before (LD), in a randomized two-block design to reduce environmental effects, while 10 plants/RIL were grown in the growth cabinet, in the same conditions as mentioned above, also in a randomized two-block design. A line with the ERECTA wild-type allele in the Ler genetic background, the two reciprocal hybrids, and both parents were included in all experiments.
Digital Imaging, Computer Analysis, and RGR Determination
The mean total leaf area (TLA) of each accession was obtained by imaging 20 to 24 plants per accession at 10 (TLA1), 15 (TLA2), and 20 (TLA3) d after transferring the seedlings to the pots. Leaf areas were determined with an image processing technique, using a Nikon digital camera (model COOLPIX 950; Nikon Corporation Imaging Products Division, Shinagawa-Ku, Tokyo), and analysis of the pictures using the computer program MetaMorph (version 4.01; Universal Imaging Corporation, West Chester, PA, www.image1.com). The mean TLA for each line of the 114 RILs was obtained by imaging five plants/line at day 10 (TLA1) and four plant/line at 15 (TLA2) and 20 (TLA3) d. The relative growth rate (RGR) was calculated according to the following equation: (lnAx − lnAy)/dt(x−y). RGR was calculated for each line based on the three measurements of rosette area, resulting in RGR2-1, RGR3-2, and RGR 3-1, referring to RGRs in the intervals 10 to 15, 15 to 20, and 10 to 20 d, respectively.
Weight, Water Content, and SLA Determinations
The mean seed weight (SW) for each accession was obtained by weighing two seed lots each of 100 seeds using a Perkin-Elmer microbalance (model AD-4 Autobalance, Boston). SW for each line of the 114 RILs was determined for one batch per line.
The mean fresh weight (FW) of the plants was determined at day 35 by harvesting and weighing the aboveground parts of two plants/accession. The mean FW for each RIL was determined at day 15 and 25, by harvesting and weighing two plants/line, one from each block. Dry weights (DW) were determined after drying the plants at 105°C for 48 h, and the water content (WC) was estimated as the relative ratio between water and dry weight using the formula [(FW-DW)/FW] × 100. The relative growth rate as based on dry weight (RGRdw) was calculated in the same way as RGR based on leaf area.
The specific leaf area (SLA) was calculated as area divided by weight (mm2 mg−1). The relation between the 22 accessions based on seed weight, fresh and dry weights, and areas at 10, 15, and 20 d was described with principle component analysis using NTSYSpc version 2.10t. (Rohlf, 2001) with standardized data, which were converted in a correlation matrix from which three eigenvectors were extracted using the EIGEN function of the NTSYS-pc program.
Chlorophyll Fluorescence Measurements
Chlorophyll fluorescence as a nondestructive means of photosynthetic capacity was measured using a MINI-PAM (S/N: 0133; WALZ Mess- und Regeltechnik, Effeltrich, Germany), with the determination of the effective quantum yield of photosynthetic energy conversion (Yield = Δ F/Fm′).
Measurement of Flowering Time and Related Traits in RILs
From the greenhouse experiment, in which 12 plants/RIL were grown in LD condition, FT for each plant was recorded as the number of days from planting until the opening of the first flower. Flowering time was also scored by counting the TLN, i.e. RL plus CL, excluding the cotyledons, since there is a close correlation between leaf number and flowering time (Koornneef et al., 1991). The following traits were also recorded: TPL, PLTS, and total number of side shoots or inflorescences (TSSN) (number of branches in the main inflorescence plus the number of side shoots from the rosette). In the phytotron experiment (SD), flowering time was scored as described above, for six plants/line. In addition, the number of rosette leaves was counted at day 24 from planting (i.e. before flowering) which refers to the leaf initiation speed (LIS).
Genetic Mapping
The mapping of the segregating population was done by using 66 molecular markers, including the morphological marker erecta, located at a distance from 1 to 15 cm on the genetic map to obtain a regular distribution among the five chromosomes. These markers were used to generate the linkage map; details are published elsewhere (Clerkx et al., 2004). This map was used for QTL analysis of the various traits.
Statistical Analysis and QTL Mapping
For each RIL, the mean value of the traits under investigation was (log10) transformed to improve normality of the distribution, except for the relative growth rates, rosette areas, and the specific leaf area. Transformed data were used for QTL analysis. The software package MapQTL version 4.0 (van Ooijen, 2000) was used to identify and locate QTL on the linkage map by using interval mapping and multiple-QTL model (MQM) mapping methods as described in its reference manual (http://www.plant.wageningen-ur.nl/products/). In a first step, putative QTLs were identified using interval mapping. Thereafter, the closest marker at each putative QTL was selected as a cofactor, and the selected markers were used as genetic background controls in the approximate multiple QTL model of MapQTL. Log of the odds (LOD) threshold values applied to declare the presence of a QTLs were estimated by performing the permutation tests implemented in MapQTL version 4.0 using at least 1,000 permutations of the original data set for each trait, resulting in a 95% LOD threshold between 2.4 and 2.6.
Two-LOD support intervals were established as 95% QTL confidence interval (van Ooijen, 1992). The estimated additive genetic effect and the percentage of variance explained by each QTL and the total variance explained by all the QTLs affecting a trait, were obtained using restricted MQM Mapping implemented with MapQTL.
Two-way interactions among the QTL identified for each trait were tested by ANOVA using the corresponding two markers as fixed factors and the trait as dependent variable, using the general linear model of the statistical package SPSS version 11.5.0. A Bonferroni correction to adjust the 0.05 threshold of significance was applied if multiple tests were performed on the same data set. Only those interactions that were significant after the Bonferroni correction are presented.
Heritabilities were calculated based on measurements on 6 to 12 plants.
This work was supported by a grant to M.E.E.-L. from the Ministry of Higher Education, Egyptian Government; by the Technology Foundation STW, Applied Science Division of NWO (project no. STW WBI4737 to E.J.M.C.); and by EU-Natural (contract no. QLG2–CT–2001–01097 to G.J.R.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.036822.
References
- Alonso-Blanco C, Blankenstijn-de Vries H, Hanhart CJ, Koornneef M (1999) Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 4710–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, El-Assal SED, Coupland G, Koornneef M (1998) Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics 149: 749–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Koornneef M (2000) Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends Plant Sci 5: 22–29 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Maloof JN, Lutes J, Dabi T, Redfern JL, Trainer GT, Werner JD, Asami T, Berry CC, Weigel D, et al. (2002) Quantitative trait loci controlling light and hormone response in two accessions of Arabidopsis thaliana. Genetics 160: 683–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto JF, Smith HG (2002) Differential genetic variation in adaptive strategies to a common environmental signal in Arabidopsis accessions; phytochrome-mediated shade avoidance. Plant Cell Environ 25: 53–63 [Google Scholar]
- Clarke JH, Mithen R, Brown JKM, Dean C (1995) QTL analysis of flowering time in Arabidopsis thaliana. Mol Gen Genet 248: 278–286 [DOI] [PubMed] [Google Scholar]
- Clerkx EJM, El-Lithy ME, Vierling E, Ruys GJ, Blankenstijn-de Vries H, Groot SPC, Vreugdenhil D, Koornneef M (2004) Analysis of natural allelic variation of Arabidopsis seed quality traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred population. Plant Physiol 135: 432–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerge RW (2002) Mapping and analysis of quantitative trait loci in experimental populations. Nat Rev Genet 3: 43–52 [DOI] [PubMed] [Google Scholar]
- Douglas SJ, Chuck G, Dengler RE, Pelecanda L, Riggs CD (2002) KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell 14: 547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans GC (1972) The Quantitative Analysis of Plant Growth. Blackwell Scientific Publications, Oxford
- Gazzani S, Gendall AR, Lister C, Dean C (2003) Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol 132: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grime JP, Hunt R (1975) Relative growth-rate: its range and adaptive significance in a local flora. J Ecol 63: 393–422 [Google Scholar]
- Jansen RC, van Ooijen JW, Stam P, Lister C, Dean C (1995) Genotype by environment interaction in genetic mapping of multiple quantitative trait loci. Theor Appl Genet 91: 33–37 [DOI] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Khurmatov KK (1982) Heterogeneity of natural populations of the Arabidopsis thaliana (Pamiro-Alay) in the flowering time. Arabid Inf Serv 19: 62–66 [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D (2004) Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol 55: 141–172 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Blankenstijn-de Vries H, Hanhart CJ, Soppe W, Peeters AJM (1994) The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild-type. Plant J 6: 911–919 [Google Scholar]
- Koornneef M, Hanhart CJ, Van der Veen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Kowalski SP, Lan TH, Feldmann KA, Paterson AH (1994) QTL mapping of naturally-occurring variation in flowering time of Arabidopsis thaliana. Mol Gen Genet 245: 548–555 [DOI] [PubMed] [Google Scholar]
- Kuittinen H, Sillanpää MJ, Savolainen O (1997) Genetic basis of adaptation: flowering time in Arabidopsis thaliana. Theor Appl Genet 95: 573–583 [Google Scholar]
- Le Corre V, Roux F, Reboud X (2002) DNA polymorphism at the FRIGIDA gene in Arabidopsis thaliana: extensive nonsynonymous variation is consistent with local selection for flowering time. Mol Biol Evol 19: 1261–1271 [DOI] [PubMed] [Google Scholar]
- Leister D, Varotto C, Pesaresi P, Niwergall A, Salamini F (1999) Large-scale evaluation of plant growth in Arabidopsis thaliana by non-invasive image analysis. Plant Physiol Biochem 37: 671–678 [Google Scholar]
- Li B, Suzuki J, Hara T (1998) Latitudinal variation in plant size and relative growth rate in Arabidopsis thaliana. Oecologia 115: 293–301 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F (2002) Bay-0 × Shahdara recombinant inbred line population: a powerful tool for genetic dissection of complex traits in Arabidopsis. Theor Appl Genet 104: 1173–1184 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Merigout P, Talbotec J, Daniel-Vedele F (2003) Quantitative trait loci analysis of nitrogen use efficiency in Arabidopsis. Plant Physiol 131: 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloof JN (2003) QTL for growth and morphology. Curr Opin Biotechnol 6: 85–90 [DOI] [PubMed] [Google Scholar]
- Maloof JN, Borevitz JO, Dabi T, Lutes J, Nehring RB, Redfern JL, Trainer GT, Wilson JM, Asami T, Berry CC, et al. (2001) Natural variation in light sensitivity of Arabidopsis. Nat Genet 29: 441–446 [DOI] [PubMed] [Google Scholar]
- McGraw JB, Garbutt K (1990) The analysis of plant growth in ecological and evolutionary studies. Trends Ecol Evol 5: 251–254 [DOI] [PubMed] [Google Scholar]
- Michaels SD, He Y, Scortecci KC, Amasino RM (2003) Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc Natl Acad Sci USA 100: 10102–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motooka S, Hayashi T, Mima Y, Konishi K (1991) Measurement of in-vitro plant growth by image processing. J Jpn Soc Hortic Sci 60: 677–684 [Google Scholar]
- Pérez-Pérez JM, Serrano-Cartagena J, Micol JL (2002) Genetic analysis of natural variations in the architecture of Arabidopsis thaliana vegetative leaves. Genetics 162: 893–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poorter H, Navas ML (2003) Plant growth and competition at elevated CO2: on winners, losers and functional groups. New Phytol 157: 175–198 [DOI] [PubMed] [Google Scholar]
- Poorter H, Remkes C (1990) Leaf area ratio and net assimilation rate of 24 wild species differing in relative growth rate. Oecologia 83: 553–559 [DOI] [PubMed] [Google Scholar]
- Rauh L, Basten C, Buckler S (2002) Quantitative trait loci analysis of growth response to varying nitrogen sources in Arabidopsis thaliana. Theor Appl Genet 104: 743–750 [DOI] [PubMed] [Google Scholar]
- Rédei GP (1992) A heuristic glance to the past of Arabidopsis genetics. In C Koncz, N Chua, J Schell, eds, Methods in Arabidopsis Research. World Scientific, Singapore, 1–15
- Rohlf FJ (2001) NTSYSpc: Numerical Taxonomy and Multivariate Analysis System, Version 2.10x. Exeter Software, Setauket, NY
- Shpak ED, Lakeman MB, Torii KU (2003) Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell 15: 1095–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Spomer LA (1987) Direct quantification of in vitro cell growth through image analysis. In Vitro Cell Dev Biol 23: 67–74 [DOI] [PubMed] [Google Scholar]
- Smith MAL, Spomer LA, Meyer MJ, McClelland MT (1989) Non-invasive image analysis evaluation of growth during plant micropropagation. Plant Cell Tissue Organ Cult 19: 91–102 [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y (1996) The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer MC, Halldorsdottir SS, Modliszewski JL, Mackay TFC, Purugganan MD (2002) Quantitative trait loci for inflorescence development in Arabidopsis thaliana. Genetics 160: 1133–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Andel J, Biere A (1990) Ecological significance of variability in growth rate and plant productivity. In H Lambers, ML Cambridge, H Konings, TL Pons, eds, Causes and Consequences of Variation in Growth Rate and Productivity of Higher Plants. SPB Publishing, The Hague, The Netherlands, 257-267
- Van Ooijen JW (1992) Accuracy of mapping quantitative trait loci in autogamous species. Theor Appl Genet 84: 803–811 [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW (2000) MapQTL (R) Version 4.0: Userfriendly Power in QTL Mapping; Addendum to the Manual of Version 3.0. Plant Research International, Wageningen, The Netherlands
- Yokoyama R, Takahashi T, Kato A, Torii KU, Komeda Y (1998) The Arabidopsis ERECTA gene is expressed in the shoot apical meristem and organ primordia. Plant J 15: 301–310 [DOI] [PubMed] [Google Scholar]