Abstract
Bread wheat (Triticum aestivum) is an allohexaploid species, consisting of three subgenomes (A, B, and D). To study the molecular evolution of these closely related genomes, we compared the sequence of a 307-kb physical contig covering the high molecular weight (HMW)-glutenin locus from the A genome of durum wheat (Triticum turgidum, AABB) with the orthologous regions from the B genome of the same wheat and the D genome of the diploid wheat Aegilops tauschii (Anderson et al., 2003; Kong et al., 2004). Although gene colinearity appears to be retained, four out of six genes including the two paralogous HMW-glutenin genes are disrupted in the orthologous region of the A genome. Mechanisms involved in gene disruption in the A genome include retroelement insertions, sequence deletions, and mutations causing in-frame stop codons in the coding sequences. Comparative sequence analysis also revealed that sequences in the colinear intergenic regions of these different genomes were generally not conserved. The rapid genome evolution in these regions is attributable mainly to the large number of retrotransposon insertions that occurred after the divergence of the three wheat genomes. Our comparative studies indicate that the B genome diverged prior to the separation of the A and D genomes. Furthermore, sequence comparison of two distinct types of allelic variations at the HMW-glutenin loci in the A genomes of different hexaploid wheat cultivars with the A genome locus of durum wheat indicates that hexaploid wheat may have more than one tetraploid ancestor.
Polyploidy has played a principal role in the evolutionary history of higher plants, with approximately 70% of flowering plants being polyploid (Masterson, 1994). Included are some of the world's most important crops, such as bread wheat, cotton, and potato. Polyploids are divided into autopolyploids and allopolypoids, depending on the nature of polyploid formation. Autopolyploids are generated from the doubling of a single species' genome, whereas allopolyploids contain two or more sets of related chromosomes that are brought together into the same nucleus, usually by interspecific fertilization, followed by chromosomal doubling. As a result of genome polyploidization, new species are created, which may show different adaptive responses to the changing environment (Wendel, 2000).
Bread wheat (Triticum aestivum) is an allohexaploid species, consisting of three sets of subgenomes (A, B, and D), each of which contains seven pairs of homoeologous chromosomes. Hexaploid wheat is one of the best-characterized examples of genome evolution through the process of allopolyploidization. The identity and evolution of hexaploid wheat's ancestral genomes have been studied extensively (Kimber and Sears, 1987; Feldman et al., 1995). Hexaploid bread wheat originated from the hybridization of a tetraploid wheat (Triticum turdigum, AABB) with a diploid progenitor, Aegilops tauschii (DD). The tetraploid wheat was derived from the hybridization of two diploid progenitors, Triticum urarta (AA) and an unconfirmed species (BB genome) related to Aegilops speltoides (SS). Recent phylogenetic studies of two plastid genes of Triticum and Aegilops species further confirmed that Triticum urartu is the A genome donor of tetraploid and hexaploid wheats and that A. tauschii is the D genome donor of hexaploid wheat. The origin of the B genome remains elusive (Huang et al., 2002).
Wheat belongs to the grass family Poaceae, which encompasses approximately 8,700 species from approximately 650 genera (Judd et al., 1999). Comparative genetic mapping in plants has provided evidence for a remarkable conservation of marker and gene order (colinearity) between related grass genomes (for reviews, see Bennetzen 2000; Devos and Gale, 2000; Schoen, 2000) and has offered the potential for the map-based cloning of agronomic traits from plant species with large genomes, such as barley (Hordeum vulgare) and wheat (Laurie and Devos, 2002; Yan et al., 2003). However, comparative mapping analyses are limited in their ability to uncover many of the important mechanisms involved in genome evolution, such as local small sequence rearrangements (like inversions and tandem duplications) and gene inactivation by point mutations and small deletions. On the other hand, comparative sequence analyses provide detailed information on the composition and organization of genomes, as well as reveal subtle forms of conservation and divergence. Recent studies have demonstrated that while gene colinearity exists among grass genomes, there are many exceptions. In addition, it has been shown that sequences in the colinear intergenic regions, which are mainly retrotransposable elements, are not conserved (for review, see Bennetzen, 2000; Gaut, 2002; Feuillet and Keller, 2002). Although retrotransposable elements are ubiquitous in plants, the number of these elements is directly correlated with the genome size (Kidwell, 2002). In many cases, retrotransposons comprise over 50% of nuclear DNA content. Retrotransposons replicate via an mRNA intermediate that is reverse transcribed into extrachromosomal DNA and integrated somewhere else in the genome (Boeke, 1989). Therefore, retrotransposons can generate mutations by inserting within or near genes. These mutations are generally stable; because they transpose via replication, the sequence at the insertion site is retained (Kumar and Bennetzen, 1999).
To date, comparative sequence studies have focused on grass genomes of distantly related species. In addition, the majority of these studies have used sequences from diploid genomes for their comparisons. Hence, the interaction of genomes in polyploid species has not been adequately addressed. A comparison of the wheat genomes provides an excellent opportunity to study genome polyploidization that has occurred in recent evolutionary history. The wheat A, B, and D genomes are estimated to have diverged between 2.5 and 4.5 million years ago (MYA) from a common ancestor (Huang et al., 2002). In comparison, the tetraploid and hexaploid wheats are young polyploids; T. turgidum evolved roughly 400,000 years ago, whereas T. aestivum has only existed for 8,000 years (Nesbitt and Samuel, 1996). Recently, several studies using amplified fragment length polyphorphisms (AFLP) have shown allopolyploidization-induced rapid genome evolution. This includes the elimination of single copy DNA, activation and silencing of specific genes, and reactivation of retrotransposons (Ozkan et al., 2001; Kashkush et al., 2002; He et al., 2003). A detailed sequence comparison of the genomes in polyploids and their diploid ancestors will allow for a better understanding of the mechanisms controlling these evolutionary events during polyploidization.
The wheat high Mr (HMW) glutenin genes are among the best studied wheat genes, primarily because they encode the wheat gluten proteins, which are critical to the physical and biochemical properties of wheat doughs (Shewry et al., 1992). Genes encoding the HMW-glutenin subunits are located at the three Glu-1 loci (Glu-1A, Glu-1B, and Glu-1D) on the long arms of the three homoeologous group 1 chromosomes of hexaploid wheat. Each locus contains two paralogous HMW-glutenin genes encoding x- and y-type HMW-glutenin subunits. We have focused our study on the characterization of the HMW-glutenin loci from polyploid wheat and its diploid ancestors in an effort to elucidate the evolution of polyploid wheat and to further understand the molecular basis of wheat dough quality. Previously, we reported the sequences of the HMW-glutenin regions of the diploid A. tauschii d-genome and tetraploid T. turgidum B genome (Anderson et al., 2003; Kong et al., 2004), two of the ancestors of hexaploid bread wheat. In addition, we sequenced the orthologous d-hordein region from the H genome of barley, which is closely related to wheat (Gu et al., 2003). In this study, we report the sequence of a 307-kb colinear region of the T. turgidum A genome. The comparative analyses of these orthologous regions provided the first view of sequence divergence on a large scale in the wheat A, B, and D genomes, and enhanced our understanding of their evolutionary relationships.
RESULTS
Gene Content and Order in the Glu-A1 Region
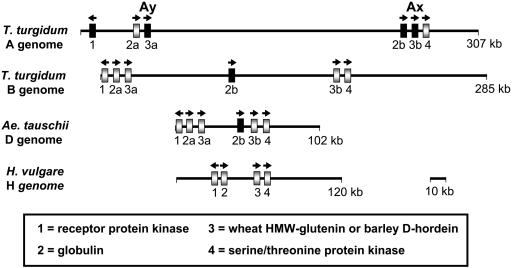
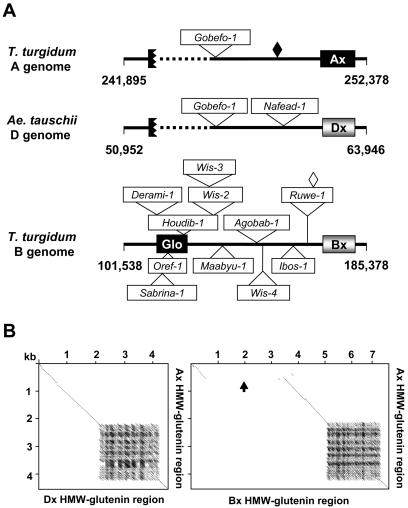
Previous sequence analyses of BAC clones containing the HMW-glutenin loci from the D genome of A. tauschii, the B genome of T. turgidum, and the barley d-hordein region revealed a general conservation of gene content and order in these orthologous regions. Six genes were identified in the wheat genomes, including duplicated sets of globulin and HMW-glutenin genes (Gu et al., 2003; Kong et al., 2004). The order of these six genes is: a Leu-rich repeat receptor kinase, a globulin, a y-type HMW-glutenin, a duplicate globulin, an x-type HMW-glutenin, and a Ser/Thr protein kinase. The duplication of the two storage protein genes is not present in the orthologous region of the barley genome (Gu et al., 2003). In the 307-kb region of the tetraploid A genome, sequences matching these six genes are present and their order is conserved as compared with those in the orthologous regions of the B and D genomes (Fig. 1; Anderson et al., 2003; Kong et al., 2004). Further analysis revealed that four of the six genes—the Leu-rich repeat receptor kinase, both copies of HMW-glutenin genes, and the second duplicated globulin genes—are inactive or disrupted in the A genome. Mechanisms causing inactivation of these genes may include retroelement insertions, coding sequence deletions, and mutations in these gene sequences (see detailed discussion below).
Figure 1.
Comparison of gene density and content in the HMW-glutenin regions among different wheat and barley genomes. Genes are represented by numbered boxes, with their corresponding names listed below. a and b denote distinct copies of the duplicated HMW-glutenin and globulin genes. Arrows indicate the direction of potential transcription of these genes. Black boxes represent inactive genes or pseudogenes. A 10-kb segment is provided as a scale reference.
The first of the two intact genes from the 5′ end in the contiguous 307-kb sequence region is a globulin gene (position 47,272–47,961). A BLASTN search of this globulin gene against the wheat expressed sequence tag (EST) database identified more than 100 matches with probability values less than e−100. These ESTs are present only in hexaploid wheat cDNA libraries constructed from endosperm tissues and whole seeds at different developmental stages, supporting an activity of the globulin gene during seed development. ESTs closely matching the globulin gene were assembled into three contigs, indicating three homoeologs from three hexaploid wheat genomes (data not shown). The globulin coding sequence identified in the tetraploid A genome differs from one of these contigs at only one base position. The other two contigs closely match the globulin genes from either the A. tauschii D (Anderson et al., 2003) or T. turgidum B (Kong et al., 2004) genomes, with sequence identities over 99%, indicating that the three globulin gene homeologs, one from each genome in hexaploid wheat, are active in the endosperm.
A Ser/Thr protein kinase gene at position 252,172 to 256,179 also appears intact in the sequenced A genome region. Although no matching ESTs were found in the wheat EST collection, this gene seems to be conserved since gene finding programs predict the same exons and the same exon/intron boundaries for the orthologous genes from other wheat and barley genomes (not shown). Whether this conserved protein kinase is expressed and has a biological function remains unknown.
Gene Inactivation in the A Genome of the Durum Wheat
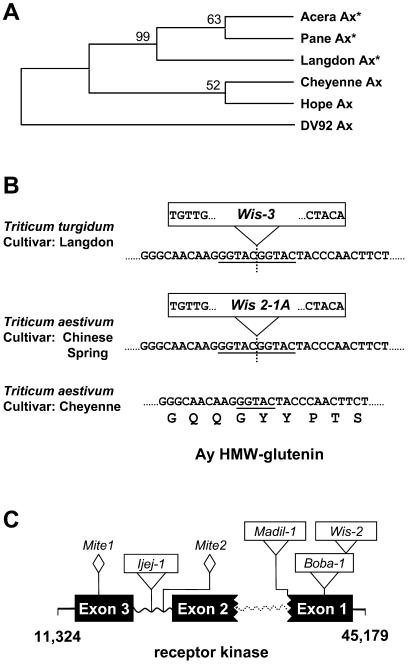
In the sequenced Glu-A1 locus of the durum wheat, both paralogous HMW-glutenins are inactive. In the x-type HMW-glutenin gene (Ax), a point mutation has changed a Gln codon (CAA) in one of the repeat domains of the HMW-glutenin subunit to an in-frame stop codon (TAA). More than 40 HMW-glutenin gene sequences isolated from different wheat species or cultivars are present in GenBank, including a complete set of the six HMW-glutenin genes from the cultivar Cheyenne, which is known for its high breadmaking quality (Anderson et al., 2002). When the nucleotide sequence of the Ax HMW-glutenin gene from durum wheat was used in a BLAST search against the National Center for Biotechnology Information database, the sequences from the top two matches (AF145590 from cultivar Pane-247, U19774 from cultivar Acera) were both found to contain the point mutation that produced the in-frame stop codon. Moreover, except for a 15-bp deletion at the 3′ end, the tetraploid Ax HMW-glutenin differs from these two hexaploid Ax HMW-glutenin genes by only two bases. The Ax HMW-glutenin genes are known to be expressed in some wheat cultivars, such as Cheyenne and Hope (Anderson and Greene, 1989; Halford et al., 1992). A comparison of the sequences of the inactive Ax HMW-glutenin genes with those of the expressed Ax HMW-glutenin genes showed that the inactive genes had five conserved base substitutions, including the in-frame stop codon (data not shown). These results suggest that the A genome from this durum wheat shares an evolutionary lineage with the A genomes from the hexaploid wheats Pane and Acera, and that the A genomes in Cheyenne and Hope were derived from a tetraploid ancestor that had diverged from the durum wheat. To further test the lineage of the A genome in hexaploid wheat cultivars, a phylogenic analysis was conducted based on sequences of the Ax HMW-glutenin coding region. The expressed and inactivated Ax HMW-glutenins are grouped in two clades (Fig. 2A). Further analysis of HMW-glutenin sequences from more wheat species will enrich our understanding of these evolutionary relationships.
Figure 2.
Gene disruption in the Glu-A1 region. A, Phylogenic analysis on Ax HMW-glutenin coding sequences from different wheats. The phylogenic tree was built by unweighted pair-group method with arithmetic mean (UPGMA) using MEGA2 software (available at www.megasoftware.net). Cheyenne, Pane, Acera, and Hope are hexaploid wheats (T. aestivum), Langdon is the tetraploid wheat (T. turgidum) analyzed in this paper, and DV92 is a diploid wheat (T. monococcum). The Ax HMW-glutenin gene from the diploid wheat was used as a outgroup. Bootstrap values are indicated. The inactive Ax HMW-glutenin genes are labeled with stars. B, Insertion of WIS element in the Ay HMW-glutenin gene. Portions of the Ay HMW-glutenin nucleotide sequences from different wheat species or cultivars are provided. The position of the Wis-3 element insertion is indicated by a vertical dashed line. The 5-bp inverted repeat sequences of the LTRs are shown in the boxed Wis retroelements. The 5-bp region that was used as a target site duplication (TSD) is underlined in the sequences. The repeat motif encoded by the given nucleotides is provided for the Cheyenne cultivar. C, Disruption of the Leu-rich receptor kinase gene. Three exons, indicated by black boxes, were predicted based on comparison with the colinear genes in the wheat D and B genomes (Anderson et al., 2003; Kong et al., 2004). The two boxes with broken borders symbolize exons that are incomplete due to a deletion event. Wavy lines represent intron regions. The dashed wavy line indicates a deleted intron. The insertion positions of retroelements are indicated.
The Ay HMW-glutenin gene in the Glu-A1 locus is disrupted by an insertion of a WIS element, Wis-3, in the repetitive domain region (Fig. 2B). A 5-bp (GGTAC) target site duplication sequence (TSD) was identified on either side of the full-length Wis-3 element. Previously, it was reported that the y-type HMW-glutenin of the A genome in the hexaploid wheat Chinese Spring is also silenced due to an insertion of a Wis retroelement (Harberd et al., 1987). A comparison of the Wis element insertions in these tetraploid and hexaploid A genomes revealed that the two Wis elements have the same insertion site, and are flanked by the same 5-bp TSD (Fig. 2B). The two retrotransposons share a 99.8% sequence identity. In addition, the sequences of the y-type HMW-glutenin from the tetraploid and hexaploid A genomes are 99% identical.
Although the Ay HMW-glutenins are always inactive in hexaploid bread wheat (Shewry et al., 1992), the mechanisms causing the inactivation vary in different cultivars. The sequence of the Ay HMW-glutenin gene from the hexaploid wheat Cheyenne does not contain the Wis retroelement insertion found in the Chinese Spring and durum wheat A genomes (Fig. 2B). Instead, there is a point mutation causing an in-frame stop codon (Forde et al., 1985). Further studies indicated that this Ay HMW-glutenin gene also contained an inactive promoter (Halford et al., 1989). It is highly unlikely that the absence of the Wis retrotransposon in cultivar Cheyenne resulted from a deletion event that exactly removed the retroelement sequence plus only one of the two 5-bp TSDs. Taken together, our results suggest that the A genome in hexaploid bread wheat has two distinct allelic variations. One is represented by the Chinese Spring A genome, whose ancestor has the same lineage as the A genome from the durum wheat in this study. The other is represented by the A genome in Cheyenne. The tetraploid ancestor contributing to the allelic variation of the A genome in Cheyenne remains to be determined.
The Leu-rich receptor kinase gene is also disrupted by repetitive DNA insertion events. As shown in Figure 2C, there are six mobile elements (three long terminal repeat [LTR] retrotransposons, one class II transposon, and two MITE insertions) present in this gene region. Two elements, Mite2 and class II transposon Ijeij-1, were inserted in the second intron regions, as revealed by comparison with the intact receptor kinase gene in the B genome (Kong et al., 2004). The other four elements are located in the coding regions. While only a single MITE (Mite1) was found in the third exon region, three LTR retrotransposons (Madil-1, Boba-1, and Wis-2) were discovered in the first exon region. The insertion of Madil-1 appears to have been accompanied by a deletion event; a region spanning the entire first intron plus the flanking coding sequences is missing at the insertion site in the receptor kinase gene of the A genome. In spite of this 3-kb deletion, the remaining receptor kinase sequence still spans a 36-kb region due to the retroelement insertions. Because of the multiple insertions of retroelements and sequence deletions in the receptor kinase region, it would be very difficult to determine the original gene structure without performing the comparative analysis with the corresponding regions of the B and D genomes (Anderson et al., 2003; Kong et al., 2004). The orthologues of the Leu-rich receptor kinase gene in the wheat D, B, and barley genomes are likely to be active since disruptions to these genes were not detected and transcripts for the barley gene are present in the EST database (Gu et al., 2003).
One common phenomenon in genome evolution is the deletion of genomic sequences, which often results in gene inactivation if it occurs in the promoter or coding sequence regions (Bennetzen and Ramakrishna, 2002). Due to such deletion events, the duplicate globulin gene at position 242,338 to 243,392 has lost most of the coding sequence as well as the 3′ untranslated region. In addition, the start codon (ATG) has been mutated to an Ile codon (ATA). Only the promoter sequence showed high identity (83%) with that of the intact globulin gene over a 500-bp region (data not shown).
Comparing Paralogous and Orthologous Regions among Wheat Genomes
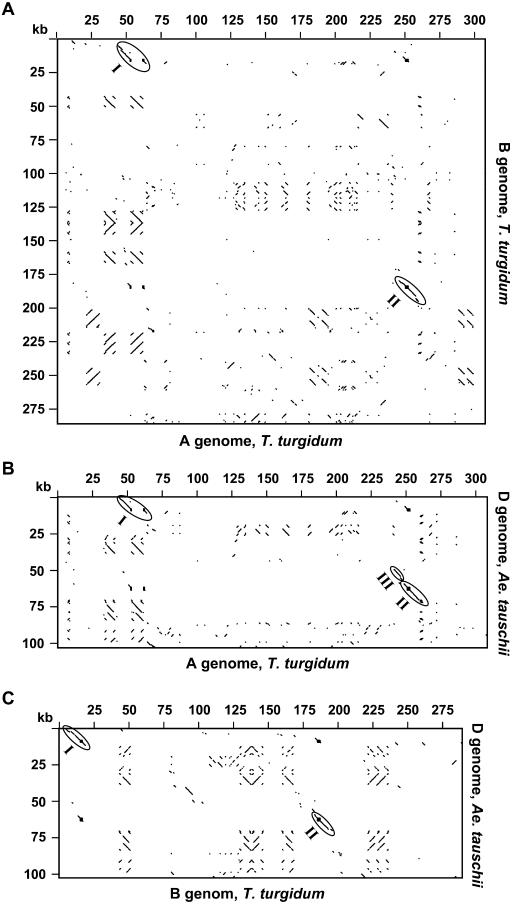
To examine sequence conservation among the three wheat genomes, we conducted pair-wise comparisons of the orthologous HMW-glutenin regions of the A, B, and D genomes. As shown in Figure 3, in only two regions are the sequences conserved among all three genomes. The intact globulin, y-type HMW-glutenin, and partial receptor kinase gene sequences are included in Region I (Fig. 3, A–C). Region II consists primarily of the sequences of the x-type HMW-glutenin and protein kinase genes. Region III, a conserved region that is composed mainly of repetitive DNAs, is only present between the A and D genomes (Fig. 3B; see discussion below). Except for these regions, the sequences are divergent among the three genomes. Although other highly related sequences were also revealed in several regions by pair-wise comparison, they are not colinear. These regions are composed of sequences of highly related LTR retrotransposons that are very abundant in the wheat genomes.
Figure 3.
Pair-wise comparisons of the orthologous HMW-glutenin regions of the A, B, and D genomes. The Dotplot analyses compared the complete available sequences in the vicinity of the Glu-A1 and Glu-B (Fig. 3A), Glu-A1 and Glu-D1 (Fig. 3B), and Glu-B1 and Glu-D1 (Fig. 3C) loci. Sequence match criteria of 60% over a 40-bp window was used in the analyses. Colinear regions are circled and numbered with I, II, and III. The gap in Region I of Figure 3, A and B is caused by the Wis-3 insertion in the Ay HMW-glutenin gene.
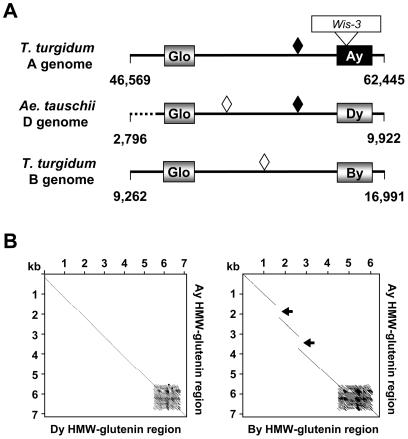
Previously, we identified that in wheat, but not in barley, a 7.2-kb segment containing a globulin and an HMW-glutenin gene was duplicated and inserted into an adjacent region, creating the y-type and x-type paralogous HMW-glutenin genes (Kong et al., 2004). To understand the evolution of the wheat genomes, we further compared the 7.2-kb orthologous regions containing the y-type HMW-glutenin from the A, B, and D genomes. It appears that the sequences in these regions are generally conserved (Fig. 4A), although several differentiating mobile elements were identified, including the Wis-3 insertion in the Ay HMW-glutenin gene as described above. Four MITE element insertions were also detected in the regions between the globulin and HMW-glutenin, one of which was shared by the D and A genomes (Fig. 4A). When the sequences from the Wis and MITE insertions were removed, dotplot analyses revealed that the three regions are significantly conserved (Fig. 4B). The A and D genome segments show 93% sequence identity. Less sequence similarity (85%) was observed when the A and B genome segments were compared. The dotplot analyses also revealed that there were either two sequence deletion events in the B genome or two insertion events in an A and D genome common ancestor. Collectively, these results suggest that the A and D genomes are more closely related to one another than either is to the B genome.
Figure 4.
Comparison of orthologous sequences in the y-type HMW-glutenin regions. A, Organization of regions containing the globulin and y-type HMW-glutenin genes. Genes are represented by shaded boxes or black box (inactive gene). Diamonds represent MITEs inserted in the intergenic regions. The two black diamonds are colinear MITEs present in the A and D genomes. The dashed line represents a region that has been deleted in the D genome. B, Dotplot analyses of y-type HMW-glutenin regions. All inserted repetitive DNA sequences were removed from the regions used in these comparisons prior to the analyses. The sequence match criteria is 60% over a 40-bp window. Arrows represent regions that are either deletions in the B genome or insertions in the A genome. The dark block-like structures indicate where repetitive motif sequences in the HMW-glutenin were found to have multiple matches within the compared sequences.
We then compared the duplicate segments containing the x-type HMW-glutenin from the three wheat genomes, as shown in Figure 5. Sequence rearrangements appear to have been more frequent in these regions than in the segments containing the y-type HMW-glutenin gene. The original 7.2-kb duplicated segment of the B genome has expanded to 84 kb due to multiple insertions of retrotransposons after the duplication event (Fig. 5A; Kong et al., 2004). When the sequences of the A and D genomes were closely examined, it was found that the duplicate globulin genes are both inactive and that exactly the same sequence region was deleted in both genomes. Moreover, a non-LTR retroelement, Gobefo-1, was identified in this region at the same insertion site in both the A and D genomes.
Figure 5.
Comparison of orthologous sequences in the x-type HMW-glutenin regions. A, Organization of regions containing the globulin and x-type HMW-glutenin genes. Genes are represented by shaded boxes or black boxes (inactive genes). Retroelements are boxed and their insertion positions indicated by either triangles or solid vertical lines. Diamonds represent MITEs. Dashed lines represent regions deleted in the A and D genomes based on comparison with the corresponding region of the B genome. The black box with a broken border represents the second globulin gene with a portion of the coding region deleted. B, Dotplot analyses of x-type HMW-glutenin regions. All inserted repetitive DNA sequences were removed from the regions used in these comparisons prior to the analyses. Sequence match criteria is 60% over a 40-bp window. The black arrow indicates a region that is either an insertion in the B genome or a deletion in the A genome.
Retroelements that distinguish the A genome from D genome were also identified. A non-LTR retroelement, Nafead-1, is present only in the D genome, while a Stowaway MITE is present only in the A genome, indicating that these insertions occurred after the divergence of the A and D genomes. Despite the differences in retrotransposon insertions in the three wheat genomes, sequence conservation is still observable when the sequences contributed by retrotransposons are removed (Fig. 5B). A dotplot analysis revealed 92% sequence identity in the 4.2-kb region between the A and D genomes, while the sequence identity between the A and B genomes is about 83%. These results are consistent with those shown in Figure 4.
We also compared the two 7.2-kb paralogous regions in the A genome. No repetitive DNA elements were found to be colinear. This suggests that either the original duplicated region contained no repetitive sequences or that elements present there have been deleted or become unrecognizable since the duplication event. In addition to the differences caused by the insertions of multiple mobile elements, there are more than 50 deletions or insertions of more than 5 bp, including the 3-kb deletion in the second globulin region. Sequence conservation is mostly located in the two HMW-glutenin regions, despite the fact that both genes are inactive. This may indicate that the inactivation of these genes occurred relatively recently, since disruption of the HMW-glutenin genes was not detected in the diploid rye (De Bustos et al., 2001), the D genome from A. tauschii (Anderson et al., 2003), nor in the Am genome from Triticum monococcum (Y. Gu, S. Heath, and O. Anderson, unpublished data), a species closely related to the hexaploid A genome donor, T. urartu (Dvorak et al., 1992). Nevertheless, the possibility that the gene inactivation occurred in the diploid ancestral A genome cannot be ruled out.
Intergenic Regions and Nested Retrotransposons
Gene spacing is dramatically different in the orthologous regions of the wheat and barley genomes, as shown in Figure 1. To further examine the composition and organization of DNA in the intergenic regions of these closely related genomes, we first compared the sequences after the y-type HMW-glutenin and before the second globulin gene, the region representing the space between the two duplicate segments. The size of this region is 41 and 85 kb in the D and B genomes, respectively (Anderson et al., 2003; Kong et al., 2004), and is 179 kb in the A genome (Fig. 6A). In these orthologous intergenic regions, retrotransposable elements comprise the majority of the sequence. Detailed examinations revealed that the sequences of three mobile elements are shared by all three genomes, suggesting that these elements inserted before the genomes diverged. One such element is a Stowaway MITE, which has identical insertion sites in all three genomes (Kong et al., 2004; Fig. 6A). In addition to this MITE, a region composed of a partial Wilma and a partial Sabrina-like element is present in both the B and D genomes (Kong et al., 2004). This structure was likely generated by a deletion event that removed the sequences of the 3′ end of the Wilma element and the 5′ end of the Sabrina-like element (Kong et al., 2004). However, in the A genome, it appears that there was a second deletion event unique to the A genome that resulted in a complete loss of the Sabrina-like sequence and left only a fragment of the Wilma 5′ LTR segment (Fig. 6A).
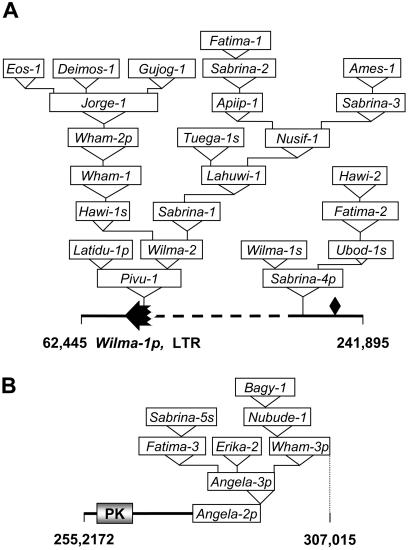
Figure 6.
Organization of retroelements in the intergenic regions of the Glu-A1 locus. A, Nested retrotransposons in the region between the y-type HMW-glutenin and the second globulin gene. The horizontal solid line represents the intergenic region. The partial Wilma LTR (black arrow) and MITE (black diamond) elements indicated along the horizontal solid line are colinear with the sequences present in D and B genomes (Kong et al., 2004). The dashed line represents the region deleted only in the A genome based on the comparison with the colinear regions of the wheat D and B genomes. Retroelements are boxed and their insertion positions indicated with either a triangle or a solid vertical line. The letters s and p following the names of the retroelements indicate solo and partial, respectively. B, Nested retrotransposons in the region after the protein kinase. The protein kinase gene is represented by a shaded box. The horizontal solid line represents the intergenic region. Retroelements are boxed and their insertion positions indicated with either a triangle or a solid vertical line.
Except for the remnants of these two retroelements and the MITE sequence, none of the other retroelements in this intergenic region appear to be conserved between the A, B, and D genomes, indicating that the remainders of the retroelement insertions happened after the divergence of the three genomes. In the A genome, a Pivu-1 element inserted into the previously described partial Wilma LTR region. This Pivu-1 element contains large blocks of nested retrotransposon insertions, with as many as 19 members (Fig. 6A). This nested structure spans a region of over 140 kb. In the largest branch of these insertions, there are eight tiers of retrotransposons nested within one another. A second nested arrangement of retrotransposons was found immediately downstream, in which four other retrotransposons are layered in a partial Sabrina element, Sabrina-4p (Fig. 6A).
In the Glu-A1 locus, another nested retrotransposon structure was identified in the region beginning after the protein kinase gene and extending to the end of the BAC. This structure is composed of eight retrotransposons and spans a total of 45 kb (Fig. 6B). More retroelements may exist in the downstream portion of this arrangement since the Angela-2p, Angela-3p, and Wham-3p sequences are interrupted because of their location at the end of the BAC contig. The colinear regions of the B and D genomes, approximately 40 kb and 100 kb, respectively, are also composed of retrotransposons (Kong et al., 2004). However, none of the retroelements were found to be shared by the other genomes, implying once again that a rapid insertion of retroelements occurred after the divergence of the three wheat genomes.
While a nested retroelement structure with multiple tiers suggests a logical order to the insertion times of their members (Fig. 6A), an actual estimate of the insertion time of a long terminal repeat (LTR) retrotransposon can be calculated based on the nucleotide substitution rate in the two LTRs. Due to the nature of the transposition process, the two LTRs of a single retrotransposon are usually identical at the time of its insertion into the new region of the host genome (Boeke, 1989). The degree of sequence divergence between two LTRs reflects the length of time since the transposition event and can be used to estimate the date of insertion (SanMiguel et al., 1998). The average base substitution rate of 6.5 × 10−9 per site per year calculated from the grass (Gramineae) adh1-adh2 region (Gaut et al., 1996) was used to predict the insertion times of 22 LTR retrotransposons in the A genome (Table I). The estimated insertion times are generally consistent with the insertion order of retrotransposons derived from the position of individual elements in the nested structure. However, a few exceptions were noted in the largest retroelement block (Fig. 6A, Table I). One such exception is that the Apiip-1 element, dated to have an insertion time of 5.5 MYA, inserted into the Nusif-1 retroelement that has existed for only 4.3 MYA. Other exceptions include the insertion of Wilma-2 (4.3 MYA) into Pivu-1 (3.7 MYA), and Nusif-1 (4.3 MYA) into Lahuwi-1 (3.4 MYA). Although the causes for these inconsistencies are unclear, a possible explanation could be that translocation events may move one nested retrotransposon structure into another, resulting in older elements existing within younger ones. Alternatively, different regions of the genome might evolve at different rates, resulting in differences in their estimated insertion times (Wicker et al., 2003). Nevertheless, apart from the Apiip-1 element, all of the other retrotransposons have predicted insertion times within the last 4.5 million years (Table I). This result is consistent with the previous observation that the diploid Triticum and Aegilops progenitors of the A, B, and D genomes in diploid, tetraploid, and hexaploid wheats all diverged between 2.5 and 4.5 MYA (Huang et al., 2002). Therefore, it appears that the divergence time of the wheat genomes was sufficiently long ago for the independent amplification of retroelements in each genome to occur, resulting in the complete differentiation of the intergenic sequences.
Table I.
Retrotransposons and the chronology of their insertions in the HMW glutenin region
| Elements | Type | Length | 5′ LTRa | 3′ LTR | Base Substitutionsb | Time |
|---|---|---|---|---|---|---|
| MYAc | ||||||
| Ames-1 | gypsy | 13,054 | 4,043 | 4,070 | 194 | 3.7 |
| Apiip-1 | gypsy | 11,017 | 970 | 971 | 70 | 5.5 |
| Bagy-1 | gypsy | 10,481 | 4,056 | 4,054 | 57 | 1.1 |
| Boba-1 | gypsy | 5,408 | 227 | 226 | 2 | 0.7 |
| Deimos-1 | LITE | 2,178 | 652 | 363* | 14 | 3.0 |
| Erika-2 | gypsy | 13,468 | 4,133 | 4,133 | 7 | 0.1 |
| Fatimah-1 | gypsy | 9,079 | 480 | 482 | 13 | 2.1 |
| Fatimah-2 | gypsy | 10,018 | 505 | 542 | 19 | 2.9 |
| Gujog-1 | gypsy | 5,790 | 485 | 485 | 13 | 2.1 |
| Hawi-2 | gypsy | 11,686 | 3,761 | 3,754 | 96 | 2.0 |
| Lahuwi-1 | gypsy | 4,628 | 1,536 | 1,469 | 65 | 3.4 |
| Madil-1 | gypsy | 9,856 | 1,593 | 1,601 | 40 | 1.9 |
| Nubude-1 | gypsy | 10,201 | 3,014 | 3,152 | 96 | 2.4 |
| Nusif-1 | gypsy | 6,256 | 886 | 911 | 50 | 4.3 |
| Pivu-1 | athila | 13,092 | 2,078 | 1,998 | 99 | 3.7 |
| Sabrina-1 | athila | 6,986 | 1,577 | 928 | 47 | 3.9 |
| Sabrina-2 | athila | 7,290 | 1,543 | 1,561 | 65 | 3.2 |
| Sabrina-3 | athila | 8,300 | 1,586 | 1,589 | 70 | 3.4 |
| Wham-1 | athila | 8,132 | 1,423 | 1,278* | 51 | 3.1 |
| Wis-2 | copia | 7,871 | 769* | 1,726 | 5 | 0.7 |
| Wis-3 | copia | 8,633 | 1,752 | 1,757 | 11 | 0.5 |
| Wilma-2 | gypsy | 10,153 | 1,528 | 1,504 | 84 | 4.3 |
Full length retrotransposons with two LTRs were used to calculate the time of insertion.
Deletions were counted as a single base substitution event.
Divergence time is calculated as K/2 × Knus (Wei et al., 2002). Knus is the base substitution rate per nucleotide per year. A Knus of 6.5 × 109 per site per year derived from the grass (Gramineae) adh1-adh2 region (Gaut et al., 1996) was employed in the calculation.
, Deletion event.
DISCUSSION
Gene Inactivation in Polyploid Wheats
Bread wheat carries triplicate chromosomes, yet many traits are inherited in a diploid fashion. Many genes are genetically diploidized, and phenotypes caused by single mutations are readily identified at the hexaploid level, suggesting that many of the triplicate loci have been inactivated. Our comparative sequence analyses of the orthologous regions in the genomes of durum wheat have allowed us to detect several gene inactivation events that occurred before the hexaploid polyploidization. We have shown that the inactivation of the Ay HMW-glutenin gene in Chinese Spring had already occurred in its tetraploid ancestor. Although the orthologous region in the diploid ancestral A genome of the durum wheat has yet to be characterized, the disruptions of these four genes in the tetraploid A genome could be associated with polyploidization. In the diploid A. tauschii D (Anderson et al., 2003) and T. monococcum Am (Y. Gu, S. Heath, and O. Anderson, unpublished data) genomes, only the duplicate globulin gene was disrupted. In the orthologous region of the barley genome, all of the genes identified were intact (Gu et al., 2003). However, it is also possible that the presence of paralogous genes tolerates mutations in the duplicate copies, e.g. the globulin genes. Characterization of a large number of the orthologous and paralogous regions from polyploid and diploid ancestral genomes will enhance our understanding of gene inactivation in polyploid species. Furthermore, we detected that three genes in the Glu-1 loci (two in the Glu-A1 and one in Glu-B1 locus) were disrupted by retrotransposon insertions, suggesting that retrotransposons play a potential role in gene inactivation in wheat. The demonstration that gene inactivation is frequent in polyploid genomes raises an interesting question. How are orthologous genes discriminated from one another? In our studies, the finding that more genes in this region of the A genome were disrupted than in the orthologous region of the B and D genomes may or may not represent a regional, random phenomenon. A more general conclusion can be drawn when orthologous regions of more loci from different genomes of allopolyploid species are analyzed.
Gene Colinearity and Density in Polyploid Genomes
Gene colinearity at different loci among grass genomes has previously been investigated at the micro-level through sequencing genomic segments of orthologous regions from rice, maize, sorghum, barley, and wheat (Bennetzen and Ramakrishna, 2002). These studies have demonstrated that gene colinearity is generally retained within grass genomes, but with many exceptions (Bennetzen, 2000; Gaut, 2002). In the Triticeae tribe, genomes of polyploid wheats and their diploid ancestors are considered to be closely related by evolution. One might expect much higher sequence conservation among the wheat genomes. Our comparative studies show that dramatic sequence changes have occurred even within the gene-containing regions. We found that although gene colinearity is retained, four of six genes are inactive in the Glu-A1 region of the durum wheat. Because of the genome polyploidization, disruption of genes could be more common in the orthologous regions of the A, B, and D genomes of polyploid wheat.
Even the gene colinearity may be not as conserved as we thought in polyploid wheat. The perturbation of gene colinearity has been recently examined through the deletion mapping of wheat ESTs into delineated chromosome bins (Akhunov et al., 2003a). Studies suggested that synteny perturbation between homoeologous chromosomes caused by locus duplications and deletions occurs more often in the distal regions, where gene density and the recombination rate are higher (Akhunov et al., 2003b).
The distance between the various genes in the colinear regions of the three wheat genomes we examined varies significantly, primarily due to the differential insertion of retroelements into these regions (Fig. 6). These insertions directly affect the gene density in these regions. The gene density in the barley genome is 1 gene/10 kb when the region from the receptor kinase and Ser/Thr protein kinase genes is taken into account. If the same regions of the D, B, and A genome are compared, the gene densities are 1 gene/11, 31, and 41 kb, respectively; approximately a 4-fold difference between the D and A genomes. Other previously sequenced BACs from gene-rich regions of the diploid wheats, T. monococcum, and A. tauschii, and barley, have shown gene densities on the order of 1 gene/5 to 40 kb, values that are at least 12 times higher than would be expected for a random gene distribution (Shirasu et al., 2000; Dubcovsky et al., 2001; Wicker et al., 2001; Brooks et al., 2002; Wei et al., 2002; Yan et al., 2003). Calculations of gene density depend on the definition used to identify a gene. If only intact or active genes are considered in the calculation, the gene density for the sequenced A genome region would increase to 1 gene/150 kb. The low gene density in the sequenced A genome region may represent a statistically insignificant local variation. On the other hand, given the observation that wheat has a large genome size and if orthologous and paralogous genes are often subject to disruption during evolution, it would be expected that the density of functional genes will be much lower than in the smaller and diploid plant genomes.
Structure and Evolution of Wheat Genomes
While model plants, like Arabidopsis and rice, have small genomes (135 and 450 Mb, respectively), hexaploid wheat is often cited as an example of a large and complex genome (16,000 Mb). The difference in genome size is due in part to genome polyploidization, but is caused mainly by the large number of repetitive sequences. In the A and B genome regions, retroelements account for over 80% of examined sequences, significantly higher than that reported for the maize genome (60%; SanMiguel et al., 1996). SanMiguel et al. (1998) estimated that all of the retrotransposon insertions found within the Adh1 region in maize occurred within the last 6 million years, and that the entire maize genome doubled in size in the last 3 million years, presumably due to this rapid amplification of retroelements. Similarly, all of the datable retrotransposons identified in the HMW-glutenin regions have estimated insertion times within the last 6 million years (Table I). The larger percentage of retroelements reported in the wheat genome as compared to the maize genome suggests that wheat has a greater rate of expansion.
Although it is still not clear why the genomes of polyploid wheat contain more retroelements than other known grass genomes, the gene redundancy conferred by polyploidization may act as a buffer against the disadvantages of various mutations that occur in gene sequences (Wendel, 2000). The insertion of retroelements into genes may contribute considerably to the rapid genome expansion of polyploid genomes. For example, the receptor kinase gene in the A genome expanded into a region of more than 30 kb. In this case, the retrotransposons inserted into the gene account for 10% of the sequenced region. Similarly, the insertion of retroelements in the second globulin gene in the B genome increased the region by 40 kb (Kong et al., 2004).
Our comparative analyses revealed that only a few repetitive DNA elements are colinear among the wheat genomes, suggesting that the majority of the retroelements were inserted into their present location after the divergence of the wheat A, B, and D genomes. Considering that retrotransposons make up over 80% of the wheat genomes, we can expect that a large portion of the genome sequence is not conserved among the three wheat genomes. Recently, Wicker et al. (2003) demonstrated that more than 90% of sequences are not conserved even in the orthologous low Mr glutenin regions of the tetraploid A and diploid Am genomes that diverged only 1 to 3 million years ago. Knowledge of sequence divergence among the wheat genomes is useful in developing strategies to construct integrated genetic and physical maps for polyploid wheat. Because of its large genome size, polyploid nature, and the uncertainty of sequence similarity among each of the subgenomes in hexaploid wheat, the development of high resolution BAC-based physical maps for common wheat is assumed to be very difficult, primarily with respect to forming contig assemblies that allow for clear separation among each of these subgenomes. Previously, we showed that HMW-glutenin BAC clones from the A and B genomes in tetraploid wheat can accurately be separated based on fingerprinting analyses and subsequent contig assembly (Kong et al., 2004). Recently, we have employed the same strategy to study the structural organization of chromosomal segments containing clusters of the gliadin gene superfamily for each of the A and B genomes (Y. Gu, C. Crossman, and O. Anderson, unpublished data). Based on these results, it should be feasible to construct separate physical maps for the A, B, and D genomes in hexaploid wheat using a global fingerprinting strategy.
Repetitive DNA and Relationships among Wheat Genomes
Despite being the most prevalent elements in grass genomes, retrotransposons have not been well exploited as tools in the study of molecular evolution for various reasons, including the fact that colinear retrotransposons are very rare in distantly related genomes (SanMiguel et al., 2002; Gu et al., 2003). However, in studies involving closely related genomes, recent transposition activities may allow us to identify retroelements that can clarify evolutionary relationships. The closely related wheat genomes in tetraploid and hexaploid wheat and their ancestral diploid genomes provide an excellent opportunity for studies of genome evolution and polyploidization. Despite many studies on wheat evolution, the origins and evolutionary relationships of these genomes are still debatable (Huang et al., 2002). The identification of the Wilma sequence present at the same position of the wheat A, B, and D genomes (Kong et al., 2004; Fig. 6), but absent in the barley genome, provides a molecular footprint that separates the barley and wheat lineages. Furthermore, identification of several transposable elements shared by the A and D genomes, but not by the B genome, indicate that the A and D genomes share a common ancestor that had previously diverged from the B genome ancestor. Several other lines of evidence discovered in this study, including a deletion event in the inactive globulin gene shared only by the A and D genomes, and higher sequence homology in the duplicated regions between the A and D genomes than the B genome, also support this conclusion.
Our analysis of the Wis insertions in the Ay HMW-glutenin genes of tetraploid and hexaploid A genomes points to more than one origin of hexaploid wheat. The analysis of the mutation in the Ax HMW-glutenin genes further supports this conclusion. It is unclear whether this sequence divergence at the A genome HMW-glutenin loci occurred in the diploid or tetraploid ancestor. If it occurred in the diploid ancestor, two distinct tetraploid polyploidization events must have occurred. The simpler explanation would be that the divergence occurred after the A and B genomes had already merged in a tetraploid species, and then that two distinct hexaploid polyploidization events occurred. Analyses of orthologous regions of the A genome from T. urartu, the wild type species of the hexaploid A genome donor (Dvorak et al., 1992), would address this question. More than one origin of hexaploid wheat has also been suggested by sequence comparison of low-copy DNA. Talbert et al. (1998) showed that two distinct D genome alleles exist in the hexaploid wheat. These alleles originated from a differentiation of the D genome pool in A. tauschii. RFLP analysis of 53 single loci with the D genome pool from different geographic regions also suggested that several A. tauschii parents contributed to the evolution of hexaploid wheat (Dvorak et al., 1998). Our finding of multiple origins of the A genome provides further evidence of genome diversity in hexaploid wheat.
MATERIALS AND METHODS
BAC Isolation and Selection
Previously, we screened a BAC library of durum wheat, Triticum turgidum subsp. durum, cv Langdon, a tetraploid species consisting of A and B genomes (Cenci et al., 2003), for large chromosome regions containing HMW-glutenin genes (Kong et al., 2004). Two contigs were formed from a total of 23 resultant positive BAC clones using a combination of BAC DNA fingerprinting and subsequent contig assembly. One contig represented the chromosomal regions from the B genome and two overlapping BAC clones from the contig spanning the Glu-B1 locus were sequenced to form a 285-kb contiguous region (Kong et al., 2004). In this report, two overlapping BAC clones from the other contig (634M12 and 110M5) were selected to sequence the HMW-glutenin regions of the A genome in tetraploid durum wheat.
BAC Sequencing
The sequencing of the two A genome BAC clones was carried out as described previously (Anderson et al., 2003; Gu et al., 2003). In brief, shotgun-sequencing libraries for selected BAC clones were first constructed from randomly sheared BAC DNAs in sizes of 3 to 6 kb. Plasmid DNAs from single colonies were purified and inserts were sequenced from both directions with T7 and T3 primers using BigDye terminator chemistry (Applied Biosystems, Foster City, CA) on an ABI3700 capillary sequencer. To complete BAC sequences, gaps, mainly resulting from the presence of G/C rich areas in the gap regions, were filled by a primer-walking approach to sequence through the difficult regions using the dGTP BigDye terminator chemistry (Applied Biosystems).
Sequence Analysis
The sequence data generated for each BAC clone was used to assemble continuous contigs using the Lasergene SeqMan module (DNAStar, www.DNAStar.com) as described previously (Anderson et al., 2003; Gu et al., 2003; Kong et al., 2004). In this module, a high stringency parameter for base calling and quality assessment was selected to generate the most accurate consensus sequence reads possible. Quality reads, with estimated 15× coverage of the insert for each BAC, were used for sequence assembly with a criteria of a 40-bp window size and a 96% match requirement. To validate the accuracy of the sequence assembly, digestion patterns of BAC DNAs with HindIII, EcoRI, and NotI were compared with the predicted restriction patterns of the computer-assembled sequences.
The A genome BAC clones 634M12 and 4B1overlap for a region of 35 kb. The combined sequence of these clones covers a region of 307,015 bp in length, spanning the Glu-A1 locus region of the tetraploid wheat A genome. The final error rate for the sequence estimated by the Seqman module is about 0.3 bp/10 kb. The sequence was deposited in GenBank with the accession number of AY494981.
For annotation, the assembled sequence of the two conjoined A genome BACs was compared with the previous annotations for the orthologous regions from the barley BAC (AY268139), D genome BAC (AF497474), and B genome BAC (AY368673). In addition, a homology search was performed against NCBI nonredundant and dbEST databases using BLASTN, BLASTX, and TBLASTX algorithms. FGENESH (http://www.softberry.com/nucleo.html) and GENESCAN (http://genemark.mit.edu/GENESCAN.htm) were used for gene prediction. DNA repetitive elements were identified with NCBI BLAST searches, with DNAStar MegAlign dotplot analysis, and by comparison with the Triticeae Repeat Sequence Database (TREP) at the GrainGenes web site at http://wheat.pw.usda.gov/ITMI/Repeats/. New retrotransposons was named according to the method described by SanMiguel et al. (2002).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY494981, AY268139, AF497474, and AY368673.
Acknowledgments
The authors are grateful to Dr. Fusheng Wei for his generous help in annotation of the sequences and to Dr. Frank You for assistance in BAC contig assembly. We also thank Drs. William R. Belknap and Roger Thilmony for critical readings of the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.038083.
References
- Akhunov ED, Akhunov AR, Linkiewicz AM, Dubcovsky J, Hummel D, Lazo G, Chao S, Anderson OD, David J, Qi L, et al. (2003. a) Synteny perturbations between wheat homoeologous chromosomes caused by locus duplications and deletions correlate with recombination rates. Proc Natl Acad Sci USA 100: 10836–10841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhunov ED, Goodyear AW, Geng S, Qi LL, Echalier B, Gill BS, Miftahudin, Gustafson JP, Lazo G, Chao S, et al. (2003. b) The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosome arms. Genome Res 13: 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson OD, Greene FC (1989) The characterization and comparative analysis of high MW glutenin genes from genomes A and B of hexaploid wheat. Theor Appl Genet 77: 689–700 [DOI] [PubMed] [Google Scholar]
- Anderson OD, Larka L, Christoffers MJ, McCue KF, Gustafson JP (2002) Comparison of orthologous and paralogous DNA flanking the wheat high molecular weight glutenin genes: sequence conservation and divergence, transposon distribution, and matrix-attachment regions. Genome 45: 367–380 [DOI] [PubMed] [Google Scholar]
- Anderson OD, Rausch C, Moullet O, Lagudah ES (2003) The wheat D-genome HMW-glutenin locus: BAC sequencing, gene distribution, and retrotransposon cluster. Funct Integr Genomics 3: 56–68 [DOI] [PubMed] [Google Scholar]
- Bennetzen JL (2000) Comparative sequence analysis of plant nuclear genomes: microcolinearity and its many exceptions. Plant Cell 12: 1021–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Ramakrishna W (2002) Numerous small rearrangements of gene content, order and orientation differentiate grass genomes. Plant Mol Biol 48: 821–827 [DOI] [PubMed] [Google Scholar]
- Boeke JD (1989) Transcription and reverse transcription of retrotransposons. Annu Rev Microbiol 43: 403–434 [DOI] [PubMed] [Google Scholar]
- Brooks SA, Huang L, Gill BS, Fellers JP (2002) Analysis of 106 kb of contiguous DNA sequence from the D genome of wheat reveals high gene density and a complex arrangement of genes related to disease resistance. Genome 45: 963–972 [DOI] [PubMed] [Google Scholar]
- Cenci A, Chantret N, Kong X, Gu YG, Anderson OD, Fahima T, Distelfeld A, Dubcovsky J (2003) Construction and characterization of a half million clone BAC library of durum wheat (Triticum turgidum ssp. durum). Theor Appl Genet 107: 931–939 [DOI] [PubMed] [Google Scholar]
- De Bustos A, Rubio P, Jouve N (2001) Characterization of two gene subunits on the 1R chromosome of rye as orthologs of each of the Glu-1 genes of hexaploid wheat. Theor Appl Genet 103: 733–742 [Google Scholar]
- Devos KM, Gale MD (2000) Genome relationship: the grass model in current research. Plant Cell 12: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Ramarkrishna W, SanMiguel P, Busso CS, Yan L, Shiloff BA, Bennetzen JL (2001) Comparative sequence analysis of colinear barley and rice bacterial artificial chromosomes. Plant Physiol 125: 1342–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J, Di Terlizzi P, Zhang H-B, Resta P (1992) The evolution of polyploid wheats: identification of the A genome donor species. Genome 36: 21–31 [DOI] [PubMed] [Google Scholar]
- Dvorak J, Luo M-C, Yang Z-L, Zhang H-B (1998) The structure of the Aegilops tauschii genepool and the evolution of hexaploid wheat. Theor Appl Genet 97: 657–670 [Google Scholar]
- Feldman M, Lupton FGH, Miller TE (1995) Wheats. In J Smartt, NW Simmonds, eds, Evolution of Crops, Ed 2. Longmam Scientific, London, pp 184–192
- Feuillet C, Keller B (2002) Comparative genomics in the grass family: molecular characterization of grass genome structure and evolution. Ann Bot (Lond) 89: 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde J, Malpica JM, Halford NG, Shewry PR, Anderson OD, Greene FC, Miflin BJ (1985) The nucleotide sequence of a HMW subunit gene located on chromosome 1A of wheat (Triticum aestivum L.) Nucl Acids Res 13: 6817–6832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut BS (2002) Evolutionary dynamics of grass genomes. New Phytol 154: 15–28 [Google Scholar]
- Gaut BS, Morton BR, McCaig BC, Clegg MT (1996) Substitution rate comparisons between grasses and palm: Synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci USA 93: 10274–10279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y-Q, Anderson OD, Londeore C, Kong X, Chibbar RN, Lazo GR (2003) Structural organization of the barley D-hordein locus in comparison with its orthologous regions of wheat genomes. Genome 46: 1084–1097 [DOI] [PubMed] [Google Scholar]
- Halford NG, Field JM, Blair H, Urwin P, Moore K, Robert L, Thompson R, Flavell RB, Tatham AS, Shewry PR (1992) Analysis of HMW-glutenin subunits encoded by chromosome 1A of bread wheat (Triticum aestivum L.) indicates quantitative effects on grain quality. Theor Appl Genet 83: 373–378 [DOI] [PubMed] [Google Scholar]
- Halford NG, Forde J, Shewry PR, Kreis M (1989) Functional analysis of the upstream regions of a silent and an expressed member of a family of wheat seed protein genes in transgenic tobacco. Plant Sci 62: 207–216 [Google Scholar]
- Harberd NP, Flavel RB, Thompson RD (1987) Identification of a transposon-like insertion in a Glu-1 allele of wheat. Mol Gen Genet 209: 326–332 [DOI] [PubMed] [Google Scholar]
- He P, Friebe BR, Gill BS, Zhou J-M (2003) Allopolyploidy alters gene expression in the highly stable hexaploid wheat. Plant Mol Biol 52: 401–414 [DOI] [PubMed] [Google Scholar]
- Huang S, Sirikhachornkit A, Su X, Faris J, Gill B, Haselkorn R, Gornicki P (2002) Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc Natl Acad Sci USA 99: 8133–8138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd WS, Campbell CD, Kellog E (1999) Plant Systematics: A Phylogenic Approach. Sinauer Associates, Sunderland, MA
- Kashkush K, Feldman M, Levy AA (2002) Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160: 1651–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG (2002) Transposable elements and the evolution of genome size in eukaryotes. Genetica 115: 49–63 [DOI] [PubMed] [Google Scholar]
- Kimber G, Sears ER (1987) Evolution in the genus Triticum and the origin of cultivated wheat. In EG Heyne, ed, Wheat and Wheat Improvement. American Society of Agronomy, Madison, WI, pp 154–164
- Kong X, Gu YQ, You FM, Dubcovsky J, Anderson OD (2004) Dynamics of the evolution of orthologous and paralogous portions of a complex locus region of the B and D genomes of wheat. Plant Mol Biol (in press) [DOI] [PubMed]
- Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33: 479–532 [DOI] [PubMed] [Google Scholar]
- Laurie DA, Devos KM (2002) Trends in comparative genetics and their potential impacts on wheat and barley research. Plant Mol Biol 48: 729–740 [DOI] [PubMed] [Google Scholar]
- Masterson J (1994) Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264: 421–423 [DOI] [PubMed] [Google Scholar]
- Nesbitt M, Samuel D (1996) From staple crop to extinction? The archaeology and history of hulled wheats, pp 41-100 In S Padulosi, K Hammer, and J Heller, eds, Hulled Wheats: Promoting the Conservation and Use of Underutilized and Neglected Crops. 4. Proceedings of the 1st International Workshop on Hulled Wheats, July 21–22, 1995. International Plant Genetic Resources Institute, Rome
- Ozkan H, Levy AA, Feldman M (2001) Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13: 1735–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL (1998) The paleontology of intergene retrotransposons of maize. Nature Genetics 20: 43–45 [DOI] [PubMed] [Google Scholar]
- SanMiguel PJ, Ramakrishna W, Bennetzen JL, Busso CS, Dubcovsky J (2002) Transposable elements, genes and recombination in a 215-kb contig from wheat chromosome 5A(m). Funct Integr Genomics 2: 70–80 [DOI] [PubMed] [Google Scholar]
- SanMiguel P, Tiknonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake Berhanm A, Springer PS, Edwards KJ, Lee M, Avramova Z, et al. (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768 [DOI] [PubMed] [Google Scholar]
- Schoen DJ (2000) Comparative genomics, marker density and statistical analysis of chromosome rearrangements. Genetics 154: 943–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry PR, Halford NG, Tatham AS (1992) High molecular weight subunits of wheat glutenin. J Cereal Sci 15: 105–120 [Google Scholar]
- Shirasu K, Schulman AH, Lahaye T, Schulze-Lefert P (2000) A contiguous 66-kb barley DNA sequence provide evidence for reversible genome expansion. Genome Res 10: 908–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert LE, Smith LY, Blake NK (1998) More than one origin of hexaploid wheat is indicated by sequence comparison of low-copy DNA. Genome 41: 402–407 [Google Scholar]
- Wei F, Wing RA, Wise RP (2002) Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell 14: 1903–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF (2000) Genome evolution in polyploids. Plant Mol Biol 42: 225–249 [PubMed] [Google Scholar]
- Wicker T, Stein N, Albar L, Feuillet C, Schlagenhauf E, Keller B (2001) Analysis of a contiguous 211-kb sequence in diploid wheat (Triticum monococcum L.) reveals multiple mechanisms of genome evolution. Plant J 26: 307–316 [DOI] [PubMed] [Google Scholar]
- Wicker T, Yahiaoui N, Guyot R, Schlagenhauf E, Liu Z-D, Dubcovsky J, Keller B (2003) Rapid genome divergence at orthologous low molecular weight glutenin loci of the A and Am genomes of wheat. Plant Cell 15: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100: 6263–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]