Abstract
The cotton (+)-δ-cadinene synthase (CAD1), a sesquiterpene cyclase, catalyzes a branch-point step leading to biosynthesis of sesquiterpene phytoalexins, including gossypol. CAD1-A is a member of CAD1 gene family, and its promoter contains a W-box palindrome with two reversely oriented TGAC repeats, which are the proposed binding sites of WRKY transcription factors. We isolated several WRKY cDNAs from Gossypium arboreum. One of them, GaWRKY1, encodes a protein containing a single WRKY domain and a putative N-terminal Leu zipper. Similar to genes encoding enzymes of cotton sesquiterpene pathway, GaWRKY1 was down-regulated in a glandless cotton cultivar that contained much less gossypol. GaWRKY1 showed a temporal and spatial pattern of expression comparable to that of CAD1-A in various aerial organs examined, including sepal, stigma, anther, and developing seeds. In suspension cells, expression of both GaWRKY1 and CAD1-A genes and biosynthesis of sesquiterpene aldehydes were strongly induced by a fungal elicitor preparation and methyl jasmonate. GaWRKY1 interacted with the 3× W-box derived from CAD1-A promoter in yeast (Saccharomyces cerevisiae) one-hybrid system and in vitro. Furthermore, in transgenic Arabidopsis plants, overexpression of GaWRKY1 highly activated the CAD1-A promoter, and transient assay in tobacco (Nicotiana tabacum) leaves demonstrated that W-box was required for this activation. These results suggest that GaWRKY1 participates in regulation of sesquiterpene biosynthesis in cotton, and CAD1-A is a target gene of this transcription factor.
In cotton plants, biosynthesis of sesquiterpene phytoalexins could be induced by fungal and bacterial infection or other environmental stimuli (Bell and Stipanovic, 1977; Bell, 1986; Essenberg et al., 1990). Formation of gossypol and related sesquiterpene aldehydes is also developmentally regulated, and these secondary metabolites accumulate in pigmented glands of aerial tissues and in epidermal and subepidermal cells of roots (Bell and Stipanovic, 1977; Halloin and Bell, 1979; Meng et al., 1999). All cotton sesquiterpene phytoalexins identified so far have a cadinene-type skeleton, and biochemical and molecular evidence has suggested that (+)-δ-cadinene is the biosynthetic precursor (Chen et al., 1995; Davila-Huerta et al., 1995; Luo et al., 2001).
(+)-δ-Cadinene synthase (CAD1), a sesquiterpene cyclase, catalyzes the first committed step in the pathway of gossypol and related sesquiterpenes (Chen et al., 1995, 1996). In Gossypium arboreum, a diploid cotton species, CAD1 is encoded by a gene family that can be divided into two subfamilies: CAD1-A with a single member and CAD1-C with several (Tan et al., 2000). Our previous reverse transcription (RT)-PCR analysis showed that CAD1-C genes were highly expressed in such aerial organs as sepal, petal, and developing seeds during early developmental stages, paralleling formation of pigment glands and accumulation of sesquiterpene aldehydes, whereas expression level of CAD1-A was low or undetectable when the same RT-PCR conditions were employed. In stem, CAD1-A transcripts were detectable only after elicitation, but those of CAD1-C were detected both before and after elicitation (Tan et al., 2000). In suspension cultured cells, both CAD1-A and CAD1-C genes were induced to express by elicitation (Chen et al., 1995, 1996). Thus, in cotton plants, genes CAD1-A and CAD1-C have different but overlapping expression patterns, and there appears to be a complex program modulating the expression of sesquiterpene cyclase genes.
WRKY proteins form a large transcription factor family with their occurrence limited to plants. There are estimated to be more than 70 WRKY proteins in Arabidopsis (Robatzek and Somssich, 2002; Singh et al., 2002; Dong et al., 2003; Kalde et al., 2003). WRKY proteins are featured by the WRKY domain, which is a 60-amino acid stretch containing a conserved amino acid sequence of WRKYGQK together with a zinc finger-like motif (Eulgem et al., 2000). There is either one or two WRKY domains in each protein; some members of the family have Leu zippers toward the N terminus (Eulgem et al., 1999; Cormack et al., 2002). Numerous in vitro and in vivo experiments have demonstrated that WRKY proteins specifically bind to the W-box (T)TGAC(C/T), a cis-acting DNA element found frequently in the promoter of defense-related genes (Rushton et al., 1996; Yang et al., 1999; Du and Chen, 2000; Yu et al., 2001).
In the past few years, there has been much progress in characterizing WRKY proteins involved in regulating plant defense responses. Many WRKY genes, such as PcWRKY1, PcWRKY4, AtWRKY18, and AtWRKY70, are involved in response to pathogen infection and elicitor treatment (Rushton et al., 1996; Chen and Chen, 2002; Cormack et al., 2002; Li et al., 2004). The parsley (Petroselinum crispum) PcWRKY1, for example, bound specifically to the W-box in PR1-1 or PR1-2 promoters, and its own expression was induced rapidly and transiently by the pathogen-derived elicitor pep25 (Rushton et al., 1996). Most of the WRKY proteins, such as AtWRKY6 and AtWRKY29, are transcription activators, whereas some WRKYs behave as repressors of transcription (Asai et al., 2002; Robatzek and Somssich, 2002). There is increasing evidence that WRKY proteins also play a diverse role in regulating other developmental and physiological processes of plants, such as trichome initiation (Johnson et al., 2002), senescence (Robatzek and Somssich, 2002; Hinderhofer and Zentgraf, 2001), and carbohydrate metabolism (Sun et al., 2003).
Here, we report that a palindrome of W-box elements is present in the CAD1-A promoter. We have isolated from G. arboreum a cDNA, GaWRKY1, coding for a Leu zipper-containing WRKY protein. GaWRKY1 interacted with the 3× W-box derived from CAD1-A promoter in yeast (Saccharomyces cerevisiae) one-hybrid system and in an electrophoretic mobility shift assay (EMSA). Expression of GaWRKY1 activated CAD1-A promoter in both transgenic Arabidopsis plants and transiently transformed tobacco (Nicotiana tabacum) leaves, and disruption of the W-box abolished the activation. In the glandless cotton cultivar, expression of both GaWRKY1 and CAD1-A genes was down-regulated, and in the glanded cultivar their temporal and spatial expression patterns were comparable. These data demonstrate that GaWRKY1 is likely a transcriptional activator of the CAD1 gene CAD1-A, participating in cotton sesquiterpene biosynthesis.
RESULTS
Isolation of GaWRKY1
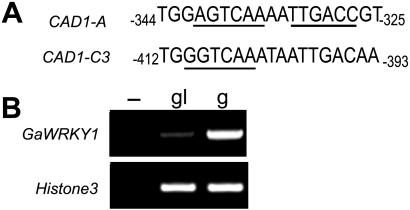
To study the transcriptional regulation of the cotton CAD1, a key enzyme in the biosynthesis pathway of gossypol and related sesquiterpenes, promoters of CAD1-A and CAD1-C3 were scanned with PLACE (http://www.dna.affrc.go.jp/htdocs/PLACE/signalscan.html). Both promoters were found to contain the typical W-box element (Fig. 1A). In CAD1-A promoter, two reversely oriented W-box cis-acting elements (AGTCAAAATTGACC) were located in the region between −340 and −323 bp, forming a palindrome (Fig. 1A). Deletion from 5′ end of the approximately 1.2-kb promoter of CAD1-A showed that the region containing the W-box was required for elicitor-induced transcription (our unpublished data). Since, as mentioned above, W-box elements are frequently found to be the binding sites of WRKY transcription factors, it is reasonable to assume that WRKY proteins are involved in transcriptional regulation of CAD1 genes of cotton.
Figure 1.
The W-box in the promoter region of CAD1-A or CAD1-C3 (A) and RT-PCR detection of GaWRKY1 transcripts in developing seeds of 25 DPA (B). The W-box is underlined. The seeds were collected from G. hirsutum L. cv Zhong-12, a glanded cultivar (g), and G. hirsutum cv GL-5, a glandless cultivar (gl). PCR was performed by 31 cycles of amplification. Histone3 transcripts were amplified as an internal control.
We then isolated WRKY cDNA fragments from G. arboreum. Expression of 10 WRKY genes in developing seeds collected at 25 d postanthesis (DPA) was examined by RT-PCR. One of them, GaWRKY1, showed a higher level of transcripts in seeds of G. hirsutum L. cv Zhong-12, a glanded cotton cultivar, than in G. hirsutum L. cv GL-5, a glandless cultivar (Fig. 1B). Expression of other nine WRKYs was either indistinguishable between the two cultivars or too low to be detected (data not shown). Previous investigations showed that aerial tissues of glandless cultivars were nearly gossypol free, and transcripts of CAD1-A and CAD1-C genes were undetectable (Liang et al., 2000; Tan et al., 2000). Therefore, GaWRKY1 was considered a candidate for transcription factors regulating the gossypol pathway.
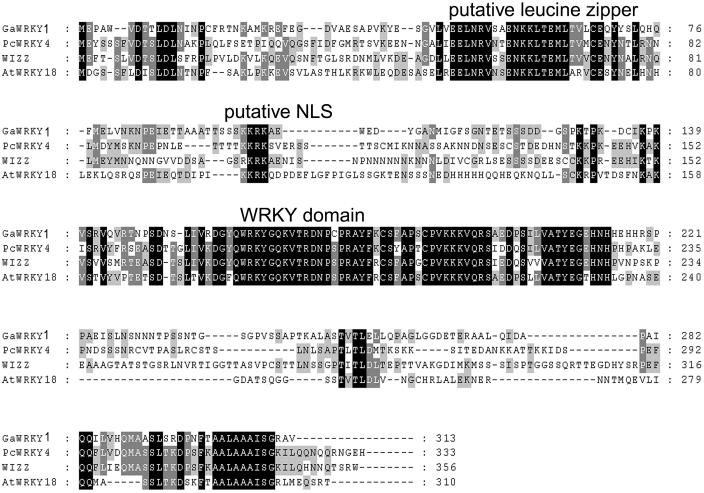
The cDNA of GaWRKY1 was isolated and found to encode a 313-amino acid protein with a predicted molecular mass of 34 kD. The deduced protein has a single WRKY domain with a zinc finger-like motif (C2H2) at its C terminus, categorizing it into Group II of the WRKY superfamily (Eulgem et al., 2000). In addition, GaWRKY1 has a putative Leu zipper in its N-terminal end (Fig. 2). GaWRKY1 exhibited high sequence identities (41%–45%) to Group IIa WRKY proteins, including PcWRKY4 of parsley (Cormack et al., 2002), AtWRKY18 of Arabidopsis (Chen and Chen, 2002), and Wizz of tobacco (Hara et al., 2000). Alignment of these protein sequences revealed that the WRKY domain, putative Leu zipper, nuclear localization site, and a C-terminal Ala-rich stretch were strictly conserved; outside these regions, however, there was little sequence conservation (Fig. 2).
Figure 2.
Alignment of GaWRKY1 with related WRKY proteins from other plant species. The deduced amino acid sequence of GaWRKY1 was aligned with PcWRKY4 (AAG35658), WIZZ (BAA87058), and AtWRKY18 (NP_567882) using ClustalW (Thompson et al., 1994) with default parameters through EMBnet (http://www.ch.embnet.org/software/ClustalW.html). Black and gray shadings, done with BOXSHADE 3.21 (http://www.ch.embnet.org/software/BOX_form.html), indicate conserved amino acid residues. Predicted domains are indicated above the sequences.
Expression Patterns of GaWRKY1 and CAD1-A
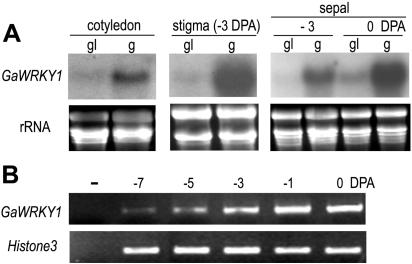
To see if GaWRKY1 expression coincides with accumulation of sesquiterpene aldehydes in other tissues, transcription levels of GaWRKY1 in the glanded and glandless cotton cultivars were further compared. RNA blot detected a significant level of GaWRKY1 mRNA in cotyledon, stigma, and sepal of the glanded cultivar, and the level was low or undetectable in the glandless cultivar (Fig. 3A). The transcripts were also detected in root, peel, petal, stigma, and anther of the glanded cotton cultivar by RNA gel blot (data not shown). In sepal, the GaWRKY1 transcript level was higher at 0 DPA than at −3 DPA. Analysis of more samples by RT-PCR confirmed that the transcript abundance of GaWRKY1 was increasing with the maturation of sepal (Fig. 3B).
Figure 3.
Expression of GaWRKY1 in G. hirsutum L. cv Zhong-12 (g) and in G. hirsutum L. cv GL-5 (gl). A, Northern analysis of GaWRKY1 transcripts in the cotyledon of 1-week-old seedlings and in stigma and sepal. B, RT-PCR analysis of GaWRKY1 transcripts in developing sepals of G. hirsutum L. cv Zhong-12. For RNA gel blot, total RNA of 40 μg was loaded on each lane. PCR was performed by 31 cycles of amplification.
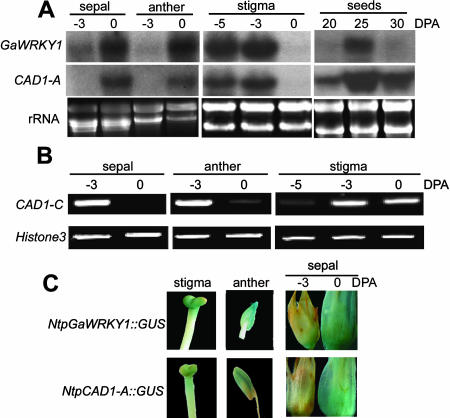
Should GaWRKY1 target to CAD1 genes, we would expect them to have similar or at least overlapping expression domains. In floral organs examined, the spatial and temporal pattern of GaWRKY1 expression was highly similar to that of CAD1-A. In sepal and anther, both GaWRK1 and CAD1-A genes had a low level of transcripts at −3 DPA, and both were up-regulated at the day of anthesis (Fig. 4A). In stigma, however, although GaWRKY1 and CAD1-A showed the same temporal expression pattern, the dynamics of transcript accumulation were different: their mRNA levels were high at −3 DPA and decreased to an undetectable level at 0 DPA (Fig. 4A). Thus, toward flower maturation, the transcript abundance of GaWRKY1 and CAD1-A was increasing in the sepal and anther but decreasing in stigma. In developing seeds, the GaWRKY1 transcript level peaked around 25 DPA, then decreased; for CAD1-A the mRNA level also increased from 20 to 25 DPA, and the level continued to be high at 30 DPA (Fig. 4A).
Figure 4.
Comparison of expression patterns of GaWRKY1 and CAD1-A. A, RNA gel blot analysis of stigma, sepal, anther, and seeds of G. hirsutum L. cv Zhong-12. B, RT-PCR analysis of CAD1-C (CAD1-C3) transcripts in developing stigma, sepal, and anther of G. hirsutum L. cv Zhong-12. PCR was performed by 23 cycles of amplification. C, Histochemical staining of GUS activities in stigma (−3 DPA), anther (0 DPA), and sepal of transgenic tobacco plants of NtpGaWRKY1::GUS and NtpCAD1-A::GUS. The tissues were stained for 24 h using X-Gluc as substrate.
As reported previously, CAD1-C3 and probably other CAD1-C genes were highly expressed in the sepal of −3 DPA, and the transcript abundance decreased at 0 DPA (Tan et al., 2000). Further analysis by RT-PCR indicated that not only in sepal but also in anther, CAD1-C3 showed opposite dynamics of mRNA accumulation to that of GaWRKY1 and CAD1-A, i.e. during flower development, the transcript level of CAD1-C3 in the sepal and anther was decreasing rather than increasing (Fig. 4B). Therefore, temporal expression pattern of CAD1-C3 was clearly different from that of GaWRKY1 in these floral organs.
To gain further insight into the relation of expression patterns between CAD1-A and GaWRKY1, promoters of both genes were fused to the β-glucuronidase (GUS) reporter gene and were transferred into tobacco plants, respectively. Histochemical staining revealed that both promoters directed GUS expression in stigma, sepal, and anther of the transgenic plants. And for the sepal, the GUS staining was stronger at 0 DPA than at −3 DPA (Fig. 4C), which was consistent with the northern results obtained from the cotton sepal (Fig. 4A). For both promoters, the expression in leaves was weak.
Induction of GaWRKY1 and CAD1-A by Elicitation
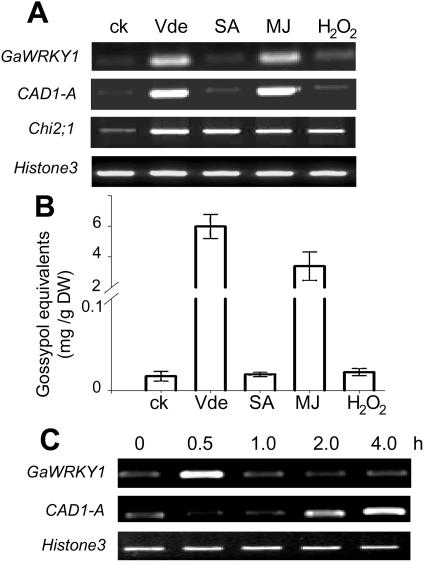
Many WRKY genes that are involved in regulating defense-related genes are themselves elicitor responsive (Rushton et al., 1996; Yang et al., 1999; Yu et al., 2001; Yoda et al., 2002; Dong et al., 2003). Because CAD1-A transcription could be activated by fungal elicitation (Chen et al., 1996; Tan et al., 2000), we would expect an inducible expression of GaWRKY1, should it be a regulator of CAD1-A. We treated the G. arboreum suspension cells with an elicitor preparation of Verticillium dahliae (Vde) and several signaling molecules, including salicylic acid (SA), methyl jasmonate (MJ), and H2O2. Examination of transcript abundance by RT-PCR revealed that Vde and MJ significantly induced the transcription of GaWRKY1 and CAD1-A, whereas SA and H2O2 exerted little effect (Fig. 5A). Coordinately, production of gossypol and related sesquiterpene aldehydes in G. arboreum cells also increased upon elicitation by Vde or MJ but not by SA neither H2O2 (Fig. 5B). Induced expression of chitinase genes in cotton plants treated with SA and the Vde elicitor has been reported (Hudspeth et al., 1996). When the chitinase gene Chi2;1 was included in RT-PCR analysis, it was induced not only by Vde but also by the three signaling molecules (Fig. 5A), indicating that the G. arboreum cells responded to all these elicitors.
Figure 5.
Induced gene expression of GaWRKY1 and CAD1-A and sesquiterpene aldehyde accumulation in G. arboreum suspension cultured cells. A, RT-PCR analysis of gene expression in G. arboreum cells treated with Vde, SA (5 mm), MJ (45 μm), and H2O2 (100 μm) for 30 min (GaWRKY1) or 4 h (CAD1-A and Chi2;1). B, Accumulation of sesquiterpene aldehydes (gossypol equivalents) in G. arboreum cells collected at 24 h postelicitation. C, Transcript levels of GaWRKY1 and CAD1-A in Vde-treated suspension cells for the time period postelicitation as indicated. PCR was performed by 30 cycles of amplification for GaWRKY1 and 29 cycles for CAD1-A.
Furthermore, in Vde-treated cells the transcript level of GaWRKY1 increased rapidly and peaked within 30 min, then declined to the original level. On the other hand, CAD1-A showed a comparatively slower rate of induction, and the transcript level was not significantly elevated until about 2 h postelicitation (Fig. 5C). Similar kinetics of transcript accumulation were also observed in MJ-treated cells (data not shown). Thus, the transcript level of GaWRKY1 increased rapidly and transiently upon elicitation, preceding the CAD1-A induction. The concomitant responses of GaWRKY1 and CAD1-A genes to different elicitor molecules and the subsequent gossypol accumulation support the assumption that GaWRKY1 participates in regulating the gossypol pathway.
Nuclear Localization and DNA-Binding Activity of GaWRKY1
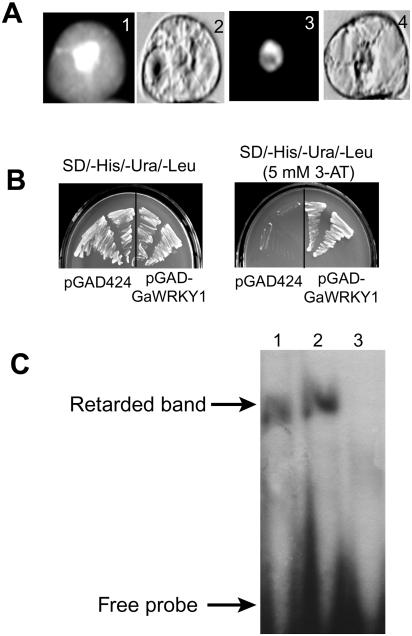
To determine subcellular localization of GaWRKY1, the open reading frame (ORF) of GaWRKY1 was in-frame fused to the green fluorescent protein (GFP) reporter gene. After introducing the construct into tobacco BY2 cells, GFP fluorescence was located exclusively in the nucleus (Fig. 6A). By contrast, BY2 cells transformed with 35S::GFP showed GFP signal throughout the cell. This indicates that GaWRKY1 is a nuclear-localized protein.
Figure 6.
Nuclear localization and DNA-binding activity of GaWRKY1. A, Subcellular localization of GFP (1 and 2) and GFP-GaWRKY1 fusion protein (3 and 4) in BY2 cells. B, Yeast one-hybrid assay using the 3×W-box as bait. Yeast cells carrying pGAD424 or pGAD-GaWRKY1 were grown for 3 d at 30°C in SD/−His/−Ura/−Leu or SD/−His/−Ura/−Leu plus 5 mm 3-amino-1,2,4-triazole (3-AT), respectively. C, EMSA performed by incubation of the purified GST-GaWRKY1 protein with 32P-labeled DNA fragment of 3×W-box. The three lanes stand for 32P-labeled 3×W-box (1), DNA fragment of the pBSK vector without the binding site as a nonspecific competitor (2), and 50-fold excess of the unlabeled 3×W-box as a competitor (3). The 3×W-box contained triple tandem copies of the W-box palindrome of CAD1-A promoter.
To determine if GaWRKY1 binds to the CAD1-A promoter, yeast one-hybrid assay was performed. A DNA fragment of 3× W-box (triple tandem repeats of the W-box palindrome of CAD1-A) was introduced into yeast cells, forming HWLW. The plasmid of pGAD-GaWRKY1, encoding the fusion protein of GAL4 binding and GaWRKY1, and the plasmid of pGAD424 were introduced into HWLW cells, respectively. We found that only the yeast clones harboring pGAD-GAWRKY1 grew on the −His−Ura−Leu synthetic dextrose (SD) base containing 5 mm 3-amino-1,2,4-triazole (Fig. 6B), indicating that GaWRKY1 bound to the 3× W-box and activated transcription in yeast. Furthermore, deletion of the GaWRKY1 showed that the zinc finger-like motif (C2H2) at the C terminus was the binding domain, and the putative Leu zipper at the N terminus significantly increased the binding affinity (data not shown).
In EMSA, the recombinant protein of glutathione S-transferase (GST)-GaWRKY1 showed binding affinities to the 3× W-box. The DNA-binding specificity was confirmed in a competition experiment with 50-fold excess of the unlabeled probe of 3×W-box (Fig. 6C). Thus, in vitro GaWRKY1 was able to specifically recognize and interact with the W-box palindrome of CAD1-A.
Activation of CAD1-A Promoter in Plants by GaWRKY1
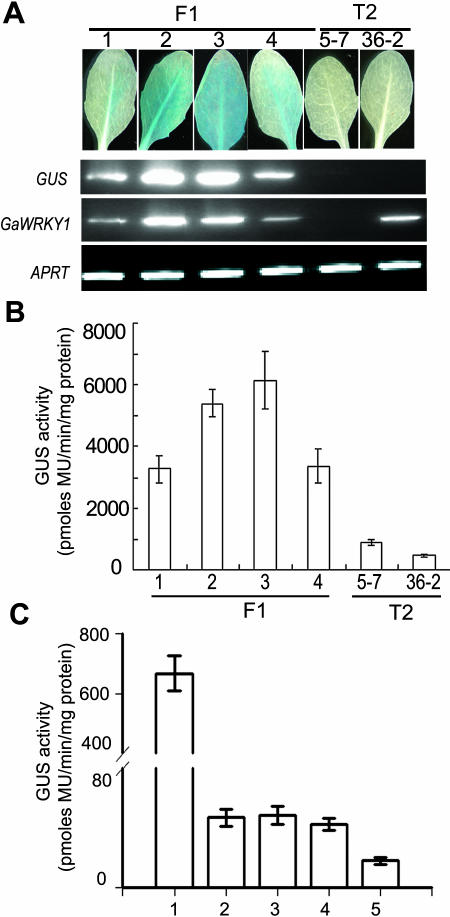
To provide evidence that GaWRKY1 protein regulates CAD1-A gene transcription, we used transgenic plants of Arabidopsis to assay their interactions. An approximately 1.2-kb promoter of CAD1-A fused with either the GUS reporter gene (pCAD1-A::GUS) or the 35S::GaWRKY1 recombinant gene was transferred into Arabidopsis plants, followed by crossing the plants' T2 lines. The GaWRKY1 transcripts were clearly detectable in a T2 line of 35S::GaWRKY1 (36-2; Fig. 7A). Plants transformed with 35S::GaWRKY1 did not show clear phenotypic changes. In the T2 plants of pCAD1-A::GUS (5-7), the GUS activity in leaves was low or too weak to be stained. However, the GUS activities became significantly higher in F1 plants (35S::GaWRKY1 × pCAD1-A::GUS) that contained a 35S::GaWRKY1 gene (Fig. 7, A and B). Analysis by RT-PCR proved that in plants transformed with pCAD1-A::GUS only, the GUS transcripts were undetectable, whereas in F1 plants, significant levels of both GaWRKY1 and GUS transcripts were present. Moreover, there was a good correlation between the transcript levels of GaWRKY1 and GUS in leaves of F1 plants (Fig. 7A). Clearly, in transgenic Arabidopsis plants, constitutive expression of GaWRKY1 strongly activated the CAD1-A promoter.
Figure 7.
Activation of CAD1-A promoter by constitutive or transient expression of GaWRKY1 in plants. A, GUS staining and RT-RCR analysis of the GUS and GaWRKY1 transcripts in mature leaves of T2 plants of 5-7 (pCAD1-A::GUS) and 36-2 (35S::GaWRKY1) lines and their F1 plants containing both constructs. B, Quantitative determination of GUS activities in mature leaves of the plants as indicated. PCR was performed by 28 cycles of amplification, and Arabidopsis Ade phosphoribosyl transferase (APRT) transcripts were amplified as an internal control. C, Transient assay of GUS activities in tobacco leaves 2 d after infiltration with Agrobacterium cells harboring pCAD1-A::GUS/35S::GaWRKY1 (1), pCAD1-A::GUS (2), mpCAD1-A::GUS/35S::GaWRKY1 (3), mpCAD1-A::GUS (4), and 35S::GaWRKY1 (5), respectively.
To investigate whether the W-box in the CAD1-A promoter was indispensable for the recognition by GaWRKY1, we generated a mutant form of pCAD1-A (mpCAD1-A), in which the W-box palindrome of AGTCAAAATTGACC was mutated to ATTCAAAATTGAAC. We then tested the interactions of GaWRKY1 with the wild-type and mutant promoters by Agrobacterium-mediated transient expression of a GUS reporter gene in tobacco leaves. The GUS activity was low in leaves expressing either pCAD1-A::GUS or mpCAD1-A::GUS alone. Coexpression of pCAD1-A::GUS with 35S::GaWRKY1 increased the GUS activity by approximately 20-fold. By contrast, 35S::GaWRKY1 did not activate mpCAD1-A (Fig. 7C). These results indicate that activation of the CAD1-A promoter by GaWRKY1 requires the W-box palindrome.
DISCUSSION
Terpenoids, the largest group of plant secondary metabolites, play a diverse role in plant-microbe, plant-herbivore, plant-plant, and plant-environment interactions (Chappell, 1995; Mahmoud and Croteau, 2002; Pichersky and Gershenzon, 2002; Aharoni et al., 2003). In cotton, gossypol and other sesquiterpene phytoalexins not only have fungistatic properties and insecticidal activities but also are toxic to mammals (Bell and Stipanovic, 1977; Davila-Huerta et al., 1995; Puckhaber et al., 2002). In previous studies, we have cloned three enzymes that catalyze consecutive steps from isopentenyl diphosphate and dimethylallyl diphosphate to 8-hydroxy-(+)-δ-cadinene of the gossypol pathway: farnesyl diphosphate synthase (Liu et al., 1999), CAD1 (Chen et al., 1995, 1996; Tan et al., 2000), and (+)-δ-cadinene-8-hydroxylase (CYP706B1; Luo et al., 2001). Genes encoding enzymes of the gossypol pathway characterized so far are all down-regulated in the glandless cultivar of G. hirsutum, as is the WRKY protein gene GaWRKY1 reported herein. As shown in Figure 1A, both CAD1-A and CAD1-C3 promoters contain W-box elements. In fact, W-boxes are also present in the promoter of CYP706B1 gene (our unpublished data). These data strongly suggest the involvement of WRKY proteins in regulation of cotton sesquiterpene biosynthesis.
In cotton suspension cultured cells, both GaWRKY1 and CAD1-A expression and sesquiterpene aldehyde biosynthesis responded to the Vde elicitor and MJ but not to SA or H2O2. The coordinated responses suggest that GaWRKY1 and CAD1-A are components of the same pathway of cotton defense reaction, which involves induced biosynthesis of sesquiterpene phytoalexins. Searching of databases revealed the presence of W-box elements in the promoter region of sesquiterpene cyclase genes of other plants, such as 5-epi-aristolochene synthase gene EAS4 of tobacco (Yin et al., 1997), putative sesquiterpene cyclase genes of Arabidopsis (At1g31950), and rice (Oryza sativa; BAC99543.1). Therefore, WRKY-W-box interactions are likely operating also in other plants in regulation of sesquiterpene biosynthesis.
Among the WRKY proteins reported so far, only a subset of members, such as ABF2, WIZZ, PcWRKY4, and PcWRKY5, have putative Leu zippers (Rushton et al., 1995; Eulgem et al., 2000; Hara et al., 2000; Cormack et al., 2002). Function of the Leu zipper has been proposed to mediate dimerization and increase the DNA-binding affinity of WRKY proteins (Cormack et al., 2002). Our results from yeast-one hybrid assay support this assumption, since removing the N-terminal Leu zipper from GaWRKY1 decreased its binding affinity to the W-box of CAD1-A promoter.
In elicitor-treated suspension cells of G. arboreum, different kinetics of induction of transcript accumulation were observed for genes encoding the transcription factor GaWRKY1, enzymes of the gossypol pathway, and a PR-10 protein. The response of GaWRKY1 to elicitation is quick and instant (Fig. 5C), whereas the induction rate of the sesquiterpene cyclase genes CAD1-A and CAD1-C3 and the cytochrome P450 gene CYP706B1 was moderate, and mRNA levels of these enzymes peaked in several hours postelicitation (Chen et al., 1995, 1996; Luo et al., 2001). For GaPR-10, which encodes a pathogenesis-related protein with in vitro ribonuclease activities, induction of gene expression was slow, and the peak level of mRNA did not appear until 12 h postelicitation (Zhou et al., 2002). Many defense-related WRKY genes, such as PcWRKY1, PcWRKY4, and AtWRKY18, also show fast and transient induction that precedes the induced expression of downstream defense genes (Rushton et al., 1996; Chen and Chen, 2002; Cormack et al., 2002). However, function of GaWRKY1 is unlikely limited to concerting defense responses, since it shows developmentally mediated expression in floral tissues. Accumulation of both GaWRKY1 and CAD1-A transcripts was increasing in the sepal and anther along with maturation, and the highest level appeared at the day of anthesis (Fig. 4, A and C). Certain senescence-associated WRKY genes, such as AtWRKY6, are expressed in floral tissues (Quirino et al., 1999), and maturation of the floral organ is also considered a senescence process (Bleecker and Patterson, 1997). Although the temporal expression pattern of GaWRKY1 in sepal and anther correlates with maturation of both organs, evidence that GaWRKY1 is involved in floral senescence is still lacking. In fact, expression of GaWRKY1 (and of CAD1-A as well) in mature stigma (0 DPA) is down- rather than up-regulated. The biological significance of differential regulation of GaWRKY1 and CAD1-A genes in different floral organs needs further investigation.
Although GaWRKY1 and CAD1-A genes showed a complex expression pattern in floral organs, their coordinated expression in various aerial organs and in suspension cultured cells indicates that regulation of CAD1-A by GaWRKY1 is spatially and temporally possible. In combination with results from DNA-binding assay and transgenic plant analyses, our data strongly suggest that CAD1-A is a target gene of GaWRKY1. The discrepancy of temporal expression patterns between CAD1-C (such as CAD1-C3) and GaWRKY1 in floral organs suggests that other WRKY proteins or bZIP-type proteins may be involved in regulation of CAD1-C genes. However, this does not exclude the possibility that GaWRKY1 may also regulate CAD1-C genes in other organs. In developing seeds, for example, genes of CAD1-A and CAD1-C showed similar temporal patterns of expression (Meng et al., 1999). Alternatively, GaWRKY1 may function as a repressor of CAD1-C genes in flower. Besides W-box palindrome, other cis-acting elements, such as those of MYB and MYC, are also present in the CAD1-A promoter. It is clear that expression of cotton CAD1 genes and biosynthesis of sesquiterpene aldehydes are mediated by both developmental and defense-related programs. Isolation of more transcription factors regulating the gossypol pathway and further elucidation of regulatory machinery of CAD1 genes will be a great help in manipulating cotton secondary metabolism and disease resistance.
MATERIALS AND METHODS
Materials
Plants of cotton (Gossypium arboreum L. cv Qingyangxiaozhi, G. hirsutum L. cv Zhong-12, and a glandless cultivar G. hirsutum cv GL-5) and tobacco (Nicotiana tabacum) were grown in a greenhouse. Fresh materials were collected as described previously (Tan et al., 2000; Luo et al., 2001). Plants of Arabidopsis (ecotype Columbia) were grown in a growth chamber at 22°C with a photoperiod of 16 h of light and 8 h of dark. G. arboreum suspension cells were cultured in liquid Murashige and Skoog medium. Elicitation was performed by treating the cells with SA (5 mm), MJ (45 μm), H2O2 (100 μm), or a crude preparation of fungal elicitor (Vde), which was obtained by precipitation of the culture fluid of Verticillium dahliae and applied at a final concentration of 1 μg of Suc equivalent per milliliter of culture, as described previously (Liu et al., 1999). Sesquiterpene aldehydes were extracted and quantitated by the phloroglucinol/HCl method (Meng et al., 1999).
Isolation of GaWRKY1 cDNA
A degenerate primer [5′-TGG(A/C) IAA(A/G) TA(C/T) GGICA(A/G) AA(A/G)-3′], corresponding to the conserved peptide sequence of WRKYGQK (Eulgem et al., 2000), together with a vector-specific T7 primer, was used in amplification of WRKY fragments from a λ-Unizap cDNA library, which was constructed from elicitor-treated G. arboreum cells (Chen et al., 1995). The PCR products were cloned into pGEM-T vector (Promega, Madison, WI) and sequenced. The cDNA of GaWRKY1 was isolated by a PCR-mediated 96-well method, with the primers GaWRKY1P0 (5′-GTCACAAGAGATAACCCCTG-3′) and GaWRKY1M0 (5′-TGATACTCTTCTCCCATTGAACCTAC-3′).
RNA Isolation and Analysis
Total RNA preparation, RT-PCR, and RNA gel blot were performed as described previously (Tan et al., 2000; Luo et al., 2001), except that in RT-PCR 1 μL of reverse transcription products was used as template without dilution. The primers used were GaWRKY1P1 (5′-ATCTTGGATCTCAACATCAACCC-3′) and GaWRKY1M1 (5′-CTATTATCAGAAGGATTGTGCGG-3′) for GaWRKY1, Histone3P (5′-GAAGCCTCATCGATACCGTC-3′) and Histone3M (5′-CTACCACTACCATCATGG-3′) for cotton Histone3 (AF024716), and Chi2P (5′-ACTCCACAGTCACCGAAACC-3′) and Chi2M (5′-ATCTTATTCCATCTCCACGG-3′) for the cotton chitinase gene Chi2;1 (Z68152). For CAD1-A, CAD1-C3 and Arabidopsis Ade phosphoribosyl transferase (APRT; NM_179383), primers were the same as reported (Payne et al., 2000; Tan et al., 2000). DNA probes of GaWRKY1 and CAD1-A were obtained by 32P-labeling of the PCR products amplified from the corresponding cDNA clones, using the same primers as for RT-PCR.
Subcellular Localization of GFP-GaWRKY
The GaWRKY1 ORF was amplified by GFPWP (5′-CGGGATCCACGTTCAGTAATG GAACCAG-3′) and GFPWM (5′-GCTCTAGACTAAATTACTTCGGCACAGAGT-3′), which contained a BamHI and XbaI site (underlined), respectively. The PCR products were digested and inserted into pGFP-JW vector, harboring an ORF encoding the GFP driven by a 35S promoter (our unpublished data), forming a chimerical gene encoding GFP-GaWRKY1. The plasmids of GFP-GaWRKY1 and pGFP-JW were then introduced into tobacco BY2 cells, respectively, via Agrobacterium-mediated transformation (Gu and Verma, 1997).
Binding Assay in Yeast One-Hybrid System
The binding assay utilized a yeast one-hybrid system (CLONTECH, Palo Alto, CA). The bait was the triple tandem copies of the CAD1-A W-box palindrome (3× W-box, 5′-TGGAGTCAAAATTGACCGTTGGAGTCAAAATTGACCGTTGGAGTCAAAATTGACCGT-3′). This fragment was inserted into pHISi and pLacZi vectors, respectively, and the two resultant plasmids were introduced into yeast cells, forming HWLW. The ORF of the GaWRKY1 was in-frame fused with the GAL4 activation domain of the one-hybrid vector pGAD424, forming pGAD-GaWRKY1. HWLW cells were then transformed with the pGAD424 and pGAD-GaWRKY1, respectively.
Electrophoretic Mobility Shift Assay
The ORF of GaWRKY1 was inserted into the expression vector pGEX-4T-1 (Amersham Pharmacia Biotech, Uppsala). GST and GST-GaWRKY1 proteins were then purified according to the instruction manual. Isolation of nuclear proteins from Vde-treated suspension cells of G. arboreum and EMSA were performed according to the published protocols (Rushton et al., 1995; Yang et al., 1999).
Plant Transformation and GUS Activity Assay
The promoters of CAD1-A (pCAD1-A) and GaWRKY1 (pGaWRKY1) were inserted into the pBI121 vector (CLONTECH), respectively, replacing the 35S promoter. These two promoter-GUS plasmids were transferred into tobacco plants by a leaf-disc method, as described previously (Liang et al., 2000), and the resulting transgenic plants were named NtpCAD1-A::GUS and NtpGaWRKY1::GUS, respectively. Arabidopsis plants were transformed with the constructs of 35S::GaWRKY1 and pCAD1-A::GUS by a floral-dip method (Clough and Bent, 1998), and the transgenic plants were named At35S::GaWRKY1 and AtpCAD1-A::GUS, respectively.
The W-box (AGTCAAAATTGACC) in pCAD1-A was mutated into ATTCAAATTGAAC by mega-primer method (Barik, 1993) with the primer mWboxP (5′-GGAAGTTGGATTCAAAATTGAA-3′); the resulting promoter was named mpCAD1-A. pCAD1-A and mpCAD1-A were inserted into the modified pBI121 vector harboring an intron-containing GUS, replacing the 35S promoter, respectively. For 35S::GaWRKY1, the ORF of GaWRKY1 was inserted into pBI121, replacing GUS. The two promoter-GUS plasmids and the 35S::GaWRKY1 were introduced into Agrobacterium strain GV3101, respectively. Transient assay using tobacco leaves was performed as described (Yang et al., 2000). The ratio of 35S::GaWRKY1 to pCAD1-A::GUS or to mpCAD1-A::GUS was adjusted 3:5 for coinfiltration.
A fluorimetric assay with 4-umbelliferyl-d-glucuronide as substrate was used to determine GUS activities, and the expression pattern was visualized with a histochemical staining using X-Gluc as substrate (Jefferson, 1987). Quantitative analysis was performed at least three times throughout this investigation.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AY507929.
Acknowledgments
We thank Professor Zongyang Wang, Dr. Hui Shen, and Mr. Bin Luo for their advice on EMSA and help on manuscript preparation.
This work was supported by the National Natural Science Foundation of China (grant nos. 30030020 and 39925005), the National 973 program (grant no. 2002CB111301), and the Chinese Academy of Sciences (grant no. KSCX2–SW–313).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.038612.
References
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Aharoni A, Giri AP, Deuerlein S, Griepink F, De Kogel WJ, Verstappen FW, Verhoeven HA, Jongsma MA, Schwab W, Bouwmeester HJ (2003) Terpenoid metabolism in wild-type and transgenic Arabidopsis plants. Plant Cell 15: 2866–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S (1993) Site-directed mutagenesis by double polymerase chain reactions. In BA White, ed, PCR Protocols. Methods in Molecular Biology, Vol 15. Humana Press, Totowa, NJ, pp 277–286 [DOI] [PubMed]
- Bell AA (1986) Physiology of secondary products. In JR Mauney, JM Stewart, eds, Cotton Physiology. The Cotton Foundation, Memphis, TN, pp 597–621
- Bell AA, Stipanovic RD (1977) The chemical composition, biological activity, and genetics of pigment glands in cotton. In Beltwide Cotton Production Research Conference, January 10-12, 1977, Atlanta
- Bleecker AB, Patterson SE (1997) Last exit: senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 9: 1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J (1995) The biochemistry and molecular biology of isoprenoid metabolism. Plant Physiol 107: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Chen Z (2002) Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol 129: 706–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XY, Chen Y, Henstein P, Davission VJ (1995) Cloning, expression and characterization of (+)-δ-cadinene synthase: a catalyst for cotton phytoalexin biosynthesis. Arch Biochem Biophys 324: 255–266 [DOI] [PubMed] [Google Scholar]
- Chen XY, Wang MS, Chen Y, Davission VJ, Heinstein P (1996) Cloning and heterologous expression of a second cotton (+)-δ-cadinene synthase. J Nat Prod 59: 944–951 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cormack RS, Eulgem T, Rushton PJ, Köchner P, Hahlbrock K, Somssich IE (2002) Leucine zipper-containing WRKY proteins widen the spectrum of immediate early elicitor-induced WRKY transcription factors in parsley. Biochim Biophys Acta 1576: 92–100 [DOI] [PubMed] [Google Scholar]
- Davila-Huerta G, Hamada H, Davis GD, Stipanovic RD, Adams CM, Essenberg M (1995) Cadinene-type sesquiterpenes induced in Gossypium cotyledons by bacterial inoculation. Phytochemistry 39: 531–536 [Google Scholar]
- Dong JX, Chen CH, Chen ZX (2003) Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol 51: 21–37 [DOI] [PubMed] [Google Scholar]
- Du L, Chen Z (2000) Identification of genes encoding receptor-like protein kinases as possible targets of pathogen- and salicylic acid-induced WRKY DNA-binding proteins in Arabidopsis. Plant J 24: 837–847 [DOI] [PubMed] [Google Scholar]
- Essenberg M, Grover PB, Jr., Cover EC (1990) Accumulation of antibacterial sesquiterpenoids in bacterially inoculated Gossypium leaves and cotyledons. Phytochemistry 29: 3107–3113 [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Schmelzer E, Hahlbrock K, Somssich IE (1999) Early nuclear events in plant defense signaling: rapid gene activation by WRKY transcription factors. EMBO J 18: 4689–4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Verma DPS (1997) Dynamics of phragmoplastin in living cells during cell plate formation and uncoupling of cell elongation from the plane of cells division. Plant Cell 9: 157–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halloin JM, Bell AA (1979) Production of nonglandular terpenoid aldehydes within diseased seeds and cotyledons of Gossypium hirsutum L. J Agric Food Chem 27: 1407–1409 [Google Scholar]
- Hara K, Yagi M, Kusano T, Sano H (2000) Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor upon wounding. Mol Gen Genet 263: 30–37 [DOI] [PubMed] [Google Scholar]
- Hinderhofer K, Zentgraf V (2001) Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 213: 469–473 [DOI] [PubMed] [Google Scholar]
- Hudspeth RL, Hobbs SL, Anderson DM, Grula JW (1996) Characterization and expression of chitinase and 1,3-beta-glucanase genes in cotton. Plant Mol Biol 31: 911–916 [DOI] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 242–250 [Google Scholar]
- Johnson SC, Kolevski B, Smyth DR (2002) TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14: 1359–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalde M, Barth M, Somssich IE, Lippok B (2003) Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol Plant Microbe Interact 16: 295–305 [DOI] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16: 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WQ, Tan XP, Chen XY, Hashimoto T, Yamada Y, Heinstein P (2000) Isolation of a (+)-δ-cadinene synthase gene CAD1-A and analysis of its expression pattern in seedlings of Gossypium arboreum L. Sci China Ser C Life Sci 30: 145–152 [DOI] [PubMed] [Google Scholar]
- Liu CJ, Heinstein P, Chen XY (1999) Expression pattern of genes encoding farnesyl diphosphate synthase and sesquiterpene cyclase in cotton suspension-cultured cells treated with fungal elicitors. Mol Plant Microbe Interact 12: 1095–1104 [DOI] [PubMed] [Google Scholar]
- Luo P, Wang YH, Wang GD, Essenberg M, Chen XY (2001) Molecular cloning and functional identification of (+)-δ-cadinene-8-hydroxylase, a cytochrome P450 monooxygenase (CYP706B1) of cotton sesquiterpene biosynthesis. Plant J 28: 95–104 [DOI] [PubMed] [Google Scholar]
- Meng YL, Jia JW, Liu CJ, Liang WQ, Heinstein P, Chen XY (1999) Coordinated accumulation of (+)-δ-cadinene synthase mRNAs and gossypol in developing seeds of Gossypium hirsutum and a new member of the cad1 family from G. arboreum. J Nat Prod 62: 248–252 [DOI] [PubMed] [Google Scholar]
- Mahmoud SS, Croteau RB (2002) Strategies for transgenic manipulation of monoterpene biosynthesis in plants. Trends Plant Sci 7: 366–373 [DOI] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Gershenzon J (2002) The formation and function of plant volatiles: perfumes for pollinator attraction and defense. Curr Opin Plant Biol 5: 237–243 [DOI] [PubMed] [Google Scholar]
- Puckhaber LS, Dowd MK, Stipanovic RD, Howell CR (2002) Toxicity of (+)- and (−)-gossypol to the plant pathogen, Rhizoctonia solani. J Agric Food Chem 50: 7017–7021 [DOI] [PubMed] [Google Scholar]
- Quirino BF, Normanly J, Amasino RA (1999) Diverse range of gene activity during Arabidopsis thaliana leaf senescence includes pathogen-independent induction of defense-related genes. Plant Mol Biol 40: 267–278 [DOI] [PubMed] [Google Scholar]
- Robatzek S, Somssich IE (2002) Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev 16: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R (1995) Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of α-Amy2 genes. Plant Mol Biol 29: 691–702 [DOI] [PubMed] [Google Scholar]
- Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE (1996) Interaction of elicitor-induced DNA binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J 15: 5690–5700 [PMC free article] [PubMed] [Google Scholar]
- Tan XY, Liang WQ, Liu CJ, Luo P, Heinstein P, Chen XY (2000) Expression pattern of (+)-δ-cadinene synthase genes and biosynthesis of sesquiterpene aldehydes in plants of Gossypium arboreum L. Planta 210: 644–651 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KB, Foley RC, Oñate-Sánchez L (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5: 430–436 [DOI] [PubMed] [Google Scholar]
- Sun C, Palmqvist S, Olsson H, Boren M, Ahlandsberg S, Jansson C (2003) A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15: 2076–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Wang Z, Fan B, Chen C, Chen Z (1999) A pathogen- and salicylic acid-induced WRKY DNA-binding activity recognizes the elicitor response element of the tobacco class I chitinase gene promoter. Plant J 18: 141–149 [Google Scholar]
- Yang Y, Li R, Qi M (2000) In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J 22: 543–551 [DOI] [PubMed] [Google Scholar]
- Yin S, Mei L, Newman J, Back K, Chappell J (1997) Regulation of sesquiterpene cyclase gene expression: characterization of an elicitor- and pathogen-inducible promoter. Plant Physiol 115: 437–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda H, Ogawa M, Yamaguchi Y, Koizumi N, Kusano T, Sano H (2002) Identification of early-responsive genes associated with the hypersensitive response to tobacco mosaic virus and characterization of a WRKY-type transcription factor in tobacco plants. Mol Genet Genomics 267: 154–161 [DOI] [PubMed] [Google Scholar]
- Yu D, Chen C, Chen Z (2001) Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13: 1527–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XJ, Lu L, Xu YH, Wang JW, Chen XY (2002) A cotton cDNA (GaPR-10) encoding a pathogenesis-related 10 protein with in vitro ribonuclease activity. Plant Sci 162: 629–636 [Google Scholar]