Abstract
In plants with crassulacean acid metabolism (CAM), dark CO2 uptake is mediated by phosphoenolpyruvate carboxylase (PEPC), an enzyme that can be regulated at transcriptional and posttranslational levels. Reversible phosphorylation of PEPC is catalyzed by a dedicated PEPC kinase, which in turn is regulated at the transcriptional level over the 24-h cycle in CAM plants. PEPC kinase controls the day/night regulation of PEPC during the CAM cycle, thus facilitating plasticity for optimizing CO2 uptake under different environmental conditions. To understand the importance of PEPC kinase in relation to its target PEPC in terms of CAM performance, the expression of the genes encoding the two enzymes was investigated in four species of Clusia that have photosynthetic patterns ranging from C3 photosynthesis to constitutive CAM. By linking changes in the expression of PEPC and PEPC kinase to day/night patterns of leaf gas exchange, organic acid, and soluble sugar contents under different environmental conditions, the genetic and metabolic limitations to CAM plasticity were assessed. The results indicate that PEPC expression is a major factor underpinning the genotypic capacity for CAM and that PEPC kinase expression does not appear to limit CAM. The day/night regulation of Ppck transcript abundance was found to be a consequence of CAM and the day/night cycling of associated metabolites, rather than the primary controlling factor for the temporal separation of carboxylation processes.
In plants with the potential for crassulacean acid metabolism (CAM), the deployment of nocturnal carboxylation, mediated by the enzyme phosphoenolpyruvate carboxylase (PEPC), permits a wide spectrum of CO2 exchange responses over the day/night cycle. Plasticity in the expression of CAM is central for optimizing carbon gain in a range of potentially limiting environments. Photosynthetic plasticity is illustrated by 24-h gas exchange profiles that can range from exclusive dark CO2 uptake, to no net CO2 uptake (but cycling of C-skeletons behind closed stomata) and from exclusive daytime CO2 uptake to continuous net uptake of CO2 over 24 h (Borland and Griffiths, 1996; Cushman and Borland, 2002). Ultimately, the expression of CAM is a reflection of both the genotype (i.e. species) and the phenotype (i.e. determined by the environment). Since PEPC activity is thought to be a major factor in limiting the magnitude of CAM, it can be hypothesized that the mechanisms that regulate PEPC activity play a key role in determining both genotypic capacity and phenotypic plasticity in CAM expression.
A number of investigations have demonstrated that PEPC activity in CAM plants is regulated at both transcriptional and posttranslational levels. Transcriptional control regulates the amount of PEPC protein, via changes in the abundance of PEPC mRNA. In the facultative species Mesembryanthemum crystallinum and Kalanchoë blossfeldiana, where CAM can be induced by salinity or dehydration, PEPC transcripts can accumulate within 2 to 3 h after stress (Schmitt, 1990; Brulfert et al., 1993; Taybi et al., 1995). Protein accumulation commences 8 to 10 h after treatment. Posttranslational control regulates the day/night activation of PEPC and ensures the temporal separation of C4 and C3 carboxylation processes that is central to the operation of CAM. PEPC is activated at night via phosphorylation of a Ser residue near the N terminus of the protein that renders the enzyme more sensitive to PEP and the positive effectors, Glc-6-P and triose-P, and less sensitive to the allosteric inhibitor, malate (Nimmo et al., 1986; Chollet et al., 1996; Vidal and Chollet, 1997). The phosphorylation state of PEPC is determined by the presence or absence of a dedicated Ca2+-independent Ser/Thr protein kinase, which in turn is regulated by a circadian oscillator (Carter et al., 1991) through day/night changes in transcript abundance (Hartwell et al., 1996). Sequence data suggest that PEPC kinase is unusual among protein kinases in that it is regulated only at the level of gene expression (Hartwell et al., 1999; Taybi et al., 2000). The circadian regulation of PEPC kinase expression can also be modified by leaf metabolic status (Borland et al., 1999). Thus, the integration of circadian and metabolic signals modulates PEPC kinase transcript abundance, which in turn controls PEPC kinase activity and the phosphorylation/activation state of PEPC over the day/night cycle (Nimmo, 2000). PEPC dephosphorylation is controlled by a protein phosphatase 2A having constant activity over the 24-h cycle (Carter et al., 1990, 1991, 1996).
The multilayered control of carbon flux through PEPC provides a mechanistic basis for the photosynthetic plasticity of CAM. However, the relative importance of PEPC and PEPC kinase genes in determining both genotypic capacity and phenotypic plasticity in CAM expression remains unclear. In M. crystallinum, both PEPC and PEPC kinase expression increase in parallel as CAM is induced (Li and Chollet, 1994; Taybi and Cushman, 1999; Taybi et al., 2000). However, other species such as the C3-CAM intermediate Clusia minor can reversibly modulate CAM expression on a day-to-day basis (Borland et al., 1996), a phenomenon that could be achieved solely on the basis of shifts in PEPC kinase expression (Borland and Griffiths, 1997). Evidence in support of a key role for PEPC kinase expression in determining CAM plasticity is provided by observations that increased expression of PEPC kinase in leaves with modified metabolic status is accompanied by increased rates of dark CO2 uptake, without any appreciable change in extractable PEPC activity (Borland et al., 1999). It is generally accepted that CAM evolved from C3 photosynthesis through increased expression of genes involved in C4 acid production and transport. A key mechanism thought to underpin CAM evolution is gene duplication followed by specific regulation of one or more isogene (Cushman and Bohnert, 1999). Different isogenes encoding PEPC have been identified in a variety of species with at least one form specialized in CAM function (Lepiniec et al., 1993; Gehrig et al., 1995, 1998). While more than one gene encodes PEPC kinase in M. crystallinum (Taybi et al., 2000), the same is also found in the C3 plant Arabidopsis (Fontaine et al., 2002) and in the C4 species Flaveria trinervia (Tsuchida et al., 2001). No specific function has been assigned to these isogenes of PEPC kinase although an isogene expressed only in root nodules was recently isolated from soybean (Xu et al., 2003).
In order to understand the importance of PEPC kinase in relation to its target PEPC in terms of CAM performance, the expression of the genes encoding the two enzymes was examined in four species of Clusia that have photosynthetic patterns ranging from C3 to CAM (Borland et al., 1998). By linking changes in the transcript abundance of PEPC and PEPC kinase to day/night patterns of leaf gas exchange, organic acid, and soluble sugar contents under different environmental conditions, the genetic and metabolic limitations to CAM plasticity were assessed. The results indicate that PEPC expression is a major factor underpinning plasticity in CAM expression and that PEPC kinase expression does not appear to limit CAM. Moreover, we demonstrate that the day/night regulation of PEPC kinase expression is a consequence of CAM and is probably mediated in response to metabolite cycling, rather than representing a major controlling factor for the temporal separation of carboxylation processes.
RESULTS AND DISCUSSION
Genotypic Capacity for CAM in Clusia
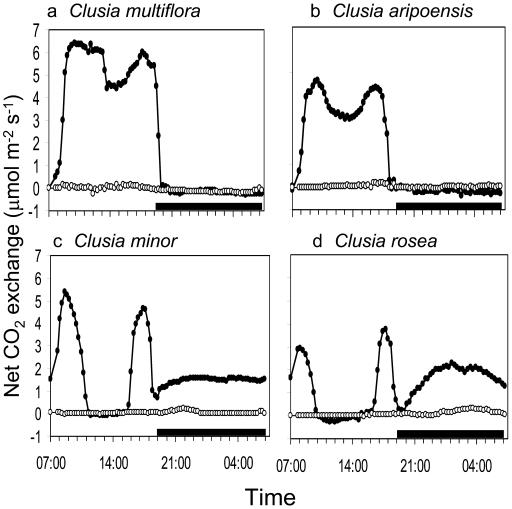
A variety of CO2 exchange profiles, ranging from C3 to constitutive CAM, were observed for the four Clusia species grown under controlled conditions with adequate water supply (Fig. 1). Clusia multiflora and Clusia aripoensis performed C3 photosynthesis, characterized by net CO2 uptake only during the day, while C. minor and Clusia rosea showed the typical 4-phase patterns associated with CAM. It has been reported previously that C. multiflora is a constitutive C3 plant and will not induce CAM in response to drought (Grams et al., 1998; Gehrig et al., 2003) while weak CAM can be induced in C. aripoensis after 7 to 10 d without water (Borland et al., 1998). Under conditions of extended drought (3 weeks), stomata remained closed over the day/night cycle in C. multiflora and C. aripoensis and only a small amount of CO2 was taken up during the dark period in both C. minor and C. rosea (Fig. 1). Upon rewatering, all plants showed full recovery from the extended drought treatment (data not shown).
Figure 1.
Day/night CO2 exchange profiles under well-watered conditions (•) and after 3 weeks without water (o) in four species of Clusia. a, C. multiflora. b, C. aripoensis. c, C. minor. d, C. rosea. The black bars on the x axes represent the periods of darkness. Each gas exchange profile is for a representative leaf from three replicate determinations.
Expression and Abundance of PEPC
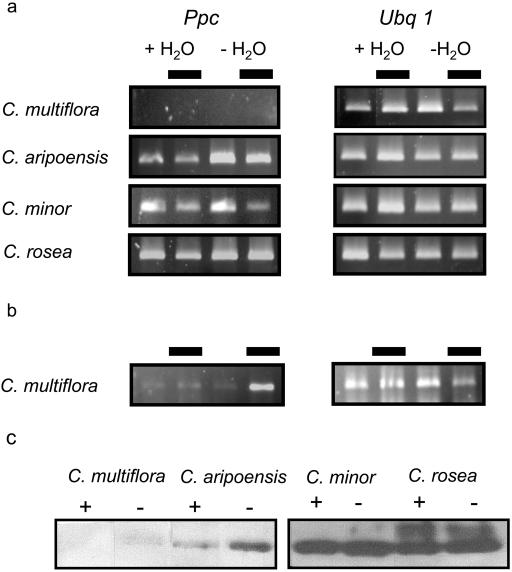
To determine if genotypic differences in the capacity for CAM were reflected by differences in the expression of PEPC, cDNA fragments of Ppc were isolated from each of the four species. At least two Ppc isogenes were found to be expressed in C. minor as suggested by RFLP of 20 clones of the 1,100-bp fragment obtained by reverse transcription (RT)-PCR (data not shown). The total abundance of Ppc transcripts in each of the four species was assessed in leaves collected during the middle of the day and night from well-watered plants and from plants deprived of water for 3 weeks (Fig. 2A). Identical RT-PCR conditions, in terms of RNA input and number of cycles, were used in order to compare the relative Ppc transcript abundance in the four species, with polyubiquitin (Ubq1) as control (Fig. 2A). Under well-watered conditions, transcript abundance of Ppc was highest in C. rosea, followed by C. minor, the two CAM-performing species, although prolonged drought had no detectable effect on the levels of Ppc transcripts in these species. In contrast, extended drought elicited an increase in Ppc transcript abundance in C. aripoensis, a species that shows C3 photosynthesis when well-watered (Fig. 1B) but that can induce limited CAM activity after mild drought (Borland et al., 1998). With these RT-PCR conditions, Ppc transcripts were undetectable in leaves of C. multiflora, but increasing the number of PCR cycles (from 30 to 40) indicated an increase in transcript abundance after prolonged drought in this constitutive C3 species (Fig. 2B). This low abundance of Ppc transcripts in C. multiflora compared to the other species resulted in a low abundance of PEPC protein (Fig. 2C). The finding that drought elicited an increase in PEPC transcript and protein abundance only in C. multiflora and C. aripoensis could be attributed to the greater water loss over the 3-week drought period from these species (30% and 26%, respectively, of leaf fresh weight) compared to the two CAM-performing species (approximately 6%). Leaf dehydration has been reported to be a key stimulus for controlling the expression of Ppc genes in both CAM (Taybi et al., 2000) and in some C3 species (Fedina and Popova, 1996; González et al., 2003).
Figure 2.
a, Steady-state levels of PEPC transcripts (Ppc) in four Clusia species measured by RT-PCR in leaf samples collected at midday and midnight (with black block representing midnight) from well-watered plants (+H2O) and after 3 weeks without water (−H2O). Ubq1 represents the control for equal RNA input and RT-PCR conditions. b, Detection of Ppc transcripts in C. multiflora after 40 cycles of PCR amplification. c, Western blots showing the abundance of PEPC protein in the same leaf samples from well-watered (+) and droughted (−) plants.
Day/night differences in Ppc transcript abundance were apparent in all the species while expression of Ubq1, the RT-PCR control, remained relatively constant. In the three species that had a capacity for CAM, Ppc transcripts appeared to be more abundant during the day (Fig. 2A). This finding is consistent with observations in CAM-performing M. crystallinum (J. Hartwell, personal communication). Day/night differences in transcript abundance were not mirrored by changes in PEPC protein that were unchanged between the day and night in all species (data not shown), indicating slow turnover of PEPC protein. It is notable that in the C3 species C. multiflora, Ppc transcripts were most abundant at night (Fig. 2B). This could imply that the Ppc transcripts in this species encoded a different isoform of PEPC. Since the abundance of Ppc transcripts and protein (Fig. 2C) were extremely low in C. multiflora in comparison with the other three species, it is possible that the transcripts isolated from this species encoded predominantly a C3 or housekeeping isoform of PEPC. This hypothesis is consistent with a recent phylogenetic cladogram of Clusia species, based on nuclear internal transcribed spacer sequences, in which C. multiflora is located on a distinct branch composed of species that are not known to perform CAM (Gehrig et al., 2003). Differences between the species in terms of relative Ppc transcript abundance were also mirrored by differences in the amount of PEPC protein, as indicated by the immunoblots (Fig. 2C). A previous investigation on C. aripoensis, C. minor, and C. rosea showed that the capacity for dark CO2 uptake was closely correlated with the amount and activity of PEPC (Borland et al., 1998). In the present work, maximum levels of malate accumulated in the four species under well-watered conditions were correlated with PEPC transcript and protein abundance. Thus, transcriptional control of PEPC abundance appears to be a key factor in determining genotypic capacity for CAM. Similar considerations on the evolution of C4 photosynthesis within the genus Flaveria have concluded that an increase in PEPC expression/amount was a cardinal event in photosynthetic diversification (Engelmann et al., 2003).
Isolation and Sequence Analysis of Ppck cDNA Fragments
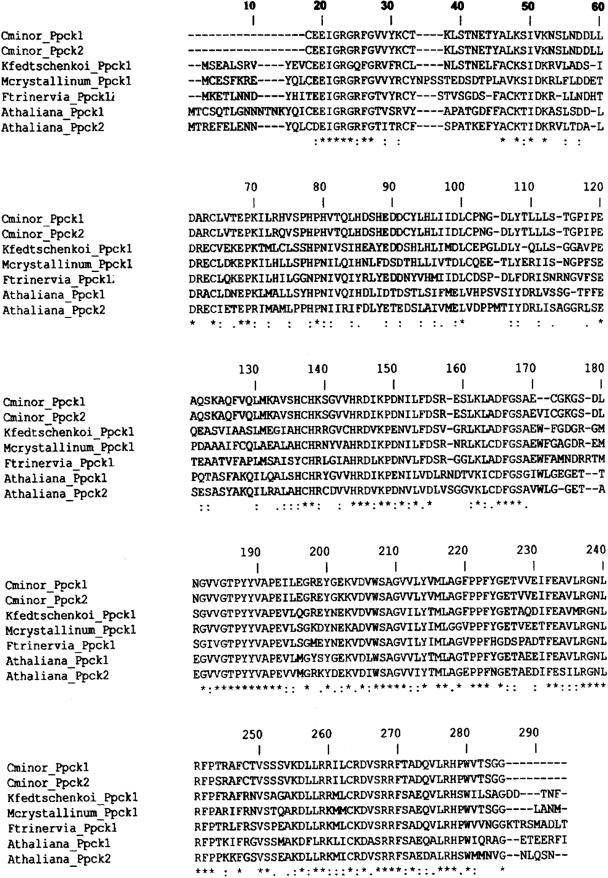
cDNA fragments for the Ppck gene were isolated by RT-PCR from each of the four Clusia species. Two distinct Ppck clones, identified by RFLP and sequencing, lacking approximately 10 amino acids at the N terminus of the PEPC kinase (in comparison with other full-length amino acid sequences) and showing high identity (approximately 98%) were isolated from each species. Figure 3 shows ClustalX amino acid sequence alignments of partial PEPC kinase 1 (AY478419) and PEPC kinase 2 (AY478420) from C. minor with the deduced PEPC kinase amino acid sequences from two CAM species, M. crystallinum (AF158091) and Kalanchoë fedtschenkoi (AF162662); the C4 plant F. trinervia (AB065100); and the C3 plant Arabidopsis (AF162660, AY040830). The conserved regions represent about 45% of the primary consensus sequence. The deduced amino acid sequences of PEPC kinase from C. minor have a shorter extension beyond the kinase domain 11 than other PEPC kinases.
Figure 3.
ClustalX amino acid sequence alignment of two partial PEPC kinases cloned from C. minor (AY478419 and AY478420) with other PEPC kinases from two CAM-plants, M. crystallinum (AF158091) and K. fedtschenkoi (AF162662); a C4 plant, F. trinervia (AB065100); and a C3 plant, Arabidopsis (AF162660, AY040830), where (.), weakly similar; (:), strongly similar; (*), identical.
There were no differences in expression patterns of the different Ppck isoforms under well-watered or droughted conditions in any of the Clusia species (data not shown), suggesting the presence of two copies of a single Ppck gene in the polyploid genome of Clusia. The existence of a small Ppck gene family has been described in species from all photosynthetic categories (i.e. C3, C4, and CAM; Nimmo, 2003). In Arabidopsis, two distinct Ppck genes (88.4% homology) are described. Both appear to be light-regulated but show contrasting expression patterns in different organs (Fontaine et al., 2002). Ppck transcripts of different sizes could be identified in the four Clusia species, which, after sequencing in C. minor, revealed the presence of an intron located within the kinase domain 11 that is similar to other Ppck genes (Nimmo, 2003). Moreover the Ppck2 cDNA contained 3′UTR of two different sizes, 150 and 288 bp, respectively. A 3′UTR of a single size (283 bp) could be detected for the Ppck1 cDNA (data not shown). The Ppck1 cDNA from C. minor contained a single (AATAAA) polyadenylation signal at 222 bp from the polyadenylation site. The Ppck2 cDNA from the same species contained a duplicated (AATAAAATAAA) polyadenylation signal at 93 and 222 bp from the polyadenylation site for the short and long 3′UTR, respectively. The 283- and 288-bp 3′UTRs of Ppck1 and Ppck2, respectively, from C. minor showed additional differences at only two positions and were shorter than the Ppck1 3′UTR from M. crystallinum (669 bp; Taybi et al., 2000). However, the presence of several canonical (GGA and GTA) subdomains in both 3′UTRs from the two Ppck genes in C. minor is similar to those described in M. crystallinum (Taybi et al., 2000). Such elements are suspected to contribute to mRNA instability in plants (Gutiérrez et al., 1999) and suggest potentially rapid turnover of Ppck transcripts. The rapid turnover of Ppck transcripts is an important aspect for the day/night regulation of PEPC kinase and PEPC phosphorylation (Nimmo, 2003) and is thought to be regulated by the 3′UTR.
Day/Night Expression of PEPC Kinase in Four Clusia Species
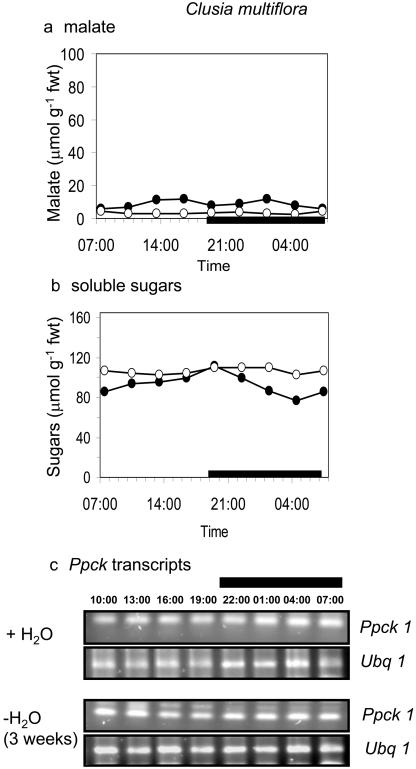
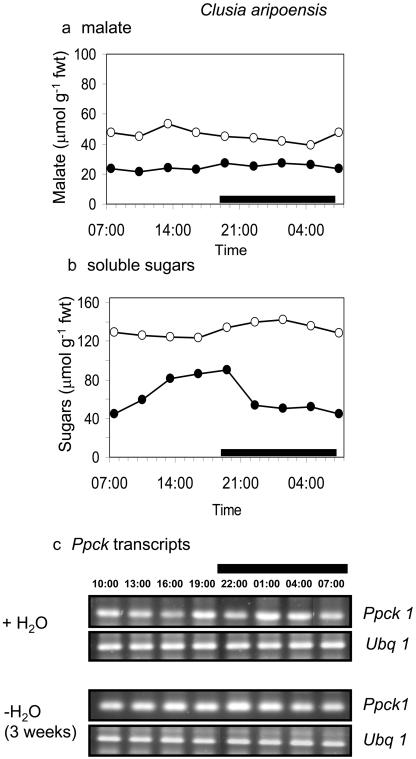
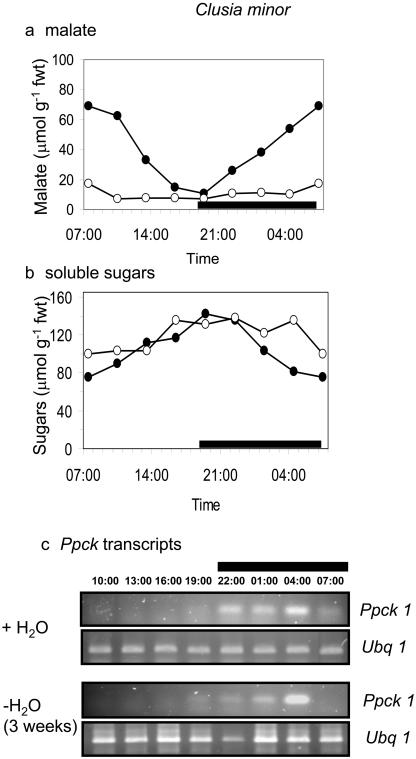
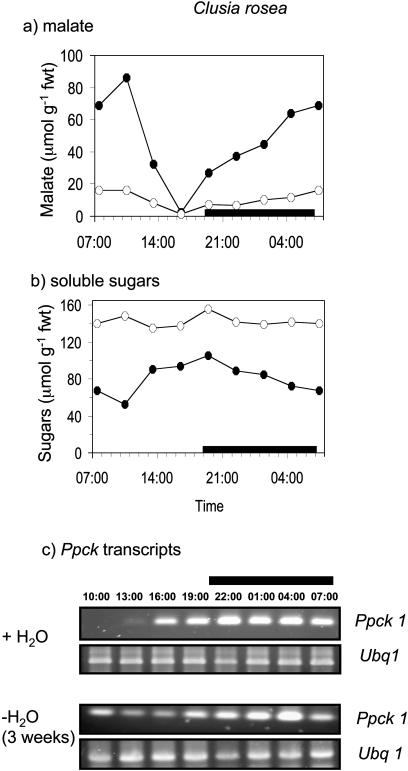
The day/night expression of the Ppck1 gene was studied in the four species under well-watered conditions and after prolonged drought (3 weeks without H2O), a treatment that limited CAM expression in both C. minor and C. rosea (Fig. 1). The rationale behind the species comparison was to determine if Ppck expression was a key factor determining the genotypic capacity for CAM. The rationale behind the two watering treatments was to determine whether Ppck expression was modulated as a direct response to drought or in response to day/night changes in the levels of metabolites, including malate and soluble sugars, the cycling of which was suppressed under severe drought stress. As shown in Figures 4 and 5, transcript levels for Ppck were comparable in both C. multiflora and C. aripoensis. Neither species displayed any CAM activity, in terms of day/night changes in malate content under well-watered or severely droughted conditions (Figs. 4A and 5A), and Ppck transcript abundance was relatively constant over the 24-h cycle (Figs. 4C and 5C). There was little discernible effect of extended drought on Ppck transcript abundance in either C. multiflora or C. aripoensis. Since drought led to an increase in Ppc transcript abundance in both of these species (Fig. 2), it appears that different signals regulate the expression of PEPC and PEPC kinase. In C. minor and C. rosea, the two species that expressed CAM under well-watered conditions as evidenced by day/night changes in malate content (Figs. 6A and 7A), day/night changes in Ppck transcript abundance were observed (Figs. 6C and 7C). In C. minor, Ppck transcripts could be detected over the first 9 to 10 h of the 24-h cycle and before the start of the dark period (Fig. 6C). Extended drought suppressed CAM expression in terms of day/night changes in malate content in C. minor (Fig. 6A). However, day/night cycling of soluble sugars was still evident (Fig. 6B) as were day/night changes in Ppck transcript abundance in this species after extended drought (Fig. 6C). In C. rosea, the species with maximal CAM expression under well-watered conditions in terms of dark CO2 uptake (Fig. 1) and day/night changes in leaf malate content (Fig. 7A), Ppck transcripts could be detected over the latter half of the light period and over the entire duration of the dark period (Fig. 7C). However, after a period of extended drought, day/night cycling of both malate and soluble sugars was suppressed in C. rosea (Fig. 7, A and B). Under these conditions, day/night changes in Ppck transcript abundance were dampened compared to well-watered plants (Fig. 7C). Specifically, down-regulation of Ppck expression at the start of the photoperiod was less apparent in severely droughted plants of C. rosea.
Figure 4.
Day/night changes in levels of a, malate; b, soluble sugars; and c, transcripts for PEPC kinase (Ppck1) and polyubiquitin (Ubq1) as control for equal RNA input and RT-PCR conditions in leaves of C. multiflora under well-watered conditions (•) and after 3 weeks without water (○). The black bars on the x axes represent the periods of darkness.
Figure 5.
Day/night changes in levels of a, malate; b, soluble sugars; and c, transcripts for PEPC kinase (Ppck1) and polyubiquitin (Ubq1) as control for equal RNA input and RT-PCR conditions in leaves of C. aripoensis under well-watered conditions (•) and after 3 weeks without water (○). The black bars on the x axes represent the periods of darkness.
Figure 6.
Day/night changes in levels of a, malate; b, soluble sugars; and c, transcripts for PEPC kinase (Ppck1) and polyubiquitin (Ubq1) as control for equal RNA input and RT-PCR conditions in leaves of C. minor under well-watered conditions (•) and after 3 weeks without water (○). The black bars on the x axes represent the periods of darkness.
Figure 7.
Day/night changes in levels of a, malate; b, soluble sugars; and c, transcripts for PEPC kinase (Ppck1) and polyubiquitin (Ubq1) as control for equal RNA input and RT-PCR conditions in leaves of C. rosea under well-watered conditions (•) and after 3 weeks without water (○). The black bars on the x axes represent the periods of darkness.
Comparison of Ppck expression profiles in the four species indicates that transcript abundance of PEPC kinase does not limit CAM expression in Clusia since the levels of maximum transcript abundance were comparable between the different species. Data obtained with other CAM species, including K. fedtschenkoi and M. crystallinum, have indicated that Ppck transcript abundance is the major factor controlling the phosphorylation of PEPC and hence nocturnal CO2 uptake (Hartwell et al., 1999; Taybi et al., 2000). Moreover, increased expression of PEPC kinase during the night in salt-stressed sorghum plants resulted in increased malate accumulation with a CAM-like pattern in this typical C4 plant (García-Mauriño et al., 2003). A constant level of Ppck expression over the 24-h cycle, as observed in C. multiflora and C. aripoensis (both operating in C3-mode), could imply that PEPC was permanently phosphorylated in these species and that PEPC abundance was the major limitation to CAM expression. The relatively high but stable levels of malate measured over the 24-h cycle in C. aripoensis, which had considerably higher levels of PEPC protein compared to C. multiflora, supports this view (Figs. 4A and 5A). However, there are reports of some situations, particularly in C3 plants, where the phosphorylation status of PEPC does not correlate with changes in PEPC kinase activity or expression (Smith et al., 1996; Osuna et al., 1999). The reported existence of a protein that reversibly inhibits PEPC kinase could represent an additional layer of control over CO2 flux through PEPC (Nimmo et al., 2001). Indeed, preliminary studies have indicated that the in vitro phosphorylation of purified PEPC is inhibited by extracts prepared from C. minor, implying the existence of this inhibitor protein in at least one of the Clusia species under investigation (J. Hartwell and A. Borland, unpublished data).
The results obtained for the four Clusia species also indicate that the day/night regulation of the Ppck gene appears to be a consequence of CAM expression and the day/night cycling of metabolites. Moreover, day/night changes in Ppck transcript abundance appear to be controlled through a down-regulation of the gene during the day rather than an up-regulation during the night. These findings contrast with results obtained with M. crystallinum in which Ppck transcripts are present in low abundance in C3 leaves and the induction of CAM is accompanied by an up-regulation of Ppck transcript abundance at night (Taybi et al., 2000). However, the results presented for Clusia are consistent with the view that the circadian control of Ppck gene expression in CAM is a consequence of metabolite transport across the tonoplast (Borland et al., 1999; Nimmo, 2000). The current working hypothesis is that cytosolic malate acts as a feedback regulator of Ppck gene expression. Thus, daytime efflux of malate from the vacuole may be a primary signal for the down-regulation of Ppck gene expression during the photoperiod in plants performing CAM (Nimmo, 2000). Clusia species can accumulate high concentrations of both malic and citric acids and background concentrations of both organic acids can be relatively high in plants performing C3 photosynthesis, as found in C. aripoensis (Fig. 5B; see also Borland et al., 1996). This contrasts with the situation in M. crystallinum where both malate and citrate contents are low in the C3 mode. Thus, in Clusia, it would appear that daytime mobilization of organic acids is a key factor regulating both the expression of CAM and the Ppck gene (Borland et al., 1996; Roberts et al., 1998). The role of other cytosolic metabolites in modulating Ppck gene expression is still unclear. It is possible that soluble sugars could play a role in this regard, as indicated by the continued day/night cycling of Ppck transcripts in droughted C. minor where malate levels remained relatively constant, yet soluble sugars continued to cycle (Fig. 6). The accumulation of soluble sugars during the day could contribute to the down-regulation of the Ppck gene. The role of soluble sugars in signaling the regulation of gene expression is well established in plants (for review, see Smeekens, 2000). To gain further insight on the regulation of Ppck expression in C. minor, a range of different treatments was used to manipulate CAM and metabolite cycling in this species.
Phenotypic Plasticity in Expression of CAM and PEPC Kinase in C. Minor
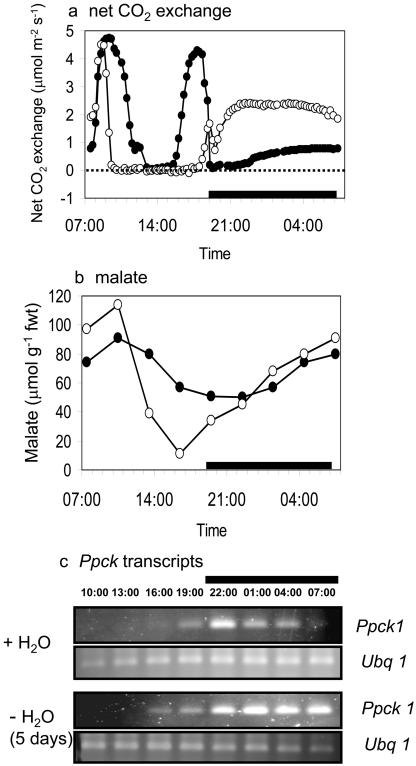
A number of studies have reported the extreme photosynthetic plasticity of C. minor under varying environmental and developmental conditions (Borland et al., 1996; Lüttge, 1996), raising the question of the role of PEPC kinase in determining phenotypic plasticity in CAM expression. Imposing mild drought stress (5 d without H2O) resulted in a substantial increase in dark CO2 uptake (Fig. 8A), together with an increase in daytime mobilization and subsequent overnight accumulation of malate (Fig. 8B) in this species. Concurrent with the increase in CAM, nocturnal Ppck transcript abundance was extended in plants subjected to mild drought (Fig. 8C). Extended nocturnal expression of Ppck, accompanied by enhanced dark CO2 uptake, has also been observed in leaves of Kalanchoë daigremontiana, in which endogenous levels of malate were reduced after exposure to an atmosphere of N2 (Borland et al., 1999). If a primary response to mild drought is enhanced sequestration of malate (or some other metabolite) in the vacuole at night, the results obtained with C. minor are consistent with the hypothesis of metabolite control of Ppck (Nimmo, 2000). A drought-induced increase in the activity of vacuolar ATPase in C. minor, as noted in M. crystallinum when CAM is induced (Barkla et al., 1999), could enhance the removal of malate from the cytosol and thus delay the down-regulation in Ppck expression.
Figure 8.
Day/night changes in a, net CO2 uptake; b, malate content; and c, transcripts for PEPC kinase (Ppck1) and polyubiquitin (Ubq1) as control for equal RNA input and RT-PCR conditions in leaves of C. minor under well-watered conditions (•) and after 5 d without water (○). The black bars on the x axes represent the periods of darkness.
A day/night difference in transcript abundance of Ppck was also found in young leaves of C. minor (Fig. 9A) that did not exhibit CAM, as indicated by the relatively low and constant levels of titratable acidity over the 14-h cycle, although significant day/night changes in soluble sugars were detected (data not shown). Mature leaves from the same plants showed similar day/night fluctuations in transcript abundance of Ppck and day/night changes in titratable acidity (Fig. 9A), and soluble sugar contents were apparent (data not shown). In contrast, subjecting plants to a low light treatment (100 μmol m−2 s−1 for 5 d) resulted in a dampening of day/night changes in metabolites as indicated by measurements of titratable acidity (Fig. 9B) and in soluble sugars, the levels of which were considerably reduced under low light (data not shown). Under these conditions, there was no apparent day/night difference in Ppck transcript abundance (Fig. 9B). Together, the results suggest interplay between organic acids and soluble sugars in the modulation of Ppck gene expression in C. minor. The marked phenotypic plasticity of plants such as C. minor for switching rapidly and reversibly between C3 photosynthesis and CAM may thus be attributed to the transport and/or enzymatic processes that regulate partitioning of metabolites such as organic acids and soluble sugars, between vacuole and cytosol.
Figure 9.
a, Abundance of transcripts for PEPC kinase (Ppck1) over the day and night in young leaves (C3 photosynthesis) and mature leaves (CAM) of C. minor with corresponding levels of titratable acidity shown for each time point. b, Abundance of Ppck1 transcripts over the day and night in mature leaves of C. minor performing C3 photosynthesis with corresponding levels of titratable acidity shown for each time point. Ubq1 transcripts are shown as control for equal RNA input and RT-PCR conditions. The black bars represent the periods of darkness.
CONCLUSIONS
Differences between species in the genotypic capacity and phenotypic plasticity of CAM expression are exemplified by tropical trees within the genus Clusia. Gas exchange profiles ranging from C3 photosynthesis through to constitutive CAM are exhibited by species from this genus. The expression and abundance of PEPC protein are major factors underpinning the genotypic capacity for CAM in Clusia. There is no evidence to indicate the existence of separate C3 and CAM-specific isoforms of PEPC kinase in Clusia. The same isoforms of Ppck genes are expressed at comparable levels in both C3 and CAM performing species, implying the existence of additional layers of control over CO2 flux through PEPC. Moreover, different signals regulate the expression of PEPC and PEPC kinase in Clusia. Day/night changes in the transcript abundance of Ppck appear to be a consequence of the day/night cycling of organic acids and soluble sugars. The changes in Ppck transcript abundance that occur over the 24-h cycle appear to be controlled through a down-regulation of the gene during the day rather than via an up-regulation during the night as reported for M. crystallinum. In Clusia, CAM-associated metabolites appear to act as signals triggering a cascade of events that control the down-regulation of the Ppck gene during the day. Thus, day/night regulation of the CAM cycle seems to be controlled at a level upstream of Ppck gene expression and PEPC phosphorylation. It is likely that metabolite flux across the vacuolar membrane will be a major controlling factor for regulating CAM and PEPC kinase expression in Clusia. Moreover, differences in the day/night expression of the Ppck gene between Clusia and M. crystallinum after CAM induction imply that the pathways linking Ppck gene expression to the well-documented circadian oscillator of CAM may have diverged between different species.
MATERIALS AND METHODS
Plant Material
All plants were propagated from cuttings; Clusia aripoensis Britt., Clusia minor, and Clusia rosea Jacq. were originally collected from various habitats on the island of Trinidad, West Indies while cuttings of Clusia multiflora were collected from northern Venezuela. Rooted cuttings were planted in commercial compost in 127–mm-diameter pots and grown up in tropical greenhouse conditions at Moorbank Botanic Gardens (University of Newcastle upon Tyne, UK). At least 2 weeks before each experiment, plants were transferred into a growth cabinet (Sanyo Gallenkamp, Leicester, UK) programmed to deliver progressive changes in light intensity and temperature at the beginning and the end of the photoperiod to mimic natural conditions. From 08:30 until 12:00, photon flux density increased to a maximum of 500 μmol m−2 s−1 at leaf height, temperature increased from 19°C to 27°C and relative humidity decreased from 80% to 60% (vapor pressure deficit increased from 1.8 to 2.9 kPa). These conditions were maintained until 16:00 when photon flux density decreased gradually until lights went off at 19:30, temperature decreased to 19°C, and relative humidity increased to 80% (vapor pressure deficit = 1.8 kPa). Over the 13-h dark period, temperature (19°C) and relative humidity (80%) remained constant.
Net CO2-Exchange Measurements
Gas exchange measurements were made continuously on a single leaf over 24-h periods with three separate runs made for each treatment. The leaf was enclosed in a porometer head that tracked the environmental conditions in the growth chamber with gas exchange parameters measured using an open infrared gas analyzer (H. Walz, Effeltrich, Germany). Rates of net CO2 exchange were calculated using DIAGAS software provided by Walz.
Organic Acid Determination
Discs were punched from replicate leaves at intervals over the day/night cycle and immediately placed in 5 mL of 80% methanol preheated to 80°C to stop all reactions. Methanol extracts were heated for 1 h to extract organic acids and soluble sugars (see below). One milliliter of methanol extract was then diluted with 2 mL of distilled water and titrated against 0.01 n NaOH solution in the presence of phenolphthalein in order to determine leaf titratable acid content. The remaining methanol extract was evaporated to dryness and subsequently taken up in 200 mm bicine, pH 7.6. The concentration of malate was determined using the enzymatic method described by Hohorst (1970).
Soluble Sugar Determination
Total soluble sugars were also measured in the methanol extracts using the phenol-sulfuric acid colorimetric assay (Dubois et al., 1956). One milliliter of 5% phenol solution was added to 1 mL of methanol extract followed by 5 mL of concentrated sulfuric acid to elicit a reaction that produced a brown precipitate in suspension. The resulting absorbance was measured in a spectrophotometer at 483 nm against a standard calibration curve established from a Glc solution of known concentration. Soluble sugar contents are expressed as μmol Glc equivalents g−1 fresh weight. All metabolite measurements were repeated, and representative data are shown.
Isolation of Partial cDNA Clones for PEPC (Ppc) and PEPC Kinase (Ppck) Genes
Isolation of partial cDNA fragments of Ppc and Ppck genes from each of the four Clusia species was achieved by RT-PCR using RNA that was bulked from extracts taken during the day and the night from unstressed and drought-stressed plants. Exactly 1 μg and 500 ng of DNase I-treated total RNA were used to amplify fragments of the two genes, respectively, by RT-PCR using degenerate primers. A 1,100-bp amplicon was obtained for the Ppc gene using degenerate primers 5′-TCHGAYTCHGGMAARGAYGC-3′ (forward primer, melting temperature (Tm) = 56°C/68°C) and 5′-GCDGCDATVCCYTTCATKG-3′ (reverse primer, Tm = 54°C/64°C), where D = A + T + G, K = T + G, M = A + C, R = A + G, Y = C + T, and V = A + C + G. A 500-bp amplicon was obtained using Ppck degenerate primers 5′-TGCGAG GAGATCGGCCGDG-3′ (forward primer, Tm = 64°C/66°C) and 5′-ACCTCCGGCGCCACGTARTAC-3′ (reverse primer, Tm = 68°C/70°C). The cDNA fragments obtained using pfu DNA polymerase (Stratagene, Amsterdam) were gel-extracted and 3′A-overhangs were added to them using Taq DNA polymerase (Bioline, London). The fragments were then cloned into PCR2.1 vector using the Topo TA cloning Kit (Invitrogen, Paisley, UK). Twenty clones for each fragment were analyzed by RFLP using four restriction enzymes: BamH1, EcoRI, XhoI, and XbaI. One clone from each restriction pattern was sequenced on an ABI 373 sequencer (Perkin-Elmer Applied Biosystems, Foster City, CA). The identity of the sequences was confirmed by searching the National Center for Biotechnology Information Network (http://www.ncbi.nlm.nih.gov) using BLAST 2.0 as search tool (Altschul et al., 1997). Multiple sequence alignments were conducted with the ClustalX multiple alignment program (Thompson et al., 1997).
The 3′ untranslated region of Ppck was recovered by 3′-RACE (Life Technologies/Gibco BRL, Paisley, UK) according to manufacturer's instructions using gene specific primers GSPa (5′-GTGTCGTTCACAGAGACAT-3′ Tm = 56°C) with two nested primers specific to each form of the Ppck gene, GSPb1 (5′-TTCGGTTCCGCTGAGTG-3′, Tm = 54°C) or GSPB2 (5′-TTCGGTTCCGCTGAGTTATT-3′, Tm = 58°C ). The anchor primers were 5′-GCCCGTCGACTAGTCTAGATTTTTTTTTTTTTTTTT-3′ and 5′-GCCCGTCGACTAGTCTAG-3′ used for reamplification. The identity of each fragment was confirmed by sequence analysis.
RNA Extraction and Semiquantitative RT-PCR
Due to high levels of phenolic compounds, Clusia tissues are particularly challenging for RNA extraction (Gehrig et al., 2000). To overcome this problem, leaves were powdered in liquid N2 and then homogenized in 0.5 m Tris buffer, pH 8, containing 0.3 m LiCl, 10 mm dithiothreitol, 10 mm EDTA, 1% (v/v), Nanonidet pyrolidone 40,000 (NP40), 5 mm urea, and 2% (w/v) polyethylene glycol 20,000. The extract was centrifuged at 13,000g for 10 min at 4°C, and the supernatant was subsequently used for RNA extraction using Tri-reagent (Helena Biosciences, Sunderland, UK) following the method previously described (Taybi and Cushman, 1999).
Semiquantitative RT-PCR assays were performed to determine the level of transcript abundance for the Ppc gene with the same degenerate primer set used for cDNA isolation and using 250 ng of DNase I-treated RNA and 30 PCR cycles. Ppck transcript abundance was assessed by RT-PCR using 250 ng of DNase I-treated RNA and two rounds of amplification of 20 cycles using specific primer sets 5′-TGCGAGGAGATCGGCCGDG-3′ (forward primer, Tm = 64°C/66°C) and 5′-GACCGAAAATTAGGAGGAT-3′ (reverse primer, Tm = 54°C) and 5′-TTAATACTCTCCACTGGGC-3′ (forward primer, Tm = 58°C) for reamplification of 1 μL of RT-PCR. As a control, transcript levels of polyubiquitin (AY478418) were determined by RT-PCR using specific primers 5′-ACAACATCCAGAAGGAGT-3′ (forward primer, Tm = 52°C) and 5′-GATCTTGGCCTTAACCAT-3′ (reverse primer, Tm = 52°C) to generate a 600-bp fragment using 250 ng of DNase I-treated RNA and 32 cycles. After amplification, the reaction products were resolved by electrophoresis on 1.2% (w/v) agarose gel and stained with ethidium bromide. Images were captured using a Gel-Doc system (Bio-Rad, Hercules, CA). All semiquantitative RT-PCR reactions were within the linear dynamic range, and all experiments were repeated twice with representative data shown. Corresponding data for CO2-exchange, malate, and soluble sugars are presented in parallel with Ppck transcript levels.
Western-Blot Analyses
To determine changes in the amount of PEPC protein in the four Clusia species, crude extracts were prepared from leaves harvested at midday and midnight from well-watered plants and plants subjected to 3 weeks of drought. Leaves were powdered in liquid N2 and then homogenized in 0.5 m Tris-HCl buffer, pH 8, containing 10 mm dithiothreitol, 10 mm EDTA, 1% (v/v) NP40, 5 mm urea and 2% (w/v) polyethylene glycol 20,000. Extracts were clarified by centrifugation at 13,000g for 10 min at 4°C. Supernatant was transferred to a fresh tube, and protein content was determined (Bradford, 1976). Exactly 20 μg of proteins from each species were resolved on 12% polyacrylamide gels, blotted onto polyvinyldenefluoride membranes (Sigma, St. Louis), and anti-PEPC polyclonal antibody was used to identify PEPC bands. Detection was achieved using the Enhanced Chemi-Luminescence (ECL) system (Amersham, Buckinghamshire, UK) following manufacturer's instructions.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AB065100, AF158091, AF162660, AF162662, AY040830, AY478418, AY478419, and AY478420.
Acknowledgments
We thank Professor Ulrich Lüttge for the gift of cuttings of C. multiflora, Dr. James Hartwell for advice in designing Ppck primers, and Professor Hans Bohnert for providing the PEPC antibody.
This work was supported by the Natural Environment Research Council, UK (grant no. GR3/12463).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.036962.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkla BJ, Vera-Estrella R, Maldonaldo-Gama M, Pantoja O (1999) Abscisic acid induction of vacuolar H+-ATPase activity in Mesembryanthemum crystallinum is developmentally regulated. Plant Physiol 120: 811–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borland AM, Griffiths H (1996) Variations in the phases of crassulacean acid metabolism and regulation of carboxylation patterns determined by carbon-isotope-discrimination techniques. In K Winter, JAC Smith, eds, Crassulacean Acid Metabolism. Biochemistry, Ecophysiology and Evolution, Ecological Studies, Vol 114. Springer-Verlag, Heidelberg, pp 230–249
- Borland AM, Griffiths H (1997) A comparative study on the regulation of C3 and C4 carboxylation processes in the constitutive crassulacean acid metabolism (CAM) plant Kalanchoë daigremontiana and the C3-CAM intermediate Clusia minor. Planta 201: 368–378 [DOI] [PubMed] [Google Scholar]
- Borland AM, Griffiths H, Maxwell C, Broadmeadow MSJ, Fordham MC (1996) CAM induction in Clusia minor L. during the transition from wet to dry season in Trinidad: the role of organic acid speciation and decarboxylation. Plant Cell Environ 19: 655–664 [Google Scholar]
- Borland AM, Hartwell J, Jenkins GI, Wilkins MB, Nimmo HG (1999) Metabolite control overrides circadian regulation of phosphoenolpyruvate carboxylase kinase and CO2 fixation in crassulacean acid metabolism. Plant Physiol 121: 889–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borland AM, Técsi LI, Leegood RC, Walker RP (1998) Inducibility of crassulacean acid metabolism (CAM) in Clusia species: physiological/ biochemical characterisation and intracellular localization of carboxylation and decarboxylation processes in three species which exhibit different degrees of CAM. Planta 205: 342–351 [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brulfert J, Güclü S, Taybi T, Pierre JN (1993) Enzymatic responses to water stress in detached leaves of the CAM plant Kalanchoë blossfeldiana Poelln., cv Tom Thumb. Plant Physiol Biochem 31: 491–497 [Google Scholar]
- Carter PJ, Fewson CA, Nimmo GA, Nimmo HG, Wilkins MB (1990) Bryophyllum fedtschenkoï protein phosphatase 2A can dephosphorylate phosphenolpyruvate carboxylase. FEBS Lett 263: 233–236 [Google Scholar]
- Carter PJ, Fewson CA, Nimmo GA, Nimmo HG, Wilkins MB (1996) Role of circadian rhythms, light and temperature in the regulation of phosphoenolpyruvate carboxylase in crassulacean acid metabolism. In K Winter, JAC Smith, eds, Crassulacean Acid Metabolism. Biochemistry, Ecophysiology and Evolution, Ecological Studies, Vol 114. Springer-Verlag, Heidelberg, pp 46–52
- Carter PJ, Nimmo HG, Fewson CA, Wilkins MB (1991) Circadian rhythms in the activity of a plant protein kinase. EMBO J 10: 2063–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chollet R, Vidal J, O'Leary MH (1996) Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 273–298 [DOI] [PubMed] [Google Scholar]
- Cushman JC, Bohnert HJ (1999) Crassulacean acid metabolism: molecular genetics. Annu Rev Plant Physiol Plant Mol Biol 50: 305–332 [DOI] [PubMed] [Google Scholar]
- Cushman JC, Borland AM (2002) Induction of crassulacean acid metabolism by water limitation. Plant Cell Environ 25: 295–310 [DOI] [PubMed] [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebus PA, Smith F (1956) Colorimetric method for the determination of sugars and related substances. Anal Chem 28: 350–356 [Google Scholar]
- Engelmann S, Bläsing OE, Gowik U, Svensson P, Westhoff P (2003) Molecular evolution of C4 phosphoenolpyruvate carboxylase in the genus Flaveria – a gradual increase from C3 to C4 characteristics. Planta 217: 717–725 [DOI] [PubMed] [Google Scholar]
- Fedina IS, Popova AV (1996) Photosynthesis, photorespiration and proline accumulation in water-stressed pea leaves. Photosynthetica (Prague) 32: 213–220 [Google Scholar]
- Fontaine V, Hartwell J, Jenkins GI, Nimmo HG (2002) Arabidopsis thaliana contains two phosphoenolpyruvate carboxylase kinase genes with different expression patterns. Plant Cell Environ 25: 115–122 [Google Scholar]
- García-Mauriño S, Monreal JA, Alvarez R, Vidal J, Echevarría C (2003) Characterization of salt stress-enhanced phosphoenolpyruvate carboxylase kinase activity in leaves of Sorghum vulgare: independence from osmotic stress, involvement of ion toxicity and significance of dark phosphorylation. Planta 216: 648–655 [DOI] [PubMed] [Google Scholar]
- Gehrig HH, Aranda J, Cushman MA, Virgo A, Cushman JC, Hammel BE, Winter K (2003) Cladogram of Panamanian Clusia based on nuclear DNA: implications for the origins of crassulacean acid metabolism. Plant Biol 5: 59–70 [Google Scholar]
- Gehrig HH, Taybi T, Kluge M, Brulfert J (1995) Identification of multiple PEPC isogenes in leaves of the facultative crassulacean acid metabolism (CAM) plant Kalanchoë blossfeldiana Poelln. cv Tom Thumb. FEBS Lett 377: 399–402 [DOI] [PubMed] [Google Scholar]
- Gehrig HH, Valentina H, Kluge M (1998) Towards a better knowledge of the molecular evolution of phosphoenolpyruvate carboxylase by comparison of partial cDNA sequences. J Mol Evol 46: 107–114 [DOI] [PubMed] [Google Scholar]
- Gehrig HH, Winter K, Cushman JC, Borland AM, Taybi T (2000) An improved RNA isolation method for succulent plant species rich in polyphenols and polysaccharides. Plant Mol Biol Report 18: 1–8 [Google Scholar]
- González M-C, Sánchez R, Cedudo FJ (2003) Abiotic stresses affecting water balance induce phosphoenolpyruvate carboxylase expression in roots of wheat seedlings. Planta 216: 985–992 [DOI] [PubMed] [Google Scholar]
- Grams TEE, Herzog B, Lüttge U (1998) Are there species in the genus Clusia with obligate C3 photosynthesis? J Plant Physiol 152: 1–9 [Google Scholar]
- Gutiérrez RA, MacIntosh GC, Green PJ (1999) Current perspectives on mRNA stability in plants: multiple levels and mechanisms of control. Trends Plant Sci 4: 429–438 [DOI] [PubMed] [Google Scholar]
- Hartwell J, Gill A, Nimmo GA, Wilkins MB, Jenkins GI, Nimmo HG (1999) Phosphoenolpyruvate carboxylase kinase is a novel protein kinase regulated at the level of transcription. Plant J 20: 333–342 [DOI] [PubMed] [Google Scholar]
- Hartwell J, Smith LH, Wilkins MB, Jenkins GI, Nimmo HG (1996) Higher plant phosphoenolpyruvate carboxylase kinase is regulated at the level of translatable mRNA in response to light or a circadian rhythm. Plant J 10: 1071–1078 [Google Scholar]
- Hohorst HJ (1970) L-malate estimation with malate dehydrogenase and NAD. In HV Bergmeyer, ed, Methods in Enzymatic Analysis. Verlag Chemie, Weinheim, pp 1544–1548
- Lepiniec L, Keryer E, Philipe H, Gadal P, Cretin C (1993) The phosphoenolpyruvate carboxylase gene family of sorghum: structure, function and molecular evolution. Plant Mol Biol 21: 487–502 [DOI] [PubMed] [Google Scholar]
- Li B, Chollet R (1994) Salt induction and the partial purification/characterisation of phosphoenolpyruvate carboxylase protein-serine kinase from an inducible crassulacean acid metabolism (CAM) plant Mesembryanthemum crystallinum L. Arch Biochem Biophys 314: 247–254 [DOI] [PubMed] [Google Scholar]
- Lüttge U (1996) Clusia: plasticity and diversity in a genus of C3/CAM intermediate tropical trees. In K Winter, JAC Smith, eds, Crassulacean Acid Metabolism. Biochemistry, Ecophysiology and Evolution, Ecological Studies, Vol 114. Springer-Verlag, Heidelberg, pp 296–311
- Nimmo GA, Nimmo HG, Hamilton ID, Fewson CA, Wilkins MB (1986) Purification of the phosphorylated night form and dephosphorylated day form of phosphoenolpyruvate carboxylase from Bryophyllum fedtshenkoï. Biochem J 239: 213–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo GA, Wilkins MB, Nimmo HG (2001) Partial purification and characterization of a protein inhibitor of phosphoenolpyruvate carboxylase kinase. Planta 213: 250–257 [DOI] [PubMed] [Google Scholar]
- Nimmo HG (2000) The regulation of phosphoenolpyruvate carboxylase in CAM plants. Trends Plant Sci 5: 75–80 [DOI] [PubMed] [Google Scholar]
- Nimmo HG (2003) Control of the phosphorylation of PEP carboxylase in higher plants. Arch Biochem Biophys 414: 189–196 [DOI] [PubMed] [Google Scholar]
- Osuna L, Pierre JN, Gonzalez MC, Alvarez R, Cejudo FJ, Echevarria C, Vidal J (1999) Evidence for a slow-turnover form of the Ca2+-independent phosphoenolpyruvate carboxylase kinase in the aleurone-endosperm tissue of germinating barley seeds. Plant Physiol 119: 511–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Borland AM, Maxwell C, Griffiths H (1998) Ecophysiology of the C3-CAM intermediate Clusia minor in Trinidad: seasonal and short-term photosynthetic characteristics of sun and shade leaves. J Exp Bot 49: 1563–1573 [Google Scholar]
- Schmitt JM (1990) Rapid concentration changes of PEPC mRNA in detached leaves of Mesembryanthemum crystallinum in response to wilting and rehydration. Plant Cell Environ 13: 845–850 [Google Scholar]
- Smeekens S (2000) Sugar-induced signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol 51: 49–81 [DOI] [PubMed] [Google Scholar]
- Smith LH, Lillo C, Nimmo HG, Wilkins MB (1996) Light regulation of phosphoenolpyruvate carboxylase in barley mesophyll protoplasts is mediated by protein synthesis and calcium, and not necessarily correlated with phosphoenolpyruvate carboxylase kinase activity. Planta 200: 174–180 [Google Scholar]
- Taybi T, Cushman JC (1999) Signalling events leading to crassulacean acid metabolism induction in the common ice plant. Plant Physiol 121: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taybi T, Patil S, Chollet R, Cushman JC (2000) A minimal serine/threonine protein kinase circadianly regulates phosphoenolpyruvate carboxylase activity in crassulacean acid metabolism-induced leaves of the common ice plant. Plant Physiol 123: 1471–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taybi T, Sotta B, Gehrig HH, Güclü S, Kluge M, Brulfert J (1995) Differential effects of abscisic acid on phosphoenolpyruvate carboxylase and CAM operation in Kalanchoë blossfeldiana. Bot Acta 198: 240–246 [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface; flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida Y, Furumoto T, Izumida A, Hata S, Izui K (2001) Phosphoenolpyruvate carboxylase kinase involved in C4 photosynthesis in Flaveria trinervia: cDNA cloning and characterization. FEBS Lett 507: 318–322 [DOI] [PubMed] [Google Scholar]
- Vidal J, Chollet R (1997) Regulatory phosphorylation of C4 PEP carboxylase. Trends Plant Sci 2: 230–237 [Google Scholar]
- Xu W, Zhou Y, Chollet R (2003) Identification and expression of a soybean nodule-enhanced PEP-carboxylase kinase gene (NE-PpcK) that shows striking up-/down-regulation in vivo. Plant J 34: 441–452 [DOI] [PubMed] [Google Scholar]