Summary
The myelodysplastic syndromes (MDS) display both haematological and biological heterogeneity with variable leukaemia potential. MicroRNAs play an important role in tumour suppression and the regulation of self-renewal and differentiation of haematopoietic progenitors. Using a microarray platform, we evaluated microRNA expression from 44 patients with MDS and 17 normal controls. We identified a thirteen microRNA signature with statistically significant differential expression between normal and MDS specimens (P < 0·01), including down-regulation of members of the leukaemia-associated MIRLET7 family. A unique signature consisting of 10 microRNAs was closely associated with International Prognostic Scoring System (IPSS) risk category permitting discrimination between lower (Low/Intermediate-1) and higher risk (Intermediate-2/High) disease (P < 0·01). Selective overexpression of MIR181 family members was detected in higher risk MDS, indicating pathogenetic overlap with acute myeloid leukaemia. Survival analysis of an independent cohort of 22 IPSS lower risk MDS patients revealed a median survival of 3·5 years in patients with high expression of MIR181 family compared to 9·3 years in patients with low MIR181 expression (P = 0·002). Our pilot study suggested that analysis of microRNA expression profile offers diagnostic utility, and provide pathogenetic and prognostic discrimination in MDS.
Keywords: microRNA, myelodysplastic syndromes, International Prognostic, Scoring System, gene expression, haematopoiesis
Myelodysplastic syndromes (MDS) comprise a heterogeneous group of haematopoietic stem cell malignancies that share a high frequency of recurrent chromosomal aberrations and a complex pathogenesis (List et al, 2004). Pathogenic mechanisms underlying abnormalities of affected haematopoietic stem cells in MDS remain poorly characterized. Senescence-associated accumulation of genetic defects is believed to play a key role in the changes in regulation of apoptosis, differentiation and proliferation potential of affected progenitors (Bernasconi, 2008). MicroRNAs are short non-coding RNAs containing 19–25 nucleotides that impair translation or induce mRNA degradation of target mRNA (Fire et al, 1998; Bartel, 2004). Over 500 microRNAs have been identified in the human genome (Bentwich et al, 2005). Analysis of microRNA expression in haematological malignancies and solid tumours has identified expression patterns that are tumour type specific with prognostic relevance (Calin et al, 2002; Debernardi et al, 2007; Garzon & Croce, 2008; Garzon et al, 2008; Jongen-Lavrencic et al, 2008; Marcucci et al, 2008a). MicroRNA expression profiling has shown that myeloblasts from patients with acute myeloid leukaemia (AML) display an expression profile that is distinct from normal bone marrow progenitors and acute lymphoblastic leukaemia (ALL) (Mi et al, 2007). Marcucci and colleagues identified a microRNA signature with independent predictive power for event-free survival in cytogenetically normal AML, which included five members of the MIR181 family that target genes involved in erythroid differentiation, homeobox genes and toll-like receptors (Marcucci et al, 2008a,b). To understand the role of microRNAs in MDS, we evaluated micro-RNA expression in bone marrow specimens from patients with MDS using a microarray platform and compared gene expression profiles to those obtained from normal donors. Gene expression was compared by International Prognostic Scoring System (IPSS) category (Greenberg et al, 1997) and test set risk signatures were confirmed in a validation set. We report microRNA class signatures with diagnostic discrimination that distinguish biological differences in MDS disease risk groups.
Methods
Patients and bone marrow specimens
Forty-four MDS and 17 normal control (NC) bone marrow mononuclear cells (BM-MNC) samples were obtained from Moffitt Cancer Center Tissue Procurement Repository. Written informed consent was obtained from all donors, and laboratory investigations were approved by the Institutional Review Board of the University of South Florida. MDS cases were characterized morphologically according to the World Health Organization (WHO) classification for myeloid malignancies (Swerdlow et al, 2008). Prognostic risk was assigned using the IPSS based on number of cytopenias in peripheral blood, percentage of bone marrow blasts and cytogenetics abnormalities (Greenberg et al, 1997). Clinical and demographic data are summarized in Table SI. BM-MNC were isolated from heparinized bone marrow aspirates on Ficoll-Hypaque gradient and cryopreserved in 20% fetal bovine serum with 10% dimethyl sulfoxide. After thawing cells were washed in phosphate-buffered saline before total RNA isolation.
RNA extraction and genome wide microRNA microarray profiling
Total RNA was isolated using QIAzol followed by RNeasy Mini kit (Qiagen, Valencia, CA, USA) according to manufacturer instructions. Genome-wide microRNA microarray profiling was performed using a microRNA microarray platform (OSU 3) consisting of 1100 microRNA probes in duplicates including 345 human and 249 mouse microRNA genes. microRNA isolated from haematopoietic cells was biotin-labelled and hybridized on microRNA chips as described elsewhere (Liu et al, 2004). Briefly, 5 µg of total RNA from each sample was reverse transcribed by using biotin end-labelled random octamers. Hybridization was carried out on the custom microRNA microarray chip (with probes length 40-b) spotted in quadruplicate with annotated active sites. The hybridized chips were washed and processed to detect biotin-containing transcripts by streptavidin-Alexa647 conjugate and scanned by using an Axon 4000B (Axon Instruments, Union City, CA, USA). Scanned images of chips were quantified by GENEPIX PRO 6·0 (Axon Instruments). Raw data were normalized using quantiles. MicroRNA was excluded when <20% of expression data had at least a 1·5-fold change in either direction from microRNA’s median value.
Statistical analysis
Statistical comparisons were made using the BRB Array tools (Simon et al, 2007). Expression data were normalized using quantiles. Class comparison between groups of arrays used random variance t tests, which provides improved estimates of gene-specific variances without assuming that all microRNAs have the same variance. The criteria for inclusion of a microRNA was a P-value less than a specified threshold value (i.e. 0·01). For constructing predictors and classifying experiments into classes based on microRNA expression levels we used class prediction in BRB Array tools. Six methods of prediction were applied: compound covariate predictor, diagonal linear discriminant analysis, k-nearest neighbour (using k = 1 and 3), nearest centroid, and support vector machines. Class prediction determined cross-validated misclassification rate and performed a permutation test to determine if the cross-validated misclassification rate was lower than it would be expected by chance. The criterion for inclusion of a gene in the predictor was a P-value less than a specified threshold value (i.e. 0·01). The output contains the result of the permutation test on the cross-validated misclassification rate, and a listing of genes that comprise the predictor, with parametric P-values for each microRNA and the CV-support percentage (percentage of times when the microRNA was used in the predictor for a leave-one-out cross-validation procedure). Heatmaps were produced using centred correlation distance and average linkage with cluster 3.0 (http://genome-www.stanford.edu) and visualized with java treeview (Volinia et al, 2010). Survival curves were generated using Kaplan–Meier method and the log-rank test to test any difference of survival curves. Holm’s method was used to adjust for the two tests.
Real time RT-PCR reverse transcription polymerase chain reaction (RT-PCR)
Single tube TaqMan MicroRNA Assays was performed to quantify mature microRNAs on Applied Biosystems Real-Time PCR instruments. All reagents, primers and probes were obtained from Applied Biosystems (Applied Biosystems, Foster City, CA, USA). RNU48 was used to normalize all RNA samples. Reverse Transcriptase Reactions and Real-Time PCR were performed according to the manufacturer’s protocols except that we used half volumes, 7·5 µl for the RT reaction and 10 µl for the PCR reaction, respectively. RNA concentrations were determined with a NanoDrop (NanoDrop Technologies, Inc, Wilmington, DE, USA). One nanogram RNA per sample was used for the assays. All RT reactions, including no-template controls and RT minus controls, were run in a GeneAmp PCR 9700 Thermocycler (Applied Biosystems). Gene expression levels were quantified using the abi prism 7900HT Sequence detection system (Applied Biosystems). Comparative real-time PCR was performed in triplicate, including no-template controls. Relative expression was calculated using the comparative Ct method (Chen et al, 2005). Wilcoxon test was used to compared values between groups, P < 0·05 was considered significant.
MicroRNA locked nucleic acid in situ hybridization (LNA-ISH) of formalin-fixed, paraffin-embedded bone marrow biopsy section
In situ hybridization (ISH) was carried out on deparaffinized bone marrow tissues using a previously published protocol (Nuovo et al, 2009), which included digestion in pepsin (1·3 mg/ml) for 30 min. The sequences of the probes containing the six dispersed locked nucleic acid (LNA) modified bases with digoxigenin conjugated to the 5′ end were: MIR181-(5′) ACCCACCGACAGCAATGAATGTT (Exiqon, Inc, Woburn, MA, USA). The probe cocktail and tissue microRNA were co-denatured at 60°C for 5 min, followed by hybridization at 37°C overnight and a low stringency wash in 0·2× saline sodium citrate (SSC) and 2% bovine serum albumin at 4°C for 10 min. The probe-target complex was seen due to the action of alkaline phosphatase on the chromogen nitroblue tetrazolium and bromochloroindolyl phosphate (NBT/BCIP). Negative controls included the use of a scrambled probe. MIR328 served as a positive control.
Results
We studied microRNA profiles in patients with MDS using microarray platform. Since a single microRNAs is able to regulate multiple genes simultaneously it is plausible that microRNAs are implicated in pathogenesis of such complex and heterogeneous disorders as MDS. We hypothesized that deregulation of microRNA expression is most probably initiated in a pluripotent MDS stem cell and subsequently affects more mature bone marrow cells represented in the fraction of bone marrow mononuclear cells. Although it would be optimal to study population of CD34+ cells this approach was not technically feasible because a microchip platform required 5 µg of total RNA for each sample analysis.
MicroRNA expression profile distinguishes MDS from normal haematopoiesis
MicroRNA expression was analysed in 44 MDS patient and 17 normal control specimens using a microRNA chip (OSU version 3). Overall, the MDS cohort included 66% patients with Low/Intermediate-1 risk IPSS categories, and 33% patients with Intermediate-2/High risk disease. WHO distribution included refractory anaemia (RA) (n = 5), refractory anaemia with ring sideroblasts (RARS) (n = 8), MDS with deletion 5q (n = 6), refractory cytopenia with multilineage dysplasia (RCMD) (n = 7), refractory anaemia with excess blasts-1 (RAEB-1) (n = 10) or RAEB-2 (n = 8), MDS unclassifiable (MDS-U) (n = 4), myelodysplastic/myeloproliferative neoplasm, unclassifiable (MDS/MPN-U) (n = 2). Overall, 290 microRNAs passed the filtering criterion; among which 42 were up-regulated and 34 down-regulated (Table SII). Fifteen microRNAs were selected among the 76 microRNAs with differential expression (P < 0·001) according to class comparison from which an expression heat map was constructed (Fig 1). Using the class prediction we found that MDS and normal controls had distinct microRNA profiles. A signature consisting of thirteen microRNAs was sufficient to discriminate between these two cohorts (Table I). Included among these are two down-regulated microRNAs that have key roles in tumourigenesis through deregulation of gene targets such as RAS, MYC, HMGA2 and CASP3 (MIRLET7 family) (Johnson et al, 2005; Park et al, 2007; Sampson et al, 2007; Tsang & Kwok, 2008) and stem/progenitor cell self-renewal and maturation potential (MIR146A, MIRLET7) (Labbaye et al, 2008; Liu et al, 2009); in addition to increased expression of microRNAs that have repressive roles in erythroid (MIR222) and megakaryocyte (MIR10A) maturation (Felli et al, 2005; Garzon et al, 2006). Of particular interest, MIR146A is encoded on the long arm of chromosome 5 within the commonly deleted region associated with the 5q- syndrome (Starczynowski et al, 2010). Two of the up-regulated microRNAs are located within homeobox gene clusters that have important roles in haematopoietic development and oncogenesis. MIR10A and MIR196A1 are located within the HOX B cluster on 17q21, whereas MIR196A2 is within the HOX C cluster at 12q13 (Calin et al, 2004).
Fig 1.
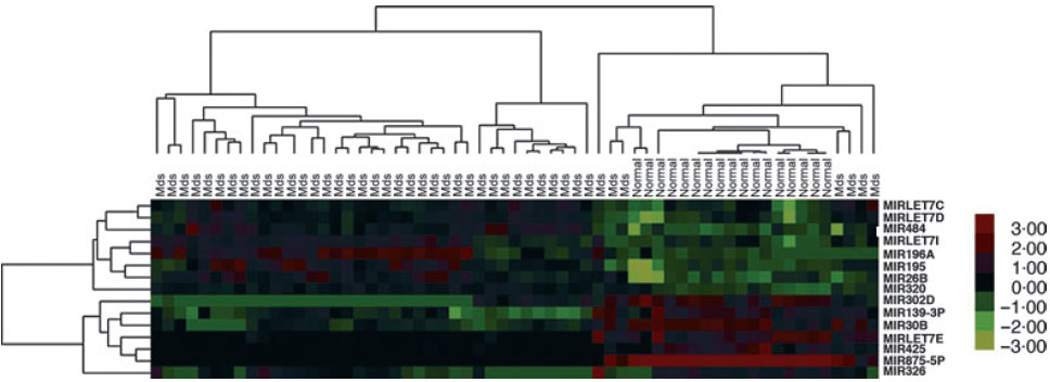
MicroRNAs can differentiate MDS and normal controls. Two-way hierarchical clustering of 44 MDS and 17 normal controls (NC) was performed after quality filtering. Hierarchical clustering showed clustering of the samples according to disease status. A heat map was generated using the expression ratios of 15 microRNAs selected out of total 73 that differed significantly (P < 0·001). Red: up-regulated microRNAs, green: downregulated microRNAs. Each column represents a MDS or NC sample, and each row represents a single microRNA. Patient samples are grouped by IPSS. The heatmap colour key gradient ranging from global minimum (green) −3 to global maximum (red) +3 is on the right side.
Table I.
MicroRNA signature differentiating MDS versus Normal Controls (NC).
| MicroRNA | Parametric P value |
Fold change |
Chromosomal location |
|---|---|---|---|
| MIR222 | 0·0020831 | 3·81 | Xp11.3 |
| MIR10A | 0·0018187 | 3·37 | 17q21.32 |
| MIR196A | 6·66E-05 | 3·04 | 17q21.32 |
| MIR320 | <1e-07 | 2·12 | 8p21.3 |
| MIR100 | 0·0298715 | 1·51 | 11q24.1 |
| MIR124 | 0·0009909 | 0·60 | 8p23.1 |
| MIR206 | 0·0093797 | 0·59 | 6p12.2 |
| MIR146A | 0·0356401 | 0·57 | 5q34 |
| MIR150 | 0·0365018 | 0·54 | 19q13.33 |
| MIR326 | 0·0001276 | 0·51 | 11q13.14 |
| MIRLET7E | <1e-07 | 0·46 | 19q13.41 |
| MIR197 | 0·000754 | 0·33 | 1p13.3 |
| MIR875-5P | <1e-07 | 0·02 | 8q22.2 |
This 13 microRNA signature was predictive (misclassification error rate after 10-fold CV, <0·01). Red: up-regulated microRNAs. Green: down-regulated microRNAs.
MicroRNA expression distinguishes higher from lower risk MDS
The microRNA expression patterns from 10 patients with higher risk (HR) MDS according IPSS score (Int-2/High) were compared to 10 patients with lower risk (LR; Low/Int-1) MDS. After normalization of data, differences in expression of 68 microRNAs were statistically significant (38 up-regulated and 30 down-regulated microRNAs, P < 0·05) (Table SIII). Thirty of these microRNAs were selected for heat map construction (Fig 2). Class prediction analysis identified a unique microRNA signature consisting of nine up-regulated and one downregulated microRNAs that discriminated the two prognostic groups (Table II). Four members of the MIR181 family were up-regulated in HR MDS specimens.
Fig 2.
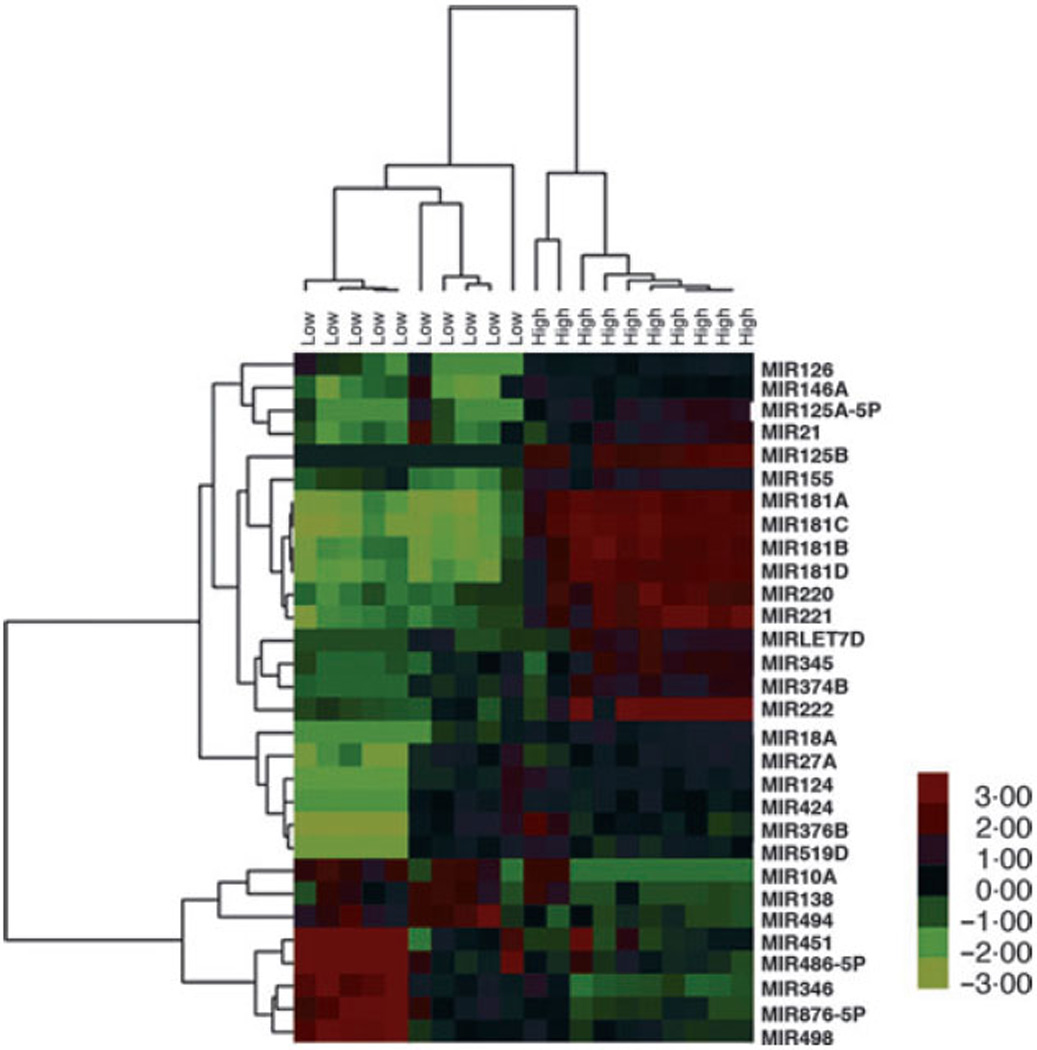
MicroRNAs can differentiate MDS according to prognosis. Hierarchical clustering of 10 high risk and 10 low risk MDS samples revealed a clustering according to prognostic group. A heat map was generated using the expression ratios of 30 microRNAs selected out of total 68 that differed significantly (P < 0·05). Red: up-regulated microRNAs, green: down-regulated microRNAs. Each column represents a patient sample, and each row represents a single microRNA. Patient samples are grouped by IPSS. The heatmap colour key gradient ranging from global minimum (green) −3 to global maximum (red) +3 is on the right side.
Table II.
MicroRNA signature differentiating HR versus LR MDS.
| MicroRNA | Parametric P value |
Fold change |
Chromosomal location |
|---|---|---|---|
| MIR181C | <1e-07 | 22·72 | 19p13.13 |
| MIR181A | <1e-07 | 19·95 | 19q33.3 |
| MIR181B | 2E-07 | 14·25 | 1q32.1 |
| MIR181D | <1e-07 | 13·88 | 19p13.13 |
| MIR221 | 4E-07 | 12·07 | Xp11.3 |
| MIR376B | 0·024925 | 4·99 | 14q32.31 |
| MIR125B | 9E-07 | 4·27 | 11q24.1 |
| MIR155 | 0·000131 | 3·62 | 21q21.3 |
| MIR130A | 0·000675 | 2·80 | 11q12.1 |
| MIR486-5P | 0·005686 | 0·14 | 8p11.21 |
The signature was predictive (misclassification error rate after 10-fold CV, <0·01). Red: up-regulated microRNAs, green: down-regulated microRNAs.
Results from the microchip platform were validated on an independent set of 18 samples (Table SIV) using quantitative RT-PCR. Expression of seven randomly selected microRNAs was evaluated (Fig 3) and confirmed the findings from the microarray chip. Next, we examined expression of MIR181B in bone marrow biopsies using in situ hybridization with locked nucleic acid (LNA)-modified probes in six bone marrow biopsies from MDS patients and two controls. MIR181B was strongly expressed in the cytoplasm of HR MDS cells (Fig 4), whereas specimens from LR MDS displayed reduced staining intensity. Although a cohort of our MDS patients was heterogeneous and included the six most common MDS subtypes according to WHO classification our results suggested that there is an overlap in molecular pathogenesis between these subtypes.
Fig 3.
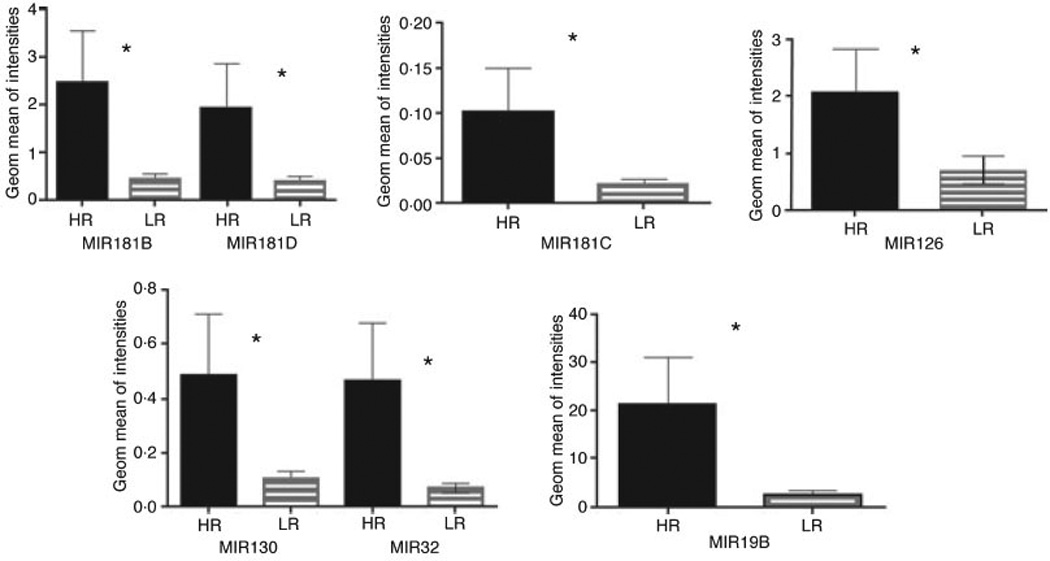
Validation of microRNA expression using independent sample set by Real -Time RT-PCR. Samples from 18 independent MDS patients (HR = 6, LR = 12) were analysed. Seven differentially expressed microRNAs were randomly selected for Real-Time RT-PCR. Significant correlation was demonstrated for all tested microRNA between microRNA microchip platform and qRT-PCR.
Fig 4.
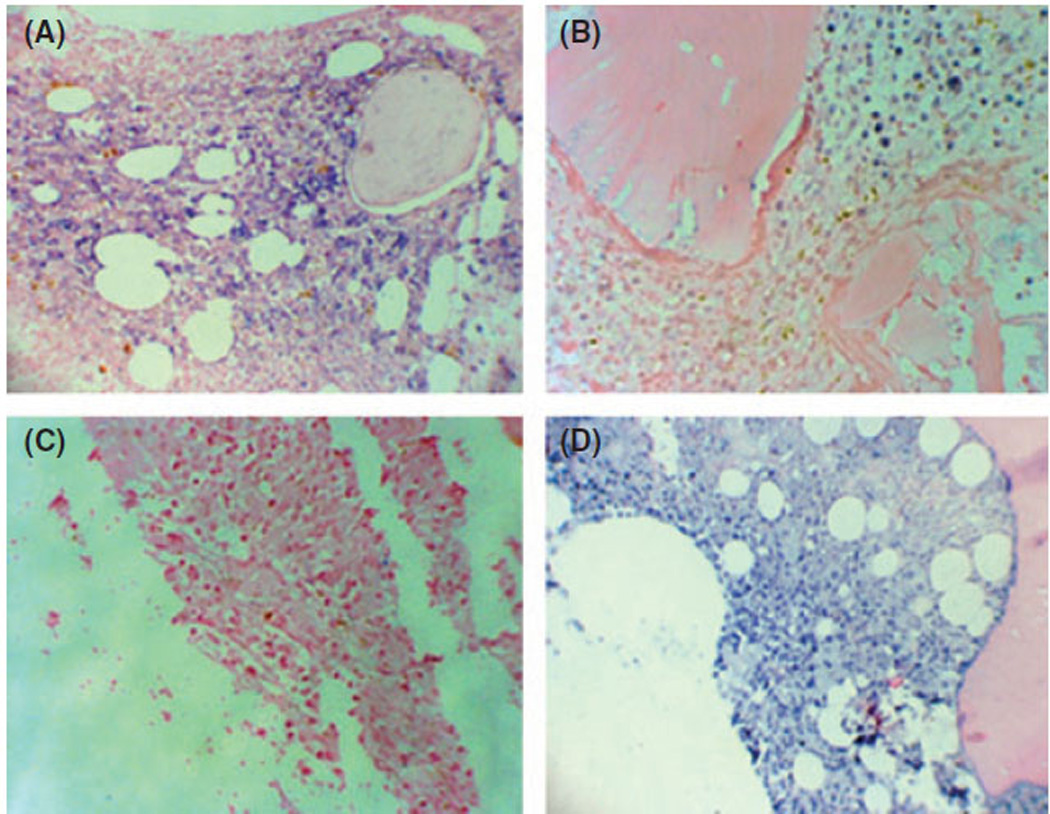
MicroRNA181B locked nucleic acid in situ hybridization (LNA-ISH) of formalin-fixed, paraffin-embedded bone marrow biopsy sections. We examined six patients with MDS and two controls. MIR181B is present in the cytoplasm of both (A) HR (+++) and (B) LR MDS (++) cells arranged in clusters on the background of normal (MIR181B negative) haematopoietic cells. (C) Negative control – scrambled microRNA probe. (D) Positive control –MIR328 (A 200×, B 400×, C 400×, D 200×).
Expression of MIR181 family correlates with survival of patients with LR MDS
We also evaluated overall survival in an independent cohort of 22 MDS patients with IPSS lower risk. Results of this analysis revealed a significant difference in survival of patients with high expression of MIR181 family, with a median survival of 3·5 years compared to 9·3 years in patients with low MIR181 expression (P = 0·002) (Fig 5). Interestingly, two of three patients in the data set that developed AML had up-regulation of the MIR181 family, whereas one did not.
Fig 5.
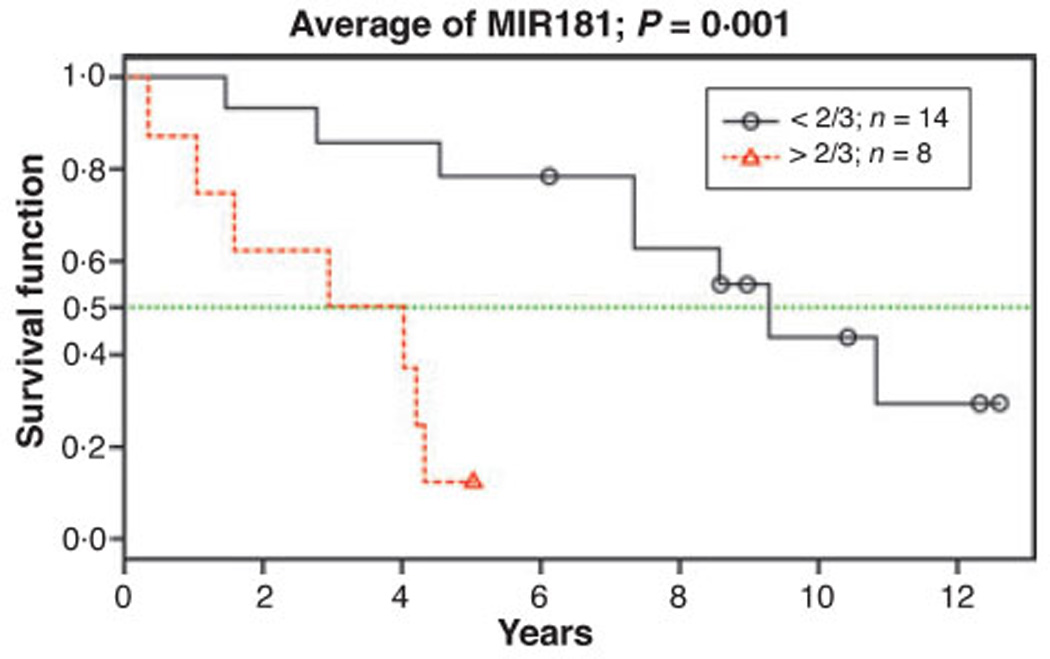
Kaplan–Meier estimate of overall survival in 22 IPSS lower-risk MDS patients by MIR181 family expression. Four microRNAs (MIR181A, MIR181B, MIR181C, MIR181D) from the 10 microRNA prognostic signature were selected due to the wide variance in gene expression (>10-fold). Since family member expression levels were highly correlated (r > 0·8), a composite score was generated by averaging four microRNAs separate standardized scores. Anticipating that about one-third of the patients should be considered as having ‘high risk’ due to high microRNA expression, we divided the patients into two groups, comparing the 14 patients with lower scores, to the eight patients with higher scores. Results showed a significant difference in survival from date of diagnosis to date of death or last visit.
Discussion
We have identified and validated two microRNA signatures that discriminate MDS from normal control subjects and patients with HR from LR MDS, respectively. A vast majority of microRNAs included in both signatures were previously found to be implicated in regulation of haematopoiesis, apoptosis and angiogenesis.
The MIR181 family plays an important role in the negative regulation of haematopoiesis including the proliferation and the differentiation of stem/progenitor cells and megakaryocytic lineage development (Georgantas et al, 2007). A putative role for MIR181 in the pathobiology of myeloid malignancies is supported by a recent report by investigators in the Cancer and Leukemia Group B (CALGB) showing that expression of five members of MIR181 family among a 12 microRNA signature was inversely associated with progression-free survival in AML patients with normal cytogenetics (Marcucci et al, 2008b).
Several microRNAs involved in stage-specific control of erythropoiesis (Choong et al, 2007; Georgantas et al, 2007) were deregulated in MDS. Expression of MIR221 and MIR222 normally decline with erythroid maturation to de-repress KIT expression and facilitate erythroblast expansion (Felli et al, 2005). Overexpression of these microRNAs, which distinguished MDS patients from controls in our study, can suppress erythroid growth and may be operative in disease-associated ineffective erythropoiesis. (Table II). The latter may be compounded by down-regulation of members of the MIR144/451 cluster in HR MDS, which are key transcriptional targets of the erythroid transcription factor GATA-1, which are normally up-regulated with late stages of differentiation (Dore et al, 2008). MIR155, a translational repressor of several myeloid transcription factors including PU.1, C/EPBβ and CSF1R (Georgantas et al, 2007; O’Connell et al, 2008), was significantly up-regulated in HR MDS. Mice transplanted with MIR155 transfected stem cells developed a myeloproliferative disorder with abnormal granulocyte morphology analogous to MDS, suggesting a role in the higher risk MDS phenotype (O’Connell et al, 2008). Recent investigations have shown that the proliferative effects of MIR155 are mediated in part through down-regulation of the phosphatase and tensin homologue (PTEN) and consequent up-regulation of the programmed cell death protein 4 (PDCD4), Src homology-2 domain-containing inositol 5-phosphatase 1 (SHIP1) and Akt phosphorylation (Yamanaka et al, 2009). Although not included in the diagnostic signatures, we found down-regulation of two MIR29 family members in MDS specimens; MIR29B in HR MDS and MIR29A in MDS compared to normal controls. Both microRNAs are located on chromosome 7q32, a region commonly deleted in MDS and therapy-related AML (Le Beau et al, 1986). These microRNAs target MCL1, a member of the BCL2 anti-apoptotic gene family that is overexpressed in AML and HR MDS (Mott et al, 2007; Economopoulou et al, 2010), as well as the DNA methytranferases DNMT-3A and -3B, and indirectly via Sp1, DNMT-1 (Fabbri et al, 2007; Garzon et al, 2009). Loss of MIR29A/B results in over-expression of MCL1 and DNMTs, with corresponding aberrant genomic methylation, suggesting that down-regulation of the MIR29 family may be a key pathogenetic feature of MDS, and as recently reported, progression of the disease to AML (Jiang et al, 2009). Megakaryocytic differentiation of common myeloid progenitors is tightly regulated by the coordinated down-regulation of MIR10A, MIR126, MIR130A and MIR155, and up-regulation of MIR150 (Liu et al, 2004; Garzon et al, 2006; Yendamuri & Calin, 2009). Induction of MIR150, which suppresses expression of c-myb, is critical for megakaryocytic lineage commitment (Barroga et al, 2008; Lu et al, 2008). Our findings of down-regulation of MIR150 accompanied by up-regulation of MIR10A in MDS patient specimens compared to normal controls suggests a possible role in dysmegakaryopoiesis and impaired thrombopoiesis in MDS. Interestingly, MIR155, MIR126 and MIR130 were overexpressed in HR-MDS, which may further suppress megakaryopoiesis and account for the higher frequency of thrombocytopenia that occurs with disease progression (Garzon et al, 2006). Further investigation is warranted to determine if these microRNA changes are associated with dysplastic megakaryocytopoiesis and severity of thrombocytopenia in MDS patients.
The elaboration of angiogenic molecules has been implicated in autocrine cytokine networks supporting myeloid progenitor self-renewal and in bone marrow microvessel density in HR MDS (Bellamy et al, 2001). Several up-regulated microRNAs in HR MDS, including MIR27B, 155, 21, 126 and 130A were previously implicated in regulation of different stages of angiogenesis (Urbich et al, 2008). MIR130A targets a site in the 3′-UTR of the anti-angiogenic homeobox gene HOXA5 to promote angiogenesis (Chen & Gorski, 2008). In a zebrafish model MIR126 was shown to control vascular integrity and micro-vessel formation by repressing negative regulators of the vascular endothelial growth factor (VEGF) signalling receptor pathway (Fish et al, 2008). Our data suggest that changes in microRNA expression may promote angiogenesis and lead to MDS disease progression. Although it is difficult to draw firm conclusions from our prognostic and diagnostic signatures in terms of a mechanism of action, our pilot study showed that putative microRNA targets obtained from published literature or from in silico prediction models are implicated in the regulation of multiple important pathogenic pathways in haematopoiesis and MDS. However, further studies are needed to determine whether selective targeting of these microRNAs might alter disease phenotype and outcome.
Conclusions
Our data reveal that changes in microRNA expression may have a fundamental role in the pathogenesis and phenotype of MDS. The vast majority of alterations in microRNA expression affected microRNAs implicated in haematopoiesis and tumourigenesis. MicroRNA expression profiling was able to discriminate between normal and MDS haematopoiesis, suggesting that molecular characterization may offer a potentially more rigorous diagnostic alternative to the reliance on morphological recognition with direct biological implications. Moreover, the identification of microRNA signatures that distinguish IPSS LR from HR MDS indicates that microRNA expression profile also offers prognostic utility that might, in the future, provide insight into appropriate therapeutic selection. Additionally, MIR181 expression may offer a molecular tool to refine discrimination of disease behaviour. Further investigations done on a large population of MDS patients are necessary to discern the possible relationship between micro-RNA profile and haematological phenotype, therapeutic response and overall survival.
Supplementary Material
Acknowledgements
This work was supported by the Harold Spielberg Research Grant received from Aplastic Anemia & MDS International Foundation. We thank Ms. Gayle Behrle for technical assistance with the manuscript editing.
Footnotes
Supporting information
Additional Supporting information may be found in the online version of this article:
Table SI. Patient Samples/Clinical Data/Microarray Platform.
Table SII. MiRNA Expression Profiling MDS versus NC.
Table SIII. MiRNAs Expression Profiling MDS HR versus LR.
Table SIV. Patient Samples/Clinical Data/Real-Time RT-PCR.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Barroga CF, Pham H, Kaushansky K. Thrombopoietin regulates c-Myb expression by modulating micro RNA 150 expression. Experimental Hematology. 2008;36:1585–1592. doi: 10.1016/j.exphem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bellamy WT, Richter L, Sirjani D, Roxas C, Glinsmann-Gibson B, Frutiger Y, Grogan TM, List AF. Vascular endothelial cell growth factor is an autocrine promoter of abnormal localized immature myeloid precursors and leukemia progenitor formation in myelodysplastic syndromes. Blood. 2001;97:1427–1434. doi: 10.1182/blood.v97.5.1427. [DOI] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nature Genetics. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Bernasconi P. Molecular pathways in myelodysplastic syndromes and acute myeloid leukemia: relationships and distinctions-a review. British Journal of Haematology. 2008;142:695–708. doi: 10.1111/j.1365-2141.2008.07245.x. [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Research. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choong ML, Yang HH, McNiece I. MicroRNA expression profiling during human cord blood-derived CD34 cell erythropoiesis. Experimental Hematology. 2007;35:551–564. doi: 10.1016/j.exphem.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Debernardi S, Skoulakis S, Molloy G, Chaplin T, Dixon-McIver A, Young BD. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21:912–916. doi: 10.1038/sj.leu.2404605. [DOI] [PubMed] [Google Scholar]
- Dore LC, Amigo JD, Dos Santos CO, Zhang Z, Gai X, Tobias JW, Yu D, Klein AM, Dorman C, Wu W, Hardison RC, Paw BH, Weiss MJ. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economopoulou C, Pappa V, Papageorgiou S, Kontsioti F, Economopoulou P, Charitidou E, Girkas K, Kapsimali V, Papasteriadi C, Tsirigotis P, Papageorgiou E, Dervenoulas J, Economopoulos T. Cell cycle and apoptosis regulatory gene expression in the bone marrow of patients with de novo myelodysplastic syndromes (MDS) Annals of Hematology. 2010;89:349–358. doi: 10.1007/s00277-009-0835-2. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, Valtieri M, Calin GA, Liu CG, Sorrentino A, Croce CM, Peschle C. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Developemental Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Current Opinion in Hematology. 2008;15:352–358. doi: 10.1097/MOH.0b013e328303e15d. [DOI] [PubMed] [Google Scholar]
- Garzon R, Pichiorri F, Palumbo T, Iuliano R, Cimmino A, Aqeilan R, Volinia S, Bhatt D, Alder H, Marcucci G, Calin GA, Liu CG, Bloomfield CD, Andreeff M, Croce CM. MicroRNA fingerprints during human megakaryocytopoiesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, Pichiorri F, Fabbri M, Coombes K, Alder H, Nakamura T, Flomenberg N, Marcucci G, Calin GA, Kornblau SM, Kantarjian H, Bloomfield CD, Andreeff M, Croce CM. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, Havelange V, Volinia S, Blum W, Rush LJ, Perrotti D, Andreeff M, Bloomfield CD, Byrd JC, Chan K, Wu LC, Croce CM, Marcucci G. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgantas RW, III, Hildreth R, Morisot S, Alder J, Liu CG, Heimfeld S, Calin GA, Croce CM, Civin CI. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, Sanz G, Sanz M, Vallespi T, Hamblin T, Oscier D, Ohyashiki K, Toyama K, Aul C, Mufti G, Bennett J. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89:2079–2088. [PubMed] [Google Scholar]
- Jiang Y, Dunbar A, Gondek LP, Mohan S, Rataul M, O’Keefe C, Sekeres M, Saunthararajah Y, Maciejewski JP. Aberrant DNA methylation is a dominant mechanism in MDS progression to AML. Blood. 2009;113:1315–1325. doi: 10.1182/blood-2008-06-163246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Lowenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111:5078–5085. doi: 10.1182/blood-2008-01-133355. [DOI] [PubMed] [Google Scholar]
- Labbaye C, Spinello I, Quaranta MT, Pelosi E, Pasquini L, Petrucci E, Biffoni M, Nuzzolo ER, Billi M, Foa R, Brunetti E, Grignani F, Testa U, Peschle C. A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nature Cell Biology. 2008;10:788–801. doi: 10.1038/ncb1741. [DOI] [PubMed] [Google Scholar]
- Le Beau MM, Albain KS, Larson RA, Vardiman JW, Davis EM, Blough RR, Golomb HM, Rowley JD. Clinical and cytogenetic correlations in 63 patients with therapy-related myelodysplastic syndromes and acute nonlymphocytic leukemia: further evidence for characteristic abnormalities of chromosomes no. 5 and 7. Journal of Clinical Oncology. 1986;4:325–345. doi: 10.1200/JCO.1986.4.3.325. [DOI] [PubMed] [Google Scholar]
- List AF, Vardiman J, Issa JP, DeWitte TM. Myelodysplastic syndromes. Hematology. American Society of Hematology Education Program. 2004:297–317. doi: 10.1182/asheducation-2004.1.297. [DOI] [PubMed] [Google Scholar]
- Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, Alder H, Bullrich F, Negrini M, Croce CM. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SP, Fu RH, Yu HH, Li KW, Tsai CH, Shyu WC, Lin SZ. MicroRNAs regulation modulated self-renewal and lineage differentiation of stem cells. Cell Transplantation. 2009;18:1039–1045. doi: 10.3727/096368909X471224. [DOI] [PubMed] [Google Scholar]
- Lu J, Guo S, Ebert BL, Zhang H, Peng X, Bosco J, Pretz J, Schlanger R, Wang JY, Mak RH, Dombkowski DM, Preffer FI, Scadden DT, Golub TR. MicroRNA-mediated control of cell fate in megakaryocyteerythrocyte progenitors. Developemental Cell. 2008;14:843–853. doi: 10.1016/j.devcel.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Maharry K, Radmacher MD, Mrozek K, Vukosavljevic T, Paschka P, Whitman SP, Langer C, Baldus CD, Liu CG, Ruppert AS, Powell BL, Carroll AJ, Caligiuri MA, Kolitz JE, Larson RA, Bloomfield CD. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B Study. Journal of Clinical Oncology. 2008a;26:5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Radmacher MD, Maharry K, Mrozek K, Ruppert AS, Paschka P, Vukosavljevic T, Whitman SP, Baldus CD, Langer C, Liu CG, Carroll AJ, Powell BL, Garzon R, Croce CM, Kolitz JE, Caligiuri MA, Larson RA, Bloomfield CD. MicroRNA expression in cytogenetically normal acute myeloid leukemia. New England Journal of Medicine. 2008b;358:1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly MB, Wang Y, Qian Z, Jin J, Zhang Y, Bohlander SK, Le Beau MM, Larson RA, Golub TR, Rowley JD, Chen J. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19971–19976. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuovo GJ, Elton TS, Nana-Sinkam P, Volinia S, Croce CM, Schmittgen TD. A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nature Protocols. 2009;4:107–115. doi: 10.1038/nprot.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. Journal of Experimental Medicine. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Shell S, Radjabi AR, Schickel R, Feig C, Boyerinas B, Dinulescu DM, Lengyel E, Peter ME. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle. 2007;6:2585–2590. doi: 10.4161/cc.6.21.4845. [DOI] [PubMed] [Google Scholar]
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a downregulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Research. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-array tools. Cancer Informatics. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- Starczynowski DT, Kuchenbauer F, Argiropoulos B, Sung S, Morin R, Muranyi A, Hirst M, Hogge D, Marra M, Wells RA, Buckstein R, Lam W, Humphries RK, Karsan A. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nature Medicine. 2010;16:49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- Swerdlow SH International Agency for Research on Cancer & World Health Organization. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: International Agency for Research on Cancer; 2008. [Google Scholar]
- Tsang WP, Kwok TT. Let-7a micro-RNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis. 2008;13:1215–1222. doi: 10.1007/s10495-008-0256-z. [DOI] [PubMed] [Google Scholar]
- Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovascular Research. 2008;79:581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A, Marchesini J, Mascellani N, Sana ME, Abu Jarour R, Desponts C, Teitell M, Baffa R, Aqeilan R, Iorio MV, Taccioli C, Garzon R, Di Leva G, Fabbri M, Catozzi M, Previati M, Ambs S, Palumbo T, Garofalo M, Veronese A, Bottoni A, Gasparini P, Harris CC, Visone R, Pekarsky Y, de la Chapelle A, Bloomston M, Dillhoff M, Rassenti LZ, Kipps TJ, Huebner K, Pichiorri F, Lenze D, Cairo S, Buendia MA, Pineau P, Dejean A, Zanesi N, Rossi S, Calin GA, Liu CG, Palatini J, Negrini M, Vecchione A, Rosenberg A, Croce CM. Reprogramming of micro RNA networks in cancer and leukemia. Genome Research. 2010;20:589–599. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka Y, Tagawa H, Takahashi N, Watanabe A, Guo YM, Iwamoto K, Yamashita J, Saitoh H, Kameoka Y, Shimizu N, Ichinohasama R, Sawada K. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood. 2009;114:3265–3275. doi: 10.1182/blood-2009-06-222794. [DOI] [PubMed] [Google Scholar]
- Yendamuri S, Calin GA. The role of microRNA in human leukemia: a review. Leukemia. 2009;23:1257–1263. doi: 10.1038/leu.2008.382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


