Abstract
The intracellular bacterial pathogen Coxiella burnetii directs biogenesis of a parasitophorous vacuole (PV) that acquires host endolysosomal components. Formation of a PV that supports C. burnetii replication requires a Dot/Icm type 4B secretion system (T4BSS) that delivers bacterial effector proteins into the host cell cytosol. Thus, a subset of T4BSS effectors are presumed to direct PV biogenesis. Recently, the PV-localized effector protein CvpA was found to promote C. burnetii intracellular growth and PV expansion. We predict additional C. burnetii effectors localize to the PV membrane and regulate eukaryotic vesicle trafficking events that promote pathogen growth. To identify these vacuolar effector proteins, a list of predicted C. burnetii T4BSS substrates was compiled using bioinformatic criteria, such as the presence of eukaryote-like coiled-coil domains. Adenylate cyclase translocation assays revealed 13 proteins were secreted in a Dot/Icm-dependent fashion by C. burnetii during infection of human THP-1 macrophages. Four of the Dot/Icm substrates, termed Coxiella vacuolar protein B (CvpB), CvpC, CvpD, and CvpE, labeled the PV membrane and LAMP1-positive vesicles when ectopically expressed as fluorescently tagged fusion proteins. C. burnetii ΔcvpB, ΔcvpC, ΔcvpD, and ΔcvpE mutants exhibited significant defects in intracellular replication and PV formation. Genetic complementation of the ΔcvpD and ΔcvpE mutants rescued intracellular growth and PV generation, whereas the growth of C. burnetii ΔcvpB and ΔcvpC was rescued upon cohabitation with wild-type bacteria in a common PV. Collectively, these data indicate C. burnetii encodes multiple effector proteins that target the PV membrane and benefit pathogen replication in human macrophages.
INTRODUCTION
Coxiella burnetii is an intracellular pathogen and the etiological agent of human Q fever. This highly infectious Gram-negative bacterium is capable of colonizing mammalian, avian, and arthropod host organisms (1). The pathogen is shed in high numbers by infected livestock and easily disseminated via aerosols (1). C. burnetii exhibits a biphasic developmental cycle in which the bacterium transitions between small cell variant (SCV) and large cell variant (LCV) forms (2–4). SCVs are 0.2 to 0.5 μm in size with densely packed chromatin and low metabolic activity. Once internalized within a host cell, SCVs differentiate into replicative LCVs of ∼1 μm in size with dispersed chromatin. The compact structure of SCVs correlates with resistance to osmotic stress, sonic disruption, and high pressure (3, 5). Therefore, the SCV is presumed to be the environmentally stable form of C. burnetii that facilitates disease transmission (3).
Successful intracellular replication of C. burnetii in mononuclear phagocytes, such as alveolar macrophages, is required for development of human Q fever, a disease that typically manifests as an acute flu-like illness (6). C. burnetii replicates within a specialized parasitophorous vacuole (PV) with characteristics of a phagolysosome (6). After internalization by a host cell, C. burnetii is sequestered within a nascent phagosome that traffics canonically through the endolysosomal system to ultimately acquire late endosomal and lysosomal markers such as Rab7, lysosome-associated membrane protein 1 (LAMP1), and cathepsin D (7). PV acquisition of acid hydrolases correlates with pronounced degradative activity that C. burnetii, by unknown mechanisms, is able to resist (7). In response to vacuole acidification, C. burnetii becomes metabolically active, resulting in the synthesis of bacterial proteins required for PV maturation (8, 9). PV biogenesis involves fusion of the vacuole with vesicles originating from endocytic, autophagic, and secretory pathways through processes regulated by multiple host factors, including Rab GTPases and soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) (10–15).
Translocation by C. burnetii of proteins directly into the host cell cytosol by a specialized type 4B secretion system (T4BSS) is required for PV formation (16–18). The C. burnetii T4BSS is homologous to the virulence-associated T4BSS of Legionella pneumophila, encoded by defect in organelle trafficking (dot) and intracellular replication (icm) genes (19). C. burnetii strains harboring Himar1 transposon (Tn) insertions in icmL or icmD (16, 17), or deletions in dotA or dotB (20), fail to secrete effector proteins and have severe defects in intracellular growth. Interestingly, a C. burnetii icmD::Tn mutant can replicate intracellularly if sequestered in a vacuole with wild-type C. burnetii that provide Dot/Icm functions in trans (16). Collectively, these data confirm that Dot/Icm T4BSS function is essential for the productive infection by C. burnetii.
Based on similar function of L. pneumophila and C. burnetii Dot/Icm T4BSSs, investigators have extensively used L. pneumophila to screen candidate C. burnetii effectors for Dot/Icm-dependent secretion using either adenylate cyclase (CyaA) or β-lactamase (BlaM) translocation assays (17, 18, 21–25). C. burnetii proteins are typically selected as candidate effectors using bioinformatic criteria that include the presence of eukaryote-like motifs (21, 23, 24), a C terminus enriched in acidic residues (22, 25–27), and/or a PmrA regulatory element upstream of the encoding gene promoter (21, 25, 28). Screening of candidate effectors has resulted in the identification of approximately 130 C. burnetii proteins that are secreted in a Dot/Icm-dependent fashion (17, 18, 21–25, 29). Recently, several groups have used new genetic tools to express proteins in C. burnetii and validate that substrates recognized by the L. pneumophila Dot/Icm system are also exported during C. burnetii infection of host cells, including six Dot/Icm substrates encoded by the QpH1 cryptic plasmid (18) and 21 substrates encoded by chromosomal genes (14, 17, 21, 25).
C. burnetii Dot/Icm effectors are predicted to remodel host endomembrane compartments for generation of the replication-permissive PV, but information is lacking on specific effector functions that promote this process. However, inactivation of C. burnetii genes is now possible, allowing direct assessment of effector requirements during C. burnetii infection of mammalian cells. From a Himar1 Tn mutant library, Weber et al. (25) identified 20 C. burnetii mutants where a Tn insertion disrupts a gene encoding a known T4BSS substrate. Five of these mutants exhibit impaired growth in J774A.1 mouse macrophages and, consequently, the affected genes are named cir genes for Coxiella effectors for intracellular replication. Martinez et al. (30) recently identified 12 C. burnetii Himar1 Tn mutants with significant growth defects in Vero epithelial cells where the Tn insertion disrupts a gene encoding a known Dot/Icm substrate. The functions of these Tn-disrupted effector-encoding genes are currently unknown (25, 30).
Recently, we functionally characterized a new T4BSS effector protein, termed CvpA for Coxiella vacuolar protein A, that was identified based on the presence of a eukaryote-like leucine-rich repeat and multiple endocytic sorting motifs (14). CvpA localizes to the PV membrane when ectopically expressed as a fusion to mCherry red fluorescent protein. A C. burnetii ΔcvpA mutant displays severe defects in intracellular replication and PV biogenesis that are rescuable by genetic complementation. In uninfected cells, mCherry-CvpA labels the membrane of endocytic recycling vesicles, an interaction mediated by the multiple endocytic sorting motifs within CvpA that bind the heterotetrameric clathrin adaptor protein complex 2 (AP2) (14, 31). Depletion of cellular AP2 or clathrin with small interfering RNA significantly inhibits C. burnetii replication. In addition, a mutated form of CvpA lacking endocytic motifs does not rescue growth of the ΔcvpA mutant. Collectively, these data suggest CvpA modulates clathrin-mediated vesicle transport events on the PV membrane that promote vacuole biogenesis (14). We predict additional C. burnetii effectors on the PV membrane regulate vesicle fusion events required for vacuole maturation and pathogen growth.
In the present study, bioinformatic criteria were used to compile a list of candidate C. burnetii T4BSS substrates that were then screened for Dot/Icm-dependent translocation during C. burnetii infection of mammalian cells. To gain insight into potential effector functions, confirmed T4BSS substrates were ectopically expressed in eukaryotic cells to identify those that traffic to the PV membrane. Four effectors localized to the PV in Coxiella-infected cells, and strains harboring targeted deletions in the encoding genes of each effector displayed significant defects in intracellular replication and PV formation. These results indicate multiple C. burnetii Dot/Icm effectors localize to the PV membrane that promote C. burnetii replication. Importantly, these findings are based on assessments of Dot/Icm substrate recognition and effector function performed directly within C. burnetii.
MATERIALS AND METHODS
Bacterial and mammalian cell culture.
C. burnetii Nine Mile RSA439 (phase II, clone 4) was cultivated axenically in ACCM-2 as previously described (32). HeLa (CCL-2; American Type Culture Collection [ATCC]) human cervical epithelial cells were cultured in Dulbecco modified Eagle medium (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (FBS) (Invitrogen) at 37°C and 5% CO2. THP-1 (TIB-202; ATCC) human monocytic leukemia cells and Vero (CCL-81; ATCC) African green monkey kidney cells were maintained in RPMI 1640 medium (Invitrogen) containing 10% FBS at 37°C and 5% CO2. THP-1 monocytes were differentiated into macrophage-like cells by overnight treatment with 200 nM phorbol-12-myristate-13-acetate (PMA) and then washed twice with phosphate-buffered saline (PBS; 1.05 mM KH2PO4, 155 mM NaCl, 2.96 mM Na2HPO4 [pH 7.2]) prior to infection.
Construction of plasmids for C. burnetii transformation.
The bacterial strains and plasmids utilized in the present study are listed in Table S1 in the supplemental material, and the primers used in plasmid construction are listed in Table S2 in the supplemental material. C. burnetii genes were amplified with Accuprime Pfx DNA polymerase (Invitrogen) from genomic DNA extracted from the Nine Mile (RSA439), Dugway (5J108-111), and K (Q154) strains of C. burnetii using gene-specific primers (Integrated DNA Technologies, Coralville, IA). For the construction of plasmids used in targeted deletion of C. burnetii genes, the 5′ and 3′ flanking sequences of cbu0021, cbu1556, cbu1818, and cbu1863 were PCR amplified using the respective primer pairs listed in Table S2 in the supplemental material. Using the In-Fusion PCR cloning system (BD Clontech, Mountain View, CA), the 5′ and 3′ amplicons were inserted into pJC-CAT linearized by BamHI/SalI digestion to generate pJC-CAT::CBU0021-5′3′, pJC-CAT::CBU1556-5′3′, pJC-CAT::CBU1818-5′3′, and pJC-CAT::CBU1863-5′3′. The kanamycin cassette was amplified from pJB-Kan (32) using either P1169-Kan-NdeI-KO-F and P1169-Kan-NdeI-KO-R or P1169-Kan-AgeI-KO-F and P1169-Kan-AgeI-KO-R and then inserted into the NdeI or AgeI site located between the 5′ and 3′ sequences within the pJC-CAT constructs to produce pJC-CAT::CBU0021-5′3′-Kan, pJC-CAT::CBU1556-5′3′-Kan, pJC-CAT::CBU1818-5′3′-Kan, and pJC-CAT::CBU1863-5′3′-Kan, which were then used in targeted gene deletion (20). For construction of Tn7 complementation plasmids, cbu1818 or cbu1863, and their upstream promoter regions were PCR amplified from genomic DNA by using the primer pairs CBU1818comp-F and CBU1818comp-R or CBU1863comp-F and CBU1863comp-R. PCR products were inserted into the EcoRI site of pMini-Tn7T-CAT (20) with In-Fusion to create pMini-Tn7T-CAT::CBU1818comp and pMini-Tn7T-CAT::CBU1863comp.
Genes conferring resistance to chloramphenicol, kanamycin, or ampicillin are approved for C. burnetii genetic transformation studies by the Rocky Mountain Laboratories Institutional Biosafety Committee and the Centers for Disease Control and Prevention, Division of Select Agents and Toxins Program.
C. burnetii transformation.
Transformation and selection of C. burnetii Δcvp mutants was conducted as described previously (20). Briefly, pJC-CAT::CBU0021-5′3′-Kan, pJC-CAT::CBU1556-5′3′-Kan, pJC-CAT::CBU1818-5′3′-Kan, and pJC-CAT::CBU1863-5′3′-Kan chromosomal integrants were selected by culturing the bacteria in ACCM-2 containing 350 μg/ml kanamycin and 3 μg/ml chloramphenicol and then subculturing the bacteria for 4 days in ACCM-2 supplemented with 1% sucrose and kanamycin to select for cointegrants that had excised the sacB-containing plasmid backbone. C. burnetii ΔcvpB, ΔcvpC, ΔcvpD, and ΔcvpE clones were isolated by limiting dilution in ACCM-2 and gene deletions confirmed by PCR (20). Transformation and selection of C. burnetii Δcvp complement strains was accomplished as previously described (20).
Construction of plasmids for expression of C. burnetii proteins in eukaryotic cells.
C. burnetii effector-coding genes were amplified by PCR using the oligonucleotide primers listed in Table S2 in the supplemental material and cloned into pENTR/D-TOPO (Invitrogen). The plasmids were transformed into E. coli Top10 (Invitrogen), purified, and then cloned genes transferred to Gateway compatible pT-Rex-DEST30/N-mCherry or pT-Rex-DEST30/N-GFP that allow anhydrotetracycline (aTet)-inducible expression of N-terminal mCherry or green fluorescent protein (GFP) fusion proteins. Plasmids were purified from E. coli TOP10 using the GenElute HP Plasmid Midiprep kit (Sigma, St. Louis, MO) and sequence confirmed.
Adenylate cyclase translocation assays.
Adenylate cyclase assays were conducted as previously described (14). Briefly, PMA-differentiated THP-1 macrophages were plated in 24-well plates (5 × 105 cells/well) and infected at a multiplicity of infection (MOI) of 50 with C. burnetii strains expressing each of the candidate proteins N-terminally fused to CyaA. After a 48 h of incubation, the cells were washed with PBS and then lysed with 200 μl of lysis buffer containing 50 mM HCl and 0.1% Triton X-100. Samples were boiled for 5 min, and 400 μl of 95% ethanol was added. The samples were dried under vacuum and resuspended in 400 μl of assay buffer (0.5 M sodium acetate [pH 6.0], 0.002% [wt/vol] bovine serum albumin). The amount of cyclic AMP (cAMP) in the samples was determined with the cAMP Biotrak enzyme immunoassay system (GE Healthcare, Piscataway, NJ) according to the nonacetylation procedure. Samples were measured in duplicate for each independent experiment (n = 3). Values are reported as the fold change in cAMP concentration versus the empty vector control (CyaA only). Proteins were deemed T4BSS substrates if the fold change in cAMP concentration was significantly greater (P > 0.05) than the CyaA only control, as judged by one-way analysis of variance (ANOVA).
Ectopic expression and immunofluorescence microscopy.
HeLa cells infected with C. burnetii for 24 h on 12-mm coverslips in a 24-well plate were transfected with 500 ng of the pT-REx-DEST30/N-GFP or pT-Rex-DEST30/N-mCherry constructs described above and 250 ng of pcDNA 6/TR (Invitrogen) using FuGENE HD (Promega, Madison, WI) as previously described (14). The cells were incubated 24 h; fresh growth medium containing 1 μg/ml aTc (Sigma) was then added to induce protein expression. The cells were fixed 24 h later with 4% paraformaldehyde and permeabilized with PBS containing 0.05% saponin and 5% FBS. For immunostaining, rabbit anti-LAMP1 polyclonal antibody (Abcam, Cambridge, England) and guinea pig anti-Coxiella serum (14) were used as primary antibodies, followed by goat anti-rabbit Alexa Fluor 594 (Invitrogen) and goat anti-guinea pig Alexa Fluor 647 (Invitrogen). Coverslips were mounted using Prolong Gold with DAPI (4′,6′-diamidino-2-phenylindole; Invitrogen) and imaged. For subcellular localization of ectopically expressed pT-Rex-DEST30/N-mCherry-CBU1556, cells were stained with a mouse antibody against transferrin receptor (Life Technologies) and goat anti-mouse Alexa Fluor 488 (Invitrogen). Imaging was conducted with a LSM710 confocal laser scanning microscope (Carl Zeiss Micro Imaging, Thornwood, NY) or a Nikon Eclipse Ti-E inverted microscope.
Quantification of C. burnetii growth and PV morphology.
THP-1 cells seeded at 5 × 105 per well in a 24-well plate were infected at an MOI of 0.5 with C. burnetii suspended in RPMI plus 10% FBS. Plates were centrifuged at 500 × g for 20 min. Infected cells were washed once with PBS and then cultured in RPMI plus 10% FBS for the remainder of the experiment. For each of three independent experiments, samples were collected in duplicate on the day of infection (day 0) and 6 days postinfection. C. burnetii growth was assessed by quantifying genomic equivalents (GE) as previously described (14).
To measure PV size, Vero cells (2 × 104) on 12-mm coverslips in a 24-well plate were infected at an MOI of 5 with C. burnetii suspended in RPMI plus 2% FBS. Plates were centrifuged at 500 × g for 20 min. Infected Vero cells were incubated 6 days and then fixed and stained with antibodies against LAMP1 and Coxiella as described above. Fluorescence micrographs were acquired, and 25 PV were measured for each C. burnetii strain in three independent experiments. For mutant complementation by coinfection, Vero cells (2 × 104) on 12-mm coverslips in a 24-well plate were infected at MOIs of 5 and 2, respectively, with the Δcvp mutant and wild-type C. burnetii expressing mCherry red fluorescent protein (16). The Δcvp mutants were visualized in PVs cohabited with wild-type C. burnetii at 6 days postinfection by immunostaining with antibodies against C. burnetii and LAMP1. Assessment of mutant PV fusion was conducted with Vero cells (2 × 104) infected at an MOI of 100 with C. burnetii in RPMI plus 2% FBS. Cells were immunostained for LAMP1 and C. burnetii at 6 days postinfection, and the number of PVs in each cell (n = 100) enumerated using fluorescence microscopy.
PV acidification.
PV acidification was examined using LysoTracker Green DND-26 (Invitrogen) according to the manufacturer's protocol. Vero cells (2 × 104) were infected at an MOI of 100 with C. burnetii in RPMI plus 2% FBS. Cells were incubated 6 days, treated with LysoTracker, and then immunostained for LAMP1 and C. burnetii.
Data analysis.
GraphPad Prism 6.0 software (San Diego, CA) was used to perform one-way ANOVA or two-way ANOVA statistical tests. All image processing and measurements were conducted using ImageJ software (W. S. Rasband, National Institutes of Health, Bethesda, MD).
RESULTS
Identification of C. burnetii T4BSS effector proteins.
Bioinformatic analysis of the C. burnetii Nine Mile (RSA493), Dugway (5J108-111), and K (Q154) genomes (33) revealed 14 genes encoding predicted proteins with characteristics of T4BSS substrates (Table 1). The identified candidate proteins contained eukaryote-like features, including coiled-coil and leucine-rich repeat domains associated with ligand recognition, and transmembrane domains predicted to mediate membrane attachment. CBU0885 contains a haloacid dehalogenase (HAD)-like domain associated with phosphatase activity (34), while CBU1457 harbors multiple Sel1 repeats known to promote protein-protein interactions (35). In addition, several genes encoding candidate proteins contained PmrA regulatory elements (28). Genes encoding each of the candidate effectors were inserted into pJB-CAT-CyaA, allowing C. burnetii expression of proteins N-terminally fused to the adenylate cyclase reporter protein CyaA (14, 18). PMA-differentiated THP-1 human macrophages were infected with wild-type C. burnetii or ΔdotA mutant strains expressing each of the CyaA constructs. THP-1 macrophages were also infected with wild-type C. burnetii transformed with the empty CyaA vector as a negative control. At 48 h postinfection, THP-1 cells were lysed, and the concentration of cytosolic cAMP measured (Fig. 1). Expression of 13 CyaA fusion proteins by C. burnetii resulted in a significant fold increase in cAMP concentration relative to C. burnetii expressing CyaA alone. No increase in cAMP concentration was detected after infection with C. burnetii ΔdotA expressing CyaA fusion proteins, indicating that secretion is Dot/Icm dependent. Collectively, these data indicated that CBU0021, CBU0534, CBU0885, CBUD0886, CBUK0790, CBU1493, CBUD0487, CBU1543, CBU1556, CBU1676, CBU1818, CBU1819, and CBU1863 are Dot/Icm substrates delivered to the host cell cytosol by the C. burnetii T4BSS. Negative translocation of CyaA-CBU1457 was not due to lack of expression since all CyaA fusion proteins were expressed as assessed by immunoblotting (data not shown).
TABLE 1.
Features of candidate Dot/Icm effectors
| Candidate effector | Size (kDa) | Features of gene or protein | Source or reference(s) |
|---|---|---|---|
| CBU0021 | 93.1 | Coiled-coil domain | 22, 30, 63 |
| CBUD0487 | 90.6 | Coiled-coil domain | This study |
| CBU0534 | 46.1 | Coiled-coil domain:, transmembrane domain | This study |
| CBUK0790 | 84.7 | PmrA regulatory element | This study |
| CBU0885 | 43.3 | Haloacid dehalogenase (HAD)-like domain | 22, 25 |
| CBUD0886 | 50.8 | Leucine-rich repeats | This study |
| CBU1457 | 78.3 | Sel1-like repeats | 21, 25 |
| CBU1493 | 60.9 | PmrA regulatory element | This study |
| CBU1543 | 22.3 | Coiled-coil domain, transmembrane domain | 21, 25 |
| CBU1556 | 64.6 | Coiled-coil domain, transmembrane domains | 21, 25 |
| CBU1676 | 41.8 | Paralog of CBU0885 | 62 |
| CBU1818 | 53.9 | Coiled-coil domain, transmembrane domains | 62 |
| CBU1819 | 42.3 | Coiled-coil domain | This study |
| CBU1863 | 33.2 | PmrA regulatory element, transmembrane domains | This study |
FIG 1.
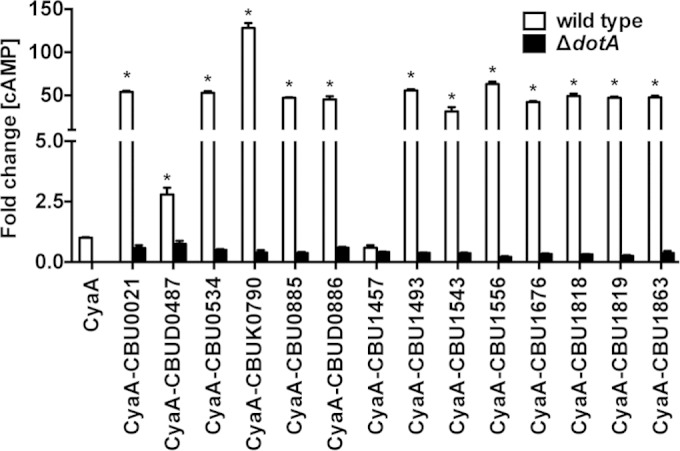
C. burnetii translocates 13 proteins via its Dot/Icm T4BSS. Cytosolic concentrations of cAMP were determined after infection of THP-1 macrophages for 48 h with wild-type C. burnetii or a ΔdotA mutant expressing CyaA fusions to candidate Dot/Icm substrates. The results are expressed as the fold change relative to wild-type C. burnetii expressing CyaA alone. Increased cytosolic cAMP concentrations indicative of protein translocation were detected for CBU0021, CBUD0487, CBU0534, CBUK0790, CBU0886, CBUD0886, CBU1493, CBU1543, CBU1556, CBU1676, CBU1818, CBU1819, and CBU1863. The results are from one experiment conducted in duplicate and are representative of three independent experiments. Error bars indicate the standard deviations from the means. Asterisks indicate a significantly greater difference (P < 0.05) compared to values for the CyaA only control as determined by one-way ANOVA.
C. burnetii effectors traffic to the PV.
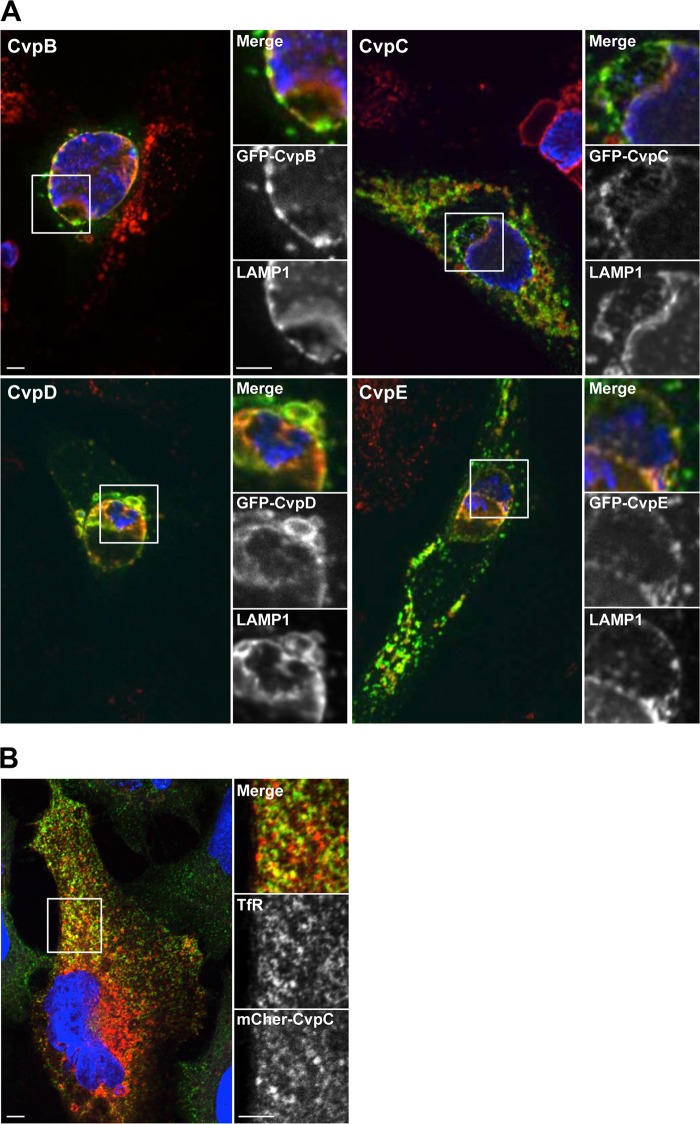
A subset of effector proteins within the repertoire of C. burnetii Dot/Icm substrates is predicted to localize to the PV membrane where they modify vesicle budding, transport, and fusion events to promote vacuole expansion. Indeed, CvpA was previously shown to localize to the PV membrane and contribute to PV biogenesis by a mechanism involving subversion of clathrin-mediated vesicular trafficking (14). To identify additional PV-localized effector proteins, the 13 validated C. burnetii Dot/Icm substrates were ectopically expressed as N-terminal fusions to GFP or mCherry fluorescent protein in HeLa cells infected with C. burnetii. At 3 days postinfection, cells were fixed and stained with antibodies against LAMP1 and C. burnetii. The ectopically expressed Dot/Icm substrates displayed subcellular localizations ranging from diffuse cytoplasmic to punctate perinuclear (Fig. 2A; see also Fig. S1 in the supplemental material). Of the 13 proteins examined, CBU0021, CBU1556, CBU1818, and CBU1863 localized to the LAMP1-positive PV membrane (Fig. 2A; see also Fig. S1 in the supplemental material), suggesting they confer effector functions associated with PV biogenesis. Consequently, these proteins were termed CvpB, CvpC, CvpD, and CvpE, respectively. To examine whether Cvp effectors also interact with the PV early after infection, HeLa cells ectopically expressing the Cvp proteins were examined at 6 h postinfection. More than 90% of C. burnetii PV labeled with ectopically expressed CvpB, CvpC, CvpD, or CvpE, suggesting the effectors target vesicular components during the initial stages of PV maturation (see Fig. S2 in the supplemental material).
FIG 2.
Four C. burnetii Dot/Icm T4BSS substrates localize to the PV membrane. (A) Representative confocal fluorescence micrographs of C. burnetii-infected HeLa cells ectopically expressing CvpB, CvpC, CvpD, or CvpE N-terminally fused to GFP. At 72 h postinfection, cells were fixed and stained with antibodies against the lysosomal membrane protein LAMP1 (red) and C. burnetii (blue). (B) CvpC-mCherry colocalizes with transferrin receptor. HeLa cells were transfected with a plasmid encoding CvpC N-terminally fused to mCherry red fluorescent protein. Cells were immunostained for transferrin receptor (green) with the DNA stained by DAPI (blue). Scale bar, 5 μm.
Although Cvp effectors traffic to the LAMP1-positive PV membrane, it remained possible that they also interact with membrane components outside the late endosomal compartment, as is the case with CvpA (14). To further examine their subcellular itineraries, each Cvp protein was ectopically expressed N-terminally fused to GFP or mCherry red fluorescent protein in uninfected HeLa cells. Transfected cells were then stained with antibodies against protein markers of early, recycling, or late endosomes, the endoplasmic reticulum (ER), the ER-Golgi intermediate compartment (ERGIC), or the cis-Golgi or trans-Golgi. As expected, all of the Cvp effectors localized with late endosomal marker LAMP1 in uninfected cells. In addition, CvpC exhibited partial localization with transferrin receptor, a marker of recycling endosomes (Fig. 2B). We have previously shown that ectopically expressed CvpA localizes to recycling endosomes and inhibits the uptake of transferrin (14). However, ectopic expression of CvpC did not alter transferrin uptake (data not shown). Moreover, none of the Cvp proteins localized with structures that labeled with antibodies against EEA1 (early endosomes), calreticulin (ER), ERGIC53 (ERGIC), giantin (cis-Golgi), or p230/golgin-245 (trans-Golgi) (data not shown). Collectively, these results suggest CvpB, CvpC, CvpD, and CvpE are C. burnetii effectors that target components of the late endosomal system, and that CvpC also targets recycling endosomes.
Cvp effectors promote intracellular replication and PV expansion.
Localization to the PV membrane suggested CvpB, CvpC, CvpD, and CvpE promote PV biogenesis and C. burnetii intracellular replication. To assess this possibility, C. burnetii cvp deletion mutants were generated and growth was assessed in THP-1 macrophages by measuring the increase in GE over 6 days. Replication of the C. burnetii ΔcvpB, ΔcvpC, ΔcvpD, and ΔcvpE mutants was significantly impaired with 49-, 276-, 160-, and 110-fold increases in GE, respectively, relative to the 713-fold increase in GE observed for wild-type C. burnetii (Fig. 3A). Defects in C. burnetii replication correlated with impaired PV biogenesis. All mutants occupied aberrantly small, tight-fitting vacuoles in Vero cells compared to the large and spacious PVs generated by wild-type C. burnetii (Fig. 3B). PVs harboring C. burnetii ΔcvpB, ΔcvpD, and ΔcvpE strains in Vero cells also exhibited impaired homotypic fusion with 57.5, 35.2, and 32.6%, respectively, containing two or more PVs compared to 16.2% of cells infected with wild-type C. burnetii (see Fig. S3A in the supplemental material). Consistent with the presence of LAMP1, PVs containing Δcvp mutants were acidified, as evidenced by the accumulation of the acidotropic fluorescent dye LysoTracker DND-26 (see Fig. S3B in the supplemental material). These acidified vacuoles supported metabolic activation of Δcvp mutants as each was capable of secreting a CyaA-CvpA fusion protein (14) (see Fig. S4 in the supplemental material). Thus, Δcvp mutant growth defects likely result from inactivation of the targeted effector.
FIG 3.
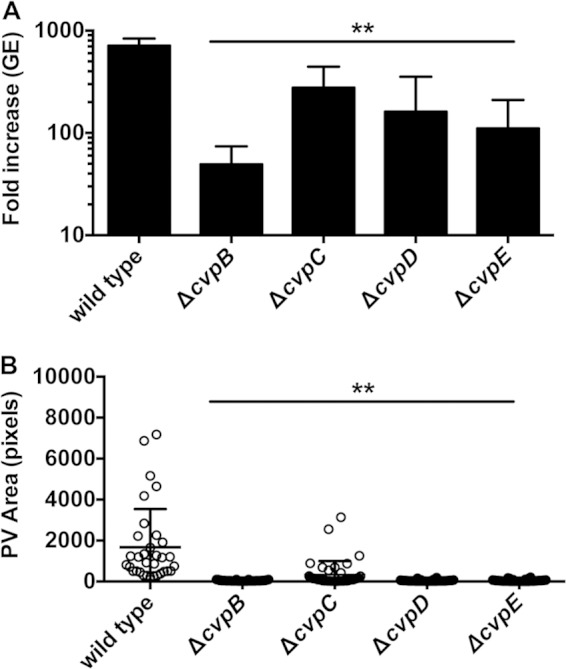
C. burnetii Δcvp mutants exhibit defects in intracellular growth and PV biogenesis. (A) Replication of wild-type C. burnetii and Δcvp mutants. Fold increases in bacterial GE at 6 days postinfection of THP-1 macrophages relative to the 0 day time point are depicted from three independent experiments, each performed in duplicate. (B) Sizes of PVs generated by wild-type C. burnetii and the Δcvp mutants 6 days postinfection in Vero cells. PV size was measured using ImageJ (n = 25), and the data are representative of three independent experiments. Error bars indicate the standard deviations from the means. Asterisks indicate a statistically significant difference (P < 0.01) compared to values for wild-type bacteria as determined by one-way ANOVA.
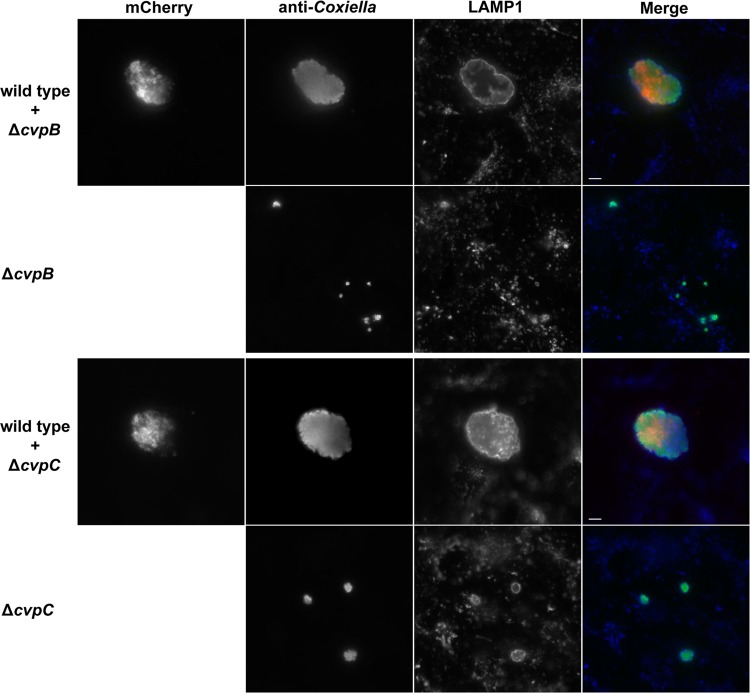
To confirm that the specific gene deletion was responsible for the intracellular growth defect of a Δcvp mutant, complementation was attempted using Tn7 to introduce a single gene copy into the chromosome under the control of a native promoter. Tn7 complementation rescued intracellular growth of the ΔcvpD and ΔcvpE mutants, as evidenced by significant increases in replication within THP-1 cells (Fig. 4A) and PV size in Vero cells (Fig. 4B). This strategy of genetic complementation failed to rescue impaired growth of C. burnetii ΔcvpB and ΔcvpC. Transformation with a multicopy plasmid encoding constitutively expressed CvpB and CvpC also failed to complement mutant growth defects (data not shown). Therefore, we used a coinfection strategy that has been previously used to show that the growth deficiencies of C. burnetii Dot/Icm mutants can be complemented by functions provided in trans by wild-type C. burnetii (16, 25). Vero cells were coinfected with wild-type C. burnetii and the ΔcvpB or ΔcvpC strain. When sequestered within a PV co-occupied by wild-type C. burnetii, the ΔcvpB and ΔcvpC mutants exhibited robust replication (Fig. 5).
FIG 4.
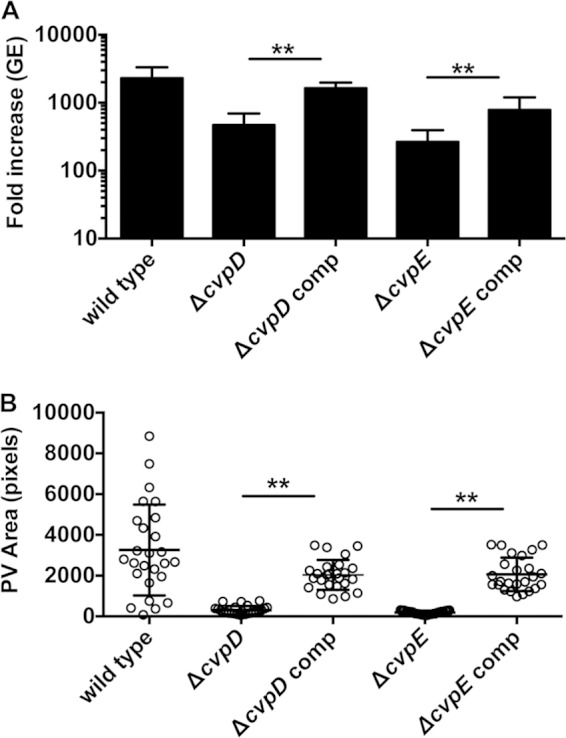
Genetic complementation of the ΔcvpD and ΔcvpE mutants rescues intracellular growth and PV biogenesis. (A) Replication of wild-type C. burnetii and complemented Δcvp mutants. Fold increases in bacterial GE at 6 days postinfection of THP-1 macrophages relative to the 0 day time point are depicted from three independent experiments, each performed in duplicate. (B) Sizes of PVs generated by the wild-type C. burnetii and complemented Δcvp mutants at 6 days postinfection of Vero cells. PV size was measured using ImageJ (n = 25), and the data are representative of three independent experiments. Error bars indicate the standard deviations from the means. Asterisks indicate a statistically significant difference (P < 0.01) compared to values for wild-type bacteria as determined by one-way ANOVA.
FIG 5.
Coinfection with wild-type C. burnetii rescues growth of ΔcvpB and ΔcvpC mutants. THP-1 macrophages were infected with the ΔcvpB or ΔcvpC mutant alone or coinfected with the ΔcvpB or ΔcvpC mutant and wild-type C. burnetii expressing mCherry red fluorescent protein. At 6 days postinfection, C. burnetii (wild type and mutant) and LAMP1 were immunostained green and blue, respectively. Robust growth of ΔcvpB and ΔcvpC mutants was only observed in PVs cohabited with wild-type C. burnetii. Scale bar, 5 μm.
DISCUSSION
The pronounced fusogenicity of the C. burnetii PV is predicted to be mediated by C. burnetii Dot/Icm effector proteins that modulate vesicle budding, transport, and fusion events on the PV membrane (6, 9, 14, 36). In support of this model is CvpA, a PV-interacting Dot/Icm effector protein that promotes PV formation (14). CvpA targets clathrin-dependent vesicular trafficking, presumably allowing C. burnetii to acquire membrane components for PV biogenesis (14). In the present study, we used genetic manipulation of C. burnetii to directly identify Dot/Icm T4BSS substrates secreted by the pathogen and to validate their importance in intracellular replication. Based on interactions with the PV membrane, four new Cvp proteins were identified among the 13 identified Dot/Icm substrates. Defects in intracellular replication and PV expansion observed for C. burnetii Δcvp mutants support the hypothesis that C. burnetii encodes a family of Dot/Icm effectors that localize to the PV membrane and modulate membrane trafficking events necessary for PV development and pathogen replication.
The Δcvp mutants occupy a LAMP1-positive, acidified PV and can secrete proteins via the T4BSS. Thus, their growth deficiencies are not related to overall failings in PV maturation and Dot/Icm function. The ΔcvpB, ΔcvpD, and ΔcvpE mutants, but not the ΔcvpC mutant, generated multi-PV in Vero cells that displayed impaired homotypic fusion. These phenotypes correlated with a severe replication defect. Rescue of mutant growth defects by Tn7 cis-complementation verified mutation of CvpD and CvpE impairs C. burnetii intracellular growth, but this strategy failed to complement growth of the ΔcvpB and ΔcvpC mutants. The reasons for lack of genetic complementation remain unclear, but may include temporal problems with gene expression, insufficient chaperone engagement, and/or altered protein levels. To bypass these issues, we and others have used coinfection with wild-type C. burnetii to rescue impaired growth of strains with mutations in the T4BSS apparatus (16, 30) and Dot/Icm effectors (25). Although coinfection rescued the growth defects of ΔcvpB and ΔcvpC mutants, we cannot rule out the possibility that the phenotypes are due to a secondary mutation that can also be complemented in trans.
The observation that cvpB, cvpC, cvpD, and cvpE are maintained as full-length genes among sequenced C. burnetii strains suggests that they modulate core host cell functions required for successful intracellular parasitism by the genus Coxiella. Polymorphisms among several previously identified C. burnetii Dot/Icm substrates (17, 24) are hypothesized to contribute to pathogenic potential (36). Of the substrates identified in the present study, cbud0487 is full length only in the Dugway (5J108-111) strain, cbud0886 is full length only in the Dugway (5J108-111) and K (Q154) strains, and cubk0790 is only full length only in the K (Q154) strain. The Nine Mile strain used here is representative of acute disease isolates, the K strain is a human chronic endocarditis isolate, and the Dugway strain is an attenuated rodent isolate (33).
Localization of bacterial effector proteins to pathogen-occupied vacuoles is commonly observed. For example, L. pneumophila translocates multiple effector proteins via its Dot/Icm T4BSS that interact with the membrane of the Legionella vacuole (LV). Dot/Icm effectors LidA, SidC, and DrrA/SidM bind specific phosphoinositides (37, 38), while LepA, LepB, LegC3, LegC2/YlfB, and LegC7/YlfA contain transmembrane domains that facilitate interaction with the LV membrane (39, 40). Similarly, Chlamydia spp. deploy a type 3 secretion system (T3SS) to deliver multiple Inc family proteins that localize to the chlamydial inclusion via a conserved bilobed hydrophobic motif (41). CvpC, CvpD, and CvpE all contain predicted transmembrane domains that may facilitate their interaction with the LAMP1-positive PV membrane. In contrast, CvpB lacks obvious transmembrane domains and therefore might target the PV membrane by binding phosphoinositides. When examined in uninfected cells, CvpB, CvpD, and CvpE consistently localize only to the late endolysosomal compartment. CvpC, in addition to LAMP1-positive structures, also localizes with transferrin receptor, an endocytic carrier protein that undergoes clathrin-dependent endocytosis and traffics through the endocytic recycling system. Thus, CvpC may traffic between late and recycling endosomal compartments, as is the case with CvpA (14).
The activities of vesicular effectors produced by other pathogens may provide clues to Cvp biochemical activities. Modulation of Rab GTPases is a prominent strategy used by intracellular pathogens to remodel their phagosome into a replication-permissive compartment (42, 43). Rab GTPases play critical roles in regulating transport of lipids and proteins that ultimately determine organelle identity (44, 45). Guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) regulate the rate of cycling between inactive GDP-bound and active GTP-bound forms of Rab GTPases (45). Pathogens secrete effectors that subvert these regulatory mechanisms to control spatial and temporal aspects of membrane transport. Of the LV-localized Dot/Icm effectors produced by L. pneumophila, multiple proteins modulate Rab1 activity to refurbish the LV with ER components and to prevent LV fusion with lysosomes. For example, LidA binds Rab1, which stabilizes its activation (46) and promotes the acquisition of ER-derived vesicles (47). Furthermore, DrrA/SidM is a GEF that posttranslationally modifies Rab1 with AMP (48–51), thereby preventing inactivation by GAP proteins (52–54). Rab1 (12), as well as the regulators of endocytic traffic Rab5 and Rab7, have been implicated in C. burnetii PV biogenesis (15, 55, 56). Thus, it is plausible that C. burnetii Cvp effectors disregulate Rab GTPases to promote maturation of its strikingly large phagolysosome-like replication compartment.
The Cvp effectors lack homology to proteins of known function, but CvpB, CvpC, and CvpD contain predicted eukaryote-like coiled-coil domains similar to bacterial effectors known to functionally mimic the activity of eukaryotic SNARE proteins (57). SNAREs regulate fusion between transport vesicles and target compartment membranes, and pathogens subvert SNARE function to promote fusion events beneficial to pathogen growth (57). The chlamydial T3SS substrate IncA harbors two SNARE-like coiled-coil motifs (58) and inhibits membrane fusion mediated by endocytic, but not exocytic, SNARE complexes (59). Expression of the L. pneumophila effector LegC3 produces a vacuolar protein sorting defect in yeast (40) linked to the disruption of trans-SNARE complexes by coiled-coil domains within the effector (60). In contrast to L. pneumophila and Chlamydia spp. that block fusion of their vacuoles with lysosomes, phagosome acidification and lysosomal fusion promotes Coxiella replication (6). Moreover, C. burnetii also requires membrane components from autophagic and secretory compartments for PV expansion (10–12, 15, 61). Several SNARE proteins, including syntaxin-8, syntaxin-17, and vesicle-associated membrane protein 7 (VAMP7), localize to the PV membrane, with syntaxin-17 and VAMP7 having demonstrated roles in vacuole expansion (11, 15). Coxiella researchers are just beginning to investigate the molecular mechanisms that control PV fusogenicity, a process that likely involves the activities of multiple Rab GTPases, SNARE proteins, and Dot/Icm effectors (11, 14–16).
When this manuscript was originally submitted, 4 of the 13 proteins we report here as substrates of the C. burnetii Dot/Icm system had also been described as L. pneumophila Dot/Icm substrates (21, 22, 25). Using a hidden semi-Markov model, Lifshitz et al. (22) identified a group of C. burnetii proteins, termed C-terminal signal for effector translocation of C. burnetii (CetCb) proteins, that have C-terminal amino acid sequences resembling those of known Dot/Icm substrates. In agreement with our findings using C. burnetii, CvpB/CetCb1 (CBU0021) and CetCb4 (CBU0885) fusions with CyaA are translocated by L. pneumophila in a Dot/Icm-dependent manner (22). Chen et al. (21) expressed multiple C. burnetii proteins in L. pneumophila fused to the β-lactamase (BlaM) reporter. These researchers concluded that CBU1457, CBU1543, and CvpC (CBU1556) are Dot/Icm substrates based on 2, 25, and 50% translocation efficiencies of the respective BlaM fusion protein (21, 25). We found that CBU1543 and CvpC fusions with CyaA are translocated by C. burnetii in a Dot/Icm-dependent fashion. In contrast, we did not detect translocation of CyaA-CBU1457 by C. burnetii at the 48-h postinfection time point. Translocation was also not detected at 72 or 96 h postinfection, nor with L. pneumophila using standard assay conditions (data not shown) (18). The reason for this disparate result is unclear but emphasizes the need to validate Dot/Icm effectors directly in C. burnetii.
Since submission of the manuscript, a study by Lifshitz and coworkers (62) demonstrated L. pneumophila Dot/Icm-dependent secretion of CBU1676 and CBU1818 (CvpD). Expression of CBU1676, or its paralog CBU0885, inhibits mitogen-activated protein kinase signaling in Saccharomyces cerevisiae. Newton et al. (63) reported that C. burnetii cig2 (cvpB) Tn insertion mutants exhibit a multi-PV growth phenotype in HeLa cells without showing an overall growth defect. We found that C. burnetii ΔcvpB replicates poorly in THP-1 human macrophages, a behavior that correlates with generation of multiple, small PVs in Vero cells. Martinez et al. (30) also found that C. burnetii strains harboring a Tn insertion in cbu0021 (cvpB) display impaired growth in Vero cells. Thus, accumulating evidence from independent laboratories indicates CvpB is a critical Dot/Icm effector that subverts cellular functions involved in PV biogenesis. Indeed, because PVs containing the cvpB mutant do not accumulate the autophagosome protein LC3, and autophagy inhibition of cells infected with wild-type bacteria produce multiple PVs similar to the C. burnetii cvpB mutant, Newton et al. (63) conclude that CvpB promotes PV interactions with autophagosomes that enhance PV maturation. Biochemical characterization of the functional activities CvpB and other Cvp effectors will provide important insight into mechanisms governing PV development.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by funding from NIH NIAID grants AI072485 (R.H.V.) and R01AI087669 (D.E.V.) and by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases (R.A.H.).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02763-14.
REFERENCES
- 1.Maurin M, Raoult D. 1999. Q fever Clin Microbiol Rev 12:518–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coleman SA, Fischer ER, Howe D, Mead DJ, Heinzen RA. 2004. Temporal analysis of Coxiella burnetii morphological differentiation. J Bacteriol 186:7344–7352. doi: 10.1128/JB.186.21.7344-7352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinzen RA, Hackstadt T, Samuel JE. 1999. Developmental biology of Coxiella burnetii. Trends Microbiol 7:149–154. doi: 10.1016/S0966-842X(99)01475-4. [DOI] [PubMed] [Google Scholar]
- 4.McCaul TF, Williams JC. 1981. Developmental cycle of Coxiella burnetii: structure and morphogenesis of vegetative and sporogenic differentiations. J Bacteriol 147:1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCaul TF, Hackstadt T, Williams JC. 1981. Ultrastructural and biological aspects of Coxiella burnetii under physical disruptions, p 267–280. In Burgdorfer W, Anacker RL (ed), Rickettsiae and rickettsial diseases. Academic Press, Inc, New York, NY. [Google Scholar]
- 6.Voth DE, Heinzen RA. 2007. Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell Microbiol 9:829–840. doi: 10.1111/j.1462-5822.2007.00901.x. [DOI] [PubMed] [Google Scholar]
- 7.Howe D, Shannon JG, Winfree S, Dorward DW, Heinzen RA. 2010. Coxiella burnetii phase I and II variants replicate with similar kinetics in degradative phagolysosome-like compartments of human macrophages. Infect Immun 78:3465–3474. doi: 10.1128/IAI.00406-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hackstadt T, Williams JC. 1981. Biochemical stratagem for obligate parasitism of eukaryotic cells by Coxiella burnetii. Proc Natl Acad Sci U S A 78:3240–3244. doi: 10.1073/pnas.78.5.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howe D, Melnicakova J, Barak I, Heinzen RA. 2003. Maturation of the Coxiella burnetii parasitophorous vacuole requires bacterial protein synthesis but not replication. Cell Microbiol 5:469–480. doi: 10.1046/j.1462-5822.2003.00293.x. [DOI] [PubMed] [Google Scholar]
- 10.Beron W, Gutierrez MG, Rabinovitch M, Colombo MI. 2002. Coxiella burnetii localizes in a Rab7-labeled compartment with autophagic characteristics. Infect Immun 70:5816–5821. doi: 10.1128/IAI.70.10.5816-5821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campoy EM, Mansilla ME, Colombo MI. 2013. Endocytic SNAREs are involved in optimal Coxiella burnetii vacuole development. Cell Microbiol 15:922–941. doi: 10.1111/cmi.12087. [DOI] [PubMed] [Google Scholar]
- 12.Campoy EM, Zoppino FC, Colombo MI. 2011. The early secretory pathway contributes to the growth of the Coxiella-replicative niche. Infect Immun 79:402–413. doi: 10.1128/IAI.00688-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howe D, Melnicakova J, Barak I, Heinzen RA. 2003. Fusogenicity of the Coxiella burnetii parasitophorous vacuole. Ann N Y Acad Sci 990:556–562. doi: 10.1111/j.1749-6632.2003.tb07426.x. [DOI] [PubMed] [Google Scholar]
- 14.Larson CL, Beare PA, Howe D, Heinzen RA. 2013. Coxiella burnetii effector protein subverts clathrin-mediated vesicular trafficking for pathogen vacuole biogenesis. Proc Natl Acad Sci U S A 110:E4770–E4779. doi: 10.1073/pnas.1309195110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDonough JA, Newton HJ, Klum S, Swiss R, Agaisse H, Roy CR. 2013. Host pathways important for Coxiella burnetii infection revealed by genome-wide RNA interference screening. mBio 4:e00606–12. doi: 10.1128/mBio.00606-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beare PA, Gilk SD, Larson CL, Hill J, Stead CM, Omsland A, Cockrell DC, Howe D, Voth DE, Heinzen RA. 2011. Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. mBio 2:e00175–11. doi: 10.1128/mBio.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carey KL, Newton HJ, Lührmann A, Roy CR. 2011. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog 7:e1002056. doi: 10.1371/journal.ppat.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voth DE, Beare PA, Howe D, Sharma UM, Samoilis G, Cockrell DC, Omsland A, Heinzen RA. 2011. The Coxiella burnetii cryptic plasmid is enriched in genes encoding type IV secretion system substrates. J Bacteriol 193:1493–1503. doi: 10.1128/JB.01359-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogel JP. 2004. Turning a tiger into a house cat: using Legionella pneumophila to study Coxiella burnetii. Trends Microbiol 12:103–105. doi: 10.1016/j.tim.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Beare PA, Larson CL, Gilk SD, Heinzen RA. 2012. Two systems for targeted gene deletion in Coxiella burnetii. Appl Environ Microbiol 78:4580–4589. doi: 10.1128/AEM.00881-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen C, Banga S, Mertens K, Weber MM, Gorbaslieva I, Tan Y, Luo ZQ, Samuel JE. 2010. Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc Natl Acad Sci U S A 107:21755–21760. doi: 10.1073/pnas.1010485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lifshitz Z, Burstein D, Peeri M, Zusman T, Schwartz K, Shuman HA, Pupko T, Segal G. 2013. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type IVB secretion signal. Proc Natl Acad Sci U S A 110:E707–E715. doi: 10.1073/pnas.1215278110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan X, Luhrmann A, Satoh A, Laskowski-Arce MA, Roy CR. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320:1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voth DE, Howe D, Beare PA, Vogel JP, Unsworth N, Samuel JE, Heinzen RA. 2009. The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J Bacteriol 191:4232–4242. doi: 10.1128/JB.01656-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber MM, Chen C, Rowin K, Mertens K, Galvan G, Zhi H, Dealing CM, Roman VA, Banga S, Tan Y, Luo ZQ, Samuel JE. 2013. Identification of Coxiella burnetii type IV secretion substrates required for intracellular replication and Coxiella-containing vacuole formation. J Bacteriol 195:3914–3924. doi: 10.1128/JB.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burstein D, Zusman T, Degtyar E, Viner R, Segal G, Pupko T. 2009. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog 5:e1000508. doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang L, Boyd D, Amyot WM, Hempstead AD, Luo ZQ, O'Connor TJ, Chen C, Machner M, Montminy T, Isberg RR. 2011. The E block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol 13:227–245. doi: 10.1111/j.1462-5822.2010.01531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zusman T, Aloni G, Halperin E, Kotzer H, Degtyar E, Feldman M, Segal G. 2007. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol Microbiol 63:1508–1523. doi: 10.1111/j.1365-2958.2007.05604.x. [DOI] [PubMed] [Google Scholar]
- 29.Maturana P, Graham JG, Sharma UM, Voth DE. 2013. Refining the plasmid-encoded type IV secretion system substrate repertoire of Coxiella burnetii. J Bacteriol 195:3269–3276. doi: 10.1128/JB.00180-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez E, Cantet F, Fava L, Norville I, Bonazzi M. 2014. Identification of OmpA, a Coxiella burnetii protein involved in host cell invasion, by multi-phenotypic high-content screening. PLoS Pathog 10:e1004013. doi: 10.1371/journal.ppat.1004013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonifacino JS, Traub LM. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 32.Omsland A, Beare PA, Hill J, Cockrell DC, Howe D, Hansen B, Samuel JE, Heinzen RA. 2011. Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl Environ Microbiol 77:3720–3725. doi: 10.1128/AEM.02826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beare PA, Unsworth N, Andoh M, Voth DE, Omsland A, Gilk SD, Williams KP, Sobral BW, Kupko JJ III, Porcella SF, Samuel JE, Heinzen RA. 2009. Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella. Infect Immun 77:642–656. doi: 10.1128/IAI.01141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeuchi H, Hirano T, Whitmore SE, Morisaki I, Amano A, Lamont RJ. 2013. The serine phosphatase SerB of Porphyromonas gingivalis suppresses IL-8 production by dephosphorylation of NF-κB RelA/p65. PLoS Pathog 9:e1003326. doi: 10.1371/journal.ppat.1003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mittl PR, Schneider-Brachert W. 2007. Sel1-like repeat proteins in signal transduction. Cell Signal 19:20–31. doi: 10.1016/j.cellsig.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 36.van Schaik EJ, Chen C, Mertens K, Weber MM, Samuel JE. 2013. Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat Rev Microbiol 11:561–573. doi: 10.1038/nrmicro3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brombacher E, Urwyler S, Ragaz C, Weber SS, Kami K, Overduin M, Hilbi H. 2009. Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate-binding effector protein of Legionella pneumophila. J Biol Chem 284:4846–4856. doi: 10.1074/jbc.M807505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ragaz C, Pietsch H, Urwyler S, Tiaden A, Weber SS, Hilbi H. 2008. The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell Microbiol 10:2416–2433. doi: 10.1111/j.1462-5822.2008.01219.x. [DOI] [PubMed] [Google Scholar]
- 39.Chen J, Reyes M, Clarke M, Shuman HA. 2007. Host cell-dependent secretion and translocation of the LepA and LepB effectors of Legionella pneumophila. Cell Microbiol 9:1660–1671. doi: 10.1111/j.1462-5822.2007.00899.x. [DOI] [PubMed] [Google Scholar]
- 40.de Felipe KS, Glover RT, Charpentier X, Anderson OR, Reyes M, Pericone CD, Shuman HA. 2008. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog 4:e1000117. doi: 10.1371/journal.ppat.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Betts HJ, Wolf K, Fields KA. 2009. Effector protein modulation of host cells: examples in the Chlamydia spp. arsenal Curr Opin Microbiol 12:81–87. doi: 10.1016/j.mib.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 42.Alix E, Mukherjee S, Roy CR. 2011. Subversion of membrane transport pathways by vacuolar pathogens. J Cell Biol 195:943–952. doi: 10.1083/jcb.201105019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brumell JH, Scidmore MA. 2007. Manipulation of rab GTPase function by intracellular bacterial pathogens. Microbiol Mol Biol Rev 71:636–652. doi: 10.1128/MMBR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Behnia R, Munro S. 2005. Organelle identity and the signposts for membrane traffic. Nature 438:597–604. doi: 10.1038/nature04397. [DOI] [PubMed] [Google Scholar]
- 45.Zerial M, McBride H. 2001. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol 2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 46.Cheng W, Yin K, Lu D, Li B, Zhu D, Chen Y, Zhang H, Xu S, Chai J, Gu L. 2012. Structural insights into a unique Legionella pneumophila effector LidA recognizing both GDP and GTP bound Rab1 in their active state. PLoS Pathog 8:e1002528. doi: 10.1371/journal.ppat.1002528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Machner MP, Isberg RR. 2006. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell 11:47–56. doi: 10.1016/j.devcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 48.Hardiman CA, Roy CR. 2014. AMPylation is critical for Rab1 localization to vacuoles containing Legionella pneumophila. mBio 5:e01035–13. doi: 10.1128/mBio.01035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ingmundson A, Delprato A, Lambright DG, Roy CR. 2007. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature 450:365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- 50.Muller MP, Peters H, Blumer J, Blankenfeldt W, Goody RS, Itzen A. 2010. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329:946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- 51.Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR. 2006. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol 8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- 52.Gazdag EM, Schobel S, Shkumatov AV, Goody RS, Itzen A. 2014. The structure of the N-terminal domain of the Legionella protein SidC. J Struct Biol 186:188–194. doi: 10.1016/j.jsb.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 53.Mishra AK, Del Campo CM, Collins RE, Roy CR, Lambright DG. 2013. The Legionella pneumophila GTPase activating protein LepB accelerates Rab1 deactivation by a non-canonical hydrolytic mechanism. J Biol Chem 288:24000–24011. doi: 10.1074/jbc.M113.470625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu Q, Hu L, Yao Q, Zhu Y, Dong N, Wang DC, Shao F. 2013. Structural analyses of Legionella LepB reveal a new GAP fold that catalytically mimics eukaryotic RasGAP. Cell Res 23:775–787. doi: 10.1038/cr.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newton HJ, McDonough JA, Roy CR. 2013. Effector protein translocation by the Coxiella burnetii Dot/Icm type IV secretion system requires endocytic maturation of the pathogen-occupied vacuole. PLoS One 8:e54566. doi: 10.1371/journal.pone.0054566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romano PS, Gutierrez MG, Beron W, Rabinovitch M, Colombo MI. 2007. The autophagic pathway is actively modulated by phase II Coxiella burnetii to efficiently replicate in the host cell. Cell Microbiol 9:891–909. doi: 10.1111/j.1462-5822.2006.00838.x. [DOI] [PubMed] [Google Scholar]
- 57.Wesolowski J, Paumet F. 2010. SNARE motif: a common motif used by pathogens to manipulate membrane fusion. Virulence 1:319–324. doi: 10.4161/viru.1.4.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Delevoye C, Nilges M, Dautry-Varsat A, Subtil A. 2004. Conservation of the biochemical properties of IncA from Chlamydia trachomatis and Chlamydia caviae: oligomerization of IncA mediates interaction between facing membranes. J Biol Chem 279:46896–46906. doi: 10.1074/jbc.M407227200. [DOI] [PubMed] [Google Scholar]
- 59.Paumet F, Wesolowski J, Garcia-Diaz A, Delevoye C, Aulner N, Shuman HA, Subtil A, Rothman JE. 2009. Intracellular bacteria encode inhibitory SNARE-like proteins. PLoS One 4:e7375. doi: 10.1371/journal.pone.0007375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bennett TL, Kraft SM, Reaves BJ, Mima J, O'Brien KM, Starai VJ. 2013. LegC3, an effector protein from Legionella pneumophila, inhibits homotypic yeast vacuole fusion in vivo and in vitro. PLoS One 8:e56798. doi: 10.1371/journal.pone.0056798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gutierrez MG, Vazquez CL, Munafo DB, Zoppino FC, Beron W, Rabinovitch M, Colombo MI. 2005. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol 7:981–993. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- 62.Lifshitz Z, Burstein D, Schwartz K, Shuman HA, Pupko T, Segal G. 2014. Identification of novel Coxiella burnetii Icm/Dot effectors and genetic analysis of their involvement in modulating a mitogen-activated protein kinase pathway. Infect Immun 82:3740–3752. doi: 10.1128/IAI.01729-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Newton HJ, Kohler LJ, McDonough JA, Temoche-Diaz M, Crabill E, Hartland EL, Roy CR. 2014. A screen of Coxiella burnetii mutants reveals important roles for Dot/Icm effectors and host autophagy in vacuole biogenesis. PLoS Pathog 10:e1004286. doi: 10.1371/journal.ppat.1004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.