Abstract
Eukaryote-like serine/threonine kinases (eSTKs) constitute an important family of bacterial virulence factors. Genome analysis had predicted putative eSTKs in Salmonella enterica serovar Typhi, although their functional characterization and the elucidation of their role in pathogenesis are still awaited. We show here that the primary sequence and secondary structure of the t4519 locus of Salmonella Typhi Ty2 have all the signatures of eukaryotic superfamily kinases. t4519 encodes a ∼39-kDa protein (T4519), which shows serine/threonine kinase activities in vitro. Recombinant T4519 (rT4519) is autophosphorylated and phosphorylates the universal substrate myelin basic protein. Infection of macrophages results in decreased viability of the mutant (Ty2Δt4519) strain, which is reversed by gene complementation. Moreover, reactive oxygen species produced by the macrophages signal to the bacteria to induce T4519, which is translocated to the host cell cytoplasm. That T4519 may target a host substrate(s) is further supported by the activation of host cellular signaling pathways and the induction of cytokines/chemokines. Finally, the role of T4519 in the pathogenesis of Salmonella Typhi is underscored by the significantly decreased mortality of mice infected with the Ty2Δt4519 strain and the fact that the competitive index of this strain for causing systemic infection is 0.25% that of the wild-type strain. This study characterizes the first eSTK of Salmonella Typhi and demonstrates its role in promoting phagosomal survival of the bacteria within macrophages, which is a key determinant of pathogenesis. This, to the best of our knowledge, is the first study to describe the essential role of eSTKs in the in vivo pathogenesis of Salmonella spp.
INTRODUCTION
Professional intracellular pathogens (PIPs), such as Salmonella, Mycobacteria, Legionella, Listeria, and Coxiella spp., frequently cause prolonged and difficult-to-treat human infections (1). Persistent survival and/or replication within phagocytes is central to the pathogenesis of these organisms. Most PIPs achieve this feat by perturbing phagosome maturation and inhibiting phagosome-lysosome (P-L) fusion, leading to evasion of killing by lysosomal hydrolases (1). A similar mechanism has been proposed for Salmonella spp. (2) but is unlikely to be a major factor promoting phagosomal survival, since Salmonella may replicate within macrophages, where mature Salmonella-containing vacuoles (SCVs) have fused with the lysosomes (3). On the other hand, P-L fusion was unaffected in RAW 264.7 macrophages infected with Salmonella enterica serovar Typhimurium LT2ΔssaV, a mutant strain that fails to translocate the type three secretion system 2 (T3SS-2) effector proteins into the host cytoplasm and is impaired in phagosomal survival and systemic virulence in mice (4). The two-component regulatory system PhoP-PhoQ plays a major role by selectively inducing a large number of survival genes, the majority of which are encoded by Salmonella pathogenicity island 2 (SPI-2), within macrophages (5, 6). It is critical to identify the prokaryotic and host genes involved in the intracellular survival and/or replication of PIPs in order to design more-efficient antimicrobial therapeutics.
Salmonella enterica serovar Typhi, a Gram-negative enterobacterium, causes a potentially lethal human disease called enteric (typhoid) fever, which is a major threat to public health in the developing world (7). However, the survival mechanism of the bacteria within macrophages remains poorly understood. Current knowledge is based on extrapolated data from a related pathogen, S. Typhimurium, which causes self-limiting gastroenteritis in humans but a typhoid-like systemic illness in mice (8). However, considerable differences exist between the genomic architectures and virulence mechanisms of the two organisms (9, 10). There is 99% homology in the housekeeping gene sequences but 11% difference in the genes encoded by the pathogenicity islands (11). Most strikingly, SPI-2 genes were found to be essential for the phagosomal survival of S. Typhimurium but redundant for S. Typhi (12). In contrast, SPI-7, which encodes the Vi polysaccharide, is present only in S. Typhi and is important for pathogenesis (13). However, SPI-7 has not been reported to promote intracellular viability.
Eukaryote-like serine/threonine kinases (eSTKs) and eukaryote-like tyrosine kinases (eTyKs) confer survival advantages on pathogens within the host by regulating phagocytosis and/or endosomal trafficking (YopO of Yersinia spp. and PknG of mycobacteria), by modulating host defense by targeting the innate signaling pathways (Legionella pneumophila LegK1 and OspG of Shigella spp.), or through unknown mechanisms (Mycobacterium tuberculosis PknH, PknI, and PknK) (14). Two S. Typhimurium eSTKs have been studied; SteC, an SPI-2 effector, promotes actin cytoskeleton rearrangement and limits the intracellular growth of the bacteria, while RdoA confers a growth advantage in nutrient-starved medium and during biofilm formation (14, 15). However, these eSTKs were not found to play a significant role in pathogenesis (16). eSTK sequences were also identified in the S. Typhi genome (10, 17). We show here that T4519 is an authentic eSTK of S. Typhi that is induced within macrophages in a reactive oxygen species (ROS)-dependent manner. The protein is translocated to the host cell cytoplasm and modulates macrophage immune responses. Finally, T4519 is required for bacterial survival within macrophages and contributes to S. Typhi pathogenesis in a mouse model.
MATERIALS AND METHODS
Computational prediction and analysis.
Homology searches for S. Typhi eSTKs were carried out by NCBI BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The template for the homology model of T4519 was used to search the Protein Data Bank (PDB) (http://www.rcsb.org). The serine/threonine kinase (STK) active-site signature {[LIVMFYC]-x-[HY]-x-D-[LIVMFY]-K-x(2)-N-[LIVMFYCT](3)} was found in the PROSITE database (http://prosite.expasy.org/) (Prosite entry PS00108).
Cells and reagents.
Cell lines and cell culture reagents were procured from the American Type Culture Collection (ATCC) and Invitrogen, respectively. Bacterial culture media were purchased from BD Difco. Molecular biology reagents were purchased as follows: DNA restriction enzymes and DNase I from New England Biolabs, cDNA synthesis reagents from Fermentas, and SYBR green master mix from ABI. Antibodies against phospho(Ser/Thr) Phe, p65, histone H3, and AKT and mitogen-activated protein (MAP) kinases were from Cell Signaling, anti-tubulin from Santa Cruz, horseradish peroxidase (HRP)-conjugated secondary antibodies from Pierce, and fluorescent-dye-conjugated secondary antibodies from Jackson. All pharmacological inhibitors were purchased from Calbiochem. All other chemicals were purchased from Sigma-Aldrich unless otherwise indicated.
Bacterial strains, plasmids, and primers.
All the bacterial strains and plasmids used in this work are listed in Table S1 in the supplemental material, while PCR primers are listed in Table S2 in the supplemental material.
Cell culture.
The human acute monocytic leukemia cell line THP-1 was maintained in RPMI 1640 medium supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS) and penicillin-streptomycin. Cells were differentiated in the presence of 100 nM phorbol 12-myristate 13-acetate (PMA) for 24 h, followed by culture in the absence of PMA for 48 h before the experiment. The mouse leukemia macrophage cell line RAW 264.7 and the human colorectal adenocarcinoma cell line HT-29 were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% FBS and penicillin-streptomycin.
Generation of Ty2 mutants and complemented strains.
Deletion mutant strains of Ty2 were generated using the suicide vector pCVD442 (18). Briefly, gene fragments were amplified, cloned into the pBlueScript SK(+) vector, subcloned into pCVD442, and transformed into SM10 λpir. Conjugation was performed between SM10 λpir and Ty2 in the absence of antibiotic selection on a Luria agar plate. Conjugants were selected in antibiotic Hektoen enteric agar plates containing streptomycin-ampicillin and were cultured in the presence of 10% sucrose in Luria-Bertani broth for several generations to remove the sacB gene. Gene manipulations were confirmed by PCR at multiple stages, and the deletions were confirmed by sequencing. For complementation, genes were cloned into the pQE60 plasmid and were electroporated into the mutant bacteria and S. Typhimurium LT2. For the translocation study, the t4519 gene was amplified, cloned into pCX340 at the 5′ end of the β-lactamase (TEM1) gene, and electroporated into the Ty2Δt4519 strain. The expression of the T4519-TEM1 fusion protein was confirmed by immunoblotting.
Preparation of human monocyte-derived macrophages.
Ten milliliters of peripheral venous blood was collected from human volunteers, diluted with an equal volume of Hanks balanced salt solution (HBSS), and overlaid on Ficoll-Paque Plus at a 3:1 (blood to Ficoll-Paque) ratio, followed by centrifugation at 1,300 rpm for 30 min at room temperature. The buffy coat containing the mononuclear cells was washed several times and was resuspended in HBSS. Monocytes were sorted using anti-human CD14+ magnetic beads as per the manufacturer's protocol (BD Biosciences). Cells (106/ml) were cultured in RPMI 1640 medium supplemented with granulocyte-macrophage colony-stimulating factor (GM-CSF) (50 ng/ml) and 20% FBS for 3 days, followed by culture for the next 3 days in the absence of GM-CSF (50 ng/ml).
Cloning, expression, and purification of bacterial proteins and generation of polyclonal antibodies.
The open reading frames (ORFs) of t4519, t4520, t2942, and t2544 or deletion mutants of t4519 were amplified from Ty2 genomic DNA using the corresponding forward and reverse primers (see Table S2 in the supplemental material), followed by cloning into the pBlueScript SK(+) vector and subcloning into the pET28a+ expression vector at the NdeI/XhoI sites. Positive clones were transformed into Escherichia coli BL21(DE3). Bacterial cultures were grown until the optical density at 600 nm (OD600) reached 0.6, and the expression of all proteins was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 5 h at 37°C. In order to obtain the purified proteins, pellets were resuspended in a denaturation buffer (0.1 M phosphate buffer, 8 M urea, 10 mM Tris-HCl, and 100 mM imidazole [pH 8.0]) and were incubated for 1 h at room temperature, followed by centrifugation at 16,000 × g for 30 min. Supernatants were collected and were passed through Ni-nitrilotriacetic acid (NTA) columns (Qiagen). The columns were washed with a wash buffer (0.1 M phosphate buffer, 8 M urea, 10 mM Tris-HCl, and 30 mM imidazole [pH 6.8]), and the bound proteins were eluted using an elution buffer (0.1 M phosphate buffer, 8 M urea, 10 mM Tris-HCl, and 30 mM imidazole [pH 5.2]). Denatured proteins were refolded by dialysis with a phosphate-urea gradient buffer (50 mM glycine, 5 mM EDTA, 10% glycerol, 10% glucose, 250 mM NaCl, 5 mM HEPES, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.1 mM dithiothreitol [DTT], 1 M Na2HPO4, and 1 M NaHPO4 [pH 7.2]). The purity and sizes of the proteins were confirmed by SDS-PAGE, and protein concentrations were quantified by a Bradford assay.
To generate anti-T4519 or anti-T2544 antiserum, male New Zealand White rabbits were injected intramuscularly with 100 μg of recombinant T4519 (rT4519) or T2544 protein emulsified with Freund's complete adjuvant (FCA), followed by three booster doses of 150 μg of protein emulsified with incomplete Freund's adjuvant (IFA) administered at intervals of 7 days. The animals were bled 7 days after the last injection; serum was separated from the blood; and the anti-T4519 or anti-T2544 IgG titer was determined by an enzyme-linked immunosorbent assay (ELISA).
Gentamicin protection assay.
THP-1 cells seeded in 24-well plates (5 × 105 cells/well) were differentiated with PMA (100 nM, 24 h). Cells were infected at a multiplicity of infection (MOI) of 10 with bacterial cultures opsonized with RPMI 1640 medium containing 10% FBS and were synchronized with the bacteria by centrifugation at 400 × g for 5 min. Infection was continued for 30 min at 37°C in the presence of 5% CO2. Extracellular bacteria were removed by repeated washing with 1× phosphate-buffered saline (PBS), and the cells were cultured in complete RPMI medium containing 100 μg/ml of gentamicin for 1 h, followed by 15 μg/ml of gentamicin until the time of the experiment. Cells were lysed with 1× PBS containing 0.25% Triton X-100, and intracellular CFU counts were carried out after the bacteria recovered from the cells were grown overnight on a Luria agar plate.
Gene expression analysis.
THP-1 or RAW 264.7 cells were seeded at a density of 2 × 106/well in 6-well plates (for bacterial mRNA expression) and were infected with bacteria as described in the preceding section. For the analysis of host mRNA expression, cells were seeded at a density of 5 × 105/well in 24-well plates and infected with bacteria as described above. Total RNA was extracted using the TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Five micrograms (for bacterial mRNA expression) or 1 μg (for host mRNA expression) of DNA-free RNA was used for cDNA synthesis using SuperScript II reverse transcriptase. Quantitative PCR (qPCR) was performed in a StepOnePlus system (ABI) using SYBR green master mix. Relative quantitation was done by the comparative threshold cycle (CT) method (19). The levels of expression of the genes of interest were normalized against that of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene (for cytokine and chemokine gene expression) or 16S rRNA (for S. Typhi gene expression) using the formula 2−ΔΔCT, where −ΔΔCT is calculated as ΔCT (sample) − ΔCT (calibrator), and ΔCT is the CT of the target gene subtracted from the CT of the housekeeping gene (GAPDH or 16S rRNA). The calibrators used in our experiments were uninfected THP-1 cells (for cytokine and chemokine gene expression). For S. Typhi gene expression, comparison was done with 0 h of infection (30 min of infection followed by 100 µg/ml of gentamicin for 1 h).
Transmission electron microscopy.
THP-1 cells seeded in 6-well plates (2 × 106 cells/well) were pulse-chased with a bovine serum albumin (BSA)-coated gold tracer (particle size, 25 nm; Electron Microscopy Sciences) for 4 h. Cells were washed with 1× PBS and were infected as described under “Gentamicin protection assay” above. Cells were fixed in 3% glutaraldehyde in phosphate buffer (pH 7.4), postfixed in 1% osmium tetroxide, dehydrated in an ascending acetone series, and embedded in Agar 100 resin (Agar Scientific). Ultrathin sections were examined under an FEI Tecnai 12 BioTwin transmission electron microscope operated at 100 kV.
In vitro kinase assay.
Kinase (rT4519, rT4519 truncated versions, T4520, T2942, or phosphorylated extracellular signal-regulated kinase [phospho-ERK]) and the substrate (myelin basic protein [MBP]) were incubated in a buffer containing 5 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 2 mM MnCl2, 1 mM DTT, 200 μM sodium orthovanadate, and 10 mM ATP for 1 h. Reactions were terminated by adding SDS sample buffer, and the proteins were subjected to immunoblot analysis.
Fluorescent microscopy.
THP-1 cells were seeded at a density at 1 × 106 per well on collagen-coated coverslips and were infected with the bacteria. After culturing as described under “Gentamicin protection assay” above, they were washed with sterile 1× PBS, fixed with 2.5% paraformaldehyde (PFA) containing 1× PBS for 10 min, washed with 1× PBS (3 times), and permeabilized with 0.1% Triton X-100 for 20 min. Fixed cells were incubated in a blocking solution (0.1% Triton containing 1× PBS supplemented with 5% [vol/vol] horse serum) for 1 h at room temperature. Cells were washed with ice-cold 1× PBS and were incubated with a rabbit polyclonal anti-Salmonella antibody (BD) overnight at 4°C, followed by fluorescein isothiocyanate (FITC)-conjugated anti-rabbit antibodies for 1 h at room temperature, and actin was stained with rhodamine-phalloidin. After a wash with 1× PBS, coverslips were mounted on clean glass slides. Stained cells were imaged under a Zeiss Axiovert fluorescence microscope, and bacteria were counted manually.
To study T4519 translocation, THP-1 cells were seeded at a density of 2 × 106 per well on collagen-coated coverslips placed in the wells of a 6-well plate and were differentiated with 100 nM PMA for 24 h. Single colonies of Salmonella strain Ty2Δt4519 harboring the TEM1 (empty vector), glutathione S-transferase (GST)-TEM1, or T4519-TEM1 expression plasmid were cultured in LB medium in the presence of appropriate antibiotics, and infections were performed as described under “Gentamicin protection assay” above. At 6 h postinfection, cells were washed three times with 1× PBS and were cultured for another 1 h in serum-free RPMI medium containing 1 μM CCF2-AM dye (Invitrogen). Cells were examined under a confocal microscope (Olympus) to detect the blue and green fluorescence emissions (∼450 nm and ∼510 nm, respectively) after excitation at 410 nm.
Cell viability assay.
THP-1 cells were seeded at a density of 5 × 104/well in 96-well plates and were infected with bacteria as described under “Gentamicin protection assay” above. The release of lactate dehydrogenase (LDH) into the culture supernatants was quantified using the In vitro Toxicology LDH assay kit (Sigma-Aldrich) according to the manufacturer's instructions. For the trypan blue dye exclusion assay, infected THP-1 cells were gently scraped, loaded with trypan blue stain (0.01%), and counted under a hemocytometer.
In vivo experiments.
All animal experiments were carried out according to the protocols approved by the Institutional Animal Ethics Committee, National Institute of Cholera and Enteric Diseases (NICED), Kolkata, India. An iron overload mouse model was used for oral S. Typhi infection as described previously (20). Briefly, BALB/c mice were intraperitoneally (i.p.) injected with Fe3+ (0.32 mg/g of body weight) and desferrioxamine (DFO; Novartis) (0.025 mg/g of body weight) 4 h prior to the bacterial challenge. Log-phase cultures of bacteria were resuspended in sterile 1× PBS and were fed orally after neutralization of gastric acidity with 5% sodium bicarbonate. In lethal-infection experiments, mice were monitored for death over the next 14 days, and mouse survival was plotted as a Kaplan-Meier graph.
For S. Typhimurium infection, BALB/c mice (6 to 8 weeks old) were infected orally with the S. Typhimurium strain LT2 (ATCC) or a T4519-expressing LT2 strain at 5 times the 50% lethal dose (LD50). The mortality of mice was monitored over next 30 days.
To study systemic infection, the liver, spleen, and heart were collected 2, 4, and 6 days after infection with sublethal doses (5 × 105 CFU) of S. Typhi strains and were macerated to make single-cell suspensions. Cell pellets were collected by centrifugation at 800 × g for 5 min and were resuspended in 1× PBS containing 0.5% Triton X-100, and serial dilutions in 1× PBS were plated in a Luria agar plate. CFU counts were normalized per gram of tissue.
Immunoblotting.
Postexperimental cells were lysed with a Nonidet P-40 lysis buffer (50 mM HEPES [pH 7.4], 100 mM NaCl, 2 mM EDTA, 0.5% Nonidet P-40, 10% glycerol, 50 mM β-glycerophosphate, 1 mM NaF, protease inhibitor cocktail). Equal amounts of total proteins in the cell lysates were separated by SDS-PAGE and were transferred to a PVDF membrane (Millipore). Blots were incubated overnight at 4°C with primary antibodies diluted in blocking buffer (Tris-buffered saline [TBS] containing 3% [wt/vol] BSA). After 3 washes with 1× TBS containing 0.1% Tween 20 (TBS-T), blots were incubated with HRP-conjugated secondary antibodies (dilution, 1:20,000) dissolved in TBS-T containing 3% (wt/vol) BSA for 1 h at room temperature. Blots were developed with the ECL Plus Western blot detection reagent (Pierce), and the chemiluminescence was transferred to autoradiographs. Signal intensity was measured by Quantity One software (Bio-Rad).
ELISA.
Cytokines and chemokines in the culture supernatants of the experimental cells and in mouse serum were measured by ELISA using an ELISA kit (eBioscience) as per the manufacturer's instructions.
p65 nuclear translocation assay.
THP-1 cells seeded in a 90-mm tissue culture petri dish (107 cells/well) were differentiated with PMA (100 nM; 24 h). Cells were infected with bacteria as described under “Gentamicin protection assay” above, and nuclear extracts were prepared as described previously (19). Briefly, THP-1 cells were harvested by scraping, resuspended in low-salt buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl), and incubated on ice for 15 min. After the addition of Nonidet P-40 (final concentration, 0.5%), the cell suspensions were passed through a 21-gauge needle 5 to 6 times. Cytoplasmic and nuclear fractions were separated by centrifugation at 14,000 rpm for 10 min at 4°C. Nuclear proteins were extracted by incubating the cell pellet in high-salt buffer C (20 mM HEPES [pH 7.9], 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol) for 30 min on ice with vigorous shaking (250 rpm). Nuclear proteins were subjected to immunoblot analysis to check p65 nuclear translocation.
Statistical analysis.
Statistical analysis was performed, and graphs were drawn using GraphPad Prism Software.
RESULTS
A putative eSTK of S. Typhi is required for survival within macrophages.
To identify S. Typhi eSTKs, which are critical for macrophage survival and pathogenesis, we first carried out an NCBI BLAST search for sequence homologies in the S. Typhi Ty2 and CT18 genomes to the reference sequence of the eukaryotic protein kinase superfamily (21). The search results identified two candidate kinases in S. Typhi (T4519/STY4822 and T4520/STY4823), which bear amino acid sequences homologous to those of the eukaryotic protein kinases over the kinase catalytic domains (58% and 63% query coverage and 23% and 25% identity, respectively). Similarity searches of T4519 and T4520 sequences with the known crystal structures in the Protein Data Bank (PDB) showed M. tuberculosis PknB heading the list. Most importantly, 11 of the 12 critical amino acid residues that define the eukaryotic protein kinase superfamily are conserved in the S. Typhi proteins (see Fig. S1A in the supplemental material). The activation loop is present between the conserved DFG (Asp-Phe-Gly) and APE (Ala-Pro-Glu) motifs. This segment includes the magnesium-binding loop, the activation loop, and the P+1 loop (22). The conserved RD (arginine-aspartate) sequence in the catalytic loop of eukaryotic kinases, which interacts with the phosphorylated Ser/Thr residues in the activation loop and stabilizes the activation loop in a conformation that allows substrate binding and catalysis, is also conserved here. In addition, one STK active-site signature {[LIVMFYC]-x-[HY]-x-D-[LIVMFY]-K-(2)-N-[LIVMFYCT](3)}, as given in the PROSITE database (Prosite entry PS00108), was identified in the T4519 sequence. The genes encoding the putative eSTKs of S. Typhi (prkX [t4519] and prkY [t4520]) are aligned in tandem with a serine/threonine (Ser/Thr) phosphatase 2C gene (prpZ [t4521]), forming the prpZ gene cluster, which is present exclusively in S. Typhi and is absent from all other Salmonella serovars. Deletion of the gene cluster has been reported to decrease the viability of S. Typhi in THP-1 cells (17).
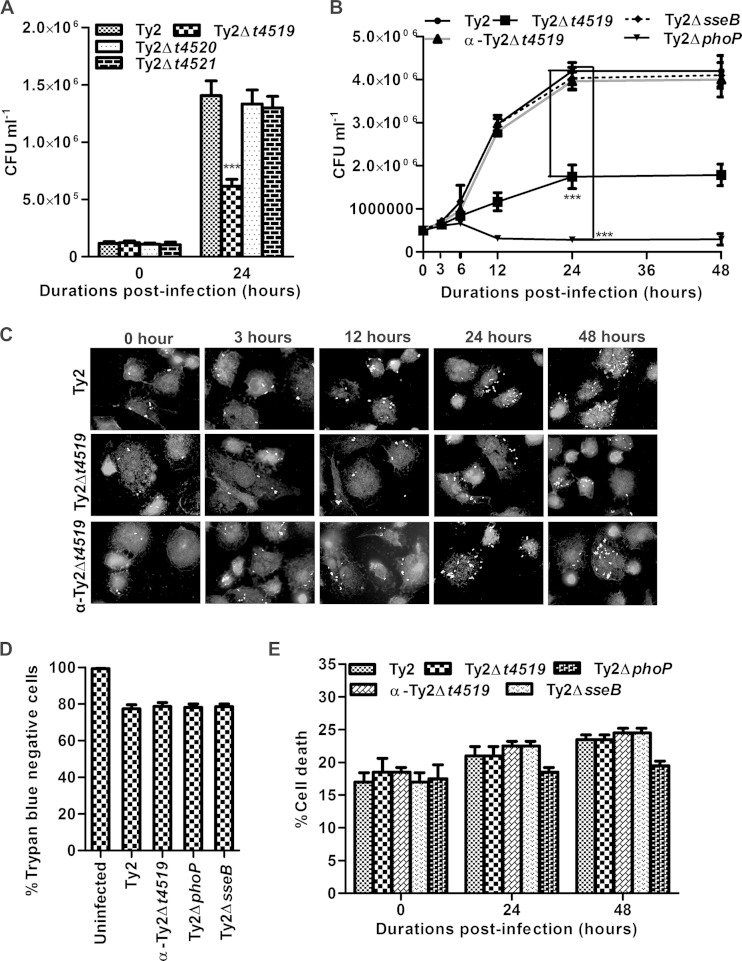
Since persistent phagosomal survival within macrophages is critical for Salmonella spp. (3), we investigated the virulence functions of the individual prpZ cluster genes by mutating one gene at a time and comparing the viability of the mutants with that of the wild-type S. Typhi strain (Ty2) within macrophages. To this end, macrophages derived from the human monocytic leukemia cell line THP-1 and macrophages of the murine cell line RAW 264.7 were infected with the bacteria at a multiplicity of infection (MOI) of 10 for 30 min, and intracellular CFU counts were analyzed 0 h and 24 h after the gentamicin protection assay. The results showed similar CFU counts of various strains at 0 h, suggesting no difference in phagocytosis. In contrast, intracellular CFU counts of the Ty2Δt4519 strain were ∼50% those of the wild-type Ty2 strain and other mutant strains after 24 h, indicating a role for T4519 in the phagosomal survival of S. Typhi (Fig. 1A). To further address this issue, macrophage cell lines were infected with the Ty2Δt4519 strain complemented with a plasmid encoding t4519 (α-Ty2Δt4519). Parallel sets of cultures were infected with Ty2ΔphoP or Ty2ΔsseB, a T3SS-2 secretion apparatus mutant strain, since the phoP and sseB genes were previously implicated in the phagosomal survival of Salmonella spp. (3). Temporal analysis of the intracellular CFU counts, as well as manual counting under the microscope, showed progressively decreasing numbers of viable Ty2Δt4519 cells relative to Ty2 cells during the replicative phase of their phagosomal life (Fig. 1B and C; see also Fig. S1B and C in the supplemental material). The compromised intracellular viability of strain Ty2Δt4519 was completely reversed by gene complementation. Deletion of phoP resulted in a marked reduction in the intracellular survival of S. Typhi, while sseB deletion had no effect on intracellular survival, confirming the dispensability of SPI-2 (12). These findings validate the integrity of the experimental system. The morphology of the intracellular bacteria was unaltered, suggesting that the t4519 mutation had no effect on the structural integrity of S. Typhi (see Fig. S1D in the supplemental material). In addition, the in vitro growth kinetics of the wild-type and mutant strains were similar in both enriched and N minimal media, indicating that T4519 is not involved in bacterial replication (see Fig. S1E and F in the supplemental material). Taken together, these results suggest that T4519 mutation impairs the survival of S. Typhi within macrophages. To rule out any effect of cell death following S. Typhi infection on intracellular CFU counts, we analyzed the infected cells by trypan blue dye exclusion and LDH release assays. In agreement with the published reports (23), we observed minimal and equivalent frequencies of death of THP-1 cells for different strains (Fig. 1D and E). Together, these results suggest that T4519 selectively promotes the phagosomal survival of S. Typhi within macrophages. While this may be critical for pathogenesis, T4519 was not found to be required for bacterial growth in epithelial cells (see Fig. S1G in the supplemental material).
FIG 1.
T4519 promotes persistent intracellular survival and replication of Salmonella Typhi in a gentamicin protection assay. PMA (100 nM, 24 h)-differentiated THP-1 cells were infected with Ty2 or a mutant strain at an MOI of 10 for 1 h. Cells were washed repeatedly and were treated with gentamicin (100 μg/ml, 1 h) to remove extracellular bacteria. Intracellular bacteria were allowed to grow for the indicated times by culturing the cells in a gentamicin-containing medium (15 μg/ml). (A and B) CFU counts of intracellular bacteria recovered after lysis of the cells. The complemented Ty2Δt4519 strain is designated α-Ty2Δt4519. Error bars, means ± standard deviations for three independent experiments. ***, P < 0.001 (by Student's t test). (C) Cells were fixed and permeabilized, and intracellular bacteria were stained with polyclonal anti-Salmonella antisera, followed by an FITC-conjugated secondary antibody. The actin cytoskeleton was stained with rhodamine-phalloidin. Representative fluorescence microscopic images from multiple fields are shown in grayscale. (D) Trypan blue-negative cell counts by hemocytometer 24 h postinfection. (E) LDH was measured in the culture supernatants of the cells.
T4519 is induced by ROS within macrophages.
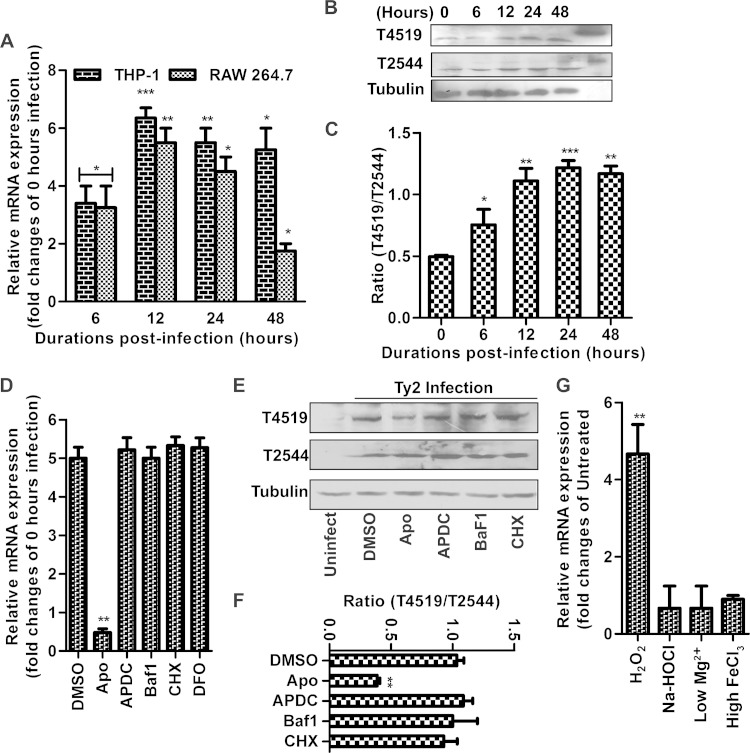
Virulence genes of intracellular pathogens may be selectively induced within macrophages (3). To investigate this issue with regard to T4519, we studied t4519 mRNA expression by the intracellular Ty2 strain. We found time-dependent induction of t4519 mRNA in THP-1 cells and RAW 264.7 macrophages, with peak expression at ∼12 h after infection (Fig. 2A). These results correlated with T4519 protein expression, which peaked at ∼24 h postinfection (Fig. 2B and C). Since the number of intracellular bacteria increased progressively with time until 24 h following infection, we adjusted for its contribution to the increase in T4519 expression by calculating the ratio of the expression of t4519 mRNA to that of the housekeeping gene 16S rRNA and the ratio of T4519 protein expression to T2544 protein expression. T2544 is a noninducible outer membrane protein expressed by various strains of S. Typhi, including Ty2 (20). However, T4519 expression was not induced in intestinal epithelial cells (IECs) (data not shown). Several host factors, such as reactive oxygen species (ROS), reactive nitrogen species (RNS), and phagosomal acidification and maturation, have been implicated in the regulation of microbial virulence genes within host macrophages. Using pharmacological inhibitors to block each of these signals, we analyzed the effects on intracellular T4519 expression (24). Cycloheximide (CHX) failed to prevent T4519 induction, indicating that the host signals to which S. Typhi responded were generated independently of new protein synthesis. Likewise, blocking of phagosomal acidification and nitrate production or chelation of iron by desferrioxamine (DFO) had no impact on T4519 expression. However, inhibition of ROS generation by pretreatment of the cells with apocyanin significantly abolished the microbial response, indicating that oxygen radicals produced by infected macrophages may be sensed by S. Typhi, leading to T4519 induction (Fig. 2D to F). To further prove this point, we studied t4519 mRNA expression after culturing S. Typhi in media containing chemical agents in order to mimic intraphagosomal conditions. Hydrogen peroxide, but not hypochlorite, N minimal medium (low pH, low Mg2+), or iron, was capable of inducing T4519, indicating that superoxide ions drive T4519 expression (Fig. 2G) (17).
FIG 2.
T4519 is an inducible Ser/Thr kinase. (A to F) PMA-differentiated THP-1 cells were infected with strain Ty2 and were subjected to a gentamicin protection assay as for Fig. 1. Error bars, means ± standard deviations for three independent experiments. Asterisks indicate significant differences by Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (A and D) Reverse transcription-qPCR results show 16S rRNA-normalized expression of t4519 mRNA at the indicated time points (A) or 12 h postinfection (D). (B and E) Immunoblots using total lysates of Ty2-infected cells. T2544 was used as a control. Results of one of three independent experiments are shown. The rightmost lane in panel B shows recombinant proteins. (C and F) Densitometry of the immunoblots. (D to F) Cells were pretreated with either the vehicle (dimethyl sulfoxide [DMSO]) (14 μM), the ROS inhibitor apocyanin (Apo) (1 mM), the RNS inhibitor ammonium pyrrolidine dithiocarbamate (APDC) (100 μM), the vacuolar H+-ATPase inhibitor bafilomycin A1 (Baf1) (50 nM), the protein translation inhibitor CHX (25 μg/ml), or the iron chelator desferrioxamine (DFO) (40 μM) for 1 h. (G) Reverse transcription-qPCR shows 16S rRNA-normalized expression of t4519 mRNA from S. Typhi either grown in LB medium and treated with H2O2 (5 mM), Na-HOCl (0.5 mM), or ferric chloride (50 mM) for 10 min or cultured in N minimal medium (low pH, low Mg2+).
T4519 is a secreted serine/threonine protein kinase.
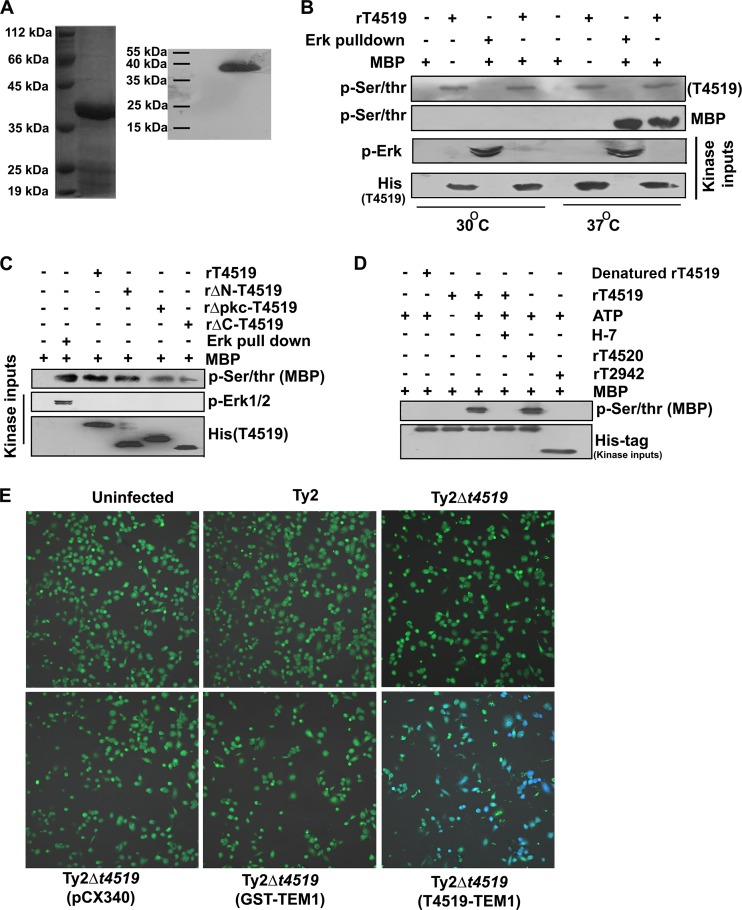
Although T4519 was predicted previously to be a Ser/Thr kinase, this was never experimentally validated (17). We used in vitro kinase assays to investigate whether T4519 is a bona fide Ser/Thr kinase. To this end, we expressed 6His-tagged T4519 in E. coli BL21(DE3) and purified the recombinant protein by Ni2+ column chromatography. Coomassie blue staining and Western blotting of the purified protein run in SDS-PAGE showed a single band of the expected size (41 kDa) (Fig. 3A). We incubated rT4519 and the active MAP kinase ERK immunoprecipitated from lipopolysaccharide (LPS)-stimulated THP-1 cells with the universal kinase substrate myelin basic protein (MBP) in a cell-free system. The equally robust phosphorylation of MBP by these two kinases suggests that T4519, like the mammalian MAP kinase ERK, is a potent Ser/Thr kinase. This was further confirmed by autophosphorylation of T4519 at the Ser/Thr residues (Fig. 3B). To map the kinase domain, the assays described above were carried out with various truncated versions of rT4519. Deletion of the C terminus (amino acids 283 to 359) or the protein kinase domain (amino acids 126 to 160) reduced the kinase activity but did not render rT4519 kinase dead, while the kinase activity of the N-terminally deleted (Δ1-54) mutant was similar to that of the wild-type protein (Fig. 3C). The specificity of the kinase functions of T4519 and the rΔN-T4519 mutant was further proved by the lack of kinase activities in the absence of ATP or denatured protein (the recombinant protein was eluted from the inclusion bodies under denaturing conditions [see Materials and Methods]) or in the presence of an outer membrane protein of S. Typhi, T2942, which is not a kinase, and by the inhibition of kinase activities by the protein kinase C (PKC) inhibitor H7 (Fig. 3D; see also Fig. S2A in the supplemental material).
FIG 3.
T4519 is a secreted serine/threonine protein kinase. (A) SDS-PAGE (left) and Western blot (right) of purified recombinant 6His-tagged T4519. (B and C) In vitro kinase assays. Full-length (B) or truncated (C) purified rT4519 proteins (2 μg) or active ERK immunoprecipitated from LPS-stimulated THP-1 cells was incubated with the substrate (MBP; 5 μg) for 1 h. (D) Recombinant proteins (2 μg) were incubated with the substrate (MBP; 5 μg) for 1 h in the presence or absence of ATP (10 mM) or H7-dihydrochloride (1 μM). Kinase activities were monitored by immunoblotting for phosphorylation of MBP and autophosphorylation of T4519. Results of one of two independent experiments are shown. (E) PMA-differentiated THP-1 cells were infected with strain Ty2 or a mutant as indicated and were subjected to a gentamicin protection assay. Some of the mutant strains expressed the empty TEM1 vector (pCX340) or a TEM1 fusion protein. Cells were cultured as described for Fig. 1 for 6 h, followed by loading with CCF2-AM (1 μM), and were analyzed by confocal microscopy after excitation at 410 nm. Emission was captured at 510 nm (green) and 450 nm (blue). Results of one of two independent experiments are shown.
Our previous results suggest that T4519 may be a secreted effector of S. Typhi (Fig. 1B). A large number of effector proteins of intracellular salmonellae are translocated to the host cytoplasm to perform their functions (3). Using the TEM1 (β-lactamase) reporter system (25), we showed that TEM1 is translocated when expressed as a T4519-TEM1 fusion protein, but not as TEM1 or GST-TEM1. This is underscored by the emission of green fluorescence by uninfected THP-1 cells, cells infected with strain Ty2 or Ty2Δt4519, or cells infected with strain Ty2Δt4519 expressing the empty vector or the TEM1-GST fusion protein upon loading with the fluorescence resonance energy transfer (FRET) dye CCF2-AM. In contrast, cells infected with the Ty2Δt4519 strain expressing the TEM1-T4519 protein emitted blue fluorescence due to the cleavage of the dye by the translocated TEM1 (β-lactamase) and the quenching of FRET (Fig. 3E). Since T4519 carries no secretion signals in its sequence, it may require a secretion system. However, S. Typhi T3SS-1 and T3SS-2 mutants (Ty2ΔinvG and Ty2ΔsseB, respectively) continued to secrete the TEM1-T4519 fusion protein, indicating that these secretion systems have no role in T4519 translocation (see Fig. S2B in the supplemental material). Taken together, these results suggest that T4519 may target a host substrate(s) to exert its role in bacterial pathogenesis.
T4519 targets a host substrate(s) to regulate the cytokine response.
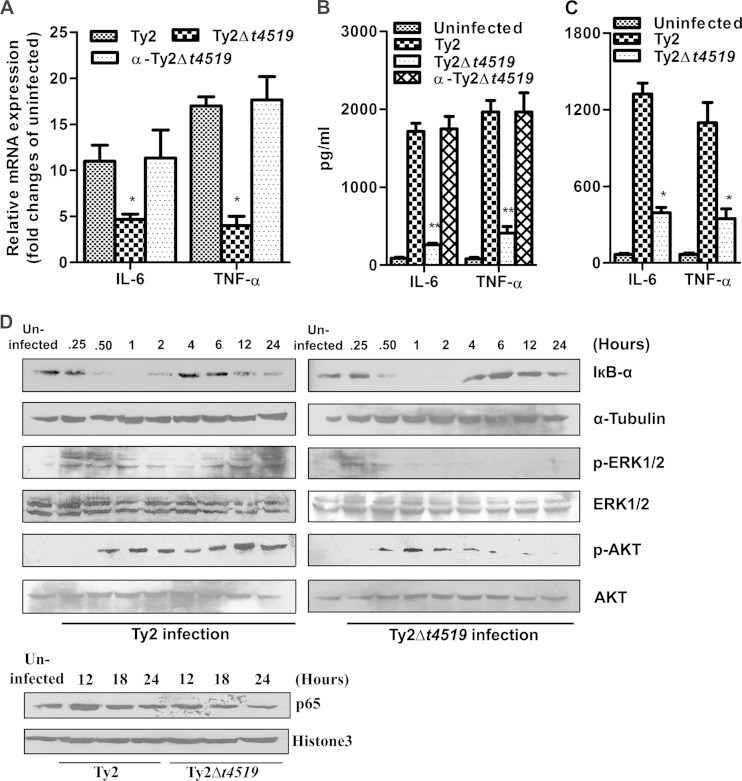
To determine whether T4519 secreted by intracellular S. Typhi targets host substrates, we studied the induction of cytokines and chemokines in infected macrophages. The results showed significantly higher induction of proinflammatory cytokines by the wild-type and complemented Ty2 strains than by strain Ty2Δt4519, suggesting that T4519 induces cytokine/chemokine responses in macrophages (Fig. 4A and B). However, cytokine/chemokine induction was not altered when the cells were infected with strain Ty2Δt4520 or Ty2Δt2942, indicating the specificity of T4519 effects (see Fig. S3 in the supplemental material). To verify the correlation of the in vivo effects with the in vitro effects of T4519, we studied serum cytokine/chemokine levels after the infection of mice with the wild-type or mutant strain. The results showed markedly elevated levels only in Ty2-infected mice 48 h postinfection, not in Ty2Δt4519-infected mice (Fig. 4C). Since the expression of cytokines/chemokines is regulated by cellular signaling pathways, we studied their activation following infection of THP-1-derived macrophages with strain Ty2 or Ty2Δt4519. Rapid activation of NF-κB (reflected by IκB-α degradation and nuclear translocation of p65), MAP kinase ERK, and phosphoinositol 3-kinase/AKT pathways was observed for both these infections at early time points. However, activation was significantly higher for Ty2 infection at later times, correlating with T4519 induction (Fig. 4D). This suggests that T4519 secreted by S. Typhi Ty2 may target host cellular proteins to exert its role.
FIG 4.
T4519 regulates cytokine/chemokine responses. (A and B) PMA-differentiated THP-1 cells were infected with the indicated bacterial strains and were cultured as described for Fig. 1. (A) Reverse transcription-qPCR shows GAPDH-normalized expression of cytokines and chemokines at 12 h postinfection. Error bars, means ± standard deviations for three independent experiments. Asterisks indicate significant differences from results for Ty2 infection by Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (B) Cytokine and chemokine levels in culture supernatants after 24 h of infection were determined by ELISA. (C) A serum ELISA was performed 48 h after infection of mice. (D) PMA-differentiated THP-1 cells were infected with strain Ty2 or Ty2Δt4519 and were cultured as described for Fig. 1 for the indicated times. (Top) Immunoblots of total-cell lysates probed for IκB-α (NF-κB activation) and for phosphorylated (indicating activation) and total (loading control) MAP kinase (ERK) and AKT. Tubulin was used as a loading control for IκB-α. (Bottom) Immunoblots of nuclear extracts from infected cells were probed for p65 (NF-κB activation) and histone 3 (loading control).
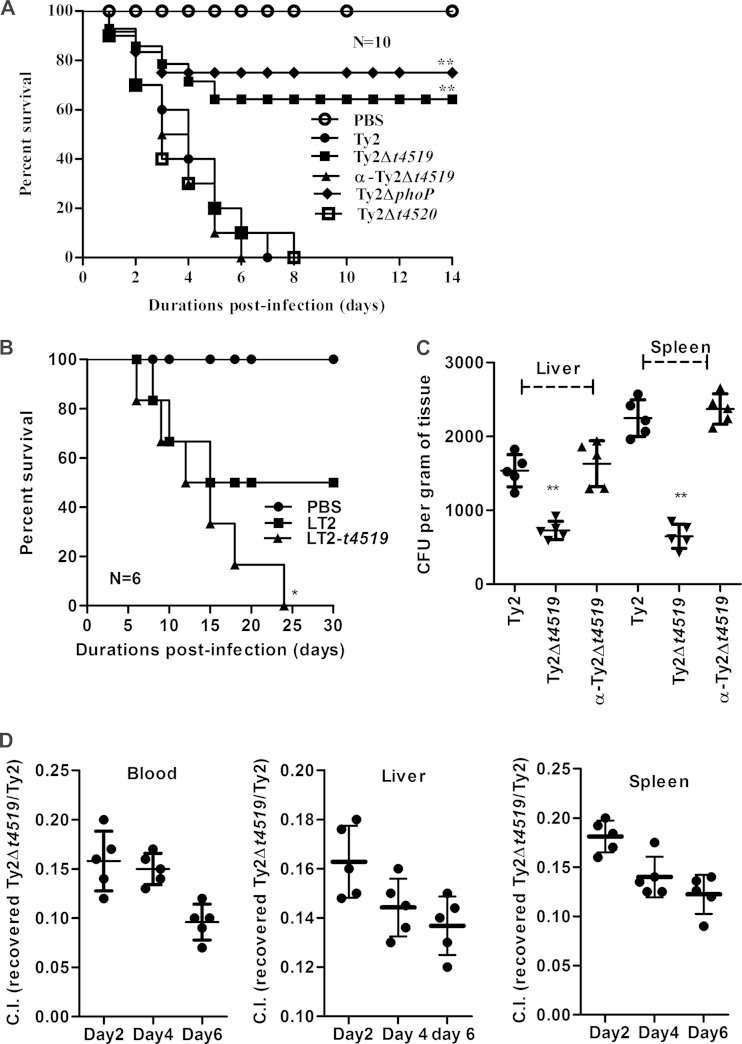
T4519 is required for S. Typhi pathogenesis in mice.
To explore the role of T4519 in bacterial pathogenesis in vivo, we used an iron overload mouse model (20). BALB/c mice infected orally with Ty2 or the complemented (α-Ty2Δt4519) strain at 5 times the LD50 showed 100% mortality within 7 days. In contrast, 65% or 75% of mice infected with an identical dose of the Ty2Δt4519 or Ty2ΔphoP strain, respectively, survived, while all the mice succumbed to Ty2Δt4520 infection (Fig. 5A). These results suggest a critical and specific role for T4519 in the pathogenesis of S. Typhi in mice. This was further supported by infection of mice with an LD50 of S. Typhimurium strain LT2 expressing T4519, which resulted in 100% lethality (Fig. 5B). The role of T4519 in systemic S. Typhi infection was underscored by the lower level of recovery of Ty2Δt4519 bacteria than of Ty2 bacteria from the visceral organs, while the recovery levels were equalized by complementation of the mutant strain (Fig. 5C). Further proof of the role of T4519 in systemic infection was provided by a competitive index assay, where oral infection with a premixed (1:1) dose of the Ty2 and Ty2Δt4519 strains resulted in significantly less recovery of the mutant strain (10 to 15% of Ty2 recovery) from the blood and visceral organs (Fig. 5D). Thus, the intracellular survival advantage conferred by T4519 promotes the pathogenesis of S. Typhi in vivo.
FIG 5.
T4519 promotes Salmonella Typhi pathogenesis in mice. (A) Kaplan-Meier survival plot following oral infection of BALB/c mice (8 to 10 weeks old) with 107 CFU of the indicated strains. (B) Survival plot for BALB/c mice after oral infection with 106 CFU of S. Typhimurium LT2 or a T4519-expressing LT2 strain. For panels A and B, asterisks indicate significant differences (**, P < 0.01) from results for Ty2 infection (A) or LT2 infection (B) by a log rank curve comparison test. (C) Groups of BALB/c mice were orally infected with sublethal doses (5 × 105) of the indicated bacterial strains. Visceral organs were harvested 2 days postinfection, and intracellular CFU counts were carried out after lysis of the cells. Horizontal bars represent mean CFU counts for all mice in one out of two independent experiments. Asterisks indicate significant differences (**, P < 0.01) from results for Ty2 infection as evaluated by the Mann-Whitney test. (D) Groups of BALB/c mice were orally infected with a premixed dose of the Ty2 and Ty2Δt4519 strains (2.5 × 105 CFU each). Blood and visceral organs were harvested at the indicated days postinfection. The intracellular CFU count of each strain was calculated, and results were plotted as a competitive index (C.I.). Data are representative of results of three independent experiments.
DISCUSSION
Combined bioinformatics and experimental approaches may be powerful tools for identifying novel virulence factors of pathogenic microorganisms and exploring the mechanisms behind pathogenesis. We demonstrated previously that an outer membrane adhesin of S. Typhi is required for binding to extracellular matrix proteins and pathogenesis in mice (20). Taking a similar approach here, we have shown that a eukaryote-like Ser/Thr protein kinase (eSTK) of S. Typhi (T4519/STY4822), which is induced within host macrophages and is secreted into the cell cytoplasm, contributes to the phagosomal survival of the bacteria. In agreement with this finding, S. Typhi Ty2Δt4519 shows significantly reduced pathogenicity in mice, a phenotype that is reversed by complementation.
eSTKs constitute an expanding family of prokaryotic proteins that allow the organisms to sense the external environment and adapt to it through various structural and metabolic changes, such as cell division, cell wall synthesis, differentiation, sporulation, biofilm formation, glucose, glycogen, and purine metabolism, secondary metabolism, and the oxidative stress response (26). eSTKs have also been shown to promote host adaptation and virulence in pathogens. Genome sequence analysis has predicted 993 eSTKs in 303 prokaryotes, but only a small fraction have been experimentally validated for kinase functions and have known substrates (22). We have shown here by in vitro kinase assays that a predicted kinase of S. Typhi indeed functions as a bona fide Ser/Thr kinase, as evidenced by autophosphorylation and phosphorylation of MBP. Autophosphorylation is common among STKs; for example, M. tuberculosis PknB is activated by autophosphorylation at residues Thr171 and Thr173 (27). The T4519 sequence has threonine residues at similar positions (Thr170, Thr173, and Thr176), which may be the sites for autophosphorylation (see Fig. S1A in the supplemental material). A kinase function of T4519 in vivo, although not demonstrated in this study, appears critical considering its role in S. Typhi survival in macrophages and pathogenesis in the mouse model. In contrast to S. aureus Stk1 (PknB), deletion of T4519 did not result in the loss of structural integrity for S. Typhi or in inhibition of its growth in vitro (28). This, coupled with its induction within macrophages, suggests that T4519 may predominantly confer a survival advantage on the bacteria during their phagosomal life. This may be critical for the persistent intracellular survival of S. Typhi in view of the functional redundancy of SPI-2 genes, which serve the same functions for S. Typhimurium. However, other prpZ cluster genes, t4520 (prkY) and t4521 (prpZ), played no role in the intracellular survival and virulence of S. Typhi. This is further supported by our finding of a 50% reduction in the phagosomal survival of S. Typhi in the gentamicin survival assay, which is similar to the results obtained in an earlier study with a strain in which the prpZ cluster genes had been deleted (17). Both studies used THP-1-derived macrophages, and we have additionally used RAW 264.7 macrophages, since primary macrophages fail to support S. Typhi replication (see Fig. S4 in the supplemental material) (23, 29). However, the results are likely to be the same for primary macrophages, considering the outcomes of the in vivo experiments.
Environmental cues may regulate the expression of eSTKs. High osmolarity induces M. tuberculosis PknD, which regulates transcription, cell wall remodeling, and virulence factor production (30). T4519 is induced by ROS within macrophages, which is further proved by its induction by H2O2 in in vitro cultures (17). This supports our observation that T4519 is expressed at low levels during the initial period of macrophage infection and fails to influence phagocytosis and early intracellular survival. A previous study, however, reported that deletion of all prpZ cluster genes increased phagocytosis (17). T4519 is not induced in epithelial cells, which produce significantly lower levels of ROS than macrophages, and thus is not required for the intraepithelial survival of S. Typhi. Unlike that of SPI-2 and SPI-3 genes, T4519 expression is not regulated by PhoP. However, T4519 failed to rescue an intracellular Ty2ΔphoP strain, perhaps due to inability to protect against H2O2- and HOCl-mediated bacterial killing (17).
eSTKs may be secreted into host cells and phosphorylate cellular substrates involved in actin rearrangements, the cell cycle, apoptosis, and host defense (14). The T3SS has been identified as the commonest secretory system in Gram-negative pathogens for the translocation of eSTKs, such as Yersinia protein kinase A (YpkA), S. Typhimurium SteC, OspG of Shigella spp. and enterohemorrhagic E. coli (EHEC) NleH1 and NleH2 (14). However, T3SS-1 and T3SS-2 were not involved in the secretion of T4519 (see Fig. S2B in the supplemental material). Other secretory systems reported for eSTKs include Legionella pneumophila Dot/Icm, a T4SS for the secretion of LegK1 to LegK4, and SecA2 for M. tuberculosis PknG (14). The Salmonella enterica serovar Heidelberg plasmid-encoded T4SS enhances the ability to invade and survive within intestinal epithelial and macrophage cell lines, perhaps through the suppression of innate host immune responses (31). In addition, the T6SS gene cluster of S. Typhimurium is required for optimal virulence in mice, perhaps due to its role in macrophage survival and replication (32). It would be important to investigate whether these secretion systems are present and functional in S. Typhi and whether they regulate T4519 translocation. Phosphorylation of cellular proteins by translocated eSTKs has been shown to facilitate bacterial persistence. This may be achieved through the modulation of host immune responses by the suppression of NF-κB activation, as observed with OspG and NleH1. SteC, on the other hand, promotes actin rearrangement by activating the MEK/ERK/myosin light-chain kinase (MLCK)/myosin IIB pathway and limits intracellular replication (14). This may be critical for the maintenance of the intracellular niche of S. Typhimurium, since increased bacterial numbers may significantly enhance the level of flagellin (FliC) translocated to the host cell cytoplasm (33), leading to cell death by inflammasome activation, as well as apoptosis induced by lysosomal membrane permeabilization (LMP). However, expression of flagellin by intracellular S. Typhi is inhibited by the regulatory protein TviA, resulting in negligible death of the infected macrophages (34).
Arrest of phagosomal maturation and inhibition of phagolysosome formation appear to be central mechanisms behind the intracellular persistence of pathogens. M. tuberculosis PknG inhibits phagosome maturation by sequestering the host cell PKCα (35). While limited data are available on S. Typhi, conflicting results have been reported regarding lysosomal fusion of S. Typhimurium-containing vacuoles (2, 3). However, no eSTKs have been suggested so far to perturb the maturation of SCVs and to inhibit their fusion with lysosomes. Functional PhoP fails to rescue the intracellular survival of the Ty2Δt4519 strain, suggesting that PhoP and T4519 may have different cellular targets in cultured macrophages. While PhoP-regulated genes primarily protect against antimicrobial peptides and oxidative damage (36), T4519 may provide protection against lysosomal degradative enzymes. A recent study reported impaired trafficking of lysosomal hydrolases by the SPI-2-encoded effector protein SifA of S. Typhimurium, leading to the formation of less potent lysosomes (37). Whether T4519 mediates similar functions during S. Typhi infection remains to be tested. Since iron is an essential trace element for which S. Typhi competes with the host, T4519 could increase intracellular survival if it helped the bacteria to scavenge iron. We ruled out this possibility by culturing infected macrophages in the presence of desferrioxamine, which had no effect on the survival of S. Typhi within macrophages (data not shown).
It is difficult to identify a mandatory role for eSTKs in in vivo pathogenesis, because of functional redundancy. We have shown here that the majority (65%) of the mice survived when infected with the Ty2Δt4519 strain at a dose (5 times the LD50 of wild-type Ty2) that is 100% lethal for infection with strain Ty2. This is the first report to shows that an eSTK is required for optimal pathogenesis of Salmonella spp. in vivo. We ruled out the possibility that iron overload of mice contributed to the difference in lethality between the Ty2 and Ty2Δt4519 strains by showing that iron plays no role in T4519 induction (Fig. 2D and G) or bacterial replication (data not shown). Moreover, T4519, which is absent from S. Typhimurium, promotes S. Typhi pathogenesis in the mouse oral infection model (Fig. 5B). It is intriguing that the lethalities of the Ty2Δt4519 and Ty2ΔphoP strains are comparable, although PhoP regulates the expression of ∼40 survival genes (38) and a PhoP mutant strain is more susceptible to phagosomal degradation by cultured macrophages (Fig. 1B). This implies that T4519 may play an additional protective role in vivo.
It is intriguing that deletion of individual SPI-2 genes often fails to generate a phenotype (3), while we observed a dominant phenotype of T4519 mutation in S. Typhi. This finding is similar to those in other experiments, where a mycobacterium in which PknG was deleted was deficient in glutamine and glutamate utilization and pathogenesis in mice (39), and deletion of the Yersinia pseudotuberculosis secreted kinase YpkA resulted in an avirulent mouse strain due to increased phagocytosis (40). However, despite the essential role of eSTKs in bacterial virulence, the mechanisms often remain obscure, as for L. pneumophila LegK2 and M. tuberculosis PknH, PknI, and PknK (14). Further studies need to be carried out to identify the host cell substrate(s) of T4519 and the mechanisms behind increased phagosomal survival and pathogenesis. It remains to be examined whether NF-κB activation by T4519 is critical for the prevention of cell death during infection, which may promote the survival of S. Typhi (41).
The activation of cellular signaling pathways regulates the production of cytokines/chemokines by macrophages. We observed increased levels of tumor necrosis factor alpha (TNF-α) and interleukin 6 (IL-6) in the culture supernatants of macrophages and in mouse serum after infection with wild-type S. Typhi Ty2, but not after Ty2Δt4519 infection. TNF-α may induce LMP, favoring intracellular survival (42). Moreover, proinflammatory cytokines may inhibit antimicrobial peptide expression, thus contributing to pathogen survival, especially at mucosal sites (43).
The present study characterizes a putative eSTK of S. Typhi and establishes it as a novel virulence factor. This will stimulate further studies to unravel the mechanisms of phagosomal persistence of S. Typhi and the pathogenesis of diseases caused by this organism. Considering that a large number of drugs targeting host kinases are already in clinical use, bacterial eSTKs and their cellular substrates may be attractive targets in the treatment of infectious diseases.
Supplementary Material
ACKNOWLEDGMENTS
This work was partly funded by extramural grants from the Department of Biotechnology, Government of India (BT/HRD/NBA/34/01/2011), and received intramural funding from the Indian Council of Medical Research, Government of India.
We thank S. Panda, NICED, for helping with the statistical analysis. We thank the Head of the Department of Biochemistry, University of Kolkata, Kolkata, India, for allowing us to use a confocal microscope at the DBT IPLS facility.
Footnotes
Atri Ta and Sayan Das contributed equally to this article.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02521-14.
REFERENCES
- 1.Flannagan RS, Cosio G, Grinstein S. 2009. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat Rev Microbiol 7:355–366. doi: 10.1038/nrmicro2128. [DOI] [PubMed] [Google Scholar]
- 2.Buchmeier NA, Heffron F. 1991. Inhibition of macrophage phagosome-lysosome fusion by Salmonella typhimurium. Infect Immun 59:2232–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haraga A, Ohlson MB, Miller SI. 2008. Salmonellae interplay with host cells. Nat Rev Microbiol 6:53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 4.Garvis SG, Beuzon CR, Holden DW. 2001. A role for the PhoP/Q regulon in inhibition of fusion between lysosomes and Salmonella-containing vacuoles in macrophages. Cell Microbiol 3:731–744. doi: 10.1046/j.1462-5822.2001.00153.x. [DOI] [PubMed] [Google Scholar]
- 5.Feng X, Walthers D, Oropeza R, Kenney LJ. 2004. The response regulator SsrB activates transcription and binds to a region overlapping OmpR binding sites at Salmonella pathogenicity island 2. Mol Microbiol 54:823–835. doi: 10.1111/j.1365-2958.2004.04317.x. [DOI] [PubMed] [Google Scholar]
- 6.García Véscovi E, Soncini FC, Groisman EA. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165–174. doi: 10.1016/S0092-8674(00)81003-X. [DOI] [PubMed] [Google Scholar]
- 7.Crump JA, Mintz ED. 2010. Global trends in typhoid and paratyphoid fever. Clin Infect Dis 50:241–246. doi: 10.1086/649541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinstein DL, O'Neill BL, Hone DM, Metcalf ES. 1998. Differential early interactions between Salmonella enterica serovar Typhi and two other pathogenic Salmonella serovars with intestinal epithelial cells. Infect Immun 66:2310–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 10.Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, Sebaihia M, Baker S, Basham D, Brooks K, Chillingworth T, Connerton P, Cronin A, Davis P, Davies RM, Dowd L, White N, Farrar J, Feltwell T, Hamlin N, Haque A, Hien TT, Holroyd S, Jagels K, Krogh A, Larsen TS, Leather S, Moule S, O'Gaora P, Parry C, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 11.Garai P, Gnanadhas DP, Chakravortty D. 2012. Salmonella enterica serovars Typhimurium and Typhi as model organisms: revealing paradigm of host-pathogen interactions. Virulence 3:377–388. doi: 10.4161/viru.21087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forest CG, Ferraro E, Sabbagh SC, Daigle F. 2010. Intracellular survival of Salmonella enterica serovar Typhi in human macrophages is independent of Salmonella pathogenicity island (SPI)-2. Microbiology 156:3689–3698. doi: 10.1099/mic.0.041624-0. [DOI] [PubMed] [Google Scholar]
- 13.Sharma A, Qadri A. 2004. Vi polysaccharide of Salmonella typhi targets the prohibitin family of molecules in intestinal epithelial cells and suppresses early inflammatory responses. Proc Natl Acad Sci U S A 101:17492–17497. doi: 10.1073/pnas.0407536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Canova MJ, Molle V. 2014. Bacterial serine/threonine protein kinases in host-pathogen interactions. J Biol Chem 289:9473–9479. doi: 10.1074/jbc.R113.529917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng J, He C, Singh VK, Martin NL, Jia Z. 2007. Crystal structure of a novel prokaryotic Ser/Thr kinase and its implication in the Cpx stress response pathway. Mol Microbiol 63:1360–1371. doi: 10.1111/j.1365-2958.2007.05611.x. [DOI] [PubMed] [Google Scholar]
- 16.Geddes K, Worley M, Niemann G, Heffron F. 2005. Identification of new secreted effectors in Salmonella enterica serovar Typhimurium. Infect Immun 73:6260–6271. doi: 10.1128/IAI.73.10.6260-6271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faucher SP, Viau C, Gros PP, Daigle F, Le Moual H. 2008. The prpZ gene cluster encoding eukaryotic-type Ser/Thr protein kinases and phosphatases is repressed by oxidative stress and involved in Salmonella enterica serovar Typhi survival in human macrophages. FEMS Microbiol Lett 281:160–166. doi: 10.1111/j.1574-6968.2008.01094.x. [DOI] [PubMed] [Google Scholar]
- 18.Philippe N, Alcaraz JP, Coursange E, Geiselmann J, Schneider D. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51:246–255. doi: 10.1016/j.plasmid.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Chakraborty K, Maity PC, Sil AK, Takeda Y, Das S. 2009. cAMP stringently regulates human cathelicidin antimicrobial peptide expression in the mucosal epithelial cells by activating cAMP-response element-binding protein, AP-1, and inducible cAMP early repressor. J Biol Chem 284:21810–21827. doi: 10.1074/jbc.M109.001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh S, Chakraborty K, Nagaraja T, Basak S, Koley H, Dutta S, Mitra U, Das S. 2011. An adhesion protein of Salmonella enterica serovar Typhi is required for pathogenesis and potential target for vaccine development. Proc Natl Acad Sci U S A 108:3348–3353. doi: 10.1073/pnas.1016180108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanks SK, Hunter T. 1995. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9:576–596. [PubMed] [Google Scholar]
- 22.Pereira SF, Goss L, Dworkin J. 2011. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol Mol Biol Rev 75:192–212. doi: 10.1128/MMBR.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwan WR, Huang XZ, Hu L, Kopecko DJ. 2000. Differential bacterial survival, replication, and apoptosis-inducing ability of Salmonella serovars within human and murine macrophages. Infect Immun 68:1005–1013. doi: 10.1128/IAI.68.3.1005-1013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arpaia N, Godec J, Lau L, Sivick KE, McLaughlin LM, Jones MB, Dracheva T, Peterson SN, Monack DM, Barton GM. 2011. TLR signaling is required for Salmonella typhimurium virulence. Cell 144:675–688. doi: 10.1016/j.cell.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charpentier X, Oswald E. 2004. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J Bacteriol 186:5486–5495. doi: 10.1128/JB.186.16.5486-5495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyagi N, Anamika K, Srinivasan N. 2010. A framework for classification of prokaryotic protein kinases. PLoS One 5:e10608. doi: 10.1371/journal.pone.0010608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boitel B, Ortiz-Lombardia M, Duran R, Pompeo F, Cole ST, Cervenansky C, Alzari PM. 2003. PknB kinase activity is regulated by phosphorylation in two Thr residues and dephosphorylation by PstP, the cognate phospho-Ser/Thr phosphatase, in Mycobacterium tuberculosis. Mol Microbiol 49:1493–1508. doi: 10.1046/j.1365-2958.2003.03657.x. [DOI] [PubMed] [Google Scholar]
- 28.Débarbouillé M, Dramsi S, Dussurget O, Nahori MA, Vaganay E, Jouvion G, Cozzone A, Msadek T, Duclos B. 2009. Characterization of a serine/threonine kinase involved in virulence of Staphylococcus aureus. J Bacteriol 191:4070–4081. doi: 10.1128/JB.01813-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strandberg KL, Richards SM, Gunn JS. 2012. Cathelicidin antimicrobial peptide expression is not induced or required for bacterial clearance during Salmonella enterica infection of human monocyte-derived macrophages. Infect Immun 80:3930–3938. doi: 10.1128/IAI.00672-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatzios SK, Baer CE, Rustad TR, Siegrist MS, Pang JM, Ortega C, Alber T, Grundner C, Sherman DR, Bertozzi CR. 2013. Osmosensory signaling in Mycobacterium tuberculosis mediated by a eukaryotic-like Ser/Thr protein kinase. Proc Natl Acad Sci U S A 110:E5069–E5077. doi: 10.1073/pnas.1321205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gokulan K, Khare S, Rooney AW, Han J, Lynne AM, Foley SL. 2013. Impact of plasmids, including those encodingVirB4/D4 type IV secretion systems, on Salmonella enterica serovar Heidelberg virulence in macrophages and epithelial cells. PLoS One 8:e77866. doi: 10.1371/journal.pone.0077866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Guo JT, Li YG, Johnston RN, Liu GR, Liu SL. 2013. The type VI secretion system gene cluster of Salmonella typhimurium: required for full virulence in mice. J Basic Microbiol 53:600–607. doi: 10.1002/jobm.201200047. [DOI] [PubMed] [Google Scholar]
- 33.Lage SL, Buzzo CL, Amaral EP, Matteucci KC, Massis LM, Icimoto MY, Carmona AK, D'Imperio Lima MR, Rodrigues MM, Ferreira LC, Amarante-Mendes GP, Bortoluci KR. 2013. Cytosolic flagellin-induced lysosomal pathway regulates inflammasome-dependent and -independent macrophage responses. Proc Natl Acad Sci U S A 110:E3321–E3330. doi: 10.1073/pnas.1305316110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atif SM, Winter SE, Winter MG, McSorley SJ, Bäumler AJ. 2014. Salmonella enterica serovar Typhi impairs CD4 T cell responses by reducing antigen availability. Infect Immun 82:2247–2254. doi: 10.1128/IAI.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walburger A, Koul A, Ferrari G, Nguyen L, Prescianotto-Baschong C, Huygen K, Klebl B, Thompson C, Bacher G, Pieters J. 2004. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science 304:1800–1804. doi: 10.1126/science.1099384. [DOI] [PubMed] [Google Scholar]
- 36.Charles RC, Harris JB, Chase MR, Lebrun LM, Sheikh A, LaRocque RC, Logvinenko T, Rollins SM, Tarique A, Hohmann EL, Rosenberg I, Krastins B, Sarracino DA, Qadri F, Calderwood SB, Ryan ET. 2009. Comparative proteomic analysis of the PhoP regulon in Salmonella enterica serovar Typhi versus Typhimurium. PLoS One 4:e6994. doi: 10.1371/journal.pone.0006994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGourty K, Thurston TL, Matthews SA, Pinaud L, Mota LJ, Holden DW. 2012. Salmonella inhibits retrograde trafficking of mannose-6-phosphate receptors and lysosome function. Science 338:963–967. doi: 10.1126/science.1227037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller SI, Mekalanos JJ. 1990. Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. J Bacteriol 172:2485–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cowley S, Ko M, Pick N, Chow R, Downing KJ, Gordhan BG, Betts JC, Mizrahi V, Smith DA, Stokes RW, Av-Gay Y. 2004. The Mycobacterium tuberculosis protein serine/threonine kinase PknG is linked to cellular glutamate/glutamine levels and is important for growth in vivo. Mol Microbiol 52:1691–1702. doi: 10.1111/j.1365-2958.2004.04085.x. [DOI] [PubMed] [Google Scholar]
- 40.Wiley DJ, Nordfeldth R, Rosenzweig J, DaFonseca CJ, Gustin R, Wolf-Watz H, Schesser K. 2006. The Ser/Thr kinase activity of the Yersinia protein kinase A (YpkA) is necessary for full virulence in the mouse, mollifying phagocytes, and disrupting the eukaryotic cytoskeleton. Microb Pathog 40:234–243. doi: 10.1016/j.micpath.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 41.Faherty CS, Maurelli AT. 2008. Staying alive: bacterial inhibition of apoptosis during infection. Trends Microbiol 16:173–180. doi: 10.1016/j.tim.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Werneburg NW, Guicciardi ME, Bronk SF, Gores GJ. 2002. Tumor necrosis factor-alpha-associated lysosomal permeabilization is cathepsin B dependent. Am J Physiol Gastrointest Liver Physiol 283:G947–G956. [DOI] [PubMed] [Google Scholar]
- 43.Arijs I, De Hertogh G, Lemaire K, Quintens R, Van Lommel L, Van Steen K, Leemans P, Cleynen I, Van Assche G, Vermeire S, Geboes K, Schuit F, Rutgeerts P. 2009. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS One 4:e7984. doi: 10.1371/journal.pone.0007984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.