Abstract
Mycobacterium tuberculosis is able to synthesize molybdopterin cofactor (MoCo), which is utilized by numerous enzymes that catalyze redox reactions in carbon, nitrogen, and sulfur metabolism. In bacteria, MoCo is further modified through the activity of a guanylyltransferase, MobA, which converts MoCo to bis-molybdopterin guanine dinucleotide (bis-MGD), a form of the cofactor that is required by the dimethylsulfoxide (DMSO) reductase family of enzymes, which includes the nitrate reductase NarGHI. In this study, the functionality of the mobA homolog in M. tuberculosis was confirmed by demonstrating the loss of assimilatory and respiratory nitrate reductase activity in a mobA deletion mutant. This mutant displayed no survival defects in human monocytes or mouse lungs but failed to persist in the lungs of guinea pigs. These results implicate one or more bis-MGD-dependent enzymes in the persistence of M. tuberculosis in guinea pig lungs and underscore the applicability of this animal model for assessing the role of molybdoenzymes in this pathogen.
INTRODUCTION
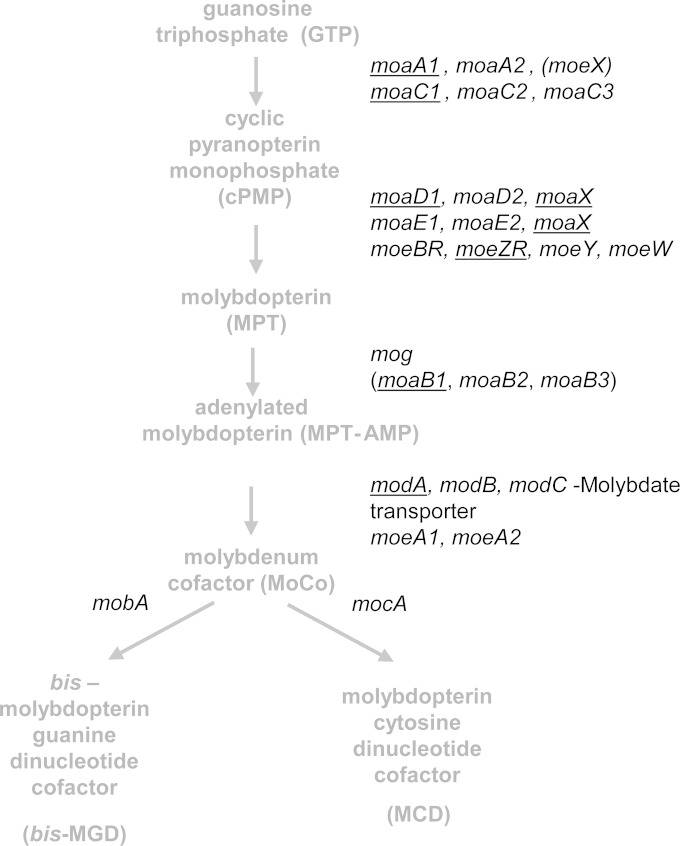
Comparative genomics suggest an association between molybdenum cofactor (MoCo) and pathogenesis in Mycobacterium tuberculosis, the causative agent of tuberculosis (TB) (1). Consistent with this notion, genes involved in MoCo biosynthesis have been identified as being essential for the growth and survival of M. tuberculosis in various models of infection (Fig. 1) (2–4). For example, transposon mutants harboring insertions in MoCo biosynthesis genes were attenuated for growth in macrophages (5–7), mice (8), and the lungs of nonhuman primates (9). Furthermore, a transposon insertion in moaD1, which encodes a subunit of molybdopterin synthase, rendered M. tuberculosis hypersensitive to oxidative stress, suggesting that the impaired macrophage invasion and phagosome maturation defects of this mutant are due to increased susceptibility to intracellular killing (10). Of particular interest is the observation that the moaA1-moaD1 gene cluster is located on a genomic island with reduced GC content and contains genetic features that are indicative of horizontal gene transfer (11). These data highlight the importance of MoCo availability during the parasitism of host macrophages (12) and potentially implicate one or more of the eight predicted MoCo-dependent enzymes in M. tuberculosis pathogenesis.
FIG 1.
MoCo biosynthetic pathway in M. tuberculosis (adapted from reference 2). MoCo biosynthesis occurs through a highly conserved, four-step pathway. Further modifications of MoCo give rise to alternative forms of the cofactor. A notable feature of this pathway is the multiplicity of homologues involved in the first two steps of the pathway. Genes identified as essential in forward genetic screens are underlined. Parentheses denote genes with weak homology. The moaX gene is reflected twice, since it contains both moaD- and moaE-like domains.
MoCo-dependent enzymes in bacteria are classified into three families according to the structure of their molybdenum centers: xanthine dehydrogenase, sulfite oxidase, and dimethyl sulfoxide (DMSO) reductase (13). The biosynthesis of MoCo involves four steps that are highly conserved across all kingdoms of life; however, certain modifications of MoCo are known to occur only in bacterial species (Fig. 1) (14–16). The bacterium-specific modification of MoCo involves the addition of nucleotides to form either molybdopterin cytosine dinucleotide (MCD) or bis-molybdopterin guanine dinucleotide (bis-MGD), which are forms of the cofactor employed by xanthine dehydrogenase and DMSO reductase-type enzymes, respectively. Biosynthesis of MCD in Escherichia coli requires MocA, a cytidylyltransferase that transfers cytosine residues to the cofactor, a process that is more efficient if Mo is bound (17). The M. tuberculosis genome contains a mocA (Rv0371c) homologue, which is located on an operon with a gene encoding CO dehydrogenase, the sole xanthine dehydrogenase-type molybdoenzyme in this organism (Table 1) (2, 18).
TABLE 1.
Putative MoCo-dependent enzymes in M. tuberculosis and their predicted/demonstrated function
| Locus designation | Gene name | Annotation | Form of cofactor requireda | Functional information and expression | Reference(s) |
|---|---|---|---|---|---|
| Rv0197 | Possible oxidoreductase | bis-MGD | Rv0197 detected in guinea pig lungs at 30 days postinfection | 33 | |
| Rv1161 | narG | Respiratory and assimilatory nitrate reductase (alpha chain) | bis-MGD | Required for the survival of M. tuberculosis under anaerobic conditions in vitro | 20 |
| Required for protection of M. tuberculosis against acid stress during hypoxia | 21 | ||||
| Reduced narGHJI expression in M. tuberculosis and M. africanum clinical isolates is associated with impaired fitness in macrophages | 46 | ||||
| Required for growth of M. tuberculosis in human macrophages | 38 | ||||
| Required for nitrite production and transcriptional reprogramming during growth of M. tuberculosis within human macrophages | 39 | ||||
| ΔnarG mutant of M. tuberculosis is more sensitive to isoniazid in human macrophages and axenic culture | 40 | ||||
| NarG detected in guinea pig lungs at 30 and 90 days postinfection | 33 | ||||
| Rv1442 | bisC | Probable biotin sulfoxide reductase | bis-MGD | ||
| Rv2900c | fdhF | Possible formate dehydrogenase H | bis-MGD | Rv2900c detected in guinea pig lungs at 30 days postinfection | 33 |
| Rv0218 | Probable conserved transmembrane protein, some similarity with sulfite oxidases | MoCo | |||
| Rv0373c | Probable carbon monoxide dehydrogenase (large chain) | MCDb/sulfurated | Implicated in the ability of M. tuberculosis H37Ra and M. bovis BCG to oxidize CO at physiologically relevant concentrations | 47, 48 | |
| CDH from Mycobacterium sp. strain JC1 demonstrated NO dehydrogenase activity | 49 | ||||
| Rv3151 | nuoG | Probable NADH dehydrogenase I (chain g) | Unknown | M. tuberculosis ΔnuoG mutant has a proapoptotic phenotype in macrophages | 28, 50 |
| NADH-ubiquinone oxidoreductase | Repressed by acid shock | 51 |
Enzymatic preferences for various cofactor forms were determined by interrogating the dbTEU database (http://gladyshevlab.org/trace_element/).
Molybdopterin cytosine dinucleotide.
M. tuberculosis also possesses a homolog of mobA (Rv2453c), which has been implicated in the production of bis-MGD in E. coli through the formation of a bis-MoCo intermediate and subsequent addition of two guanine residues on the terminal phosphate groups (15, 19). Of the eight molybdoenzymes identified through bioinformatic analysis of the M. tuberculosis proteome, four are predicted to utilize bis-MGD: Rv0197, Rv1161 (NarG), Rv1442 (BisC), and Rv2900c (FdhF) (2) (Table 1). Three of these proteins form parts of multisubunit enzyme complexes. The best studied of them is the narGHI-encoded nitrate reductase (NR), which is required for survival of M. tuberculosis under anaerobic conditions in vitro (20) and for protection against acid stress during hypoxia (21). The NarGHI respiratory complex from M. tuberculosis has both respiratory and assimilatory NR functions (22). Although the loss of NarGHI function impaired the ability of Mycobacterium bovis BCG to persist in organs of immunocompetent as well as immune-deficient mice (23, 24), the functional inactivation of this enzyme had no effect on growth or survival of M. tuberculosis in mice (20). The reason for this discrepancy is unclear but may be due to the differential abundance of the narGHJI transcript (25) and/or the fact that the lesions produced in mice are insufficiently hypoxic to manifest in a survival phenotype of an narG mutant of M. tuberculosis (20). The bisC gene encodes a probable biotin sulfoxide reductase (Table 1) which may be involved in reducing spontaneous oxidation products of biotin (26), a cofactor required by M. tuberculosis during infection in mice (27). Little is known about the roles of FdhF (a subunit of formate dehydrogenase) and Rv0197 (a putative oxidoreductase) in the physiology of M. tuberculosis. Of the remaining molybdoenzymes, NuoG, a subunit of the type I NADH dehydrogenase (NDH-1), has been implicated in inhibiting macrophage apoptosis through neutralization of NOX2-derived reactive oxygen species (28). The inhibition is proposed to allow for distribution of M. tuberculosis between the various subclasses of immune cells in the lung and prevent trafficking of bacilli to the draining lymph node (29). The role of carbon monoxide dehydrogenase in M. tuberculosis has not been investigated.
In this study, we employed a genetic approach to further explore the role of bis-MGD in M. tuberculosis. Our results confirm the functionality of mobA and its essentiality for NR activity in M. tuberculosis. We also show that loss of mobA function specifically impairs the ability of M. tuberculosis to persist in the lungs of guinea pigs, thereby implicating one or more bis-MGD-dependent enzymes in mycobacterial persistence in this animal model.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and mutant construction.
Bacterial strains and plasmids are detailed in Table S1 in the supplemental material. All Escherichia coli strains were grown in Luria-Bertani broth or on Luria agar and incubated at 37°C. Mycobacterium tuberculosis strains were cultured standing in Middlebrook 7H9 medium supplemented with 0.2% glycerol, Middlebrook oleic acid-albumin-dextrose-catalase (OADC) enrichment, and 0.05% Tween 80. Nitrate assimilation experiments and nitrate accumulation assays were performed as previously described (3). Briefly, for assimilation experiments, strains were grown aerobically in MB media supplemented with nitrate, while DTA-nitrate media (Dubos broth base containing 5% glycerol, 0.5% albumin, 0.75% dextrose, 0.05% Tween 80, and 5 mM NaNO3) was used for nitrite accumulation assays. Respiratory NR activity was measured by monitoring nitrite accumulation with the Griess assay. CFUs were determined by plating appropriate dilutions on Middlebrook 7H10 medium supplemented with 0.5% glycerol and Middlebrook OADC enrichment. Where appropriate, ampicillin (Ap), kanamycin (Km), and hygromycin (Hyg) were used in E. coli cultures at 200, 50, and 150 μg/ml, respectively. In mycobacterial cultures, Hyg, Km, and gentamicin (Gm) were used at 50, 25, and 10 μg/ml, respectively.
Construction of mutant and complemented strains.
The primers listed in Table S1 in the supplemental material were used to amplify upstream (TmobUF and TmobUR) and downstream (TmobDF and TmobDR) homologous sequences and construct the suicide plasmid, p2mobA. This plasmid was used to generate a ΔmobA mutant by two-step allelic exchange as previously described (30). This approach resulted in a mutant carrying an in-frame deletion in mobA spanning from codons 45 to 182, where the resulting protein lacked 137 amino acids. The genotype of the mutant strain was confirmed by PCR and Southern hybridization (see Fig. S1A). A complementation vector (pMobA) carrying M. tuberculosis mobA fused to the promoter region upstream of Rv2455c was constructed by PCR amplification and cloning of the resulting amplicon into pMV306H.
Analysis of PDIM production in M. tuberculosis.
Two μCi of [14C]propionic acid was added to log-phase cultures of M. tuberculosis, which were incubated for a further 48 h. The cultures then were harvested and the pellet resuspended in 5 ml of 10:1 methanol-0.3% NaCl. The lipid fraction was extracted into petroleum ether and separated by thin-layer chromatography on silica plates. Labeled lipids were visualized autoradiographically by exposing the dried plates to Kodak Biomax imaging film at room temperature for 10 to 12 days.
Human monocyte infection.
Peripheral blood-derived human monocytes were isolated and infected with wild-type or mutant strains of M. tuberculosis at a multiplicity of infection of 1 bacillus per monocyte (1:1) in RPMI medium as previously described (31). At each time point, 500 μl of supernatant for cytokine analysis was removed from each well of infected cells and stored at −70°C until use. Triton X-100 was added to a final concentration of 0.1% and incubated for 5 min to lyse monocytes. Supernatants from infected monocytes were analyzed using a Bio-Plex Th1/Th2 human cytokine panel assay (Bio-Rad, Hercules, CA) according to the manufacturer's instructions {cytokines analyzed: interleukin-2 [IL-2], IL-4, IL-5, IL-10, IL-12(p70), IL-13, granulocyte-macrophage colony-stimulating factor [GM-CSF], gamma interferon [IFN-γ], and tumor necrosis factor alpha [TNF-α]}. The results were read using a Bio-Plex 200 system (Bio-Rad, Hercules, CA), and data and statistical analyses were carried out using GraphPad Prism, version 4 (GraphPad, San Diego, CA). Using this analysis, we obtained robust data only for TNF-α secretion and relatively low levels of the other cytokines assessed. The protocol for these studies was approved by Stellenbosch University's Health Research Ethics Committee (N13/09/126).
Histology.
Right caudal lung lobes from each guinea pig were fixed with 4% paraformaldehyde in phosphate-buffered saline. Paraffin-embedded sections from these tissues were cut to 5 μm, mounted on glass slides, deparaffinized, and stained using hematoxylin and eosin. Differences in pathology were determined by visual assessment of the number and extent of lesions in sections from each animal.
Murine and guinea pig infections.
Six- to 8-week-old female C57BL/6 mice from Jackson Laboratories (Bar Harbor, ME) were infected with the H37Rv, ΔmobA, or ΔmobA attB::pMobA strain through the respiratory route. Approximately 5 to 10 organisms were implanted in the lungs of each mouse, as confirmed by plating lung homogenates 3 h after infection. Bacterial loads (CFU) in the lungs, liver, and spleen of infected mice were assessed at selected time points over a period of 90 days. For guinea pig infections, thawed aliquots of frozen cultures of the three strains were diluted in sterile water to an inoculum concentration of 106 CFU/ml in 10 ml. A Madison chamber aerosol generation device was used to expose the animals to M. tuberculosis. This device was calibrated to deliver approximately 20 bacilli into the lungs. Lung bacterial loads were determined by plating serial dilutions of tissue homogenates on Middlebrook 7H11 agar. The protocols for these studies were approved by the IACUC at the Public Health Research Institute, New Jersey Medical School, Rutgers University, New Brunswick, New Jersey (mouse infections), and the Department of Microbiology, Immunology and Pathology, Colorado State University, Fort Collins (guinea pig infections), Colorado.
Statistical analysis.
The unpaired t test with Welch's correction was used to assess statistical significance of pairwise comparisons using GraphPad Prism Software.
RESULTS
The mobA gene is dispensable for growth of M. tuberculosis in vitro but is required for NR activity.
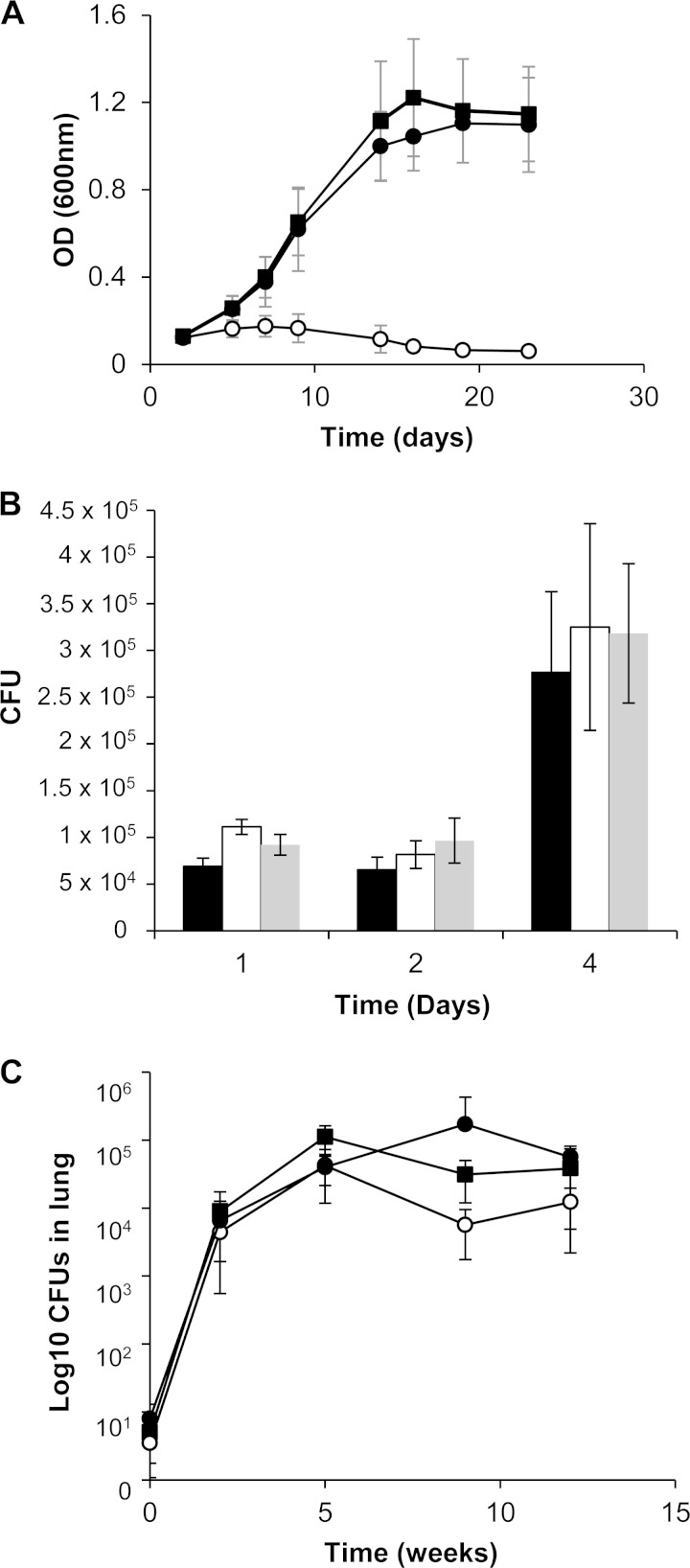
To assess the role of bis-MGD in pathogenicity, we constructed a mutant strain of M. tuberculosis H37Rv carrying an in-frame, unmarked deletion in the mobA gene. The mutant displayed no growth defect in Middlebrook 7H9, DTA, or DTA-nitrate (data not shown), confirming that mobA is dispensable for growth under these conditions. In prior work, we demonstrated the utility of NarGHI-catalyzed NR activity as a readout for MoCo production in M. tuberculosis (3). Since NarGHI belongs to the DMSO reductase family of enzymes that utilize the bis-MGD form of the cofactor, we reasoned that its activity likewise would provide a means of assessing the production of bis-MGD. The determination of the level of nitrite produced by the wild-type, ΔmobA, and ΔmobA attB::pMobA strains yielded 2,547 ± 319, 12 ± 17, and 3,088 ± 57 mmol nitrite/ml, respectively, consistent with an association between mobA function and respiratory NR activity. Furthermore, deletion of mobA completely abrogated nitrate assimilation, as evidenced by the inability of the ΔmobA mutant to grow on nitrate as the sole nitrogen source (Fig. 2A). Both NR-associated defects were fully rescued by genetic complementation, confirming that the phenotypes observed were attributable to the loss of mobA function. These data confirm the essentiality of mobA for NarGHI-dependent growth of M. tuberculosis on nitrate as the sole nitrogen source and confirm that this gene is the sole guanylyltransferase responsible for producing bis-MGD in this organism.
FIG 2.
Nitrate assimilation and survival of M. tuberculosis strains in human monocytes and mice. (A) Strains were cultured in MB media with nitrate as the sole nitrogen source for NR-dependent assimilation and growth. The results shown are the means and standard deviations from three independent experiments. OD, optical density. (B) Growth of the ΔmobA mutant in human monocytes. The solid bar represents the wild type, open bars represent the ΔmobA mutant, and gray bars represent the genetically complemented derivative. Data are the means from experiments with 6 independent donors. (C) Survival of the ΔmobA mutant in the lungs of infected mice. Four animals per strain were used at day 0, and 5 animals per strain were used at all other time points. ■, wild type; ○, ΔmobA mutant; ●, ΔmobA attB::pMobA mutant.
Deletion of mobA has no effect on growth or survival of M. tuberculosis in human monocytes.
MoCo-dependent enzymes or genes required for MoCo biosynthesis in M. tuberculosis previously have been implicated in the colonization/apoptosis of host macrophages (2). To investigate the effect of abrogating bis-MGD production on intracellular growth and survival of M. tuberculosis, human monocytes from 6 healthy volunteers were infected with the wild-type, mutant, and complemented strains. No differences were observed in the ability of the mutant to grow and survive (Fig. 2B). Furthermore, secretion of tumor necrosis factor alpha from monocytes was indistinguishable between cells infected with the wild-type and mutant strains (data not shown).
bis-MGD is not required for growth and persistence of M. tuberculosis in mice but is required for persistence in guinea pig lungs.
The role of bis-MGD in pathogenesis then was assessed by monitoring the growth and survival of the three strains in the murine model of TB infection. Since mutations that affect the production of phthiocerol dimycocerosate (PDIM) are known to attenuate the virulence of M. tuberculosis in mice, the strains first were assessed for PDIM production and shown to be PDIM+ (see Fig. S1B in the supplemental material). C57BL/6 mice then were infected by the aerosol route, and the progress of the infection was monitored over a period of 90 days. All three strains achieved and maintained comparable lung bacillary loads over this time course (Fig. 2C). Similarly, no differences in bacterial dissemination to the spleen and liver were noted (data not shown). It has been demonstrated that an NR-deficient mutant of M. tuberculosis does not display growth defects in mice (20), and our data now confirm that the remaining three bis-MGD-dependent enzymes also are dispensable for virulence in the murine model of TB infection.
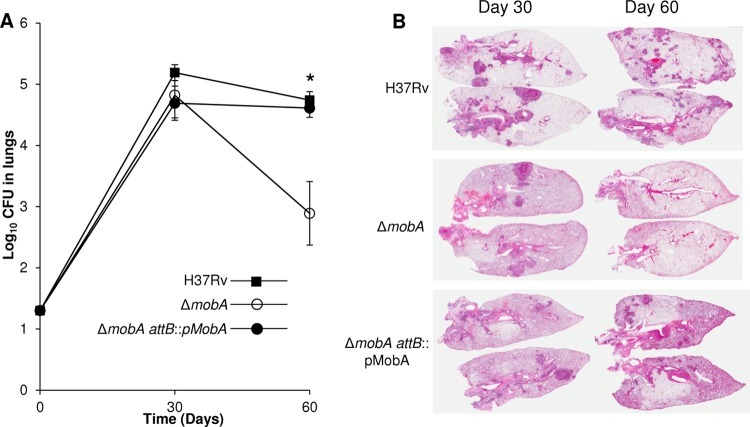
In light of these findings, we reasoned that an animal model that more closely recapitulates the pathology of human tuberculosis is required in order to investigate the impact of simultaneously crippling all bis-MGD enzymes, including NarGHI, on the virulence of M. tuberculosis through the loss of MobA function. Therefore, we turned to the guinea pig model in which hypoxic tuberculous granulomas are formed (32). Female outbred Hartley guinea pigs were infected by the aerosol route, and the bacterial burden in the lungs was determined at 30 and 60 days postinfection (Fig. 3A). No significant difference was observed in the lung bacillary load of animals infected with the wild-type, ΔmobA mutant, or complemented strain 30 days postinfection, arguing against any role for bis-MGD-dependent enzymes during acute infection. However, at 60 days, the bacterial burden in animals infected with the mutant strain was significantly lower than that of the wild-type and complemented strains (P = 0.014). Histological examination of the lung tissue at 30 days revealed inflammation and granuloma formation in animals infected with all three strains (Fig. 3B). However, at the later time point, the inflammation seen in animals infected with the mutant strain was reduced compared to the significant lung involvement observed in animals infected with the wild-type and complemented strains. These findings specifically implicate MobA, and, as a result, one or more bis-MGD-dependent enzymes in the persistence of M. tuberculosis in the lungs of infected guinea pigs.
FIG 3.
Growth and survival of M. tuberculosis strains in guinea pigs. (A) Survival of the wild-type strain, ΔmobA strain, and ΔmobA attB::pMobA strain in lungs of infected guinea pigs over 60 days. Five animals were sacrificed per time point. *, P = 0.014. (B) Pathology in infected guinea pig lungs.
DISCUSSION
The detection of NarG, NarH, and FdhF in guinea pigs during pulmonary infection (33) confirms that these proteins are expressed by M. tuberculosis in this animal model (32). Drug treatment of guinea pigs infected with M. tuberculosis results in the resolution of secondary lesions but has a reduced effect on primary lesions, where some bacteria are found in the necrotic zone, and the majority of the persisting organisms are located in the acellular rim of the lesion (34). Importantly, this particular area of the granuloma is hypoxic. The adaptive response of M. tuberculosis to hypoxia has been the subject of intense investigation and has been shown to comprise both transient and enduring components (35). The former is associated with the induction of an ∼47-gene regulon, controlled by the DosR/DosS/DosT two-component system, which also is induced by CO and NO (36). A key feature of the DosR response is the upregulation of the narK2-encoded nitrate transporter, which results in elevated NR activity under hypoxic conditions (37). M. tuberculosis has two bacterial NR homologues, NarGHI and NarX (2); the former is a bona fide molybdoenzyme, whereas the latter lacks the MoCo binding domain (Pfam domain PF01568) and most likely is not a bis-MGD-dependent enzyme. The transcript for narX has been detected in bacteria derived from sputum of TB patients, but unlike NarGHI, whose role in nitrate reduction is well established, the role of NarX in the physiology and metabolism of M. tuberculosis remains unclear (37). Thus, the essentiality of MobA for NarGHI activity suggests that the persistence defect of the mobA mutant under the hypoxic conditions that prevail in the lesions of infected guinea pigs are due, at least in part, to the loss of NarGHI function. Recent work has shown that nitrite production by NarGHI enhances the survival of M. tuberculosis in human macrophages (38, 39). The production of nitrite by M. tuberculosis when cultured at physiologic oxygen tension in human macrophages was shown to enable the adaptation of the organism to the host environment through the cessation of growth, decreased ATP consumption, and the differential expression of approximately 120 genes (39). These findings suggest a dual role for NarGHI in energy metabolism and in producing the mediator of an adaptive response to the intracellular environment. Endogenous nitrite production by M. tuberculosis reduces its sensitivity to isoniazid both within macrophages and in axenic culture, implicating NarGHI in intrinsic resistance to this antibiotic (40). However, the implications of these findings for pulmonary TB in humans remain unknown. As outlined above, we observed no survival defect of the mobA mutant in primary human monocytes. The reasons for this discrepancy presently are unclear but may be due to differences in cell type (monocytes versus macrophages).
In E. coli, the bisC-encoded biotin sulfoxide reductase is able to reduce both biotin sulfoxide and methionine sulfoxide to biotin and methionine, respectively (41). This dual activity for BisC also was recently demonstrated in Salmonella enterica serovar Typhimurium and linked to the survival of this organism under conditions of oxidative stress. In this case, the reduction of methionine sulfoxide by BisC was important for survival in the presence of oxidative stress in vitro, whereas biotin sulfoxide reduction was required for defense against host-derived oxidative stress generated in activated macrophages and mice (42). Given the high degree of homology between the M. tuberculosis and E. coli/Salmonella enterica serovar Typhimurium BisC homologues (40% or 39% homology between M. tuberculosis and E. coli or Salmonella enterica serovar Typhimurium), it is tempting to speculate that BisC plays an analogous role in methionine and/or biotin regeneration in M. tuberculosis, which similarly encounters significant oxidative stress during the course of infection. The remaining bis-MGD dependent enzymes have not been functionally characterized in M. tuberculosis. As such, their individual and collective contributions to the persistence defect of the mobA mutant in guinea pigs have yet to be established.
A study comparing the survival of a pool of 80 M. tuberculosis mutants in mice and guinea pigs demonstrated that while a high degree of concordance exists between the two models, some genes were differentially required for survival (43). The dispensability of mobA for the growth and survival of M. tuberculosis in mice suggests that none of the bis-MGD-dependent molybdoenzymes is required for virulence in this model. In the case of NarGHI, these data are consistent with a previous study which found no virulence defect for a ΔnarG mutant in mice (20). This likely is due to the lack of hypoxia in mouse granulomas, unlike those found in guinea pigs, rabbits, and nonhuman primates, which are hypoxic (20, 32). Thus, our results emphasize the importance of using animal models that more closely recapitulate conditions that prevail in human infection (44) in order to discern the role of certain M. tuberculosis enzymes or enzyme families.
With respect to the predicted essentiality of MoCo biosynthetic genes in various forward genetic screens, our results also suggest that the attenuation in mice of mutants of M. tuberculosis harboring transposon insertions in MoCo biosynthetic genes is unrelated to bis-MGD-dependent enzyme function. This, in turn, implicates other MoCo-dependent enzymes in pathogenesis and/or points to a role for MoCo biosynthetic enzymes in other pathways. The majority of MoCo biosynthetic genes identified as being required for the growth and survival of M. tuberculosis in the intracellular environment or in the organism of infected mice map to the first two steps of the biosynthetic pathway, where the highest degree of gene expansion also occurs (2). This may reflect the involvement of MoCo biosynthetic genes in other metabolic pathways. For example, the second step of the MoCo biosynthetic pathway has been linked to cysteine metabolism through moeZR, which is able to thiocarboxylate both MoaD1/MoaD2 as well as CysO, which acts as a sulfide donor for a cysteine biosynthetic pathway in M. tuberculosis (45). Therefore, the phenotypes of mutants harboring insertions in the molybdopterin synthase subunits or sulfurtransferases may result from perturbations in cysteine and sulfur biosynthesis.
In conclusion, we demonstrate that the mobA gene is required for bis-MGD production and associated NR activity in M. tuberculosis. Our data also confirm that bis-MGD is required for persistence in the guinea pig, but not the murine model of TB infection. These observations confirm a role for one or more of the enzymes that depend on this form of MoCo for prolonged survival in the hypoxic primary lesions that accumulate in the lungs of infected guinea pigs.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Research Foundation, the South African Medical Research Council (to V.M.), and the Howard Hughes Medical Institute (Senior International Research Scholars grant to V.M. and an International Early Career Scientist grant to B.D.K). M.J.W. also was supported by the Columbia University-Southern African Fogarty AITRP and PHRI-AURUM-Global Infectious Diseases Research Programme. G.K. was supported by NIAID grant AI54338. I.M.O. was supported by NIAID grant AI092002.
We acknowledge the generous support of Paul van Helden, the MRC Centre for Tuberculosis Research, and the Stellenbosch University node of the Centre of Excellence for Biomedical TB Research.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02722-14.
REFERENCES
- 1.McGuire AM, Weiner B, Park ST, Wapinski I, Raman S, Dolganov G, Peterson M, Riley R, Zucker J, Abeel T, White J, Sisk P, Stolte C, Koehrsen M, Yamamoto RT, Iacobelli-Martinez M, Kidd MJ, Maer AM, Schoolnik GK, Regev A, Galagan J. 2012. Comparative analysis of Mycobacterium and related Actinomycetes yields insight into the evolution of Mycobacterium tuberculosis pathogenesis. BMC Genomics 13:120. doi: 10.1186/1471-2164-13-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams M, Mizrahi V, Kana BD. 2014. Molybdenum cofactor: a key component of Mycobacterium tuberculosis pathogenesis? Crit Rev Microbiol 40:18–29. doi: 10.3109/1040841X.2012.749211. [DOI] [PubMed] [Google Scholar]
- 3.Williams MJ, Kana BD, Mizrahi V. 2011. Functional analysis of molybdopterin biosynthesis in mycobacteria identifies a fused molybdopterin synthase in Mycobacterium tuberculosis. J Bacteriol 193:98–106. doi: 10.1128/JB.00774-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi T, Xie J. 2011. Molybdenum enzymes and molybdenum cofactor in mycobacteria. J Cell Biochem 112:2721–2728. doi: 10.1002/jcb.23233. [DOI] [PubMed] [Google Scholar]
- 5.Brodin P, Poquet Y, Levillain F, Peguillet I, Larrouy-Maumus G, Gilleron M, Ewann F, Christophe T, Fenistein D, Jang J, Jang M-S, Park S-J, Rauzier J, Carralot J-P, Shrimpton R, Genovesio A, Gonzalo-Asensio JA, Puzo G, Martin C, Brosch R, Stewart GR, Gicquel B, Neyrolles O. 2010. High content phenotypic cell-based visual screen identifies Mycobacterium tuberculosis acyltrehalose-containing glycolipids involved in phagosome remodeling. PLoS Pathog 6:e1001100. doi: 10.1371/journal.ppat.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacGurn JA, Cox JS. 2007. A genetic screen for Mycobacterium tuberculosis mutants defective for phagosome maturation arrest identifies components of the ESX-1 secretion system. Infect Immun 75:2668–2678. doi: 10.1128/IAI.01872-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosas-Magallanes V, Stadthagen-Gomez G, Rauzier J, Barreiro LB, Tailleux L, Boudou F, Griffin R, Nigou J, Jackson M, Gicquel B, Neyrolles O. 2007. Signature-tagged transposon mutagenesis identifies novel Mycobacterium tuberculosis genes involved in the parasitism of human macrophages. Infect Immun 75:504–507. doi: 10.1128/IAI.00058-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camacho LR, Ensergueix D, Perez E, Gicquel B, Guilhot C. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol Microbiol 34:257–267. doi: 10.1046/j.1365-2958.1999.01593.x. [DOI] [PubMed] [Google Scholar]
- 9.Dutta NK, Mehra S, Didier PJ, Roy CJ, Doyle LA, Alvarez X, Ratterree M, Be NA, Lamichhane G, Jain SK, Lacey MR, Lackner AA, Kaushal D. 2010. Genetic requirements for the survival of tubercle bacilli in primates. J Infect Dis 201:1743–1752. doi: 10.1086/652497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mestre O, Hurtado-Ortiz R, Dos Vultos T, Namouchi A, Cimino M, Pimentel M, Neyrolles O, Gicquel B. 2013. High throughput phenotypic selection of Mycobacterium tuberculosis mutants with impaired resistance to reactive oxygen species identifies genes important for intracellular growth. PLoS One 8:e53486. doi: 10.1371/journal.pone.0053486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becq J, Gutierrez MC, Rosas-Magallanes V, Rauzier J, Gicquel B, Neyrolles O, Deschavanne P. 2007. Contribution of horizontally acquired genomic islands to the evolution of the tubercle bacilli. Mol Biol Evol 24:1861–1871. doi: 10.1093/molbev/msm111. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Behr MA. 2014. Building a better bacillus: the emergence of Mycobacterium tuberculosis. Front Microbiol 5:139. doi: 10.3389/fmicb.2014.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hille R. 1996. The mononuclear molybdenum enzymes. Chem Rev 96:2757–2816. doi: 10.1021/cr950061t. [DOI] [PubMed] [Google Scholar]
- 14.Mendel RR. 2013. The molybdenum cofactor. J Biol Chem 288:13165–13172. doi: 10.1074/jbc.R113.455311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iobbi-Nivol C, Leimkuhler S. 2013. Molybdenum enzymes, their maturation and molybdenum cofactor biosynthesis in Escherichia coli. Biochim Biophys Acta 1827:1086–1101. doi: 10.1016/j.bbabio.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Schwarz G, Mendel RR, Ribbe MW. 2009. Molybdenum cofactors, enzymes and pathways. Nature 460:839–847. doi: 10.1038/nature08302. [DOI] [PubMed] [Google Scholar]
- 17.Neumann M, Mittelstädt G, Seduk F, Iobbi-Nivol C, Leimkühler S. 2009. MocA is a specific cytidylyltransferase involved in molybdopterin cytosine dinucleotide biosynthesis in Escherichia coli. J Biol Chem 284:21891–21898. doi: 10.1074/jbc.M109.008565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 19.Reschke S, Sigfridsson KG, Kaufmann P, Leidel N, Horn S, Gast K, Schulzke C, Haumann M, Leimkuhler S. 2013. Identification of a bis-molybdopterin intermediate in molybdenum cofactor biosynthesis in Escherichia coli. J Biol Chem 288:29736–29745. doi: 10.1074/jbc.M113.497453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aly S, Wagner K, Keller C, Malm S, Malzan A, Brandau S, Bange FC, Ehlers S. 2006. Oxygen status of lung granulomas in Mycobacterium tuberculosis infected mice. J Pathol 210:298–305. doi: 10.1002/path.2055. [DOI] [PubMed] [Google Scholar]
- 21.Tan MP, Sequeira P, Lin WW, Phong WY, Cliff P, Ng SH, Lee BH, Camacho L, Schnappinger D, Ehrt S, Dick T, Pethe K, Alonso S. 2010. Nitrate respiration protects hypoxic Mycobacterium tuberculosis against acid- and reactive nitrogen species stresses. PLoS One 5:e13356. doi: 10.1371/journal.pone.0013356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malm S, Tiffert Y, Micklinghoff J, Schultze S, Joost I, Weber I, Horst S, Ackermann B, Schmidt M, Wohlleben W, Ehlers S, Geffers R, Reuther J, Bange F-C. 2009. The roles of the nitrate reductase NarGHJI, the nitrite reductase NirBD and the response regulator GlnR in nitrate assimilation of Mycobacterium tuberculosis. Microbiology 155:1332–1339. doi: 10.1099/mic.0.023275-0. [DOI] [PubMed] [Google Scholar]
- 23.Fritz C, Maass S, Kreft A, Bange F-C. 2002. Dependence of Mycobacterium bovis BCG on anaerobic nitrate reductase for persistence is tissue specific. Infect Immun 70:286–291. doi: 10.1128/IAI.70.1.286-291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber I, Fritz C, Ruttkowski S, Kreft A, Bange FC. 2000. Anaerobic nitrate reductase (narGHJI) activity of Mycobacterium bovis BCG in vitro and its contribution to virulence in immunodeficient mice. Mol Microbiol 35:1017–1025. doi: 10.1046/j.1365-2958.2000.01794.x. [DOI] [PubMed] [Google Scholar]
- 25.Sohaskey CD, Modesti L. 2009. Differences in nitrate reduction between Mycobacterium tuberculosis and Mycobacterium bovis are due to differential expression of both narGHJI and narK2. FEMS Microbiol Lett 290:129–134. doi: 10.1111/j.1574-6968.2008.01424.x. [DOI] [PubMed] [Google Scholar]
- 26.Pierson DE, Campbell A. 1990. Cloning and nucleotide sequence of bisC, the structural gene for biotin sulfoxide reductase in Escherichia coli. J Bacteriol 172:2194–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woong Park S, Klotzsche M, Wilson DJ, Boshoff HI, Eoh H, Manjunatha U, Blumenthal A, Rhee K, Barry CE III, Aldrich CC, Ehrt S, Schnappinger D. 2011. Evaluating the sensitivity of Mycobacterium tuberculosis to biotin deprivation using regulated gene expression. PLoS Pathog 7:e1002264. doi: 10.1371/journal.ppat.1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Velmurugan K, Chen B, Miller JL, Azogue S, Gurses S, Hsu T, Glickman M, Jacobs WR Jr, Porcelli SA, Briken V. 2007. Mycobacterium tuberculosis nuoG is a virulence gene that inhibits apoptosis of infected host cells. PLoS Pathog 3:e110. doi: 10.1371/journal.ppat.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blomgran R, Desvignes L, Briken V, Ernst JD. 2012. Mycobacterium tuberculosis inhibits neutrophil apoptosis, leading to delayed activation of naive CD4 T cells. Cell Host Microbe 11:81–90. doi: 10.1016/j.chom.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parish T, Stoker NG. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969–1975. [DOI] [PubMed] [Google Scholar]
- 31.Manca C, Paul S, Barry CE III, Freedman VH, Kaplan G. 1999. Mycobacterium tuberculosis catalase and peroxidase activities and resistance to oxidative killing in human monocytes in vitro. Infect Immun 67:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Via LE, Lin PL, Ray SM, Carrillo J, Allen SS, Eum SY, Taylor K, Klein E, Manjunatha U, Gonzales J, Lee EG, Park SK, Raleigh JA, Cho SN, McMurray DN, Flynn JL, Barry CE. 2008. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun 76:2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kruh NA, Troudt J, Izzo A, Prenni J, Dobos KM. 2010. Portrait of a pathogen: the Mycobacterium tuberculosis proteome in vivo. PLoS One 5:e13938. doi: 10.1371/journal.pone.0013938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenaerts AJ, Hoff D, Aly S, Ehlers S, Andries K, Cantarero L, Orme IM, Basaraba RJ. 2007. Location of persisting mycobacteria in a guinea pig model of tuberculosis revealed by r207910. Antimicrob Agents Chemother 51:3338–3345. doi: 10.1128/AAC.00276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rustad TR, Sherrid AM, Minch KJ, Sherman DR. 2009. Hypoxia: a window into Mycobacterium tuberculosis latency. Cell Microbiol 11:1151–1159. doi: 10.1111/j.1462-5822.2009.01325.x. [DOI] [PubMed] [Google Scholar]
- 36.Chim N, Johnson PM, Goulding CW. 2014. Insights into redox sensing metalloproteins in Mycobacterium tuberculosis. J Inorg Biochem 133:118–126. doi: 10.1016/j.jinorgbio.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sohaskey CD, Wayne LG. 2003. Role of narK2X and narGHJI in hypoxic upregulation of nitrate reduction by Mycobacterium tuberculosis. J Bacteriol 185:7247–7256. doi: 10.1128/JB.185.24.7247-7256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jung JY, Madan-Lala R, Georgieva M, Rengarajan J, Sohaskey CD, Bange FC, Robinson CM. 2013. The intracellular environment of human macrophages that produce nitric oxide promotes growth of mycobacteria. Infect Immun 81:3198–3209. doi: 10.1128/IAI.00611-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cunningham-Bussel A, Zhang T, Nathan CF. 2013. Nitrite produced by Mycobacterium tuberculosis in human macrophages in physiologic oxygen impacts bacterial ATP consumption and gene expression. Proc Natl Acad Sci U S A 110:E4256–E4265. doi: 10.1073/pnas.1316894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cunningham-Bussel A, Bange FC, Nathan CF. 2013. Nitrite impacts the survival of Mycobacterium tuberculosis in response to isoniazid and hydrogen peroxide. Microbiologyopen 2:901–911. doi: 10.1002/mbo3.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ezraty B, Bos J, Barras F, Aussel L. 2005. Methionine sulfoxide reduction and assimilation in Escherichia coli: new role for the biotin sulfoxide reductase BisC. J Bacteriol 187:231–237. doi: 10.1128/JB.187.1.231-237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Denkel LA, Rhen M, Bange FC. 2013. Biotin sulfoxide reductase contributes to oxidative stress tolerance and virulence in Salmonella enterica serovar Typhimurium. Microbiology 159:1447–1458. doi: 10.1099/mic.0.067256-0. [DOI] [PubMed] [Google Scholar]
- 43.Jain SK, Hernandez-Abanto SM, Cheng QJ, Singh P, Ly LH, Klinkenberg LG, Morrison NE, Converse PJ, Nuermberger E, Grosset J, McMurray DN, Karakousis PC, Lamichhane G, Bishai WR. 2007. Accelerated detection of Mycobacterium tuberculosis genes essential for bacterial survival in guinea pigs, compared with mice. J Infect Dis 195:1634–1642. doi: 10.1086/517526. [DOI] [PubMed] [Google Scholar]
- 44.Young D. 2009. Animal models of tuberculosis. Eur J Immunol 39:2011–2014. doi: 10.1002/eji.200939542. [DOI] [PubMed] [Google Scholar]
- 45.Voss M, Nimtz M, Leimkühler S. 2011. Elucidation of the dual role of mycobacterial MoeZR in molybdenum cofactor biosynthesis and cysteine biosynthesis. PLoS One 6:e28170. doi: 10.1371/journal.pone.0028170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Homolka S, Niemann S, Russell DG, Rohde KH. 2010. Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog 6:e:1000988. doi: 10.1371/journal.ppat.1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.King GM. 2003. Uptake of carbon monoxide and hydrogen at environmentally relevant concentrations by mycobacteria. Appl Environ Microbiol 69:7266–7272. doi: 10.1128/AEM.69.12.7266-7272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park SW, Hwang EH, Park H, Kim JA, Heo J, Lee KH, Song T, Kim E, Ro YT, Kim SW, Kim YM. 2003. Growth of Mycobacteria on carbon monoxide and methanol. J Bacteriol 185:142–147. doi: 10.1128/JB.185.1.142-147.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park SW, Song T, Kim SY, Kim E, Oh J-I, Eom C-Y, Kim YM. 2007. Carbon monoxide dehydrogenase in mycobacteria possesses a nitric oxide dehydrogenase activity. Biochem Biophys Res Commun 362:449–453. doi: 10.1016/j.bbrc.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 50.Miller JL, Velmurugan K, Cowan MJ, Briken V. 2010. The type I NADH dehydrogenase of Mycobacterium tuberculosis counters phagosomal NOX2 activity to inhibit TNF-alpha-mediated host cell apoptosis. PLoS Pathog 6:e:1000864. doi: 10.1371/journal.ppat.1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fisher MA, Plikaytis BB, Shinnick TM. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J Bacteriol 184:4025–4032. doi: 10.1128/JB.184.14.4025-4032.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.