Abstract
Amyloids are proteins with cross-β-sheet structure that contribute to pathology and inflammation in complex human diseases, including Alzheimer's disease, Parkinson's disease, type II diabetes, and secondary amyloidosis. Bacteria also produce amyloids as a component of their extracellular matrix during biofilm formation. Recently, several human amyloids were shown to activate the NLRP3 inflammasome, leading to the activation of caspase 1 and production of interleukin 1β (IL-1β). In this study, we investigated the activation of the NLRP3 inflammasome by bacterial amyloids using curli fibers, produced by Salmonella enterica serovar Typhimurium and Escherichia coli. Here, we show that curli fibers activate the NLRP3 inflammasome, leading to the production of IL-1β via caspase 1 activation. Investigation of the underlying mechanism revealed that activation of Toll-like receptor 2 (TLR2) by curli fibers is critical in the generation of IL-1β. Interestingly, activation of the NLRP3 inflammasome by curli fibers or by amyloid β of Alzheimer's disease does not cause cell death in macrophages. Overall, these data identify a cross talk between TLR2 and NLRP3 in response to the bacterial amyloid curli and generation of IL-1β as a product of this interaction.
INTRODUCTION
Amyloid proteins are produced by both bacteria and humans. In humans, more than 60 amyloidogenic proteins are produced throughout the body (1). Amyloids accumulate, forming deposits, during several complex diseases, such as Alzheimer's disease (AD), Parkinson's disease, type II diabetes, and secondary amyloidosis. Although it was initially thought that amyloids were only misfolded proteins causing disease pathology, it is becoming more apparent that the proteins have a function in the human body (2, 3). For instance, Pmel17, involved in melanin production, prevents melanocyte cytotoxicity (4–6), and secretory hormones in the endocrine system are stored in a cross-beta-sheet-rich structure in secretory granules (7). Furthermore, it has recently been proposed that the amyloid β peptide, found in the senile plaques of Alzheimer's disease patients, binds specific DNA regions and participates in gene regulation (8).
Bacteria produce amyloids as a component of their extracellular matrix (ECM) to build multicellular communities termed biofilms (9). Biofilms are characterized by their resistant nature in response to environmental insults, including chemical treatments, antibiotics, and the immune system (10). It is thought that the amyloids act as a shield to protect bacteria in biofilms due to their highly resistant nature against chemicals and proteolytic enzymes. Although it is estimated that up to 40% of bacterial species produce amyloids in their biofilms (11), most of these proteins remain uncharacterized. Curli fibers, amyloids produced in the biofilms of Escherichia coli and Salmonella enterica serovar Typhimurium, are the most studied bacterial amyloid to date. Curli fibers are encoded by the csg gene cluster formed by two operons, csgBAC and csgDEFG (12–14). CsgA, the major subunit of the curli fiber, requires CsgB for nucleation into fibers around the bacterial cell. CsgA contains 5 imperfect repeats that are important for β-sheet formation in the amyloid. Similarly, CsgB contains repeats that drive the β-sheet structure required for nucleation and anchoring of the curli fibers to the cell outer membrane (9, 15–17). Curli fibers are detected by the immune system during infections, including urinary tract infections, sepsis, and gastrointestinal inflammation caused by E. coli or S. Typhimurium, leading to the expression of important cytokines and chemokines (18–21).
Although bacterial and human amyloids do not share primary amino acid sequence homology, all amyloids are characterized by their conserved cross-β-sheet quaternary structure. Recently, we discovered that curli fibers are recognized by Toll-like receptor 2 (TLR2), a pattern recognition receptor of the immune system (20). Intriguingly, human amyloid β (22), islet amyloid polypeptide (IAPP) (23), and serum amyloid A (SAA) (24) are also recognized by TLR2, suggesting that the conserved structure of these proteins serves as a target for the immune surveillance of potential pathogenic proteins. Consistent with this idea, when the amyloid structure of either curli or amyloid β is disrupted by a single-amino-acid substitution or by scrambling the amino acid sequence, these molecules are no longer recognized by TLR2 (25).
Interleukin 1β (IL-1β) is a key cytokine that has been implicated in the pathogenesis of several inflammatory diseases, including bacterial infections (26). Generation of IL-1β has also been implicated in neuroinflammation and Alzheimer's disease (27). IL-1β is produced as a biologically inactive precursor form, pro-IL-1β, via the activation of membrane-bound TLRs. Cleavage of pro-IL-1β to active IL-1β by caspase 1 (IL-1β converting enzyme [ICE]) is controlled by inflammasomes, cytosolic danger signal receptors (28). NLRP3 is the best-characterized inflammasome; it can be activated by a wide range of danger signals and uses apoptosis-associated speck-like protein containing a CARD (ASC) as an adaptor molecule (26). Recently, human amyloid β, islet amyloid polypeptide, and SAA have all been demonstrated to activate the NLRP3 inflammasome, resulting in the production of IL-1β (29–31).
In this study, we investigated whether the bacterial amyloid curli triggers IL-1β production through a mechanism similar to that of known human amyloids. We determined that curli fibers activate caspase 1 via activation of the NLRP3 inflammasome. Furthermore, we show that the recognition of curli fibers by both TLR2 and NLRP3 is required for the generation of IL-1β.
MATERIALS AND METHODS
Bacterial strains.
S. Typhimurium IR715 is a fully virulent, nalidixic acid-resistant strain derived from the ATCC strain 14028 (32). An S. Typhimurium msbB mutant (RPW3) and a fliC fljB mutant (EHW26) were described previously (33). A derivative of IR715 carrying an unmarked deletion of csgBA has been described elsewhere (21). S. Typhimurium expressing green fluorescent protein (GFP) under the control of the curli promoter was obtained by cloning the csgBA promoter from IR715 using primer 5′ GGAATTCGAGACGTGGCATTAACCTGGACAGCACAA3′ and reverse primer 5′ GGGATCCGCTGTCACCCTGGACCTGGTCGTACATAGC 3′. The resulting PCR product was cloned upstream of a gene encoding GFP on plasmid pDW6 (34), kindly provided by Brad Cookson from University of Washington, yielding plasmid pCT125 (PcsgBA::gfp), which was then electroporated into S. Typhimurium IR715. S. Typhimurium IR715 harboring pDW5 (34), which expresses GFP constitutively under the control of the tetA promoter (PtetA::gfp), was also used. Bacterial strains were grown in LB medium with nalidixic acid (50 μg/ml) or carbenicillin (100 μg/ml) where necessary.
Tissue culture media and reagents.
RPMI medium (catalog number 11875085) was purchased from Life Technologies. The Cytotox 96 Non-Radioactive Cytotoxicity Assay kit was purchased from Promega (catalog number G1780). Western blot lysis buffer consisted of 20 mM HEPES (pH 7.5), 150 mM NaCl, 10% glycerol, 1% Triton X-100, and 1 mM EDTA in water. Protease Inhibitor Cocktail Set III, EDTA-Free (Calbiochem; catalog number 539134) was added to the lysis buffer immediately before use. Ultrapure LPS-SM was purchased from Invivogen (catalog number tlrl-smlps). Recombinant human amyloid β 1-42 peptide was purchased from Anaspec (catalog number 24224). The mouse IL-1β ELISA Max Standard kit was purchased from BioLegend (catalog number 432602). The mouse IL-6 Ready-Set-GO! ELISA kit was purchased from eBioscience (catalog number 88-7064). Hexafluoro-2-propanol (HFIP) was purchased from Sigma-Aldrich. The specific caspase 1 inhibitor z-YVAD-fmk was purchased from Bachem (catalog number N-1330). Cytochalasin D (catalog number C2618) and glyburide (catalog number G2539) were both purchased from Sigma-Aldrich and dissolved in dimethyl sulfoxide (DMSO).
Curli purification.
Curli fibers were purified from the S. Typhimurium msbB mutant (RPW3) utilizing the established protocol, as described previously (35). Briefly, contaminating cell macromolecules were removed from sonicated cell extracts by enzymatic digestion and preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Insoluble fimbriae were recovered from the well of the gel.
Mouse strains.
C57BL/6 mice were purchased from Jackson Laboratories. TLR2-deficient (B6.129-TLR2tm1Kir/J) mice were purchased from Jackson Laboratories and bred in the Temple University School of Medicine Animal Facility. Nlrp3−/−, ASC−/−, and Casp1−/− Casp11−/− mice were maintained at the University of Pennsylvania Veterinary School Animal Facilities. All procedures were approved by the Temple University and University of Pennsylvania Institutional Animal Care and Use Committees.
Amyloid β fibrillization.
Amyloid polymerization was conducted as previously described (36). One milligram of human amyloid β 1-42 was dissolved in 400 μl of HFIP. The sample was incubated for 1 h with rocking and vortexed for 1 min. HFIP solution (200 μl) was aliquoted into two separate 1.5-ml Eppendorf tubes, and the HFIP was evaporated using a Speed-Vac system (Sorvall) at room temperature. The dried amyloid β was then dissolved in 22.1 μl of DMSO, and 265 μl of 100 mM HEPES buffer, pH 7.3, was added to the DMSO solution. The amyloid β was allowed to incubate for 7 days at 37°C. During the 7-day incubation, samples were briefly vortexed daily. By day 7, the fibrils appear in the solution as a precipitate. The samples were centrifuged at 15,000 rpm for 10 min, the HEPES buffer was removed, and the samples were washed twice with sterile water. A bicinchoninic acid (BCA) assay (Calbiochem) was conducted to determine the final protein concentration of amyloid β fibrils.
BMDMs.
Bone marrow-derived macrophages (BMDMs) from 6- to 8-week-old C57BL/6, TLR2-deficient, NLRP3-deficient, ASC-deficient, and caspase 1/caspase 11-deficient mice were differentiated as previously described (20). Briefly, femurs of mice were flushed, and a single-cell suspension of the bone marrow was prepared in RPMI medium. The suspension was centrifuged at 1,000 rpm for 10 min to pellet the cells. The cells were then resuspended in bone marrow macrophage medium. The bone marrow macrophage differentiation medium prepared with conditioned L929 cell media was changed on the fourth day of culture. On the seventh day of culture, the cells were seeded into the wells of a 24-well plate at a density of 500,000 cells/well.
The cells were stimulated by dose titration of purified curli fibers (10 to 0.5 μg/ml), lipopolysaccharide (LPS) (50 ng/ml), or fibrillized amyloid β (50 μg/ml). The cells were primed for 3 h with LPS (50 ng/ml) before infection with S. Typhimurium IR715 (multiplicity of infection [MOI], 1:20) grown to exponential phase for maximal inflammasome activation (37) or grown under curli-inducing conditions as described previously (20). When grown under exponential-growth conditions, an overnight starter culture was first grown in LB medium with continuous shaking at 37°C. The bacteria were then diluted 1 to 40 in LB medium containing 300 mM NaCl and grown further under static conditions at 37°C for 3 h. The cells were stimulated for 4 or 24 h, and the supernatants were collected to assay for IL-6 and IL-1β by enzyme-linked immunosorbent assay (ELISA). The cell supernatants and lysates were kept for lactate dehydrogenase (LDH) assay. IL-6 and IL-1β ELISAs were conducted according to the manufacturer's instructions.
For inhibitor experiments, BMDMs were treated with cytochalasin D (2.5 μM), YVAD (20 μM), or glyburide (50 μM) 1 h prior to stimulation; 5 μg/ml of curli fibers was used to stimulate the BMDMs for 24 h, and the supernatants were collected for IL-1β ELISA measurement.
LDH assay.
The Cytotox 96 Non-Radioactive Cytotoxicity Assay (Promega) was completed according to the manufacturer's instructions. Briefly, supernatants from cells stimulated for 24 h were collected to determine LDH release into the supernatant during stimulation. Lysis buffer (10×) was added to the wells to a 1× final concentration in the medium. The cells were incubated for 45 min at 37°C to lyse the cells. The 24-well plate was centrifuged for 4 min at 200 rpm, and medium was then collected for total-LDH determination. Assay buffer (12 ml) was mixed with one vial of assay substrate, and 50 μl of supernatant or 50 μl of lysate was placed in a 96-well plate and mixed with 50 μl of the assay substrate. The plate was incubated for 15 min while protected from light, and 50 μl of Stop Solution was added to each well. The absorbance of the wells was read at 490 nm on a BMG Labtech Polarstar plate reader. Cell death was calculated as the ratio of LDH activity in the supernatant alone over the maximum LDH activity of lysed cells.
RESULTS
Curli fibers induce secretion of IL-1β.
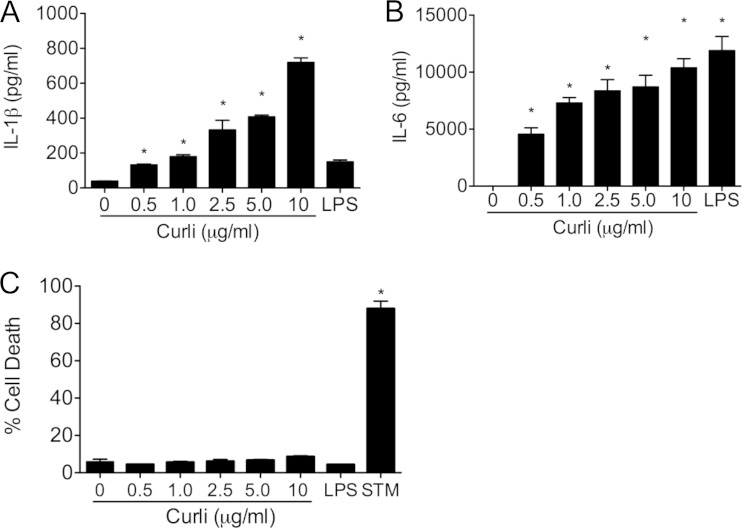
Curli fibers play a pivotal role as a structural component of the ECM of enteric biofilms (38). While curli fibers differ in primary sequence from human amyloids, all amyloids form similar β-sheet quaternary structures. Recent work has shown that the amyloid β found in the plaques present in the brains of AD patients activates the NLRP3 inflammasome, leading to the production of IL-1β (39, 40). Further studies also determined that two other human amyloids, IAPP and SAA, are also recognized by NLRP3 (29–31). In order to determine if the bacterial amyloid curli could also induce secretion of IL-1β, BMDMs from wild-type C57BL/6 mice were stimulated for 24 h with a dose titration (0 to 10 μg/ml) of curli fibers. It is important to note that the curli fibers were purified from an msbB mutant that makes a tetra-acylated LPS that does not activate TLR4. Stimulation of BMDMs with purified curli induced secretion of IL-1β in a dose-dependent fashion. As expected, LPS did not induce secretion of IL-1β (Fig. 1A). Curli also induced IL-6 production by BMDMs, as reported previously (36, 41). LPS caused robust secretion of IL-6, likely through the activation of TLR4 (Fig. 1B).
FIG 1.
Purified curli fibers elicit IL-1β secretion in macrophages. (A and B) C57BL/6 bone marrow-derived macrophages were stimulated with a dose titration (0, 0.5, 1, 2.5, 5, and 10 μg/ml) of purified curli fibers and a single dose of LPS (50 ng/ml). After 24 h, the supernatants were collected, and IL-1β (A) and IL-6 (B) levels were determined by ELISA. (C) An LDH assay was used to determine percent cytotoxicity. S. Typhimurium (STM) infection at an MOI of 20 was utilized as a positive control for the cytotoxicity assay. Shown are average means and standard errors (SE) from three independent experiments. *, P < 0.05.
During infections of macrophages, IL-1β release is concomitant with cell death and the release of LDH into the supernatant (42). To determine whether curli amyloids were causing cytotoxicity and cell death, an LDH assay was conducted on the supernatants and cell lysates from the BMDMs stimulated with the dose titration of purified curli fibers. Interestingly, while infection of BMDMs with S. Typhimurium IR715 caused cell death and LDH release, stimulation of BMDMs with curli fibers failed to cause cell death, even though IL-1β was released after 24 h of stimulation, suggesting that curli fibers are not cytotoxic to the cells even at the highest concentration of 10 μg/ml. LPS was used as a negative control and did not induce any cell death (Fig. 1C). These data provided evidence that purified curli fibers trigger IL-1β production without causing cytotoxicity and LDH release.
IL-1β secretion depends on the activation of TLR2.
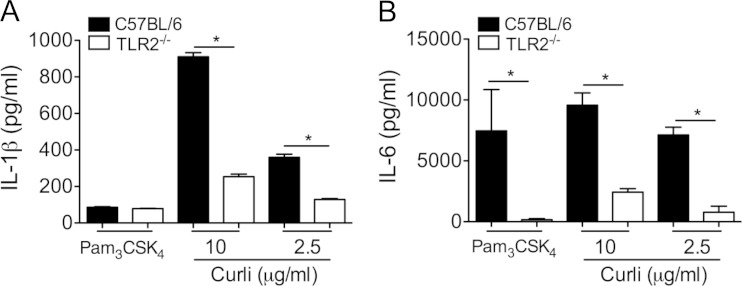
While inflammasome activation provides the necessary enzymatic activity for the cleavage of pro-IL-1β to IL-1β, an additional signal is required for the transcription and translation of pro-IL-1β. One of the commonest signals for the production of pro-IL-1β is TLR activation. In fact, many inflammasome-activating molecules require priming of immune cells with TLR ligands like LPS in order to initiate the production of IL-1β (43, 44). We have shown that curli amyloids, as well as amyloid β, signal through TLR2 to cause the production of cytokines, including IL-6 (25). Since curli activates TLR2, we hypothesized that curli generates the first necessary pro-IL-1β signal for mature IL-1β production through TLR2 activation. In order to test the requirement for TLR2 in IL-1β production in response to curli, C57BL/6 and TLR2-deficient BMDMs were stimulated with 10 μg/ml or 2.5 μg/ml of curli, and IL-1β was measured in the supernatants by ELISA. We determined that wild-type BMDMs produced IL-1β in response to both doses of purified curli fibers. Interestingly, TLR2-deficient BMDMs had reduced production of IL-1β (Fig. 2A). Consistent with the previous reports, IL-6 secretion by BMDMs was significantly abrogated in the absence of TLR2 (Fig. 2B). Intriguingly, treatment of wild-type or TLR2-deficient BMDMs with the synthetic ligand for the TLR2/TLR1 heterocomplex, Pam3CSK4, did not result in the production of IL-1β (Fig. 2A). In contrast the BMDMs produced high levels of IL-6 in a TLR2-dependent manner (Fig. 2B). Collectively, these data suggest that the loss of TLR activation in TLR2-deficient BMDMs impedes the production of pro-IL-1β, resulting in lower levels of IL-1β in response to curli fibers. Furthermore, curli activates the inflammasome pathways, leading to the generation of mature IL-1β.
FIG 2.
Curli induces IL-1β and IL-6 secretion in a TLR2-dependent manner. C57BL/6 and TLR2-deficient macrophages were stimulated with Pam3CSK4 (100 ng/ml) and purified curli fibers (2.5 and 10 μg/ml). Twenty-four hours after stimulation, the supernatants were collected and assayed for IL-1β (A) and IL-6 (B) production by ELISA. Shown are average means and SE from three independent experiments. *, P < 0.05.
Curli fibers activate the NLRP3 inflammasome.
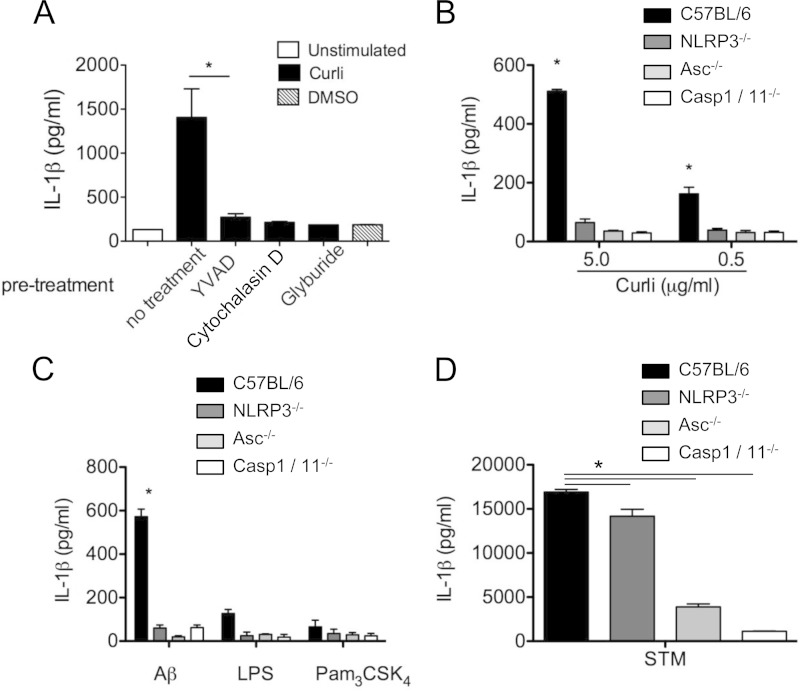
Caspase 1 is a cysteine protease that enzymatically cleaves pro-IL-1β to make mature IL-1β, which becomes active through the formation of a protein complex called the inflammasome. The inflammasome complex recognizes conserved microbial molecular patterns in the cytosol. Both the NLRC4 and the NLRP3 inflammasomes are activated in response to S. Typhimurium infection. Detection of flagellin during S. Typhimurium infection leads to the activation of caspase 1, secretion of IL-1β, and macrophage death via the NLRC4 inflammasome (45). Recently, several human amyloids have been shown to activate NLRP3. Since the recognition of amyloids by TLR2 is dependent on their conserved quaternary structure, we hypothesized that curli amyloids would also activate the NLRP3 inflammasome. Initially, we tested our hypothesis by utilizing inhibitors of NLRP3 inflammasome pathways on BMDMs from wild-type mice. We pretreated BMDMs with the specific NLRP3 inhibitor glyburide (50 μM) or the caspase 1 inhibitor YVAD (20 μM) for 1 h prior to stimulation. The BMDMs were stimulated with 5 μg/ml of curli fibers for 24 h. We found that IL-1β secretion was strongly induced by the purified curli fibers. Furthermore, each of the inhibitors caused inhibition of IL-1β secretion. This evidence pointed to NLRP3 and caspase 1 dependence for curli-induced secretion of IL-1β. Cytochalasin D, an inhibitor of phagocytosis, also blocked IL-1β secretion in BMDMs treated with curli, suggesting that internalization of curli fibers is essential for the induction of IL-1β secretion by BMDMs (Fig. 3A).
FIG 3.
IL-1β secretion in response to curli and amyloid β is dependent on caspase 1 activation. (A) C57BL/6 macrophages were pretreated with cytochalasin D (2.5 μM), YVAD (20 μM), or glyburide (50 μM) 1 h prior to stimulation. The cells were then stimulated with 5 μg/ml of curli or DMSO; 24 h poststimulation, the supernatants were collected and assayed by ELISA for IL-1β secretion. (B to D) C57BL/6, NLRP3-deficient, ASC-deficient, and caspase 1 (Casp1)-deficient bone marrow-derived macrophages were stimulated with purified curli (0.5 or 5.0 μg/ml) (B); polymerized recombinant amyloid β 1-42 (50 μg/ml) (Aβ), LPS (50 ng/ml), and Pam3CSK4 (100 ng/ml) (C); or S. Typhimurium infection at an MOI of 20 (D). The supernatants were collected 24 h after stimulation and assayed for IL-1β by ELISA. Shown are average means and SE from three independent experiments. *, P < 0.05.
In order to confirm our findings that curli induces IL-1β via the activation of NLRP3 and caspase 1, we tested BMDMs from C57BL/6 wild-type mice, as well as NLRP3-, ASC-, and caspase 1/caspase 11-deficient mice. These BMDMs were treated with a high dose (5.0 μg/ml) and a low dose (0.5 μg/ml) of purified curli fibers, fibrillar amyloid β (50 μg/ml), and LPS (50 ng/ml) for 24 h. As a positive control, the BMDMs were also infected with S. Typhimurium. As expected, both the low and high doses of purified curli fibers triggered IL-1β secretion in wild-type BMDMs (Fig. 3B). Amyloid β fibers induced IL-1β in wild-type BMDMs, while LPS failed to induce IL-1β, as expected (Fig. 3C). NLRP3-, ASC-, and caspase 1/caspase 11-deficient BMDMs failed to secrete IL-1β in response to purified curli fibers or amyloid β fibers (Fig. 3B and C). S. Typhimurium infection eliciting of IL-1β was partially dependent on NLRP3, as flagellin activates the NLRC4 inflammasome for IL-1β secretion (46). As expected, IL-1β production was significantly abrogated in both ASC- and caspase 1/caspase 11-deficient BMDMs 24 h postinfection by S. Typhimurium (Fig. 3D).
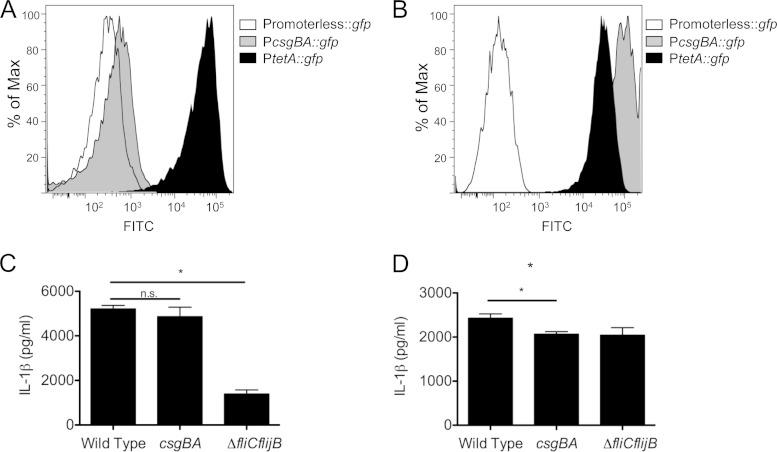
In order to determine whether curli fibers are expressed in S. Typhimurium grown under conditions optimal for inflammasome activation, we used an S. Typhimurium strain carrying a plasmid in which the csgBA promoter drives expression of a GFP reporter construct (PcsgBA::gfp). S. Typhimurium strains carrying a plasmid that constitutively expresses GFP via the tetA promoter (PtetA::gfp) or a promoterless gfp were used as positive and negative controls, respectively. Expression of gfp was determined by flow cytometry. When grown under conditions optimal for inflammasome activation, curli fibers were expressed by only a small fraction of the bacterial population (Fig. 4A). However, when the strain was grown under conditions optimal for curli expression, gfp expression was detectable in the entire population by flow cytometry (Fig. 4B). When BMDMs were infected with wild-type S. Typhimurium, its curli mutant (csgBA), or the flagellin mutant (fliC fljB) grown under conditions optimal for inflammasome activation (Fig. 4C) or curli expression (Fig. 4D) for 4 h, the curli mutant was significantly attenuated in its ability to elicit IL-1β only when the bacterial strains were grown under curli-inducing conditions (Fig. 4D). There was no significant difference in the IL-1β produced by the wild-type S. Typhimurium and the curli mutant grown under conditions optimal for inflammasome activation, while the flagellin mutant was severely attenuated in its ability to elicit IL-1β (Fig. 4C).
FIG 4.
S. Typhimurium curli fibers lead to IL-1β production in BMDMs. (A and B) Curli expression was determined in S. Typhimurium bacteria that were grown under conditions optimal for inflammasome activation (A) or curli expression (B) using S. Typhimurium carrying PcsgBA::gfp on a plasmid. S. Typhimurium carrying a plasmid containing promoterless gfp was used as a negative control. S. Typhimurium carrying a plasmid that constitutively expresses GFP via PtetA::gfp was used as a positive control. (C and D) C57BL/6 macrophages pretreated with LPS (50 ng/ml) were infected at an MOI of 20 with S. Typhimurium grown under conditions that were optimal for either inflammasome activation (C) or curli expression (D). The supernatants were collected 4 h after infection and assayed by ELISA for IL-1β. Shown are average means and SE from three independent experiments. *, P < 0.05; n.s., not significant.
NLRP3 activation by amyloids does not cause cytotoxicity.
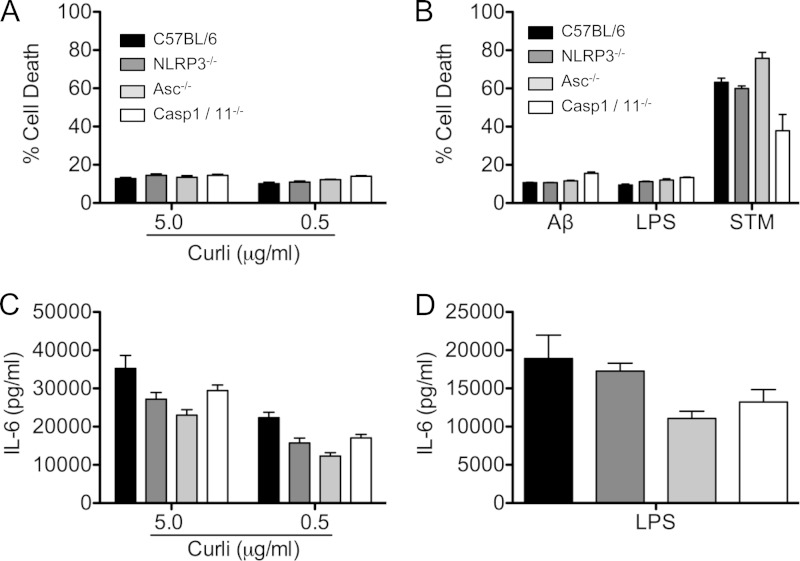
The process of pyroptosis leads to the release of the powerful proinflammatory cytokine IL-1β. In this process, cell death occurs and LDH is released through the permeable cell membrane, which has lost its integrity. Activation of inflammasomes can lead to pyroptosis via caspase 1 activation (46, 47). To determine whether the bacterial amyloid, curli, elicits pyroptosis resulting from NLRP3 activation, BMDMs from wild-type, NLRP3-deficient, ASC-deficient, and caspase 1/caspase 11-deficient mice were treated with curli at 5.0- and 0.5-μg/ml concentrations. While these concentrations of curli did induce IL-1β secretion from BMDMs (Fig. 3B), no cytotoxicity was detected above baseline in any of the macrophage genotypes using an LDH assay (Fig. 5A). Similarly, amyloid β also failed to produce any cytotoxicity in the BMDMs. As expected, LPS did not cause any cytotoxicity, and infection with S. Typhimurium IR715 caused over 50% cytotoxicity in the BMDMs from all mouse genotypes (Fig. 5B). NLRP3, ASC, and caspase 1 deficiency did not cause any significant differences in IL-6 production in response to curli or LPS (Fig. 5C and D).
FIG 5.
Curli fibers do not trigger cell death. (A and B) C57BL/6, NLRP3-deficient, ASC-deficient, and caspase 1-deficient bone marrow-derived macrophages were stimulated with purified curli (0.5 or 5.0 μg/ml) (A) or polymerized recombinant amyloid β 1-42 (50 μg/ml), LPS (50 ng/ml), or S. Typhimurium infection at an MOI of 20 (B). Cell death was determined by LDH assay. (C and D) IL-6 secretion was determined by ELISA. Shown are average means and SE from three independent experiments.
DISCUSSION
A variety of structurally different molecules, including bacterial toxins, ATP, and crystals, activate the NLRP3 inflammasome (28, 44). Several insoluble human proteins, which form amyloid fibers found in various complex diseases, have also been shown to activate the NLRP3 inflammasome. They include amyloid β, SAA, and IAPP (29–31, 40). One of the major hallmarks of Alzheimer's disease is the accumulation of amyloid β in the brains of patients. Prolonged exposure to amyloid β leads to persistent activation of microglia. IL-1β production and caspase 1 activation are observed in diseased tissues and in microglia exposed to amyloid β (48, 49). Intriguingly, loss of NLRP3 or caspase 1 in mouse models of Alzheimer's disease resulted in the protection of mice from disease symptoms, including spatial memory (39). Subsequent studies showed that amyloid β indeed activates NLRP3 and caspase 1, leading to the production inflammatory cytokines, including IL-1β (40).
Here, we show that curli fibers, the insoluble amyloid fibers found in the ECMs of E. coli and S. Typhimurium biofilms, also elicit IL-1β production in macrophages (Fig. 1). Similar to human amyloids, we determined that the IL-1β production elicited by curli fibers was also mediated by the NLRP3 inflammasome, as macrophages lacking the components of the inflammasome, NLRP3 or ASC, failed to produce IL-1β (Fig. 3B). This was dependent on the activation of caspase 1, since macrophages treated with a caspase 1 inhibitor, YVAD, or macrophages lacking caspase 1 and caspase 11 also failed to respond to curli fibers (Fig. 3A and B). Consistent with previous findings, amyloid β also triggered IL-1β production through the NLRP3 inflammasome (Fig. 3C). We used S. Typhimurium as a control to show the activation of inflammasome complexes. We determined that S. Typhimurium activated NLRP3. Although IL-1β production was significantly reduced in NLRP3-deficient macrophages, large quantities of IL-1β were still produced by these macrophages (Fig. 3D). The flagellin mutant was severely attenuated in its ability to elicit IL-1β production (Fig. 4C). Since flagellin activates the NLRC4 inflammasome (45), we think that the large quantities of IL-1β in NLRP3-deficient macrophages were due to the activation of the NLRC4 inflammasome by flagellin. Curli fibers expressed by S. Typhimurium elicited IL-1β production only when the strains were grown under curli-inducing conditions (Fig. 4D). Consistent with our findings, a recent study reported that the NLRP3 inflammasome was activated by the mitochondrial superoxide induced during S. Typhimurium infection of BMDMs (50). Under curli-inducing conditions, the flagellin mutant did not have a profound effect on IL-1β production (Fig. 4D).
It is interesting that the activation of the NLRP3 inflammasome with either curli or amyloid β did not result in cell death in macrophages (Fig. 1C and 5A and B). Although a large body of evidence points to the toxic effect of amyloid β on neuronal cells, macrophages seem to be resistant to the toxic nature of amyloid β. Amyloid β-mediated neuronal cell death has been linked to the generation of reactive oxygen species, as well as caspase 2 (51, 52). Interestingly, a recent study showed that when neuronal cells were cultured with caspase 1-deficient microglial cells, amyloid β did not cause neuronal cell death. However, caspase 1-deficient cells failed to upregulate chemokines, suggesting that the activation of the NLRP3 inflammasome contributes to the secondary chemotactic and neurotoxic effects of amyloid β mediated by microglia. These findings suggest that the activation of the NLRP3 inflammasome by bacterial amyloids in macrophages could have profound effects on surrounding immune cells.
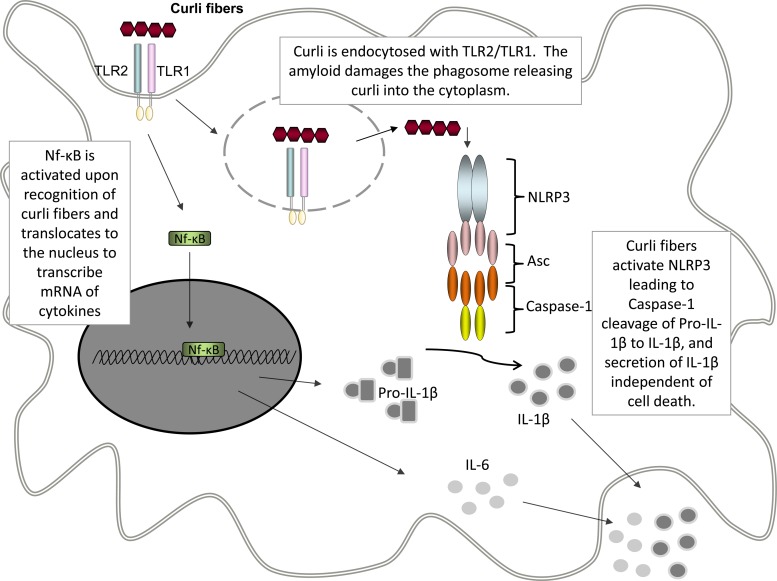
The immune system recognizes curli fibers through a mechanism similar to that observed with human amyloid fibers. The fibrillar amyloid structure of both curli and human amyloids activates the TLR2/TLR1 receptor complex, leading to the activation of the immune system (22, 25). Here, we have demonstrated that macrophages deficient in TLR2 failed to produce IL-1β (Fig. 2). In addition, treatment of macrophages with cytochalasin D, an inhibitor of phagocytosis, disrupts IL-1β production (Fig. 3A). These results indicate that internalization upon TLR2 activation by curli is critical in NLRP3 inflammasome activation. It has previously been reported that amyloid β causes damage to the lysosomal compartment (40). We think that this could be a common mechanism used by amyloid proteins, leading to the fibers' access to the cytosol, where they could be detected by the NLRP3 inflammasome (Fig. 6). It is important to note that the cooperation between TLR2 and NLRP3 is not common to all TLR2 ligands, since the synthetic TLR2/TLR1 ligand Pam3CSK4 does not activate the NLRP3 complex (Fig. 2A). Currently, we are working out the details of how curli fibers access the cytosol and activate the NLRP3 inflammasome.
FIG 6.
Proposed model of curli-induced IL-1β secretion. Curli fibers stimulate the TLR2/TLR1 heterocomplex, which causes activation of NF-κB and expression of proinflammatory cytokines, including pro-IL-1β and IL-6. Upon activation, TLR2/TLR1 are internalized, leading to endocytosis of the curli fibers, where they can activate NLRP3 and caspase 1. Caspase 1 then cleaves pro-IL-1β to IL-1β, which is secreted into the extracellular space.
In addition to enteric pathogens, curli-like amyloid fibers have been identified in several human pathogens, including Staphylococcus aureus and Mycobacterium tuberculosis, and human commensals, such as E. coli Nissle 1917 (53, 54). Therefore, future studies on how bacterial amyloids interact with the immune system will not only shed light on the pathogeneses of several human diseases, but will also provide insight into how the commensal bacteria are detected at the mucosal sites. Interestingly, decreased expression of NLRP3 has been linked to susceptibility to Crohn's disease, and NLRP3 has been shown to participate in the regulation of the epithelial barrier (55, 56). Recently, it was demonstrated that the detection of curli fibers in the gastrointestinal mucosa leads to reinforcement of the epithelial barrier (57). Since curli fibers are recognized by the receptors that have been implicated in barrier regulation in the gut, TLR2 and NLRP3, the detection of microbiota-associated amyloids may play a pivotal role in the regulation of the mucosal barrier and intestinal homeostasis. In addition, elucidating the interactions of bacterial amyloids with the immune system and determining the commonalities between human amyloids may provide clues to why bacteria express molecules that are structurally similar to human amyloids present in human diseases.
ACKNOWLEDGMENTS
Work in C.T.'s laboratory was supported by National Institutes of Health, National Institute of Allergy and Infectious Diseases, grants 1R03AI107434 and 1R21AI105370. Work in I.E.B.'s laboratory was supported by National Institutes of Health, National Institute of Allergy and Infectious Diseases, grant 1R21AI105346-01.
REFERENCES
- 1.Mok KH, Pettersson J, Orrenius S, Svanborg C. 2007. HAMLET, protein folding, and tumor cell death. Biochem Biophys Res Commun 354:1–7. doi: 10.1016/j.bbrc.2006.12.167. [DOI] [PubMed] [Google Scholar]
- 2.Otzen D. 2010. Functional amyloid: turning swords into plowshares. Prion 4:256–264. doi: 10.4161/pri.4.4.13676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiti F, Dobson CM. 2006. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 4.Theos AC, Truschel ST, Raposo G, Marks MS. 2005. The Silver locus product Pmel17/gp100/Silv/ME20: controversial in name and in function. Pigment Cell Res 18:322–336. doi: 10.1111/j.1600-0749.2005.00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leonhardt RM, Vigneron N, Rahner C, Van den Eynde BJ, Cresswell P. 2010. Endoplasmic reticulum export, subcellular distribution, and fibril formation by Pmel17 require an intact N-terminal domain junction. J Biol Chem 285:16166–16183. doi: 10.1074/jbc.M109.097725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfefferkorn CM, McGlinchey RP, Lee JC. 2010. Effects of pH on aggregation kinetics of the repeat domain of a functional amyloid, Pmel17. Proc Natl Acad Sci U S A 107:21447–21452. doi: 10.1073/pnas.1006424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, Rissman RA, Singru PS, Nilsson KP, Simon R, Schubert D, Eisenberg D, Rivier J, Sawchenko P, Vale W, Riek R. 2009. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325:328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maloney B, Lahiri DK. 2011. The Alzheimer's amyloid beta-peptide (Abeta) binds a specific DNA Abeta-interacting domain (AbetaID) in the APP, BACE1, and APOE promoters in a sequence-specific manner: characterizing a new regulatory motif. Gene 488:1–12. doi: 10.1016/j.gene.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hufnagel DA, Tukel C, Chapman MR. 2013. Disease to dirt: the biology of microbial amyloids. PLoS Pathog 9:e1003740. doi: 10.1371/journal.ppat.1003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D, Nielsen PH. 2007. Amyloid adhesins are abundant in natural biofilms. Environ Microbiol 9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- 12.Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu Rev Microbiol 60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hung C, Zhou Y, Pinkner JS, Dodson KW, Crowley JR, Heuser J, Chapman MR, Hadjifrangiskou M, Henderson JP, Hultgren SJ. 2013. Escherichia coli biofilms have an organized and complex extracellular matrix structure. mBio 4:e00645–00613. doi: 10.1128/mBio.00645-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammer ND, Schmidt JC, Chapman MR. 2007. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc Natl Acad Sci U S A 104:12494–12499. doi: 10.1073/pnas.0703310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Smith DR, Jones JW, Chapman MR. 2007. In vitro polymerization of a functional Escherichia coli amyloid protein. J Biol Chem 282:3713–3719. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans ML, Chapman MR. 2014. Curli biogenesis: order out of disorder. Biochim Biophys Acta 1844:1551–1558. doi: 10.1016/j.bbamcr.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bian Z, Brauner A, Li Y, Normark S. 2000. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J Infect Dis 181:602–612. doi: 10.1086/315233. [DOI] [PubMed] [Google Scholar]
- 19.Kai-Larsen Y, Luthje P, Chromek M, Peters V, Wang X, Holm A, Kadas L, Hedlund KO, Johansson J, Chapman MR, Jacobson SH, Romling U, Agerberth B, Brauner A. 2010. Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog 6:e1001010. doi: 10.1371/journal.ppat.1001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tukel C, Raffatellu M, Humphries AD, Wilson RP, Andrews-Polymenis HL, Gull T, Figueiredo JF, Wong MH, Michelsen KS, Akcelik M, Adams LG, Baumler AJ. 2005. CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol Microbiol 58:289–304. doi: 10.1111/j.1365-2958.2005.04825.x. [DOI] [PubMed] [Google Scholar]
- 21.Nishimori JH, Newman TN, Oppong GO, Rapsinski GJ, Yen JH, Biesecker SG, Wilson RP, Butler BP, Winter MG, Tsolis RM, Ganea D, Tukel C. 2012. Microbial amyloids induce interleukin 17A (IL-17A) and IL-22 responses via Toll-like receptor 2 activation in the intestinal mucosa. Infect Immun 80:4398–4408. doi: 10.1128/IAI.00911-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S, Liu Y, Hao W, Wolf L, Kiliaan AJ, Penke B, Rube CE, Walter J, Heneka MT, Hartmann T, Menger MD, Fassbender K. 2012. TLR2 is a primary receptor for Alzheimer's amyloid beta peptide to trigger neuroinflammatory activation. J Immunol 188:1098–1107. doi: 10.4049/jimmunol.1101121. [DOI] [PubMed] [Google Scholar]
- 23.Westwell-Roper C, Ehses JA, Verchere BC. 2012. Activation of Toll-like receptor 2 by islet amyloid polypeptide: a trigger for islet inflammation in type 2 diabetes? Can J Diabetes 36(Suppl):S18. doi: 10.1016/j.jcjd.2012.07.076. [DOI] [Google Scholar]
- 24.Cheng N, He R, Tian J, Ye PP, Ye RD. 2008. Cutting edge: TLR2 is a functional receptor for acute-phase serum amyloid A. J Immunol 181:22–26. doi: 10.4049/jimmunol.181.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tukel C, Wilson RP, Nishimori JH, Pezeshki M, Chromy BA, Baumler AJ. 2009. Responses to amyloids of microbial and host origin are mediated through Toll-like receptor 2. Cell Host Microbe 6:45–53. doi: 10.1016/j.chom.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brodsky IE, Monack D. 2009. NLR-mediated control of inflammasome assembly in the host response against bacterial pathogens. Semin Immunol 21:199–207. doi: 10.1016/j.smim.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Shaftel SS, Griffin WS, O'Banion MK. 2008. The role of interleukin-1 in neuroinflammation and Alzheimer disease: an evolving perspective. J Neuroinflammation 5:7. doi: 10.1186/1742-2094-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrilli V, Papin S, Tschopp J. 2005. The inflammasome. Curr Biol 15:R581. doi: 10.1016/j.cub.2005.07.049. [DOI] [PubMed] [Google Scholar]
- 29.Westwell-Roper C, Nackiewicz D, Dan M, Ehses JA. 2014. Toll-like receptors and NLRP3 as central regulators of pancreatic islet inflammation in type 2 diabetes. Immunol Cell Biol 92:314–323. doi: 10.1038/icb.2014.4. [DOI] [PubMed] [Google Scholar]
- 30.Ather JL, Ckless K, Martin R, Foley KL, Suratt BT, Boyson JE, Fitzgerald KA, Flavell RA, Eisenbarth SC, Poynter ME. 2011. Serum amyloid A activates the NLRP3 inflammasome and promotes Th17 allergic asthma in mice. J Immunol 187:64–73. doi: 10.4049/jimmunol.1100500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niemi K, Teirila L, Lappalainen J, Rajamaki K, Baumann MH, Oorni K, Wolff H, Kovanen PT, Matikainen S, Eklund KK. 2011. Serum amyloid A activates the NLRP3 inflammasome via P2X7 receptor and a cathepsin B-sensitive pathway. J Immunol 186:6119–6128. doi: 10.4049/jimmunol.1002843. [DOI] [PubMed] [Google Scholar]
- 32.Stojiljkovic I, Baumler AJ, Heffron F. 1995. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol 177:1357–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raffatellu M, Chessa D, Wilson RP, Dusold R, Rubino S, Baumler AJ. 2005. The Vi capsular antigen of Salmonella enterica serotype Typhi reduces Toll-like receptor-dependent interleukin-8 expression in the intestinal mucosa. Infect Immun 73:3367–3374. doi: 10.1128/IAI.73.6.3367-3374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cummings LA, Wilkerson WD, Bergsbaken T, Cookson BT. 2006. In vivo, fliC expression by Salmonella enterica serovar Typhimurium is heterogeneous, regulated by ClpX, and anatomically restricted. Mol Microbiol 61:795–809. doi: 10.1111/j.1365-2958.2006.05271.x. [DOI] [PubMed] [Google Scholar]
- 35.Collinson SK, Emody L, Muller KH, Trust TJ, Kay WW. 1991. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J Bacteriol 173:4773–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rapsinski GJ, Newman TN, Oppong GO, van Putten JP, Tukel C. 2013. CD14 protein acts as an adaptor molecule for the immune recognition of Salmonella curli fibers. J Biol Chem 288:14178–14188. doi: 10.1074/jbc.M112.447060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee CA, Falkow S. 1990. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci U S A 87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zogaj X, Bokranz W, Nimtz M, Romling U. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect Immun 71:4151–4158. doi: 10.1128/IAI.71.7.4151-4158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heneka MT, Kummer MP, Stutz A, Delekate A, Schwartz S, Vieira-Saecker A, Griep A, Axt D, Remus A, Tzeng TC, Gelpi E, Halle A, Korte M, Latz E, Golenbock DT. 2013. NLRP3 is activated in Alzheimer's disease and contributes to pathology in APP/PS1 mice. Nature 493:674–678. doi: 10.1038/nature11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T, Fitzgerald KA, Latz E, Moore KJ, Golenbock DT. 2008. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol 9:857–865. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tukel C, Nishimori JH, Wilson RP, Winter MG, Keestra AM, van Putten JP, Baumler AJ. 2010. Toll-like receptors 1 and 2 cooperatively mediate immune responses to curli, a common amyloid from enterobacterial biofilms. Cell Microbiol 12:1495–1505. doi: 10.1111/j.1462-5822.2010.01485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Denes A, Lopez-Castejon G, Brough D. 2012. Caspase-1: is IL-1 just the tip of the ICEberg? Cell Death Dis 3:e338. doi: 10.1038/cddis.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moretti J, Blander JM. 2014. Insights into phagocytosis-coupled activation of pattern recognition receptors and inflammasomes. Curr Opin Immunol 26:100–110. doi: 10.1016/j.coi.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Latz E, Xiao TS, Stutz A. 2013. Activation and regulation of the inflammasomes. Nat Rev Immunol 13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Broz P, Newton K, Lamkanfi M, Mariathasan S, Dixit VM, Monack DM. 2010. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J Exp Med 207:1745–1755. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergsbaken T, Fink SL, Cookson BT. 2009. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol 7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ting JP, Willingham SB, Bergstralh DT. 2008. NLRs at the intersection of cell death and immunity. Nat Rev Immunol 8:372–379. doi: 10.1038/nri2296. [DOI] [PubMed] [Google Scholar]
- 48.Akama KT, Van Eldik LJ. 2000. Beta-amyloid stimulation of inducible nitric-oxide synthase in astrocytes is interleukin-1beta- and tumor necrosis factor-alpha (TNFalpha)-dependent, and involves a TNFalpha receptor-associated factor- and NFkappaB-inducing kinase-dependent signaling mechanism. J Biol Chem 275:7918–7924. doi: 10.1074/jbc.275.11.7918. [DOI] [PubMed] [Google Scholar]
- 49.Griffin WS, Stanley LC, Ling C, White L, MacLeod V, Perrot LJ, White CL III, Araoz C. 1989. Brain interleukin 1 and S-100 immunoreactivity are elevated in Down syndrome and Alzheimer disease. Proc Natl Acad Sci U S A 86:7611–7615. doi: 10.1073/pnas.86.19.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wynosky-Dolfi MA, Snyder AG, Philip NH, Doonan PJ, Poffenberger MC, Avizonis D, Zwack EE, Riblett AM, Hu B, Strowig T, Flavell RA, Jones RG, Freedman BD, Brodsky IE. 2014. Oxidative metabolism enables Salmonella evasion of the NLRP3 inflammasome. J Exp Med 211:653–668. doi: 10.1084/jem.20130627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kadowaki H, Nishitoh H, Urano F, Sadamitsu C, Matsuzawa A, Takeda K, Masutani H, Yodoi J, Urano Y, Nagano T, Ichijo H. 2005. Amyloid beta induces neuronal cell death through ROS-mediated ASK1 activation. Cell Death Differ 12:19–24. doi: 10.1038/sj.cdd.4401528. [DOI] [PubMed] [Google Scholar]
- 52.Troy CM, Rabacchi SA, Friedman WJ, Frappier TF, Brown K, Shelanski ML. 2000. Caspase-2 mediates neuronal cell death induced by beta-amyloid. J Neurosci 20:1386–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alteri CJ, Xicohtencatl-Cortes J, Hess S, Caballero-Olin G, Giron JA, Friedman RL. 2007. Mycobacterium tuberculosis produces pili during human infection. Proc Natl Acad Sci U S A 104:5145–5150. doi: 10.1073/pnas.0602304104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwartz K, Syed AK, Stephenson RE, Rickard AH, Boles BR. 2012. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog 8:e1002744. doi: 10.1371/journal.ppat.1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Villani AC, Lemire M, Fortin G, Louis E, Silverberg MS, Collette C, Baba N, Libioulle C, Belaiche J, Bitton A, Gaudet D, Cohen A, Langelier D, Fortin PR, Wither JE, Sarfati M, Rutgeerts P, Rioux JD, Vermeire S, Hudson TJ, Franchimont D. 2009. Common variants in the NLRP3 region contribute to Crohn's disease susceptibility. Nat Genet 41:71–76. doi: 10.1038/ng.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zaki MH, Boyd KL, Vogel P, Kastan MB, Lamkanfi M, Kanneganti TD. 2010. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oppong GO, Rapsinski GJ, Newman TN, Nishimori JH, Biesecker SG, Tukel C. 2013. Epithelial cells augment barrier function via activation of the Toll-like receptor 2/phosphatidylinositol 3-kinase pathway upon recognition of Salmonella enterica serovar Typhimurium curli fibrils in the gut. Infect Immun 81:478–486. doi: 10.1128/IAI.00453-12. [DOI] [PMC free article] [PubMed] [Google Scholar]