Abstract
Chlamydia trachomatis is an obligate intracellular epitheliotropic bacterial pathogen of humans. Infection of the eye can result in trachoma, the leading cause of preventable blindness in the world. The pathophysiology of blinding trachoma is driven by multiple episodes of reinfection of conjunctival epithelial cells, producing an intense chronic inflammatory response resulting in submucosal tissue remodeling and scarring. Recent reports have shown that infection with trachoma organisms lacking the cryptic chlamydial plasmid is highly attenuated in macaque eyes, a relevant experimental model of human trachoma infection. To better understand the molecular basis of plasmid-mediated infection attenuation and the potential modulation of host immunity, we conducted transcriptional profiling of human epithelial cells infected with C. trachomatis plasmid-bearing (A2497) and plasmid-deficient (A2497P−) organisms. Infection of human epithelial cells with either strain increased the expression of host genes coding for proinflammatory (granulocyte-macrophage colony-stimulating factor [GM-CSF], macrophage colony-stimulating factor [MCSF], interleukin-6 [IL-6], IL-8, IL-1α, CXCL1, CXCL2, CXCL3, intercellular adhesion molecule 1 [ICAM1]), chemoattraction (CCL20, CCL5, CXCL10), immune suppression (PD-L1, NFKB1B, TNFAIP3, CGB), apoptosis (CASP9, FAS, IL-24), and cell growth and fibrosis (EGR1 and IL-20) proteins. Statistically significant increases in the levels of expression of many of these genes were found in A2497-infected cells compared to the levels of expression in A2497P−-infected cells. Our findings suggest that the chlamydial plasmid plays a focal role in the host cell inflammatory response to infection and immune avoidance. These results provide new insights into the role of the chlamydial plasmid as a chlamydial virulence factor and its contributions to trachoma pathogenesis.
INTRODUCTION
Chlamydia trachomatis is an obligate epitheliotropic pathogen of mucosal surfaces. Isolates exist as multiple serovariants that cause trachoma and sexually transmitted infections (STIs), both of which are important human health problems globally. Trachoma is caused by serovars A, B, Ba, and C, whereas STIs are caused by serovars D to L3. Trachoma, a neglected tropical disease (1) of the developing world, is the leading cause of preventable infectious blindness; WHO estimates that as many as 2.3 million people are blind or suffer serious vision impairment because of the disease (2). Chlamydial infections of the urogenital mucosae are the most common bacterial cause of sexually transmitted diseases in both industrialized nations and developing countries (3, 4). Chlamydial infection of the female genital tract can result in serious sequelae, such as salpingitis, tubal factor infertility, and ectopic pregnancy (5).
The pathophysiology of chronic chlamydial diseases is unknown but is thought to be the result of persistent or repeated bouts of reinfection that drive chronic inflammatory responses, producing fibrosis and scarring of the conjunctival surface and fallopian tubes. Two general immunologically based hypotheses have been proposed to describe the function of chlamydial reinfection in mediating chronic damaging inflammation: (i) a chlamydial antigen-specific Th1 cell-mediated delayed-type hypersensitivity (DTH) response (6–9) and (ii) an epithelial cellular response to infection (8). In the DTH hypothesis, following reactivation from persistence or reinfection, antigen-specific Th1 cells producing proinflammatory cytokines are recruited to the site of infection with sufficient ferocity and numbers to cause damaging disease sequelae. In contrast, the cellular response theorizes that it is the infected epithelial cell itself, through production of inflammatory cytokines, chemokines, and growth factors, that is responsible for producing the damaging disease sequelae following infection (8). It is likely that both of these mechanisms contribute to chlamydial inflammatory disease, but it is unclear which one might predominate in eliciting damaging pathological immunity. Regardless, what role chlamydial genes play in orchestrating infection-mediated pathology is unknown, particularly in the case of the cellular hypothesis of inflammatory disease.
The chlamydial plasmid has been shown to be an important chlamydial virulence factor in both murine (10, 11) and nonhuman primate (12) models of infection, as plasmid-deficient organisms produce attenuated infections with decreased organism loads, resulting in reduced or no postinfection pathology. The molecular basis for this attenuation is unclear but could involve plasmid-regulated genes (13) that function as Toll-like receptor 2 (14) or tumor necrosis factor alpha (TNF-α) receptor antagonists (15, 16). On the other hand, the basis of plasmid-mediated pathology might be a direct host-pathogen relationship caused by infection with virulent plasmid-bearing organisms.
To our knowledge, no studies have examined the proinflammatory response of cultured human epithelial cells infected with a C. trachomatis plasmid-deficient strain. We hypothesize that if the chlamydial plasmid is playing an important role in inflammatory pathogenesis and the cellular paradigm is a primary contributing factor in this host-pathogen interaction, cells infected with a plasmidless strain might exhibit a reduced ability to secrete proinflammatory cytokines compared to their plasmid-bearing parental strain.
Here, we describe the transcriptome analysis of HeLa host cells infected with either a virulent trachoma plasmid-bearing organism (A2497) or its isogenic plasmid-deficient strain (A2497P−). Our results indicate that, compared to the response of mock-infected cells, both the virulent A2497 and the A2497P− strains elicit a transcriptome proinflammatory cytokine and T cell chemokine response that is similar and comparable to what others have described. However, when A2497 was compared to A2497P−, significant increases in the levels of expression of many inflammation, chemoattraction, cell growth, or fibrosis genes in A2497 relative to the levels of expression in A2497P− were observed. These results support a role, at least in part, for the host cellular response as a source of damaging proinflammatory cytokines and implicate the chlamydial plasmid as a virulence factor in mediating this response.
MATERIALS AND METHODS
Cell culture and chlamydial strains.
Chlamydia trachomatis strains A2497 and A2497P− were previously described as being chromosomally isogenic, with the only genomic difference being the presence (A2497) or absence (A2497P−) of the cryptic plasmid (13). Human epithelial (HeLa 229, ATCC CCL-2.1) cells were grown at 37°C with 5% CO2 in high-glucose-containing Dulbecco's modified Eagle medium (DMEM) (Mediatech, Inc.) supplemented with 10% fetal bovine serum (DMEM-10). Six 6-well plates containing HeLa 229 cell monolayers (1.5 × 106 cells/well) were infected with A2497 or A2497P− (multiplicity of infection [MOI] = 1) or mock infected as previously described (13) in the absence of cycloheximide, with six replicates being used for each infection condition and postinfection (p.i.) harvest time point. Supernatants were aspirated, and the monolayers were harvested by suspension into 1 ml of RLT buffer (Qiagen). RNA and genomic DNA extraction was conducted according to the manufacturer's specifications.
Phase microscopy and glycogen staining.
Monolayers of HeLa 229 cells grown in 24-well tissue culture plates were infected at an MOI of 0.5 with either C. trachomatis A2497 or A2497P−. Chlamydiae were cultured in DMEM-10 without cycloheximide. At 0, 4, 12, 24, 48, and 72 h p.i., cultures were imaged by phase microscopy before and after staining with 200 μl of a 1:10 dilution of Lugol's iodine (5% iodine, 10% potassium iodide in 50% ethanol) in phosphate-buffered saline (PBS).
RNA and DNA extraction and microarray target preparation.
Eighteen 6-well plates were seeded with HeLa 229 cells, which were grown to confluence and infected with either the A2497 or A2497P− strain or mock infected (medium alone). A total of 108 wells representing three infection treatments, six time points, and six biological replicates were assigned to culture plates using a randomized block design to avoid confounding of the results by treatment and time. Samples were processed by the Genomics Unit of the Research Technologies Section at RML, NIAID, NIH, and were assigned to each well as follows. Samples from each time point were allocated on three 6-well plates. The three infection conditions were represented by two biological replicates on each plate. Two replicates on each plate were selected from six available replicates and randomly assigned to a well on the plate. Wells were seeded, infected, and harvested in the same order starting from well A1 on plate 1 through well B3 on plate 18. RNA and genomic DNA were extracted and quality controlled as described previously (13). Mock-infected samples were also used as a way to verify that neighboring culture wells were not cross-contaminated. Four replicate A2497- or A2497P−-infected HeLa 229 cell RNAs and three mock-infected cell RNAs were randomly selected from six replicates for DNA microarray target preparation. DNA microarray target synthesis and quality control were performed as described previously (17).
qPCR genome copy analysis and qRT-PCR array validation.
Quantitative PCR (qPCR) for determination of genome copy numbers was performed as described by Song et al. (13). Quantitative reverse transcription-PCR (qRT-PCR) validation for the human genes CCL5, IL6, IL8 (CXCL8), IFIT2, TNFAIP3, CD274 (PD-L1), and CSF2 was performed as previously described (17), with small differences. The human peptidylprolyl isomerase A gene (PPIA) was used as a reference gene (see Table S3 in the supplemental material) on the basis of the high signal intensity, the low level of change in expression across all conditions, and the uniqueness of the probe set. Thirty-six RNAs representing A2497- and A2497P−-infected and mock-infected samples harvested at 24, 48, and 72 h postinfection were used for qRT-PCR validation. qRT-PCR oligonucleotides for IFIT2 and IL8 genes were purchased from Life Technologies (Carlsbad, CA), and the remainder of the oligonucleotides were purchased from Biosearch Technologies (Petaluma, CA). Due to the low level of expression of PD-L1, qRT-PCR was performed directly against the cDNA for the biological replicates, while for all other genes, a 0.1 dilution of the cDNAs was used. A Pearson correlation analysis between array and qRT-PCR data was performed, and the results are reported when the P values, determined using a t test across time, were greater than 0.5 and less than 0.05. Determination of significance (P ≤ 0.05) for differential expression, determined by qRT-PCR, of the seven genes mentioned above in comparisons of A2497 and A2497P− was performed temporally in GraphPad software using a t test.
Microarray chip processing and analysis were performed essentially as described previously (18), with a few exceptions, as follows. In brief, hybridization, fluidics, and scanning were performed according to standard Affymetrix protocols. Command Console (CC) software (v3.1; Affymetrix) was used to convert the image files to cell intensity data (cel files). All cel files, which represented individual samples, were normalized using the trimmed mean scaling method within the Expression Console (EC; v1.2, Affymetrix) to produce the analyzed cel (chp files) and report files. The cel files were input into Partek Genomics Suite software (Partek, Inc., St. Louis, MO) and quantile normalized to produce a principal components analysis (PCA) graph and dendrogram (data not shown). An analysis of variance (ANOVA) was performed within Partek Genomics Suite software to obtain multiple-test-corrected P values at the 0.05 significance level, using the false discovery rate (FDR) method (19). The P values were combined with fold change values, signal confidence (the level above the background), and call consistency (as a percentage), calculated using custom Excel templates for each comparison of interest. The resulting data were analyzed for the 48-h time point using Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems), generating the networks, functional analyses, and pathway construction.
Microarray data accession number.
The array data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (GEO) database (20) and are accessible through GEO series accession number GSE56168.
RESULTS
Inclusion morphology and glycogen accumulation differences between A2497 and A2497P−.
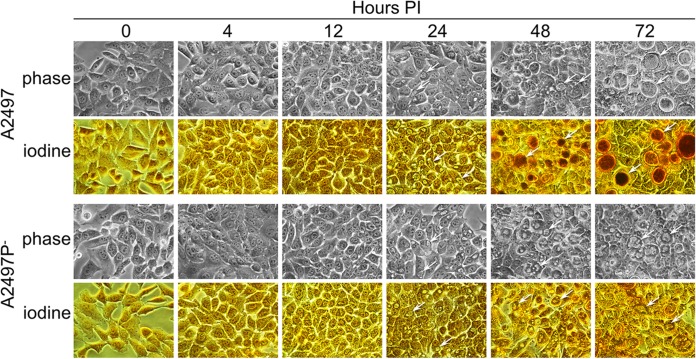
Monolayers of HeLa 229 cells infected with either C. trachomatis A2497 or A2497P− were imaged at 0, 4, 12, 24, 48, and 72 h p.i. by phase microscopy before and after staining with iodine (Fig. 1). No inclusions of either strain were detected by phase microscopy at 4 and 12 h p.i. At 24 h p.i., the inclusions of both the A2497 and A2497P− strains were comparable in size and were glycogen negative. At 48 and 72 h p.i., distinct differences in both inclusion morphology and glycogen staining were evident between the A2497 and A2497P− strains. The A2497 strain produced active inclusions and stained strongly for glycogen, while the A2497P− strain produced abnormal inclusions with a donut-like morphology and failed to stain for glycogen.
FIG 1.
Iodine-stained HeLa 229 cells infected with C. trachomatis A2497 or A2497P−. Chlamydia-infected HeLa 229 cell cultures were photographed unfixed by using phase microscopy at 0, 4, 12, 24, 48, and 72 h p.i. At each time point, supernatants were removed and stained with Lugol's iodine solution in PBS to visualize glycogen accumulation. Arrows, inclusions. Both A2497 and A2497P− inclusions are visible by 24 h p.i. Glycogen (dark brown) is detectable in A2497 inclusions only at 48 and 72 h p.i. The atypical donut inclusion morphology is evident in the A2497P− strain at 48 and 72 h p.i. Magnifications, ×400.
Host transcriptome analysis of A2497- and A2497P−-infected cells compared to mock-infected cells.
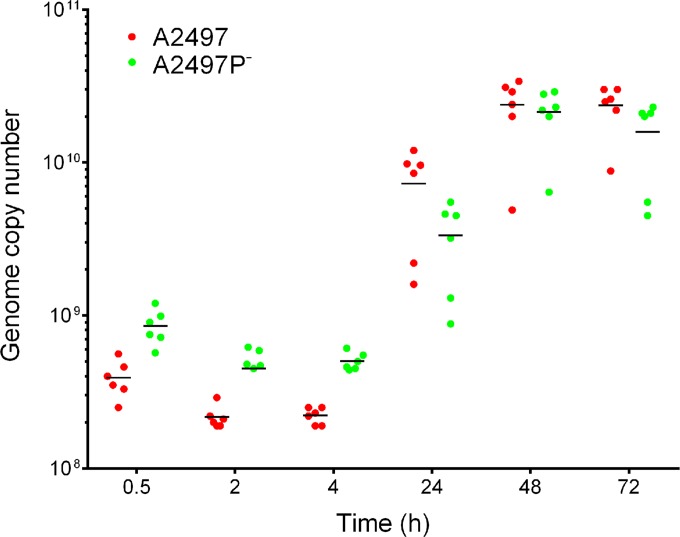
RNAs and DNAs were isolated from infected and uninfected HeLa 229 cells at 30 min, 2 h, 4 h, 24 h, 48 h, and 72 h p.i., and the DNA was analyzed by qPCR to determine chlamydial genomic equivalents of A2497 and A2497P− (Fig. 2). Chlamydial genome copy numbers increased at 24 h p.i. and plateaued for both strains at 48 and 72 h p.i. Figure 2 demonstrates that the genome copy numbers were approximately equivalent over time for both A2497 and A2497P−.
FIG 2.
qPCR analysis of genome copy number during infection with A2497 and A2497P− chlamydiae. Six biological replicates per time point are shown for A2497 and A2497P−. Horizontal lines, average values for the biological replicates per time point p.i.
Following microarray scanning, replicate data sets were combined and expression values were averaged. An ANOVA comparing A2497- and A2497P−-infected cells versus mock-infected controls was performed to obtain multiple-test-corrected P values using the false discovery rate (FDR) method (19). This comparison identified genes exhibiting differential expression levels passing a 0.05 significance level (see Materials and Methods). For the time points 30 min, 2 h, and 4 h p.i., no significant transcriptional difference in host gene expression (P ≤ 0.05) was found between either A2497- or A2497P−-infected cells and uninfected host cells.
At 24 h p.i., 416 (A2497) and 43 (A2497P−) genes demonstrated significant differential expression in infected cells. At 48 h p.i., these numbers increased to 10,347 and 7,175 genes for A2497- and A2497P−-infected cells, respectively. Finally, at 72 h p.i., 12,878 (A2497) and 12,911 (A2497P−) genes were significantly differentially expressed. These results clearly demonstrate that A2497 influences host gene expression more than A2497P− at 24 and 48 h p.i., but by 72 h p.i., both strains affect the expression of a similar number of genes. In total, 19,248 host genes, or 34% of the array, demonstrated significant differential expression (P ≤ 0.05) when A2497- or A2497P−-infected cells were compared to mock-infected cells. These data are presented in Table S1 in the supplemental material.
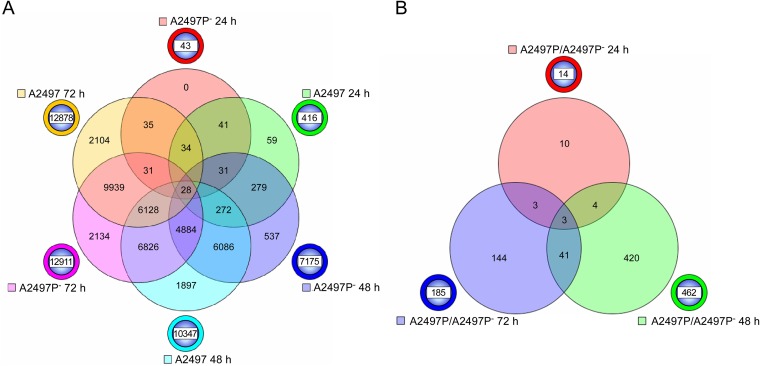
A Venn diagram was constructed to show gene numbers, unique or shared, across the three later time points for comparisons of A2497- or A2497P−-infected cells versus mock-infected cells (Fig. 3A). At 24 h p.i., 41 of the 43 genes (96%) identified in the A2497P−-infected cells were also observed to be included among the 416 genes of the A2497-infected cells. At 48 h p.i., 6,086 genes of the 7,175 genes (86%) expressed by A2497P− were shared with those expressed by A2497-infected cells (10,347 genes). Finally, at 72 h p.i., 9,939 of the 12,911 genes (78%) in A2497P−-infected cells were common to the A2497-infected cells (12,878 genes). These results demonstrate that over time the majority of statistically significant, differentially expressed genes were the same across all three time points (24, 48, and 72 h) for both A2497- and A2497P−-infected cells.
FIG 3.
Six-way and three-way Venn diagrams of genes significantly differentially expressed in A2497- and A2497P−-infected cells. (A) A six-way Venn diagram of the total number of genes that are significantly differentially expressed (P ≤ 0.05) at 24, 48, and 72 h p.i. between A2497- and A2497P−-infected cells when cells infected with each strain are compared to mock-infected controls. The chlamydial strain and time p.i. are indicated next to the circle beside the Venn diagram of a color similar to the color in the Venn diagram. The numbers of genes that are unique across time points for each chlamydial strain are shown in the circle beside the Venn diagram. The numbers of genes shared in common are indicated at the intersections of the circles in the Venn diagram. (B) Three-way Venn diagram of the total number of genes significantly differentially expressed (P ≤ 0.05) between A2497- and A2497P−-infected cells at 24, 48, and 72 h p.i. The chlamydial strain and time p.i. are indicated next to the circle beside the Venn diagram of a color similar to the color in the Venn diagram. The numbers of genes that are unique across time points for each chlamydial strain are shown in the circle beside the Venn diagram. The numbers of genes shared in common are indicated at the intersections of the circles in the Venn diagram.
To view similarities or differences in gene expression between A2497- and A2497P−-infected cells and mock-infected cells, we constructed a heat map grouping gene families by inflammation, apoptosis, cell growth or fibrosis, metabolism, as well as other annotations that appeared to dominate the array data set (see Fig. S1 in the supplemental material). Similar expression directionality and fold change levels for A2497- and A2497P−-infected cells were observed for these genes. Briefly, significant upregulation of expression of genes encoding inflammatory proteins (CSF1, CSF2, CXCL1, intercellular adhesion molecule 1 [ICAM1], interleukin-1α [IL-1α], IL-1β, IL-6, IL-8, IL-7 receptor [IL-7R], OASL, TNFAIP6, TNFRSF12A), interferon-induced proteins (IFIT1 to IFIT3), chemoattractant proteins (CCL20, CCL5, CCR5, CXCL10, CXCL2, CXCL3), apoptosis-inducing proteins (CASP1, CASP9, CGB, FAS, IL-24, IL-20, PHLDA1, NFKBIB, TRAF4), proteins involved in fibrosis or cell growth (ANGPTL4, COL5A1, COL8A1, IGFBP3, IL-11, MUC13, PEX7, RARRES1, SER PINB3, SERPINB4, TGFB1), and proteins involved in metabolism and stress was observed for both A2497- and A2497P−-infected cells.
A2497 induces increased inflammatory and immune responses compared to A2497P−.
The expression level differences observed between A2497- and A2497P−-infected cells compared to mock-infected cells led us to perform direct comparisons of the signal intensities of the 19,248 genes differentially expressed between A2497- and A2497P−-infected cells. At 24, 48, and 72 h p.i., 14, 462, and 185 genes, respectively, were significantly differentially expressed (P < 0.05) between A2497- and A2497P−-infected cells, providing a total of 616 different genes. All 616 genes, along with their fold change values, are shown in Table S2 in the supplemental material. Figure 3B shows a Venn diagram depicting the number of shared or unique genes present in this comparison of A2497- and A2497P−-infected cells. Forty-one genes were found to be common between the samples obtained at 48 and 72 h p.i., while three genes were shared at all three time points. The three common genes encode proteins CCL5 (a chemokine), HERC5 (a ubiquitin ligase known to assist the conjugation of ISG15 to viral proteins), and SOD2 (an enzyme which converts superoxide to H2O2). Four genes were common in the comparison of samples obtained at 24 and 48 h p.i., with the 4th gene encoding the interferon-induced protein IFIT3. The numbers of genes unique to the 48- and 72-h-p.i. time points were 420 and 144, respectively (Fig. 3B).
All 616 differentially expressed genes identified in the A2497-versus-A2497P− comparisons were analyzed using NCBI database searches and significant categories identified in IPA. As shown in Table 1, proinflammatory genes with increased expression in A2497-infected cells relative to A2497P−-infected cells were predominant, with 45 of those exhibiting a significant fold change of ≥1.5, and their functional annotation suggested an association with chlamydial immunopathogenesis. Fold change values ranged from 1.50 to 11.45, and those found in the significant IPA categories of infiltration of leukocytes, inflammatory response, proliferation of connective tissue cells, or angiogenesis are indicated (Table 1). The gene encoding CSF2 (granulocyte-macrophage colony-stimulating factor [GM-CSF]), a cytokine controlling the production, differentiation, and function of granulocytes and macrophages, was one of the most differentially expressed immunomodulatory genes in our data set, and its expression was 11.45-fold higher in A2497-infected cells than A2497P−-infected cells at 72 h p.i. Other interesting genes demonstrating increased expression in A2497-infected cells encoded three GRO chemokines, CXCL1, CXCL2, and CXCL3 (GROα, GROβ, and GRO3, respectively). In addition, the genes for EDN1 (a peptide capable of vasoconstriction), FPR1 (a formylated peptide receptor important for neutrophil infiltration), ICAM (a surface glycoprotein that binds to integrins of type CD11a/CD18), and the pleiotropic cytokines IL-1α, IL-6, IL-8, and PENK (a peptide implicated in immune cell response, proliferation, and migration) demonstrated elevated levels of expression in A2497-infected cells (Table 1).
TABLE 1.
Increased expression of proinflammatory molecules in A2497-infected versus A2497P−-infected HeLa cellsa
| UniGene designation | Gene | Gene title | Involvement in: |
Fold change at: |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Infiltration of leukocytes | Inflammatory response | Proliferation of connective tissue cells | Angiogenesis | 24 h | 48 h | 72 h | |||
| Hs.480042 | AREG | Amphiregulin/amphiregulin B | X | 1.89 | |||||
| Hs.201398 | C1QTNF1 | C1q and tumor necrosis factor-related protein 1 | 1.55 | ||||||
| Hs.2490 | CASP1 | Caspase 1, apoptosis-related cysteine peptidase | X | X | 1.55 | ||||
| Hs.75498 | CCL20 | Chemokine (C-C motif) ligand 20 | 3.25 | ||||||
| Hs.514821 | CCL5b (RANTES) | Chemokine (C-C motif) ligand 5 | X | X | X | 1.56 | |||
| Hs.172944 | CGB | Chorionic gonadotropin beta polypeptide | X | 5.06 | |||||
| Hs.1349 | CSF2b (GM-CSF) | Colony-stimulating factor 2 (granulocyte-macrophage) | 11.45 | ||||||
| Hs.789 | CXCL1 (GROα) | Chemokine (C-X-C motif) ligand 1 | X | X | 1.90 | 2.96 | |||
| Hs.632586 | CXCL10 (C7) | Chemokine (C-X-C motif) ligand 10 | 1.56 | ||||||
| Hs.719458 | CXCL2 (GROβ) | Chemokine (C-X-C motif) ligand 2 | X | X | 1.66 | 2.25 | |||
| Hs.617230 | CXCL3 (GRO3) | Chemokine (C-X-C motif) ligand 3 | X | X | X | 1.64 | 2.63 | ||
| Hs.348883 | CYR61 (GIG1) | Cysteine-rich angiogenic inducer 61 | X | X | 1.84 | ||||
| Hs.190622 | DDX58 (RIGI) | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | 1.75 | ||||||
| Hs.713645 | EDN1 | Endothelin 1 | X | X | X | 1.89 | |||
| Hs.655559 | EGR1 | Early growth response 1 | 1.78 | ||||||
| Hs.736403 | FOXC1 | Forkhead box C1 | X | 1.63 | |||||
| Hs.753 | FPR1 | Formyl peptide receptor 1 | X | 3.82 | |||||
| Hs.26663 | HERC5 | HECT, RLD-containing E3 ubiquitin ligase 5 | 1.57 | ||||||
| Hs.8867 | HRG | Histidine-rich glycoprotein | X | 3.90 | |||||
| Hs.643447 | ICAM1 | Intercellular adhesion molecule 1 | X | X | X | 1.62 | |||
| Hs.163173 | IFIH1 | Interferon induced with helicase C domain 1 | 1.92 | ||||||
| Hs.20315 | IFIT1 | Interferon-induced protein with tetratricopeptide receptor 1 | 1.59 | ||||||
| Hs.437609 | IFIT2b | Interferon-induced protein with tetratricopeptide receptor 2 | 1.68 | ||||||
| Hs.744072 | IFIT3 | Interferon-induced protein with tetratricopeptide receptor 3 | 1.58 | 1.56 | |||||
| Hs.1722 | IL1A | Interleukin-1α | X | X | X | X | 1.59 | ||
| Hs.708393 | IL20 | Interleukin-20 | 2.10 | ||||||
| Hs.654458 | IL6b | Interleukin-6 (interleukin-β2) | X | X | X | X | 2.45 | 2.64 | |
| Hs.624 | IL8b | Interleukin-8 | 1.81 | 2.81 | |||||
| Hs.458485 | ISG15 | ISG15 ubiquitin-like modifier | X | 1.62 | |||||
| Hs.459265 | ISG20 | Interferon-stimulated exonuclease gene of 20 kDa | 1.56 | ||||||
| Hs.272373 | MUC13 | Mucin 13, cell surface associated | 2.15 | ||||||
| Hs.487046 | NCOA6 | Nuclear receptor coactivator 6 | X | 1.83 | |||||
| Hs.5940 | NEDD9 | Neural precursor cell expressed | X | 1.55 | |||||
| Hs.9731 | NFKBIB | Nuclear factor kappa gene enhancer in B cell inhibitor, beta | 1.72 | ||||||
| Hs.118633 | OASL | 2′-5′-Oligoadenylate synthetase-like | 1.60 | ||||||
| Hs.37982 | PAPPA2 | Pappalysin 2 | 9.60 | ||||||
| Hs.521989 | PD-L1b | CD274 molecule | 2.11 | ||||||
| Hs.104920 | PENK | Proenkephalin | 2.46 | ||||||
| Hs.187284 | RARRES1 | Retinoic acid receptor responder | 1.74 | ||||||
| Hs.131269 | SERPINB3 | Serpin peptidase inhibitor, clade B, member 3 | 2.48 | ||||||
| Hs.227948 | SERPINB4 | Serpin peptidase inhibitor, clade B, member 4 | 1.71 | ||||||
| Hs.123035 | SOD2 | Superoxide dismutase 2, mitochondrial | 2.67 | 1.85 | 1.92 | ||||
| Hs.517033 | TGM2 | Transglutaminase 2 | X | X | X | 1.50 | |||
| Hs.211600 | TNFAIP3b | Tumor necrosis factor alpha-induced protein 3 | 1.78 | 2.58 | |||||
| Hs.437322 | TNFAIP6 | Tumor necrosis factor alpha-induced protein 6 | X | 2.20 | |||||
Gene expression in HeLa 229 cells infected with strain A2497 was compared to that in cells infected with A2497P− at 24, 48, and 72 h p.i. The common gene symbol along with alternate symbols for each gene is shown. For all genes, only fold changes above 1.5-fold and with a P value of <0.05 (by ANOVA) for changes in expression between A2497- and A2497P−-infected HeLa 229 cells are shown. The absence of a number in the fold change columns indicates that the comparisons did not meet these criteria. X, presence of the gene in the IPA category shown.
qRT-PCR confirmation was performed for this gene (Fig. 4).
Increased levels of expression of genes encoding chemoattractants, such as CCL20, CCL5 (RANTES), and CXCL10 (C7; CXCL10 is a chemokine and ligand for the CXCR3 receptor), were also found in the A2497-infected host cells (Table 1). Eight proteins that function as innate antiviral effectors and whose genes were significantly upregulated in A2497-infected cells, specifically, DDX58 (RIGI), IFIH1 (MDA5), IFIT1, IFIT2, IFIT3, ISG15, ISG20, and OASL, were observed. IFIT1, IFIT2, and IFIT3 are interferon inducible, and their proteins act as inhibitors of cellular and viral processes. ISG15, ISG20, and OASL are also interferon-inducible genes and are antiviral in function (Table 1).
The expression of anti-inflammatory genes was increased in A2497-infected cells at fold change levels of 1.72 to 5.06 (Table 1). Of note, the genes encoding PD-L1 (CD274; which is involved in immunological tolerance and immunity), CGB (a chemoattractant for T-regulatory cells), as well as two inhibitors of NF-kappa B activation (NFKBIB and TNFAIP3) exhibited elevated gene expression levels in A2497-infected cells.
We also found numerous genes whose annotation is associated with angiogenesis, cell growth, connective tissue, or fibrosis to be upregulated in expression in A2497-infected cells. The proteins encoded by those genes exhibiting the highest expression differential were pappalysin 2 (PAPPA2; a regulator of insulin-like growth factor), HRG (a prothrombotic protein), MUC13 (an epithelial cell-secreted surface glycoprotein), and IL-20 (an IL-10 family member involved in promoting angiogenesis) (Table 1). Finally, genes encoding proteins functioning in an antiapoptotic manner, which are associated with aberrant epithelial cell proliferation, showed a 2.48-fold increase in expression at 48 h p.i. (SERPINB3) and a modest 1.71-fold increase at 72 h p.i. (SERPINB4) in A2497-infected cells.
qRT-PCR validation of microarray data.
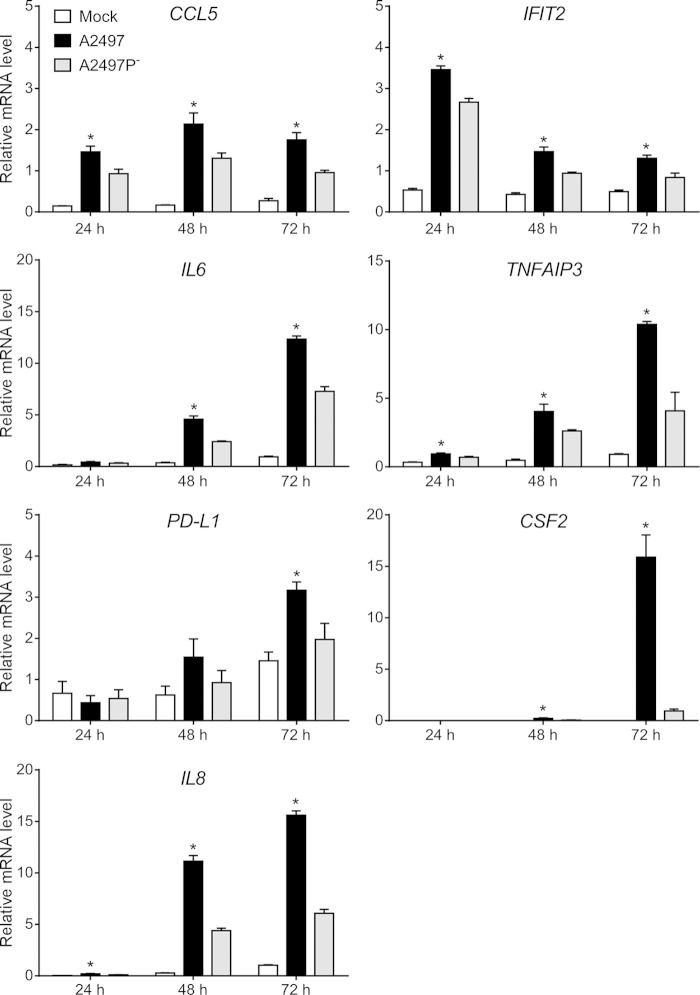
Seven gene transcripts exhibiting significant expression differentials between A2497- and A2497P−-infected cells were identified and used as qRT-PCR validation targets to corroborate our microarray results. The target mRNA transcripts were CCL5, IFIT2, IL6, TNFAIP3, PD-L1 (CD274), CSF2, and IL8 (CXCL8). A constitutively expressed housekeeping gene, the peptidylprolyl isomerase A (PPIA) gene, was chosen for statistical comparison of the results. qRT-PCRs were performed on the same RNAs used for the microarrays, and the results are plotted in Fig. 4. Normalized expression levels of all genes at all times p.i. were significantly different (P < 0.05) (see Table S4 in the supplemental material), with the exceptions being IL6 at 24 h p.i., PD-L1 at 24 and 48 h p.i., and CSF2 at 24 h p.i. (Fig. 4). When the qRT-PCR data were compared to the microarray data for these genes, high correlation values and their associated P values were obtained (see Table S4 in the supplemental material), thus supporting our microarray data.
FIG 4.
Confirmation of microarray data by qRT-PCR. Normalized signals for CCL5, IFIT2, IL6, TNDAIP3, PD-L1, CSF2, and IL8 human mRNA were analyzed in a subset of microarray samples. The mean relative mRNA level for each treatment group (the A2497-, A2497P−-, and mock-infected groups) was plotted, and mRNA level variation between the 6 replicate samples for each group is expressed as the standard error of the mean. All mRNAs were present at a lower copy number in strain A2497P−-infected samples than A2497-infected samples at 24, 48, and 72 h p.i., except for PD-L1 at 24 h. All copy number differences between A2497- and A2497P−-infected cells, with the exception of IL6 at 24 h p.i., PD-L1 at 24 and 48 h p.i., and CSF2 at 24 h p.i., were statistically significant (t test, P ≤ 0.05) (see Table S4 in the supplemental material), as indicated by asterisks above the bars for A2497.
IPA.
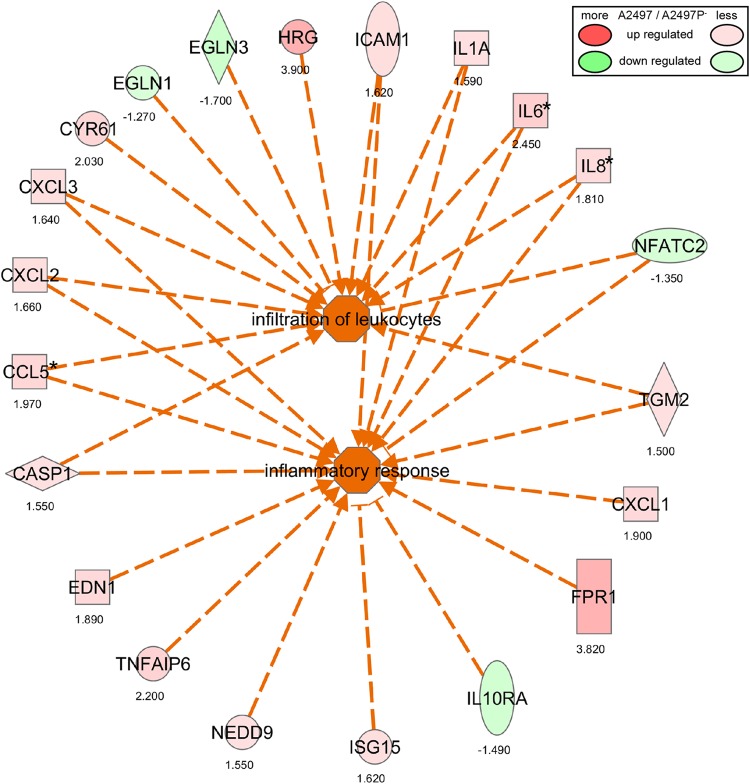
Ingenuity Pathway Analysis (IPA) allows integration of data from different platforms and provides insight into molecular and chemical interactions as well as cellular phenotypes and potential disease processes. Therefore, the interconnectivity of the 462 genes differentially expressed between A2497- and A2497P−-infected cells at 48 h p.i. was analyzed using Ingenuity Pathway Analysis, and a number of significant categories and pathways were identified (data not shown). We combined two high-ranking (according to their P values and activation z-scores) and overlapping categories, specifically, the leukocyte migration category and the inflammatory response category, creating the combined pathway depicted in Fig. 5. This combined pathway demonstrates, through differential gene expression and presumed coordinated function at the protein level, that a greater leukocyte migration and inflammatory response appears to occur in A2497-infected cells than A2497P−-infected cells.
FIG 5.
Infiltration of leukocytes and inflammatory response pathways for A2497- versus A2497P−-infected cells at 48 h p.i., as determined by IPA analysis. Two significant IPA categories (infiltration of leukocytes and inflammatory response) (P ≤ 0.05) and the interconnectivity between them are shown. Function and directionality are indicated by the key on the right. Numbers below the genes indicate the fold change in the levels of expression of the gene in A2497-infected versus A2497P−-infected cells. *, the results were confirmed by qRT-PCR.
Several other high-ranking categories were also identified, in particular, proliferation of connective tissue as well as angiogenesis, both of which are shown diagrammatically in Fig. S2 and S3 in the supplemental material, respectively. Five genes (EDN1, IL6, IL8, IL1A, and IGFBP3) were found to be common between both pathways, suggesting that similar yet different genes and mechanisms may be involved (see Fig. S2 and S3 in the supplemental material). The pathway results shown in Fig. S2 and S3 in the supplemental material demonstrate that many of the genes described in Table 1 and Table S2 in the supplemental material for angiogenesis and proliferation of connective tissue are connected to each other by their expression profiles, again suggesting a greater function or involvement of these pathways in A2497-infected cells.
DISCUSSION
The strong tendency for naturally occurring chlamydial isolates to maintain their cryptic plasmid is a compelling argument for the importance of the plasmid to the pathogenicity of chlamydial infection (10, 21–23). Clearly, recent studies have shown that plasmid-deficient strains are attenuated in mouse and nonhuman primate infection models (11, 12, 24). The molecular basis for this attenuation remains largely unanswered. However, there is a direct relationship between the attenuation phenotype and the plasmid's ability to regulate specific plasmid and chromosomal genes (10, 13, 24). A major attenuating phenotype of the plasmid-deficient trachoma strain A2497P− is the spontaneous resolution of A2497P− infection in macaque eyes without the production of a clinical ocular pathology (12). In contrast, infection with A2497 plasmid-bearing organisms is protracted and evokes an intense ocular inflammatory response. These distinctly different pathological outcomes for in vivo A2497 and A2497P− infections suggest that the plasmid is contributing to sustained infection and the induction of an inflammatory pathology. However, it is unclear from these in vivo studies what the role of the plasmid might be in eliciting inflammatory processes. Possibilities are (i) a direct interaction with infected nonimmune conjunctival epithelial cells, the cellular paradigm of pathogenesis (8), or (ii) induction of host adaptive immune mechanisms. To investigate a potential role for the plasmid in the generation of a nonimmune cellular basis of pathogenesis, we conducted a comparative transcriptome analysis of human epithelial cells infected with the A2497 or A2497P− strain.
HeLa cells infected with either strain generated differential transcriptional profiles for genes encoding IL-1α, IL-1β, IL-6, IL-7R, OASL, IL-8, macrophage colony-stimulating factor (MCSF), RANTES, interferon-inducible antiviral proteins (IFIH1, IFIT1, IFIT2, IFIT3), GROA, CASP1, IL-11, CXCL2, and CXCL3 that were similar to each other, relative to the mock-infected controls at 24 to 48 h p.i. The absence of an early host transcriptional response indicates that chlamydial attachment, entry, or early elementary body (EB)-to-reticulate body (RB) differentiation and growth were stealthy in nature and that neither outer membrane proteins nor lipopolysaccharide (LPS) engaged cognate cytoplasmic membrane signaling receptors to trigger these host proinflammatory responses. In the case of chlamydial LPS, this lack of response is consistent with the molecule's unique carbohydrate and lipid structure, which has been shown to be significantly less active in the induction of proinflammatory cytokines than LPSs of typical Gram-negative organisms (25). Conversely, induction of host genes encoding mRNAs for proinflammatory cytokines was dramatically increased at late times p.i. (24 to 48 h). This delayed host response to infection implicates stress or chlamydial secreted proteins in the induction of proinflammatory genes. The temporal kinetics of our transcriptional findings are consistent with what others have reported for the secretion of proinflammatory cytokines by C. trachomatis-infected HeLa cells (26–28).
Despite the similar transcription profiles of cells infected with A2497 and A2497P−, there were distinct differences in the levels of mRNA expression between the strains. Genes significantly upregulated in A2497-infected cells compared to their level of regulation in A2497P−-infected cells encoded the proinflammatory proteins and chemokines IL-1α, IL-6, IL-8, CXCL1, CXCL2, CXCL3, ICAM1, and RANTES, as well as the chemoattractants CCL20, CCL5, and gamma Interferon-inducible protein-10 (IP-10). The gene encoding CCL5 (RANTES) was previously shown to be upregulated in human prostate epithelial cells (29) and in patients with late-stage blinding trachoma (30, 31).
Lastly, and very importantly, the elevated expression of PD-L1 mRNAs in A2497-infected cells relative to A2497P−-infected cells has direct implications for CD8+ T cell-mediated immunity to chlamydial infection. The protein PD-L1 has recently been shown to limit the mucosal CD8+ T cell response to murine chlamydial infection of the female genital tract (32). Collectively, these findings suggest that plasmid-deficient chlamydiae may lose this immune avoidance ability and therefore become more readily controlled by chlamydia-specific CD8+ T cells. Consistent with this conclusion are the recent findings of Olivares-Zavaleta et al. (33), who demonstrated that macaques vaccinated with A2497P− organisms elicit a strongly protective anti-CD8-mediated immunity. Moreover, the level of expression of the gene encoding CGB, an immunosuppressive chemoattractant for T regulatory cells (34), was increased 5.06-fold in A2497-infected cells compared to the level of expression in A2497P−-infected cells. Thus, a pattern of gene expression changes that appear to subvert protective CD8+ T cell immunity emerges in A2497-infected epithelial cells.
Collectively, our results support, at least in part, the cellular paradigm of chlamydial pathogenesis proposed by Stephens (8), which is that chlamydial infection of nonimmune epithelial cells alone is capable of generating a damaging pleiotropic inflammatory immunity. We further conclude that these chlamydia-host cell interactions might also serve to evade adaptive host defense immune mechanisms and contribute to fibrosis. In each case, however, our findings show that plasmid-deficient organisms are less capable of provoking the cellular responses that exacerbate inflammation, prolong infection, promote fibrosis-like effects, and evade adaptive immune mechanisms.
Alternatively, one could make the intuitive argument that the level of expression of proinflammatory cytokine and chemokine genes by virulent A2497 organisms is, in fact, marginal, and its biological significance is thus questioned. In fact, Buckner et al. (35) reported that infection of human polarized endocervical cells by virulent chlamydiae resulted in only a modest increase in proinflammatory cytokines, leading these investigators to propose that chlamydiae may in fact be stealth pathogens using evasion strategies to circumvent proinflammatory cytokines and T cell-activating chemokines. Considering that natural chlamydial infection of humans is caused predominantly by plasmid-bearing organisms and results in reinfection or persistent infection, an accumulative proinflammatory and immunomodulatory host response might be sufficient to produce an enhanced damaging inflammatory response and a concomitant decreased ability to immunologically control infection in time.
In summary, a global picture begins to emerge from our findings to suggest that the chlamydial plasmid functions to (i) evoke an inflammatory response by infected epithelial cells, (ii) induce the expression of host genes that downregulate CD8+ T cell responses to mucosal infection, and (iii) provoke altered epithelial expression of cell growth-, cell growth control-, fibrosis-, and connective tissue-related genes, the resultant phenotypic hallmarks of chronic chlamydial diseases. In contrast, plasmid-deficient chlamydiae have a reduced ability to control or influence the expression of these host genes, which could therefore help explain their strong in vivo attenuation phenotype (12) and unique ability to induce protective CD8+ T cell-mediated immunity (34).
Supplementary Material
ACKNOWLEDGMENTS
We thank Anita Mora for graphical assistance and Kelly Haynes for manuscript preparation.
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02764-14.
REFERENCES
- 1.Hotez PJ, Kamath A. 2009. Neglected tropical diseases in sub-Saharan Africa: review of their prevalence, distribution, and disease burden. PLoS Negl Trop Dis 3:e412. doi: 10.1371/journal.pntd.0000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Resnikoff S, Pascolini D, Etya'ale D, Kocur I, Pararajasegaram R, Pokharel GP, Mariotti SP. 2004. Global data on visual impairment in the year 2002. Bull World Health Organ 82:844–851. [PMC free article] [PubMed] [Google Scholar]
- 3.Bébéar C, de Barbeyrac B. 2009. Genital Chlamydia trachomatis infections. Clin Microbiol Infect 15:4–10. doi: 10.1111/j.1469-0691.2008.02647.x. [DOI] [PubMed] [Google Scholar]
- 4.Brunham RC, Rey-Ladino J. 2005. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat Rev Immunol 5:149–161. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- 5.Vasilevsky S, Grueb G, Nardelli-Haefliger D, Baud D. 2014. Genital Chlamydia trachomatis: understanding the roles of innate and adaptive immunity in vaccine research. Clin Microbiol Rev 27:346–370. doi: 10.1128/CMR.00105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison RP, Belland RJ, Lyng K, Caldwell HD. 1989. Chlamydial disease pathogenesis. The 57-kD chlamydial hypersensitivity antigen is a stress response protein. J Exp Med 170:1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morrison RP, Lyng K, Caldwell HD. 1989. Chlamydial disease pathogenesis. Ocular hypersensitivity elicited by a genus-specific 57-kD protein. J Exp Med 169:663–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stephens RS. 2003. The cellular paradigm of chlamydial pathogenesis. Trends Microbiol 11:44–51. doi: 10.1016/S0966-842X(02)00011-2. [DOI] [PubMed] [Google Scholar]
- 9.Watkins NG, Hadlow WJ, Moos AB, Caldwell HP. 1986. Ocular delayed hypersensitivity: a pathogenetic mechanism of Chlamydial conjunctivitis in guinea pigs. Proc Natl Acad Sci U S A 83:7480–7484. doi: 10.1073/pnas.83.19.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlson JH, Whitmire WM, Crane DD, Wicke L, Virtaneva K, Sturdevant DE, Kupko JJ, Porcella SF, Martinez-Orengo N, Heinzen RA, Kari L, Caldwell HD. 2008. The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect Immun 76:2273–2283. doi: 10.1128/IAI.00102-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Connell CM, Ingalls RR, Andrews CW Jr, Scurlock AM, Darville T. 2007. Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J Immunol 179:4027–4034. doi: 10.4049/jimmunol.179.6.4027. [DOI] [PubMed] [Google Scholar]
- 12.Kari L, Whitmire WM, Olivares-Zavaleta N, Goheen MM, Taylor LD, Carlson JH, Sturdevant GL, Lu C, Bakios LE, Randall LB, Parnell MJ, Zhong G, Caldwell HD. 2011. A live-attenuated chlamydial vaccine protects against trachoma in nonhuman primates. J Exp Med 208:2217–2223. doi: 10.1084/jem.20111266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song L, Carlson JH, Whitmire WM, Kari L, Virtaneva K, Sturdevant DE, Watkins H, Zhou B, Sturdevant GL, Porcella SF, McClarty G, Caldwell HD. 2013. Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence-associated genes. Infect Immun 81:636–644. doi: 10.1128/IAI.01305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darville T, O'Neill JM, Andrews CWJ, Nagarajan UM, Stahl L, Ojjcius DM. 2003. Toll-like receptor-2, but not Toll-like receptor-4, is essential for development of oviduct pathology in chlamydial genital tract infection. J Immunol 171:6187–6197. doi: 10.4049/jimmunol.171.11.6187. [DOI] [PubMed] [Google Scholar]
- 15.Dong X, Liu Y, Chang X, Lei L, Zhong G. 2014. Signaling via tumor necrosis factor receptor 1 but not Toll-like receptor 2 contributes significantly to hydrosalpinx development following Chlamydia muridarum infection. Infect Immun 82:1833–1839. doi: 10.1128/IAI.01668-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murthy AK, Li W, Chaganty BK, Kamalakaran S, Guentzel MN. 2011. Tumor necrosis factor alpha production from CD8+ T cells mediates oviduct pathological sequelae following primary genital Chlamydia muridarum infection. Infect Immun 79:2928–2935. doi: 10.1128/IAI.05022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackey-Lawrence NM, Guo X, Sturdevant DE, Virtaneva K, Hernandez MM, Houpt E, Sher A, Porcella SF, Petri WAJ. 2013. Effect of the leptin receptor Q223R polymorphism on the host transcriptome following infection with Entamoeba histolytica. Infect Immun 81:1460–1470. doi: 10.1128/IAI.01383-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Lai Y, Villaruz AE, Cha DJ, Sturdevant D, Otto M. 2007. Gram-positive three-component antimicrobial peptide-sensing system. Proc Natl Acad Sci U S A 104:9469–9474. doi: 10.1073/pnas.0702159104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and proven approach to multiple testing. J R Stat Soc Series B Stat Methods 57:289–300. [Google Scholar]
- 20.Edgar R, Domrachev M, Lash AE. 2002. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Comanducci M, Ricci S, Cevenini R, Ratti G. 1990. Diversity of the Chlamydia trachomatis common plasmid in biovars with different pathogenicity. Plasmid 23:149–154. doi: 10.1016/0147-619X(90)90034-A. [DOI] [PubMed] [Google Scholar]
- 22.Rockey DD. 2011. Unraveling the basic biology and clinical significance of the chlamydial plasmid. J Exp Med 208:2159–2162. doi: 10.1084/jem.20112088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell M, Darville T, Chandra-Kuntal K, Smith B, Andrews CWJ, O'Connell CM. 2011. Infectivity acts as in vivo selection for maintenance of the chlamydial cryptic plasmid. Infect Immun 79:98–107. doi: 10.1128/IAI.01105-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sigar IM, Schripsema JH, Wang Y, Clarke IN, Cutcliffe LT, Seth-Smith HM, Thomson NR, Bjartling C, Unemo M, Persson K, Ramsey KH. 2014. Plasmid deficiency in urogenital isolates of Chlamydia trachomatis reduces infectivity and virulence in a mouse model. Pathog Dis 70:61–69. doi: 10.1111/2049-632X.12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heine H, Muller-Loennies S, Brade L, Lindner B, Brade H. 2003. Endotoxic activity and chemical structure of lipopolysaccharides from Chlamydia trachomatis serotypes E and L2 and Chlamydophila psittaci 6BC. Eur J Biochem 270:440–450. doi: 10.1046/j.1432-1033.2003.03392.x. [DOI] [PubMed] [Google Scholar]
- 26.Buchholz KR, Stephens RS. 2006. Activation of the host cell proinflammatory interleukin-8 response by Chlamydia trachomatis. Cell Microbiol 8:1768–1779. doi: 10.1111/j.1462-5822.2006.00747.x. [DOI] [PubMed] [Google Scholar]
- 27.Rasmussen SJ, Eckmann L, Quayle AJ, Shen L, Zhang YX, Anderson DJ, Fierer J, Stephens RS, Kagnoff MF. 1997. Secretion of proinflammatory cytokines by epithelial cells in response to Chlamydia infection suggests a central role for epithelial cells in chlamydial pathogenesis. J Clin Invest 99:77–87. doi: 10.1172/JCI119136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia M, Bumgarner RE, Lampe MF, Stamm WE. 2003. Chlamydia trachomatis infection alters host cell transcription in diverse cellular pathways. J Infect Dis 187:424–434. doi: 10.1086/367962. [DOI] [PubMed] [Google Scholar]
- 29.Sellami H, Said-Sadier N, Znazen A, Gdoura R, Ojcius DM, Hammami A. 2014. Chlamydia trachomatis infection increases the expression of inflammatory tumorigenic cytokines and chemokines as well as components of the Toll-like receptor and NF-κB pathways in human prostate epithelial cells. Mol Cell Probes 28:147–154. doi: 10.1016/j.mcp.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Burton MJ, Rajak SN, Bauer J, Weiss HA, Tolbert SB, Shoo A, Habtamu E, Manjurano A, Emerson PM, Mabey DC, Holland MJ, Bailey RL. 2011. Conjunctival transcriptome in scarring trachoma. Infect Immun 79:499–511. doi: 10.1128/IAI.00888-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Natividad A, Freeman TC, Jeffries D, Burton MJ, Mabey DC, Bailey RL, Holland MJ. 2010. Human conjunctival transcriptome analysis reveals the prominence of innate defense in Chlamydia trachomatis infection. Infect Immun 78:4895–4911. doi: 10.1128/IAI.00844-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fankhauser SC, Starnbach MN. 2014. PD-L1 limits the mucosal CD8+ T cell response to Chlamydia trachomatis. J Immunol 192:1079–1090. doi: 10.4049/jimmunol.1301657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olivares-Zavaleta N, Whitmire BW, Kari L, Sturdevant GL, Caldwell HD. 2014. CD8+ T cells define an unexpected role in live-attenuated vaccine protective immunity against Chlamydia trachomatis infection in macaques. J Immunol 192:4648–4654. doi: 10.4049/jimmunol.1400120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schumacher A, Heinze K, Witte J, Poloski E, Linzke N, Woidacki K, Zenclussen AC. 2013. Human chorionic gonadotropin as a central regulator of pregnancy immune tolerance. J Immunol 190:2650–2658. doi: 10.4049/jimmunol.1202698. [DOI] [PubMed] [Google Scholar]
- 35.Buckner LR, Lewis ME, Greene SJ, Foster TP, Quayle AJ. 2013. Chlamydia trachomatis infection results in a modest pro-inflammatory cytokine response and a decrease in T cell chemokine secretion in human polarized endocervical epithelial cells. Cytokine 63:151–165. doi: 10.1016/j.cyto.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.