Abstract
Mycobacterium abscessus is a pathogenic, rapidly growing mycobacterium involved in pulmonary and cutaneo-mucous infections worldwide, to which cystic fibrosis patients are exquisitely susceptible. The analysis of the genome sequence of M. abscessus showed that this bacterium is endowed with the metabolic pathways typically found in environmental microorganisms that come into contact with soil, plants, and aquatic environments, where free-living amoebae are frequently present. M. abscessus also contains several genes that are characteristically found only in pathogenic bacteria. One of them is MAB_0555, encoding a putative phospholipase C (PLC) that is absent from most other rapidly growing mycobacteria, including Mycobacterium chelonae and Mycobacterium smegmatis. Here, we report that purified recombinant M. abscessus PLC is highly cytotoxic to mouse macrophages, presumably due to hydrolysis of membrane phospholipids. We further showed by constructing and using an M. abscessus PLC knockout mutant that loss of PLC activity is deleterious to M. abscessus intracellular survival in amoebae. The importance of PLC is further supported by the fact that M. abscessus PLC was found to be expressed only in amoebae. Aerosol challenge of mice with M. abscessus strains that were precultured in amoebae enhanced M. abscessus lung infectivity relative to M. abscessus grown in broth culture. Our study underlines the importance of PLC for the virulence of M. abscessus. Despite the difficulties of isolating M. abscessus from environmental sources, our findings suggest that M. abscessus has evolved in close contact with environmental protozoa, which supports the argument that amoebae may contribute to the virulence of opportunistic mycobacteria.
INTRODUCTION
The recognition of the role of Mycobacterium abscessus in human pathology has taken several decades, due to confusion in many studies between this mycobacterium and the very closely related species Mycobacterium chelonae. It was only in 1992 that these two species were distinguished and M. abscessus elevated to the rank of species (1).
These two phylogenetically closely related, rapidly growing mycobacteria (RGM), which have identical 16S ribosomal rRNA gene sequences, are distinguished by different pathogenicity patterns. M. chelonae, generally less pathogenic than M. abscessus, is implicated in skin and soft tissue infections and only occasionally involved in lung infections. M. abscessus is currently the most frequently isolated RGM in human pathology and the main RGM involved in lung infections (2, 3), with a particular link to cystic fibrosis (CF) patients (4–6). M. abscessus is also the main RGM responsible for iatrogenic infections in humans (postinjection abscesses, cardiac surgery infections, and plastic surgery infections) (7–9).
The environmental source of M. abscessus that might serve as a reservoir for human infection is currently unknown (10). Although the gene pool of M. abscessus (11) suggests that this bacterium has evolved in an aquatic environment at the interface with plants, as shown by the presence of genes coding for resistance to arsenic, i.e., cysteine desulfurases, which are found mainly in environmental organisms (11), some other genes of M. abscessus indicate that this bacterium tends to specialize in intracellular parasitism (12). The hypothesis that M. abscessus has evolved in an aquatic environment has been strengthened by a recent study showing that it can replicate and survive within zebrafish embryos at 28°C, where it can be pathogenic and capable of inducing lethal infections (13).
Amoebae are an integral part of this aquatic and telluric environment, and several reports have already shown an association of mycobacteria with free-living amoebae in water networks (14–17), although some species, including M. abscessus, were not recovered at all (18, 19), mainly due to aggressive methods of decontamination (20). Mycobacteria can grow in amoebae (21–25), and amoebic coculture has been successfully used to isolate Mycobacterium massiliense (26), a member of the M. abscessus complex. M. abscessus was also described as being able to multiply in trophozoites and to survive in amoeba cysts, the persistent stage of amoebae (12, 23), supporting the idea that factors other than rapid growth may be involved in mycobacterium-amoeba interactions.
Comparative genomic analyses of M. abscessus, M. chelonae, and Mycobacterium smegmatis genomes has allowed the confirmation of differences observed between these RGM in terms of pathogenicity (27) and intracellular behavior (28; A.-L. Roux, T. Deramaudt, R. Simeone, A. Viljoen, A. Bernut, A. Bah, N. Dulphy, M. Rottman, A. Toubert, J.-L. Gaillard, L. Tailleux, L. Kremer, I. Vergne, C. de Chastellier, L. Majlessi, R. Brosch, and J.-L. Herrmann, unpublished data) by highlighting several M. abscessus key genes encoding virulence factors (11). Interestingly, these genes seem to have been acquired by horizontal gene transfer (HGT) mainly from aquatic and telluric pathogenic bacteria, including those playing a major role in patients with CF: Pseudomonas spp. and Burkholderia spp. (11). One key determinant acquired by HGT is phospholipase C (PLC), encoded by the plcC gene (MAB_0555) (11). PLC was reported to be involved in the intracellular survival of Mycobacterium tuberculosis (29) and is absent from both M. chelonae and M. smegmatis. However, neither its role in the pathogenicity of M. abscessus nor its interaction with eukaryotic cells has been investigated yet.
Bacterial PLCs are known to play important roles in bacterial pathogenesis, increasing bacterial survival by inducing inappropriate host cellular signaling mechanisms and direct cytotoxicity or by impairing lung inflammatory responses (for reviews, see references 30, 31, and 32). In mycobacteria, PLC (and sphingomyelinase) activity seems to be associated with the most virulent species (33).
Association of PLC activity with virulent species prompted us to initiate a detailed molecular characterization of the M. abscessus PLC activities. Here, we describe the biological activities of the M. abscessus PLC. Data were obtained from experiments with purified recombinant PLC, as well as from analysis of its role in three different eukaryotic infection models, for which we employed a PLC knockout mutant of M. abscessus and its complemented derivative that were both constructed in this study.
MATERIALS AND METHODS
Mycobacterial and amoeba strains, reagents, and antibodies.
Smooth M. abscessus CIP104536TS (CIP-S) and the recombinant strain M. smegmatis mc2155 groEL1ΔC (34) were grown aerobically at 37°C in Middlebrook 7H9 medium supplemented with 0.2% glycerol. Acanthamoeba castellanii (ATCC 30010) was grown at 28°C without CO2 in PYG broth (35). p-Nitrophenylphosphorylcholine (p-NPPC) and the unlabeled phospholipids phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylinositol (PI) were from Sigma-Aldrich. Radiolabeled 1-palmitoyl-2-[14C]palmitoyl-glycerophosphocholine (DPPC) and [1-14C]palmitic acid were from GE Healthcare. Bacillus cereus PC-PLC enzyme was from Sigma-Aldrich. Polyclonal mouse anti-PLC antibodies were obtained after three DNA immunizations of mice with a plasmid containing the PLC sequence under the control of a cytomegalovirus (CMV) promoter ((V. Le Moigne, M. Rottman, C. Goulard, B. Barteau, I. Poncin, N. Soismier, S. Canaan, B. Pitard, J.-L. Gaillard, and J.-L. Herrmann, unpublished results).
Cloning of PLC (MAB_0555).
MAB_0555, encoding M. abscessus PLC with its predicted Tat signal sequence, was amplified by PCR (see Table S1 in the supplemental material), gel purified, and cloned after ligation into pCR2.1-TOPO (Life Technologies, France) (pTOPO-MAB_0555). MAB_0555 was again amplified from pTOPO-MAB_0555 with a second set of primers (see Table S1 in the supplemental material), which includes restriction sites for HindIII and NcoI. Amplified products were then digested, purified, and cloned into pMyC (pMyC-MAB_0555) as previously described (36). pMyc-MAB_0555 was transformed into the recombinant strain M. smegmatis mc2155 groEL1ΔC, which was further used for protein purification of PLC.
PLC purification, enzymatic activity, and cell experiments with purified recombinant M. abscessus PLC.
PLC was purified from a single transformed colony of M. smegmatis::pMyc-MAB_0555 as described previously for M. tuberculosis PLCs (36). Recombinant M. abscessus PLC (rPLCMa) was concentrated to 1 mg/ml, analyzed by MALDI-TOF (matrix-assisted laser desorption ionization–time of flight) mass spectrometry and N-terminal sequencing, and stored at −80°C. Phospholipase C activity was measured with p-NPPC, PC, or sphingomyelin (SM) as the substrate, as previously described (36), using the fluorescent Amplex red PC-PLC kit assay or the fluorescent Amplex red sphingomyelinase kit assay (Molecular Probes, Life Technologies) as described by the supplier. Competition assays between PC and PE (or PI) (30 mg/ml and 25 mg/ml in chloroform, respectively) were performed using the Amplex red PC-PLC assay for substrate preference. Inhibition assays were performed using the D609 compound (9.38 mM final concentration in water) (Sigma), at molecular inhibitor/enzyme ratios of 200 and 600. The residual activity was measured using the Amplex red PC-PLC kit as described above. The hemolytic and cytotoxic effects of purified rPLCMa were evaluated as previously described (36). Incorporation of labeled fatty acids ([1-14C] palmitic acid) into macrophages and rPLCMa activity on radiolabeled macrophages were evaluated as described previously (36, 37).
Construction of the PLC KO (MAB_0555) mutant in M. abscessus.
The M. abscessus PLC knockout (KO) mutant was obtained by allelic exchange in M. abscessus CIP-S using the strategy previously reported (38). Briefly, the zeocin cassette (Streptoalloteichus hindustanus ble) was inserted into the HindIII-ClaI region spanning the 3′ end of MAB_0554 and into MAB_0555 (nucleotides 731 to 801 in MAB_0554 to nucleotides 1 to 590 in MAB_0555). The overall fragment was cloned into pMVZ261 and further restricted by PvuII-HpaI for purification and electroporation in M. abscessus CIP-S bearing the recombineering plasmid pJV53 (39). Homologous recombination was checked by a first PCR screen using forward and reverse primers outside the deleted region (see Table S1 in the supplemental material) and then by Southern blotting using a zeocin probe and a 532-bp probe matching the 3′ end of MAB_0555 and prepared by amplification using forward and reverse primers (see Table S1 in the supplemental material). To complement the M. abscessus PLC KO mutant, pTOPO-MAB_0555 was digested and MAB_0555 was cloned under the control of the hsp60 promoter into the integrative plasmid pMVZ361-Kan-Zeo. The plasmid was then electroporated into the wild-type (WT) and PLC KO strains. In vitro growth of WT, PLC KO, and PLC-complemented M. abscessus strains was monitored at 600 nm.
TLC and mass spectrometry comparative analysis of the WT and PLC KO mutant of M. abscessus.
Mycobacterial wet cells were sequentially extracted with CHCl3-CH3OH (1:2, vol/vol), with CHCl3-CH3OH (1:1, vol/vol) and then three times with CHCl3-CH3OH (2:1, vol/vol). The organic phases were pooled, extensively washed with water, and evaporated to dryness. Lipids were analyzed by thin-layer chromatography (TLC) on silica gel 60-precoated plates (0.25-mm thickness; Merck) developed with CHCl3-CH3OH (90:10 [vol/vol]) for glycolipids or CHCl3-CH3OH-H2O (60:35:8) for phospholipids. Sugar-containing compounds were visualized by spraying plates with 0.2% anthrone in concentrated sulfuric acid, followed by heating, whereas the Dittmer-Lester reagent and ninhydrin were used to detect phosphorus- and amino group-containing substances, respectively.
For mass spectrometry (MS) experiments, total lipids were extracted from bacterial cell pellets with methanol (MeOH)-CHCl3 (2:1, vol/vol) overnight at room temperature (RT). Supernatants were filtered and then poured into new tubes for evaporation under nitrogen flow. CHCl3-MeOH (2:1, vol/vol) was then added to cell pellets and incubated at RT for 24 h. After incubation, the contents of the glass tubes were filtered on glass pipettes and poured into the corresponding tube containing the previously evaporated materials. After solvent evaporation, H2O-CHCl3 (1:1, vol/vol) was added, and the tube was incubated for 24 h. After water-lipid separation, water was removed until the organic phase was limpid. Lipids extracts were evaporated and dissolved in isopropanol-methanol (70:30, vol/vol), 0.02% (mass/vol) formic acid, 0.01% (mass/vol) ammonium hydroxide. Electrospray ionization quadrupole time-of-flight mass spectrometry was performed as previously described (40). Briefly, lipid extracts were injected by infusion into the MS. Ionization was maintained at 325°C with a 5-liter/min drying gas flow, a 200,000-Pa nebulizer pressure, and 5,500 V. Spectra were collected in positive-ion mode from m/z 200 to 3,000 at 1 spectrum/s. Spectrometer was calibrated in positive-ion mode with a sodium iodide solution (NaI at 2 μg/ml in 50% isopropanol). Collision-induced dissociation (CID) MS was performed with energy of 30 V. Data were collected and processed through Analyst QS 1.1 software from AB-MDS-Sciex.
Coculture of M. abscessus strains and murine macrophages.
Bone marrow-derived murine macrophages (BMDMs) were prepared as previously described (41). BMDMs were grown in RPMI 1640 medium containing 10% fetal calf serum (FCS) at 37°C with 5% CO2. Coculture experiments were performed as previously described (36, 41, 42) at a multiplicity of infection (MOI) of one bacterium per macrophage. After 3 h of incubation, the cells were washed 3 times with RPMI to remove extracellular bacteria and incubated with amikacin (250 μg/ml) to kill the remaining extracellular bacteria. Fresh medium containing 50 μg/ml of amikacin was then added. The number of CFU/ml was determinate at days 0, 1, 3, and 6 of the culture after cold-water lysis of macrophages.
Coculture of M. abscessus strains and A. castellanii.
For amoeba infection assays, M. abscessus cultures were washed 3 times in 30 ml of Page's modified Neff's amoeba saline (PAS), which contains no source of carbon or azote (35). The mycobacterial inoculum was thoroughly mixed, and mycobacteria were then dispersed by 10 passages of the bacterial suspension through a 25-gauge needle attached to a 5-ml syringe followed by 10 passages through a 29.5-gauge needle attached to a 1-ml syringe. Mycobacterial suspensions were then adjusted in PAS buffer to a concentration of 2.5 × 107 bacteria per ml by measuring the optical density at 600 nm (OD600). CFU counts were also confirmed on the inocula. Five hundred microliters of the A. castellanii suspension was washed three times in PAS buffer and dispatched into a 48-well plate. Following 1 h of incubation at 32°C, the amoeba monolayer was inoculated with 200 μl of a bacterial suspension (MOI, 25 bacteria/amoeba). After 3 h of incubation at 32°C, extracellular mycobacteria were removed by three thorough washings in PAS buffer, followed by one supplementary 2-h incubation in the presence of 100 μg/ml of amikacin, in order to kill all extracellular mycobacteria. PAS (500 μl) was the added. Every 24 h, 50 μl (107) heat-inactivated Escherichia coli (70°C for 60 min) was added to each well to slow the transition from trophozoite to cyst. The number of CFU/ml was determined for each M. abscessus strain after lysis of the A. castellanii monolayer with 1% SDS for 30 min at 32°C, at 1, 2, 3, and 5 days of coculture.
Western blotting for PLC expression.
M. abscessus strains, either grown in 7H9 or cocultured 1 to 2 days with amoebae, were lysed by sonication on ice (three times for 30 s each) with proteases inhibitors (Complete Mini; Roche) plus E64 (20 μM final concentration) and leupeptin (20 μM final concentration). Thirty micrograms of cell lysates was separated on SDS-PAGE and transferred onto nitrocellulose membranes, which were then incubated with murine anti-PLC antibodies diluted 1/300. After addition of a rabbit anti-mouse antibody linked to peroxidase, the signal was revealed using the Sirius chemiluminescent substrate (Advansta, USA).
RT-PCR for PLC mRNA expression.
For total mRNA extraction of mycobacteria from macrophage cocultures, macrophages were infected for 5 days with M. abscessus (MOI = 1) in F75 flasks. Each day, the culture medium was discarded, and the infected monolayer was washed with 1× phosphate-buffered saline (PBS) and then resuspended in 10 ml of guanidine thiocyanate solution (4 M) to lyse macrophages. The lysates were then centrifuged at 2,500 × g for 30 min to concentrate intracellular bacteria. The pellet of intracellular mycobacteria or pellet of mycobacteria cultivated at 30°C or 37°C was then resuspended in TRIzol, and total-mRNA extraction was performed using TRIzol in the presence of zirconia/silica beads and after bead beating at maximum speed for 30 s twice. After a chloroform-and-isopropanol precipitation, RNA samples were treated twice with DNase I Amp Grade (Invitrogen) (1 U/μg of RNA). Total RNA integrity and concentration were assessed with the Experion automated electrophoresis system (Bio-Rad). One microgram of total RNA was used for a reverse transcription reaction with oligo(dT)12–18 primers and SuperScript II reverse transcriptase (SuperScript first-strand synthesis system for reverse transcription-PCR [RT-PCR]; Invitrogen, Carlsbad, CA). Negative controls were made by replacing the reverse transcriptase with diethyl pyrocarbonate-treated water. Diluted cDNA was combined with primer/probe sets (see Table S1 in the supplemental material) and SYBR green I master mix (Roche) according to the manufacturer's recommendations. Samples were normalized internally using the average cycle threshold (CT) of sigA as the reference (42). sigA was used as the constitutive gene as previously described (43). The concentration ratio (target/sigA mRNA) was calculated using ReLQuant Roche software and expressed in arbitrary units.
Mouse model of M. abscessus aerosol infection.
BALB/c mice were challenged with aerosolized M. abscessus using an aerosol generator under agreement number B92-033-01. This apparatus used a Micro Mist small-volume nebulizer (Hudson RCI-Teleflex Medical, Research Triangle Park, NC, USA) containing 6 ml of mycobacterial solution at various concentrations. Presleeping mice (isoflurane; Abbott, Rungis, France) were anesthetized with 200 μl of Hypnomidate (etomidate; Janssen-Cilag, France) and placed in an open 50-ml syringe fixed on top of a closed compartment containing the nebulizer. The nebulization in this device lasted 15 min, the time necessary to vaporize all the bacterial solution. Aerosol infections were performed with fresh aliquots of M. abscessus strains grown on 7H9 as described previously (41), to achieve an inoculum of 1 × 108 mycobacteria. When mice were infected by aerosolized M. abscessus strains cocultured with amoebae, the infected amoebae were prepared by rapping the flasks vigorously and centrifuging at 1,000 × g for 5 min. The resulting pellet was suspended in 10 ml of PBS and adjusted to a concentration of 2 × 106 CFU/ml after SDS lysis of amoebae. Lungs, livers, and spleens were collected in sterile distilled water and homogenized, and 10-fold serial dilutions were then plated on VCA3 plates (vancomycin, colimycin, and amphotericin B; bioMérieux, France) for CFU enumeration. Plates were incubated at 37°C up to 5 days. Results were expressed as the mean log10 CFU per organ. The minimum detection limit per organ was 20 CFU (or 1.3 log10 CFU) per lung, spleen, or liver. A two-way analysis of variance (ANOVA) with a Tukey posttest were performed using GraphPad prism program version 5 for statistical comparison.
Statistical analysis.
Fisher's exact test and Student's t test were used. A P value of <0.05 was considered significant.
RESULTS
M. abscessus PLC resembles the PLC from CF pathogens.
Ripoll et al. (11) reported the presence of a PLC-encoding gene (MAB_0555) in the M. abscessus genome. MAB_0555 has a length of 1,440 bp and encodes a 479-amino-acid protein (11). Inspection of the syntenic genomic regions and comparison with other mycobacterial genomes suggest that MAB_0555 was inserted just downstream of MAB_0554, which encodes a potential hydrolase/lipase (11) that is conserved in a wide range of fast-growing and slowly growing mycobacteria (see Fig. S1 in the supplemental material). Phylogenetic analysis demonstrated a nonmycobacterial origin of MAB_0555, and more recent protein comparison performed for this study confirmed this trait (Table 1). The strongest identities were observed with several recently discovered Actinomycetes but more importantly in several major CF pathogens (Table 1): Burkholderia cenocepacia PLC (43% identity), Pseudomonas aeruginosa nonhemolytic and hemolytic PLCs (45% and 41% identity, respectively), Burkholderia multivorans PLC (41% identity), and Stenotrophomonas maltophilia putative nonhemolytic PLC (41% identity). Of note, a 29% identity was also observed with PLC of another CF pathogen, Aspergillus fumigatus putative phosphatidylglycerol PLC. By comparison, the percent identity with the 4 PLCs present in M. tuberculosis varied between 38 and 42% (Table 1).
TABLE 1.
Maximal percentage of identity between the different PLC amino acid sequences from M. abscessus and M. tuberculosis or nonmycobacterial microorganismsa
| Organism | Uniprot no. | Protein name | Size (aa) | Maximal % identity | Length of homology region (aa) |
|---|---|---|---|---|---|
| Nonmycobacterial bacteria | |||||
| Pseudomonas aeruginosa | P15713* | Nonhemolytic PLC | 692 | 45 | 503 |
| Burkholderia cenocepacia | A0KBL6 | PLC | 723 | 43 | 538 |
| Ralstonia pickettii | B2UDZ8 | PLC, phosphocholine specific | 700 | 42 | 521 |
| Burkholderia cenocepacia | A0B2U2 | PLC | 704 | 42 | 519 |
| Pseudomonas aeruginosa | P06200 | Hemolytic PLC | 730 | 42 | 515 |
| Bordetella avium | Q2KUR2 | Nonhemolytic PLC | 693 | 42 | 509 |
| Stenotrophomonas maltophilia | B2FL44 | Putative nonhemolytic PLC | 706 | 41 | 525 |
| Burkholderia cenocepacia | A0K5Z1 | PLC | 714 | 41 | 516 |
| Burkholderia multivorans | B3CZ34 | PLC | 718 | 41 | 515 |
| Burkholderia multivorans | A9ADA7 | PLC, phosphocholine specific | 752 | 41 | 496 |
| Bordetella avium | Q2L0W2 | Nonhemolytic PLC | 723 | 40 | 530 |
| Ralstonia pickettii | B2UCT8 | PLC, phosphocholine specific | 719 | 40 | 520 |
| Burkholderia multivorans | A9ALB9 | PLC | 771 | 39 | 519 |
| Bordetella bronchiseptica | Q7WH05 | Putative phospholipase | 627 | 39 | 425 |
| Burkholderia cenocepacia | A0B3P1 | PLC | 777 | 38 | 559 |
| Pseudomonas fluorescens | Q4KC01 | PLC, phosphocholine specific | 715 | 38 | 546 |
| Ralstonia pickettii | B2UJE7 | PLC | 474 | 30 | 456 |
| Burkholderia multivorans | B3CZQ1 | PLC | 528 | 30 | 214 |
| Burkholderia multivorans | B3D7M3 | PLC | 554 | 30 | 204 |
| Burkholderia multivorans | A9AKI0 | PLC | 556 | 29 | 182 |
| Burkholderia cenocepacia | A0B0A3 | PLC | 557 | 29 | 180 |
| Mycobacteria | |||||
| Mycobacterium tuberculosis | P9WIA8* | PLC 4 | 514 | 42 | 465 |
| Mycobacterium tuberculosis | P9WIB0* | PLC 3 | 517 | 40 | 496 |
| Mycobacterium tuberculosis | P9WIB2* | PLC 2 | 521 | 39 | 504 |
| Mycobacterium tuberculosis | P9WIB4* | PLC 1 | 520 | 38 | 503 |
| Fungi | |||||
| Aspergillus fumigatus | B0XWP7 | Phosphatidylglycerol specific phospholipase, putative | 492 | 29 | 392 |
| Aspergillus fumigatus | B0YCK0 | Phosphatidylglycerol specific PLC, putative | 509 | 26 | 406 |
| Aspergillus fumigatus | B0XPD6 | Phosphoesterase superfamily protein | 456 | 26 | 396 |
Alignments were performed with Basic Local Alignment Search Tool (BLAST) program at http://blast.ncbi.nlm.nih.gov, using the protein BLAST algorithm. aa, amino acids. *, PLC was biochemically characterized.
M. abscessus purified recombinant PLC hydrolyzes eukaryotic cells. (i) Expression in M. smegmatis and purification.
rPLCMa was purified after acetamide induction. The ionic detergent Sarkosyl (1%) in the lysis buffer allowed solubilization and purification of the recombinant enzyme using immobilized metal ion affinity chromatography and fast protein liquid chromatography (IMAC-FPLC). Under these conditions, 10 to 15 mg of pure rPLCMa was obtained per liter of culture. SDS-PAGE and MALDI-TOF analysis of purified PLC showed that the apparent molecular mass of recombinant protein was compatible (Fig. 1A) with the expected theoretical molecular mass (52 kDa) based on the amino acid sequence, including the full-length TAT signal peptide (noncleaved), as previously noticed for M. tuberculosis PLCs (36). We then used approaches similar to those described previously (36) for the evaluation of M. abscessus PLC.
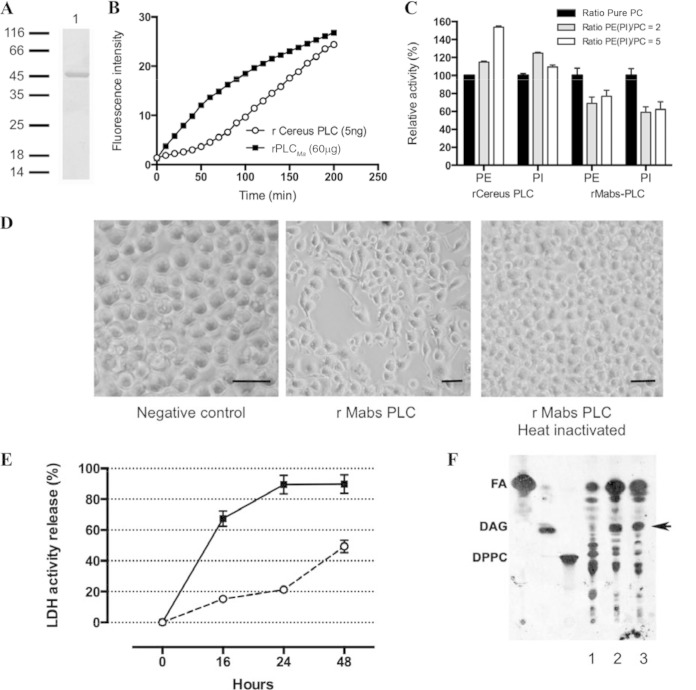
FIG 1.
(A, B, and C) Biochemical characterization of rPLCMa. (A) Purified rPLCMa was loaded on 12% SDS-PAGE. Molecular weight standards are on the left. Lane 1 contained purified rPLCMa (10 μg). The sample was loaded under reducing conditions, and the gel was then stained with Coomassie brilliant blue. (B) Time course hydrolysis of phosphatidylcholine (PC) by 60 μg of rPLCMa (■) or 5 ng of recombinant B. cereus PC-PLC (○). The release of phosphocholine was measured indirectly by the fluorescence measurement of resorufin released using the Amplex red phosphatidylcholine kit and continuously monitored at a λexc of 510 nm and λem of 590 nm, (C) Substrate preference of rPLCMa and recombinant B. cereus PC-PLC. Competition assays between PC and phosphatidylethanolamine (PE) (or phosphatidylinositol [PI]) were carried out using different phospholipid ratios (PE [or PI]/PC = 2 or 5) and pure PC. A final PC quantity of 0.1 μmol was used in the pure-PC assay, and 0.2 μmol and 0.5 μmol of PE (or PI) were used for the other PE (PI)/PC ratios. The PC-PLC activity was continuously measured using the Amplex red PC-PLC kit. The relative activity (percent) of PC hydrolysis was calculated from the ratio of PC activity in the presence of PE (or PI) over PC activity in the absence of PE (or PI). (D, E, and F) Cytotoxic effects of rPLCMa on mouse macrophages. (D) Macrophage cellular state after 24 h of incubation, with no recombinant PLC (buffer only [negative control]), with 50 μg of purified rPLCMa (rPLCMa), and with 50 μg of heat-inactivated purified rPLCMa (rPLCMa, heat inactivated), as shown by light microscopy (magnification, ×200; bar, 25 μm). (E) Purified rPLCMa (50 μg; ■) and 15 μg of PC-PLC from B. cereus (○) were incubated with 1 × 106 RAW264.7 mouse macrophages. The values are shown as percent lysis, in which the amount of LDH released in wells with macrophages incubated with enzymes was compared to total LDH released in control wells in which all of the macrophages had been deliberately lysed. LDH released was quantified at 16, 24, and 48 h. The values are the means for triplicate samples. (F) Autoradiography of TLC plate showing the release of radiolabeled DAG, the product of phospholipid hydrolysis (indicated by the black arrow), after incubation of rPLCMa (50 μg, lane 2) and PC-PLC from B. cereus (15 μg, lane 3) with radiolabeled macrophages. For the negative control (lane 1), only the buffer without any pure enzyme was added in the incubation medium. Abbreviations: DAG; diacylglycerol; FA, free fatty acid; DPPC, dipalmitoyl-glycerophosphocholine.
(ii) Biochemical hydrolytic activity of rPLCMa.
rPLCMa was able to hydrolyze p-NPPC with turnover and specific activity (5.68 mmol min−1 mg−1) similar to those of B. cereus and C. perfringens PLCs (7.75 mmol min−1 mg−1 and 3.1 mmol min−1 mg−1, respectively) (see Fig. S2A in the supplemental material). Comparatively, rPLCMa specific activity (0.1 nmol min−1 mg−1) measured using PC as the substrate (Fig. 1B) was far below the specific activity observed for B. cereus PC-PLC (165 nmol min−1 mg−1). It is noteworthy that both B. cereus PC-PLC and rPLCMa hydrolyze the same substrate; however, the catalytic mechanisms differ between the proteins, since the effect of D609 inhibitor totally abolished the activity of B. cereus PLC, while it had no effect on rPLCMa activity (data not shown).
rPLCMa was active within a large range of temperatures (25 to 55°C), its optimal temperature being 37°C, with an optimal pH of 7 to 7.5 (see Fig. S2B and C in the supplemental material). This activity was stable for the first 48 h but was completely abolished upon heat inactivation.
No sphingomyelinase activity was observed for rPLCMa (data not shown). However, the rPLCMa and B. cereus PC-PL substrate preferences and the activities of both enzymes were determined in the presence of different ratios of PI/PC or PE/PC (PE [or PI]/PC ratios = 2 and 5) (Fig. 1C). Under these experimental conditions, the hydrolysis of only the PC can be detected, and the substrate preference was deduced by the increasing or decreasing activity on PC. The PC-PLC activity of the B. cereus enzyme increased significantly (by a factor of 1.5) in the presence of PE, suggesting a better PC substrate presentation in the presence of PE. In contrast, the PC-PLC activity of rPLCMa decreased by a factor of 1.5 in the presence of PE (Fig. 1C). Similar results were obtained in the presence of PI (Fig. 1C), suggesting no substrate preference. Finally, when a mixture of phospholipids was used as the substrate (Fig. 1C), the behavior of both enzymes can vary, again suggesting a different hydrolysis mechanism between B. cereus PC-PLC and rPLCMa.
(iii) rPLCMa lyses eukaryotic cells.
rPLCMa did not lyse erythrocytes, unlike B. cereus or C. perfringens PLCs (data not shown). Comparatively, rPLCMa exhibited a strong cytotoxic effect on mouse macrophages (Fig. 1D) and on the amount of lactate dehydrogenase (LDH) released (Fig. 1E). A level of macrophage lysis of 17% was detected after 16 h of incubation and increased over time to reach 55% after 48 h, while the cell monolayer was not affected in the presence of heat-inactivated rPLCMa.
We next evaluated rPLCMa macrophage cytotoxicity with [1-14C]palmitic acid incorporation into membrane phospholipids, and measuring the amount of radiolabeled membrane phospholipids released after membrane degradation. Purified rPLCMa released radiolabeled diacylglycerol (DAG), unlike the control sample (Fig. 1F). Altogether, we demonstrated that rPLCMa is active and possesses cytotoxic activity against macrophages, mainly by degrading the phospholipids of the cell membrane.
M. abscessus PLC as a virulence factor in eukaryotic cell infection models. (i) Construction of an M. abscessus PLC KO mutant.
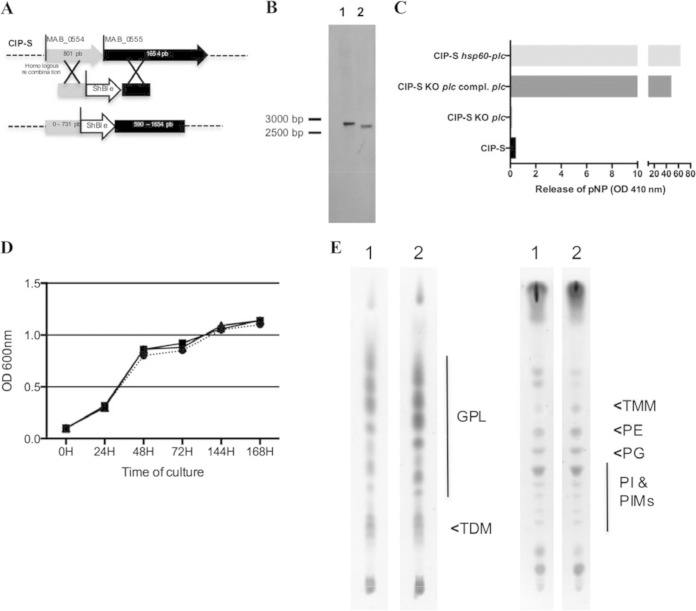
An M. abscessus PLC KO mutant was obtained by allelic exchange using the recombineering system (38). For the construction of the mutant, a zeocin cassette was inserted between the 3′ end of MAB_0554 and the 5′ end of MAB_0555, thereby completely disrupting the PLC-encoding gene MAB_0555 (Fig. 2A). Southern blot analysis allowed us to confirm that double crossing-over and disruption of MAB_0555 had occurred (Fig. 2B). We then constructed a PLC-complemented version by inserting a pMVZ361-hsp60pro-MAB_0555 plasmid into the KO mutant. For control purposes, we also inserted the same plasmid into the WT M. abscessus strain. Measurement of the PLC activity of the parental CIP-S strain, the PLC KO mutant, the complemented mutant, and the WT strain carrying the additional copy of PLC supplied by pMVZ361-hsp60pro-MAB_0555 revealed low and no PLC activity for the WT strain and the PLC M. abscessus KO mutant, respectively. These findings argue for low expression of PLC under in vitro conditions and confirmed the disruption of MAB_0555 in the KO mutant (Fig. 2C). In contrast, expression of PLC in either of the pMVZ361-hsp60pro-MAB_0555-carrying strains was associated with high and comparable activity (Fig. 2C). Of note, in vitro growth characteristics of the WT, the PLC M. abscessus KO mutant, and its complemented version were similar (Fig. 2D). In addition, lipid extract comparison by TLC (Fig. 2E) and MS (data not shown) between the WT strain and PLC M. abscessus KO mutant did not highlight qualitative or quantitative differences in parietal lipid composition between both strains.
FIG 2.
Construction of the M. abscessus plc KO mutant by homologous recombination (HR). (A) MAB_0554 and MAB_0555 are separated by 4 bp. The M. abscessus plc KO mutant was thus constructed by amplifying a nearly 2-kbp M. abscessus fragment encompassing this region and cloning the zeocin resistance gene (S. hindustanus ble) into the HindIII (position 731 in MAB_0554)-ClaI (position 590 in MAB_0555) sites. The entire fragment was then electroporated into M. abscessus CIP-S containing the pJV53 plasmid inserted by HR into the M. abscessus chromosome. (B) Southern blotting analysis was performed after genomic DNA restriction by KpnI and gel electrophoresis. A 532-bp probe targeting the 3′ conserved region of MAB_0555 was used for hybridization with DNA fragments: a 2,704-bp band is observed with the WT strain (lane 1), and a 2,635-bp band is observed with the M. abscessus KO mutant (lane 2). (C) Respective PLC activity of the different constructed M. abscessus strains. Phospholipase C activity was measured with p-NPPC for the CIP-S, CIP-S plc KO, CIP-S plc KO plc-complemented, and CIP-S-hsp60-plc (M. abscessus pMVZ361-hsp60pro-MAB_0555, used as a control for PLC activity) strains. (D) In vitro growth curves estimated by spectrophotometry (OD600) of CIP-S (circles), CIP-S plc KO (squares), and CIP-S plc KO plc-complemented (triangles) strains. (E) Glycolipid and phospholipid patterns of the different constructed M. abscessus strains. Total lipid contents of the WT M. abscessus strain (lanes 1) and the plc KO M. abscessus mutant (lanes 2) were analyzed by TLC using CHCl3-CH3OH (90:10, vol/vol) (left) and CHCl3-CH3OH-H2O (60:35:8, vol/vol/vol) (right) as the solvent systems and anthrone revelation (GPLs, glycopeptidolipids; TDM, trehalose dimycolate; TMM, trehalose monomycolate; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PIMs, phosphatidylinositol mannosides).
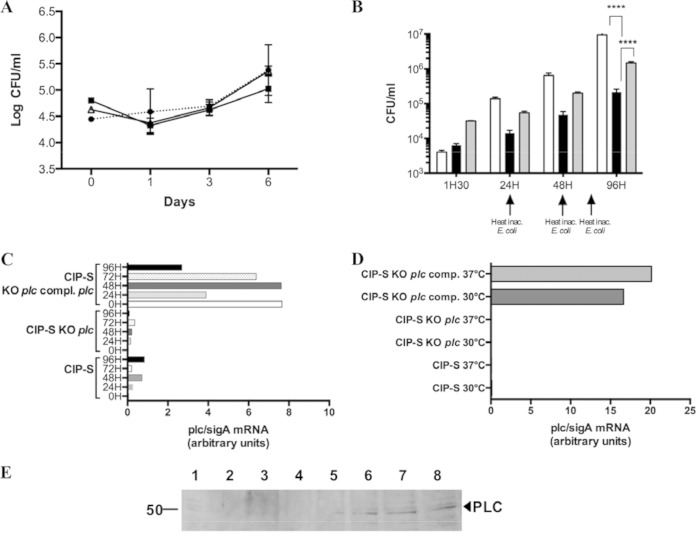
(ii) Survival of the PLC M. abscessus KO mutant in BMDMs.
To investigate the role of PLC in intracellular survival of M. abscessus, BMDMs were cocultivated with the WT strain, the PLC M. abscessus KO mutant, and the complemented PLC M. abscessus KO mutant. The absence of MAB_0555 (PLC) failed to alter the growth of the WT strain in BMDMs, as the WT, PLC mutant, and complemented strains exhibited similar growth throughout the experiment (Fig. 3A). The comparable intracellular growth rate strongly suggests that the PLC activity does not confer any supplementary advantage to M. abscessus within murine macrophages.
FIG 3.
Survival of plc M. abscessus mutant in eukaryotic cells. (A) Growth of mycobacterial strains within BMDMs recorded by CFU evaluation after 1, 3, and 6 days of coculture. The CIP-S (circles), CIP-S plc KO (squares), and CIP-S plc KO plc-complemented (triangles) strains were used. (B) Growth of mycobacterial strains within amoebae recorded by CFU evaluation after 1.5 h and 1, 2, and 4 days of coculture. CIP-S (white bars), CIP-S plc KO (black bars), and CIP-S plc KO plc-complemented (gray bars) strains were used. Experiments were repeated five times in triplicate at different times for both panels A and B (***, P < 0.001). (C) mRNA plc/sigA ratio (in arbitrary units) for the CIP-S, CIP-S plc KO, and CIP-S plc KO plc-complemented strains cocultivated with macrophages for 5 days. (D) mRNA plc/sigA ratio (in arbitrary units) for the CIP-S, CIP-S plc KO, and CIP-S plc KO plc-complemented strains cultivated in rich medium (7H9) at 30°C or 37°C. The results are representative of two independent experiments (C and D). (E) Western blot analysis of PLC expression during coculture of mycobacterial strains with A. castellanii. Lane 1, total extract (30 μg) of CIP-S cultivated in 7H9 medium; lane 2, total extract (30 μg) of amoebae cultivated for 96 h in PAS buffer in the absence of mycobacteria; lanes 3 to 7, total extract (30 μg) of amoebae cocultivated for 3 h (lane 3), 24 h (lane 4), 48 h (lane 5), 72 h (lane 6), or 96 h (lane 7) in PAS buffer in the presence of CIP-S; lane 8, total extract (30 μg) of amoebae cocultivated for 48 h in PAS buffer in the presence of the CIP-S plc KO plc-complemented strain. This picture is representative of three independent experiments.
(iii) Survival of the PLC M. abscessus KO mutant in A. castellanii.
A. castellanii and M. abscessus strains were cocultured in order to evaluate the PLC contribution to M. abscessus survival in amoebae (23). As shown in Fig. 3B, WT M. abscessus was able to replicate and survive inside amoebae, although it was unable to grow in the PAS amoeba medium used during the coculture (data not shown). By comparison, the M. abscessus PLC KO mutant was greatly impaired and unable to survive throughout the course of the experiment (Fig. 3B). The lack of PLC seems to have been directly involved in this result, since the complemented PLC M. abscessus KO mutant strain was able to grow (although less than the WT) and to survive inside amoebae (Fig. 3B). Of note, the PLC M. abscessus KO mutant strain complemented only with the MAB_0554 gene behaved like the PLC M. abscessus KO mutant strain, confirming that the observed defect was not due to the 3′ loss of MAB_0554 when the PLC KO mutant was being constructed (data not shown). Finally, we could not evaluate if growth in amoebae affects the production of the mycobacterial glycolipids, as the number of mycobacteria obtained after coculturing is insufficient to carry out a glycolipid analysis by TLC. In addition, the smooth morphotype was very stable after coculturing in amoebae, as previously demonstrated (13, 41, 43).
The different behavior of the mutant in amoebae or BMDMs suggested the putative occurrence of de novo synthesis of PLC when M. abscessus is cocultivated with A. castellanii, which is of particular importance given the very low PLC production in in vitro-grown M. abscessus (Fig. 2C). We therefore further assessed this phenomenon by analyzing and comparing PLC expression at a transcriptional and translational level.
Expression of PLC is induced when M. abscessus is cocultured in A. castellanii.
MAB_0555 mRNA expression was quantified by RT-PCR after intramacrophage coculture (Fig. 3C) and in vitro growth (30°C and 37°C) of the different strains (Fig. 3D). Under both ex vivo and in vitro conditions, specific MAB_0555 transcriptional expression was observed only in the complemented strains carrying the pMVZ361::MAB_0555 driving PLC expression under the control of the constitutive hsp60 promoter (Fig. 3C and D). Absence of specific MAB_0555 mRNA expression was observed for the wild-type and the PLC-KO M. abscessus strains in macrophages (Fig. 3C) and between the two growth temperatures in vitro (Fig. 3D).
In order to confirm the presence of the expressed PLC by the wild-type strain in amoebae, Western blot (WB) analysis using polyclonal antibodies was preferred to RT-PCR. WB clearly revealed the presence of PLC when bacteria were cocultured in amoebae compared to mycobacteria grown in 7H9 rich medium, despite a strong amoeba proteolytic activity during preparation of the protein extract (Fig. 3E). A 47-kDa protein, which corresponds to the native molecular mass of PLC (without the TAT signal peptide), was detected only in the presence of amoebae after 48 h until 96 h of coculture with either the WT (Fig. 3E, lanes 5 to 7) or the complemented strains (Fig. 3E, lane 8).
In vivo behavior of the PLC M. abscessus KO mutant in the aerosol mouse model of infection.
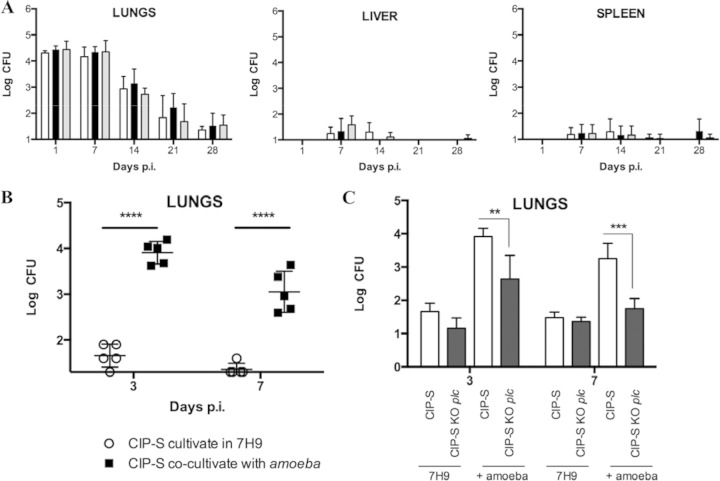
All three M. abscessus strains were independently used to infect mice by aerosol for 15 min using an inoculum of 1 × 108 mycobacteria. CFU were enumerated in lungs, livers, and spleens at days 1, 7, 14, 21, and 28. A similar decline was observed for WT strain, the PLC M. abscessus KO mutant, and the complemented PLC M. abscessus KO mutant (Fig. 4A), in agreement with results from the BMDM infection studies.
FIG 4.
Effect of coculture of mycobacterial strains within amoebae on virulence in mice. (A) BALB/c mice were aerosolized with the CIP-S (white bars), CIP-S plc KO (black bars), and CIP-S plc KO plc-complemented (gray bars) strains cultivated in 7H9 medium. Mice were challenged with 108 mycobacteria and sacrificed at days 1, 7, 14, 21, and 28. Lungs, livers, and spleens were collected, homogenized, diluted (1/1, 1/5, 1/25, and 1/125), and cultured on VCA3 agar plates. CFU were counted after 5 days of growth. Twenty-five mice per group were challenged. (B) BALB/c mice were aerosolized with CIP-S cultivated in 7H9 medium (○) or obtained after 3 days of coculture in amoebae (■). Mice were sacrificed at days 3 and 7. Lungs were collected, diluted (1/1, 1/5, 1/25, and 1/125), and cultured on VCA3 agar plates. CFU were counted for the different dilutions after 5 days of growth. Ten mice per group were challenged. (****, P < 0.0001). (C) BALB/c mice were aerosolized with the CIP-S (empty bars) or CIP-S plc KO (black bars) strain cultivated in 7H9 medium or obtained after 3 days of coculture within amoebae. Mice were sacrificed at days 3 and 7. Lungs were collected, diluted (1/1, 1/5, 1/25, and 1/125), and cultured on VCA3 agar plates. CFU were counted after 5 days of growth. Ten mice per group were challenged. (**, P < 0.01; ***, P < 0.001).
Since coculture of M. abscessus and A. castellanii significantly induces the expression of PLC in M. abscessus, we therefore addressed the question of whether a previous coculture of M. abscessus with A. castellanii would impact the M. abscessus virulence phenotype in the mouse model of infection. As shown in Fig. 3B, we recovered from cocultures of M. abscessus in amoebae only 2 × 106 mycobacteria, which was 100 times less than the in vitro-prepared inoculum. At this infectious dose, CFU counts only in the lungs were performed, as no mycobacteria could be recovered from livers and spleens (Le Moigne et al., unpublished). However, after only one expansion in amoebae, we obtained significantly higher bacterial loads in the lungs at 3 and 7 days postinfection (CFU per lungs) than with the same strain grown in vitro in rich 7H9 medium (Fig. 4B). Amoeba lysate itself added to M. abscessus before aerosol challenge was ineffective in increasing M. abscessus bacterial load in the lungs compared to coculturing M. abscessus within amoebae (data not shown).
The role of PLC expression in the enhanced survival in the lungs of immunocompetent mice was then explored by testing the phenotype of the PLC-deficient isogenic mutant cocultured with A. castellanii. Experiments demonstrated that the bacterial burden achieved by the M. abscessus plc KO mutant was significantly lower at 3 days (P < 0.01) and at 7 days (P < 0.001) postinfection than the one achieved by the WT M. abscessus strain (Fig. 4C). Taken together, these results indicate that growth of M. abscessus in Acanthamoeba increases virulence of M. abscessus in mice, most likely by inducing the expression of PLC.
DISCUSSION
Phospholipases are considered key virulence factors and are synthesized by bacterial species causing disparate infectious disease, from infection causing massive tissue destruction to food-borne diseases. Some well-studied PLCs involved in virulence are the α toxin of Clostridium perfringens, the PLC-H and PLC-N of P. aeruginosa, and the two PI-PLCs of Listeria monocytogenes. Cytolysis is the most common characteristic attributed to bacterial phospholipase virulence factors (31, 32). We demonstrated that M. abscessus synthesizes a PLC with lytic activity against eukaryotic cells, with membrane phospholipid degradation and DAG production. We were not able to demonstrate a hemolytic activity for M. abscessus PLC, indicating that it resembles P. aeruginosa PLC-N, which also lacks hemolytic activity (44). This aspect might be crucial for degradation and penetration of the mucus layer present in the lungs, as observed for P. aeruginosa (31, 32). As with eukaryotic PLC, the hydrolysis of phospholipids by M. abscessus PLC leads to the production of DAG, a well-known lipid second messenger. DAG has been shown to activate protein kinase C, which is known to modulate the activation of neutrophils and macrophages (45). This activity further emphasizes the inflammatory response observed in CF lungs and participates in the increased virulence of M. abscessus in CF patients compared to other mycobacteria (46).
Apart from playing a role in virulence, PLCs are thought to function in phosphate and carbon source acquisition (32). Synthesis of both P. aeruginosa PLC-H and PLC-N is regulated by inorganic phosphate at the transcriptional level by the positive regulator PhoB (47). Induction of PI-PLC synthesis was also shown for other bacteria in such a limited-resource environment. PI-PLC activity of Lactobacillus rhamnosus markedly depends on the amount of carbohydrate in the culture medium (48). Similar reports showed that PI-PLC-producing bacteria, including B. cereus, Bacillus thuringiensis, L. monocytogenes, and Staphylococcus aureus, required glucose-free medium for PLC activity to be detected (49–51). We demonstrated the absence of growth of M. abscessus in low-nutrient medium like PAS (data not shown) and correlatively a growth in the presence of amoebae, indicating that amoebae helped M. abscessus survive in a low-nutrient and/or eukaryotic environment. More importantly, amoeba-cocultured M. abscessus expressed virulence factors to be able to infect more aggressively the host as shown by the de novo production of PLC, as observed with the increased virulence of M. abscessus in mice when cocultured with Acanthamoeba compared to culture in 7H9 medium. Several mycobacteria were shown to survive in amoebae (12), but increased virulence after passage on amoebae was only shown for M. avium (21). PLC expression was stimulated when M. abscessus was grown intracellularly in Acanthamoeba but not in macrophages, thus pointing out the possible existence of regulatory networks similar to those observed in low-nutrient environments. In contrast, not much is presently known about induction of M. tuberculosis PLCs (29) and the role of the PhoP/PhoR regulatory network in the synthesis of PLC (52).
From the present data, we conclude that M. abscessus does not exhibit a clear substrate preference, as it was not possible to distinguish the phospholipids (PE, PI, or PC) preferentially hydrolyzed by M. abscessus PLC. An advantage conferred by the M. abscessus PLC activity might thus be the local degradation in the lungs of the dipalmitoyl-phosphatidylcholine, a major component of the surfactant, which would provide nutrients and/or osmoprotectants (47) necessary for M. abscessus survival in lung tissue. In CF patients, the thickening of the surfactant may thereby promote colonization and implementation of bacteria benefiting from PLC activity (Table 1). Colonizing CF lungs with already de novo-synthesized PLC would represent a major advantage for M. abscessus and might explain its specific tropism compared to other closely related RGM, such as M. chelonae. It remains to be established whether M. abscessus expresses PLC activity in infected patients. However, we have preliminary serological data confirming an anti-PLC antibody response in M. abscessus-infected CF patients (Le Moigne et al., unpublished).
Of importance was the choice of three different ex vivo and in vivo models to decipher the role of the PLC in M. abscessus virulence, as it helped us define the eukaryotic environment, where PLC activity was crucial for M. abscessus survival. Amoeba-bacterium interactions have recently been well defined (53), notably for mycobacteria (54, 55), and helped us in this study to consider the role of amoebae in shaping virulence of the bacteria toward a pathogenic phenotype (18, 53, 56, 57). Exchange and acquisition of genetic material have helped M. abscessus in gaining key virulence factors compared to other RGM. These de novo-synthesized virulence factors might explain the peculiar link between M. abscessus and CF lungs. Finally, it should be mentioned that identification of an environmental source where M. abscessus might come into contact with environmental amoebae would be very important for learning more about the potential contamination risks for CF patients. We are presently seeking such hidden sources of M. abscessus infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Laurent Kremer for critical reading of the manuscript.
We thank Vaincre La Mucoviscidose (VLM IC1014 and RF20120600689) and the Région Ile-de-France (Domaine d'Intérêt Majeur Maladies Infectieuses et Emergentes) for funding the postdoctoral fellowship to V.L.M. and a Ph.D. program for F.L.C. We are also grateful for the support from the Centre National de la Recherche Scientifique (CNRS).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02032-14.
REFERENCES
- 1.Kusunoki S, Ezaki T. 1992. Proposal of Mycobacterium peregrinum sp. nov., nom. rev., and elevation of Mycobacterium chelonae subsp. abscessus (Kubica et al.) to species status: Mycobacterium abscessus comb. nov. Int J Syst Bacteriol 42:240–245. doi: 10.1099/00207713-42-2-240. [DOI] [PubMed] [Google Scholar]
- 2.Griffith DE, Girard WM, Wallace RJ. 1993. Clinical features of pulmonary disease caused by rapidly growing mycobacteria. An analysis of 154 patients. Am Rev Respir Dis 147:1271–1278. [DOI] [PubMed] [Google Scholar]
- 3.Brown-Elliott BA, Wallace RJ. 2002. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin Microbiol Rev 15:716–746. doi: 10.1128/CMR.15.4.716-746.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olivier KN, Weber DJ, Wallace RJ, Faiz AR, Lee J-H, Zhang Y, Brown-Elliot BA, Handler A, Wilson RW, Schechter MS, Edwards LJ, Chakraborti S, Knowles MR. 2003. Nontuberculous mycobacteria. I: multicenter prevalence study in cystic fibrosis. Am J Respir Crit Care Med 167:828–834. doi: 10.1164/rccm.200207-678OC. [DOI] [PubMed] [Google Scholar]
- 5.Jönsson BE, Gilljam M, Lindblad A, Ridell M, Wold AE, Welinder-Olsson C. 2007. Molecular epidemiology of Mycobacterium abscessus, with focus on cystic fibrosis. J Clin Microbiol 45:1497–1504. doi: 10.1128/JCM.02592-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roux A-L, Catherinot E, Ripoll F, Soismier N, Macheras E, Ravilly S, Bellis G, Vibet M-A, Le Roux E, Lemonnier L, Gutierrez C, Vincent V, Fauroux B, Rottman M, Guillemot D, Gaillard J-L. 2009. Multicenter study of prevalence of nontuberculous mycobacteria in patients with cystic fibrosis in France. J Clin Microbiol 47:4124–4128. doi: 10.1128/JCM.01257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wallace RJ, Brown BA, Griffith DE. 1998. Nosocomial outbreaks/pseudo-outbreaks caused by nontuberculous mycobacteria. Annu Rev Microbiol 52:453–490. doi: 10.1146/annurev.micro.52.1.453. [DOI] [PubMed] [Google Scholar]
- 8.Viana-Niero C, Lima KVB, Lopes ML, da Silva Rabello MC, Marsola LR, Brilhante VCR, Durham AM, Leão SC. 2008. Molecular characterization of Mycobacterium massiliense and Mycobacterium bolletii in isolates collected from outbreaks of infections after laparoscopic surgeries and cosmetic procedures. J Clin Microbiol 46:850–855. doi: 10.1128/JCM.02052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zelazny AM, Root JM, Shea YR, Colombo RE, Shamputa IC, Stock F, Conlan S, McNulty S, Brown-Elliott BA, Wallace RJ, Olivier KN, Holland SM, Sampaio EP. 2009. Cohort study of molecular identification and typing of Mycobacterium abscessus, Mycobacterium massiliense, and Mycobacterium bolletii. J Clin Microbiol 47:1985–1995. doi: 10.1128/JCM.01688-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, Inns T, Reacher M, Haworth CS, Curran MD, Harris SR, Peacock SJ, Parkhill J, Floto RA. 2013. Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381:1551–1560. doi: 10.1016/S0140-6736(13)60632-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ripoll F, Pasek S, Schenowitz C, Dossat C, Barbe V, Rottman M, Macheras E, Heym B, Herrmann J-L, Daffé M, Brosch R, Risler J-L, Gaillard J-L. 2009. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One 4:e5660. doi: 10.1371/journal.pone.0005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamrabet O, Merhej V, Pontarotti P, Raoult D, Drancourt M. 2012. The genealogic tree of mycobacteria reveals a long-standing sympatric life into free-living protozoa. PLoS One 7:e34754. doi: 10.1371/journal.pone.0034754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernut A, Herrmann J-L, Kissa K, Dubremetz J-F, Gaillard J-L, Lutfalla G, Kremer L. 2014. Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation. Proc Natl Acad Sci U S A 111:E943–E952. doi: 10.1073/pnas.1321390111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falkinham JO. 2009. Surrounded by mycobacteria: nontuberculous mycobacteria in the human environment. J Appl Microbiol 107:356–367. doi: 10.1111/j.1365-2672.2009.04161.x. [DOI] [PubMed] [Google Scholar]
- 15.Thomas V, Herrera-Rimann K, Blanc DS, Greub G. 2006. Biodiversity of amoebae and amoeba-resisting bacteria in a hospital water network. Appl Environ Microbiol 72:2428–2438. doi: 10.1128/AEM.72.4.2428-2438.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von Reyn CF, Waddell RD, Eaton T, Arbeit RD, Maslow JN, Barber TW, Brindle RJ, Gilks CF, Lumio J, Lähdevirta J. 1993. Isolation of Mycobacterium avium complex from water in the United States, Finland, Zaire, and Kenya. J Clin Microbiol 31:3227–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montecalvo MA, Forester G, Tsang AY, du Moulin G, Wormser GP. 1994. Colonisation of potable water with Mycobacterium avium complex in homes of HIV-infected patients. Lancet 343:1639. doi: 10.1016/S0140-6736(94)93093-7. [DOI] [PubMed] [Google Scholar]
- 18.Greub G, Raoult D. 2004. Microorganisms resistant to free-living amoebae. Clin Microbiol Rev 17:413–433. doi: 10.1128/CMR.17.2.413-433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feazel LM, Baumgartner LK, Peterson KL, Frank DN, Harris JK, Pace NR. 2009. Opportunistic pathogens enriched in showerhead biofilms. Proc Natl Acad Sci U S A 106:16393–16399. doi: 10.1073/pnas.0908446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radomski N, Cambau E, Moulin L, Haenn S, Moilleron R, Lucas FS. 2010. Comparison of culture methods for isolation of nontuberculous mycobacteria from surface waters. Appl Environ Microbiol 76:3514–3520. doi: 10.1128/AEM.02659-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cirillo JD, Falkow S, Tompkins LS, Bermudez LE. 1997. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect Immun 65:3759–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinert M, Birkness K, White E, Fields B, Quinn F. 1998. Mycobacterium avium bacilli grow saprozoically in coculture with Acanthamoeba polyphaga and survive within cyst walls. Appl Environ Microbiol 64:2256–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adékambi T, Ben Salah S, Khlif M, Raoult D, Drancourt M. 2006. Survival of environmental mycobacteria in Acanthamoeba polyphaga. Appl Environ Microbiol 72:5974–5981. doi: 10.1128/AEM.03075-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas V, McDonnell G. 2007. Relationship between mycobacteria and amoebae: ecological and epidemiological concerns. Lett Appl Microbiol 45:349–357. doi: 10.1111/j.1472-765X.2007.02206.x. [DOI] [PubMed] [Google Scholar]
- 25.Mba Medie F, Ben Salah I, Henrissat B, Raoult D, Drancourt M. 2011. Mycobacterium tuberculosis complex mycobacteria as amoeba-resistant organisms. PLoS One 6:e20499. doi: 10.1371/journal.pone.0020499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adékambi T, Reynaud-Gaubert M, Greub G, Gevaudan M-J, La Scola B, Raoult D, Drancourt M. 2004. Amoebal coculture of “Mycobacterium massiliense” sp. nov. from the sputum of a patient with hemoptoic pneumonia. J Clin Microbiol 42:5493–5501. doi: 10.1128/JCM.42.12.5493-5501.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rottman M, Catherinot E, Hochedez P, Emile J-F, Casanova J-L, Gaillard J-L, Soudais C. 2007. Importance of T cells, gamma interferon, and tumor necrosis factor in immune control of the rapid grower Mycobacterium abscessus in C57BL/6 mice. Infect Immun 75:5898–5907. doi: 10.1128/IAI.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Byrd TF, Lyons CR. 1999. Preliminary characterization of a Mycobacterium abscessus mutant in human and murine models of infection. Infect Immun 67:4700–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raynaud C, Guilhot C, Rauzier J, Bordat Y, Pelicic V, Manganelli R, Smith I, Gicquel B, Jackson M. 2002. Phospholipases C are involved in the virulence of Mycobacterium tuberculosis. Mol Microbiol 45:203–217. doi: 10.1046/j.1365-2958.2002.03009.x. [DOI] [PubMed] [Google Scholar]
- 30.Titball RW. 1993. Bacterial phospholipases C Microbiol Rev 57:347–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Songer JG. 1997. Bacterial phospholipases and their role in virulence. Trends Microbiol 5:156–161. doi: 10.1016/S0966-842X(97)01005-6. [DOI] [PubMed] [Google Scholar]
- 32.Schmiel DH, Miller VL. 1999. Bacterial phospholipases and pathogenesis. Microbes Infect 1:1103–1112. doi: 10.1016/S1286-4579(99)00205-1. [DOI] [PubMed] [Google Scholar]
- 33.Johansen KA, Gill RE, Vasil ML. 1996. Biochemical and molecular analysis of phospholipase C and phospholipase D activity in mycobacteria. Infect Immun 64:3259–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noens EE, Williams C, Anandhakrishnan M, Poulsen C, Ehebauer MT, Wilmanns M. 2011. Improved mycobacterial protein production using a Mycobacterium smegmatis groEL1ΔC expression strain. BMC Biotechnol 11:27. doi: 10.1186/1472-6750-11-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rowbotham TJ. 1983. Isolation of Legionella pneumophila from clinical specimens via amoebae, and the interaction of those and other isolates with amoebae. J Clin Pathol 36:978–986. doi: 10.1136/jcp.36.9.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakala N′goma JC, Schué M, Carrière F, Geerlof A, Canaan S. 2010. Evidence for the cytotoxic effects of Mycobacterium tuberculosis phospholipase C towards macrophages. Biochim Biophys Acta 1801:1305–1313. doi: 10.1016/j.bbalip.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Garcia JR, Curi R, Martins EF, Carpinelli AR. 2001. Macrophages transfer [14C]-labelled fatty acids to pancreatic islets in culture. Cell Biochem Funct 19:11–17. doi: 10.1002/cbf.887. [DOI] [PubMed] [Google Scholar]
- 38.Medjahed H, Reyrat J-M. 2009. Construction of Mycobacterium abscessus defined glycopeptidolipid mutants: comparison of genetic tools. Appl Environ Microbiol 75:1331–1338. doi: 10.1128/AEM.01914-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Kessel JC, Marinelli LJ, Hatfull GF. 2008. Recombineering mycobacteria and their phages. Nat Rev Microbiol 6:851–857. doi: 10.1038/nrmicro2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Layre E, Collmann A, Bastian M, Mariotti S, Czaplicki J, Prandi J, Mori L, Stenger S, De Libero G, Puzo G, Gilleron M. 2009. Mycolic acids constitute a scaffold for mycobacterial lipid antigens stimulating CD1-restricted T cells. Chem Biol 16:82–92. doi: 10.1016/j.chembiol.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Catherinot E, Clarissou J, Etienne G, Ripoll F, Emile J-F, Daffé M, Perronne C, Soudais C, Gaillard J-L, Rottman M. 2007. Hypervirulence of a rough variant of the Mycobacterium abscessus type strain. Infect Immun 75:1055–1058. doi: 10.1128/IAI.00835-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roux A-L, Ray A, Pawlik A, Medjahed H, Etienne G, Rottman M, Catherinot E, Coppée J-Y, Chaoui K, Monsarrat B, Toubert A, Daffé M, Puzo G, Gaillard J-L, Brosch R, Dulphy N, Nigou J, Herrmann J-L. 2011. Overexpression of proinflammatory TLR-2-signalling lipoproteins in hypervirulent mycobacterial variants. Cell Microbiol 13:692–704. doi: 10.1111/j.1462-5822.2010.01565.x. [DOI] [PubMed] [Google Scholar]
- 43.Pawlik A, Garnier G, Orgeur M, Tong P, Lohan A, Le Chevalier F, Sapriel G, Roux A-L, Conlon K, Honoré N, Dillies M-A, Ma L, Bouchier C, Coppée J-Y, Gaillard J-L, Gordon SV, Loftus B, Brosch R, Herrmann JL. 2013. Identification and characterization of the genetic changes responsible for the characteristic smooth-to-rough morphotype alterations of clinically persistent Mycobacterium abscessus. Mol Microbiol 90:612–629. doi: 10.1111/mmi.12387. [DOI] [PubMed] [Google Scholar]
- 44.Ostroff RM, Vasil AI, Vasil ML. 1990. Molecular comparison of a nonhemolytic and a hemolytic phospholipase C from Pseudomonas aeruginosa. J Bacteriol 172:5915–5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishizuka Y. 1992. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science 258:607–614. doi: 10.1126/science.1411571. [DOI] [PubMed] [Google Scholar]
- 46.Catherinot E, Roux A-L, Vibet M-A, Bellis G, Ravilly S, Lemonnier L, Le Roux E, Bernède-Bauduin C, Le Bourgeois M, Herrmann J-L, Guillemot D, Gaillard J-L. 2013. Mycobacterium avium and Mycobacterium abscessus complex target distinct cystic fibrosis patient subpopulations. J Cyst Fibros 12:74–80. doi: 10.1016/j.jcf.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 47.Shortridge VD, Lazdunski A, Vasil ML. 1992. Osmoprotectants and phosphate regulate expression of phospholipase C in Pseudomonas aeruginosa. Mol Microbiol 6:863–871. doi: 10.1111/j.1365-2958.1992.tb01537.x. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez AV, Baigorí MD, Alvarez S, Castro GR, Oliver G. 2001. Phosphatidylinositol-specific phospholipase C activity in Lactobacillus rhamnosus with capacity to translocate. FEMS Microbiol Lett 204:33–38. doi: 10.1111/j.1574-6968.2001.tb10858.x. [DOI] [PubMed] [Google Scholar]
- 49.Goldfine H, Knob C. 1992. Purification and characterization of Listeria monocytogenes phosphatidylinositol-specific phospholipase C Infect Immun 60:4059–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Low MG. 1981. Phosphatidylinositol-specific phospholipase C from Staphylococcus aureus. Methods Enzymol 71:741–746. doi: 10.1016/0076-6879(81)71087-5. [DOI] [PubMed] [Google Scholar]
- 51.Griffith OH, Volwerk JJ, Kuppe A. 1991. Phosphatidylinositol-specific phospholipases C from Bacillus cereus and Bacillus thuringiensis. Methods Enzymol 197:493–502. doi: 10.1016/0076-6879(91)97175-X. [DOI] [PubMed] [Google Scholar]
- 52.Gonzalo Asensio J, Maia C, Ferrer NL, Barilone N, Laval F, Soto CY, Winter N, Daffé M, Gicquel B, Martín C, Jackson M. 2006. The virulence-associated two-component PhoP-PhoR system controls the biosynthesis of polyketide-derived lipids in Mycobacterium tuberculosis. J Biol Chem 281:1313–1316. doi: 10.1074/jbc.C500388200. [DOI] [PubMed] [Google Scholar]
- 53.Gomez-Valero L, Buchrieser C. 2013. Genome dynamics in Legionella: the basis of versatility and adaptation to intracellular replication. Cold Spring Harb Perspect Med 3:a009993. doi: 10.1101/cshperspect.a009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hagedorn M, Soldati T. 2007. Flotillin and RacH modulate the intracellular immunity of Dictyostelium to Mycobacterium marinum infection. Cell Microbiol 9:2716–2733. doi: 10.1111/j.1462-5822.2007.00993.x. [DOI] [PubMed] [Google Scholar]
- 55.Hagedorn M, Rohde KH, Russell DG, Soldati T. 2009. Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts. Science 323:1729–1733. doi: 10.1126/science.1169381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cosson P, Soldati T. 2008. Eat, kill or die: when amoeba meets bacteria. Curr Opin Microbiol 11:271–276. doi: 10.1016/j.mib.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 57.Boritsch EC, Supply P, Honoré N, Seeman T, Stinear TP, Brosch R. 2014. A glimpse into the past and predictions for the future: the molecular evolution of the tuberculosis agent. Mol Microbiol 93:835–852. doi: 10.1111/mmi.12720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.