Abstract
MicroRNAs (miRNAs) have important roles in the initiation and progression of human cancer, but their role in head and neck cancer development and progression is not well defined. We aimed to determine whether specific miRNAs and their target mRNAs contribute to head and neck cancer pathogenesis and progression. To identify miRNAs associated with head and neck squamous cell carcinomas (HNSCCs), we analyzed HNSCC cell lines, normal head and neck tissues and normal keratinocytes by miRNA profiling; a group of differentially expressed miRNAs was identified, which includes miR-125b. Decreased expression of miR-125b is known to occur in epithelial cancers and many target mRNAs for this miR have been reported. We found decreased expression of miR-125b-1 and hypermethylation of its promoter in HNSCC compared with its non-malignant counterpart. The TACSTD2 (also known as TROP2) gene was identified and validated as a direct target of miR-125b-1. Abnormal expression of TACSTD2 cell-surface glycoprotein has been reported in most epithelial tumors, and the overexpressions of this mRNA and protein product has been considered a useful tumor marker. We report that miR-125b-1 causes mitogen-activated protein kinase pathway dysfunction through regulation of TACSTD2 expression. Thus, loss of miR-125b-1 may have a key role in the pathogenesis and progression of squamous cell carcinomas of head and neck and possibly of other tumors.
Keywords: miRNA, MAPK, TACSTD2, HNSCC, miR-125b
INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common neoplasm worldwide, and includes tumors of the oral cavity, larynx and pharynx.1 Although diagnosis and treatment of HNSCC have improved, the survival rate has not improved substantially in the past 40 years.2,3 In this study, we focus on the relationship between HNSCC and microRNA (miRNA) dysregulation.
miRNAs are 19–24 nucleotide noncoding RNAs that regulate the translation and degradation of target mRNAs,4,5 and a large fraction of protein-coding genes are under the control of miRNAs. miRNAs have a fundamental role in the regulation of important cellular functions, such as proliferation, apoptosis, death, stress response, differentiation and development, and dysregulation of miRNA is associated with a variety of disorders, in particular cancers, including HNSCC.6–8 Recent studies have demonstrated that miR-125b is dysregulated in multiple types of cancer, including breast,9 oral,10 bladder11 and anaplastic thyroid carcinomas,12 suggesting that miR-125b could have a role in tumor development. miR-125b-1 and miR-125b-2 originated from independent precursors located on different chromosome loci, but their mature sequences and targets are identical. In this study, we found that expression of miR-125b-1 is suppressed in HNSCC cell lines, in association with TACSTD2 overexpression. TACSTD2 is a 36-kDa cell-surface glycoprotein overexpressed in a variety of late stage epithelial carcinomas with low-to-no expression in normal tissues;13–17 it is overexpressed in a variety of human cancers, both metastatic and invasive tumors. Analysis of tumor tissues from cancer patients revealed association of increased TACSTD2 expression with poor prognosis and higher frequency of metastasis.13,14,16,17
In this study, the TACSTD2 gene was identified as a direct and functional target of miR-125b-1 in squamous cell carcinogenesis. Upmodulation of miR-125b-1 expression inhibited cell invasion, colony formation and anchorage-independent growth of HNSCC cells. We further showed that miR-125b-1 may regulate the extracellular signal–regulated kinase (ERK)–MAPK (mitogen-activated protein kinase) signaling pathway in HNSCC. Thus, our findings reveal novel roles of miR-125b-1 in the pathogenesis and progression of epithelial tumors.
RESULTS
miRNA expression signature differentiates HNSCCs from nonneoplastic head and neck cells or tissue
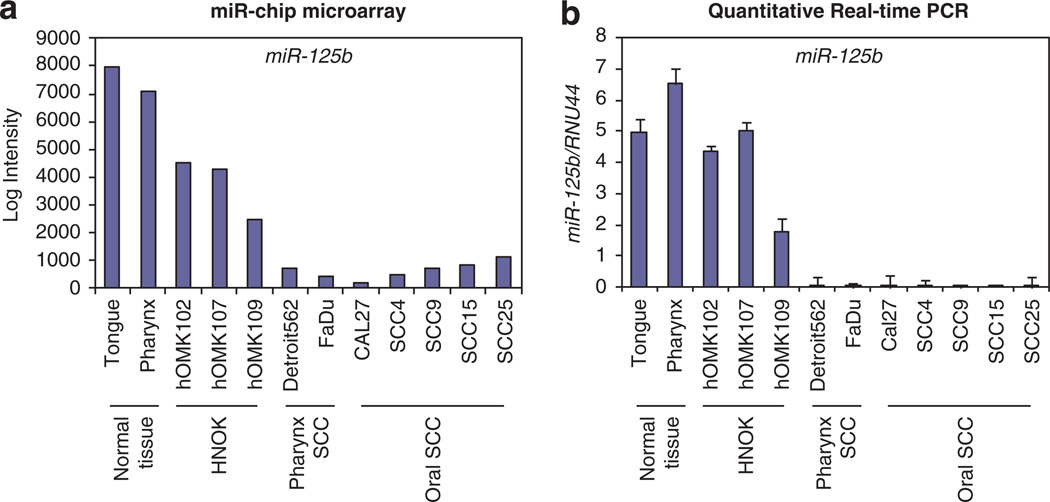
We analyzed the miRNA expression profiles of seven human HNSCC-derived cell lines, three normal primary oral keratinocyte cells and head and neck tissues from two healthy subjects, using a miRNA microarray platform for genome-wide assessment of expression levels of human miRNAs.18,19 Marked differences in expression were observed in 292 human miRNAs of HNSCCs vs non-cancer tissues or cell lines. Using twofold expression difference as a cutoff level, we identified 72 upregulated and 43 downregulated miRNAs. Expression of 20 miRNAs was altered more than fourfold in HNSCC samples (fold change: > 4, false discovery rate: <0.05). Among these, miR-125b was the most significantly downregulated miRNA (−100-fold), (Table 1, Figure 1a) and its downregulation was validated by quantitative real-time PCR (qPCR; Figure 1b).
Table 1.
Differential miR expressed in the HNSCC comparing with the non-HNSCC
| MicroRNA | Chromosomal location | Fold change (HNSCC/ non-HNSCC) |
P-value |
|---|---|---|---|
| Upregulated in HNSCC | |||
| miR-146b | 10q24.32 | 171 | 3.77E-06 |
| miR-340-5p | 5q35.3 | 57 | 2.78E-10 |
| miR-15b-3p | 3q25.33 | 54 | 5.72E-11 |
| miR-135a-5p | 3q21.1 | 50 | 7.46E-12 |
| miR-16-5p | 13q14.2 | 35 | 4.35E-09 |
| miR-15b | 3q25.33 | 33 | 4.99E-08 |
| miR-486 | 8p11.21 | 25 | 7.11E-06 |
| miR-148b | 12q13.13 | 23 | 9.16E-10 |
| miR-182-5p | 7q32.2 | 7.6 | 1.36E-06 |
| miR-21 | 17q23.1 | 5.7 | 6.60E-07 |
| Downregulated in HNSCC | |||
| miR-125b-1 | 11q24.1 | 0.01 | 6.38E-11 |
| miR-145 | 5q32 | 0.01 | 5.64E-08 |
| miR-619 | 12q24.11 | 0.01 | 1.20E-14 |
| miR-618 | 12q21.31 | 0.01 | 1.23E-14 |
| miR-616 | 12q13.3 | 0.01 | 8.63E-07 |
| miR-770 | 14q32.2 | 0.01 | 2.46E-14 |
| miR-302d | 4q25 | 0.02 | 1.98E-11 |
| miR-372 | 19q13.42 | 0.03 | 1.33E-10 |
| miR-643 | 19q13.41 | 0.03 | 2.52E-09 |
| miR-340 | 5q35.3 | 0.04 | 2.03E-09 |
Abbreviations: HNSCC, head and neck squamous cell carcinoma; miR, microRNA.
List of differentially expressed miRNAs in HNSCC compared with normal tissues and normal keratinocytes. miRNAs are sorted by P-value of the univariate test (BRB tools). The first 10 genes are significant at the nominal 0.01 level of the univariate test. False discovery rate or q-value is the expected percentage of genes identified by chance.
Figure 1.
Expression of miR-125b in HNSCC. Expression of miR-125b in HNSCC cell lines, normal tissues and normal keratinocytes. (a) Microarray data for miR-125b. (b) qPCR validation of miR-125b expression.
miR-125b-1 is hypermethylated in SCC
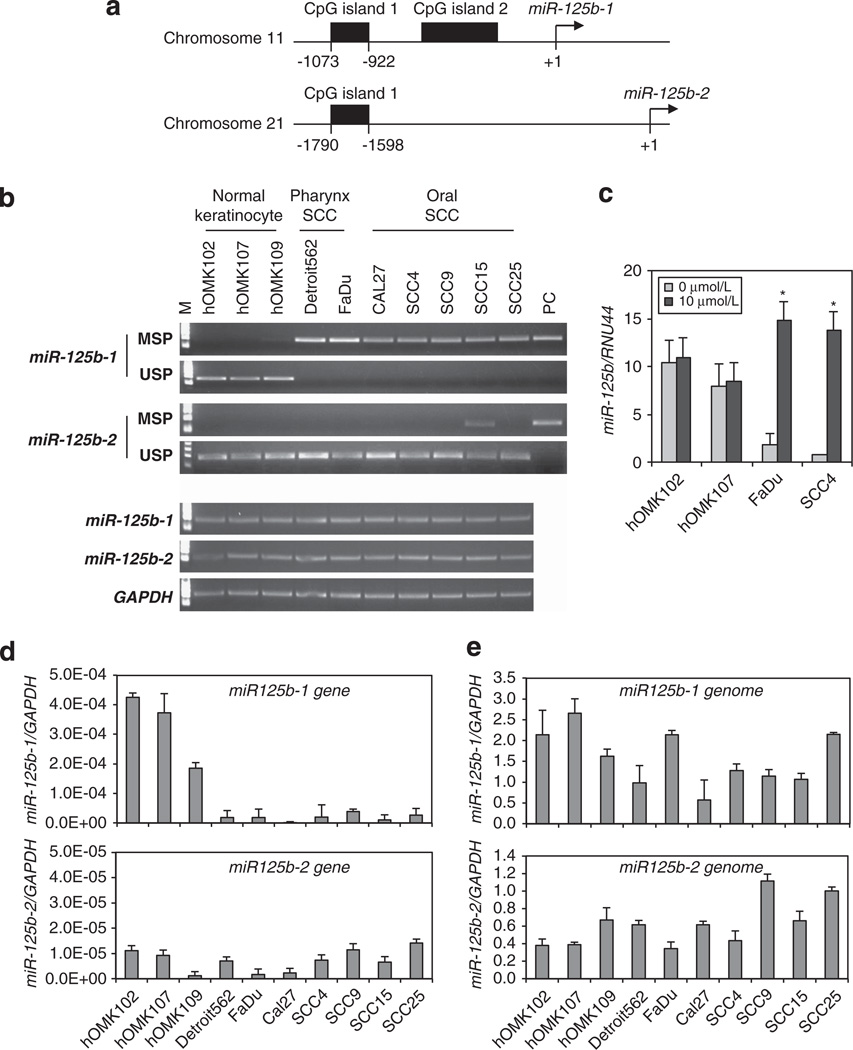
To examine mechanisms of downregulation of miR-125b expression in HNSCC, we assessed the deletion and methylation status of the locus. We selected 2000 bps of sequence upstream of miR-125b-1 and miR-125b-2 (Figure 2a) for examination. Two CpG-rich regions upstream of miR-125b-1 and one CpG-rich region upstream of miR-125b-2 were found by using the MethPrimer online tool (http://www.urogene.org/methprimer/). CpG island region 1 upstream of miR-125b-1 was found to be hypermethylated in all HNSCC cells, while non-cancer cells showed absence of methylation. The CpG island region upstream of miR-125b-2 was not methylated in the HNSCC cells (Figure 2b upper). Expression of the hypermethylated miR-125b was restored after treatment with the demethylating agent, 5-aza-2′-deoxycytidine (5-aza-CdR), for 5 days (Figure 2c). To assess the correlation between DNA methylation status of miR-125b-1, 2 and expression of the two loci, we performed real-time PCR analysis of precursor miR-125b-1, 2. We found that the gene expression in hypermethylated miR-125b-1 locus, but not miR-125b-2, was repressed in all HNSCC cells (Figure 2d). By using real-time or end-point PCR analysis, we searched for deletion of the miR-125b-1, 2. In this result, we found some differences of copy number. However, there is no significant difference between normal keratinocytes and HNSCC cells, and we did not observe any loss at the genomic region of either of the miR-125b-1, 2 loci (Figures 2b and e lower panel). These results suggest that the loss of expression of miR-125b-1 was caused by promoter methylation and matured miR-125b expression reflects it.
Figure 2.
DNA methylation or deletion status of miR-125b-1 and miR-125b-2 in HNSCC-derived cells. (a) Identification of CpG island enriched sites of the miR-125b-1 and miR-125b-2 regulatory regions using MethPrimer program. (b) Using semiquantitative PCR analysis, methylation and deletion status of miR-125b-1, 2 alleles were defined. Upper panel, lane MSP (methylation-specific PCR) and USP (unmethylation-specific PCR) lanes corresponded to amplification methylated and unmethylated DNA, respectively. Universal methylated human DNA standard having all CpG sites methylated (performed as per instructions provided in the EZ DNA methylation kit, Zymo Research) was used as positive control (PC). Lower panel, genomic DNA was PCR-amplified using semiquantitative RT–PCR analysis for detection of deletion of miR-125b-1 and miR-125b-2 genomic loci. (c) Expression of miR-125b using qRT–PCR analysis in normal keratinocytes cells hOMK102, hOMK107 and SCC cell lines FaDu, SCC4 after treatment with 10 µmol/l 5-aza-CdR. Vertical bar above each column indicate ± s.d. as determined by triplicated assays. *P < 0.001. (d) qRT–PCR analysis to determine the expression of precursor miR-125b-1, 2 in HNSCC cells and non-cancer cells. Values represent mean ± s.d. as determined by triplicated assays (n = 3). (e) qRT–PCR analysis was used to assess the allele status of the miR-125b-1, 2 loci. Values represent mean ± s.d. as determined by triplicated assays (n = 3).
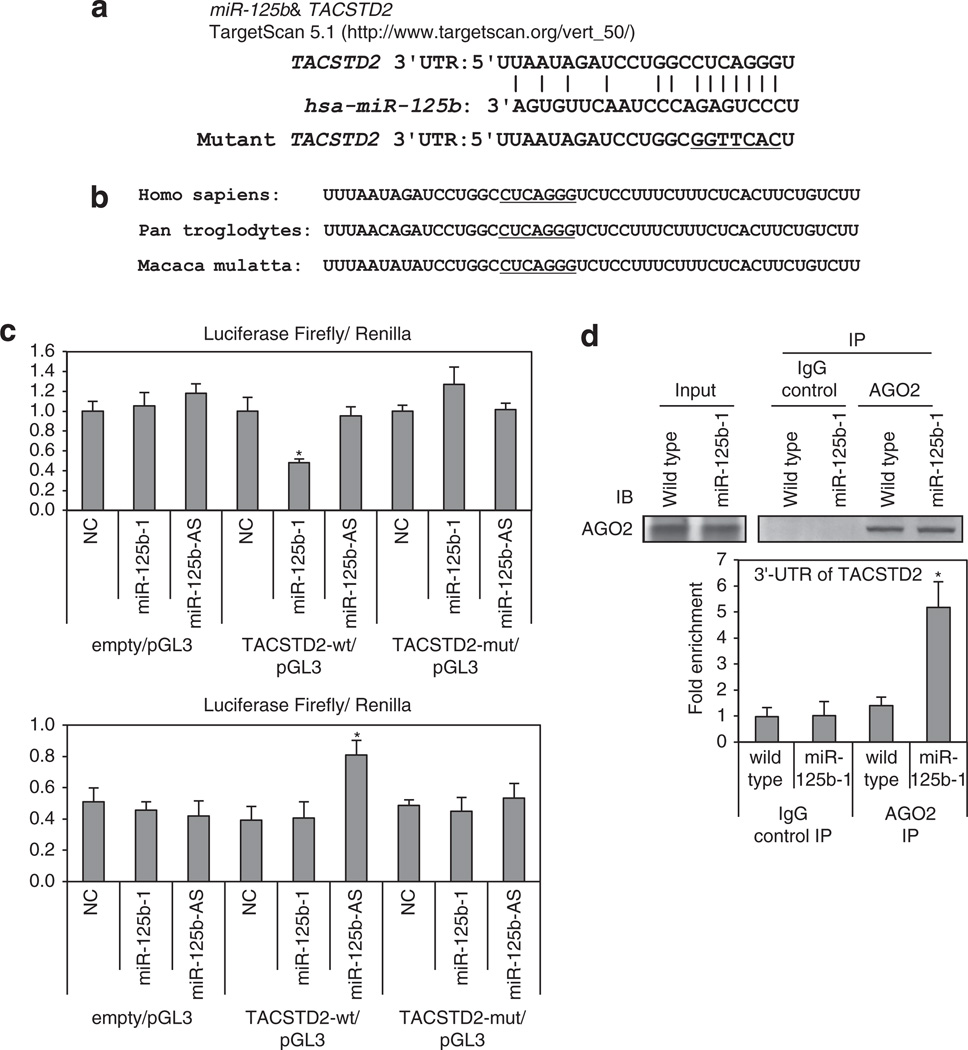
TACSTD2 is a target of SCC-deregulated miR-125b-1 by miRNA target prediction
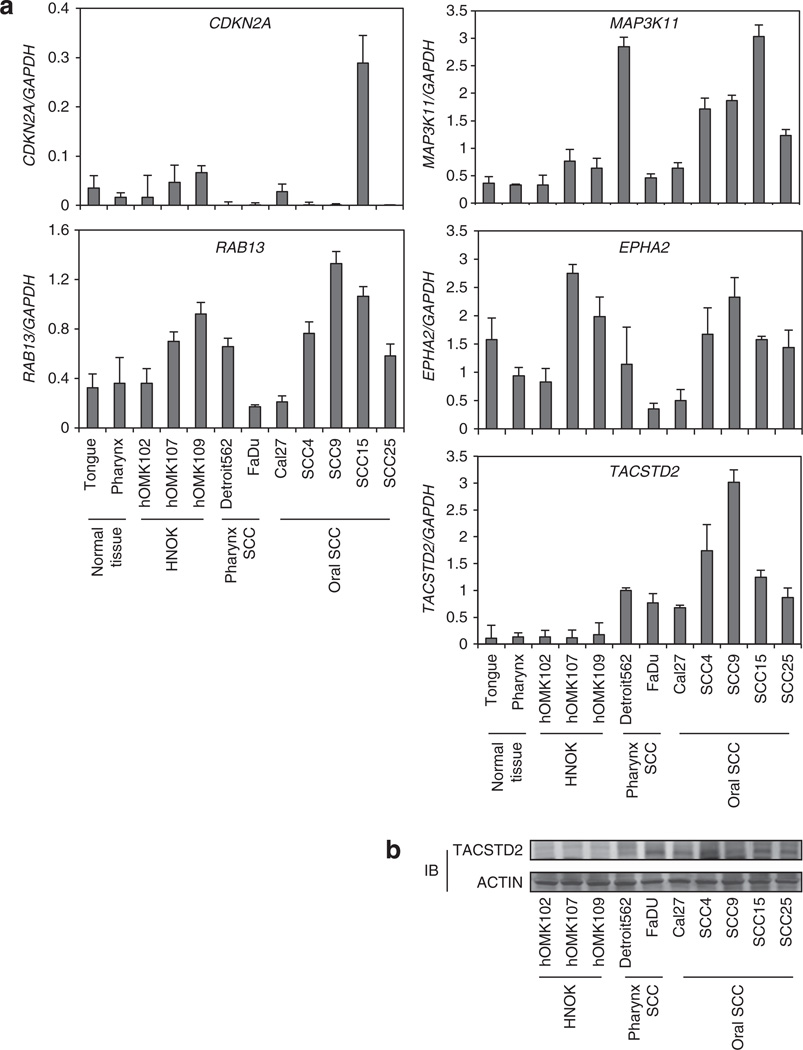
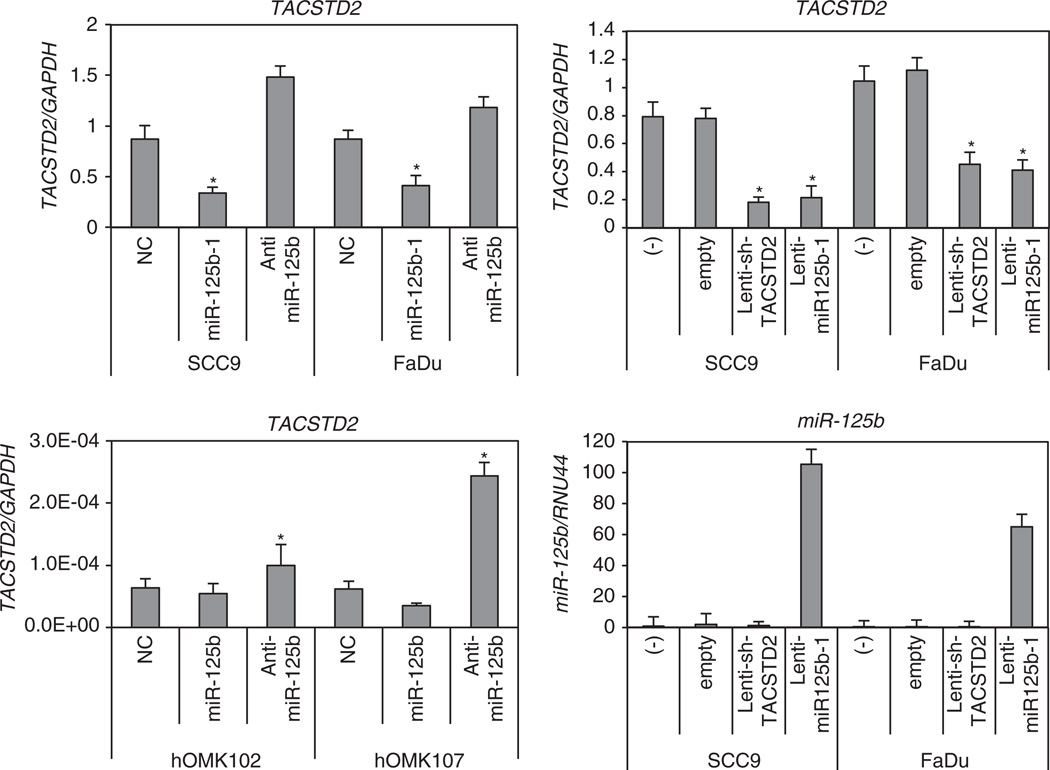
Identification of miRNA-regulated gene targets is a necessary step to understand miRNA functions. We searched for additional miR-125b-1 targets using computer-aided miRNA target prediction programs, MicroCosm Targets (http://www.ebi.ac.uk/enrightsrv/microcosm/htdocs/targets/v5/), TargetScan (http://www. targetscan.org/), PicTar (http://pictar.mdc-berlin.de/), miRBase targets and miRanda (microrna.org and miRbase), and > 1000 candidate downstream targets of miR-125b were identified, including many putative miR-125b-1 target genes that might have a role in cell invasion, such as EPHA2, RAB13, MAP3K11, CDKN2A and TACSTD2. We performed qPCR analysis in HNSCC cells compared with normal samples. Among these candidate genes, we found that TACSTD2 mRNA and protein levels were overexpressed in HNSCCs and showed an inverse correlation with miR-125b-1 expression (Figures 3a and b). TACSTD2 was selected as a promising candidate of the target. The TACSTD2 gene product is a cell-surface glycoprotein belonging to the TACSTD gene family and is highly overexpressed by a variety of epithelial carcinomas with low or no expression in normal tissues.13–17 We found that overexpression of miR-125b-1 using precursor miRNA oligo and miRNA-expressing lentivirus in both SCC9 and FaDu cells reduced the mRNA level of TACSTD2 > 50% (*P < 0.05). Furthermore, transfection of a miR-125b inhibitor antisense miRNA into SCC9 FaDu, hOMK102 and hOMK107 cells led to markedly increased expression of TACSTD2 (Figure 4). MiRanda analysis indicated that TACSTD2 contains one miR-125b-binding site in its 3’ untranslated region (UTR; Figure 5a), a site highly conserved across primates species (chimpanzee, rhesus macaque and human) but not rodents (Figure 5b). Therefore, we constructed vectors containing wild type or mutant 3′-UTR of human TACSTD2 fused downstream of the firefly luciferase gene. The wild-type or mutant vector was co-transfected into HEK-293T cells with miR-125b-1 expression construct or vector control. The transfection efficacy was normalized by co-transfection with renilla reporter vector. As shown in Figure 5c upper part, miR-125b-1 expression led to significantly decreased relative luciferase activity of wild-type TACSTD2 3′-UTR (> 50%), whereas reduction of luciferase activity with mutant TACSTD2 3′-UTR was not detected. We also performed luciferase assay using endogenous miR-125b-1 expressing normal hOMK107 cell and the luciferase activity was increased by antisense miR-125b (Figure 5c lower part), suggesting that miR-125b-1 could bind to the 3′-UTR of TACSTD2. In addition, we immunoprecipitated Argonaute 2 (AGO2), a core component of RNA-induced silencing complex (RISC), and demonstrated the direct binding of miR-125b-1 to endogenous TACSTD2 mRNA (Figure 5d). Taken together, these findings indicate that TACSTD2 is a target of miR-125b-1.
Figure 3.
miR-125b downstream candidate genes in HNSCC CDKN2A, RAB13, MAP3K11, EPHA2 and TACSTD2 mRNA level using qRT–PCR analysis in HNSCC cell lines compared with normal tissues and normal keratinocytes. TACSTD2 protein level is elevated in HNSCC cell lines compared with normal keratinocytes. IB, immunoblot.
Figure 4.
miR-125b-1 expression downmodulates TACSTD2 mRNA in HNSCC cells. qPCR analysis of TACSTD2 and miR-125b expression is shown. The expression level of TACSTD2 was downregulated by miR-125b-1 and upregulated after anti-miR125b treatment in HNSCC cell lines (left upper). TACSTD2 was induced by anti-miR-125b in normal keratinocytes (left lower). The expression level of TACSTD2 was also downregulated by lentiviral short-hairpin RNA of TACSTD2 and lenti-miR-125b-1 in HNSCC cell lines (right upper). Infection with miR-125b-1 expression was confirmed in right lower figure. Vertical bar above each column indicates ± s.d. as determined for triplicated assays. *P < 0.05.
Figure 5.
miR-125b targets TACSTD2 mRNA at 3′-UTR. (a) A predicted miR-125b target site on TACSTD2 3′-UTR was identified by TargetScan 5.1 software. In the figure the alignment of TACSTD2 3′-UTR with miR-125b is shown. The site of target mutagenesis is indicated in underline. (b) Potential binding site of miR-125b on TACSTD2 3′-UTR in different species. The seed sequence is underlined. (c) Luciferase reporter plasmid pGL3 carrying the 3′-UTR of TACSTD2, mutant 3′-UTR or empty was transfected into HEK-293T (upper panel) or hOMK107 (lower panel) cells with miR-125b-1, antisense miR-125b or negative control of miR (NC). The cells were harvested 24 h later for luciferase assays. Data are mean ± s.d. of three independent experiments and each measured in triplicate (*P < 0.05). (d) Lysates from FaDu cells expressing miR-125b-1 or wild type were immunoprecipitated with anti-AGO2 antibody or nonspecific IgG; RNA was isolated from the IP fractions and real-time PCR used to demonstrate the significant enrichment for 3′-UTR TACSTD2 in the immunoprecipitates of miR-125b-1 loaded RISC. Data shown are mean ± s.d. of TACSTD2 relative abundance in the various AGO2-IPs from three independent experiments. RISC-associated GAPDH was used for normalization (*P < 0.01). Upper, western blot analysis of immunoprecipitate of AGO2-containing RISC (right) and input protein (left) demonstrate equal pulldown and expression of this protein in wild-type or miR-125b-1-expressed cells. IgG control confirms the specificity of the IP.
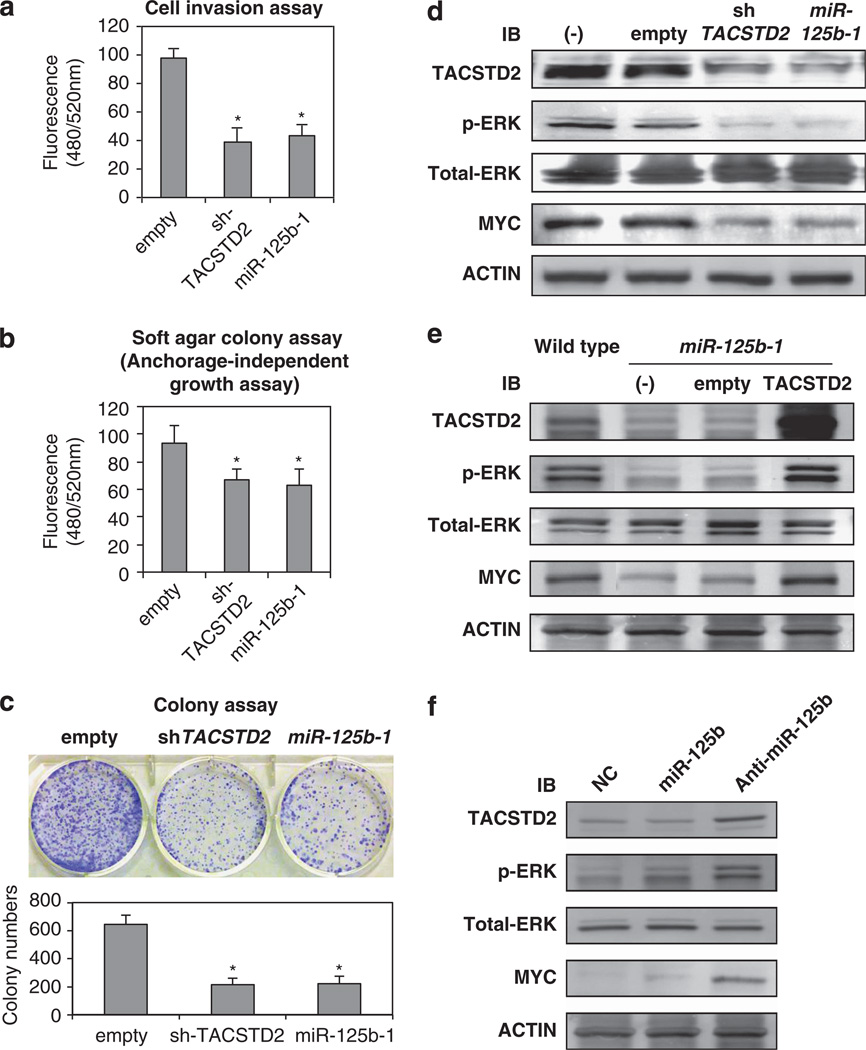
miR-125b-1 impairs clonogenicity and invasiveness of HNSCC cells
Increased invasion and proliferation is a characteristic of aggressive cancer cells. To determine whether the effect of miR-125b-1 and TACSTD2 could result in increased cell phenotype changes, we performed cell invasion and colony-formation assays. As shown in Figure 6a, FaDu cells with stable miR-125b-1 overexpression or TACSTD2 suppression showed a significant reduction in invasion ability compared with FaDu cells stably expressing vector control. Moreover, colony-formation assay for anchorage-independent and -dependent growth demonstrated that enhanced expression of miR-125b-1 and inhibition of TACSTD2 significantly impaired the clonogenicity of FaDu cells when compared with control cells (Figures 6b and c).
Figure 6.
miR-125b-1 expression inhibits cell invasion, anchorage-independent growth and colony formation by downmodulating the TACSTD2-MAPK pathway. (a–c) Invasion and growth suppression by ectopically expressed sh-TACSTD2 and miR-125b in FaDu cells. Colonies grown from FaDu cells stably transfected with Lenti-empty, Lenti-sh-TACSTD2 or Lenti-miR125b-1 were counted. The experiments were performed three times using triplicate samples, and average scores are indicated with error bars on the histogram (*P < 0.05). (d) Repression of MAPK pathway by overexpressed miR-125b-1. Western blot analysis of FaDu cells with knocked down TACSTD2 or overexpressing miR-125b-1. Western blots of total protein extracted from wild type (−), control (empty), knockdown TACSTD2 cell pools (sh-TACSTD2) or miRNAoverexpressing cell pools (miR-125b-1) were probed for TACSTD2, phospho-ERK1/2 (p-ERK), total ERK1/2 (total-ERK), MYC and ACTIN. (e) Rescue of MAPK pathway by overexpression of TACSTD2. Western blot analysis of FaDu cells with overexpressing miR-125b-1 indicated recovery pERK and MYC by TACSTD2 overexpression. (f) Overexpression of TACSTD2, pERK and MYC in hOMK107 cells transfected with antimiR- 125b. IB, immunoblot.
miR-125b-1 inhibits activation of the MAPK pathway through modulation of TACSTD2 expression
Next we focused on identification of signal pathways that may be affected by miR-125b-1 because a recent report shows that murine Tacstd2 increases the level of phosphorylated ERK1/2.20 We determined that phosphorylation of ERK and the downstream effector, MYC, were significantly decreased in FaDu cells stably expressing short-hairpin RNA for TACSTD2 (sh-TACSTD2); miR-125-1 overexpression also led to reduced levels of phospho-ERK (p-ERK) and MYC (Figure 6d). These reduced MAPK pathways were rescued by overexpression of TACSTD2 in LV-miR-125b-1-FaDu cells (Figure 6e) or by suppression of miR125b in keratinocyte hOMK107 (Figure 6f).
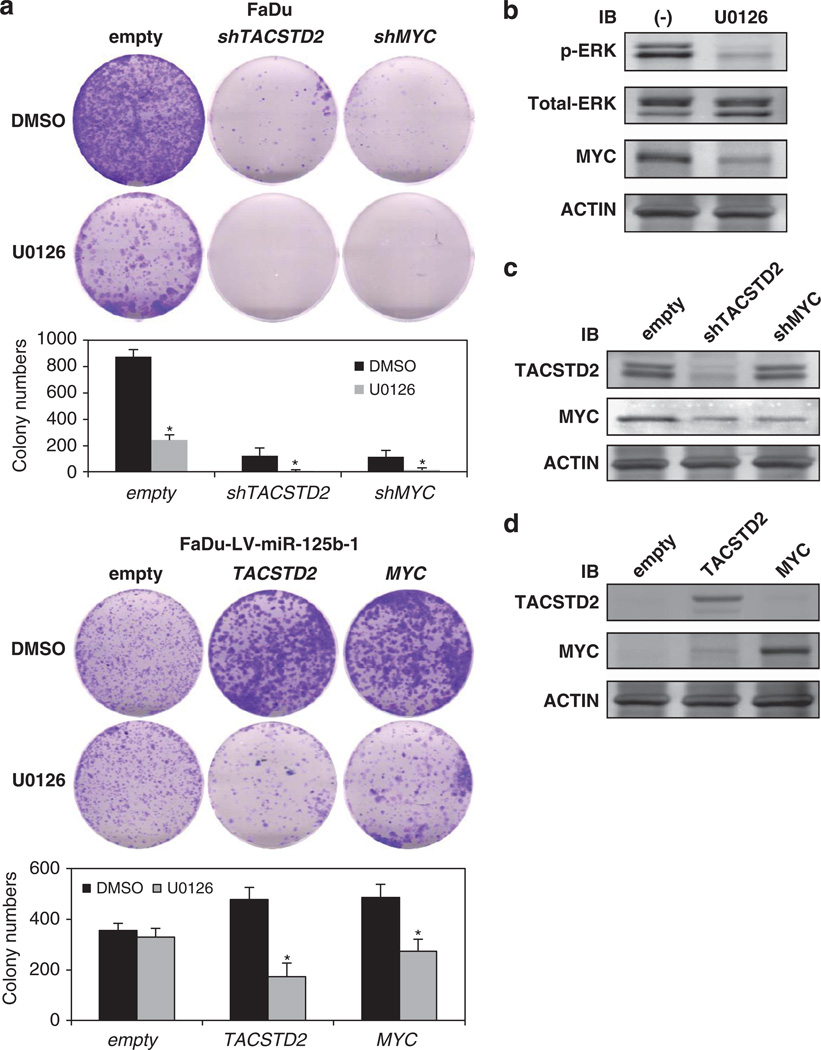
An inhibitor of ERK phosphorylation, U0126, reduces cell proliferation of HNSCC cells
To ascertain the biochemical effect of inhibition of ERK phosphorylation, we examined the effect of U0126, an inhibitor of ERK phosphorylation21 by colony-formation assay. Colony formation was inhibited by suppression of TACSTD2 and MYC using short-hairpin RNAs, and the suppression has enhanced by U0126 (Figure 7a upper part). Moreover, U0126 suppressed colonyformation ability by transfection with TACSTD2 and MYC in FaDu-LV-miR-125b-1 cells (Figure 7a lower part). ERK and the downstream effector, MYC with effect of U0126, effect of transfection of shTACSTD2, shMYC, TACSTD2 and MYC were determined by western blot in FaDu or FaDu-LV-miR-125b-1 cells (Figures 7b–d). The results showed inhibition of cell proliferation by U0126 and demonstrated that HNSCC cell proliferation depended on expression of TACSTD2 and ERK phosphorylation.
Figure 7.
U0126, specific inhibitor of ERK phosphorylation, inhibits growth of HNSCC. (a) Growth suppression in FaDu or FaDu-LV-miR-125b-1 cells transfected with shTACSTD2, shMYC, TACSTD2 or MYC and treated with U0126 at a concentration of 1 µm. The experiments were performed three times using triplicate samples, and average scores are indicated with error bars on the histogram (*P < 0.05). (b) Downmodulation of phospho-ERK and MYC in FaDu cells treated with U0126. (c) Suppression of TACSTD2 and MYC transfected with shTACSTD2 and shMYC in FaDu cells. (d) Overexpression of TACSTD2 and MYC in FaDu-LV-mir-125b-1 cells transfected with TACSTD2 and MYC. IB, immunoblot.
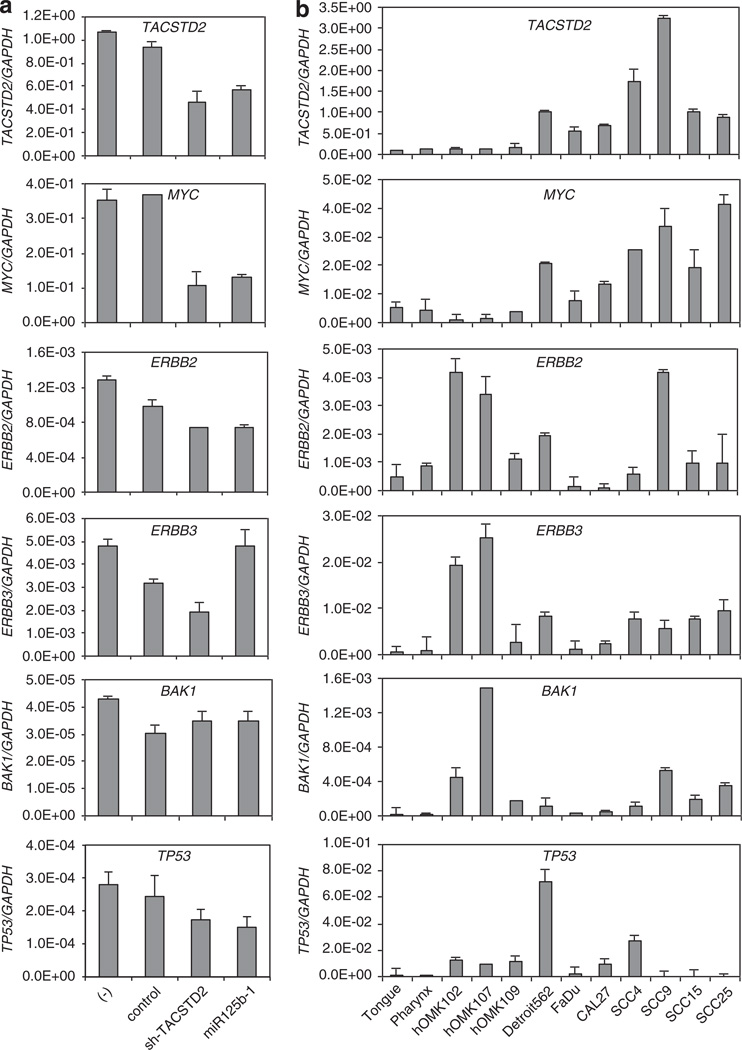
Inverse correlation of expression of miR-125b and miR-125b target genes
Many targets of miR-125b have been reported, but their evaluation in HNSCC had not been reported. In this study, we assessed expression of the reported targets of miR-125b in HNSCC-derived cells. We found that the levels of TACSTD2 and MYC expression were downregulated by shTACSTD2 and miR-125b-1 (Figure 8a), and these reductions were confirmed at the mRNA and protein expression levels (Figure 6d). TACSTD2 and MYC expression correlated inversely with miR-125b expression level and was high in HNSCC cell line samples compared with normal tissues or normal keratinocytes, while ERBB2, 3, BAK1 and TP53 expression were not (Figure 8b).
Figure 8.
Differential expression and validation of miR-125b and downstream genes in HNSCC. (a) qRT–PCR analysis of TACSTD2, MYC mRNA and reported miR-125b downstream genes in FaDu cells infected with lenti-shTACSTD2 or lenti-miR-125b. (b) Expression of TACSTD2, MYC mRNA and reported miR-125b downstream genes in HNSCC cell lines, normal keratinocytes and normal tissues by qPCR validation. Values represent mean ± s.d. as determined by triplicated assays.
DISCUSSION
miRNA dysregulation has been found in all human cancers.4 Although aberrant expression of miR-489, miR-21, miR-205, miR-204 and miR-125b has been observed in head and neck cancers by miRNA profiling studies,22–26 their functional involvement in cancer pathogenesis and progression had not been assessed. We focused on functional studies of miR-125b and showed that miR-125b-1 has a tumor-suppressive role in HNSCC. This result was validated by the inhibition of tumorigenesis after overexpression of miR-125b-1 in HNSCC cells by invasion and clonogenicity analyses.
Decrease of miR-125b expression relative to normal cell controls is the one of the most commonly reported miRNA changes in a variety of cancers, and there are several reports of the possible targets of this miRNA.27–31 miR-125b-1 and miR-125b-2, which are precursors of miR-125b, are located on chromosome 11q24.1 and 21q21.1, respectively, regions involved in loss of heterozygosity in oral, lung, breast and ovarian cancer.32–35 However, the mechanism of regulation of miR-125b-1, 2 expressions had not been defined, because the mature sequences of the two miRNAs are identical, and microarray and qPCR methods used did not allow discrimination between miR-125b-1 and miR-125b-2. Recent studies have shown that miRNAs can also be dysregulated by promoter methylation in human malignancies.36,37 In this study, we carried out deletion analysis and examined the methylation status of miR-125b-1, 2 to clarify the regulation of this miRNA in HNSCC. We found that none of the genomic regions that included miR-125b-1 and miR-125b-2 were deleted. On the other hand, we found that the promoter of miR-125b-1 but not of miR-125b-2 was hypermethylated in all HNSCC-derived cells we tested and that the methylation status of miR-125b-1 was inversely correlated with the expression of miR-125b-1 precursor RNA. These data suggested that miR-125b-1 is downregulated by methylation.
Using bioinformatics tools and database searches, > 1000 candidate targets of miR-125b were identified. We analyzed expression of some of them and selected for study TACSTD2 gene and the protein, with known alterations of expression in epithelial cancer, including HNSCC.13–17 Our study, using luciferase assay and AGO2 containing RISC immunoprecipitation (IP), showed that the TACSTD2 gene is directly targeted by miR-125b-1. TACSTD2 is highly expressed in several types of cancers13–17 and expression of TACSTD2 has been correlated with increased metastatic disease and decreased overall survival.13,15,17 We showed that knocking down TACSTD2 expression by short-hairpin RNA in HNSCC cells resulted in a 50–70% inhibition of colony formation in anchorage-dependent and -independent assays and > 50% inhibition in the cell invasion assay. Thus, our results suggest that TACSTD2 may be a key player in HNSCC by functioning as an oncogene. A very recent report suggests that murine Tacstd2 expression increases the level of phosphorylated ERK, suggesting its possible involvement in murine pancreatic cancer.20 The functional role of TACSTD2 in cancer is not well understood and its physiological ligand is still unknown. By contrast, activation of the MAPK pathway by membrane protein and cell-surface receptor is well known.27,38 Our study demonstrated that miR125b-1 suppressed MAPK by regulating the targeted TACSTD2 and the suppressed MAPK pathway recovered with re-overexpression of TACSTD2. To investigate the clinical relevance of the MAPK pathway, we tested the effects of this pathway inhibition using U0126 as an ERK phosphorylation inhibitor. U0126 is a highly potent inhibitor of ERK1/2 (IC50 = 0.1–20 µm), a protein kinase with important roles in cell proliferation.21 In response to treatment with U0126, cell proliferation was decreased in parallel with inhibition of phosphorylation of ERK.
TP53 and BAK1 were also reported as targets of miR-125b in prostate cancer and neuroblastoma cells, consistently with the oncogenic function of miR-125b.30,39 These reports contradict tumor-suppressor function of miR-125b, and the role of miR-125b in malignancy has been somewhat controversial. In our study, the assessment of the expression levels of miR-125b-1 and of several reported targets of miR-125b, ERBB2, ERBB3, BAK1 and TP53 did not show inverse correlation in HNSCCs. On the other hand, mRNA expression levels of TACSTD2 and MYC clearly showed inverse correlation. MYC is known to be a downstream target of phospho-ERK.40
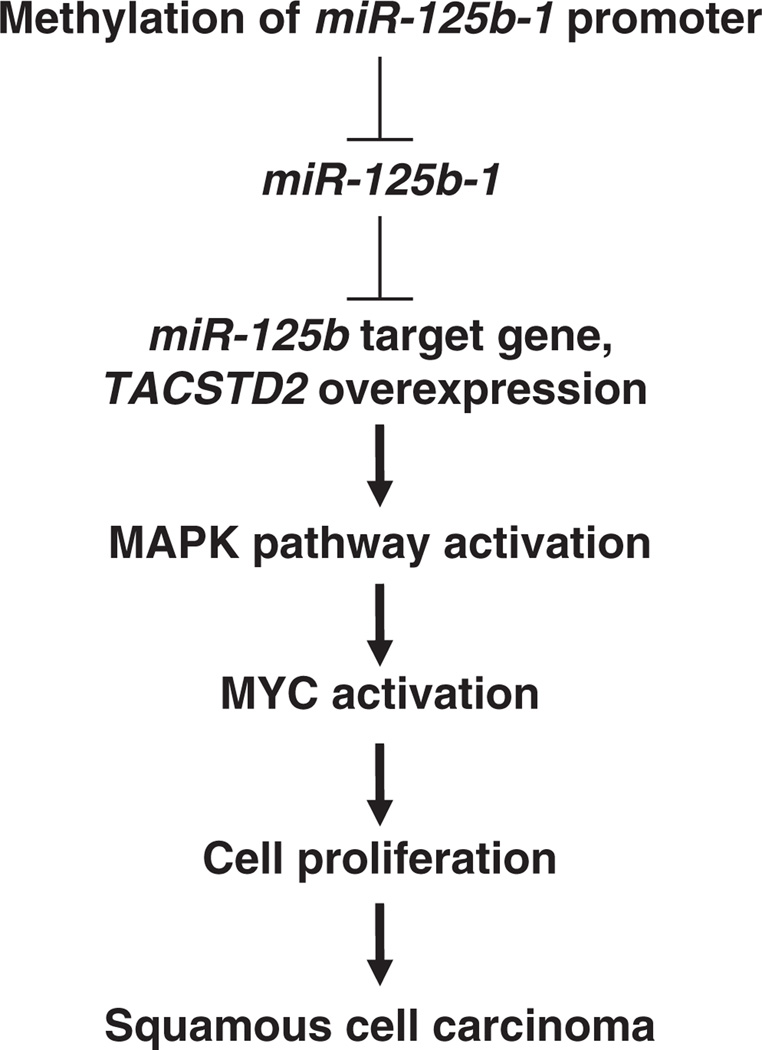
Our investigation is the first to show that miR-125b-1 directly regulates TACSTD2 expression and contributes to MAPK pathway activation in HNSCC (Figure 9). This pathway could be targeted by drugs, providing novel opportunities for rational treatment of HNSCC.
Figure 9.
Proposed dysregulated miR-125b-1 signal pathway involved in SCC.
MATERIALS AND METHODS
Cell lines and tissue samples
Human squamous carcinoma-derived cell lines, including pharyngeal cancers (FaDu, Detroit562) and oral cancers (Cal 27, SCC4, SCC9, SCC15, SCC25), were obtained from ATCC (Manassas, VA, USA). SCC4, SCC9, SCC15 and SCC25 cell lines were maintained in DMEM (Dulbecco’s modified eagle’s medium) plus Ham’s 12 (1:1) medium supplemented with 10% fetal calf serum. Detroit 562, FaDu and CAL 27 cell lines were cultured as monolayers in DMEM, supplemented with 10% fetal bovine serum, 0.075% sodium bicarbonate, 0.6 mg/ml l-glutamine and kept at 37 1C in humidified atmosphere with 5% CO2 in air. Human primary keratinocytes from normal oral mucosa (hOMK102, hOMK107, hOMK109) were obtained from Cell Research Corporation (International Plaza, Singapore) and maintained in Epilife basal medium with 1% epilife-defined growth supplement and 1% antibiotics and used between passages 4 and 8. Human normal tissue, tongue and pharynx were obtained from BioChain (Newark, CA, USA). Human papillomavirus testing was performed in all HNSCC cell lines, primary keratinocytes and normal tissues evaluating by PCR using consensus primers (Takara, Mountain View, CA, USA), and all samples were confirmed negative for presence of Human papillomavirus (Supplementary Figure S1).
miRNA microarrays-based miRNA expression profiling assay and data analysis
Total RNA from cell lines and tissues was isolated by TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Microarray analysis was performed as described.38 Briefly, 5 µg of total RNA was used for hybridization on miRNA microarray chips. These chips contain genespecific 40-mer oligonucleotide probes, spotted and covalently attached to a polymeric matrix. The microarrays were hybridized in 6 × SSPE (0.9 m NaCl, 60mm NaH2PO4 · H2O, 8mm EDTA, pH 7.4)/30% formamide at 25 °C for 18 h, washed in 0.75 × TNT (Tris-HCl/NaCl/Tween 20) at 37 °C for 40 min and processed by using a method of direct detection of the biotin-containing transcripts by streptavidin-Alexa Fluor 647 conjugate. Processed slides were scanned by using a microarray scanner, with the laser set to 635 nm, at fixed photomultiplier tube (PMT) setting, and a scan resolution of 10 mm.
Bisulfite conversion of genomic DNA and methylation-specific PCR
Bisulfite conversion of genomic DNA was performed as described in the EZ DNA methylation kit (Zymo Research, Irvine, CA, USA) to create a template for methylation-specific PCR. For bisulfite sequencing, the sodium bisulfite-modified DNA was subjected to PCR with primers for miR-125b-1 (Gene ID: 406911) designed to amplify nucleotides from 922 to 1073 relative to the transcription start point and primers for miR-125b-2 (Gene ID: 262205364) designed to amplify nucleotides from 1598 to 1790 relative to the transcription start point.
5-aza-CdR treatment for demethylating
Normal human keratinocytes hOMK102, 107 and SCC cell lines FaDu, SCC4 were seeded 1 × 106 per well in six-well plates and cultured with 10 µmol/l 5-aza-CdR (Sigma-Aldrich, St Louis, MO, USA) for 5 days. The medium containing agent was replaced every 24 h. RNA was isolated and quantitative reverse transcriptase–PCR (qRT–PCR) was carried out to evaluate the restoration of miR-125b expression after 5-aza-CdR treatment.
qPCR and end-point PCR for detection of mature, precursor or genome of miR-125b and candidate genes
To detect mature miR-125b expression, we used Trizol reagent to isolate total RNA, which was then amplified by TaqMan stem-loop RT–PCR method.23 TaqMan miRNA assays used the Human Panel-Early Access Kit (ABI, Forest City, CA, USA), which includes 10 human cells as well as 3 negative controls. We also used individual miR-125b-specific primer sets and TaqMan probe from ABI to detect miR-125b expression in patient specimens and cell lines. In all, 5 µg of total RNA was subjected to cDNA synthesis by using SuperScript III First-Strand Synthesis System (Invitrogen) for detection of miR-125b1, 2 precursor and mir-125b downstream gene. At the end of the reaction, 40 µl of TE (10mm Tris, 1mm EDTA, pH7.6) was added and 1 µl of the reaction (1/60 volume) was used as a template for PCR. TACSTD2 or candidate target genes transcripts and miR-125b1, 2 precursor were quantified by using QuantiTect SYBR Green RT–PCR kit (QIAGEN, Valencia, CA, USA) and iCycler real-time PCR detection system (Bio-Rad, Hercules, CA, USA) for the detection of PCR product. Primer sequences are shown in Supplementary Table S1. Cycle threshold (Ct) value of the tested cDNA was converted to weight according to the standard curve. For detection of loss of genomic region of miR-125b-1, 2, genomic DNA was extracted and quantified by using Quantitect SYBR RT– PCR kit and iCycler real-time PCR detection system (Bio-Rad).
Construction for reporter assay
A reporter plasmid was constructed by cloning into pGL3 control vector (Promega, Madison, WI, USA), a 200-bp genomic sequence corresponding to part of the 3′-UTR region (TACSTD2 #1), using PCR primers, 5′-TG CTCTAGAGATTTCGGTATCGTCCCAGA-3′ and 5′-TGCTCTAGAGGACCGAAAG GGGATACATT-3′. The PCR product was then digested with XbaI and cloned into the reporter plasmid pGL3 control (Promega) downstream of the luciferase gene. As the negative control, mutations into the miR-125bbinding site of the TACSTD2 3′-UTR were introduced using mutant assay for GeneTailor (Invitrogen). The primers used for the miR-binding site mutagenesis are as follows and mutation sequences are indicated by noncapitalized letters: TACSTD2-mutF 5′-TTAATAGATCCTGGCggttcacTCTCCT TTCTT-3′ and TACSTD2-mutR 5′-GCCAGGATCTATTAAACCTGGTGTG-3′. All plasmid constructs were confirmed by DNA sequencing.
Luciferase target assay
HEK-293T and hOMK107 cells were counted, 4 × 105 cells seeded in 12- well plates and co-transfected using Lipofectamine 2000 (Invitrogen) with 2 µg of TACSTD2#1/pGL3-control vector and miR-125b with 2 µg of pRL-TK vector (Promega, San Luis Obispo, CA, USA). At 24 h after transfection, the cells were lysed in 500 µl of a passive lysis buffer. Cell lysates were directly used in the luciferase assay. Quantification of luciferase activities and calculation of relative ratios were carried out manually with a luminometer. Firefly activity was normalized to Renilla activity to control for the transfection efficiency. Data shown are mean ± s.d. of the ratio of luciferase activity (miR-125b vs control oligo) of a representative assay; experiments were performed in triplicate and repeated three times.
IP of AGO2-containing RISC
Wild-type and Lenti-miR-125b-1-expressing FaDu cells were washed once with phosphate-buffered saline and lysed for 30 min in buffer containing 25mm Tris-HCl, pH 7.4, 150mm KCl, 5mm EDTA, 0.5% Nonidet P-40, 5mm dithiothreitol, protease inhibitors and 100 units/ml of RNase inhibitor (Invitrogen). To form the AGO2/anti-AGO2 antibody complexes, 1mg of lysate was incubated with 5 µg of anti-AGO2 antibody (Abnova, Walnut, CA, USA) or normal mouse IgG (immunoglobulin G) for 3 h, followed by 50 µl of protein A/G agarose beads for 2 h. Subsequently, the immunocomplexes were washed three times and used for RNA isolation (9/10 of the IP) or immunoblots (1/10 of the IP). cDNA was synthesized with 50 ng of RNA using SuperScript III cDNA synthesis kit (Invitrogen), and qRT–PCR was performed to determine TACSTD2 enrichment in the RISC; RISC-associated GAPDH (glyceraldehyde 3-phosphate dehydrogenase) was used for normalization.
miR and gene transfection and lentivirus overexpression
For overexpression of pre-miR-125-b-1, anti-miR-125b and pre-miR-control (Applied Biosystems, Foster City, CA, USA) or TACSTD2 and MYC (GeneCopoeia, Rockville, MD, USA), each component was transfected into FaDu and SCC9 using Lipofectamine 2000 (Invitrogen). For exogenous miR- 125b-1 stable expression, lentivirus vector and empty vector (System Biosciences, Mountain View, CA, USA) with a green fluorescent protein tag were used to prepare empty vector or vector carrying the miR-125b insert for co-transfection with helper plasmids into 293FT cell to produce lentiviruses. The stable cell lines were established by lentiviral infection.
RNA interference
Short hairpins (Santa Cruz Biotech, Dallas, TX, USA) targeting human TACSTD2 and MYC were cloned into the lentiviral vector. Lentivirus was generated as previously described.20 Infected cells were selected in puromycin to generate pooled stable knockdown populations.
Western blot analysis
Whole-cell lysates were prepared from FaDu and SCC9 cells 16 h after transfection or each condition and subjected to western blot analysis. Proteins were subjected to SDS–PAGE and transferred to PVDF membrane. After treatment with blocking reagent (PIERCE, Rockford, IL, USA), the membrane was incubated with goat polyclonal antiserum against TACSTD2 (R&D, Minneapolis, MN, USA), rabbit polyclonal antisera against ERK1/2, phospho-ERK1/2 (Thr202/Tyr204) and MYC obtained from Cell Signaling (Danvers, MA, USA).
Cell invasion assay
Effect of miR-125b on the invasion ability of SCC9 and FaDu cells was determined using QCM 96-well Cell Invasion Assay kit (Chemicon, Temecula, CA, USA). Cells infected with either miR-125b or control lentiviral vectors were seeded into inserts at 2 × 104 per insert in serumfree medium and then transferred to wells filled with culture medium containing 10% fetal bovine serum. The plates with cells were incubated for 16 h at 37 °C in a CO2 incubator. After incubation, non-invading cells on the top of the membrane were removed by scraping. Migrating cells on the bottom of the insert were incubated with 150 µl of prewarmed cell detachment buffer for 30 min, dissociated from the membrane and detected by CyQuant GR dye. The fluorescence plate was read using a 480/520nm filter set.
Soft agar colony-formation assay agar (0.6%; bottom agar) was added to six-well plates and allowed to solidify. Cells (1 × 105) were resuspended in 1ml of 0.3% agar (top agar) and overlayed onto the bottom agar. Cells were fed with top agar every 3 days, and after 4 weeks, colonies in agar were solubilized and measured by fluorescence plate reader.
Cell colony-forming assay
FaDu cells infected with miR-125b, shTACSTD2, shMYC or control lentiviral vectors or transfected with TACSTD2, MYC or control vectors were seeded at 1 × 103 per 3ml medium in six-well plates. Two weeks later, colonies were counted using the QuantityOne software (Bio-Rad). The mean ± s.d. of three independent experiments performed in triplicate was reported.
U0126 treatment for ERK inhibition
Cells for various experimental conditions were plated and U0126 (Cell Signaling Technology) was added into the culture medium at a concentration of 1 µm. For a single treatment, three identical plates were treated and cultured for 24 h.
Statistical analysis
The mean ± s.d. was calculated for each data point. Differences between groups were analyzed by the Student’s t-test or analysis of variance (Dunnett’s multiple comparison test). A P-value < 0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Kay Huebner (Ohio State University) for careful editing of this manuscript and Sharon Palko (Ohio State University) for administrative support.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Author contributions: HN and CMC designed research; HN, RC, and TK performed research; HN, CT, JP, CFC, SV, and CMC analyzed data; HN and CMC wrote the paper. All authors critically reviewed and edited the paper.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Forastiere A, Koch W, Trotti A, Sidransky D. Head and neck cancer. N Eng J Med. 2001;345:1890–1900. doi: 10.1056/NEJMra001375. [DOI] [PubMed] [Google Scholar]
- 3.Mao L, Hong WK, Papadimitrakopoulou VA. Focus on head and neck cancer. Cancer Cell. 2004;5:311–316. doi: 10.1016/s1535-6108(04)00090-x. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 5.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu CJ, Tsai MM, Hung PS, Kao SY, Liu TY, Wu KJ, et al. miR-31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma. Cancer Res. 2010;70:1635–1644. doi: 10.1158/0008-5472.CAN-09-2291. [DOI] [PubMed] [Google Scholar]
- 7.Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 8.Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer. 2007;6:5. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 10.Henson BJ, Bhattacharjee S, O’Dee DM, Feingold E, Gollin SM. Decreased expression of miR-125b and miR-100 in oral cancer cells contributes to malignancy. Genes Chromosomes Cancer. 2009;48:569–582. doi: 10.1002/gcc.20666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345–352. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 12.Visone R, Pallante P, Vecchione A, Cirombella R, Ferracin M, Ferraro A, et al. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2007;26:7590–7595. doi: 10.1038/sj.onc.1210564. [DOI] [PubMed] [Google Scholar]
- 13.Fong D, Moser P, Krammel C, Gostner JM, Margreiter R, Mitterer M, et al. High expression of TROP2 correlates with poor prognosis in pancreatic cancer. Br J Cancer. 2008;99:1290–1295. doi: 10.1038/sj.bjc.6604677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fong D, Spizzo G, Gostner JM, Gastl G, Moser P, Krammel C, et al. TROP2: a novel prognostic marker in squamous cell carcinoma of the oral cavity. Mod Pathol. 2008;21:186–191. doi: 10.1038/modpathol.3801001. [DOI] [PubMed] [Google Scholar]
- 15.Mühlmann G, Spizzo G, Gostner J, Zitt M, Maier H, Moser P, et al. TROP2 expression as prognostic marker for gastric carcinoma. J Clin Pathol. 2009;62:152–158. doi: 10.1136/jcp.2008.060590. [DOI] [PubMed] [Google Scholar]
- 16.Nakashima K, Shimada H, Ochiai T, Kuboshima M, Kuroiwa N, Okazumi S, et al. Serological identification of TROP2 by recombinant cDNA expression cloning using seram of patients with esophageal squamous cell carcinoma. Int J Cancer. 2004;112:1029–1035. doi: 10.1002/ijc.20517. [DOI] [PubMed] [Google Scholar]
- 17.Ohmachi T, Tanaka F, Mimori K, Inoue H, Yanaga K, Mori M. Clinical significance of TROP2 expression in colorectal cancer. Clin Cancer Res. 2006;12:3057–3063. doi: 10.1158/1078-0432.CCR-05-1961. [DOI] [PubMed] [Google Scholar]
- 18.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 20.Cubas R, Zhang S, Li M, Chen C, Yao Q. Trop2 expression contributes to tumor pathogenesis by activating the ERK MAPK pathway. Mol Cancer. 2010;9:25321. doi: 10.1186/1476-4598-9-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 22.Kikkawa N, Hanazawa T, Fujimura L, Nohata N, Suzuki H, Chazono H, et al. miR-489 is a tumour-suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC) Br J Cancer. 2010;103:877–884. doi: 10.1038/sj.bjc.6605811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura S, Naganuma S, Susuki D, Hirono Y, Yamaguchi A, Fujieda S, et al. Expression of microRNAs in squamous cell carcinoma of human head and neck and the esophagus: miR-205 and miR-21 are specific markers for HNSCC and ESCC. Oncol Rep. 2010;6:1625–1633. doi: 10.3892/or_00000804. [DOI] [PubMed] [Google Scholar]
- 24.Lee Y, Yang X, Huang Y, Fan H, Zhang Q, Wu Y, et al. Network modeling identifies molecular functions targeted by miR-204 to suppress head and neck tumor metastasis. PLoS Comput Biol. 2010;6:e1000730. doi: 10.1371/journal.pcbi.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hui AB, Lenarduzzi M, Krushel T, Waldron L, Pintilie M, Shi W, et al. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clin Cancer Res. 2010;16:1129–1139. doi: 10.1158/1078-0432.CCR-09-2166. [DOI] [PubMed] [Google Scholar]
- 26.Avissar M, Christensen BC, Kelsey KT, Marsit CJ. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clin Cancer Res. 2009;15:2850–2855. doi: 10.1158/1078-0432.CCR-08-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC. Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem. 2007;282:1479–1486. doi: 10.1074/jbc.M609383200. [DOI] [PubMed] [Google Scholar]
- 28.Xia HF, He TZ, Liu CM, Cui Y, Song PP, Jin XH, et al. MiR-125b expression affects the proliferation and apoptosis of human glioma cells by targeting Bmf. Cell Physiol Biochem. 2009;23:347–358. doi: 10.1159/000218181. [DOI] [PubMed] [Google Scholar]
- 29.Liang L, Wong CM, Ying Q, Fan DN, Huang S, Ding J, et al. MicroRNA-125b suppressesed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology. 2010;52:1731–1740. doi: 10.1002/hep.23904. [DOI] [PubMed] [Google Scholar]
- 30.Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu M, et al. An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci USA. 2007;104:19983–19988. doi: 10.1073/pnas.0706641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang L, Luo J, Cai Q, Pan Q, Zeng H, Guo Z, et al. MicroRNA-125b suppresses the development of bladder cancer by targeting E2F3. Int J Cancer. 2011;128:1758–1769. doi: 10.1002/ijc.25509. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto N, Uzawa K, Miya T, Watanabe T, Yokoe H, Shibahara T, et al. Frequent allelic loss/imbalance on the long arm of chromosome 21 in oral cancer: evidence for three discrete tumor suppressor gene loci. Oncol Rep. 1999;6:1223–1227. doi: 10.3892/or.6.6.1223. [DOI] [PubMed] [Google Scholar]
- 33.Yamada H, Yanagisawa K, Tokumaru S, Taguchi A, Nimura Y, Osada H, et al. Detailed characterization of a homozygously deleted region corresponding to a candidate tumor suppressor locus at 21q11-21 in human lung cancer. Genes Chromosomes Cancer. 2008;47:810–818. doi: 10.1002/gcc.20582. [DOI] [PubMed] [Google Scholar]
- 34.Negrini M, Rasio D, Hampton GM, Sabbioni S, Rattan S, Carter SL, et al. Definition and refinement of chromosome 11 regions of LOH in breast cancer: identification of a new region at 11q23-q24. Cancer Res. 1995;55:3003–3007. [PubMed] [Google Scholar]
- 35.Rasio D, Negrini M, Manenti G, Dragani T, Croce CM. Loss of heterozygosity at chromosome 11q in lung adenocarcinoma: identification of three independent regions. Cancer Res. 1995;55:3988–3991. [PubMed] [Google Scholar]
- 36.Zhang Y, Yan LX, Wu QN, Du ZM, Chen J, Liao DZ, et al. miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res. 2011;71:3552–3562. doi: 10.1158/0008-5472.CAN-10-2435. [DOI] [PubMed] [Google Scholar]
- 37.Bueno MJ, Pérez de Castro I, Gómez de Cedrón M, Santos J, Calin GA, Cigudosa JC, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;6:496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 38.Nakanishi H, Nakamura T, Canaani E, Croce CM. ALL1 fusion proteins induce deregulation of EphA7 and ERK phosphorylation in human acute leukemias. Proc Natl Acad Sci USA. 2007;104:14442–14447. doi: 10.1073/pnas.0703211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gupta S, Davis RJ. MAP kinase binds to the NH2-terminal activation domain of c-Myc. FEBS Lett. 1994;353:281–285. doi: 10.1016/0014-5793(94)01052-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.