Abstract
MicroRNAs (miRNAs) play a critical role in development and progression of cancers. Deregulation of MicroRNA-9 (miR-9) has been documented in many types of cancers but their role in the development of esophageal squamous cell carcinoma (ESCC) has not been studied. This study aimed to investigate the effect of miR-9 in esophageal cancer metastasis. The up-regulation of miR-9 was frequently detected in primary ESCC tumor tissue, which was significantly associated with clinical progression (P = 0.022), lymph node metastasis (P = 0.007) and poor overall survival (P < 0.001). Functional study demonstrated that miR-9 promoted cell migration and tumor metastasis, which were effectively inhibited when expression of miR-9 was silenced. Moreover, we demonstrated that miR-9 interacted with the 3′-untranslated region of E-cadherin and down-regulated its expression, which induced β-catenin nuclear translocation and subsequently up-regulated c-myc and CD44 expression. In addition, miR-9 induced epithelial-mesenchymal transition (EMT) in ESCC, a key event in tumor metastasis. Taken together, our study demonstrates that miR-9 plays an important role in ESCC metastasis by activating β-catenin pathway and inducing EMT via targeting E-cadherin. Our study also suggests miR-9 can be served as a new independent prognostic marker and/or as a novel potential therapeutic target for ESCC.
Keywords: miR-9, ESCC, E-cadherin, metastasis, β-catenin
INTRODUCTION
Esophageal cancer is one of the most common solid malignancies in the world and ranks as the sixth-leading cause of cancer-related mortality [1]. Esophageal squamous cell carcinoma (ESCC) is the predominant histologic type in East Asia, especially in the high-risk areas in northern China [2, 3]. Despite the great advances achieved in diagnosis and multimodality therapies recently, the prognosis of ESCC is still poor with a dismal 5-year survival rate of 20–30% [4]. The high probability of metastasis and recurrence is still the major reason of grim prognosis, yet the precise molecular mechanism of metastatic dissemination is still not completely clear [3]. Therefore, understanding the factors involved in ESCC metastasis is required for the identification of new prognostic biomarkers and therapeutic targets.
In the last few years, growing body of evidences indicate that microRNAs (miRNAs) are involved in multiple cellular processes as posttranscriptional regulators and particularly in cancer development and progression [5, 6]. miRNAs are a diverse class of 20–24 nucleotide that plays important roles in gene regulation by pairing to the 3′-untranslated region (3′-UTR) of target mRNAs to direct their posttranscriptional repression [7]. Deregulation of miRNAs has been reported to play roles in ESCC metastasis by acting as activators or inhibitors [8–10]. In our recent study, microarray strategy was applied to identify differentially expressed miRNAs in ESCC cells by comparing miRNA profiles between tumor and paired non-tumor tissues [11]. MiR-9 was investigated in this study because its deregulation has been reported in several types of cancers, including breast cancer [12], colorectal cancer [13] and melanoma [14]. However, the role of miR-9 in the development and progress of ESCC remains unclear. In the present study, overexpression of miR-9 was frequently detected in primary ESCC cases, which was associated with clinical progression, lymph node metastasis and poor overall survival. Functional study found that miR-9 increased cell motility in vitro and tumor metastasis in vivo. Although E-cadherin has been documented as a target of miR-9 in breast cancer [12], we further demonstrated that miR-9 directly targeted the 3′-UTR of E-Cadherin and activated the β-catenin signaling pathway in ESCC.
RESULTS
Up-regulation of miR-9 is frequently detected in ESCC
To evaluate expression situation of miR-9 in clinical ESCC specimens, quantitative real-time PCR (qRT-PCR) was used to compare expression levels of miR-9 between tumor and corresponding non-tumor esophageal mucosa in 67 ESCCs. Up-regulation of miR-9 was detected in 35/67 (52.24%) of ESCC tumors, compared with corresponding nontumorous tissues (defined as >2-fold increase). The average miR-9 expression was significantly higher in tumor tissues than in their normal counterparts (P = 0.0016) (Figure 1A). Expression level of miR-9 in 9 human ESCC cell lines was also detected by qRT-PCR and the result showed that up-regulation of miR-9 could be detected in 8/9 cell lines (except EC109) compared with a pool of 5 nontumorous tissues as a normal control (Figure 1B).
Figure 1. miR-9 was frequently up-regulated in primary ESCC cases and cell lines.
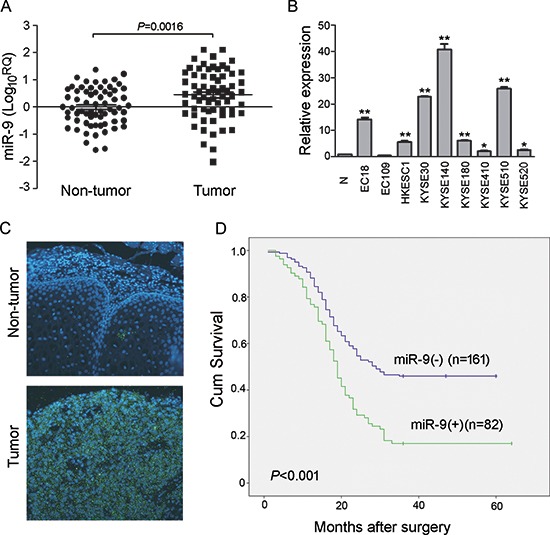
(A) qRT-PCR shows that miR-9 was frequently up-regulated in 67 primary ESCC tissues compared with their adjacent non-tumor tissues. (P = 0.0016, independent t test). Expression of miR-9 was shown in log10 scale and normalized to U6. (B) Up-regulation of miR-9 was detected in all tested ESCC cell lines except EC109 compared with pool of non-tumor tissues (N). U6 was set as an endogenous control. *P < 0.05; **P < 0.001. (C) Representative, of miR-9 expression (green signals) in a pair of ESCC tumor tissue and corresponding non-tumor tissue detected by MISH. Nuclei were conterstained by DAPI (blue color). Original magnification, 40 × objective. (D) Kaplan-Meier analysis indicates up-regulation of miR-9 was significantly associated with poorer overall survival rates of ESCC patients (P < 0.001).
Up-regulation of miR-9 is associated with ESCC metastasis and poor prognosis
To investigate the clinical significance of miR-9 up-regulation in ESCC, expression of miR-9 was evaluated by microRNA in situ hybridization (MISH) in a tissue microarray containing 300 pairs of primary ESCCs and their paired non-tumor samples. Informative results were observed in 243 pairs of ESCC cases, while non-informative cases included lost samples and samples with limited number of cells. The overexpression of miR-9, defined as its fluorescent signals in tumor tissue obviously more and stronger than that in the corresponding non-tumor tissue, was detected in 82/243 (33.74%) of informative ESCC tissues (Figure 1C). The clinical association analysis found that overexpression of miR-9 was significantly associated with advanced clinical stage (P = 0.022) and lymph node metastasis (P = 0.007, Table 1). Kaplan-Meier analysis indicated that overexpression of miR-9 was significantly associated with poorer overall survival (log-rank test, P < 0.001, Figure 1D). Further, multivariate Cox regression analysis revealed that overexpression of miR-9 is an independent prognostic factor for poor survival of patients with ESCC (P = 0.009, Table 2).
Table 1. Clinicopathological correlation of miR-9 expression in ESCC.
| Feature | miR-9 expression level | |||
|---|---|---|---|---|
| All | Normal | Upregulated | P | |
| Age | 0.069 | |||
| ≥60 years | 139 | 98(70.5) | 41(29.5) | |
| >60 years | 104 | 63(60.6) | 41(39.4) | |
| Differentiation | 0.093 | |||
| Well | 60 | 33(55.0) | 27(45.0) | |
| Moderate | 156 | 108(69.2) | 48(30.8) | |
| Poor | 27 | 20(74.1) | 7(25.9) | |
| Tumer invasion | 0.705 | |||
| T1 | 18 | 12(66.7) | 6(33.3) | |
| T2 | 18 | 10(55.6) | 8(44.4) | |
| T3 | 45 | 32(71.1) | 13(28.9) | |
| T4 | 162 | 107(66.0) | 55(34.0) | |
| Lymph node metastasis | 0.007* | |||
| N0 | 138 | 101(73.2) | 37(26.8) | |
| N1 | 105 | 60(57.1) | 45(42.9) | |
| Clinical stage | 0.022* | |||
| Stage I-II | 161 | 115(71.4) | 46(28.6) | |
| Stage III-IV | 82 | 46(56.1) | 36(43.9) | |
Significant difference
Table 2. Cox proportional hazard regression analyses for overall survival.
| Clinicopathological Features | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| HR(95%Cl) | P | HR (95%Cl) | P | |
| miR-9 UPregulation | 1.913(1.391–2.632) | <0.001* | 1.543 (1.112–2.140) | 0.009* |
| Gender | 0.785(0.568–1.085) | 0.142 | ||
| Age | 1.216(0.886–1.669) | 0.225 | ||
| Differentiation | 1.574(1.192–2.079) | <0.001* | 1.465(1.114–1.928) | 0.006* |
| Tumor invasion | 1.481(1.197–1.831) | <0.001* | 1.267(1.012–1.586) | 0.039* |
| Clinical stage | 2.483(1.802–3.420) | <0.001* | 1.545 (0.799–2.990) | 0.196 |
| LN metastasis | 2.138(1.556–2.938) | <0.001* | 1.218 (0.654–2.268) | 0.534 |
CI = confidence interval; HR = hazard ratio;
Statistical significance (p < 0.05) is shown in bold.
miR-9 promotes cell migration and tumor metastasis
To investigate the oncogenic function of miR-9, miR-9 was cloned into a lentiviral vector and stably transfected into ESCC cell lines HKESC1 and KYSE410 (Figure 2A). Empty vector-transfected cells were used as controls. Tumorigenic effect of miR-9 was studied by XTT cell growth assay, foci formation assay and tumor formation in nude mice. Unexpectedly, no significant difference was detected by XTT assay between miR-9 transfected cells and control cells (data not shown). Foci formation and tumor formation in nude mice showed that miR-9 could increase number of foci formed and promote tumor growth in tested mice in KYSE410 cells, but not in HKESC1 cells (Figure 2B and 2C). Since overexpression of miR-9 has been significantly associated with ESCC metastasis, its role in cell migration and invasion was then investigated by both in vitro and in vivo assays. Cell migration assay showed that miR-9 could significantly increase cell motility in HKESC1 and KYSE410 cells compared with the empty vector-transfected cells (P < 0.01, Figure 3A). When endogenous miR-9 was silenced by a siRNA against miR-9 in KYSE30 and KYSE510 cells (Figure 3B), the number of migrated cells decreased significantly compared with scramble siRNA-transfected control cells (P < 0.01, Figure 3C).
Figure 2. Functional study of miR-9.
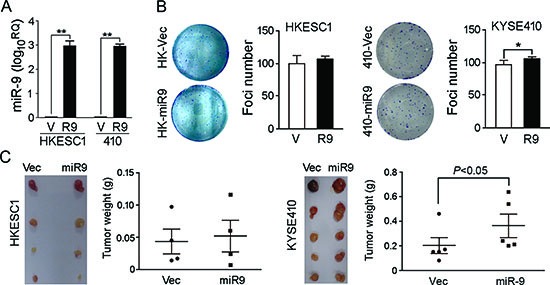
(A) Relative expression of miR-9 was detected by qRT-PCR in miR-9-transfected (R9) HKESC1 and KYSE410 cells compared with empty vector-transfected cells (V). **P < 0.001. (B) Foci formation assay was performed to compare frequency of foci formation between miR-9- and empty vector-transfected cells. Results are expressed as mean ±S.E.M. of three independent experiments. *P < 0.05. (C) Representative images of xenografts and summary of tumor weight in tumor formation in nude mice. miR-9- and empty vector-transfected cells were inoculated subcutaneously to the flanks of nude mice. Xenografts were isolated and weighted after 4 weeks.
Figure 3. miR-9 promotes cell migration and tumor metastasis.
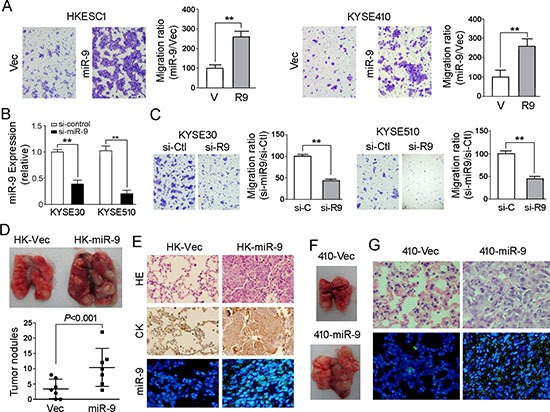
(A) Representative images and summary of cell migration assay. Compared with empty vector-transfected cells, miR-9-transfected cells could promote cell migration in HKESC1 and KYSE410 cells. The results are expressed as mean ± S.E.M. of three independent experiments. **P < 0.01. (B) Down-regulation of miR-9 was detected by qRT-PCR when miR-9 was silenced by a siRNA in KYSE30 and KYSE510 cells. Cells treated with scramble siRNA were used as control cells. Results are expressed as mean ± S.E.M. of three independent experiments. **P < 0.01. (C) Representative images and summary of migrated cells between siRNA-miR-9 and scramble siRNA treated KYSE30 and KYSE510 cells. The results are expressed as mean ± S.E.M. of three independent experiments. **P < 0.01. (D) miR-9 could promote ESCC metastasis in vivo. Representative images of lungs derived from mice injected with HK-miR-9 and control cells (upper). Visible tumor nodules were counted and summarized (bottom). (E) H&E staining (upper) and IHC staining of CK (middle) was performed on pulmonary sections derived from mice. Original magnification: 20 × objective. Expression of miR-9 (green signals) was detected by MISH in pulmonary sections (bottom). Original magnification: 40 × objective. (F) Representative image of lung derived from mice injected with 410-miR-9 and control cells. (G) H&E staining (upper) was performed on pulmonary sections derived from mice injected with 410-miR-9 and control cells. Original magnification: 40 × objective. Expression of miR-9 was detected by MISH in pulmonary sections (bottom). Original magnification: 20 × objective.
To further validate the effect of miR-9 on tumor metastasis, in vivo metastasis assay was performed in nude mice. miR-9 expressing HKESC1 (HK-miR-9) and KYSE410 (410-miR-9) cells were injected into nude mice via tail vein, respectively. Empty vector trasfected cells were used as controls (HK-Vec and 410-Vec). Eight weeks later, tested mice were sacrificed and the metastatic modules formed on the lungs and livers were counted. No visible metastatic nodule was observed in livers of mice except one HK-miR-9 mouse (data not shown). Pulmonary metastatic nodules induced by HK-miR-9 cells and HK-Vec cells were detected in 7/7 and 5/7 of tested mice, respectively. As shown in Figure 3D, the number of pulmonary metastatic nodules induced by HK-miR-9 cells was significantly higher than that induced by control cells (P < 0.001, independent student's t test). Histological study with H&E stained paraffin block sections confirmed that the pulmonary nodules were metastatic cancers (Figure 3E). IHC staining with anti-cytokeratin antibody was performed to validate that the cells originated from injected tumor cells (Figure 3E). MISH demonstrated that miR-9 was overexpressed in metastatic nodule induced by miR-9 overexpressed cells compared with control cells (Figure 3E). The pulmonary metastatic nodules induced by 410-miR-9 cells could be only observed in one mouse whereas no visible tumor nodule was observed in 410-Vec mice (Figure 3F). Histological and MISH studies confirmed the metastatic nodule was cancer with miR-9 overexpression (Figure 3G).
E-cadherin is a putative target of miR-9 in ESCC
By searching the targets of miR-9 with TargetScan and miRanda, one potential target E-cadherin was selected for further study because it has been associated with tumor metastasis [15]. 3′-UTR of E-cadherin mRNA contains one complementary site for miR-9. To examine whether E-cadherin is regulated by miR-9 in ESCC, we tested the expression of E-cadherin in miR-9 overexpressed cell lines HKESC1 and KYSE410. qRT-PCR and western blotting results showed that expression of E-cadherin was significantly reduced in HKESC1 and KYSE410 when miR-9 was stably transfected into these cells compared with empty vector-transfected control cells (P < 0.01, Figure 4A). When miR-9 was silenced in KYSE30 and KYSE510 cells, expression of E-cadherin was increased significantly (P < 0.01, Figure 4B).
Figure 4. miR-9 down-regulates E-cadherin in ESCC cells.
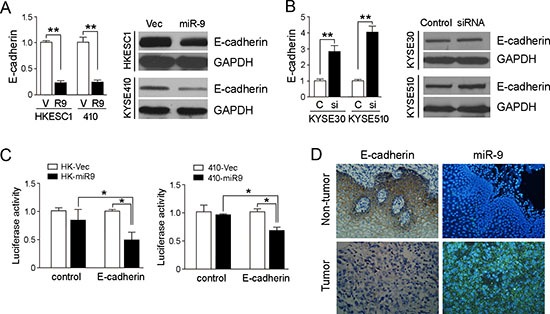
(A) qRT-PCR and Western blotting results showed that miR-9 could effectively down-regulate E-cadherin expression in HK-miR-9 and 410-miR-9 cells compared with control cells. qRT-PCR results were expressed as mean ± S.E.M. of three independent experiments. **P < 0.001. GAPDH was set as an internal control in Western blot analysis. (B) Expressions of E-cadherin in both mRNA and protein levels were increased when miR-9 was silenced in KYSE30 and KYSE510 cells by a siRNA against miR-9, compared with scramble siRNA-treated cells. C, control scramble siRNA; si, siRNA against miR-9. **P < 0.01. (C) Luciferase assay was performed to confirm that miR-9 could target E-cadherin. miR-9 overexpressed cells and control cells were co-transfected with pMIR-REPORT-CDH1(3′-UTR)/empty vector with pRL-TK, and relative luciferase activity was detected. *P < 0.05. (D) Representative pictures of E-cadherin staining (by IHC) and miR-9 (by MISH) in a pair of ESCC tumor tissue and corresponding non-tumor tissue. Results showed that E-cadherin was down-regulated in miR-9 overexpressed tumor tissue. Original magnification: 20 × objective.
A luciferase reporter assay was then performed to verify whether E-cadherin is the direct target of miR-9. The potential miR-9 binding sequence in the 3′-UTR region of E-cadherin was cloned into a luciferase reporter vector and transiently transfected into the miR-9 overexpressed cells and vector control cells. Decreased luciferase activity in miR-9 overexpressed cells was observed compared with the control cell (Figure 4C), indicating that E-cadherin was a downstream target of miR-9.
miR-9 level is negatively correlated with expression of E-cadherin in ESCC
To investigate the correlation between expressions of miR-9 and E-cadherin in clinical samples, IHC was performed using a TMA containing 300 pairs of primary ESCCs. Informative results were observed in 237 pairs (both tumor and non-tumor tissues) of ESCCs. Non-informative cases were mainly caused by tissue loss in either tumor or paired non-tumor tissues. E-cadherin staining was calculated by adding the scores for the percentage and intensity of positive staining of E-cadherin. Since the staining index of E-cadherin in non-tumor samples was ≥5, down-regulation of E-cadherin was defined as staining index <5. Down-regulation of E-cadherin was detected in 103/237 (43.5%) of informative ESCC tissues compared with their paired non-tumor tissues (Figure 4D). Moreover, the correlation between miR-9 and E-cadherin was analyzed by Pearson χ2 test in 216 cases with both miR-9 and E-cadherin information, a negative correlation was detected between miR-9 and E-cadherin (P = 0.02, Supplementary Table 1).
miR-9 promotes tumor metastasis via nuclear translocation of β-catenin
E-cadherin/β-catenin complex is known for connecting the actin cytoskeleton to the adherens junctions [16]. When the cytoplasmic E-cadherin decreased, β-catenin is released from the complex. Stabilized “free” β-catenin goes to the nucleus, binds T-cell factor/lymphoid enhancer factor (TCF/Lef) and activates transcription of target genes such as c-myc, cyclin D1, CD44 and VEGF, which are responsible for cell proliferation and metastasis [17]. To explore the molecular mechanism of miR-9/E-cadherin in promoting metastasis, subcellular localization of β-catenin was detected by immunofluorescence (IF) analysis. IF result showed that β-catenin translocation from the cell membrane to nucleus was observed in both miR-9-transfected HKESC1 and KYSE410 cells, whereas β-catenin was mainly located on the membrane in the empty vector-transfected cells (Figure 5A). To determine whether the nuclear translocation of β-catenin can upregulate its downstream target genes, western blot analysis was used to test expressions of c-myc, cyclin D1, CD44 and VEGF. Increased expressions of c-myc, CD44 and VEGF were detected in HK-miR-9 and 410-miR-9 cells compared with their control cells (Figure 5B). No significant change was observed on cyclin D1 protein level. When miR-9 was knocked down in KYSE30 and KYSE510 cells, the protein levels of c-myc, CD44 and VEGF were decreased (Figure 5C).
Figure 5. miR-9 activates β-catenin pathway and induces EMT.
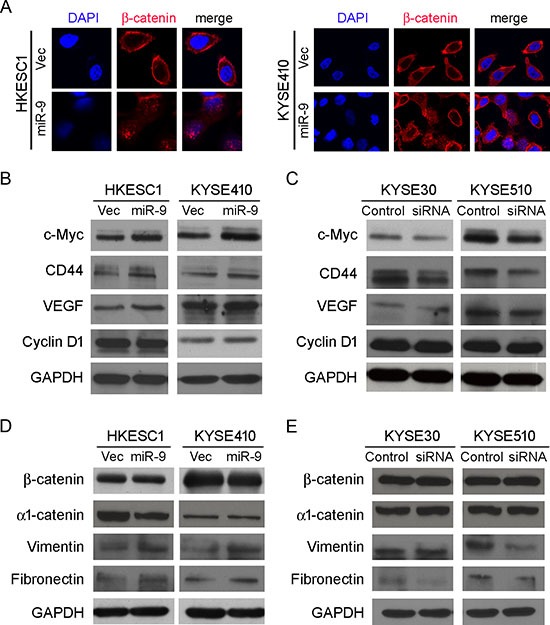
(A) IF showed that nuclear translocation of β-catenin (red) happened in miR-9-transfected cells while β-catenin located mainly on the membrane in empty vector-transfected cells. Nuclei were counterstained with DAPI. Original magnification: 40 × objective. (B-E) Western blot analysis was used to compare protein expression levels between miR-9- and empty vector-transfected cells (B and D), or between cells treated with siRNA against miR-9 and scramble siRNA (C and E). GAPDH was set as internal control.
miR-9 induces epithelial-mesenchymal transition in ESCC
As epithelial-mesenchymal transition (EMT) is one of the key events involved in invasion and metastasis of tumor cells, several epithelial markers (β-catenin and α1-catenin) and mesenchymal markers (Vimentin and Fibronectin) were tested by Western blot analysis. The results found that down-regulations of epithelial markers were not obvious except decrease of α1-catenin in HK-miR-9 cells (Figure 5D). Upregulation of Vimentin and Fibronectin could be detected in both HK-miR-9 and 410-miR-9 cells (Figure 5D). In miR-9 silenced KYSE30 and KYSE510 cells, mesenchymal markers were down-regulated except Vimentin in KYSE30 cells (Figure 5E). These data suggest that miR-9 could induce partial EMT that promoted cancer cell invasion and metastasis.
DISCUSSION
Previous studies have reported that miRNAs, such as oncomiRs or anti-oncomiRs, play important roles in cancer development and progression by acting as activators or inhibitors [18]. More recently, several specialized miRNAs termed metastamirs have been implicated in the regulation of tumor metastasis [19]. Metastamirs always play important roles in various steps of metastasis rather than tumorigenesis.[12, 19, 20] Characterization of metastamirs and their targets involved in the progression of ESCC may lead to the identification of novel prognostic markers and therapeutic targets. miR-9, a highly conserved microRNA, was reported to be expressed predominantly in the central nervous system of the developing embryo exhibiting a prodifferentiation function [21]. In addition to its involvement in neurogenesis, miR-9 was first found to be up-regulated in primary brain tumors [22]. Up-regulation of miR-9 has been also reported in breast cancer [12], colorectal cancer [13] and melanoma [14]. Interestingly, down-regulation of miR-9 has been reported in gastric [23] and ovarian cancers [24], suggesting the diversity role of miR-9 in different cancers.
Here we report that miR-9 was frequently up-regulated in primary ESCC tumors, which was significantly associated with advanced clinical stage (P = 0.022), lymph node metastasis (P = 0.007) and poor survival rate (P < 0.001) of ESCC. Moreover, multivariate Cox regression analysis revealed that up-regulation of miR-9 is an independent prognostic factor for poor survival of patients with ESCC. Functional study found that miR-9 could increase cell migration and metastasis, implying that miR-9 is a metastamiR in ESCC, and it could be used as a prognostic biomarker in ESCC.
As reported by many studies, informatics analysis indicated that E-cadherin might be the downstream target of miR-9 [12, 25]. In the present study, E-cadherin was down-regulated in miR-9 overexpressed cells and up-regulated when endogenous miR-9 was silenced. Luciferase assay also confirmed that miR-9 could target E-cadherin directly. E-cadherin is a crucial protein mediated cell-cell adhesion to hold the epithelial cells tight together. Loss of E-cadherin decreases the cellular adhesion, resulting in an increase in cellular motility [26]. It has been reported that down-regulation of E-cadherin in esophageal carcinoma was associated with an increased invasive and metastatic potential [15]. Moreover, E-cadherin can sequester β-catenin as a complex to connect the actin cytoskeleton to the adherent junctions [26]. Upon loss of E-cadherin expression, β-catenin was released from the complex. Stabilized “free” β-catenin goes to the nucleus and modulates b-catenin/Tcf-mediated gene expression to promote cell invasion and metastasis [27]. It was also reported that loss of E-cadherin activates EGFR-MEK/ERK signaling and promotes invasion in non-small cell lung cancer [28]. Recently, Ma et al. reported that miR-9 could target E-cadherin and allow liberation of β-catenin, which then activates the VEGF to promote metastasis in breast cancer [12]. Our results confirmed miR-9 to be involved in regulation of cell metastasis by targeting E-cadherin in ESCC. In the ESCC primary tumor tissues, miR-9 is negatively correlated with the expression of E-cadherin (R = −0.163, P < 0.05). Although miR-9 could not cause significant change on β-catenin protein level, the nuclear translocation of β-catenin was observed in miR-9 overexpressed cells. Western blotting analysis demonstrated that the downstream targets of β-catenin, including VEGF, c-myc and CD44 were up-regulated in miR-9 overexpressed cells, suggesting that these genes may participate in the tumor metastasis in ESCC.
Since epithelial-mesenchymal transition is one of the key events involved in tumor invasion and metastasis [27], the effect of miR-9 on EMT was also investigated in the study. As we mentioned earlier, no significant change on β-catenin protein level was observed. α1-catenin decreased in HK-miR-9 cells and increased in miR-9 silenced KYSE30 cells. Changes of 2 mesenchymal markers (Vimentin and Fibronectin) were also detected in miR-9 overexpressed and silenced cells, suggesting that the metastatic effect of miR-9 was via inducing EMT in ESCC. In summary, miR-9 was frequently overexpressed in ESCC specimens and played a crucial role in ESCC metastasis through targeting E-cadherin, promoting β-catenin nuclear translocation and subsequently inducing EMT.
MATERIALS AND METHODS
Cell lines and primary tumor tissues
Six ESCC cell lines (KYSE30, KYSE140, KYSE180, KYSE410, KYSE510, and KYSE520) were obtained from DSMZ (Braunschweig, Germany), the German Resource Centre for Biological Material [29]. Three Chinese ESCC cell lines (EC18, EC109, and HKESC1) were kindly provided by our fellows at the University of Hong Kong (Professors G. Srivastava and G.S. Tsao) [30]. Sixty seven pairs of primary ESCCs and corresponding nontumorous tissues were collected directly after surgical resection at Linzhou Cancer Hospital (Henan, China). A total of 300 formalin-fixed and paraffin-embedded ESCCs and their adjacent nontumorous tissue samples were also kindly provided by Linzhou Cancer Hospital. All patients recruited in the study did not receive preoperative treatment. Clinical samples used in this study were approved by the Committees for Ethical Review of Research at Zhengzhou University (Zhengzhou, China) and Sun Yat-Sen University Cancer Center (Guangzhou, China).
RNA isolation and quantitative real-time PCR
Total RNA was extracted with TRIzol (Invitrogen, Calsbad, CA) according to the manufacturer's instructions. Reverse transcription was performed using the PrimeScriptRT reagent Kit (Promega, Madison, WI). Real-time PCR was carried out using SYBR Green SuperMix (Roche, Basel, Switzerland) and ABI7900HT Fast Real-Time PCR system (Applied Biosystems, Foster City, CA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 was used as internal control. The primers of miR-9 and U6 were purchased from HAPK Biotechnonology (Shenzhen, China). Other primer sequences used in the study were listed in supporting data.
Tissue microarray (TMA) and immunohistochemical staining (IHC)
Tissue microarray containing 300 pairs of primary ESCC cases (tumor and corresponding non-tumor tissues) was constructed as described previously [31]. None of the patients in the study has received follow-up radiation or chemotherapy. IHC was performed using a standard streptavidin-biotin-peroxidase complex method. In brief, a TMA section was deparaffinized and rehydrated. Endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide for 15 minutes. For antigen retrieval, the TMA slide was microwave-treated in 10 mM citrate buffer (pH 6.0) for 10 minutes. The slide was then incubated with anti-E-cadherin monoclonal antibody (Cell Signalling Technology, Danvers, MA). The nucleus was counterstained using Meyer's hematoxylin. E-cadherin immunoreactivity was calculated by adding the scores for the percentage of E-cadherin -positive cells (<5%, 0; 5%–25%, 1; 25%–50%, 2; 50%–75%, 3; >75%, 4) and the intensity of E-cadherin-positive staining (negative, 0; weak, 1; moderate, 2; or strong, 3). Informative results were observed in 237 pairs of ESCC cases, while non-informative samples included lost samples and inappropriate staining.
microRNA in situ hybridization (MISH)
An oligonucleotide probe (5′-TCATACAGCTAGATAACCAAAGA-3′) complementary to hsa-miR-9-5p was purchased from the Exonbio Lab (Guangzhou, China). This oligonucleotide contains three 2′-fluoro-modified RNA residues (2′-F RNA) at the 4th, 10th and 20th bases, which can increase melting temperature and result in enhanced hybridization stability. Both 5′ and 3′ ends were labeled by digoxin (DIG). A scramble probe 5′-TCTTACACCTAGATAAGCAAAGA-3′ was used as a negative control. MISH was performed as described previously [32]. Informative results were observed in 243 pairs of ESCCs. Non-informative cases included lost samples and samples with limited number of cells.
Establishment of miR-9 overexpression ESCC cell lines
Lentiviral construct containing pre-miR-9 (GeneCopoeia, China) was packaged using the ViraPower Mix (Invitrogen, Carlsbad, CA) in 293FT cells. pre-miR-9-expressing lentivirus was used to stably transfect ESCC cells (KYSE410 and HKESC1) to establish miR-9-overexpressing cells. Empty vector-transfected cells were established as controls. A siRNA against miR-9 (Genepharma, China) was transfected into KYSE30 and KYSE510 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The cells transfected with scrambled inhibitor (NC) (Genepharma, China) were used as negative controls.
Cell growth assay, foci formation assay and in vivo tumor formation assay
For cell growth assay, 1 × 103 cells were plated in 96-well plates and the cell growth rate was assayed by CCK-8 kit (Dojindo, Japan) according to the manufacturer's instruction. For foci formation assay, 1 × 103 cells were plated in 6-well plates, After 2 weeks culture, colonies consisted of >50 cells were counted with 1% crystal violet staining. Triplicate independent experiments were performed.
Animal experiments were performed in compliance with the guidelines for the Welfare of Experimental Animals in Sun Yat-sen University Cancer Center. For in-vivo study, HK-miR-9 (2 × 106), 410-miR-9 (3 × 106) and empty vector transfected control cells were subcutaneously injected into the dorsal flanks of nude mice, respectively (4 for HKESC1 group and 5 for KYSE410 group). After 4 weeks, tumor formation in nude mice was examined by measuring tumor weight.
Cell motility assay
Migration assay was performed with migration chamber (BD Biosciences, Flanklin Lakes, NJ) following the manufacturer's instructions. Cells located on the lower side of the chamber were fixed, stained, and counted under a microscope. Triplicate independent experiments were performed.
In vivo metastasis assay
For the in vivo metastasis assays, 7 × 105 cells were injected intravenously through the tail vein into 4- to 5-week-old nude mouse (7 for HKESC1 group and 6 for KYSE410 group), respectively. The mice were sacrificed 8 weeks later and the intrahepatic and pulmonary metastatic nodules were carefully examined and counted. The livers and lungs were fixed for further study. The tissue blocks were cut into 5 μm sections and stained with haematoxylin-eosin.
Antibodies and Western blotting
Quantified protein lysates were resolved on SDS-PAGE gel, transferred onto PVDF membrane (Millipore, Billerica, MA), and immunoblotted with anti-human antibodies E-cadherin, β-catenin, Vimentin, α1-catenin, cyclin D1, c-myc (Cell Signalling Technology, Danvers, MA), GAPDH and CD44 (Santa Cruz, CA), VEGF (BD Biosciences, Franklin Lakes, NJ), and cytokeratin (Maixin, China). Blots were visualized with enhanced chemiluminescence (Amersham Biosciences, Piscataway, NJ).
Luciferase assay
An 819-bp fragment of CDH1 3′-UTR containing the putative binding site of miR-9 was amplified by PCR (Supporting data) and cloned into pMIR-REPORT vector (Ambion, Carlsbad, CA). The miR-9 overexpressed cells and the vector control cells were transfected with the reporter constructs containing the target binding sequence of CDH1, the pMIR-REPORT vector was used as control. Renilla luciferase vector (pRL-TK) was cotransfected into KYSE410 and HKESC1 cells as a normalizing transfection control, Dual luciferase signals were measured 48 hours later by the Dual-luciferase assay kit (Promega, Madison, WI).
Immunofluorescence (IF)
Immunofluorescence analysis was performed as described previously [33]. Briefly, cells were fixed and incubated with the primary antibody β-catenin (Immunofluorescence Preferred; Cell Signaling Technology, Danvers, MA) overnight at 4°C. After washing with PBS, cells were then incubated with fluorescence-conjugated secondary antibody (Invitrogen, Calsbad, CA). Images were captured after stained with Anti-fade DAPI solution.
Statistical analysis
SPSS standard V.16.0 (SPSS, Inc., IL) was used for Statistical analysis, Student's t tests was used to analysis the significance of difference. Survival analysis was performed using Kaplan-Meier plots and log-rank tests. The correlation between miR-9 expression and clinicopathological characteristics was analyzed by Pearson χ2 test. Univariable and multivariable Cox proportional hazard regression model was used to assess the survival hazard. Differences were considered significant when P < 0.05.
SUPPLEMENTARY DATA AND TABLE
Acknowledgments
This work was supported by China National Basic Research Program (2012CB967001), Hong Kong RGC General Research Fund (HKU7668/11M), the National Natural Science Foundation of China (81172338 and 81272416), the National Key Sci-Tech Special Project of Infectious Diseases (2013ZX10002-011-005) and Young Talent Teachers Plan of SYSU (11ykpy58, 13ykzd26), GDDESTIP (2013KJCX0001).
Footnotes
Conflicts of interests
The authors declared that no conflict of interest exists.
REFERENCES
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA: a cancer journal for clinicians. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Yang CS. Research on esophageal cancer in China: a review. Cancer Res. 1980;40:2633–2644. [PubMed] [Google Scholar]
- 3.Enzinger PC, Mayer RJ. Esophageal cancer. The New England journal of medicine. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA: a cancer journal for clinicians. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 5.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nature reviews Genetics. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Guo Y, Chen Z, Zhang L, Zhou F, Shi S, Feng X, Li B, Meng X, Ma X, Luo M, Shao K, Li N, Qiu B, Mitchelson K, Cheng J, He J. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68:26–33. doi: 10.1158/0008-5472.CAN-06-4418. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, Iwatsuki M, Watanabe M, Baba H. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer. 2013;119:1159–1167. doi: 10.1002/cncr.27895. [DOI] [PubMed] [Google Scholar]
- 10.Wang K, Li J, Guo H, Xu X, Xiong G, Guan X, Liu B, Li J, Chen X, Yang K, Bai Y. MiR-196a binding-site SNP regulates RAP1A expression contributing to esophageal squamous cell carcinoma risk and metastasis. Carcinogenesis. 2012;33:2147–2154. doi: 10.1093/carcin/bgs259. [DOI] [PubMed] [Google Scholar]
- 11.Kong KL, Kwong DL, Chan TH, Law SY, Chen L, Li Y, Qin YR, Guan XY. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 2012;61:33–42. doi: 10.1136/gutjnl-2011-300178. [DOI] [PubMed] [Google Scholar]
- 12.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, Westermann F, Speleman F, Vandesompele J, Weinberg RA. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu L, Chen H, Zhou D, Li D, Bai R, Zheng S, Ge W. MicroRNA-9 up-regulation is involved in colorectal cancer metastasis via promoting cell motility. Medical oncology. 2012;29:1037–1043. doi: 10.1007/s12032-011-9975-z. [DOI] [PubMed] [Google Scholar]
- 14.Shiiyama R, Fukushima S, Jinnin M, Yamashita J, Miyashita A, Nakahara S, Kogi A, Aoi J, Masuguchi S, Inoue Y, Ihn H. Sensitive detection of melanoma metastasis using circulating microRNA expression profiles. Melanoma research. 2013 doi: 10.1097/CMR.0b013e328363e485. [DOI] [PubMed] [Google Scholar]
- 15.Shimada Y, Hashimoto Y, Kan T, Kawamura J, Okumura T, Soma T, Kondo K, Teratani N, Watanabe G, Ino Y, Sakamoto M, Hirohashi S, Imamura M. Prognostic significance of dysadherin expression in esophageal squamous cell carcinoma. Oncology. 2004;67:73–80. doi: 10.1159/000080289. [DOI] [PubMed] [Google Scholar]
- 16.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nature reviews Molecular cell biology. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 17.Ceteci F, Ceteci S, Karreman C, Kramer BW, Asan E, Gotz R, Rapp UR. Disruption of tumor cell adhesion promotes angiogenic switch and progression to micrometastasis in RAF-driven murine lung cancer. Cancer cell. 2007;12:145–159. doi: 10.1016/j.ccr.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 18.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nature reviews Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 19.Hurst DR, Edmonds MD, Welch DR. Metastamir: the field of metastasis-regulatory microRNA is spreading. Cancer Res. 2009;69:7495–7498. doi: 10.1158/0008-5472.CAN-09-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 21.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 22.Nass D, Rosenwald S, Meiri E, Gilad S, Tabibian-Keissar H, Schlosberg A, Kuker H, Sion-Vardy N, Tobar A, Kharenko O, Sitbon E, Lithwick Yanai G, Elyakim E, Cholakh H, Gibori H, Spector Y, et al. MiR-92b and miR-9/9* are specifically expressed in brain primary tumors and can be used to differentiate primary from metastatic brain tumors. Brain pathology. 2009;19:375–383. doi: 10.1111/j.1750-3639.2008.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo H, Zhang H, Zhang Z, Zhang X, Ning B, Guo J, Nie N, Liu B, Wu X. Down-regulated miR-9 and miR-433 in human gastric carcinoma. Journal of experimental & clinical cancer research : CR. 2009;28:82. doi: 10.1186/1756-9966-28-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laios A, O'Toole S, Flavin R, Martin C, Kelly L, Ring M, Finn SP, Barrett C, Loda M, Gleeson N, D'Arcy T, McGuinness E, Sheils O, Sheppard B, J OL. Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Molecular cancer. 2008;7:35. doi: 10.1186/1476-4598-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu MH, Huang CC, Pan MR, Chen HH, Hung WC. Prospero homeobox 1 promotes epithelial-mesenchymal transition in colon cancer cells by inhibiting E-cadherin via miR-9. Clin Cancer Res. 2012;18:6416–6425. doi: 10.1158/1078-0432.CCR-12-0832. [DOI] [PubMed] [Google Scholar]
- 26.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nature reviews Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 28.Bae GY, Choi SJ, Lee JS, Jo J, Lee J, Kim J, Cha HJ. Loss of E-cadherin activates EGFR-MEK/ERK signaling, which promotes invasion via the ZEB1/MMP2 axis in non-small cell lung cancer. Oncotarget. 2013;4:2512–2522. doi: 10.18632/oncotarget.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimada Y, Imamura M, Wagata T, Yamaguchi N, Tobe T. Characterization of 21 newly established esophageal cancer cell lines. Cancer. 1992;69:277–284. doi: 10.1002/1097-0142(19920115)69:2<277::aid-cncr2820690202>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 30.Wong ML, Tao Q, Fu L, Wong KY, Qiu GH, Law FB, Tin PC, Cheung WL, Lee PY, Tang JC, Tsao GS, Lam KY, Law S, Wong J, Srivastava G. Aberrant promoter hypermethylation and silencing of the critical 3p21 tumour suppressor gene, RASSF1A, in Chinese oesophageal squamous cell carcinoma. Int J Oncol. 2006;28:767–773. [PubMed] [Google Scholar]
- 31.Xie D, Sham JS, Zeng WF, Lin HL, Che LH, Wu HX, Wen JM, Fang Y, Hu L, Guan XY. Heterogeneous expression and association of beta-catenin, p16 and c-myc in multistage colorectal tumorigenesis and progression detected by tissue microarray. Int J Cancer. 2003;107:896–902. doi: 10.1002/ijc.11514. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Li X, Li Y, Yang H, Wang L, Qin Y, Liu H, Fu L, Guan XY. Cell-specific detection of miR-375 downregulation for predicting the prognosis of esophageal squamous cell carcinoma by miRNA in situ hybridization. PLoS One. 2013;8:e53582. doi: 10.1371/journal.pone.0053582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L, Chan TH, Yuan YF, Hu L, Huang J, Ma S, Wang J, Dong SS, Tang KH, Xie D, Li Y, Guan XY. CHD1L promotes hepatocellular carcinoma progression and metastasis in mice and is associated with these processes in human patients. J Clin Invest. 2010;120:1178–1191. doi: 10.1172/JCI40665. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


