Abstract
Auxin is a versatile plant hormone that plays an essential role in most aspects of plant growth and development. Auxin regulates various growth processes by modulating gene transcription through a SCFTIR1/AFB-Aux/IAA-ARF nuclear signaling module. Recent work has generated clues as to how multiple layers of regulation of the auxin signaling components may result in diverse and specific response outputs. In particular, interaction and structural studies of key auxin signaling proteins have produced novel insights into the molecular basis of auxin-regulated transcription and may lead to a refined auxin signaling model.
Introduction
The plant hormone auxin, primarily indole-3-acetic acid (IAA), has a central role in nearly all aspects of plant growth and development, including hypocotyl elongation, embryogenesis, organogenesis, organ polarity establishment and tropic responses. One of the most intriguing questions in plant biology is how such a simple molecule can have so many diverse and context-specific roles. Extensive studies on different aspects of auxin biology have suggested that local auxin concentration, which is determined by auxin metabolism and auxin transport, as well as auxin signaling contribute to the diversity and specificity of auxin response. In addition, positive and negative feedback between auxin response, metabolism and transport all contribute to the robustness of the auxin network [EJ Chapman and M Estelle [1],2–7]. In the last ten years, studies in Arabidopsis thaliana and other plant species have identified a major auxin-signaling pathway. Auxin is perceived by a co-receptor complex consisting of an F-box protein from the TRANSPORT INHIBITOR RESPONSE 1/AUXIN SIGNALING F-BOX PROTEINS (TIR1/AFBs) family and a member of the Auxin/INDOLE ACETIC ACID (Aux/IAA) family of transcriptional repressors [8–14]. Transcription is directly regulated by a third family of proteins called the AUXIN RESPONSE FACTORs (ARFs) [15–17]. The TIR1/AFB proteins are subunits of a SKP1–Cul1–F-box (SCF)-type E3 ligase called SCFTIR1/AFB. When auxin levels are low, Aux/IAA proteins bind to ARFs, and repress their transcriptional activity. This repression involves recruitment of a co-repressor called TOPLESS (TPL). Auxin acts as a “molecular glue” to promote interaction between the two components of the auxin co-receptor, TIR1/AFB and Aux/IAA, leading to ubiquitination and 26S proteasome-mediated degradation of the latter (Fig. 1B) [10,13]. Therefore, a rise in auxin level releases Aux/IAA repression on ARFs, thus activating the transcriptional response. This review will focus on recent advances in our understanding of auxin perception and signaling through the well-established TIR1/AFB-Aux/IAA-ARF pathway, with an emphasis on the mechanisms that will potentially result in diversity and specificity of auxin.
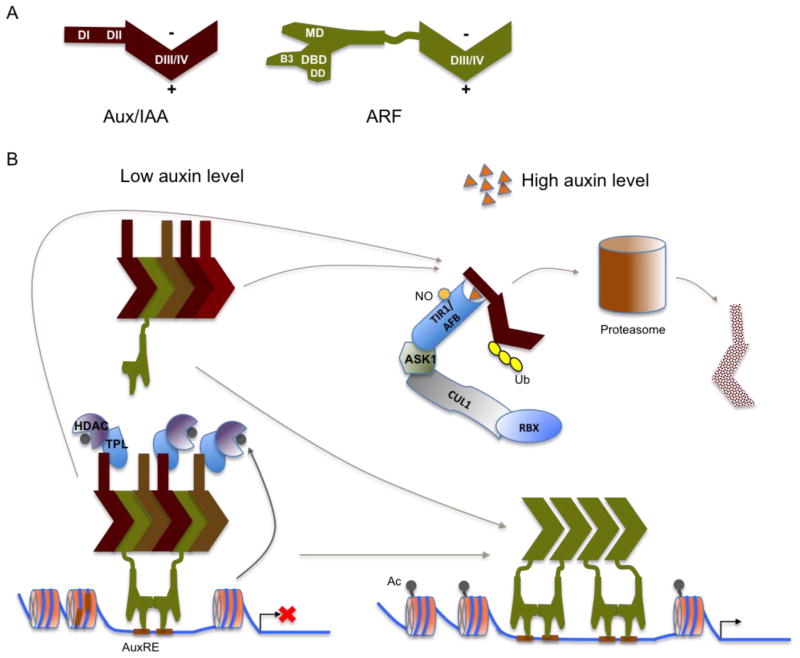
Figure 1. A model for the TIR1/AFB-mediated auxin signaling pathway.
A. DIII/IV regions of Aux/IAAs and ARFs are homologous and the DIII/IV of most Aux/IAAs and ARFs form a PB1 domain that comprises both an acidic (+) and a basic (−) faces.
B. When auxin level is low, Aux/IAA proteins and ARFs form multimers through directional interaction between the acidic and basic interfaces of their DIII/IV regions. Aux/IAAs in the multimers recruit co-repressor complexes, which at least consist of TPLs and HDACs and repress transcription of target promoter through removing acetyls (Ac) from local chromatin. In addition, Aux/IAAs in the multimers may block ARFs from efficient binding to AuxREs in their target promoters. When auxin level is high, auxin promotes ubiquitination and degradation of Aux/IAAs through a SCFTIR1/AFB-proteosome module, and the released ARFs form dimers and even higher-order complexes that activate expression of target genes.
The TIR1/AFB-Aux/IAA co-receptor perceives the auxin signal
A combination of biochemical and genetic approaches lead to the discovery that TIR1 and the related AFB proteins function as long-sought auxin receptors [8,9 ]. Instead of causing a substrate modification, commonly required for substrate recognition by many other cullin-based E3 ligases, auxin enhances the interaction between SCFTIR1/AFB and Aux/IAAs by directly binding to TIR1. Subsequent structural studies further revealed that auxin enhances the TIR1-Aux/IAA interaction by filling in the bottom of a hydrophobic pocket on the surface of TIR1 and that an Aux/IAA substrate peptide occupies the rest of the pocket on top of auxin [10]. Strikingly, auxin binding does not alter the conformation of TIR1. Rather the hormone acts as a “molecular glue” to stabilize the interaction between TIR1 and the Aux/IAA protein. Therefore, TIR1/AFB and Aux/IAA proteins work together as a co-receptor for auxin.
In Arabidopsis, the TIR1/AFB auxin receptor family comprises 6 members: TIR1 and AFB1 through AFB5, which fall into 3 phylogenetic clades [11,18]. When auxin response was assessed in the root, mutations in TIR1, but not the other members of the family, cause a significant decrease in response. However, defects in auxin response were drastically enhanced in higher-order mutants (e.g., tir1afb2afb3), indicating the existence of functional redundancy among the TIR1/AFB genes [11,18 ]. Further studies demonstrate that different TIR1/AFBs exhibit divergent properties. Genetic analysis of different combinations of tir1, afb1, afb2 and afb3 mutants revealed that TIR1 and AFB2 play more important roles than the others at least during seedling development [18]. The pattern of TIR1, AFB1, AFB2 and AFB3 mRNA accumulation is very broad and in general quite similar. However, the pattern of TIR1, AFB2, and AFB3 protein accumulation is highly restricted to growing regions of the plant suggesting a posttranscriptional regulatory mechanism. In contrast, AFB1 protein accumulation is similar to the pattern of transcription. Promoter swap experiments indicate that the difference between TIR1 and AFB1 regulation resides with the coding sequence of the protein [18].
In vitro pull-down experiments and yeast-two-hybrid-based protein interaction studies revealed that different TIR1/AFBs showed different affinity for the same Aux/IAA [13,18], suggesting that combinations of different TIR1/AFB receptors may partially account for the diversity of auxin response. Furthermore, AFB5 (and probably also AFB4) binds the synthetic auxin picloram with much higher affinity than TIR1 [13,19,20]. The different affinity that different receptor proteins exhibit for picloram may result from amino acid substitutions within the auxin-binding pocket. However, variation in other amino acids should also be considered since recent results showed that amino acids outside the auxin-binding pocket also contribute to auxin-mediated assembly of the co-receptor system [21]. In addition, Havens et al. systemically characterized the degradation of different Aux/IAA proteins in combination with TIR1 and AFB2 using a synthetic system in yeast [14]. In this system, exogenous auxin promotes interaction between the TIR1/AFB and Aux/IAA-YFP proteins resulting in degradation of the Aux/IAA-YFP fusion protein through the yeast ubiquitination system. By monitoring the loss of Aux/IAA-YFP, the authors discovered that TIR1 and AFB2 have very different activities even with the same Aux/IAA protein. This result further supports the notion that selection of TIR1/AFBs may contribute to the diversity and specificity of auxin response Given that, in different contexts, the available concentrations of different TIR1/AFB receptors could vary significantly, many quantitatively different combinations of TIR1/AFB proteins may form, and result in various signaling outputs, thus enhancing the tunability of auxin response. Taken together, the variation among the TIR1/AFBs could be an important contributor to the diversity and specificity of auxin response.
In addition to the TIR1/AFB family, the Aux/IAA family also comprises multiple members and has been shown to contribute to the diversity and specificity of auxin response. The Arabidopsis genome encodes 29 Aux/IAA proteins. Mis-expression and promoter-swap studies have showed that variation in protein sequence is important for conferring distinct functions to different Aux/IAAs [17,22,23]. Most Aux/IAA proteins have four conserved modular domains (Fig. 1A) [24]. The N-terminal domain I (DI) is required for transcriptional repression and recruits co-repressor proteins including TOPLESS (TPL) [25,26]. The C-terminal domain (CTD, also called DIII/IV, or separately as DIII and DIV) mediates interaction with the ARF transcription factors. Domain II (DII) is located between DI and DIII/IV and is responsible for protein instability through direct interaction with auxin and the TIR1/AFBs.
DII contains a 13 amino acid degron motif, which is required for the characteristic instability of Aux/IAAs [8,9,27 ]. Studies on the structure of a TIR1-auxin-IAA7 degron complex showed that the degron directly contacts TIR1 and auxin [10]. Mutations in the amino acid sequences of the DII, especially the highly conserved Gly-Trp-Pro-Pro-Val (GWPPV) motif stabilize Aux/IAAs by abolishing its interaction with the SCFTIR1/AFB complexes [8,9,28]. Based on the degron nature of DII, an auxin sensor has been developed by fusing a fragment of IAA28 containing DII with a florescence protein [29]. An increase in auxin level triggers degradation of the fusion protein. Although DII is both necessary and sufficient for auxin-mediated degradation of Aux/IAAs [8–10,27], data published by several labs demonstrate that sequences outside of DII also contribute to the instability of Aux/IAAs by affecting their affinity for auxin and TIR1/AFBs [12–14]. The DII sequences of most Aux/IAAs are conserved, but variations exist [13]. Using yeast two hybrid and quantitative IAA binding assays, Calderón Villalobos et al. showed that different combinations of TIR1 and Aux/IAA proteins form co-receptor complexes with a wide range of auxin-binding affinities [13]. Furthermore, degradation assays of Aux/IAAs using an in planta approach and the synthetic yeast system both showed that different Aux/IAAs exhibit different degradation rates [12,14]. Therefore, various combinations of TIR1/AFB and Aux/IAA proteins may be the molecular basis for the large and dynamic range of auxin responses and the diversity of processes in which auxin acts.
TPL-Aux/IAA complexes repress auxin-responsive gene expression through affecting chromatin conformation
Aux/IAAs repress transcriptional activity of ARFs by recruiting co-repressors including TPL and TOPLESS RELATED (TPR) proteins through an Ethylene response factor-Associated amphiphilic Repression (EAR) motif in the DI domain (Fig. 1B) [26]. Genetic, pharmacological and biochemical evidence demonstrates that one mechanism of TPL/TPRs-mediated transcriptional repression is through recruiting histone deacetylases (HDACs) [25,30,31 ]. HDACs remove acetylation from histone tails of nucleosomes, and hypoacetylation of histones correlates with chromatin status that favors transcriptional repression (Fig. 1B) [32]. Histone acetylation has been shown to play an important role in modulating auxin-responsive gene expression [33]. Interestingly, different TPL/TPRs may interact with different transcription factors, and the HDACs recruited by TPL/TPRs may also vary [25,30,31,34], suggesting additional layers of regulation may exist through combinations of different Aux/IAAs, TPL/TPRs and HDACs.
Aux/IAA-ARF multimerization regulates target gene expression
The interaction between the ARF transcription factors and the Aux/IAA co-repressors is a key aspect of auxin regulation [35]. DIII/IV regions of Aux/IAA proteins and ARF transcription factors are homologous and mediate Aux/IAA-ARF interaction. Similar to Aux/IAAs, ARFs also consist of modular domains. In addition to the DIII/IV, ARFs contain an amino-terminal DNA-binding domain (DBD) and a middle domain (MD) that acts as either an activation or repression domain. There are 23 ARF genes in Arabidopsis. Based on transient assays in transfected protoplasts, five Arabidopsis ARFs, (ARF5, ARF6, ARF7, ARF8 and ARF19) were characterized as transcriptional activators. The other ARFs were classified as repressors based on either the transient assays or the putative repression motif carried in their MD [15]. Multiple lines of evidence indicate that DIII/IV is required for interaction between Aux/IAAs and ARFs and also homo- or hetero-interaction between Aux/IAAs themselves [36,37 ]. It has been shown that most Aux/IAAs can interact with many Aux/IAAs and ARFs and the interactions between Aux/IAAs are largely indiscriminant, at least in the yeast-2-hybrid context [37]. Several different interaction assays have shown that some ARFs interact with themselves or other ARFs [38–42]. Therefore, in addition to ARF-Aux/IAA dimerization, DIII/IV has been thought to mediate ARF-ARF dimerization as well. However, in a systematic yeast-two-hybrid-based screen for interactions among Aux/IAA and ARF proteins that utilized just the DIII/IV domain, only a few ARF-ARF interactions were observed, whereas many ARF-Aux/IAA and Aux/IAA-Aux/IAA interacting pairs were identified [37]. These results suggest that either DIII/IV is not the major player for ARF-ARF interactions or that DIII/IV is not sufficient to mediate efficient ARF-ARF interaction in yeast (see below).
It has been thought for many years that Aux/IAAs repress transcriptional activity of ARFs by forming Aux/IAA-ARF dimers through the conserved DIII/IV regions. However, recent studies suggest that Aux/IAA can also form multimers [42–44]. Bioinformatics analyses suggested that the DIII/IV region of most Aux/IAAs (except for IAA29, IAA31, IAA32 and IAA33) and ARFs (except for ARF10, ARF14, ARF15, ARF16, ARF20 and ARF21) forms a type I/II Phox and Bem1p (PB1) protein-protein interaction domain (Fig. 1A) [45]. A PB1 domain can comprise an acidic surface (type I), a basic surface (type II), or both (type I/II). For type I/II PB1 domains the presence of acidic and basic surfaces on opposite faces of the domain facilitates directional interactions resulting in the formation of polymers. Structural studies of DIII/IV of ARF5 and ARF7 demonstrate that multimerization between ARFs and Aux/IAAs can occur and that multiple Aux/IAAs may be required for efficient repression (Fig. 1B)[42,44]. In agreement with this notion, the auxin response defects of transgenic plants overexpressing iaa16-1 (an auxin-insensitive stabilized mutant) were reverted by mutations of the conserved interface residues on either one of the two electrostatic interfaces of IAA16 PB1 domain. These mutations abolish Aux/IAA-ARF or Aux/IAA-Aux/IAA multimerization but not dimerization since the remaining interface (type I or II) can still form dimers with the oppositely-charged interface (type II or I) of an Aux/IAA or ARF[42]. When in vitro pulldown assays were conducted to test the interaction between ARF5 and IAA12, mutations in either interface of the PB1 domain of IAA12 significantly reduced the interaction, whereas mutations in both interfaces abolished the interaction [44]. Furthermore, in plants, mutations in one surface of the overexpressed ARF7 significantly increased the response of the DR5 promoter to auxin. In contrast, when both surfaces were mutated ARF7 induced DR5 activity independently of auxin [44]. These results support the notion that multimerization between Aux/IAAs and ARFs is required for efficient repression of the transcriptional activity of ARFs. It is still not clear if ARF7 forms DIII/IV-based homo-multimers in physiological conditions. Most interaction studies using DIII/IV of ARFs and Aux/IAAs have shown that the heterotypic ARF-Aux/IAA interactions are much stronger than homotypic ARF-ARF interactions [37,42 ]. Therefore, it is likely that in physiological conditions, when Aux/IAAs are present, ARF-Aux/IAA complexes are formed with many more Aux/IAAs than ARFs (Fig. 1B). Of course it is also possible that ARFs form higher-order complexes, especially when Aux/IAAs are degraded at high auxin levels. Taken together, these data refine the classical Aux/IAA-ARF-dimerization-based auxin-signaling model (Fig. 1). The inclusion of more than one Aux/IAA in a transcription-regulatory complex may enhance the tunability of auxin response since different Aux/IAAs involved will allow more inputs of developmental and environmental signals. Simultaneously, the new findings confront auxin researchers with many new questions. Do Aux/IAA-ARF complexes contain different Aux/IAA or ARF proteins and what is the composition of the complex in different cellular contexts? Does multimerization affect the affinity of Aux/IAAs for other signaling components? Does mutimerization affect binding of ARFs to target promoters? Do IAA29, IAA31, IAA32 and IAA33 disturb mutimerization since they only contain one electrostatic interface in their PB1 domain according to the bioinformatics prediction [45]?. More structural studies on different Aux/IAAs and ARFs multimers may help to answer these questions and gain further insight into the impact of multimerization of Aux/IAAs and ARFs on diversity and specificity of auxin response.
It is still not clear how repressor ARFs regulate gene repression or whether their function is auxin responsive. Yeast-two-hybrid assays have identified very few interactions between DIII/IV of repressor ARFs and that of Aux/IAAs [37]. Three hypotheses have been offered concerning the mechanism of ARF repression [6]. First, repressor ARFs may repress the transcriptional activities of activator ARFs by forming heterodimers. Second, repressor ARFs may directly recruit co-repressors TPL/TPRs through the EAR domains that have been identified in some repressor ARFs [34]. Alternatively, repressor ARFs may compete with activator ARFs for AuxREs in target promoters. Actually, all these three mechanisms may exist simultaneously, and more structural and functional analyses are needed to shed light on the detailed mechanisms.
ARFs regulate auxin responsive expression of genes by binding to AuxREs in their promoters
It has long been recognized that ARFs directly bind to auxin-response elements (AuxREs) in the promoters of auxin responsive genes through their DBD [16,46–48]. Surprisingly, a recent structural study of ARF5 and ARF1 DNA binding domains (DBD) revealed that a subdomain within the DBD, called dimerization domain or DD (Figure 1A), mediates homodimerization in both solution and in crystals [49]. Substitution of the conserved residues on the dimerization interface of ARF5 DD attenuated or even abolished ARF5 homodimerization. Importantly, the mutations also affected ARF5 function in plants, suggesting that DBD-mediated dimerization is required for ARF function. When dimerization was tested in a transient protoplast expression system, deletion of DIII/IV only slightly reduced homo-dimerization of ARF5, suggesting that DBD is sufficient for dimerization in vivo, but that interactions through domain III/IV may help to stabilize dimers [49]. The residues that contribute to ARF-ARF binding are highly conserved among different ARFs. Furthermore, the same mechanism of DBD-mediated homodimerization was also observed for ARF1, which is phylogenetically distinct from ARF5. Therefore, it is likely that most, if not all, ARFs homodimerize through their DBD.
In addition, these authors showed that homodimerization of the ARF DBD generated cooperative DNA binding to a inverted repeat of two AuxREs, although weaker monomeric ARF binding was also detected [49]. The canonical AuxRE TGTCTC was originally identified as an enriched motif in the promoter of auxin response genes [46], and was later shown to be sufficient for ARF binding [48]. By co-crystalizing ARF1 with AuxRE DNA, Boer et al. reveal that ARFs bind AuxREs through the B3 subdomain within the DBD (Figure 1A) [49]. Strikingly, protein binding microarray studies showed that both ARF1 and ARF5 prefer the same DNA binding site despite the fact that the two ARFs have very different biological functions. Quantitative assays reveal that ARF5 binding to the AuxRE is stronger than that of ARF1. Further studies showed that ARF5 is more tolerant of changes in the spacing between adjacent AuxREs than is ARF1. This suggests a model in which ARF dimers bind two AuxREs as molecular calipers with ARF-specific preferences with respect to AuxRE spacing. However, these results were obtained from in vitro studies using purified DBDs. It is possible that in physiological conditions, sequence degeneration of AuxREs may still be an important factor that determines binding preference of different ARFs. Moreover, ARFs that bind to AuxREs with non-optimal spacing for DBD-mediated ARF dimerization may also play some roles in regulating gene expression. In certain conditions, these ARFs may form multimers through DIII/IV with each other or ARF dimers that bind to AuxREs with preferred spacing. In turn, the multimerization of ARFs may affect local chromatin structure, thereby modulating the promoter activity. Therefore, both the sequence and the distribution pattern of AuxREs in target promoters may contribute to diversity and specificity of auxin responsive gene expression.
Cho et al. recently showed that auxin treatment promotes binding of ARF7 and ARF19 to their target promoters, which correlates with the enhancement of transcriptional activity of the ARFs [50]. Similarly, Walcher et al. reported previously that auxin treatment enhances ARF5 binding to its target promoters [51]. These findings suggest that in addition to repressing the transcriptional activity of ARFs, the Aux/IAA co-repressor complexes may block ARFs from efficient binding to their target promoters. The complexes may directly affect the accessibility of the DBD for dimerization or binding to AuxREs. Alternatively, the Aux/IAA co-repressor complexes that bind DNA-binding ARFs may maintain the chromatin in a ‘tight’ state, thus preventing efficient access of more ARFs.
Post-translational modifications of the core auxin components affect auxin signaling
Like many other proteins involved in signal transduction, the components of auxin signaling can be subjected to post-translational modifications. Terrile et al. showed that nitric oxide (NO) modulates auxin signaling through S-nitrilation of TIR1 (Fig. 1B) [52]. TIR1 S-nitrilation enhances TIR1-Aux/IAA interaction, promoting degradation of Aux/IAA proteins by the SCFTIR1/AFB complex. Furthermore, ARFs have recently been shown to undergo phosphorylation through a TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR (TDIF)-TDIF RECEPTOR (TDR)-BRASSINOSTEROID-INSENSITIVE2 (BIN2) module [50]. During the process of lateral root initiation, the secreted peptide TDIF binds its receptor TDR, and the TDIF-initiated TDR signaling directly acts to promote BIN2-mediated phosphorylation of ARF7 and ARF19. ARF phosphorylation suppresses their interaction with Aux/IAAs, thus enhancing DNA binding and transcriptional activity. Similarly, a previous report showed that BIN2 also phosphorylates ARF2 [53]. However, in this case, phosphorylation leads to loss of DNA binding independent of Aux/IAA binding. In any case, these data suggest that post-translational modifications of ARFs may constitute another layer of regulation of auxin signaling outputs.
Other transcription factors may modulate auxin signaling by interacting with Aux/IAAs and ARFs
In addition to the interaction between Aux/IAAs and ARFs, both groups of proteins have been reported to interact with other transcriptional regulators. A recent study showed that a sunflower Aux/IAA, HaIAA27 binds a heat shock transcription factor HaHSFA9, and further demonstrated that HaIAA27 regulates seed longevity by repressing HaHSFA9 activity [54]. Moreover, a MYB transcription factor MYB77 interacts with both IAA19 and ARFs including ARF1, ARF2, ARF5, ARF6 and ARF7 [55]. In addition, it has been shown that a bHLH transcription factor BIGPETALp (BPEp) interacts with ARF8 [56], and that a transcriptional co-regulator BREVIS RADIX (BRX) interacts with ARF5 [57]. In most of these cases, DIII/IV has been shown to be required for the interactions. These findings are especially interesting since they suggest that other signaling pathways may modulate auxin signaling through direct protein-protein interactions with Aux/IAAs or ARFs. These interactions may lead to integration of signals from multiple signaling pathways.
Conclusion
Although both the structure of the auxin molecule and the core SCFTIR1/AFB-Aux/IAA-ARF auxin perception and signaling module are quite simple, multiple layers of regulation allow the occurrence of diverse and dynamic auxin responses. As described in this review, different combinations of auxin signaling components, formation of transcriptional regulatory complexes, post-translational modifications, and interaction of core auxin signaling components with factors involved in other signaling pathways may all contribute to the diversity and specificity of auxin response. Furthermore, when these aspects of regulation are combined with regulation of transcription [18,23,37,58,59], small RNA-mediated post-transcriptional regulation of gene expression [60], auxin metabolism [2,4 ], directional transport [3] and feedbacks existing between different processes [61], a multidimensional regulatory matrix will form. In addition to the SCFTIR1/AFB-Aux/IAA-ARF module, another auxin signaling pathway involving AUXIN BINDING PROTEIN1 (ABP1) has been shown to mediate non-transcriptional auxin responses and play a critical role in plant development [62,63]. Moreover, a SKP2A-mediated pathway may affect auxin-regulated cell cycle control [64]. Therefore, it will be intriguing (although challenging) to both dissect the details and understand the integrity of the network underlying the fine-tuning, plasticity and specificity of auxin response that integrates developmental and environmental cues. In the future, alongside with genetic, genomics, cellular biological and biochemical studies, structural studies on complexes involved in auxin signaling (e.g., the Aux/IAA-ARF multimers) will help gain insights into molecular basis of auxin signaling. In addition, computational approaches may open new avenues to understand the complex network of auxin response.
Highlights.
The TIR1/AFB-Aux/IAA-ARF module has a key role in auxin signaling
Combinatorial protein interactions contribute to the specificity of auxin signaling.
Multimerization of Aux/IAAs and ARFs mediates transcriptional regulation by auxin.
Acknowledgments
Research in the authors’ laboratory is supported by grants from the National Institutes of Health (GM43644), the Department of Energy (De-FG02-09ER16007), the Howard Hughes Medical Institute, and the Gordon and Betty Moore Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet. 2009;43:265–285. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- 2.Zhao Y. Auxin Biosynthesis and Its Role in Plant Development. Annual Review of Plant Biology. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrasek J, Friml J. Auxin transport routes in plant development. Development. 2009;136:2675–2688. doi: 10.1242/dev.030353. [DOI] [PubMed] [Google Scholar]
- 4.Ludwig-Müller J. Auxin conjugates: their role for plant development and in the evolution of land plants. Journal of Experimental Botany. 2011;62:1757–1773. doi: 10.1093/jxb/erq412. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi K-i. The interaction and integration of auxin signaling components. Plant and Cell Physiology. 2012 doi: 10.1093/pcp/pcs035. [DOI] [PubMed] [Google Scholar]
- 6.Pierre-Jerome E, Moss BL, Nemhauser JL. Tuning the auxin transcriptional response. Journal of Experimental Botany. 2013;64:2557–2563. doi: 10.1093/jxb/ert100. [DOI] [PubMed] [Google Scholar]
- 7.Peer WA. From perception to attenuation: auxin signalling and responses. Current Opinion in Plant Biology. 2013;16:561–568. doi: 10.1016/j.pbi.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 9.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 10.Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 11.Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jurgens G, Estelle M. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Dreher KA, Brown J, Saw RE, Callis J. The Arabidopsis Aux/IAA Protein Family Has Diversified in Degradation and Auxin Responsiveness. The Plant Cell Online. 2006;18:699–714. doi: 10.1105/tpc.105.039172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13**.Calderon Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol. 2012;8:477–485. doi: 10.1038/nchembio.926. This study showed that different Aux/IAAs exhibit different affinity to TIR1/AFBs and auxin, suggesting that combinations of Aux/IAA and TIR1/AFBs may result in broad sensitivity to auxin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14**.Havens KA, Guseman JM, Jang SS, Pierre-Jerome E, Bolten N, Klavins E, Nemhauser JL. A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol. 2012;160:135–142. doi: 10.1104/pp.112.202184. The authors developed a synthetic auxin response system, that can be used to study pairwise interactions between the auxin receptors TIR1/AFBs and the repressors Aux/IAAs. Taking advantage of this system, the authors systematically characterized the diversity of degradation bahaviors among TIR1/AFB-Aux/IAA pairs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guilfoyle TJ, Hagen G. Auxin response factors. Curr Opin Plant Biol. 2007;10:453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15:533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weijers D, Benkova E, Jager KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jurgens G. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 2005;24:1874–1885. doi: 10.1038/sj.emboj.7600659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M. Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci U S A. 2009;106:22540–22545. doi: 10.1073/pnas.0911967106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenham K, Santner A, Castillejo C, Mooney S, Sairanen I, Ljung K, Estelle M. The AFB4 auxin receptor is a negative regulator of auxin signaling in seedlings. Curr Biol. 2011;21:520–525. doi: 10.1016/j.cub.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Lee S, Sundaram S, Armitage L, Evans JP, Hawkes T, Kepinski S, Ferro N, Napier RM. Defining Binding Efficiency and Specificity of Auxins for SCFTIR1/AFB-Aux/IAA Co-receptor Complex Formation. ACS Chemical Biology. 2014;9:673–682. doi: 10.1021/cb400618m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu H, Moss BL, Jang SS, Prigge M, Klavins E, Nemhauser JL, Estelle M. Mutations in the TIR1 auxin receptor that increase affinity for auxin/indole-3-acetic acid proteins result in auxin hypersensitivity. Plant Physiol. 2013;162:295–303. doi: 10.1104/pp.113.215582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knox K, Grierson CS, Leyser O. AXR3 and SHY2 interact to regulate root hair development. Development. 2003;130:5769–5777. doi: 10.1242/dev.00659. [DOI] [PubMed] [Google Scholar]
- 23.Muto H, Watahiki MK, Nakamoto D, Kinjo M, Yamamoto KT. Specificity and Similarity of Functions of the Aux/IAA Genes in Auxin Signaling of Arabidopsis Revealed by Promoter-Exchange Experiments among MSG2/IAA19, AXR2/IAA7, and SLR/IAA14. Plant Physiology. 2007;144:187–196. doi: 10.1104/pp.107.096628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lokerse AS, Weijers D. Auxin enters the matrix--assembly of response machineries for specific outputs. Curr Opin Plant Biol. 2009;12:520–526. doi: 10.1016/j.pbi.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 25.Long JA, Ohno C, Smith ZR, Meyerowitz EM. TOPLESS Regulates Apical Embryonic Fate in Arabidopsis. Science. 2006;312:1520–1523. doi: 10.1126/science.1123841. [DOI] [PubMed] [Google Scholar]
- 26.Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319:1384–1386. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- 27.Ramos JA, Zenser N, Leyser O, Callis J. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell. 2001;13:2349–2360. doi: 10.1105/tpc.010244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell. 2001;13:2809–2822. doi: 10.1105/tpc.010289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29*.Brunoud G, Wells DM, Oliva M, Larrieu A, Mirabet V, Burrow AH, Beeckman T, Kepinski S, Traas J, Bennett MJ, et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482:103–106. doi: 10.1038/nature10791. In this paper, the authors described a novel auxin sensor that was made by fusing a peptide containing domain II (DII, which is responsible for protein degradation) of IAA28 to a florescence protein VENUS. Since the abundance of the DII-VENUS fusion protein is auxin dependent, it can be used to monitor auxin levels in living cells. [DOI] [PubMed] [Google Scholar]
- 30.Krogan NT, Hogan K, Long JA. APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development. 2012;139:4180–4190. doi: 10.1242/dev.085407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Kim J, Somers DE. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proceedings of the National Academy of Sciences. 2013;110:761–766. doi: 10.1073/pnas.1215010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anzola JM, Sieberer T, Ortbauer M, Butt H, Korbei B, Weinhofer I, Müllner AE, Luschnig C. Putative Arabidopsis Transcriptional Adaptor Protein (PROPORZ1) is required to modulate histone acetylation in response to auxin. Proceedings of the National Academy of Sciences. 2010;107:10308–10313. doi: 10.1073/pnas.0913918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Causier B, Ashworth M, Guo W, Davies B. The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 2012;158:423–438. doi: 10.1104/pp.111.186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guilfoyle TJ, Hagen G. Auxin response factors. Current Opinion in Plant Biology. 2007;10:453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 36.Kim J, Harter K, Theologis A. Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci U S A. 1997;94:11786–11791. doi: 10.1073/pnas.94.22.11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol. 2011;7:508. doi: 10.1038/msb.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hardtke CS, Ckurshumova W, Vidaurre DP, Singh SA, Stamatiou G, Tiwari SB, Hagen G, Guilfoyle TJ, Berleth T. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development. 2004;131:1089–1100. doi: 10.1242/dev.00925. [DOI] [PubMed] [Google Scholar]
- 39.Ouellet F, Overvoorde PJ, Theologis A. IAA17/AXR3: biochemical insight into an auxin mutant phenotype. Plant Cell. 2001;13:829–841. doi: 10.1105/tpc.13.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J-FBJ, Xiong Y, Li L, McCormack M. Large-Scale Protein-Protein Interaction Analysis in Arabidopsis Mesophyll Protoplasts by Split Firefly Luciferase Complementation. PLoS ONE. 2011;6:e27364. doi: 10.1371/journal.pone.0027364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muto H, Nagao I, Demura T, Fukuda H, Kinjo M, Yamamoto KT. Fluorescence Cross-Correlation Analyses of the Molecular Interaction between an Aux/IAA Protein, MSG2/IAA19, and Protein–Protein Interaction Domains of Auxin Response Factors of Arabidopsis Expressed in HeLa Cells. Plant and Cell Physiology. 2006;47:1095–1101. doi: 10.1093/pcp/pcj080. [DOI] [PubMed] [Google Scholar]
- 42**.Korasick DA, Westfall CS, Lee SG, Nanao MH, Dumas R, Hagen G, Guilfoyle TJ, Jez JM, Strader LC. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proceedings of the National Academy of Sciences. 2014 doi: 10.1073/pnas.1400074111. This study, together with Nanao et al. (2014) used a combination of bioinformatics, structural and genetic studies demonstrate that multimerization between Aux/IAAs and ARFs may be required for efficient Aux/IAA repression of transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43*.Guilfoyle TJ, Hagen G. Getting a Grasp on Domain III/IV Responsible for Auxin Response Factor-IAA Protein Interactions. Plant Science. 2012 doi: 10.1016/j.plantsci.2012.04.003. In this review, bioinformatics analyses predicted that the DIII/IV regions of most ARFs and Aux/IAAs form a type I/II PB1 domain that may mediate oligomerization of these factors through directional interations. [DOI] [PubMed] [Google Scholar]
- 44**.Nanao MH, Vinos-Poyo T, Brunoud G, Thévenon E, Mazzoleni M, Mast D, Lainé S, Wang S, Hagen G, Li H, et al. Structural basis for oligomerization of auxin transcriptional regulators. Nat Commun. 2014;5 doi: 10.1038/ncomms4617. This study, together with Korasick et al. (2014) used a combination of bioinformatics, structural and genetic studies demonstrate that multimerization between Aux/IAAs and ARFs may be required for efficient Aux/IAA repression of transcription. [DOI] [PubMed] [Google Scholar]
- 45.Guilfoyle TJ, Hagen G. Getting a grasp on domain III/IV responsible for Auxin Response Factor–IAA protein interactions. Plant Science. 2012;190:82–88. doi: 10.1016/j.plantsci.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 46.Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ. Composite structure of auxin response elements. Plant Cell. 1995;7:1611–1623. doi: 10.1105/tpc.7.10.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ulmasov T, Hagen G, Guilfoyle TJ. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- 49**.Boer DR, Freire-Rios A, van den Berg Willy AM, Saaki T, Manfield Iain W, Kepinski S, López-Vidrieo I, Franco-Zorrilla Jose M, de Vries Sacco C, Solano R, et al. Structural Basis for DNA Binding Specificity by the Auxin-Dependent ARF Transcription Factors. Cell. 2014;156:577–589. doi: 10.1016/j.cell.2013.12.027. The authors studied the crystal structures of the DNA binding domains (DBD) of two functionally divergent ARFs, ARF1 and ARF5, and discovered that DBD mediates both DNA binding and dimerization through distict subdomains. Furthermore, the authors showed that dimerization results in cooperative DNA binding, which is critical for the in vivo function of ARFs. In vitro experiments revealed that the spacing tolerated between two binding sites of ARF1 and ARF5 dimers is different. These findings will lead to novel insights into the mechanisms of transcriptional control by auxin signaling. [DOI] [PubMed] [Google Scholar]
- 50*.Cho H, Ryu H, Rho S, Hill K, Smith S, Audenaert D, Park J, Han S, Beeckman T, Bennett MJ, et al. A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat Cell Biol. 2014;16:66–76. doi: 10.1038/ncb2893. The authors demonstrated that during lateral root development, a TDIF-TDR module-controlled BIN2-mediated phospharylation of ARF7 and ARF19. Phosphorylation suppressed their interaction with Aux/IAA repressors, thus potentiating their transcriptional activity. [DOI] [PubMed] [Google Scholar]
- 51.Walcher CL, Nemhauser JL. Bipartite promoter element required for auxin response. Plant Physiol. 2012;158:273–282. doi: 10.1104/pp.111.187559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Terrile MC, Paris R, Calderon-Villalobos LI, Iglesias MJ, Lamattina L, Estelle M, Casalongue CA. Nitric oxide influences auxin signaling through S-nitrosylation of the Arabidopsis TRANSPORT INHIBITOR RESPONSE 1 auxin receptor. Plant J. 2012;70:492–500. doi: 10.1111/j.1365-313X.2011.04885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci U S A. 2008;105:9829–9834. doi: 10.1073/pnas.0803996105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carranco R, Espinosa JM, Prieto-Dapena P, Almoguera C, Jordano J. Repression by an auxin/indole acetic acid protein connects auxin signaling with heat shock factor-mediated seed longevity. Proceedings of the National Academy of Sciences. 2010;107:21908–21913. doi: 10.1073/pnas.1014856107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell. 2007;19:2440–2453. doi: 10.1105/tpc.107.050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varaud E, Brioudes F, Szécsi J, Leroux J, Brown S, Perrot-Rechenmann C, Bendahmane M. AUXIN RESPONSE FACTOR8 Regulates Arabidopsis Petal Growth by Interacting with the bHLH Transcription Factor BIGPETALp. The Plant Cell Online. 2011;23:973–983. doi: 10.1105/tpc.110.081653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scacchi E, Salinas P, Gujas B, Santuari L, Krogan N, Ragni L, Berleth T, Hardtke CS. Spatio-temporal sequence of cross-regulatory events in root meristem growth. Proceedings of the National Academy of Sciences. 2010;107:22734–22739. doi: 10.1073/pnas.1014716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rademacher E, Moller B, Lokerse AS, Llavata-Peris CI, van den Berg W, Weijers D. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J. 2011 doi: 10.1111/j.1365-313X.2011.04710.x. [DOI] [PubMed] [Google Scholar]
- 59.Rademacher EH, Lokerse AS, Schlereth A, Llavata-Peris CI, Bayer M, Kientz M, Freire Rios A, Borst JW, Lukowitz W, Jürgens G. Different Auxin Response Machineries Control Distinct Cell Fates in the Early Plant Embryo. Dev Cell. 2012;22:211–222. doi: 10.1016/j.devcel.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 60.Plavskin Y, Timmermans MCP. Small RNA-Regulated Networks and the Evolution of Novel Structures in Plants. Cold Spring Harbor Symposia on Quantitative Biology. 2012;77:221–233. doi: 10.1101/sqb.2013.77.014878. [DOI] [PubMed] [Google Scholar]
- 61.Kieffer M, Neve J, Kepinski S. Defining auxin response contexts in plant development. Curr Opin Plant Biol. 2010;13:12–20. doi: 10.1016/j.pbi.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 62.Sauer M, Kleine-Vehn J. AUXIN BINDING PROTEIN1: The Outsider. The Plant Cell Online. 2011;23:2033–2043. doi: 10.1105/tpc.111.087064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu T, Dai N, Chen J, Nagawa S, Cao M, Li H, Zhou Z, Chen X, De Rycke R, Rakusová H, et al. Cell Surface ABP1-TMK Auxin-Sensing Complex Activates ROP GTPase Signaling. Science. 2014;343:1025–1028. doi: 10.1126/science.1245125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.del Pozo JC, Manzano C. Auxin and the ubiquitin pathway. Two players–one target: the cell cycle in action. Journal of Experimental Botany. 2013 doi: 10.1093/jxb/ert363. [DOI] [PubMed] [Google Scholar]