SUMMARY
Monoclonal antibodies derived from blood plasma cells of acute HIV-1-infected individuals are predominantly targeted to the HIV Env gp41 and cross-reactive with commensal bacteria. To understand this phenomenon, we examined anti-HIV responses in ileum B cells using recombinant antibody technology and probed their relationship to commensal bacteria. The dominant ileum B cell response was to Env gp41. Remarkably, a majority (82%) of the ileum anti-gp41 antibodies cross-reacted with commensal bacteria, and of those, 43% showed non-HIV-1 antigen polyreactivity. Pyrosequencing revealed shared HIV-1 antibody clonal lineages between ileum and blood. Mutated immunoglobulin G antibodies cross-reactive with both Env gp41 and microbiota could also be isolated from the ileum of HIV-1 uninfected individuals. Thus, the gp41 commensal bacterial antigen cross-reactive antibodies originate in the intestine, and the gp41 Env response in HIV-1 infection can be derived from a preinfection memory B cell pool triggered by commensal bacteria that cross-react with Env.
INTRODUCTION
The plasma cell and memory B cell pools in intestine contain a normal subset of B cells reactive with intestinal commensal bacteria (Benckert et al., 2011). In acute HIV-1 infection (AHI), virus replication is prominent in the gastrointestinal tract, with early depletion of CD4+ T cells (Brenchley et al., 2004; Guadalupe et al., 2003; Mehandru et al., 2006; Pope and Haase, 2003; Veazey et al., 1998; 2001) as well as early destruction of B cell germinal centers (Levesque et al., 2009). Initial plasma (Tomaras et al., 2008) and mucosal fluid (Yates et al., 2013) antibody response in AHI is targeted to HIV-1 Env gp41. The AHI gp41 antibody response is nonneutralizing and does not select viral escape mutants (Tomaras et al., 2008). Rather, it is the initial autologous gp120 neutralizing antibody response that is the first Env antibody shown to select viral escape mutants (Moore et al., 2009; Richman et al., 2003; Wei et al., 2003).
Recombinant monoclonal antibodies (mAbs) isolated from blood plasmablasts and/or plasma cells (hereafter termed plasma cells) of individuals with AHI were predominantly targeted to Env gp41 and were polyreactive with both host and environmental antigens including commensal bacteria (Liao et al., 2011). These observations raised the hypothesis that a component of the peripheral blood HIV-1 Env gp41 response in blood originates from polyreactive memory B cells activated prior to transmission by environmental antigens (Liao et al., 2011).
Here we have used single B cell sorting and recombinant antibody technology to probe the plasma cell and memory B cell repertoire of the terminal ileum in early and chronic HIV-1 infection. We found that the terminal ileum plasma cell and memory B cell repertoire was comprised of predominantly polyclonally activated, non-HIV-1-reactive antibodies, and the dominant early HIV-1 B cell response in the terminal ileum was targeted to Env gp41. Remarkably, 82% of HIV-1 gp41-reactive terminal ileum antibodies cross-reacted with intestinal commensal bacterial antigens, and mutated antibodies cross-reactive with Env gp41 and intestinal commensal bacteria were isolated from HIV-1 un-infected individuals. Thus, the antibody response to HIV-1 may be shaped by intestinal B cells stimulated by microbiota to develop a preinfection pool of memory B cells cross-reactive with HIV-1 gp41.
RESULTS
HIV-1 gp41-Reactive Antibodies in Terminal Ileum in Early and Chronic HIV-1 Infection Individuals
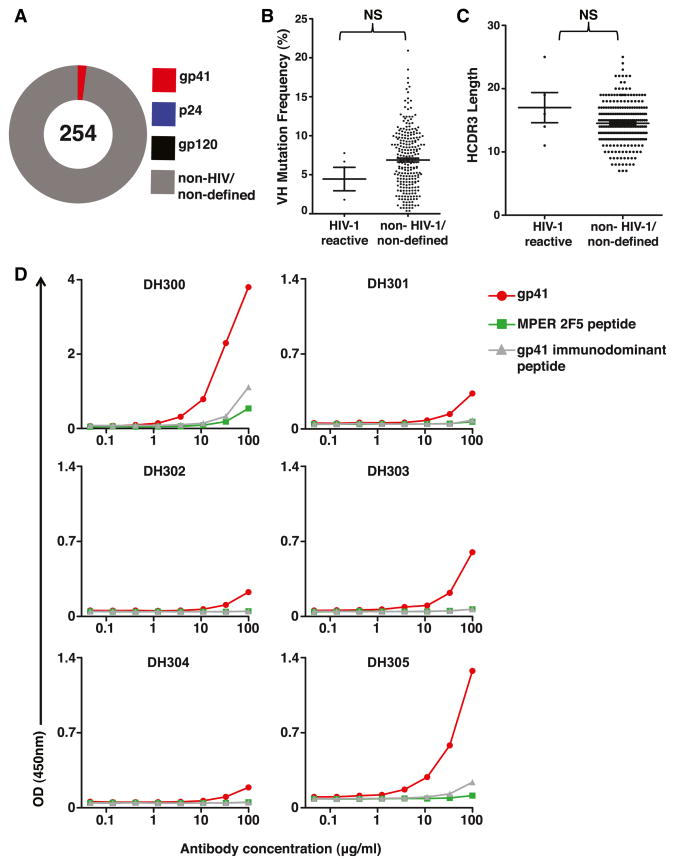
We investigated the plasma cell response to HIV-1 infection within the terminal ileum of six early HIV-1 infection (EHI) individuals (Table S1). We expressed 114 mAbs from plasma cells and 140 mAbs from memory B cells recovered from terminal ileum. Of the 254 total mAbs isolated from EHI individuals, only 5 (2.0%) reacted with gp41 and none (0.0%) with gp120 (Figure 1 and Table S2). HIV-1-reactive mAbs primarily utilized heavy-chain variable gene segments from VH family 3. VH mutation frequencies ranged from 0.0% to 10.4%, and HCDR3 lengths ranged from 11 to 25 amino acids. There were no statistical differences between the mean VH mutation frequencies and HCDR3 lengths of the HIV-1-reactive antibodies compared to non-HIV-1-reactive antibodies isolated from terminal ileum plasma cells from EHI individuals (Figures 1B and 1C). All recombinant HIV-1 mAbs were expressed with an immunoglobulin G1 (IgG1) backbone; their original isotypes were IgA1, IgA2, and IgG3 (Table S2). IgA2 and IgG3 only made up 6.7% and 5.1% of total terminal ileum mAbs isolated from EHI, respectively (Table S3). Four of the five gp41-reactive mAbs were low affinity, with effective antibody binding 50% concentrations (EC50s) of >100 μg/ml. DH300 had the highest apparent affinity, with an EC50 of only >25 μg/ml (Figure 1D and Table S2). With a VH mutation frequency of 10.4%, the heavy chain of DH300 was also the most mutated of the EHI terminal ileum HIV-1-reactive mAbs isolated (Table S2). These HIV-1-reactive mAbs were tested for neutralization against the easy-to-neutralize (tier 1) viruses, ADA, MN, and SF162, and the difficult-to-neutralize virus (tier 2) DU156, and all were nonneutralizing when assayed in the TZM-bl pseudovirus infection assay. Thus, the plasma cell and memory B cell response in EHI was polyclonal, and the HIV-1-reactive mAbs were targeted to Env gp41 and were nonneutralizing.
Figure 1. Characteristics of Antibodies Isolated from Terminal Ileum Plasma Cells and Memory B Cells of EHI Individuals.
(A) The total number of mAbs generated from wells with one VHDHJH and one VLJL gene isolated is indicated in the center of the pie chart. The percentages of mAbs binding to gp41, gp120, p24, and non-HIV-1 antigens are indicated by colors.
(B) Frequency of somatic mutations in VH gene segments of HIV-1-reactive antibodies compared to non-HIV-1-reactive or nondefined mAbs from terminal ileum plasma cells and memory B cells of six EHI individuals. Mean and SEM are indicated by lines.
(C) The HCDR3 lengths of HIV-1-reactive mAbs compared to non-HIV-1-reactive or nondefined antibodies isolated from terminal ileum B cells, with plasma cells and memory B cells pooled. Mean and SEM are indicated by lines.
(D) Six recombinant mAbs (DH300, DH301, DH302, DH303, DH304, and DH305) produced in a rIgG1 backbone were evaluated for reactivity with HIV-1 rgp41, SP62 = 2F5 MPER gp41 epitope peptide (QQEKNEQELLELDKWASLWN), and sp400 = gp41 immunodominant peptide (RVLAVERYLRDQQLLG IWGCSGKLICTTAVPWNASWSNKSLNK) by ELISA. They were tested in 3-fold dilutions ranging from 100 to 0.05 μg/ml (x axis).
See also Tables S1, S2, and S3.
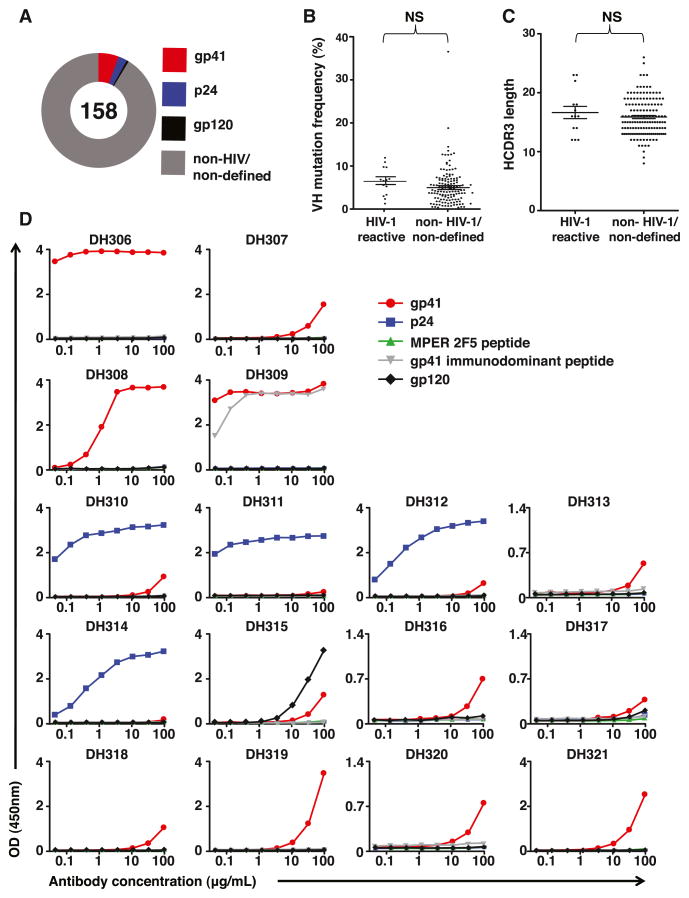
We next characterized the plasma cell response in the terminal ileum of three chronically HIV-1-infected (CHI) individuals, 038-7, 004-0, and 071-8 (Table S1). From these individuals, we expressed 158 mAbs from terminal ileum plasma cells; 14 (8.8%) of these mAbs reacted with HIV-1 antigens, 9 (5.7%) with Env gp41, 4 (2.5%) with HIV-1 capsid protein (p24), and 1 (0.6%) with Env gp120 (Figure 2 and Table S2). Similar to the gp41 antibodies from EHI individuals, the HIV-1-reactive mAbs isolated from CHI individual 071-8 predominantly used VH family 3 gene segments; mutation frequencies ranged from 1.3% to 7.0%, and the majority of the original mAbs were of IgA isotype (Table S2). In contrast, 8 of the 9 (89%) HIV-1-reactive mAbs from plasma cells of CHI individual 004-0 used VH1-69; all these VH1-69 antibodies were originally IgG1 (Table S2). Both individuals, 071-8 and 004-0, have three genomic copies of VH1-69 as determined by digital PCR (Table S4). The number of B cells utilizing the gene segment VH1-69 has been reported to be proportional to the gene copy number of certain VH1-69 alleles (Sasso et al., 1996). However, this was not seen at the terminal ileum single B cell level in our study, where both 071-8 and 004-0 had three genomic copies of VH1-69, yet only 004-0 predominately used VH1-69 to respond to HIV-1 infection (Table S2).
Figure 2. Characteristics of Antibodies Isolated from Terminal Ileum Plasma Cells of CHI Individuals.
(A) The total number of mAbs generated is indicated in the center of the pie chart. The percentages of mAbs binding to gp41, gp120, p24, and non-HIV-1/nondefined antigens are indicated by colors.
(B) Frequency of somatic mutations in VH gene segments of HIV-1-reactive antibodies compared to non-HIV-1- or nondefined reactive antibodies from terminal ileum plasma cells and memory B cells of three CHI subjects. Mean and SEM are indicated by lines.
(C) The HCDR3 lengths of HIV-1-reactive antibodies compared to non-HIV-1- or nondefined reactive antibodies isolated from terminal plasma cells of three CHI individuals. Mean and SEM are indicated by lines.
(D) A total of 16 recombinant mAbs (DH306, DH307, DH308, DH309, DH310, DH311, DH312, DH313, DH314, DH315, DH316, DH317, DH318, DH319, DH320, and DH321) produced in a rIgG1 backbone were evaluated for reactivity with HIV-1 rgp41, p24, sp62 = 2F5 MPER gp41 epitope peptide (QQEKNEQELLELDKWASLWN) and sp400 = gp41 immunodominant peptide (RVLAVERYLRD QQLLGIWGCSGKLICTTAVPWNASWSNKSLNK), and MN gp120 gD- by ELISA. They were tested in 3-fold dilutions ranging from 100 to at least 0.05 μg/ml (x axis).
See also Tables S1, S2, S4, and S5.
The VH mutation frequencies of antibodies from individual 004-0 ranged from 3.3% to 11.9%, and HCDR3 lengths ranged from 12 to 23 amino acids (Table S2). There were no statistical differences between the mean VH mutation frequencies and HCDR3 lengths of the HIV-1-reactive mAbs compared to non-HIV-1-reactive mAbs isolated from terminal ileum plasma cells from CHI individuals (Figures 2B and 2C). The estimated EC50s for gp41 binding of these antibodies ranged from <0.1 μg/ml to >100 μg/ml (Table S2). DH306 and DH309 had high apparent affinities to gp41 (EC50s of <0.1 μg/ml). DH310, DH311, DH312, and DH314 had high affinities to Gag p24 (EC50s of <1 μg/ml −<0.1) (Figure 2D and Table S2). These HIV-1-reactive mAbs were also tested for neutralization against viruses, ADA, MN, SF162, and DU156 in TZM-bl assays and were nonneutralizing.
Because the HIV-1 antigen-specific terminal ileum mAbs account for such a small proportion of the plasma cell and memory B cell response as measured by single-cell sorting, we next quantified the Env-specific memory B cell pool by an alternative method. We assayed paired peripheral blood mononuclear cells (PBMCs) and terminal ileum samples from four CHI individuals (078-2, 067-8, 072-3, and 076-4) (Table S1) by flow cytometry analysis of HIV-1 Env-specific memory B cells with a fluorescent-labeled consensus group M gp140 Env, consensus-S (CON-S) previously shown to bind to clade B-reactive antibodies (Liao et al., 2006; Tomaras et al., 2008). We found means of 0.04% ± 0.02%, 0.26% ± 0.24%, and 0.20% ± 0.29% IgM, IgG, and IgA CON-S gp140-reactive memory B cells, respectively, in blood (Table S5). The mean percentage of IgM, IgG, and IgA CON-S gp140-reactive memory B cells in terminal ileum were 0.01% ± 0.02%, 0.05% ± 0.1%, and 0.03% ± 0.06%, respectively (Table S5). Thus, by flow cytometry with a fluorophor-labeled Env, there was also a relative dearth of HIV-1 Env-reactive memory B cells in terminal ileum compared to blood in CHI.
Terminal Ileum HIV-1-Reactive Antibodies Were Cross-Reactive with Commensal Bacterial Antigens
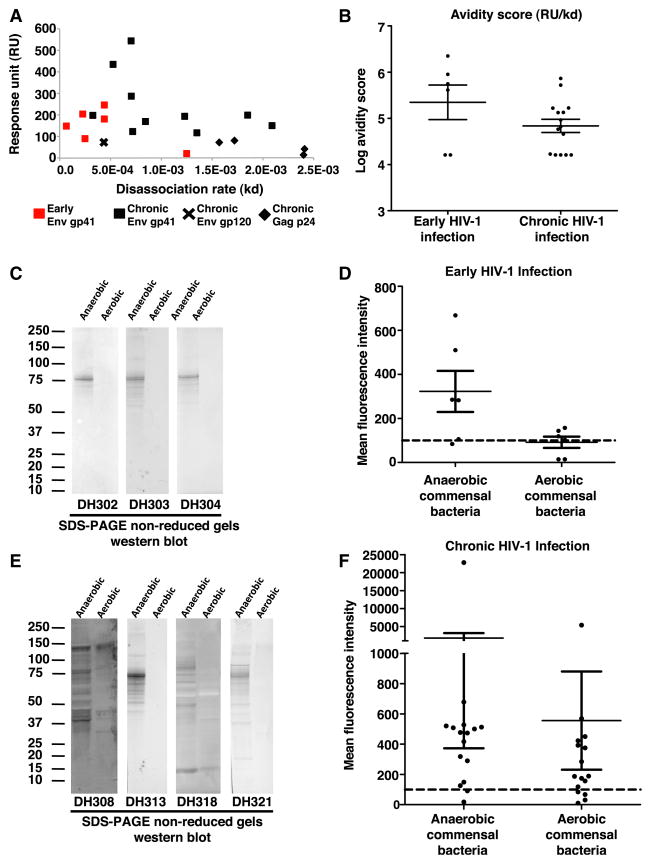
We tested HIV-1-reactive mAbs isolated from terminal ileum of EHI for reactivity to antigens in anaerobic commensal bacteria whole-cell lysates (WCLs) by surface plasmon resonance (SPR) and to both anaerobic and aerobic commensal bacteria WCLs by western blot analysis. Of the six gp41-reactive antibodies from EHI, all were reactive to anaerobic intestinal commensal bacteria by both SPR and western blot (Figures 3A–3C and S1 and Table S6). Similarly, 11 of the 16 HIV-1-reactive mAbs isolated from the terminal ileum of CHI cross-reacted with anaerobic commensal bacteria by SPR and western blot (Figures 3A, 3B, 3E, and S1 and Table S6). Antibody reactivity to aerobic and anaerobic commensal bacteria was also tested in Luminex-based binding antibody multiplex assays (BAMAs) (Figure S2A). Of 17 antibodies positive in western blot and SPR, 14 could also be confirmed in BAMA (Figures 3D, 3F, S1, and S2 and Table S6).
Figure 3. Commensal Bacteria Cross-Reactivity of HIV-1-Reactive Antibodies Isolated from Terminal Ileum Plasma Cells and Memory B Cells of EHI and CHI Individuals.
(A) HIV-1-reactive mAbs isolated from terminal ileum plasma cells and memory B cells of EHI and CHI individuals were tested for reactivity to anaerobic commensal bacteria by SPR. The response unit and off rate for each antibody that reacted with anaerobic commensal bacteria in this assay is plotted. Antibodies isolated from EHI individuals are indicated in red, and antibodies isolated from CHI individuals are indicated in black. The HIV-1 reactivity of the antibodies is indicated by shapes.
(B) The avidity score (response unit [RU] / off rate [kd]) of the terminal ileum HIV-1-reactive antibodies binding to anaerobic WCL by SPR.
(C) HIV-1-reactive mAbs isolated from EHI terminal ileum B cells were also tested for reactivity to anaerobic and aerobic commensal bacteria WCL by SDS-PAGE western blot. A total of 100 μg of each anaerobic and aerobic WCL was loaded in individual lanes, and mAbs were tested at 20 μg/ml in both nonreducing and reducing conditions. Three representative westerns under nonreducing conditions are shown.
(D) HIV-1-reactive mAbs isolated from terminal ileum plasma cells and memory B cells of EHI individuals were tested for reactivity to anaerobic and aerobic commensal bacteria by BAMA at 100 μg/ml. Mean and SEM are indicated by lines.
(E) HIV-1-reactive mAbs isolated from CHI terminal ileum B cells were also tested for reactivity to anaerobic and aerobic commensal bacteria WCLs by SDS-PAGE western blot. A total of 100 μg of anaerobic and aerobic WCLs was loaded in individual lanes, and mAbs were tested at 20 μg/ml in both nonreducing and reducing conditions. Four representative westerns under nonreducing conditions are shown.
(F) HIV-1-reactive mAbs isolated from terminal ileum plasma cells and memory B cells of CHI individuals were also tested for reactivity to anaerobic and aerobic commensal by BAMA at 100 μg/ml. Mean and SEM are indicated by lines. See also Figures S1–S3 and Tables S2 and S6.
To determine if HIV-1 and commensal bacteria cross-reactive mAbs were polyreactive/autotreactive, we tested the HIV-1-reactive mAbs in Luminex AtheNA ANA II and HEp-2 immunofluorescence ANA assays. Four of the six gp41-commensal bacteria cross-reactive mAbs from EHI terminal ileum were not reactive with additional antigens by these assays (Figure S1B and Table S6). Of the 11 HIV-1 and commensal bacteria cross-reactive mAbs isolated from CHI terminal ileum plasma cells, six were not reactive in either assay (Figure S2B and Table S6).
In addition to HIV-reactive mAbs, we produced and purified 19 terminal ileum mAbs that did not bind HIV-1 epitopes by ELISA or BAMA (Table S2). Of these, four antibodies (21%) were reactive with intestinal bacterial WCLs by both western blot and BAMA (Figures S1A and S3 and Table S6). Three of these four antibodies were not reactive in AtheNA ANA II or HEp-2 ANA assays (Table S6). Therefore, not all commensal bacteria-reactive antibodies from intestine were cross-reactive with gp41.
Affinity Maturation of Commensal Bacteria Cross-Reactive Antibodies to Autologous Envelope
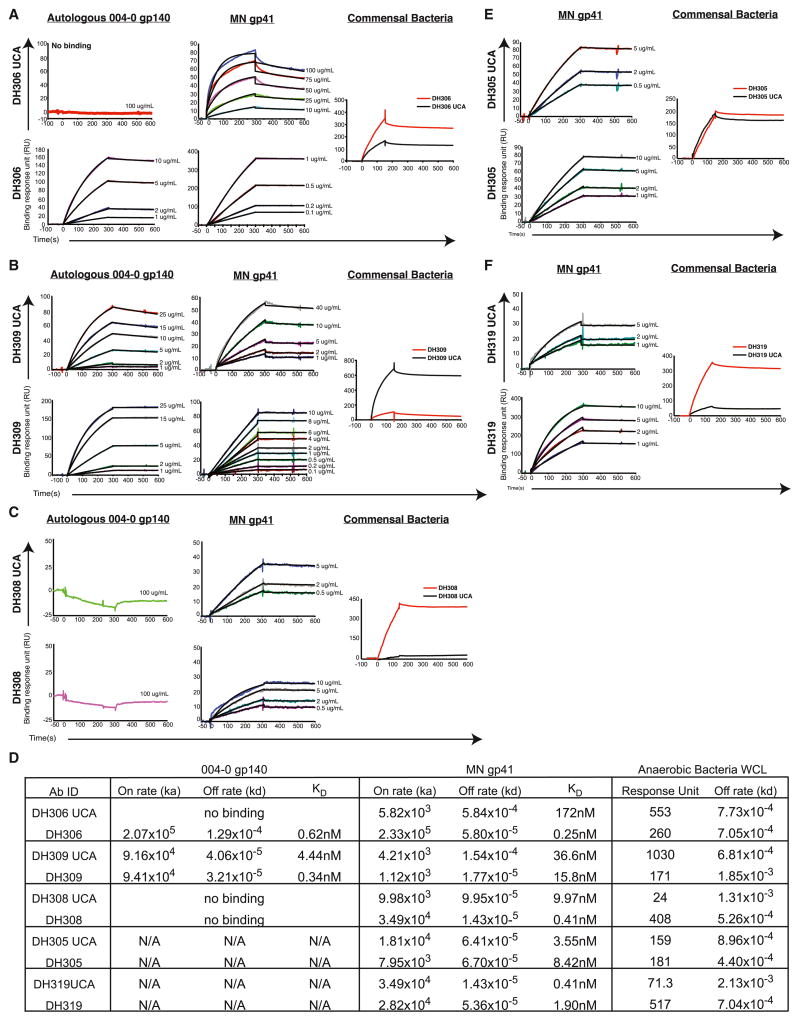
To determine if HIV-1 gp41-reactive antibodies that were cross-reactive with commensal bacteria underwent affinity maturation to gp41, we inferred the heavy- and light-chain unmutated common ancestors (UCAs) of five gp41-reactive mAbs, DH306, DH309, DH308, DH305, and DH319, and produced their UCAs, termed DH306 UCA, DH309 UCA, DH308 UCA, DH305 UCA, and DH319 UCA, respectively. For mAbs isolated from 004-0, DH306, DH309, and DH308, we determined UCA and mature antibody affinities to autologous HIV-1 004-0 gp140 and heterologous HIV-1 MN gp41, as well as relative binding to commensal bacterial antigens. Affinity for the autologous Env increased from undetectable binding to 0.62 nM when comparing the UCA DH306 UCA and the mature antibody DH306 and similarly increased from 4.44 nM to 0.34 nM for DH309 UCA and DH309 (Figures 4A, 4B, and 4D). The mature antibody DH306 also had a greater reactivity to commensal bacteria compared to its UCA (Figure 4A). Binding to the 004-0 T/F gp140 was undetectable for DH308 UCA and DH308; however, affinity to MN gp41 increased from 9.97 nM to 0.41 nM (Figures 4C and 4D). In contrast, DH305 UCA and DH319 UCA had high affinities of 3.55 nM and 0.41 nM to MN gp41, respectively, and affinity did not increase upon accumulation of mutations in the mature mAbs, DH305 and DH319 (Figures 4E and 4F). Therefore, in three commensal bacterial antigen cross-reactive gp41 clonal lineages, affinity maturation to gp41 could be demonstrated from UCAs to mature antibodies.
Figure 4. HIV-1 gp41 and Commensal Bacteria Cross-Reactive Antibodies from the Terminal Ileum Show Affinity Maturation to Autologous and Heterologous HIV-1 Envelope.
(A–C) SPR binding curves of UCA and mature mAbs immobilized with an anti-Fc receptor antibody binding to titrations of autologous HIV-1 gp140 from individual 004-0 and to MN gp41. Relative binding to commensal bacteria was also determined by SPR. (A) DH306 UCA and DH306; (B) DH309 UCA and DH309; and (C) DH308 UCA and DH308.
(D) Table of the on rates, off rates, and apparent dissociation constant (KD) for each mature mAb and UCA pair binding to the HIV-1 Envs tested and the response unit and off rate of mAb binding to anaerobic bacteria WCL. N/A: this antibody was not tested for binding to 004-0 gp140.
(E and F) SPR binding curves of UCA and mature mAbs immobilized with an anti-Fc receptor antibody binding to titrations of MN gp41. Autologous Env was not available for these two mAbs. Relative binding to commensal bacteria was also determined by SPR. (E) DH305 UCA and DH305. (F) DH319 UCA and DH319.
HIV-1 gp41 Commensal Bacterial Cross-Reactive Antibodies Isolated from the Terminal Ileum of Uninfected Individuals
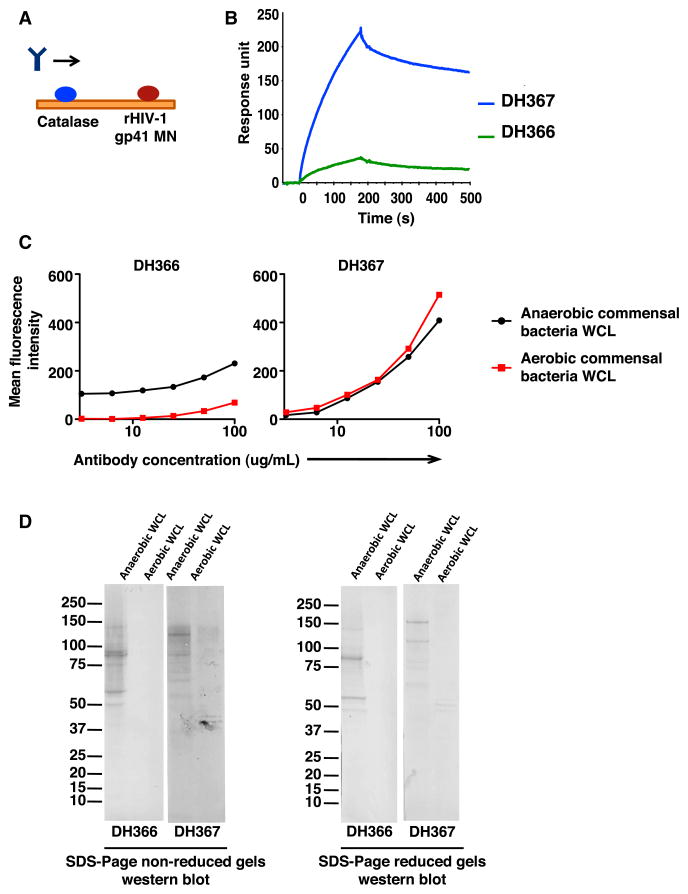
If preinfection terminal ileum antibodies cross-reactive with intestinal commensal bacteria and gp41 are responsible for the initial antibody response to HIV-1 Env gp41 following HIV-1 infection, mutated gp41 and gut flora cross-reactive antibodies should exist in the terminal ileum of uninfected individuals. To investigate this hypothesis, we sorted single plasma cells and memory B cells from three HIV-1 uninfected individuals (Table S1) and identified two low-affinity gp41-reactive antibodies, DH366 and DH367, both of which also reacted with intestinal commensal bacteria (Figures 5 and S1B and Table S2). Both antibodies used VH gene segments from family 3 and were class-switched to IgG; the VH mutation frequencies of these antibodies were 5.2% and 9.7% (Table S2). Therefore, commensal bacteria-reactive mutated B cells that are cross-reactive with Env gp41 can be found in the intestinal B cell repertoire of HIV-1 uninfected individuals, supporting the notion that the initial gp41 antibody response to HIV-1 derived from preexisting commensal bacterial cross-reactive memory B cells.
Figure 5. HIV-1-Reactive Antibodies Isolated from Terminal Ileum Plasma Cells and Memory B Cells of Uninfected Individuals React to Both HIV-1 gp41 and Commensal Bacteria.
(A) SPR strategy used to confirm HIV-1 reactivity of mAbs isolated from the terminal ileum of uninfected individuals. Signal generated by antibody binding to catalase was subtracted from signal generated by antibody binding to recombinant MN gp41.
(B) Antibodies DH366 and DH367 isolated from terminal ileum plasma cells that were natural IgG1 and IgG3 antibodies, respectively, were produced in a rIgG1 backbone and were evaluated for reactivity with HIV-1 MN gp41 by SPR. The VHs of DH366 and DH367 were mutated 6.6% and 11.8%, respectively.
(C) DH366 and DH367 were tested for reactivity to anaerobic and aerobic commensal bacteria by BAMA. Dilutions were 2-fold, ranging from 100–3.1 μg/ml (x axis).
(D) Reactivity to anaerobic and aerobic commensal bacteria under nonreducing and reducing conditions by SDS-PAGE western blot.
E. coli RNA Polymerase Is One Intestinal Bacterial Antigen Cross-Reactive with HIV-1 gp41 Antibodies
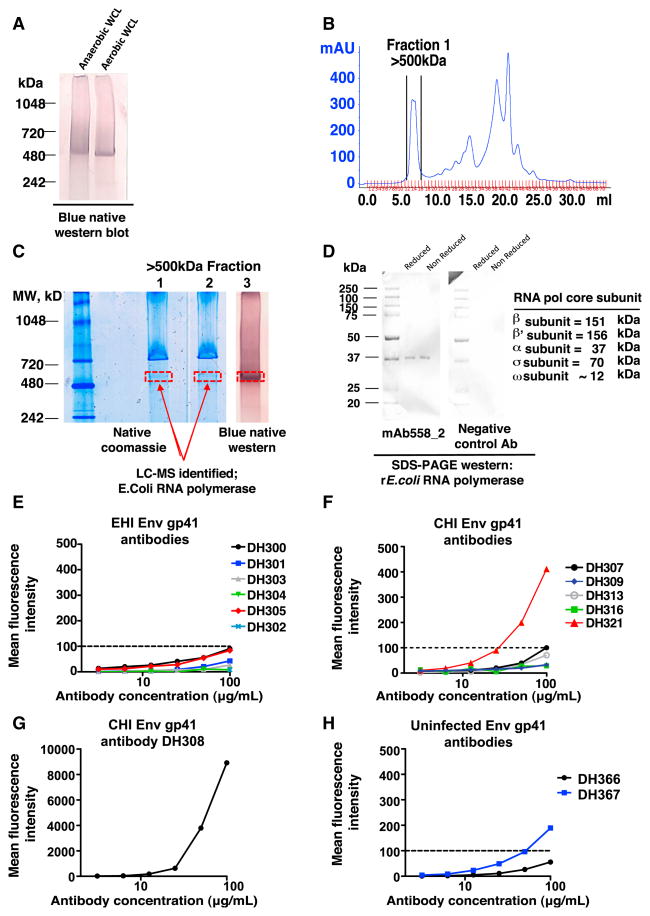
To identify antigens in commensal bacteria cross-reactive with gp41 mAbs, we used the AHI blood-derived HIV-1 gp41, gut bacterial WCL-reactive antibody 558_2 previously reported to bind to an ~520 kDa band of both aerobic and anaerobic commensal bacteria WCLs (Liao et al., 2011) (Figure 6A). The large molecular weight fraction of bacterial WCL was isolated by size exclusion chromatography (SEC) (Figure 6B), and isoelectric zoom fractionation showed that the protein reactive with mAb558_2 migrated to the gel compartment with pH 7–10 (Figure S4A). E. coli RNA polymerase subunits β, β′, and α were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) of the 520 kDa excised bands from two lanes of the SEC-enriched >500 kDa fraction analyzed on a Native-PAGE gel (Figures 6B, 6C, and S4B–S4D). We determined that mAb558_2 binding was specific for the core enzyme of E. coli RNA polymerase (Figures 6D, S4E, and S4F). By western blot, we mapped the specificity of mAb558_2 to the 37 kDa α subunit of recombinant E. coli RNA polymerase (Figure 6D).
Figure 6. Identification of E. coli RNA Polymerase as One Cross-Reactive Commensal Bacterial Antigen Recognized by HIV-1 gp41 Antibodies.
(A) Western blot analysis following NativePAGE gel run showing that mAb 558_2 (Liao et al., 2011) binds to an ~520 kDa protein band in anaerobic and aerobic intestinal bacterial WCL.
(B). Protein fractions from bacterial WCL with molecular weight of ~500 kDa were collected following size exclusion chromatography (SEC).
(C) The ~500 kDa fraction shows enrichment of the 520 kDa protein by 1D native Coomassie blue (lanes 1 and 2) and blue native western blotting with mAb558_2 (lane 3). The ~520 kDa bands from two gels, identified in two red boxes, were excised from the gel and determined to be E. coli RNA polymerase by LC-MS/MS.
(D) Recombinant E. coli RNA polymerase core protein was run on a denaturing SDS-PAGE gel under both reducing and nonreducing conditions and blotted with mAb558_2 and a hemagglutinin (HA) flu-reactive antibody Ab1248 as a negative control.
(E–H) Reactivity of terminal ileum Env gp41 commensal bacteria cross-reactive antibodies with rE. coli RNA polymerase was determined by BAMA. Dilutions were 2-fold, ranging from 100–3.1 μg/ml (x axis). (E) HIV-1 gp41 commensal bacteria cross-reactive mAbs isolated from the terminal ileum of EHI individuals. (F) HIV-1 gp41 commensal bacteria cross-reactive mAbs isolated from the terminal ileum of CHI individuals. DH309 and DH316 are representative of the other lowest binders, DH317 and DH318. (G) H308, a gp41 mAb isolated from a terminal ileum plasma cell from CHI individual 004-0, is the strongest rE. coli RNA polymerase binder. DH308 used the VH gene segment 1–69, which was 8.6% mutated and was naturally IgG1. (H) HIV-1 gp41 commensal bacteria cross-reactive mAbs isolated from the terminal ileum of uninfected individuals. See also Figure S4.
We found that 2 of 14 (14.3%) Env gp41 and intestinal commensal bacteria cross-reactive EHI and CHI terminal ileum antibodies also reacted with rRNA polymerase by BAMA. (Figures 6E–6G) Moreover, the gp41 commensal bacterial cross-reactive antibody DH367 isolated from the terminal ileum of an uninfected individual also reacted with rRNA polymerase by BAMA (Figure 6H).
Terminal Ileum HIV-1-Reactive Antibody Clonal Lineage Members Shared by Terminal Ileum and Peripheral Blood Compartments
We next asked if HIV-1 and commensal bacteria cross-reactive B cells recirculate in the terminal ileum and peripheral blood. We studied paired blood samples of three of the individuals (042-8, 004-0, and 071-8) from whom we had isolated terminal ileum plasma cell and memory B cell mAbs (Table S1). We sorted single plasma cells and memory B cells from PBMCs and identified 13 antibodies with HIV-1 reactivity (Figure S5 and Table S2). By single-cell PCR, we were able to identify four clonal lineages within the terminal ileum and one clonal lineage within the blood (Table S7). However, by these methods we were unable to identify any clonal lineages with members shared between the terminal ileum and blood.
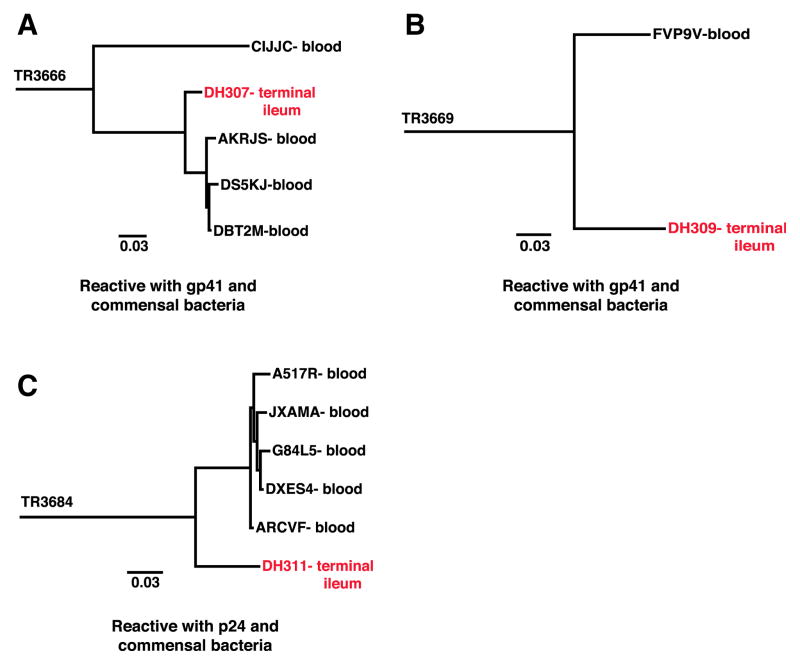
We next conducted pyrosequencing of genomic DNA isolated from PBMCs taken at the same time as terminal ileum samples from chronically infected individuals 004-0 and 071-8 and searched the sequences for VHDHJH members clonally related to the 149 terminal ileum VHDHJH sequences isolated from these same two individuals by single-cell sorting. By this method we identified a total of 18 clonal lineages that had members in both terminal ileum and blood compartments (Figures 7 and S6 and Table S7). Thus, 12% of terminal ileum B cells isolated by single-cell PCR had cross-compartment clonal lineage members in the blood. Of these 18 cross-compartment clonal lineages, we determined that two clonal lineages were cross-reactive with Env gp41 and intestinal commensal bacteria, and one lineage was cross-reactive with HIV-1 Gag p24 and commensal bacteria (Figures 1D, 2D, 7, and S6 and Table S2).
Figure 7. Phylogenetic Trees of Ig Heavy-Chain Clonal Lineages with Members Derived from Blood B Cells and Terminal Ileum B Cells, with Known Antibody Reactivity.
(A–C) Trees are rooted on the inferred UCA. Nodes are labeled with the antibody or sequence ID and sample they were isolated from. Red nodes indicate B cells isolated by single-cell PCR from terminal ileum B cells that were produced in large scale and screened for reactivity to HIV-1 antigens and commensal bacteria WCLs. The reactivity of the terminal ileum mAb is noted below each tree. Black nodes indicate VHDHJH sequences identified by pyrosequencing of time-matched peripheral PBMCs. Tree IDs are located to the left of each tree: TR3666 (A), TR3669 (B), and TR3684 (C). See also Figures S5–S7 and Table S7.
To determine if the cross-compartmentalization of B cell clonal lineages identified in 004-0 and 071-8 was due to contamination of the terminal ileum tissue biopsies with blood B cells trafficking through the ileum vasculature without entering the tissue, we performed quantitative image analysis of B cells in the terminal ileum of HIV-1-infected individuals and found that of the 12 terminal ileum biopsies studied, only 0.2% of the CD20+ cells within the tissue samples were found within blood vessels (Figure S7). Thus, blood contamination of the biopsy could not explain the 12% of terminal ileum B cells isolated by single-cell PCR as contaminating B cells from the blood compartment.
DISCUSSION
In this study we have demonstrated that the dominant plasma cell antibody population to HIV-1 in both EHI and CHI in the terminal ileum was nonneutralizing, directed to Env gp41, and cross-reactive with intestinal commensal bacterial antigens. One such bacterial antigen identified was the α subunit of E. coli RNA polymerase. Similar specificities of gp41 commensal bacteria cross-reactive mutated antibodies could be isolated from HIV-1 uninfected individuals. Moreover, we demonstrated sharing of terminal ileum clonal lineage members with the blood compartment, providing support for the hypothesis that blood B cells cross-reactive with intestinal bacteria and gp41 are derived from the intestinal tract.
The preponderance of gp41 antibodies in terminal ileum plasma cell and memory B cell pools now potentially explains the mechanism of induction of gp41 antibody immunodominance in plasma and mucosal fluid studies (Tomaras et al., 2008; Yates et al., 2013). Liao et al. (2011) showed a predominance of blood gp41 antibodies from HIV-1 plasma cell-derived mAbs from AHI and found them to be a minority of the plasma cell pool 17–46 days after HIV-1 transmission. The polyclonal pool of non-HIV-1-reactive B cells is likely due to the massive cytokine storm that occurs early on after HIV-1 transmission (Stacey et al., 2009) and prompted us to ask if the plasma cell and memory B cell pools in terminal ileum would be a location of a more robust HIV-1 Env antibody response. Instead, we found in both EHI and CHI that terminal ileum contained primarily non-HIV-1-reactive polyclonal plasma and memory B cells, and the few HIV-1-reactive B cells that were present were targeted to Env gp41.
Host-specific bacterial colonization of the gastrointestinal tract is required for normal development of the intestinal immune system (Chung et al., 2012; Erturk-Hasdemir and Kasper, 2013; Hooper et al., 2012). Germ-free mice have numerous immunological deficiencies, including small Peyer’s patches and mesenteric lymph nodes, reduced secretory IgA, fewer plasma cells, CD4+ T cells and CD8+ T cells, and diminished antimicrobial peptide production (Erturk-Hasdemir and Kasper, 2013; Hooper et al., 2012; Round and Mazmanian, 2009). Recolonization of germ-free mice with host-specific commensal bacteria ameliorates these defects (Chung et al., 2012; Smith et al., 2007). The presence of intestinal commensal bacteria induces immune maturation that is not only required for gut homeostasis, but helps generate a pool of mature adaptive immune cells prepared to protect the host from infections. The pre-HIV-1 infection presence of B cells within the intestine cross-reactive with both bacterial antigens and HIV-1 gp41 is evidence of molecular mimicry between HIV-1 antigens and bacteria antigens and suggests an explanation for why the initial antibody response to AHI in the plasma and mucosal fluids is to gp41 (Fujinami et al., 1983; Liao et al., 2011; Oldstone, 1998; Srinivasappa et al., 1986; Tomaras et al., 2008; Yates et al., 2013).
Isolation of mutated gp41 and gut flora cross-reactive antibodies from terminal ileum HIV-1 uninfected individuals directly suggests that commensal or pathogenic bacteria or other cross-reactive environmental antigens can trigger gp41 cross-reactive responses before HIV-1 infection. These data provide evidence in support of the hypothesis that the dominant HIV-1 gp41 antibody response after HIV-1 transmission is mediated by previously activated memory B cells that are present before HIV-1 infection and cross-reactive with intestinal bacteria. Once HIV-1 infection occurs, then gp41 would begin to trigger the previously activated bacterial-driven lineages toward affinity maturation to gp41-specific antibodies. A critical test of this notion would be to demonstrate that reactivity in commensal bacteria-gp41 lineage begins with a gut flora-reactive UCA followed by acquisition of gp41 reactivity upon affinity maturation. In the present study, we provide three examples of gp41-reactive antibodies, DH306, DH308, and DH309, that showed affinity maturation to autologous and/or heterologous Env (Figure 4). In the case of antibody DH306, reactivity of the UCA with gp41, but not the T/F Env gp140, may well be an example of cross-reactive stimulation of the UCA by an environmental gp41 cross-reactive antigen before transmission that gave rise to the affinity mature antibody that, after infection, reacted with the autologous T/F Env. Both DH308 and DH308 UCA bound to MN gp41 with nanomolar affinity but did not bind to the autologous T/F gp140 (Figure 4). It is important to note that antibody DH308 was isolated from individual 004-0 3 years into infection. Thus, it is likely that a T/F Env variant selected by antibodies over time initiated the DH308 lineage, given the high level of affinity maturation to gp41 from ~10 nM in DH308 UCA to 0.4 M in the mature antibody DH308 (Figure 4D). DH305 UCA and DH319 UCA are examples of naturally paired, unmutated VHDHJH and VLJL with high affinities for viral antigens and B cell clonal lineages reaching an affinity ceiling prior to accumulation of the mutations found in the mature mAbs as previously described (Batista and Neuberger, 1998). We have also previously shown that in a reconstructed blood gp41 clonal lineage, the UCA and the first intermediate antibody in the lineage were commensal bacteria reactive, but not gp41-reactive (Liao et al., 2011). Instead, gp41 reactivity only occurred later in clonal lineage development, and after gp41 reactivity occurred, there was affinity maturation to HIV-1 env gp41 in the clonal lineage. The presence of CD4+ memory T cells cross-reactive with both HIV-1 antigens and microbial peptides in uninfected adults (Campion et al., 2014; Su et al., 2013), and the 5.2% and 11.9% VH mutation frequencies of DH306 and DH309, suggested that the affinity maturation of gp41 commensal bacteria cross-reactive B cells to gp41 is T cell dependent.
We now directly demonstrate the intestinal tract origin for commensal bacteria-gp41 cross-reactive antibodies found in the blood. Moreover, we demonstrated that 21% of commensal bacteria-reactive B cells were not gp41 reactive, adding additional support to the idea that, in HIV-1-infected individuals, the gp41-reactive plasma cells and memory B cells represented a response to HIV-1. The proportion of these control non-HIV-1-reactive antibodies isolated from the terminal ileum of HIV-1-infected individuals that reacted with gut flora (21%) is greater than the ~12% of plasma cells from the terminal ileum of HIV-1 uninfected individuals determined to be reactive with specific gut flora by Benckert et al. (2011). Microbial translocation that occurs in HIV-1 infection may account for this higher level of commensal bacteria-reactive terminal ileum B cells in our study (Brenchley et al., 2006).
A critical test of the hypothesis that blood gp41 commensal bacteria-reactive B cells arise in the intestine was to determine if commensal bacteria-gp41 clonal lineages shared members with blood B cells. Indeed, we have now found evidence for three such intestinal commensal bacteria-gp41 clonal lineages shared by both terminal ileum and peripheral blood compartments (Figures 7 and S6 and Table S7).
In summary, these data provide evidence for the hypothesis that the postinfection B cell response to HIV-1 is shaped by the preinfection B cell repertoire to environmental antigens. Env gp41 antibodies cross-react with human intestinal commensal bacteria, suggesting that commensal bacteria play critical roles in shaping the preinfection response to HIV-1, and demonstrate a major role for the memory B cell pool in contributing to the initial antibody response to HIV-1. These data also raise the hypothesis that the human B cell response to a wide variety of other infectious agents may similarly be affected by cross-reactivity to environmental antigens.
EXPERIMENTAL PROCEDURES
Study Subjects
Terminal ileum, blood, and bone marrow samples were collected from six EHI individuals 47–200 days after transmission, ten CHI individuals greater than 200 days after transmission, as estimated from patient history and Fiebig classification (Fiebig et al., 2003), and three HIV-1 uninfected individuals (Table S1). All individuals studied were from the United States. Table S1 shows the clinical characteristics of the individuals studied. All work related to human subjects was carried out with the informed consent of trial participants and in compliance with Institutional Review Board protocols approved by Duke University Medical Center and the University of North Carolina Medical Center.
Flow Cytometry Analysis of Terminal Ileum and Blood B Cells
Terminal ileum mononuclear cells were isolated from gut tissues, and a single-cell suspension was formed by passing cells through 100 μM cell strainer (Fisher Scientific). The cells were then labeled with a panel of fluorochrome-conjugated mAbs to label distinct B cell subsets in blood and terminal ileum. A detailed protocol is included in the Supplemental Experimental Procedures.
PCR Amplification of Plasma Cell and Memory B Cell Immunoglobulin VH and VL Genes
The Ig VHDHJH and VLJL genes of the sorted plasma cell and memory B cells were amplified by RT and nested PCR using the method as reported (Liao et al., 2009; Tiller et al., 2008; Wardemann et al., 2003; Wrammert et al., 2008).
Sequencing, Sequence Annotation, Quality Control, and Data Management of Ig VHDHJH and VLJL Sequences
These methods were completed as previously described (Liao et al., 2011), and a detailed explanation is included in Supplemental Experimental Procedures.
High-Throughput DNA Sequencing of Ig V(D)J Gene Segments
These methods were completed as previously described (Boyd et al., 2009), and a detailed explanation is included in Supplemental Experimental Procedures.
Identification of Clone Members and Inference of UCA
Clonal relatedness of VHDHJH and VLJL sequences was determined as described (Kepler, 2013; Liao et al., 2013).
Expression of VHDHJH and VLJL as Full-Length IgG1 Recombinant mAbs
PCR was used to assemble linear full-length Ig heavy- and light-chain gene expression cassettes using the Ig VHDHJH and VL gene pairs as previously described (Liao et al., 2009, 2011, 2013). We determined that, in most cases, isolation of one heavy chain and two light chains is an artifact of sorting two B cells into one well, and the heavy-chain and light-chain pairing could not be precisely determined. These antibodies were not included in statistical analysis of frequencies of total antibodies, but such heavy-chain sequences were used for analysis of clonal relationships, and VHDHJH and VL pairs that reacted with HIV-1 antigens were included in the total list of HIV-1 antibodies. Additional explanation is included in the Supplemental Experimental Procedures.
Assays for Antibody Reactivity
The recombinant mAbs expressed in small-scale and large-scale transfections were assayed for antibody reactivity to HIV-1 antigens and a panel of non-HIV-1 antigens by ELISA and BAMA as previously described (Liao et al., 2009, 2011). Antibodies produced in large scale and protein A purified were titrated at concentrations ranging from 100 μg/ml to 0.046 μg/ml at 3-fold dilutions for ELISA assays. Positivity cutoffs for reactivity were set at 3-fold above background and an optical density (OD) of 0.130 at 100 μg/ml. An antigen list is included in the Supplemental Experimental Procedures.
The apparent affinity of HIV-1-reactive antibodies was calculated in molar concentration from EC50 values using a four-parametric sigmoid curve-fitting analysis. Antibodies were titrated by 3-fold dilutions at concentrations ranging from 100 μg/ml to 0.046 μg/ml titrations. Antibodies were considered to have high affinity if the EC50 was less than 1 μg/ml, midrange affinity if the EC50 ranged between 1 and 50 μg/ml, and low affinity if the EC50 was greater than 50 μg/ml.
As previously described (Tomaras et al., 2008), BAMA assays are conducted with carboxylated fluorescent beads (Luminex) covalently coupled to small quantities (25 μg) of antigen and are incubated with antibody from small-scale transfection or after column purification and binding is detected with biotin-labeled mouse anti-human IgG (SouthernBiotech). Further explanation and an antigen list are included in the Supplemental Experimental Procedures.
For indirect immunofluorescence on HEp-2 cells, all antibodies grown in large scale were assayed for reactivity to HEp-2 cells at 50 μg/ml and 25 μg/ml (Inverness Medical Professional Diagnostics) by indirect immunofluorescence staining (Haynes et al., 2005). Antibody reactivity to autoantigens was also determined by antibody multiplex AtheNA Multi-Lyte ANA II test (Wampole Laboratories) (Haynes et al., 2005). Antibodies were studied in a dose dilution starting at 50 μg/ml and determined to be reactive when binding antibody multiplex assay scores were 225 MFI units or greater (Haynes et al., 2005).
For western blot analysis of commensal bacteria reactivity, 100 μg of both aerobic and anaerobic lysates were run on 4%–12% Tris-Bis SDS-PAGE (Life Technologies) for 1 hr 29 min at 150 V in both reduced and nonreduced conditions. NuPAGE sample reducing agent at 1× was used for reducing conditions (Life Technologies). Antigens were transferred to nitrocellulose using Life Technologies iBlot Gel Transfer system. Antibody binding was tested at 20 μg/ml for all antibodies, and the Anti-Human IgG (whole molecule)-Alkaline Phosphatase antibody produced in goat (Sigma) was used at a 1:5,000 dilution. Detection occurred directly on the nitrocellulose using Western Blue (Promega).
Surface Plasmon Resonance
To confirm the reactivity to gp41 of the antibodies isolated from the terminal ileum of uninfected individuals, SPR binding assays were performed on a Biacore 3000 (GE Healthcare) maintained at 25°C. Recombinant catalase (GE Healthcare) and HIV-1 gp41 MN were immobilized on a CM5 sensor chip by standard amine coupling, as previously described (Alam et al., 2008, 2009).
To determine the reactivity of terminal ileum HIV-1-reactive mAbs to anaerobic intestinal commensal bacteria, SPR binding assays were performed on a Biacore 4000 (Biacore). HIV-1-reactive mAbs were immobilized on CM5 sensor chips by standard amine coupling, and reactivity was determined by double reference subtraction. Response generated by nonspecific binding of anaerobic commensal bacteria WCLs to control antibody palivizumab (anti-RSV IgG1 mAb) (Johnson et al., 1997) was subtracted from signal generated by antigen binding to HIV-1-reactive mAbs. The positivity cutoff was calculated as three times the response generated by antigen binding to a second negative control antibody Ab1248 (anti-influenza/hemagglutinin). Rate constants were measured using 1:1 Langmuir equation. Additional explanation of measuring rate constants for mAb binding to bacteria WCL is included in the Supplemental Experimental Procedures. Glycine-HCL (pH 2.0) was used as the regeneration buffer.
To determine the affinity of mature mAbs and their respective UCAs, Env SPR binding titrations were performed using Biacore 3000 at 25°C. Autologous Env, 040 gp140 isolated from individual 004-0 (Table S1) (Bar et al., 2012; Liao et al., 2013), and heterologous Env MN gp41 were used. Antibodies were captured on a CM5 sensor chip coupled with anti-human Fc antibody. Rate constants were measured using global curve fitting to binding curves obtained from Env titrations.
Preparation of Intestinal Anaerobic and Aerobic Commensal Bacteria Lysates
Two separate preparations of bacteria were inoculated from stool specimens from 4–5 individuals and grown on agar plates under anaerobic and aerobic conditions at 30°C. For each preparation the individual stool samples were pooled, but aerobic and anaerobic extracts were prepared separately. A detailed protocol is included in the Supplemental Experimental Procedures.
Identification of gp41 mAb-Reactive Protein in Intestinal Bacterial Lysate
Anaerobe gut lysate (total protein ~4 mg) was fractionated on a Superdex S200 (GE Healthcare) size exclusion column, and protein fractions with molecular size > 500 kDa were pooled and concentrated. SPR binding and western blot analysis (~520 kDa band) confirmed that the high molecular weight fraction from size exclusion chromatography was gp41 mAb276 reactive. The size-enriched and mAb276-reactive fraction comprised about 10%–20% of the total gut lysate proteins. About 180 μg of size-fractionated lysate protein was loaded on two adjacent lanes of a blue native gel, and ~520 kDa bands that blotted with Mab 276 were cut out of the Coomassie-stained blue native gel. The cut out protein band was subjected to trypsin and chymotrypsin digestion, and protein identification was performed by LC-MS/MS.
IgG VH1-69 Copy Number Assay
Primers and a probe designed for VH1-69 gene segments were used in the QX100 Droplet Digital PCR system by Bio-Rad. A more detailed protocol is included in the Supplemental Experimental Procedures.
Statistical Analysis
All analysis data sets were compiled and completed with SAS v.9.2 (SAS Institute). To compare VH mutation frequency and HCDR3 lengths, a mixed model was performed to account for multiple observations taken from individual patients.
Quantitative Image Analysis
Quantitative image analysis was performed on terminal ileum tissue B cells and blood vessels as described (Levesque et al., 2009).
Supplementary Material
Acknowledgments
The authors are grateful for technical assistance from Marietta Gustilo for processing terminal ileum tissue; Josh Amos for processing blood and performing flow cytometry experiments; Michele J. Donathan, Judith Lucas, William Williams, Jessica Peel, and Robert Meyerhoff for help with design and quality control of antibody binding assays; Dana Momeyer and Glenn Overman for genomic DNA isolation; Jamie Pritchett for plasmid preparation; Feng Yan for his contributions to isolating RNA polymerase from anaerobic bacteria WCLs; and Shelley Stewart and Kara Anasti for running SPR experiments. We also thank Jennifer Kirchherr and Caroline Cockrell for program management support, Michael Root for gp41 5-helix bundle recombinant protein, Jeff Lifson for AT-2-inactivated HIV-1 virions, and Andrew Z. Fire and Garnett Kelsoe for valuable discussion.
Research reported in this publication was supported by the National Institute of Allergy and Infectious Disease of the National Institutes of Health, by the Center of HIV/AIDS Vaccine Immunology, grant number U19-AI067854, and by the Center for HIV/AIDS Vaccine Immunology- Immunogen Discovery grant number UM1-AI100645-01 from the NIH, NIAID, Division of AIDS, and by a Viral Oncology Training Grant, grant number T32-CA009111 from the NCI, NIH.
Footnotes
ACCESSION NUMBERS
The GenBank accession numbers for isolated antibody sequences reported in this paper are as follows: DH300 (KM067693 and KM067694), DH301 (KM067695 and KM067696), DH302 (KM067697 and KM067698), DH303 (KM067699 and KM067700), DH304 (KM067701 and KM067702), DH305 (KM067703 and KM067704), DH306 (KM067705 and KM067706), DH307 (KM067707 and KM067708), DH308 (KM067709 and KM067710), DH309 (KM067711 and KM067712), DH310 (KM067713 and KM067714), DH311 (KM067715 and KM067716), DH312 (KM067717 and KM067718), DH313 (KM067719 and KM067720), DH314 (KM067721 and KM067722), DH315 (KM067723 and KM067724), DH316 (KM067725 and KM067726), DH317 (KM067727 and KM067728), DH318 (KM067729 and KM067730), DH319 (KM067731 and KM067732), DH320 (KM067733 and KM067734), DH321 (KM067735 and KM067736), DH322 (KM067737 and KM067738), DH323 (KM067739 and KM067740), DH324 (KM067741 and KM067742), DH325 (KM067743 and KM067744), DH326 (KM067745 and KM067746), DH327 (KM067747 and KM067748), DH328 (KM067749 and KM067750), DH329 (KM067751 and KM067752), DH330 (KM067753 and KM067754), DH331 (KM067755 and KM067756), DH332 (KM067757 and KM067758), DH333 (KM067759 and KM067760), DH334 (KM067761 and KM067762), DH335 (KM067763 and KM067764), DH336 (KM067765 and KM067766), DH337 (KM067767 and KM067768), DH338 (KM067769 and KM067770), DH339 (KM067771 and KM067772), DH340 (KM067773 and KM067774), DH341 (KM067775 and KM067776), DH342 (KM067777 and KM067778), DH343 (KM067779 and KM067780), DH344 (KM067781 and KM067782), DH345 (KM067783 and KM067784), DH346 (KM067785 and KM067786), DH347 (KM067787 and KM067788), DH348 (KM067789 and KM067790), DH349 (KM067791 and KM067792), DH350 (KM067793 and KM067794), DH351 (KM067795 and KM067796), DH352 (KM067797 and KM067798), DH353 (KM067799 and KM067800), DH354 (KM067801 and KM067802), DH355 (KM067803 and KM067804), DH366 (KM067805 and KM067806), and DH367 (KM067807 and KM067808).
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.chom.2014.07.003.
References
- Alam SM, Scearce RM, Parks RJ, Plonk K, Plonk SG, Sutherland LL, Gorny MK, Zolla-Pazner S, Vanleeuwen S, Moody MA, et al. Human immunodeficiency virus type 1 gp41 antibodies that mask membrane proximal region epitopes: antibody binding kinetics, induction, and potential for regulation in acute infection. J Virol. 2008;82:115–125. doi: 10.1128/JVI.00927-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SM, Morelli M, Dennison SM, Liao HX, Zhang R, Xia SM, Rits-Volloch S, Sun L, Harrison SC, Haynes BF, Chen B. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci USA. 2009;106:20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar KJ, Tsao CY, Iyer SS, Decker JM, Yang Y, Bonsignori M, Chen X, Hwang KK, Montefiori DC, Liao HX, et al. Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLoS Pathog. 2012;8:e1002721. doi: 10.1371/journal.ppat.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista FD, Neuberger MS. Affinity dependence of the B cell response to antigen: a threshold, a ceiling, and the importance of off-rate. Immunity. 1998;8:751–759. doi: 10.1016/s1074-7613(00)80580-4. [DOI] [PubMed] [Google Scholar]
- Benckert J, Schmolka N, Kreschel C, Zoller MJ, Sturm A, Wiedenmann B, Wardemann H. The majority of intestinal IgA+ and IgG+ plasmablasts in the human gut are antigen-specific. J Clin Invest. 2011;121:1946–1955. doi: 10.1172/JCI44447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd SD, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med. 2009;1:12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, Douek DC. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- Campion SL, Brodie TM, Fischer W, Korber BT, Rossetti A, Goonetilleke N, McMichael AJ, Sallusto F. Proteome-wide analysis of HIV-specific naive and memory CD4+ T cells in unexposed blood donors. J Exp Med. 2014;211:1273–1280. doi: 10.1084/jem.20130555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erturk-Hasdemir D, Kasper DL. Resident commensals shaping immunity. Curr Opin Immunol. 2013;25:450–455. doi: 10.1016/j.coi.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, Heldebrant C, Smith R, Conrad A, Kleinman SH, Busch MP. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17:1871–1879. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- Fujinami RS, Oldstone MB, Wroblewska Z, Frankel ME, Koprowski H. Molecular mimicry in virus infection: crossreaction of measles virus phosphoprotein or of herpes simplex virus protein with human intermediate filaments. Proc Natl Acad Sci USA. 1983;80:2346–2350. doi: 10.1073/pnas.80.8.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadalupe M, Reay E, Sankaran S, Prindiville T, Flamm J, McNeil A, Dandekar S. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol. 2003;77:11708–11717. doi: 10.1128/JVI.77.21.11708-11717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S, Oliver C, Prince GA, Hemming VG, Pfarr DS, Wang SC, Dormitzer M, O’Grady J, Koenig S, Tamura JK, et al. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J Infect Dis. 1997;176:1215–1224. doi: 10.1086/514115. [DOI] [PubMed] [Google Scholar]
- Kepler TB. Reconstructing a B-cell clonal lineage. I Statistical inference of unobserved ancestors. F1000Res. 2013;2:103. doi: 10.12688/f1000research.2-103.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MC, Moody MA, Hwang KK, Marshall DJ, Whitesides JF, Amos JD, Gurley TC, Allgood S, Haynes BB, Vandergrift NA, et al. Polyclonal B cell differentiation and loss of gastrointestinal tract germinal centers in the earliest stages of HIV-1 infection. PLoS Med. 2009;6:e1000107. doi: 10.1371/journal.pmed.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Sutherland LL, Xia SM, Brock ME, Scearce RM, Vanleeuwen S, Alam SM, McAdams M, Weaver EA, Camacho Z, et al. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology. 2006;353:268–282. doi: 10.1016/j.virol.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Levesque MC, Nagel A, Dixon A, Zhang R, Walter E, Parks R, Whitesides J, Marshall DJ, Hwang KK, et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J Virol Methods. 2009;158:171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Chen X, Munshaw S, Zhang R, Marshall DJ, Vandergrift N, Whitesides JF, Lu X, Yu JS, Hwang KK, et al. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. J Exp Med. 2011;208:2237–2249. doi: 10.1084/jem.20110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehandru S, Poles MA, Tenner-Racz K, Jean-Pierre P, Manuelli V, Lopez P, Shet A, Low A, Mohri H, Boden D, et al. Lack of mucosal immune reconstitution during prolonged treatment of acute and early HIV-1 infection. PLoS Med. 2006;3:e484. doi: 10.1371/journal.pmed.0030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Gray ES, Morris L. Specificity of the autologous neutralizing antibody response. Curr Opin HIV AIDS. 2009;4:358–363. doi: 10.1097/COH.0b013e32832ea7e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldstone MB. Molecular mimicry and immune-mediated diseases. FASEB J. 1998;12:1255–1265. doi: 10.1096/fasebj.12.13.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope M, Haase AT. Transmission, acute HIV-1 infection and the quest for strategies to prevent infection. Nat Med. 2003;9:847–852. doi: 10.1038/nm0703-847. [DOI] [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasso EH, Johnson T, Kipps TJ. Expression of the immunoglobulin VH gene 51p1 is proportional to its germline gene copy number. J Clin Invest. 1996;97:2074–2080. doi: 10.1172/JCI118644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Srinivasappa J, Saegusa J, Prabhakar BS, Gentry MK, Buchmeier MJ, Wiktor TJ, Koprowski H, Oldstone MB, Notkins AL. Molecular mimicry: frequency of reactivity of monoclonal antiviral antibodies with normal tissues. J Virol. 1986;57:397–401. doi: 10.1128/jvi.57.1.397-401.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey AR, Norris PJ, Qin L, Haygreen EA, Taylor E, Heitman J, Lebedeva M, DeCamp A, Li D, Grove D, et al. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol. 2009;83:3719–3733. doi: 10.1128/JVI.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4(+) memory-phenotype T cells are abundant in unexposed adults. Immunity. 2013;38:373–383. doi: 10.1016/j.immuni.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras GD, Yates NL, Liu P, Qin L, Fouda GG, Chavez LL, Decamp AC, Parks RJ, Ashley VC, Lucas JT, et al. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol. 2008;82:12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, Rosenzweig M, Johnson RP, Desrosiers RC, Lackner AA. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- Veazey RS, Marx PA, Lackner AA. The mucosal immune system: primary target for HIV infection and AIDS. Trends Immunol. 2001;22:626–633. doi: 10.1016/s1471-4906(01)02039-7. [DOI] [PubMed] [Google Scholar]
- Wardemann H, Yurasov S, Schaefer A, Young JW, Meffre E, Nussenzweig MC. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates NL, Stacey AR, Nolen TL, Vandergrift NA, Moody MA, Montefiori DC, Weinhold KJ, Blattner WA, Borrow P, Shattock R, et al. HIV-1 gp41 envelope IgA is frequently elicited after transmission but has an initial short response half-life. Mucosal Immunol. 2013;6:692–703. doi: 10.1038/mi.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.