Highlights
-
•
The expression of matrilin VWA domains was improved.
-
•
The proper folding was monitored by NMR and CD spectroscopy.
-
•
Binding properties were analyzed by surface plasmon resonance.
-
•
Differences between bacterially and eukaryotically expressed domains were detected.
-
•
We recommend eukaryotic expression for structural analysis.
Keywords: Matrilin, Extracellular matrix, von Willebrand factor A domain
Abstract
VWA domains are the predominant independent folding units within matrilins and mediate protein–protein interactions. Mutations in the matrilin-3 VWA domain cause various skeletal diseases. The analysis of the pathological mechanisms is hampered by the lack of detailed structural information on matrilin VWA domains. Attempts to resolve their structures were hindered by low solubility and a tendency to aggregation. We therefore took a comprehensive approach to improve the recombinant expression of functional matrilin VWA domains to enable X-ray crystallography and nuclear magnetic resonance (NMR) studies. The focus was on expression in Escherichia coli, as this allows incorporation of isotope-labeled amino acids, and on finding conditions that enhance solubility. Indeed, circular dichroism (CD) and NMR measurements indicated a proper folding of the bacterially expressed domains and, interestingly, expression of zebrafish matrilin VWA domains and addition of N-ethylmaleimide yielded the most stable proteins. However, such proteins did still not crystallize and allowed only partial peak assignment in NMR. Moreover, bacterially expressed matrilin VWA domains differ in their solubility and functional properties from the same domains expressed in eukaryotic cells. Structural studies of matrilin VWA domains will depend on the use of eukaryotic expression systems.
Introduction
The matrilins form a four-member family of extracellular matrix proteins and belong to the superfamily of von Willebrand factor A (VWA)1-containing proteins. Matrilins are expressed in cartilage and many other forms of extracellular matrix, where they are part of fibrillar or filamentous structures and mediate interactions between collagen-containing fibrils and other matrix components. They are modular, multi-subunit proteins with most subunits consisting of two VWA-domains interconnected by a variable number of epidermal growth factor (EGF)-like domains (for review see [1]). Only matrilin-3 lacks the second VWA-domain. The C-terminal α-helical coiled-coil-domain enables the oligomerisation of single subunits to trimers (matrilin-1 and -4) and tetramers (matrilin-2 and -3). N-terminal of the coiled-coil-domain matrilin-2 carries a unique domain and at the N-terminus matrilin-2 and -3 have short sequences with a clustering of positively charged side chains.
Matrilins act as adaptor proteins in the extracellular matrix and this function can be modulated by structural heterogeneity. Alternative splicing, the formation of homo- and heteroligomers and proteolytic processing lead to variability in subunit and domain composition and can decrease or increase the binding avidity. Matrilins interact with a variety of extracellular matrix proteins such as aggrecan, collagen I, II and IX, cartilage oligomeric matrix protein (COMP) and the small leucine rich repeat proteoglycans decorin and biglycan [2–8]. Genetic deletion of matrilin-1 and -3 in mouse leads to moderate changes in cartilage collagen fibril volume density [9]. Mutations in matrilins, especially in matrilin-3, are associated with various skeletal diseases, e.g. multiple epiphyseal dysplasia (MED), bilateral hereditary micro-epiphyseal dysplasia, spondylo-epi-metaphyseal dysplasia, and osteoarthritis [10–13]. Most of the mutations in matrilin-3 that cause MED are located in the β-sheet of the VWA domain [14] and often lead to retention of matrilin-3 in the endoplasmic reticulum (ER) resulting in ER-stress response [15–18]. However, other mutations were identified where the mutated form of matrilin-3 is secreted and affects the collagen fibril organization [8].
The interactions of matrilins with other proteins are mediated by their VWA domains. VWA domains are modules of about 200 amino acids which are present in a large number of intra- and extracellular proteins [19]. VWA domains show a classical Rossmann-fold [20] with a central β-sheet and surrounding amphipathic α-helices. In many VWA domains a well-conserved metal ion dependent adhesion site (MIDAS) motif is involved in ligand binding [21]. The MIDAS motif is composed of five amino acid residues (DXSXSXnTXnD) present in three different loops at the C-terminal end of the β-sheet, the top face of the domain. Many VWA domains, e.g. the matrilin-3 VWA domain, contain an imperfect MIDAS motif where single residues are substituted by similar amino acids. The N- and C-termini are in close proximity at the bottom face of the domain where the structure is often stabilized by a disulfide bridge formed by the two terminal cysteine residues. An exception is the collagen VI α3N5 domain that has N- and C-termini at opposite sides of the domain [22].
The structures of several VWA domains have been solved, with the best characterized being the integrin I domains and the complement factor VWA domains (for review see [23]). The VWA domains of collagens and matrilins form two neighboring phylogenetic subbranches [24], but no VWA domain structure of the matrilin subbranch is known. The identity between the VWA domains of matrilins is highest between matrilin-1 VWA1 and matrilin-3 VWA1 (62.6%) and lowest between matrilin-1 VWA1 and matrilin-2 VWA2, as well as between matrilin-4 VWA1 and matrilin-4 VWA2 or matrilin-1 VWA2 (all 37.2%). The identity between matrilin VWA domains and the previously structurally characterized homologs is lower and only the identities between the matrilin-1 VWA2 domain and the VWA domain (I-domain) of the α subunit of integrin α1β1 (31.4%) and between the matrilin-2 VWA1 the N5 domain of the collagen VI α3 chain (30.1%) is higher than 30%. The recombinant expression of single matrilin VWA domains in eukaryotic cells has been described [5–7]. However, in this study we took a comprehensive approach to the recombinant expression of functional VWA domains of all matrilins to enable studies of structure and interactions by X-ray crystallography and nuclear magnetic resonance (NMR). The focus was on expression in Escherichia coli as this would allow the incorporation of isotope-labeled amino acids, needed for NMR studies, and was expected to give high protein yields. However, we found that bacterially expressed matrilin VWA domains, unlike VWA domains from some other proteins, differ in their solubility and functional properties from such domains expressed in eukaryotic cells.
Materials and methods
Protein expression and purification
All constructs were generated by PCR using the primers listed in Supplemental Table 1. The primers introduced sequences for restriction sites that were used for digestion of the PCR product and subsequent ligation into a modified pET 15b expression vector (Novagen) for expression in E. coli or a modified pCEP-Pu-vector [25] for expression in human 293EBNA cells. Both vectors contain an N-terminal One-STrEP-tag. The pCEP-Pu-vector in addition carries a BM40 signal sequence to allow secretion of the recombinant protein. For the expression of a codon-optimized VWA domain the vector pJexpress (DNA2.0) was used. Constructs encoding fusion proteins with the B1 domain from streptococcal protein G and with the chaperone SlyD were generated by inserting the PCR-fragment into the vector GEV2 [26] or a modified SlyD fusion expression vector [27].
The matrilin constructs for bacterial expression were used for transformation of E. coli BL21 Rosetta™ cells (Novagen). Single colonies were cultured overnight at 37 °C in 10 ml LB-medium and 50 μg/ml ampicillin for selection. One liter LB-medium (or M9-minimal medium containing 15N-ammonium chloride and 13C-glucose for isotope labeling) was inoculated with the overnight culture. The cultures were grown with a flask shaking rate of 220 rpm in the presence of 50 μg/ml ampicillin to A600 = 0.4−0.6 at 37 °C. After adding isopropyl-β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM, the cultures were incubated overnight at 28 °C. The harvested cells were then resuspended in 50 ml of the buffer needed for subsequent experiments and lysed by sonication. After centrifugation at 15,000 rpm for 20 min, the supernatant was used for affinity purification.
The construct for eukaryotic expression was used for transfection of human 293EBNA cells (Invitrogen) using the FuGENE HD transfection reagent (Promega) according to the manufacturer’s instructions. 293EBNA cells transfected with constructs encoding COMP T3 + TC domains [5] or the decorin core protein lacking its glycosaminoglycan attachment site in a His-tag containing vector were kindly provided by Frank Zaucke. Transfected cells were selected with puromycin (0.5 μg/ml). Cells were cultured in DMEM/F-12 + GlutaMAX-I cell culture medium (Gibco) containing 10% (v/v) fetal calf serum (FCS).
Bacterial supernatant or cell culture medium was applied to a StrepTactin column (IBA). Subsequently the column was washed with buffer containing the appropriate additives (see Table 2). 10 mM Tris–HCl, 150 mM NaCl, pH 7.4, or 20 mM Hepes, 150 mM NaCl, 2 mM MgCl2, pH 7.4, was used when the protein was expressed for circular dichroism spectroscopy or surface plasmon resonance experiments, respectively. For NMR experiments 50 mM KH2PO4/K2HPO4, 150 mM NaCl, pH 7.4, was used or, when a lower conductivity was needed, 20 mM Bis–Tris, 150 mM KCl, pH 7.0. For elution of the recombinant protein 2.5 mM desthiobiotin was added to the washing buffer. His-tag containing proteins were loaded on a TALON metal affinity resin column (Clontech), washed with Tris buffered saline (TBS) and eluted with a serial dilution of imidazole in TBS (5–300 mM).
Table 2.
Buffer additives, temperature conditions and constructs used in attempts to increase the solubility of the matrilin-4 VWA2 domain.
| Buffer additives | 50 mM Arginine + glutamic acid | [37] | + |
| binding partner COMP T3 + TC (molar ratio 1:1) | [31] | + | |
| 5 mM Iodoacetamide | + | ||
| 0.06% Polyethylene glycol (PEG)-400 und PEG-4000 | [31] | + | |
| 1 mM N-ethylmaleimide | + | ||
| 0.5 M Trehalose | [38] | + | |
| 2 M Urea | [39] | + | |
| 5 mM CHAPS | [31] | − | |
| 0.5% Citraconic acid anhydride | − | ||
| 1% Cyclodextrin | iFOLD | − | |
| 0.5 M and 1 M NDSB-256 | iFOLD | − | |
| 5 mM and 10 mM Dithiothreitol | [31] | − | |
| 10% Glycerol | [40] | − | |
| 0.5 M Lactose | [31] | − | |
| 0.1% and 1% Octyl glucoside | [31] | − | |
| 1.5 M Sorbitol | iFOLD | − | |
| Temperature | Purification at 4 °C | − | |
| Purification at 23 °C | − | ||
| Purification at 37 °C | − | ||
| Heat shock at 42 °C (prior to adding IPTG) | − | ||
| Constructs | Matrilin-4 VWA2 without cysteines | − | |
| Fusion protein with the chaperone SlyD | [27] | − | |
| Fusion protein with the B1 domain of the streptococcal protein G | [26] | Protein in inclusion bodies | |
+ = protein solubility increased, − = protein solubility not increased, iFOLD = iFOLD Protein Refolding System 2 (Novagen).
Protein solubility testing
To test protein solubility, buffer additives were introduced in the washing and elution buffer during the affinity purification of the matrilin-4 VWA2 domain. After elution, the protein was stored overnight at 4 °C, 23 °C or 37 °C and time was measured until the protein visibly precipitated. If the protein did not precipitate overnight, it was concentrated using an Amicon Ultra-4 Centrifugal Filter Unit (Millipore) to 2–10 mg/ml. Subsequently, time was measured until visible protein precipitation occurred. An increase in solubility was noted if the time until precipitation increased.
SDS–polyacrylamide gel electrophoresis
Affinity-purified proteins were subjected to SDS–polyacrylamide gel electrophoresis on a 15% (w/v) polyacrylamide gel and stained with colloidal Coomassie staining solution [28].
Mass spectrometry
The MALDI-TOF measurement was carried out by the Central Bioanalytic Core Unit in the Center for Molecular Medicine Cologne, University of Cologne. The protein was desalted on a C4 Zip Tip (Millipore) and measured in Super DHB (a mixture of 2,5-dihydroxybenzoic acid (2,5-DHB) and 2-hydroxy-5-methoxybenzoic acid) at a mass range of 10–80 kDa.
Circular dichroism spectroscopy
Circular dichroism spectra were recorded in a Jasco J-715 spectropolarimeter using a thermostated 1 mm path length quartz cell (Hellma). Proteins were dissolved in 10 mM Tris, 150 mM NaCl, pH 7.4, at a concentration of 0.2 mg/ml. Far-ultraviolet spectra (180–250 nm) were recorded at 20 °C. The buffer background was subtracted and the data converted to mean molar residue ellipticity.
Surface plasmon resonance spectroscopy
All measurements were performed on a BIAcore 2000 instrument at 25 °C. COMP T3 + TC and the decorin core protein were immobilized on a CM5 chip. The chip surface was activated by a 1:1 mixture of 100 mM N-hydroxysuccinimide (NHS) and 400 mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC). Immobilization was carried out at a flow rate of 5 μl/min for 20 min in 25 mM sodium acetate, pH 4.5, or pH 5.0. The remaining free NHS esters were inactivated with 1 M ethanolamine–HCl, pH 8.5. Non-bound proteins were removed with 2 M NaCl in 25 mM sodium acetate, pH 5.0. For kinetic measurements the analyte was passed over the chip surface in a serial dilution in running buffer (10–3000 nM). The analyte was injected at a flow rate of 30 μl/min with an association period of 300 s and a dissociation period of 500 s. Remaining bound proteins were removed with 2 M NaCl in running buffer. The baseline resulting from passing the analyte through a blank flow cell was subtracted from the experimental data.
Nuclear magnetic resonance
A 15N 13C labeled 100 μM mouse matrilin-4 VWA2 domain was prepared in in 20 mM Bis–Tris, 150 mM KCl, 1 mM NEM, pH 7 and a 15N 13C labeled 300 μM zebrafish matrilin-4 VWA2 domain was prepared in 20 mM Bis–Tris, 150 mM KCl, pH 7, respectively. NMR experiments with the murine domain were performed at 25 °C on a Varian Inova 600 MHz (1H) instrument equipped with a z-gradient triple resonance room temperature probe. 2D 1H–15N and 1H–13C HSQC spectra incorporating gradient enhanced coherence selection and water flip back were recorded with acquisition times of 63 ms (t1, 15N) and 64 ms (t2, NH) and 16 ms (t1, 13C) and 51 ms (t2, H), respectively, using standard pulse sequences as implemented in BioPack (Varian, Palo Alto, US). NMR-experiments with the zebrafish VWA domain were performed at the Medical Research Council in Mill Hill (London, UK) at a Bruker Avance 700 MHz (1H) instrument equipped with a z-gradient cryoprobe. 2D 1H–15N HSQS spectra were recorded with acquisition times 104 ms (1H) and 47 ms (15N).
Data were processed using water suppression and mild resolution enhancement using NMRPipe [29] and were analyzed with NMRView [30].
Results
Expression and stability testing of matrilin VWA domains
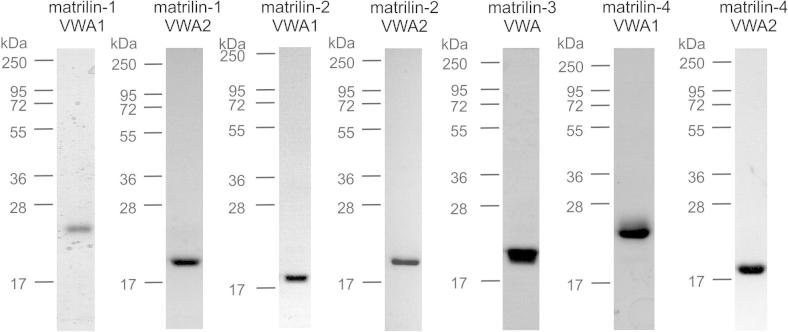
To be able to study structure and interactions of the matrilin VWA domains, we systematically optimized the recombinant expression of these. For use with methods that require high protein concentrations or isotope labeling, e.g. X-ray crystallography and nuclear magnetic resonance (NMR), the proteins were expressed in a bacterial system (Table 1). Constructs encoding all murine matrilin VWA domains were expressed in E. coli Rosetta™ cells. After cultivation, VWA domain expressing cells were resuspended in the required buffer and sonicated. After centrifugation, the supernatant was used for affinity purification of the VWA domain via a One-STrEP-tag (Fig. 1). However, bacterially expressed and purified matrilin VWA domains always precipitated when the protein concentration was raised to 2 mg/ml or higher. The highest solubility was achieved for the matrilin-3 VWA1 and the matrilin 4 VWA2 domain.
Table 1.
List of experiments performed with bacterially expressed proteins.
| VWA domain construct | Performed experiments |
|---|---|
| Matrilin-1 VWA1a | Solubility testing |
| Matrilin-1 VWA2a | Solubility testing, CD |
| Matrilin-2 VWA1a | Solubility testing, CD |
| Matrilin-2 VWA2a | Solubility testing |
| Matrilin-3 VWAa | Solubility testing, CD, NMR |
| Matrilin-4 VWA1a | Solubility testing, CD |
| Matrilin-4 VWA2a | Solubility testing, CD, SPR, NMR, X-ray crystallography |
| Matrilin-4 VWA2a without cysteine residues | Solubility testing |
| Matrilin-4 VWA2a fused to SlyD | Solubility testing |
| Matrilin-4 VWA2a fused to B1 domain of streptococcal protein G | Solubility testing |
| Matrilin-3a VWAb | Solubility testing |
| Matrilin-4 VWA1b | Solubility testing |
| Matrilin-4 VWA2b | Solubility testing, CD, NMR, X-ray crystallography |
Mouse.
Zebrafish.
Fig. 1.
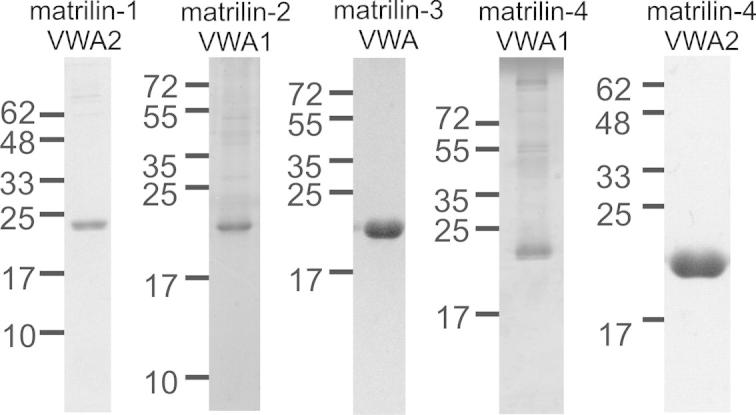
Purified murine matrilin VWA domains expressed in E. coli. After expression, the VWA domains were affinity purified via a One-STrEP-tag. The elution fraction was subjected to electrophoresis on a SDS–polyacrylamide gel (15%) and stained with Coomassie. Matrilin-1 VWA1 and matrilin-2 VWA2 could not be expressed in sufficient amounts.
To increase solubility, buffer additives were tested by introducing these in the washing and elution buffers during the affinity purification of the matrilin-4 VWA2 domain, chosen as a prototype for solubility testing. The agents used were detergents (e.g. CHAPS and octyl glucoside), osmolytes (e.g. amino acids, polyols and sugars) and alkylating agents (NEM and iodoacetamide) (overview see [31]). Some of these additives increased the solubility of the matrilin-4 VWA2 domain while others did not (Table 2). The highest solubility was achieved after adding 1 mM N-ethylmaleimide (NEM). However, even in the presence of solubility increasing additives the VWA domain eventually precipitated, although at a later time point.
To determine if temperature influences the solubility, the matrilin-4 VWA2 domain was purified and stored at different temperatures (Table 2). Purification and storage at 4 °C or at 37 °C did not improve the solubility compared to room temperature. A heat shock was applied for 30 min prior to induction of protein expression to release chaperones that should facilitate folding. However, the solubility of the matrilin-4 VWA2 domain was not increased. A fusion protein of the matrilin-4 VWA2 domain and the chaperone SlyD was generated to facilitate proper folding of the domain and thereby decreasing aggregation [27]. This fusion protein was however only poorly expressed. A second fusion protein that connects matrilin-4 VWA2 to the B1 domain of the streptococcal protein G [26] to increase solubility yielded more protein in inclusion bodies.
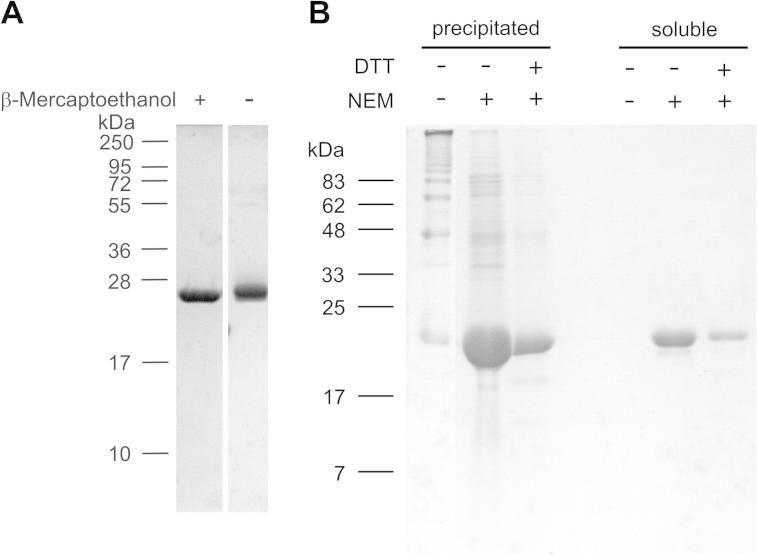
All matrilin VWA domains contain two cysteine residues that may form disulfide bonds to stabilize the structure. Such proteins often migrate differently on SDS-polyacrylamide gels under reducing and non-reducing conditions. However, the matrilin-4 VWA2 domain expressed in E. coli showed similar migration under both conditions (Fig. 2A). The disulfide bonds may not be correctly formed during protein expression in bacteria. To test this possibility, precipitated material was subjected to SDS–polyacrylamide gel electrophoresis under non-reducing conditions (Fig. 2B). A pattern with many bands representing higher aggregates was obtained. The presence of NEM or DTT in the buffer used for purification markedly decreased the proportion of aggregates. Remaining soluble material gave a single band representing the monomeric domain. To eliminate all disulfide exchange, a construct was generated for expression of a matrilin-4 VWA2 domain that lacks the flanking cysteine residues. However, the resulting protein showed an increased tendency for aggregation and precipitation. Thus, the cysteine residues are necessary for protein stability, while disulfide bond exchange enhances protein aggregation which also leads to precipitation.
Fig. 2.
Analysis of disulfide bonding of the matrilin-4 VWA2 domain expressed in E. coli. (A) The matrilin-4 VWA2 domain was subjected to electrophoresis on a 15% (w/v) SDS–polyacrylamide gel with and without prior reduction with β-mercaptoethanol. (B) Matrilin-4 VWA2 was purified without buffer additives and in the presence of 1 mM NEM or 1 mM NEM and 10 mM DTT, respectively. After visible protein precipitation in the elution fractions, precipitated and remaining soluble protein were subjected to SDS–PAGE without reduction on a 15% polyacrylamide gel and stained with Coomassie. Left: molecular mass marker in kDa.
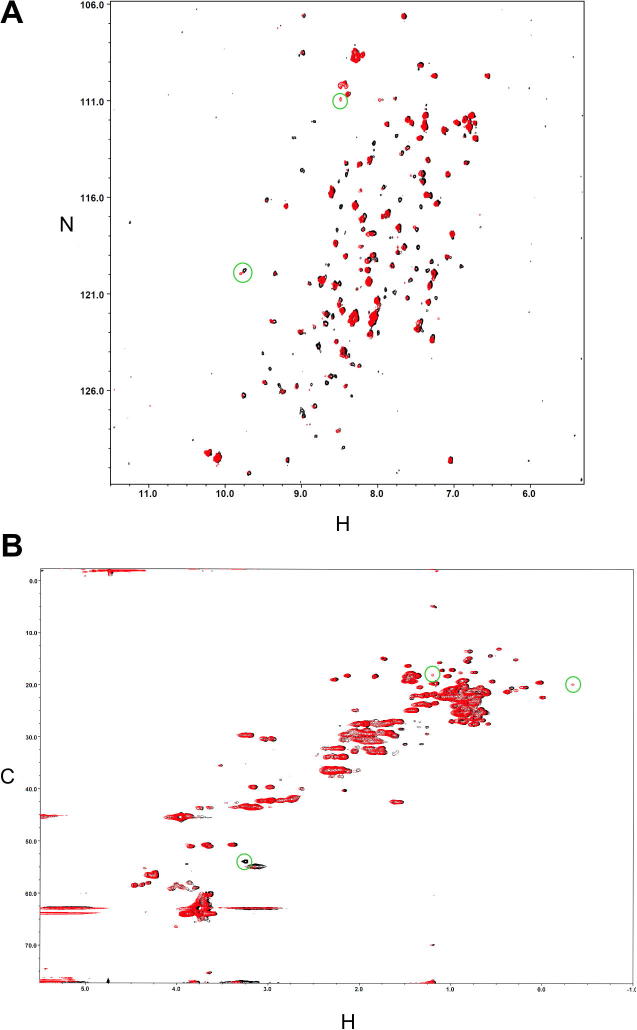
The matrilin VWA domains showed decreased solubility in the presence of EDTA. Divalent cations may bind to the MIDAS motif and cause a conformational change of the VWA domain that favors solubility. To test for divalent cation binding, 1H–15N-HSCQ- and 1H–13C-HSCQ-spectra were recorded of a 15N–13C-labeled matrilin-4 VWA2 domain in the presence of EDTA or Mg2+ (Fig. 3). Both spectra were well dispersed, indicating a distinct folding of the protein. Most of the resonances remained in the same position in both conditions with only a small number of peaks that were exclusively seen in one of the two conditions. This indicates the overall structure of the protein stays the same and that Mg2+ binding only causes a local structural perturbation. It was also noted that overall the peak intensities in the presence of EDTA are somewhat lower leading to an apparent loss of resonances at the chosen plotting threshold in Fig. 3. There are a number of possible contributing factors such as enhanced aggregation or line broadening due to increased internal μs-ms dynamics of the protein in the presence of EDTA. Also the addition of EDTA triggered a slight protein precipitation during the NMR measurement. Both effects are consistent with a less stable protein in the absence of Mg2+ indicating that divalent cations can stabilize the structure of this VWA domain.
Fig. 3.
Comparison of the solution structures of bacterially expressed mouse matrilin-4 VWA2 domain in the presence of Mg2+ or EDTA by high resolution NMR. 1H–15N-HSQC-spectra (A) and 1H–13C-HSQC-spectra (B) of the 15N–13C-labeled mouse matrilin-4 VWA2 domain ((100 μM, 20 mM Bis–Tris, 150 mM KCl 1 mM NEM, at pH = 7 and at a temperature of 25 °C)) were recorded after adding 2 mM MgCl2 (black) or 2 mM EDTA (red), respectively. Chemical shift differences in the spectra are highlighted with green circles. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Comparison of different matrilin VWA domains
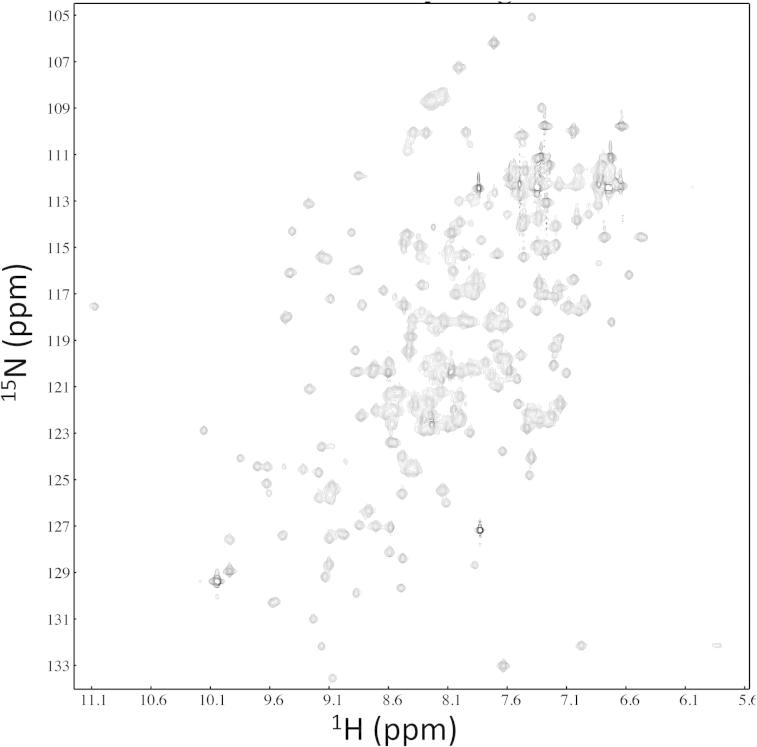
In addition to the murine matrilin VWA domains the zebrafish matrilin-3a and matrilin-4 VWA domains were expressed in E. coli. Interestingly, the highest protein solubility among all matrilin VWA domains expressed in bacteria was obtained for a matrilin-4 VWA2 domain from zebrafish. This domain could be concentrated to 10 mg/ml without visible protein precipitation. A codon-optimized version (DNA2.0) of the zebrafish matrilin-4 VWA2 domain was affinity purified from E. coli supernatants by use of a One-STrEP-tag (Fig. 4). The protein was mainly in the monomeric form and only slight amounts of dimers and higher aggregates were detected. A high resolution 1H–15N HSQC spectrum of the 15N–13C labeled domain was recorded to assess the folding state (Fig. 5). The spectrum is well dispersed and has lineshapes consistent with the size of a monomeric protein sample. Discounting NH2 resonances arising from side chains, approximately 180 NH resonances could be observed. While this is broadly consistent with the number of signals expected for this construct there are still in the order of 10% of signals missing, suggesting that there are some regions of the protein that have broad lineshapes potentially due to rapid solvent exchange or μs–ms time scale dynamics. Overall the spectrum suggests that this domain is folded. However, collection of triple resonance data for backbone assignments was impeded by the limited lifetime at the required sample concentrations for these experiments. The zebrafish matrilin-4 VWA2 domain was also used for crystallization attempts, but the structure could not be determined due to lacking crystallization.
Fig. 4.
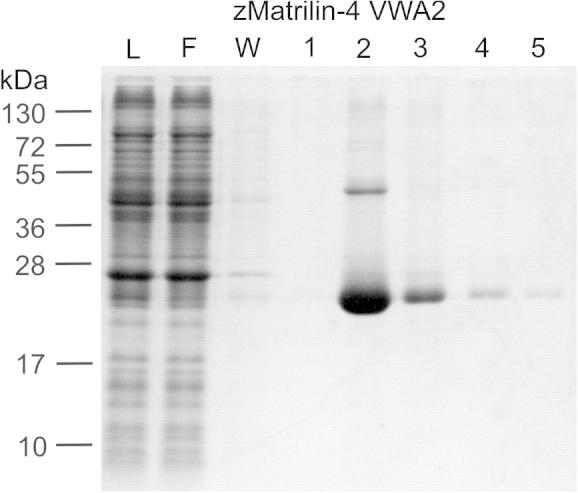
Purification of the bacterially expressed zebrafish matrilin-4 VWA2 domain. The matrilin-4 VWA2 domain was affinity purified via a One-STrEP-tag. Bacterial lysate (L), flow through (F), washing fraction (W) and the first four elution fractions (1–4) were subjected to a SDS–polyacrylamide gel (15%) without reduction and stained with Coomassie. Left: molecular mass marker in kDa.
Fig. 5.
Solution structure of the bacterially expressed zebrafish matrilin-4 VWA2 domain. 1H–15N HSQC spectrum of bacterially expressed zebrafish matrilin-4 VWA2 domain from zebrafish (M4zA2) recorded at 25 °C in 20 mM Bis–Tris, 150 KCl, pH 7, at 600 MHz. The spectrum is well dispersed and shows approximately the correct number of NH peaks indicating a well-folded and structurally homogeneous protein construct.
To determine if bacterially expressed murine matrilin VWA domains have similar properties as eukaryotically expressed domains, all such domains were also expressed in human 293EBNA cells (Fig. 6). All these VWA domains were better soluble than the corresponding bacterially expressed proteins. For example, at 2 mg/ml the eukaryotically expressed murine matrilin-4 VWA2 domain remained in solution whereas the murine protein expressed in E. coli precipitated. A proper formation of the disulfide bond in the protein expressed in human cells is indicated by a mobility shift in SDS–polyacrylamide gels after reduction (Fig. 7) that is in contrast to the VWA domain expressed in E. coli.
Fig. 6.
Purified murine matrilin VWA domains expressed in 293EBNA cells. After expression, the VWA domains were affinity purified via a One-STrEP-tag and subjected to electrophoresis on a SDS-polyacrylamide gel (15%) and stained with Coomassie.
Fig. 7.
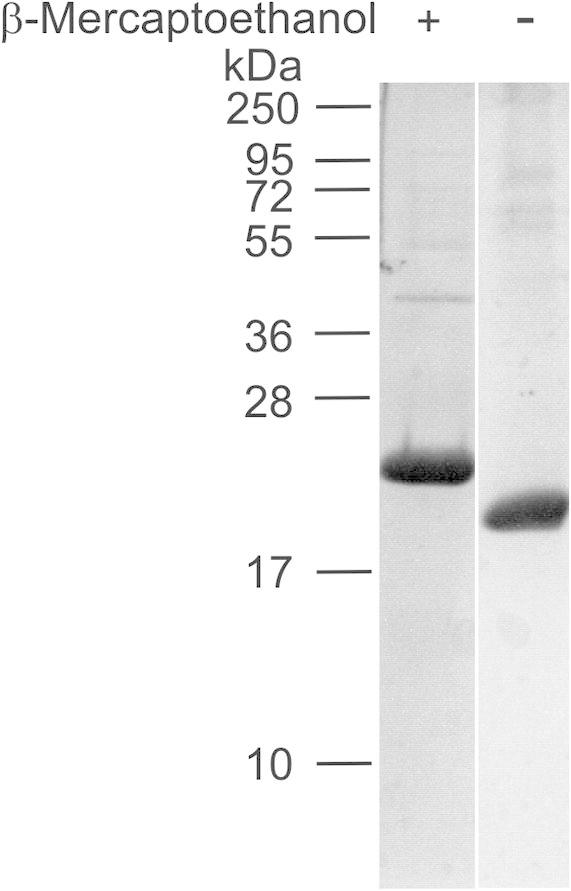
Analysis of the disulfide bond present in matrilin-4 VWA2 domain expressed in human 293EBNA cells. The matrilin-4 VWA2 domain was affinity-purified via a One-STrEP-tag and subjected to electrophoresis on a 15% (w/v) SDS–polyacrylamide gel with and without prior reduction with β-mercaptoethanol. Proteins were stained with Coomassie. Left: molecular mass marker in kDa.
Comparison of folding and interactions of matrilin VWA domains expressed in E. coli and eukaryotes
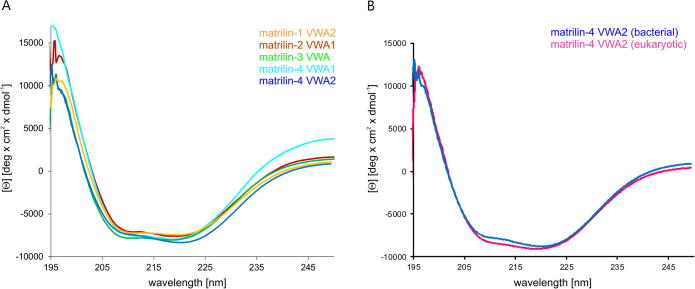
To determine if the bacterially expressed matrilin VWA domains were properly folded, circular dichroism (CD) measurements were performed (Fig. 8A). Matrilin-1 VWA1 and matrilin-2 VWA2 could not be purified in sufficient amounts. All other matrilin VWA domains showed far-UV spectra that are consistent with that of a typical Rossmann fold [32] with minima at 209 and 222 nm. Thus, the bacterially expressed protein domains contain the secondary structure elements present in a Rossmann fold. Together with the well-dispersed NMR spectra of the matrilin-4 VWA2 domain shown above (Fig. 3) the data suggest that the bacterially expressed domains overall have the correct three dimensional structure. Direct comparison of the CD-spectra of matrilin-4 VWA2 domains expressed either in E. coli or in human 293EBNA cells showed a great similarity (Fig. 8B). Thus, the bacterially expressed matrilin VWA domains adopt a native like structure.
Fig. 8.
Analysis of the folding of bacterially expressed matrilin VWA domains by circular dichroism spectroscopy. (A) Far-UV CD spectroscopy of bacterially expressed matrilin-1 VWA2 (orange), matrilin-2 VWA1 (red), matrilin-3 (green), matrilin-4 VWA1 (light blue) and matrilin-4 VWA2 (dark blue) shows spectra typical for the Rossmann fold occurring in VWA domains. (B) The spectra of bacterially (blue) und eukaryotically (pink) expressed matrilin-4 VWA2 domain are closely similar indicating the same content of different secondary structure elements. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
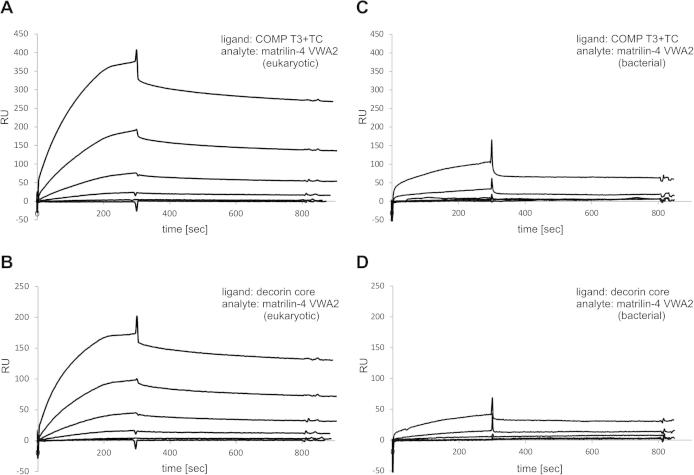
The matrilin-4 VWA2 domain was investigated more closely by comparing the binding properties of bacterially and eukaryotically expressed constructs to COMP [5,6] and decorin [5,33], two well characterized binding partners of matrilins. Surface plasmon resonance measurements were performed using COMP T3 + TC, representing the matrilin binding domains of COMP [5], and the decorin core protein as ligands (Fig. 9). While the matrilin-4 VWA2 domain expressed in 293EBNA cells strongly interacted with both binding partners, the VWA domain expressed in E. coli showed much weaker binding.
Fig. 9.
Binding of bacterially and eukaryotically expressed matrilin-4 VWA2 to COMP T3 + TC and decorin core protein as measured by surface plasmon resonance. The eukaryotically expressed matrilin-4 VWA2 domain binds in a concentration dependent manner to the immobilized COMP T3 + TC domains (A) and to the immobilized decorin core protein (B). The binding of the bacterially expressed domain to both ligands was markedly reduced (C and D). Matrilin-4 VWA2 was used as an analyte in a serial dilution (10 nM, 30 nM, 100 nM, 300 nM, 1000 nM, 3000 nM). RU = resonance units.
It can be excluded that glycosylation, only present on a eukaryotic protein, is necessary for the binding. Mass spectrometry (MALDI-TOF) showed that the measured mass (21121.0 m/z) was consistent with the calculated mass (21122.8 m/z) of the unmodified VWA domain.
Discussion
Disulfide exchange leads to stability problems after expression in E. coli
Matrilin VWA domains were initially expressed in bacteria as several successfully crystallized VWA domains from other proteins have earlier been expressed in E. coli. This is a cost-effective system, which yields large amounts of protein in a short time and allows isotope labeling. However, bacterially expressed matrilin VWA domains showed a stronger tendency to aggregation and a lower solubility, although CD and NMR spectroscopy indicated that they were appropriately folded. For structure determination by either X-ray crystallography or NMR protein concentrations of several mg/ml are needed. This could not overcome by using a number of stabilizing buffer additives, as the bacterially expressed matrilin VWA domains were not stable in solution. Furthermore, engineering solubility enhancing tags onto the proteins designed did not yield stable solutions of matrilin VWA domains.
All matrilin VWA domains expressed in human cells showed an increased stability compared to the same protein domains expressed in E. coli. The eukaryotic expression of recombinant extracellular proteins allows representative posttranslational modifications, such as glycosylation and it has indeed been shown that the presence of glycan chains on the VWA domains of matrilin-1 affects their structure and thermal stability [7]. However, this does not explain the better solubility of the eukaryotically expressed matrilin-4 VWA2 domain that was characterized in this study, as it does not carry such modifications. Nevertheless, in a functional assay only the eukaryotically expressed VWA domains interacted strongly with their physiological binding partners, whereas the bacterially expressed domains displayed much weaker binding. The problems encountered when expressing this domain in bacteria probably arose from incorrectly formed disulfide bonds. In several VWA domains disulfide bonds serve to stabilize the structure [15], but the matrilin-4 VWA2 domain was shown to form insoluble aggregates due to disulfide exchange. The extent of shuffling could be decreased by the addition of alkylating agents like N-ethylmaleimide or iodoacetamide. However, even with these additives stable protein solutions could not be obtained. Eukaryotic expression allowed a correct formation of disulfide bonds indicated by a mobility shift in SDS–polyacrylamide gels after reduction and yielded protein solutions that were stable for several weeks. Both eukaryotic and bacterial cells have reducing conditions in their cytoplasm, but eukaryotic cells allow the formation of disulfide bonds under oxidative conditions in the ER. As bacterial cells lack this compartment, recombinant proteins with disulfide bonds in their native structure are often misfolded [34]. With the exception of the VWA domains of the anthrax toxin receptor and the von Willebrand factor A1 and A3 domains, those VWA domains that were previously expressed in bacteria and crystallized carried no terminal cysteine residues or contained either no cysteine residue or a single one in the middle of the protein sequence. In contrast, all matrilin VWA domains contain cysteine residues at the C- and N-terminus, which most likely form an intramolecular disulfide bond. A construct of the matrilin-4 VWA2 domain that lacks the terminal cysteine residues showed a strong tendency to aggregation. Thus, the disulfide bond is important for stabilization of the fold. Hence, the data suggest that structural studies of matrilin VWA domains would benefit from an eukaryotic expression system. Still crystallization attempts with the eukaryotically expressed matrilin-4 VWA2 and matrilin-3 VWA domains were not yet successful (data not shown) indicating that correct disulfide bond formation may not be sufficient for crystal formation and may be a result of an intrinsic tendency of these domains to aggregate. Unspecific “stickiness” seems to be more pronounced for matrilin VWA domains than for other VWA domains. In electron microscopy the VWA domains of matrilin-1 and -2 show a self-interaction [35,36]. This may well be a result of the role of the matrilins as adaptor proteins in the extracellular matrix and the fact that their VWA domains provide the sites of interaction. Aggregation of matrilin VWA domains may be physiologically relevant and could contribute to the supramolecular assembly of matrilins.
Acknowledgements
The research leading to these results has received funding from the European Community’s Seventh Framework Programme under grant agreement n°602300 (SYBIL) and by the Boehringer Ingelheim Fonds. J.M.W. would like to thank the Wellcome Trust for NMR equipment funds at the Southampton NMR center (Grant No. 090658/Z/09/Z) and Stuart Findlow for technical support. We thank the MRC for providing access to the Biomedical NMR Centre at Mill Hill (Grant No. U117533887). We would also like to acknowledge the use of the crystallization facilities at the Centre for Biological Sciences at the University of Southampton in the Life Science Building.
Footnotes
Abbreviations used: CD, circular dichroism; COMP, cartilage oligomeric matrix protein; DTT, dithiothreitol; EBNA, Epstein Barr nuclear antigen; EDC, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide; ER, endoplasmic reticulum; HSQC, heteronuclear single quantum coherence; MALDI-TOF, matrix-assisted laser desorption/ionization-time of flight; MED, multiple epiphyseal dysplasia; MIDAS, metal ion dependent adhesion site; NEM, N-ethylmaleimide; NHS, N-hydroxysuccinimide; NMR, nuclear magnetic resonance; VWA, von Willebrand factor A.
Appendix A. Supplementary data
References
- 1.Klatt A.R., Becker A.-K.A., Neacsu C.D., Paulsson M., Wagener R. The matrilins: modulators of extracellular matrix assembly. Int. J. Biochem. Cell Biol. 2011;43:320–330. doi: 10.1016/j.biocel.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 2.Paulsson M., Heinegård D. Matrix proteins bound to associatively prepared proteoglycans from bovine cartilage. Biochem. J. 1979;183:539–545. doi: 10.1042/bj1830539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piecha D., Wiberg C., Mörgelin M., Reinhardt D.P., Deák F., Maurer P. Matrilin-2 interacts with itself and with other extracellular matrix proteins. Biochem. J. 2002;367:715–721. doi: 10.1042/BJ20021069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiberg C., Klatt A.R., Wagener R., Paulsson M., Bateman J.F., Heinegård D. Complexes of matrilin-1 and biglycan or decorin connect collagen VI microfibrils to both collagen II and aggrecan. J. Biol. Chem. 2003;278:37698–37704. doi: 10.1074/jbc.M304638200. [DOI] [PubMed] [Google Scholar]
- 5.Mann H.H., Ozbek S., Engel J., Paulsson M., Wagener R. Interactions between the cartilage oligomeric matrix protein and matrilins. Implications for matrix assembly and the pathogenesis of chondrodysplasias. J. Biol. Chem. 2004;279:25294–25298. doi: 10.1074/jbc.M403778200. [DOI] [PubMed] [Google Scholar]
- 6.Fresquet M., Jowitt T.A., Ylöstalo J., Coffey P., Meadows R.S., Ala-Kokko L. Structural and functional characterization of recombinant matrilin-3 A-domain and implications for human genetic bone diseases. J. Biol. Chem. 2007;282:34634–34643. doi: 10.1074/jbc.M705301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fresquet M., Jowitt T.A., Stephen L.A., Ylöstalo J., Briggs M.D. Structural and functional investigations of Matrilin-1 A-domains reveal insights into their role in cartilage ECM assembly. J. Biol. Chem. 2010;285:34048–34061. doi: 10.1074/jbc.M110.154443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otten C., Hansen U., Talke A., Wagener R., Paulsson M., Zaucke F. A matrilin-3 mutation associated with osteoarthritis does not affect collagen affinity but promotes the formation of wider cartilage collagen fibrils. Hum. Mutat. 2010;31:254–263. doi: 10.1002/humu.21182. [DOI] [PubMed] [Google Scholar]
- 9.Nicolae C., Ko Y.-P., Miosge N., Niehoff A., Studer D., Enggist L. Abnormal collagen fibrils in cartilage of matrilin-1/matrilin-3-deficient mice. J. Biol. Chem. 2007;282:22163–22175. doi: 10.1074/jbc.M610994200. [DOI] [PubMed] [Google Scholar]
- 10.Chapman K.L., Mortier G.R., Chapman K., Loughlin J., Grant M.E., Briggs M.D. Mutations in the region encoding the von Willebrand factor A domain of matrilin-3 are associated with multiple epiphyseal dysplasia. Nat. Genet. 2001;28:393–396. doi: 10.1038/ng573. [DOI] [PubMed] [Google Scholar]
- 11.Mostert A.K., Dijkstra P.F., Jansen B.R.H., van Horn J.R., de Graaf B., Heutink P. Familial multiple epiphyseal dysplasia due to a matrilin-3 mutation: further delineation of the phenotype including 40 years follow-up. Am. J. Med. Genet. A. 2003;120A:490–497. doi: 10.1002/ajmg.a.20034. [DOI] [PubMed] [Google Scholar]
- 12.Stefánsson S.E., Jónsson H., Ingvarsson T., Manolescu I., Jónsson H.H., Olafsdóttir G. Genomewide scan for hand osteoarthritis: a novel mutation in matrilin-3. Am. J. Hum. Genet. 2003;72:1448–1459. doi: 10.1086/375556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borochowitz Z.U., Scheffer D., Adir V., Dagoneau N., Munnich A., Cormier-Daire V. Spondylo-epi-metaphyseal dysplasia (SEMD) matrilin 3 type: homozygote matrilin 3 mutation in a novel form of SEMD. J. Med. Genet. 2004;41:366–372. doi: 10.1136/jmg.2003.013342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fresquet M., Jackson G.C., Loughlin J., Briggs M.D. Novel mutations in exon 2 of MATN3 affect residues within the alpha-helices of the A-domain and can result in the intracellular retention of mutant matrilin-3. Hum. Mutat. 2008;29:330. doi: 10.1002/humu.9518. [DOI] [PubMed] [Google Scholar]
- 15.Cotterill S.L., Jackson G.C., Leighton M.P., Wagener R., Mäkitie O., Cole W.G. Multiple epiphyseal dysplasia mutations in MATN3 cause misfolding of the A-domain and prevent secretion of mutant matrilin-3. Hum. Mutat. 2005;26:557–565. doi: 10.1002/humu.20263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otten C., Wagener R., Paulsson M., Zaucke F. Matrilin-3 mutations that cause chondrodysplasias interfere with protein trafficking while a mutation associated with hand osteoarthritis does not. J. Med. Genet. 2005;42:774–779. doi: 10.1136/jmg.2004.029462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leighton M.P., Nundlall S., Starborg T., Meadows R.S., Suleman F., Knowles L. Decreased chondrocyte proliferation and dysregulated apoptosis in the cartilage growth plate are key features of a murine model of epiphyseal dysplasia caused by a matn3 mutation. Hum. Mol. Genet. 2007;16:1728–1741. doi: 10.1093/hmg/ddm121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nundlall S., Rajpar M.H., Bell P.A., Clowes C., Zeeff L.A.H., Gardner B. An unfolded protein response is the initial cellular response to the expression of mutant matrilin-3 in a mouse model of multiple epiphyseal dysplasia. Cell Stress Chaperones. 2010;15:835–849. doi: 10.1007/s12192-010-0193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whittaker C.A., Hynes R.O. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol. Biol. Cell. 2002;13:3369–3387. doi: 10.1091/mbc.E02-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rossmann M.G., Moras D., Olsen K.W. Chemical and biological evolution of nucleotide-binding protein. Nature. 1974;250:194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- 21.Lee J.O., Rieu P., Arnaout M.A., Liddington R. Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18) Cell. 1995;80:631–638. doi: 10.1016/0092-8674(95)90517-0. [DOI] [PubMed] [Google Scholar]
- 22.Becker A.-K.A., Mikolajek H., Paulsson M., Wagener R., Werner J.M. A structure of a collagen VI VWA domain displays N and C termini at opposite sides of the protein. Struct. Lond. Engl. 2014;1993(22):199–208. doi: 10.1016/j.str.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Springer T.A. Complement and the multifaceted functions of VWA and integrin I domains. Struct. Lond. Engl. 2006;1993(14):1611–1616. doi: 10.1016/j.str.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumánovics A., Lindahl K.F. G7c in the lung tumor susceptibility (Lts) region of the Mhc class III region encodes a von Willebrand factor type A domain protein. Immunogenetics. 2001;53:64–68. doi: 10.1007/s002510100297. [DOI] [PubMed] [Google Scholar]
- 25.Kohfeldt E., Maurer P., Vannahme C., Timpl R. Properties of the extracellular calcium binding module of the proteoglycan testican. FEBS Lett. 1997;414:557–561. doi: 10.1016/s0014-5793(97)01070-3. [DOI] [PubMed] [Google Scholar]
- 26.Huth J.R., Bewley C.A., Jackson B.M., Hinnebusch A.G., Clore G.M., Gronenborn A.M. Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci. Publ. Protein Soc. 1997;6:2359–2364. doi: 10.1002/pro.5560061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scholz C., Schaarschmidt P., Engel A.M., Andres H., Schmitt U., Faatz E. Functional solubilization of aggregation-prone HIV envelope proteins by covalent fusion with chaperone modules. J. Mol. Biol. 2005;345:1229–1241. doi: 10.1016/j.jmb.2004.10.091. [DOI] [PubMed] [Google Scholar]
- 28.Kang D., Gho Y., Suh M., Kang C. Highly sensitive and fast protein detection with coomassie brilliant blue in sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Bull. Korean Chem. Soc. 2002;23:1511–1512. [Google Scholar]
- 29.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 30.Johnson B.A. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 31.Bagby S., Tong K.I., Ikura M. Optimization of protein solubility and stability for protein nuclear magnetic resonance. Methods Enzymol. 2001;339:20–41. doi: 10.1016/s0076-6879(01)39307-2. [DOI] [PubMed] [Google Scholar]
- 32.Coinçon M., Heitz A., Chiche L., Derreumaux P. The betaalphabetaalphabeta elementary supersecondary structure of the Rossmann fold from porcine lactate dehydrogenase exhibits characteristics of a molten globule. Proteins. 2005;60:740–745. doi: 10.1002/prot.20507. [DOI] [PubMed] [Google Scholar]
- 33.Wiberg C., Hedbom E., Khairullina A., Lamandé S.R., Oldberg A., Timpl R. Biglycan and decorin bind close to the n-terminal region of the collagen VI triple helix. J. Biol. Chem. 2001;276:18947–18952. doi: 10.1074/jbc.M100625200. [DOI] [PubMed] [Google Scholar]
- 34.de Marco A. Strategies for successful recombinant expression of disulfide bond-dependent proteins in Escherichia coli. Microb. Cell Factories. 2009;8:26. doi: 10.1186/1475-2859-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hauser N., Paulsson M. Native cartilage matrix protein (CMP). A compact trimer of subunits assembled via a coiled-coil alpha-helix. J. Biol. Chem. 1994;269:25747–25753. [PubMed] [Google Scholar]
- 36.Piecha D., Muratoglu S., Mörgelin M., Hauser N., Studer D., Kiss I. Matrilin-2, a large, oligomeric matrix protein, is expressed by a great variety of cells and forms fibrillar networks. J. Biol. Chem. 1999;274:13353–13361. doi: 10.1074/jbc.274.19.13353. [DOI] [PubMed] [Google Scholar]
- 37.Golovanov A.P., Hautbergue G.M., Wilson S.A., Lian L.-Y. A simple method for improving protein solubility and long-term stability. J. Am. Chem. Soc. 2004;126:8933–8939. doi: 10.1021/ja049297h. [DOI] [PubMed] [Google Scholar]
- 38.Singer M.A., Lindquist S. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol. Cell. 1998;1:639–648. doi: 10.1016/s1097-2765(00)80064-7. [DOI] [PubMed] [Google Scholar]
- 39.Bhuyan A.K. Protein stabilization by urea and guanidine hydrochloride. Biochemistry (Mosc.) 2002;41:13386–13394. doi: 10.1021/bi020371n. [DOI] [PubMed] [Google Scholar]
- 40.Vagenende V., Yap M.G.S., Trout B.L. Mechanisms of protein stabilization and prevention of protein aggregation by glycerol. Biochemistry (Mosc.) 2009;48:11084–11096. doi: 10.1021/bi900649t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.