Background: The molecular mechanism by which α-conotoxin RegIIA inhibits α3β4, α3β2, and α7 nAChRs is unknown.
Results: Alanine scanning mutagenesis and molecular dynamic simulations of RegIIA revealed Asn11 and Asn12 confer improved selectivity at α3β4 nAChR.
Conclusion: We synthesized the [N11A,N12A]RegIIA analog that selectively inhibits α3β4.
Significance: These findings could be used to develop α3β4-selective drugs to treat lung cancer.
Keywords: Electrophysiology, Molecular Dynamics, Nicotinic Acetylcholine Receptors (nAChR), Oocyte, Peptides, α-conotoxin, Xenopus Oocyte, Alanine Scanning Mutagenesis
Abstract
Activation of the α3β4 nicotinic acetylcholine receptor (nAChR) subtype has recently been implicated in the pathophysiology of various conditions, including development and progression of lung cancer and in nicotine addiction. As selective α3β4 nAChR antagonists, α-conotoxins are valuable tools to evaluate the functional roles of this receptor subtype. We previously reported the discovery of a new α4/7-conotoxin, RegIIA. RegIIA was isolated from Conus regius and inhibits acetylcholine (ACh)-evoked currents mediated by α3β4, α3β2, and α7 nAChR subtypes. The current study used alanine scanning mutagenesis to understand the selectivity profile of RegIIA at the α3β4 nAChR subtype. [N11A] and [N12A] RegIIA analogs exhibited 3-fold more selectivity for the α3β4 than the α3β2 nAChR subtype. We also report synthesis of [N11A,N12A]RegIIA, a selective α3β4 nAChR antagonist (IC50 of 370 nm) that could potentially be used in the treatment of lung cancer and nicotine addiction. Molecular dynamics simulations of RegIIA and [N11A,N12A]RegIIA bound to α3β4 and α3β2 suggest that destabilization of toxin contacts with residues at the principal and complementary faces of α3β2 (α3-Tyr92, Ser149, Tyr189, Cys192, and Tyr196; β2-Trp57, Arg81, and Phe119) may form the molecular basis for the selectivity shift.
Introduction
Nicotinic acetylcholine receptors (nAChR)4 are ligand-gated ion channels expressed in the central nervous system (CNS) and peripheral nervous system (1). They are pentameric receptors composed of a combination of α subunits (α2–10) and β subunits (β2–4). Heteromeric (for example, α3β2 and α4β2) and homomeric isoforms (only α7 and α9 subunits) of nAChRs exhibit diverse structural and functional heterogeneity (2). Their physiological role in modulating pre- and post-synaptic transmission in the CNS, and visceral and somatic sensory transmission in the peripheral nervous system are well understood (3, 4).
nAChRs have been implicated in the pathophysiology of a number of health conditions, including Alzheimer disease, schizophrenia, tobacco addiction, and lung cancer (5). Our understanding of isoform distribution and neurophysiological roles of individual receptor subtypes in these conditions is limited by a lack of adequate isoform-specific probes (6).
An initial report suggesting that nAChRs may regulate cancer cell growth (7) was followed by a number of studies investigating the role of nAChRs in cancer development and progression (reviewed in Ref. 8). Two studies identified tobacco-specific nitrosamines as potent nAChR agonists and inhibitors of cancer cell apoptosis (9). In addition, genome-wide studies identified associations between lung cancer and several single nucleotide polymorphisms within the gene cluster encoding the α3, α5, and β4 nAChR subunits (10). Additionally, activation of the α3β4 nAChR, a predominant subtype expressed in sympathetic and parasympathetic neurons of mammalian autonomic ganglia (11–13), is known to be associated with nicotine addiction and drug abuse (14, 15). However, our understanding of the physiological relationship is limited.
Conotoxins are bioactive peptides isolated from the venom of cone snails of the genus Conus (16). α-Conotoxins are a specific class of short, disulfide-constrained peptides with a conserved Cys-framework, CCXnCXmC. Xn and Xm represent the number of amino acids and are used to subclassify peptides such as α4/3, α4/4, and α4/7, which specifically target various nAChR isoforms (17). Thus they represent excellent molecular probes to elucidate the physiological roles of nAChR subtypes in normal and disease states (18). The structural and functional properties of a number of α-conotoxins have been characterized (16–19). Although α-conotoxins such as ImII and RgIA exhibit selective inhibitory activity at α7 (20) and α9α10 (21) nAChR subtypes, respectively, most known α-conotoxins target multiple nAChR subtypes (16, 17). α-Conotoxin AuIB is the only known peptide that selectively targets the α3β4 nAChR subtype albeit with low potency (IC50 = 2.5 μm) (22, 23).
Mutagenesis experiments have become an important tool to improve selectivity and potency of receptor inhibitors (see for example, Ref. 24). Previously we reported the discovery and isolation from Conus regius venom of the α4/7-conotoxin RegIIA (25). RegIIA potently inhibits ACh-evoked currents of α3β4, α3β2, and α7 nAChR isoforms. Given the pathophysiological association of the α3β4 nAChR subtype with various disorders such as lung cancer and nicotine addiction, we have now employed mutagenesis with the aim of improving the selectivity profile of RegIIA. Using alanine scanning mutagenesis and modeling studies, we also identified critical α-conotoxin RegIIA residues that interact with α3β2, α3β4, and α7 nAChR ACh-binding sites.
EXPERIMENTAL PROCEDURES
Peptide Synthesis
All of the peptide analogs were assembled on rink amide methylbenzhydrylamine resin (Novabiochem; 0.7 mmol g−1) using o-benzotriazole-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HBTU)-mediated manual solid-phase peptide synthesis, with an in situ neutralization procedure for N-(9-fluorenyl)methoxycarbonyl (Fmoc) chemistry. Each cycle consisted of Fmoc deprotection with 20% piperidine in dimethylformamide, followed by Fmoc amino acid coupling using HBTU and N,N-diisopropylethylamine in dimethylformamide. A 2-fold excess of Fmoc amino acids was used in the coupling reactions. All peptides were synthesized in globular conformation (I-III and II-IV disulfide connectivity) through incorporation of Fmoc-Cys acetamidomethyl (Acm)–OH at positions 2 and 8 of the amino acid sequence (I-III disulfide bond). The efficiency of the coupling reactions were checked using the Kaiser Ninhydrin test.
Peptides were cleaved from the dried resin (0.4 g) by treatment with 100 ml of TFA, triisopropylsilane, and water as scavengers (95:2.5:2.5, TFA:triisopropylsilane:water, v/v/v). The reaction was allowed to proceed at room temperature (20–23 °C) for 2.5 h. The TFA was then evaporated, and the peptide was precipitated with ice-cold ether, filtered, dissolved in 50% buffer A/B (buffer A: H2O, 0.05% TFA; buffer B: 90% CH3CN, 10% H2O, 0.045% TFA) and lyophilized. Crude peptides were purified by reversed phase-high performance liquid chromatography (RP-HPLC) on a Phenomenex C18 column using a gradient of 0–80% buffer B for 80 min and the elutant was monitored at 215/280 nm. Unless otherwise stated, the same conditions were used in subsequent purification steps. Electrospray-mass spectroscopy (ESI-MS) confirmed the molecular mass of the fractions collected.
Fractions displaying the correct molecular mass for linear peptide were pooled and lyophilized for oxidation. Linear peptides were oxidized in two steps. First, they were dissolved in 0.1 m NH4HCO3 (pH 8.2) at a concentration of 0.3 mg/ml. Stirring overnight at room temperature formed the II-IV disulfide bond. The acetamidomethyl (Acm) protecting groups were stable under these conditions. Second, iodine (0.01–0.1 m) was added for 5 min at 37 °C, and excess iodine was destroyed by adding sodium ascorbate. This formed the I-III disulfide bond. The oxidized peptides were purified by RP-HPLC using a gradient of 0–80% buffer B over 160 min. Analytical RP-HPLC and ESI-MS confirmed the purity and molecular mass of the synthesized peptides.
NMR Spectroscopy
Nuclear magnetic resonance (NMR) data for all peptides were recorded on Bruker Avance 500- and 600-MHz spectrometers, with samples dissolved in 90% H2O, 10% D2O. Two-dimensional NMR experiments included TOCSY (total correlation spectroscopy) and NOESY (nuclear Overhauser effect spectroscopy) recorded at 280 K. Spectra were analyzed using Topspin 1.3 (Bruker) and Sparky software. Unless specified, spectra were recorded at pH 3.5.
Two-electrode Voltage Clamp Electrophysiological Recordings of nAChRs Expressed in Xenopus Oocytes
Stage V-VI oocytes were harvested from mature female Xenopus laevis anesthetized with 0.1% tricaine, following protocols approved by the RMIT Animal Ethics Committee. RNA and Xenopus oocytes were prepared, then nAChR subtypes were expressed in oocytes as described previously (26). Briefly, cDNAs encoding the rat α3, β2, and β4 subunits, and human α7 subunit were subcloned into oocyte expression vector pT7TS and the α6/α3 chimera plasmid was kindly provided by Dr. Michael McIntosh (University of Utah). mRNA corresponding to each subunit was prepared using the mMESSAGE mMACHINE Kit (Ambion®, Invitrogen). Oocytes were injected with 5 ng of cRNA for each subtype in a 1:1 ratio. However, for the expression of α6/α3 receptors, 15–20 ng of cRNA for each subunit was injected. The injected oocytes were then incubated for 2–5 days at 18 °C in ND96 buffer (96 mm NaCl, 2 mm KCl, 1 mm CaCl2, 1 mm MgCl2, and 5 mm HEPES, pH 7.4) supplemented with 50 mg/liter of gentamicin and 100 μg/units/ml of penicillin-streptomycin, before recording. Membrane currents from Xenopus oocytes were recorded at room temperature (20–23 °C), using a bath solution of ND96 as described above. A two-electrode voltage clamp (virtual ground circuit) with either a GeneClamp 500B amplifier (Molecular Devices, Sunnyvale, CA) or an automated work station with eight channels in parallel, including drug delivery and online analysis (OpusXpressTM 6000A, Axon Instruments Inc.) was used.
All recordings were made using voltage recording and current-injecting electrodes were pulled from borosilicate glass (GC150T-7.5, Harvard Apparatus Ltd., Holliston, MA) and had resistances of 0.3–1.5 MΩ when filled with 3 m KCl. Oocytes were voltage clamped at a holding potential of −80 mV. During recordings, oocytes were perfused continuously at a rate of 2 ml/min, with ACh (200 μm for α7 and 50 μm for all other nAChR subtypes) applied for 2 s at 2 ml/min. A 180- to 240-s interval washout period was used between every ACh application. The inhibitory effect of each peptide at the respective concentration represents the ratio of ACh-evoked peak amplitude evoked before and following 300-s incubation with peptide. Data were filtered at 10 Hz and sampled at 500 Hz (27).
Data Analysis
Concentration-response curves for antagonists were fitted by unweighted nonlinear regression to the logistic equation,
where Ex is the response, X is the antagonist concentration, Emax is the maximal response, nH is the slope factor, and IC50 is the antagonist concentration giving 50% inhibition of the maximal response. All electrophysiological data were pooled (n = 4–8 for each data point) and represent arithmetic mean ± S.E. of the fit. Computation was done using GraphPad Prism 6.03 (GraphPad Software, Inc., La Jolla, CA).
Homology Modeling
Homology models of the extracellular ligand-binding domain of rat (α3)2(β2)3 and (α3)2(β4)3 nAChRs bound to RegIIA or [N11A,N12A]RegIIA were constructed using the crystallographic coordinates of Aplysia californica acetylcholine-binding protein, co-crystallized with the double mutant α-conotoxin PnIA[A10L,D14K] (Protein Data Bank accession code 2BR8) (28) as a template. RegIIA and double mutant peptides were modeled bound to the α(+)β(−) receptor binding sites using the geometry of the PnIA mutant in the acetylcholine-binding protein crystal structure as a template. This provided a suitable 4/7 α-conotoxin-bound conformation of the receptor for subsequent molecular dynamics (MD) simulations and analyses.
Rat α3, β2, and β4 sequences were obtained from the Swiss-Prot database (codes P04757, P12390, and P12392, respectively), and aligned with the template sequence using the ClustalW server. BLOSUM was used as the scoring matrix. Using the multiple alignment as input, 10 models each of the RegIIA-α3β2, RegIIA-α3β4, [N11A,N12A]RegIIA-α3β2, and [N11A,N12A]RegIIA-α3β4 complexes were generated using Modeler9v6 (29). The top ranking models were selected and validated using PROCHECK (30). MD simulations were then done on the top model of each of the four receptor complexes. The overall fold of each homology model complex is qualitatively similar.
MD Simulations
Each of the four receptor complexes was put in a separate cubic simulation box, with an edge length of 100 × 100 × 100 Å, and solvated with 27,229 (for α3β2) or 27,194 (α3β4) TIP3P water molecules. To neutralize charge and maintain an ionic concentration of ∼150 mm, 115 (for α3β2) or 88 (α3β4) Na+ and 70 Cl− ions were added to the solvent. All simulations were performed using GROMACS version 4.5.5 (31, 32), with CHARMM27 force-field version 2.0 (with cmap) (33).
Before undergoing MD simulations, the complexes were energy minimized using the steepest descent algorithm and an energy gradient convergence criterion of 0.01 kcal/mol/Å. All MD simulations were performed using a constant particle number, pressure, and temperature ensemble, with temperature maintained at 300 K using the v-rescale temperature coupling algorithm (34), and pressure maintained at 1 bar using the Parrinello-Rahman pressure coupling algorithm (35). Time steps of 2 fs were used to integrate all simulations.
Solvent equilibration simulations of 100 ps lengths were performed. In these simulations, the non-hydrogen atoms of the receptor and toxins were positionally restrained so the solvent and ions could undergo motions to reach equilibrium from an initially energetically unfavorable state, without disturbing the protein. Subsequent simulations of all four complexes were then performed with all atoms free of the system to undergo dynamics.
To improve conformational sampling, we performed 10 independent simulations for each complex, using different random seeds to assign initial particle velocities. Each simulation was performed for 20 ns (i.e. 200 ns of trajectory per complex). To reduce bias from initial homology model conformations, all analyses were performed on the final 10 ns of the trajectories.
Unless indicated, all data were taken as an average over the 10 independent simulations. Molecular graphics were produced using VMD version 1.9.2 (36). All analyses were performed using a combination of VMD, GROMACS analysis software suite, and in-house scripts.
The interatomic contact difference plot (ΔN) was calculated by determining the total number of toxin contacts within 4.5 Å of each receptor residue for wild-type and mutant RegIIA, averaged over 10 independent simulations and both α(+)β(−) interfaces (20 data points per receptor residue). To obtain ΔN values, the wild-type contact number was subtracted from that of the double mutant.
RESULTS
Synthesis of RegIIA Analogs
We used an alanine scan mutagenesis approach to elucidate the molecular mechanism underlying inhibition of the α3β4 nAChR subtype by RegIIA. This technique is well established, and in conjunction with atomistic MD simulations, has enabled significant advances in molecular pharmacology (see for example Refs. 37 and 38). α-Conotoxin RegIIA belongs to the α4/7 subclass having the conserved Cys-framework CCXnCXmC. Due to its I-III and II-IV disulfide connectivity, native RegIIA exhibits a classical helical, globular structure (25). This globular conformation balances the shape, charge, and polarity of the peptide. As α-conotoxins contain four cysteines there are two additional disulfide isomers that can form during oxidative folding or disulfide reshuffling, the ribbon (I-IV and II-III disulfide bonds) or bead (I-II and III-IV disulfide bonds) isomers. Changes in the disulfide connectivity are reflected in the peptide conformation and can have a significant impact on α-conotoxin potency and specificity at nAChRs (39, 40).
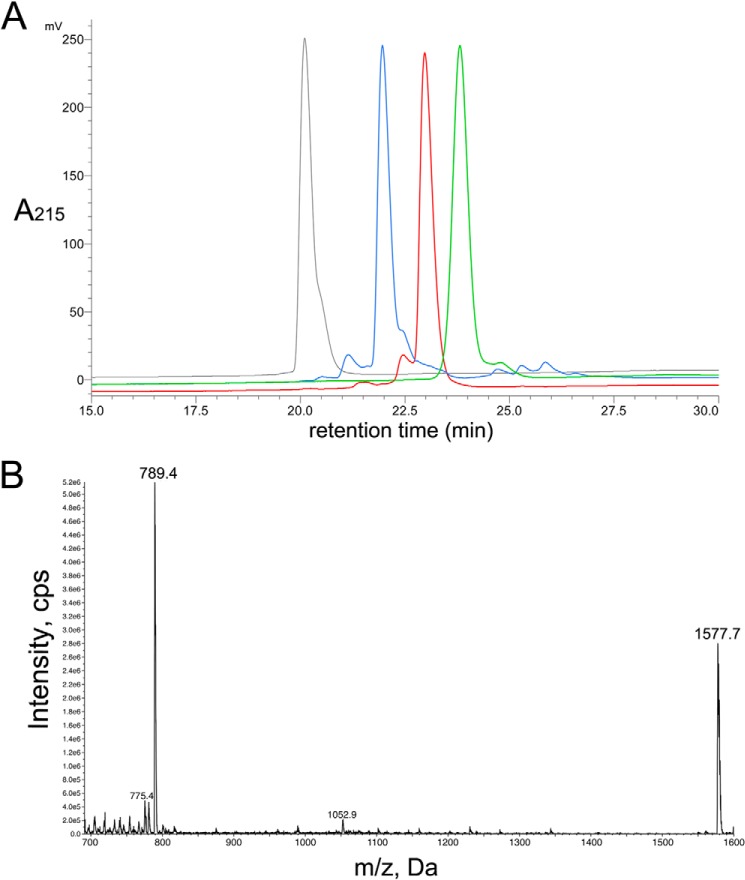
We used regioselective disulfide bond formation with Acm-protected cysteine residues incorporated at positions 1 and 3, and a two-step oxidation procedure, to produce the alanine mutant peptides in a globular conformation (I-III and II-IV disulfide bonds). The two-step oxidation process was confirmed by RP-HPLC, ESI-MS (Fig. 1), and two-dimensional NMR. Fig. 2 show the negative 1H shift values between amino acid positions 3 and 7 indicating the presence of an α-helix secondary structure.
FIGURE 1.
HPLC and ESI-MS profile of [N11A,N12A]RegIIA. A, an overlay of the analytical RP-HPLC traces for RegIIA (black), reduced and Acm-protected [N11A,N12A]RegIIA (red), oxidized and Acm-protected [N11A,N12A]RegIIA (blue), and [N11A,N12A]RegIIA (green). Peptides were analyzed with a Phenomenex C18 Jupiter 300 column (150 × 2 mm) using a solvent gradient from 5 to 50% buffer B for 35 min. B, ESI-MS data of [N11A,N12A]RegIIA.
FIGURE 2.
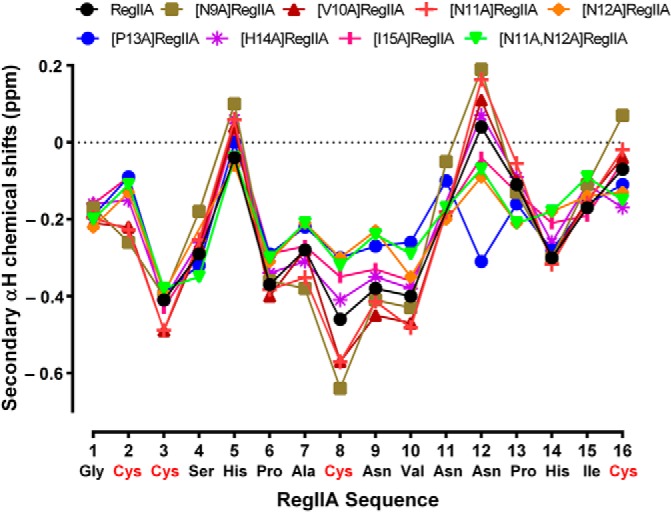
Secondary αH shifts of RegIIA and alanine analogs at 280 K. The secondary shift values of the peptide backbone (residues 2–12) for most of the analogs correlate well with RegIIA.
Alanine Mutagenesis Reveals Key Residues for RegIIA-nAChR Activity
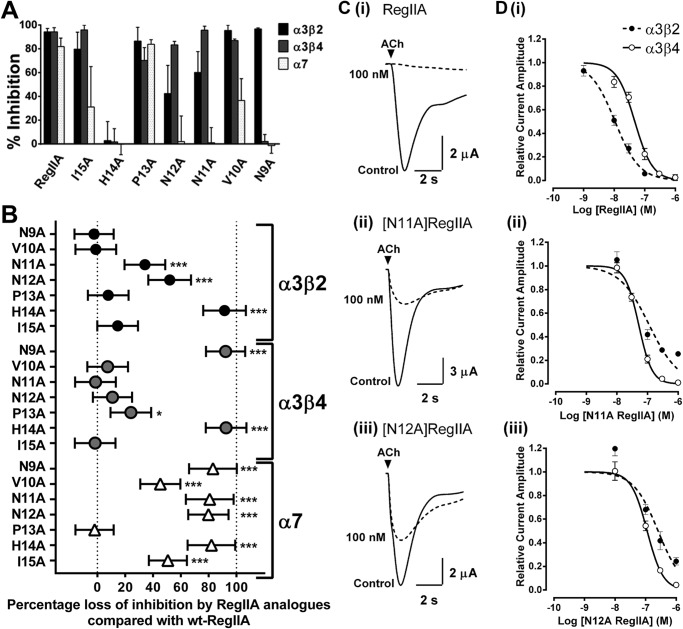
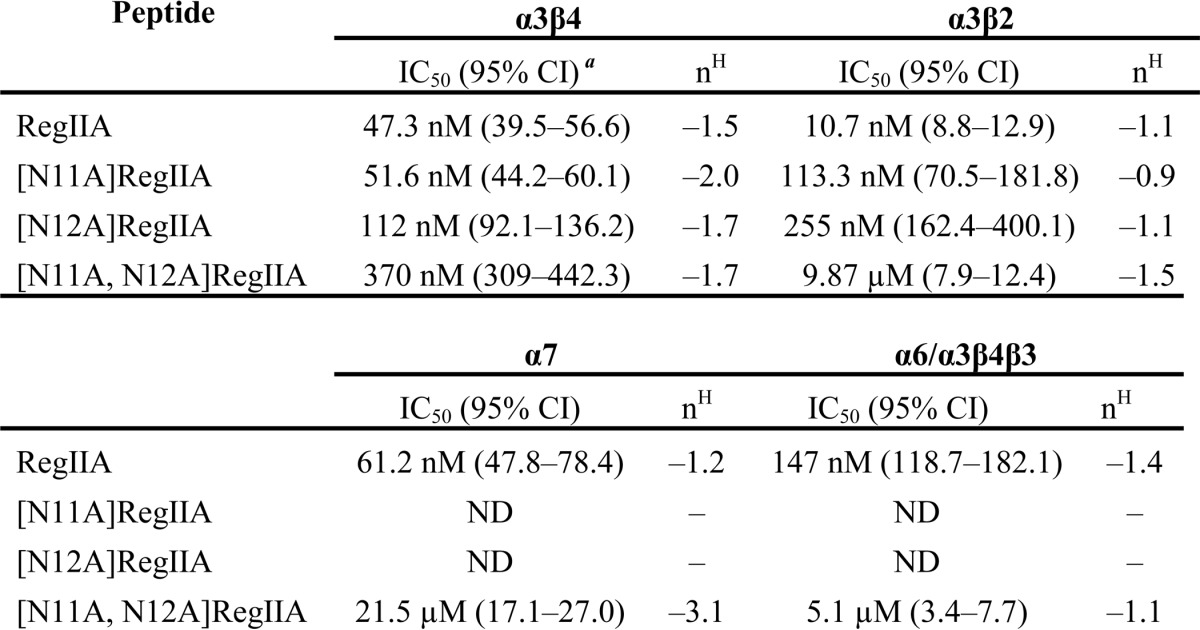
To understand the structure-activity relationship of RegIIA at nAChRs, we tested loop 2 alanine analogs of RegIIA (at a concentration of 300 nm) on α3β2, α3β4, and α7 nAChR subtypes (Fig. 3A). [H14A]RegIIA showed complete loss of activity at all three nAChR subtypes. All RegIIA analogs, except [P13A]RegIIA, showed significantly reduced or complete loss in activity at the α7 nAChR subtype (Fig. 3B). [N9A]RegIIA was the only analog that showed no activity at the α3β4 and α7 nAChR subtype, whereas maintaining inhibition at α3β2 subtype. In contrast, we observed improved selectivity of [N11A]RegIIA and [N12A]RegIIA for α3β4 with significantly reduced (∼50%) inhibition of the α3β2 nAChR subtype (Fig. 3, B and C). This was apparent from the shift in concentration-response curves for [N11A]RegIIA and [N12A]RegIIA (Fig. 3D). IC50 values for [N11A]RegIIA and [N12A]RegIIA at the α3β2 nAChR subtype were 113.3 (95% Cl 70.6–181.8; nH = −0.9) and 255 nm (95% Cl 162.4–400.1; nH = −1.1), respectively. At the α3β4 nAChR subtype, the IC50 values were 51.6 nm (95% Cl 44.2–60.1; nH = −2.0) and 112 nm (95% Cl 92.1–136.2; nH = −1.7), respectively (Table 1).
FIGURE 3.
Inhibition of nAChR subtypes expressed in Xenopus oocytes by RegIIA and alanine analogs. A, bar graph of inhibition of nAChR subtypes by RegIIA and its analogs. B, two-way analysis of variance scatter plot illustrating the loss of activity of RegIIA analogs (300 nm) relative to wild-type RegIIA at various nAChR subtypes. [H14A]RegIIA completely lost its activity at α3β2, α3β4, and α7 nAChRs. [N9A]RegIIA was more selective for the α3β2 subtype than RegIIA. [N11A]RegIIA and [N12A]RegIIA selectivity for the α3β4 nAChR subtype significantly improved. All analogs, except [P13A]RegIIA, significantly lost activity at the α7 nAChR subtype. ***, p < 0.001; *, p < 0.05; n = 4–6. C, superimposed traces showing inhibition of α3β2 nAChR-mediated ACh-evoked currents by 100 nm RegIIA (i), [N11A]RegIIA (ii), and [N11A]RegIIA (iii). D, concentration-response curves for RegIIA (i), [N11A]RegIIA (ii) and [N12A]RegIIA (iii) inhibition of the α3β4 (black line, open symbols) and α3β2 nAChR subtypes (dash line, closed symbols). [N11A]RegIIA and [N12A]RegIIA shifted the curve to the right for the α3β2 nAChR subtype, giving an IC50 value of 116 and 278 nm, respectively. All data represents mean ± S.E., n = 4–6.
TABLE 1.
RegIIA and analog inhibition of nAChR subtypes
IC50 values with 95% confidence interval (CI). Hill slope (nH) obtained from concentration-response curves for RegIIA and analogues at α3β2, α3β4, α7 and α6/α3β4β3 nAChR subtypes. All data represent mean of n = 4–6 experiments. ND, not determined.
[N11A,N12A]RegIIA: a Selective α3β4 nAChR Antagonist
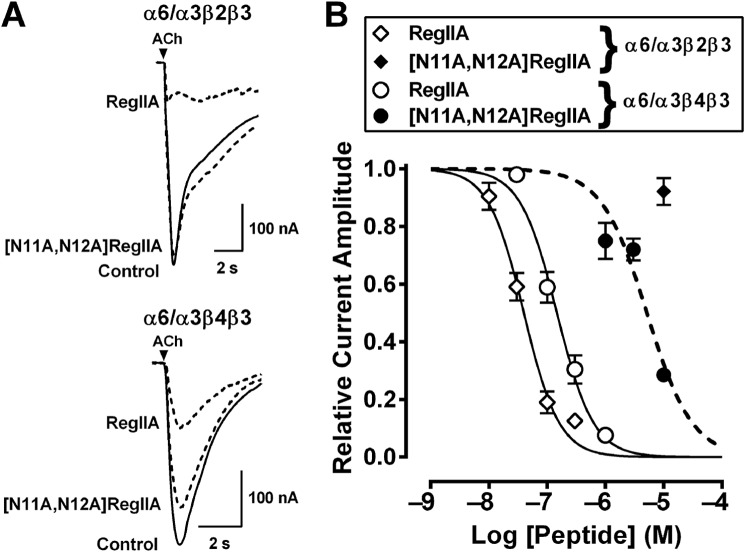
We synthesized the double mutant [N11A,N12A]RegIIA to better understand the cumulative effect of the two residues on nAChR activity. [N11A,N12A]RegIIA inhibited the α3β4 nAChR subtype with an IC50 of 370 nm (95% Cl 3.09–442.3; nH = −1.7), a 7-fold less potency than native RegIIA (Fig. 4). However, at the α3β2 and α7 nAChR subtypes, potency decreased by 1,000- (IC50 = 9.9 μm) and 360-fold (IC50 = 21.5 μm), respectively (Table 1). This indicates a decrease in selectivity for the α3β2 and α7 nAChR subtypes. Furthermore, the RegIIA double mutation led to a significant loss in activity at α6-containing receptors (Fig. 5). In comparison, wild-type RegIIA potently inhibited α6/α3β2β3 and α6/α3β4β3 nAChR subtypes, with IC50 values of 40 and 147 nm, respectively (Fig. 5). [N11A,N12A]RegIIA had ∼36-fold less potency at the α6/α3β4β3 nAChR subtype than RegIIA (Fig. 5 and Table 1), and no activity at the α6/α3β2β3 nAChR subtype at a concentration of 10 μm (Fig. 5). Inhibition of α3β4 by 100 nm RegIIA and [N11A,N12A]RegIIA was reduced by greater than 40%, with a 10-fold (30 to 300 μm) increase in the ACh concentration (data not shown). This shift in the concentration-response relationship clearly indicates that RegIIA and [N11A,N12A]RegIIA are competitive antagonists of α3β4 nAChRs.
FIGURE 4.
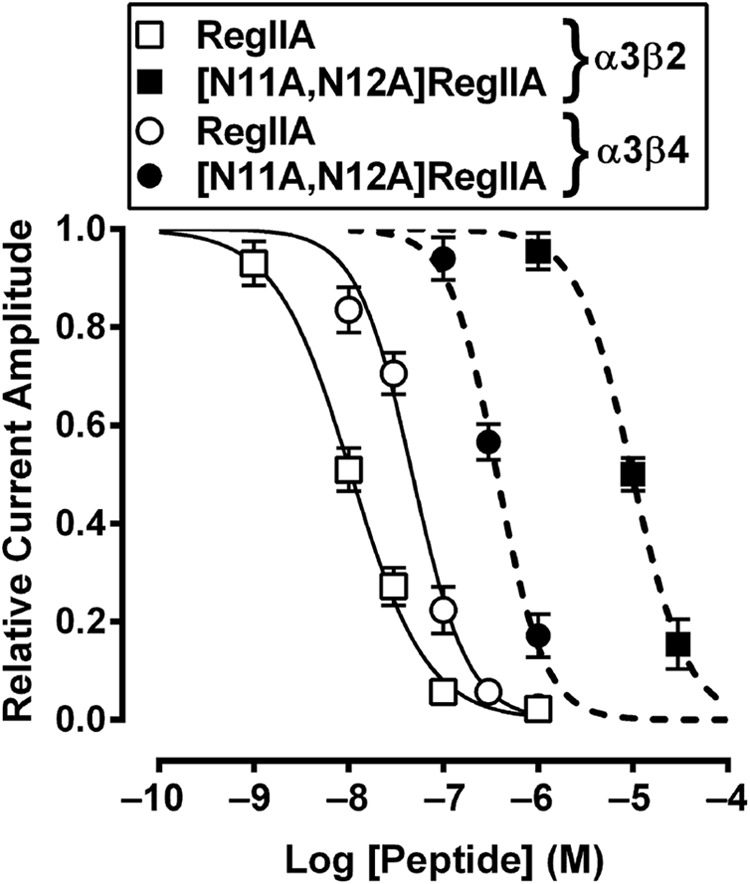
[N11A,N12A]RegIIA inhibition of α3β2 and α3β4 nAChR compared with wild-type RegIIA inhibition of these receptors. Concentration-response curve for [N11A,N12A]RegIIA gave IC50 values of 370 nm and 9.9 μm at α3β4 (▴) and α3β2 (■) receptors, respectively, with an approximate 27-fold increased selectivity for α3β4 nAChR compared with RegIIA. Data represent mean ± S.E., n = 4–6.
FIGURE 5.
[N11A,N12A]RegIIA inhibition of α6/α3β2β3 and α6/α3β4β3 nAChR compared with wild-type RegIIA inhibition of these receptors. A, superimposed traces showing ACh-evoked current inhibition of α6/α3β2β3 and α6/α3β4β3 nAChR subtypes by RegIIA and [N11A,N12A]RegIIA. B, wild-type RegIIA was active at α6-containing receptors with IC50 values of 40 and 147 nm at α6/α3β2β3 (♢) and α6/α3β4β3 (○) receptors, respectively. Concentration-response curve for [N11A,N12A]RegIIA gave an IC50 value of 5.1 μm at α6/α3β4β3 (●), with an approximate 35-fold decrease in potency compared with RegIIA. [N11A,N12A]RegIIA showed no activity at α6/α3β2β3 (♦) when tested at a concentration of 10 μm. Data represents mean ± S.E., n = 3–6.
Key Toxin-Receptor Interactions Revealed by Homology Modeling and MD Simulations
To understand the higher selectivity of [N11A,N12A]RegIIA for α3β4 than α3β2, we have used homology models and atomistic MD simulations of wild-type RegIIA and [N11A,N12A]RegIIA bound to either α3β2 or α3β4 (Fig. 6A). The initial homology models of wild-type RegIIA bound to the two nAChR subtypes suggest that Asn11 and Asn12 of RegIIA make contact predominantly with the α3(+) face, although Asn11 also makes contact with β2-Arg81 and Lys79 (β4-Arg79 and Ile77) (Fig. 6B).
FIGURE 6.
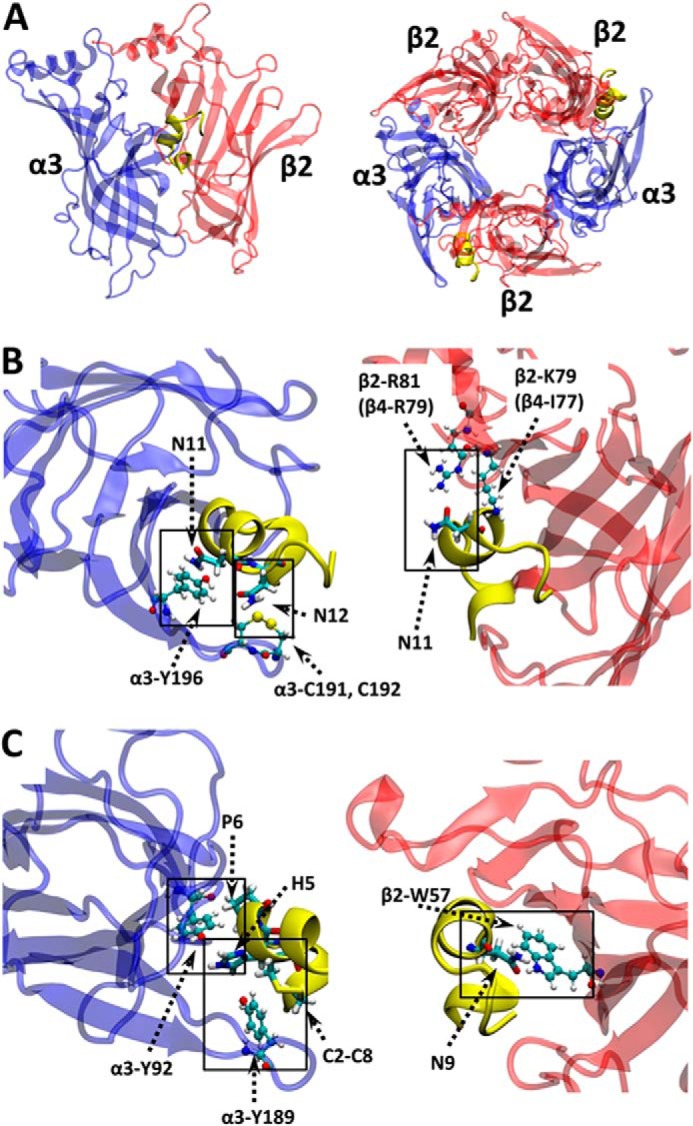
Homology model and MD simulation of RegIIA bound to α3β2. A, illustration of the initial structure (homology model) of RegIIA toxin bound to the presumed inter-subunit C-loop pockets are shown in yellow. B, MD simulation snapshot (at 20 ns) of RegIIA bound with α3β2 showing interactions between Asn11, Asn12 and receptor residues at the α3(+) and β2(−) faces; C, and of pairwise interactions that are substantially reduced at α3β2 compared with α3β4 after the [N11A,N12A] mutation. Conotoxin side chains of interest are in licorice form. Receptor side chains are shown as ball and stick. Atoms are color coded as: C = cyan, H = white, N = blue, O = red, and S = yellow.
Because the interactions between nAChR and RegIIA-Asn11/Asn12 are similar for both α3β2 and α3β4, it is difficult to rationalize the substantial selectivity change after double mutation by employing “static” homology models alone. We therefore employed MD simulations, which take into account solvent, temperature, and protein dynamics, to compare changes in atomic contacts at the wild-type and mutated receptors. In particular, we examined the effects of the [N11A,N12A] mutation on contacts at sites distant from these positions (including the β(−) face, which differs substantially between β2 and β4), which may help explain the basis of RegIIA selectivity.
Fig. 7 shows the change in number of toxin atoms (y axis) residing within 4.5 Å of the molecular surface of each receptor residue (x axis) at the α(+)β(−) interfaces (“ΔN profiles”). This is a measure of loss (negative values on the y axis) or gain (positive values) of toxin contacts as a result of the [N11A,N12A] double mutation. The ΔN profiles for α3β2 and α3β4 reveal that the double mutation generally reduces toxin-receptor contacts at α(+) face residues. However, the effect at the β(−) face is mixed, with reduced or increased number of contacts for different residues. As expected, mutation of Asn11 and Asn12 leads to decreased contact with receptor residues in the immediate proximity of these (RegIIA) positions. Contact between the Asn11 position with α3-Tyr196/β2-Arg81 (β4-Arg79), and the Asn12 position with α3-Cys192, are reduced compared with that of wild-type RegIIA (Fig. 6B).
FIGURE 7.
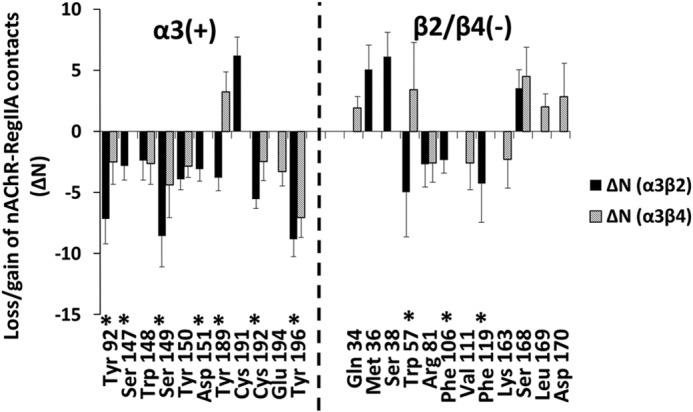
Change in number of toxin atoms (ΔN) that lie within 4.5 Å of receptor residues (x axis) due to [N11A,N12A] double mutation of RegIIA. Asterisks indicate residues with substantially fewer contacts for α3β2 compared with α3β4. Residues from the β2 subunit are shown on the x axis. Data from α3β4 is at the homologous positions. Error bars are mean ± S.E. calculated over α(+)/β(−) interfaces for 10 independent simulations (20 data points in total).
The [N11A,N12A] mutation also markedly reduces toxin-receptor contacts at regions distant from the mutation sites. Of particular interest and to explain the selectivity of the mutant for α3β4, are receptor residues with substantially fewer toxin contacts for α3β2 than α3β4 nAChR subtypes. Some of the pairwise interactions involving these residues are illustrated in Fig. 6C and are also marked with asterisks in Fig. 7. The most prominent residues include α3-Tyr92, Ser149, Tyr189, Cys192, and Tyr196 at the principal face, and β2-Trp57 (β4-Trp55) and β2-Phe119 (β4-Gln117) at the complementary face. These residues may be essential for RegIIA inhibition of α3β2. In particular, the marked loss of contact at α3-Cys192 in both α3β2 and α3β4 nAChR subtypes is consistent with similar observations from previous studies of α-conotoxin analogs and modifications. van Lierop et al. (41) showed dicarba modification of the C2-C8 disulfide bond in Vc1.1 resulted in loss of activity at α9α10. Their MD simulations showed reduced contact between the modified toxin and Cys-loop disulfide atoms. Grishin et al. (38) found [F9A]AuIB lost its activity at α3β4 with the MD simulations showing reduced contact between the toxin and Cys-loop sulfur atoms due to this mutation. Our current data supports the crucial role of Cys-loop sulfur atoms in conotoxin inhibition of neuronal nAChRs.
Double mutation also caused a loss of contact between Asn9/Pro6 and β2-Trp57, whereas slightly increasing the contact between Asn9/Pro6 and the homologous position at β4-Trp59 (Fig. 7). This, in addition to functional data for [N9A]RegIIA showing complete loss in activity at α3β4 (Fig. 3), suggests interaction between Asn9 and/β4-Trp59 might be important for inhibition of the α3β4 subtype. This loss of toxin contact at α3β2, but not α3β4, may also contribute to the marked selectivity change of the double mutant [N11A,N12A]RegIIA toward the α3β4 nAChR subtype.
DISCUSSION
Since the discovery, in the worm-hunting cone snail Conus imperialis, of α-conotoxin ImI and its action on neuronal nAChRs, numerous α-conotoxins have been identified and functionally characterized (16–19). A number of peptides have been identified from the venom of C. regius, a Western Atlantic worm-hunting cone snail species. These peptides belong to various superfamilies with eight being part of the A-superfamily that is distinguished by cysteine framework I (42). The conotoxin composition of C. regius venom is clinically relevant because it contains the α-conotoxin RegIe and RegIIA both of which have been identified as potential therapeutics. RgIA (free carboxyl C terminus form of [O6P]RegIe) selectively targets pain transmission by modulation of the α9α10 nAChR subtype and high voltage-activated N-type calcium channel currents, via GABAB receptor activation (43). α-Conotoxin RegIIA potently inhibits activity of the α3β4 nAChR subtype (25) that has been implicated in the pathophysiology of lung cancer.
RegIIA exhibits homology with a number of peptides (Table 2) having a conserved SHPA sequence in loop 1 and a NNP motif in loop 2. The Ser and Pro residues of the SHPA motif are highly conserved in peptides of different subclasses that inhibit various nAChR subtypes (Table 2). Sequence variations in loop 2 contribute to the unique nAChR subtype selectivity of α-conotoxins (44). However, the NNP motif is conserved in peptides such as α-conotoxins OmIA, EpI, PnIA, TxIA, and ArIB that specifically inhibit α3β2 and α7 nAChR subtypes (16). This observation is consistent with our finding that alanine mutation of the NNP motif of RegIIA significantly affected inhibition at α3β2 and α7 nAChR subtypes (Fig. 3A). Alanine mutations at other positions provided further information about the structure-function relationship between α-conotoxins and neuronal nAChRs. Asparagine (RegIIA position nine) to alanine mutation completely abolished inhibition of the α7 and α3β4 nAChRs.
TABLE 2.
Sequence alignment of α-conotoxins targeting various nAChR subtypes
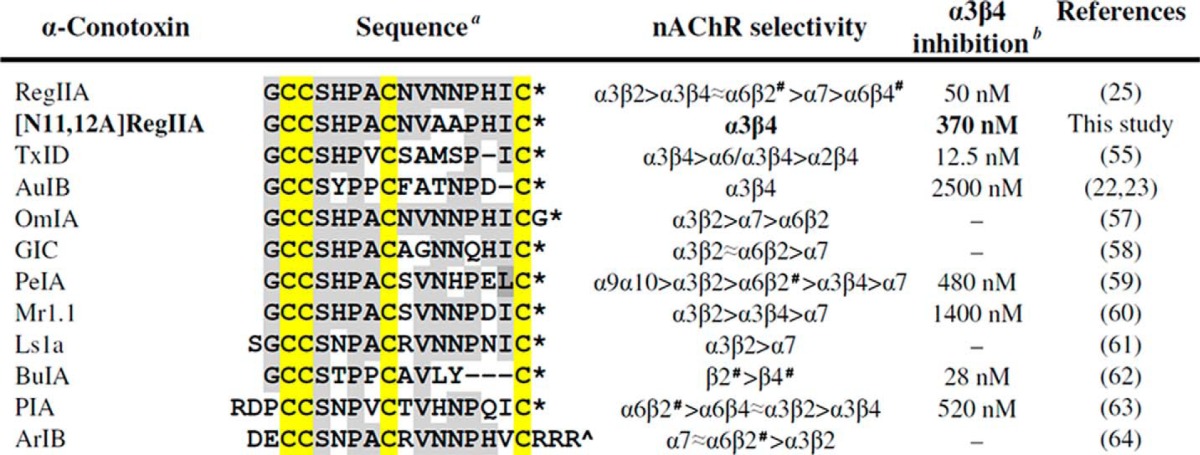
a Amino acids homologous to RegIIA are labeled with a grey background. The conserved cysteine framework is highlighted in yellow. * indicates an amidated C terminus; ^ free carboxyl C terminus; and # presence of additional subunits.
b Peptides inhibiting the α3β4 nAChR subtype and their corresponding IC50 values are shown.
The marked shifts in selectivity observed by the analogs for either α3β2 or α3β4 subtype is intriguing given that both receptors share a common principal subunit face at the presumed conotoxin binding site (Fig. 6A). Previous x-ray studies of α-conotoxin-acetylcholine-binding protein complexes, with support from synthetic analogs, have improved our understanding of ligand-receptor interactions (45–47). The extracellular N-terminal domains of the β2 and β4 nAChR subunits bind α-conotoxins and exhibit 70% sequence homology. Recent MD simulation studies also reveal well preserved structural topology of α3β2 and α3β4 nAChRs. However, the ACh-binding pocket interface between the α and β subunits was larger in α3β2 than α3β4 nAChR subtypes (48). Even though, both Asn11 and Asn12 primarily interact with the α3(+) interface, this difference could explain the shift in selectivity of [N11A] and [N12A]RegIIA for α3β4 over α3β2.
Our findings are also supported by data from a recent molecular docking study of α-conotoxin GIC with human α3β2 and α3β4 nAChR subtypes. The study revealed all three subunits have residues that interact with GIC (α3 subunit: Tyr92, Tyr150, Tyr189, and Tyr196; β2 subunit: Trp57, Val111, Phe119, and Leu121; and β4 subunit: Trp57, Ile111, Leu119 (Gln119 in rat β4) and Leu121). Interestingly, the α3-Tyr196 and β2-Phe119 residues of the α3β2-GIC model were more closely located than the α3-Tyr196 and β4-Leu119 residues of the α3β4-GIC model (48). A comprehensive receptor mutagenesis study of the various α-conotoxins (MII, GID, and PnIA) that inhibit the α3β2 nAChR subtype indicates that the β2 subunit pharmacophore comprising Thr59, Glu61, Val111, Phe119, and Leu121 residues, also has a significant role in ligand binding (49). It is important to note that, whereas both of the above studies identified similar residues interacting with the conotoxins, increasing literature on the pharmacological difference between rat and human nAChR receptors is emerging (50, 51). The current study was carried out using rat nAChR subunit clones, however, the activity of RegIIA and [N11A,N12A]RegIIA may differ at human nAChR subtypes.
The selective α3β4 antagonist, AuIB, expanded our knowledge of the distribution and physiological functions of this nAChR subtype in various tissues, including dopaminergic signaling in medial habenula (52), glutamatergic neurotransmission in cardiac vagal neurons (53), and keratinocyte chemokinesis (54). However, the low potency of AuIB at α3β4 and its off-target effect on Cav2.2 channel modulation via GABAB receptor activation has hampered its use in in vivo studies. Recent studies in development of selective and potent α3β4 antagonists led to the discovery and synthesis of two novel α-conotoxins: TxID, a novel α4/6-conotoxin from Conus textile that potently blocks α3β4 (IC50 = 12.5 nm) (55) and TP-2212-59, a synthetic analog of α4/4-conotoxin BuIA (IC50 = 2.3 nm) (56). Although both peptides are potent inhibitors of the α3β4 nAChR subtype, TxID also inhibits α6/α3β4 with only 7.5-fold less potency (IC50 = 94.1 nm) (55) and the activity of TP-2212-59 at other nAChR subtypes such as α6-containing receptors is yet to be tested (56).
We report the successful synthesis of [N11A,N12A]RegIIA, an α3β4 nAChR subtype-selective antagonist. [N11A,N12A]RegIIA is ∼4-fold more potent than AuIB and could be used to decipher the physiological role of α3β4 nAChR in pathological states. MD simulations reveal a direct loss in pairwise contacts between positions 11 and 12 and the principal face of the receptor, including destabilization in other toxin-receptor contacts at the complementary face. This is qualitatively consistent with the 1000-fold decrease in [N11A,N12A]RegIIA inhibition of the α3β2 nAChR subtype. Our homology models and MD simulations also suggest that at the β2 subunit, Asn11 is close to two basic residues (β2-Arg81 and Lys79); whereas at the β4 subunit, Asn11 is close to one basic (β4-Arg79) and one non-polar residue (β4-Ile77). This has implications for rational toxin modification. Based on the receptor environment surrounding Asn11, it is possible that a N11K mutant would enhance selectivity for α3β4, because a Lys at position 11 would introduce electrostatic repulsion with β2-Arg81 and β2-Lys79. There would likely also be a reduction in affinity at α3β4. However, the repulsion between N11K and β4-Arg79 might be partially offset by favorable contacts between the CH2 groups of N11K and β4-Ile77. Further efforts to examine and optimize the selectivity of RegIIA are presently ongoing.
Our study increases understanding of the interactions of RegIIA with various nAChR subtypes. It also identifies key residues such as Asn9, Asn11, and Asn12 involved in toxin-receptor interaction. This information will be valuable in the design and development of potent, α3β4-selective drugs to treat lung cancer and nicotine addiction.
Acknowledgments
We thank Prof. Norelle Daly for help with NMR spectral assignment, Dr. Han-Shen Tae for carrying out the competitive antagonism experiments of RegIIA and [N11A,N12A]RegIIA, and Dr. Hartmut Cuny for helpful comments on a draft of the manuscript.
This work was supported in part by an Australian Research Council (ARC) Discovery Project Grant (to D. J. A. and F. M.) and the Victorian Life Sciences Computation Initiative (VLSCI) for provision of computational resources (project grants VR0009 and NCq75 to A. H. and D. J. A.).
- nAChR
- nicotinic acetylcholine receptor
- ACh
- acetylcholine
- RP-HPLC
- reverse phase-HPLC
- ESI
- electrospray ionization
- MD
- molecular dynamics
- HBTU
- o-benzotriazole-N,N,N′,N′-tetramethyluronium hexafluorophosphate
- Acm
- acetamidomethyl
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- NMR
- nuclear magnetic resonance.
REFERENCES
- 1. Albuquerque E. X., Pereira E. F., Alkondon M., Rogers S. W. (2009) Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol. Rev. 89, 73–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gotti C., Clementi F., Fornari A., Gaimarri A., Guiducci S., Manfredi I., Moretti M., Pedrazzi P., Pucci L., Zoli M. (2009) Structural and functional diversity of native brain neuronal nicotinic receptors. Biochem. Pharmacol. 78, 703–711 [DOI] [PubMed] [Google Scholar]
- 3. Huh K. H., Fuhrer C. (2002) Clustering of nicotinic acetylcholine receptors: from the neuromuscular junction to interneuronal synapses. Mol. Neurobiol. 25, 79–112 [DOI] [PubMed] [Google Scholar]
- 4. Dani J. A., Bertrand D. (2007) Nicotinic acetylcholine receptors and nicotinic cholinergic mechanisms of the central nervous system. Annu. Rev. Pharmacol. Toxicol. 47, 699–729 [DOI] [PubMed] [Google Scholar]
- 5. Hurst R., Rollema H., Bertrand D. (2013) Nicotinic acetylcholine receptors: from basic science to therapeutics. Pharmacol. Ther. 137, 22–54 [DOI] [PubMed] [Google Scholar]
- 6. Gotti C., Moretti M., Gaimarri A., Zanardi A., Clementi F., Zoli M. (2007) Heterogeneity and complexity of native brain nicotinic receptors. Biochem. Pharmacol. 74, 1102–1111 [DOI] [PubMed] [Google Scholar]
- 7. Schuller H. M. (1989) Cell type specific, receptor-mediated modulation of growth kinetics in human lung cancer cell lines by nicotine and tobacco-related nitrosamines. Biochem. Pharmacol. 38, 3439–3442 [DOI] [PubMed] [Google Scholar]
- 8. Thunnissen F. B. (2009) Acetylcholine receptor pathway and lung cancer. J. Thorac. Oncol. 4, 943–946 [DOI] [PubMed] [Google Scholar]
- 9. Improgo M. R., Scofield M. D., Tapper A. R., Gardner P. D. (2010) The nicotinic acetylcholine receptor CHRNA5/A3/B4 gene cluster: dual role in nicotine addiction and lung cancer. Prog. Neurobiol. 92, 212–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tournier J. M., Birembaut P. (2011) Nicotinic acetylcholine receptors and predisposition to lung cancer. Curr. Opin. Oncol. 23, 83–87 [DOI] [PubMed] [Google Scholar]
- 11. Skok V. I. (2002) Nicotinic acetylcholine receptors in autonomic ganglia. Auton. Neurosci. 97, 1–11 [DOI] [PubMed] [Google Scholar]
- 12. Wang N., Orr-Urtreger A., Korczyn A. D. (2002) The role of neuronal nicotinic acetylcholine receptor subunits in autonomic ganglia: lessons from knockout mice. Prog. Neurobiol. 68, 341–360 [DOI] [PubMed] [Google Scholar]
- 13. Park K. S., Cha S. K., Kim M. J., Kim D. R., Jeong S. W., Lee J. W., Kong I. D. (2006) An α3β4 subunit combination acts as a major functional nicotinic acetylcholine receptor in male rat pelvic ganglion neurons. Pflügers Arch. 452, 775–783 [DOI] [PubMed] [Google Scholar]
- 14. Stoker A. K., Markou A. (2013) Unraveling the neurobiology of nicotine dependence using genetically engineered mice. Curr. Opin. Neurobiol. 23, 493–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Muldoon P. P., Jackson K. J., Perez E., Harenza J. L., Molas S., Rais B., Anwar H., Zaveri N. T., Maldonado R., Maskos U., McIntosh J. M., Dierssen M., Miles M. F., Chen X., De Biasi M., Damaj M. I. (2014) The α3β4* nicotinic ACh receptor subtype mediates physical dependence to morphine: mouse and human studies. Br. J. Pharmacol. 171, 3845–3857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Azam L., McIntosh J. M. (2009) α-Conotoxins as pharmacological probes of nicotinic acetylcholine receptors. Acta Pharmacol. Sin. 30, 771–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lebbe E. K., Peigneur S., Wijesekara I., Tytgat J. (2014) Conotoxins targeting nicotinic acetylcholine receptors: an overview. Mar. Drugs 12, 2970–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Essack M., Bajic V. B., Archer J. A. (2012) Conotoxins that confer therapeutic possibilities. Mar. Drugs 10, 1244–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Akondi K. B., Muttenthaler M., Dutertre S., Kaas Q., Craik D. J., Lewis R. J., Alewood P. F. (2014) Discovery, synthesis, and structure-activity relationships of conotoxins. Chem. Rev. 114, 5815–5847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ellison M., McIntosh J. M., Olivera B. M. (2003) α-Conotoxins ImI and ImII. Similar α7 nicotinic receptor antagonists act at different sites. J. Biol. Chem. 278, 757–764 [DOI] [PubMed] [Google Scholar]
- 21. Vincler M., Wittenauer S., Parker R., Ellison M., Olivera B. M., McIntosh J. M. (2006) Molecular mechanism for analgesia involving specific antagonism of α9α10 nicotinic acetylcholine receptors. Proc. Natl. Acad. Sci. U.S.A. 103, 17880–17884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luo S., Kulak J. M., Cartier G. E., Jacobsen R. B., Yoshikami D., Olivera B. M., McIntosh J. M. (1998) α-Conotoxin AuIB selectively blocks α3β4 nicotinic acetylcholine receptors and nicotine-evoked norepinephrine release. J. Neurosci. 18, 8571–8579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grishin A. A., Wang C. I., Muttenthaler M., Alewood P. F., Lewis R. J., Adams D. J. (2010) α-Conotoxin AuIB isomers exhibit distinct inhibitory mechanisms and differential sensitivity to stoichiometry of α3β4 nicotinic acetylcholine receptors. J. Biol. Chem. 285, 22254–22263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Halai R., Clark R. J., Nevin S. T., Jensen J. E., Adams D. J., Craik D. J. (2009) Scanning mutagenesis of α-conotoxin Vc1.1 reveals residues crucial for activity at the α9α10 nicotinic acetylcholine receptor. J. Biol. Chem. 284, 20275–20284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Franco A., Kompella S. N., Akondi K. B., Melaun C., Daly N. L., Luetje C. W., Alewood P. F., Craik D. J., Adams D. J., Marí F. (2012) RegIIA: An α4/7-conotoxin from the venom of Conus regius that potently blocks α3β4 nAChRs. Biochem. Pharmacol. 83, 419–426 [DOI] [PubMed] [Google Scholar]
- 26. Hogg R. C., Hopping G., Alewood P. F., Adams D. J., Bertrand D. (2003) α-Conotoxins PnIA and [A10L]PnIA stabilize different states of the α7-L247T nicotinic acetylcholine receptor. J. Biol. Chem. 278, 26908–26914 [DOI] [PubMed] [Google Scholar]
- 27. Nevin S. T., Clark R. J., Klimis H., Christie M. J., Craik D. J., Adams D. J. (2007) Are α9α10 nicotinic acetylcholine receptors a pain target for α-conotoxins? Mol. Pharmacol. 72, 1406–1410 [DOI] [PubMed] [Google Scholar]
- 28. Celie P. H., Kasheverov I. E., Mordvintsev D. Y., Hogg R. C., van Nierop P., van Elk R., van Rossum-Fikkert S. E., Zhmak M. N., Bertrand D., Tsetlin V., Sixma T. K., Smit A. B. (2005) Crystal structure of nicotinic acetylcholine receptor homolog AChBP in complex with an α-conotoxin PnIA variant. Nat. Struct. Mol. Biol. 12, 582–588 [DOI] [PubMed] [Google Scholar]
- 29. Sali A., Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 30. Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 31. Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. (2005) GROMACS: fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 [DOI] [PubMed] [Google Scholar]
- 32. Lindahl E., Hess B., van der Spoel D. (2001) GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model 7, 306–317 [Google Scholar]
- 33. Bjelkmar P., Larsson P., Cuendet M. A., Hess B., Lindahl E. (2010) Implementation of the CHARMM force field in GROMACS: Analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J. Chem. Theory Comput. 6, 459–466 [DOI] [PubMed] [Google Scholar]
- 34. Bussi G., Donadio D., Parrinello M. (2007) Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 [DOI] [PubMed] [Google Scholar]
- 35. Parrinello M., Rahman A. J. (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 [Google Scholar]
- 36. Humphrey W., Dalke A., Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 37. Hogg R. C., Miranda L. P., Craik D. J., Lewis R. J., Alewood P. F., Adams D. J. (1999) Single amino acid substitutions in α-conotoxin PnIA shift selectivity for subtypes of the mammalian neuronal nicotinic acetylcholine receptor. J. Biol. Chem. 274, 36559–36564 [DOI] [PubMed] [Google Scholar]
- 38. Grishin A. A., Cuny H., Hung A., Clark R. J., Brust A., Akondi K., Alewood P. F., Craik D. J., Adams D. J. (2013) Identifying key amino acid residues that affect α-conotoxin AuIB inhibition of α3β4 nicotinic acetylcholine receptors. J. Biol. Chem. 288, 34428–34442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Armishaw C. J. (2010) Synthetic α-conotoxin mutants as probes for studying nicotinic acetylcholine receptors and in the development of novel drug leads. Toxins 2, 1471–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dutton J. L., Bansal P. S., Hogg R. C., Adams D. J., Alewood P. F., Craik D. J. (2002) A new level of conotoxin diversity, a non-native disulfide bond connectivity in α-conotoxin AuIB reduces structural definition but increases biological activity. J. Biol. Chem. 277, 48849–48857 [DOI] [PubMed] [Google Scholar]
- 41. van Lierop B. J., Robinson S. D., Kompella S. N., Belgi A., McArthur J. R., Hung A., MacRaild C. A., Adams D. J., Norton R. S., Robinson A. J. (2013) Dicarba α-conotoxin Vc1.1 analogues with differential selectivity for nicotinic acetylcholine and GABAB receptors. ACS Chem. Biol. 8, 1815–1821 [DOI] [PubMed] [Google Scholar]
- 42. Franco A., Pisarewicz K., Moller C., Mora D., Fields G. B., Marí F. (2006) Hyperhydroxylation: a new strategy for neuronal targeting by venomous marine molluscs. Prog. Mol. Subcell. Biol. 43, 83–103 [DOI] [PubMed] [Google Scholar]
- 43. Callaghan B., Haythornthwaite A., Berecki G., Clark R. J., Craik D. J., Adams D. J. (2008) Analgesic α-conotoxins Vc1.1 and RgIA inhibit N-type calcium channels in rat sensory neurons via GABAB receptor activation. J. Neurosci. 28, 10943–10951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Millard E. L., Daly N. L., Craik D. J. (2004) Structure-activity relationships of α-conotoxins targeting neuronal nicotinic acetylcholine receptors. Eur. J. Biochem. 271, 2320–2326 [DOI] [PubMed] [Google Scholar]
- 45. Galzi J. L., Revah F., Black D., Goeldner M., Hirth C., Changeux J. P. (1990) Identification of a novel amino acid α-tyrosine 93 within the cholinergic ligands-binding sites of the acetylcholine receptor by photoaffinity labeling: additional evidence for a three-loop model of the cholinergic ligand-binding sites. J. Biol. Chem. 265, 10430–10437 [PubMed] [Google Scholar]
- 46. Dellisanti C. D., Yao Y., Stroud J. C., Wang Z. Z., Chen L. (2007) Crystal structure of the extracellular domain of nAChR α1 bound to α-bungarotoxin at 1.94-Å resolution. Nat. Neurosci. 10, 953–962 [DOI] [PubMed] [Google Scholar]
- 47. Ulens C., Hogg R. C., Celie P. H., Bertrand D., Tsetlin V., Smit A. B., Sixma T. K. (2006) Structural determinants of selective α-conotoxin binding to a nicotinic acetylcholine receptor homolog AChBP. Proc. Natl. Acad. Sci. U.S.A. 103, 3615–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee C., Lee S. H., Kim D. H., Han K. H. (2012) Molecular docking study on the α3β2 neuronal nicotinic acetylcholine receptor complexed with α-conotoxin GIC. BMB Rep. 45, 275–280 [DOI] [PubMed] [Google Scholar]
- 49. Dutertre S., Nicke A., Lewis R. J. (2005) β2 subunit contribution to 4/7 α-conotoxin binding to the nicotinic acetylcholine receptor. J. Biol. Chem. 280, 30460–30468 [DOI] [PubMed] [Google Scholar]
- 50. Shiembob D. L., Roberts R. L., Luetje C. W., McIntosh J. M. (2006) Determinants of α-conotoxin BuIA selectivity on the nicotinic acetylcholine receptor β subunit. Biochemistry 45, 11200–11207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Azam L., McIntosh J. M. (2012) Molecular basis for the differential sensitivity of rat and human α9α10 nAChRs to α-conotoxin RgIA. J. Neurochem. 122, 1137–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McCallum S. E., Cowe M. A., Lewis S. W., Glick S. D. (2012) α3β4 nicotinic acetylcholine receptors in the medial habenula modulate the mesolimbic dopaminergic response to acute nicotine in vivo. Neuropharmacology 63, 434–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kamendi H. W., Cheng Q., Dergacheva O., Gorini C., Jameson H. S., Wang X., McIntosh J. M., Mendelowitz D. (2009) Abolishment of serotonergic neurotransmission to cardiac vagal neurons during and after hypoxia and hypercapnia with prenatal nicotine exposure. J. Neurophysiol. 101, 1141–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chernyavsky A. I., Arredondo J., Marubio L. M., Grando S. A. (2004) Differential regulation of keratinocyte chemokinesis and chemotaxis through distinct nicotinic receptor subtypes. J. Cell Sci. 117, 5665–5679 [DOI] [PubMed] [Google Scholar]
- 55. Luo S., Zhangsun D., Zhu X., Wu Y., Hu Y., Christensen S., Harvey P. J., Akcan M., Craik D. J., McIntosh J. M. (2013) Characterization of a novel α-conotoxin TxID from Conus textile that potently blocks rat α3β4 nicotinic acetylcholine receptors. J. Med. Chem. 56, 9655–9663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chang Y. P., Banerjee J., Dowell C., Wu J., Gyanda R., Houghten R. A., Toll L., McIntosh J. M., Armishaw C. J. (2014) Discovery of a potent and selective α3β4 nicotinic acetylcholine receptor antagonist from an α-conotoxin synthetic combinatorial library. J. Med. Chem. 57, 3511–3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Talley T. T., Olivera B. M., Han K. H., Christensen S. B., Dowell C., Tsigelny I., Ho K. Y., Taylor P., McIntosh J. M. (2006) α-Conotoxin OmIA is a potent ligand for the acetylcholine-binding protein as well as α3β2 and α7 nicotinic acetylcholine receptors. J. Biol. Chem. 281, 24678–24686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McIntosh J. M., Dowell C., Watkins M., Garrett J. E., Yoshikami D., Olivera B. M. (2002) α-Conotoxin GIC from Conus geographus, a novel peptide antagonist of nicotinic acetylcholine receptors. J. Biol. Chem. 277, 33610–33615 [DOI] [PubMed] [Google Scholar]
- 59. McIntosh J. M., Plazas P. V., Watkins M., Gomez-Casati M. E., Olivera B. M., Elgoyhen A. B. (2005) A novel α-conotoxin, PeIA, cloned from Conus pergrandis, discriminates between rat α9α10 and α7 nicotinic cholinergic receptors. J. Biol. Chem. 280, 30107–30112 [DOI] [PubMed] [Google Scholar]
- 60. Peng C., Chen W., Sanders T., Chew G., Liu J., Hawrot E., Chi C. (2010) Chemical synthesis and characterization of two α4/7-conotoxins. Acta Biochim. Biophys. Sin. 42, 745–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Inserra M. C., Kompella S. N., Vetter I., Brust A., Daly N. L., Cuny H., Craik D. J., Alewood P. F., Adams D. J., Lewis R. J. (2013) Isolation and characterization of α-conotoxin LsIA with potent activity at nicotinic acetylcholine receptors. Biochem. Pharmacol. 86, 791–799 [DOI] [PubMed] [Google Scholar]
- 62. Azam L., Dowell C., Watkins M., Stitzel J. A., Olivera B. M., McIntosh J. M. (2005) α-Conotoxin BuIA, a novel peptide from Conus bullatus, distinguishes among neuronal nicotinic acetylcholine receptors. J. Biol. Chem. 280, 80–87 [DOI] [PubMed] [Google Scholar]
- 63. Dowell C., Olivera B. M., Garrett J. E., Staheli S. T., Watkins M., Kuryatov A., Yoshikami D., Lindstrom J. M., McIntosh J. M. (2003) α-Conotoxin PIA is selective for α6 subunit-containing nicotinic acetylcholine receptors. J. Neurosci. 23, 8445–8452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Whiteaker P., Christensen S., Yoshikami D., Dowell C., Watkins M., Gulyas J., Rivier J., Olivera B. M., McIntosh J. M. (2007) Discovery, synthesis, and structure activity of a highly selective α7 nicotinic acetylcholine receptor antagonist. Biochemistry 46, 6628–6638 [DOI] [PubMed] [Google Scholar]