Background: Rubisco, a key photosynthetic enzyme of eight large and eight small subunits, is phylogenetically divided into green and red types.
Results: The small subunits of red-type Rubisco contain a C-terminal β-hairpin extension that mediates efficient assembly of the holoenzyme.
Conclusion: The C-terminal β-hairpin renders red-type Rubisco independent of specialized assembly chaperones.
Significance: These findings can help in bioengineering red-type Rubisco into crop plants.
Keywords: Molecular Chaperone; Photosynthesis; Protein Assembly; Protein Folding; Ribulose-1,5-bisphosphate Carboxylase/Oxygenase (RuBisCO)
Abstract
Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) is the key enzyme involved in photosynthetic carbon fixation, converting atmospheric CO2 to organic compounds. Form I Rubisco is a cylindrical complex composed of eight large (RbcL) subunits that are capped by four small subunits (RbcS) at the top and four at the bottom. Form I Rubiscos are phylogenetically divided into green- and red-type. Some red-type enzymes have catalytically superior properties. Thus, understanding their folding and assembly is of considerable biotechnological interest. Folding of the green-type RbcL subunits in cyanobacteria is mediated by the GroEL/ES chaperonin system, and assembly to holoenzyme requires specialized chaperones such as RbcX and RAF1. Here, we show that the red-type RbcL subunits in the proteobacterium Rhodobacter sphaeroides also fold with GroEL/ES. However, assembly proceeds in a chaperone-independent manner. We find that the C-terminal β-hairpin extension of red-type RbcS, which is absent in green-type RbcS, is critical for efficient assembly. The β-hairpins of four RbcS subunits form an eight-stranded β-barrel that protrudes into the central solvent channel of the RbcL core complex. The two β-barrels stabilize the complex through multiple interactions with the RbcL subunits. A chimeric green-type RbcS carrying the C-terminal β-hairpin renders the assembly of a cyanobacterial Rubisco independent of RbcX. Our results may facilitate the engineering of crop plants with improved growth properties expressing red-type Rubisco.
Introduction
The enzyme Rubisco3 catalyzes the carboxylation of the five-carbon sugar ribulose 1,5-bisphosphate in the dark reactions of photosynthesis, thereby mediating the conversion of atmospheric CO2 into usable sugars. In its most abundant form (form I), Rubisco is a hexadecameric complex composed of eight large (RbcL, ∼50 kDa) and eight small subunits (RbcS, ∼12–15 kDa) (1, 2). The RbcL subunits form a cylindrical tetramer of anti-parallel dimers that is capped by four RbcS subunits at the top and four RbcS subunits at the bottom. Based on the alignment of RbcL sequences, form I Rubisco is phylogenetically divided into green and red branches (3–5). Green-type Rubisco occurs in cyanobacteria, green algae and plants, whereas the red type is found in photosynthetic bacteria, non-green algae and phytoplankton.
Despite its central role in the global carbon cycle, Rubisco is a slow catalyst. Moreover, it discriminates poorly between CO2 and O2 (6). Although the fixation of CO2 results in the synthesis of usable sugars, the competing reaction with O2 (photorespiration) decreases the efficiency of carbon fixation (7, 8). Extensive efforts have focused on engineering a Rubisco enzyme with improved specificity for CO2 (1, 9–12). Interestingly, compared with their green-type homologs, several of the red-type enzymes display a >2-fold higher specificity for CO2 coupled with a lower Michaelis constant for CO2 (13). Specificity for CO2 over O2 (Sc/o) determines preferential carboxylation over oxygenation of ribulose-1,5-bisphosphate during Rubisco catalysis. A comparison of the kinetic properties of different forms of Rubisco suggests an inverse relation between Sc/o and turnover rate. So far, the highest specificity factor (Sc/o, 167) is reported for the Rubisco from the filamentous red algae Griffithsia monilis, which has a turnover rate of 2.6 s−1 (13). By comparison, the bacterial red-type enzyme from Rhodobacter sphaeroides possess a similar CO2 turnover rate (∼2.0 to 2.9 s−1), but a significantly lower Sc/o (∼55) (12, 14, 15). It has been proposed that transplantation of the G. monilis Rubisco into C3 plant chloroplasts would raise crop carbon gain by up to ∼30% without any increase in the amount of Rubisco per unit leaf area at ambient conditions by improving the efficiency of light, water and nitrogen usage (11, 12). However, when the rbcLS operon from the red algae Galdieria sulfuraria (Gs) or the diatom Phaeodactylum tricornutum (Pt) was inserted into the tobacco plastid genome, the heterologous Rubisco subunits were expressed abundantly but failed to assemble into an active holoenzyme (13), suggesting incompatibility with the chloroplast chaperones involved in the folding and assembly of green-type Rubisco. Folding of the RbcL subunit is mediated by the chaperonin system: GroEL/ES in proteobacteria and cyanobacteria and Cpn60/Cpn10/Cpn20 in algae and plants (16–18). Moreover, several factors have recently been reported to be required for the efficient assembly of the RbcL8S8 complex. These factors include the assembly chaperone, RbcX, and the Rubisco accumulation factors 1 and 2 (RAF1; RAF2), which are structurally unrelated but conserved in most organisms harboring green-type Rubisco (19–25). A combination of GroEL/ES and RbcX allowed the in vitro reconstitution of cyanobacterial Rubisco from Synechococcus sp. PCC 6301 (Syn6301) (20). Interestingly, organisms containing red-type Rubisco lack the rbcX or RAF1 genes (26–29), suggesting an alternative mode of assembly.
Here, we analyzed the chaperone requirement for the in vitro folding and assembly of a prokaryotic red-type form I Rubisco from the proteobacterium Rhodobacter sphaeroides (Rs). We show that the folding of the RbcL subunit requires the GroEL/ES chaperonin, but the assembly to holoenzyme is independent of accessory factors. Instead, assembly is mediated by the RbcS subunits in a manner dependent on red-type specific C-terminal β-hairpin extensions. The extensions of the four RbcS subunits that cap the RbcL8 complex form an eight stranded β-barrel structure, partially occupying the central solvent channel of the holoenzyme (2, 30, 31). Mutational analysis and the finding that red-type RbcS can mediate green-type Syn6301RbcL assembly in the absence of RbcX, suggest that the red-type RbcS fulfills the role of an assembly factor.
EXPERIMENTAL PROCEDURES
Molecular Biology
The rbcL and rbcS genes from R. sphaeroides were amplified from the plasmid pET30bRsrbcLS (15) using restriction sites NdeI/BamHI sites to generate the plasmids pET30bRsrbcL and pET30bRsrbcS, respectively. A C-terminally truncated version of the rbcS gene comprising residues 1–106 was also generated (pET30bRsrbcSΔEF). The QuikChange protocol (Stratagene) was employed to introduce point mutations in RsRbcS. P. tricornutum rbcS was amplified from the total genomic DNA and cloned into pET11a plasmid using the NdeI/BamHI restriction sites resulting in pET11aPtrbcS. G. sulfuraria rbcS was cloned into pET11a from the pGs plasmid comprising the plastome fragment encoding rbcL-rbcS-cbbX. A hybrid version of Syn6301RbcS containing the C-terminal extension of RsRbcS (residues 106–129) was generated as follows: the three penultimate amino acids Gly-109, Arg-110, and Tyr-111 of Syn6301RbcS in pET11aSyn6301rbcS (19) were deleted using QuikChange protocol (Stratagene) to generate the plasmid pET11aSyn6301rbcSΔGRY. Primers were designed to introduce the RsRbcS C-terminal extension resulting in pET11aSyn6301rbcSΔGRY-RsrbcSEF. The plasmids for expression of the cyanobacterial proteins Syn6301RbcL, Syn6301RbcS, Syn7002RbcS, and N-terminally His6-tagged AnaCARbcX are described by Saschenbrecker et al. (19). Coding regions of all constructs were verified by DNA sequencing.
Protein Purification
Unless stated otherwise, all purification procedures were performed at 4 °C. Concentrations of pure proteins were determined spectrophotometrically at 280 nm. E. coli GroEL and GroES were purified as described previously (32, 33). The Syn6301RbcL8 was purified as described by Saschenbrecker et al. (19). N-terminally tagged AnaCARbcX was purified according to the protocol of Liu et al. (20). Expression and purification of Rubisco from R. sphaeroides was performed as described by Mueller-Cajar et al. (15). Purification of denatured RsRbcL and Syn6301RbcL from inclusion bodies was performed essentially as described previously (19, 20). The proteins were stored in buffer (40 mm Tris, pH 8.0, 250 mm sucrose, 10 mm EDTA, 6 m GuHCl) at −80 °C. RbcS proteins were also purified from inclusion bodies but were refolded during dialysis against 25 mm Tris, pH 8.0, 50 mm NaCl (19). Purified RbcS proteins (see Fig. 4A) were concentrated and stored in buffer containing 25 mm Tris pH 8.0, 50 mm NaCl, 10% (v/v) glycerol, and 5 mm DTT.
FIGURE 4.
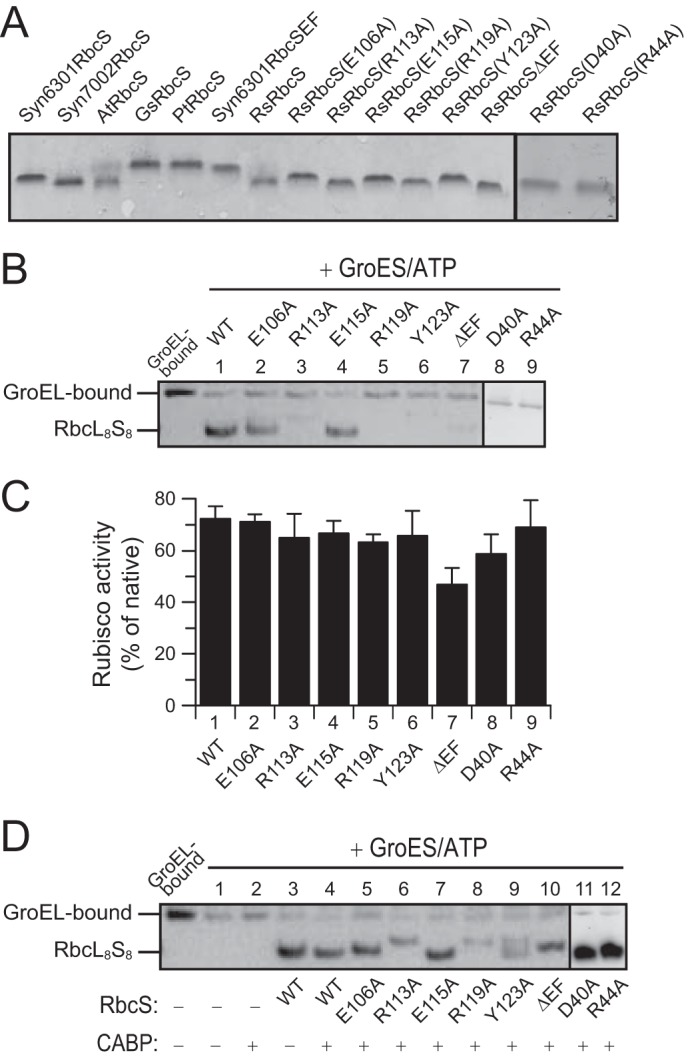
Requirement for assembly of residues in the C-terminal β-hairpin and the βAB loop of R. sphaeroides RbcS. A, purified and refolded RbcS proteins used in this study. Shown is an analysis by SDS-PAGE and Coomassie staining. B, reconstitution of RsRubisco assembly with WT and mutant RbcS subunits as indicated analyzed by native PAGE and immunoblotting against RbcL. 90-min end points are shown (see Fig. 2A). GroEL-bound RsRbcL from a reaction lacking ATP is shown as control. C, Rubisco activities are shown in reactions as described in B. Error bars indicate S.D. of at least three independent experiments. D, reconstitution reactions as described in B in the presence or absence of 0.1 mm CABP.
Rubisco Refolding
GroEL-assisted refolding of GuHCl-denatured RsRbcL was performed at 25 °C as described previously (20) with minor modifications. Denatured RsRbcL was diluted to 0.5 μm final concentration into ice-cold refolding buffer (20 mm MOPS-KOH, pH 7.5, 100 mm KCl, 5 mm Mg(OAc)2, 10 mm DTT, 0.5 mg ml−1 BSA) containing 1 μm GroEL. Final concentration of GuHCl in the refolding assay was maintained at <30 mm. RbcL not captured by GroEL and aggregated was removed by centrifugation at 16,200 × g for 5 min at 4 °C followed by addition of 2 μm GroES. Refolding was initiated by the addition of 10 mm ATP and stopped by addition of apyrase (0.25 units/μl). Addition of RbcS and other proteins was as specified in the figure legends. CABP was added at a concentration of 0.1 mm when indicated. Reactions were analyzed by 6% native PAGE or NuPAGE® Novex® 3–8% Tris acetate (Invitrogen) or SDS-PAGE followed by immunoblotting with anti-RbcL antibody where indicated. RsRbcL was detected with antibody from Agrisera and Syn6301RbcL with antibodies raised in rabbits (20). Rubisco activity was analyzed as described previously with modifications (20, 34). Activities of in vitro reconstituted enzyme are expressed in percent of native control, which refers to the activity of recombinant, purified holoenzyme.
CABP-binding Assay
RsRbcL was expressed in E. coli with or without overexpression of GroEL/ES. Soluble cell lysate was obtained by centrifugation of crude cell lysate at 16,200 × g for 15 min at 4 °C. Equivalent amounts of lysate containing RsRbcL were incubated with purified RsRbcS and holoenzyme formation was allowed to proceed for 15 min at 25 °C. The concentration of Rubisco active sites was measured by stoichiometric binding of the tight binding inhibitor [2-14C]carboxyarabinitol-P2 after preincubation with 25 mm NaHCO3 and 15 mm MgCl2, as described previously (35).
Thermal Stability Assay
The apparent melting temperature (Tm) of RsRubisco (RsRbcL8S8) and mutant variants was determined as described (10, 36). Equal concentrations of soluble protein from E. coli lysates expressing RsRbcL were incubated with wild-type or mutant RbcS for 30 min at 25 °C to allow formation of holoenzyme. Rubisco activities were measured as above (at 25 °C) after incubation of lysate containing holoenzyme at increasing temperatures from 30 to 70 °C for 10 min, followed by incubation on ice for 5 min. Activities measured at 30 °C were set to 100%.
RESULTS
RsRbcL Is Expressed in E. coli in a Soluble, Assembly-competent Form
We have observed previously that green-type cyanobacterial RbcL is generally highly insoluble upon expression in E. coli, even when GroEL/ES is overexpressed (19). In contrast, red-type RbcL from R. sphaeroides (RsRbcL) was 16 ± 3% soluble upon expression in E. coli cells containing endogenous GroEL/ES levels, and 33 ± 2% soluble protein was recovered when GroEL/ES was transiently overexpressed from an arabinose-inducible plasmid before induction of RsRbcL with IPTG (Fig. 1A). While at higher concentration (when GroEL/ES was overexpressed), the soluble RsRbcL migrated on native-PAGE as a smear of poorly resolved species consistent with a dynamic ensemble of oligomeric states, it was hardly detectable at lower concentration (Fig. 1B). The soluble RsRbcL was assembly-competent and formed the RbcL8S8 holoenzyme upon addition of purified RsRbcS to the cell lysate (Fig. 1B). The RbcL8S8 complex was enzymatically active as reflected by its ability to bind the substrate analog [2-14C]carboxyarabinitol-P2 (35) (Fig. 1C). Overexpression of GroEL/ES resulted in a ∼2.6-fold increase in holoenzyme protein and a ∼3.4-fold increase in Rubisco active sites.
FIGURE 1.
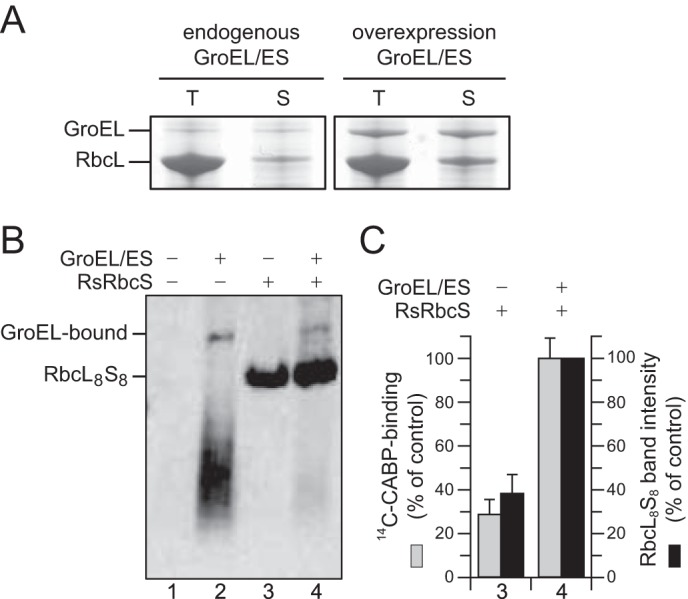
Requirement of GroEL/ES chaperonin for R. sphaeroides Rubisco folding upon expression in E. coli. A, RsRbcL was expressed in E. coli with or without overexpression of GroEL/ES. Total (T) and soluble (S) fractions of cell lysates were analyzed by SDS-PAGE and Coomassie staining. B, soluble fractions were analyzed by native PAGE with or without supplementation of purified RsRbcS when indicated, followed by immunoblotting against RbcL. C, Rubisco active sites in reactions 3 and 4 from B were quantified by [2-14C]carboxyarabinitol-P2 (14C-CABP) binding assay. Error bars indicate S.D. of three independent experiments with the values obtained in the presence of GroEL/ES and RsRbcS set to 100%. Band intensities of RbcL8S8 were quantified by densitometry, and the band intensity obtained with GroEL/ES overexpression was set to 100%.
To obtain more detailed insight into RsRubisco folding and assembly, we performed reconstitution experiments using purified proteins in vitro. Upon dilution of denatured RsRbcL from 6 m GuHCl into GroEL containing refolding buffer in the absence of ATP, RsRbcL bound stably to GroEL and migrated as a high molecular weight complex on native PAGE (Fig. 2A, panel i). Addition of GroES and ATP to start the GroEL/ES reaction cycle resulted in the time-dependent release of the folded RsRbcL subunit (Fig. 2A, panel ii). This RsRbcL remained soluble (Fig. 2B), but was enzymatically inactive and not detectable by native-PAGE as a defined species (Fig. 2, A, panel ii, and C). Thus, RsRbcL differs from the behavior of green-type Syn6301RbcL which retains affinity for GroEL after folding, unless RbcX is present (20). When RsRbcS was added during GroEL/ES-mediated folding, enzymatically active RsRbcL8S8 complex formed at an apparent half-time of ∼20 min and with a high yield of ∼80% (Fig. 2, A, panel iii, and C). As the GroEL/ES reaction was stopped with apyrase at different times to deplete ATP, this time course reflects the kinetics of RsRbcL subunit folding and release from GroEL. Folded RsRbcL remained assembly competent as demonstrated by the addition of RsRbcS after refolding (Fig. 2A, panel iv, and C). These results show that RsRubisco assembly is independent of a separate auxiliary assembly factor. The folding properties of RsRbcL differ from that of Syn6301RbcL. Moreover, RsRbcS appears to have a critical role in shifting a dynamic ensemble of RbcL oligomers to the well defined holoenzyme complex.
FIGURE 2.
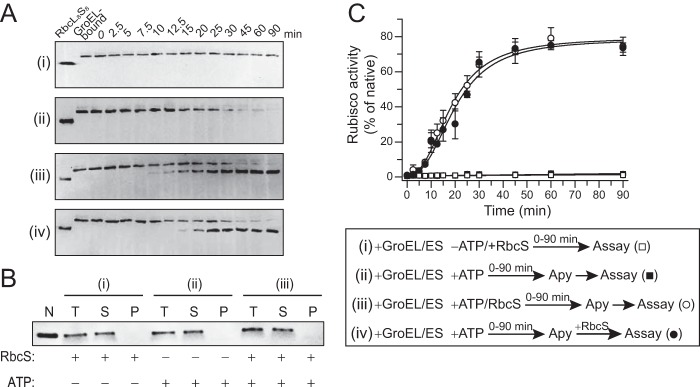
In vitro reconstitution of R. sphaeroides Rubisco with purified components. A, time course of RsRbcL folding and assembly analyzed by native PAGE and immunoblotting against RbcL. GuHCl denatured RsRbcL was diluted into GroEL/ES containing buffer and incubated either in the absence of ATP but presence of RsRbcS (panel i) or presence of ATP (panels ii–iv) without RsRbcS (panel ii), with RsRbcS (panel iii), or RsRbcS added after GroEL/ES cycling was stopped with apyrase (Apy) (panel iv). B, solubility of RsRbcL from reactions (panel i) to (panel iii) analyzed by fractionation into total (T), soluble (S), and pellet (P) fractions by centrifugation, followed by SDS-PAGE and immunoblotting against RbcL. C, Rubisco activities in reconstitution reactions as described in A. See legend for sequence of addition. Activities are expressed as percent of native enzyme control (kcat ∼2 CO2 s−1) (15). Error bars indicate S.D. of at least three independent experiments.
The C-terminal β-Hairpin Extension of RsRbcS Mediates Assembly and Stability
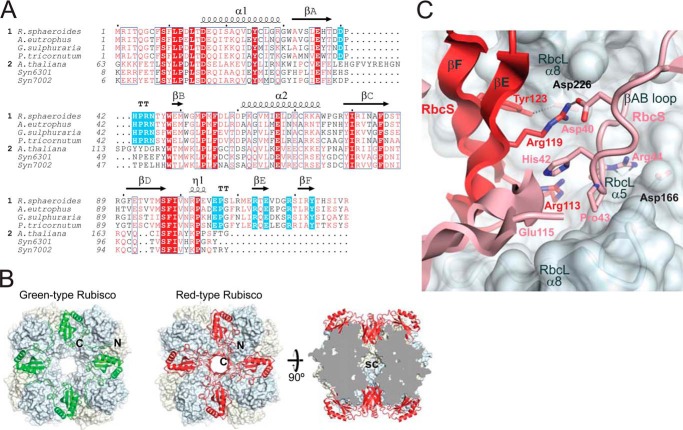
In the assembly of Syn6301RbcL, RbcX functions in stabilizing antiparallel RbcL dimers and facilitates formation of the RbcL8 complex. RbcS binding then results in a conformational change in the RbcL subunits, thereby causing the displacement of RbcX (21). In the case of RsRubisco, the RsRbcS subunits might play a similar role in assembly by linking RsRbcL dimers. All red-type RbcS subunits possess a ∼25-amino acid C-terminal extension that forms a β-hairpin structure, consisting of β-strands βE and βF (Fig. 3A). As illustrated by the structure of the red-type Rubisco of Alcaligenes eutrophus, the βEF hairpins from four adjacent RbcS subunits are assembled to an eight-stranded β-barrel that protrudes into the central solvent channel of the holoenzyme (Fig. 3B). The RbcS β-barrel engages in extensive hydrogen bond and salt bridge contacts with the RbcL subunits (2, 30, 31, 37). Tyr-123 in βF is able to form a hydrogen bond with Asp-226 in helix α8 of RbcL (Fig. 3C). Furthermore, Arg-113, Glu-115, and Arg-119 of the βEF hairpin stabilize the conformation of the conserved βAB loop (residues 40–45) (Fig. 3A) in the adjacent RbcS subunit through a hydrogen bond between Arg-119 and Asp-40 and π-stacking interactions between the Arg-113 guanidinium group and the imidazole ring of His-42 (Fig. 3C). The βAB loop in turn packs against the RbcL subunit of the adjacent RbcL dimer (at helices α5 and α8) involving a hydrogen bond of Arg-44 (conserved in red-type RbcS) to the C-terminal end of RbcL helix α5 at residue Asp-163. In addition, the guanidinium moiety of Arg-44 makes electrostatic interactions with the helix α5 dipole and Asp-166 of RbcL. Thus, the RbcS β-barrel formed by βEF hairpins is expected to confer additional stability to the red-type Rubisco relative to the green-type holoenzyme. The RsRbcS subunits may either first interact with the RsRbcL anti-parallel dimer, followed by holoenzyme formation, or they may first form a tetramer platform on to which RbcL assembles.
FIGURE 3.
Structural comparison of red- and green-type Rubisco small subunits. A, alignment of representative RbcS sequences from red-type (1) (Uniprot accession codes P27998, Rhodobacter sphaeroides; P09658, Alcaligenes eutrophus; P23756, Galdieria sulfuraria; A0T0E2, Phaeodactylum tricornutum) and green-type organisms (2) (Uniprot accession codes P10785, Arabidopsis thaliana; P04716, Synechococcus sp. PCC 6301; Q44178, Synechococcus sp. PCC 7002) using Clustal-X. Similar residues are shown in red, and identical residues were shown in white on red background. Secondary structure elements are indicated above the sequences. Residues of red-type RbcS implicated in Rubisco assembly are highlighted in cyan. B, the RbcS subunit arrangements in the crystal structures of green- and red-type Rubiscos from Synechococcus sp. PCC 6301 and Alcaligenes eutrophus (PDB codes 1RBL and 1BXN, respectively) (30, 47). The left and middle panels show top views of Syn6301 and A. eutrophus RbcL8S8, respectively; the RbcS subunits are shown in ribbon representation, and the RbcL octameric core is shown in surface representation. The right panel shows a cross-section along the 4-fold axis through the complex of A. eutrophus. N, N terminus; C, C terminus; sc, solvent channel. C, interactions at the interface between small and large subunits in red-type Rubisco from A. eutrophus (PDB 1BXN) (30). The interacting regions of adjacent RbcS subunits are shown in red and pink ribbon representations. The RbcL subunits are shown in surface representation. The side chains of critical residues are highlighted in stick representation.
To investigate the contribution of the C-terminal β-hairpin of RbcS to assembly, we either truncated the C terminus of RsRbcS by 23 residues to generate RsRbcSΔEF or made point mutations in the highly conserved residues Asp-40, Arg-44, Glu-106, Arg-113, Glu-115, Arg-119, and Tyr-123 to alanine. The mutant proteins were purified from inclusion bodies and refolded upon removal of denaturant by dialysis (Fig. 4A). When added to an in vitro reconstitution reaction, the mutants of the βEF hairpin (R113A, R119A, Y123A, and ΔEF) failed to mediate the formation of holoenzyme detectable on native PAGE, whereas complex formation was preserved with mutants E106A and E115A (Fig. 4B). Similarly, mutation of the conserved residues Asp-40 and Arg-44 in the βAB loop (Fig. 3A) impaired holoenzyme formation (Fig. 4B). Surprisingly, the RbcS mutants were nevertheless able to reconstitute RsRubisco activity to wild-type level, except for RsRbcSΔEF, which resulted in a ∼30% lower yield (Fig. 4C).
The presence of Rubisco activity, despite the absence of detectable RbcL8S8 complex on native PAGE, may be explained by a stabilizing effect of the sugar substrate ribulose-1,5-bisphosphate, which was absent during reconstitution and native PAGE. Note that the antiparallel RbcL dimer has two sugar binding sites formed by the subunit interface, and substrate binding has been reported to stabilize the dimer (2, 38, 39). To test this possibility, we added the high affinity substrate analog 2-carboxyarabinitol-1,5-bisphosphate (CABP) during reconstitution (Fig. 4D). In the presence of CABP, the effect of mutations in the C-terminal β-hairpin and the βAB loop were compensated to varying degrees, and RsRbcL8S8 complexes were clearly observed on native-PAGE (Fig. 4D), confirming a stabilizing effect of the sugar substrate. Note that the migration behavior of some of the mutant complexes varied relative to wild-type, which may reflect differences in charge and shape properties.
We next analyzed the thermal stability of Rubisco holoenzyme containing wild-type (WT) or mutant RbcS. RsRbcL was expressed in E. coli and holoenzyme reconstituted by adding RbcS subunits. The lysates were then incubated for 10 min at increasing temperatures up to 70 °C, followed by Rubisco activity assay at 25 °C. The apparent melting temperature (Tm) of RsRbcL8SΔEF8 (∼47 °C) was reduced by ∼6 °C compared with WT RsRubisco (∼53 °C) (Fig. 5A and Table 1), confirming a substantial stabilizing effect of the β-hairpin. Moreover, in vitro reconstitution showed that the kinetics of Rubisco assembly with RbcSΔEF was ∼3.5-fold slower than with WT RsRbcS (Fig. 5B). The RbcS point mutations in the βAB loop and the C-terminal β-hairpin also reduced the melting temperature (Table 1), except for E106A (∼53 °C) and E115A (∼54 °C), consistent with the ability of these mutants to produce stable RbcL8S8 on native-PAGE in the absence of CABP (Fig. 4B). In summary, the RbcL8S8 complex can form in the absence of the β-hairpin loop, but presence of the βEF region strongly accelerates the assembly process and stabilizes the holoenzyme.
FIGURE 5.
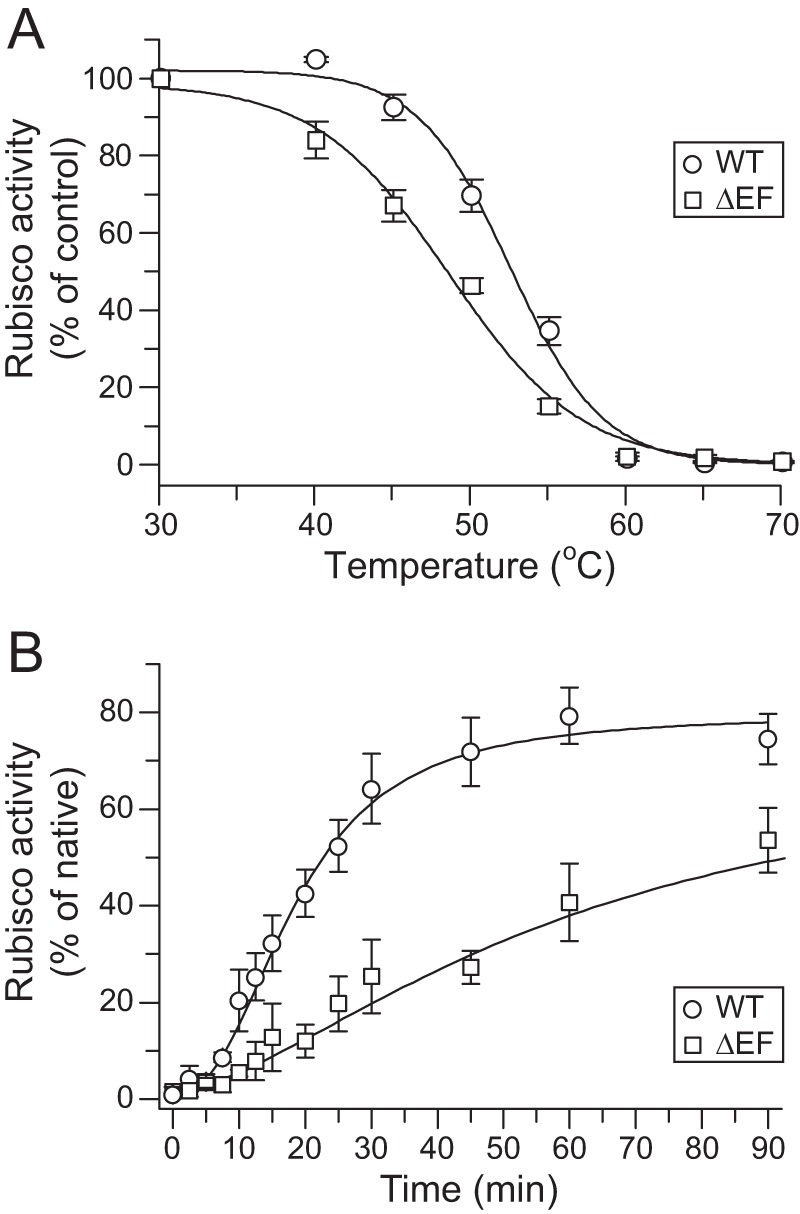
Role of the C-terminal β-hairpin of R. sphaeroides RbcS in Rubisco stability and assembly. A, thermal stability of Rubisco containing WT RsRbcS or RsRbcSΔEF. RbcL and RbcS were co-expressed in E. coli, and lysates were incubated at the indicated temperatures for 10 min, followed by cooling on ice and activity assay at 25 °C. Activities measured after incubation at 30 °C were set to 100%. Shown is S.D. from at least three independent experiments. B, carboxylation activity during in vitro reconstitution of RsRubisco with WT RsRbcS or RsRbcSΔEF. Experiments were performed essentially described as in Fig. 2C (panel iii).
TABLE 1.
Apparent melting temperatures (Tm) of wild-type RsRubisco (WT) and variants
| Sample (Tm) | Tm (°C) |
|---|---|
| WT | 53.25 ± 1.35 |
| RsRbcSE106A | 52.67 ± 0.75 |
| RsRbcSR113A | 48.25 ± 2.43 |
| RsRbcSE115A | 53.99 ± 0.36 |
| RsRbcSR119A | 50.03 ± 1.86 |
| RsRbcSY123A | 48.56 ± 1.16 |
| RsRbcSΔEF | 47.42 ± 1.43 |
| RsRbcSD40A | 51.13 ± 0.62 |
| RsRbcSR44A | 44.80 ± 2.23 |
Heterologous Red-type RbcS Mediates Rubisco Assembly but Does Not Confer Catalytic Activity
To investigate whether RbcL8S8 assembly correlates with catalytic activity, we used various red- and green-type RbcS subunits with RsRbcL in the in vitro reconstitution assay. As analyzed by native PAGE, the folded RsRbcL assembled with the RbcS subunits from the red algae G. sulfuraria and the red-type diatom P. tricornutum (Pt) but with none of the tested green-type subunits (Fig. 6A), consistent with the requirement of the C-terminal β-hairpin region for assembly. Notably, the heterologous RbcL8S8 complexes were essentially enzymatically inactive (Fig. 6B). This indicates that heterologous red-type RbcS is competent for the assembly but not sufficient for activity. RbcS proteins are generally more divergent (∼25–30% identity) in sequence than RbcL subunits (∼50–60% identity) and are apparently critical for holoenzyme activity through long range interactions between RbcS and the active site of the cognate RbcL dimer. Thus, structural competence for assembly does not correlate with the capacity to generate catalytically active enzyme.
FIGURE 6.
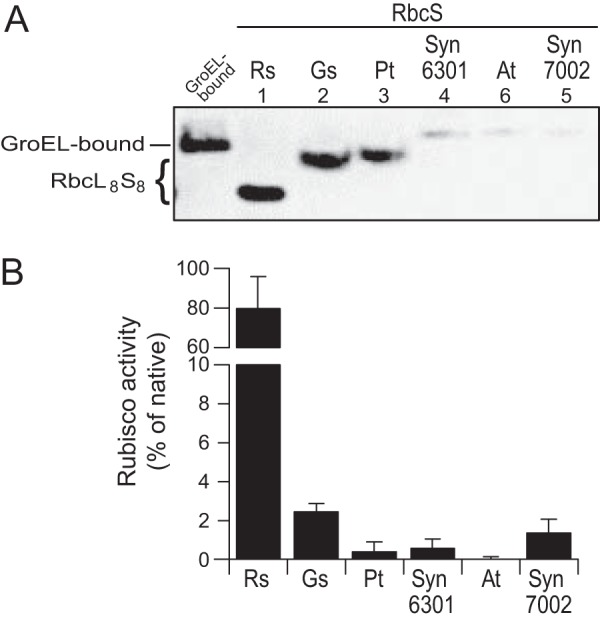
Heterologous red-type RbcS subunits allow assembly but do not generate active enzyme. A, assembly of RsRbcL with RsRbcS or RbcS subunits from G. sulfuraria (Gs), P. tricornutum (Pt), Syn6301, A. thaliana (At) or Syn7002. Reconstitution reactions were performed and analyzed as in Fig. 4B. B, Rubisco activities in reactions as described in A. Error bars indicate S.D. of at least three independent experiments.
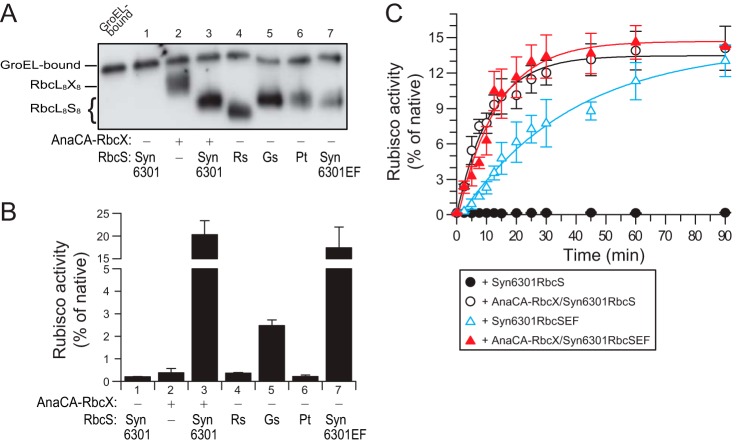
Red-type RbcS Subunits Can Overcome the Requirement of Green-type RbcL for the RbcX Assembly Factor
Based on the results above, we speculated that the absence of the β-hairpin region in green-type RbcS may explain the requirement of certain cyanobacterial Rubiscos for the RbcX assembly chaperone (19–21). We tested this using green-type RbcL from Syn6301, which requires RbcX for assembly with RbcS in vitro and remains bound to GroEL in the absence of RbcX (20). In vitro reconstitution confirmed that the formation of enzymatically active Syn6301RbcL8S8 complex was dependent on RbcX (Fig. 7, A and B). Note that the yield of this reaction is limited to ∼20% due to the inefficient release of Syn6301RbcL from GroEL (20). RbcX-independent assembly was detected by native PAGE with the red-type RbcS subunits from R. sphaeroides, G. sulfuraria, and P. tricornutum (Fig. 7A), but very little activity was measurable (Fig. 7B). Strikingly, a C-terminal fusion of Syn6301RbcS with the βEF region of RsRbcS (Syn6301RbcSEF) was able to produce active holoenzyme with similar final yield as Syn6301RbcS in the presence of RbcX (Fig. 7B). However, the apparent rate at which active enzyme was produced was substantially slower than that achieved with AnaCARbcX and Syn6301RbcS (Fig. 7C). This is probably due to a lower affinity of Syn6301RbcSEF for the folded RbcL subunits that tend to rebind to chaperonin. Combination of Syn6301RbcSEF and RbcX again resulted in rapid assembly (Fig. 7C). Thus, the C-terminal β-hairpin of red-type RbcS can substitute for the green-type assembly chaperone during in vitro reconstitution but with reduced assembly kinetics.
FIGURE 7.
Assembly of Syn6301RbcL Rubisco enzymes using heterologous red-type small subunits or green/red hybrid RbcS. A, assembly of Syn6301RbcL with Syn6301RbcS or RbcS subunits from R. sphaeroides, G. sulfuraria, or P. tricornutum or Syn6301RbcSEF with or without AnaCARbcX as indicated. Reconstitution reactions were performed and analyzed as in Fig. 4B. B, Rubisco activities in reactions as described in A. Error bars indicate S.D. of at least three independent experiments.
DISCUSSION
The RbcS subunits of green- and red-type Rubiscos are distinct in that the latter contain a C-terminal β-hairpin extension. The β-hairpins of 4 RbcS subunits form an eight-stranded β-barrel structure that protrudes into the central solvent channel of the RbcL8 core. Here we have shown for the Rubisco of R. sphaeroides that this structure confers to the enzyme the ability to assemble the RbcL8S8 complex independent of assembly chaperones such as RbcX. Although the β-hairpin is not essential for the assembly of red-type Rubisco and its enzymatic activity, it strongly enhances the rate of assembly and also increases the stability of the holoenzyme. Attachment of the C-terminal β-hairpin extension of RsRbcS to the green-type Syn6301RbcS allowed the assembly of Syn6301RbcL8SEF8 without RbcX, albeit with slower kinetics.
In all organisms containing red-type Rubisco, the rbcS gene follows the rbcL gene in an operon (3, 40). In eukaryotes with red-type Rubisco (non-green algae and diatoms), both genes are always plastid-encoded and linked (13). Such an organization would ensure that subunit stoichiometry is maintained independently of the overall rates of transcription and translation. In contrast, the genes encoding green-type rbcL and rbcS in cyanobacteria are generally in an operon with the assembly chaperone rbcX located in the intergenic space (41). In green algae and plants, the rbcL is chloroplast encoded, whereas the rbcS and rbcX genes are nuclear encoded (1). The operon linkage of red-type rbcL and rbcS genes, and thus their common regulation, may be a reflection of the function of RbcS in assembly. Given the finding that some red-type Rubiscos have higher CO2 specificities than the green-type enzymes (30, 42, 43), it is of interest to ask why organisms with green-type Rubiscos do not utilize an RbcS-assisted assembly mechanism but instead have evolved specialized assembly chaperones (19–25, 44). One possible reason is that the assembly factors, by providing independence of the rbcL-rbcS operon structure, may have facilitated the transfer of rbcS to the nuclear genome, an organization that presumably provided superior metabolic regulation and outweighed the advantage of RbcS-assisted assembly. Notably, co-expression of RbcL and RbcS from the red algae G. sulfuraria or the diatom P. tricornutum in tobacco chloroplasts failed to result in assembled red-type Rubisco (13), although these proteins would be expected to be independent of the green-type assembly factors. One possible explanation for the failed assembly is that the green-type RbcS or an assembly factor(s) may have exerted a dominant negative effect. It is also possible that the general chloroplast chaperone systems may be unable to mediate the folding of the red-type Rubisco subunits, considering that the non-green algal plastome also encodes its own Hsp70 (DnaK) and chaperonin (GroEL) (45, 46).
The exact mechanism by which the β-hairpins mediate assembly is not yet clear. Our reconstitution experiments did not provide evidence that red-type RbcS first forms a stable tetramer platform on to which RbcL dimers assemble. Neither do the red-type RbcL subunits assemble independent of RbcS to stable octameric core complexes, in contrast to green-type cyanobacterial RbcL (20, 21). Our data indicate that upon GroEL/ES-mediated folding, the RbcL subunits form a range of soluble oligomeric states which are in dynamic equilibrium (Fig. 1B). Addition of RbcS rapidly shifts this equilibrium toward holoenzyme formation: The RbcS β-barrel engages in extensive hydrogen bond and salt bridge contacts with the RbcL subunits (Fig. 3C). Furthermore, the βEF hairpin of one RbcS (residues 106–129) stabilizes the conserved βAB loop (residues 40–45) in the adjacent RbcS subunit (Fig. 3A and C), which in turn packs against the other RbcL subunit of the RbcL dimer. Thus, the RbcS β-barrel stabilizes the assembly at the level of antiparallel RbcL dimers and holds the dimers together. The RbcL dimer can be stabilized to some extent by sugar substrate binding, allowing assembly to occur with C-terminally truncated RbcS (Fig. 4D). However, only formation of the β-barrel drives rapid holoenzyme assembly (Fig. 5B). Our findings should facilitate future efforts to increase the yield of crop plants by expressing catalytically superior forms of red-type Rubisco in chloroplasts (6, 13).
Acknowledgments
We thank Prof. S. Whitney (Australian National University, Canberra, Australia) for the G. sulfuraria plastome fragment encoding rbcL-rbcS-cbbX. We thank K. Vasudeva Rao for the initial cloning of G. sulfuraria and A. thaliana RbcS and A. Sharma for the synthesis of CABP. We thank A. Bracher for preparing Fig. 3.
Footnotes
- Rubisco
- ribulose 1,5-bisphosphate carboxylase/oxygenase
- At
- Arabidopsis thaliana
- Gs
- Galdieria sulphuraria
- GuHCl
- guanidinium hydrochloride
- CABP
- 2-carboxyarabinitol-1,5-bisphosphate
- Pt
- Phaeodactylum tricornutum
- RbcL
- Rubisco large subunit
- RbcS
- Rubisco small subunit
- Rs
- Rhodobacter sphaeroides
- Syn6301
- Synechococcus sp. PCC 6301
- Syn7002
- Synechococcus sp. PCC 7002.
REFERENCES
- 1. Spreitzer R. J., Salvucci M. E. (2002) RUBISCO: structure, regulatory interactions, and possibilities for a better enzyme. Annu. Rev. Plant Biol. 53, 449–475 [DOI] [PubMed] [Google Scholar]
- 2. Andersson I., Backlund A. (2008) Structure and function of Rubisco. Plant Physiol. Biochem. 46, 275–291 [DOI] [PubMed] [Google Scholar]
- 3. Tabita F. R. (1999) Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a different perspective. Photosynth. Res. 60, 1–28 [Google Scholar]
- 4. Tabita F. R., Satagopan S., Hanson T. E., Kreel N. E., Scott S. S. (2008) Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J. Exp. Bot. 59, 1515–1524 [DOI] [PubMed] [Google Scholar]
- 5. Badger M. R., Bek E. J. (2008) Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot. 59, 1525–1541 [DOI] [PubMed] [Google Scholar]
- 6. Parry M. A.., Andralojc P. J., Scales J. C., Salvucci M. E., Carmo-Silva A. E., Alonso H., Whitney S. M. (2013) Rubisco activity and regulation as targets for crop improvement. J. Exp. Bot. 64, 717–730 [DOI] [PubMed] [Google Scholar]
- 7. Andrews T. J., Lorimer G. H. (1987) Rubisco: structure, mechanisms, and prospects for improvement. In The Biochemistry of Plants: A Comprehensive Treatise (Hatch M. D., Boardman N. K., eds), pp. 131–218, Academic Press, New York [Google Scholar]
- 8. Gutteridge S., Gatenby A. A. (1995) Rubisco synthesis, assembly, mechanism, and regulation. Plant Cell 7, 809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. John Andrews T., Whitney S. M. (2003) Manipulating ribulose bisphosphate carboxylase/oxygenase in the chloroplasts of higher plants. Arch. Biochem. Biophys. 414, 159–169 [DOI] [PubMed] [Google Scholar]
- 10. Spreitzer R. J., Peddi S. R., Satagopan S. (2005) Phylogenetic engineering at an interface between large and small subunits imparts land-plant kinetic properties to algal Rubisco. Proc. Natl. Acad. Sci. U.S.A. 102, 17225–17230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Long S. P., Zhu X. G., Naidu S. L., Ort D. R. (2006) Can improvement in photosynthesis increase crop yields? Plant Cell Environ. 29, 315–330 [DOI] [PubMed] [Google Scholar]
- 12. Whitney S. M., Houtz R. L., Alonso H. (2011) Advancing our understanding and capacity to engineer nature's CO2-sequestering enzyme, Rubisco. Plant Physiol. 155, 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Whitney S. M., Baldet P., Hudson G. S., Andrews T. J. (2001) Form I Rubiscos from non-green algae are expressed abundantly but not assembled in tobacco chloroplasts. Plant J. 26, 535–547 [DOI] [PubMed] [Google Scholar]
- 14. Horken K. M., Tabita F. R. (1999) Closely related form I ribulose bisphosphate carboxylase/oxygenase molecules that possess different CO2/O2 substrate specificities. Arch. Biochem. Biophys. 361, 183–194 [DOI] [PubMed] [Google Scholar]
- 15. Mueller-Cajar O., Stotz M., Wendler P., Hartl F. U., Bracher A., Hayer-Hartl M. (2011) Structure and function of the AAA+ protein CbbX, a red-type Rubisco activase. Nature 479, 194–199 [DOI] [PubMed] [Google Scholar]
- 16. Hill J. E., Hemmingsen S. M. (2001) Arabidopsis thaliana type I and II chaperonins. Cell Stress Chaperones 6, 190–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weiss C., Bonshtien A., Farchi-Pisanty O., Vitlin A., Azem A. (2009) Cpn20: siamese twins of the chaperonin world. Plant Mol. Biol. 69, 227–238 [DOI] [PubMed] [Google Scholar]
- 18. Tsai Y. C., Mueller-Cajar O., Saschenbrecker S., Hartl F. U., Hayer-Hartl M. (2012) Chaperonin cofactors, Cpn10 and Cpn20, of green algae and plants function as hetero-oligomeric ring complexes. J. Biol. Chem. 287, 20471–20481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saschenbrecker S., Bracher A., Rao K. V., Rao B. V., Hartl F. U., Hayer-Hartl M. (2007) Structure and function of RbcX, an assembly chaperone for hexadecameric Rubisco. Cell 129, 1189–1200 [DOI] [PubMed] [Google Scholar]
- 20. Liu C., Young A. L., Starling-Windhof A., Bracher A., Saschenbrecker S., Rao B. V., Rao K. V., Berninghausen O., Mielke T., Hartl F. U., Beckmann R., Hayer-Hartl M. (2010) Coupled chaperone action in folding and assembly of hexadecameric Rubisco. Nature 463, 197–202 [DOI] [PubMed] [Google Scholar]
- 21. Bracher A., Starling-Windhof A., Hartl F. U., Hayer-Hartl M. (2011) Crystal structure of a chaperone-bound assembly intermediate of form I Rubisco. Nat. Struct. Mol. Biol. 18, 875–880 [DOI] [PubMed] [Google Scholar]
- 22. Feiz L., Williams-Carrier R., Wostrikoff K., Belcher S., Barkan A., Stern D. B. (2012) Ribulose-1,5-bis-phosphate carboxylase/oxygenase accumulation factor1 is required for holoenzyme assembly in maize. Plant Cell 24, 3435–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wheatley N. M., Sundberg C. D., Gidaniyan S. D., Cascio D., Yeates T. O. (2014) Structure and identification of a pterin dehydratase-like protein as a RuBisCO assembly factor in the α-carboxysome. J. Biol. Chem. 289, 7973–7981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kolesinski P., Belusiak I., Czarnocki-Cieciura M., Szczepaniak A. (2014) Rubisco accumulation factor 1 from Thermosynechococcus elongatus participates in the final stages of ribulose-1,5-bisphosphate carboxylase/oxygenase assembly in Escherichia coli cells and in vitro. FEBS J. 281, 3920–3932 [DOI] [PubMed] [Google Scholar]
- 25. Feiz L., Williams-Carrier R., Belcher S., Montano M., Barkan A., Stern D. B. (2014) RAF2 - A novel Rubisco biogenesis factor in maize. Plant J. 10.1111/tpj.12686 [DOI] [PubMed] [Google Scholar]
- 26. Armbrust E. V., Berges J. A., Bowler C., Green B. R., Martinez D., Putnam N. H., Zhou S., Allen A. E., Apt K. E., Bechner M., Brzezinski M. A., Chaal B. K., Chiovitti A., Davis A. K., Demarest M. S., Detter J. C., Glavina T., Goodstein D., Hadi M. Z., Hellsten U., Hildebrand M., Jenkins B. D., Jurka J., Kapitonov V. V., Kröger N., Lau W. W., Lane T. W., Larimer F. W., Lippmeier J. C., Lucas S., Medina M., Montsant A., Obornik M., Parker M. S., Palenik B., Pazour G. J., Richardson P. M., Rynearson T. A., Saito M. A., Schwartz D. C., Thamatrakoln K., Valentin K., Vardi A., Wilkerson F. P., Rokhsar D. S. (2004) The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86 [DOI] [PubMed] [Google Scholar]
- 27. Bowler C., Allen A. E., Badger J. H., Grimwood J., Jabbari K., Kuo A., Maheswari U., Martens C., Maumus F., Otillar R. P., Rayko E., Salamov A., Vandepoele K., Beszteri B., Gruber A., Heijde M., Katinka M., Mock T., Valentin K., Verret F., Berges J. A., Brownlee C., Cadoret J. P., Chiovitti A., Choi C. J., Coesel S., De Martino A., Detter J. C., Durkin C., Falciatore A., Fournet J., Haruta M., Huysman M. J., Jenkins B. D., Jiroutova K., Jorgensen R. E., Joubert Y., Kaplan A., Kröger N., Kroth P. G., La Roche J., Lindquist E., Lommer M., Martin-Jézéquel V., Lopez P. J., Lucas S., Mangogna M., McGinnis K., Medlin L. K., Montsant A., Oudot-Le Secq M. P., Napoli C., Obornik M., Parker M. S., Petit J.-L., Porcel B. M., Poulsen N., Robison M., Rychlewski L., Rynearson T. A., Schmutz J., Shapiro H., Siaut M., Stanley M., Sussman M. R., Taylor A. R., Vardi A., Dassow P. v., Vyverman W., Willis A., Wyrwicz L. S., Rokhsar D. S., Weissenbach J., Armbrust E. V., Green B. R., van de Peer Y., Grigoriev I. V. (2008) The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244 [DOI] [PubMed] [Google Scholar]
- 28. Kontur W. S., Schackwitz W. S., Ivanova N., Martin J., Labutti K., Deshpande S., Tice H. N., Pennacchio C., Sodergren E., Weinstock G. M., Noguera D. R., Donohue T. J. (2012) Revised sequence and annotation of the Rhodobacter sphaeroides 2.4.1 genome. J. Bacteriol. 194, 7016–7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bhattacharya D., Price D. C., Chan C. X., Qiu H., Rose N., Ball S., Weber A. P. M., Cecilia Arias M., Henrissat B., Coutinho P. M., Krishnan A., Zäuner S., Morath S., Hilliou F., Egizi A., Perrineau M. M., Yoon H. S. (2013) Genome of the red alga Porphyridium purpureum. Nat. Commun. 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hansen S., Vollan V. B., Hough E., Andersen K. (1999) The crystal structure of rubisco from Alcaligenes eutrophus reveals a novel central eight-stranded β-barrel formed by β-strands from four subunits. J. Mol. Biol. 288, 609–621 [DOI] [PubMed] [Google Scholar]
- 31. Spreitzer R. J. (2003) Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase. Arch. Biochem. Biophys. 414, 141–149 [DOI] [PubMed] [Google Scholar]
- 32. Hayer-Hartl M. K., Weber F., Hartl F. U. (1996) Mechanism of chaperonin action: GroES binding and release can drive GroEL-mediated protein folding in the absence of ATP hydrolysis. EMBO J. 15, 6111–6121 [PMC free article] [PubMed] [Google Scholar]
- 33. Brinker A., Pfeifer G., Kerner M. J., Naylor D. J., Hartl F. U., Hayer-Hartl M. (2001) Dual function of protein confinement in chaperonin-assisted protein folding. Cell 107, 223–233 [DOI] [PubMed] [Google Scholar]
- 34. Goloubinoff P., Christeller J. T., Gatenby A. A., Lorimer G. H. (1989) Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfolded state depends on two chaperonin proteins and Mg-ATP. Nature 342, 884–889 [DOI] [PubMed] [Google Scholar]
- 35. Mueller-Cajar O., Whitney S. M. (2008) Evolving improved Synechococcus Rubisco functional expression in Escherichia coli. Biochem. J. 414, 205–214 [DOI] [PubMed] [Google Scholar]
- 36. Karkehabadi S., Peddi S. R., Anwaruzzaman M., Taylor T. C., Cederlund A., Genkov T., Andersson I., Spreitzer R. J. (2005) Chimeric small subunits influence catalysis without causing global conformational changes in the crystal structure of ribulose-1,5-bisphosphate carboxylase/oxygenase. Biochemistry 44, 9851–9861 [DOI] [PubMed] [Google Scholar]
- 37. Sugawara H., Yamamoto H., Shibata N., Inoue T., Okada S., Miyake C., Yokota A., Kai Y. (1999) Crystal structure of carboxylase reaction-oriented ribulose 1, 5-bisphosphate carboxylase/oxygenase from a thermophilic red alga, Galdieria partita. J. Biol. Chem. 274, 15655–15661 [DOI] [PubMed] [Google Scholar]
- 38. Alonso H., Blayney M. J., Beck J. L., Whitney S. M. (2009) Substrate-induced assembly of Methanococcoides burtonii d-ribulose-1,5-bisphosphate carboxylase/oxygenase dimers into decamers. J. Biol. Chem. 284, 33876–33882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Lun M., van der Spoel D., Andersson I. (2011) Subunit interface dynamics in hexadecameric Rubisco. J. Mol. Biol. 411, 1083–1098 [DOI] [PubMed] [Google Scholar]
- 40. Hartman F. C., Harpel M. R. (1994) Structure, function, regulation, and assembly of d-ribulose-1,5-bisphosphate carboxylase/oxygenase. Annu. Rev. Biochem. 63, 197–234 [DOI] [PubMed] [Google Scholar]
- 41. Larimer F. W., Soper T. S. (1993) Overproduction of Anabaena 7120 ribulose-bisphosphate carboxylase/oxygenase in Escherichia coli. Gene 126, 85–92 [DOI] [PubMed] [Google Scholar]
- 42. Read B. A., Tabita F. R. (1992) A hybrid ribulosebisphosphate carboxylase/oxygenase enzyme exhibiting a substantial increase in substrate specificity factor. Biochemistry 31, 5553–5560 [DOI] [PubMed] [Google Scholar]
- 43. Kanevski I., Maliga P., Rhoades D. F., Gutteridge S. (1999) Plastome engineering of ribulose-1,5-bisphosphate carboxylase/oxygenase in tobacco to form a sunflower large subunit and tobacco small subunit hybrid. Plant Physiol. 119, 133–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kolesiński P., Piechota J., Szczepaniak A. (2011) Initial characteristics of RbcX proteins from Arabidopsis thaliana. Plant Mol. Biol. 77, 447–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reith M. (1995) Molecular-biology of rhodophyte and chromophyte plastids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 549–575 [Google Scholar]
- 46. Allen J. F., de Paula W. B., Puthiyaveetil S., Nield J. (2011) A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci. 16, 645–655 [DOI] [PubMed] [Google Scholar]
- 47. Newman J., Branden C. I., Jones T. A. (1993) Structure determination and refinement of ribulose 1,5-bisphosphate carboxylase/oxygenase from Synechococcus PCC6301. Acta Crystallogr. D Biol. Crystallogr. 49, 548–560 [DOI] [PubMed] [Google Scholar]