Background: Synthesis of the E. coli O9a polysaccharide requires a multidomain polymerase and a terminator protein.
Results: C-terminal deletions in the polymerase compromise fidelity or activity.
Conclusion: Complex interactions between polymerase domains and the terminator are essential for accurate synthesis.
Significance: These properties determine serotype specificity and may be exploited in glycoengineering to produce glycans with novel structures and applications.
Keywords: Carbohydrate Biosynthesis, Enzyme Structure, Glycobiology, Glycosyltransferase, Lipopolysaccharide (LPS), Microbiology, Polysaccharide, Protein Complex
Abstract
The Escherichia coli O9a O-polysaccharide (O-PS) is a prototype for bacterial glycan synthesis and export by an ATP-binding cassette transporter-dependent pathway. The O9a O-PS possesses a tetrasaccharide repeat unit comprising two α-(1→2)- and two α-(1→3)-linked mannose residues and is extended on a polyisoprenoid lipid carrier by the action of a polymerase (WbdA) containing two glycosyltransferase active sites. The N-terminal domain of WbdA possesses α-(1→2)-mannosyltransferase activity, and we demonstrate in this study that the C-terminal domain is an α-(1→3)-mannosyltransferase. Previous studies established that the size of the O9a polysaccharide is determined by the chain-terminating dual kinase/methyltransferase (WbdD) that is tethered to the membrane and recruits WbdA into an active enzyme complex by protein-protein interactions. Here, we used bacterial two-hybrid analysis to identify a surface-exposed α-helix in the C-terminal mannosyltransferase domain of WbdA as the site of interaction with WbdD. However, the C-terminal domain was unable to interact with WbdD in the absence of its N-terminal partner. Through deletion analysis, we demonstrated that the α-(1→2)-mannosyltransferase activity of the N-terminal domain is regulated by the activity of the C-terminal α-(1→3)-mannosyltransferase. In mutants where the C-terminal catalytic site was deleted but the WbdD-interaction site remained, the N-terminal mannosyltransferase became an unrestricted polymerase, creating a novel polymer comprising only α-(1→2)-linked mannose residues. The WbdD protein therefore orchestrates critical localization and coordination of activities involved in chain extension and termination. Complex domain interactions are needed to position the polymerase components appropriately for assembly into a functional complex located at the cytoplasmic membrane.
Introduction
Many different macromolecules containing complex carbohydrates (glycoconjugates) are found on the surfaces of living cells. These structures play crucial roles in the interactions between the cell and its environment or in communications and interactions between one cell and another. Glycoconjugates on the surfaces of bacteria are important in survival against factors in the environment or against the protective responses of mammalian or plant hosts. The function of a particular glycoconjugate is dictated by its structure, and the precise order of sugars and linkages in a glycan are dictated by the specificities of glycosyltransferase (GT)5 enzymes. Although 97 GT families are recognized in the Carbohydrate-Active Enzyme (CAZy) database (1) based on bioinformatics or structural criteria (2), the features dictating acceptor/donor specificity are often unknown and cannot be inferred from either catalytic fold or GT family (3). Most GTs catalyze the formation of a single glycosidic linkage, but increasing numbers of polymerases are being identified. These enzymes sequentially add multiple residues to an acceptor molecule, creating complex glycans of varying lengths without a template. Examples are found in prokaryotes and eukaryotes, and some form highly important products such as cellulose, chitin, hyaluronic acid, chondroitin, and polysialic acid (4, 5). Depending on the enzyme and the complexity of the product, polymerases may contain one or more GT active site(s). Polymerases operate by either “processive” or “distributive” mechanisms (6, 7). Processive enzymes catalyze multiple rounds of glycose transfer to a given acceptor while maintaining the growing glycan in a single active site. In contrast, strictly distributive enzymes possess one or more active sites and release the glycan product after each addition. The structural principles that guide efficiency and fidelity of distributive multidomain polymerase enzymes are largely unknown. The polymerase involved in the biosynthesis of the lipopolysaccharide (LPS) O9a O-polysaccharide (O-PS) antigen in Escherichia coli provides a prototype for addressing these questions.
The O9a system is a representative of the widespread ATP-binding cassette transporter-dependent assembly pathway (8, 9). The pathway (see Fig. 1A) begins with the transfer of N-acetylglucosamine 1-phosphate to a 55-carbon polyisoprenoid lipid acceptor (undecaprenol phosphate) catalyzed by WecA (10, 11). Undecaprenol diphosphate-GlcpNAc is then committed to O9a biosynthesis by the action of WbdC and WbdB, two GDP-mannose (GDP-Manp)-dependent mannosyltransferases that add a trimannose adapter region (12). These enzymes and the resulting structure are conserved in E. coli serotypes O9a, O9, and O8, which produce structurally related O-PSs differing in the type and sequence of linkages in their repeat units (9). A serotype-specific multidomain polymerizing mannosyltransferase (WbdA) extends this adapter to create the precise repeating unit O-PS (12, 13). In E. coli O9a, the peripheral membrane protein WbdD terminates polymerization by adding a methyl phosphate to the non-reducing end of the nascent O9a polymer (14–16). This terminal modification is required for recognition and export of the completed O-PS across the cytoplasmic membrane by its cognate ATP-binding cassette transporter. The recognition event is mediated by the nucleotide-binding domain polypeptide of the transporter, which possesses a serotype-specific carbohydrate-binding module that only binds terminated O-PS chains (17, 18). The WbdD terminator plays an additional pivotal structural role in recruiting WbdA to the membrane (19). The stoichiometry of WbdA:WbdD in active complexes is a critical factor in establishing the chain length distribution of the resulting glycans, and the “variable geometry” model has been developed to explain this process (20). The key element in the model is that complexes with different stoichiometries have different physical sizes that generate a range of glycan chain lengths. Implicit in this model is a structural element that serves as a molecular ruler to establish chain length. This role is fulfilled by an extended α-helical bundle that holds the WbdD trimer together and maintains a critical distance between the catalytic sites of the terminator and its site of interaction with the WbdA polymerase (21). WbdD is therefore a central player in a sophisticated quality control system that dictates the distribution of chain lengths and marks those chains with a terminal export tag. After export, the O-PS is transferred to the lipid A-core portion of LPS, translocated to the cell surface, and inserted into the outer membrane (22, 23).
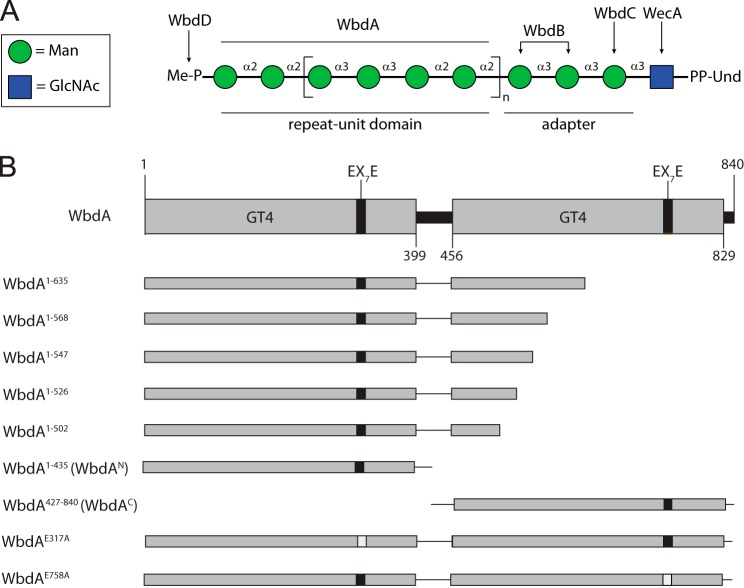
FIGURE 1.
Biosynthesis of the O9a antigenic polysaccharide in E. coli and a schematic representation of the WbdA constructs used in this study. In A, the structure of the undecaprenol diphosphate (Und-PP)-linked biosynthetic intermediate is shown. Sugars are represented using the nomenclature established by the Consortium for Functional Glycomics. The enzymes responsible for the formation of each part of the glycan are identified. The polymerase, WbdA, is solely responsible for the extension of the repeat unit region of the glycan, extending a trisaccharide adapter. B describes the WbdA active site mutants, domains, and truncated proteins whose function was examined. Domain predictions were identified using the NCBI Conserved Domain Database (33). This allowed the separation of WbdAN and WbdAC, which retain activity and synthesize authentic O9a polysaccharide when coexpressed in E. coli ΔwbdA (13).
The WbdA polymerase possesses a modular design comprising two identifiable GT(family)4 domains as defined by the CAZy classification system (1, 2). The two GT4 active sites correlate with the presence of α-(1→2) and α-(1→3) linkages in the O9a tetrasaccharide repeat unit (13) (Fig. 1A). Coexpression of the two domains (with coding sequences cloned in separate plasmids) restores O9a biosynthesis in an E. coli wbdA deletion mutant, but neither domain shows any in vivo activity when expressed alone (13). Surprisingly, when examined in vitro with synthetic oligosaccharide acceptors, the purified N-terminal domain (WbdAN) transfers multiple Manp residues to the synthetic acceptor to create α-(1→2)-linked polymannose, which is not seen in the authentic O9a glycan population (13). The same product can be generated in vivo in cells expressing a full-length WbdA polypeptide possessing a defect in the EX7E motif in the C-terminal catalytic site (13). This motif is involved in hydrogen bonding to GDP-Manp substrate (24) and is essential for catalysis in each of the mannosyltransferase domains of WbdA. The collective data indicate that WbdAN is an α-(1→2)-Manp GT but that fidelity of its function in O9a biosynthesis requires the presence of the C-terminal domain (WbdAC). In contrast, constructs containing only WbdAC show no activity in vivo or in vitro (13). The purpose of this study was to rationalize the mechanistic basis for the difference in in vivo and in vitro activities of the two GT4 domains.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are described in Table 1. Bacteria were grown in lysogeny broth (LB) (25) at 37 °C unless otherwise stated. Where appropriate, media contained one or more of the following supplements: d-glucose (0.4%, w/v), d-mannose (0.1%, w/v), l-arabinose (0.2%, w/v), kanamycin (50 μg/ml), ampicillin (100 μg/ml), chloramphenicol (34 μg/ml), 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal) (40 μg/ml), and isopropyl β-d-thiogalactopyranoside (0.5 mm).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or property | Source or Ref. |
|---|---|---|
| E. coli | ||
| Top10 | F−, mcrA, Δ(mrr-hsdRMS-mcrBC), φ80, lacZΔM15, ΔlacX74, deoR, nupG, recA1, araD139, Δ(ara-leu)7697, galU, galK, rpsL(Strr), endA1 | Invitrogen |
| BL21 | B F− dcm ompT hsdS(rB− mB−) gal [malB+]K-12(λS) | Novagen |
| BTH101 | cya-854, recA1, endA1, gyrA96, thi1, hsdR17, spoT1, rfbD1, glnV44(AS); Nalr | 54 |
| CWG1105 | O9a:K−; trp his lac rpsL cpsK30, manA, ΔwbdA; Smr; Tcr | 12 |
| Plasmid | ||
| pBAD24 | Plasmid vector with l-arabinose-inducible promoter; Apr | 26 |
| pKT25 | Bacterial two-hybrid vector with sequence encoding the T25 fragment of B. pertussis adenylate cyclase upstream of a multiple cloning site; Kmr | 54 |
| pUT18 | Bacterial two-hybrid vector with sequence encoding the T18 fragment of B. pertussis adenylate cyclase downstream of a multiple cloning site; Apr | 54 |
| pUT18C | Bacterial two-hybrid vector with sequence encoding the T18 fragment of B. pertussis adenylate cyclase upstream of a multiple cloning site; Apr | 54 |
| pKT25-Zip | pKT25 derivative encoding the CyaA T25 fragment fused to the leucine zipper domain of the yeast GCN4 protein; Kmr | 54 |
| pUT18C-Zip | pUT18C derivative encoding the CyaA T18 fragment fused to the leucine zipper domain of the yeast GCN4 protein; Apr | 54 |
| pUT18-Zip | pUT18 derivative encoding the CyaA T18 fragment fused to the leucine zipper domain of the yeast GCN4 protein; Apr | 54 |
| pWQ284 | pBAD24 derivative containing a chloramphenicol resistance cassette; Cmr | 18 |
| pWQ486 | pKT25 derivative encoding T25-WbdD; Kmr | 19 |
| pWQ487 | pUT18C derivative encoding T18-WbdA; Apr | 19 |
| pWQ492 | pBAD24 derivative encoding WbdA-His10; Apr | 19 |
| pWQ589 | pBAD24 derivative containing a kanamycin resistance cassette; Kmr | 13 |
| pWQ590 | pBAD24 derivative encoding His10-WbdA1–435; Apr | 13 |
| pWQ591 | pWQ284 derivative encoding His10-WbdA427–840; Cmr | 13 |
| pWQ630 | pWQ631 derivative encoding WbdAE758A; Kmr | 13 |
| pWQ631 | pWQ589 derivative encoding WbdD475–708-WbdA; Kmr | 13 |
| pWQ765 | pWQ631 derivative containing WbdD475–708-WbdA (frameshift mutation after amino acid 635, now encodes S636RPRCQRTVHWHAG-); Kmr | This study |
| pWQ766 | pWQ284 derivative encoding His10-WbdA1–435; Cmr | This study |
| pWQ769 | pWQ492 derivative encoding WbdAE317A-His10; Apr | This study |
| pWQ770 | pWQ631 derivative encoding WbdD475–708-WbdA1–635; Kmr | This study |
| pWQ771 | pWQ631 derivative encoding WbdD475–708-WbdA1–568; Kmr | This study |
| pWQ772 | pWQ631 derivative encoding WbdD475–708-WbdA1–547; Kmr | This study |
| pWQ773 | pWQ631 derivative encoding WbdD475–708-WbdA1–526; Kmr | This study |
| pWQ774 | pWQ631 derivative encoding WbdD475–708-WbdA1–502; Kmr | This study |
| pWQ775 | pUT18C derivative encoding T18-WbdA1–635; Apr | This study |
| pWQ776 | pUT18C derivative encoding T18-WbdA1–568; Apr | This study |
| pWQ777 | pUT18C derivative encoding T18-WbdA1–547; Apr | This study |
| pWQ778 | pUT18C derivative encoding T18-WbdA1–526; Apr | This study |
| pWQ779 | pUT18C derivative encoding T18-WbdA1–502; Apr | This study |
| pWQ780 | pUT18C derivative encoding T18-WbdA1–435; Apr | This study |
| pWQ781 | pUT18 derivative encoding WbdA427–840-T18; Apr | This study |
DNA Methods
Pwo DNA polymerase (Roche Applied Science) was used to PCR amplify DNA fragments used in cloning. The oligonucleotide primers (Sigma) used for PCR introduced restriction endonuclease sites and epitope tags; their particular features are described in supplemental Table S1. DNA fragments were purified using the PureLink PCR Purification kit (Invitrogen). The PureLink Quick Plasmid Miniprep kit (Invitrogen) was used to purify plasmid DNA. Restriction endonucleases (Invitrogen and New England Biolabs) and T4 DNA ligase (New England Biolabs) were used as described by the manufacturers. PfuUltra high fidelity DNA polymerase (Stratagene) was used for site-directed mutagenesis of plasmids according to the QuikChange method (Stratagene). All DNA constructs were assessed by restriction endonuclease digestion and verified by DNA sequencing. Sequencing reactions were performed by the Genomics Facility at the Advanced Analysis Center (University of Guelph).
Generation of WbdA Constructs
Nested C-terminal truncations of WbdA (see Fig. 1B) were generated in pWQ631 using complementary oligonucleotide primers containing base changes that encoded a stop codon and a unique restriction site for screening purposes. PCR products were digested using DpnI and transformed into E. coli Top10. For complementation experiments, each of the plasmids encoding truncated WbdA derivatives also contained sequences encoding the C-terminal fragment of WbdD (residues 475–708). This fragment has been used in previous work to increase the quantity of O-PS synthesized by WbdA constructs (13) and was used here for the same purpose. For protein expression and purification, DNA fragments encoding WbdA (and its various derivatives) were cloned behind the pBAD arabinose-inducible promoter (26). To assess protein-protein interactions, DNA fragments were cloned into pUT18 and pUT18C to create N- and C-terminal fusions of WbdA with the T18 fragment of the Bordetella pertussis adenylate cyclase.
Expression of WbdA and Its C-terminally Truncated Derivatives
5-ml cultures of E. coli CWG1105 (ΔwbdA) transformed with pWQ631-based plasmids were grown in LB medium at 37 °C until the A600 reached 0.3. Growth was continued at 20 °C until the A600 reached 0.5 after which wbdA expression was induced with l-arabinose (0.01%, w/v) and incubation was continued for 16 h. Culture volumes containing equivalent amounts of cells (as determined by optical density) were subjected to centrifugation at 5000 × g for 10 min, and the pellets were resuspended in SDS-PAGE loading buffer (27).
Purification of Decahistidine-tagged WbdA Derivatives
This protocol was adapted from methods described previously (13). In brief, protein was purified from exponential phase 500-ml cultures of E. coli BL21 transformed with plasmids pWQ492, pWQ769, and pWQ590 encoding WbdA-His10, WbdAE317A-His10, and His10-WbdA1–435 (i.e. His10-WbdAN), respectively. These plasmids are all pBAD derivatives (26), and gene expression was induced overnight at 20 °C with l-arabinose as described above. Cells were collected by centrifugation and resuspended in 100 ml of buffer A (20 mm BisTris, 250 mm NaCl, 5% (w/v) glycerol, pH 7.0) supplemented with Complete Mini EDTA-free protease inhibitor tablets (Roche Applied Science). Cells were lysed using an EmulsiFlex homogenizer (Avestin), and the lysate was cleared by centrifugation at 5000 × g for 10 min. The cell-free lysate was treated with 5 μl of Benzonase nuclease (Novagen) for 15 min at room temperature and then centrifuged at 100,000 × g for 60 min to remove cell membranes. Purification was performed using an ÄKTA Explorer system (GE Healthcare) and a 5-ml HiTrap chelating HP column charged with Ni2+ (GE Healthcare). Individual lysates containing His10-tagged WbdA derivatives were loaded at 4 ml/min, and the column was washed sequentially with 5 column volumes of buffer A containing 0, 50, 75, 125, and 250 mm imidazole. Fractions containing the His10-tagged WbdA derivatives were pooled, exchanged into Buffer B (20 mm BisTris, 50 mm NaCl, pH 7.0), concentrated using a Vivaspin column (50,000 molecular weight cutoff; Sartorius Biolab Products), and stored for subsequent use in in vitro enzyme reactions. The concentrations of WbdA-His10 (30 mg/ml), WbdAE317A-His10 (4.5 mg/ml), and His10-WbdAN (5 mg/ml) were estimated from A280 values, and the theoretical extinction coefficients were predicted by ProtParam (28).
Size Exclusion Chromatography
Purified WbdA-His10 in Buffer A containing 250 mm imidazole was dialyzed with Buffer C (50 mm BisTris, 150 mm NaCl, pH 7.0). WbdA-His10 (0.2 mg at 2 mg/ml) was eluted through a Superose 6 HR 10/30 size exclusion column (GE Healthcare) with Buffer C at 0.5 ml/min. Molecular mass was estimated from a standard curve derived from the standards β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), ovalbumin (44 kDa), and carbonic anhydrase (29 kDa).
Bacterial Two-hybrid Interactions
E. coli BTH101 was transformed with the appropriate two-hybrid plasmid combinations. The transformed cell suspensions were serially diluted in LB and plated on LB agar containing X-Gal, antibiotics, isopropyl β-d-thiogalactopyranoside, and l-arabinose (if required). Plates were incubated at 30 °C for 48 h before two-hybrid interactions were assessed visually for the presence of blue colonies. For quantitative β-galactosidase assays, cultures of E. coli BTH101 containing recombinant two-hybrid plasmids were grown for 16 h at 37 °C in LB containing glucose and antibiotics. Each starter culture was subcultured (1:50) into 5 ml of LB supplemented with antibiotics and isopropyl β-d-thiogalactopyranoside (and l-arabinose if required), and the cultures were grown with shaking at 30 °C for 24 h. The optical density (at 600 nm) of each culture was measured, and 1:10 dilutions in LB were prepared. Aliquots (70 μl) of the diluted cultures were assayed for β-galactosidase activity using the Pierce β-galactosidase assay kit according to the stopped microplate plate assay protocol with a reaction time of 7.5 min. Bacterial Protein Extraction Reagent (BPER-II; Pierce) was used in the place of Yeast Protein Extraction Reagent provided with the kit. Each two-hybrid combination was measured from at least three independent cultures. Activity is reported in Miller units (29).
In Vitro Mannosyltransferase Reactions Using Synthetic Acceptors
In vitro biosynthesis was performed using synthetic fluorescein-tagged oligosaccharide acceptors as described previously (12, 13). Acceptor A (α-Manp-(1→2)-α-Manp-(1→2)-α-Manp-(1→3)-α-Manp) represents the repeat unit of the O9a antigen, whereas acceptor B (α-Manp-(1→3)-α-Manp-(1→3)-β-GlcpNAc) represents the conserved reducing terminal trisaccharide of the O9a antigens. Standard reactions were performed for 30 min in 250-μl reaction volumes with 10 μm enzyme, 0.5 mm acceptor, and 5 mm GDP-Manp in Buffer D (50 mm HEPES, 20 mm MgCl2, 1 mm dithiothreitol, pH 7.5). Product generation was initially verified by thin layer chromatography using AL SIL G TLC plates (Whatman) developed in ethyl acetate:water:1-butanol:acetic acid (5:4:4:2.5). Fluorescent reaction products were detected with a hand-held UV lamp. Reaction mixtures were diluted in 1 ml of water and loaded onto a C18 Sep-Pak cartridge (Waters). The cartridge was washed extensively with water, and the products were eluted in 2 ml of 60% (v/v) aqueous acetonitrile and concentrated using a SpeedVac concentrator. MALDI-TOF mass spectra of the reaction products were obtained on a Bruker ultrafleXtreme MALDI-TOF/TOF in negative ion mode. All NMR spectra were acquired in D2O at 27 °C on an Agilent VNMRS 700-MHz spectrometer equipped with a cryoprobe. The spectra were referenced to an external standard of acetone (2.22 ppm for 1H and 31.07 ppm for 13C at 27 °C).
Protein and LPS PAGE
For routine monitoring of proteins, cells were solubilized in SDS-PAGE loading buffer (27). LPS samples were prepared by digestion of these whole-cell lysates with proteinase K (30). Both protein and LPS samples were heated at 100 °C for 10 min prior to analysis by SDS-PAGE in Tris-glycine buffer (27). Silver staining was used to visualize LPS (31), and Simply Blue (Invitrogen) was used to visualize protein. For Western immunoblotting, protein and LPS samples were transferred to nitrocellulose membranes (Protran, PerkinElmer Life Sciences). LPS immunoblots were probed with O9a-specific rabbit antiserum (14) and alkaline phosphatase-conjugated goat anti-rabbit secondary antibody (Cedarlane Laboratories) and developed using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Roche Applied Science) substrates. Protein immunoblots were probed with WbdA-specific rabbit antiserum (19) or mouse pentahistidine-specific monoclonal antibody (Qiagen), and the secondary antibodies were either horseradish peroxidase-conjugated goat anti-rabbit (Sigma) or horseradish peroxidase-conjugated goat anti-mouse (Jackson ImmunoResearch Laboratories). Protein immunoblots were developed using Luminata Classico (Millipore) chemiluminescent substrate.
Bioinformatics Analyses
Molecular modeling of WbdAC was performed using the Phyre2 server in the intensive mode (32). Conserved domains were identified by the NCBI conserved domain database (13, 33).
RESULTS
In Vitro Activity of the C-terminal GT4 Domain
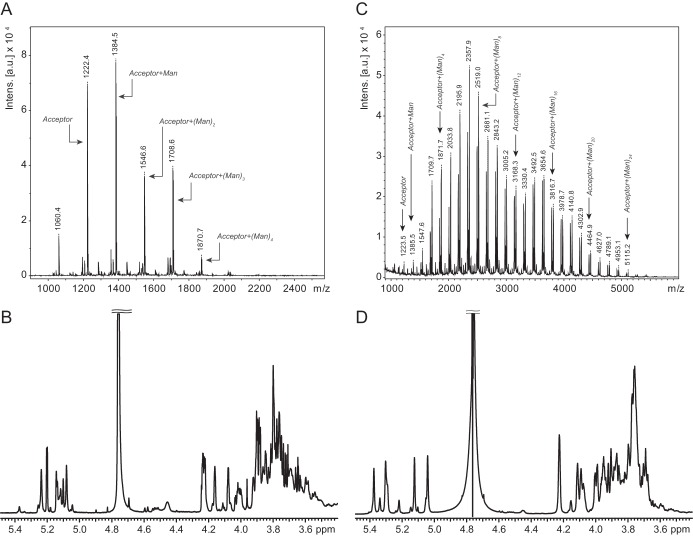
Previously, we identified the N-terminal GT4 domain (WbdAN) as the source of α-(1→2)-mannosyltransferase activity (12). However, no in vitro mannosyltransferase activity was detected with the C-terminal (putative) GT4 domain (WbdAC) in either the presence or absence of its N-terminal partner. The observation that a full-length protein bearing a mutation in the EX7E motif of the putative C-terminal GT4 active site (WbdAE758A; Fig. 1B) could synthesize α-(1→2)-linked polymannose led us to test the prediction that the C-terminal GT4 domain possessed α-(1→3)-mannosyltransferase activity. We therefore investigated the activity of the corresponding mutant (WbdAE317A), inactivated in the N-terminal EX7E motif, using synthetic oligosaccharide acceptors. This construct showed activity with both acceptors (data not shown) as was found with wild type WbdA, but only acceptor B was examined in depth because its structure (α-Manp-(1→3)-α-Manp-(1→3)-β-GlcpNAc) lacks α-(1→2) linkages, simplifying the analysis. The MS spectrum following reaction with WbdAE317A revealed the acceptor (m/z 1223) and multiple products with increments of m/z 162 expected for one Manp residue; products with up to four added mannose residues were detected (Fig. 2A). The reaction products were analyzed by 1H NMR spectroscopy (Fig. 2B). The major peaks in the spectrum are derived from a compound consisting of the acceptor extended by two α-(1→3)-Manp residues as expected from MS data. This compound generated major anomeric peaks at 5.24, 5.14, 5.12, and 5.08 ppm. These signals could be assigned to Manp with α-(1→3) linkages through two-dimensional NMR spectra, including COSY, TOCSY, and tROESY spectra (data not shown). Specifically in the tROESY spectrum, the signal at 5.14 ppm showed a correlation to H3 of the ring with an anomeric signal at 5.12 ppm, the signal at 5.12 ppm showed a correlation to H3 of the ring with an anomeric signal at 5.08 ppm, and the signal at 5.08 ppm showed a correlation to H3 of the ring with an anomeric signal at 5.24 ppm. No signals indicative of α-(1→2) linkages were observed. These results were consistent with the values predicted by the CASPER NMR analysis program (34–36) (accessed October 28, 2014) (Table 2). We then performed a reaction with the same acceptor incubated with WbdAE317A and WbdAN. When analyzed in isolation, WbdAN extends a synthetic acceptor to generate α-(1→2)-linked polymannose (13), but in combination, these two constructs reconstituted authentic O9a biosynthesis. The 1H NMR spectrum of these products revealed anomeric signals at 4.99, 5.08, 5.25, and 5.33 ppm (Fig. 2D). These values are consistent with the predictions from the CASPER program for the O9a repeat unit (Table 2), the published values for the authentic O9a glycan (37), and the in vitro products obtained with the same acceptor and full-length WbdA (12). The MS spectrum revealed a series of products, each differing by a single mannose residue (m/z 162). The largest products of significant quantity showed 24 added residues, equivalent to six (tetrasaccharide) repeat units of the authentic O9a glycan, but additional peaks with intensities near the background level suggest that larger products are possible (Fig. 2C).
FIGURE 2.
Characterization of the products of in vitro biosynthesis reactions. Reactions were performed with acceptor B (α-Manp-(1→3)-α-Manp-(1→3)-β-GlcpNAc; m/z 1223) and WbdAE317A (A and B) or WbdAE317A plus WbdAN (C and D). Products were analyzed by MALDI MS (A and C) and 1H NMR spectroscopy (B and D). Intens., intensity; a.u., arbitrary units.
TABLE 2.
Comparison of the chemical shifts for the anomeric protons of the products generated in in vitro reactions with those predicted by the CASPER database for the structures shown
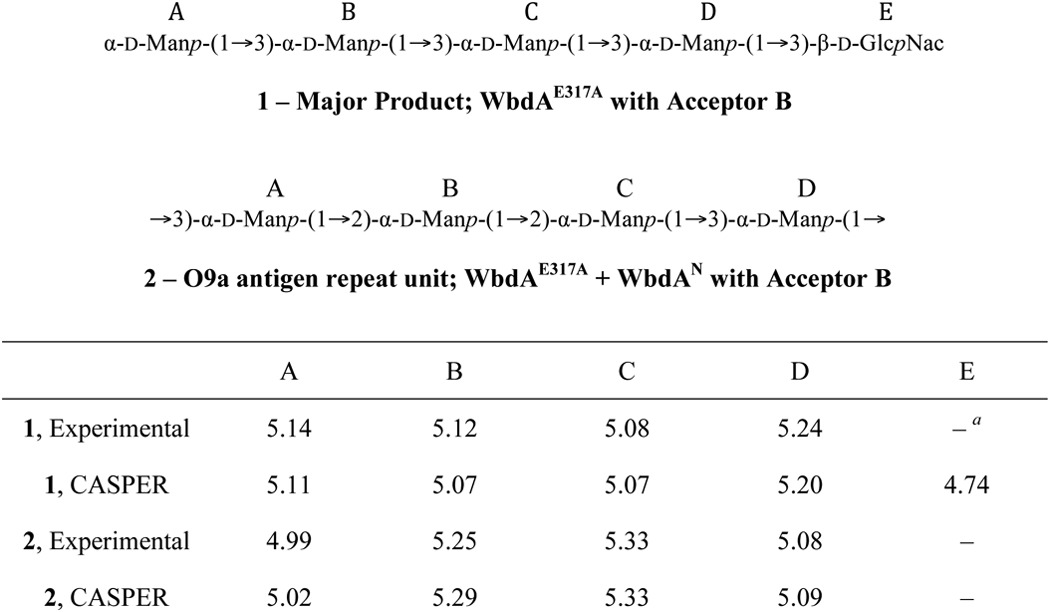
a The HOD peak at 4.76 ppm is too large to see whether the anomeric signal for β-d-GlcpNAc is present.
A Region within the C-terminal Part of WbdA Is Required for in Vivo Activity of the N-terminal GT4 Domain
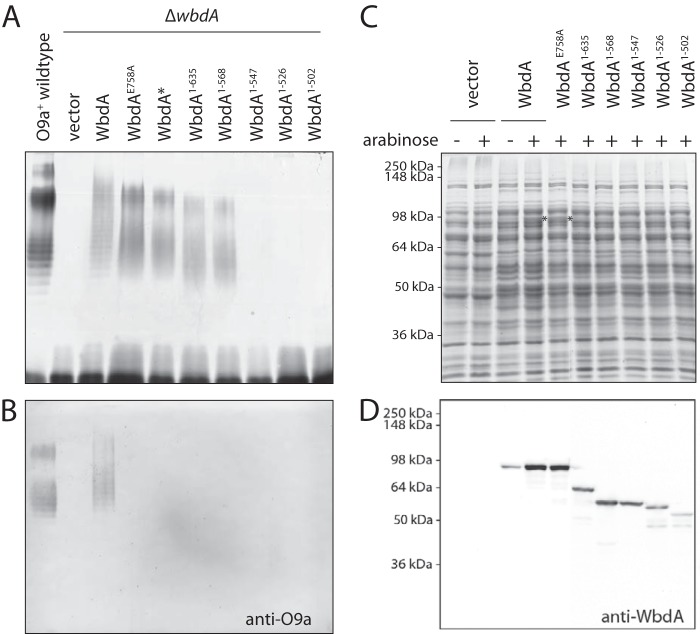
With the activities of the two GT4 domains now assigned, we turned our attention to the molecular basis for the lack of detectable in vivo activity of WbdAN despite its ability to restore authentic O9a production in cells co-transformed with plasmids expressing WbdAC (13). Furthermore, when examined in vitro, WbdAN showed robust poly-α-(1→2)-mannosyltransferase activity, and the same activity was replicated in vivo in cells expressing WbdAE758A, which possesses a mutation in the catalytically essential EX7E motif in the C-terminal GT4 active site (13). As reported previously, lysates of cells expressing WbdAE758A lack the typical O9a LPS banding pattern (reflecting size increments of a tetrasaccharide O-PS repeat unit), and LPS molecules with the aberrant α-(1→2)-linked O-PS no longer reacted with anti-O9a antibodies in Western immunoblots (Fig. 3, A and B). The same LPS phenotype was evident in a strain expressing a wbdA construct where a stop codon was inadvertently introduced, creating a protein (designated WbdA*) that possessed a frameshift mutation at residue 635 and terminated prematurely at residue 649 (Fig. 3, A and B). The collective results suggest that in vivo poly-α-(1→2)-mannosyltransferase activity of the N-terminal GT4 domain is dependent on a region of the C-terminal domain and that this is missing in WbdAN.
FIGURE 3.
Capacity for production of α-(1→2)-mannan by WbdA proteins with varying C-terminal deletions. A shows LPS profiles revealed by silver-stained SDS-PAGE of lysates from cells containing WbdA and its derivatives. B shows the corresponding Western immunoblot using antibodies specific for the O9a antigen. C and D show expression of the proteins in cell lysates following induction with 0.01% l-arabinose. Duplicate samples were separated by SDS-PAGE and stained to show equivalent sample loading (C) or probed in Western immunoblots with antibodies specific for WbdA (D). The asterisks in C identify overexpressed WbdA protein visible in the stained protein profile. Full-length WbdA has a predicted size of 95.3 kDa but always runs faster in SDS-PAGE with an observed apparent size of 84.5 kDa in lysates or following purification. The truncated derivatives also showed smaller apparent sizes: WbdA1–635, 71.2/67.3 kDa; WbdA1–568, 63.5/58.7 kDa; WbdA1–547, 61.3/58.1 kDa; WbdA1–526, 58.9/56.1 kDa; WbdA1–502, 56.2/52.2 kDa. All plasmid constructs were transformed into E. coli ΔwbdA (CWG1105).
To determine the minimal portion of WbdA required for in vivo poly-α(1→2)-mannosyltransferase activity, a series of C-terminal truncations of WbdA was constructed guided by WbdA* as a starting point and the location of the N-terminal GT4 domain as an end point (Fig. 1B). E. coli ΔwbdA cells expressing the WbdA1–635 construct (equivalent to WbdA*) produced the α-(1→2)-polymannose glycan as expected. The shortest functional construct was WbdA1–568, whereas WbdA1–547 and proteins with more extensive C-terminal deletions were all unable to restore any O-PS synthesis. All of the constructs generated WbdA proteins (Fig. 3, C and D), indicating that inactivity did not simply reflect selective proteolytic degradation of some truncated versions of WbdA. The expression level of the truncated products was slightly less than those of the full-length proteins based on Western immunoblotting. This does not necessarily correlate with protein amount due to loss of epitopes (through truncation) recognized by the rabbit anti-WbdA antibodies, but it is consistent with the absence of visible bands in the stained SDS-polyacrylamide gel for any construct shorter than the full-length protein. A reduction in the amount of WbdA may explain the slight downward shift in the size distribution of the α-(1→2)-polymannose glycans they generate (Fig. 3A) because alterations in stoichiometry of WbdA:WbdD are known to effect chain length (14, 20).
In Vivo Poly-α-(1→2)-mannosyltransferase Activity Correlates with the Ability of WbdD to Recruit the Truncated WbdA Derivatives
Previously, we demonstrated that WbdA activity was dependent on protein-protein interactions with WbdD, which recruits the polymerase to a functional complex on the cytoplasmic membrane (19). The C-terminal region of WbdD involved in this interaction was localized to a predicted 30-residue C-terminal helix, but the region of WbdA involved was not investigated. The in vivo activities for the truncated WbdA proteins could be explained by an essential interacting region in the C-terminal part of WbdA that is deleted in some constructs, and bacterial two-hybrid results proved this hypothesis to be correct. Interactions were measured between the T25-WbdD fusion protein and WbdA variants fused to an N-terminal T18 fragment. This was based on the high level of β-galactosidase activity seen with the full-length proteins (Table 3), consistent with data published previously (19). With the full-length proteins, these positions of the CyaA fragments had no effect on WbdA or WbdD activity (19). Positive interactions were detected between T25-WbdD and T18-WbdA1–635 and between T25-WbdD and T18-WbdA1–568, whereas none of the constructs unable to synthesize the poly-α-(1→2)-mannan demonstrated any significant interaction with WbdD.
TABLE 3.
Protein-protein interactions defined by bacterial two-hybrid experiments
| Bacterial two-hybrid combination | β-Galactosidase activity | S.E. |
|---|---|---|
| Miller units | ||
| T25-WbdD + T18-WbdA | 7777 | 149 |
| T25-WbdD + WbdA-T18 | 2714 | 89 |
| T25-WbdD + T18-WbdA1–635 | 742 | 18 |
| T25-WbdD + T18-WbdA1–568 | 1403 | 8 |
| T25-WbdD + T18-WbdA1–547 | 279 | 11 |
| T25-WbdD + T18-WbdA1–526 | 284 | 20 |
| T25-WbdD + T18-WbdA1–502 | 198 | 10 |
| T25-WbdD + T18-WbdAN | 232 | 32 |
| T25-WbdD + WbdAC-T18 | 193 | 42 |
| T25-WbdD + T18-WbdAN + WbdAC | 1840 | 646 |
| T25-WbdD + WbdAC-T18 + WbdAN | 557 | 249 |
| T18-WbdAN + WbdAC-T25 | 279 | 25 |
| pKT25 + pUT18C (negative control) | 314 | 15 |
| pKT25-Zip + pUT18C-Zip (positive control) | 2549 | 105 |
Interactions between WbdD and the C-terminal WbdA Domain Require the Participation of the N-terminal Domain
The two-hybrid data reinforce the need for a productive association between WbdA and WbdD for polymerase activity. Because co-expression of WbdAN and WbdAC restored O9a biosynthesis in an E. coli ΔwbdA mutant (13), an interaction between WbdAC and WbdD was anticipated. Unexpectedly, this combination showed no interaction (Table 3), so two-hybrid experiments were performed in which WbdAN was introduced together with WbdAC-T18 and T25-WbdD. When WbdAN was present, an interaction between WbdAC-T18 and T25-WbdD was evident. In these transformants, the three-plasmid system led to some instability and larger variation in measured β-galactosidase activities (Table 3), but the colonies on X-Gal plates consistently showed a blue color indicative of the positive interaction. Significantly, when WbdAC was included, a robust signal was detected, reflecting strong interaction between T18-WbdAN (which lacks the site of interaction) and T25-WbdD. Therefore, although the interaction site appears to reside within WbdAC, its ability to associate with WbdD shows a strict dependence on WbdAN. WbdAN therefore appears to be critical for creating a conformational situation that allows WbdAC and WbdD to interact. The WbdAC-dependent interaction between WbdAN and WbdD could reflect an altered affinity of WbdAN for WbdD in the presence of WbdAC. Alternatively, it may require WbdAC as an intermediary with no direct binding between WbdAN and WbdD. Given the observed complex interactions and dependences, this question cannot be resolved by two-hybrid approaches and will require a solved structure for the heterocomplex.
Restoration of O9a Antigen Production by Introduction of the C-terminal GT4 Domain
These complex interactions led us to examine the poly-α-(1→2)-mannosyltransferase activities of the C-terminal truncated proteins when co-expressed with a complete (catalytically active) WbdAC domain. LPS profiles and reactivity with anti-O9a antibodies were examined in lysates from E. coli ΔwbdA cells containing WbdAC and each one of the C-terminal truncations (Fig. 4, A and B). The control containing WbdAN and WbdAC recapitulates the restoration of O9a O-PS production reported previously (13). Similar results were obtained with WbdA1–502, WbdA1–526, and WbdA1–527. In contrast, no significant O9a O-PS was detected in cells expressing WbdA1–568 and WbdA1–635; i.e. those constructs capable of synthesizing the α-(1→2)-polymannose glycan continued to synthesize that product regardless of the presence of a catalytically active WbdAC. The same result was obtained with cells expressing WbdAE758A (EX7E mutation in the C-terminal GT4 active site) and WbdAC (Fig. 4, C and D). In contrast, no O-PS was produced in cells containing WbdAE317A (N-terminal EX7E mutation) and WbdAN. There are two possible interpretations for these data. In one, access of the catalytically active C-terminal GT4 domain into an active complex is blocked by occupancy of WbdD with a WbdA construct that retains its interaction site but possesses no C-terminal GT4 activity. Alternatively, the region of WbdAN that is essential for coordinating the interaction of WbdAC with WbdD is already occupied in the truncated versions with its linked but inactive WbdAC and is no longer available to interact with the separate active WbdAC domain. It is also conceivable that both scenarios contribute to the observed phenotype.
FIGURE 4.
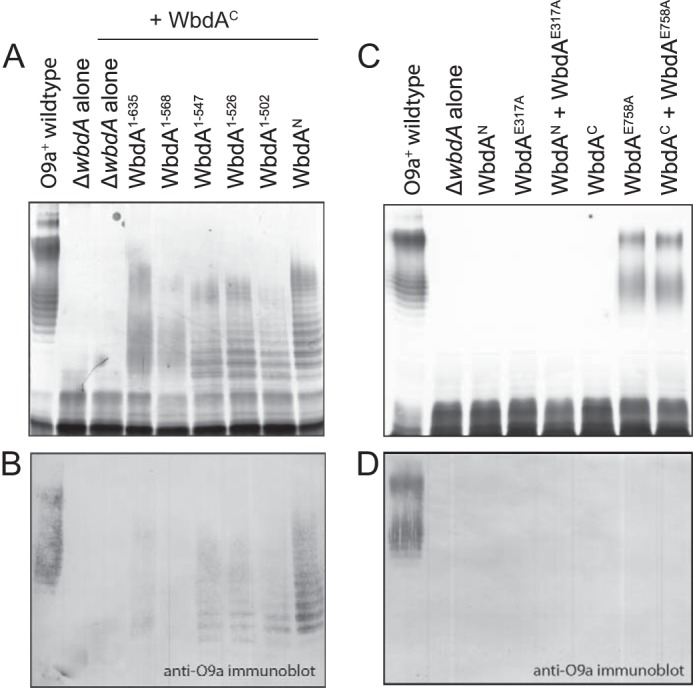
Active synthesis of α-(1→2)-polymannose precludes restoration of O9a with an active WbdAC α-(1→3)-mannosyltransferase domain in trans. E. coli CWG1105 cells were co-transformed with compatible plasmids encoding the identified WbdA C-terminal deletions and WbdAC (A and B). C and D show the results of complementation experiments where plasmids encoding derivatives with EX7E catalytic site mutations were co-transformed with plasmids expressing the individual domains. The LPS products in whole-cell lysates were revealed by silver-stained SDS-PAGE (A and C) and Western immunoblotting with antibodies specific for the O9a antigen (B and D).
WbdA Exists as a Monomer
Although WbdA and WbdD clearly interact and WbdD is a trimer (16), the possibility existed of further homotypic interactions involving WbdA. In two-hybrid analyses, no interactions could be detected regardless of the locations of the CyaA fragment reporters (data not shown). To confirm this, WbdA-His10 was purified and examined by size exclusion chromatography. The majority of the protein was eluted as a monomer with only a (variable) trace of aggregate evident in the profile (Fig. 5).
FIGURE 5.
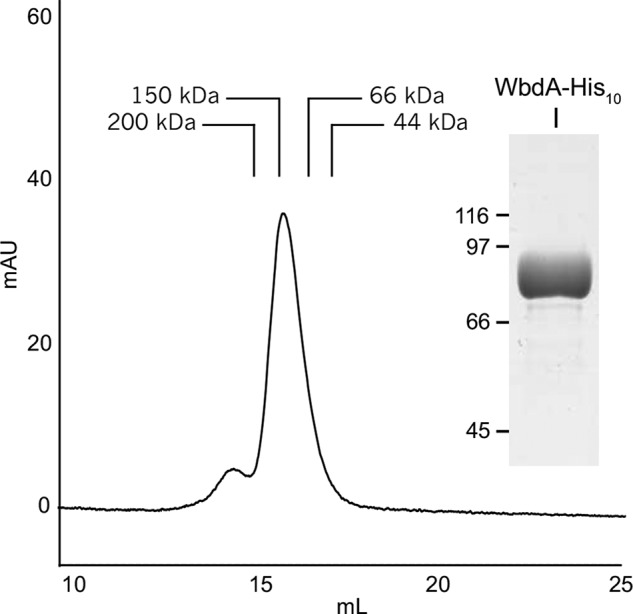
Size exclusion chromatography of purified WbdA-His10. WbdA-His10 was purified (inset) and eluted through a Superose 6 HR 10/30 column. The predominant peak corresponded with an estimated molecular mass of 120 kDa consistent with monomeric WbdA-His10 (theoretical molecular mass, 96.1 kDa). A minor amount of the protein eluted before the 200-kDa standard. The molecular masses of the protein standards are as follows: β-amylase, 200 kDa; alcohol dehydrogenase, 150 kDa; bovine serum albumin, 66 kDa; ovalbumin, 44 kDa. mAU, milli-absorbance units.
DISCUSSION
WbdA is a two-domain polymerizing GT responsible for the synthesis of a glycan containing a tetrasaccharide repeat unit. The N-terminal GT4 catalytic site has α-(1→2)-mannosyltransferase activity, and we now can assign α-(1→3)-mannosyltransferase activity to the C-terminal GT4 site. The in vitro product profiles obtained using WbdA at different substrate concentrations are consistent with WbdA being a distributive enzyme, releasing product after each mannosyl transfer reaction (12). Reproducible production of a tetrasaccharide in which two α-(1→2)-linked Manp residues are always followed by two α-(1→3)-linked Manp residues represents an interesting challenge for such enzymes. Enzymes exist where a single GT site sequentially adds a defined number of residues to an acceptor. For example, PglH is a UDP-GalpNAc-dependent GT involved in N-glycosylation of proteins in Campylobacter jejuni (38), and it is proposed that its ability to reliably add three residues is precisely controlled by the relative binding affinities of the enzyme for the growing acceptor (39). An involvement of the active sites of WbdA in its fidelity is suggested by analysis of the WbdA protein from serotype O9a and its homolog from serotype O9 with the latter O-PS containing a pentasaccharide repeat unit (three α-(1→2)-linked Manp residues followed by two α-(1→3)-linked Manp residues). A mutation (C80R) in a residue residing in the modeled active site of WbdAN (13) changes specificity of the WbdA protein from the O9 pentasaccharide repeat unit to the O9a tetrasaccharide (40). However, the distributive nature (at least in vitro) and the observation that WbdAN activity becomes unconstrained in the absence of functional WbdAC suggest a more complex situation. In WbdA, the interplay between the two functional catalytic sites is essential for fidelity with WbdAC acting as a regulatory constraint on the WbdAN mannosyltransferase.
Multidomain, bifunctional polymerases with two active sites have been described in the biosynthesis of hyaluronan (41, 42), chondroitin (42, 43), and heparosan (44–46). However, how the sites interact is unclear. With WbdA, modeling of individual sites can be done with confidence based on homologs (13), but the relative juxtaposition of the domains cannot be resolved without a structure for the complete protein. The only crystal structure available for any multidomain polymerase is KfoC, the chondroitin synthase involved in the biosynthesis of the capsular polysaccharide in E. coli K4 (47). In this enzyme, two almost identical active sites add single GlcpNAc and glucuronic acid (GlcpA) residues in the disaccharide repeat unit. The active sites are located at the opposite sides of the protein separated by 60 Å. A 71-residue (largely disordered) region from the N terminus wraps around the surface of the C-terminal GlcpA-transferase domain, but its importance in catalysis or organization of the overall structure is uncertain. Nevertheless, interaction between the domains is essential for polymerase activity because N-terminal deletion constructs still possess the ability to transfer single GlcpNAc or GlcpA residues to an acceptor, but the protein no longer acts as a polymerase (48). The structure also reveals hydrophobic interactions between two hexapeptides, one in each domain, that influence the conformation of the binding pocket of the C-terminal GlcpA-transferase, creating an unusual orientation of the substrate in the catalytic site. The significance of this feature is also unknown. Clearly, KfoC and WbdA both demonstrate complex domain interactions essential for function, but additional crystal structures are needed to determine the mechanistic consequences and establish whether any generic architectural principles are involved.
Organization of WbdA into a functional active biosynthesis complex requires its interaction with WbdD; there is no known terminator component for KfoC (19). This process presumably engages WbdA at the membrane with the natural undecaprenol diphosphate-linked oligosaccharide acceptor and creates the proper structural context for chain termination (14–16, 20). Previously, the interaction domain in WbdD was localized to a 30-residue α-helix at the C terminus (19). In analysis of the C-terminal deletions of WbdA, interaction was still possible with WbdA1–568 but was lost in WbdA1–547. This region can be modeled using other GT4 enzymes as a scaffold, and the structure of mycobacterial PimA, a GDP-Manp-dependent GT4 mannosyltransferase that is involved in the biosynthesis of phosphoinositol mannosides (49) can be used to predict substrate binding locations (Fig. 6). The region of interaction identified by deletion analysis extends from residues Val548–Ile568 (highlighted in red) and encompasses a surface-exposed α-helix located between Val548 and Ala556 that represents an excellent candidate for interaction. Interestingly, the interactions between WbdA and WbdD are complex. Although the WbdAC domain possesses the interaction site and shows correct catalytic activity when expressed with its WbdAN partner, interactions between WbdAC and WbdD were dependent on the presence of WbdAN. As expected, WbdAN could only interact with WbdD when WbdAC provided the interaction site. Productive interactions were compromised when inactivated GT domains were present. The results indicate that the separated GT domains are able to recreate the correct configuration for authentic O9a polymerase activity in vivo. However, the same is not true in vitro as the same combination can only generate the α-(1→2)-polymannose glycan (13). In this situation, WbdAC is not appropriately positioned to participate in polymerization and properly constrain the activity of WbdAN presumably because WbdD is absent. These results presumably highlight the critical scaffold and orientation role played by WbdD in vivo and will only be fully appreciated if the structure of the WbdA-WbdD heterocomplex can be solved. Although in vitro O9a synthesis was restored by combining WbdAN and WbdAE317A, the same combination did not work in vivo. The environment in vitro (high enzyme and substrate concentrations and a soluble acceptor) is radically different from the in vivo situation, making it impossible to directly relate the in vivo and in vitro data. However, the results do reinforce the need to consider both in vivo and in vitro situations when assigning functions to individual GT active sites in multidomain polymerases.
FIGURE 6.
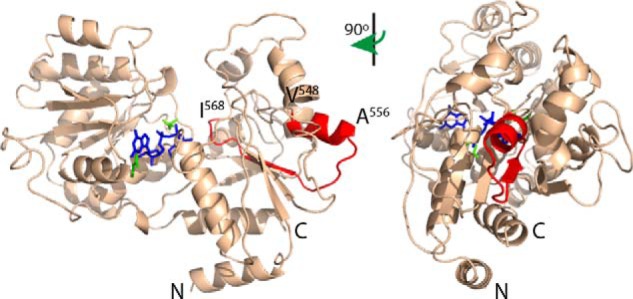
Structural model of WbdAC highlighting the region of interaction with WbdD. Residues 440–832 of WbdA were modeled with high confidence using Phyre2 in the intensive mode. The region of interaction identified by deletion analysis (highlighted in red) corresponds to a surface-exposed α-helix located between Val548 and Ala556. GDP-Manp (colored blue) was modeled into the active site through structural alignment with mycobacterial PimA (Protein Data Bank code 2GEJ), a GDP-Manp-dependent GT4 mannosyltransferase that is involved in the biosynthesis of phosphoinositol mannosides (49), that was crystallized in the presence of GDP-Manp. Glutamate residues of the WbdA EX7E motif are colored green.
GTs have applications in emerging technologies, including glycoengineering in bacteria (50), and bacterial GTs that use sugar nucleotide donors are important in these efforts because of the vast range of unique specificities as well as their potential to build human glycan mimics. Polymerases are being investigated for various roles in enzymatic (and chemoenzymatic) syntheses (e.g. (4, 51–53) where the goal is to produce “glyco-products” with defined chemical and physical characteristics. Understanding the structure and function of multidomain polymerases will be essential for their successful application in glycoengineering efforts.
Supplementary Material
This work was supported in part by grants from the Natural Sciences and Engineering Research Council (to C. W. and T. L. L.) and by the Alberta Glycomics Centre (to T. L. L.).

This article contains supplemental Table S1.
- GT
- glycosyltransferase
- O-PS
- O-polysaccharide
- GlcpNAc
- N-acetylglucosamine
- Manp
- mannose
- LB
- lysogeny broth
- BisTris
- [bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- TOCSY
- total correlation spectroscopy
- tROESY
- transverse rotating frame Overhauser enhancement spectroscopy
- GalpNAc
- N-acetylgalactosamine
- Glcp
- glucose
- GlcpA
- glucuronic acid
- X-Gal
- 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside.
REFERENCES
- 1. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Coutinho P. M., Deleury E., Davies G. J., Henrissat B. (2003) An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328, 307–317 [DOI] [PubMed] [Google Scholar]
- 3. Lairson L. L., Henrissat B., Davies G. J., Withers S. G. (2008) Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 [DOI] [PubMed] [Google Scholar]
- 4. DeAngelis P. L., Liu J., Linhardt R. J. (2013) Chemoenzymatic synthesis of glycosaminoglycans: re-creating, re-modeling and re-designing nature's longest or most complex carbohydrate chains. Glycobiology 23, 764–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Keys T. G., Fuchs H. L., Galuska S. P., Gerardy-Schahn R., Freiberger F. (2013) A single amino acid toggles Escherichia coli polysialyltransferases between mono- and bifunctionality. Glycobiology 23, 613–618 [DOI] [PubMed] [Google Scholar]
- 6. Breyer W. A., Matthews B. W. (2001) A structural basis for processivity. Protein Sci. 10, 1699–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Levengood M. R., Splain R. A., Kiessling L. L. (2011) Monitoring processivity and length control of a carbohydrate polymerase. J. Am. Chem. Soc. 133, 12758–12766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cuthbertson L., Kos V., Whitfield C. (2010) ABC transporters involved in export of cell surface glycoconjugates. Microbiol. Mol. Biol. Rev. 74, 341–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Greenfield L. K., Whitfield C. (2012) Synthesis of lipopolysaccharide O-antigens by ABC transporter-dependent pathways. Carbohydr. Res. 356, 12–24 [DOI] [PubMed] [Google Scholar]
- 10. Jann K., Goldemann G., Weisgerber C., Wolf-Ullisch C., Kanegasaki S. (1982) Biosynthesis of the O9 antigen of Escherichia coli. Initial reaction and overall reaction. Eur. J. Biochem. 127, 157–164 [DOI] [PubMed] [Google Scholar]
- 11. Rick P. D., Hubbard G. L., Barr K. (1994) Role of the rfe gene in the synthesis of the O8 antigen in Escherichia coli K-12. J. Bacteriol. 176, 2877–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Greenfield L. K., Richards M. R., Li J., Wakarchuk W. W., Lowary T. L., Whitfield C. (2012) Biosynthesis of the polymannose lipopolysaccharide O-antigens from Escherichia coli serotypes O8 and O9a requires a unique combination of single- and multiple-active site mannosyltransferases. J. Biol. Chem. 287, 35078–35091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greenfield L. K., Richards M. R., Vinogradov E., Wakarchuk W. W., Lowary T. L., Whitfield C. (2012) Domain organization of the polymerizing mannosyltransferases involved in synthesis of the Escherichia coli O8 and O9a lipopolysaccharide O antigens. J. Biol. Chem. 287, 38135–38149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clarke B. R., Cuthbertson L., Whitfield C. (2004) Nonreducing terminal modifications determine the chain length of polymannose O antigens of Escherichia coli and couple chain termination to polymer export via an ATP-binding cassette transporter. J. Biol. Chem. 279, 35709–35718 [DOI] [PubMed] [Google Scholar]
- 15. Clarke B. R., Richards M. R., Greenfield L. K., Hou D., Lowary T. L., Whitfield C. (2011) In vitro reconstruction of the chain termination reaction in biosynthesis of the Escherichia coli O9a O-polysaccharide: the chain-length regulator, WbdD, catalyzes the addition of methyl phosphate to the non-reducing terminus of the growing glycan. J. Biol. Chem. 286, 41391–41401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hagelueken G., Huang H., Clarke B. R., Lebl T., Whitfield C., Naismith J. H. (2012) Structure of WbdD: a bifunctional kinase and methyltransferase that regulates the chain length of the O antigen in Escherichia coli O9a. Mol. Microbiol. 86, 730–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cuthbertson L., Powers J., Whitfield C. (2005) The C-terminal domain of the nucleotide-binding domain protein Wzt determines substrate specificity in the ATP-binding cassette transporter for the lipopolysaccharide O antigens in Escherichia coli serotypes O8 and O9a. J. Biol. Chem. 280, 30310–30319 [DOI] [PubMed] [Google Scholar]
- 18. Cuthbertson L., Kimber M. S., Whitfield C. (2007) Substrate binding by a bacterial ABC transporter involved in polysaccharide export. Proc. Natl. Acad. Sci. U.S.A. 104, 19529–19534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clarke B. R., Greenfield L. K., Bouwman C., Whitfield C. (2009) Coordination of polymerization, chain termination, and export in assembly of the Escherichia coli O9a antigen in an ABC transporter-dependent pathway. J. Biol. Chem. 284, 30662–30672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. King J. D., Berry S., Clarke B. R., Morris R. J., Whitfield C. (2014) Lipopolysaccharide O antigen size distribution is determined by a chain extension complex of variable stoichiometry in Escherichia coli O9a. Proc. Natl. Acad. Sci. U.S.A. 111, 6407–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hagelueken G., Clarke B. R., Huang H., Tuukkanen A., Danciu I., Svergun D. I., Liu H., Whitfield C., Naismith J. H. (2014) A coiled-coil structural domain acts as a molecular ruler in LPS chain length regulation. Nat. Struct. Mol. Biol. 10.1038/nsmb.2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Raetz C. R., Whitfield C. (2002) Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Whitfield C., Trent M. S. (2014) Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128 [DOI] [PubMed] [Google Scholar]
- 24. Guerin M. E., Kordulakova J., Schaeffer F., Svetlikova Z., Buschiazzo A., Giganti D., Gicquel B., Mikusova K., Jackson M., Alzari P. M. (2007) Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria. J. Biol. Chem. 282, 20705–20714 [DOI] [PubMed] [Google Scholar]
- 25. Miller J. H. (1972) Experiments in Molecular Genetics, pp. 432–433, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 26. Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 28. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wlikins M. R., Appel R. D., Bairoch A. (2005) in The Proteomics Protocols Handbook (Walker J., ed) pp. 571–707, Humana Press, Totowa, NJ [Google Scholar]
- 29. Miller J. H. (1972) Experiments in Molecular Genetics, pp. 352–355, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 30. Hitchcock P. J., Brown T. M. (1983) Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsai C. M., Frasch C. E. (1982) A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119, 115–119 [DOI] [PubMed] [Google Scholar]
- 32. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 33. Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C., Gonzales N. R., Gwadz M., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Lu F., Marchler G. H., Mullokandov M., Omelchenko M. V., Robertson C. L., Song J. S., Thanki N., Yamashita R. A., Zhang D., Zhang N., Zheng C., Bryant S. H. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jansson P. E., Stenutz R., Widmalm G. (2006) Sequence determination of oligosaccharides and regular polysaccharides using NMR spectroscopy and a novel Web-based version of the computer program CASPER. Carbohydr. Res. 341, 1003–1010 [DOI] [PubMed] [Google Scholar]
- 35. Lundborg M., Widmalm G. (2011) Structural analysis of glycans by NMR chemical shift prediction. Anal. Chem. 83, 1514–1517 [DOI] [PubMed] [Google Scholar]
- 36. Roslund M. U., Säwén E., Landström J., Rönnols J., Jonsson K. H., Lundborg M., Svensson M. V., Widmalm G. (2011) Complete 1H and 13C NMR chemical shift assignments of mono-, di-, and trisaccharides as basis for NMR chemical shift predictions of polysaccharides using the computer program casper. Carbohydr. Res. 346, 1311–1319 [DOI] [PubMed] [Google Scholar]
- 37. Parolis L. A., Parolis H., Dutton G. G. (1986) Structural studies of the O-antigen polysaccharide of Escherichia coli O9a. Carbohydr. Res. 155, 272–276 [DOI] [PubMed] [Google Scholar]
- 38. Glover K. J., Weerapana E., Imperiali B. (2005) In vitro assembly of the undecaprenylpyrophosphate-linked heptasaccharide for prokaryotic N-linked glycosylation. Proc. Natl. Acad. Sci. U.S.A. 102, 14255–14259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Troutman J. M., Imperiali B. (2009) Campylobacter jejuni PglH is a single active site processive polymerase that utilizes product inhibition to limit sequential glycosyl transfer reactions. Biochemistry 48, 2807–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kido N., Kobayashi H. (2000) A single amino acid substitution in a mannosyltransferase, WbdA, converts the Escherichia coli O9 polysaccharide into O9a: generation of a new O-serotype group. J. Bacteriol. 182, 2567–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jing W., DeAngelis P. L. (2000) Dissection of the two transferase activities of the Pasteurella multocida hyaluronan synthase: two active sites exist in one polypeptide. Glycobiology 10, 883–889 [DOI] [PubMed] [Google Scholar]
- 42. Jing W., DeAngelis P. L. (2003) Analysis of the two active sites of the hyaluronan synthase and the chondroitin synthase of Pasteurella multocida. Glycobiology 13, 661–671 [DOI] [PubMed] [Google Scholar]
- 43. Sobhany M., Kakuta Y., Sugiura N., Kimata K., Negishi M. (2008) The chondroitin polymerase K4CP and the molecular mechanism of selective bindings of donor substrates to two active sites. J. Biol. Chem. 283, 32328–32333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kane T. A., White C. L., DeAngelis P. L. (2006) Functional characterization of PmHS1, a Pasteurella multocida heparosan synthase. J. Biol. Chem. 281, 33192–33197 [DOI] [PubMed] [Google Scholar]
- 45. Chavaroche A. A., van den Broek L. A., Springer J., Boeriu C., Eggink G. (2011) Analysis of the polymerization initiation and activity of Pasteurella multocida heparosan synthase PmHS2, an enzyme with glycosyltransferase and UDP-sugar hydrolase activity. J. Biol. Chem. 286, 1777–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chavaroche A. A., van den Broek L. A., Boeriu C., Eggink G. (2012) Synthesis of heparosan oligosaccharides by Pasteurella multocida PmHS2 single-action transferases. Appl. Microbiol. Biotechnol. 95, 1199–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Osawa T., Sugiura N., Shimada H., Hirooka R., Tsuji A., Shirakawa T., Fukuyama K., Kimura M., Kimata K., Kakuta Y. (2009) Crystal structure of chondroitin polymerase from Escherichia coli K4. Biochem. Biophys. Res. Commun. 378, 10–14 [DOI] [PubMed] [Google Scholar]
- 48. Sobhany M., Kakuta Y., Sugiura N., Kimata K., Negishi M. (2012) The structural basis for a coordinated reaction catalyzed by a bifunctional glycosyltransferase in chondroitin biosynthesis. J. Biol. Chem. 287, 36022–36028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Korduláková J., Gilleron M., Mikusova K., Puzo G., Brennan P. J., Gicquel B., Jackson M. (2002) Definition of the first mannosylation step in phosphatidylinositol mannoside synthesis. PimA is essential for growth of mycobacteria. J. Biol. Chem. 277, 31335–31344 [DOI] [PubMed] [Google Scholar]
- 50. Baker J. L., Çelik E., DeLisa M. P. (2013) Expanding the glycoengineering toolbox: the rise of bacterial N-linked protein glycosylation. Trends Biotechnol. 31, 313–323 [DOI] [PubMed] [Google Scholar]
- 51. Lindhout T., Iqbal U., Willis L. M., Reid A. N., Li J., Liu X., Moreno M., Wakarchuk W. W. (2011) Site-specific enzymatic polysialylation of therapeutic proteins using bacterial enzymes. Proc. Natl. Acad. Sci. U.S.A. 108, 7397–7402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Keys T. G., Fuchs H. L., Ehrit J., Alves J., Freiberger F., Gerardy-Schahn R. (2014) Engineering the product profile of a polysialyltransferase. Nat. Chem. Biol. 10, 437–442 [DOI] [PubMed] [Google Scholar]
- 53. Fiebig T., Berti F., Freiberger F., Pinto V., Claus H., Romano M. R., Proietti D., Brogioni B., Stummeyer K., Berger M., Vogel U., Costantino P., Gerardy-Schahn R. (2014) Functional expression of the capsule polymerase of Neisseria meningitidis serogroup X: a new perspective for vaccine development. Glycobiology 24, 150–158 [DOI] [PubMed] [Google Scholar]
- 54. Karimova G., Pidoux J., Ullmann A., Ladant D. (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.