Background: The ubiquitin-conjugating enzyme Cdc34 rapidly associates with ubiquitin ligase SCF to achieve processive ubiquitination.
Results: Protein cross-linking was used to capture native Cdc34-SCF complexes.
Conclusion: The acidic tail domain of Cdc34 interacts with the basic canyon region on SCF in multiple conformations.
Significance: Multiple conformations between Cdc34 and SCF enable their rapid association.
Keywords: E3 Ubiquitin Ligase, Protein-Protein Interaction, Ubiquitin, Ubiquitin-conjugating Enzyme (E2 Enzyme), Ubiquitylation (Ubiquitination)
Abstract
In the ubiquitin-proteasome system, protein substrates are degraded via covalent modification by a polyubiquitin chain. The polyubiquitin chain must be assembled rapidly in cells, because a chain of at least four ubiquitins is required to signal for degradation, and chain-editing enzymes in the cell may cleave premature polyubiquitin chains before achieving this critical length. The ubiquitin-conjugating enzyme Cdc34 and ubiquitin ligase SCF are capable of building polyubiquitin chains onto protein substrates both rapidly and processively; this may be explained at least in part by the atypically fast rate of Cdc34 and SCF association. This rapid association has been attributed to electrostatic interactions between the acidic C-terminal tail of Cdc34 and a feature on SCF called the basic canyon. However, the structural aspects of the Cdc34-SCF interaction and how they permit rapid complex formation remain elusive. Here, we use protein cross-linking to demonstrate that the Cdc34-SCF interaction occurs in multiple conformations, where several residues from the Cdc34 acidic tail are capable of contacting a broad region of the SCF basic canyon. Similar patterns of cross-linking are also observed between Cdc34 and the Cul1 paralog Cul2, implicating the same mechanism for the Cdc34-SCF interaction in other members of the cullin-RING ubiquitin ligases. We discuss how these results can explain the rapid association of Cdc34 and SCF.
Introduction
Ubiquitin-mediated proteolysis is responsible for controlling the half-lives of thousands of proteins in human cells. Polyubiquitin chains are assembled onto proteins through the concerted action of ubiquitin-conjugating enzymes (E2s)2 and ubiquitin ligases (E3s). The E2 forms a covalent intermediate with ubiquitin, a conserved 76-amino acid protein, whereas the E3 recruits both the E2∼ubiquitin and the protein substrate targeted for ubiquitination (1). Ubiquitin is typically transferred directly from E2 to the protein substrate, although in some cases an E3∼ubiquitin intermediate is established prior to substrate ubiquitination (2, 3). Polyubiquitinated proteins are recognized and degraded by the 26 S proteasome, a large, multisubunit protease.
Much effort has gone into uncovering the mechanism of action of the cullin-RING ubiquitin ligase (CRL) E3s that collectively control ∼20% of ubiquitin-mediated proteolysis in human cells (4, 5). The Skp1-cullin-F-box (SCF) E3s are the archetypal CRLs and arguably the most intensively studied E3 to date (6). The SCF is a modular multisubunit enzyme containing four subunits: a substrate binding subunit that contains a conserved F-box domain; Rbx1, which contains a RING (really interesting new gene) domain responsible for recruiting the E2∼ubiquitin; Cul1, which acts as a scaffold by simultaneously binding to both Rbx1 and the F-box subunit; and Skp1, which helps tether the F-box protein to Cul1.
Polyubiquitin chain formation occurs on human SCF-bound substrates through the sequential action of two E2 enzymes, UbcH5 and Cdc34 (7). UbcH5 transfers the initial ubiquitin to the protein substrate, followed by elongation of the polyubiquitin chain by Cdc34. Specifically, Cdc34∼ubiquitin is recruited to the SCF-protein substrate complex, which stimulates the discharge of ubiquitin from Cdc34 to the substrate. Cdc34 then dissociates from the SCF, allowing for recruitment of another Cdc34∼ubiquitin. The cycling of Cdc34∼ubiquitin for spent Cdc34 occurs rapidly, resulting in the stepwise addition of ubiquitins to SCF-bound substrates in a processive manner (8, 9).
Cdc34 contains both a canonical E2 catalytic domain and a highly acidic C-terminal extension called the acidic tail. The Cdc34 acidic tail is both phylogenetically conserved and essential for Cdc34 function. For instance, deletion of the tail results in a massive loss of Cdc34 activity in vitro (10), and a tail-deleted cdc34 mutant was unable to complement a yeast strain harboring a temperature-sensitive mutation at the endogenous locus (11). The acidic tail promotes the binding of Cdc34 to SCF through its direct interaction with a conserved basic region on the Cul1 subunit called the basic canyon. This electrostatic interaction enables extremely fast association rates between Cdc34 and SCF and at least partially explains why ubiquitin chains are processively assembled onto SCF-bound substrates (8).
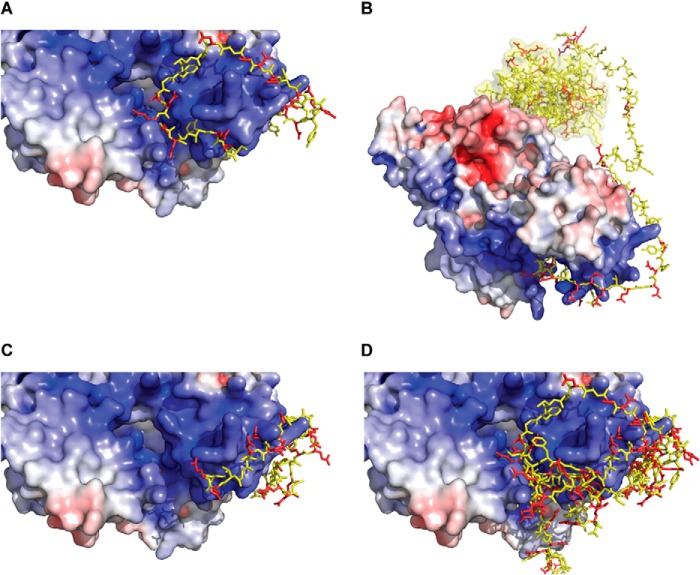
In our previous work, molecular modeling was used to determine whether the Cul1 basic canyon can accommodate the Cdc34 acidic tail (8). This procedure resulted in numerous models of the Cdc34-SCF complex where residues from the acidic tail nestled into the basic canyon; however, the precise location of acidic tail residues in relation to the basic canyon varied considerably (Fig. 1, A–D). Nevertheless, it remains possible that the Cdc34 acidic tail may interact with the Cul1 basic canyon in a single conformation; thus, we wanted to use experimental methods to elucidate the mode of binding. Using a cysteine-based cross-linking approach, we demonstrate that cysteine residues placed along multiple positions within the Cdc34 acidic tail can cross-link to a broad region of the Cul1 basic canyon, in particular near the conserved Cul1 basic canyon residue Lys 679. Our results are most consistent with a model whereby the Cdc34 acidic tail binds to Cul1 in multiple conformations, which explains how the tail promotes the rapid association of Cdc34 with SCF.
FIGURE 1.
Rosetta molecular models showing the interaction of the Cdc34 acidic tail with the Cul1 basic canyon. Four distinct models from some 30,000 computational trajectories have been selected as representative models (8). A, the electrostatic surface potential of Cul1 is shown, where blue indicates the highly positively charged surface of the basic canyon. Cdc34 acidic tail residues are shown in ball-and-stick representations, where acidic residues are red and non-acidic ones are yellow. Notice how the acidic residues are all contacting the positively charged surface. B, identical model as in A, except the view has been expanded to include the Cdc34 catalytic domain (yellow surface). Notice that the intervening tail region between the docked distal acidic tail and the Cdc34 catalytic domain is sufficiently long to allow interaction of the catalytic domain with Rbx1. C, same as in A except showing a different model from Rosetta. D, superposition of the acidic tail conformations from four representative molecular models. Notice that no two conformations are identical.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
All proteins were expressed in either Escherichia coli or Hi5 insect cells. All Cdc34 proteins were expressed in the Rosetta (DE3) host strain (EMD Millipore) and purified by virtue of their N-terminal histidine tag using nickel-nitrilotriacetic acid-agarose beads (Qiagen) and gel filtration chromatography using a Superdex 200 10/300 column (GE Healthcare). The histidine tags were cleaved from the Cdc34 N terminus using tobacco etch virus protease. Human Cul1-Rbx1 proteins were expressed and purified using the “Split-n-Coexpress” protocol (12). Briefly, the Cul1 subunit is co-expressed as two fragments, an N-terminal domain and a C-terminal one, along with the Rbx1 subunit that has been fused to glutathione S-transferase (GST) at its N terminus. This permits the high level expression of soluble Cul1-Rbx1 proteins in the BL21(DE3) E. coli host strain. After initial purification of the complexes on glutathione-Sepharose 4B beads (GE Healthcare), the purification tags were cleaved using thrombin protease (Sigma), followed by cation exchange chromatography and gel filtration using a Superdex 200 10/300 column. Human Cul2-Rbx1 proteins were expressed and purified as previously described (13). Human E1 protein was expressed in Hi5 insect cells and purified as previously described (14). Human βTrCP-Skp1 complex was expressed in Hi5 insect cells and purified as described (8, 15). The purification of neddylated Cul1-Rbx1 complexes has been described previously (15). All proteins were gel-filtered into storage buffer (30 mm Tris, pH 7.5, 100 mm NaCl, 1 mm DTT, 10% glycerol), divided into aliquots, and snap-frozen in liquid nitrogen for later use. Human ubiquitin was purchased from Boston Biochem. Ub-β-catenin peptide substrate (New England Peptide) was prepared and labeled with 32P as described previously (8, 15).
Process for Selecting Residues in the Cul1 BC for Cys Replacement
A multiple sequence alignment of both human Cul1 paralogs and orthologs has been described previously (8). This alignment was used to identify positively charged residues located in the basic canyon region that are also phylogenetically conserved. It was reasoned that these residues are candidates for forming favorable ionic interactions with the negatively charged Cdc34 acidic tail during Cdc34-SCF complex formation. The x-ray structure of human Cul1 was analyzed to identify residues from the culled list that are also highly exposed to bulk solvent (16). This produced the following Cul1 residues: Lys-431, Lys-432, Lys-435, Lys-472, Arg-473, Lys-515, Lys-522, Lys-645, Lys-647, Lys-678, Lys-679, Arg-681, and Lys-689. From this list, Lys-435, Lys-472, Lys-515, Lys-645, and Lys-679 were chosen as final candidates for Cys replacement because their locations collectively sample the entire basic canyon region.
Multiturnover Ubiquitination Assay
Cdc34 1C, the Cdc34 2C proteins, and all Cul1-Rbx1 complexes were tested for activity using standard multiturnover conditions. First, 1 μm E1, 60 μm ubiquitin, and 10 μm Cdc34 were incubated in a reaction buffer containing 30 mm Tris, pH 7.5, 100 mm NaCl, 5 mm MgCl2, 2 mm DTT, and 2 mm ATP at room temperature for 1 min. Both Cul1-Rbx1 and βTrCP-Skp1 complexes were added (0.1 μm final), followed by an additional incubation period of 2 min. The reaction was initiated by adding Ub-β-catenin peptide (3 μm final) and quenched with reducing SDS-PAGE sample buffer. Unmodified substrate and ubiquitinated products were separated by SDS-PAGE, followed by autoradiography and quantitation using ImageQuant software (GE Healthcare). Note that the choice of Ub-β-Catenin peptide as substrate bypasses the UbcH5-dependent substrate priming step such that these ubiquitination reactions only require Cdc34 for E2 enzyme.
Diubiquitin Synthesis Assay
1 μm E1 was incubated with 5 μm 32P-labeled K48R ubiquitin and 5 μm wild-type (WT) Cdc34 protein in a buffer containing 30 mm Tris, pH 7.5, 100 mm NaCl, 5 mm MgCl2, 2 mm DTT, and 2 mm ATP at room temperature for 1 min (14). Reactions were then supplemented with either buffer or cullin-Rbx1 complexes (WT Cul1-Rbx1, WT Cul2-Rbx1, or R648C Cul2-Rbx1) to a final concentration of 0.5 μm. The reactions were incubated for an additional minute, followed by initiation of product formation with 70 μm Asp-77 ubiquitin (further details can be found on the use of K48R and Asp-77 ubiquitin in this assay in Ziemba et al. (14). Reactions were quenched at various time points with reducing SDS-PAGE buffer. Both ubiquitin substrates and diubiquitin products were resolved by SDS-PAGE, followed by autoradiography and quantitation using ImageQuant software (GE Healthcare).
Labeling of Cdc34 Proteins
Cdc34 proteins were first dialyzed against a buffer containing 30 mm Tris, pH 7.5, 100 mm NaCl, and 5% glycerol using a Slide-A-Lyzer MINI dialysis unit, 7,000 molecular weight cut-off (Pierce). A fresh aliquot of bismaleimidoethane (BMOE; Pierce) was dissolved in DMSO to generate a 2.4 mm stock solution, followed by assembly of the labeling reaction with 120 μm BMOE and 60 μm Cdc34 protein in labeling buffer (PBS containing 5 mm EDTA). After a 10-min incubation at room temperature, the reactions were desalted to remove excess BMOE using Zeba spin desalting columns, 7,000 molecular weight cut-off (Pierce). Alternatively, a fresh aliquot of 2,2′-dipyridyldisulfide (DPS; Sigma) was dissolved in 50% DMSO to generate a 50 mm stock solution, followed by assembly of the labeling reaction with 2.5 mm DPS and 60 μm Cdc34 protein in labeling buffer (30 mm Tris, pH 7.5, 100 mm NaCl, 5% glycerol). After a 15-min incubation at room temperature, the reactions were desalted to remove excess DPS. All labeled proteins were quickly divided into aliquots and snap-frozen with liquid nitrogen for later use.
Cross-linking Reactions, Visualization, and Quantitation
Cross-linking reactions induced with BMOE were assembled as follows. 10 μm Cdc34 was added to 1 μm Cul1-Rbx1 in a reaction buffer containing PBS and 5 mm EDTA. The reactions were quenched using reducing SDS-PAGE sample buffer supplemented with 10 mm DTT after a 10- or 60-s incubation period. Cross-linking reactions induced with DPS were assembled as follows. 10 μm Cdc34 was added to 1 μm Cul1-Rbx1 in a reaction buffer containing 30 mm Tris, pH 7.5, 100 mm NaCl, and 5% glycerol. The reactions were quenched using non-reducing SDS-PAGE sample buffer after 10 or 60 s, followed immediately by SDS-PAGE and Western blotting. Western blots were probed using antibodies against either Cul1 (1:5,000 dilution factor; Invitrogen (catalog no. 322400)), Cdc34 (1:10,000 dilution factor; BD Biosciences (catalog no. 610249)), or Cul2 (1:2,500 dilution factor; Invitrogen (catalog no. 511800)). Protein levels were detected using Western Bright ECL (Bio Express) on a Molecular Imager ChemiDoc XRS+ system (Bio-Rad) and quantitated using Image Lab software (Bio-Rad). Note that the anti-Cul1 and anti-Cul2 antibodies recognize epitopes at the C-terminal end of Cul1 and Cul2, and because these proteins are expressed as fragments, the Cul1 and Cul2 Western blots correspond to the protein levels for the C-terminal domains.
RESULTS
A cysteine-based cross-linking approach was developed to uncover the mode of binding between the Cdc34 acidic tail (AT) and the Cul1 basic canyon (BC) on SCF. Specifically, single cysteine residues were systematically introduced along the distal Cdc34 AT region (which was previously shown to be the critical part of the tail for functional binding to SCF) (17). Single cysteine residues were also introduced at structural positions sampling the entire Cul1 BC region that contained basic residues conserved in both human Cul1 orthologs and paralogs. Our premise is that the percentage of cysteine residues in both the Cdc34 AT and the Cul1 BC that can form Cdc34-Cul1 cross-links will provide insight into how the Cdc34 AT docks to the Cul1 BC. For instance, the binding of the Cdc34 AT to the Cul1 BC in a single, unique conformation would result in the close proximity of only a limited number of residues from both Cdc34 and Cul1 in Cdc34-SCF complexes, restricting the cross-linking of cysteines in the Cul1 BC to Cdc34. On the other hand, the binding of the Cdc34 AT with the Cul1 BC through multiple conformations would probably result in the proximity of a greater number of residues from both the Cdc34 AT and the Cul1 BC and more promiscuous Cdc34-Cul1 cross-linking.
Eight Cdc34 and five Cul1 cysteine mutants were produced, enabling the detection of 40 potential Cdc34-Cul1 cross-links. Because human Cdc34 naturally contains a conserved cysteine residue in the active site as well as two naturally occurring cysteine residues in the AT region, the two cysteines in the AT were first mutated to serines to generate Cdc34 1C. Cysteine residues were then introduced at evenly spaced intervals along the protein primary structure by replacing existing residues with a single cysteine (Fig. 2A). Thus, these Cdc34 proteins contain only two cysteines, including the active site one; they are referred to as Cdc34 2C. The human Cul1 amino acid sequence contains 12 naturally occurring cysteine residues. Because our attempt to express a cysteineless version was unsuccessful, single cysteine residues were introduced in the Cul1 BC without perturbing the naturally occurring ones (Fig. 2, B and C).
FIGURE 2.
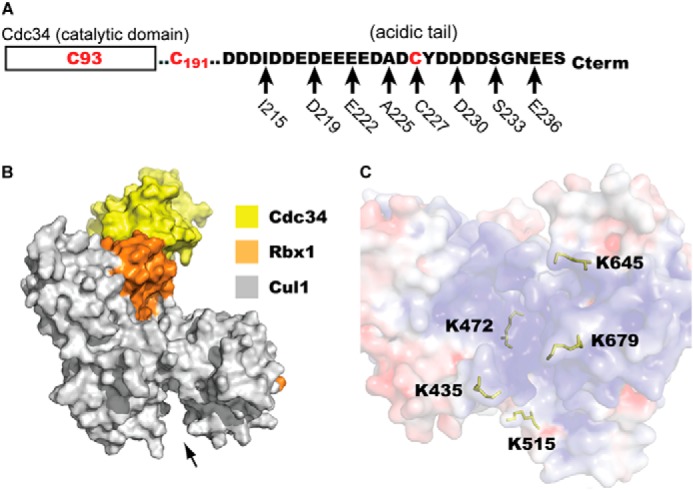
Probing the interaction of the Cdc34 acidic tail with SCF using a cysteine cross-linking strategy. A, domain organization of human Cdc34 highlighting the amino acid sequence of the acidic tail region responsible for binding to SCF. Three naturally occurring cysteine residues exist in wild-type Cdc34: the active site residue Cys-93, Cys-191, and Cys-227. The mutation of Cys-191 and Cys-227 to serines produces Cdc34 1C. Using Cdc34 1C as a template, eight positions within the acidic tail served as sites for the introduction of a single cysteine residue in the acidic tail. These constructs are collectively referred to as the Cdc34 2C proteins. B, surface representation of a model of Cdc34 docked onto Cul1-Rbx1. The arrow shows the location of the Cul1 basic canyon. C, close-up view of the Cul1 basic canyon. The electrostatic surface potential of Cul1 is shown, where blue indicates the highly positively charged surface of the basic canyon. The positions of five Cul1 residues that have been individually replaced with a cysteine are highlighted as yellow ball-and-stick representations.
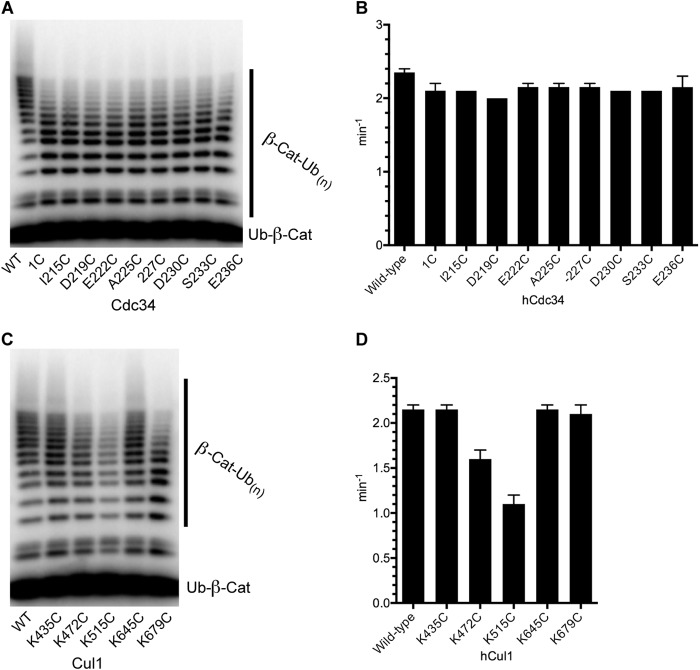
The functional integrity of the Cdc34 proteins and the SCF ligases used in this investigation were confirmed using an established SCF-dependent in vitro ubiquitination assay (15, 18). First, Cdc34 1C and the Cdc34 2C proteins equally showed only a modest decrease in the average number of ubiquitins attached to substrate compared with WT Cdc34 (Fig. 3, A and B). The SCF complexes containing either K435C or K645C Cul1 were identical in activity to SCF complexes containing WT Cul1, and the activities of SCF enzymes containing K472C or K679C Cul1 were only slightly decreased (Fig. 3, C and D). Finally, SCF containing K515C Cul1 was more substantially affected by the mutation; however, the activity of this enzyme still resulted in polyubiquitin chains that extended to as many as 16 ubiquitin protomers on substrate. These results demonstrate that the cysteine-engineered proteins can form functional complexes.
FIGURE 3.
Cdc34 1C and 2C proteins as well as the Cul1-Rbx1 cysteine mutants retain ubiquitination activities within 2-fold of the WT enzymes. A, Ub-β-catenin peptide ubiquitination reactions containing either WT Cdc34, Cdc34 1C, or the Cdc34 2C proteins and WT SCFβTrCP. Ubiquitination reactions were incubated for 5 min, followed by quenching and SDS-PAGE. The rate of the reaction is defined as the fraction of Ub-β-catenin peptide that is modified by one or more ubiquitins, normalized to the ratio of the substrate and enzyme concentration and divided by the time of incubation. B, graph plotting the rates of ubiquitination with WT Cdc34, Cdc34 1C, or Cdc34 2C proteins in the presence of WT SCFβTrCP. Error bars, S.E. of measurement from duplicate data points. C, same as in A except the reactions contain WT Cdc34 and either WT or cysteine-containing mutant Cul1 subunits of SCFβTrCP. D, graph plotting the rates of ubiquitination with WT or cysteine mutant Cul1-containing SCFβTrCP complexes. Error bars, S.E. of measurement from duplicate data points.
The homobifunctional cross-linking reagent BMOE was initially used to induce cross-linking between Cdc34 proteins in complex with SCF. BMOE was chosen because the cysteine-reactive maleimide groups are joined by an 8-Å alkyl spacer arm, which affords some flexibility on how close the cysteine in Cdc34 must get to the cysteine in Cul1 for cross-linking to occur.
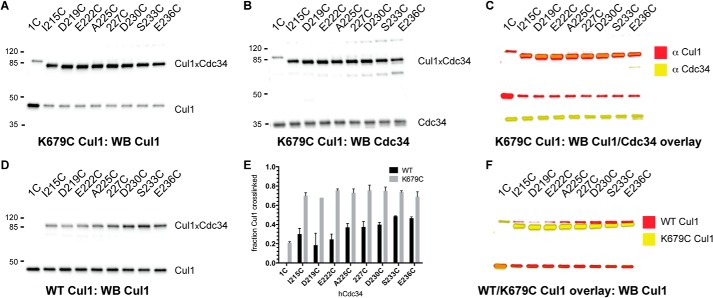
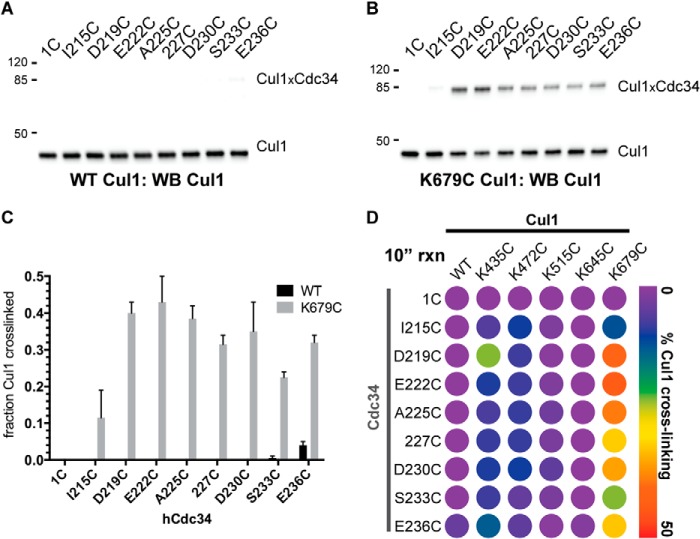
We began our investigation by detecting cross-linking between either Cdc34 1C or the Cdc34 2C proteins and K679C Cul1-Rbx1. A saturating concentration of Cdc34 protein that had been preactivated with BMOE was incubated with K679C Cul1-Rbx1 for 1 min followed by quenching. Notably, all eight reactions containing K679C Cul1 and a Cdc34 2C protein resulted in the formation of single bands that were both cross-reactive to anti-Cul1 and anti-Cdc34 antibodies (Fig. 4, A–C). The amount of Cul1 converted into Cdc34-Cul1 also did not significantly differ for all eight Cdc34 2C proteins (Fig. 4E). A small amount of Cdc34 1C formed cross-links to K679C Cul1 (this is probably nonspecific because the Cdc34 active site is not known to interact with the Cul1 BC); however, the amount was ∼4-fold less than for the eight Cdc34 2C proteins.
FIGURE 4.
The Cdc34 acidic tail forms cross-links to multiple residues in the Cul1 basic canyon. A, Cul1 Western blot of reactions between either Cdc34 1C or Cdc34 2C proteins and K679C Cul1-Rbx1 in the presence of BMOE. Note that very little product is formed between Cdc34 1C and Cul1, whereas almost all Cul1 is cross-linked to the Cdc34 2C proteins. B, Cdc34 Western blot of the identical reactions shown in A. C, color overlay of the Cul1 (red) and Cdc34 (yellow) Western blots, demonstrating that cross-linked products consist of both Cdc34 and Cul1. D, Cul1 Western blot of reactions between either Cdc34 1C or Cdc34 2C proteins and WT Cul1-Rbx1. E, graph showing the fraction of either WT or K679C Cul1 subunits that cross-linked to either Cdc34 1C or Cdc34 2C proteins. Error bars, S.E. of measurement from duplicate data points. F, color overlay of reactions between WT (red) or K679C (yellow) Cul1-Rbx1 and Cdc34 1C or Cdc34 2C proteins. Note the difference in electrophoretic mobilities for WT Cul1-Cdc34 and K679C Cul1-Cdc34 products.
Surprisingly, cross-linking was observed between all eight Cdc34 2C proteins and WT Cul1, although the efficiencies of cross-linking were substantially less than for reactions containing K679C Cul1-Rbx1 (Fig. 4, D and E). Furthermore, no cross-linking was observed in the control reaction containing Cdc34 1C and WT Cul1. Overlaying of the Western blots corresponding to the cross-linking reactions for WT and K679C Cul1-Rbx1 indicated a significant difference in the electrophoretic mobilities of the cross-linked products (Fig. 4F). We speculated that this was probably due to the fact that the cross-linking between K679C Cul1 and Cdc34 was specific to Cys 679 (which is not present in WT Cul1); however, we wanted to rigorously demonstrate this by identifying the cysteines in WT Cul1 that are responsible for cross-linking to Cdc34.
An analysis of the Cul1-Rbx1 structure identified a cysteine residue, Cys-551, which is in close proximity to the BC region on Cul1 (Fig. 5A). To test whether Cys-551 is responsible for forming cross-links between WT Cul1 and Cdc34, C551S Cul1-Rbx1 was generated and tested in cross-linking reactions with the Cdc34 2C proteins. This mutation resulted in the near elimination of cross-linking with Cdc34 2C proteins (Fig. 5B), and the subsequent mutation of Lys-679 into a cysteine residue in C551S Cul1 restored cross-linking to Cdc34 (Fig. 5C).
FIGURE 5.
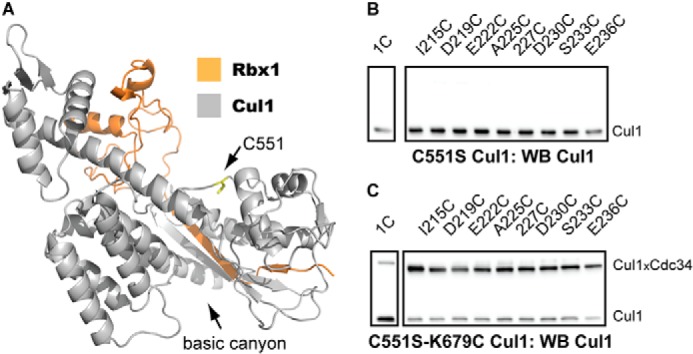
Native cysteine residue Cys-551 in WT Cul1 forms cross-links to the Cdc34 2C proteins. A, ribbon diagram of the C-terminal domain of Cul1 (gray) and Rbx1 (orange). The Cys-551 side chain is shown in a ball-and-stick representation (yellow), and its location is highlighted by the black arrow. The structure is from Protein Data Bank entry 1LDK (16). B, Cul1 Western blot of reactions between either Cdc34 1C or Cdc34 2C proteins and C551S Cul1-Rbx1 in the presence of BMOE. Notice that no Cdc34-Cul1 cross-links are observed. C, Cul1 Western blot of reactions between either Cdc34 1C or Cdc34 2C proteins and C551S/K679C Cul1-Rbx1 in the presence of BMOE. Notice that the introduction of a cysteine residue at Lys-679 in C551S Cul1-Rbx1 restores cross-linking to the Cdc34 2C proteins. The time of incubation for all reactions was 1 min.
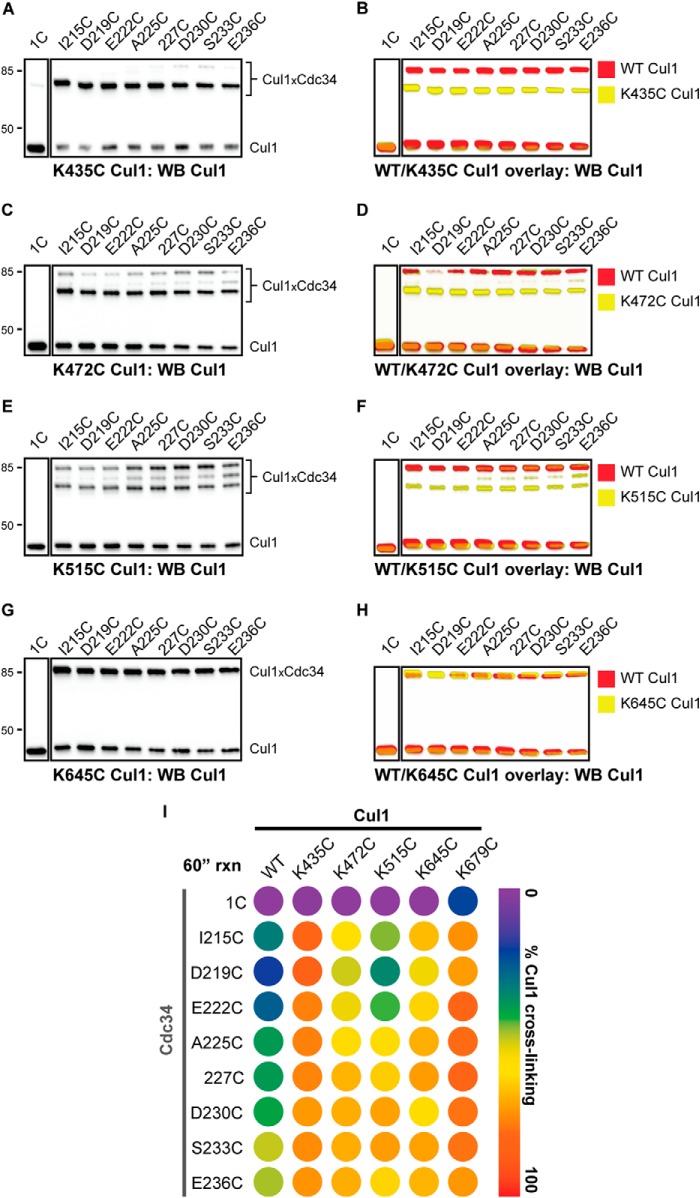
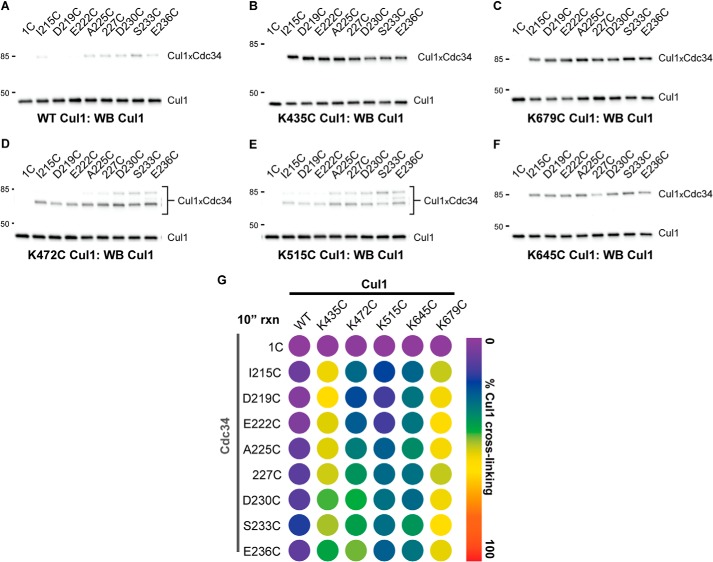
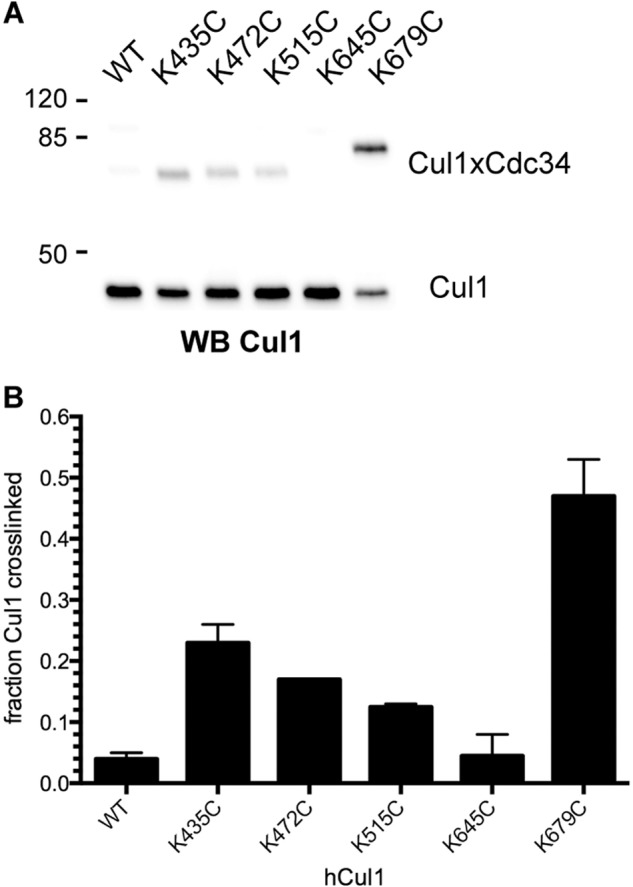
Cross-linking reactions were next performed with the Cdc34 proteins and the remaining four Cul1 proteins containing a unique cysteine in the BC. Similar to K679C Cul1, all of the Cdc34 2C proteins showed robust cross-linking to K435C Cul1, whereas Cdc34 1C showed almost no cross-linking (Fig. 6, A, B, and I). Cross-linking was also observed between the Cdc34 2C proteins and K472C Cul1; however, note that two distinct Cdc34-Cul1 product bands were observed in the Cul1 Western blots (Fig. 6C). It is likely that one of these bands corresponds to a cross-linked product between the native Cul1 residue Cys-551 and a Cdc34 2C protein because the electrophoretic mobility of this band is identical to that for the WT Cul1-Cdc34 product (Fig. 6D), and the other product band would then correspond to a Cys-472 Cul1-Cdc34 2C cross-link. Note that the latter product is more abundant than the Cys-551 Cul1-Cdc34 one, indicating a preference for cross-linking to Cys-472, which is more centrally located within the Cul1 BC than Cys-551. Reactions between K515C Cul1 and the Cdc34 2C proteins also showed partitioning of the Cdc34 cross-linking to either Cys-551 or Cys-515 on Cul1, and in this case, the amounts of these products were comparable (Fig. 6, E and F). Finally, cross-linking was observed between the Cdc34 2C proteins and K645C Cul1 (Fig. 6, G and H). (Unfortunately, the electrophoretic mobilities of Cdc34-Cul1 cross-links from reactions with WT or K645C Cul1-Rbx1 are identical; however, the increase in signal for Cdc34-Cul1 cross-links from reactions with K645C Cul1-Rbx1 suggests that cross-linking is occurring between Cys-645 and Cdc34.) In summary, a broad region of the Cul1 BC can cross-link to the entire Cdc34 AT region tested, which suggests a large degree of conformational flexibility for the interaction of the Cdc34 AT with the Cul1 BC (Fig. 6I).
FIGURE 6.
The Cdc34 acidic tail forms cross-links to multiple positions in the Cul1 basic canyon. Cross-linking was performed in the presence of BMOE-activated Cdc34 proteins for 60 s prior to quenching. A, Cul1 Western blot of reactions between either Cdc34 1C or Cdc34 2C proteins and K435C Cul1-Rbx1 in the presence of BMOE. Note that very little product is formed between Cdc34 1C and Cul1, whereas almost all Cul1 is cross-linked to the Cdc34 2C proteins. B, color overlay of reactions between WT (red) or K435C (yellow) Cul1-Rbx1 and Cdc34. Note the difference in electrophoretic mobilities for WT Cul1-Cdc34 and K435C Cul1-Cdc34 products, indicating that these cross-links probably involve different cysteine residues on Cul1. C and D, same as in A and B, except with K472C Cul1-Rbx1. E and F, same as in A and B, except with K515C Cul1-Rbx1. G and H, same as in A and B, except with K645C Cul1-Rbx1. Notice that the electrophoretic mobilities of the Cul1-Cdc34 products are identical in the WT and K645C Cul1 color overlay. However, there is an increase in signal for the reactions containing K645C Cul1-Rbx1 compared with reactions containing WT Cul1-Rbx1 (see Fig. 4D), suggesting that at least some cross-linking is occurring between Cys-645 and the Cdc34 2C proteins. I, heat map for the values of the fraction of Cul1 converted to Cdc34-Cul1 cross-links for WT and all five cysteine mutant Cul1-Rbx1 proteins. In cases where more than one Cdc34-Cul1 product is observed (e.g. K472C Cul1 and K515C Cul1), the values for the fraction converted have been combined. Values represent the average from duplicate measurements.
It is well known that caution should be taken when interpreting results from BMOE-induced cross-linking reactions because nonspecific cross-linking can be problematic. Although all Cul1 proteins were tested for cross-linking to Cdc34 1C (which serves as a negative control because the Cdc34 active site is not known to interact with the Cul1 BC), two additional controls were performed to further substantiate the results. First, previous studies had demonstrated that the interaction of the Cdc34 AT with the Cul1 BC is stabilized by electrostatic forces, and thus the affinity of Cdc34 for SCF is salt-dependent (8). Encouragingly, the efficiency of cross-linking for 227C Cdc34 2C to K679C Cul1 was dramatically reduced in the presence of increasing salt concentrations (Fig. 7, A and B). Second, cysteine residues were engineered at exposed positions on the Cdc34 catalytic domain (Ser-36 and Lys-167) in a tail-deleted Cdc34 mutant. The deletion of the AT domain from Cdc34 should eliminate interaction of Cdc34 with the Cul1 BC, and both S36C and K167C Cdc34 Δ190 showed almost no cross-linking to K679C Cul1-Rbx1 (Fig. 7C). Thus, the BMOE-induced cross-linking between Cdc34 and Cul1 probably reflects the formation of a bona fide Cdc34-SCF complex.
FIGURE 7.
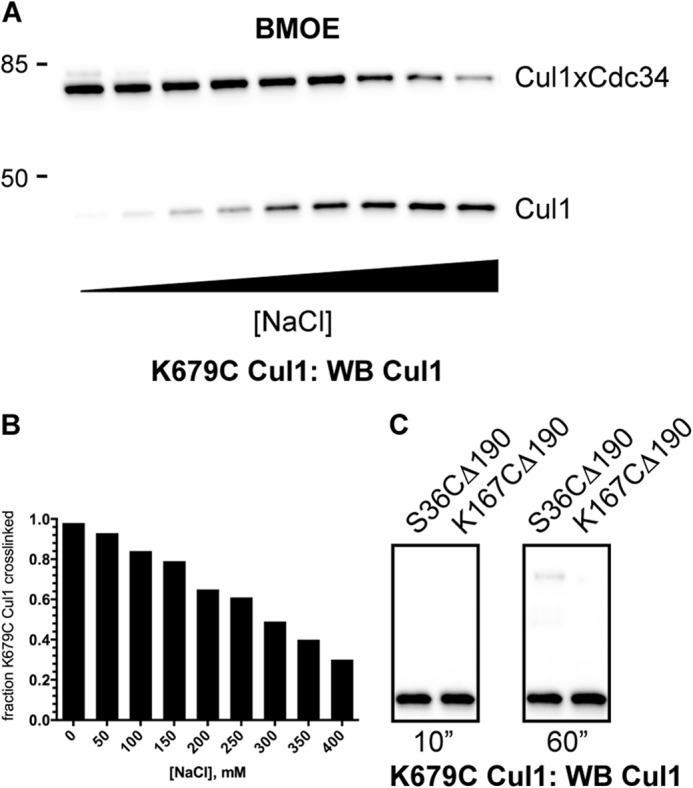
Cross-linking between K679C Cul1-Rbx1 and BMOE-activated Cdc34 proteins is affected by the ionic strength of the reaction buffer and is dependent on the presence of the Cdc34 acidic tail. A, Cul1 Western blot of reactions between 227C Cdc34 2C and K679C Cul1-Rbx1 with increasing concentrations of NaCl in the presence of BMOE. Note that K679C Cul1 was chosen because Lys-679 is centrally located in the basic canyon region. B, graph showing the fraction of K679C Cul1 that was converted into Cdc34-Cul1 products as a function of the concentration of NaCl in the reaction buffer. C, cross-linking between K679C Cul1-Rbx1 and either S36C Δ190 or K167C Δ190 Cdc34 2C proteins. Both S36C Δ190 and K167C Δ190 Cdc34 were incubated in the presence of BMOE followed by desalting. BMOE-activated Cdc34 proteins were then incubated with K679C Cul1-Rbx1 for the indicated amounts of time, followed by SDS-PAGE and detection of proteins by Cul1 Western blotting.
Because product formation for the cross-linking reactions between Cdc34 2C proteins and either K679C or K435C Cul1-Rbx1 had approached completion during the 1-min incubation period, the cross-linking reactions were repeated for all pairwise Cdc34-Cul1 combinations and quenched after only 10 s. Notably, the cross-linking between Cdc34 2C proteins and WT Cul1-Rbx1 was negligible; however, both K679C and K435C Cul1 subunits were still efficiently converted into Cdc34-Cul1 cross-linked products (Fig. 8, A–C and G). Product formation for K472C, K515C, and K645C Cul1-Rbx1 complexes was still apparent at 10 s; however, the levels were significantly reduced compared with the reactions with 1-min incubation periods (Fig. 8, D–F and G). Two key points can be made from these results: 1) cross-linking formation between the Cdc34 AT and the Cul1 BC is highly tolerant of the position of the cysteine residue in the Cdc34 AT region, and 2) although cross-linking was observed between cysteines in the Cdc34 AT and a broad region of the Cul1 BC, it was most efficient when cysteines were introduced at either Lys-679 or Lys-435.
FIGURE 8.
Conducting the BMOE-induced cross-linking reactions with a shorter incubation period reveals the greater reactivity of K679C and K435C Cul1-Rbx1 complexes with the Cdc34 2C proteins in comparison with the other Cul1 mutants. Cross-linking was performed in the presence of BMOE-activated Cdc34 proteins for 10 s prior to quenching. A, Cul1 Western blot of reactions between either Cdc34 1C or Cdc34 2C proteins and WT Cul1-Rbx1 in the presence of BMOE. Note that very little product is formed between the Cdc34 2C proteins and WT Cul1 (compare with Fig. 4D). B, same as A, except with K435C Cul1-Rbx1. C, same as A, except with K679C Cul1-Rbx1. D, same as A, except with K472C Cul1-Rbx1. E, same as A, except with K515C Cul1-Rbx1. F, same as A, except with K645C Cul1-Rbx1. G, heat map for the values of the fraction of Cul1 converted to Cdc34-Cul1 cross-links. Similar to Fig. 6, in cases where more than one Cdc34-Cul1 product is observed, the values for the fraction converted have been combined. Values represent the average from duplicate measurements.
To gain greater insight into the interaction of the Cdc34 AT and the Cul1 BC, cross-linking reactions were performed between the Cul1-Rbx1 complexes and Cdc34 1C and 2C proteins that were activated for cross-linking with DPS. Here, cross-linking occurs when a cysteine residue modified by DPS is attacked by a thiolate anion from an adjacent cysteine, resulting in disulfide bond formation. This method places a much greater stringency on the proximities of cysteines from Cdc34 and Cul1, because the sulfur atoms must be within ∼2 Å for disulfide bonding to occur, whereas they can be separated by as much as 10 Å for cross-linking with BMOE. Consistent with this, no cross-linking was observed between Cdc34 1C or the Cdc34 2C proteins and WT Cul1-Rbx1 or between Cdc34 1C and K679C Cul1-Rbx1 during the 10-s incubation period (Fig. 9, A–C). However, product formation was observed between the Cdc34 2C proteins and K679C Cul1-Rbx1 (Fig. 9B). Notably, I215C Cdc34 2C showed a substantial reduction in the level of Cdc34-Cul1 cross-linking as compared with the other Cdc34 2C proteins (note that Ile-215 is the most proximal position in the Cdc34 AT that was tested). Less product formation was observed in reactions between the Cdc34 2C proteins and both K435C and K472C Cul1-Rbx1 complexes, and cross-linking was nearly absent from reactions containing either K515C or K645C Cul1-Rbx1 complexes (Fig. 9D). Similar to the cross-linking reactions that were induced with BMOE, the DPS-induced cross-linking showed a strict salt dependence, and both S36C and K167C Cdc34 Δ190 activated with DPS showed almost no cross-linking to K679C Cul1-Rbx1 (Fig. 10).
FIGURE 9.
The Cdc34 acidic tail shows a preference for disulfide cross-linking to Cys-679 in the Cul1 basic canyon. A, Cul1 Western blot of reactions between either Cdc34 1C or Cdc34 2C proteins that had been activated with DPS and WT Cul1-Rbx1. B, same as in A, except K679C Cul1-Rbx1 was used. C, graph showing the fraction of either WT or K679C Cul1 subunits that cross-linked to either Cdc34 1C or Cdc34 2C proteins. Error bars, S.E. of measurement from duplicate data points. D, heat map showing the fraction of Cul1 converted to Cdc34-Cul1 cross-links for WT and all five cysteine mutant Cul1-Rbx1 proteins. Cdc34 proteins had been activated with DPS prior to their incubation with the Cul1-Rbx1 proteins. Values represent the average from duplicate measurements.
FIGURE 10.
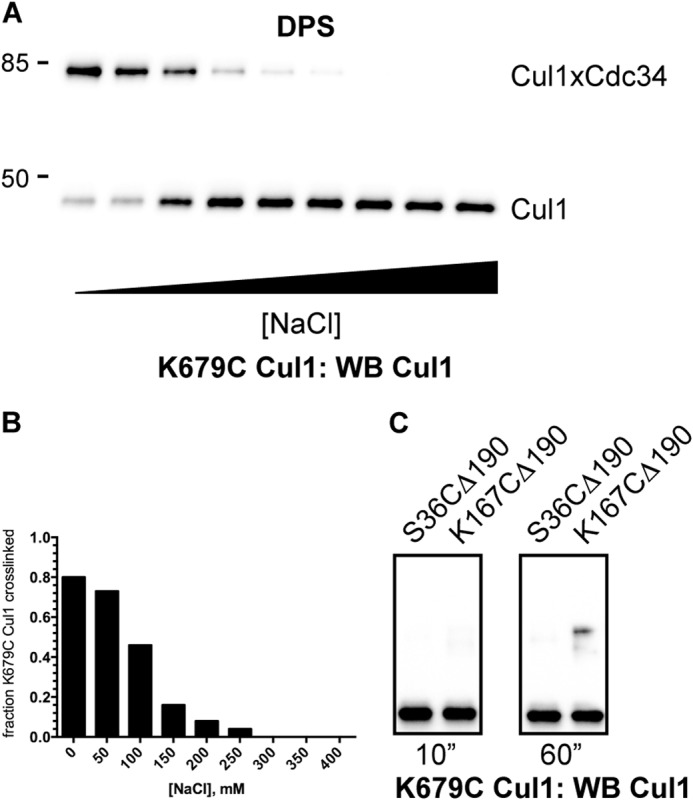
Cross-linking between K679C Cul1-Rbx1 and DPS-activated Cdc34 proteins is affected by the ionic strength of the reaction buffer and is dependent on the presence of the Cdc34 acidic tail. A, Cul1 Western blot of reactions between 227C Cdc34 2C and K679C Cul1-Rbx1 with increasing concentrations of NaCl in the presence of DPS. Note that K679C Cul1 was chosen because Lys-679 is centrally located in the basic canyon region. B, graph showing the fraction of K679C Cul1 that was converted into Cdc34-Cul1 products as a function of the concentration of NaCl in the reaction buffer. C, cross-linking between K679C Cul1-Rbx1 and either S36C Δ190 or K167C Δ190 Cdc34 2C proteins. DPS-activated Cdc34 proteins were incubated with K679C Cul1-Rbx1 for the indicated amounts of time, followed by SDS-PAGE and detection of proteins by Cul1 Western blotting.
Cross-linking reactions between cysteine residues induced by DPS are dependent on the deprotonation of the attacking thiol group (in this case, the cysteine located in the Cul1 BC), and thus their pKa values can affect the rates of cross-linking. To uncover potential differences in the pKa values of the Cul1 cysteine residues that participated in cross-linking, DPS-induced cross-linking reactions between 227C Cdc34 2C and WT or mutant Cul1-Rbx1 complexes were repeated at pH 9 (note that the previous reactions were incubated at pH 7.5). As expected, the fraction of Cul1 converted into Cdc34-Cul1 cross-links increased for the Cul1-Rbx1 complexes for which product formation was observable at pH 7.5 (Fig. 11). Nevertheless, cross-linked product formation was still considerably greater for K679C Cul1-Rbx1 as compared with the other SCF complexes, indicating that the results for the DPS-induced cross-linking reactions are not significantly influenced by differences in the cysteine pKa values.
FIGURE 11.
Cross-linking between 227C Cdc34 and K679C Cul1 is more robust than for the other Cul1 cysteine-containing mutants, even at high pH. A, Cul1 Western blot of reactions between 227C Cdc34 and either WT Cul1-Rbx1 or the five Cul1-Rbx1 cysteine mutant complexes in the presence of DPS and a reaction buffer composed of 30 mm Tris, pH 9.0, 100 mm NaCl, and 5% glycerol (note that the pH of the reaction buffer was 7.5 for all previous reactions). Reactions were incubated for 10 s prior to quenching with non-reducing SDS-PAGE buffer. B, graph showing the fraction of Cul1 proteins that cross-linked to 227C Cdc34. Error bars, S.E. of measurement from duplicate data points.
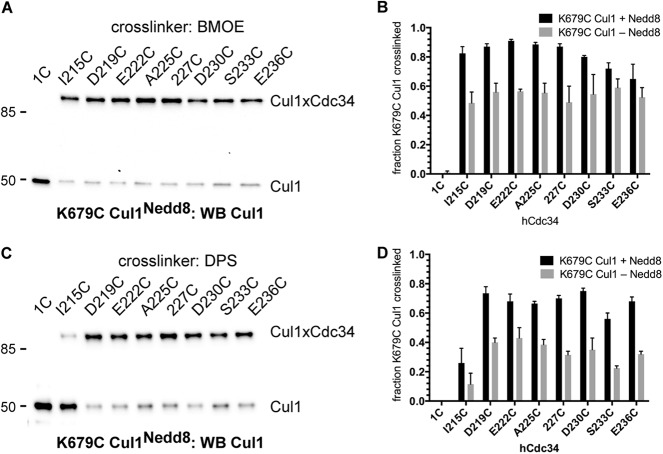
The activities of SCF ubiquitin ligases are controlled in cells by a process called neddylation (15, 19–21). Here, the ubiquitin-like protein, Nedd8, is conjugated to a lysine residue on Cul1, which induces conformational changes affecting numerous biochemical activities of SCF. To explore how neddylation affects the interaction of the Cdc34 AT with the Cul1 BC, K679C Cul1-Rbx1 was neddylated and used in cross-linking reactions with either the Cdc34 1C or Cdc34 2C proteins. The neddylation of K679C Cul1 resulted in a modest increase in the amount of cross-linking to the Cdc34 2C proteins, regardless of whether BMOE or DPS was used to induce cross-linking (Fig. 12). Furthermore, each Cdc34 2C protein still formed similar amounts of Cdc34-Cul1 products (one exception is I215C Cdc34 2C in cross-linking reactions induced by DPS). The modest increase in cross-linking induced by the neddylation of Cul1 may potentially be explained by a positive electrostatic surface on Nedd8 that faces the cullin BC region and may attract the negatively charged Cdc34 AT (19). Although it may at first seem surprising that neddylation has only a modest effect on Cdc34 cross-linking to SCF, these results are consistent with previous studies showing that the neddylation of Cul1 only minimally increases the affinity of Cdc34 for SCF (15) and that, although neddylation causes substantial conformational changes in the SCF structure, the BC region is to a large extent not affected (8).
FIGURE 12.
The neddylation of SCF stimulates the cross-linking of Cdc34 to Cul1. A, Cul1 Western blot of reactions between either Cdc34 1C or Cdc34 2C proteins that had been activated with BMOE and neddylated K679C Cul1-Rbx1. B, graph showing the fraction of either neddylated (black bars) or unneddylated (gray bars) K679C Cul1 subunits that cross-linked to either Cdc34 1C or Cdc34 2C proteins. Error bars, S.E. of measurement from duplicate data points. C and D, same as A and B, except the Cdc34 1C and 2C proteins had been activated with DPS. Note that K679C Cul1 was chosen for these experiments because Lys-679 is centrally located in the basic canyon region.
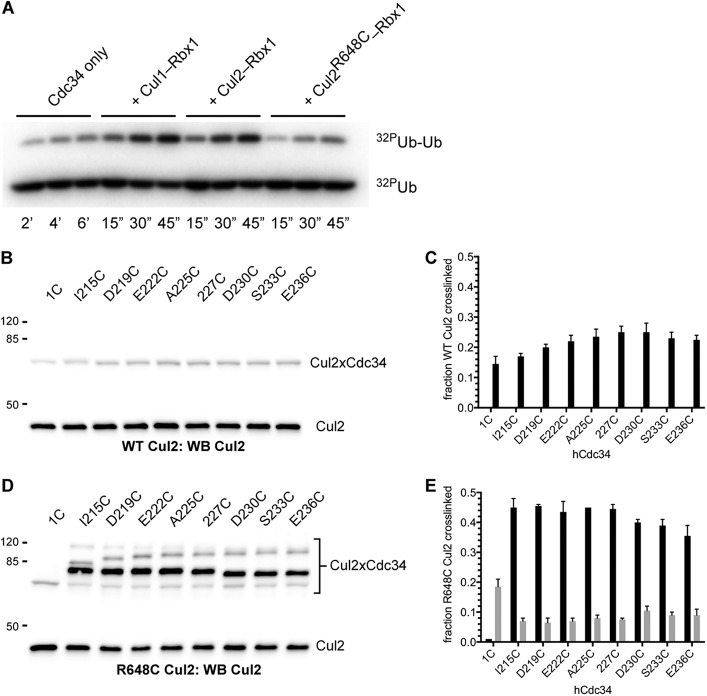
SCF ubiquitin ligases are members of the larger CRL family, and many of the positively charged Cul1 BC residues are also conserved in Cul1 paralogs. This suggests that other members of the CRLs may also contain basic canyons that interact with the Cdc34 AT in a similar manner as with the Cul1 BC. To determine whether this is the case, we used our cysteine-based cross-linking approach, wherein a cysteine was introduced in the Cul1 homolog Cul2 at Arg-648. (Note that this position is structurally equivalent to Lys-679 in Cul1.) Both WT and R648C Cul2-Rbx1 were functional in a Cdc34 activation assay (Fig. 13A), although the R648C mutation in Cul2 resulted in a modest loss of activity that was similar to the effect of the K679C mutation in Cul1 on SCF activity.
FIGURE 13.
The Cdc34 acidic tail forms cross-links to a residue located in the Cul2 basic canyon. A, time course for a diubiquitin synthesis assay for WT Cdc34 alone or in the presence of either WT Cul1-Rbx1, WT Cul2-Rbx1, or R648C Cul2-Rbx1. Notice that both WT Cul1-Rbx1 and WT Cul2-Rbx1 stimulate the Cdc34-dependent formation of diubiquitin by ∼20-fold, and the reduction in the activity of R648C Cul2-Rbx1 as compared with WT Cul2-Rbx1 is only ∼3-fold. B, Cul2 Western blot of reactions between either Cdc34 1C or Cdc34 2C proteins that had been activated with BMOE and WT Cul2-Rbx1. C, graph showing the fraction of Cul2 protein that cross-linked to either Cdc34 1C or the Cdc34 2C proteins. Error bars, S.E. of measurement from duplicate data points. D and E, same as in B and C, except cross-linking reactions contained R648C Cul2-Rbx1. Quantitation of the levels for the fastest migrating Cdc34-Cul2 product species is shown as gray bars; product levels for the next fastest species are shown as black bars.
Cross-linking reactions between BMOE-activated Cdc34 1C and Cdc34 2C proteins were first performed in the presence of WT Cul2-Rbx1. A minor fraction of Cul2 was converted into Cdc34-Cul2 products in reactions containing Cdc34 1C or the Cdc34 2C proteins (Fig. 13, B and C). Note that products with identical electrophoretic mobilities were observed in reactions containing R648C Cul2-Rbx1 as well; however, the amounts of this product were substantially reduced for the Cdc34 2C proteins compared with Cdc34 1C (Fig. 13, D and E). Furthermore, additional Cdc34-Cul2 products were observed but only between the Cdc34 2C proteins and R648C Cul2. Moreover, these products were in much greater abundance than the products observed between Cdc34 1C or the Cdc34 2C proteins and WT Cul2. Similar to cross-linking reactions between the Cdc34 2C proteins and the Cul1-Rbx1 complexes, the amounts of Cdc34-Cul2 products did not significantly deviate when comparing each of the Cdc34 2C proteins. These results suggest that the Cdc34 AT interacts with the Cul2 BC region similarly to how it interacts with the Cul1 BC.
DISCUSSION
Cysteine-based cross-linking is a powerful approach for identifying residues located at the interfaces of interacting proteins and has been useful in the characterization of both E1-E2 and E2-E3 interactions (22–24). The interactions between the Cdc34 AT region and the Cul1 and Cul2 BCs were probed by cross-linking, and the following observations can be made from the results. 1) Cross-linking was observed between all eight residues tested within the Cdc34 AT and six sites within the Cul1 BC region (including the native Cul1 residue Cys-551, located in close proximity to the BC). 2) Cross-linking was most robust when a cysteine was introduced at Lys-679 in the Cul1 BC. 3) Cross-linking efficiencies were similar for all Cdc34 2C proteins that had been activated with BMOE; however, I215C Cdc34 2C showed a substantial reduction in cross-linking to K679C Cul1 in reactions with DPS-activated Cdc34, suggesting that Ile-215 represents a boundary for the portion of the Cdc34 AT that interacts with the Cul1 BC (note that this result is in agreement with previous investigations that mapped the Cul1-binding region of the Cdc34 AT using either direct binding or ubiquitination assays (8, 17)). 4) Cross-linking occurred between each of the Cdc34 2C proteins and the Cul2 BC with similar levels of product formation, suggesting that the Cdc34 AT may interact with the BCs of the CRLs through a conserved mechanism involving multiple conformations.
We propose the following model to explain how interaction of the Cdc34 AT with the Cul1 BC enables the rapid association of Cdc34 with SCF. Typically, when two separated proteins are capable of forming an interaction and collide in solution, a productive encounter only occurs during the off-chance that the interacting surfaces are in close alignment (25). Exceptions to this rule are made possible through complementary electrostatic interactions between the proteins that increase the probability of productive encounters (26). In the case of Cdc34 and SCF, functional binding occurs when the Cdc34 catalytic domain docks onto the Rbx1 subunit of SCF, which stimulates the discharge of ubiquitin from Cdc34 to SCF-bound substrate. Prior to this, the Cdc34 AT interacts with the Cul1 BC, which serves to increase the effective concentration of the Cdc34 catalytic domain and Rbx1. Furthermore, the precise alignment of the Cdc34 AT with the Cul1 BC is unnecessary, because the intervening polypeptide chain between the distal AT and the Cdc34 catalytic domain is sufficiently long to allow interaction of the catalytic domain with Rbx1 even when the AT is docked onto different surfaces on the BC (this has been verified through molecular modeling of the Cdc34-SCF complex; see Fig. 1B). Thus, the collision of Cdc34 and SCF results in productive Cdc34-SCF complexes more often than for typical protein-protein interactions at least in part because the docking of the Cdc34 AT onto the Cul1 BC is possible in multiple conformations.
Nonspecific electrostatic interactions have probably evolved to promote fast association rates between proteins in many biological systems. For instance, many eukaryotic transcription factors contain highly acidic domains that can be transferred between transcription factors without disrupting their function (27, 28). Indeed, this is reminiscent of an engineered Cdc34 protein where the distal AT residues were replaced with only acidic ones without compromising Cdc34 function in vitro (17). As another potential example, consider the disordered C-terminal domain of RNA polymerase II that is dynamically phosphorylated during transcription. These phosphorylation events can signal for the binding of scores of proteins to the transcription complex (29). It is tempting to speculate that at least some of these proteins may associate rapidly with RNA polymerase II through electrostatic interactions similar to that of Cdc34 and SCF.
Additional details, such as whether the Cdc34 AT binds to the basic canyon and changes conformations prior to its dissociation from SCF, await a more thorough structural characterization. The results presented here pave the way for these and other studies, and the production of highly specific, cross-linked Cdc34-SCF complexes may prove useful toward the elucidation of an activated Cdc34∼ubiquitin x-ray structure in the presence of SCF.
Acknowledgments
We thank Casey Hall and Shirley Shen (University of Nevada, Las Vegas, Genomics Center) for DNA sequencing, care of critical instruments, and expert advice. We thank Chris Lima for helpful discussion.
This work was supported, in whole or in part, by National Institutes of Health, NIGMS, Grant P20 GM103440.
- E2
- ubiquitin-conjugating enzyme
- E1
- ubiquitin-activating enzyme
- E3
- ubiquitin ligase
- SCF
- Skp1-cullin-F-box ligase
- CRL
- cullin-RING ligase
- AT
- acidic tail
- BC
- basic canyon
- Ub
- ubiquitin
- BMOE
- bismaleimidoethane
- DPS
- 2,2′-dipyridyldisulfide
- WB
- Western blot.
REFERENCES
- 1. Metzger M. B., Pruneda J. N., Klevit R. E., Weissman A. M. (2014) RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 1843, 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kamadurai H. B., Qiu Y., Deng A., Harrison J. S., Macdonald C., Actis M., Rodrigues P., Miller D. J., Souphron J., Lewis S. M., Kurinov I., Fujii N., Hammel M., Piper R., Kuhlman B., Schulman B. A. (2013) Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. eLife 2, e00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kee Y., Huibregtse J. M. (2007) Regulation of catalytic activities of HECT ubiquitin ligases. Biochem. Biophys. Res. Commun. 354, 329–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 5. Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., Brownell J. E., Burke K. E., Cardin D. P., Critchley S., Cullis C. A., Doucette A., Garnsey J. J., Gaulin J. L., Gershman R. E., Lublinsky A. R., McDonald A., Mizutani H., Narayanan U., Olhava E. J., Peluso S., Rezaei M., Sintchak M. D., Talreja T., Thomas M. P., Traore T., Vyskocil S., Weatherhead G. S., Yu J., Zhang J., Dick L. R., Claiborne C. F., Rolfe M., Bolen J. B., Langston S. P. (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 [DOI] [PubMed] [Google Scholar]
- 6. Petroski M. D., Deshaies R. J. (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 7. Wu K., Kovacev J., Pan Z. Q. (2010) Priming and extending: a UbcH5/Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol. Cell 37, 784–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kleiger G., Saha A., Lewis S., Kuhlman B., Deshaies R. J. (2009) Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell 139, 957–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pierce N. W., Kleiger G., Shan S. O., Deshaies R. J. (2009) Detection of sequential polyubiquitylation on a millisecond timescale. Nature 462, 615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu K., Chen A., Tan P., Pan Z. Q. (2002) The Nedd8-conjugated ROC1-CUL1 core ubiquitin ligase utilizes Nedd8 charged surface residues for efficient polyubiquitin chain assembly catalyzed by Cdc34. J. Biol. Chem. 277, 516–527 [DOI] [PubMed] [Google Scholar]
- 11. Mathias N., Steussy C. N., Goebl M. G. (1998) An essential domain within Cdc34p is required for binding to a complex containing Cdc4p and Cdc53p in Saccharomyces cerevisiae. J. Biol. Chem. 273, 4040–4045 [DOI] [PubMed] [Google Scholar]
- 12. Li T., Pavletich N. P., Schulman B. A., Zheng N. (2005) High-level expression and purification of recombinant SCF ubiquitin ligases. Methods Enzymol. 398, 125–142 [DOI] [PubMed] [Google Scholar]
- 13. den Besten W., Verma R., Kleiger G., Oania R. S., Deshaies R. J. (2012) NEDD8 links cullin-RING ubiquitin ligase function to the p97 pathway. Nat. Struct. Mol. Biol. 19, 511–516, S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ziemba A., Hill S., Sandoval D., Webb K., Bennett E. J., Kleiger G. (2013) Multimodal mechanism of action for the Cdc34 acidic loop: a case study for why ubiquitin-conjugating enzymes have loops and tails. J. Biol. Chem. 288, 34882–34896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Saha A., Deshaies R. J. (2008) Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 32, 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zheng N., Schulman B. A., Song L., Miller J. J., Jeffrey P. D., Wang P., Chu C., Koepp D. M., Elledge S. J., Pagano M., Conaway R. C., Conaway J. W., Harper J. W., Pavletich N. P. (2002) Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709 [DOI] [PubMed] [Google Scholar]
- 17. Kleiger G., Hao B., Mohl D. A., Deshaies R. J. (2009) The acidic tail of the Cdc34 ubiquitin-conjugating enzyme functions in both binding to and catalysis with ubiquitin ligase SCFCdc4. J. Biol. Chem. 284, 36012–36023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu G., Xu G., Schulman B. A., Jeffrey P. D., Harper J. W., Pavletich N. P. (2003) Structure of a β-TrCP1-Skp1-β-catenin complex: destruction motif binding and lysine specificity of the SCF(β-TrCP1) ubiquitin ligase. Mol. Cell 11, 1445–1456 [DOI] [PubMed] [Google Scholar]
- 19. Duda D. M., Borg L. A., Scott D. C., Hunt H. W., Hammel M., Schulman B. A. (2008) Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134, 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamoah K., Oashi T., Sarikas A., Gazdoiu S., Osman R., Pan Z. Q. (2008) Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1's C-terminal tail. Proc. Natl. Acad. Sci. U.S.A. 105, 12230–12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scott D. C., Sviderskiy V. O., Monda J. K., Lydeard J. R., Cho S. E., Harper J. W., Schulman B. A. (2014) Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell 157, 1671–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Calabrese M. F., Scott D. C., Duda D. M., Grace C. R., Kurinov I., Kriwacki R. W., Schulman B. A. (2011) A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases. Nat. Struct. Mol. Biol. 18, 947–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaiser S. E., Mao K., Taherbhoy A. M., Yu S., Olszewski J. L., Duda D. M., Kurinov I., Deng A., Fenn T. D., Klionsky D. J., Schulman B. A. (2012) Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7-Atg3 and Atg7-Atg10 structures. Nat. Struct. Mol. Biol. 19, 1242–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Olsen S. K., Lima C. D. (2013) Structure of a ubiquitin E1-E2 complex: insights to E1-E2 thioester transfer. Mol. Cell 49, 884–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fersht A. (1985) Enzyme Structure and Mechanism, 2nd Ed., pp. 147–148, W. H. Freeman, New York [Google Scholar]
- 26. Schreiber G., Fersht A. R. (1996) Rapid, electrostatically assisted association of proteins. Nat. Struct. Biol. 3, 427–431 [DOI] [PubMed] [Google Scholar]
- 27. Gill G., Ptashne M. (1987) Mutants of GAL4 protein altered in an activation function. Cell 51, 121–126 [DOI] [PubMed] [Google Scholar]
- 28. Gill G., Sadowski I., Ptashne M. (1990) Mutations that increase the activity of a transcriptional activator in yeast and mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 87, 2127–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Phatnani H. P., Greenleaf A. L. (2006) Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20, 2922–2936 [DOI] [PubMed] [Google Scholar]