Background: Synthesis of bacteriochlorophylls is essential for the photosynthetic capture of solar energy.
Results: Chlorophyllide a oxidoreductase (COR) performs nitrogenase-related redox catalysis using two cysteine-ligated [4Fe-4S] clusters.
Conclusion: A reductive trans protonation mechanism and an identical substrate binding mode for nitrogenase-like enzymes was deduced.
Significance: Energy transduction of COR is relevant for related systems involved in the reduction of chemically stable multibonds.
Keywords: Electron Paramagnetic Resonance (EPR), Electron Transport, Iron-Sulfur Protein, Nitrogenase, Photosynthetic Pigment, COR, DPOR, Chlorophyllide Oxidoreductase, Dark-operative Protochlorophyllide Oxidoreductase
Abstract
Bacteriochlorophyll a biosynthesis requires the stereo- and regiospecific two electron reduction of the C7-C8 double bond of chlorophyllide a by the nitrogenase-like multisubunit metalloenzyme, chlorophyllide a oxidoreductase (COR). ATP-dependent COR catalysis requires interaction of the protein subcomplex (BchX)2 with the catalytic (BchY/BchZ)2 protein to facilitate substrate reduction via two redox active iron-sulfur centers. The ternary COR enzyme holocomplex comprising subunits BchX, BchY, and BchZ from the purple bacterium Roseobacter denitrificans was trapped in the presence of the ATP transition state analog ADP·AlF4−. Electron paramagnetic resonance experiments revealed a [4Fe-4S] cluster of subcomplex (BchX)2. A second [4Fe-4S] cluster was identified on (BchY/BchZ)2. Mutagenesis experiments indicated that the latter is ligated by four cysteines, which is in contrast to the three cysteine/one aspartate ligation pattern of the closely related dark-operative protochlorophyllide a oxidoreductase (DPOR). In subsequent mutagenesis experiments a DPOR-like aspartate ligation pattern was implemented for the catalytic [4Fe-4S] cluster of COR. Artificial cluster formation for this inactive COR variant was demonstrated spectroscopically. A series of chemically modified substrate molecules with altered substituents on the individual pyrrole rings and the isocyclic ring were tested as COR substrates. The COR enzyme was still able to reduce the B ring of substrates carrying modified substituents on ring systems A, C, and E. However, substrates with a modification of the distantly located propionate side chain were not accepted. A tentative substrate binding mode was concluded in analogy to the related DPOR system.
Introduction
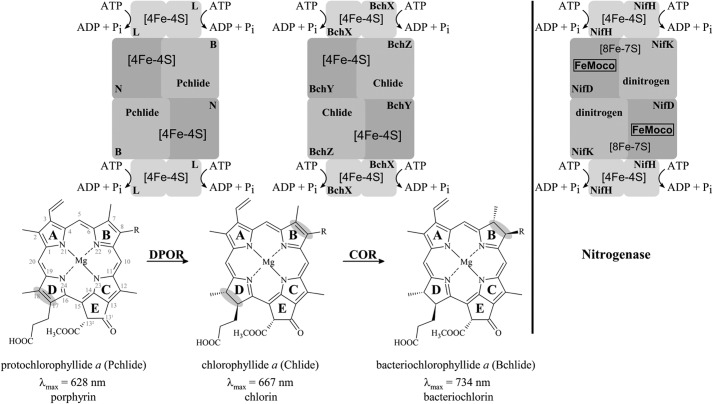
The energy supply of the global biosphere by photosynthesis vitally depends on the biosynthesis of (bacterio)chlorophylls, a process yielding several billion tons of these molecules seasonally (1). Another essential process of life on earth is the reduction of atmospheric dinitrogen to ammonia. The two processes share closely related enzymes. The sophisticated biochemistry of a nitrogenase-type reductase plays in many photosynthetic organisms a fundamental role for the formation of the chlorin and bacteriochlorin ring system of chlorophylls (Chls)2 and bacteriochlorophylls (Bchls) (Fig. 1) (2, 3). The biosynthesis of both Chls and Bchls requires the stereospecific reduction of the C17-C18 double bond of ring D of the porphyrin protochlorophyllide a (Pchlide), which can be catalyzed by the nitrogenase-like enzyme, dark-operative protochlorophyllide a oxidoreductase (DPOR) (Fig. 1, left) (4, 5). The resulting chlorin molecule is termed chlorophyllide a (Chlide). It requires a second stereospecific reduction step of the C7-C8 double bond of ring B to obtain the characteristic bacteriochlorin ring structure of Bchls. This formation of bacteriochlorophyllide a (Bchlide) is catalyzed by a second nitrogenase-like enzyme, termed chlorophyllide a oxidoreductase (COR) (Fig. 1, center) (6). DPOR and COR mediate significant changes of the respective pigment absorption properties, which improves light capturing and energy transduction processes of photosynthesis (compare Fig. 1) (7).
FIGURE 1.
Reduction of the aromatic ring system of Pchlide and Chlide by DPOR and COR. ATP-dependent dark-operative protochlorophyllide a oxidoreductase (DPOR) catalyzes the stereospecific reduction of ring D (highlighted in gray) of protochlorophyllide a (Pchlide). The DPOR reaction cycle includes the transient formation of an octameric L2(NB)2L2 protein complex (top, left). Bacteriochlorophyll a biosynthesis requires a second stereospecific ring reducing step on ring B (highlighted in gray) which is catalyzed by chlorophyllide a oxidoreductase (COR). ATP-dependent COR catalysis also includes the transient formation of an (BchX)2(BchY/BchZ)2(BchX)2 complex (top, center) for the reduction of chlorophyllide a (Chlide). R is either a vinyl or an ethyl group. The homologous nitrogenase complex (NifH)2(NifD/NifK)2(NifH)2 is indicated on the right. Involved redox-active [Fe-S] clusters and the iron- and molybdenum-containing cofactor (FeMoco) are indicated. Orthologous subunits of DPOR, COR, and nitrogenase are indicated by gray shading.
The COR enzyme is composed of three protein subunits, BchY, BchZ, and BchX, whereas the related subunits of DPOR are designated N, B, and L (gene names chlN, chlB, and chlL in Chl synthesizing and bchN, bchB, and bchL in Bchl synthesizing organisms) (4, 7–9). All these subunits share amino acid sequence homology to the related subunits of nitrogenase termed NifD, NifK, and NifH, respectively (compare Fig. 1) (7). Although subunits BchX, L, and NifH share a sequence identity at the amino acid level of ∼33%, subunits BchY, N, NifD and BchZ, B, NifK show lower identity values of ∼15% (7, 10). Three-dimensional protein structures of DPOR and nitrogenase revealed an oligomeric architecture consisting of heterotetrameric core complexes (N/B)2 and (NifD/NifK)2, respectively, which require the dynamic interplay with the respective homodimeric subcomplexes L2 and (NifH)2 (compare Fig. 1) (11, 12).
To mediate the ATP-driven electron transfer and substrate reduction process, both subcomplexes of DPOR carry redox active [Fe-S] clusters (13, 14): a symmetric intersubunit [4Fe-4S] cluster located on L2 and an uncommon (N/B)2 [4Fe-4S] cluster, which is asymmetrically ligated by three cysteinyl residues of subunit N and an unusual aspartate ligand from subunit B (Cys-17, Cys-42, Cys-103, and Asp-36 Prochlorococcus marinus numbering).
The schematic comparison in Fig. 1 highlights the involved redox-active components of DPOR and the related metallo-centers found in nitrogenase: a closely related [4Fe-4S] cluster on (NifH)2 and the highly unusual [8Fe-7S] (P cluster) and the Mo-7Fe-9S-X-homocitrate cluster (FeMoco) located on (NifD/NifK)2 (MoFe-protein) (12, 15–17). Accordingly, the final [4Fe-4S] cluster-dependent Pchlide reduction and the specific substrate recognition in the active site of (N/B)2 of DPOR are unrelated to nitrogenase catalysis. In contrast, identical mechanisms have been elucidated for the initial steps of DPOR and nitrogenase catalysis (2, 11).
For DPOR (and nitrogenase), L2-dependent ((NifH)2-dependent) ATP hydrolysis triggers the transient association and dissociation of subcomplexes L2 and (N/B)2 (of subcomplexes (NifH)2 and (NifD/NifK)2). Thereby, the appropriate timing of the accompanying electron transfer processes is controlled (11, 16).
In analogy to DPOR and nitrogenase, a catalytic (BchY/BchZ)2 complex in combination with a homodimeric subcomplex (BchX)2 was proposed for the COR enzyme (10). Sequence alignments of BchX proteins with the closely related L subunits of DPOR revealed a strong conservation of both cysteinyl ligands responsible for [4Fe-4S] cluster formation and also for key residues involved in ATP binding and hydrolysis (7).
To some extent these theoretical findings are supported by a recent Rhodobacter sphaeroides study where a weak EPR signal was tentatively ascribed to a single [4Fe-4S]1+ cluster on (BchX)2 or alternatively to [2Fe-2S]1+ clusters located on each BchX protomer (18). The individual BchY protein and also the co-purified (BchY/BchZ)2 protein complex from R. sphaeroides showed a weak EPR signal indicative for a [3Fe-4S]1+ cluster (18). However, this cluster type is often found under non-physiological conditions, and a more detailed interpretation would also require the careful analysis of the oligomeric subunit structure of the respective proteins.
In vitro assays for COR, DPOR, and nitrogenase make use of the artificial electron donor dithionite in the presence of high concentrations of ATP (6, 19, 20). For DPOR and nitrogenase it was proposed that the consumption of ATP is required to overcome the kinetic barrier imminent to Pchlide or dinitrogen reduction (4, 21).
To date no experimental data concerning COR substrate recognition are available in the literature. Structural investigations for DPOR from P. marinus revealed that the Pchlide ring system is completely buried in a hydrophobic cavity. This binding mode requires substantial conformational rearrangements for the channeling of the substrate into this pocket (11, 14). A detailed mechanism for the regio- and stereospecific protonation of Pchlide was elucidated on the basis of biochemical and structural data. The C18 protonation is mediated via a water molecule that is adequately positioned by combined interaction with residue His-394 and the propionate moiety of the Pchlide substrate, whereas Asp-290 is directly responsible for the protonation at C17 (P. marinus numbering of subunit B).
In this study we establish a recombinant system for COR from Roseobacter denitrificans. Based on individually purified COR subcomplexes, the ligand architecture of the respective [Fe-S] clusters is resolved in a combined biochemical and spectroscopic approach. Furthermore, the transient COR subcomplex could be trapped. Subsequently, a series of chemically modified substrate molecules is employed to identify specific determinants for Chlide substrate recognition. The obtained functional substrates allow us to propose the localization of the COR reaction step within the bacteriochlorophyll biosynthetic pathway. Besides this, a tentative model for the reductive protonation of the COR substrate is proposed and compared with the DPOR mechanism.
EXPERIMENTAL PROCEDURES
Production and Purification of R. denitrificans COR
Genes bchY and bchZ from R. denitrificans (OCh 114, German Collection of Microorganisms and Cell Cultures (DSMZ)) were PCR-amplified using primers CTATAGGATCCATGATCAAGGGTCACCCGC and CTATACTCGAGCTAAATCATCTCCTGCGCTTTG and primers CTATACTCGAGAATTTCACACAGGAAACAGTATTCATGTTAGTCACAGATCATG and CTATTGCGGCCGCTCATGTCTGATCCTCCAAATC, respectively (the implemented Escherichia coli-specific ribosomal binding site upstream of bchY is marked in bold letters, and employed restriction sites are in italics). Both genes were ligated into the BamHI and NotI sites of pET32a to yield plasmid pET-bchYZ. Subsequently, a PreScissionTM protease binding site sequence (bold letters) was implemented upstream of bchY. For this purpose bchY and bchZ were amplified from pET-bchYZ using primers TATATAGAGCTCGCTGGAAGTTCTGTTCCAGGGGCCCCTGCAGATGATCAAGGGTC and TATATATAGCGGCCGCTAAATCATCTCC and primers CAGCAGCATATGTTAGTCACAGATC and CAGCAGCTCGAGTCATGTCTGATC and cloned into the NdeI/XhoI and SacI/NotI sites of the pACYCDuet-1 vector, respectively. Plasmid pET-bchYZ and the resulting plasmid pACYC-bchYZ (variations detailed in parentheses) were transformed into E. coli BL21(DE3) Codon Plus RIL (E. coli BL21(DE3)). Cells were grown aerobically at 17 °C (25 °C) in 500 ml of LB medium containing 1 mm Fe(III) citrate and 1 mm l-cysteine. Protein production was induced at an A578 of 0.5 by the addition of 50 μm isopropyl-β-d-thiogalactopyranoside. After 16 h (4 h) of cultivation, the respective cultures were supplemented with 0.15 g (0 g) sodium dithionite and incubated for 2 h (45 min) at 17 °C without agitation in an anaerobic chamber (Coy Laboratories, Grass Lake, MI) to allow [Fe-S] cluster formation. All subsequent steps were performed under anaerobic conditions (95% N2, 5% H2, <1 ppm O2) using N2-saturated buffers. Cells were harvested by centrifugation, and the bacterial cell pellet was resuspended in 15 ml of buffer 1 (100 mm HEPES-NaOH (pH 7.5), 10 mm MgCl2, 500 mm NaCl) and disrupted by a single passage through a French press at 16,000 p.s.i. into an anaerobic bottle. After centrifugation for 60 min at 110,000 × g at 4 °C, the supernatant was applied to 500 μl (1 ml) of a Ni2+-loaded chelating Sepharose FF (GE Healthcare) equilibrated with buffer 1. After 2 (3) washing steps with 20 ml (5 ml) of buffer 1 and a pre-elution step using 10 ml (3 ml) of buffer 1 containing 30 mm (20 mm) imidazole, the recombinant fusion protein BchY in complex with BchZ was eluted using 2 ml (1.5 ml) of buffer 1 containing 200 mm imidazole (200 mm imidazole and 10% (w/v) glycerol). Alternatively, the BchY/BchZ protein complex was liberated from the resin by thrombin (Sigma) cleavage (for construct pET-bchYZ). Protein fractions containing the (BchY/BchZ)2 complex were identified by SDS-PAGE.
The overproduction and purification of BchX using plasmid pET-bchX was essentially performed as described elsewhere (10) in the presence of plasmid pISC (22), which encodes for proteins of the isc-operon to ensure appropriate [Fe-S] clusters assembly in E. coli (protein production for 21 h in the presence of 300 μm isopropyl-β-d-thiogalactopyranoside and 1 mm l-cysteine followed by anaerobic incubation for 45 min without agitation).
Determination of Native Molecular Mass
Analytical gel permeation chromatography for (BchY/BchZ)2 was performed using a Superdex 200 HR 10/30 column (GE Healthcare), equilibrated with buffer 1 containing 25 mm NaF and 500 mm AlCl3. The native molecular mass of (BchX)2 was analyzed using a Superdex 200 HR 26/60 column (GE Healthcare) in the presence of buffer 2 (buffer 1 containing 150 mm NaCl). Columns were calibrated with protein standards (molecular weight marker kit MWGF 1000; Sigma) at a flow rate of 0.5 ml min−1 (BchY/BchZ)2 and 1 ml min−1 for (BchX)2, respectively (10).
Site-directed Mutagenesis of COR
Up to two nucleotides of plasmid pACYC-bchYZ were exchanged using the QuikChangeTM site-directed mutagenesis kit (Stratagene). Mutations in the bchY gene were introduced using the following oligonucleotides (exchanged nucleotides are in bold): C62A, CAAACCACAAAGCATGGCTCCGGCGTTCGGGTC and GACCCGAACGCCGGAGCCATGCTTTGTGGTTTG; C86A, GAGCGGGTCGGCGGCCTGCGTCTATGGTC and GACCATAGACGCAGGCCGCCGACCCGCT; C87A, GCGGGTCGGCGTGCGCCGTCTATGGTCTG and CAGACCATAGACGGCGCACGCCGACCCGC; C145A, GTGGTGACAAACCTTGCTGTGCCGACAGCC and GGCTGTCGGCACAGCAAGGTTTGTCACCAC. Oligonucleotides for the exchange of codons in bchZ were: D30C, CAGGTCGTGATCTGCGGCCCCGTGGGCTG and CAGCCCACGGGGCCGCAGATCACGACCTG; C35A, GGCCCCGTGGGCGCCGAAAACCTGCCCGTC and GACGGGCAGGTTTTCGGCGCCCACGGGGCC; C35S, GGCCCCGTGGGCAGCGAAAACCTGCCCGTC and GACGGGCAGGTTTTCGCTGCCCACGGGGCC; C35D, GGCCCCGTGGGCGACGAAAACCTGCCCGTC and GACGGGCAGGTTTTCGTCGCCCACGGGGCC; C35D/S94C, GTGGTCACCGGGTGCATTGCCGAGATGATCG and CGATCATCTCGGCAATGCACCCGGTGACCAC and the primers for C35D (see above).
Determination of Protein Concentration
The concentration of purified (BchY/BchZ)2 and (BchX)2 proteins was determined using the Bradford reagent (Sigma) according to the manufacturer's instructions with bovine serum albumin as a standard.
N-terminal Amino Acid Sequence Determination
Automated Edman degradation was used to confirm the identity of purified proteins and to quantify the amounts of the individual subunits in the protein complex (BchY/BchZ)2.
UV-visible Light Absorption Spectroscopy
UV-visible absorption spectra of purified (BchY/BchZ)2 and (BchX)2 complexes were recorded using a V-650 spectrometer (Jasco) under anaerobic conditions.
Iron Determination Method
The iron content of purified protein complexes was determined as described in Huberman and Pérez (23).
In Vitro Iron-Sulfur Cluster Reconstitution of (BchX)2
The in vitro reconstitution of [4Fe-4S] clusters was performed as previously described (24) using 4.5 mol of ammonium iron citrate and 4.5 mol of lithium sulfide/mol of protein complex in buffer 2 containing 200 mm imidazole and 10% (w/v) glycerol (12 h incubation at 4 °C). The reconstituted protein was desalted by NAP-25 size-exclusion chromatography (GE Healthcare) according to the manufacturer's instructions.
Preparation of EPR Samples
Purified proteins were concentrated up to 200 μm using an Amicon stirred ultracentrifugation cell (Millipore) equipped with a 30,000-Da cut-off membrane. Sodium dithionite was added to a final concentration of 12.5 mm. After 15 min of incubation, samples were transferred into quartz EPR tubes and frozen in liquid nitrogen.
EPR Spectroscopy
X-Band EPR spectra were recorded on a Bruker Elexsys E-500 CW X-band spectrometer. The sample tubes of 5-mm diameter were placed in a standard TE102 resonator. Low temperature measurements were obtained using an Oxford ESR 900 helium flow cryostat (3–300 K). Baseline corrections were performed by subtracting a background spectrum, obtained under the same experimental conditions from an empty tube.
Protochlorophyllide Preparation
Pchlide was isolated from the bchL-deficient Rhodobacter capsulatus strain ZY5 as described earlier (20, 25).
Chlorophyllide Preparation
Chlide was isolated from 5 liters of cell culture of the bchF- and bchZ-deficient R. capsulatus strain CB1200 (9) as described in Gough et al. (26). Purified Chlide was dissolved in 100 μl of DMSO containing 30 mm Tricine-NaOH (pH 8.0) and stored at −20 °C.
Substrate Analogs
Synthesis and/or isolation of the respective pigments was described elsewhere (27, 28).
Coupled DPOR/COR Activity Assay
Coupled DPOR/COR activity measurements were performed in 250-μl assays containing 100 mm HEPES-NaOH (pH 7.5), 10 mm MgCl2, 150 mm NaCl, 2 mm ATP, 2.7–3.5 μm Pchlide, 5 mm 1,4-dithio-d,l-threitol, and 0.7 mm sodium dithionite (as electron donor) and an ATP-regenerating system containing creatine phosphate (20 mm) and creatine phosphokinase (0.1 units/μl). The standard DPOR/COR activity assay was supplemented with 40 μl of a cell-free E. coli extract containing the overproduced N, B, and L subunits of Chlorobaculum tepidum DPOR (2) to provide the natural Chlide substrate of COR. Then, 40–1,100 pmol of the purified (BchY/BchZ)2 complex and 0.25–40 μl of a cell-free E. coli extract containing the overproduced (BchX)2 protein were added. All assays were incubated in the dark for 30 or 60 min at 34 °C. The reaction was stopped by adding 500 μl of acetone. After two identical centrifugation steps (10 min, 12,100 × g) Pchlide, Chlide, and Bchlide formation was determined by UV-visible spectroscopy using the following extinction coefficients: Pchlide ϵ626 = 30.4 mm−1cm−1 (20), Chlide ϵ665 = 74.9 mm−1cm−1 (29), Bchlide ϵ734 = 44.7 mm−1cm−1 (6). In all cases assays were completed by control experiments in which the (BchX)2 protein or the (BchY/BchZ)2 complex was omitted.
Chlide Reduction Assay
COR activity was measured in 125-μl assays under conditions described for the coupled DPOR/COR activity assay, omitting the C. tepidum DPOR substrate generating system. Instead, purified Chlide (or modified substrates) was added at a concentration of 10 μm. Reactions were stopped by adding 500 μl of acetone. After centrifugation (30 min, 12,000 × g) samples were extracted with 1 volume of hexane. The lower aqueous acetone phase was analyzed by UV-visible spectroscopy. Chlide and Bchlide were quantified as described above. Assays were completed by control experiments in which the (BchX)2 protein or the (BchY/BchZ)2 complex was omitted.
Formation of the Ternary COR Complex
1.3 nmol of the purified (BchX)2 protein was immobilized on 100 μl of S-agarose (Merck Millipore) on a 10-ml gravity-flow column (Bio-Rad). Then formation of MgADP·AlF4− was initiated by the addition of 10 mm MgADP and 2 mm AlCl3 to a solution of buffer 2 containing 50 mm NaF (trapping buffer). 1 ml of this solution was supplemented with 3.45 nmol of the (BchY/BchZ)2 complex in the presence of 63 μm Chlide. This mixture was incubated for 1 h with the immobilized (BchX)2 protein to allow ternary complex formation. The column was washed with 500 μl of trapping buffer. The affinity matrix was then suspended in 400 μl of trapping buffer and subsequently removed from the column. After a centrifugation step, all immobilized proteins were liberated from the S-agarose with 100 μl of SDS sample buffer. The denatured protein samples (10 min, 95 °C) were then subjected to SDS-PAGE analysis. COR ternary complex investigations were always completed by parallel control experiments in which the (BchX)2 or the (BchY/BchZ)2 protein was omitted.
Purification of Chlide-saturated (BchY/BchZ)2
73 nmol of His-tagged (BchY/BchZ)2 were immobilized on 1 ml of Ni2+ loaded chelating Sepharose FF. After washing with 2 ml of buffer 1 containing 20 mm imidazole (buffer 3) the column was incubated for 10 min with 1 ml of buffer 3 containing 365 μm Chlide. After two washing steps with 2 ml of buffer 1 containing 25 mm imidazole the Chlide-saturated COR complex was eluted from the resin using 1.5 ml of buffer 1 containing 200 mm imidazole and 10% (w/v) glycerol. The amount of protein-bound Chlide was determined by UV-visible spectroscopy.
Sequence Analysis of BchX
The structure based sequence alignment of the L protein from P. marinus and R. sphaeroides was performed as described elsewhere (11). Then, sequence-based alignments of 15 representative BchX and L proteins, respectively, were calculated using ClustalW (30, 31). Both types of sequence comparisons were manually assembled, and the degree of conservation was highlighted. Structurally conserved amino acid residues located at the dimer interface of L2, residues responsible for the L2/(N/B)2 protein-protein interaction and all key residues for the dynamic switch mechanism of L2 are indicated (see Fig. 4) (11). The secondary structure prediction for BchX was calculated with SOPMA (32).
FIGURE 4.
Sequence and/or structure based alignment of BchX and L subunits. The structure based alignment of the L subunits from P. marinus and R. sphaeroides was calculated as described under “Experimental Procedures” using PDB coordinates 2YNM and 3FWY. α-Helices (dark blue) and β-strands (light blue) are indicated. The prediction of the secondary structure of R. denitrificans BchX was performed using the SOPMA program (dark blue and light blue shading). Sequence-based alignments of representative BchX (Cons 1), L (Cons 3), and BchX/L (Cons 2) sequences were calculated. Conserved residues and those with similar properties are indicated by an asterisk (*), colon (:), or period (.). This sequence-based conservation pattern and the structure-based alignment of the two L proteins was manually assembled. Differences in the structural and primary sequence alignment are indicated (−). Key residues for the dynamic protein-protein interaction of L2 and (NB)2 are indicated in yellow. Residues of the L2 dimer interface are marked in black boxes (11). Ligands of the inter-subunit [4Fe-4S] cluster of L2 proteins are marked in red. Rd, R. denitrificans; Pm, P. marinus; Rs, R. sphaeroides.
RESULTS
Purification and Biochemical Characterization of R. denitrificans COR
The genes coding for subunits BchY and BchZ from R. denitrificans were cloned into a standard E. coli expression vector. The plasmid pET-bchYZ encodes for an N-terminal thioredoxin/His/S-tag fusion protein of BchY and for the untagged BchZ protein. Efficient translation of the bchY gene was fostered by the vector encoded ribosomal binding site, whereas the bchZ gene was actively fused to an E. coli-specific ribosomal binding site sequence to allow for high level production of BchZ. Alternatively, equimolar overproduction of both COR subunits was accomplished by using the pACYC-bchYZ construct containing individual promoters for the N-terminal His-tagged BchY protein and for subunit BchZ.
COR subcomplex (BchX)2 was produced with a separate production system as an N-terminal thioredoxin/His/S-tag fusion protein. The employed E. coli BL21(DE3) host was additionally carrying plasmid pISC (coding for genes of the isc-operon) to ensure efficient iron-sulfur cluster biogenesis under BchX overproducing conditions (22). Starting with cell harvesting and disruption, all COR proteins were maintained under anaerobic conditions throughout purification and all other subsequent experiments.
BchY fusion proteins (as thioredoxin/His/S-tagged or His-tagged protein) were purified using Ni2+-chelating Sepharose (GE Healthcare). Either the intact fusion protein was eluted in the presence of 200 mm imidazole or the native protein was liberated from the resin by on-column cleavage with thrombin. SDS-PAGE analyses confirmed the relative molecular mass of thioredoxin/His/S-BchY (Mr = 71,000), BchY (Mr = 53,000), or His-BchY (Mr = 56,000; compare Fig. 3A, lane 2). Both purification strategies always revealed stoichiometric amounts of a second protein (Mr = 53,000; compare Fig. 3A, lane 2) as judged by SDS-PAGE. The co-purified protein was identified as subunit BchZ (N-terminal sequence MLVTDH), and a molar ratio of 1:1.2 for His-BchY versus BchZ was determined from the Edman degradation experiments.
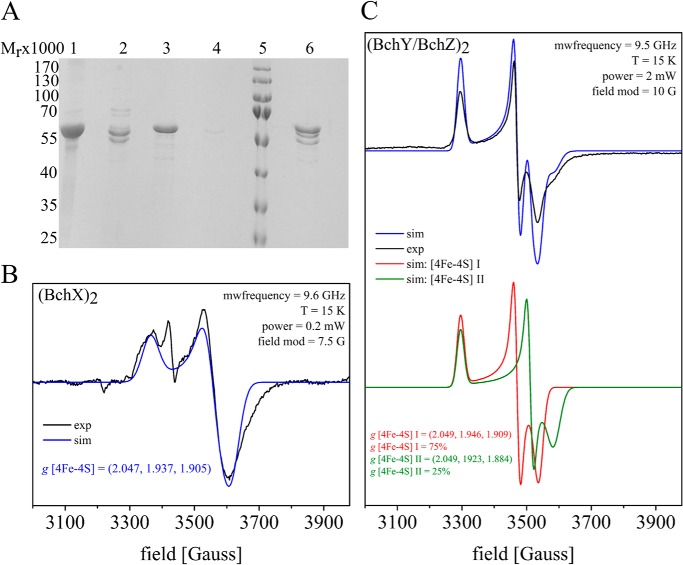
FIGURE 3.
EPR spectra of (BchY/BchZ)2 and (BchX)2 proteins and ternary complex formation of COR. A, SDS-PAGE analyses of purified recombinant COR subcomplexes and trapping of the ternary COR complex. Lane 1, affinity-purified thioredoxin/His/S-BchX after elution with imidazole; lane 2, affinity-purified His-BchY/BchZ after imidazole elution; lanes 3–6, ternary COR complex formation analysis in the presence of MgADP·AlF4−. The purified thioredoxin/His/S-BchX protein was immobilized on S-agarose and subsequently used as bait in the presence of MgADP·AlF4− and (BchY/BchZ)2. After an extensive washing step, all immobilized proteins were liberated due to SDS sample buffer treatment. Lanes 3 and 4, control experiments in the absence of (BchY/BchZ)2 and (BchX)2, respectively. Lane 5, molecular mass markers, relative molecular mass (×1000) are indicated. Lane 6, trapped COR complex containing subunits BchX, BchY, and BchZ. B and C, EPR spectra of COR subcomplexes after dithionite reduction (black) in combination with the corresponding simulations (blue, red, and green). The respective microwave frequency, temperature, microwave power, and modulation amplitude is indicated. All g values are calculated from the respective simulations. B, the spectra of reconstituted (BchX)2 indicates an intersubunit [4Fe-4S] cluster. mwfrequency, microwave frequency. C, the spectra of (BchY/BchZ)2 exhibited a superposition of two non-identical [4Fe-4S] clusters.
(BchX)2 was analogously purified via Ni2+-chelating Sepharose. SDS-PAGE analysis revealed the relative molecular mass of thioredoxin/His/S-BchX (Mr = 53,000; see Fig. 3A, lane 1) and BchX (Mr = 40,000).
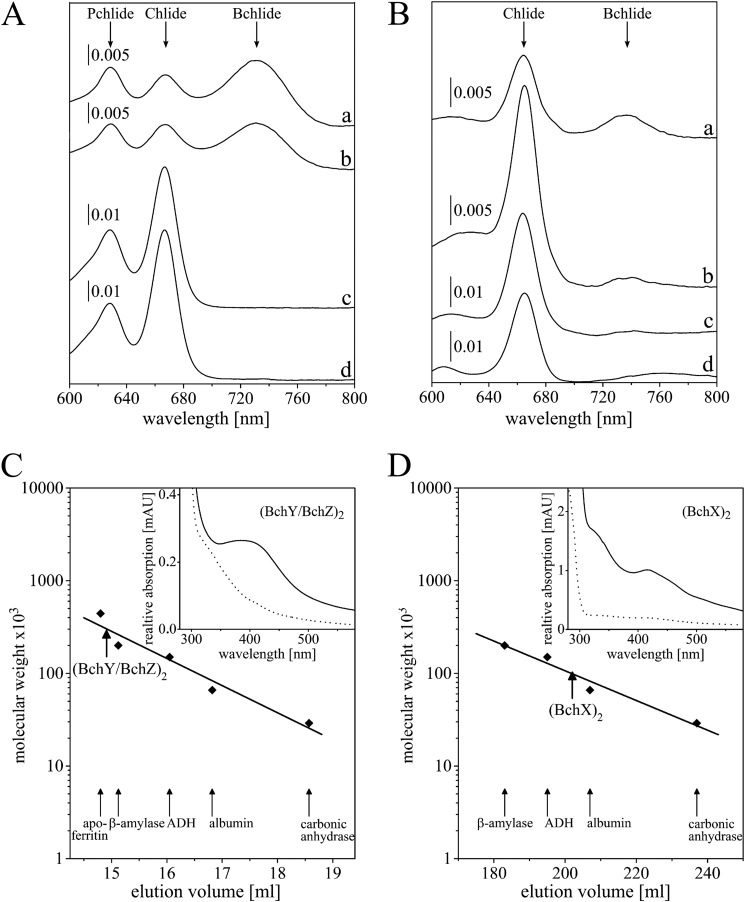
Size exclusion chromatography revealed a native molecular mass of 280,000 Da for the purified BchY/BchZ complex that is indicative for a heterotetrameric subunit architecture (calculated molecular weights of His-BchY and BchZ were 56,370 and 53,380, respectively). Accordingly, the catalytic COR complex of R. denitrificans was denoted as (BchY/BchZ)2.
Identical experiments revealed a relative molecular weight of 85,000 for the purified BchX subunit (calculated monomer mass, 39,969 Da). Therefore, a dimeric (BchX)2 subcomplex of COR was concluded in agreement with a recent study for C. tepidum BchX (10) and also as observed for the related L2 and (NifH)2 complexes of DPOR and nitrogenase (10, 33). Obviously, the quaternary structure of both COR subcomplexes parallels the respective stoichiometry of the related DPOR and nitrogenase systems (compare Fig. 1) (2, 20, 33, 34).
Reconstitution of COR Activity in a Coupled DPOR/COR Assay
Enzymatic activity for R. denitrificans COR was reconstituted by using a well established DPOR system (2) in the presence of the easily available Pchlide molecule (20, 25) to provide the COR substrate Chlide. For the reconstitution of the COR enzyme, 285 pmol of the purified (BchY/BchZ)2 complex were supplemented with the overproduced (BchX)2 protein (as a cell-free extract) in the presence of 3.5 μm Pchlide (20, 25), 5 mm 1,4-dithio-d,l-threitol, 0.7 mm sodium dithionite (as artificial electron donor), and 2 mm ATP in combination with an ATP-regenerating system. UV-visible spectroscopic analyses of the acetone-extracted pigments clearly revealed the efficient conversion of Pchlide (λmax 628 nm) (20, 25) into Chlide (λmax 667 nm) (26, 29), which then results in the formation of Bchlide (λmax 734 nm) (Fig. 2A, spectrum a) (6). The ATP-dependent R. denitrificans COR catalysis requires the presence of both subcomplexes as no Bchlide formation was observed in the absence of either (BchY/BchZ)2 or (BchX)2 (Fig. 2A, spectra c and d).
FIGURE 2.
Enzymatic activity of R. denitrificans COR and characterization of subcomplexes (BchY/BchZ)2 and (BchX)2. A, UV-visible absorption spectra of a coupled DPOR/COR activity assay for 1 h at 34 °C after acetone pigment extraction. Absorption maxima of the Pchilde substrate (628 nm), Chlide (667 nm), and Bchlide (734 nm) are indicated. Trace a, the coupled standard DPOR/COR activity assay containing 285 pmol (BchY/BchZ)2; trace b, coupled activity assay using 237 pmol of the mutant complex (BchYC86A/BchZ)2; traces c and d, negative controls in the absence of COR subunits (BchY/BchZ)2 and (BchX)2, respectively. B, UV-visible absorption spectra of COR activity assays in the presence of Chlide. Trace a, COR assay containing 100 pmol of (BchY/BchZ)2; trace b, analogous assay using the substrate analog Zn-BPheid c (5); traces c and d, control reactions devoid of COR subunit (BchY/BchZ)2 and (BchX)2, respectively (with Chlide as substrate). C, analytical gel permeation chromatography of purified (BchY/BchZ)2 (32 μm) using a Superdex 200 HR 10/300 gel permeation column under anaerobic conditions at a flow rate of 0.5 ml−1 min−1 (absorbance determination at 280 nm). Protein standards were apoferritin (Mr = 443,000), β-amylase (Mr = 200,000), alcohol dehydrogenase (ADH; Mr = 150,000), albumin (Mr = 66,000), and carbonic anhydrase (Mr = 29,000). The elution volume of (BchY/BchZ)2 is indicated (bold arrow). mAU, absorbance units. D, analytical gel permeation chromatography of (BchX)2. Purified (BchX)2 (0.12 mm) was analyzed using a Superdex 200 HR 26/60 column under anaerobic conditions at a flow rate of 1 ml−1 min−1 as described above. Insets of C and D, UV-visible absorption analysis of purified (BchY/BchZ)2 and (BchX)2 at concentrations of 9.4 μm and 0.22 mm, respectively. Spectra were recorded under anaerobic conditions (solid line) and after aerobic sample incubation (dotted line).
The experiment devoid of (BchX)2 also indicates that the electron transferring L2 component of DPOR cannot substitute for the COR specific reductase (BchX)2 in the employed DPOR/COR assay. Only recently a related experiment revealed enzymatic DPOR activity in the presence of a chimeric enzyme composed of subunit (BchX)2 from R. denitrificans and (N/B)2 from C. tepidum (10).
R. denitrificans (BchX)2 Coordinates an Intersubunit [4Fe-4S] Cluster
Concentrated (BchX)2 protein fractions showed a brownish color, and UV-visible spectroscopy revealed an absorption maximum at 428 nm indicative of a [4Fe-4S] cluster (Fig. 2D, inset, solid line). As already published (10), the iron content of 3.4 mol of iron/mol of (BchX)2 suggests the presence of a single intersubunit [4Fe-4S] center as also described for the related L2 and (NifH)2 proteins of DPOR and nitrogenase (4, 16). Exposure to atmospheric oxygen rapidly eliminated the absorption signal at 428 nm (Fig. 2D, inset, dotted line), analogously to the surface-exposed [4Fe-4S] centers of L2 and (NifH)2 (4, 35). The protein sample was subjected to EPR measurements to confirm the proposed [4Fe-4S] cluster of R. denitrificans (BchX)2 (Fig. 3B). A rhombic S = 1/2 signal was obtained with g values of g1 = 2.047, g2 = 1.937, and g3 = 1.905 located in a region characteristic for [4Fe-4S]1+ clusters (36, 37). The observed data are in agreement with related EPR experiments for the L2 protein of DPOR (4, 38) for (NifH)2 of Azotobacter vinelandii and also for (BchX)2 from R. sphaeroides (g1 = 2.02, g2 = 1.93 and g3 = 1.92) (18, 39). Similar to what was reported for (NifH)2 of A. vinelandii (39), no evidence for spin states larger than S = 1/2 (e.g. S = 3/2; S = 5/2) was obtained for the reconstituted (BchX)2 sample. It should, however, be noted that the spin count of the S = 1/2 signal is around 10%. Therefore, the involvement of higher spin states (with EPR signals broadened beyond detection) cannot be completely excluded. The additional signal at g = 2.002 probably is indicative of the presence of minor amounts of a partially reconstituted [3Fe-4S] cluster.
Our biophysical COR experiments suggest a [4Fe-4S] cluster-dependent redox catalysis of (BchX)2 that is analogous to the related L2 and (NifH)2 proteins (11, 16). These DPOR and nitrogenase subcomplexes make use of a nucleotide-dependent switch mechanism to mediate ternary protein complex formation and the subsequent electron transfer step. Accordingly, potential protein-protein interaction of COR components (BchX)2 and (BchY/BchZ)2 was investigated.
Substrate Binding of (BchY/BchZ)2
His-tagged (BchY/BchZ)2 was immobilized on a Ni2+-chelating Sepharose column, and a 5-fold molar excess of Chlide was added. After an extensive washing step, the substrate-saturated (BchY/BchZ)2 protein was eluted and subsequently analyzed by UV-visible absorption spectroscopy. The Chlide binding experiments revealed the presence of 1.4 mol of Chlide/mol (BchY/BchZ)2 complex.
Trapping the Ternary COR Complex
COR catalysis requires the interaction of (BchX)2 and (BchY/BchZ)2 to facilitate substrate reduction. In this study an ATP analog, (MgADP·AlF4−) was used for trapping the ternary complex of COR. MgADP in combination with the inorganic compound AlF4− mimics the ATP molecule in the transition state of nucleotide hydrolysis as previously shown for nitrogenase and DPOR (11, 12). The S-tagged (BchX)2 protein was specifically immobilized on an S-agarose column and incubated with a 2.5-fold molar excess of (BchY/BchZ)2 in the presence of MgADP·AlF4− and 63 μm Chlide. After a subsequent washing step, all column-bound proteins were eluted under denaturing conditions and subsequently analyzed by SDS-PAGE. Fig. 3A, lane 6, reveals the “bait” protein (BchX)2 with a relative molecular weight of 53,000 in the presence of COR subunits BchY (56,000) and BchZ (53,000) (compare Fig. 3A, lane 6 with lanes 2 and 3). All proteins were identified by N-terminal sequencing, and a molar ratio of 1.2:0.5:0.3 for BchX versus BchY versus BchZ was determined from the Edman degradation. The column material did not reveal any unspecific binding of (BchY/BchZ)2 (Fig. 3A, lane 4).
These results are indicative for a closely related nucleotide-dependent switch mechanism for COR, DPOR, and nitrogenase, which links ATP hydrolysis to conformational changes. From structural investigations, the overall rearrangements of L2 and (NifH)2 have been described as a rotation of the nucleotide-bound monomers toward the dimer interface (11, 16). The sequence analysis depicted in Fig. 4 clearly reveals a striking degree of sequence conservation for BchX residues that are tentatively located at the dimer interface of (BchX)2 (related residues of the L2 structures are highlighted in the black boxes). Besides this, all key residues for ATP hydrolysis (Lys-37 and Asp-155), the “P-loop” (Gly-36–Thr-44), and the residues responsible for the nucleotide-dependent signal transduction (“switch I region” Gly-64–Thr-72, “docking loop” Ile-84–Glu-96, and also the “switch II region” Asp-151–Phe-161 including cluster ligand Cys-158) are also conserved in BchX proteins (P. marinus numbering). The overall sequence analysis shown in Fig. 4 clearly supports the proposed dynamic switch mechanism of (BchX)2 as an initial step of COR catalysis.
In contrast to this, the theoretical analysis of the subsequent substrate reduction process reveals an unclear picture with respect to the involved redox active [Fe-S] cluster of (BchY/BchZ)2. Sequence analysis for subunits N versus BchY and B versus BchZ only revealed the presence of three conserved iron-sulfur cluster-coordinating cysteine residues in the sequence of BchY (10), whereas no equivalent was evident for the unusual aspartate ligand from DPOR subunit B.
(BchY/BchZ)2 Coordinates Two Non-Identical [4Fe-4S] Clusters
Concentrated fractions of purified (BchY/BchZ)2 showed a brownish color, and the UV-visible spectrum revealed an absorption maximum at 428 nm characteristic for [4Fe-4S] clusters (Fig. 2C, inset, solid line). This absorption peak was bleached upon long term oxygen exposure (Fig. 2C, inset, dashed line), and 6 mol of iron/mol of (BchY/BchZ)2 were determined. By using a standard extinction coefficient of 15 mm−1 cm−1 at 410 nm (40, 41), a content of 1.4 mol of [4Fe-4S] cluster/mol of (BchY/BchZ)2 was detected. Both methods are indicative for the presence of two [4Fe-4S] clusters on (BchY/BchZ)2. Standard methods (42) did not reveal any flavin cofactor.
The (BchY/BchZ)2 protein was further characterized by EPR experiments. After reduction with 12.5 mm sodium dithionite, a complex S = 1/2 spin signal indicative for the presence of a [4Fe-4S]+1 cluster type was observed. Simulation of the obtained EPR spectrum was possible by superposition of two signals, [4Fe-4S] I and [4Fe-4S] II, with different g values in an intensity ratio of 75%:25% (Fig. 3 C). The g values estimated for the first cluster from the simulation were g1 = 2.049, g2 = 1.946, and g3 = 1.909. The second cluster [4Fe-4S] II, exhibited g values of g1 = 2.049, g2 = 1.923, and g3 = 1.884. No evidence for S = 3/2 or S = 5/2 high spin states were observed from the analyzed (BchY/BchZ)2 sample. The measured data are in a range where reduced [4Fe-4S] clusters are expected (36, 37). The differing g values of [4Fe-4S] I and [4Fe-4S] II clusters might indicate subtle differences in the specific cluster coordination. A similar, non-identical [4Fe-4S] cluster coordination was also observed for the DPOR (N/B)2 complex from P. marinus (4).
Mutagenesis of Potential Ligands of the [4Fe-4S] Cluster of (BchY/BchZ)2
The observed [4Fe-4S] characteristics in combination with theoretical sequence analysis (10) are indicative for a differing ligation pattern in (BchY/BchZ)2 when compared with (N/B)2 of DPOR. Careful inspection of the sequence alignment indicates three conserved cysteine residues Cys-62, Cys-87, and Cys-145 of BchY, which might mirror cluster ligands Cys-17, Cys-41, and Cys-103 of DPOR subunit N (P. marinus numbering) (10, 11). However, neither the unusual fourth aspartate ligand nor the stabilizing Cys-95 of DPOR is present in the sequence of subunit BchZ (10, 13). The conserved residues Cys-86 of BchY and residues Asp-30 and Cys-35 of BchZ were, therefore, considered as alternative cluster ligands in the subsequent mutagenesis study (10).
Cys-62, Cys-87, Cys-145 of BchY and Cys-35 of BchZ Participate in Iron-Sulfur Cluster Coordination
All conserved cysteines of BchY were individually replaced by alanine residues in a site-directed mutagenesis approach. The purified variant protein complexes C62A, C87A, and C145A did not reveal any detectable activity (Table 1), and the respective iron contents were substantially reduced as indicated by values of 63, 21, and 26% (as related to the wild type; compare Table 1). By contrast, for the mutant complex C86A, a moderately reduced specific activity of 50% and an iron content of 100% in comparison to the wild-type protein were observed (Fig. 2A, spectrum b, and Table 1). The related EPR analysis of this mutant clearly indicated characteristic [4Fe-4S]+1 EPR S = 1/2 signals (simulated values [4Fe-4S] I: g1 = 2.048, g2 = 1.937, g3 = 1.919; [4Fe-4S] II: g1 = 2.048, g2 = 1.937, and g3 = 1.882; data not shown) analogous to the wild type. From these mutational data the critical involvement of only Cys-62, Cys-87, and Cys-145 of BchY in COR cluster formation and redox catalysis was concluded.
TABLE 1.
Characterization of mutant protein complexes
Table of enzymatic activities and iron content of different mutant COR enzymes. The [4Fe-4S] cluster of subcomplex (BchY/BchZ)2 is coordinated by four cysteine ligands. The enzymatic activity and the iron content of the wild-type R. denitrificans COR was set as 100%. Activities below the detection limit of the employed assay are indicated (n.d.). Conservation of the related amino acid residues in COR, DPOR, and nitrogenase is indicated (+).
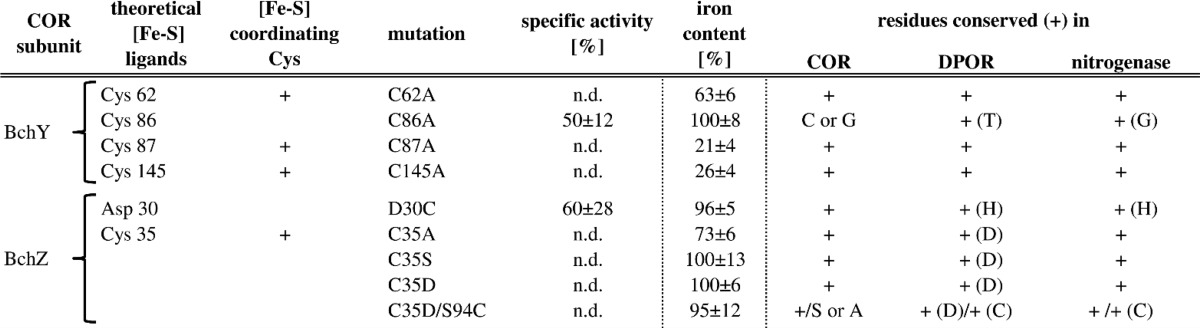
According to this, the highly conserved Asp-30 and Cys-35 residues located on subunit BchZ were considered as an additional [4Fe-4S] cluster ligand. The analysis of the D30C mutant complex indicated a moderately reduced activity of 60%. However, no decrease of the iron content was observed in comparison to the wild type (Table 1), and characteristic S = 1/2 [4Fe-4S]+1 cluster signals like the wild type were obtained (simulated values [4Fe-4S] I: g1 = 2.047, g2 = 1.940, g3 = 1.919; [4Fe-4S] II: g1 = 2.047, g2 = 1.940, and g3 = 1.895; Fig. 5A).
FIGURE 5.
EPR spectra of different (BchY/BchZ)2 mutants. A and B, EPR spectra of mutated COR (BchY/BchZ)2 subcomplexes after dithionite reduction (black) in combination with the corresponding simulations (blue and green). The respective microwave frequency (mwfrequency), temperature, microwave power, and modulation amplitude are indicated. All g values are calculated from the respective simulations. A, the spectra of (BchY/BchZD30C)2 exhibited a superposition of two non-identical [4Fe-4S] clusters. The spectra of (BchY/BchZC35S)2 indicated a [4Fe-4S]1+ cluster. B, the spectra of (BchY/BchZC35D)2 and (BchY/BchZC35D/S94C)2 indicate an intersubunit [4Fe-4S] cluster.
By contrast, (BchY/BchZ)2 complexes containing a C35A or C35S mutation were enzymatically inactive. The C35A mutation indicated a clear decrease of the iron content (73%), whereas the C35S mutant showed an identical iron content as the wild type. It was proposed that the isosteric C35S mutation still allows for a serine ligation of the overall [4Fe-4S] cluster of COR. Subsequent EPR experiments revealed characteristic S = 1/2 [4Fe-4S]+1 cluster signals as observed for the wild type protein (simulated values [4Fe-4S] g1 = 2.046, g2 = 1.93 and g3 = 1.88; Fig. 5A) Such artificial cluster formation due to a cysteine/serine ligand exchange is well known (2, 14, 43).
According to these results, Cys-35 was concluded as the fourth ligand of (BchY/BchZ)2. Obviously, COR catalysis is based on a four cysteine-ligated [4Fe-4S] of (BchY/BchZ)2, whereas DPOR (N/B)2 makes use of an unusual three cysteine/one aspartate ligation pattern.
Conversion of COR Ligands into DPOR-like Cluster Ligands
To investigate the closely related redox chemistry of COR and DPOR, Cys-35 of BchY was substituted by an aspartate ligand in analogy to the catalytic (N/B)2 complex of DPOR. This mutant complex revealed an identical iron content as observed for the wild type protein; however, no enzymatic activity was determined in the reconstitution assay. The EPR characterization of this C35D mutant protein complex revealed a rather weak S = 1/2 [4Fe-4S]+1 cluster signal (spin count ∼10%) with g parameters similar to those observed for the wild type protein (simulated values [4Fe-4S] g1 = 2.046, g2 = 1.93, g3 = 1.88; Fig. 5B). These spectroscopic data might be indicative for a three-cysteine/one-aspartate ligation pattern for the [4Fe-4S] cluster of (BchY/BchZ)2.
Biochemical and structural investigations of DPOR revealed an important function of Cys-95 for the stabilization of the [4Fe-4S] cluster of (N/B)2 (2, 11, 14). On the basis of sequence comparisons, Ser-94 of BchZ was identified as a potential substitute for Cys-95 of subunit B (P. marinus numbering). Ser-94 was subsequently mutagenized into a cysteine residue, and the resultant double mutant C35D/S94C was analyzed enzymatically and EPR spectroscopically. The EPR signal for this double mutant clearly shows the presence of a [4Fe-4S]+1 cluster as indicated by S = 1/2 g values of g1 = 2.05, g2 = 1.934, and g3 = 1.899 (Fig. 5B). These results might indicate a three-cysteine/one-aspartate ligation pattern of (BchY/BchZ)2 as in the related DPOR structures. However, no enzymatic activity for this cluster mutant was observed in COR activity assays. These results might indicate differing redox properties of the respective [4Fe-4S] clusters of DPOR and COR; minor differences in the ligand geometry might account for the inactive C35D/S94C mutant complex.
Table 1 summarizes the mutagenesis data and relates the positions of critical amino acids to the respective subunits of DPOR and nitrogenase. Interestingly, the Cys-35 ligand of BchZ has a direct counterpart in the respective subunit of nitrogenase (13). Cys-95 of NifK from A. vinelandii functions as a direct ligand of the unusual [8Fe-7S] cluster (P-cluster) of nitrogenase, and the related cysteine to serine amino acid exchange also resulted in a complete loss of nitrogenase activity. In analogy to the herein presented EPR data, this nitrogenase cluster variant revealed an “intact” [8Fe-7S] cluster (43).
Substrate Recognition of (BchY/BchZ)2
For the analysis of R. denitrificans COR substrate recognition, an “uncoupled” Chlide reduction assay was established. COR activity was reconstituted in vitro using recombinant protein components (BchY/BchZ)2 and (BchX)2 in the presence of an ATP-regenerating system, a 10 μm concentration of substrate Chlide and a 0.7 mm concentration of the artificial electron donor, sodium dithionite (compare UV-visible measurement Fig. 2B, spectrum a). The specific activity of this assay (42 pmol mg−1 min−1) was set as 100%, and all further kinetic experiments were related to this standard Chlide assay. Measurements were always completed by control experiments in the absence of either (BchY/BchZ)2 or (BchX)2 (compare Fig. 2B, spectra c and d).
Overall, seven magnesium- or zinc-containing chlorin compounds with altered substituents on ring systems A, E, and/or D were analyzed as potential substrates of COR. These molecules 2-8 in combination with the respective spectroscopic characteristics are depicted in Fig. 6. The use of zinc derivatives instead of magnesium-coordinating compounds has been well established for the reconstitution of photosynthetic reaction centers (44) as well as for the analysis of (bacterio)chlorophyll biosynthetic enzymes (4, 28, 45, 46).
FIGURE 6.
Analysis of artificial substrate utilization of COR. A, the natural Chlide substrate and zinc or magnesium containing analogs with modifications on pyrrole rings A and E. B, zinc derivatives with modifications on pyrrole rings A, E, and D. The enzymatic activity of R. denitrificans COR in the presence of Chlide was set as 100%, and all other values were related to this. Characteristics of the Qy band absorption of the respective pigments are indicated. *, absorption maxima determined in the present investigation. #, absorption maxima of Bchlide according to Nomata et al. (6).
Substrate Analogs with Altered Substituents on Rings A and E
Activity assays in the presence of Zn-3-Acetyl-Pheid a (2) revealed a relative activity of 30% when compared with the COR assay using the natural Chlide substrate. Obviously, the presence of a zinc central atom together with a more bulky and more polar acetyl group on C3 is tolerated in the active site of the COR enzyme. By contrast, the related DPOR enzyme does not accept an acetyl group on C3 (4). This result might indicate that different substrates comprising either a 3-vinyl or alternatively a 3-acetyl group might be accepted as a COR substrate in vivo.
Assays using a magnesium-containing compound Pyro-Chlide a (3) and also using the related zinc-containing molecule Zn-Pyro-Pheid a (4) indicated significant COR activities of 30 and 50%, respectively. Both molecules are devoid of the methoxycarbonyl substituent at the C132 position. Accordingly, the C132 substituent of Chlide is not a determinant for COR substrate recognition.
Zn-BPheid c (5) is also devoid of the C132-methoxycarbonyl group on ring E and possesses an additional methyl group at the C20 position. Besides this, compound (5) is also carrying a hydroxyl group at C31, as does 31-hydroxy Chlide, which has been described as an intermediate of the Bchl biosynthetic pathway (47). The obtained activity of 30% (Fig. 2B, spectrum b) for this substrate again indicates that the modification of the C132 and the C31 position together with the additional C20 methyl group is tolerated in the active site of COR. Due to the observed activities for Zn-3-Acetyl-Pheid a (2) and Zn-BPheid c (5), the hydroxylation of the C31-vinyl position and the subsequent conversion into a 3-acetyl group must also be considered as enzymatic steps in the pathway that potentially precedes the COR-catalyzed reduction on ring B.
Substrate Analogs with Altered Substituents on Ring A and Ring E
Zn-31-132-di-OH-Pheid a (6) is again carrying a C31-hydroxyl group on ring A. Furthermore, this zinc compound contains an additional C132 hydroxyl group on ring E. This molecule revealed an identical activity as observed for the natural Chlide substrate. Again, this result supports the idea of a broadened substrate specificity of R. denitrificans COR that would also include 31-hydroxy Chlide as a “natural” substrate (47). Moreover, this result reconfirms that substrate recognition of COR does not require the E ring substituents of Chlide on C13. Obviously, the known aggregation tendency of Zn-31-132-di-OH-Pheid a (6) in aqueous solution (48) does not interfere with its recognition.
Substrate Analogs with Altered Substituents on Ring A, D, and E
Activity assays using Zn-3-formyl-Me-Pheid a (7) or Zn-3-formyl-Pyro-Me-Pheid a (8) did not result in any detectable COR activity. Because the presence or absence of the 132 substituent does not interfere with the reaction (see above), a lack of activity is likely due to 3-formyl substituent or to methylation of the propionic acid side chain at ring D. The C3-formyl group is of similar size as the C3-vinyl group of the natural Chlide substrate so that a steric effect due to the substitution on ring A was excluded. The methyl ester modification on ring D might then be responsible for the discrimination of compounds 7 and 8 as a COR substrate. In this context it is noteworthy that the propionate group of the related DPOR substrate Chlide acts as a key determinant of substrate recognition and catalysis. The C17-propionate group of the Chlide substrate of COR is, however, distant from the catalytic target on ring B. The observed discrimination of compounds 7 and 8 might indicate that the overall chlorin ring system of Chlide is relevant for the substrate recognition of COR. According to this, the binding of the Chlide substrate in a buried binding pocket analogously as described for the related DPOR system (4) would account for the results of the Chlide binding and substrate recognition experiments of the present investigation.
DISCUSSION
Due to the conjugated nature of the Chlide molecule, an unusually negative reduction potential for the trans-hydrogenation is expected. Therefore, the COR catalyzed two-electron reduction of the C7-C8 double bond is linked to the hydrolysis of ATP molecules. The nucleotide-dependent switch protein (BchX)2 triggers the ATP-dependent transfer of electrons via a [4Fe-4S] cluster onto a second [4Fe-4S] cluster located on (BchY/BchZ)2. The interaction of these subcomplexes (BchX)2 and (BchY/BchZ)2 was efficiently trapped after incubation with the non-hydrolyzable ATP analog, ADP·AlF4−. These experiments clearly indicate that the initial electron transfer steps of COR catalysis resemble DPOR and nitrogenase catalysis. By contrast, the subsequent substrate reduction process differs for the individual systems. COR and DPOR catalysis is not based on equivalents of the unusual P-cluster or molybdenum-containing cofactor of nitrogenase. Instead, COR and DPOR make use of a more conventional [4Fe-4S] cluster for the direct transfer of electrons onto the substrate. However, with respect to the differing reduction state of the COR and DPOR substrate (chlorin versus porphyrin), a four cysteine versus a three-cysteine/one-aspartate ligation pattern might be required to appropriately modulate the redox potential of (BchY/BchZ)2 and (NB)2, respectively.
The presented EPR data for (BchY/BchZ)2 indicated an almost quantitative spin count for S = 1/2 (with no evidence for higher spin e.g. S = 3/2, S = 5/2) analogously as also described for the (NB)2 protein from P. marinus (4). By contrast, an S = 3/2 ground spin state was reported for the (NB)2 protein from R. capsulatus (49). Minor structural rearrangements of the cluster ligands might influence the interaction between the iron atoms, thereby altering the electronic ground state of the respective [4Fe-4S] cluster. With respect to this, the presence of a tightly bound substrate and the presence of an artificial cluster ligand (mutation D36C in the related R. capsulatus study) (49) or a solvent-dependent spin crossover between the low spin and high state also must be considered (39, 50–52). The EPR experiments of the present study did not reveal evidence for high spin ground states of mutant proteins C35S, C35D, and C35D/S94C of (BchY/BchZ)2. These data might mirror the outcome of a mutant study for a four-cysteine-ligated cluster of a ferredoxin from A. vinelandii, which revealed the same S = 1/2 spin signal before and after cysteine versus aspartate replacement (53).
The substrate orientation in the three-dimensional structure of the catalytic DPOR complex revealed that the electrons for the D ring reduction enter the conjugated ring system of Pchlide via the remotely located ring systems A and/or B. Subsequently, the reductive trans protonation mechanism of DPOR is based on a hydrophobic substrate binding cavity devoid of water molecules so that the substrate protonation then relies on the stereospecific addition of two protons via precisely positioned amino acid molecules. Alignments of the respective DPOR and COR sequences did not reveal any of the highly conserved proton donor residues in the sequence of COR (10). Furthermore, the propionate group located on C17 of ring D has been identified as an important determinant for DPOR and COR substrate recognition. Based on these initial findings, an identical binding mode (with respect to the ring orientation) for the Chlide substrate in the active site of COR might be the evolutionary clue for enzymatic systems with different active site specificities. Such identical ring orientation in the active site of COR would solely require two alternative proton donors for the specific reduction of ring B after formation of an a π-anion radical by a conserved electron insertion path via ring A and B (54). In the active site of ferredoxin-dependent bilin reductases, an identical tetrapyrrole ring orientation also facilitates the synthesis of different reaction products (55).
Only recently a new specificity for the COR enzyme from the Bchl b-producing organism, Blastochloris viridis was identified. This enzyme is not only capable of the reduction of the C7-C8 double bond of Chlide. It also catalyzes the reduction of the diene system (comprised of the C7/C8 double bond and the C8 vinyl group) to a C8 ethylidene group (56). This new catalytic activity of COR is based on the 1,4-hydrogenation of the C7 and C82 positions of Chlide (instead of the well known 1,2-hydrogenation of C7 and C8). This new activity of an orthologous COR enzyme might solely require the spatial rearrangement of a single proton donor (C82 instead of C8) for the synthesis of a Bchl compound with significantly altered spectroscopic properties.
These findings might indicate the plasticity of the herein proposed substrate binding and protonation mechanism of COR. The catalytic differences of COR, DPOR, and nitrogenase might have implications for related multiprotein complexes that are involved in the reduction of chemically stable double and/or triple bonds. Future experiments should be focused on the structural investigation of COR.
Acknowledgments
We gratefully thank Simone Virus and Beate Jaschok-Kentner for technical assistance.
This work was supported by Deutsche Forschungsgemeinschaft Grant JA 470/9-2.
- Chl
- chlorophyll
- Bchl
- bacteriochlorophyll
- Pchlide
- protochlorophyllide
- DPOR
- protochlorophyllide oxidoreductase
- Chlide
- chlorophyllide
- Bchlide
- bacteriochlorophyllide
- COR
- chlorophyllide oxidoreductase
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Hendry G. A. F., Houghton J. D., Brown S. B. (1987) The degradation of chlorophyll: a biological enigma. New Phytol. 107, 255–302 [DOI] [PubMed] [Google Scholar]
- 2. Bröcker M. J., Virus S., Ganskow S., Heathcote P., Heinz D. W., Schubert W. D., Jahn D., Moser J. (2008) ATP-driven reduction by dark-operative protochlorophyllide oxidoreductase from Chlorobium tepidum mechanistically resembles nitrogenase catalysis. J. Biol. Chem. 283, 10559–10567 [DOI] [PubMed] [Google Scholar]
- 3. Burke D. H., Alberti M., Hearst J. E. (1993) The Rhodobacter capsulatus chlorin reductase-encoding locus, bchA, consists of three genes, bchX, bchY, and bchZ. J. Bacteriol. 175, 2407–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bröcker M. J., Wätzlich D., Uliczka F., Virus S., Saggu M., Lendzian F., Scheer H., Rüdiger W., Moser J., Jahn D. (2008) Substrate recognition of nitrogenase-like dark operative protochlorophyllide oxidoreductase from Prochlorococcus marinus. J. Biol. Chem. 283, 29873–29881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fujita Y., Matsumoto H., Takahashi Y., Matsubara H. (1993) Identification of a nifDK-like gene (ORF467) involved in the biosynthesis of chlorophyll in the cyanobacterium Plectonema boryanum. Plant Cell Physiol. 34, 305–314 [PubMed] [Google Scholar]
- 6. Nomata J., Mizoguchi T., Tamiaki H., Fujita Y. (2006) A second nitrogenase-like enzyme for bacteriochlorophyll biosynthesis: reconstitution of chlorophyllide a reductase with purified X-protein (BchX) and YZ-protein (BchY-BchZ) from Rhodobacter capsulatus. J. Biol. Chem. 281, 15021–15028 [DOI] [PubMed] [Google Scholar]
- 7. Burke D. H., Hearst J. E., Sidow A. (1993) Early evolution of photosynthesis: clues from nitrogenase and chlorophyll iron proteins. Proc. Natl. Acad. Sci. U.S.A. 90, 7134–7138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Suzuki J. Y., Bollivar D. W., Bauer C. E. (1997) Genetic analysis of chlorophyll biosynthesis. Annu. Rev. Genet. 31, 61–89 [DOI] [PubMed] [Google Scholar]
- 9. Bollivar D. W., Suzuki J. Y., Beatty J. T., Dobrowolski J. M., Bauer C. E. (1994) Directed mutational analysis of bacteriochlorophyll a biosynthesis in Rhodobacter capsulatus. J. Mol. Biol. 237, 622–640 [DOI] [PubMed] [Google Scholar]
- 10. Wätzlich D., Bröcker M. J., Uliczka F., Ribbe M., Virus S., Jahn D., Moser J. (2009) Chimeric nitrogenase-like enzymes of (bacterio)chlorophyll biosynthesis. J. Biol. Chem. 284, 15530–15540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moser J., Lange C., Krausze J., Rebelein J., Schubert W. D., Ribbe M. W., Heinz D. W., Jahn D. (2013) Structure of ADP-aluminium fluoride-stabilized protochlorophyllide oxidoreductase complex. Proc. Natl. Acad. Sci. U.S.A. 110, 2094–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schindelin H., Kisker C., Schlessman J. L., Howard J. B., Rees D. C. (1997) Structure of ADP x AIF4(−)-stabilized nitrogenase complex and its implications for signal transduction. Nature 387, 370–376 [DOI] [PubMed] [Google Scholar]
- 13. Bröcker M. J., Schomburg S., Heinz D. W., Jahn D., Schubert W. D., Moser J. (2010) Crystal structure of the nitrogenase-like dark operative protochlorophyllide oxidoreductase catalytic complex (ChlN/ChlB)2. J. Biol. Chem. 285, 27336–27345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Muraki N., Nomata J., Ebata K., Mizoguchi T., Shiba T., Tamiaki H., Kurisu G., Fujita Y. (2010) X-ray crystal structure of the light-independent protochlorophyllide reductase. Nature 465, 110–114 [DOI] [PubMed] [Google Scholar]
- 15. Schmid B., Einsle O., Chiu H. J., Willing A., Yoshida M., Howard J. B., Rees D. C. (2002) Biochemical and structural characterization of the cross-linked complex of nitrogenase: comparison to the ADP-AlF4−-stabilized structure. Biochemistry 41, 15557–15565 [DOI] [PubMed] [Google Scholar]
- 16. Tezcan F. A., Kaiser J. T., Mustafi D., Walton M. Y., Howard J. B., Rees D. C. (2005) Nitrogenase complexes: multiple docking sites for a nucleotide switch protein. Science 309, 1377–1380 [DOI] [PubMed] [Google Scholar]
- 17. Hu Y., Ribbe M. W. (2011) Biosynthesis of nitrogenase FeMoco. Coord. Chem. Rev. 255, 1218–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim E. J., Kim J. S., Lee I. H., Rhee H. J., Lee J. K. (2008) Superoxide generation by chlorophyllide a reductase of Rhodobacter sphaeroides. J. Biol. Chem. 283, 3718–3730 [DOI] [PubMed] [Google Scholar]
- 19. Wherland S., Burgess B. K., Stiefel E. I., Newton W. E. (1981) Nitrogenase reactivity: effects of component ratio on electron flow and distribution during nitrogen fixation. Biochemistry 20, 5132–5140 [DOI] [PubMed] [Google Scholar]
- 20. Fujita Y., Bauer C. E. (2000) Reconstitution of light-independent protochlorophyllide reductase from purified bchl and BchN-BchB subunits. In vitro confirmation of nitrogenase-like features of a bacteriochlorophyll biosynthesis enzyme. J. Biol. Chem. 275, 23583–23588 [DOI] [PubMed] [Google Scholar]
- 21. Kurnikov I. V., Charnley A. K., Beratan D. N. (2001) From ATP to electron transfer: electrostatics and free-energy transduction in nitrogenase. J. Phys. Chem. 105, 5359–5367 [Google Scholar]
- 22. Takahashi Y., Nakamura M. (1999) Functional assignment of the ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster involved in the assembly of Fe-S clusters in Escherichia coli. J. Biochem. 126, 917–926 [DOI] [PubMed] [Google Scholar]
- 23. Huberman A., Pérez C. (2002) Nonheme iron determination. Anal. Biochem. 307, 375–378 [DOI] [PubMed] [Google Scholar]
- 24. Flühe L., Knappe T. A., Gattner M. J., Schäfer A., Burghaus O., Linne U., Marahiel M. A. (2012) The radical SAM enzyme AlbA catalyzes thioether bond formation in subtilosin A. Nat. Chem. Biol. 8, 350–357 [DOI] [PubMed] [Google Scholar]
- 25. Heyes D. J., Ruban A. V., Wilks H. M., Hunter C. N. (2002) Enzymology below 200 K: the kinetics and thermodynamics of the photochemistry catalyzed by protochlorophyllide oxidoreductase. Proc. Natl. Acad. Sci. U.S.A. 99, 11145–11150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gough S. P., Rzeznicka K., Peterson Wulff R., Francisco Jda C., Hansson A., Jensen P. E., Hansson M. (2007) A new method for isolating physiologically active Mg-protoporphyrin monomethyl ester, the substrate of the cyclase enzyme of the chlorophyll biosynthetic pathway. Plant Physiol. Biochem. 45, 932–936 [DOI] [PubMed] [Google Scholar]
- 27. Klement H., Helfrich M., Oster U., Schoch S., Rüdiger W. (1999) Pigment-free NADPH:protochlorophyllide oxidoreductase from Avena sativa L. Purification and substrate specificity. Eur. J. Biochem. 265, 862–874 [DOI] [PubMed] [Google Scholar]
- 28. Helfrich M., Schoch S., Lempert U., Cmiel E., Rüdiger W. (1994) Chlorophyll synthetase cannot synthesize chlorophyll a′. Eur. J. Biochem. 219, 267–275 [DOI] [PubMed] [Google Scholar]
- 29. McFeeters R. F., Chichester C. O., Whitaker J. R. (1971) Purification and properties of chlorophyllase from Ailanthus altissima (Tree-of-Heaven). Plant Physiol. 47, 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 31. Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Combet C., Blanchet C., Geourjon C., Deléage G. (2000) NPS@: network protein sequence analysis. Trends Biochem. Sci. 25, 147–150 [DOI] [PubMed] [Google Scholar]
- 33. Nomata J., Swem L. R., Bauer C. E., Fujita Y. (2005) Overexpression and characterization of dark-operative protochlorophyllide reductase from Rhodobacter capsulatus. Biochim. Biophys. Acta 1708, 229–237 [DOI] [PubMed] [Google Scholar]
- 34. Igarashi R. Y., Seefeldt L. C. (2003) Nitrogen fixation: the mechanism of the Mo-dependent nitrogenase. Crit. Rev. Biochem. Mol. Biol. 38, 351–384 [DOI] [PubMed] [Google Scholar]
- 35. Wiig J. A., Lee C. C., Fay A. W., Hu Y., Ribbe M. W. (2011) Purification of nitrogenase proteins. Methods Mol. Biol. 766, 93–103 [DOI] [PubMed] [Google Scholar]
- 36. Mouesca J.-M., Lamotte B. (1998) Iron-sulfur clusters and their electronic and magnetic properties. Coord. Chem. Rev. 178–180, 1573–1614 [Google Scholar]
- 37. Guigliarelli B., Bertrand P. (1999) Application of EPR spectroscopy to the structural and functional study of iron-sulfur proteins. Adv. Inorg. Chem. 47, 421–497 [Google Scholar]
- 38. Nomata J., Kitashima M., Inoue K., Fujita Y. (2006) Nitrogenase Fe protein-like Fe-S cluster is conserved in L-protein (BchL) of dark-operative protochlorophyllide reductase from Rhodobacter capsulatus. FEBS Lett. 580, 6151–6154 [DOI] [PubMed] [Google Scholar]
- 39. Lindahl P. A., Day E. P., Kent T. A., Orme-Johnson W. H., Münck E. (1985) Mossbauer, EPR, and magnetization studies of the Azotobacter vinelandii Fe protein. Evidence for a [4Fe-4S]1+ cluster with spin S = 3/2. J. Biol. Chem. 260, 11160–11173 [PubMed] [Google Scholar]
- 40. Hänzelmann P., Hernández H. L., Menzel C., García-Serres R., Huynh B. H., Johnson M. K., Mendel R. R., Schindelin H. (2004) Characterization of MOCS1A, an oxygen-sensitive iron-sulfur protein involved in human molybdenum cofactor biosynthesis. J. Biol. Chem. 279, 34721–34732 [DOI] [PubMed] [Google Scholar]
- 41. Shen G., Balasubramanian R., Wang T., Wu Y., Hoffart L. M., Krebs C., Bryant D. A., Golbeck J. H. (2007) SufR coordinates two [4Fe-4S]2+, 1+ clusters and functions as a transcriptional repressor of the sufBCDS operon and an autoregulator of sufR in cyanobacteria. J. Biol. Chem. 282, 31909–31919 [DOI] [PubMed] [Google Scholar]
- 42. Latimer M. T., Painter M. H., Ferry J. G. (1996) Characterization of an iron-sulfur flavoprotein from Methanosarcina thermophila. J. Biol. Chem. 271, 24023–24028 [DOI] [PubMed] [Google Scholar]
- 43. May H. D., Dean D. R., Newton W. E. (1991) Altered nitrogenase MoFe proteins from Azotobacter vinelandii. Analysis of MoFe proteins having amino acid substitutions for the conserved cysteine residues within the β-subunit. Biochem. J. 277, 457–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frank H. A., Chynwat V., Posteraro A., Hartwich G., Simonin I., Scheer H. (1996) Triplet state energy transfer between the primary donor and the carotenoid in Rhodobacter sphaeroides R-26.1 reaction centers exchanged with modified bacteriochlorophyll pigments and reconstituted with spheroidene. Photochem. Photobiol. 64, 823–831 [DOI] [PubMed] [Google Scholar]
- 45. Griffiths W. T. (1980) Substrate-specificity studies on protochlorophyllide reductase in barley (Hordeum vulgare) etioplast membranes. Biochem. J. 186, 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rüdiger W., Böhm S., Helfrich M., Schulz S., Schoch S. (2005) Enzymes of the last steps of chlorophyll biosynthesis: modification of the substrate structure helps to understand the topology of the active centers. Biochemistry 44, 10864–10872 [DOI] [PubMed] [Google Scholar]
- 47. Biel A. J., Marrs B. L. (1983) Transcriptional regulation of several genes for bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulata in response to oxygen. J. Bacteriol. 156, 686–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brody S. S. (1968) Low-temperature fluorescence excitation spectra for long-wavelength emission as a function of greening in Euglena gracilis and chlorophyll A concentration in vitro: a mathematical model to describe both systems. Biophys. J. 8, 210–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kondo T., Nomata J., Fujita Y., Itoh S. (2011) EPR study of 1Asp-3Cys ligated 4Fe-4S iron-sulfur cluster in NB-protein (BchN-BchB) of a dark-operative protochlorophyllide reductase complex. FEBS Lett. 585, 214–218 [DOI] [PubMed] [Google Scholar]
- 50. Conover R. C., Kowal A. T., Fu W. G., Park J. B., Aono S., Adams M. W., Johnson M. K. (1990) Spectroscopic characterization of the novel iron-sulfur cluster in Pyrococcus furiosus ferredoxin. J. Biol. Chem. 265, 8533–8541 [PubMed] [Google Scholar]
- 51. Onate Y. A., Finnegan M. G., Hales B. J., Johnson M. K. (1993) Variable temperature magnetic circular dichroism studies of reduced nitrogenase iron proteins and [4Fe-4S]+ synthetic analog clusters. Biochim. Biophys. Acta 1164, 113–123 [DOI] [PubMed] [Google Scholar]
- 52. Hagen W. R., Eady R. R., Dunham W. R., Haaker H. (1985) A novel S = 3/2 EPR signal associated with native Fe-proteins of nitrogenase. FEBS Lett. 189, 250–254 [DOI] [PubMed] [Google Scholar]
- 53. Jung Y. S., Bonagura C. A., Tilley G. J., Gao-Sheridan H. S., Armstrong F. A., Stout C. D., Burgess B. K. (2000) Structure of C42D Azotobacter vinelandii FdI. A Cys-X-X-Asp-X-X-Cys motif ligates an air-stable [4Fe-4S]2+/+ cluster. J. Biol. Chem. 275, 36974–36983 [DOI] [PubMed] [Google Scholar]
- 54. Nomata J., Kondo T., Mizoguchi T., Tamiaki H., Itoh S., Fujita Y. (2014) Dark-operative protochlorophyllide oxidoreductase generates substrate radicals by an iron-sulphur cluster in bacteriochlorophyll biosynthesis. Sci. Rep. 4, 5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Busch A. W., Reijerse E. J., Lubitz W., Frankenberg-Dinkel N., Hofmann E. (2011) Structural and mechanistic insight into the ferredoxin-mediated two-electron reduction of bilins. Biochem. J. 439, 257–264 [DOI] [PubMed] [Google Scholar]
- 56. Tsukatani Y., Yamamoto H., Harada J., Yoshitomi T., Nomata J., Kasahara M., Mizoguchi T., Fujita Y., Tamiaki H. (2013) An unexpectedly branched biosynthetic pathway for bacteriochlorophyll b capable of absorbing near-infrared light. Sci. Rep. 3, 1217. [DOI] [PMC free article] [PubMed] [Google Scholar]