Background: Myocyte enhancer factor 2 (MEF2) proteins are essential for skeletal muscle development and regeneration, but their diverse roles in differentiation have not been defined.
Results: Individual MEF2 proteins regulate distinct gene programs in skeletal muscle.
Conclusion: Certain genes are preferentially sensitive to a specific MEF2 isoform.
Significance: These findings provide opportunities to modulate MEF2 isoform-sensitive genes in skeletal muscle health and disease.
Keywords: Differentiation, Myogenesis, RNA Interference (RNAi), Transcription Factor, Transcriptomics
Abstract
Skeletal muscle differentiation requires precisely coordinated transcriptional regulation of diverse gene programs that ultimately give rise to the specialized properties of this cell type. In Drosophila, this process is controlled, in part, by MEF2, the sole member of an evolutionarily conserved transcription factor family. By contrast, vertebrate MEF2 is encoded by four distinct genes, Mef2a, -b, -c, and -d, making it far more challenging to link this transcription factor to the regulation of specific muscle gene programs. Here, we have taken the first step in molecularly dissecting vertebrate MEF2 transcriptional function in skeletal muscle differentiation by depleting individual MEF2 proteins in myoblasts. Whereas MEF2A is absolutely required for proper myoblast differentiation, MEF2B, -C, and -D were found to be dispensable for this process. Furthermore, despite the extensive redundancy, we show that mammalian MEF2 proteins regulate a significant subset of nonoverlapping gene programs. These results suggest that individual MEF2 family members are able to recognize specific targets among the entire cohort of MEF2-regulated genes in the muscle genome. These findings provide opportunities to modulate the activity of MEF2 isoforms and their respective gene programs in skeletal muscle homeostasis and disease.
Introduction
Formation of multinucleated, contractile skeletal myotubes from muscle precursor cells involves precise integration of numerous muscle gene programs that are regulated by the cooperative activity of muscle-specific and broadly expressed transcription factors (1, 2). The gene regulatory network that drives skeletal muscle formation in Drosophila is centered on MEF2,2 the exclusive member of this evolutionarily conserved transcription factor in these animals, whose activity is essential for muscle differentiation and the control of a spectrum of genes throughout all stages of muscle development (3–5). Whereas Drosophila possess a single Mef2 gene, vertebrates have evolved to encode multiple isoforms of MEF2, adding to the complexity of the transcriptional circuitry required to govern this biological process. Thus, to properly understand skeletal muscle differentiation in vertebrates, it is important to dissect the mechanisms by which individual members of the MEF2 transcription factor family regulate muscle gene programs.
The vertebrate MEF2 family of transcription factors is encoded by four genes: Mef2a, -b, -c, and -d, which display differences in their temporal and tissue expression patterns but are co-expressed throughout developing and adult skeletal muscle (6). Furthermore, vertebrate MEF2 proteins share substantial amino acid similarity in their DNA-binding domains and bind to similar cis-acting sequences, thereby obscuring the mechanisms through which these factors regulate distinct target genes (3). The lack of conservation between the four individual MEF2 isoforms in their carboxyl-terminal transactivation domain and the dramatically different phenotypes of the various MEF2-deficient vertebrate models suggests that individual MEF2 isoforms regulate distinct gene programs in muscle. MEF2 transcriptional function is further complicated by complex tissue-specific and temporal alternative splicing of mRNA transcripts produced from each of the four Mef2 genes (3, 7, 8).
There is growing evidence that the MEF2 family of transcription factors function nonredundantly in mammalian skeletal muscle. Skeletal muscle-specific MEF2C knock-out mice display widespread myofiber structural abnormalities perinatally, and a subset of these mice survives to adulthood with alterations in skeletal myofiber type (9, 10). Conversely, mice with a global deletion of MEF2A or skeletal muscle-specific deletion of MEF2D display normal skeletal muscle development (9). Interestingly, adult MEF2D knock-out mice also exhibit alterations in skeletal myofiber type distribution (10). We and others have previously shown that MEF2A deficiency results in impaired differentiation of C2C12 skeletal myoblasts and primary myoblasts isolated from injured skeletal muscle of adult MEF2A knock-out mice (11, 12). In contrast, differentiation is not impaired when MEF2A is deleted specifically in adult satellite cells in the context of muscle injury but is perturbed when MEF2A, -C, and -D are deleted simultaneously in these cells (13). These results suggest that some of the transcriptional functions of mammalian MEF2 proteins in skeletal muscle overlap but that distinct regulatory activities emerge in certain biological settings.
Because no study to date has comprehensively analyzed the individual transcriptomes of the four mammalian MEF2 proteins in muscle, we sought to gain a better understanding of their diverse regulatory roles in skeletal muscle differentiation. Toward this end, we developed isoform-specific short hairpin RNAs to knock down the expression of each MEF2 protein in C2C12 myoblasts, followed by phenotypic analysis of myotube formation along with global gene expression profiling. With the exception of MEF2A, we found that knockdown of MEF2B, -C, or -D either individually or in combination did not impair C2C12 myotube formation, indicating that MEF2B, -C, and -D, are dispensable for this process. Remarkably, dysregulated gene expression analysis revealed that individual mammalian MEF2 proteins regulate numerous nonoverlapping genes in C2C12 cells. Finally, computational analysis of MEF2 isoform-dependent target genes failed to show differences in the predicted MEF2 DNA-binding sites, but each gene set displayed dramatically different enriched transcription factor-binding site motifs located within their proximal upstream regulatory regions.
EXPERIMENTAL PROCEDURES
Cell Culture
C2C12 and COS cells were cultured as described previously (14). Specificity of the Mef2b and Mef2d shRNAs was tested by co-transfection of COS cells with MEF2-FLAG and pENTR shRNA constructs. For modulating MEF2 expression levels in myoblasts, C2C12 cells were transduced either with adenoviruses harboring shRNAs targeting individual Mef2 isoforms or in combination with adenoviruses overexpressing full-length MEF2 cDNAs. Cells were allowed to differentiate for 72 h before being analyzed.
Plasmids
MEF2A-FLAG and MEF2C-FLAG plasmids were generated by cloning full-length mouse cDNAs into pCMV-tag4 (Invitrogen). MEF2B-FLAG and MEF2D-FLAG (human) were kind gifts from T. Gulick (Sanford Burnham Medical Research Institute, Orlando, FL). shRNA sequences targeting either Mef2b or Mef2d transcripts were cloned into the pENTRTM/U6 RNAi entry vector according to the manufacturer's instructions (Invitrogen).
shRNA Design and Knockdown
Adenoviruses carrying shRNAs were generated as described previously (15). Briefly, shRNA sequences targeting either Mef2b or Mef2d transcripts were generated using the BLOCK-iT RNAiTM designer system (Invitrogen). Human, mouse, and rat mRNA sequences were analyzed for conservation of the proposed shRNA sequences. shRNAs that targeted multiple Mef2 isoforms or that were found within alternatively spliced exons were excluded. The Mef2a shRNA adenovirus was used at a multiplicity of infection of 25 for all assays. The Mef2b, -c, and -d shRNA adenoviruses were used at a multiplicity of infection of 50 for all assays. β-gal, MEF2A, MEF2C, MEF2D, and MEF2-VP16 overexpression adenoviruses were generously provided by Jeff Molkentin (Children's Hospital, Cincinnati, OH) and Ken Walsh (Boston University Medical School) and were used at a multiplicity of infection of 50.
Microarray
Seventy-two hours after induction of differentiation, total RNA from shlacZ (n = 6), shMef2a (n = 6), shMef2b (n = 6), shMef2c (n = 6), and shMef2d (n = 6) C2C12 myotubes was prepared by TRIzol® isolation (Invitrogen). Samples were pooled in sets, for a total of three biological replicates per condition. Samples were hybridized to the Mouse GeneChip® Gene 1.0 ST array (Affymetrix) at the Boston University Microarray Facility (n = 3 per shRNA; 15 arrays total). Microarray data are available in GEO (NCBI) with accession number GSE63798.
Construction of Isoform Sensitive Gene Sets
Microarray data were annotated with Entrez ID numbers. Genes were analyzed for statistical significance and sorted into groups based on shared dysregulation among the treatment groups.
Quantitative RT-PCR
RNA from C2C12 MEF2 knockdown experiments (n = 3) was used to synthesize cDNA using reverse transcriptase (Moloney murine leukemia virus) with random hexamers (Promega). Quantitative RT-PCR was performed in triplicate wells using Power SYBR® Green Master Mix (Applied Biosystems) with the 7900HT sequence detection system (Applied Biosystems). The primers used were: Gapdh forward, 5′-TGGCAAAGTGGAGATTGTTGCC, and reverse, 5′-AAGATGGTGATGGGCTTCCCG; Cdkn1c forward, 5′-CCAATGCGAACGACTTCTTCGC, and reverse, 5′-AACTAACTCATCTCAGACGTTTGCGC; Sept4 forward, 5′-TACACTCATGGTGGCAGGAGAATCTG, and reverse, 5′-CACTCTGTGTTGTTGACTGCATCC; Hspb7 forward, 5′-GCTGAGAAGCTGGCAGCTGATG, and reverse, 5′-ATCTCAGTCCGGAAGGTCTGCTG; Myom1 forward, 5′-CTACTCTGGACGGCAAGTGCAC, and reverse, 5′-GTGGTCCGTTTGGAGGTTGC; Stc2 forward, 5′-CTGCAGAACACAGCGGAGATCC, and reverse, 5′-CTGGGCATCGAATTTTCCAGCGT; Tex16 forward, 5′-CTTCTTGCCCTTTCAAGGTGT, and reverse, 5′-TACCTGTTTGGAGTCTGAGCTGAA; Selp forward, 5′-TACACAGCCTCCTGCCAGGA, and reverse, 5′-CTGAAGGTGCACTGTGAGTTGAAGG; C1ql1 forward, 5′-GGTCACCAACCTAGGCAACAACTAC, and reverse, 5′-CTCCATCCAGCTTGATGAAGACCTC; Bace2 forward, 5′-ACTCAGAGAGCTCCAGCACATACC, and reverse, 5′-GCCAAAGCAGCATAAGCAAGTCC; Pi16 forward, 5′-CTGCAGATGAGGTGGGATG, and reverse, 5′-GCCGTGCTGAAATTGTAATACTC; Themis forward, 5′-CTACGGACGACCTTTTTGAAAT, and reverse, 5′-CTAAGATCCTCGAAGCCTGGTA; Glipr1 forward, 5′-ACTCAGGTTGTTTGGGCAGACAG, and reverse, 5′-TGCAGAGACTGTTGAGACACTTGTCA; Cpa4 forward, 5′-GTACACGCAAAGCCAGAACC, and reverse, 5′-CCATGGTACACTTCAGAGCAAG; Fam78a forward, 5′-AGCAGGGCATGTCTAGCTGG, and reverse, 5′-CACGTGGTGAAGCTCTGGTC; and Ppp1r3a forward, 5′-GCTAGACTTGATGATAAACCAACGG, and reverse, 5′-CCCATGAACAAGTCAGTGTTGA.
Western Blot Analysis
Western blots were performed as previously described (12). Antibodies included: anti-GAPDH (1:1,000; Santa Cruz), anti-MEF2 (1:1,000; Santa Cruz), anti-MEF2C (1:1,000; Sparrow Biosciences), anti-MEF2D (1:1,000: BD Biosciences), anti-FLAG (1:10,000; Sigma), anti-α-tubulin (1:1,000; Sigma), and anti-MF-20 (1:200; supernatant, DSHB). Blots were incubated with horseradish peroxidase-conjugated secondary antibodies (1:10,000; Sigma) and reacted with Western Lightning chemiluminescent reagent (PerkinElmer Life Sciences).
Computational Pathway Analysis
Statistically distinct gene sets sensitive to individual MEF2 isoforms were analyzed using three independent pathway analysis algorithms. Gene Ontology term and Kyoto Encyclopedia of Genes and Genomes pathway analyses were performed through the DAVID bioinformatics database (16, 17). Ingenuity Pathway Analysis (Ingenuity® systems) was used to determine the canonical cellular pathways associated with each uniquely sensitive gene set.
MEF2-binding Site Variation Analysis
MEF2-binding site comparisons were performed by extracting putative MEF2-binding sites from the proximal promoter regions (5 kb upstream of the putative transcriptional start site) of gene sets uniquely sensitive to each MEF2 isoform using the Find Individual Motif Occurrences (FIMO) tool from the Multiple Em for Motif Elicitation (MEME) suite (18). Extraction was performed by scoring 10 base pair motifs against the MEF2 motif stored in the JASPAR database (MA0052.1) using a p value threshold of less than 0.0001. The output position-weight matrices were then used to compile a sequence logo using the WebLogo (19).
De Novo Motif Discovery
Transcription factor-binding motif enrichment analysis was performed on the proximal promoter region of genes preferentially sensitive to each MEF2 isoform using MatInspector from the Genomatix software suite (20). A default background composed of a cross-section of genomic promoter sequences was used to discriminate between enriched features and nonspecific promoter regions. The resulting transcription factor motifs were then sorted by Z score. Motifs with a Z score of greater than or equal to 2 were considered to be enriched. Additional data for each enriched motif was extracted from the Genomatix MatBase and NCBI databases.
Statistical Analysis
All numerical quantification is representative of the mean ± S.E. of at least three independently performed experiments. Statistically significant differences between two populations of data were determined using Student's t test. p values of ≤0.05 were considered to be statistically significant. The technical quality of the arrays was evaluated using relative log expressions and normalized unscaled standard error. Relative log expressions and normalized unscaled standard error values >0.1 and >1.05, respectively, are considered out of normal limits. All arrays had median values within the limits of these tests. Microarray data were normalized using the robust multiarray average algorithm and were log2 transformed by default. Knockdown efficiency was determined by calculating a fold change for each MEF2 isoform knockdown relative to the shlacZ control. Significant dysregulation of gene expression was determined using a one-way analysis of variance. The Benjamini-Hochberg false discovery rate correction was then applied to obtain corrected q values, and a q value threshold of less than or equal to 0.05 was used to determine significant dysregulation. Tukey's honest significant difference post hoc test was performed to identify significantly dysregulated genes and correct for multiple testing error across all intergroup comparisons. A corrected q value of 0.05 was used to determine statistically significant gene dysregulation among groups.
RESULTS
MEF2 Isoform-specific Short Hairpin RNA Adenovirus
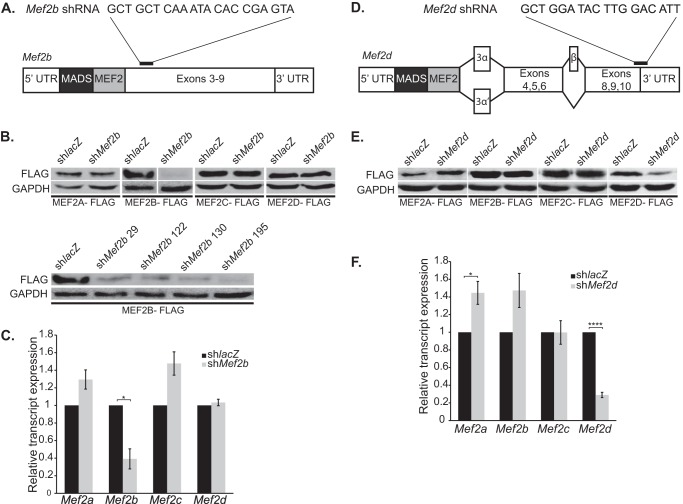
To address the roles of the mammalian MEF2 transcription factors in skeletal muscle differentiation, we depleted each protein in C2C12 myoblasts using isoform-specific shRNA adenoviruses. These shRNAs were designed to target all mRNA transcripts generated from each Mef2 gene. Previously, we described the specific and robust knockdown of MEF2A and MEF2C using shRNA adenoviruses (11, 15). We subsequently generated shRNAs to specifically target MEF2B (Fig. 1A) and MEF2D (Fig. 1D). The efficacy of these newly designed MEF2 shRNAs was examined in COS cells co-transfected with each shRNA along with either MEF2B-FLAG or MEF2D-FLAG. These shRNAs robustly knocked down the expression of the respective MEF2 protein without affecting the expression of the other MEF2 family members, demonstrating the specificity of these shRNAs (Fig. 1, B and E). The MEF2B and MEF2D shRNAs were subsequently packaged into adenovirus for transduction in C2C12 cells. Transduction of MEF2B- and MEF2D-specific shRNA adenoviruses robustly knocked down the expression of the respective endogenous MEF2 factor but did not deplete the expression of the other MEF2 family members (Fig. 1, C and F).
FIGURE 1.
Robust and specific knockdown of MEF2 proteins using shRNA adenoviruses. A and D, schematic representations of Mef2b (A) and Mef2d (D) transcripts. shRNA adenoviruses were generated to target the carboxyl-terminal region of the Mef2 transcripts (black bar) for knockdown in C2C12 myoblasts. The target sequences excluded regions containing alternatively spliced exons and were selected based on homology between mouse, human, and rat Mef2b or Mef2d sequences. B and E, Western blot analysis of shMef2b (B) and shMef2d (E) specificity against overexpressed MEF2 constructs in COS cells. Extracts for shMef2b knockdown of MEF2B-FLAG Western blot were cropped for image clarity (B, upper panel) but analyzed on the same gel (B, lower panel). shMef2b 195 (sequence shown in A) was used to generate adenovirus. C and F, quantitative RT analysis of endogenous Mef2b (C) and Mef2d (F) knockdown in C2C12 myotubes at differentiation day 3. The data are means ± S.E. *, p < 0.05; ****, p < 0.0001.
Knockdown of MEF2 Proteins in C2C12 Myoblasts
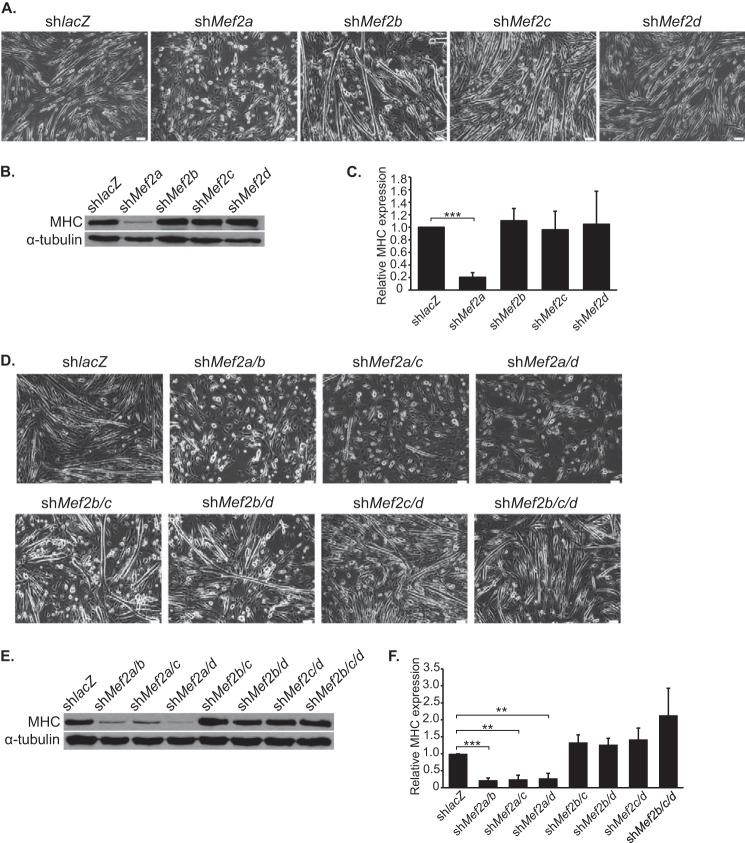
The four mammalian MEF2 factors display different temporal expression patterns in C2C12 differentiation (11, 21). Therefore, we focused our analysis of the individual MEF2 isoform knockdowns on differentiation day 3, because this reflects the time at which the four MEF2 proteins are co-expressed in myotubes. Proliferating C2C12 myoblasts were transduced with the shRNA adenoviruses, and on differentiation day 3, MEF2-depleted myotubes were evaluated for gross morphological defects in the formation of multinucleated myotubes. As previously reported by us and others, MEF2A depletion resulted in impaired myotube formation and differentiation (Fig. 2, A–C) (11, 12). By contrast, individual depletion of the other MEF2 proteins failed to show any obvious impairment in myotube formation or differentiation (Fig. 2, A–C). These results suggest that, with the exception of MEF2A, the remaining MEF2 proteins are dispensable for C2C12 myogenic differentiation.
FIGURE 2.
shRNA-mediated knockdown of MEF2 proteins in C2C12 myoblasts. C2C12 myoblasts were transduced with adenoviruses harboring shRNAs targeting Mef2a, -b, -c, or -d, or with an shRNA against lacZ as a negative control. A–C, knockdown of Mef2a (A), but not Mef2b, -c, or -d, resulted in impaired myotube formation and differentiation as shown by Western blot analysis of the muscle-specific marker myosin heavy chain (MHC) (B and C). D, combinatorial knockdown of Mef2a/b, Mef2a/c, and Mef2a/d failed to modulate the differentiation defect observed in the Mef2a knockdown alone. Additionally, simultaneous knockdown of Mef2b/c, Mef2b/d, Mef2c/d, and Mef2b/c/d failed to produce any overt morphological defects in C2C12 differentiation. E and F, Western blot analysis of myosin heavy chain (E) and accompanying densitometry (F) for the combinatorial knockdowns show that differentiation is only affected in MEF2A depleted cells. The data are means ± S.E. **, p < 0.01; ***, p < 0.001.
We then asked whether depletion of MEF2B, -C, or -D could modulate the impaired differentiation phenotype of MEF2A-deficient C2C12 cells. As shown in Fig. 2 (D–F), individual depletion of the other MEF2 proteins in the MEF2A-deficient C2C12 myoblast background resulted in a phenotype similar to MEF2A deficiency alone (Fig. 2D, upper panels). We subsequently knocked down MEF2B, -C, and -D in combination to investigate the potential redundancy of these proteins in C2C12 differentiation. Knockdown of MEF2B and -C, MEF2B and -D, MEF2C and -D, or all three MEF2 proteins did not adversely affect C2C12 differentiation (Fig. 2, D, lower panels; E; and F). These results demonstrate that MEF2A is sufficient for C2C12 differentiation and that it may play a dominant role in this context.
MEF2 Overexpression in Wild Type and MEF2A-depleted C2C12 Cells
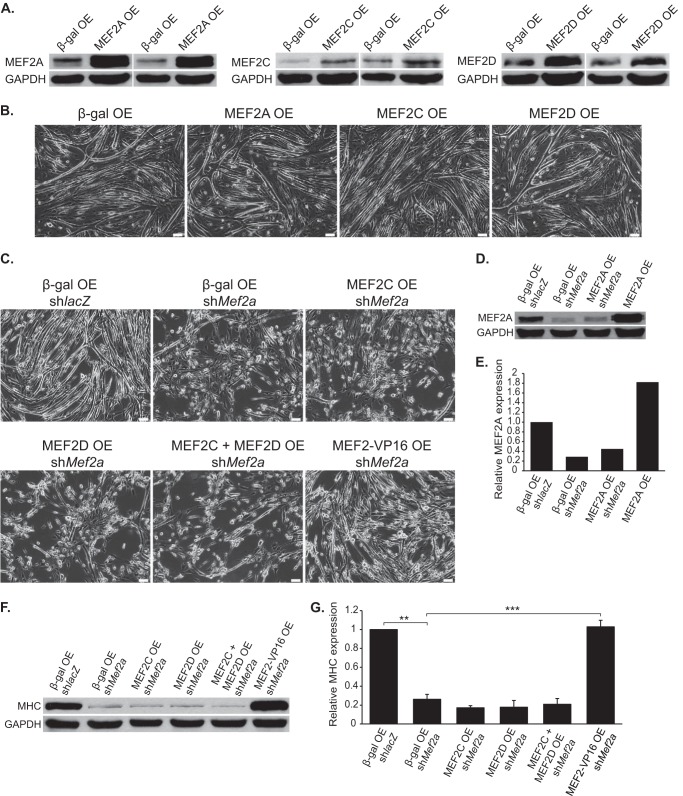
In a complementary set of experiments, we asked whether overexpression of each MEF2 protein in C2C12 myoblasts is sufficient to induce a morphological phenotype in these cells. For these experiments, we used adenoviruses harboring MEF2A, -C, and -D cDNAs, which have been previously described (22). Transduction of these MEF2 viruses individually in C2C12 myoblasts resulted in an increase of the respective MEF2 protein over endogenous levels (Fig. 3A). However, overexpression of the individual MEF2 isoforms in proliferating C2C12 myoblasts followed by differentiation failed to trigger an overt morphological phenotype in these cells (Fig. 3B). Curiously, contrary to a recent report (8), acute overexpression of MEF2D did not appear to enhance myotube formation in C2C12 cells, even though this adenovirus encodes the muscle-specific α2 isoform described in that study.
FIGURE 3.
Overexpression and rescue of MEF2 in C2C12 myoblasts. A, Western blot analyses of C2C12 cells transduced with MEF2 isoform adenoviruses confirm an increase in MEF2 protein levels, relative to the β-gal control. OE, overexpression. B, overexpression of MEF2A, -C, or -D did not overtly modulate C2C12 myotube formation. Interestingly, overexpression of the MEF2D muscle-specific isoform did not result in enhanced myotube formation, as previously described. C, overexpression of MEF2-VP16, but not overexpression of MEF2C or -D alone or in combination, was able to rescue the differentiation defect observed in MEF2A depleted C2C12 myotubes. D, overexpression of MEF2A was unable to restore MEF2A to wild type levels, because the MEF2A cDNA encoded in this adenovirus is knocked down by shMef2a. E, quantification of MEF2A Western blot analysis. F, Western blot analysis of myosin heavy chain (MHC) expression demonstrates overexpression of MEF2-VP16, but not MEF2C or -D alone or in combination, was able to rescue impaired differentiation in MEF2A deficient cells. G, quantification of the myosin heavy chain expression Western blot analysis. The data are means ± S.E. **, p < 0.01; ***, p < 0.001.
We next determined whether overexpression of MEF2A, -C, or -D was capable of rescuing the differentiation defect in MEF2A-deficient C2C12 cells. Transduction of MEF2C or -D adenoviruses failed to rescue myotube formation or differentiation in MEF2A-depleted C2C12 cells (Fig. 3, C, F, and G). By contrast, transduction of MEF2A-depleted C2C12 cells with MEF2-VP16 adenovirus (22), consisting of only the MEF2C DNA-binding and the VP16 transactivation domains, effectively promoted myotube formation and differentiation (Fig. 3, C, F, and G). Importantly, the rescue by MEF2-VP16 strongly suggests that impaired C2C12 differentiation is caused by the specific knockdown of MEF2 and not secondary off target effects caused by the shRNA. It is worth noting that MEF2A overexpression was unable to rescue the MEF2A-deficient phenotype because the MEF2A cDNA encoded in this adenovirus is knocked down by this shRNA (Fig. 3, D and E).
Gene Expression Profiles and Comparative Analysis of Individual MEF2 Knockdown in C2C12 Cells
The lack of readily observable phenotypes in the individual and combinatorial knockdowns of MEF2B, -C, and -D suggested that these proteins function redundantly in C2C12 myotube formation. Furthermore, the inability of MEF2C and -D overexpression to rescue impaired myotube formation in MEF2A-depleted C2C12 cells demonstrates that they are not functionally redundant with MEF2A in this process. Therefore, to determine what sets of genes and cellular processes are regulated by individual MEF2 proteins in skeletal myotubes, we performed global gene expression profiling for each knockdown.
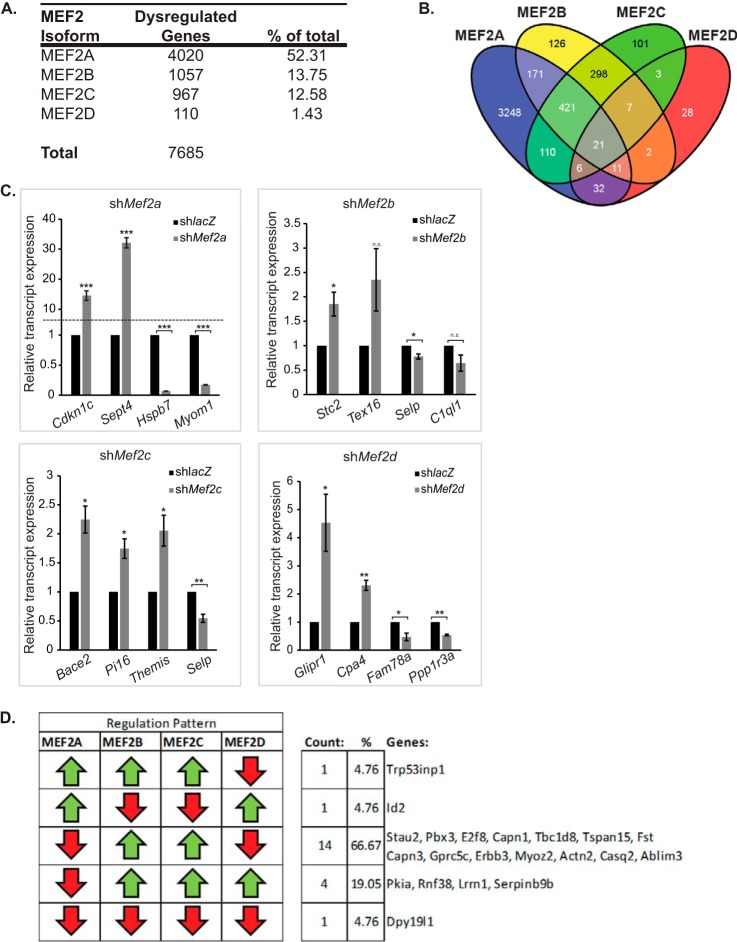
Microarray analysis of C2C12 myotubes depleted of individual MEF2 proteins (n = 3 arrays for each MEF2 shRNA) resulted in a range of significantly dysregulated genes (Fig. 4, A and B), as determined by a one-way analysis of variance using the Benjamini-Hochberg false discovery rate correction and a threshold q value less than or equal to 0.05. Using these stringent criteria, the most striking difference in the total number of dysregulated genes was observed for MEF2A and -D knockdowns. As shown in Fig. 4 (A and B), depletion of MEF2A revealed 4,020 significantly dysregulated genes, whereas MEF2D deficiency resulted in only 110 dysregulated genes. A wide disparity was also noted when comparing the MEF2A knockdown with that of MEF2B and -C, suggesting that MEF2A plays a major transcriptional function in C2C12 cells. The microarray results were subsequently validated by quantitative RT-PCR analysis on a subset of the top dysregulated genes (up- and down-regulated) from each individual knockdown. Most of the genes examined displayed the expected dysregulation (Fig. 4C).
FIGURE 4.
Comparative analysis of MEF2 knockdown gene sets. Microarray analysis reveals that C2C12 cells are differentially sensitive to depletion of the four MEF2 isoforms. A, summary of total significantly dysregulated (q < 0.05) genes in each MEF2 isoform shRNA knockdown. B, a composite Venn diagram incorporating all overlapping gene sets as determined by the Tukey's honest significant difference post hoc test (q < 0.05). C, quantitative RT-PCR analysis of a subset of genes dysregulated in the Mef2 knockdown microarrays. Cdkn1c, cyclin-dependent kinase inhibitor 1c; Sept4, septin 4; Hspb7, heat shock protein family member 7; Myom1, myomesin 1; Stc2, stanniocalcin 2; Tex16, testis expressed gene 16; Selp, selectin, platelet; C1ql1, compliment component 1, q subcomponent-like 1; Bace2, beta-site app-cleaving enzyme 2; Pi16, peptidase inhibitor 16; Themis, thymocyte selection associated; Glipr1, GLI pathogenesis-related 1 (glioma); Cpa4, carboxypeptidase a4; Fam78a, family with sequence similarity 78; Ppp1r3a, protein phosphatase 1, regulatory (inhibitor) subunit 3a. The primer sequences are listed under “Experimental Procedures.” D, only five of the possible patterns of dysregulation are represented in the commonly dysregulated gene set. Of these patterns, the most prevalent group (66%) were genes that were down-regulated in MEF2A or MEF2D depletion and up-regulated in MEF2B or MEF2C depletion. Additionally, only a single gene, Dpy19l1 (DumPY19-like 1), was dysregulated in the same direction upon individual knockdown of each MEF2 factor. The data are means ± S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant.
To identify genes sensitive to a given MEF2 isoform, we compared the various dysregulated gene sets to determine the extent of overlapping genes. MEF2-dependent genes were designated as isoform-sensitive (or nonoverlapping) if they were significantly different from each other using the Tukey's honest significant difference post hoc test. Based on these strict statistical criteria, a subset of genes in each MEF2 knockdown was clearly found to be significantly more sensitive to the respective MEF2 isoform compared with the other family members. As shown in the Venn diagram (Fig. 4B), these nonoverlapping dysregulated groups included 3,248 genes (81% of the total dysregulated by MEF2A) for MEF2A, 126 genes (12%) for MEF2B, 101 genes (10%) for MEF2C, and 28 genes (25%) for MEF2D. This comparative analysis also revealed that many (75–90%) of the genes dysregulated in the MEF2B, -C, and -D knockdowns were dysregulated in one or more of the other MEF2 depletions. Furthermore, the overlap of dysregulated genes in the pairwise combinations of MEF2D and either B or C gene sets was quite low (two and three genes, respectively) in comparison with the extent of overlap seen in any of the other combinations, such as MEF2A and -B (171 overlapping genes), MEF2A and -C (110 genes), MEF2A and -D (32 genes), and MEF2B and -C (298 genes). Finally, there were only 21 of over 7,000 dysregulated genes (<0.003%) shared by all four MEF2 knockdowns. These results suggest that only a small fraction of MEF2-dependent genes in C2C12 cells can be regulated by all four MEF2 proteins, but most can be regulated by two or three of these transcription factors, either as homodimers or combinations of heterodimers.
Analysis of the dysregulated patterns, i.e. up- or down-regulated in each MEF2 gene set, of the 21 genes revealed that only five of the potential twenty-four different patterns were represented (Fig. 4D). Interestingly, genes that were down-regulated upon depletion of MEF2A or MEF2D were up-regulated in MEF2B or MEF2C depletion and were the most prevalent group, comprising two-thirds of the dysregulated patterns. Additionally, only a single gene, Dpy19l1 (DumPY19-like 1), was down-regulated in each of the MEF2 isoform depletions. Dpy19l1 is the apparent ortholog of a Caenorhabditis elegans gene, dpy-19, that encodes a transmembrane protein with C-mannosyltransferase activity involved in neuroblast migration (23, 24). Dpy19l1 may also function upstream of genes involved in muscle development in worms, making it an exciting MEF2-dependent gene to further investigate (WormBase).
Classification of Cellular Processes in MEF2 Knockdown Gene Sets
To gain insight into the distinct roles of the MEF2 family in C2C12 myotubes, nonoverlapping dysregulated genes in each subgroup were categorized into cellular processes using three independent functional pathway analysis algorithms: Ingenuity Pathway Analysis (IPA), Gene Ontology, and Kyoto Encyclopedia of Genes and Genomes. IPA of MEF2-sensitive gene sets revealed that the preferentially dysregulated target genes from each MEF2 knockdown function in vastly different cellular processes (Table 1). Many genes distinctly sensitive to MEF2A play roles in calcium signaling and actin cytoskeletal rearrangement. Genes preferentially regulated by MEF2B are associated with hepatic fibrotic pathways, ovarian cancer signaling, and human stem cell pluripotency, whereas genes preferentially regulated by MEF2C are involved in control of the G1/S cell cycle checkpoint, eicosanoid signaling, and estrogen-mediated S phase entry. Genes regulated by MEF2D appear to be involved in JAK2-like signaling and hypoxic response. Other canonical pathways in the MEF2D gene set, such as atherosclerosis signaling and AMPK signaling, did not reach statistical significance. Interestingly, IPA also revealed that some MEF2 family members regulate similar cellular processes, even though the isoform-sensitive genes do not overlap and code for different proteins. For example, the canonical pathways related to cancer was shared by MEF2A (molecular mechanisms of cancer) and MEF2C (ovarian cancer signaling), and Rho signaling was shared by MEF2A (Rho family GTPases) and MEF2D (RhoA signaling). These results likely reflect distinct proteins belonging to the same pathway. IPA was also performed on the 21 dysregulated genes common to all four MEF2 proteins (Table 1). Of the top five canonical pathways, three appear to be highly relevant to muscle function. Integrin and FAK signaling have been shown to play a role in adhesion and signaling at the myofiber periphery (26), and calpain proteases are calcium-regulated proteases important in skeletal muscle homeostasis (27).
TABLE 1.
Analysis of canonical pathways associated with each MEF2 isoform
Each preferentially sensitive MEF2 gene set, including the 21 commonly dysregulated genes, were analyzed using Ingenuity Pathway Analysis software. The top five most significantly regulated canonical pathways are provided. These pathways were analyzed for statistically significant association with each unique MEF2-sensitive gene set, and all were found to be significant (p < 0.05) with the exception of the MEF2D-associated canonical pathways, likely because of the small number of MEF2D-sensitive genes dysregulated in the expression analysis. Also provided is the amount of genes dysregulated in each MEF2 factor knockdown in relation to the accepted number of genes associated with each canonical pathway (ratio).
| Canonical pathway | p value | Ratio |
|---|---|---|
| MEF2A | ||
| Molecular mechanisms of cancer | 2.40E-08 | 92/387 (0.238) |
| Calcium signaling | 2.26E-07 | 55/217 (0.253) |
| Germ cell-Sertoli cell junction signaling | 6.20E-06 | 46/169 (0.272) |
| Actin cytoskeletal signaling | 1.63E-05 | 57/242 (0.236) |
| Signaling by Rho family GTPases | 1.96E-05 | 60/262 (0.229) |
| MEF2B | ||
| Hepatic fibrosis/hepatic stellate cell activation | 1.31E-03 | 5/155 (0.032) |
| Ovarian cancer signaling | 7.57E-03 | 4/152 (0.026) |
| Human embryonic stem cell pluripotency | 7.77E-03 | 4/161 (0.025) |
| Ceramide signaling | 1.13E-02 | 3/91 (0.033) |
| Apoptosis signaling | 1.47E-02 | 3/100 (0.03) |
| MEF2C | ||
| Cell cycle: G1/S checkpoint regulation | 3.14E-03 | 3/68 (0.044) |
| Eicosanoid signaling | 3.14E-03 | 3/85 (0.035) |
| Molecular mechanisms of cancer | 5.49E-03 | 6/387 (0.016) |
| Estrogen-mediated S phase entry | 5.48E-03 | 2/28 (0.071) |
| Sulfate activation for sulfonation | 9.42E-03 | 1/8 (0.125) |
| MEF2D | ||
| Role of JAK2 in hormone-like cytokine signaling | 4.03E-02 | 1/37 (0.027) |
| Hypoxia signaling in the cardiovascular system | 7.69E-02 | 1/68 (0.015) |
| RhoA signaling | 1.32E-01 | 1/122 (0.008) |
| Artherosclerosis signaling | 1.37E-01 | 1/138 (0.007) |
| AMPK signaling | 1.50E-01 | 1/180 |
| All MEF2 | ||
| Regulation of cellular mechanics by calpain protease | 3.20E-05 | 3/73 (0.041) |
| nNOS signaling in neurons | 1.20E-03 | 2/52 (0.038) |
| Integrin signaling | 1.24E-03 | 3/208 (0.014) |
| Amyloid processing | 1.41E-03 | 2/61 (0.033) |
| FAK signaling | 4.05E-03 | 2/106 (0.019) |
Like the IPA, both Gene Ontology and Kyoto Encyclopedia of Genes and Genomes algorithms revealed that the various MEF2 isoform target genes function in different cellular processes, and in most instances, these algorithms predicted pathways similar to those predicted by IPA (supplemental Tables S1 and S2). It is worth noting that some overlap in basic skeletal gene programs is observed among the MEF2 family, particularly between MEF2A and -C. Taken together, these computational analyses suggest that the four mammalian MEF2 factors regulate partially overlapping but predominantly distinct gene programs in C2C12 differentiation.
Identification of Distinct TF-binding Sites Associated with Each MEF2 Knockdown Gene Set
To mechanistically understand the preferential sensitivity of target genes to MEF2 isoforms, we reasoned that sensitivity to a given MEF2 protein is determined by minor variations in the MEF2 DNA-binding site consensus sequence, with the assumption that a number of these genes are direct targets. Based on FIMO analysis of the proximal promoter regions, no significant variations were observed in the consensus MEF2 DNA-binding sequence, CTA(A/T)4TAG (data not shown).
In the absence of any variations in MEF2-binding site sequence, we reasoned that the isoform sensitivity of dysregulated genes in each MEF2 group results from a distinct combination of transcription factor (TF)-binding sites in the various promoters. To identify TF motifs enriched in gene sets associated with a single MEF2 factor knockdown, we performed de novo motif discovery using the MatInspector transcription factor-binding site enrichment analysis (Genomatix). Motifs were considered significantly enriched in a set of promoters if the Z score was greater than or equal to 2. As shown in Fig. 5A, over 90% of the overrepresented binding site motifs were shared by two or more MEF2 factors, and less than 10% of enriched motifs were found in genes regulated by a single MEF2 protein. De novo motifs were then sorted into groups based on their enrichment in gene sets associated with each of the four MEF2 isoforms. This analysis showed that 14% of MEF2A-, 1% of MEF2B-, 0% of MEF2C-, and 5% of MEF2D-sensitive genes harbored motifs that were preferentially enriched in proximal promoter regions in the respective MEF2 knockdown (Fig. 5B). Similar to the disparity in the number of dysregulated genes in each MEF2 knockdown, the number of de novo motifs associated with each MEF2 gene set varied greatly. As shown in Fig. 5B, a total of 15 DNA-binding site motifs were identified specifically in the MEF2A-sensitive gene set and were not shared by the other MEF2 isoform gene sets. This was followed by one distinct motif in the MEF2B-sensitive gene set and two distinct motifs in the MEF2D-sensitive gene set. Remarkably, the MEF2C-sensitive dysregulated genes did not harbor DNA-binding site motifs specific to this data set. This analysis indicates that all of the de novo motifs in MEF2C-sensitive genes were found in one or more of the other MEF2 isoform gene sets.
FIGURE 5.
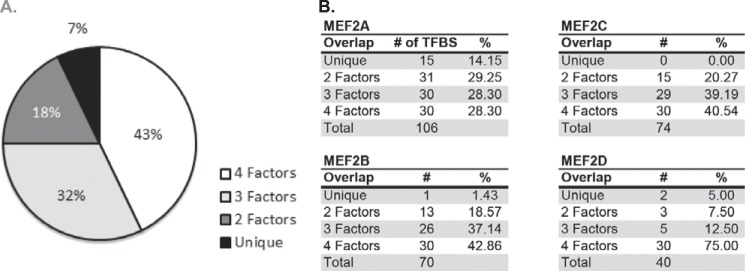
Identification of distinct TF-binding sites associated with each MEF2 knockdown gene set. A, transcription factor-binding motif enrichment analysis was performed on the proximal promoter regions of each MEF2 gene set. Approximately 43% of binding motifs were shared by all four gene sets, 32% were shared by three MEF2 gene sets, 18% were shared by two MEF2 gene sets, and 7% are enriched only in genes regulated by a single MEF2 factor. B, breakdown of motif distribution by MEF2 isoform. MEF2A had the highest percentage of uniquely enriched motifs, and no unique motifs were identified for MEF2C-sensitive genes. TFBS, transcription factor binding site.
Another intriguing result of this analysis was the finding that not a single DNA-binding sequence motif was preferentially associated with the overlapping genes from the MEF2B- and -C-, MEF2B- and -D-, and MEF2C- and -D-sensitive promoter sets (supplemental Table S3). All of the motifs found in these overlapping gene sets were also found in the MEF2A sensitive gene set. Thus, there may be a common mechanism through which a subset of MEF2-dependent genes is regulated by the individual MEF2 proteins.
Regardless of the MEF2 isoform-sensitive gene set, most of the transcription factors predicted to bind to these DNA sequence motifs are broadly expressed, and the vast majority encode C2H2 zinc finger and homeodomains (Table 2). These particular binding domains are among the most prevalent in transcription factor superfamilies (28–30). Moreover, they can function both as activators and repressors and regulate a broad spectrum of target genes and cellular processes, including differentiation. Although these are large transcription factor families, this analysis clearly identified specific transcription factors (Table 2).
TABLE 2.
Transcription factor-binding site enrichment analysis
Binding site enrichment analysis was performed on gene sets preferentially regulated by each MEF2 factor using the Genomatix software suite. Fifteen motifs were uniquely enriched in the MEF2A-sensitive gene set, two were enriched in the MEF2B set, no motifs were enriched in the MEF2C set, and two were enriched in the MEF2D set. The table provides the description of each enriched motif with a calculated Z score (Z > 2 was considered significantly enriched). The known binding factors and their relevant binding domains are included. Finally, a summary of the function of each of these binding factors is provided.
| TF module | Module description | Z score | Known factor | Binding domain | Function |
|---|---|---|---|---|---|
| Mef2a | |||||
| V$SF1F | Vertebrate steroidogenic factor | 13.53 | Nr5a1 | C4 zinc finger domain | Essential for embryonic sex determination |
| V$DLXF | Distal-less homeodomain transcription factors | 12.93 | Dlx1–Dlx6 | Homeodomain | Repressor, important of embryonic development (shared function) |
| V$IKZF | Ikaros family zinc finger 5 | 10.16 | Ikzf5 | C2H2 zinc finger domain | Repressor, lymphocyte development |
| V$BARB | Barbiturate-inducible element box | 9.68 | Not characterized | Not characterized | |
| V$ZF05 | C2H2 zinc finger transcription factor 5 | 9.49 | Zfp410 | C2H2 zinc finger domain | ECM remodeling |
| V$IKRS | Ikaros zinc finger family | 8.8 | Ikzf1–Ikzf4 | C2H2 zinc finger domain | Repressor, lymphocyte development (shared function) |
| V$THAP | THAP domain containing protein | 6.72 | Thap1 | THAP domain | Cell cycle progression downstream of Rb-E2F signaling |
| V$HEAT | Heat shock factors | 6.34 | Hsf1, Hsf2, Hsf4 | HSF-HTH | Heat-sensitive activator, embryonic development |
| V$ATBF | AT-binding transcription factor | 6.15 | Zfhx3 | C2H2 zinc finger domain, C2HC zinc finger domain, homeodomain | Repressor, promotes neuronal and myogenic differentiation |
| V$OVOL | OVO homolog-like transcription factor | 6.15 | Ovol1, Ovol2 | C2H2 zinc finger domain | Ovol1: repressor, contributes to epithelial development |
| Ovol2: repressor, embryonic cardiovascular development | |||||
| V$GTBX | GT box | 4.38 | Zfp628 | C2H2 zinc finger domain | Uncharacterized transcriptional regulator |
| V$PTF1 | Pancreas transcription factor 1, heterotrimeric transcription factor | 4.33 | Ptf1a, Rbpj, Rbpjl | Basic helix-loop-helix | Ptf1a: involved in pancreatic and neuronal development |
| Rbpj: involved in cardiac and hematopoeitic development | |||||
| Rbpjl: uncharacterized transcriptional regulator | |||||
| V$RU49 | Zinc finger transcription factor RU49, zinc finger proliferation 1-Zipro1 | 3.4 | Zscan21 | C2H2 zinc finger domain | Uncharacterized transcriptional regulator |
| V$HZIP | Homeodomain-leucine zipper transcription factors | 2.21 | Homez | Homeodomain | Uncharacterized transcriptional regulator |
| V$CHOP | C/EBP homologous protein (CHOP) | 2.12 | Ddit3 | bZIP | Stress sensitive transcriptional regulator, negative regulator of myogenic differentiation |
| Mef2b | |||||
| V$BTBF | BTB/POZ (broad complex, TramTrack, Bric-a-brac/pox viruses and zinc fingers) | 2.56 | Zbtb33 | BTB-POZ C2H2 zinc fingers | Repressor of Wnt target genes; activator of an unrelated set of genes |
| Mef2c | |||||
| Mef2d | |||||
| V$TALE | TALE homeodomain class recognizing TG motifs | 3.94 | Meis1, Meis2, Meis3, Pknox1, Pknox2, Tgif1, Tgif2 | TALE class homeodomain | Meis1: angiogenic and hematopoeitic development |
| Meis2: mammalian eye/neuronal development | |||||
| Meis3: activator, downstream of PKB signaling | |||||
| Pknox1: activator, hematopoeitic and angiogenic development | |||||
| Pknox2: associated with actin cytoskeleton | |||||
| Tgif1: Downstream of TGF-β signaling, embryonic development | |||||
| Tgif2: uncharacterized regulator downstream of TGF-β signaling | |||||
| V$DICE | Downstream immunoglobulin | 2.31 | Gtf2i, Gtf2ird1 | GTF2I repeat domain | Gtf2i: embryonic development, negative regulator of angiogenesis |
| Control element, critical for B cell activity and specificity | Gtf2ird1: modulates cell cycle progression, skeletal muscle differentiation | ||||
We next performed an extensive literature search to determine the cellular function of these predicted transcription factors (Table 2) and focus on those that were likely to play a role in skeletal muscle differentiation. Four of the TF-binding sites enriched in the MEF2-sensitive gene sets bind factors that may play a role in skeletal muscle differentiation. In the MEF2A sensitive module, the distal-less homeobox transcription factors Dlx1–6 were among the most significantly enriched TFs. Dlx factors are important for limb and craniofacial development. Although few studies exist on their role in muscle development, a related family member, Msx1, is associated with maintenance of an undifferentiated state during migration of proliferative skeletal muscle precursors (25). Another significantly enriched MEF2A-associated TF module identified Zfhx3/Atbf1, a well characterized inhibitor of myogenic differentiation. It acts primarily as a transcriptional repressor through the obstruction of E-box motifs on muscle-specific promoters. Although Zfhx3 functions as a repressor, one splice variant, Zfhx3-B, acts as a transcriptional activator that contributes to myogenic differentiation (31).
Two families of transcription factors were associated with MEF2D-sensitive genes in our de novo motif analysis. Meis1 and Pknox2 are members of a small family of transcription factors that function in skeletal muscle as pioneer transcription factors to stabilize the MyoD/E12 DNA interaction on suboptimal E box sites, such as the E box found on the myogenin promoter (32–34). Although little is known about the function of Pknox2, it is similar in homology to Meis1 and is highly expressed in skeletal muscle tissue (35).
The final transcription factor associated with MEF2D-sensitive genes is Gtf2ird1/Gtf3. Gtf3 shares high homology with TFII-1, suggesting an important role in the integration of muscle-specific transcription factor activity and the general transcriptional machinery of the cell (36). Gtf3 has 23 known splice variants, 11 of which are only detectable in skeletal muscle. Interestingly, the human Gtf3 ortholog is localized to a region of the genome that is deleted in Williams-Beuren syndrome, a disease associated with muscle weakness and atrophy. Of the genes deleted in this region, Gtf3 is the only one associated with skeletal muscle function, suggesting an important role for Gtf3 activity in human skeletal muscle (37).
The aforementioned transcription factors represent interesting candidates to further investigate the mechanisms of MEF2 isoform-specific transcriptional regulation. Although we highlighted transcription factors that may function in muscle, the computational analysis suggests that specificity in gene regulation by MEF2 isoforms is primarily established by unique combinations of broadly expressed transcription factors. The potential role of these transcription factors in muscle and their ability to mediate specificity to MEF2 isoform-dependent gene programs remains to be seen.
DISCUSSION
The molecular mechanisms by which MEF2 controls gene expression and how its activity is regulated by signaling pathways in muscle have been intensely investigated. Previous studies have used dominant negative and genome wide ChIP approaches to report the role of MEF2 in skeletal myoblast differentiation in vitro (38–40). However, these studies did not explore potential differences in MEF2 isoforms, and a carefully designed systematic analysis into the distinct transcriptional functions of the individual vertebrate MEF2 proteins in this process has not been performed. Here, we have used acute isoform-specific knockdown of the four mammalian MEF2 proteins in a well defined muscle differentiation context followed by genome-wide expression profiling to examine the requirement of each MEF2 isoform and its downstream pathways in this process.
Individual knockdown of mammalian MEF2 family members in C2C12 cells revealed three key findings. First, that MEF2A is absolutely required for C2C12 differentiation and that the remaining MEF2 proteins, MEF2B, -C, and -D, were dispensable for this process. Second, that there was a surprisingly broad spectrum in the number of genes sensitive to a given MEF2 isoform. Third, although depletion of MEF2B, -C, or -D either individually, in combination, or altogether did not cause an overt phenotype, we discovered that deficiency of individual MEF2 isoforms resulted in significant gene dysregulation. Interestingly, many of these genes did not overlap and were sensitive to a particular mammalian MEF2 isoform.
Along these lines, our transcriptome results show varying degrees of overlap with previously published MEF2 ChIP studies. Using a custom CpG island array and a pan-MEF2 antibody, 20 genes were identified in C2C12 cells whose promoters were enriched for MEF2 binding (39). Of these, only nine overlapped with our microarray data and were found exclusively in the MEF2A gene set, suggesting that they may be direct targets of MEF2A. Another study used a standardized microarray 4 days postdifferentiation (C2C12 cells) to identify 22 genes enriched for MEF2 binding (40), of which 16 were significantly dysregulated in our study. These 16 genes were distributed among the MEF2 knockdown gene sets. A more recent study (8) looked at the binding enrichment of two MEF2D splice variants in C2C12 cells using ChIP sequencing. These assays yielded 160 genes with significant MEF2D binding on differentiation day 5, of which 126 were found in our MEF2 knockdown gene sets. Curiously, we found many of their MEF2D-specific genes to be sensitive to the knockdown of other MEF2 factors. These results suggest that genes bound by MEF2D may also be regulated by other MEF2 isoforms, depending on context or are regulated by specific combinations of MEF2D heterodimers. Finally, while this work was in revision, an RNA sequencing analysis of MEF2A siRNA knockdown in C2C12 myotubes (day 2) was performed (41). Of the 1,207 dysregulated genes described in that study, ∼50% of the genes were found to overlap with our MEF2A dysregulated gene set. The relatively modest overlap of these gene sets may reflect the different time points, knockdown method, or procedure used to analyze gene expression profiles.
Overexpression of MEF2C, MEF2D, or both in the MEF2A-deficient C2C12 background was unable to rescue the impaired differentiation defect in these myoblasts. However, the MEF2-VP16 adenovirus was able to restore myotube formation and expression of MHC to wild type levels. This difference may be explained by the ability of the MEF2-VP16 protein to function as a potent transcriptional activator and overcome any encoded specificity within the regulatory regions of MEF2A-dependent genes. Additionally, because MEF2-VP16 contains the MEF2C DNA-binding domain, which is highly conserved among the mammalian MEF2 proteins and has only a single amino acid difference between this and the MEF2A DNA-binding domain, these results suggest that target gene selectivity is directed by the carboxyl-terminal transactivation domain of the MEF2 proteins. Therefore, an interesting area of investigation in the future would be to generate chimeric MEF2 proteins harboring different combinations of the DNA-binding and carboxyl-terminal domains to determine whether any of these MEF2 variants are capable of rescuing the differentiation defect or altering target gene preferences.
Given the profound differentiation defect in MEF2A-deficient C2C12 cells, we searched the MEF2A dysregulated gene set for genes that may help explain the phenotype. Although the MEF2A gene set has numerous dysregulated genes that may explain the phenotype, we were intrigued by four of these genes. The first gene that caught our attention was MyoD. MyoD is a master muscle regulatory transcription factor in myogenic differentiation, and among its myriad roles in muscle gene regulation, its knockdown prevents myoblast differentiation (42). We also noted the down-regulation of myoferlin (Myof). Myoferlin is responsible for sarcolemmal repair at the site of myoblast fusion, and a deficiency prevents myoblast fusion (43). The final two gene candidates, palladin (Palld) and formin-binding protein (Fnbp1l), are actin-associating remodeling molecules that play integral roles in actin-mediated signaling required for myogenic differentiation and myotube fusion (44, 45). Loss of either of these proteins leads to a fusion deficient phenotype in C2C12 differentiation. Although expression of none of these genes is completely abrogated in the MEF2A knockdown, the collective down-regulation of these genes may contribute to defective C2C12 differentiation.
The cellular pathways dependent on individual MEF2 isoforms in C2C12 myoblasts were found to vary considerably, which is not surprising given the extent of nonoverlapping genes. MEF2A was predicted to regulate genes involved in actin cytoskeletal rearrangement, consistent with its regulation of structural genes encoding proteins localized to the costamere in cardiac muscle (15, 46). It is intriguing that regulation of the actin cytoskeleton was not identified among the top canonical pathways in the MEF2B, -C, or -D dysregulated gene sets, considering these MEF2 proteins have been shown to regulate muscle structural genes in vitro and in vivo (46). Although expression profiling clearly identified this category of genes in all of the MEF2 knockdowns, other pathways were found to be more significantly dysregulated in the MEF2B, -C, or -D gene sets. Based on the extensive overlap of the MEF2B, -C, and -D dysregulated genes, it appears that regulation of muscle structural genes can be compensated for by any one of the remaining MEF2 proteins and/or that regulation of these genes is more dependent on MEF2A. Given the importance of cytoskeletal proteins in muscle function, it is possible that these genes have evolved less stringent transcriptional mechanisms relating to their regulation by the various MEF2 isoforms.
The computational analysis of predicted MEF2-binding sites in each of the MEF2-sensitive dysregulated gene sets did not identify any significant differences in this consensus sequence. This is not entirely surprising because the binding to different MEF2 consensus sequences in an isoform-specific manner has not been reported. One caveat of using FIMO to identify MEF2 cis-acting sequences is that this algorithm is based exclusively on published MEF2-binding sites, thereby restricting the MEF2 motif analysis to this limited data set. Nevertheless, the computational analysis revealed a significant difference in the transcription factor modules of overrepresented DNA-binding sites associated with each of the various MEF2 isoform gene sets. An interesting finding that emerged in this analysis is the observation that not a single, predicted transcription factor-binding site motif was uniquely associated with the MEF2C-sensitive cohort. This likely resulted from complete overlap with analogous motifs found in the other MEF2 isoform-regulated genes. These findings suggest that mechanisms beyond distinct co-regulators in the proximal promoter are necessary for MEF2C-dependent gene regulation in skeletal muscle. Perhaps transcription of MEF2C-sensitive genes by MEF2C requires a specific arrangement of common DNA-binding site motifs rather than unique sequences in these regulatory regions.
Another intriguing finding was the lack of E-box motifs, which are recognized by basic helix-loop-helix proteins such as MyoD. It is firmly established that MEF2 and the MyoD family form an important cooperative interaction in the regulation of muscle genes (47). In our computational analysis, E-box motifs were actually found to be significantly underrepresented. Our interpretation of this result is that E-box motifs are prevalent throughout the mouse genome (48) and consequently, using proximal promoters as the background from which motifs are subtracted, result in significantly reduced frequency of this binding site in the MEF2-sensitive gene sets.
Our findings reveal isoform-specific differences in the regulation of genes by the mammalian MEF2 transcription factors in C2C12 differentiation. It is important to note, although C2C12 myoblasts are a widely accepted in vitro model of skeletal muscle differentiation, that the MEF2-dependent transcriptome in these cells may differ from that in primary skeletal myoblasts. Additionally, our computational data support the model whereby MEF2-dependent transcriptional regulation is modulated by co-factor interactions and not through differences in the MEF2 consensus sequence. The present study has taken this notion a step further by demonstrating that the target gene selectivity of a given MEF2 isoform may be dictated by one or more distinct co-regulatory partners. Our results pave the way for future studies of MEF2-dependent regulation of isoform-sensitive genes in muscle by focusing on the in vivo occupancy of specific dimeric combinations of MEF2 proteins in the context of chromatin.
Supplementary Material
Acknowledgments
We thank members of the Naya laboratory for critical reading of the manuscript. We are grateful to Jeff Molkentin (Cincinnati Children's Hospital) for providing the MEF2 overexpression adenoviruses and to Tod Gulick (Sanford Burnham Medical Research Institute) for the MEF2-FLAG expression plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grant HL73304 (to F. J. N.) and diversity supplement HL73304-S1 (to N. L. E.). This work was also supported by funds from the Beckman Scholars Program (to S. E. N.), a Boston University Undergraduate Research Opportunities Program (UROP) fellowship (to Y. M.), and Boston University Bioinformatics Clinical and Translational Science Institute Grant U54-TR001012.
Microarray data are available in GEO(NCBI) with accession number GSE63798.

This article contains supplemental Tables S1–S3.
- MEF2
- myocyte enhancer factor 2
- IPA
- Ingenuity Pathway Analysis
- TF
- transcription factor
- FIMO
- Find Individual Motif Occurrences
- MEME
- Multiple Em for Motif Elicitation.
REFERENCES
- 1. Buckingham M., Rigby P. W. (2014) Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell 28, 225–238 [DOI] [PubMed] [Google Scholar]
- 2. Braun T., Gautel M. (2011) Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol. 12, 349–361 [DOI] [PubMed] [Google Scholar]
- 3. Black B. L., Olson E. N. (1998) Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 14, 167–196 [DOI] [PubMed] [Google Scholar]
- 4. Potthoff M. J., Olson E. N. (2007) MEF2: a central regulator of diverse developmental programs. Development. 134, 4131–4140 [DOI] [PubMed] [Google Scholar]
- 5. Sandmann T., Jensen L. J., Jakobsen J. S., Karzynski M. M., Eichenlaub M. P., Bork P., Furlong E. E. (2006) A temporal map of transcription factor activity: Mef2 directly regulates target genes at all stages of muscle development. Dev. Cell 10, 797–807 [DOI] [PubMed] [Google Scholar]
- 6. Edmondson D. G., Lyons G. E., Martin J. F., Olson E. N. (1994) Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 120, 1251–1263 [DOI] [PubMed] [Google Scholar]
- 7. Zhu B., Ramachandran B., Gulick T. (2005) Alternative pre-mRNA splicing governs expression of a conserved acidic transactivation domain in myocyte enhancer factor 2 factors of striated muscle and brain. J. Biol. Chem. 280, 28749–28760 [DOI] [PubMed] [Google Scholar]
- 8. Sebastian S., Faralli H., Yao Z., Rakopoulos P., Palii C., Cao Y., Singh K., Liu Q. C., Chu A., Aziz A., Brand M., Tapscott S. J., Dilworth F. J. (2013) Tissue-specific splicing of a ubiquitously expressed transcription factor is essential for muscle differentiation. Genes Dev. 27, 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Potthoff M. J., Arnold M. A., McAnally J., Richardson J. A., Bassel-Duby R., Olson E. N. (2007) Regulation of skeletal muscle sarcomere integrity and postnatal muscle function by Mef2c. Mol. Cell Biol. 27, 8143–8151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Potthoff M. J., Wu H., Arnold M. A., Shelton J. M., Backs J., McAnally J., Richardson J. A., Bassel-Duby R., Olson E. N. (2007) Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J. Clin. Invest. 117, 2459–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Snyder C. M., Rice A. L., Estrella N. L., Held A., Kandarian S. C., Naya F. J. (2013) MEF2A regulates the Gtl2-Dio3 microRNA mega-cluster to modulate WNT signaling in skeletal muscle regeneration. Development 140, 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seok H. Y., Tatsuguchi M., Callis T. E., He A., Pu W. T., Wang D. Z. (2011) miR-155 inhibits expression of the MEF2A protein to repress skeletal muscle differentiation. J. Biol. Chem. 286, 35339–35346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu N., Nelson B. R., Bezprozvannaya S., Shelton J. M., Richardson J. A., Bassel-Duby R., Olson E. N. (2014) Requirement of MEF2A, C, and D for skeletal muscle regeneration. Proc. Natl. Acad. Sci. U.S.A. 111, 4109–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McCalmon S. A., Desjardins D. M., Ahmad S., Davidoff K. S., Snyder C. M., Sato K., Ohashi K., Kielbasa O. M., Mathew M., Ewen E. P., Walsh K., Gavras H., Naya F. J. (2010) Modulation of angiotensin II mediated cardiac remodeling by the MEF2A target gene Xirp2. Circ. Res. 106, 952–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ewen E. P., Snyder C. M., Wilson M., Desjardins D., Naya F. J. (2011) The Mef2A transcription factor coordinately regulates a costamere gene program in cardiac muscle. J. Biol. Chem. 286, 29644–29653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang D. W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 17. Huang D. W., Sherman B. T., Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grant C. E., Bailey T. L., Noble W. S. (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics. 27, 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crooks G. E., Hon G., Chandonia J.-M., Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Quandt K., Frech K., Karas H., Wingender E., Werner T. (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23, 4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ramachandran B., Yu G., Li S., Zhu B., Gulick T. (2008) Myocyte enhancer factor 2A is transcriptionally autoregulated. J. Biol. Chem. 283, 10318–10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu J., Gong N. L., Bodi I., Aronow B. J., Backx P. H., Molkentin J. D. (2006) Myocyte enhancer factors 2A and 2C induce dilated cardiomyopathy in transgenic mice. J. Biol. Chem. 281, 9152–9162 [DOI] [PubMed] [Google Scholar]
- 23. Buettner F. F., Ashikov A., Tiemann B., Lehle L., Bakker H. (2013) C. elegans DPY-19 is a C-mannosyltransferase glycosylating thrombospondin repeats. Mol. Cell 50, 295–302 [DOI] [PubMed] [Google Scholar]
- 24. Watanabe K., Takebayashi H., Bepari A. K., Esumi S., Yanagawa Y., Tamamaki N. (2011) Dpy19l1, a multi-transmembrane protein, regulates the radial migration of glutamatergic neurons in the developing cerebral cortex. Development 138, 4979–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bendall A. J., Abate-Shen C. (2000) Roles for Msx and Dlx homeoproteins in vertebrate development. Gene 247, 17–31 [DOI] [PubMed] [Google Scholar]
- 26. Mayer U. (2003) Integrins: redundant or important players in skeletal muscle? J. Biol. Chem. 278, 14587–14590 [DOI] [PubMed] [Google Scholar]
- 27. Sorimachi H., Ono Y. (2012) Regulation and physiological roles of the calpain system in muscular disorders. Cardiovasc. Res. 96, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Messina D. N., Glasscock J., Gish W., Lovett M. (2004) An ORFeome-based analysis of human transcription factor genes and the construction of a microarray to interrogate their expression. Genome Res. 14, 2041–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu J., Stormo G. D. (2008) Context-dependent DNA recognition code for C2H2 zinc-finger transcription factors. Bioinformatics 24, 1850–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Svingen T., Tonissen K. F. (2006) Hox transcription factors and their elusive mammalian gene targets. Heredity 97, 88–96 [DOI] [PubMed] [Google Scholar]
- 31. Berry F. B., Miura Y., Mihara K., Kaspar P., Sakata N., Hashimoto-Tamaoki T., Tamaoki T. (2001) Positive and negative regulation of myogenic differentiation of C2C12 cells by isoforms of the multiple homeodomain zinc finger transcription factor ATBF1. J. Biol. Chem. 276, 25057–25065 [DOI] [PubMed] [Google Scholar]
- 32. Heidt A. B., Rojas A., Harris I. S., Black B. L. (2007) Determinants of myogenic specificity within MyoD are required for noncanonical E box binding. Mol. Cell. Biol. 27, 5910–5920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grade C. V., Salerno M. S., Schubert F. R., Dietrich S., Alvares L. E. (2009) An evolutionarily conserved Myostatin proximal promoter/enhancer confers basal levels of transcription and spatial specificity in vivo. Dev. Genes Evol. 219, 497–508 [DOI] [PubMed] [Google Scholar]
- 34. Knoepfler P. S., Bergstrom D. A., Uetsuki T., Dac-Korytko I., Sun Y. H., Wright W. E., Tapscott S. J., Kamps M. P. (1999) A conserved motif N-terminal to the DNA-binding domain of myogenic bHLH transcription factors mediates cooperative DNA binding with Pbx-Meis1/Prep1. Nucleic Acids Res. 27, 3752–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Imoto I., Sonoda I., Yuki Y., Inazawa J. (2001) Identification and characterization of human PKNOX2, a novel homeobox-containing gene. Biochem. Biophys. Res. Commun. 287, 270–276 [DOI] [PubMed] [Google Scholar]
- 36. O'Mahoney J. V., Guven K. L., Lin J., Joya J. E., Robinson C. S., Wade R. P., Hardeman E. C. (1998) Identification of a novel slow-muscle-fiber enhancer binding protein, MusTRD1. Mol. Cell Biol. 18, 6641–6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tassabehji M., Carette M., Wilmot C., Donnai D., Read A. P., Metcalfe K. (1999) A transcription factor involved in skeletal muscle gene expression is deleted in patients with Williams syndrome. Eur. J. Hum. Genet. 7, 737–747 [DOI] [PubMed] [Google Scholar]
- 38. Ornatsky O. I., Andreucci J. J., McDermott J. C. (1997) A dominant-negative form of transcription factor MEF2 inhibits myogenesis. J. Biol. Chem. 272, 33271–33278 [DOI] [PubMed] [Google Scholar]
- 39. Paris J., Virtanen C., Lu Z., Takahashi M. (2004) Identification of MEF2-regulated genes during muscle differentiation. Physiol. Genomics 20, 143–151 [DOI] [PubMed] [Google Scholar]
- 40. Blais A., Tsikitis M., Acosta-Alvear D., Sharan R., Kluger Y., Dynlacht B. D. (2005) An initial blueprint for myogenic differentiation. Genes Dev. 19, 553–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wales S., Hashemi S., Blais A., McDermott J. C. (2015) Global MEF2 target gene analysis in cardiac and skeletal muscle revealsnovel regulation of DUSP6 by p38MAPK-MEF2 signaling. Nucleic Acids Res. 42, 11349–11362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Megeney L. A., Kablar B., Garrett K., Anderson J. E., Rudnicki M. A. (1996) MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 10, 1173–1183 [DOI] [PubMed] [Google Scholar]
- 43. Doherty K. R., Cave A., Davis D. B., Delmonte A. J., Posey A., Earley J. U., Hadhazy M., McNally E. M. (2005) Normal myoblast fusion requires myoferlin. Development 132, 5565–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nguyen N. U., Liang V. R., Wang H.-V. (2014) Actin-associated protein palladin is required for migration behavior and differentiation potential of C2C12 myoblast cells. Biochem. Biophys. Res. Commun. 452, 728–733 [DOI] [PubMed] [Google Scholar]
- 45. George B., Jain N., Fen Chong P., Hou Tan J., Thanabalu T. (2014) Myogenesis defect due to Toca-1 knockdown can be suppressed by expression of N-WASP. Biochim. Biophys. Acta 1843, 1930–1941 [DOI] [PubMed] [Google Scholar]
- 46. Estrella N. L., Naya F. J. (2014) Transcriptional networks regulating the costamere, sarcomere, and other cytoskeletal structures in striated muscle. Cell Mol. Life Sci. 71, 1641–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Molkentin J. D., Olson E. N. (1996) Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc. Natl. Acad. Sci. U.S.A. 93, 9366–9373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cao Y., Yao Z., Sarkar D., Lawrence M., Sanchez G. J., Parker M. H., MacQuarrie K. L., Davison J., Morgan M. T., Ruzzo W. L., Gentleman R. C., Tapscott S. J. (2010) Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell 18, 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.