Abstract
A detailed understanding of the processes controlling adipogenesis is instrumental in the fight against the obesity epidemic. Adipogenesis is controlled by a transcriptional cascade composed of a large number of transcriptional factors, among which CCAAT/enhancer-binding protein (C/EBP) β plays an essential role. During 3T3-L1 adipocyte differentiation, C/EBPβ is induced early to transactivate the expression of C/EBPα and peroxisome proliferator-activated receptor γ (PPARγ), two master transcription factors for terminal adipocyte differentiation. Studies in recent years have revealed many new target genes of C/EBPβ, implicating its participation in many other processes during adipogenesis, such as mitotic clonal expansion, epigenetic regulation, unfolded protein response, and autophagy. Moreover, the function of C/EBPβ is highly regulated by post-translational modifications, which are crucial for the proper activation of the adipogenic program. Advances toward elucidation of the function and roles of the post-translational modification of C/EBPβ during adipogenesis will greatly improve our understanding of the molecular mechanisms governing adipogenesis.
Keywords: adipogenesis, Autophagy, CCAAT-Enhancer-binding Protein (C/EBP), Cell Proliferation, Epigenetics, Post-translational Modification (PTM), Unfolded Protein Response (UPR)
Introduction
Adipose tissue is not only a key depot for energy storage but is also involved in the dynamic regulation of metabolism (1). The upsurge of adipose tissue mass plays a central role in obesity-related complications such as type 2 diabetes, hypertension, hyperlipidemia, and arteriosclerosis (2). Both the increase of adipocyte size (hypertrophy) and the increase of adipocyte number (hyperplasia) are major contributors to the development of obesity (3). Thus, a tight control of adipocyte development and function is critical in maintaining whole body energy homeostasis, and a full understanding of the mechanisms regulating adipose formation would provide precious information on the way to control of obesity.
Much of our knowledge of adipocyte differentiation has been obtained by studying adipocyte cell culture models. The 3T3-L1 cell line is one of the best studied cellular models (4). Upon the treatment with differentiation inducers (a combination of 3-isobutyl-1-methylxanthine, dexamethasone, and insulin), growth-arrested 3T3-L1 preadipocytes re-enter the cell cycle, a process referred to as mitotic clonal expansion (MCE),2 which contributes to the hyperplasia of adipocytes. The adipogenic gene expression program is initiated during and after 2–3 rounds of MCE, ultimately leading to terminal adipocyte differentiation (5).
The adipogenic program requires a cascade of multiple transcription factors (6), among which is CCAAT/enhancer-binding protein β (C/EBPβ), an important transcriptional factor belonging to the leucine zipper family. Knockdown of C/EBPβ in 3T3-L1 preadipocytes blocks adipogenesis (7, 8), whereas its overexpression is sufficient to induce 3T3-L1 adipocyte differentiation without the hormonal inducers normally required (9). The functional importance of C/EBPβ during adipocyte development has also been demonstrated in vivo. Disruption of the C/EBPβ gene in mice caused decreased fat mass because of impaired development of adipose tissue (10). Thus, C/EBPβ plays a crucial role during adipocyte differentiation.
As an important early factor of adipogenesis, C/EBPβ is induced rapidly after the addition of adipogenic stimuli and is responsible for inducing the expression of C/EBPα and peroxisome proliferator-activated receptor γ (PPARγ), two master adipogenic transcription factors, by binding to their promoters (11). In this way, C/EBPβ promotes terminal adipocyte differentiation. Besides the abovementioned function, a number of studies have illuminated additional roles of C/EBPβ during adipogenesis, through its transcriptional regulation of many new target genes. Furthermore, the function of C/EBPβ is elaborately regulated by post-translational modifications (PTMs), including phosphorylation, acetylation, methylation, O-GlcNAcylation, ubiquitination, and SUMOlation. Herein, the new functions and PTMs of C/EBPβ during adipogenesis will be reviewed.
Role and Mechanism of C/EBPβ in Mitotic Clonal Expansion
Despite some controversy (12), multiple studies indicate that MCE is a necessary step for the terminal adipocyte differentiation of 3T3-L1 preadipocytes. The extracellular signal-regulated kinase kinase (MEK) inhibitor U0126 and cyclin-dependent kinase inhibitor roscovitine, which inhibit the cell cycle at different points, block MCE as well as adipogenesis (5, 13). The DNA synthesis inhibitor aphidicolin and the anti-proliferation reagent rapamycin also block MCE and 3T3-L1 preadipocyte differentiation (14, 15). Moreover, knockdown of histone acetyltransferase binding to ORC1 (HBO1), a positive regulator for the initiation of DNA replication, impairs the ability of 3T3-L1 preadipocytes to differentiate into mature adipocytes by inhibiting DNA replication and MCE (16). It is hypothesized that DNA replication during MCE increases the accessibility of promoter or enhancer elements to factors required for transcription of genes involved in the initiation of differentiation (17).
Several lines of evidence have shown that C/EBPβ is involved in MCE. When subjected to the same differentiation protocol as 3T3-L1 preadipocytes, a subset of mouse embryo fibroblasts undergoes MCE and terminal differentiation into adipocytes. Mouse embryo fibroblasts from C/EBPβ−/− mice, however, neither undergo MCE nor differentiate into adipocytes (5). Furthermore, knockdown of C/EBPβ by siRNA in 3T3-L1 preadipocytes prevents MCE as well as adipocyte differentiation (7). Additionally, overexpression of a dominant-negative C/EBPβ (A-C/EBP) that blocks C/EBPβ DNA binding activity by dimerizing through its leucine zipper (18) also disrupts MCE and adipogenesis in 3T3-L1 cells (19). Intriguingly, C/EBPβ takes part in the proliferation of certain other cell types such as lobuloalveolar cells, osteoblasts, and keratinocytes (20–22), further supporting an important role of C/EBPβ in cell proliferation.
To understand how C/EBPβ promotes MCE, a promoter-wide ChIP-on-chip analysis combined with gene expression microarrays was performed to identify the potential target genes of C/EBPβ at the early stage of 3T3-L1 adipocyte differentiation (8). Four cell cycle genes (Cdc45l, Mcm3, Gins1, and Cdc25c) and the chromatin assembly gene histone H4 were identified as C/EBPβ target genes. Mcm3 is a component of MCM2–7 (mini-chromosome maintenance proteins 2–7) complex, whereas Gins1 is a subunit of GINS (go-ichi-ni-san) complex. Cdc45l, MCM2–7, and GINS form a large complex referred to as CMG, which is involved in the regulation of eukaryotic chromosomal DNA replication (23). Cdc25c is a phospho-tyrosine phosphatase that contributes to S-phase and M-phase entry of the cells (24). Histone H4 is the most highly conserved and strictly cell cycle-regulated nucleosomal protein critical for normal progression of S phase (7). Knockdown of these four cell cycle genes and histone H4 significantly impaired MCE, whereas ectopic expression of these genes together significantly reverses the inhibitory effect of C/EBPβ siRNA on MCE, indicating that these genes are important downstream effectors of C/EBPβ to promote MCE (8).
Growing lines of evidence have indicated that epigenetics play an essential role in adipogenesis (25–27). Kdm4b is a Jmjc-domain-containing histone demethylase for H3K9me3 (28). Studies have shown that Kdm4b is required for estrogen receptor α (ERα)-regulated breast cancer progression and mammary epithelial cell proliferation (29). Interestingly, Kdm4b has been shown to be required for MCE (8). It is identified as a target gene of C/EBPβ and functions as a co-factor of C/EBPβ to demethylate H3K9me3 in the regulatory regions of C/EBPβ-regulated cell cycle genes and chromatin assembly gene as mentioned above, thereby promoting their expression and MCE (8). Thus, a profound role for C/EBPβ in the epigenetic control is revealed, suggesting a novel feed forward mechanism involving C/EBPβ and Kdm4b in the regulation of MCE. It should be noted that the function of C/EBPβ and Kdm4b is specific to MCE because knockdown of C/EBPβ or Kdm4b neither affects the expression of these cell cycle genes and histone H4 nor impairs cell proliferation in pre-confluent 3T3-L1 preadipocytes (8). It is possible that additional co-factors or a specific PTM of C/EBPβ, which might be absent in pre-confluent 3T3-L1 cells, are required for C/EBPβ and Kdm4b to ensure MCE. In addition, it is noteworthy that deletion of C/EBPβ results in retarded proliferation of mammary epithelial cells and severe inhibition of lobuloalveolar development (20), which is similar to the phenotype of conditional deletion of Kdm4b in mammary epithelium (8). Therefore, it would be interesting to investigate the potential collaboration between C/EBPβ and Kdm4b during mammary gland development.
C/EBPβ activates the expression of PPARγ and C/EBPα by directly binding to their promoters. Although C/EBPβ is induced very early in adipocyte differentiation, the expression of PPARγ and C/EBPα occurs much later (11). This lag appears necessary because PPARγ and C/EBPα are both anti-mitotic, and their premature expression would otherwise prevent MCE, a required step for adipocyte differentiation. G9a is an important euchromatic methyltransferase that is responsible for the majority of H3K9me2 in the cells (30). Recent evidence suggests that G9a-mediated H3K9me2 mainly associates with transcriptional silencing (31). G9a plays important roles in various biological processes and has been shown to be a repressor of adipogenesis (27). In 3T3-L1 cells, a transient induction of G9a by C/EBPβ was detected during MCE (32). Then, G9a inhibited PPARγ and C/EBPα expression through H3K9 dimethylation of their promoters. Hence, C/EBPβ up-regulates G9a that delays the transactivation of PPARγ and C/EBPα so as to guarantee MCE, providing another line of evidence for the participation of C/EBPβ in epigenetic regulation.
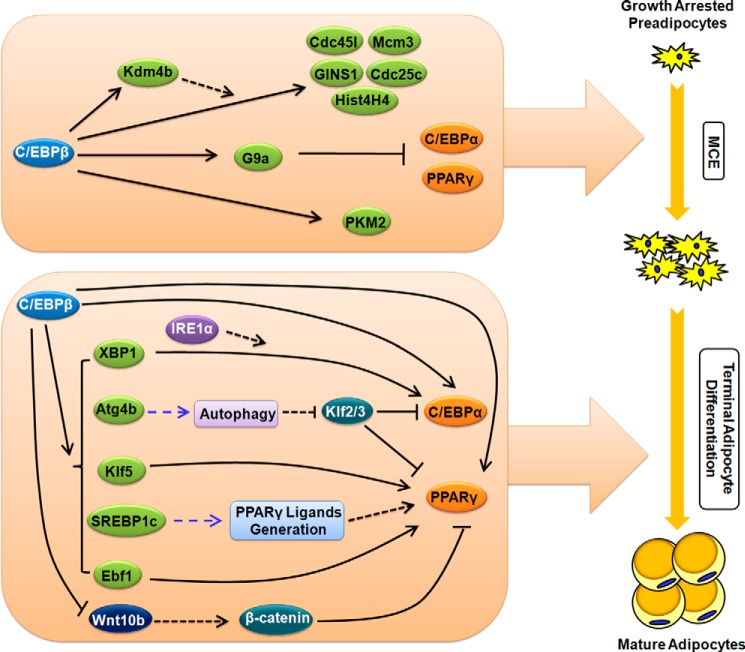
The embryonic M2 isoform of pyruvate kinase (PKM2) has attracted much attention because of its critical role in aerobic glycolysis of tumor cells, namely the Warburg effect (33). Instead of PKM1, tumor cells commonly express PKM2, which may contribute to the metabolism shift from oxidative phosphorylation to aerobic glycolysis and tumorigenesis (34). Besides, PKM2 has also been reported to promote tumor growth via regulating cell cycle progression and oncogene expression (35, 36). Of interest, PKM2 expression is elevated during the early stage of 3T3-L1 adipogenesis, and knockdown of PKM2 compromises MCE (37). Further studies, however, are needed to investigate the mechanism of PKM2 in MCE. Importantly, PKM2 is identified as a target gene of C/EBPβ during MCE (37). Consequently, transactivation of PKM2 by C/EBPβ contributes to facilitating MCE. Collectively, these findings (Fig. 1) provide new clues to understanding the action of C/EBPβ in the proliferation of certain specific cell types.
FIGURE 1.
Multiple roles of C/EBPβ during adipogenesis. Besides its well known function in the direct transactivation of C/EBPα and PPARγ, many new roles of C/EBPβ during adipogenesis have been revealed in the past decade. At the early stage of 3T3-L1 adipocyte differentiation, C/EBPβ transactivates the expression of multiple cell cycle-related genes to facilitate MCE, a required step for terminal adipocyte differentiation. A novel feed forward mechanism involving C/EBPβ and Kdm4b in the regulation of MCE is illustrated. Moreover, C/EBPβ transiently transactivates the expression of G9a, which delays the expression of C/EBPα and PPARγ, two anti-proliferation factors, so as to ensure MCE. The transactivation of Kdm4b (a histone demethylase) and G9a (a histone methyltransferase) by C/EBPβ provides evidence for the epigenetic control of MCE by C/EBPβ. At the late stage of 3T3-L1 adipocyte differentiation, C/EBPβ is involved in the activation of UPR and autophagy, through the transactivation of Xbp1 and Atg4b, respectively. In addition, C/EBPβ activates the expression of some other transcriptional factors and inhibits the expression of Wnt10b, an anti-adipogenic factor. Together, these effects ultimately lead to the activation or up-regulation of C/EBPα and PPARγ, thereby promoting terminal adipocyte differentiation. Black solid lines with arrowheads or blunt ends indicate transcriptional regulation of gene expression. Black dashed lines with arrowheads indicate promotion of activity. A black dashed line with a blunt end indicates inhibition of protein stability. Blue dashed lines with arrowheads indicate promotion of biological processes.
Role of C/EBPβ in Terminal Adipocyte Differentiation
C/EBPβ is an important factor to initiate the transcriptional cascades that culminate in the expression of two essential adipogenic factors, PPARγ and C/EBPα (38). Apart from its well established role in activating the expression of PPARγ and C/EBPα, studies in recent years have brought to light a number of new targets of C/EBPβ, which extends our knowledge of its role in terminal adipocyte differentiation.
Unfolded protein response (UPR) is a complex signaling cascade activated by the perturbations in endoplasmic reticulum homeostasis to coordinate multiple signaling pathways and control a variety of physiologies (39). Among the three branches of UPR, the inositol-requiring enzyme 1α (IRE1α)/X-box binding protein 1 (XBP1) pathway, which plays a crucial role in glucose and lipid metabolism as well as in insulin function, is the most conserved branch (40). Because dramatic transformations take place during the differentiation from preadipocytes to mature adipocytes, it is hypothesized that adipocytes might exhibit increased level of UPR so as to relieve the stress burden on the endoplasmic reticulum imposed by the increased biosynthesis of protein and lipids (41). A recent study demonstrates that adipogenesis is associated with the increase of UPR and that the IRE1a-XBP1 pathway is indispensable for adipogenesis (42). Knockdown of IRE1α or XBP1 in 3T3-L1 cells significantly inhibits adipogenesis, and XBP1 could directly transactivate the expression of C/EBPα, a master gene of adipogenesis, to promote adipocyte differentiation. Intriguingly, C/EBPβ is responsible for the induction of XBP1 by binding to its proximal promoter region (42). Thus, through regulating the expression of XBP1, C/EBPβ participates in the activation of UPR, a required process for adipogenesis.
Autophagy is a cellular process that delivers cytosolic components to lysosomes for degradation (43). It is involved in a variety of physiological and pathophysiological processes, such as nutrient starvation, immune responses, tumor suppression, cell death, and so on (44). Recent studies have demonstrated that autophagy is required for cell differentiation of certain cell types, including adipocyte differentiation (45, 46). Autophagy was induced during adipogenesis, promoting the degradation of Klf2 and Klf3, two negative regulators of adipocyte differentiation, which is mediated by the adaptor protein p62/SQSTM1 (47). In 3T3-L1 cells, C/EBPβ has been identified as an activator of autophagy through the transactivation of Atg4b, an important autophagy gene that exposes glycine from LC3 precursor at its C terminus to form LC3-I and is essential for autophagosome formation (47). Of interest, C/EBPβ has been shown to regulate circadian autophagy rhythm in the liver (48). These findings highlight an important role of C/EBPβ in controlling the program of autophagy gene expression during some biological processes, including adipogenesis.
Many other target genes of C/EBPβ have been reported and shown to be important for terminal adipocyte differentiation. For instance, C/EBPβ transactivates the expression of Klf5, sterol-responsive element-binding protein 1c (SREBP1c), and early B-cell factor 1 (Ebf1). Klf5 is a key transcription factor for adipogenesis through promoting PPARγ expression (49). SREBP1c is an important pro-adipogenic transcriptional factor that regulates the expression of many lipid metabolism genes and contributes to the generation of endogenous PPARγ ligands (50). Ebf1 promotes adipogenesis by activating PPARγ transcription (51). On the other hand, C/EBPβ is involved in suppression of Wnt/β-catenin signaling through transcriptional inhibition of the expression of Wnt10b, a major Wnt ligand that inhibits adipogenesis (52). Taken together, these findings shed light on the multiple roles of C/EBPβ in terminal adipogenic differentiation (Fig. 1).
Post-translational Modifications (PTMs) of C/EBPβ during Adipogenesis
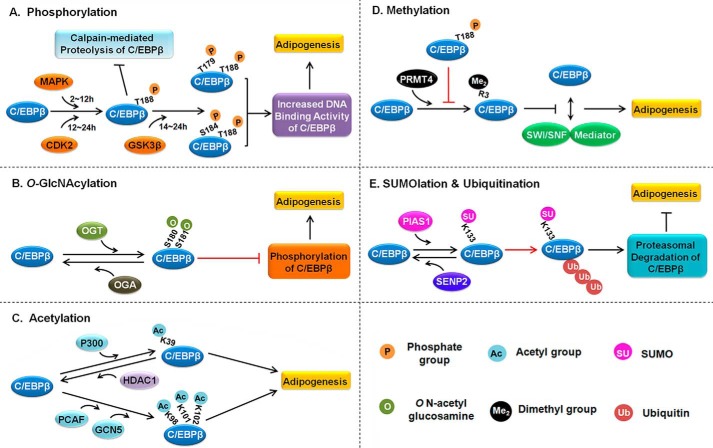
Because of the important role of C/EBPβ in triggering the adipogenic program, it is necessary to gain mechanistic insights into the regulation of C/EBPβ so as to better understand the process controlling adipogenesis. The regulation of C/EBPβ during adipogenesis occurs at multiple levels, including transcriptional regulation, translational regulation, and PTM. Studies on the PTMs of C/EBPβ, including phosphorylation, O-GlcNAcylation, acetylation, methylation, ubiquitination, and SUMOlation, have progressed a lot in recent years, and this progress will be discussed in detail here (Fig. 2). Some of the studies that did not investigate cells during adipogenesis will also be discussed, which might help us better understand the PTMs of C/EBPβ.
FIGURE 2.
The PTMs of C/EBPβ during adipogenesis. A, phosphorylation. C/EBPβ is phosphorylated on Thr-188 by MAPK (2–12 h after adipogenic induction) and by CDK2 (12–24 h after adipogenic induction), followed by GSK3β-mediated phosphorylation on Ser-184 or Thr-179. This dual phosphorylation induces conformational changes in C/EBPβ, which activates its DNA binding and facilitates adipogenesis. B, O-GlcNAcylation. The modification of O-GlcNAc on Ser-180 and Ser-181 of C/EBPβ prevents its phosphorylation on Thr-188, Ser-184, and Thr-179, thus suppressing its DNA binding activity. OGA, β-N-acetylglucosaminidase; OGT, β-N-acetylglucosaminyltransferase. C, acetylation. In general, acetylation of C/EBPβ increases its transcriptional activity to promote adipogenesis. D, methylation. PRMT4/CARM1 dimethylates C/EBPβ on Arg-3, which interferes with the interaction between C/EBPβ and SWI/SNF and inhibits adipogenesis. MAPK/CDK2-mediated phosphorylation on Thr-188 could block PRMT4/CARM1-mediated dimethylation of C/EBPβ on Arg-3. E, SUMOlation and ubiquitination. PIAS1-mediated SUMOlation of C/EBPβ on Lys-133 promotes its ubiquitination and proteasomal degradation, thereby suppressing adipogenesis. SUMO-specific protease SENP2 reverses the SUMOlation of C/EBPβ to promote adipogenesis. The cross-talks between different types of PTMs are indicated by red solid lines with arrowheads or blunt ends. The black solid lines with arrowheads at both ends indicate protein interaction.
Regulation of signaling transduction depends not only on the identity of phospho-sites but also on when phosphorylation events occur. Studies have shown that sequential phosphorylation of C/EBPβ is critical for 3T3-L1 adipocyte differentiation (53). C/EBPβ is expressed rapidly after adipogenic induction (≤4 h) and phosphorylated on Thr-188 by MAPK. After 10–12 h of induction, 3T3-L1 cells re-enter the cell cycle, and the activity of MAPK falls off. When the cells enter S-phase, the rising activity of CDK2/cyclin A keeps maintaining the C/EBPβ phosphorylation on Thr-188 (13). At the onset of S-phase, GSK3β translocates into the nucleus and C/EBPβ is phosphorylated by GSK3β on Ser-184 or Thr-179 (53). Phosphorylation of Thr-188 appears to prime C/EBPβ for the subsequent phosphorylation on Ser-184 or Thr-179. Studies indicate that this dual phosphorylation induces a conformational change in C/EBPβ that allows dimerization through its C-terminal leucine zipper domain. Dimerization brings the adjacent basic regions into position to hold the C/EBP regulatory elements of its target genes in a “scissor-like” grip (54). All these actions of the dual phosphorylated C/EBPβ facilitate its DNA binding and transcriptional regulatory activities. Recent studies showed that phosphorylation also contributes to the stability of C/EBPβ. Both ex vivo and in vitro experiments indicated that phosphorylation on Thr-188 by MAPK or CDK2/cyclin A protected C/EBPβ from calpain-mediated proteolysis (55).
Protein O-GlcNAc glycosylation is a widespread PTM for both nuclear protein and cytoplasmic protein. It is different from the classical glycosylation of secreted proteins and membrane protein, but is similar to phosphorylation on some level. Both O-GlcNAcylation and phosphorylation can take place on serine and threonine residues. It has been demonstrated that C/EBPβ could be modified by O-GlcNAcylation on Ser-180 and Ser-181, which are very close to its phosphorylation sites (Thr-188, Ser-184, Thr-179) (56). Studies have proved that the O-GlcNAcylation of these two sites suppressed the phosphorylation on the adjacent sites, thereby delaying 3T3-L1 adipocyte differentiation (56). Thus, O-GlcNAcylation of C/EBPβ modulates its phosphorylation and transcription activity through the adjacent sites-mediated competition.
Protein acetylation contributes to the protein interaction with DNA and/or other proteins, like co-activators and other transcriptional regulators. C/EBPβ has a plurality of acetylation sites, whose acetylation can modulate its function. Cesena et al. (57) discovered that the nuclear co-activator p300 possesses acetyltransferase activity and modifies C/EBPβ on multiple lysine residues. The acetylation on Lys-39 of C/EBPβ is critical for its transcriptional activity. Furthermore, Lys-39 deacetylation mediated by HDAC1 down-regulates its activity during adipogenesis (58). Wiper-Bergeron et al. (59) reported that, in the process of glucocorticoid-induced preadipocyte differentiation, acetylase GCN5 and p300/CBP-associated factor (PCAF) mediate C/EBPβ acetylation on Lys-98, Lys-101, and Lys-102, and this acetylation acts as a molecular switch repressing the interaction of C/EBPβ with HDAC1 and reducing the affinity between C/EBP β and the corepressor mSin3a. In some cases, however, HDAC1 can strengthen the function of C/EBPβ. Xu et al. (60) reported that acetylation on Lys-215 and Lys-216 decreases the binding activity of C/EBPβ to the promoter of ID1 (inhibitor of DNA-binding protein), and HDAC1-mediated deacetylation can activate this transcription.
Methylation modifies not only DNA and histone, but also some transcription factors. Pless et al. (61) found that Lys-39 of C/EBPβ could be modified by histone methyltransferase G9a and that this modification could inhibit its transcriptional activity. Moreover, the interaction of C/EBPβ with G9a could be inhibited by C/EBPβ phosphorylation (61). Kowenz-Leutz et al. (62) showed the relationship between C/EBPβ phosphorylation and arginine methylation. Protein arginine methyltransferase 4 (PRMT4/CARM1) interacts with C/EBPβ and dimethylates it on Arg-3. The phosphorylation of C/EBPβ by Ras/MAPK, however, blocks the interaction between C/EBPβ and PRMT4/CARM1 and inhibits the methylation on Arg-3. The Arg-3 methylation constrains the interaction between C/EBPβ and SWI/SNF and inhibits adipocyte differentiation. Consequently, C/EBPβ phosphorylation by Ras signaling pathway and arginine methylation reciprocally regulates the interaction between C/EBPβ and epigenetic complexes during adipocyte differentiation.
The lysine residue is not only the substrate of acetylation and methylation but is also modified by ubiquitin. Through the sequential action of E1-activating enzyme, E2-binding enzyme, and E3 ligase, the ubiquitin polymers are connected to the target proteins. Hattori et al. (63) found that the C/EBP family proteins are degraded by the ubiquitin-proteasome pathway. In the process of C/EBP protein ubiquitination, ubiquitin ligases or modifying enzymes specifically recognize the monomer form of C/EBP proteins, so as to remove the transcriptionally inactive monomer of C/EBP proteins and maintain a basal level of C/EBP proteins in cells. The homologous dimerization of C/EBPβ or the heterologous dimerization of C/EBPβ with other C/EBP family proteins can make the protein itself stable (63).
With in-depth study of ubiquitination, the small ubiquitin-like modifications (SUMOlation), have attracted more and more attention. SUMOlation is a reversible PTM that regulates the protein subcellular localization, nucleocytoplasmic transportation, protein stability, and interaction, by the conjugation of the small ubiquitin related modifier (SUMO) to target proteins (64). Kim et al. (65) reported that C/EBP family proteins, including C/EBPα, C/EBPβ, C/EBPδ, and C/EBPϵ, are modified by SUMO. There is a conserved motif containing 5 amino acids ((I/V/L)KXEP) in C/EBP family proteins, and the lysine residue in this motif is specific to SUMO modification (65). C/EBPβ is a SUMO target, and SUMO modification controls its transcriptional activity. Eaton and Sealy (66) found that SUMO is conjugated to Lys-173 residue of C/EBPβ, and blocking SUMOlation on Lys-173 by Lys to Ala mutation relieves its repression on cyclin D1 promoter. Subramanian et al. (67) found that SUMO modification in the synergy control motifs of multiple C/EBP molecules could limit their transcriptional activity. Berberich-Siebelt et al. (68) also found that SUMO could be conjugated to the lysine residue of C/EBPβ in the conserved motif Ile/Val-Lys-X-Glu of the central regulatory domain, which weakened the inhibitory effect of C/EBPβ on c-Myc in murine T cells. Interestingly, this SUMOlation promoted the location of C/EBPβ around the centrosome of heterochromatin, which suggests that SUMO could regulate C/EBPβ function by changing its subnuclear localization (68). It was recently reported that PIAS1, a SUMO E3 ligase, could interact with C/EBPβ and SUMOylate it on Lys-133, leading to increased ubiquitination and degradation of C/EBPβ (69). Consequently, PIAS1 is a negative regulator in adipogenesis by promoting the SUMOlation and degradation of C/EBPβ. Conversely, the SUMO-specific protease Sentrin/SUMO-specific protease 2 (SENP2) plays a critical role in promoting adipogenesis by de-SUMOlation and stabilization of C/EBPβ (70).
In summary, the modification of C/EBPβ, involving the cross-talks of different types of PTMs, finely tunes its function. Considering the fact that the current studies on C/EBPβ PTMs are performed in vitro, knock-in mice expressing PTM-related C/EBPβ mutants are needed to dissect the physiological relevance of C/EBPβ PTMs in regard to adipose tissue development.
Conclusion
Much progress has been made in the past decade in defining the role of C/EBPβ during adipogenesis. The expression and activity of C/EBPβ play a profound role in modulating a wide array of target genes that are important in facilitating adipogenesis. Also, the identification and characterization of C/EBPβ target genes have provided critical information for understanding the function of C/EBPβ in adipogenesis. Meanwhile, the PTM controlling C/EBPβ function has been intensively explored. It should be noted, however, that some studies on the role and regulation of C/EBPβ are mainly based on murine cell models in vitro, which heightens the need for further verifying these findings in vivo and translating them from mouse to human. As our knowledge of the multifaceted nature of C/EBPβ during adipogenesis increases, it is believed that C/EBPβ and factors regulating its function will provide potential targets for the treatment of obesity-related disorders.
Acknowledgments
The Department of Biochemistry and Molecular Biology is supported by Shanghai Leading Academic Discipline Project B110 and 985 Project 985 III-YFX0302.
This work was supported, in whole or in part, by National Key Basic Research Project Grant 2011CB910201 and 2013CB530601, State Key Program of National Natural Science Foundation Grant 31030048C120114, Shanghai Key Science and Technology Research Project 10JC1401000 (to Q. Q. T.), National Natural Science Foundation Grant 30870510 (to X. L.), National Natural Science Foundation Grants 31000603 and 31370027 (to L. G.).
This work is dedicated to the memory of Professor M. Daniel Lane, our friend and mentor and a pioneer in understanding mechanisms of adipocyte differentiation.
- MCE
- mitotic clonal expansion
- C/EBPβ
- CCAAT/enhancer-binding protein β
- C/EBPα
- CCAAT enhancer binding protein α
- IRE1α
- inositol-requiring enzyme 1α
- PKM2
- M2 isoform of pyruvate kinase
- PIAS1
- protein inhibitor of activated STAT1
- PTM
- post-translational modification
- PPARγ
- peroxisome proliferator-activated receptor γ
- SUMO
- small ubiquitin-like modifier
- UPR
- unfolded protein response
- XBP1
- X-box binding protein 1
- PRMT4/CARM1
- protein arginine methyltransferase 4
- CBP
- CREB-binding protein
- CREB
- cAMP-response element-binding protein.
REFERENCES
- 1. Rosen E. D., Spiegelman B. M. (2006) Adipocytes as regulators of energy balance and glucose homeostasis. Nature 444, 847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Haslam D. W., James W. P. (2005) Obesity. Lancet 366, 1197–1209 [DOI] [PubMed] [Google Scholar]
- 3. Shepherd P. R., Gnudi L., Tozzo E., Yang H., Leach F., Kahn B. B. (1993) Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J. Biol. Chem. 268, 22243–22246 [PubMed] [Google Scholar]
- 4. Green H., Kehinde O. (1975) An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell 5, 19–27 [DOI] [PubMed] [Google Scholar]
- 5. Tang Q. Q., Otto T. C., Lane M. D. (2003) Mitotic clonal expansion: a synchronous process required for adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 100, 44–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Farmer S. R. (2006) Transcriptional control of adipocyte formation. Cell Metab. 4, 263–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Y. Y., Li X., Qian S. W., Guo L., Huang H. Y., He Q., Liu Y., Ma C. G., Tang Q. Q. (2011) Transcriptional activation of histone H4 by C/EBPβ during the mitotic clonal expansion of 3T3-L1 adipocyte differentiation. Mol. Biol. Cell 22, 2165–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guo L., Li X., Huang J. X., Huang H. Y., Zhang Y. Y., Qian S. W., Zhu H., Zhang Y. D., Liu Y., Liu Y., Wang K. K., Tang Q. Q. (2012) Histone demethylase Kdm4b functions as a co-factor of C/EBPβ to promote mitotic clonal expansion during differentiation of 3T3-L1 preadipocytes. Cell Death Differ. 19, 1917–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yeh W. C., Cao Z., Classon M., McKnight S. L. (1995) Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 9, 168–181 [DOI] [PubMed] [Google Scholar]
- 10. Tanaka T., Yoshida N., Kishimoto T., Akira S. (1997) Defective adipocyte differentiation in mice lacking the C/EBPβ and/or C/EBPδ gene. EMBO J. 16, 7432–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tang Q. Q., Lane M. D. (1999) Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev. 13, 2231–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qiu Z., Wei Y., Chen N., Jiang M., Wu J., Liao K. (2001) DNA synthesis and mitotic clonal expansion is not a required step for 3T3-L1 preadipocyte differentiation into adipocytes. J. Biol. Chem. 276, 11988–11995 [DOI] [PubMed] [Google Scholar]
- 13. Li X., Kim J. W., Grønborg M., Urlaub H., Lane M. D., Tang Q. Q. (2007) Role of cdk2 in the sequential phosphorylation/activation of C/EBPβ during adipocyte differentiation. Proc. Natl. Acad. Sci. U.S.A. 104, 11597–11602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reichert M., Eick D. (1999) Analysis of cell cycle arrest in adipocyte differentiation. Oncogene 18, 459–466 [DOI] [PubMed] [Google Scholar]
- 15. Yeh W. C., Bierer B. E., McKnight S. L. (1995) Rapamycin inhibits clonal expansion and adipogenic differentiation of 3T3-L1 cells. Proc. Natl. Acad. Sci. U.S.A. 92, 11086–11090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johmura Y., Osada S., Nishizuka M., Imagawa M. (2008) FAD24 acts in concert with histone acetyltransferase HBO1 to promote adipogenesis by controlling DNA replication. J. Biol. Chem. 283, 2265–2274 [DOI] [PubMed] [Google Scholar]
- 17. Cornelius P., MacDougald O. A., Lane M. D. (1994) Regulation of adipocyte development. Annu. Rev. Nutr. 14, 99–129 [DOI] [PubMed] [Google Scholar]
- 18. Greenwel P., Tanaka S., Penkov D., Zhang W., Olive M., Moll J., Vinson C., Di Liberto M., Ramirez F. (2000) Tumor necrosis factor α inhibits type I collagen synthesis through repressive CCAAT/enhancer-binding proteins. Mol. Cell Biol. 20, 912–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang J. W., Tang Q. Q., Vinson C., Lane M. D. (2004) Dominant-negative C/EBP disrupts mitotic clonal expansion and differentiation of 3T3-L1 preadipocytes. Proc. Natl. Acad. Sci. U.S.A. 101, 43–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seagroves T. N., Krnacik S., Raught B., Gay J., Burgess-Beusse B., Darlington G. J., Rosen J. M. (1998) C/EBPβ, but not C/EBPα, is essential for ductal morphogenesis, lobuloalveolar proliferation, and functional differentiation in the mouse mammary gland. Genes Dev. 12, 1917–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iyer V. V., Kadakia T. B., McCabe L. R., Schwartz R. C. (2004) CCAAT/enhancer-binding protein-β has a role in osteoblast proliferation and differentiation. Exp. Cell Res. 295, 128–137 [DOI] [PubMed] [Google Scholar]
- 22. Zhu S., Yoon K., Sterneck E., Johnson P. F., Smart R. C. (2002) CCAAT/enhancer binding protein-β is a mediator of keratinocyte survival and skin tumorigenesis involving oncogenic Ras signaling. Proc. Natl. Acad. Sci. U.S.A. 99, 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aparicio T., Ibarra A., Méndez J. (2006) Cdc45-MCM-GINS, a new power player for DNA replication. Cell Div. 1, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Turowski P., Franckhauser C., Morris M. C., Vaglio P., Fernandez A., Lamb N. J. (2003) Functional cdc25C dual-specificity phosphatase is required for S-phase entry in human cells. Mol. Biol. Cell 14, 2984–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wakabayashi K., Okamura M., Tsutsumi S., Nishikawa N. S., Tanaka T., Sakakibara I., Kitakami J., Ihara S., Hashimoto Y., Hamakubo T., Kodama T., Aburatani H., Sakai J. (2009) The peroxisome proliferator-activated receptor γ/retinoid X receptor α heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol. Cell Biol. 29, 3544–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang L., Jin Q., Lee J. E., Su I. H., Ge K. (2010) Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 7317–7322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang L., Xu S., Lee J. E., Baldridge A., Grullon S., Peng W., Ge K. (2013) Histone H3K9 methyltransferase G9a represses PPARγ expression and adipogenesis. EMBO J. 32, 45–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fodor B. D., Kubicek S., Yonezawa M., O'Sullivan R. J., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T. (2006) Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 20, 1557–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kawazu M., Saso K., Tong K. I., McQuire T., Goto K., Son D. O., Wakeham A., Miyagishi M., Mak T. W., Okada H. (2011) Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS One 6, e17830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tachibana M., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Shinkai Y. (2002) G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16, 1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shinkai Y., Tachibana M. (2011) H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 25, 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li S. F., Guo L., Qian S. W., Liu Y., Zhang Y. Y., Zhang Z. C., Zhao Y., Shou J. Y., Tang Q. Q., Li X. (2013) G9a is transactivated by C/EBPβ to facilitate mitotic clonal expansion during 3T3-L1 preadipocyte differentiation. Am. J. Physiol. Endocrinol. Metab. 304, E990–E998 [DOI] [PubMed] [Google Scholar]
- 33. Luo W., Semenza G. L. (2012) Emerging roles of PKM2 in cell metabolism and cancer progression. Trends Endocrinol. Metab. 23, 560–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Christofk H. R., Vander Heiden M. G., Harris M. H., Ramanathan A., Gerszten R. E., Wei R., Fleming M. D., Schreiber S. L., Cantley L. C. (2008) The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 [DOI] [PubMed] [Google Scholar]
- 35. Yang W., Xia Y., Ji H., Zheng Y., Liang J., Huang W., Gao X., Aldape K., Lu Z. (2011) Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature 480, 118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang W., Xia Y., Hawke D., Li X., Liang J., Xing D., Aldape K., Hunter T., Alfred Yung W. K., Lu Z. (2012) PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 150, 685–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jiang Y., Guo L., Xie L. Q., Zhang Y. Y., Liu X. H., Zhang Y., Zhu H., Yang P. Y., Lu H. J., Tang Q. Q. (2014) Proteome profiling of mitotic clonal expansion during 3T3-L1 adipocyte differentiation using iTRAQ-2DLC-MS/MS. J. Proteome Res. 13, 1307–1314 [DOI] [PubMed] [Google Scholar]
- 38. Tang Q. Q., Lane M. D. (2012) Adipogenesis: from stem cell to adipocyte. Annu. Rev. Biochem. 81, 715–736 [DOI] [PubMed] [Google Scholar]
- 39. Lee J., Ozcan U. (2014) Unfolded protein response signaling and metabolic diseases. J. Biol. Chem. 289, 1203–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Park S. W., Ozcan U. (2013) Potential for therapeutic manipulation of the UPR in disease. Semin. Immunopathol. 35, 351–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gregor M. F., Hotamisligil G. S. (2007) Thematic review series: Adipocyte Biology. Adipocyte stress: the endoplasmic reticulum and metabolic disease. J. Lipid Res. 48, 1905–1914 [DOI] [PubMed] [Google Scholar]
- 42. Sha H., He Y., Chen H., Wang C., Zenno A., Shi H., Yang X., Zhang X., Qi L. (2009) The IRE1α-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab. 9, 556–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mizushima N. (2007) Autophagy: process and function. Genes Dev. 21, 2861–2873 [DOI] [PubMed] [Google Scholar]
- 44. Cecconi F., Levine B. (2008) The role of autophagy in mammalian development: cell makeover rather than cell death. Dev. Cell 15, 344–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Singh R., Xiang Y., Wang Y., Baikati K., Cuervo A. M., Luu Y. K., Tang Y., Pessin J. E., Schwartz G. J., Czaja M. J. (2009) Autophagy regulates adipose mass and differentiation in mice. J. Clin. Invest. 119, 3329–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Baerga R., Zhang Y., Chen P. H., Goldman S., Jin S. (2009) Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy 5, 1118–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guo L., Huang J. X., Liu Y., Li X., Zhou S. R., Qian S. W., Liu Y., Zhu H., Huang H. Y., Dang Y. J., Tang Q. Q. (2013) Transactivation of Atg4b by C/EBPβ promotes autophagy to facilitate adipogenesis. Mol. Cell Biol. 33, 3180–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ma D., Panda S., Lin J. D. (2011) Temporal orchestration of circadian autophagy rhythm by C/EBPβ. EMBO J. 30, 4642–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Oishi Y., Manabe I., Tobe K., Tsushima K., Shindo T., Fujiu K., Nishimura G., Maemura K., Yamauchi T., Kubota N., Suzuki R., Kitamura T., Akira S., Kadowaki T., Nagai R. (2005) Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 1, 27–39 [DOI] [PubMed] [Google Scholar]
- 50. Payne V. A., Au W. S., Lowe C. E., Rahman S. M., Friedman J. E., O'Rahilly S., Rochford J. J. (2010) C/EBP transcription factors regulate SREBP1c gene expression during adipogenesis. Biochem. J. 425, 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jimenez M. A., Akerblad P., Sigvardsson M., Rosen E. D. (2007) Critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol. Cell Biol. 27, 743–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chung S. S., Lee J. S., Kim M., Ahn B. Y., Jung H. S., Lee H. M., Kim J. W., Park K. S. (2012) Regulation of Wnt/β-catenin signaling by CCAAT/enhancer binding protein β during adipogenesis. Obesity (Silver Spring) 20, 482–487 [DOI] [PubMed] [Google Scholar]
- 53. Tang Q. Q., Grønborg M., Huang H., Kim J. W., Otto T. C., Pandey A., Lane M. D. (2005) Sequential phosphorylation of CCAAT enhancer-binding protein β by MAPK and glycogen synthase kinase 3β is required for adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 102, 9766–9771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kim J. W., Tang Q. Q., Li X., Lane M. D. (2007) Effect of phosphorylation and S–S bond-induced dimerization on DNA binding and transcriptional activation by C/EBPβ. Proc. Natl. Acad. Sci. U.S.A. 104, 1800–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang Y. Y., Li S. F., Qian S. W., Zhang Y. Y., Liu Y., Tang Q. Q., Li X. (2012) Phosphorylation prevents C/EBPβ from the calpain-dependent degradation. Biochem. Biophys. Res. Commun. 419, 550–555 [DOI] [PubMed] [Google Scholar]
- 56. Li X., Molina H., Huang H., Zhang Y. Y., Liu M., Qian S. W., Slawson C., Dias W. B., Pandey A., Hart G. W., Lane M. D., Tang Q. Q. (2009) O-Linked N-acetylglucosamine modification on CCAAT enhancer-binding protein β: role during adipocyte differentiation. J. Biol. Chem. 284, 19248–19254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ceseña T. I., Cardinaux J. R., Kwok R., Schwartz J. (2007) CCAAT/enhancer-binding protein (C/EBP) β is acetylated at multiple lysines: acetylation of C/EBPβ at lysine 39 modulates its ability to activate transcription. J. Biol. Chem. 282, 956–967 [DOI] [PubMed] [Google Scholar]
- 58. Ceseña T. I., Cui T. X., Subramanian L., Fulton C. T., Iñiguez-Lluhí J. A., Kwok R. P., Schwartz J. (2008) Acetylation and deacetylation regulate CCAAT/enhancer binding protein β at K39 in mediating gene transcription. Mol. Cell Endocrinol. 289, 94–101 [DOI] [PubMed] [Google Scholar]
- 59. Wiper-Bergeron N., Salem H. A., Tomlinson J. J., Wu D., Haché R. J. (2007) Glucocorticoid-stimulated preadipocyte differentiation is mediated through acetylation of C/EBPβ by GCN5. Proc. Natl. Acad. Sci. U.S.A. 104, 2703–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xu M., Nie L., Kim S. H., Sun X. H. (2003) STAT5-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBPβ. EMBO J. 22, 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pless O., Kowenz-Leutz E., Knoblich M., Lausen J., Beyermann M., Walsh M. J., Leutz A. (2008) G9a-mediated lysine methylation alters the function of CCAAT/enhancer-binding protein-β. J. Biol. Chem. 283, 26357–26363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kowenz-Leutz E., Pless O., Dittmar G., Knoblich M., Leutz A. (2010) Crosstalk between C/EBPβ phosphorylation, arginine methylation, and SWI/SNF/Mediator implies an indexing transcription factor code. EMBO J. 29, 1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hattori T., Ohoka N., Inoue Y., Hayashi H., Onozaki K. (2003) C/EBP family transcription factors are degraded by the proteasome but stabilized by forming dimer. Oncogene 22, 1273–1280 [DOI] [PubMed] [Google Scholar]
- 64. Yeh E. T. (2009) SUMOylation and De-SUMOylation: wrestling with life's processes. J. Biol. Chem. 284, 8223–8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kim J., Cantwell C. A., Johnson P. F., Pfarr C. M., Williams S. C. (2002) Transcriptional activity of CCAAT/enhancer-binding proteins is controlled by a conserved inhibitory domain that is a target for sumoylation. J. Biol. Chem. 277, 38037–38044 [DOI] [PubMed] [Google Scholar]
- 66. Eaton E. M., Sealy L. (2003) Modification of CCAAT/enhancer-binding protein-β by the small ubiquitin-like modifier (SUMO) family members, SUMO-2 and SUMO-3. J. Biol. Chem. 278, 33416–33421 [DOI] [PubMed] [Google Scholar]
- 67. Subramanian L., Benson M. D., Iñiguez-Lluhí J. A. (2003) A synergy control motif within the attenuator domain of CCAAT/enhancer-binding protein α inhibits transcriptional synergy through its PIASy-enhanced modification by SUMO-1 or SUMO-3. J. Biol. Chem. 278, 9134–9141 [DOI] [PubMed] [Google Scholar]
- 68. Berberich-Siebelt F., Berberich I., Andrulis M., Santner-Nanan B., Jha M. K., Klein-Hessling S., Schimpl A., Serfling E. (2006) SUMOylation interferes with CCAAT/enhancer-binding protein β-mediated c-myc repression, but not IL-4 activation in T cells. J. Immunol. 176, 4843–4851 [DOI] [PubMed] [Google Scholar]
- 69. Liu Y., Zhang Y. D., Guo L., Huang H. Y., Zhu H., Huang J. X., Liu Y., Zhou S. R., Dang Y. J., Li X., Tang Q. Q. (2013) Protein inhibitor of activated STAT 1 (PIAS1) is identified as the SUMO E3 ligase of CCAAT/enhancer-binding protein β (C/EBPβ) during adipogenesis. Mol. Cell Biol. 33, 4606–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chung S. S., Ahn B. Y., Kim M., Choi H. H., Park H. S., Kang S., Park S. G., Kim Y. B., Cho Y. M., Lee H. K., Chung C. H., Park K. S. (2010) Control of adipogenesis by the SUMO-specific protease SENP2. Mol. Cell Biol. 30, 2135–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]