Background: The calcium-activated chloride channel ANO1 regulates multiple physiological processes.
Results: We identified residues that when mutated affected channel activity, intracellular trafficking, or localization and report the first structure-function map of ANO1.
Conclusion: The re-entrant loop mediates calcium/voltage sensitivity and activation of ANO1.
Significance: We provide new tools for studying ANO1 function in biological systems and its potential as a therapeutic target.
Keywords: Chloride Channel, Ion Channel, Mutagenesis, Protein Structure, Site-directed Mutagenesis, ANO1, TMEM16A, Variomics
Abstract
The calcium-activated chloride channel ANO1 regulates multiple physiological processes. However, little is known about the mechanism of channel gating and regulation of ANO1 activity. Using a high-throughput, random mutagenesis-based variomics screen, we generated and functionally characterized ∼6000 ANO1 mutants and identified novel mutations that affected channel activity, intracellular trafficking, or localization of ANO1. Mutations such as S741T increased ANO1 calcium sensitivity and rendered ANO1 calcium gating voltage-independent, demonstrating a critical role of the re-entrant loop in coupling calcium and voltage sensitivity of ANO1 and hence in regulating ANO1 activation. Our data present the first unbiased and comprehensive study of the structure-function relationship of ANO1. The novel ANO1 mutants reported have diverse functional characteristics, providing new tools to study ANO1 function in biological systems, paving the path for a better understanding of the function of ANO1 and its role in health and diseases.
Introduction
Despite their critical functions in cells, the molecular identity of calcium-activated chloride channels (CaCCs)2 had been enigmatic for many years, but in 2008, three independent laboratories identified ANO1 (anoctamin 1, also known as TAOS2, DOG1, ORAOV2, and TMEM16A) as the first CaCC (1–4). ANO1 is widely expressed in secretory epithelia (5–8), as well as in a variety of other cell types, including smooth muscles and sensory neurons (6, 9, 10). ANO1 exhibits an important function in regulating airway fluid secretion, gut motility, secretory functions of exocrine glands, renal function, (vascular) smooth muscle contraction, and nociception (8, 10, 11). The physiological relevance of ANO1 is further underscored by several disease states associated with ANO1 dysfunction, including asthma, gastroparesis, hypertension, rota-virus-induced diarrhea, polycystic kidney disease, as well as its high expression and amplification in various cancers (12–17).
ANO1 belongs to the family of anoctamins that includes 10 members (ANO1–10) (18). Besides ANO1, only ANO2 and ANO6 have conclusively been shown to evoke the appearance of calcium-activated chloride current (19, 20). Anoctamins do not share any significant sequence homology with other known ion channel proteins, and in the absence of a crystal structure the prediction of the topology is limited to the methods of bioinformatics. Hydropathy analysis predicts an eight-transmembrane structure with cytoplasmic N and C termini (21), although this model has recently been questioned (4, 22–24).
In addition to the lack of information about ANO1's topology and structure, the mechanism of ANO1 activation is even less understood. Although it is well established that ANO1 is activated by calcium, it is not clear whether calcium directly binds to the channel or whether auxiliary calcium-binding proteins (i.e. calmodulin) are needed (24–27). ANO1 lacks “classical” calcium-binding motifs found in other channels. Recently, residues in the third intracellular loop of ANO1 have been proposed to mediate calcium sensitivity of ANO1 (23, 24), and reconstitution of ANO1 in proteoliposomes has been shown to be sufficient to recapitulate the biophysical properties of ANO1 (28), suggesting a direct activation by calcium. However, it remains unclear how calcium binding regulates the activity of the channel. In the absence of a crystal structure for ANO1, alternative ways to study the structure-function of ANO1 are needed.
Here, we report the results of an unbiased approach to investigate the structure-function relationship of ANO1. By using a high-throughput, random mutagenesis-based variomics screen, we generated and functionally characterized a library of ∼6000 mutants of ANO1. We identified several residues in ANO1 that when mutated significantly changed the functional properties of ANO1 in multiple ways, by causing the channel to be inactive at high calcium levels, by inducing activity even at low calcium concentrations, or by affecting the intracellular trafficking and localization. In summary, our data provide the first unbiased and comprehensive study of the structure-function relationship of ANO1. The newly identified, functionally diverse group of ANO1 mutants provides new tools for the study of ANO1 function in biological systems, which will lead to a better understanding of the function of ANO1, its physiological function, and therapeutic potential.
EXPERIMENTAL PROCEDURES
Error-prone PCR-mediated Mutagenesis
Error-prone PCR-mediated mutagenesis was performed using the GeneMorph II random mutagenesis kit (Stratagene). Briefly, 500–1000 ng of plasmid DNA (pcDNA3.1-ANO1(abc)-His) was mutagenized by PCR according to the manufacturer's protocol using the following primers: ANO1-N-fw, 5′-GGGAGACCCAAGCTGGCTAGTTAAGC-3′, and ANO1-N-rev, 5′-CGCCACTGTGCTGGATATCTGCAG-3′; ANO1-C-fw, 5′-GCAGATGCGGCTGAACTACAGATGG-3′, and ANO1-C-rev, 5′-TCTTCCTCGAAGCCGGTCAGGTC-3′. PCR products were separated by gel electrophoresis and purified using a gel extraction kit (Qiagen). After restriction enzyme digest (HindIII/PpuMI for N-terminal and PpuMI/EcoRI for C-terminal library), the DNA was purified with a PCR purification kit (Qiagen) and cloned into the parental vector at equivalent positions (pcDNA3.1-ANO1(abc)-His). Mutagenized DNA was transformed into Escherichia coli DH5α maximum efficiency (Invitrogen), and two libraries with around 3000 mutants each were prepared as described previously (29). The 96-well plates containing normalized mutant DNA (40 ng/μl) were re-arrayed into 384-deep well bar-coded bioassay plates (Greiner Bio-One) by transferring 62.5 μl (2.5 μg of DNA) of mutant plasmid with a MiniTrak robot (PerkinElmer Life Sciences) into the assigned 384-well plate coordinates. Each 96-well plate was stamped four times on a 384-well plate to allow for the parallel testing of multiple assay conditions on the same plate. In total, 66 unique 384-well plates (33 plates per library, ∼6000 mutants total) containing an individual clone within each well were generated. All plates were stored at −20 °C.
Next Generation Sequencing
All library constructs were pooled, and the genomic library was prepared, sequenced, and analyzed as described previously (29). Nonbiased assessment of codon usage was calculated by dividing the total mutations across the coding sequence by the potential mutations. All potential codons were then determined based on the three alternative bases at each location, assuming one mutation per codon. The data have been deposited in the NCBI SRA database, bioproject ID PRJNA261442.
Cell Culture and Transfection
CHO cells (Invitrogen) were cultured in DMEM/F-12; HEK293TN (Invitrogen) and HEK293 cells stably expressing the H148Q/I152L variant of YFP (generated as described in Ref. 30) were maintained in DMEM supplemented with fetal bovine serum (10% v/v) at 37 °C, 5% CO2. For screening, cells were harvested using trypsin/EDTA buffer and seeded on multiwell plates. Forward transfection of cells with the mutant library was performed in poly-d-lysine-coated 384-well black clear bottom plates (BD Biosciences) using FuGENE6 (Promega) according to the manufacturer's instructions. Briefly, 1 μl (40 ng) of cDNA library or wild-type ANO1, mixed with 0.12 μl of FuGENE 6 in a total of 20 μl, was added to the each well of cells using a Biomek robotic workstation. Cells were then incubated for a total of 2 days at 37 °C, 5% CO2. After 1 day, the media from the plates were replaced with 65 μl of fresh growth media. Follow-up experiments were performed in a 96-well format. Cells in collagen-coated 96-well black clear bottom plates (Corning Glass) were transfected as described above using 160 ng of DNA and 0.48 μl of FuGENE 6 in a total volume of 20 μl of OptiMEM per well. For the analysis of protein expression, cells were seeded and transfected in 6-well plates using 1 μg of DNA and 3 μl of FUGENE 6.
Generation of Stable Cell Lines
The sequence for human ANO1 (abc-splice form) was cloned into an HIV-based lentiviral vector with puromycin resistance gene (pEZ-ANO1(abc)-Lv105, GeneCopoeia). For virus packaging, pEZ-ANO1(abc)-Lv105 was cotransfected with vira-power lentiviral packaging mix (Invitrogen) into HEK293TN cells according to the manufacturer's protocol. CHO cells were infected with lentivirus in the presence of 10 μg/ml Polybrene (Applied Bioanalytical). Cells stably expressing ANO1 were selected with 10 μg/ml puromycin. Stable cell pools were seeded into 96-well plates aiming for single cells by serial dilution, and two identical plates were prepared by splitting the cells after ∼10 days. One plate was fixed and stained for ANO1 using an anti-His antibody as described. The clones with the highest expression of ANO1 were selected from the second plate, expanded, and established as a stable cell line.
Compounds
CaCCinh-A01 was synthesized in-house. EGTA-AM was purchased from Calbiochem, and all other chemicals were purchased from Sigma.
YFP-quench Assay
For the screening of the mutant libraries, transiently transfected HEK293-YFP-cells were washed once in wash buffer (2 mm KCl, 1 mm CaCl2, 1 mm MgCl2, 135 mm NaCl, 10 mm d-glucose, 20 mm HEPES, pH 7.4), and 40 μl of wash buffer was added to each well (Biomek robotic workstation). The YFP baseline signal was recorded for 30 s on an FDSS platform using a GFP filter set (excitation 480 nm/emission 540 nm), and 10 μl of assay solution (2 mm KCl, 1 mm CaCl2, 1 mm MgCl2, 350 mm NaI, 10 mm d-glucose, 10 mm HEPES, pH 7.4) was added containing 500 μm carbachol (final concentration 100 μm carbachol (EC100)) or an equal amount of DMSO (unstimulated), and YFP fluorescence was recorded for 2 min (1 measurement/s). Kinetics of YFP-quench were analyzed manually, and mutants were classified as described in the text. For follow up of single mutants, transiently transfected cells in 96-well plates were washed once in wash buffer, and 80 μl of wash buffer (containing compound or DMSO as indicated) was added to each well. After 10 min, the plates were analyzed on an EnVision plate reader with a fluid handling system module and a GFP bottom read filter module (excitation 485 nm/band width 14 nm, emission 535 nm/band width 25 nm, bottom mirror D505 FITC). Baseline YFP signal was measured for 3 s before 20 μl of assay solution containing 500 μm carbachol (final concentration 100 μm carbachol (EC100)), or an equal amount of DMSO (unstimulated) was injected per well, and YFP fluorescence was recorded for 30 s (1 measurement/s). YFP fluorescence recordings were normalized for the initial background-subtracted average fluorescence. The relative quench was calculated as the difference between the initial fluorescence and the residual YFP fluorescence at the end of the measurement (30 s) and was used to compare different mutants (relative activity).
Site-directed Mutagenesis
Single amino acid mutants were generated by site-directed mutagenesis (Q5 mutagenesis kit, New England Biolabs) according to the manufacturer's protocol using pBM-ANO1(abc)-His or pEZ-ANO1(abc)-Lv105 as a template. Primers were designed using the NEBaseChanger on-line tool (New England Biolabs) and were synthesized by Integrated DNA Technologies. Sequences are available upon request. Maxi-preps (Qiagen) were performed for all mutants. The introduced mutation was confirmed by Sanger sequencing of the complete coding region of ANO1 (GeneWiz).
High-Content Image Analysis of ANO1 Expression Level
Screening plates were fixed with 4% paraformaldehyde immediately after analysis and were stored at 4 °C. Plates were washed twice in PBS and were incubated in blocking solution (3% (w/v) BSA, 0.3% Triton X-100 in PBS) for 1 h at room temperature. Primary antibody (anti-His, D3I1O, Cell Signaling) was added, and plates were incubated overnight at 4 °C. Plates were washed twice in PBS, and Alexa660-conjugated secondary antibody was added in blocking buffer for 1 h at room temperature. Nuclei were stained with DAPI. Imaging was conducted using a WiScan automated microscope (Idea Bio-Medical) with a 10 × 0.4 NA UPLSAPO objective (Olympus), recording 16 fields/well. Image analysis has been performed as described previously (31). In brief, fluorescence intensities were quantified on a cell by cell basis. For this, a mask defining the nuclear and cellular regions was applied. For the analysis of the screening plates, individual staining intensities were normalized to the average staining intensity of each plate, and the three inactive clones with the highest expression (>75% of the average staining intensity of the plate) were picked.
Confocal Microscopy
Cells were grown on collagen-coated glass coverslips, fixed with 4% paraformaldehyde, and stained as described above and using an Alexa594-conjugated secondary antibody. Coverslips were mounted in VectaShield (Vector Laboratories), and cells were acquired on a laser scanning confocal microscope (LSM 510 Axiovert 200 M, Carl Zeiss). A 63 × 1.4 NA oil immersion objective was used for imaging with the pinhole diameter set for 1-μm optical z-sections. 512 × 512 pixel images were collected with the ZEN software package (Carl Zeiss) with an x,y resolution of 0.28 μm/pixel. z-stacks were recorded with a size increment of 0.5 μm. For each set of samples, the laser intensity and detector gain were set for the most intensely stained specimen such that there was no saturation, and all other samples were imaged at the same settings. Images were exported to Adobe Photoshop for figure preparation. Images were cropped to show a representative field of cells using the middle section of each z-stack, and they originated from the same respective experiment. All focus, contrast, and brightness settings remained constant during imaging preparation.
Western Blotting
Immunoblotting was performed as described previously (32). ANO1 was detected using an anti-ANO1 antibody (SP31, Abcam). Tubulin served as a loading control.
Immunoprecipitation
3 × 106 HEK293-YFP cells were plated per 10-cm dish, allowed to adhere overnight, and were transfected with a total of 6 μg of DNA using 18 μl of FuGENE 6. Cells were harvested 48 h after transfection and lysed in RIPA buffer (Cell Signaling). ∼1 mg of lysate was incubated with 40 μl of anti-FLAG M2 beads (Sigma) or 40 μl of Ni-NTA-coated beads (Invitrogen) for 1 h at 4 °C. Proteins bound to the beads after washing were eluted in Laemmli buffer for 10 min at 70 °C and analyzed by immunoblotting.
Electrophysiology
Whole-cell voltage-clamp recordings were made using the QPatch planar patch clamp system (Sophion BioSciences, operated according to the manufacturer's specification) by testing cells stably expressing the indicated lentiviral constructs or cells transiently infected with BacMam virus (33), as described previously (32). Trypsin-harvested cells were resuspended in serum-free medium containing 1 mg/ml trypsin inhibitor. Gigaseal formation, whole-cell configuration, and series resistance compensation were established at room temperature (21–23 °C) using QPatch software. The extracellular recording solution contained the following (in mm): N-methyl-d-glucamine (130), EGTA (10), HEPES (10), MgCl2 (1), CaCl2 (6), sucrose (45), pH 7.3 (adjusted with HCl). The intracellular solution contained the following (in mm): N-methyl-d-glucamine (130), EGTA (20), HEPES (10), MgCl2 (1), 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (10), Mg-ATP (2), CaCl2 (varying levels), pH 7.25 (adjusted with HCl; osmolarity adjusted to 320 with sucrose). CaCl2 additions were calculated to achieve the free [Ca2+]i levels indicated in the figures as described previously (29). CaCCinh-A01 was used from 30 mm stock in 100% DMSO and diluted in extracellular solution (1.1–100 μm) maintaining constant 0.3% DMSO throughout. For evaluating the whole-cell conductance (G), voltage steps of 1-s duration were given from a holding potential of −70 mV to voltages between −70 and +70 mV or −100 and +260 mV in 10- and 20-mV steps, respectively, followed by a step to −70 mV to observe tail currents. The extended voltage range was necessary to approach maximal conductances and enabled us to fit the obtained data. The conductance was calculated from tail currents at −70 mV as G = I/(Vm − ECl), where I is the peak tail current; Vm is the pulse voltage, and ECl is the equilibrium potential for chloride for our recording conditions. To analyze the [Ca2+]i dependence of ANO1 activation at different voltages, we have varied the [Ca2+]i from 30 nm up to 10 μm. The dependence of G on [Ca2+]i was fitted at each voltage by the Hill equation: G = Gmax[Ca2+]inH/([Ca2+]inH + EC50nH), where EC50 is the [Ca2+]i at which half-maximal activation occurred and nH is the Hill coefficient. To analyze the voltage dependence of ANO1 activation, the normalized conductance (G/Gmax) at a given [Ca2+]i was plotted against the voltage of the pre-pulse, and a Boltzmann equation was fitted to the data: G/Gmax = 1/(1 + exp(z(V½ − V)F/RT)), where z is the equivalent gating charge associated with voltage-dependent channel opening; V is the membrane potential; V½ is the membrane potential producing half-maximal activation; F is the Faraday constant; R is the gas constant; and T is the absolute temperature. The maximum conductance Gmax was estimated from the fit of the voltage dependence of G at 1 μm free [Ca2+]i, and G at each [Ca2+]i was then normalized to the same Gmax. To evaluate the deactivation kinetics, the voltage protocol included a single 1-s pre-pulse at +100 mV followed by 4-s long voltage steps between −120 and +80 mV in 20-mV increments. Single exponential functions, A + B·exp(−t/τdeact) (where A and B are parameters representing the plateau and the span of the current decay, and τdeact is the time constant describing the deactivation time course), were fitted to tail currents at each potential. For CaCCinh-A01 experiments, a 1.5-s duration pulse at +70 mV from and returning to a holding potential of −70 mV was combined with a voltage ramp protocol that changed the membrane voltage continuously from −90 to +70 mV over 1.5 s. Current amplitudes recorded at the end of the depolarizing step at +70 mV before and after cumulative applications of increasing concentrations of CaCCinh-A01 were normalized and plotted to evaluate the CaCCinh-A01-mediated inhibition of ANO1. Data are presented as means ± S.E., with n indicating the number of cells. Student's unpaired two-tailed t test was used for statistical analysis, and p < 0.05 was considered significant.
RESULTS
Functional Characterization of a High-Throughput Random Mutagenesis Library for ANO1 Identifies Inactive and Constitutively Active Mutants
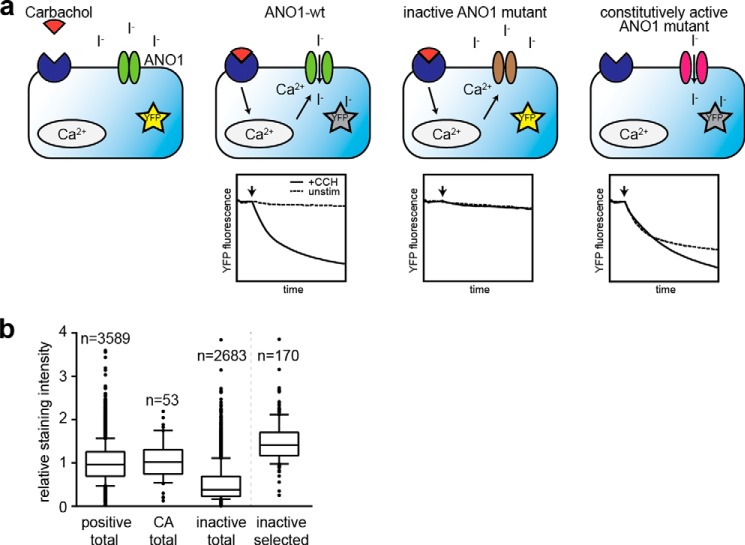
Using high-throughput random mutagenesis, we aimed to identify amino acids critical for ANO1 function. We generated two libraries with a total of ∼6000 mutants of ANO1 (abc-splice form) by error-prone PCR targeting the N-terminal half or the C-terminal half of the coding region for ANO1 (∼3000 clones were picked for each library). The quality, coverage, and mutation rate of our mutagenesis libraries were analyzed by next generation sequencing (29). The data revealed a homogeneous coverage of the complete coding region of ANO1 and an overall mutation rate of 0.78 mutations/kb (Table 1). A YFP-quench assay was used to measure the activity of the cloned ANO1 mutants. With this assay, the influx of anions (iodide) in cells expressing a halide-sensitive mutant of YFP (YFP H148Q/I152L) can be monitored by measuring the quenching of YFP fluorescence (Fig. 1a) (34, 35). HEK293 cells stably expressing YFP-(H148Q/I152L) were transfected with the ANO1 mutants, and the rate of YFP quenching was compared with cells transfected with wild-type ANO1 (ANO1-WT). Because ANO1 activity depends on the presence of intracellular calcium (1, 3, 4), we stimulated the cells with the muscarinic receptor agonist carbachol (CCH) and screened for ANO1 mutants that failed to show CCH-induced quenching of YFP (inactive mutants). To find mutations that render ANO1 more active at low calcium levels (herein conferred to as constitutively active mutants), we measured ANO1 activity in the absence of CCH and compared the rate of YFP quenching to ANO1-WT transfected cells. By testing ∼6000 mutants under unstimulated and CCH-stimulated conditions, we identified a total of 2683 inactive mutants and 53 constitutively active ANO1 mutants. Only 3589 mutants showed wild-type activity of ANO1 (active mutants). To exclude inactive mutants that were caused by nonsense mutations, truncations, or mutations that prevent ANO1 from being expressed, we stained the screening plates after completion of the YFP assay for ANO1 using a C-terminal His tag, and expression of ANO1 was quantified by high-content imaging analysis (Fig. 1b). The 170 inactive mutants of ANO1 with the highest expression and all 53 constitutively active mutants were sequenced and compared with the wild-type sequence for ANO1. Alignment of the sequences of the 170 inactive mutants identified 132 distinct inactive mutants with a total of ∼360 amino acid changes targeting 268 amino acid positions, including mutations of residues previously reported to be critical for ANO1 function (Arg-537, Leu-575, Glu-727, Glu-756, and Asp-760) (1, 23, 24, 36). Furthermore, 25 mutants were found to have deletions in the coding region for ANO1 and were excluded from the analysis. Analysis of the sequences for the 53 constitutively active mutant clones identified 49 distinct constitutively active mutants for ANO1, with a total of 95 amino acid changes, targeting 68 amino acid positions. These data highlight the merit of high-throughput random mutagenesis, coupled to a functional cell-based characterization, to discover novel mutations with divergent effects on ANO1 protein function in a cellular system.
TABLE 1.
Characteristics of the ANO1 mutant library as determined by next generation sequencing
| % | % | ||
|---|---|---|---|
| Transition | |||
| A→G | 51.89 | A→G, T→C | 52.63 |
| T→C | 53.33 | ||
| G→A | 57.33 | G→A, C→T | 58.27 |
| C→T | 59.20 | ||
| Transversion | |||
| A→T | 40.78 | A→T, T→A | 37.80 |
| T→A | 35.03 | ||
| A→C | 7.34 | A→C, T→G | 9.57 |
| T→G | 11.65 | ||
| G→C | 10.88 | G→C, C→G | 10.24 |
| C→G | 9.61 | ||
| G→T | 31.79 | G→T, C→A | 31.49 |
| C→A | 31.19 | ||
| Bias indicator | |||
| Transition/Transversion | 1.24 | ||
| AT→GC/GC→AT | 0.76 | ||
| A→N, T→N | 52.37 | ||
| G→N, C→N | 47.63 | ||
FIGURE 1.
Functional characterization of a library of ∼6000 ANO1 mutants. a, schematic summary of the principle of the YFP-quench assay. HEK293 cells stably expressing a halide-sensitive variant of YFP (YFP H148Q/I152L) were transiently transfected with ANO1, and the ANO1-mediated influx of iodide was analyzed by measuring YFP fluorescence before and after stimulation of intracellular calcium release with CCH. Exemplary measurements are shown. b, relative staining intensity of ANO1 (normalized to the mean staining intensity of the active ANO1 mutants) in the three experimental groups (positive, wild-type like activity; CA, constitutive activity; inactive, no activity) and the subset of inactive mutants selected for further characterization in subsequent experiments. Each box represents the median and 25th/75th percentiles; whiskers depict the minimum and maximum.
Identification of Novel Mutations That Cause Inactivity of ANO1
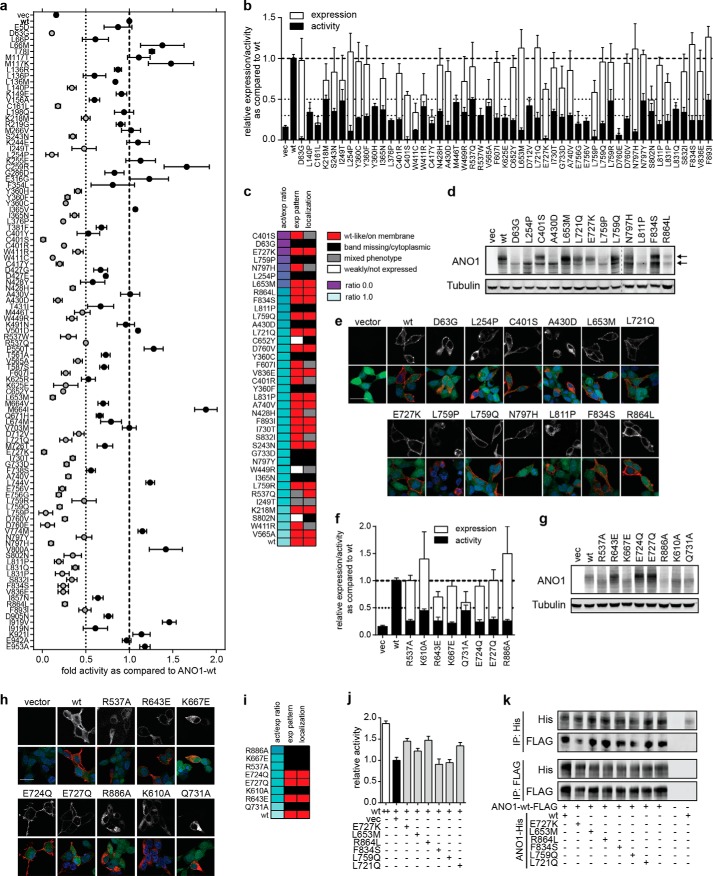
Most of the mutant constructs for ANO1 that scored in our primary screen showed multiple mutations. To identify the causative mutation, 177 ANO1 mutants (103/74 mutants for inactive and constitutively active mutations, respectively) with single amino acid changes were generated by site-directed mutagenesis, and their function was tested in the YFP-quench assay. We identified 52 single amino acid mutations that caused a decrease in CCH-induced ANO1 activity of >50% as compared with ANO1-WT (Fig. 2a). To analyze whether the mutations had any effect on the expression level of ANO1, we quantified the expression of all 52 inactive ANO1 mutants by using high-content imaging analysis (Fig. 2b). Fourteen mutants were found to be only weakly expressed and were excluded from further analysis. Calculating the ratio of the activity and expression level for each mutation allowed us to rank order the mutants to identify the mutants with the most significant decrease in relative ANO1 activity (Fig. 2c). Furthermore, the expression of all mutants was analyzed by Western blotting as exemplarily shown in Fig. 2d for selected inactive mutants. ANO1-WT is glycosylated and is detected as a double band, corresponding to the mature and immature form of ANO1 (18, 32). Fourteen out of the 38 inactive mutants with confirmed ANO1 expression lacked the expression of the upper band for ANO1, indicative of a lack of maturation likely caused by a mislocalization of ANO1 in the cell, thus providing an explanation for the lack of activity in the YFP assay. In agreement with this hypothesis, we analyzed the localization of all ANO1 mutants by high-content imaging, which showed a mainly intracellular localization of all 14 mutants with an altered expression pattern in the Western blot (Fig. 2c). Seventeen mutants were found to be expressed on the plasma membrane. In addition, we analyzed the subcellular localization for our top 13 inactive mutants with the lowest activity/expression ratio of ANO1 (ratio <0.25) by immunofluorescence in HEK293-YFP cells using confocal microscopy (Fig. 2e). Consistent with the expression pattern on the Western blot, 8/13 inactive ANO1 mutants were expressed primarily on the plasma membrane, whereas ANO1-D63G, L254P, A430D, L759P, and L811P were mainly intracellularly localized, indicating accumulation of ANO1 in the endoplasmic reticulum/Golgi network of the cells with little or no expression of ANO1 on the plasma membrane. Taken together, we report 38 inactive mutants for ANO1 with high expression, among which 17 mutants showed localization to the plasma membrane. These mutants are of particular interest when compared with the previously reported inactive mutants of ANO1. Although we were able to confirm expression and a >50% decrease in ANO1 activity for all tested mutants, only R643E, E724Q, and E727Q showed a double band pattern of ANO1 on a Western blot (Fig. 2, f, g, and i). We found ANO1-R537A, K667E, R886A, K610A, and Q731A to be localized in intracellular compartments of the cells, whereas only ANO1-R643E, E724Q, and E727Q were expressed on the plasma membrane (Fig. 2, h and i). Our data suggest that mutations of ANO1 are directly affecting the biochemical activity of the channel or the subcellular localization of the protein, both of which result in loss of ANO1 activity on the plasma membrane. These results highlight the need for a careful characterization of inactive ANO1 mutants to distinguish the lack of activity caused by mutations affecting critical residues for ANO1 function from mutations causing mislocalization of ANO1.
FIGURE 2.
Characterization of inactive ANO1 mutants. a, activity of selected single amino mutants of ANO1 measured in a YFP-quench assay after stimulation with CCH. Data were normalized to the activity of ANO1-wild type (wt) mean ± S.E. n ≥ 4. b, comparison of the activity (as determined in a) and expression level (measured by high-content imaging) of 52 inactive ANO1 mutants. Data were normalized to the respective signal of ANO1-WT (mean ± S.E., n ≥ 4). c, summary of the characterization of inactive mutants. The activity/expression ratio was calculated by dividing the relative activity with the relative expression level. The expression pattern was analyzed by Western blotting and the localization by high-content imaging. d, Western blot analysis of selected inactive ANO1 mutants (act/expression ratio <0.25). Arrows indicate the two distinct bands detected for ANO1. Tubulin served as a loading control. Representative images are shown. Image has been cropped at indicated position (dashed line). e, immunofluorescence analysis of ANO1 expression and localization (anti-His, red) in HEK293-YFP cells (YFP expression, green), nuclear stain (DAPI, blue). Representative confocal images are shown. The upper panel depicts the signal for ANO1 in gray scale. Scale bar, 20 μm. f, comparison of the activity (YFP-quench assay) and expression level (high-content imaging) of previously described inactive ANO1 mutants. Data were normalized to the respective signal of ANO1-WT and are presented as the mean ± S.E. of ≥4 experiments. g, Western blot analysis of inactive ANO1 mutants previously described in the literature. vec, vector. h, immunofluorescence analysis of ANO1 expression and localization of previously described inactive ANO1 mutants as described in e. i, summary of the characterization of inactive mutants previously reported in the literature. j, activity of ANO1-WT in the YFP-quench assay when coexpressed with equal amounts of vector, wild type, or the indicated inactive mutants. Data were normalized to the activity of ANO1-WT cotransfected with equal amounts of empty vector and are presented as the mean ± S.E. of ≥8 measurements. k, coimmunoprecipitation (IP) analysis of the hetero-oligomerization of ANO1-WT and inactive mutants of ANO1. HEK293-YFP cells were transfected with equal amounts of the indicated mutants (His-tagged) and ANO1-WT (FLAG-tagged), and ANO1 was precipitated from the lysate using Ni-NTA or anti-FLAG antibody-coated beads, and the amount of copurified ANO1-WT or inactive mutant, respectively, was determined by Western blotting.
Using Inactive Mutants to Probe the Functional Structure of ANO1
ANO1's quaternary structure has recently been described as a homodimer, and perturbation of dimerization has been shown to interfere with ANO1's activity (37–39). However, it remains unclear whether both ANO1 monomers contribute to the formation of the active pore in the dimer (one-pore model) or whether each monomer in the dimer on its own can form an active pore (two-pore model). To address this question, we cotransfected HEK293-YFP cells with ANO1-WT and equal amounts of inactive, membrane-localized ANO1 mutants (ANO1-IA) and measured ANO1 activity in the YFP-quench assay. In the one-pore model where both ANO1 units contribute to the active pore, the coexpression of inactive ANO1 should result in a decrease in ANO1 activity, i.e. a dominant-negative effect, because a heterodimer between ANO1-WT and ANO1-IA is expected to be inactive. On the contrary, in the two-pore model, coexpression of inactive ANO1 is not expected to result in a decrease in ANO1 activity given that the ANO1-WT part in an ANO1-WT/ANO1-IA heterodimer can still form an active pore. Cell were either transfected with equal amounts of WT plasmid and vector or with equal amounts WT plasmid and ANO1-IA. As shown in Fig. 2j, coexpression of ANO1-IA did not result in a decrease in ANO1 activity hence supporting a two-pore model. Coimmunoprecipitation experiments showed that the inactive mutants dimerized and interacted with ANO1-WT at similar levels as the WT (Fig. 2k). Hence, our results support a two-pore quaternary structure model for ANO1, where each molecule of ANO1 functions as an independent pore.
Functional Characterization of Constitutively Active Mutants of ANO1
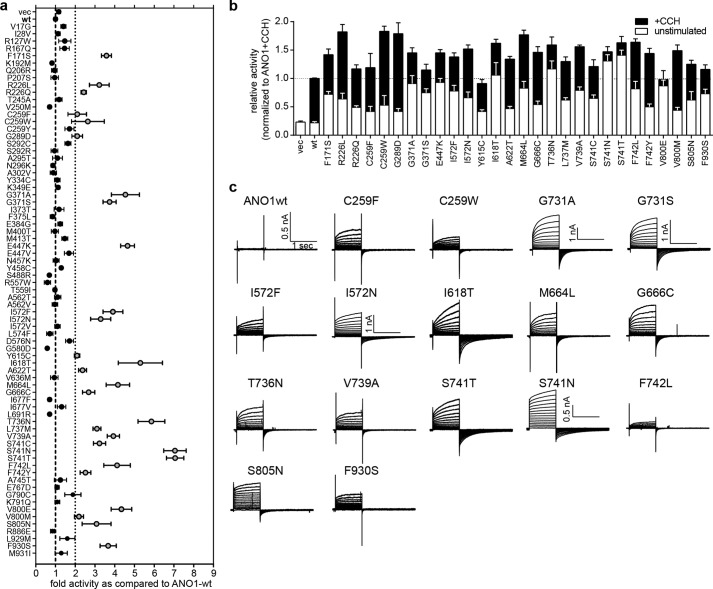
Our primary screen identified 49 mutants (with a total of 95 mutations) of ANO1 with significant activity under nonstimulated (low calcium) conditions. Out of the 74 single amino acid mutants we generated by site-directed mutagenesis, 28 mutants showed a significant increase in activity in the YFP-quench assay under unstimulated conditions as well as after stimulation with CCH as compared with ANO1-WT (Fig. 3a). Four of the mutants (ANO1-I618T, T736N, S741N, and S741T) demonstrated an activity level under unstimulated conditions that was above that of ANO1-WT at maximal stimulation, representing a hyperactive, constitutively active phenotype of ANO1 (Fig. 3b). All 28 mutants were expressed at similar levels as the wild-type and showed the characteristic double band pattern for ANO1 on a Western blot and localized to the plasma membrane (data not shown). Notably, 8 of the 28 mutants mapped to a predicted transmembrane domain of ANO1 (transmembrane domain 6, TM6), suggesting an important role of this region for the regulation of ANO1 activity. Whole-cell recordings of the chloride conductance of an exemplary subset of our constitutively active mutants at 180 nm free intracellular calcium concentrations further demonstrated the high activity of the mutant channel under low intracellular calcium conditions, which is not sufficient to activate the WT channel under physiological conditions (Fig. 3c).
FIGURE 3.
Characterization of constitutively active ANO1 mutants. a, activity of selected single amino mutants of ANO1 measured in an YFP-quench assay in the absence of CCH (unstimulated). Data were normalized to the activity of ANO1-WT (mean ± S.E., n ≥ 4). b, comparison of the activity (YFP-quench assay) of 28 constitutively active ANO1 mutants in the absence (unstimulated) and presence of CCH (+CCH). Data were normalized to the respective signal of ANO1-WT (mean ± S.E., n ≥ 4). c, whole-cell voltage clamp recordings of ANO1-mediated chloride currents of the indicated mutants at 180 nm free intracellular calcium. Voltage steps of 1-s duration were given from a holding potential of −70 mV to voltages between −70 and +70 mV in 10-mV increments, followed by a step to −70 mV. If not otherwise indicated, the scale shown for ANO1-wt applies for all panels.
Using Constitutively Active Mutants to Study the Mechanism of Action of the ANO1 Inhibitor CaCCinh-A01
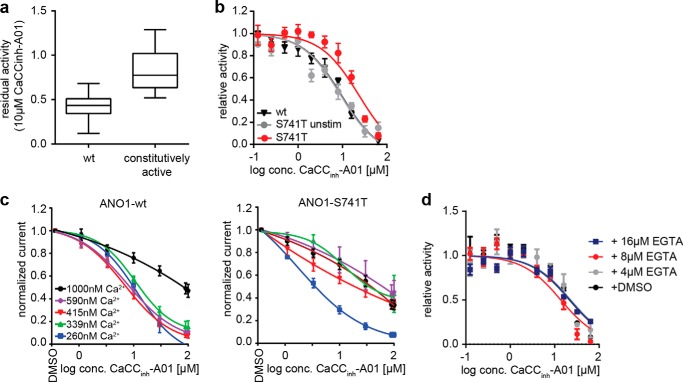
CaCCinh-A01 inhibits ANO1-dependent chloride conductance. However, direct binding of CaCCinh-A01 to ANO1 has not been shown, and its mechanism of action remains elusive. We tested the effect of CaCCinh-A01 on our constitutively active mutants and found that CaCCinh-A01 was less effective in inhibiting ANO1 mutants with constitutive activity as compared with the wild-type channel (Fig. 4, a and b). Both channel types demonstrated a reduction in inhibitor efficacy and potency at higher levels of intracellular calcium (Fig. 4c). We hypothesized that the reduction in inhibitor efficacy and potency is more pronounced in the mutants due to their increased calcium sensitivity and thus higher activity, rather than mutation-induced disruption to the inhibitor-binding site. If CaCCinh-A01 is less effective in inhibiting the active channel, an increase in the fraction of ANO1 present in the inactive conformation should result in an increased sensitivity to CaCCinh-A01. To test this hypothesis, we compared the IC50 value of CaCCinh-A01 for inhibiting the activity of ANO1-S741T under unstimulated (inactive + active conformation) and stimulated conditions (active conformation). ANO1-S741T showed an ∼4-fold increase in sensitivity to inhibition by CaCCinh-A01 under unstimulated conditions, corroborating our hypothesis that CaCCinh-A01 preferentially binds and stabilizes the inactive form of ANO1 (Fig. 4b). The decrease in sensitivity of ANO1-S741T under stimulated conditions was not caused by competition with calcium, as co-treatment with EGTA did not affect the sensitivity of ANO1-S741T to inhibition by CaCCinh-A01 (Fig. 4d). Whole-cell voltage clamp measurements of ANO1-dependent chloride currents after treatment with CaCCinh-A01 showed that ANO1-S741T was not completely inhibited by CaCCinh-A01 in the tested concentration range (Fig. 4c). These data are in full agreement with the hypothesis that CaCCinh-A01 binds preferentially to a less active channel state.
FIGURE 4.
CaCCinh-A01 sensitivity of ANO1 mutants. a, residual activity of all 28 constitutively active single amino mutants of ANO1 as compared with WT after treatment with 10 μm CaCCinh-A01 measured in a YFP-quench assay in the presence of CCH. Data were normalized to the activity of the DMSO-treated ANO1-WT (wt). Each box represents the median and 25th/75th percentiles of ≥4 measurements; whiskers depict the minimum and maximum. b, activity of ANO1-WT and ANO1-S741T in the YFP-quench assay after treatment with the indicated concentrations of CaCCinh-A01. Data present mean ± S.E. of ≥3 measurements. c, CaCCinh-A01 concentration-dependent inhibition of ANO1 currents at the indicated range of calcium concentrations, determined by whole-cell voltage clamp recordings (+70 mV). Data were normalized to basal currents in the absence of inhibitor (mean ± S.E. of ≥3). d, activity of ANO1-S741T in the YFP-quench assay after treatment with the indicated concentrations of CaCCinh-A01 and EGTA-AM or DMSO. Data present mean ± S.E. of ≥3 measurements.
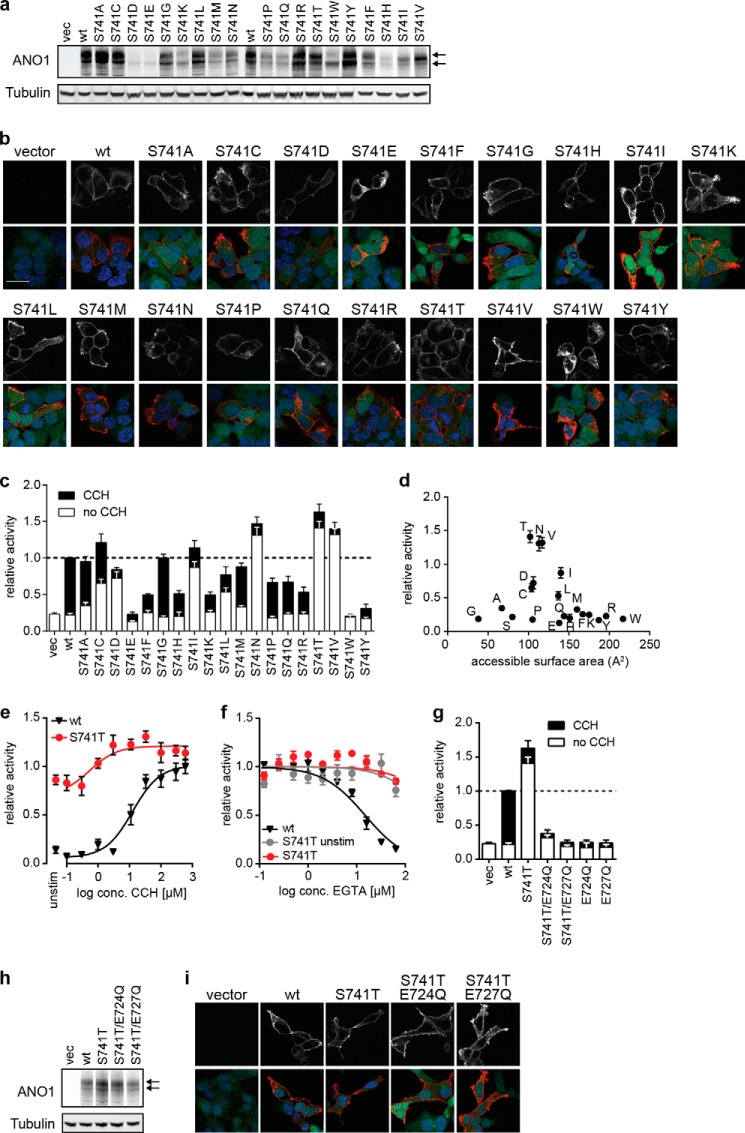
ANO1-S741T Shows an Increased Sensitivity to Calcium
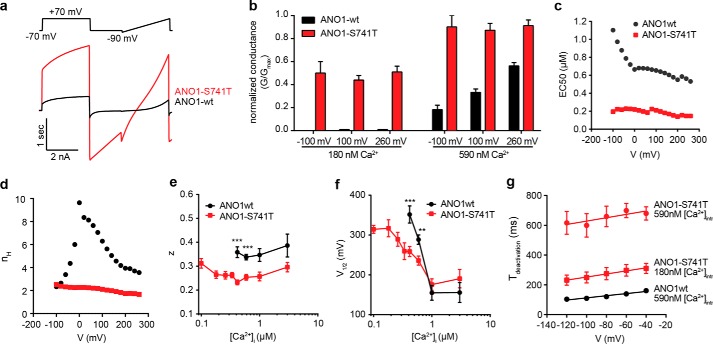
Mutation of Ser-741 to Thr (ANO1-S741T) resulted in the strongest activation of ANO1 in the YFP assay under unstimulated conditions with only a minor additional increase in activity after stimulation with CCH, suggesting an increase in sensitivity to intracellular calcium (Fig. 3b). Intrigued by the significance of the effect caused by this rather conservative amino acid substitution, we investigated the effect of all possible amino acid substitutions at Ser-741. With the exception of S741E and S741W, all mutants were expressed and localized at the plasma membrane (Fig. 5, a and b). S741E and S741W showed a significant intracellular accumulation of ANO1 accompanied by a decrease in the expression level for S741E, providing an explanation for the observed inactivity of these mutants. In addition to ANO1-S741T and S741N, we found that substitution of Ser-741 with isoleucine or valine (S741I and S741V) caused a similar constitutive activity of ANO1 under nonstimulated conditions (Fig. 5c). Correlation of the observed phenotype under unstimulated conditions with the chemical properties of the substituting amino acid residues showed that the constitutively active phenotype was dependent on the size of the side chain, although there was no correlation to the charge, hydrophobicity, or polarity of the side chain, suggesting that Ser-741 plays a critical role in the activation of ANO1 and when mutated can facilitate conformational changes required to open the channel, for example by fostering binding of calcium (Fig. 5d). To test whether ANO1-S741T enhances the activation of the channel by free intracellular calcium, we recorded the activity of ANO1-S741T after stimulation with increasing concentrations of CCH in the YFP-quench assay. Although ANO1-WT showed a robust dose-dependent increase in activity with increasing concentrations of CCH, ANO1-S741T showed only a minor increase in activity, with already ∼90% of wild-type activity under unstimulated conditions (Fig. 5e). Consistent with an increased calcium sensitivity of ANO1-S741T, treatment with EGTA-AM led to only a minor decrease in activity under both stimulated and unstimulated conditions, even at concentrations of EGTA-AM that blocked the activity of ANO1-WT (Fig. 5f). These data strengthen the hypothesis that ANO1-S741T exhibits an increased affinity for or even complete independence on intracellular calcium. To test whether the activity of S741T is independent of calcium, we mutated two of the proposed calcium-binding sites of ANO1. Mutation of either Glu-727 or Glu-724 to glutamine completely diminished the activity of S741T even under stimulated conditions, suggesting that S741T still requires calcium to be activated (Fig. 5, g and i). Consistent with our hypothesis, the whole-cell current recording of ANO1-dependent currents over a large range of membrane voltage potentials (Vm) and intracellular free calcium concentrations showed a profound increase in calcium sensitivity for ANO1-S741T (Fig. 6, a–c). Although there was no activation in the absence of intracellular free calcium, calcium concentrations as small as 100 nm activated robust levels of S741T currents that were reached by the wild-type channel only in the presence of ∼5-fold higher Ca2+ concentrations (ANO1-S741T: 0.60 ± 0.07 nS/pF, n = 32; ANO1-WT: 0.37 ± 0.06 nS/pF and 1.03 ± 0.11 nS/pF at 415 and 590 nm, respectively, n = 31 and 44, Vm = 60 mV). This occurred despite the ANO1-S741T channel showing smaller overall maximal conductance density than the ANO1-WT, 10.8 ± 0.8 nS/pF, n = 21, and 23.1 ± 2.7 nS/pF, n = 10, respectively. Determination of the EC50 value of calcium-dependent activation of ANO1 at different holding potentials showed a left shift of the curves for S741T as compared with ANO1-WT (Fig. 6c). Remarkably, although ANO1-WT showed the characteristic voltage-dependent increase in calcium sensitivity, the EC50 value of calcium remained constant for ANO1-S741T for all tested Vm values, indicating a decoupling of calcium sensitivity from voltage dependence in ANO1-S741T. Consistent with this hypothesis, we found that in contrast to the wild-type channel, the Hill coefficient (nH) for ANO1-S741T, indicative of the cooperativity of Ca2+ binding, was not dependent on voltage (Fig. 6d). Thus, the mutation renders the Ca2+ binding of ANO1-S741T voltage-independent. Conductance of ANO1-S741T was still modulated by Vm values, although this dependence was significantly altered when compared with ANO1-WT. The S741T mutation caused a left shift in the normalized conductance G/Gmax versus Vm curves at low Ca2+ concentrations and surprisingly, altered the slope (Fig. 6, e and f). We looked at the two parameters resulting from the Boltzmann fit of the G-V relations, V½, the membrane potential producing half-maximal activation representing the conformational energy associated with voltage-independent channel opening, and z, the equivalent gating charge associated with voltage-dependent channel opening (Fig. 6, e and f). ANO1-S741T showed a decrease in V½, suggesting an increased ability of Ca2+ to reduce the activation energy required for the channel to open. The Vm sensitivity (z) of ANO1-S741T was calcium-independent as reported previously for ANO1-WT (40), but it was reduced by one-fourth as compared with the wild type, suggesting the mutation disrupts either the moving charge or the electrical field in which the charge that confers voltage sensitivity is moving. Furthermore, we found that S741T showed a drastic elongation of the deactivation time (Fig. 6g). These data are consistent with S741T facilitating the active conformation of the channel and Ser-741 regulating the conformational changes coupling calcium binding and voltage sensitivity of ANO1.
FIGURE 5.
Mutation of ANO1-Ser-741 to Thr facilitates activation of ANO1. a, Western blot analysis Ser-741-ANO1 mutants. The arrows indicate the two distinct bands detected for ANO1. Tubulin served as a loading control. Representative images are shown. b, immunofluorescence analysis of Ser-741-ANO1 mutant expression and localization (anti-His, red) in HEK293-YFP cells (YFP expression, green), nuclear stain (DAPI, blue). Representative confocal images are shown. The upper panel depicts the signal for ANO1 in gray scale. Scale bar, 20 μm. c, comparison of the activity (YFP-quench assay) of ANO1 mutants with alternative amino acid changes at Ser-741 in the absence (unstimulated) and presence of CCH (+CCH). Data were normalized to the respective signal of ANO1-WT (mean ± S.E., n ≥ 4). vec, vector. d, relative activity of each Ser-741 mutant is plotted as a function of side chain surface area. Letters indicate the substituted amino acid. e, comparison of the activity of ANO1-WT and ANO1-S741T at different concentrations of CCH (YFP-quench assay). Data were normalized to the maximal activity of ANO1-WT (mean ± S.E., n ≥ 4). f, sensitivity of ANO1-WT and ANO1-S741T to inhibition with EGTA. Activity of ANO1-WT/-S741T in the YFP-quench assay was measured after treatment with the indicated concentrations of EGTA-AM in the presence or absence of CCH. Data were normalized to the activity after treatment with DMSO and are presented as the mean ± S.E. of ≥4 experiments. g, relative activity of indicated double mutant of ANO1-S741T in the YFP-quench assay as compared with ANO1-S741T and ANO1-WT. h, Western blot analysis of ANO1-S741T mutants. The arrows indicate the two distinct bands detected for ANO1. Tubulin served as a loading control. Representative images are shown. i, immunofluorescence analysis of ANO1-S741T mutants as described in b.
FIGURE 6.
Electrophysiological characterization of ANO1-S741T. a, representative whole-cell current traces recorded for ANO1-WT and ANO1-S741T at 415 nm intracellular free calcium. Voltage protocol as shown in the figure. b, normalized conductance (G/Gmax) of ANO1-WT and ANO1-S741T. G has been normalized to the individual maximal conductances at the specified voltage. c, EC50 value of the calcium-dependent activation of ANO1-WT and ANO1-S741T at the indicated membrane potentials. EC50 values were obtained from fitting of the mean [Ca2+]i − whole-cell conductance response relationship with the Hill-Langmuir equation. The mean conductance at a given potential and [Ca2+]i was obtained by averaging conductances measured in n = 4–50 and n = 4–64 cells for ANO1-WT and ANO1-S741T, respectively. d, Hill coefficient across a range of membrane potentials from the same fit as described in a. e, plot of the estimated membrane potential producing half-maximal activation (V½) of ANO1-WT/S741T versus the concentration of intracellular free calcium. V½ was obtained from fitting a Boltzmann equation to the voltage dependence of normalized whole-cell conductance at a given [Ca2+]i. The maximum conductance Gmax was estimated from the fit of the voltage dependence of G at 1 μm free [Ca2+]i, and G at each [Ca2+]i was then normalized to the same Gmax. n = 9–45. f, plot of the estimated gating charge (z) associated with voltage-dependent channel opening of ANO1-WT/S741T versus the concentration of intracellular free calcium. Fitting of data as described in e. g, kinetic analysis of the deactivation time of ANO1-WT/S741T. Tail currents were fitted to single exponentials, and the time constants were plotted versus membrane potential. n = 5–15. **, p < 0.01; ***, p < 0.001.
DISCUSSION
N Terminus of ANO1 Regulates Trafficking to the Plasma Membrane and Affects Channel Activity
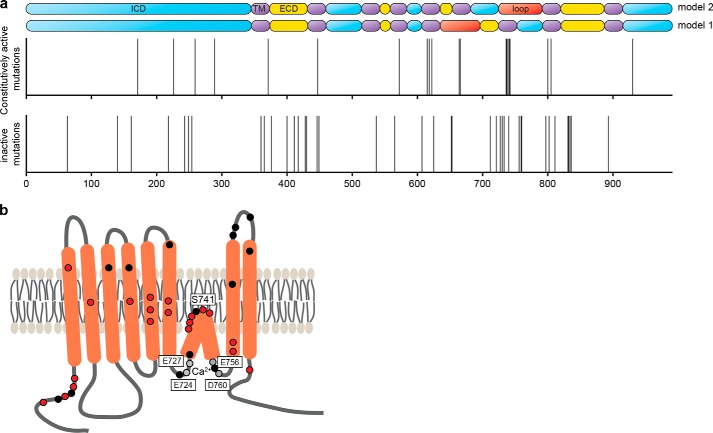
Using an unbiased, high-throughput random mutagenesis approach, we have identified several novel single amino acid mutations in ANO1 that affected ANO1 activity, trafficking, and subcellular localization (Fig. 7). Among the inactive mutants of ANO1 with confirmed expression of ANO1, we found multiple mutations to interfere with the localization of ANO1 to the plasma membrane. Point mutations in the intracellular domains of ANO1 can point to the existence of trafficking motifs. The N terminus of ANO1 has been described to be critical for the expression and trafficking of ANO1 to the plasma membrane (41–44). We identified four mutations in the N terminus that prevented ANO1 from being trafficked to the plasma membrane, opening up the possibility that these residues contribute to a noncanonical trafficking motif in ANO1 and that additional regulatory proteins bind the N terminus and are involved in regulating trafficking of the channel. Our data support the hypothesis that the N terminus of ANO1 is critical for localization of ANO1 to the plasma membrane. Furthermore, we identified two inactive mutants with mutations in the N terminus of ANO1 that were expressed on the plasma membrane. Mutations in the same N-terminal region of ANO1 caused constitutive activity of the channel and suggest that besides its role in ANO1 trafficking the N terminus of ANO1 is also involved in the regulation of channel gating.
FIGURE 7.
Functional map of ANO1. a, pictogram showing the localization of all identified inactive (lower panel) and constitutively active (middle panel) mutants of ANO1 (x axis, amino acid position of ANO1). Each line represents the position of a mutant. The upper panel depicts the predicted topologies for ANO1 (model 1 is based on Refs. 1, 4 and model 2 is based on Ref. 23). Blue, intracellular domain; purple, transmembrane domain; yellow, extracellular domain; red, loop region with proposed re-entrant loop. b, schematic illustrating the predicted topology of ANO1-abc (23, 24). The proposed calcium-binding sites of ANO1 are indicated by gray dots, and the location of the newly identified constitutively active mutants and inactive mutants with membrane localization are highlighted with red and black dots, respectively.
Mutations in the Transmembrane Domains and C Terminus of ANO1
With the exception of mutations in the C and N terminus, all identified constitutively active mutations map to the predicted transmembrane domains of ANO1, pointing to a contribution of all transmembrane domains to the formation of an active channel. A high number of inactive mutations in the transmembrane domains of ANO1 identified in our screen also showed a defect in channel trafficking and expression. This observation demonstrates the technical difficulties in studying functional domains that are highly sensitive to misfolding and highlight the value of our gain-of-function and constitutively active mutants of ANO1 as a new tool to study the processes leading to ANO1 activation.
Interestingly, we found a cluster of inactive mutations in the last extracellular domain of ANO1, a region that has not been implicated in regulation of channel activity before. We did not find these mutations to affect the glycosylation or localization of ANO1, suggesting residues 831–836 directly affect channel activation. Further studies are needed to explore the function of the last extracellular domain of ANO1 in channel activation.
ANO1 Mutants as a Tool to Study the Function of ANO1 in Biological Systems
The variety of functional mutations in ANO1 reported in this study introduces novel tools to study the function of ANO1 in biological systems. Here, we used our newly identified mutants to probe the functional structure of ANO1 and to investigate the mechanisms of action of a small molecule inhibitor of ANO1. ANO1 has been described to form homodimers, and dimerization has been found to be critical for ANO1 function (37–39). By using inactive plasma membrane-expressed mutants of ANO1, we demonstrate a 2-pore model for ANO1, in which each monomer of ANO1 in a dimer constitutes a separate pore, similar to the quaternary structure of CLC channels (45, 46). Furthermore, using our constitutively active mutants as a tool, we showed that CaCCinh-A01 inhibits ANO1 by interacting with and potentially stabilizing the inactive state of ANO1. Alternatively, the conformational changes leading to activation of ANO1 induced by calcium or mutations could prevent CaCCinh-A01 from binding. The data presented in this study are consistent with both proposed models. Without the precise knowledge of the binding site of CaCCinh-A01 in ANO1, further studies are necessary to elucidate the mechanism of CaCCinh-A01-dependent inhibition of ANO1 in more detail. These results highlight the value of ANO1 mutants as a tool for investigating ANO1 channel function and its role in biological systems.
ANO1 Mutants Support the Elucidation of the Function of Anoctamins in Health and Disease
Members of the anoctamin gene family have been linked to human disease. Although no disease-causing mutations in ANO1 have yet been identified, mutations in ANO3, ANO5, and ANO10 have been associated with human genetic diseases. Mutation of Trp-490 in ANO3 has been identified to cause a dominant form of craniometrical dystonia (47). Our screen identified a mutation of the corresponding conserved residue in ANO1 (ANO1-W449R), which leads to a loss of function accompanied by a decrease in expression and mislocalization of ANO1. ANO5 was found to be mutated at Asp-81 (D81G) in a patient with limb girdle muscular dystrophy type 2L, and mutations of ANO5-Ile-865 was reported in a patient with anoctaminopathy (48, 49). Mutations at the homolog residues in ANO1 (Asp-63/Ile-919) caused a loss of function and mislocalization of ANO1. Similarly, mutation of ANO10 (L510R) has been identified to cause an autosomally recessive form of cerebellar ataxia (50), and we found mutations at the corresponding highly conserved residue in ANO1 (Leu-737) to cause increased constitutive activity of ANO1. Although we are only beginning to understand the function of anoctamins in disease, it is possible that mutations of these highly conserved residues across the ANO1 family may translate to similar functional defects as observed for ANO1, which may result in the clinical phenotype. Our results highlight the potential of unbiased characterization of ANO1's structure-function to support the elucidation of the function of other anoctamins in health and disease. Activation of ANO1 has been discussed to be potentially useful to treat cystic fibrosis by compensating for the loss of function of cystic fibrosis transmembrane conductance regulator and by improving mucociliary clearance (13). Current approaches to investigate this hypothesis are limited to overexpression of ANO1-WT paired with stimulation of calcium signaling to activate the channel or a novel class of small molecules that can temporarily potentiate ANO1 activity (51). The constitutively active mutants identified in this study present novel tools for investigating the function and a potential therapeutic benefit of ANO1 activation in cystic fibrosis. Moreover, inhibition of ANO1 activation might be potentially beneficial in the treatment of cancer, asthma, or diarrhea (6, 16, 32). Recent screening approaches have identified several small molecule inhibitors of ANO1 (35, 52, 53). However, these approaches rely on the activation of ANO1 activity by secondary stimuli to increase intracellular calcium concentration, and direct binding of these molecules to ANO1 has not been shown. Using the constitutively active mutants reported in this study in such screening experiments would eliminate the need to engage secondary pathways to activate ANO1 and could aid in reducing the number of false-positive hits due to inhibition of secondary pathways.
Newly Identified Mutations Support a Revised Topology of ANO1
Because of the lack of a crystal structure, predictions of ANO1's topology are limited on bioinformatic approaches. Hydropathy analysis predicts an eight-transmembrane structure with cytoplasmic N and C termini (21). Whereas most models consistently predict transmembrane domains 1–5 and 7–8, the predictions for the proposed structure of the region between transmembrane domains 5–7 are ambiguous (23). Yang et al. (4) proposed the region following transmembrane domains 5 to form a re-entrant loop that faces the extracellular environment (Fig. 7a, model 1). Mutations of three highly conserved, positively charged residues in this region drastically changed the ion selectivity of ANO1, prompting the authors to suggest the region around the re-entrant loop (amino acids 642–672) to constitute the pore of the channel (4). This model has recently been questioned. Yu et al. (23) proposed an alternative model for the topology of ANO1, based on epitope accessibility experiments, in which the region between amino acids 653 and 670 completely crosses the membrane to form transmembrane domain 6 followed by an intracellular loop with a partial intercalation into the membrane (Fig. 7a, model 2). In support of model 2, residues Glu-724/727 and Glu-756/Asp-760 have been proposed as the calcium-binding sites of ANO1 (23, 24), pointing to an intracellular location. In this model, amino acids 728–752 are proposed to form a short re-entrant loop (24). The location of ANO1's pore, however, remains elusive. Here, we identified a hot spot of mutations in the region preceding transmembrane domain 7 of ANO1, consisting of a distinct peak of constitutively active mutations surrounded by a broader accumulation of inactive mutants. Our data are consistent with a role of this region in calcium binding (inactive mutations) and point to a critical function of the loop region (amino acids 728–752 in model 2) in the regulation of channel activity (constitutively active mutants). In contrast, we identified only two inactive mutations mapping to the proposed pore region in model 1 (amino acids 642–672), only one of which was expressed at the membrane. Given the confirmed coverage of this region in our mutagenesis approach, the low number of functional mutations points to only a minor function of this region in regulating ANO1 activity. Furthermore, we found previously identified mutations used to map the pore region of ANO1 to exhibit a severe defect in trafficking and expression, making the analysis of the functional data challenging. Taken together, our data suggest an important role of amino acids 721–727 and 756–760 in calcium sensing and hence an intracellular location. Furthermore, our data are consistent with residues 728–752 forming a re-entrant loop and suggest an important role of the loop region for the activity of ANO1. Thus, our findings corroborate the revised topology model of ANO1 (model 2). Our data are consistent with the possibility that amino acids 728–752 are involved in forming the pore of ANO1; however, further structural information is needed to validate this model.
Notably, we did not identify any mutations with functional effects in regions predicted to constitute a possible binding site for calmodulin (26, 27, 54), suggesting only a minor role of calmodulin in the regulation of ANO1 activity in our system, as already proposed by other studies (25, 28).
S741T Facilitates ANO1 Activation and Renders Its Calcium Sensitivity Independent of Voltage
Residues Glu-724/727 and Glu-756/Asp-760 have been proposed as the calcium-binding sites of ANO1 (23, 24), suggesting a model in which calcium is sandwiched between those residues, creating a loop between amino acids 728 and 752 that enters the membrane. Strikingly, the majority of our constitutively active mutants mapped to the predicted re-entrant loop, with Ser-741 mapping half-way between the two proposed calcium-binding sites. This finding proposes a model, in which calcium binding to residues Glu-756/Asp-760 and Glu-727/Glu-724 causes a conformational change in the loop domain that results in the activation of the channel and in which the identified constitutively active mutants of ANO1 mapping in the loop region, all on top S741T, facilitate the calcium-induced conformational changes and thus the activation of the channel at low calcium concentrations. Characterization of the electrophysiological properties of ANO1-S741T showed that the mutation resulted in an uncoupling of the characteristic voltage and calcium dependence of ANO1, with calcium inducing channel activation with the same potency and cooperativity across the tested voltage range, thus transforming the channel from a calcium/voltage-gated channel to a ligand-gated channel. Moreover, S741T facilitated the voltage-independent step that translates the calcium binding into channel opening as indicated by the V½ shifts and increased the open probability of the channel by decreasing its deactivation time. The voltage dependence of the calcium sensitivity has been described to result from either voltage-dependent Ca2+ binding to the closed state or from voltage-independent Ca2+ binding to a channel conformation that itself is voltage-dependent (55, 56). The observed voltage-independent activation of ANO1-S741T, accompanied by an increase in calcium sensitivity and the decrease in V½ and the gating charge, supports the existence of membrane-potential independent calcium-binding sites outside of the electrical field and supports the model that the S741T mutation causes conformational changes enhancing the accessibility of these binding sites. The effect of mutations at residue Ser-741 was not dependent on the polarity, charge, or hydrophobicity of the side chain, but it showed a dependence on a very narrow size limit of the side chain. Together with the predicted location of Ser-741 in the center of the re-entrant loop, this observation argues against a direct involvement of Ser-741 in calcium binding and rather support for an allosteric effect of residue Ser-741 on regulating the conformational changes coupling calcium binding and voltage sensitivity of ANO1.
In summary, our comprehensive study of the structure-function of ANO1 has identified several residues critical for ANO1 function, localization, and expression. Our study supports the revised topology model of ANO1, demonstrates a critical role of the re-entrant loop for the activation of the channel, and provides novel mechanistic insight into the calcium-dependent gating of ANO1. The collection of functionally characterized mutations described in this study will aid the investigation of the molecular mechanisms underlying the activation of ANO1 and its cellular function.
Acknowledgments
We thank Ned Kirkpatrick (Novartis Institutes for Biomedical Research) for technical assistance with the confocal microscope and Anne-Lise Glotin for technical assistance with the YFP-quench assay.
All authors are employees of Novartis Institutes for Biomedical Research.
- CaCC
- calcium-activated chloride channel
- CCH
- carbachol
- pF
- picofarad.
REFERENCES
- 1. Caputo A., Caci E., Ferrera L., Pedemonte N., Barsanti C., Sondo E., Pfeffer U., Ravazzolo R., Zegarra-Moran O., Galietta L. J. (2008) TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science 322, 590–594 [DOI] [PubMed] [Google Scholar]
- 2. Duran C., Thompson C. H., Xiao Q., Hartzell H. C. (2010) Chloride channels: often enigmatic, rarely predictable. Annu. Rev. Physiol. 72, 95–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schroeder B. C., Cheng T., Jan Y. N., Jan L. Y. (2008) Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell 134, 1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang Y. D., Cho H., Koo J. Y., Tak M. H., Cho Y., Shim W. S., Park S. P., Lee J., Lee B., Kim B. M., Raouf R., Shin Y. K., Oh U. (2008) TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature 455, 1210–1215 [DOI] [PubMed] [Google Scholar]
- 5. Gomez-Pinilla P. J., Gibbons S. J., Bardsley M. R., Lorincz A., Pozo M. J., Pasricha P. J., Van de Rijn M., West R. B., Sarr M. G., Kendrick M. L., Cima R. R., Dozois E. J., Larson D. W., Ordog T., Farrugia G. (2009) Ano1 is a selective marker of interstitial cells of Cajal in the human and mouse gastrointestinal tract. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G1370–G1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang F., Zhang H., Wu M., Yang H., Kudo M., Peters C. J., Woodruff P. G., Solberg O. D., Donne M. L., Huang X., Sheppard D., Fahy J. V., Wolters P. J., Hogan B. L., Finkbeiner W. E., Li M., Jan Y. N., Jan L. Y., Rock J. R. (2012) Calcium-activated chloride channel TMEM16A modulates mucin secretion and airway smooth muscle contraction. Proc. Natl. Acad. Sci. U.S.A. 109, 16354–16359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang F., Rock J. R., Harfe B. D., Cheng T., Huang X., Jan Y. N., Jan L. Y. (2009) Studies on expression and function of the TMEM16A calcium-activated chloride channel. Proc. Natl. Acad. Sci. U.S.A. 106, 21413–21418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hwang S. J., Blair P. J., Britton F. C., O'Driscoll K. E., Hennig G., Bayguinov Y. R., Rock J. R., Harfe B. D., Sanders K. M., Ward S. M. (2009) Expression of anoctamin 1/TMEM16A by interstitial cells of Cajal is fundamental for slow wave activity in gastrointestinal muscles. J. Physiol. 587, 4887–4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Manoury B., Tamuleviciute A., Tammaro P. (2010) TMEM16A/anoctamin 1 protein mediates calcium-activated chloride currents in pulmonary arterial smooth muscle cells. J. Physiol. 588, 2305–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cho H., Yang Y. D., Lee J., Lee B., Kim T., Jang Y., Back S. K., Na H. S., Harfe B. D., Wang F., Raouf R., Wood J. N., Oh U. (2012) The calcium-activated chloride channel anoctamin 1 acts as a heat sensor in nociceptive neurons. Nat. Neurosci. 15, 1015–1021 [DOI] [PubMed] [Google Scholar]
- 11. Faria D., Rock J. R., Romao A. M., Schweda F., Bandulik S., Witzgall R., Schlatter E., Heitzmann D., Pavenstädt H., Herrmann E., Kunzelmann K., Schreiber R. (2014) The calcium-activated chloride channel Anoctamin 1 contributes to the regulation of renal function. Kidney Int. 85, 1369–1381 [DOI] [PubMed] [Google Scholar]
- 12. Zhang C. H., Li Y., Zhao W., Lifshitz L. M., Li H., Harfe B. D., Zhu M. S., ZhuGe R. (2013) The TMEM16A Ca2+-activated Cl- channel in airway smooth muscle contributes to airway hyperresponsiveness. Am. J. Respir. Crit. Care Med. 187, 374–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sondo E., Caci E., Galietta L. J. (2014) The TMEM16A chloride channel as an alternative therapeutic target in cystic fibrosis. Int. J. Biochem. Cell Biol. 187, 73–76 [DOI] [PubMed] [Google Scholar]
- 14. Britschgi A., Bill A., Brinkhaus H., Rothwell C., Clay I., Duss S., Rebhan M., Raman P., Guy C. T., Wetzel K., George E., Popa M. O., Lilley S., Choudhury H., Gosling M., Wang L., Fitzgerald S., Borawski J., Baffoe J., Labow M., Gaither L. A., Bentires-Alj M. (2013) Calcium-activated chloride channel ANO1 promotes breast cancer progression by activating EGFR and CAMK signaling. Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.1217072110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Forrest A. S., Joyce T. C., Huebner M. L., Ayon R. J., Wiwchar M., Joyce J., Freitas N., Davis A. J., Ye L., Duan D. D., Singer C. A., Valencik M. L., Greenwood I. A., Leblanc N. (2012) Increased TMEM16A-encoded calcium-activated chloride channel activity is associated with pulmonary hypertension. Am. J. Physiol. Cell Physiol. 303, C1229–C1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ousingsawat J., Mirza M., Tian Y., Roussa E., Schreiber R., Cook D. I., Kunzelmann K. (2011) Rotavirus toxin NSP4 induces diarrhea by activation of TMEM16A and inhibition of Na+ absorption. Pflugers Arch. 461, 579–589 [DOI] [PubMed] [Google Scholar]
- 17. Tanaka T., Nangaku M. (2014) ANO1: an additional key player in cyst growth. Kidney Int. 85, 1007–1009 [DOI] [PubMed] [Google Scholar]
- 18. Pedemonte N., Galietta L. J. (2014) Structure and function of TMEM16 proteins (Anoctamins). Physiol. Rev. 94, 419–459 [DOI] [PubMed] [Google Scholar]
- 19. Pifferi S., Dibattista M., Menini A. (2009) TMEM16B induces chloride currents activated by calcium in mammalian cells. Pflugers Arch. 458, 1023–1038 [DOI] [PubMed] [Google Scholar]
- 20. Martins J. R., Faria D., Kongsuphol P., Reisch B., Schreiber R., Kunzelmann K. (2011) Anoctamin 6 is an essential component of the outwardly rectifying chloride channel. Proc. Natl. Acad. Sci. U.S.A. 108, 18168–18172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Galindo B. E., Vacquier V. D. (2005) Phylogeny of the TMEM16 protein family: some members are overexpressed in cancer. Int. J. Mol. Med. 16, 919–924 [PubMed] [Google Scholar]
- 22. Das S., Hahn Y., Walker D. A., Nagata S., Willingham M. C., Peehl D. M., Bera T. K., Lee B., Pastan I. (2008) Topology of NGEP, a prostate-specific cell:cell junction protein widely expressed in many cancers of different grade level. Cancer Res. 68, 6306–6312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu K., Duran C., Qu Z., Cui Y. Y., Hartzell H. C. (2012) Explaining calcium-dependent gating of anoctamin-1 chloride channels requires a revised topology. Circ. Res. 110, 990–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tien J., Peters C. J., Wong X. M., Cheng T., Jan Y. N., Jan L. Y., Yang H. (2014) A comprehensive search for calcium binding sites critical for TMEM16A calcium-activated chloride channel activity. Elife 10.7554/eLIFE.02772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yu K., Zhu J., Qu Z., Cui Y. Y., Hartzell H. C. (2014) Activation of the Ano1 (TMEM16A) chloride channel by calcium is not mediated by calmodulin. J. Gen. Physiol. 143, 253–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tian Y., Kongsuphol P., Hug M., Ousingsawat J., Witzgall R., Schreiber R., Kunzelmann K. (2011) Calmodulin-dependent activation of the epithelial calcium-dependent chloride channel TMEM16A. FASEB J. 25, 1058–1068 [DOI] [PubMed] [Google Scholar]
- 27. Vocke K., Dauner K., Hahn A., Ulbrich A., Broecker J., Keller S., Frings S., Möhrlen F. (2013) Calmodulin-dependent activation and inactivation of anoctamin calcium-gated chloride channels. J. Gen. Physiol. 142, 381–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Terashima H., Picollo A., Accardi A. (2013) Purified TMEM16A is sufficient to form Ca2+-activated Cl- channels. Proc. Natl. Acad. Sci. U.S.A. 110, 19354–19359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bill A., Rosethorne E. M., Kent T. C., Fawcett L., Burchell L., van Diepen M. T., Marelli A., Batalov S., Miraglia L., Orth A. P., Renaud N. A., Charlton S. J., Gosling M., Gaither L. A., Groot-Kormelink P. J. (2014) High throughput mutagenesis for identification of residues regulating human prostacyclin (hIP) receptor expression and function. PLoS One 9, e97973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qiu Z., Dubin A. E., Mathur J., Tu B., Reddy K., Miraglia L. J., Reinhardt J., Orth A. P., Patapoutian A. (2014) SWELL1, a plasma membrane protein, is an essential component of volume-regulated anion channel. Cell 157, 447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paran Y., Lavelin I., Naffar-Abu-Amara S., Winograd-Katz S., Liron Y., Geiger B., Kam Z. (2006) Development and application of automatic high resolution light microscopy for cell-based screens. Methods Enzymol. 414, 228–247 [DOI] [PubMed] [Google Scholar]
- 32. Bill A., Hall M. L., Borawski J., Hodgson C., Jenkins J., Piechon P., Popa O., Rothwell C., Tranter P., Tria S., Wagner T., Whitehead L., Gaither L. A. (2014) Small molecule facilitated degradation of ANO1–a new targeting approach for anticancer therapeutics. J. Biol. Chem. 289, 11029–11041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McPate M., Bhalay G., Beckett M., Fairbrother S., Gosling M., Groot-Kormelink P. J., Lane R., Kent T., Van Diepen M. T., Tranter P., Verkuyl J. M. (2014) The development of automated patch clamp assays for canonical transient receptor potential channels TRPC3, -6, and -7. Assay Drug Dev. Technol. 12, 282–292 [DOI] [PubMed] [Google Scholar]
- 34. Jayaraman S., Haggie P., Wachter R. M., Remington S. J., Verkman A. S. (2000) Mechanism and cellular applications of a green fluorescent protein-based halide sensor. J. Biol. Chem. 275, 6047–6050 [DOI] [PubMed] [Google Scholar]
- 35. De La Fuente R., Namkung W., Mills A., Verkman A. S. (2008) Small-molecule screen identifies inhibitors of a human intestinal calcium-activated chloride channel. Mol. Pharmacol. 73, 758–768 [DOI] [PubMed] [Google Scholar]
- 36. Scudieri P., Sondo E., Caci E., Ravazzolo R., Galietta L. J. (2013) TMEM16A-TMEM16B chimaeras to investigate the structure-function relationship of calcium-activated chloride channels. Biochem. J. 452, 443–455 [DOI] [PubMed] [Google Scholar]
- 37. Fallah G., Romer T., Detro-Dassen S., Braam U., Markwardt F., Schmalzing G. (2011) TMEM16A(a) /anoctamin-1 shares a homodimeric architecture with CLC chloride channels. Mol. Cell. Proteomics 10.1074/mcp.M110.004697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sheridan J. T., Worthington E. N., Yu K., Gabriel S. E., Hartzell H. C., Tarran R. (2011) Characterization of the oligomeric structure of the Ca2+-activated Cl− channel Ano1/TMEM16A. J. Biol. Chem. 286, 1381–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tien J., Lee H. Y., Minor D. L., Jr., Jan Y. N., Jan L. Y. (2013) Identification of a dimerization domain in the TMEM16A calcium-activated chloride channel (CaCC). Proc. Natl. Acad. Sci. U.S.A. 110, 6352–6357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xiao Q., Yu K., Perez-Cornejo P., Cui Y., Arreola J., Hartzell H. C. (2011) Voltage- and calcium-dependent gating of TMEM16A/Ano1 chloride channels are physically coupled by the first intracellular loop. Proc. Natl. Acad. Sci. U.S.A. 108, 8891–8896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suzuki T., Suzuki J., Nagata S. (2014) Functional swapping between transmembrane protein TMEM16A and TMEM16F. J. Biol. Chem. 289, 7438–7447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sondo E., Scudieri P., Tomati V., Caci E., Mazzone A., Farrugia G., Ravazzolo R., Galietta L. J. (2014) Non-canonical translation start sites in the TMEM16A chloride channel. Biochim. Biophys. Acta 1838, 89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maass K., Fischer M. A., Seiler M., Temmerman K., Nickel W., Seedorf M. (2009) A signal comprising a basic cluster and an amphipathic α-helix interacts with lipids and is required for the transport of Ist2 to the yeast cortical ER. J. Cell Sci. 122, 625–635 [DOI] [PubMed] [Google Scholar]
- 44. Fischer M. A., Temmerman K., Ercan E., Nickel W., Seedorf M. (2009) Binding of plasma membrane lipids recruits the yeast integral membrane protein Ist2 to the cortical ER. Traffic 10, 1084–1097 [DOI] [PubMed] [Google Scholar]
- 45. Ludewig U., Pusch M., Jentsch T. J. (1996) Two physically distinct pores in the dimeric ClC-0 chloride channel. Nature 383, 340–343 [DOI] [PubMed] [Google Scholar]
- 46. Jentsch T. J., Stein V., Weinreich F., Zdebik A. A. (2002) Molecular structure and physiological function of chloride channels. Physiol. Rev. 82, 503–568 [DOI] [PubMed] [Google Scholar]
- 47. Charlesworth G., Plagnol V., Holmström K. M., Bras J., Sheerin U. M., Preza E., Rubio-Agusti I., Ryten M., Schneider S. A., Stamelou M., Trabzuni D., Abramov A. Y., Bhatia K. P., Wood N. W. (2012) Mutations in ANO3 cause dominant craniometrical dystonia: ion channel implicated in pathogenesis. Am. J. Hum. Genet. 91, 1041–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sarkozy A., Hicks D., Hudson J., Laval S. H., Barresi R., Hilton-Jones D., Deschauer M., Harris E., Rufibach L., Hwang E., Bashir R., Walter M. C., Krause S., van den Bergh P., Illa I., Pénisson-Besnier I., De Waele L., Turnbull D., Guglieri M., Schrank B., Schoser B., Seeger J., Schreiber H., Gläser D., Eagle M., Bailey G., Walters R., Longman C., Norwood F., Winer J., Muntoni F., Hanna M., Roberts M., Bindoff L. A., Brierley C., Cooper R. G., Cottrell D. A., Davies N. P., Gibson A., Gorman G. S., Hammans S., Jackson A. P., Khan A., Lane R., McConville J., McEntagart M., Al-Memar A., Nixon J., Panicker J., Parton M., Petty R., Price C. J., Rakowicz W., Ray P., Schapira A. H., Swingler R., Turner C., Wagner K. R., Maddison P., Shaw P. J., Straub V., Bushby K., Lochmüller H. (2013) ANO5 gene analysis in a large cohort of patients with anoctaminopathy: confirmation of male prevalence and high occurrence of the common exon 5 gene mutation. Hum. Mutat. 34, 1111–1118 [DOI] [PubMed] [Google Scholar]
- 49. Bolduc V., Marlow G., Boycott K. M., Saleki K., Inoue H., Kroon J., Itakura M., Robitaille Y., Parent L., Baas F., Mizuta K., Kamata N., Richard I., Linssen W. H., Mahjneh I., de Visser M., Bashir R., Brais B. (2010) Recessive mutations in the putative calcium-activated chloride channel Anoctamin 5 cause proximal LGMD2L and distal MMD3 muscular dystrophies. Am. J. Hum. Genet. 86, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vermeer S., Hoischen A., Meijer R. P., Gilissen C., Neveling K., Wieskamp N., de Brouwer A., Koenig M., Anheim M., Assoum M., Drouot N., Todorovic S., Milic-Rasic V., Lochmüller H., Stevanin G., Goizet C., David A., Durr A., Brice A., Kremer B., van de Warrenburg B. P., Schijvenaars M. M., Heister A., Kwint M., Arts P., van der Wijst J., Veltman J., Kamsteeg E. J., Scheffer H., Knoers N. (2010) Targeted next-generation sequencing of a 12.5 Mb homozygous region reveals ANO10 mutations in patients with autosomal-recessive cerebellar ataxia. Am. J. Hum. Genet. 87, 813–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Namkung W., Yao Z., Finkbeiner W. E., Verkman A. S. (2011) Small-molecule activators of TMEM16A, a calcium-activated chloride channel, stimulate epithelial chloride secretion and intestinal contraction. FASEB J. 25, 4048–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kumar S., Namkung W., Verkman A. S., Sharma P. K. (2012) Novel 5-substituted benzyloxy-2-arylbenzofuran-3-carboxylic acids as calcium activated chloride channel inhibitors. Bioorg. Med. Chem. 20, 4237–4244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Namkung W., Phuan P. W., Verkman A. S. (2011) TMEM16A inhibitors reveal TMEM16A as a minor component of calcium-activated chloride channel conductance in airway and intestinal epithelial cells. J. Biol. Chem. 286, 2365–2374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jung J., Nam J. H., Park H. W., Oh U., Yoon J. H., Lee M. G. (2013) Dynamic modulation of ANO1/TMEM16A HCO3(−) permeability by Ca2+/calmodulin. Proc. Natl. Acad. Sci. U.S.A. 110, 360–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Arreola J., Melvin J. E., Begenisich T. (1996) Activation of calcium-dependent chloride channels in rat parotid acinar cells. J. Gen. Physiol. 108, 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kuruma A., Hartzell H. C. (2000) Bimodal control of a Ca2+-activated Cl− channel by different Ca2+ signals. J. Gen. Physiol. 115, 59–80 [DOI] [PMC free article] [PubMed] [Google Scholar]