Background: Collagen-binding proteins regulate tissue-specific extracellular matrices.
Results: CHADL is enriched in cartilage, binds collagen, and modulates the chondrocyte phenotype.
Conclusion: CHADL regulates the chondrocyte microenvironment.
Significance: Characterizing novel collagen-associated proteins is crucial to understand the constitution and function of specialized extracellular matrices.
Keywords: Cartilage, Chondrocyte, Chondrogenesis, Collagen, Connective Tissue
Abstract
The constitution and biophysical properties of extracellular matrices can dramatically influence cellular phenotype during development, homeostasis, or pathogenesis. These effects can be signaled through a differentially regulated assembly of collagen fibrils, orchestrated by a family of collagen-associated small leucine-rich proteins (SLRPs). In this report, we describe the tissue-specific expression and function of a previously uncharacterized SLRP, chondroadherin-like (CHADL). We developed antibodies against CHADL and, by immunohistochemistry, detected CHADL expression mainly in skeletal tissues, particularly in fetal cartilage and in the pericellular space of adult chondrocytes. In situ hybridizations and immunoblots on tissue lysates confirmed this tissue-specific expression pattern. Recombinant CHADL bound collagen in cell culture and inhibited in vitro collagen fibrillogenesis. After Chadl shRNA knockdown, chondrogenic ATDC5 cells increased their differentiation, indicated by increased transcript levels of Sox9, Ihh, Col2a1, and Col10a1. The knockdown increased collagen II and aggrecan deposition in the cell layers. Microarray analysis of the knockdown samples suggested collagen receptor-related changes, although other upstream effects could not be excluded. Together, our data indicate that the novel SLRP CHADL is expressed in cartilaginous tissues, influences collagen fibrillogenesis, and modulates chondrocyte differentiation. CHADL appears to have a negative regulatory role, possibly ensuring the formation of a stable extracellular matrix.
Introduction
To gain a comprehensive understanding of connective tissue biology, it is crucial to study proteins that regulate tissue- and function-specific collagen fibril assembly. Over the past three decades, several homologous small leucine-rich proteins (SLRPs)2 have been identified and functionally evaluated. Many of these proteins are extracellularly associated with collagen and can influence collagen fibrillogenesis in vitro (1), for example decorin, biglycan, fibromodulin, and lumican. SLRP knockout mouse phenotypes reveal that the lack of a given SLRP cannot be compensated for by another SLRP. Collagen fibrils in specific knockout mouse tissues appear to assemble in a disordered manner. This leads to tissue-specific phenotypes. Decorin-deficient mice have fragile skin (2), lumican-deficient mice have opaque corneas (3), biglycan-deficient mice have osteoporotic bones (4), and fibromodulin-deficient mice have mechanically weak tendons with increased collagen cross-linking (5, 6). Compound SLRP deficiency further aggravates the abnormal collagen fibril phenotype, suggesting concerted action of SLRPs during collagen fibrillogenesis (5, 7, 8). Therefore, the tissue-specific or even temporospecific expression of SLRPs modulates the architecture and cross-linking of the growing collagen fibers. Some SLRPs can even inhibit binding to collagen of each other, e.g. fibromodulin and lumican (9–11) or asporin and decorin (12, 13), which contributes to another level of collagen fibrillogenesis regulation.
Not all SLRPs have been characterized. One that is still undescribed is chondroadherin-like (CHADL). CHADL resides on chromosome 22 and is 19% homologous with chondroadherin. Chondroadherin is a collagen- and integrin α2β1-binding SLRP expressed in cartilage and bone (14–17) whose deficiency in mice leads to thinner cortical bone and a longer proliferative growth plate zone (18). The conspicuous difference between CHADL and other SLRPs is its size. Twice as large as most SLRPs, the CHADL gene appears to have arisen by tandem duplication of an entire single SLRP gene, the middle gap having been joined by a proline- and arginine-rich linker domain. Also, the integrin-binding site of chondroadherin is not well conserved in CHADL, and, unlike other SLRPs, CHADL features several interspersed cysteine residues besides the conserved SLRP-characteristic cysteine loops in the N- and C-terminal flanking (LRRNT and LRRCT) domains (Fig. 1A).
FIGURE 1.
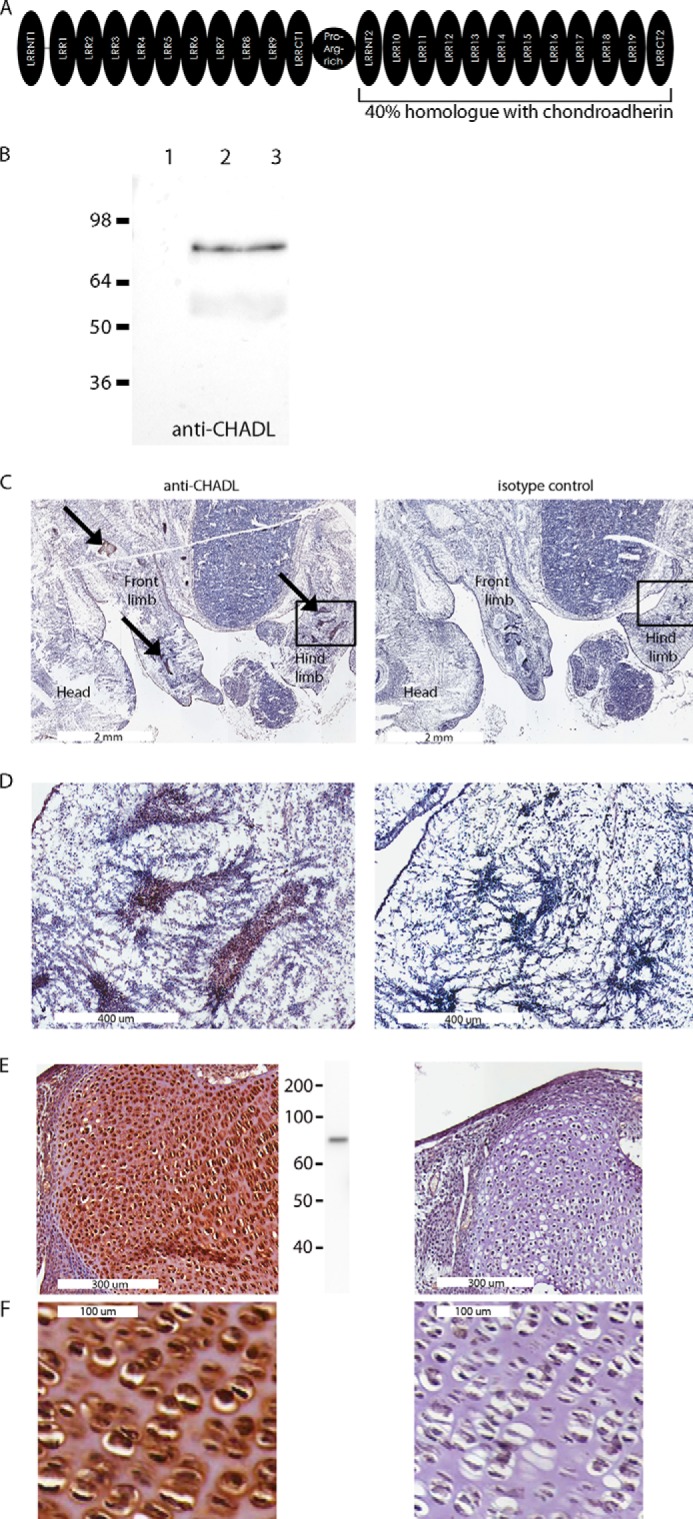
A, domain distribution in CHADL. Each half of CHADL contains several leucine-rich domains (LRR) flanked by LRRNT and LRRCT domains. Both halves are linked by a proline- and arginine-rich domain. B, characterization of anti-CHADL antibody. Immunoblots performed on medium collected from control non-transfected cells (lane 1), CHADL-expressing cells transfected with CHADL-pCEP4 expression vector (lane 2), and cell lysate from chondrogenic ATDC5 cells (lane 3). C, whole-mount mouse E14.5 embryos stained with anti-CHADL (left panel) and isotype negative controls (right panel). Stainings were most prominent in mesenchymal condensations (arrows). Mesenchymal condensations in the marked areas are magnified in D. E, stained 2-month-old knee joint cartilage. Center panel, immunoblot of cartilage extract probed with anti-CHADL. F, magnified image of E to demonstrate the pericellular distribution of CHADL.
Here we evaluated the expression profile and function of CHADL to assess the role of CHADL in connective tissue biology. We detected CHADL primarily in extracellular matrices of cartilage tissues and found it to be associated with collagen and influence chondrocyte differentiation in vitro.
EXPERIMENTAL PROCEDURES
Reagents
Insulin, transferrin, sodium selenite, glucose oxidase, catalase, and hygromycin were from Sigma. Nuclease-free water, PBS, SSC, Tris, and Lipofectamine were from Invitrogen. α Minimum Eagle's medium, DMEM/F12, and 293 expression medium were from Thermo Scientific. The ATDC5 and HFL1 cell lines were from Sigma. The Chadl Stellaris probes were from Biosearch Technologies. The Ni-NTA affinity purification cartridge was from Qiagen. Antibodies were from Abcam (anti-collagen I, catalog no. ab34710; anti-collagen III, catalog no. ab7778), Pierce (anti-aggrecan, catalog no. PA1-1745), Genscript (anti-actin), or in-house (anti-collagen II).
Antibody against CHADL
A rabbit polyclonal antibody was made against the peptide FPSDTQLLDLRRNH, covering amino acids 423–436 of the human CHADL protein. The corresponding mouse Chadl sequence is FPNDTQLLDLRRNH. The antiserum was purified on protein A-Sepharose, and the specificity was confirmed by immunoblotting against cell medium containing recombinant CHADL or against medium from non-transfected control cells.
Immunohistochemistry
Sections of frozen mouse embryos, ranging from E10-E17, and 2-month-old mouse knee joints were fixed in 4% formalin in PBS for 5 min, rinsed in TBS, and incubated in 0.3% hydrogen peroxide for 15 min. The slides were then incubated with hyaluronidase and chondroitinase ABC for 15 min. After rinsing with TBS, the slides were blocked with 10% goat serum in TBS for 1 h and then incubated with anti-CHADL diluted to 1 μg/ml in TBS with 1% goat serum. The slides were then washed with TBS and stained with the ultra-sensitive ABC rabbit IgG staining kit (Pierce) and diaminobenzidine peroxidase substrate kit (Vector Labs).
In Situ Hybridization
Frozen sections of mouse embryos or 2-week old mouse articular cartilage were fixed with phosphate-buffered 4% formaldehyde for 10 min, washed twice with PBS, and permeabilized for 5 h in 70% ethanol. The slides were equilibrated in wash buffer (2× SSC, 10% formamide) twice for 3 min and hybridized overnight at 37 °C with 1 μm Stellaris Chadl antisense probes diluted in hybridization buffer (10% dextran sulfate, 2× SSC, and 10% formamide). Slides were then incubated in wash buffer for 30 min at 37 °C and once again under the same conditions but with 5 ng/ml DAPI. The slides were then resuspended in 2× SSC, equilibrated with anti-fade GLOX buffer (2× SSC, 0.4% glucose, and 10 mm Tris-HCl (pH 8.0)), and incubated with GLOX buffer including glucose oxidase (1:100 v/v) and catalase (1:100 v/v). The slides were imaged in a fluorescence microscope.
Cell Culture
ATDC5 cells were cultured in DMEM/F12 1:1 medium with 5% fetal bovine serum (HyClone), 30 nm sodium selenite, and 10 μg/ml transferrin in a 5% CO2 atmosphere. For differentiation, 10 μg/ml insulin and 35 μg/ml ascorbate was added. HFL1 cells were cultured in α minimum Eagle's medium with 10% fetal bovine serum and 50 μg/ml ascorbate.
Immunoblotting
PVDF membranes with human tissue lysates were purchased from Zyagen. Mouse cartilage and other connective tissue lysates were obtained by boiling the tissues in 1% SDS. Cell layer homogenates were made by solubilizing the cells in TRIzol reagent (Invitrogen) and purifying proteins according to the instructions of the manufacturer. The cell medium was mixed with SDS-PAGE loading buffer. Protein amounts were quantified using Micro-BCA assays (Pierce). 30 μg of total protein was run on 4–12% BisTris SDS-PAGE reducing gels (Genscript). Proteins were transferred to a nitrocellulose membrane. The membranes were immunoblotted using 5% milk in TBS for blocking, TBS with 0.5% Tween 20 for washing, and washing buffer with 0.5% milk as antibody diluent. All antibodies were used at 1 μg/ml, and secondary HRP-conjugated donkey anti-rabbit or anti-mouse antibodies (Dako) were diluted 1:15,000. Blots were developed with SuperSignal West Dura (Pierce) and imaged using a charge-coupled device camera.
Recombinant Protein Expression
cDNA for human CHADL was synthesized and cloned into the pCEP4 vector with His6 tag-encoding sequences added to both flanks. This construct was transfected into 293T cells using Lipofectamine 2000, and CHADL-expressing cells were cloned and selected using 250 μg/ml hygromycin. After expansion, cells were grown in 293 expression medium supplemented with 50 μg/ml hygromycin. Protein was purified from the collected medium using Ni-NTA cartridges coupled to an Äkta chromatograph. The protein identity was confirmed using mass spectrometry.
Proximity Ligation Assay
The assay reagent Duolink® was purchased from Sigma. HFL1 cells were seeded on coverslips at 10,000 cells/well and transfected with the CHADL-pCEP construct using Lipofectamine 2000. For the negative control, cells were transfected with a His-tagged, non-collagen-binding domain of fibromodulin (LRR domains 1–3) in the pCEP vector. This fibromodulin domain is partially homologous with the CHADL LRR domains, but does not bind collagen, as shown previously (10. After 1 day, the cells were briefly fixed with paraformaldehyde and processed for the proximity ligation assay according to the instructions of the manufacturer. Interactions were detected using antibodies against the His tag (Abcam, catalog no. ab18184) and against procollagen (LF42, a gift from Dr. Larry Fisher).
Collagen Fibrillogenesis Functional Assay
The assay is described in Ref. 19. Briefly, 100 μg/ml of acid-extracted pepsinated collagen type I was neutralized into HEPES buffer with 0.15 m NaCl and incubated with or without recombinant CHADL protein at 5 μg/ml. This ratio amounts to a 5-fold molar excess of collagen. Samples were incubated at 37 °C in a spectrometer that continuously measured absorbance at 400 nm.
Cosedimentation Assay
100 μg/ml collagen type I and 5 μg/ml CHADL (i.e. a 5-fold molar excess of collagen) were incubated in PBS buffer (pH 7.4) at 37 °C for 8 h. The mixture was centrifuged at 20,000 × g. The supernatant and pellet were mixed separately with SDS-PAGE loading buffer, run on a 4–12% BisTris reducing gel, and transferred to a nitrocellulose membrane. Immunoblots for collagen and CHADL were performed using rabbit anti-collagen and rabbit anti-CHADL antibodies.
Cell-binding Assay
The assay is described in Ref. 17. A 48-well plate was precoated with CHADL, collagen II, or BSA at 5 μg/ml in PBS. 105kc cells (chondrosarcoma) were seeded in PBS at 50,000 cells/well, incubated for 1 h, and then the non-bound cells were washed off. The bound cells were quantified using a lysosomal N-acetylglucosaminidase assay (20).
shRNA Knockdown of Chadl Expression in ATDC5 Cells
27-mer shRNA duplexes were purchased from Origene. The only effective knockdown was obtained from the construct ACAAGGUAGAGAAACAAAGAGACCA. A universal scrambled negative control duplex (Origene) was used as a negative control. ATDC5 cells were transfected with 30 nm shRNA using Lipofectamine 2000 according to the protocol of the manufacturer. After 16 h of incubation, the medium was changed to differentiation medium that was replaced every 2 days for the remaining course of the experiment. Knockdown of the Chadl transcript was confirmed by qPCR using TaqMan assays (Invitrogen) run on an Applied Biosystems 7900 HT detection system. Cell cultures were used for microarray studies and for assessing gene expression by qPCR and immunoblotting.
Alcian Blue Staining
Cells were rinsed with PBS three times and fixed with 95% ethanol for 5 min at −20 °C. Then the cells were stained with 0.1% Alcian blue 8GX in 0.1 m HCl overnight and washed three times with water. The staining was quantified by dissolving the dye in 6 m guanidine HCl overnight, and absorbance was read at 650 nm.
Microarrays
ATDC5 cells were transfected with Chadl shRNA and differentiated for 5 days. RNA was isolated using a Quick-RNA kit (Zymo Research). Amplified RNA was generated, hybridized onto an Affymetrix MG-430 PM array strip, and processed according to the instructions of the manufacturer. Two control samples and two different knockdown samples were used. The raw data were normalized using robust multi-array analysis (21), in which raw intensities are background-corrected, log2-transformed, and then normalized using quantiles, as provided by R and Bioconductor. A linear model was fitted to the data using the linear modeling of microarray data (22) package to obtain an expression value for each probe set. Probe sets with a log2 -fold change >0.5 and p < 0.05 were considered differentially expressed. To functionally characterize the resulting gene lists, the Database for Annotation, Visualization and Integrated Discovery (23) and Ingenuity Pathway Analysis (Ingenuity Systems®) were employed.
RT-PCR and Real-time PCR
Total RNA was extracted from cells with TRIzol reagent (Invitrogen). 100 ng of RNA was used for reversed transcription using the First Strand cDNA synthesis system (Origene). Real-time PCR was performed with a TaqMan RNA-to-CT 1-step kit (Invitrogen) using TaqMan probes and an Applied Biosystems 7900 HT detection system. Gene expression was normalized to the Actb transcript.
RESULTS
Tissue Expression Profiling of Chadl
To test the specificity of our anti-CHADL antibody, we performed immunoblotting on medium collected from 293 cells transfected with the CHADL expression vector. As a negative control, we used medium collected from non-transfected cells. Only the cells expressing CHADL produced protein, detected as an 80-kDa band with the anti-CHADL antibody (Fig. 1B). Preimmune antiserum did not recognize recombinant CHADL (data not shown).
To evaluate where Chadl is expressed in mice, we performed immunohistochemistry on whole-mount mouse embryos using the anti-CHADL antibody. We could not detect Chadl in whole-mount embryos before E13.5 (data not shown), at which stage, and in later developmental stages, the staining was prominent only in mesenchymal condensations and in cartilaginous tissues and not present in any other tissues. Fig. 1, C and D, shows a typical staining in chondrogenic condensations at E14.5. We also detected Chadl in 2-month-old articular knee cartilage (Fig. 1E), where the protein was localized to the extracellular space in the area immediately surrounding the chondrocytes (Fig. 1F), and no apparent staining appeared in other limb tissues (data not shown). This finding was in concert with immunoblotting of mouse cartilage extract with anti-CHADL. An 80-kDa Chadl band could be detected in articular cartilage (Fig. 1E, center panel). We also performed immunoblots of other tissue extracts but could not detect Chadl in any of the following: brain, stomach, intestine, colon, liver, lung, kidney, heart, ovary, skeletal muscle, spleen, testis, thymus, placenta, and pancreas. We did not detect Chadl in other connective tissues, including skin, ligament, tendon, meniscus, and bone (data not shown).
In situ hybridization using Chadl antisense probes on whole-mount mouse embryos also revealed a high Chadl transcript level in the mesenchymal condensations and in cartilage, and we also detected Chadl transcripts in articular cartilage (Fig. 2). Together, data from the different tissue profiling methods suggest that CHADL is expressed preferentially in embryonic developing skeleton and in extracellular matrix of adult cartilage.
FIGURE 2.
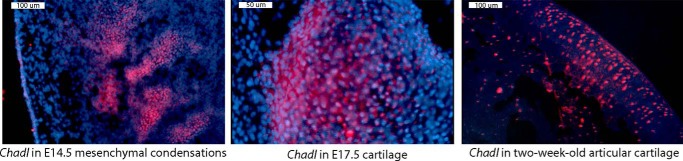
In situ hybridization of Chadl transcripts in embryonic chondrogenic tissues and in 2-week-old articular cartilage. Frozen tissue sections were briefly fixed, permeabilized with ethanol, and probed with a mix of antisense Chadl Stellaris red fluorescent probes. Sections were counterstained with DAPI (blue nuclei).
Functional Relevance of Chadl in Collagen Biology
One half of CHADL is 40% homologous with the collagen-binding protein chondroadherin. To investigate whether CHADL also associates with collagen, we used proximity ligation assays on collagen-producing HFL1 cells transfected with the His tag-CHADL-pCEP expression vector. The assay revealed a strong association of CHADL with collagen, which was not observed in cells transfected with a non-collagen-binding His-tagged fibromodulin fragment (Fig. 3A).
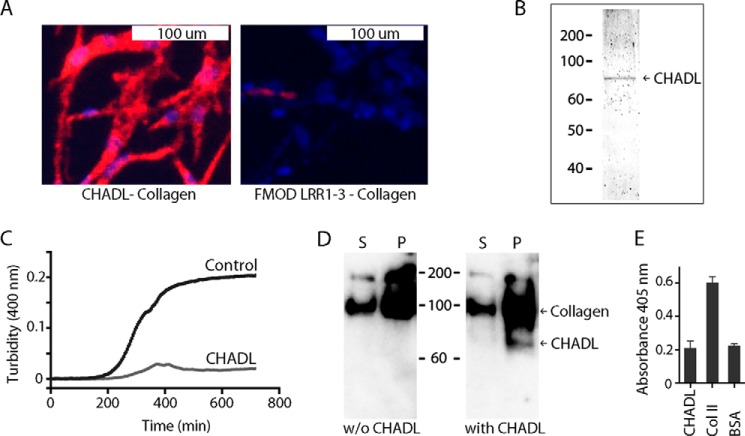
FIGURE 3.
Interaction of CHADL with collagen. A, proximity ligation assay on HFL1 fibroblasts transfected with the expression vector containing His-tagged CHADL. Cells were fixed, incubated with mouse anti-His and rabbit anti-procollagen antibodies, and then processed for proximity ligation assay using secondary antibodies and reagents. Protein interactions are detected by red fluorescence. Cells were counterstained with DAPI (blue nuclei). Right panel, negative control. Cells were transfected with a non-collagen-binding His-tagged fibromodulin fragment containing the LRR1–3 domains. B, Coomassie-stained gel of Ni-NTA purified recombinant His-tagged CHADL expressed in human 293 cells. C, collagen fibrillogenesis in vitro assay. Pepsin-extracted collagen was neutralized in HEPES-buffered saline buffer and supplemented with recombinant CHADL at a molar ratio of 5:1. The control sample was without CHADL. Samples were incubated at 37 °C, and absorbance at 400 nm was continuously recorded to follow fibrillogenesis in real time. D, cosedimentation assay. 3 μg of collagen was incubated with or without (w/o) 150 ng of CHADL in PBS (molar ratio collagen:CHADL, 5:1), incubated at 37 °C, and centrifuged after 8 h to separate the formed fibrils from soluble collagens. The two collagen fractions (pellet (P) and supernatant (S)) were run on SDS-PAGE and immunoblotted for collagen and CHADL. E, cell-binding assay. Chondrocytes were added to wells precoated with CHADL, collagen II (positive control), or BSA (negative control). After 1 h, the non-bound cells were removed, and the bound cells were quantified using lysosomal N-acetylglucosaminidase assay. Error bars show mean ± S.D. (n = 3).
We also investigated whether recombinant CHADL (Fig. 3B) could influence collagen fibrillogenesis in vitro. Although the control reaction progressed through a steady log phase, eventually reaching a plateau, supplementing the reaction with CHADL at a collagen:CHADL molar ratio of 5:1 (5-fold molar excess of collagen) reduced the extent of fibrillogenesis by over 90% (Fig. 3C). In a similar experiment, we mixed collagen and CHADL at a 5-fold molar excess of collagen, allowed fibrils to form, and then centrifuged the sample to spin down the fibrils. The pellet and the supernatant were separately analyzed by immunoblotting for collagen and CHADL. CHADL was present exclusively in the pelleted fraction of collagens (Fig. 3D).
Because the homologue chondroadherin interacts with chondrocytes via integrins, we also tested whether CHADL can influence cell binding. However, we could not detect any interactions between chondrocytes and CHADL (Fig. 3E).
Effect of Chadl Knockdown on Differentiating Chondrogenic Cells
Because the tissue profiling data suggest a specific function of Chadl in chondrocyte biology, we hypothesized that Chadl knockdown in a chondrogenic cell would influence specific cellular functions pertinent to cartilage biology. Therefore, we studied the effects of Chadl knockdown in chondrogenic ATDC5 cells.
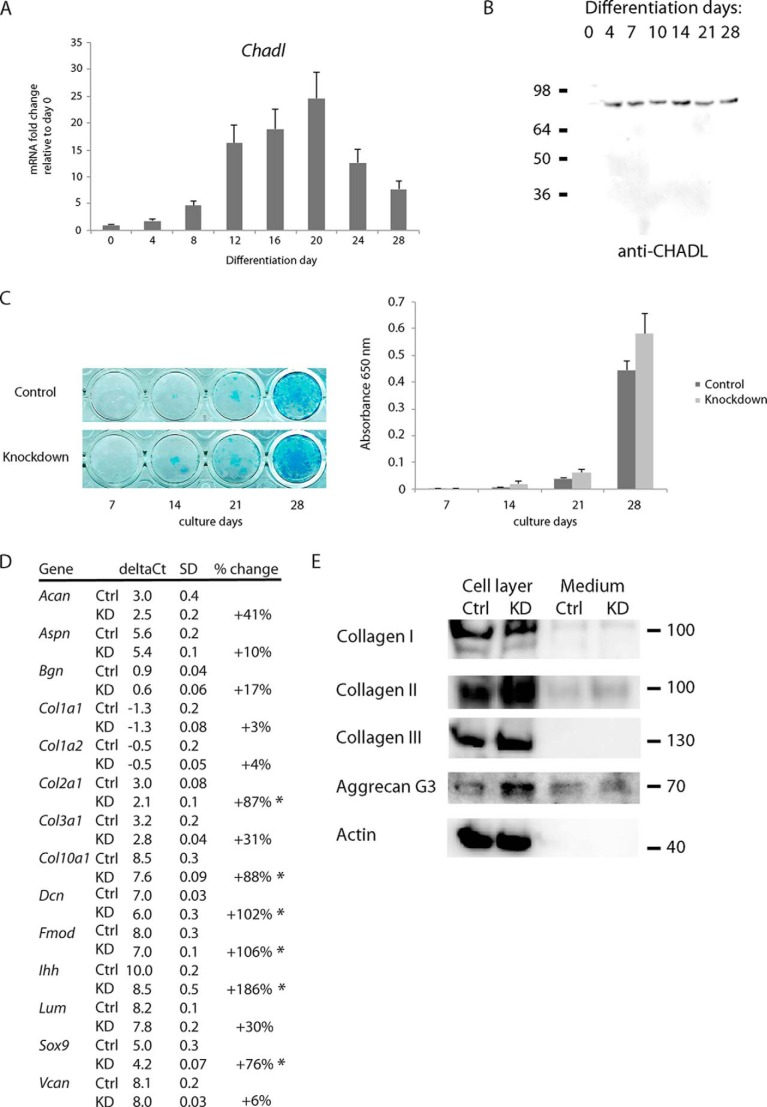
First we examined the temporal Chadl expression pattern in the differentiating ATDC5 cells using both qPCR and immunoblotting. Chadl transcripts increased in early stages of differentiation and persisted until day 20, after which they decreased (Fig. 4A). Evaluated by immunoblotting, Chadl protein levels varied only slightly during the entire differentiation period. Notably, Chadl appears to be absent in the prechondrogenic, non-differentiating, stage (Fig. 4B).
FIGURE 4.
A, expression of Chadl in differentiating chondrogenic ATDC5 cells measured by qPCR using TaqMan assays (n = 3). Transcript levels were normalized to levels in the predifferentiation stage (day 0). B, immunoblotting of differentiating ATDC5 cells. Cells were solubilized in TRIzol on different differentiation days. Proteins were extracted and run on reducing SDS-PAGE, transferred to a nitrocellulose membrane, and blotted for CHADL. C, Alcian blue staining and quantification of differentiating ATDC5 cells after Chadl shRNA knockdown or negative control shRNA. Cells were fixed at the indicated differentiation days and stained for proteoglycans using Alcian blue. Staining was quantified by measuring absorbance at 650 nm after solubilizing the stained samples (n = 4) with guanidine. D, qPCR on differentiation markers (Ihh, Col10a1, Sox9, Col2a1) and extracellular matrix genes in Chadl shRNA knockdown-treated or control shRNA-treated differentiating ATDC5 cells (n = 3) at differentiation day 14. Transcript levels were normalized to Actb. For clarity, ΔCt values and S.D. values were rounded off. The percent change was calculated from ΔΔCt (data not shown) using original values. *, p < 0.05 calculated using unpaired Student's t test. Ctrl, control; KD, knockdown sample. E, immunoblots on collagens and aggrecan using cell layer material extracted with TRIzol or cell medium from cultures used in D. Proteins were run on a 4–12% BisTris reduced SDS-PAGE, transferred to a nitrocellulose membrane, and immunoblotted for collagens I, II, and III, the aggrecan G3 domain (here cleaved off the full-length aggrecan), and actin.
Next, we analyzed the effects of Chadl shRNA-mediated knockdown on the differentiating ATDC5 cells. The shRNA construct induced over 75% Chadl knockdown between days 3 and 7 post-transfection, after which the Chadl levels started to increase (evaluated by qPCR, data not shown). We analyzed whether the knockdown of Chadl during early differentiation induced long-term effects on the cartilaginous phenotype of ATDC5 cells. In general, Chadl knockdown enhanced the differentiation of the cells. More intense Alcian blue staining indicated increased cartilage matrix proteoglycan production in cells treated with Chadl shRNA (Fig. 4C). Although not statistically significant, data from three independent experiments showed a similar trend. More dramatic changes were observed by qPCR. At day 14 post-transfection, the knockdown samples had markedly increased levels of Sox9 and Ihh, both associated with increased chondrocyte differentiation. We also detected a general increase of transcripts of many matrix-related genes, including collagens and some SLRPs (Fig. 4D), along with increased deposition of collagen II and aggrecan in the cell layers but with minor or no changes in the amount of collagens I and III (Fig. 4E). We did not observe any nonspecific interferon response or cell stress response induced by the shRNAs, as evaluated by mRNA microarray. We did not detect any changes in expression of the interferon-response genes Oas1, Oas2, Ifitm1, Mx1, or of endoplasmic reticulum stress-associated protein disulfide isomerase, calreticulin, or autophagy-associated LC3.
Effects of Chadl Knockdown Assessed by Microarrays
We used microarrays to assess the signaling and transcriptional profiles following Chadl knockdown. We analyzed transcripts from day 4 after shRNA transfection, when the knockdown effect was strongest. Hierarchical clustering of the top 300 genes by variance showed that Chadl knockdown resulted in a contrasting transcriptional response (24). Using differential expression analysis with a log2 -fold change cutoff of 1.0, 56 non-redundant genes were up-regulated, whereas 46 genes were down-regulated. With a lower cutoff of 0.5, 293 up-regulated and 199 down-regulated genes were detected. Diseases and functional categories predicted to be activated were related to growth and proliferation (z score >2.0), with the transcriptome profiles being common in angiogenesis, growth of neuritis, proliferation of connective tissue cells, and formation of cellular protrusions (Fig. 5A). The relevant genes were the up-regulated insulin-like growth factor binding protein 3 (IGFBP3), vascular cell adhesion molecule 1 (VCAM1), and MET proto-oncogene, and the down-regulated histone deacetylase 9 (Hdac9) and cyclin-dependent kinase inhibitor 1A (CDKN1A) (Fig. 5B). Utilizing the clustering function of the Database for Annotation, Visualization, and Integrated Discovery (on the basis of Gene Ontology terms), the top categories were consistent and comprised positive regulation of developmental process and cell differentiation and proliferation. Genes associated with extracellular structure organization, ossification, and skeletal system development were also enriched.
FIGURE 5.
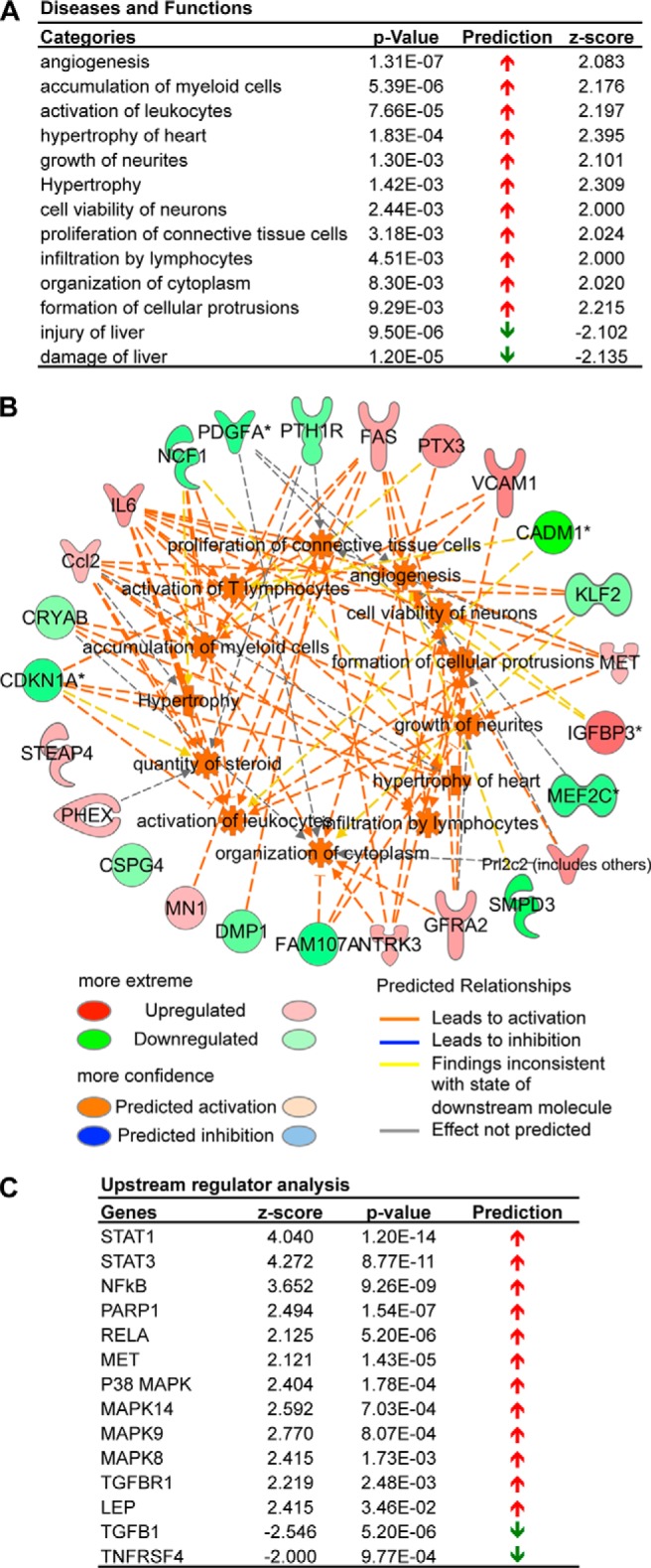
Transcriptomics analysis of Chadl shRNA knockdown in ATDC5 cells. A, diseases and biological functions for the differentially expressed genes with their respective activation p values, prediction states, and activation z scores. B, a network indicating the effects of the differentially regulated genes (outer circle) on individual diseases and functions categories (inner circle). C, identification of regulator cascades upstream of the observed transcriptional changes by upstream regulator analysis.
Using upstream regulator analysis to identify upstream regulator cascades, the predictions were consistent with a phenotype showing enhanced differentiation. RelA (p65) was predicted to be activated (activation z score of 2.125) and is known to up-regulate Sox9 (25), the master regulator of chondrogenesis. In addition, other differentiation regulators were predicted to be activated: NF-κB, STAT1, STAT3, and p38 MAPK (activation z-scores of 3.652, 4.040, 4.272, and 2.414, respectively) (Fig. 5C). Therefore, these findings are consistent with the functional role of Chadl in chondrogenesis.
DISCUSSION
Influence of extracellular matrices on cellular behavior depends significantly on the structure of the collagenous matrix. This structure can be modulated by a family of collagen-binding SLRPs, whose temporospatially regulated expression contributes to the fine regulation of the growing collagen fibrils (1–3, 5–7, 11, 13). To develop a comprehensive understanding of this process, it is necessary to identify and characterize the function of SLRPs. In this report, we profiled the expression and evaluated the function of a previously undescribed SLRP, CHADL. We observed CHADL to be expressed preferentially in embryonic cartilaginous tissues as well as in developing and in adult cartilage (Figs. 1 and 2). Interestingly, the expression in embryonic tissues did not appear until E13.5, correlating with the onset of extracellular matrix production and chondrogenic differentiation. Using our antibodies and in situ hybridization probes, we could not detect CHADL in other major organs or other connective tissues. Of course, we cannot exclude CHADL being expressed in other contexts, e.g. under specific pathological conditions, or being present in amounts below the detection limit of our methods. In cartilage, the protein is deposited in the extracellular matrix and in the pericellular matrix but not in the interterritorial space. This could allude to its potential function of regulating early collagen fibrillogenesis in cartilage. Furthermore, the expression of CHADL appeared to persist in mesenchymal tissues and cartilage from E13.5 on to adulthood, suggesting its role in maintaining homeostasis in the cartilage matrix. Together, this apparent tissue specificity could render CHADL useful as a biomarker for cartilage-related disorders, e.g. arthritis. The presence of CHADL in synovial fluid or blood samples and correlation with specific joint disorders deserves further attention.
What is the function of CHADL in cartilage? Because many SLRPs are collagen-associated, we investigated whether CHADL also interacts with and influences collagen fibrillogenesis. Proximity ligation assays showed that CHADL indeed associates with collagen, and that recombinant CHADL, at low molarity, reduces in vitro collagen fibrillogenesis and binds to collagen fibrils (Fig. 3). Interestingly, chondroadherin, although generally sharing the tissue-specific expression pattern of CHADL and interacting with collagen, does not change the course of fibrillogenesis.3 This can be due to the more bulky nature of CHADL. Being twice the size of chondroadherin, the potential for steric hindrance of collagen fibril assembly is improved. CHADL could also contain other collagen-binding sites targeted for a very effective restraining of the fibril assembly. Such a function could be important for maintaining a proper collagen structure near the surface of a chondrocyte, where the collagen matrix is less dense than farther from the cell, in the interterritorial space.
In addition, CHADL does not share the integrin-mediated cell-binding properties of chondroadherin (16), making it functionally disparate from its close homologue (Fig. 3E). Indeed, for this and other unique features of CHADL mentioned earlier, the name chondroadherin-like is misleading when considering its functional properties.
Because of its effect on collagen fibrillogenesis, CHADL could influence collagen receptor-mediated cellular response. After Chadl knockdown, microarray analysis indicated the activation of STAT1/3 and p38 MAPK. STAT1/3 and p38 MAPK can be regulated by collagen-binding integrin α2β1 and discoidin domain receptor collagen receptor signaling (26, 27). In addition, p65 - inducer of Sox9 in early chondrocyte differentiation (25) - was predicted to be activated (Fig. 5). The knockdown also accelerated longer term chondrocyte differentiation. Several days after the knockdown, chondrocyte differentiation markers Col2a1, Sox9, Col10a1 and Ihh were up-regulated, along with the synthesis of collagen and proteoglycans (Fig. 4). These transcripts were unaffected at day 4, probably because of low basal expression at the early stage of differentiation. Taken together, chondrocyte differentiation was accelerated after Chadl knockdown, some of which may have been triggered by an altered collagen structure.
In summary, the novel SLRP chondroadherin-like (CHADL) is an extracellular matrix protein, expressed in developing cartilaginous tissues, and in young and adult cartilage. CHADL is sequestered to pericellular space of the chondrocytes, and associates with collagen, modulating collagen fibrillogenesis. This regulatory role in chondrocyte's collagenous microenvironment appears to influence cell differentiation, evidenced by our Chadl knockdown studies. Indeed, CHADL appears to have a negative regulatory role during chondrocyte differentiation, possibly ensuring the formation of a stable extracellular matrix. Further animal studies are required to determine the impact of CHADL in vivo, mainly to what extent CHADL is restricted to chondrogenesis of articular cartilage, cartilage maturation and degradation, or if CHADL can function during fracture healing or tendon/ligament injuries. It also remains to be investigated whether CHADL can be a useful biomarker in specific joint and skeletal disorders.
Acknowledgments
We thank Prof. Dick Heinegard for discussions and for support and initiation of this project. We also thank Prof. Catharina Svanborg for supporting this work and for critical reading of the manuscript.
This study was supported by grants from the Swedish Research Council, the Crafoord Foundation, the Magnus Bergvall Foundation, the Åke Wiberg Foundation, the Greta and Johan Kock Foundation, and the Alfred Österlund Foundation.
The microarray data reported in this paper have been submitted to the Gene Expression Omnibus Repository with accession number GSE57740.
V. Tillgren, unpublished observation.
- SLRP
- small leucine-rich repeat protein
- LRR
- leucine-rich repeat
- CHADL
- chondroadherin-like
- Ni-NTA
- nickel-nitrilotriacetic acid
- E10
- embryonic day 10
- qPCR
- quantitative PCR.
REFERENCES
- 1. Kalamajski S., Oldberg A. (2010) The role of small leucine-rich proteoglycans in collagen fibrillogenesis. Matrix Biol. 29, 248–253 [DOI] [PubMed] [Google Scholar]
- 2. Danielson K. G., Baribault H., Holmes D. F., Graham H., Kadler K. E., Iozzo R. V. (1997) Targeted disruption of decorin leads to abnormal collagen fibril morphology and skin fragility. J. Cell Biol. 136, 729–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chakravarti S., Petroll W. M., Hassell J. R., Jester J. V., Lass J. H., Paul J., Birk D. E. (2000) Corneal opacity in lumican-null mice: defects in collagen fibril structure and packing in the posterior stroma. Invest. Ophthalmol. Vis. Sci. 41, 3365–3373 [PMC free article] [PubMed] [Google Scholar]
- 4. Xu T., Bianco P., Fisher L. W., Longenecker G., Smith E., Goldstein S., Bonadio J., Boskey A., Heegaard A. M., Sommer B., Satomura K., Dominguez P., Zhao C., Kulkarni A. B., Robey P. G., Young M. F. (1998) Targeted disruption of the biglycan gene leads to an osteoporosis-like phenotype in mice. Nat. Genet. 20, 78–82 [DOI] [PubMed] [Google Scholar]
- 5. Jepsen K. J., Wu F., Peragallo J. H., Paul J., Roberts L., Ezura Y., Oldberg A., Birk D. E., Chakravarti S. (2002) A syndrome of joint laxity and impaired tendon integrity in lumican- and fibromodulin-deficient mice. J. Biol. Chem. 277, 35532–35540 [DOI] [PubMed] [Google Scholar]
- 6. Kalamajski S., Liu C., Tillgren V., Rubin K., Oldberg Å., Rai J., Weis M., Eyre D. R. (2014) Increased C-telopeptide cross-linking of tendon type I collagen in fibromodulin-deficient mice. J. Biol. Chem. 289, 18873–18879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ezura Y., Chakravarti S., Oldberg A., Chervoneva I., Birk D. E. (2000) Differential expression of lumican and fibromodulin regulate collagen fibrillogenesis in developing mouse tendons. J. Cell Biol. 151, 779–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang G., Chen S., Goldoni S., Calder B. W., Simpson H. C., Owens R. T., McQuillan D. J., Young M. F., Iozzo R. V., Birk D. E. (2009) Genetic evidence for the coordinated regulation of collagen fibrillogenesis in the cornea by decorin and biglycan. J. Biol. Chem. 284, 8888–8897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Svensson L., Närlid I., Oldberg A. (2000) Fibromodulin and lumican bind to the same region on collagen type I fibrils. FEBS Lett. 470, 178–182 [DOI] [PubMed] [Google Scholar]
- 10. Kalamajski S., Oldberg A. (2007) Fibromodulin binds collagen type I via Glu-353 and Lys-355 in leucine-rich repeat 11. J. Biol. Chem. 282, 26740–26745 [DOI] [PubMed] [Google Scholar]
- 11. Kalamajski S., Oldberg A. (2009) Homologous sequence in lumican and fibromodulin leucine-rich repeat 5–7 competes for collagen binding. J. Biol. Chem. 284, 534–539 [DOI] [PubMed] [Google Scholar]
- 12. Kalamajski S., Aspberg A., Oldberg A. (2007) The decorin sequence SYIRIADTNIT binds collagen type I. J. Biol. Chem. 282, 16062–16067 [DOI] [PubMed] [Google Scholar]
- 13. Kalamajski S., Aspberg A., Lindblom K., Heinegård D., Oldberg A. (2009) Asporin competes with decorin for collagen binding, binds calcium and promotes osteoblast collagen mineralization. Biochem. J. 423, 53–59 [DOI] [PubMed] [Google Scholar]
- 14. Larsson T., Sommarin Y., Paulsson M., Antonsson P., Hedbom E., Wendel M., Heinegård D. (1991) Cartilage matrix proteins: a basic 36-kDa protein with a restricted distribution to cartilage and bone. J. Biol. Chem. 266, 20428–20433 [PubMed] [Google Scholar]
- 15. Neame P. J., Sommarin Y., Boynton R. E., Heinegård D. (1994) The structure of a 38-kDa leucine-rich protein (chondroadherin) isolated from bovine cartilage. J. Biol. Chem. 269, 21547–21554 [PubMed] [Google Scholar]
- 16. Camper L., Heinegård D., Lundgren-Akerlund E. (1997) Integrin α2β1 is a receptor for the cartilage matrix protein chondroadherin. J. Cell Biol. 138, 1159–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haglund L., Tillgren V., Addis L., Wenglén C., Recklies A., Heinegård D. (2011) Identification and characterization of the integrin α2β1 binding motif in chondroadherin mediating cell attachment. J. Biol. Chem. 286, 3925–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hessle L., Stordalen G. A., Wenglén C., Petzold C., Tanner E., Brorson S. H., Baekkevold E. S., Önnerfjord P., Reinholt F. P., Heinegård D. (2013) The skeletal phenotype of chondroadherin deficient mice. PloS ONE 8, e63080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hedbom E., Heinegård D. (1989) Interaction of a 59-kDa connective tissue matrix protein with collagen I and collagen II. J. Biol. Chem. 264, 6898–6905 [PubMed] [Google Scholar]
- 20. Landegren U. (1984) Measurement of cell numbers by means of the endogenous enzyme hexosaminidase: applications to detection of lymphokines and cell surface antigens. J. Immunol. Methods 67, 379–388 [DOI] [PubMed] [Google Scholar]
- 21. Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., Speed T. P. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wettenhall J. M., Smyth G. K. (2004) LimmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics 20, 3705–3706 [DOI] [PubMed] [Google Scholar]
- 23. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3. [PubMed] [Google Scholar]
- 24. Edgar R., Domrachev M., Lash A. E. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Caron M. M., Emans P. J., Surtel D. A., Cremers A., Voncken J. W., Welting T. J., van Rhijn L. W. (2012) Activation of NF-κB/p65 facilitates early chondrogenic differentiation during endochondral ossification. PloS ONE 7, e33467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu H., Bihan D., Chang F., Huang P. H., Farndale R. W., Leitinger B. (2012) Discoidin domain receptors promote α1β1- and α2β1-integrin mediated cell adhesion to collagen by enhancing integrin activation. PloS ONE 7, e52209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu L., Peng H., Glasson S., Lee P. L., Hu K., Ijiri K., Olsen B. R., Goldring M. B., Li Y. (2007) Increased expression of the collagen receptor discoidin domain receptor 2 in articular cartilage as a key event in the pathogenesis of osteoarthritis. Arthritis Rheum. 56, 2663–2673 [DOI] [PubMed] [Google Scholar]