Background: ERBB4 plays an important role in the etiology and progression of lung cancer.
Results: miR-193a-3p suppressed proliferation and invasion and promoted apoptosis in lung cancer cells and xenograft mice by negatively regulating ERBB4.
Conclusion: miR-193a-3p exerted an anti-tumor effect by negatively regulating ERBB4 in lung cancer.
Significance: This study may open new avenues for future lung cancer therapies.
Keywords: Cancer, Cancer Biology, Epidermal Growth Factor Receptor (EGFR), Lung Cancer, MicroRNA (miRNA)
Abstract
ERBB4, one of four ErbB receptor tyrosine kinase family members, plays an important role in the etiology and progression of lung cancer. In this study, we found that the ERBB4 protein levels were consistently up-regulated in lung cancer tissues, whereas the mRNA levels varied randomly, suggesting that a post-transcriptional mechanism was involved in regulating ERBB4 expression. Because microRNAs are powerful post-transcriptional regulators of gene expression, we used bioinformatic analyses to search for microRNAs that can potentially target ERBB4. We identified specific targeting sites for miR-193a-3p in the 3′-UTR of ERBB4. We further identified an inverse correlation between miR-193a-3p levels and ERBB4 protein levels, but not mRNA levels, in lung cancer tissue samples. By overexpressing or knocking down miR-193a-3p in lung cancer cells, we experimentally confirmed that miR-193a-3p directly recognizes the 3′-UTR of the ERBB4 transcript and regulates ERBB4 expression. Furthermore, the biological consequences of the targeting of ERBB4 by miR-193a-3p were examined in vitro via cell proliferation, invasion, and apoptosis assays and in vivo using a mouse xenograft tumor model. We demonstrated that the repression of ERBB4 by miR-193a-3p suppressed proliferation and invasion and promoted apoptosis in lung cancer cells and that miR-193a-3p exerted an anti-tumor effect by negatively regulating ERBB4 in xenograft mice. Taken together, our findings provide the first clues regarding the role of miR-193a-3p as a tumor suppressor in lung cancer through the inhibition of ERBB4 translation.
Introduction
Lung cancer is the most commonly diagnosed cancer and the leading cause of cancer-related deaths worldwide (1). Non-small cell lung cancer (NSCLC)5 is the predominant type of lung cancer, accounting for 75–80% of all cases (2). According to data from the International Association for the Study of Lung Cancer, the 5-year overall survival rate remains 15% (2, 3). The majority of lung cancer patients exhibit advanced disease, and chemotherapeutic agents are of limited use in patients who have relapsed tumors and who present metastatic disease (2, 3). Therefore, greater knowledge of the molecular mechanisms underlying lung carcinogenesis is of major importance and might provide novel strategies to improve survival and quality of life in lung cancer patients.
Following substantial advances in our understanding of tumor biology, key signaling pathways involved in mediating lung cancer growth and progression have been identified (4). Dominant oncogenes and tumor suppressor genes involved in the pathogenesis of lung cancer have attracted substantial interest, and their central roles and fundamental contribution to the misbehavior of lung cancer cells have become clear (5). One example is the ErbB family of receptor tyrosine kinases, whose overexpression is thought to be important in the development of a variety of human cancers, including lung cancer (6, 7). The ErbB family consists of four closely related members, including EGFR (epidermal growth factor receptor)/ERBB1 (v-Erb-b2 avian erythroblastic leukemia viral oncogene homolog 1)/HER1, ERBB2/HER2, ERBB3/HER3, and ERBB4/HER4 (8). To date, EGFR and ERBB2 have garnered the most attention, with therapeutics targeting EGFR in lung cancer and ERBB2 in breast cancer approved for clinical use (8). However, ERBB4 is emerging as an increasingly important player in the genesis and progression of human cancers. ERBB4 is frequently detected in a wide range of human cancers, including carcinomas of the colon, prostate, lung, ovary, pancreas, endometrium, bronchus, cervix, stomach, thyroid, and astrocytoma (9–20). Clinical and experimental data suggest that ERBB4 may play a significant role in the growth and survival of different human tumors (21). For example, it was reported that ERBB4 increases the proliferation potential of human lung cancer cells (22). In addition, analyses of ERBB4 expression or mutation may be of prognostic or predictive value (14). For example, ERBB4 expression is reported to be correlated with metastatic potential and patient survival in NSCLC (22). On the other hand, despite the fact that the applicability of ERBB4 as a drug target is still uncertain, several patents involving the utilization of ERBB4 have recently been issued (23). Some studies also provide evidence that ERBB4 plays a critical role in human lung cancer and may serve as a molecular target for anticancer therapy (22). Thus, manipulating the functions of ERBB4 may be therapeutically beneficial in cancer. However, despite these recent advances in our understanding of the important roles of ERBB4 in tumorigenesis, the precise molecular mechanism through which ERBB4 contributes to lung cancer progression remains largely unknown, highlighting the need for further investigations.
Over the past decade, a class of small, non-coding, single-stranded RNAs known as microRNAs (miRNAs) have emerged as major regulators of the initiation and progression of human cancers, including lung cancer (24, 25). The up-regulation of oncogenic miRNAs (targeting tumor suppressor genes) and the down-regulation of tumor-suppressive miRNAs (targeting oncogenes) lead to the dysfunction of cancer cells, including malignant proliferation, invasion, and metastasis (26–28). Among the miRNAs correlated with carcinogenesis, miR-193a-3p is one of the most important. Dysregulation of miR-193a-3p has been reported in various types of cancer, such as NSCLC (29), prostate cancer (30), breast cancer (31), head and neck squamous cell carcinoma (32), colorectal cancer (33), myeloid leukemia (34), and Wilms tumor (35). The carcinogenic impact of miR-193a-3p has been attributed to its repression of c-Kit (34) and the PTEN/PI3K signaling pathway in acute myeloid leukemia (34); of KRAS and PLAU in colon cancer (36); of PLAU (37) and EGFR-driven cell cycle network proteins (38) in breast cancer; of ARHGAP19, CCND1, ERBB4, KRAS, and Mcl-1 in epithelial ovarian cancer (39); of PLAU in hepatocellular carcinoma (40); and of Mcl-1 in NSCLC (41). Thus, miR-193a-3p functions as a tumor suppressor in human cancers.
In this study, we predicted that ERBB4 is a target of miR-193a-3p. After measuring the expression levels of miR-193a-3p and ERBB4 in human lung cancer tissues and paired noncancerous tissues, we detected an inverse correlation between miR-193a-3p expression and ERBB4 protein levels, but not mRNA levels, in human lung cancer tissues. The direct inhibition of ERBB4 translation by miR-193a-3p and the potential role of miR-193a-3p as a tumor suppressor in lung carcinogenesis have been experimentally validated in vitro and in vivo.
MATERIALS AND METHODS
Cells and Human Tissues
The human lung cancer cell lines A549, HCC827, and H1975 were purchased from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). A549-Luc, A549-Luc-miR-193a-3p, and A549-Luc-ERBB4 cells were produced by Genscript (Nanjing, China). All cells were cultured in DMEM supplemented with 10% fetal bovine serum (Invitrogen) and incubated in 5% CO2 at 37 °C in a water-saturated atmosphere. The lung tumors and paired normal adjacent tissues were derived from patients undergoing a surgical procedure at the Drum Tower Hospital affiliated with Medical School of Nanjing University (Nanjing, China). All of the patients or their guardians provided written consent, and the Ethics Committee from Nanjing University approved all aspects of this study. Tissue fragments were immediately frozen in liquid nitrogen at the time of surgery and stored at −80 °C. The clinical features of the patients are listed in Table 1.
TABLE 1.
Clinical features of lung cancer patients
| Age | Gender | Tumor subtype | Pathological stage | Smoking history | |
|---|---|---|---|---|---|
| Case 1 | 58 | Male | Adenocarcinoma | IIIA | Smoker |
| Case 2 | 67 | Male | Adenocarcinoma | IIB | Smoker |
| Case 3 | 48 | Male | Squamous cell carcinoma | IIIA | Smoker |
| Case 4 | 55 | Male | Squamous cell carcinoma | IIA | Never smoker |
| Case 5 | 70 | Female | Adenocarcinoma | IA | Never smoker |
| Case 6 | 62 | Female | Adenocarcinoma | IIB | Smoker |
RNA Isolation and Quantitative RT-PCR
Total RNA was extracted from the cultured cells and human tissues using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Assays to quantify miRNAs were performed using Taqman miRNA probes (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Briefly, 1 μg of total RNA was reverse-transcribed to cDNA using avian myeloblastosis virus reverse transcriptase (TaKaRa, Dalian, China) and a stem-loop RT primer (Applied Biosystems). The reaction conditions were as follows: 16 °C for 30 min, 42 °C for 30 min, and 85 °C for 5 min. Real-time PCR was performed using a TaqMan PCR kit on an Applied Biosystems 7300 sequence detection system. The reactions were incubated in a 96-well optical plate at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. All of the reactions were run in triplicate. After the reaction, the cycle threshold (CT) data were determined using fixed threshold settings, and the mean CT of the triplicate PCRs was determined. A comparative CT method was used to compare each condition with the controls. The relative levels of the miRNAs in cells and tissues were normalized to U6. The amount of miRNA relative to the internal control U6 was calculated using the 2−ΔΔCT equation, in which ΔΔCT = (CT miRNA − CT U6)target − (CT miRNA − CT U6)control.
To quantify ERBB4 mRNA, 1 μg of total RNA was reverse-transcribed to cDNA using oligo(dT) and avian myeloblastosis virus reverse transcriptase (TaKaRa) in the reaction, which was performed with the following conditions: 42 °C for 60 min and 70 °C for 10 min. Next, real-time PCR was performed using the RT product SYBER Green dye (Invitrogen) and specific primers for ERBB4 and GAPDH. The sequences of the primers were as follows: ERBB4 (sense), 5′-AATTGGTGGGCTCTTCATTC-3′; ERBB4 (antisense), 5′-CGTGGACATACTCCAACAGG-3′; GAPDH (sense), 5′-GATATTGTTGCCATCAATGAC-3′; and GAPDH (antisense), 5′-TTGATTTTGGAGGGATCTCG-3′. The reactions were incubated at 95 °C for 5 min, followed by 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. After the reactions were complete, the CT values were determined by setting a fixed threshold. The relative amount of ERBB4 mRNA was normalized to GAPDH.
Overexpression and Knockdown of miR-193a-3p
Synthetic pre-miR-193a-3p, anti-miR-193a-3p, and scrambled negative control RNAs (pre-scramble and anti-scramble) were purchased from Ambion (Austin, TX). Cells were seeded in 6-well plates or 60-mm dishes and were transfected using Lipofectamine 2000 (Invitrogen) the following day when the cells were ∼70% confluent. In each well, an equal amount of pre-miR-193a-3p, anti-miR-193a-3p, or scrambled negative control RNA were used. The cells were harvested 24 h after transfection for quantitative RT-PCR and Western blotting.
Luciferase Reporter Assay
To test the direct binding of miR-193a-3p to the target gene ERBB4, a luciferase reporter assay was performed as described previously (42). The entire 3′-untranslated region (3′-UTR) of human ERBB4 was PCR-amplified from human genomic DNA. The PCR products were inserted into the p-MIR-reporter plasmid (Ambion), and the insertion was confirmed by sequencing. To test the binding specificity, the sequences that interacted with the miR-193a-3p seed sequence were mutated (all three binding positions were mutated), and the mutant ERBB4 3′-UTR was inserted into an equivalent luciferase reporter. For luciferase reporter assays, A549 cells were cultured in 24-well plates, and each well was transfected with 1 μg of firefly luciferase reporter plasmid; 1 μg of a β-galactosidase (β-gal) expression plasmid (Ambion); and an equal amount (100 pmol) of pre-miR-193a-3p, anti-miR-193a-3p, or the scrambled negative control RNA using Lipofectamine 2000 (Invitrogen). The β-gal plasmid was used as a transfection control. Twenty-four hours after transfection, the cells were assayed using a luciferase assay kit (Promega, Madison, WI).
Plasmid Construction and siRNA Interference Assay
The siRNA sequence targeting the human ERBB4 cDNA was designed and synthesized by GenePharma (Shanghai, China). The siRNA sequence was 5′-CGGGAAUCUCAUCUUUCUU-3′. A scrambled siRNA was included as a negative control. A mammalian expression plasmid encoding the human ERBB4 open reading frame (pReceiver-M02-ERBB4) was purchased from GeneCopoeia (Germantown, MD). An empty plasmid served as a negative control. The ERBB4 expression vector and ERBB4 siRNA were transfected into A549 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Total RNA and protein were isolated 24 h post-transfection. The ERBB4 mRNA and protein expression levels were assessed by quantitative RT-PCR and Western blotting.
Construction of the miR-193a-3p Overexpression Lentiviral Vector
Lentivirus for miR-193a-3p overexpression was purchased from Invitrogen. Lentivirus was added to A549 cells at 70% confluence in 6-well plates or 100-mm dishes at a multiplicity of infection of 10 together with Polybrene at a final concentration of 5 μg/ml according to the manufacturer's instructions. Cells were then harvested for quantitative RT-PCR, Western blotting, or animal experiments.
Protein Extraction and Western Blotting
All cells were rinsed with PBS (pH 7.4) and lysed on ice for 30 min in radioimmune precipitation assay lysis buffer (Beyotime, Haimen, China) supplemented with a protease and phosphatase inhibitor mixture (Thermo Scientific catalog no. 78440). The tissue samples were frozen solid with liquid nitrogen, ground into powder, and lysed on ice for 30 min in radioimmune precipitation assay lysis buffer containing the protease and phosphatase inhibitor mixture. When necessary, sonication was used to facilitate lysis. Cell lysates or tissue homogenates were centrifuged for 10 min (12,000 × g, 4 °C). The supernatant was collected, and the protein concentration was calculated using the Pierce BCA protein assay kit (Thermo Scientific). The protein levels were analyzed via Western blot using the corresponding antibodies. The protein levels were normalized by probing the same blots with a GAPDH antibody. The following antibodies were purchased from the corresponding sources: anti-ERBB4 (C-7) (Santa Cruz Biotechnology, Inc., catalog no. sc-8050) and anti-GAPDH (Santa Cruz Biotechnology, catalog no. sc-365062). Protein bands were analyzed using the ImageJ software.
Cell Viability Assay
To assess cell viability, A549 cells were seeded in triplicate in 96-well plates at a density of 5 × 103 cells/well in 100 μl of culture medium. The cell proliferation index was measured using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Sigma), which was performed 12, 24, 36, and 48 h after transfection according to the manufacturer's instructions.
Cell Invasion Assay
The invasion ability of A549 cells transfected with the pre-miR-193a-3p, anti-miR-193a-3p, or ERBB4 overexpression plasmid was tested in a Transwell Boyden chamber (6.5 mm; Costar). The polycarbonate membranes (8-μm pore size) on the bottom of the upper compartment of the Transwells were coated with 1% human fibronectin (R&D Systems catalog no. 1918-FN). The cells were harvested 24 h after transfection and suspended in FBS-free DMEM culture medium. Then cells were added to the upper chamber (4 × 104 cells/well). At the same time, 0.5 ml of DMEM containing 10% FBS was added to the lower compartment, and the Transwell-containing plates were incubated for 12 h in a 5% CO2 atmosphere saturated with H2O. After incubation, cells that had entered the lower surface of the filter membrane were fixed with 4% paraformaldehyde for 25 min at room temperature, washed three times with distilled water, and stained with 0.1% crystal violet in 0.1 m borate and 2% ethanol for 15 min at room temperature. Cells remaining on the upper surface of the filter membrane (non-migrant) were gently scraped off using a cotton swab. Images of the lower surfaces (with migrant cells) were captured by a photomicroscope (5 fields/chamber) (BX51 Olympus, Japan), and the cells were counted blindly.
Apoptosis Assays
The apoptosis of A549 cells transfected with the pre-miR-193a-3p, anti-miR-193a-3p, or ERBB4 overexpression plasmid was tested using an annexin V-FITC/propidium iodide staining assay. A549 cells were cultured in 12-well plates and transfected with pre-miR-193a-3p, anti-miR-193a-3p, or ERBB4 overexpression plasmid to induce apoptosis. Pre-miR-control and control siRNAs served as negative controls. Cells were cultured overnight in both serum-containing complete medium and serum-depleted medium, and the attached and floating cells were then harvested. Flow cytometry analysis of apoptotic cells was carried out using an annexin V-FITC/propidium iodide staining kit (BD Biosciences, CA). After washing with cold PBS, the cells were resuspended in binding buffer (100 mm HEPES, pH 7.4, 100 mm NaCl, and 25 mm CaCl2) followed by staining with annexin V-FITC/propidium iodide at room temperature for 15 min in the dark. Apoptotic cells were then evaluated by gating propidium iodide and annexin V-positive cells on a fluorescence-activated cell-sorting (FACS) flow cytometer (BD Biosciences). All experiments were performed in triplicate.
Establishment of Tumor Xenografts in Mice
All protocols involving animal experiments were approved by the Ethics Committee of Nanjing University. Fifteen female SCID (severe combined immune deficiency) mice (nu/nu; each 5–6 weeks old) were purchased from the Model Animal Research Centre of Nanjing University and were anesthetized by continuous flow of 2–3% isoflurane. Then the chest area was sterilized three times with a 70% alcohol pad prior to air drying. Next, 2 × 106 A549-Luc, A549-Luc-miR-193a-3p, or A549-Luc-ERBB4 cells suspended in 100 μl of Dulbecco's PBS were injected into the mediastinum (5 mice/group). The needle was inserted on the right side of the sternum, midway down, 5 mm deep, and at a 45° angle with respect to the chest wall. Tumor volume was assessed by bioluminescent imaging (BLI) (Genogen IVISTM 200 system) starting on day 3. The exposure time was set at 5 s for consistent analysis during follow-up scans. A more detailed protocol can be found in the published mouse model (43). Tumor volumes were assessed by BLI on days 3, 12, 22, and 35. The photon counts from the five mice of each group were averaged. The -fold increase in BLI was calculated as the proportion of post-treatment BLI to baseline BLI (on the first day of treatment). On day 35, the mouse lung tumors were removed, fixed in 10% formalin for 24 h, and then further processed for hematoxylin and eosin (H&E) staining and immunohistochemical staining for Ki-67 and ERBB4.
Statistical Analysis
All Western blot images are representative of at least three independent experiments. Quantitative RT-PCR, luciferase reporter assays, and cell viability and apoptosis assays were performed in triplicate, and each experiment was repeated several times. The data shown are the mean ± S.E. of at least three independent experiments. The differences were considered statistically significant at p < 0.05 using Student's t test.
RESULTS
Up-regulation of ERBB4 Protein but Not mRNA in NSCLC Tissues
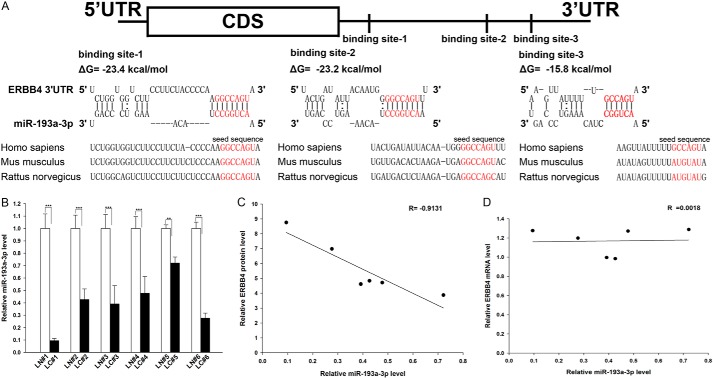
We first determined the expression patterns of ERBB4 in NSCLC tissues. After measuring the levels of ERBB4 protein in six pairs of NSCLC tissues and corresponding noncancerous tissues (the clinical features of these tissue samples are listed in Table 1) via Western blotting, we found that ERBB4 protein levels were significantly higher in the cancer tissues (Fig. 1, A and B). Subsequently, we performed quantitative RT-PCR to measure the levels of ERBB4 mRNA in the same six pairs of cancerous and noncancerous tissues. We found that ERBB4 mRNA levels did not differ significantly between the cancerous and noncancerous tissues (Fig. 1C). This disparity between ERBB4 protein and mRNA levels in NSCLC tissues strongly suggests that a post-transcriptional mechanism is involved in the regulation of ERBB4.
FIGURE 1.
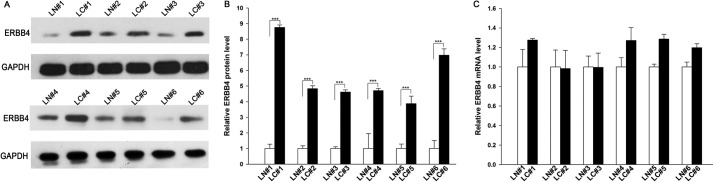
ERBB4 protein and mRNA expression levels in lung cancer tissues. A and B, Western blotting analysis of the expression levels of the ERBB4 protein in six pairs of lung cancer (LC) and normal adjacent tissue (LN) samples. A, representative image; B, quantitative analysis. C, quantitative RT-PCR analysis of the relative expression levels of ERBB4 mRNA in six pairs of LC and LN samples. ***, p < 0.001. Error bars, S.E.
Identification of Conserved miR-193a-3p Target Sites within the 3′-UTR of ERBB4
One important mode of post-transcriptional regulation is the repression of mRNA transcripts by miRNAs. miRNAs are therefore likely to play a biologically relevant role in regulating ERBB4 expression in NSCLC. Three computational algorithms, including TargetScan (44), miRanda (45), and PicTar (46), were used in combination to identify potential miRNAs that can target ERBB4. Using this approach, only three miRNAs (miR-193a-3p, miR-130, and miR-301) were identified as candidate regulators of ERBB4. miR-130 and miR-301 were excluded because they were reported to be up-regulated in lung tumors (47, 48), which is not in agreement with the concept that miRNAs should have expression patterns that are opposite to those of their targets (49–51). In contrast, miR-193a-3p was confirmed to be down-regulated in lung tumors (29, 41) and was therefore retained for further analysis. The predicted interactions between miR-193a-3p and the targeting sites within the 3′-UTR of ERBB4 are illustrated in Fig. 2A. Three predicted hybridizations were observed between miR-193a-3p and the 3′-UTR of ERBB4. The minimum free energy values of the three hybridizations between miR-193a-3p and ERBB4 are −15.8, −23.2, and −23.4 kcal/mol, which are well within the range of genuine miRNA-target pairs. Moreover, there is perfect base pairing between the seed region (the core sequence that encompasses the first 2–8 bases of the mature miRNA) and the cognate targets. Furthermore, the miR-193a-3p binding sequences in the ERBB4 3′-UTR are highly conserved across species.
FIGURE 2.
Detection of an inverse correlation between miR-193a-3p and ERBB4 levels in lung cancer tissue samples. A, schematic depicting the hypothetical duplexes formed by the interactions between the binding sites in the ERBB4 3′-UTR (top) and miR-193a-3p (bottom). The predicted free energy value of each hybrid is indicated. The seed recognition sites are denoted, and all nucleotides in these regions are highly conserved across species, including human, mouse, and rat. B, quantitative RT-PCR analysis of the expression levels of miR-193a-3p (in the form of the miRNA/U6 ratio) in six pairs of LC and LN samples. C, Pearson's correlation scatter plot of the -fold change of the levels of miR-193a-3p and ERBB4 protein in human lung cancer tissues. D, Pearson's correlation scatter plot of the -fold change of the levels of miR-193a-3p and ERBB4 mRNA in human lung cancer tissues. **, p < 0.01; ***, p < 0.001. Error bars, S.E.
Detection of an Inverse Correlation between miR-193a-3p and ERBB4 Levels in NSCLC Tissues
We next investigated whether miR-193a-3p was inversely correlated with ERBB4 in NSCLC. After determining the levels of miR-193a-3p in the same six pairs of NSCLC tissues and corresponding noncancerous tissues, we found that miR-193a-3p levels were indeed down-regulated in NSCLC tissues (Fig. 2B). The inverse correlation between miR-193a-3p and ERBB4 protein levels (Fig. 2C) and the disparity between the miR-193a-3p and ERBB4 mRNA levels (Fig. 2D) were further illustrated using Pearson's correlation scatter plots. Because animal miRNAs are generally believed to block translational processes without affecting transcript levels, the results strongly indicated the involvement of an miRNA-mediated post-transcriptional regulatory mechanism in ERBB4 repression. Thus, ERBB4 was deduced to be an miR-193a-3p target not only by computational prediction but also by noting the inverse correlation between miR-193a-3p and ERBB4 protein levels, but not mRNA levels, in human NSCLC tissues.
Validation of ERBB4 as a Direct Target of miR-193a-3p
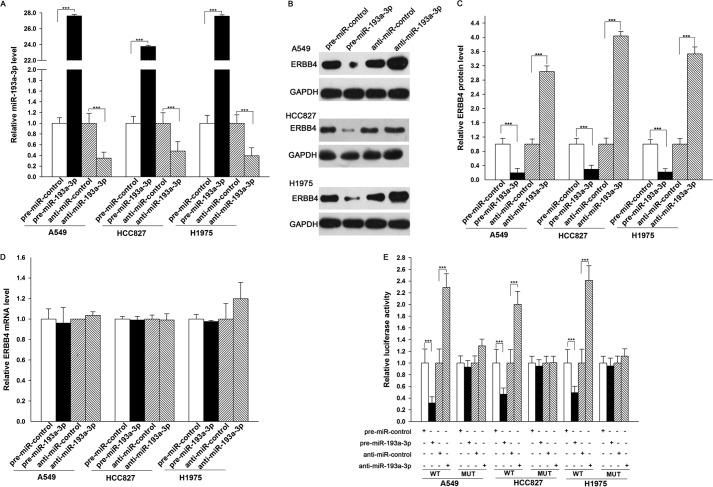
The correlation between miR-193a-3p and ERBB4 was further examined by evaluating ERBB4 expression in the human lung cancer cell lines A549, HCC827, and H1975 after miR-193a-3p overexpression or knockdown. In these experiments, overexpression was achieved by transfecting cells with pre-miR-193a-3p, which is a synthetic RNA oligonucleotide that mimics the miR-193a-3p precursor. miR-193a-3p knockdown was achieved by transfecting cells with anti-miR-193a-3p, which is a chemically modified antisense oligonucleotide designed to specifically target mature miR-193a-3p. The efficient overexpression and knockdown of miR-193a-3p in A549, HCC827, and H1975 cells are shown in Fig. 3A. Cellular miR-193a-3p levels were increased ∼20-fold when A549, HCC827, and H1975 cells were transfected with pre-miR-193a-3p, and these levels dropped to ∼30% of the normal level when A549, HCC827, and H1975 cells were treated with anti-miR-193a-3p. As anticipated, overexpressing miR-193a-3p significantly suppressed the ERBB4 protein levels in A549, HCC827, and H1975 cells, whereas miR-193a-3p knockdown had the opposite effect on ERBB4 expression in these cells (Fig. 3, B and C). To determine the regulatory level at which miR-193a-3p influenced ERBB4 expression, we repeated the above experiments and examined the expression of ERBB4 mRNA after transfection. Neither overexpression nor knockdown of miR-193a-3p affected ERBB4 mRNA levels in A549, HCC827, and H1975 cells (Fig. 3D). These results demonstrated that miR-193a-3p specifically regulates ERBB4 expression at the post-transcriptional level, which is the most common mechanism for animal miRNAs.
FIGURE 3.
Direct post-transcriptional regulation of ERBB4 expression by miR-193a-3p. A, quantitative RT-PCR analysis of the miR-193a-3p levels in A549, HCC827, and H1975 cells treated with pre-miR-control, pre-miR-193a-3p, anti-miR-control, or anti-miR-193a-3p. B and C, Western blot analysis of ERBB4 protein levels in A549, HCC827, and H1975 cells treated with pre-miR-control, pre-miR-193a-3p, anti-miR-control, or anti-miR-193a-3p. B, representative image; C, quantitative analysis. D, quantitative RT-PCR analysis of ERBB4 mRNA levels in A549, HCC827, and H1975 cells treated with pre-miR-control, pre-miR-193a-3p, anti-miR-control, or anti-miR-193a-3p. E, firefly luciferase reporters containing wild-type (WT) or mutant (MUT) miR-193a-3p binding sites in the ERBB4 3′-UTR were co-transfected into A549, HCC827, and H1975 cells along with pre-miR-control, pre-miR-193a-3p, anti-miR-control, or anti-miR-193a-3p. Twenty-four hours post-transfection, the cells were assayed using a luciferase assay kit. ***, p < 0.001. Error bars, S.E.
To determine whether the negative regulatory effects of miR-193a-3p on ERBB4 expression were mediated through the binding of miR-193a-3p to the presumed sites in the 3′-UTR of the ERBB4 mRNA, the full-length 3′-UTR of ERBB4 containing the three presumed miR-193a-3p binding sites was placed downstream of the firefly luciferase gene in a reporter plasmid. The resulting plasmid was transfected into A549, HCC827, and H1975 cells along with pre-miR-193a-3p, anti-miR-193a-3p, or scrambled negative control RNAs. As expected, luciferase activity was markedly reduced in the cells transfected with pre-miR-193a-3p and increased in the cells transfected with anti-miR-193a-3p (Fig. 3E). Furthermore, we introduced point mutations into the corresponding complementary sites in the 3′-UTR of ERBB4 to eliminate the predicted miR-193a-3p binding sites (all three binding positions were mutated). This mutated luciferase reporter was unaffected by either overexpression or knockdown of miR-193a-3p (Fig. 3E). This finding suggested that the binding sites strongly contribute to the interaction between miR-193a-3p and ERBB4 mRNA. In conclusion, our results demonstrated that miR-193a-3p directly recognizes and binds to the 3′-UTR of the ERBB4 mRNA transcript and inhibits ERBB4 translation in lung cancer cells.
The Role of miR-193a-3p in Regulating ERBB4 in Lung Cancer Cells
We next analyzed the biological consequences of the miR-193a-3p-driven repression of ERBB4 expression in lung cancer cells. Because ERBB4 is known to be involved in the regulation of cell proliferation, invasion, and apoptosis, we investigated whether miR-193a-3p would regulate ERBB4 to modulate cell proliferation, invasion, and apoptosis in lung cancer cells.
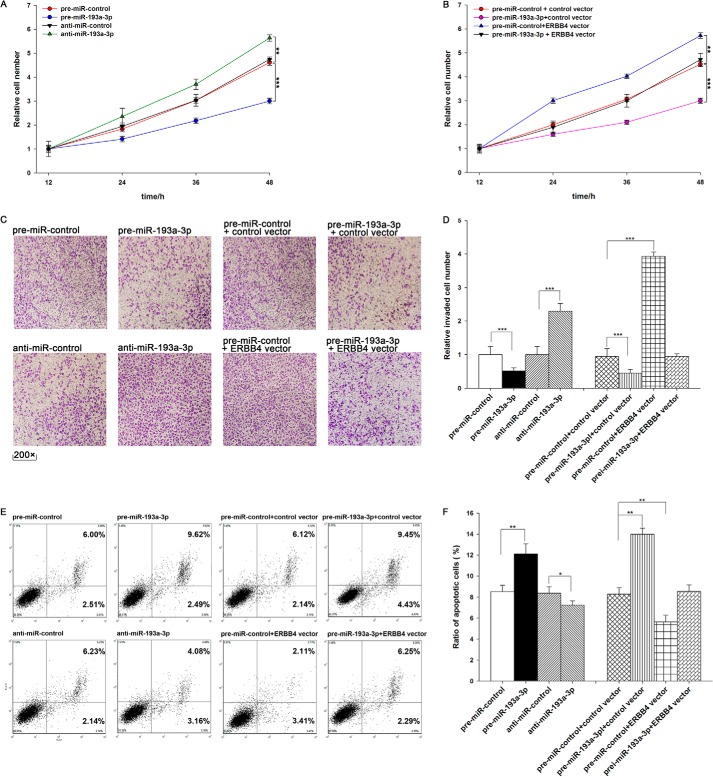
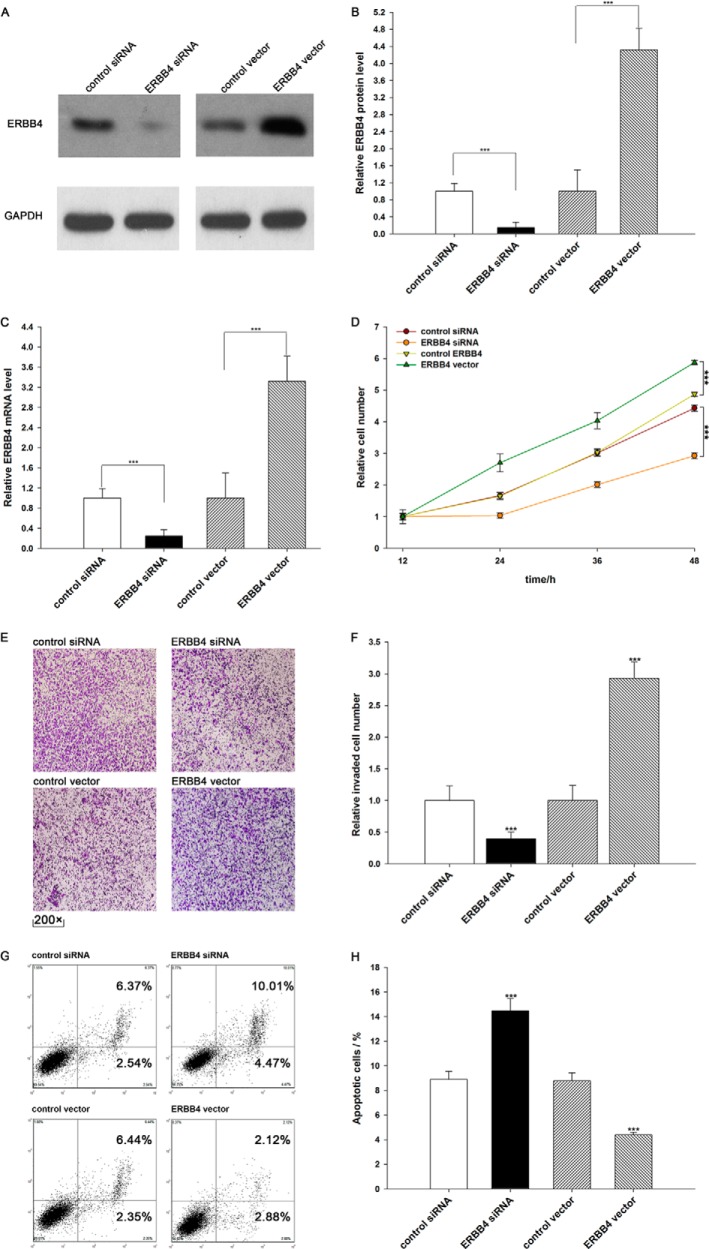
We evaluated the effects of miR-193a-3p on the proliferation of A549 cells using the MTT assay. As expected, A549 cells transfected with pre-miR-193a-3p showed decreased proliferation; in contrast, knocking down miR-193a-3p had the opposite effect on cell proliferation (Fig. 4A). Subsequently, we investigated whether overexpression or knockdown of ERBB4 would have an impact on cell proliferation. To knock down ERBB4, an siRNA targeting ERBB4 was designed and then transfected into A549 cells. To overexpress ERBB4, an expression plasmid designed to specifically express the full-length open reading frame (ORF) of ERBB4 without the miR-193a-3p-responsive 3′-UTR was also constructed and transfected into A549 cells. Efficient knockdown and overexpression of ERBB4 in A549 cells are shown in Fig. 5, A–C. A549 cells transfected with ERBB4 siRNA showed inhibited cell proliferation; in contrast, transfection with the ERBB4 overexpression plasmid had the opposite effect on cell proliferation (Fig. 5D). Moreover, compared with cells transfected with pre-miR-193a-3p alone, those transfected with both pre-miR-193a-3p and the ERBB4 overexpression plasmid exhibited significantly higher proliferation rates (Fig. 4B), suggesting that miR-193a-3p-resistant ERBB4 is sufficient to rescue the suppression of ERBB4 by miR-193a-3p and attenuate the anti-proliferative effect of miR-193a-3p on lung cancer cells. Taken together, these results indicate that miR-193a-3p might inhibit cell proliferation by silencing ERBB4.
FIGURE 4.
The role of miR-193a-3p targeting of ERBB4 in the regulation of proliferation, invasion, and apoptosis in lung cancer cells. A, the MTT viability assay was performed 12, 24, 36, and 48 h after the transfection of A549 cells with pre-miR-control, pre-miR-193a-3p, anti-miR-control, or anti-miR-193a-3p. B, the MTT viability assay was performed 12, 24, 36, and 48 h after the transfection of A549 cells with pre-miR-control plus control vector, pre-miR-193a-3p plus control vector, pre-miR-control plus ERBB4 vector, or pre-miR-193a-3p plus ERBB4 vector. C and D, Transwell analysis of invaded A549 cells treated with equal doses of pre-miR-control, pre-miR-193a-3p, anti-miR-control, anti-miR-193a-3p, pre-miR-control plus control vector, pre-miR-193a-3p plus control vector, pre-miR-control plus ERBB4 vector, or pre-miR-193a-3p plus ERBB4 vector. C, representative image; D, quantitative analysis. E and F, A549 cells were transfected with equal doses of pre-miR-control, pre-miR-193a-3p, anti-miR-control, anti-miR-193a-3p, pre-miR-control plus control vector, pre-miR-193a-3p plus control vector, pre-miR-control plus ERBB4 vector, or pre-miR-193a-3p plus ERBB4 vector. Cell apoptosis profiles were analyzed by flow cytometry. The biparametric histogram shows cells in early (bottom right quadrant) and late apoptotic states (top right quadrant). Viable cells are double negative (bottom left quadrant). E, representative image; F, quantitative analysis. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Error bars, S.E.
FIGURE 5.
Down-regulation of ERBB4 by siRNA and up-regulation of ERBB4 by an overexpression vector in lung cancer cells. A and B, Western blotting analysis of ERBB4 protein levels in A549 cells treated with control siRNA, ERBB4 siRNA, control vector, or ERBB4 vector. A, representative image; B, quantitative analysis. C, quantitative RT-PCR analysis of ERBB4 mRNA levels in A549 cells treated with control siRNA, ERBB4 siRNA, control vector, or ERBB4 vector. D, the MTT viability assay was performed 12, 24, 36, and 48 h after the transfection of A549 cells with control siRNA, ERBB4 siRNA, control vector, or ERBB4 vector. E and F, Transwell analysis of invaded A549 cells treated with equal doses of control siRNA, ERBB4 siRNA, control vector, or ERBB4 vector. E, representative image; F, quantitative analysis. G and H, A549 cells were transfected with equal doses of control siRNA, ERBB4 siRNA, control vector, or ERBB4 vector. Cell apoptosis profiles were analyzed by flow cytometry. The biparametric histogram shows cells in early (bottom right quadrant) and late apoptotic states (top right quadrant). Viable cells are double negative (bottom left quadrant). G, representative image; H, quantitative analysis. ***, p < 0.001. Error bars, S.E.
We also evaluated the effects of miR-193a-3p on the invasion ability of A549 cells using Transwell invasion assays. The Transwell invasion assay showed that the percentage of invaded cells was significantly lower in A549 cells transfected with pre-miR-193a-3p and higher in cells transfected with anti-miR-193a-3p (Fig. 4, C and D). Additionally, the transfection of ERBB4 siRNA markedly reduced the number of A549 cells that passed through the Transwell chamber, whereas transfection of the ERBB4 overexpression plasmid significantly stimulated cell invasion (Fig. 5, E and F). However, when A549 cells were co-transfected with pre-miR-193a-3p and the ERBB4 overexpression plasmid, ERBB4 dramatically attenuated the suppressive effect of miR-193a-3p on cell invasion (Fig. 4, C and D). These results indicate that miR-193a-3p might inhibit cell invasion by silencing ERBB4.
Finally, we investigated cell apoptosis using flow cytometry analysis. The results showed that the percentage of apoptotic cells was significantly higher in A549 cells transfected with pre-miR-193a-3p and lower in cells transfected with anti-miR-193a-3p (Fig. 4, E and F). In addition, transfecting ERBB4 siRNA markedly increased the percentage of apoptotic cells when compared with cells transfected using control siRNA, whereas transfecting the ERBB4 overexpression plasmid decreased cell apoptosis (Fig. 5, G and H). Furthermore, when A549 cells were simultaneously transfected with pre-miR-193a-3p and the ERBB4 overexpression plasmid, ERBB4 dramatically attenuated the pro-apoptotic effect of miR-193a-3p (Fig. 4, E and F). These results indicate that miR-193a-3p might modulate cell apoptosis by down-regulating ERBB4 in lung cancer cells. Taken together, our results suggest that ERBB4 is crucial to the proliferation, invasion, and apoptosis of lung cancer cells and that miR-193a-3p might inhibit proliferation and invasion and promote apoptosis in lung cancer cells by silencing ERBB4.
The Effect of miR-193a-3p and ERBB4 on the Growth Rate of Lung Cancer Cells in Vivo
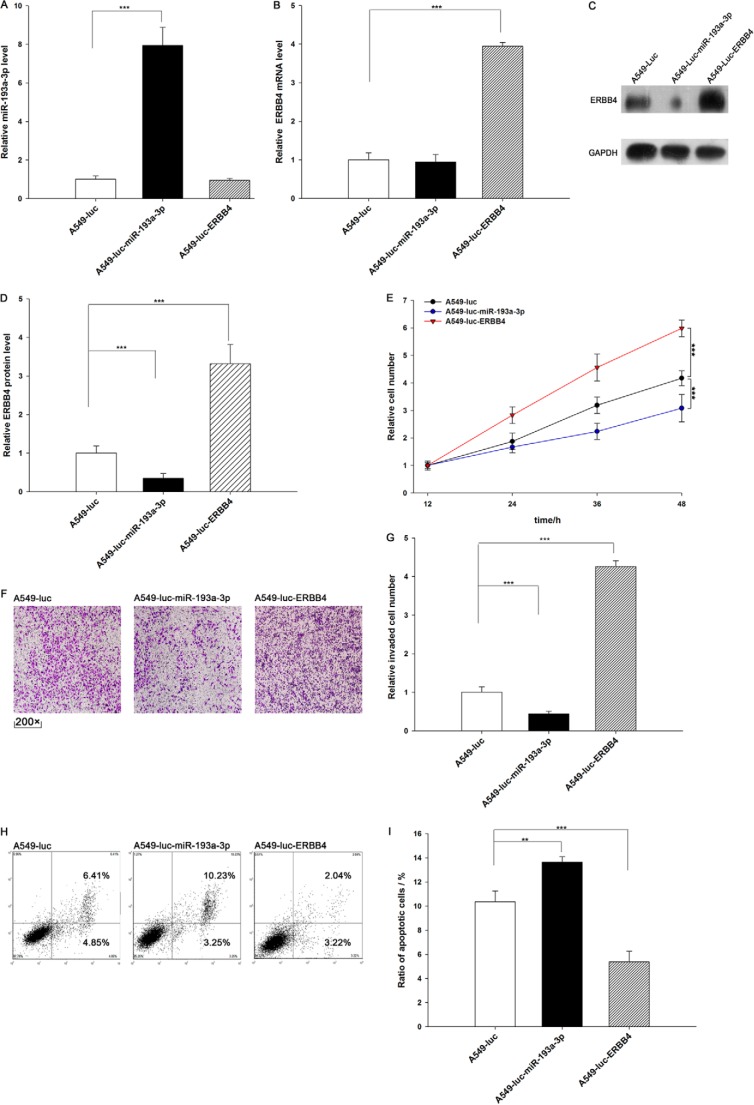
We evaluated the effects of miR-193a-3p and ERBB4 on the growth of lung cancer cell xenografts in mice. We designed and produced three types of A549 lines. A549-Luc cells contain a knocked in luciferase gene. A549-Luc-miR-193a-3p cells contain a knocked in luciferase and pre-miR-193a-3p gene, and this type of cell can express both luciferase and miR-193a-3p (Fig. 6A). A549-Luc-ERBB4 cells contain a knocked in luciferase and ERBB4 gene, and this type of cell can express both luciferase and ERBB4 (Fig. 6, B–D). We analyzed cell proliferation, invasion, and apoptosis in these three cell lines. As expected, A549-Luc-miR-193a-3p cells showed decreased proliferation and invasion and increased apoptosis compared with A549-Luc cells (Fig. 6, E–I). In contrast, A549-Luc-ERBB4 cells displayed higher proliferation and invasion rates and a lower apoptosis rate compared with A549-Luc cells (Fig. 6, E–I). These results again reveal that miR-193a-3p inhibits proliferation and invasion and promotes apoptosis in lung cancer cells and that ERBB4 produces the opposite effects.
FIGURE 6.
Effects of miR-193a-3p and ERBB4 on cell proliferation, invasion, and apoptosis in A549-Luc, A549-Luc-miR-193a-3p, and A549-Luc-ERBB4 cells. A, quantitative RT-PCR analysis of the miR-193a-3p levels in A549-Luc, A549-Luc-miR-193a-3p, and A549-Luc-ERBB4 cells. B, quantitative RT-PCR analysis of ERBB4 mRNA levels in A549-Luc, A549-Luc-miR-193a-3p, and A549-Luc-ERBB4 cells. C and D, Western blotting analysis of protein levels in A549-Luc, A549-Luc-miR-193a-3p, and A549-Luc-ERBB4 cells. C, representative image; D, quantitative analysis. E, the MTT viability assay was performed after 12, 24, 36, and 48 h in A549-Luc, A549-Luc-miR-193a-3p, and A549-Luc-ERBB4 cells. F and G, Transwell analysis of invaded A549-Luc, ERBB4 A549-Luc-miR-193a-3p, and A549-Luc-ERBB4 cells. F, representative image; G, quantitative analysis. H and I, cell apoptosis profiles of A549-Luc, A549-Luc-miR-193a-3p, and A549-Luc-ERBB4 cells by flow cytometry. The biparametric histogram shows cells in early (bottom right quadrant) and late apoptotic states (top right quadrant). Viable cells are double negative (bottom left quadrant). H, representative image; I, quantitative analysis. **, p < 0.01; ***, p < 0.001. Error bars, S.E.
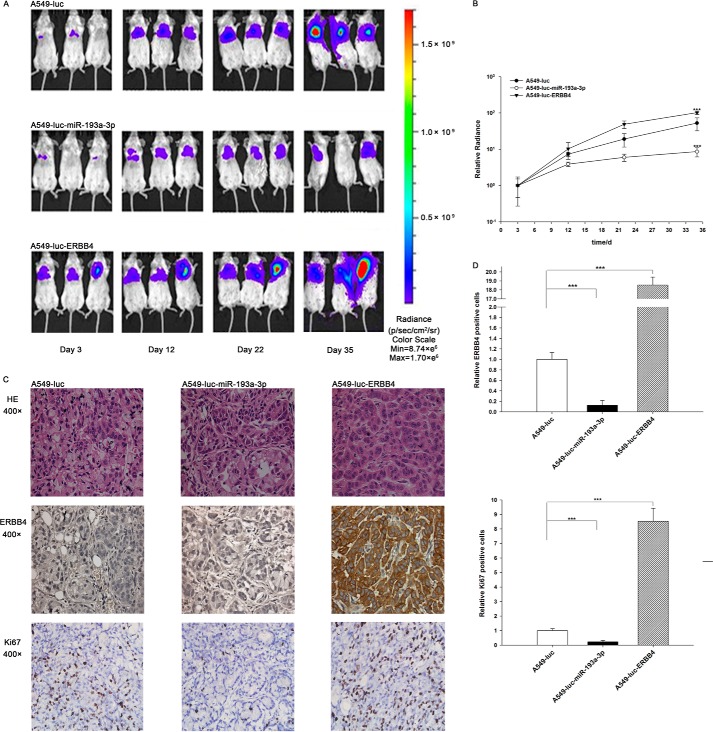
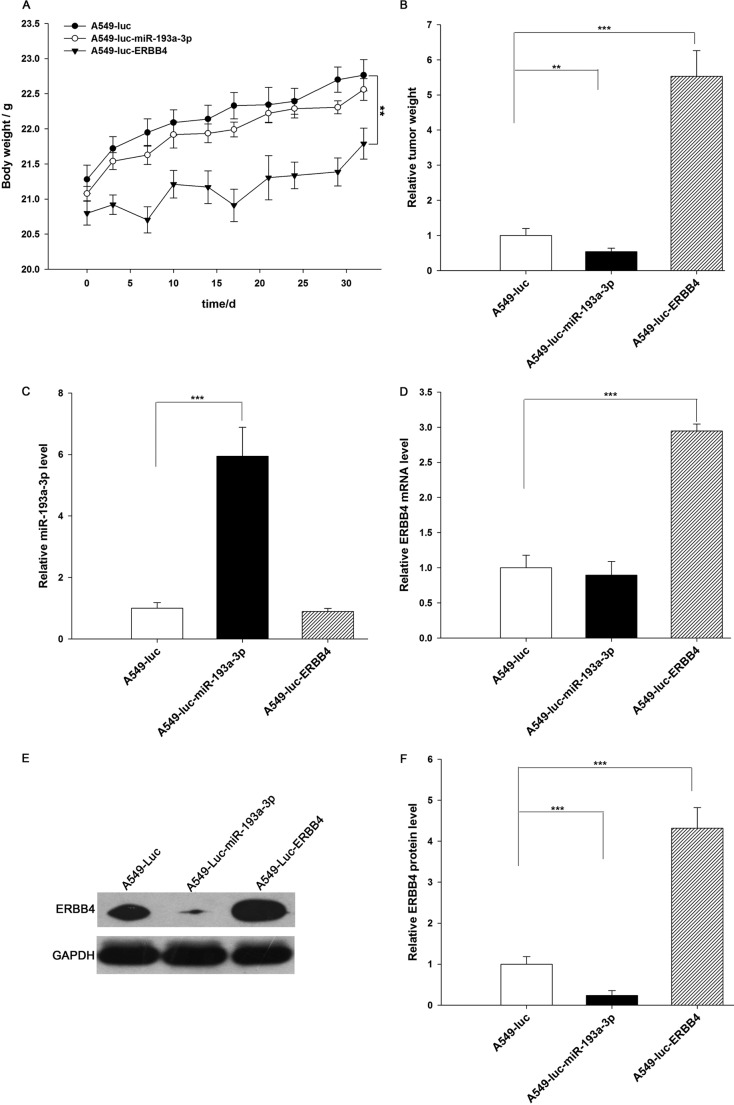
Next, these three types of cells were implanted into mice, and tumor growth was evaluated on days 3, 12, 22, and 35 after implantation. Throughout the whole implantation process, the group implanted with A549-Luc-miR-193a-3p cells had a lower rate of xenograft tumor growth compared with the A549-Luc-implanted group, whereas the group implanted with A549-Luc-ERBB4 cells had a higher rate of xenograft tumor growth (Fig. 8, A and B). As a result, the group implanted with A549-Luc-ERBB4 cells showed significant body weight loss compared with the groups implanted with either A549-Luc or A549-Luc-miR-193a-3p cells (Fig. 7A). After 35 days of xenograft growth in vivo, mice were sacrificed, and the weight of the tumors was measured. A significant suppression in the weights of the tumors was observed in the group implanted with A549-Luc-miR-193a-3p cells, whereas the weight of the tumors in the group implanted with A549-Luc-ERBB4 cells was dramatically increased (Fig. 7B). Total RNA and protein were also extracted from each xenograft and used to evaluate the expression of miR-193a-3p and ERBB4. The group implanted with A549-Luc-miR-193a-3p cells showed a significant increase in the expression of miR-193a-3p compared with the group implanted with A549-Luc (Fig. 7C). Likewise, ERBB4 mRNA levels were increased in the group implanted with A549-Luc-ERBB4 cells (Fig. 7D). Compared with the group implanted with A549-Luc, the group implanted with A549-Luc-miR-193a-3p cells displayed reduced ERBB4 protein levels, whereas the group implanted with A549-Luc-ERBB4 cells showed elevated ERBB4 protein levels (Fig. 7, E and F). In addition, immunohistochemical studies revealed the presence of lower levels of ERBB4 in the group implanted with A549-Luc-miR-193a-3p cells and higher levels of ERBB4 in the group implanted with A549-Luc-ERBB4 cells (Fig. 8, C and D). H&E staining of the xenograft tissues also showed more cell mitosis in the group implanted with A549-Luc-ERBB4 cells compared with the groups implanted with either A549-Luc or A549-Luc-miR-193a-3p cells (Fig. 8C). Finally, the proliferative activity of the tumor cells was assessed via immunocytochemistry using the mouse monoclonal antibody Ki-67. The tumor cell proliferation rate, as measured by the percentage of Ki-67-positive tumor cells, was decreased in the group implanted with A549-Luc-miR-193a-3p cells and increased in the group implanted with A549-Luc-ERBB4 cells (Fig. 8, C and D). These results were consistent with the findings of the in vitro assays, which firmly validated the role of miR-193a-3p in suppressing tumorigenesis through the targeting of ERBB4.
FIGURE 8.
Effects of miR-193a-3p or ERBB4 overexpression on the growth of lung cancer cell xenografts in mice. A and B, mice were separated into three groups: those implanted with A549-Luc (control), those with A549-Luc-miR-193a-3p, and those with A549-Luc-ERBB4. The first BLI was performed on day 3 after injection and repeated on days 12, 22, and 35. The intensity of BLI is represented by the color. A, representative image; B, -fold increase in BLI calculated as the proportion of BLI to baseline BLI (day 3). Photon counts of mice from each group were averaged. C and D, the tumors from mice implanted with A549-Luc, A549-Luc-miR-193a-3p, or A549-Luc-ERBB4 were subjected to H&E staining and immunohistochemical staining for Ki-67 and ERBB4. C, representative image; D, quantitative analysis. ***, p < 0.001.
FIGURE 7.
Effects of miR-193a-3p and ERBB4 on the growth of lung cancer cell xenografts in mice. A, body weights of mice implanted with A549-Luc, A549-Luc-miR-193a-3p, or A549-Luc-ERBB4 were performed at 0, 3, 10, 14, 17, 21, 24, 29, and 32 days. B, the tumor weight of mice implanted with A549-Luc, A549-Luc-miR-193a-3p, or A549-Luc-ERBB4 mice. C, quantitative RT-PCR analysis of miR-193a-3p levels in tumors from mice implanted with A549-Luc, A549-Luc-miR-193a-3p, or A549-Luc-ERBB4. D, quantitative RT-PCR analysis of ERBB4 mRNA levels in tumors from mice implanted with A549-Luc, A549-Luc-miR-193a-3p, or A549-Luc-ERBB4. E and F, Western blotting analysis of ERBB4 protein levels in tumors from mice implanted with A549-Luc, A549-Luc-miR-193a-3p, or A549-Luc-ERBB4. E, representative image; F, quantitative analysis. **, p < 0.01; ***, p < 0.001. Error bars, S.E.
DISCUSSION
Members of the ErbB subfamily of receptor tyrosine kinases are important regulators of normal physiology as well as pathologies such as cancer. EGFR and ERBB2 were among the first clinically validated targeted therapies. In contrast, relatively little is known about the biological significance of ERBB4 signaling in cancer. However, recent advances in our understanding of the biological role of ERBB4 in carcinogenesis, as well as the potential clinical relevance of ERBB4 in cancer therapy, have underlined the urgent need to fully elucidate the mechanisms that govern ERBB4 regulation. In this study, we found that silencing ERBB4 expression using siRNA could inhibit proliferation and invasion and promote apoptosis in lung cancer cells, whereas overexpressing ERBB4 induced opposing effects, validating its role as an essential oncogene during lung tumorigenesis. Interestingly, we identified a discordance between ERBB4 protein and mRNA levels in human lung cancer tissues. The results suggest a post-transcriptional regulation mechanism involved in ERBB4 repression. One centrally important mode of post-transcriptional regulation is the repression of mRNA transcripts by miRNAs. Therefore, we searched for miRNAs that can target ERBB4 and identified miR-193a-3p as a candidate. In addition, by overexpressing or knocking down miR-193a-3p in lung cancer cells, we experimentally validated the direct inhibition of ERBB4 translation by miR-193a-3p. Finally, we showed that miR-193a-3p inhibited ERBB4 expression and consequently inhibited proliferation and invasion and promoted apoptosis in cultured lung cancer cells and facilitated tumor growth in xenograft mice. The results delineate a novel regulatory network employing miR-193a-3p and ERBB4 to fine tune cell proliferation, invasion, and apoptosis. Interestingly, a recent study has shown that miR-193a-3p can suppress the metastasis of human NSCLC by down-regulating the ERBB4 signaling pathway (52). These studies, combined with ours, reveal the importance of miR-193a-3p targeting ERBB4 as a novel regulatory pathway in lung cancer progression.
miRNAs are aberrantly expressed in cancer and can function as oncogenes or tumor suppressor genes (26, 53). In this study, we found that the levels of miR-193a-3p were lower in lung cancer tissues than in normal, noncancerous tissues. These results suggest that miR-193a-3p may be involved in the pathogenesis of lung cancer as a tumor suppressor. Indeed, miR-193a-3p has been reported in many studies to be down-regulated in cancers (31, 33, 38, 52, 54, 55). On the other hand, it is well known that a single miRNA can target multiple genes, whereas multiple miRNAs can target a single gene. Thus, miR-193a-3p may have multiple different mRNA targets other than ERBB4, and these additional targets may also play important roles in carcinogenesis. For example, it has been reported that miR-193a-3p could repress the expression of the c-kit proto-oncogene and function as a methylation-silenced tumor suppressor in acute myeloid leukemia (34). Therefore, at this stage, the most important question is to investigate how critical this new pathway would be in the web of lung carcinogenesis. In this study, we found that overexpressing miR-193a-3p could inhibit proliferation and invasion and promote apoptosis in lung cancer cells and that ERBB4 reduction can mimic miR-193a-3p induction. Interestingly, we observed that the restoration of ERBB4 expression can successfully attenuate the anti-proliferative, anti-invasion, and pro-apoptotic effects of miR-193a-3p on lung cancer cells, although miR-193a-3p has many other targets. These results suggest that the targeting of ERBB4 is a major mechanism by which miR-193a-3p exerts its tumor-suppressive function. Therefore, the modulation of ERBB4 by miR-193a-3p might explain, at least in part, why the down-regulation of miR-193a-3p during lung carcinogenesis can promote cell growth and invasion and accelerate lung cancer formation.
Intense efforts have been directed at inhibiting the activities of the ErbB family in cancer patients by designing antibodies against the ligand binding domains and small molecules against the tyrosine kinase domains. Both approaches have shown considerable clinical promise. However, increasing evidence suggests that the majority of cancer patients do not respond to these therapies, and those who do show an initial response but are ultimately refractory to treatment (56). Recently, several patents pertaining to the utilization of ERBB4 have been issued, and some studies have also provided evidence that ERBB4 may serve as a molecular target for anticancer therapy (22). Thus, ERBB4 is likely to become a new target for lung cancer treatment. On the other hand, given the dysregulation of miRNAs in cancer development, the correction of cellular miRNA levels has emerged as a potential therapeutic strategy. The overexpressed miRNAs can be silenced using “antagomirs,” and the re-expression of miRNAs that are lost in cancers can be achieved by overexpressing miRNA mimics. In this study, treatment with miR-193a-3p showed an anti-tumor effect both in vitro and in vivo through the negative regulation of ERBB4. Thus, it is hypothesized that a replacement treatment with miR-193a-3p mimics may be a promising strategy for cancers characterized by miR-193a-3p down-regulation. In summary, as important emerging modulators in cellular pathways, miR-193a-3p and ERBB4 may provide attractive, novel therapeutic targets for lung cancer treatment. In future studies, treatments with both miR-193a-3p mimics and ERBB4-targeted drugs may offer a viable strategy for lung cancer therapy.
Taken as a whole, this study delineates a novel regulatory network employing miR-193a-3p and ERBB4 to fine tune proliferation, invasion, and apoptosis in lung cancer cells. This study may open new avenues for future lung cancer therapies.
This work was supported by National Basic Research Program of China (973 Program) Grant 2014CB542300; National Natural Science Foundation of China Grants 81101330, 31271378, and 81250044; Natural Science Foundation of Jiangsu Province Grant BK2012014; and Research Special Fund for Public Welfare Industry of Health Grant 201302018. This work was also supported by the program for New Century Excellent Talents in University from the Ministry of Education, China, Grant NCET-12-0261.
- NSCLC
- non-small cell lung cancer
- miRNA
- microRNA
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- BLI
- bioluminescent imaging
- EGFR
- EGF receptor.
REFERENCES
- 1. Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. (2011) Global cancer statistics. CA Cancer J. Clin. 61, 69–90 [DOI] [PubMed] [Google Scholar]
- 2. Ramalingam S. S., Owonikoko T. K., Khuri F. R. (2011) Lung cancer: new biological insights and recent therapeutic advances. CA Cancer J. Clin. 61, 91–112 [DOI] [PubMed] [Google Scholar]
- 3. Heist R. S., Engelman J. A. (2012) SnapShot: non-small cell lung cancer. Cancer Cell 21, 448 e442 [DOI] [PubMed] [Google Scholar]
- 4. Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 5. Zochbauer-Muller S., Gazdar A. F., Minna J. D. (2002) Molecular pathogenesis of lung cancer. Annu. Rev. Physiol. 64, 681–708 [DOI] [PubMed] [Google Scholar]
- 6. Harpole D. H., Jr., Marks J. R., Richards W. G., Herndon J. E., 2nd, Sugarbaker D. J. (1995) Localized adenocarcinoma of the lung: oncogene expression of erbB-2 and p53 in 150 patients. Clin. Cancer Res. 1, 659–664 [PubMed] [Google Scholar]
- 7. Schneider P. M., Praeuer H. W., Stoeltzing O., Boehm J., Manning J., Metzger R., Fink U., Wegerer S., Hoelscher A. H., Roth J. A. (2000) Multiple molecular marker testing (p53, C-Ki-ras, c-erbB-2) improves estimation of prognosis in potentially curative resected non-small cell lung cancer. Br. J. Cancer 83, 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bublil E. M., Yarden Y. (2007) The EGF receptor family: spearheading a merger of signaling and therapeutics. Curr. Opin. Cell Biol. 19, 124–134 [DOI] [PubMed] [Google Scholar]
- 9. Furger C., Fiddes R. J., Quinn D. I., Bova R. J., Daly R. J., Sutherland R. L. (1998) Granulosa cell tumors express erbB4 and are sensitive to the cytotoxic action of heregulin-β2/PE40. Cancer Res. 58, 1773–1778 [PubMed] [Google Scholar]
- 10. Gilmour L. M., Macleod K. G., McCaig A., Gullick W. J., Smyth J. F., Langdon S. P. (2001) Expression of erbB-4/HER-4 growth factor receptor isoforms in ovarian cancer. Cancer Res. 61, 2169–2176 [PubMed] [Google Scholar]
- 11. Graber H. U., Friess H., Kaufmann B., Willi D., Zimmermann A., Korc M., Büchler M. W. (1999) ErbB-4 mRNA expression is decreased in non-metastatic pancreatic cancer. Int. J. Cancer 84, 24–27 [DOI] [PubMed] [Google Scholar]
- 12. Haugen D. R., Akslen L. A., Varhaug J. E., Lillehaug J. R. (1996) Expression of c-erbB-3 and c-erbB-4 proteins in papillary thyroid carcinomas. Cancer Res. 56, 1184–1188 [PubMed] [Google Scholar]
- 13. Lyne J. C., Melhem M. F., Finley G. G., Wen D., Liu N., Deng D. H., Salup R. (1997) Tissue expression of neu differentiation factor/heregulin and its receptor complex in prostate cancer and its biologic effects on prostate cancer cells in vitro. Cancer J. Sci. Am. 3, 21–30 [PubMed] [Google Scholar]
- 14. Merimsky O., Staroselsky A., Inbar M., Schwartz Y., Wigler N., Mann A., Marmor S., Greif J. (2001) Correlation between c-erbB-4 receptor expression and response to gemcitabine-cisplatin chemotherapy in non-small-cell lung cancer. Ann. Oncol. 12, 1127–1131 [DOI] [PubMed] [Google Scholar]
- 15. Merimsky O., Issakov J., Bickels J., Kollender Y., Flusser G., Soyfer V., Schwartz I., Inbar M., Meller I. (2002) ErbB-4 expression in limb soft-tissue sarcoma: correlation with the results of neoadjuvant chemotherapy. Eur. J. Cancer 38, 1335–1342 [DOI] [PubMed] [Google Scholar]
- 16. Srinivasan R., Benton E., McCormick F., Thomas H., Gullick W. J. (1999) Expression of the c-erbB-3/HER-3 and c-erbB-4/HER-4 growth factor receptors and their ligands, neuregulin-1 α, neuregulin-1 β, and betacellulin, in normal endometrium and endometrial cancer. Clin. Cancer Res. 5, 2877–2883 [PubMed] [Google Scholar]
- 17. Gilbertson R. J., Perry R. H., Kelly P. J., Pearson A. D., Lunec J. (1997) Prognostic significance of HER2 and HER4 coexpression in childhood medulloblastoma. Cancer Res. 57, 3272–3280 [PubMed] [Google Scholar]
- 18. Gilbertson R. J., Clifford S. C., MacMeekin W., Meekin W., Wright C., Perry R. H., Kelly P., Pearson A. D., Lunec J. (1998) Expression of the ErbB-neuregulin signaling network during human cerebellar development: implications for the biology of medulloblastoma. Cancer Res. 58, 3932–3941 [PubMed] [Google Scholar]
- 19. Gilbertson R., Hernan R., Pietsch T., Pinto L., Scotting P., Allibone R., Ellison D., Perry R., Pearson A., Lunec J. (2001) Novel ERBB4 juxtamembrane splice variants are frequently expressed in childhood medulloblastoma. Genes Chromosomes Cancer 31, 288–294 [DOI] [PubMed] [Google Scholar]
- 20. Gilbertson R. J., Bentley L., Hernan R., Junttila T. T., Frank A. J., Haapasalo H., Connelly M., Wetmore C., Curran T., Elenius K., Ellison D. W. (2002) ERBB receptor signaling promotes ependymoma cell proliferation and represents a potential novel therapeutic target for this disease. Clin. Cancer Res. 8, 3054–3064 [PubMed] [Google Scholar]
- 21. Klapper L. N., Kirschbaum M. H., Sela M., Yarden Y. (2000) Biochemical and clinical implications of the ErbB/HER signaling network of growth factor receptors. Adv. Cancer Res. 77, 25–79 [PubMed] [Google Scholar]
- 22. Starr A., Greif J., Vexler A., Ashkenazy-Voghera M., Gladesh V., Rubin C., Kerber G., Marmor S., Lev-Ari S., Inbar M., Yarden Y., Ben-Yosef R. (2006) ErbB4 increases the proliferation potential of human lung cancer cells and its blockage can be used as a target for anti-cancer therapy. Int. J. Cancer 119, 269–274 [DOI] [PubMed] [Google Scholar]
- 23. Paatero I., Elenius K. (2008) ErbB4 and its isoforms: patentable drug targets? Recent Pat. DNA Gene Seq. 2, 27–33 [DOI] [PubMed] [Google Scholar]
- 24. Kang S. M., Lee H. J. (2014) MicroRNAs in human lung cancer. Exp. Biol. Med. 239, 1505–1513 [DOI] [PubMed] [Google Scholar]
- 25. Guz M., Rivero-Müller A., Okoń E., Stenzel-Bembenek A., Polberg K., Słomka M., Stepulak A. (2014) MicroRNAs: role in lung cancer. Dis. Markers 2014, 218169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Calin G. A., Croce C. M. (2006) MicroRNA signatures in human cancers. Nat. Rev. Cancer 6, 857–866 [DOI] [PubMed] [Google Scholar]
- 27. Ma L., Weinberg R. A. (2008) Micromanagers of malignancy: role of microRNAs in regulating metastasis. Trends Genet. 24, 448–456 [DOI] [PubMed] [Google Scholar]
- 28. Nicoloso M. S., Spizzo R., Shimizu M., Rossi S., Calin G. A. (2009) MicroRNAs: the micro steering wheel of tumour metastases. Nat. Rev. Cancer 9, 293–302 [DOI] [PubMed] [Google Scholar]
- 29. Heller G., Weinzierl M., Noll C., Babinsky V., Ziegler B., Altenberger C., Minichsdorfer C., Lang G., Döme B., End-Pfützenreuter A., Arns B. M., Grin Y., Klepetko W., Zielinski C. C., Zöchbauer-Müller S. (2012) Genome-wide miRNA expression profiling identifies miR-9-3 and miR-193a as targets for DNA methylation in non-small cell lung cancers. Clin. Cancer Res. 18, 1619–1629 [DOI] [PubMed] [Google Scholar]
- 30. Walter B. A., Valera V. A., Pinto P. A., Merino M. J. (2013) Comprehensive microRNA profiling of prostate cancer. J. Cancer 4, 350–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tahiri A., Leivonen S. K., L−uders T., Steinfeld I., Ragle Aure M., Geisler J., Mäkelä R., Nord S., Riis M. L. H., Yakhini Z., Kleivi Sahlberg K., Børresen-Dale A. L., Perälä M., Bukholm I. R. K., Kristensen V. N. (2014) Deregulation of cancer-related miRNAs is a common event in both benign and malignant human breast tumors. Carcinogenesis 35, 76–85 [DOI] [PubMed] [Google Scholar]
- 32. Chen D., Cabay R. J., Jin Y., Wang A., Lu Y., Shah-Khan M., Zhou X. (2013) MicroRNA deregulations in head and neck squamous cell carcinomas. J. Oral Maxillofac. Res. 4, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yong F. L., Law C. W., Wang C. W. (2013) Potentiality of a triple microRNA classifier: miR-193a-3p, miR-23a and miR-338-5p for early detection of colorectal cancer. BMC Cancer 13, 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gao X. N., Lin J., Li Y. H., Gao L., Wang X. R., Wang W., Kang H. Y., Yan G. T., Wang L. L., Yu L. (2011) MicroRNA-193a represses c-kit expression and functions as a methylation-silenced tumor suppressor in acute myeloid leukemia. Oncogene 30, 3416–3428 [DOI] [PubMed] [Google Scholar]
- 35. Watson J. A., Bryan K., Williams R., Popov S., Vujanic G., Coulomb A., Boccon-Gibod L., Graf N., Pritchard-Jones K., O'Sullivan M. (2013) miRNA profiles as a predictor of chemoresponsiveness in Wilms' tumor blastema. PLoS One 8, e53417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iliopoulos D., Rotem A., Struhl K. (2011) Inhibition of miR-193a expression by Max and RXRα activates K-Ras and PLAU to mediate distinct aspects of cellular transformation. Cancer Res. 71, 5144–5153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Noh H., Hong S., Dong Z., Pan Z. K., Jing Q., Huang S. (2011) Impaired microRNA processing facilitates breast cancer cell invasion by upregulating urokinase-type plasminogen activator expression. Genes Cancer 2, 140–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Uhlmann S., Mannsperger H., Zhang J. D., Horvat E. Á., Schmidt C., Küblbeck M., Henjes F., Ward A., Tschulena U., Zweig K., Korf U., Wiemann S., Sahin O. (2012) Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol. Syst. Biol. 8, 570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nakano H., Yamada Y., Miyazawa T., Yoshida T. (2013) Gain-of-function microRNA screens identify miR-193a regulating proliferation and apoptosis in epithelial ovarian cancer cells. Int. J. Oncol. 42, 1875–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Salvi A., Conde I., Abeni E., Arici B., Grossi I., Specchia C., Portolani N., Barlati S., De Petro G. (2013) Effects of miR-193a and sorafenib on hepatocellular carcinoma cells. Mol. Cancer 12, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang J., Yang B., Han L., Li X., Tao H., Zhang S., Hu Y. (2013) Demethylation of miR-9-3 and miR-193a genes suppresses proliferation and promotes apoptosis in non-small cell lung cancer cell lines. Cell Physiol. Biochem. 32, 1707–1719 [DOI] [PubMed] [Google Scholar]
- 42. Chen X., Guo X., Zhang H., Xiang Y., Chen J., Yin Y., Cai X., Wang K., Wang G., Ba Y., Zhu L., Wang J., Yang R., Zhang Y., Ren Z., Zen K., Zhang J., Zhang C. Y. (2009) Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 28, 1385–1392 [DOI] [PubMed] [Google Scholar]
- 43. Li B., Blanc J. M., Sun Y., Yang L., Zaorsky N. G., Giacalone N. J., Torossian A., Lu B. (2014) Assessment of M867, a selective caspase-3 inhibitor, in an orthotopic mouse model for non-small cell lung carcinoma. Am. J. Cancer Res. 4, 161–171 [PMC free article] [PubMed] [Google Scholar]
- 44. Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. (2003) Prediction of mammalian microRNA targets. Cell 115, 787–798 [DOI] [PubMed] [Google Scholar]
- 45. John B., Enright A. J., Aravin A., Tuschl T., Sander C., Marks D. S. (2004) Human MicroRNA targets. PLoS Biol. 2, e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. (2005) Combinatorial microRNA target predictions. Nat. Genet. 37, 495–500 [DOI] [PubMed] [Google Scholar]
- 47. Wang X. C., Tian L. L., Wu H. L., Jiang X. Y., Du L. Q., Zhang H., Wang Y. Y., Wu H. Y., Li D. G., She Y., Liu Q. F., Fan F. Y., Meng A. M. (2010) Expression of miRNA-130a in nonsmall cell lung cancer. Am. J. Med. Sci. 340, 385–388 [DOI] [PubMed] [Google Scholar]
- 48. Du L., Schageman J. J., Irnov, Girard L., Hammond S. M., Minna J. D., Gazdar A. F., Pertsemlidis A. (2010) MicroRNA expression distinguishes SCLC from NSCLC lung tumor cells and suggests a possible pathological relationship between SCLCs and NSCLCs. J. Exp. Clin. Cancer Res. 29, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ambros V. (2004) The functions of animal microRNAs. Nature 431, 350–355 [DOI] [PubMed] [Google Scholar]
- 50. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 51. He L., Hannon G. J. (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531 [DOI] [PubMed] [Google Scholar]
- 52. Yu T., Li J., Yan M., Liu L., Lin H., Zhao F., Sun L., Zhang Y., Cui Y., Zhang F., He X., Yao M. (2014) MicroRNA-193a-3p and -5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene, in press [DOI] [PubMed] [Google Scholar]
- 53. Esquela-Kerscher A., Slack F. J. (2006) Oncomirs: microRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269 [DOI] [PubMed] [Google Scholar]
- 54. Kwon J. E., Kim B. Y., Kwak S. Y., Bae I. H., Han Y. H. (2013) Ionizing radiation-inducible microRNA miR-193a-3p induces apoptosis by directly targeting Mcl-1. Apoptosis 18, 896–909 [DOI] [PubMed] [Google Scholar]
- 55. Ma K., He Y., Zhang H., Fei Q., Niu D., Wang D., Ding X., Xu H., Chen X., Zhu J. (2012) DNA methylation-regulated miR-193a-3p dictates resistance of hepatocellular carcinoma to 5-fluorouracil via repression of SRSF2 expression. J. Biol. Chem. 287, 5639–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Aurisicchio L., Marra E., Luberto L., Carlomosti F., De Vitis C., Noto A., Gunes Z., Roscilli G., Mesiti G., Mancini R., Alimandi M., Ciliberto G. (2012) Novel anti-ErbB3 monoclonal antibodies show therapeutic efficacy in xenografted and spontaneous mouse tumors. J. Cell. Physiol. 227, 3381–3388 [DOI] [PubMed] [Google Scholar]