Background: In Bombyx mori, the genes responsible for silk biosynthesis appear to be controlled by hormones.
Results: Bmdimm is specifically expressed in silk glands and directly regulates the expression of fibroin H-chain (fib-H) by the juvenile hormone (JH)-Met-Kr-h1 pathway.
Conclusion: JH is involved in synthesis of fib-H through Bmdimm.
Significance: This pathway provides new insights into hormonal regulation of silk protein genes.
Keywords: Basic Helix-Loop-Helix (bHLH) Transcription Factor, Gene Expression, Gene Regulation, Molecular Biology, Transcription Factor, Bmdimm, Bombyx mori, Silk Gland, Fibroin H-chain, Juvenile Hormone
Abstract
The genes responsible for silk biosynthesis are switched on and off at particular times in the silk glands of Bombyx mori. This switch appears to be under the control of endogenous and exogenous hormones. However, the molecular mechanisms by which silk protein synthesis is regulated by the juvenile hormone (JH) are largely unknown. Here, we report a basic helix-loop-helix transcription factor, Bmdimm, its silk gland-specific expression, and its direct involvement in the regulation of fibroin H-chain (fib-H) by binding to an E-box (CAAATG) element of the fib-H gene promoter. Far-Western blots, enzyme-linked immunosorbent assays, and co-immunoprecipitation assays revealed that Bmdimm protein interacted with another basic helix-loop-helix transcription factor, Bmsage. Immunostaining revealed that Bmdimm and Bmsage proteins are co-localized in nuclei. Bmdimm expression was induced in larval silk glands in vivo, in silk glands cultured in vitro, and in B. mori cell lines after treatment with a JH analog. The JH effect on Bmdimm was mediated by the JH-Met-Kr-h1 signaling pathway, and Bmdimm expression did not respond to JH by RNA interference with double-stranded BmKr-h1 RNA. These data suggest that the JH regulatory pathway, the transcription factor Bmdimm, and the targeted fib-H gene contribute to the synthesis of fibroin H-chain protein in B. mori.
Introduction
The Bombyx mori silk gland is a terminally differentiated silk-producing organ divided into anterior (ASG),3 middle (MSG), and posterior (PSG) silk gland regions. The yield of silk closely depends on the developmental stage of the silk glands and associates with the synthesis and secretion of silk proteins (1). Fibroin, the major silk protein component, is produced by the PSG and composed of three proteins as follows: fibroin heavy chain (fib-H), light chain (fib-L), and p25 proteins. These three proteins form a hexameric complex with a ratio of 6:6:1 of fib-H/fib-L/p25 in the silk assemblage (2). The mRNA levels from these genes and the corresponding fibroin protein accumulation in silk glands vary depending on the developmental stage (3, 4). Identification and characterization of transcription factors that regulate the tissue-specific expression of fibroin genes is a key step toward understanding the molecular mechanism of silk protein synthesis in B. mori.
Several transcription factors involved in transcriptional regulation of silk genes have been reported, including Bombyx Fkh/SGF-1 (5), a homolog of the protein encoded by the Drosophila melanogaster region-specific homeotic forkhead gene (6); SGF-2 (7); POU-M1/SGF-3 (8, 9), a homolog of Drosophila Cf1-a; and FMBP-1 (10). The gene expression profiles of these factors in silk glands have been characterized individually (10, 11) or by genome-wide analysis (12). A basic helix-loop-helix (bHLH) transcription factor, Bmsage, which is a homolog of Drosophila Sage (13, 14), is involved in the regulation of the fib-H gene via interaction with SGF1 (15). The bHLH domain is ∼60 amino acids and has a basic DNA-binding region of 15 amino acids followed by two α-helices separated by a variable loop region (16). The binding sites of a bHLH protein feature a conserved E-box-binding sequence motif, CANNTG, in which the central 2 bp are unique to the protein (17). The DNA-binding specificity of the protein to the center of the E-box motif is influenced by a single basic residue, which is arginine in Max, Myc, and other bHLH-zipper proteins (18). In addition to DNA binding activity, the bHLH domain also promotes dimerization, allowing the formation of homodimer or heterodimer complexes of transcription factors (19, 20). In Drosophila, expression of the bHLH transcription factor dimmed is highly restricted to neurosecretory cells (21). Dimmed activates expression of the peptidylglycine-a-hydroxylating monooxygenase by forming a homodimer that binds to the E-box element of the target gene (22). In B. mori, 52 bHLH genes have been identified in 39 bHLH families (23). However, no study has reported on their function in direct regulation of silk protein synthesis.
Juvenile hormone (JH) is a “status quo” hormone that is necessary for maintaining larval nature during insect life (24). Kruppel homolog 1 (Kr-h1), a C2H2 zinc finger-type transcription factor, is an early gene inducible by JH that represses metamorphic differentiation of the adult abdominal epidermis in Drosophila (25). Recent studies demonstrate that JH binds to a candidate receptor, methoprene-tolerant (Met) (26–28), forming a functional JH receptor complex with another bHLH-PAS transcription factor, steroid receptor co-activator (FISC/Taiman) (29, 30). The JH-Met-steroid receptor co-activator complex binds to the JH-response element and activates transcription of Kr-h1 (31, 32). The JH-Met-Kr-h1 cascade is conserved in the larval-pupal transition in holometabolous insects and the nymphal-adult transition in hemimetabolous insects (33, 34). In B. mori, Kr-h1 is involved in the repression of metamorphosis, and silk glands are enlarged in transgenic silkworms that overexpress Kr-h1 (35).
In B. mori silk glands, the genes responsible for silk biosynthesis are switched on and off at particular times (3). This switch appears to be under the control of endogenous and exogenous hormones. The larval period is prolonged, and silk synthesis is increased when JH compounds are applied before 96 h of the fifth instar stage (36). The DNA content in silk glands of JH-treated larvae is increased two times over controls (37), and RNA synthesis in silk glands is suppressed and then increased after JH analog (JHA) application (38). However, the mechanism by which JH regulates silk gland development and silk protein synthesis and whether Kr-h1 plays a direct role in regulating silk gene expression remains to be elucidated.
In this study, the bHLH transcription factor gene Bmdimm was identified, and its function in regulation of fib-H synthesis was analyzed. The results suggest that this gene is critical for silk protein synthesis in B. mori.
MATERIALS AND METHODS
Experimental Insects and Cells
Wild type strain Dazao (normal yield silk strain) and strain 872 (high yield silk strain) were obtained from the Gene Resource Library of Domesticated Silkworm, Southwest University, China. Both the percent of larvae spinning cocoons (cocoon shelling rate) and silk protein production were higher for strain 872 than for strain Dazao. Larvae were reared on fresh mulberry leaves or an artificial diet at 25 °C under a photoperiod of 12 h light/12 h dark with 75% relative humidity. The B. mori cell line BmE (39), originally derived from embryo cells, expresses endogenous Bmdimm and fib-H and was maintained at 27 °C in Grace's medium supplemented with 10% FBS (HyClone). The BmN cells (maintained in our laboratory), which originated from ovarian tissues, were maintained at 27 °C in TC100 medium supplemented with 10% FBS (HyClone).
Bioinformatic Analysis
Screening and identification of candidate genes were carried out by analyzing the silkworm microarray database. Candidate genes were verified by domain prediction using SMART. Prediction of open reading frames and translated amino acid sequences were performed by using ExPASy. Phylogenetic analysis was conducted with bHLH superfamily genes from B. mori, D. melanogaster, Mus musculus, Scyliorhinus canicula, Danio rerio, Ciona savignyi, Pediculus humanus corporis, Rhipicephalus pulchellus, and Takifugu rubripes (Table 1) using Clustal X with default parameters (40) and using a neighbor-joining method with MEGA version 5.0 (41). Sequences of fib-H and Bmdimm promoters were from SILKDB. Potential cis-response elements in the fib-H and Bmdimm promoters were analyzed using MATINSPECTOR.
TABLE 1.
Name of gene and accession number for phylogenetic analysis from NCBI
| Genes | Species | Accession no. |
|---|---|---|
| Bmdimm | B. mori | KC820643 |
| dimmed | D. melanogaster | NP_523611.1 |
| dimmed | T. rubripes | XP_003961540.1 |
| dimmed | Tribolium castaneum | XP_971229.1 |
| mist1 | P. humanus corporis | EEB19867.1 |
| dimmed | Rhipicephalus pulchellus | JAA56849.1 |
| Mist1 | D. rerio | ABJ97073.1 |
| Mist1 | Rattus norvegicus | AAF17707.1 |
| MIST1 | M. musculus | AAD51766.1 |
RT-PCR and qRT-PCR
Total RNA was prepared using an E.Z.N.A. Total RNA kit II according to the protocol provided by the manufacturer (Omega, Norcross, GA). Reverse transcription PCR (RT-PCR) was as described previously (15). Primers for amplifying Bmdimm, fib-H, BmKr-h1, BmMet2, BmMet1, BmSRC, and BmRpl3 are listed in Table 2. Template DNA was denatured at 95 °C for 5 min, followed by 28–35 cycles of 95 °C for 10 s, 60 °C for 15 s, and 72 °C for 30 s. PCR products were separated on 1% agarose gels and stained with ethidium bromide. The silkworm housekeeping gene for ribosomal protein L3 (BmRpl3) was used as an internal control for normalization of RNA.
TABLE 2.
Primer sequences used in this study
F indicates forward, and R indicates reverse.
| Assay | Gene | Primer sequences (5′ to 3′) |
|---|---|---|
| RT-PCR | fib-H | F, ATACGCTTGGTCGTCAAAATCTG |
| R, TCTGTGTCATCTGCTTCATCTCG | ||
| RT-PCR | Bmdimm | F, ATGCCACACTGGGTAAC |
| R, GAAGAAACCTTGCGTCCG | ||
| RT-PCR | BmMet1 | F, AATCTTGCCACCAACAGC |
| R, ACCCAACGCACATCTTCT | ||
| RT-PCR | BmMet2 | F, ACGGCCATTAAATCCTTG |
| R, GGTAAACCCTCACGACAC | ||
| RT-PCR | BmSRC | F, TCAAACGAGTCAAATAGGGTCA |
| R, GCGGTCGGTGGTAGGGTT | ||
| RT-PCR | BmKr-h1 | F, TCACAACCTACGCCAACA |
| R, CGCTCCTCGTCACCTATC | ||
| RT-PCR | BmRpl3 | F, TCGTCATCGTGGTAAGGTCAA |
| R, TTTGTATCCTTTGCCCTTGGT | ||
| qRT-PCR | Bmsage | F, AGCAATCACGAAGGTCCGC |
| R, CGTATCGTGGTTGGAGTCGT | ||
| qRT-PCR | Bmdimm | F, CGTGGAACCCGCATTTGTA |
| R, AACCTCGGCAATCCAGTCG | ||
| qRT-PCR | fib-H | F, TATCCAGGACGAAGTAAGAAACAA |
| R, TCTGTGTCATCTGCTTCATCTCG | ||
| qRT-PCR | BmMet1 | F, AATCTTGCCACCAACAGC |
| R, ACCCAACGCACATCTTCT | ||
| qRT-PCR | BmMet2 | F, ACGGCCATTAAATCCTTG |
| R, GGTAAACCCTCACGACAC | ||
| qRT-PCR | BmMet2-ORF | F, ATGAAGCGTCCAAATAAC |
| R, TGACATCGTCCAAGAAAC | ||
| qRT-PCR | BmSRC | F, TCAAACGAGTCAAATAGGGTCA |
| R, GCGGTCGGTGGTAGGGTT | ||
| qRT-PCR | BmKr-h1 | F, AACCCATACTGGCGAGCG |
| R, ATACGACGGTGTACTTGC | ||
| qRT-PCR | BmKr-h1-ORF | F, ATGATAGGTGACGAGGAGC |
| R, CCAGTATGGGTTCGGTAG | ||
| qRT-PCR | SGF1 | F, CCTTTCTACAGACAAAACCAGC |
| R, GTCAGGATGTAGCGTCCAAAA | ||
| qRT-PCR | Brc-Z2 | F, TGGACAGTCAGACGCAACA |
| R, TAAGAACGGCGGACGAG | ||
| qRT-PCR | Brc-Z4 | F, TATGGCCCTTCCAACCCTGAT |
| R, AGGTGTTGCTGCTCCGTGTG | ||
| qRT-PCR | BmRpl3 | F, TTCGTACTGGCTCTTCTCGT |
| R, CAAAGTTGATAGCAATTCCCT | ||
| ChIP-qPCR | fibH-E-box | F, TGGACAGATTTGGCTTTG |
| R, CACTAGAGGAACGGGACA | ||
| ChIP-qPCR | Bmdimm-Kr CRE | F, GTCAGTCCTACGGCATAT |
| R, GATAGTGCCACAACATAAAA |
For quantitative real time PCR (qRT-PCR), SYBR Green kits were used according to the manufacturer (TaKaRa Co., Japan) with primers in Table 2 and the following conditions: denaturation at 95 °C for 10 min followed by 40 cycles at 95 °C for 10 s, 60 °C for 30 s, and 72 °C for 35 s with an ABI7500 real time PCR machine (Applied Biosystems) using FastStart Universal SYBR Green Master. Relative mRNA levels of target genes were calculated with the 2−ΔΔCt method (42), where the target gene expression level was normalized to the expression of the internal marker gene BmRpl3. Three independent replicates were performed.
Fluorescent in Situ Hybridization
Fresh silk glands were dissected from B. mori larvae on day 3 of the fifth instar and fixed with 4% (v/v) formaldehyde after washing with PBS, pH 7.4. Samples for hybridizing with digoxigenin (DIG)-labeled DNA probes were prepared by the standard paraffin procedure. DIG-labeled DNA probes were synthesized using a DIG DNA Labeling Mix (Roche Applied Science). Pre-hybridization, hybridization, immune reaction, and sample mounting were performed as described (43).
Production of Recombinant Protein and Western Blots
The coding region of Bmdimm was amplified with specific primers (Table 3) and cloned into pET28a vector (Novagen, Germany). Positive clones were transformed into Escherichia coli strain BL21 (DE3) cells (TransGen, Beijing, China) to express Bmdimm recombinant protein, which was purified as described by Zhao et al. (15). Purified protein was injected into New Zealand White rabbits for polyclonal antibody preparation.
TABLE 3.
Primers for DNA constructs in this study
| Plasmid name | PCR template | Primer sequences (5′ to 3′) |
|---|---|---|
| pET-28a/Bmdimm | cDNA | F, CGCGGATCCATGCGCCCAAGACGCGCT |
| R, CCCAAGCTTCAGAAGAAACCTTGCG | ||
| fib-H865-Luc | Genomic DNA | F, CGGGGTACCAAGCTTGTTGTACAAAACTG |
| R, CTAGCTAGCGCTGATTTGAAAAAGTTGAA | ||
| fib-H400-Luc | Genomic DNA | F, CGGGGTACCATTTACCCATCCAAGGCATTC |
| R, CTAGCTAGCGCTGATTTGAAAAAGTTGAA | ||
| fib-H37-Luc | Genomic DNA | F, CGGGGTACCATTTTCAGTATAAAAAG |
| R, CTAGCTAGCGCTGATTTGAAAAAGTTGAA | ||
| pGEX-4T-Bmdimm | cDNA | F, CGCGGATCCATGCCACACTGGGTAACT |
| R, ATTTGCGGCCGCTCAGAAGAAACCTTGCG | ||
| pGEX-4T-Bmdimm-Mut-E108T | pGEX-4T-Bmdimm | F, GTCTCGAGAGCAATACAAGAGAAAGAATGCGA |
| R, GCATTCTTTCTCTTGTATTGCTCTCGAGACGA | ||
| pGEX-4T-Bmdimm-Mut-R109T | pGEX-4T-Bmdimm | F, GTCTCGAGAGCAATGAAACAGAAAGAATGCGA |
| R, ATTCGCATTCTTTCTGTTTCATTGCTCTCGAG | ||
| pGEX-4T-Bmdimm-Mut-R113T | pGEX-4T-Bmdimm | F, AAAGAGAAAGAATGACAATGCATTCCCTCAAC |
| R, CGGTTGAGGGAATGCATTGTCATTCTTTCTCT | ||
| pGEM-T-BmKr-h1 | cDNA | F, ATGATAGGTGACGAGGAGCGAG |
| R, CTATGATTCTGTAGCTGGCG | ||
| 1180-BmKr-h1 | cDNA | F, CGCGGATCCATGTACCCATACGATGTTCCAGATTACGCTATAGGTGACGAGGAGCGAG |
| R, ATTTGCGGCCGCCTATGATTCTGTAGCTGGCG | ||
| 1180-Bmdimm | pGEX-4T-Bmdimm | F, CGCGGATCCATGGAACAAAAACTCATCTCAGAAGAGGATCTGCCACACTGGGTAACT |
| R, ATTTGCGGCCGCTCAGAAGAAACCTTGCG | ||
| 1180-Bmsage | pGEX-4T-Bmsage | F, CGCGGATCCATGGATTACAAGGATGACGACGATAAGTACAATCAAACATAC |
| R, ATTTGCGGCCGCTTAGTATCTCTGTTGACGC | ||
| 1180-BmMet2 | cDNA | F, CGCGGATCCATGGATTACAAGGATGACGACGATAAGGCTGATTGGTCTCTG |
| R, ATTTGCGGCCGCTTATTGTTGTTGGTTTTTCT |
Protein extracts were isolated from Malpighian tubules, fat bodies, heads, midguts, ASG, MSG, PSG, epidermises, and gonads from day 3 fifth instar larvae. Western blots used Bmdimm antibody as described in Zhao et al. (15).
DNA Transfection and Hormone Treatment
Three 5′-truncated fragments (−865 to +1, −400 to +1, and −37 to +1) of the fib-H promoter were generated by PCR from genomic DNA using specific primers (Table 3). Amplified fragments were cloned into the luciferase reporter plasmid, pGL3 basic vector (Promega, Madison, WI), at the N-terminal end between the KpnI and NheI restriction sites to generate fib-H865-Luc, fib-H400-Luc, and fib-H37-Luc. Primers for amplifying the ORFs of Bmsage, Bmdimm, BmKr-h1, and BmMet2 are listed in Table 3. Target fragments were obtained by gel purification and cloned into an 1180 (Hrs1000-BmAct4-LUC-Ser1PA) expression vector (maintained in our laboratory) between BamHI and NotI sites. Highly purified plasmid DNA was prepared using Qiagen Plasmid Midi kits (Qiagen, Germany). Transfection vectors were used to transfect BmE cells using X-tremeGENE HP DNA transfection reagent (Roche Applied Science) as described by the manufacturer. An EGFP transfection vector (maintained in our laboratory) was used as a control. For hormone treatment, a JH analog (Sigma) was dissolved in DMSO and applied to the culture medium at final concentrations of 0.1, 1, or 10 μm for 6, 12, or 24 h, and cells were collected for RNA extraction.
Far-Western Blot and ELISA
A Bmdimm-glutathione S-transferase (GST) fusion protein expression vector was constructed by subcloning full-length Bmdimm cDNA into the GST gene fusion vector pGEX-4T-1 (Amersham Biosciences). Expression and purification of recombinant Bmdimm-GST were as described by Zhao et al. (15). Site-directed mutants were made using QuikChange site-directed mutagenesis kits (Stratagene, La Jolla, CA). Primers for producing mutants E108T, R109T, and R113T are listed in Table 3.
Far-Western blots were as described by Wu et al. (44). Purified protein (1 μg) was separated by SDS-PAGE with 15% (w/v) polyacrylamide gels as prey protein. Following electrophoresis, proteins were transferred to PVDF membranes, denatured, and renatured. Membranes were blocked using BSA and probed with purified bait protein. GST protein was used as a negative control.
To confirm interactions between Bmsage and Bmdimm, Bmsage-Bmdimm binding was detected by ELISA (45). Purified Bmdimm-GST and BSA (0.5 μg) were dissolved in 50 mm Tris-HCl, pH 7.4, 12.7 mm EDTA and fixed in wells of 96-well polystyrene plates overnight at 4 °C. Plates was washed three times with PBS containing 0.05% Tween 20, pH 7.4, and blocked with 1% (w/v) BSA in PBS for 2 h at 37 °C. After three washes with PBS, plates were incubated with Bmsage-GST in PBS for 2 h at 37 °C and probed with rabbit polyclonal antibody against Bmsage and secondary antibody conjugated with horseradish peroxidase at 37 °C. After three washes with PBS, 3,3′,5,5′-tetramethylbenzidine (Beyotime, Beijing, China) was added to plates as a chromogenic reagent, and plates were kept in the dark for 15 min. Absorption at 450 nm was detected by Microplate Reader (Bio-Rad).
Immunostaining and Co-immunoprecipitation
Immunostaining of Bmsage and Bmdimm was as described in Liu et al. (46). For immunostaining, BmE cells were grown on glass coverslips in Grace's medium supplemented with 10% FBS (HyClone). Cells were transfected with expression plasmids using X-tremeGENE HP DNA Transfection Reagent (Roche Applied Science) as described above. After transfection for 48 h, cells were fixed for 10 min at room temperature with 4% (v/v) formaldehyde in PBS and blocked for 30 min in PBS containing 0.1% (w/v) BSA and 5% (v/v) goat serum. Samples were treated with primary antibody (anti-FLAG monoclonal M2 mouse (Sigma), anti-Myc monoclonal rabbit (Sigma), or anti-Bmsage rabbit) for 1 h before incubation with secondary antibody (anti-mouse IgG FITC or anti-rabbit Alexa 488) for 30 min at room temperature. Samples were mounted using a mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI) and photographed using confocal microscopy (Olympus FV1000, Japan).
To further confirm whether Bmdimm protein interacted with Bmsage protein, co-immunoprecipitation was conducted with nuclear extracts from BmE cells overexpressing FLAG-Bmsage and Myc-Bmdimm. Antibody (10 μg) diluted in 200 μl of lysate/washing buffer (25 mm Tris, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 5% glycerol, 0.25 mm phenylmethylsulfonyl fluoride) was added to 50 μl (1.5 mg) of 5% (w/v) bovine serum albumin (BSA)-blocked Dynabeads (Beyotime, Beijing, China) and incubated at room temperature with rotation for 10 min. Supernatant was collected by centrifugation. Bead-antibody complexes were washed twice in 200 μl of washing buffer, and 200 μl of nuclear extract (1 mg of protein) was added. Mixtures were incubated with rotation for 2 h at 4 °C followed by centrifugation. Precipitates were washed five times with washing buffer, and the immunoprecipitated complexes were suspended in SDS sample buffer and analyzed by SDS-PAGE followed by Western blot using the indicated antibodies as described in Lin et al. (47).
Electrophoretic Mobility Shift Assay
To test protein binding to regulatory elements, electrophoretic mobility shift assays (EMSA) were performed according to Kethidi et al. (48). The potential E-box element sequence at −464 to −438 (5′-TTGATTTACAAATGTTTTTTTGGTG-3′) was used as a probe. Oligonucleotides were labeled using Cy3 from the 5′-end and annealed to produce a double-stranded probe. DNA-binding reactions were in 10 μl containing 10 μg of nuclear protein extract or 1 μg of purified recombinant protein and 2 μl of 5× binding buffer (Beyotime, Beijing, China). Labeled probe (5 μm) was added after incubation for 20 min at 25 °C, and incubation was continued for 25 min. For competition assays, unlabeled double-stranded probe was added to the reaction at the same time as labeled probe. Mixtures were loaded onto 5% (w/v) polyacrylamide gels and electrophoresed in 1× TBE buffer (45 mm Tris borate, 1 mm EDTA, pH 8.3). After electrophoresis, gels were scanned and photographed with a TYPHOON scanner (Amersham Biosciences).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed according to the manufacturer's instructions (Upstate/Millipore, Temecula, CA). DNA-protein complexes were sheared by sonication, and 1% was used as input. Anti-Bmdimm, anti-HA, and normal rabbit IgG were used for immunoprecipitation. PCR was performed using primers listed in Table 2, and the products were run in 2% agarose gels. Enrichment of promoter binding levels was analyzed by real time PCR in triplicate and expressed as a percentage over input. Quantified results are presented as means ± S.D.
Application of Methoprene
For in vivo hormone treatment, JH analog (Sigma) was dissolved in acetone and diluted to 1 μg/μl. JHA was topically applied to larvae along the dorsal surface at 1 μg per g of fresh body weight as described in previous studies (36). Silk glands were dissected and processed for RNA isolation 24 h after treatment.
For in vitro treatment, silk glands were dissected under sterile conditions in ice-cold insect Ringer (130 mm NaCl, 5 mm KCl, 0.1 mm CaCl2, and 1 mm PMSF) and rinsed in 500 μl of TC-100 insect culture medium (Sigma) supplemented with streptomycin sulfate (50 μg/ml) (49). Tissues were incubated in culture medium with or without JHA for different time periods in 95% relative humidity, 5% CO2, and 25 °C. Silk glands treated with 0.05% acetone were used as controls. After incubation, tissues were rinsed in ice-cold Ringer and processed for RNA isolation.
Double-stranded RNA Synthesis and RNA Interference
Template DNA fragments of BmKr-h1 for synthesis of double-stranded RNA (dsRNA) were amplified by PCR with primers listed in Table 3 and cloned into pGEM-T simple vector, from which dsRNA for BmKr-h1 was synthesized using RiboMAX SP6 and T7 large scale RNA production systems according to the manufacturer's instructions (Promega). dsRNA for EGFP was used as a control. BmE cells (5 × 105 cells) were transfected with 5 μg of dsRNA using X-tremeGENE HP DNA transfection reagent (Roche Applied Science). Cells were treated at 12 h post-transfection with 10 μm JHA for an additional 12 h and harvested for RNA isolation.
Statistical Analysis
All data were statistically analyzed by independent sample t test. Asterisks indicate significant differences (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
RESULTS
Bioinformatic Analysis of the Bmdimm Gene
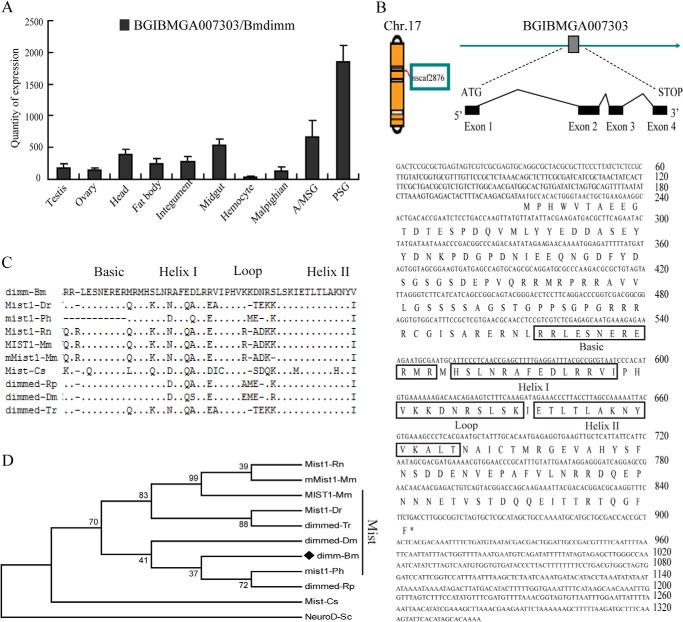
A gene (ID BGIBMGA007303) with a higher expression level in silk glands compared with other tissues was identified using whole silkworm genome tissue microarray data (Fig. 1A). This gene was located on chromosome 17 and consisted of four exons spanning a 4.6-kb fragment in nscaf2876 (Fig. 1B). The open reading frame was 636 bp and encoded a protein of 211 amino acids with a bHLH domain (amino acids 107–160) (Fig. 1B). A BLAST search with the deduced amino acid sequence revealed that the protein was highly similar to several closely related members of the Mist subfamily of the bHLH family (dimmed and Mist1 proteins) in vertebrate and invertebrate species (Fig. 1C), and was named Bmdimm. The bHLH domain of Bmdimm was 74% identical to D. melanogaster dimmed (NP_523611.1) and 72.5% identical to M. musculus Mist (AAD51766.1) (Fig. 1D), suggesting that Bmdimm belonged to the Mist-related subfamily of the bHLH family.
FIGURE 1.
Bioinformatic analysis. A, expression of Bmdimm in silkworm tissues on day 3 of fifth larval instar based on microarray database information. B, location of B. mori dimm in the silkworm genome (left), gene structure (exon-intron diagram), and nucleotide and deduced amino acid sequences. The boxes are amino acid sequences of bHLH. C, bHLH motif in Bmdimm compared with other bHLH proteins, including the Mist subfamily of vertebrates, Dmdimm, and Rpdimm of invertebrates. Dots, identical amino acids; dashes, gaps introduced to maximize alignment. D, phylogenetic tree of bHLH transcription factors. The MEGA5 program was used with a neighbor-joining algorithm. GenBank accession numbers are listed in Table 1.
Bmdimm Is Expressed Specifically in Silk Glands and Its Expression Is Closely Related to Expression of the Fib-H Gene
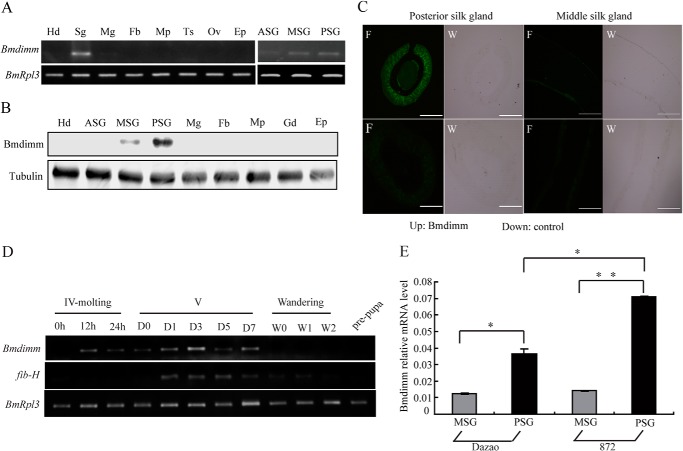
To verify the tissue specificity of Bmdimm expression, gene expression was examined in different B. mori larval tissues at day 3 of the fifth instar using RT-PCR. Bmdimm transcripts were mainly detected in silk glands (Fig. 2A). Western blot analysis showed that expression of Bmdimm mainly occurred in MSG and PSG cells with a much higher level of protein in the PSG (Fig. 2B). To further specify the location of Bmdimm expression in silk glands, in situ hybridization assays were performed using a DIG-labeled probe. The results showed that Bmdimm mRNA staining was high in the PSG and only faintly visible in the MSG (Fig. 2C). The location and patterns of Bmdimm expression were consistent with that of the fib-H gene in PSG cells (2, 15), implying that Bmdimm might be related to the fib-H gene.
FIGURE 2.
Expression pattern of Bmdimm in B. mori silk glands. A, expression of Bmdimm in different B. mori tissues on day 3 of fifth larval instar assayed by RT-PCR. BmRpl3 expression is shown as a control. Tissues are as follows: Hd, head; Sg, silk gland; Mg, midgut; Fb, fat body; Mp, Malpighian; Ts, testis; Ov, ovary; Ep, epidermis; ASG, anterior silk gland; MSG, middle silk gland; PSG, posterior silk gland. B, protein level of Bmdimm in different B. mori tissues on day 3 of fifth larval instar. Tubulin is shown as a control. Tissues are as follows: Hd, Head; ASG, anterior silk gland; MSG, middle silk gland; PSG, posterior silk gland; Mg, midgut; Fb, fat body; Mp, Malpighian; Gd, gonad; Ep, epidermis. C, expression of Bmdimm mRNA in MSG and PSG detected by fluorescent in situ hybridization. DIG-labeled DNA probes were used for in situ hybridization. MSG, longitudinal section; PSG, cross-section; control groups used sense DNA probes. F, fluorescence; W, white light. Scale bar, 300 μm. D, expression of Bmdimm and fib-H in different stages assayed by RT-PCR. BmRpl3 expression was used as a control. Developmental stages are as follows: IV, fourth instar; V, fifth instar (0, 1, 3, 5, and 7 days); wandering (0, 1, and 2 days), and pre-pupa. E, expression of Bmdimm in MSG and PSG assayed by qRT-PCR. BmRpl3 expression was used as a control. Dz, Dazao, low silk strain; 872, high silk strain. Results are expressed as means ± S.D. of three independent experiments; *, p < 0.05; **, p < 0.01.
To explore the relationship between Bmdimm and fib-H, the temporal expression patterns of Bmdimm and fib-H mRNA in silk glands from fourth instar molting larvae to pharate adults were analyzed using RT-PCR. Bmdimm mRNA was detected at a very low level in larvae at the fourth molt, but mRNA increased gradually, peaking at day 5 of fifth instar, followed by a decrease to low levels during the wandering to pupation stages (Fig. 2D). This expression pattern was similar to the expression of the fib-H gene (Fig. 2D). Both the percent of larvae spinning cocoons (cocoon shelling rate) and silk protein production were higher in strain 872 than in strain Dazao, the standard strain used for the silkworm genome sequence, and the expression level of fib-H was 2.0-fold higher in strain 872 than in strain Dazao at day 3 of the fifth instar (15). Bmdimm mRNA was significantly higher in PSG than MSG in both Dazao and 872, and additively, the mRNA level of Bmdimm in PSG but not in MSG was 2.0-fold higher in 872 than in Dazao (Fig. 2E). These results suggested that Bmdimm might be involved in the regulation of fib-H in the PSG.
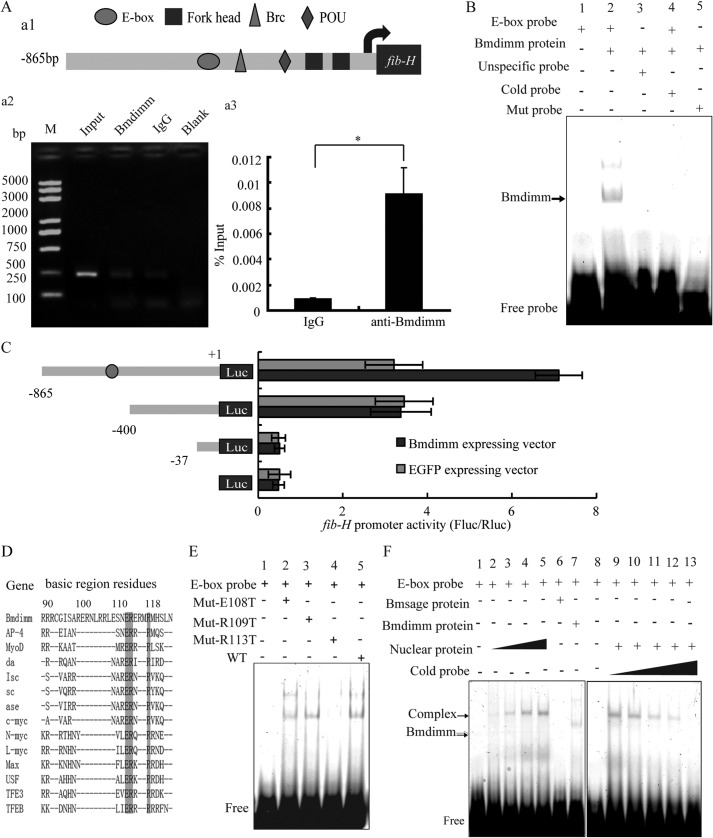
Bmdimm Regulates the Transcription of Fib-H by Binding to Its E-box Element
To explore whether the expression of fib-H was regulated directly by Bmdimm, the relationship between the regulatory sequence of fib-H and Bmdimm was studied. An 865-bp fragment from the 5′-upstream region of fib-H was cloned. Prediction of potential regulatory elements in this promoter region revealed several possible transcription factor-binding elements such as forkhead, POU, Brc, and E-box sequences (Fig. 3A, panel a1). We focused on the E-box element (CANNTG) in this region of the fib-H promoter because it might bind to transcription factors containing a bHLH domain. To test whether or not Bmdimm bound to the E-box element, chromatin immunoprecipitation (ChIP) assays were performed using a specific antibody against Bmdimm. A positive band that corresponded to the fib-H promoter was detected by PCR when we used the anti-Bmdimm antibody; few or no bands were detected in the negative controls (Fig. 3A, panel a2). To quantify the amount of precipitated DNA, qRT-PCR was performed after the ChIP assay using a primer for the fib-H promoter containing E-box element regions. The results indicated significant enrichment of DNA from the E-box element region compared with the IgG control (Fig. 3A, panel a3).
FIGURE 3.
Bmdimm regulates the expression of fib-H in B. mori. A, predicted cis-elements and ChIP analysis. Panel a1, potential cis-elements predicted in the promoter of B. mori fib-H using the MATINSPECTOR program. Panels a2 and a3, ChIP assays for Bmdimm binding to the E-box element using anti-Bmdimm and normal rabbit IgG. PCR products were analyzed on 2% agarose gels. Enrichment of promoter levels was analyzed by qRT-PCR in triplicate and expressed as a percentage over input. B, binding of Bmdimm protein to the E-box element in vitro analyzed by EMSA. C, effect of recombinant Bmdimm protein on expression of a luciferase reporter under control of the fib-H promoter. Relative luciferase activity is presented as a ratio of firefly luciferase activity to Renilla luciferase activity. Experiments were repeated three times independently, and average expression is expressed as a mean ± S.D.; *, p < 0.05. D, amino acid sequences for Bmdimm and other bHLH proteins aligned using Clustal X. E, Bmdimm sequences for glutamic acid and arginine were mutated to generate Mut-E108T, Mut-R109T, and Mut-R113T by site-directed mutagenesis and analyzed by EMSA. F, binding of Bmdimm, Bmsage, and nuclear extracts to the E-box element in vitro by EMSA.
To confirm that Bmdimm protein bound directly to the E-box element, recombinant GST-Bmdimm protein was expressed and purified, and after removal of the GST tag using thrombin, the recombinant protein was used for electrophoretic mobility shift assays (EMSA) with a labeled E-box probe. Bmdimm protein bound to the E-box probe but not to a nonspecific probe (Fig. 3B, lanes 2 and 3), and the binding of Bmdimm to the labeled probe was competitively suppressed by unlabeled cold probe (Fig. 3B, lane 4). Mutating the E-box probe from CAAATG to TAAACT resulted in no binding of Bmdimm (Fig. 3B, lane 5), further suggesting that Bmdimm protein bound specifically to the E-box element in vitro.
To determine whether the E-box sequence was a cis-element for regulation of fib-H transcription, three truncated promoter sequences of fib-H (−865 to +1, −400 to +1, and −37 to +1) were cloned into a pGL3-basic luciferase reporter vector for analysis of promoter activity. Constructs were co-transfected into BmE cells with a Bmdimm-expressing vector or an EGFP-expressing vector, and promoter activity was measured at 48 h post co-transfection using a dual-luciferase reporter system. A strong expression signal was detected when the fib-H865-LUC vector was co-transfected with the Bmdimm-expressing vector compared with co-transfection with the control EGFP vector; however, the promoter activity of the fib-H400-LUC vector was not significantly different from the control (Fig. 3C). These results suggested that the −865- to −400-bp region was critical for transcription activation of the fib-H gene through Bmdimm binding to the E-box element.
Binding specificity at the E-box center is influenced by a single basic residue in bHLH proteins (18). The glutamic acid at position 108 and arginine at 109 and 113 in the basic region of Bmdimm are conserved between bHLH proteins (Fig. 3D). The glutamic acid and arginine in Bmdimm were mutated individually to threonine by site-directed mutagenesis to generate Mut-E108T, Mut-R109T, and Mut-R113T proteins. By EMSA, Mut-R113T did not bind to the E-box probe (Fig. 3E, lane 4), but Mut-E108T and Mut-R109T bound similarly to Bmdimm protein (wild type, WT) (Fig. 3E, lanes 2, 3, and 5), indicating that Arg-113 was a key amino acid for Bmdimm binding to the E-box element, whereas Glu-108 and Arg-109 were not.
To characterize further the binding activity of Bmdimm to the E-box of the fib-H promoter, nuclear extracts were isolated from B. mori PSG, incubated in vitro with the E-box probe, and analyzed by EMSA. A protein in the nuclear extracts bound to the E-box probe in a dose-dependent manner (Fig. 3F, lanes 2–5). The band was competitively inhibited by unlabeled probe (Fig. 3F, lanes 9–13). However, the size of the bound protein in nuclear extracts appeared to be larger than Bmdimm (Fig. 3F, lane 7), implying that Bmdimm might form a complex with another protein when binding to the E-box probe. The Bmsage protein, another bHLH transcription factor expressed specifically in silk glands (15), did not bind the probe alone (Fig. 3F, lane 6), suggesting that it was not the additional protein in nuclear extracts that bound to the E-box probe.
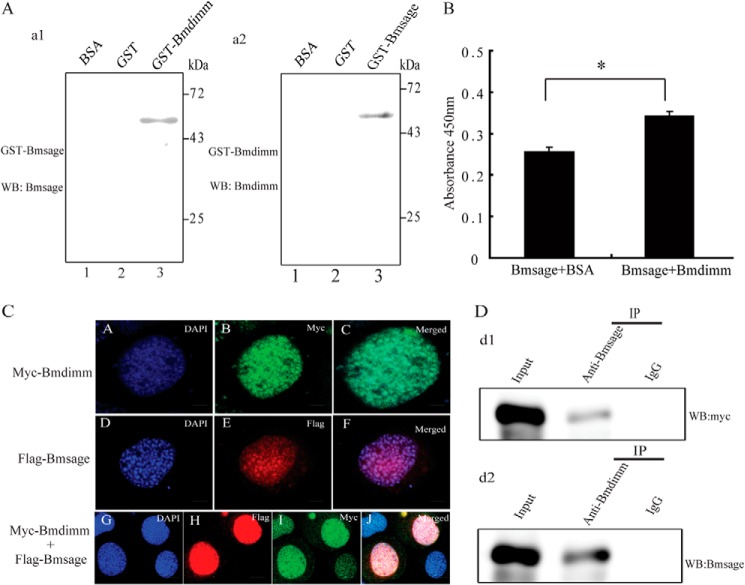
Interaction between Bmdimm and Bmsage
To determine whether Bmdimm and Bmsage interacted, far-Western blot assays were carried out with purified Bmdimm-GST and Bmsage-GST fusion proteins. The results indicated that Bmdimm bound to Bmsage on membranes (Fig. 4A, panel a1, lane 3), and vice versa, Bmsage bound to Bmdimm (Fig. 4A, panel a2, lane 3). Enzyme-linked immunosorbent assays (ELISA) showed that the absorbance of Bmsage/Bmdimm at 450 nm was significantly higher than the absorbance of Bmsage/BSA (Fig. 4B), suggesting Bmsage bound to Bmdimm in vitro. To examine further whether Bmdimm and Bmsage interacted, Myc-tagged Bmdimm and FLAG-tagged Bmsage were expressed in BmE cells. Immunostaining revealed that Bmdimm and Bmsage were co-localized in nuclei (Fig. 4C, panels A–J). Nuclear extracts from BmE cells overexpressing both FLAG-tagged Bmsage and Myc-tagged Bmdimm were used for immunoprecipitation with anti-Bmsage or anti-Bmdimm, followed by Western blots using Myc antibody or Bmsage antibody. The results indicated that Bmdimm protein was present in anti-Bmsage immunoprecipitates (Fig. 4D, panel d1), and vice versa, Bmsage protein was present in anti-Bmdimm immunoprecipitates (Fig. 4D, panel d2). These results demonstrated that Bmdimm protein interacted with Bmsage protein in the nucleus.
FIGURE 4.
Bmdimm protein interacts with Bmsage. A, interaction of Bmsage with Bmdimm. Panel a1, purified Bmdimm-GST used as prey protein was incubated with purified Bmsage-GST as bait protein and detected by anti-Bmsage. Panel a2, purified Bmsage-GST used as prey protein was incubated with Bmdimm-GST as bait protein and detected by anti-Bmdimm. Purified GST and BSA proteins were used as negative controls. B, binding of Bmdimm and Bmsage detected by ELISA. BSA was used as a negative control. Results are expressed as means ± S.D. of three independent experiments; *, p < 0.05. C, immunostaining of Bmdimm and Bmsage in BmE cells. Scale bar, 5 μm. D, nuclear extracts immunoprecipitated (IP) with anti-Bmsage or anti-Bmdimm followed by Western blot (WB) using Myc antibody (panel d1) or Bmsage antibody (panel d2).
Effect of JH on Fib-H Is Mediated by Bmdimm
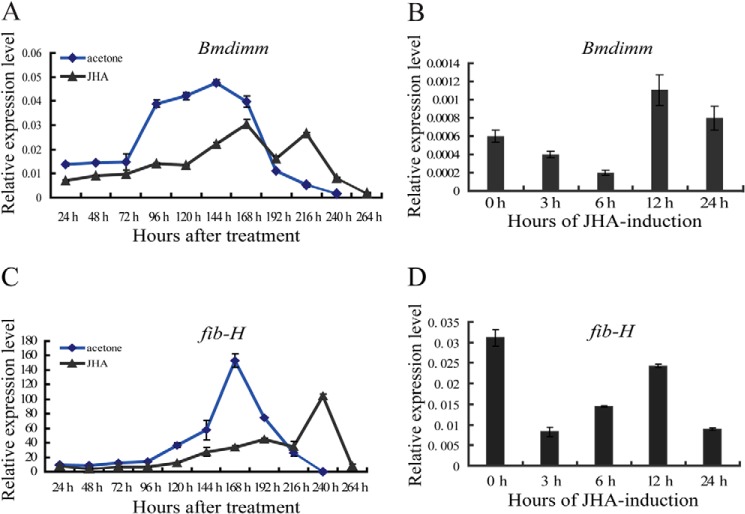
Previous studies showed that JH is involved in the synthesis of silk proteins (50–52). To examine the effect of JH on fib-H expression, 1 μg/μl JHA was topically applied to 1-day-old fifth instar larvae. Not surprisingly, larval development was prolonged for 2–3 days by treatment with JHA compared with control larvae treated with acetone. The levels of Bmdimm transcripts after JHA treatment were determined by qRT-PCR. In silk glands of control silkworms treated with acetone, expression of Bmdimm increased with larval development, peaking around 144 h and then gradually decreasing near the end of the pupal stage. By contrast, in silk glands of silkworms treated with JHA, expression of Bmdimm was lower than in control silkworms until 180 h and then reached a second expression peak that was delayed by about 24 h compared with the control (Fig. 5A). These results indicated that Bmdimm responded to exposure to JHA in vivo.
FIGURE 5.
Bmdimm responds to JHA in silk glands in vivo and in vitro. A and C, expression of Bmdimm and fib-H in larvae in vivo after JHA induction assayed by qRT-PCR. JHA was dissolved in acetone, diluted to 1 μg/μl, and topically applied to larvae along the dorsal surface at 1 μg per g of fresh body weight. Acetone was used for the control. B and D, expression of Bmdimm and fib-H in silk glands in vitro after JHA induction assayed by qRT-PCR. Silk glands were dissected from the 1st day (V0) of the fifth larval instar and incubated with JHA for 0, 3, 6, 12, or 24 h. Results are expressed as means ± S.D. of three independent experiments. BmRpl3 expression is shown as a control.
Using isolated silk glands, we investigated whether Bmdimm was also regulated by JH in vitro. Silk glands were isolated on the 1st day (V0) of the fifth instar, when larvae expressed Bmdimm during normal development (Fig. 2D). Silk glands were preincubated in hormone-free culture medium for 1 h and transferred to medium containing 0.1 μm JHA for 0, 3, 6, 12, or 24 h before RNA analysis. Expression of Bmdimm decreased until 6 h post-JHA treatment and then peaked at 12 h (Fig. 5B). These results indicated that even in isolated silk glands, Bmdimm responded to JH induction.
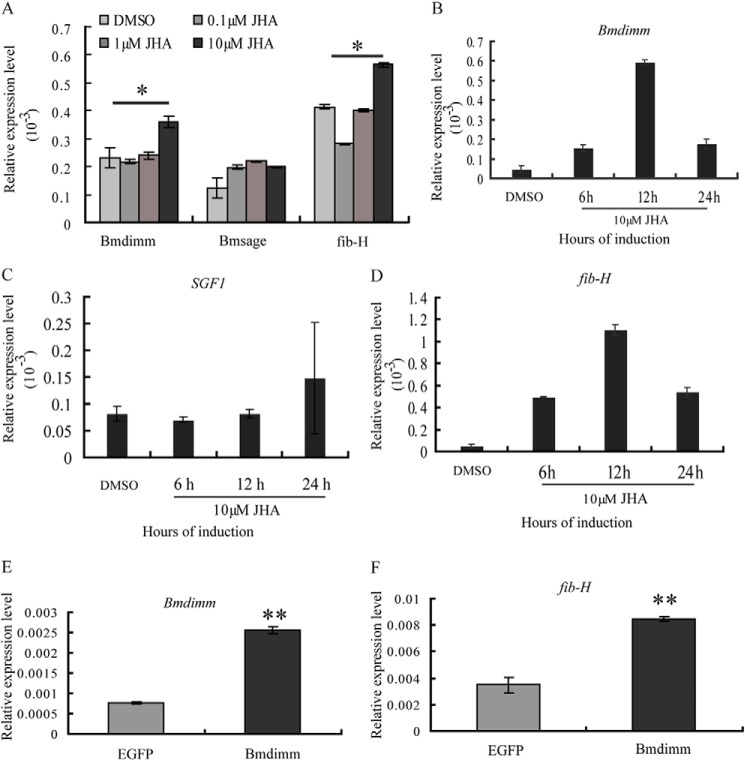
To determine whether fib-H is induced by the increased Bmdimm expression, the expression patterns of Bmdimm and fib-H were first compared in vivo and in isolated silk glands. The results showed that in the intact silkworm, the expression pattern of fib-H was basically consistent with that of Bmdimm, although its expression peak was delayed about 24 h compared with Bmdimm (Fig. 5, A versus C). In silk glands cultured in vitro, the expression of fib-H decreased initially and then increased until 12 h post-JHA treatment (Fig. 5D). Bmdimm was further investigated by qRT-PCR after treatment with different concentrations of JHA for 12 h in BmE cell lines. Its expression slightly declined in 0.1 μm JHA but increased gradually in higher levels of JHA (Fig. 6A). A significant increase happened from 6 to 12 h in 10 μm JHA (Fig. 6B). However, expression of Bmsage and silk gland factor SGF1 was not affected significantly by JHA treatment (Fig. 6, A and C). These results indicated that Bmdimm expression was time-dependently and dose-dependently induced by JHA in BmE cells.
FIGURE 6.
Effect of JH on Bmdimm in BmE cells. A, expression of Bmdimm, Bmsage, and fib-H assayed by qRT-PCR. BmRpl3 expression was used as a control. Dose-related response to JHA administration. BmE cells were cultured in the presence of 0, 0.1, 1, or 10 μm JHA for 12 h. Results are expressed as means ± S.D. of three independent experiments; *, p < 0.05. B–D, expression of Bmdimm (B), SGF1 (C), and fib-H (D) assayed by qRT-PCR. BmRpl3 expression was used as a control. Time-related response to JHA administration. BmE cells were cultured in culture medium with 10 μm JHA solution for 6, 12, or 24 h. E, expression of Bmdimm assayed by qRT-PCR after overexpression of Bmdimm in BmE cells. BmRpl3 expression was used as a control. F, effect of overexpression of Bmdimm on fib-H expression in BmE cells assayed by qRT-PCR. BmRpl3 expression was used as a control. Results are expressed as means ± S.D. of three independent experiments; **, p < 0.01.
Similarly, in BmE cells, fib-H expression declined slightly in 0.1 μm JHA but increased gradually with increasing JHA (Fig. 6A). At 10 μm, JHA treatment for 6, 12, or 24 h, fib-H expression gradually increased from 6 to 12 h (Fig. 6D), which was also consistent with that of Bmdimm (Fig. 5B). Furthermore, endogenous fib-H mRNA was significantly increased by overexpressing Bmdimm (Fig. 6, E and F). Together, all the above results indicated that the JH-Bmdimm-fib-H pathway is involved in synthesis of fib-H in B. mori.
Regulation of Bmdimm by JH Is Mediated by the JH-Met-Kr-h1 Signaling Pathway
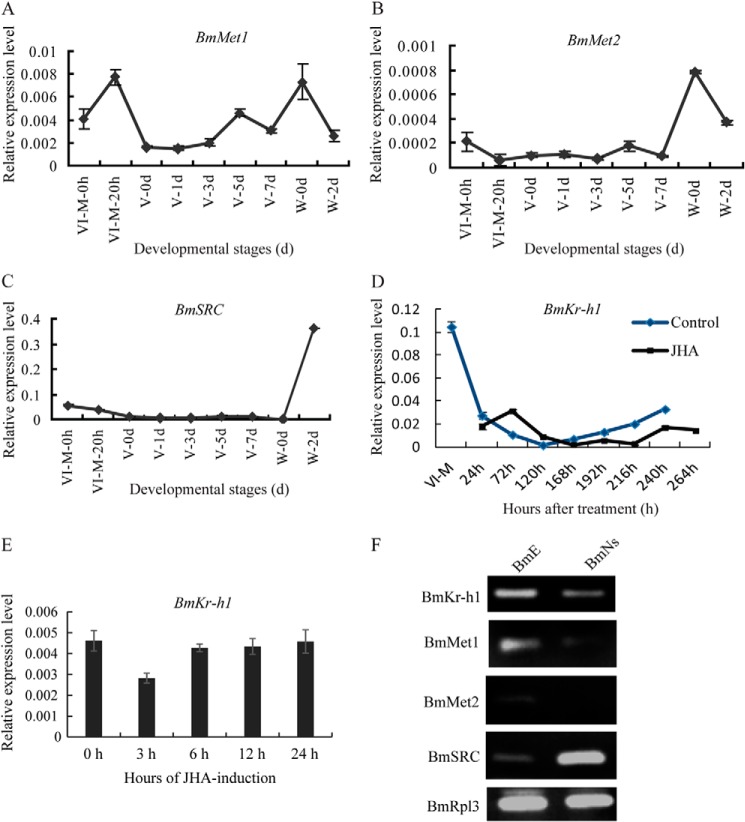
To determine how JH regulates the expression of Bmdimm, developmental changes in the expression of JH receptors and early inducible genes in silk glands of B. mori were determined by qRT-PCR (Fig. 7). The BmMet1 was highly expressed in the fourth molting stage, peaking at 20 h of this stage (VI-M-20 h), lowly expressed during the fifth instar, then increased at early spinning, and decreased again during late wandering stage (W-2 d) (Fig. 7A). Although the level of BmMet1 was slightly different from that of BmMet2 in the fourth molting stage, their overall expression patterns were very similar (Fig. 7B). BmSRC expression was similar to that of BmMet2 in the fourth molting and fifth instar stage but slightly different in the wandering stage (Fig. 7C). The expression pattern of the JH early inducible gene, BmKr-h1, was closely similar to those of BmMet and BmSRC in control silkworms from the fourth molting to spinning stage (Fig. 7D), which was correlated with the titer of JH in the hemolymph of B. mori (35), but its expression was prolonged in JHA-treated silkworms (Fig. 7D). In silk glands cultured in vitro, the expression of BmKr-h1 slightly declined in 3 h but increased gradually with prolonged treatment (Fig. 7E), further confirming a consistency with dependence of Bmdimm expression on BmKr-h1 expression after JHA treatment.
FIGURE 7.
Expression of JH receptor and BmKr-h1 in silk glands in vivo, in vitro, and in cultured B. mori cells. A–C, BmMet1 (A), BmMet2 (B), and BmSRC (C) transcript levels determined by qRT-PCR in normally developing silkworms without JHA treatment. D, expression of BmKr-h1 in silk glands from silkworms treated with JHA or acetone (control). E, expression of BmKr-h1 in silk glands cultured in vitro. BmRpl3 expression was used as a control. Results are expressed as means ± S.D. of three independent experiments. F, expression of BmMet1, BmMet2, BmSRC, and BmKr-h1 in BmE and BmN cells analyzed by RT-PCR. BmRpl3 expression was used as a control.
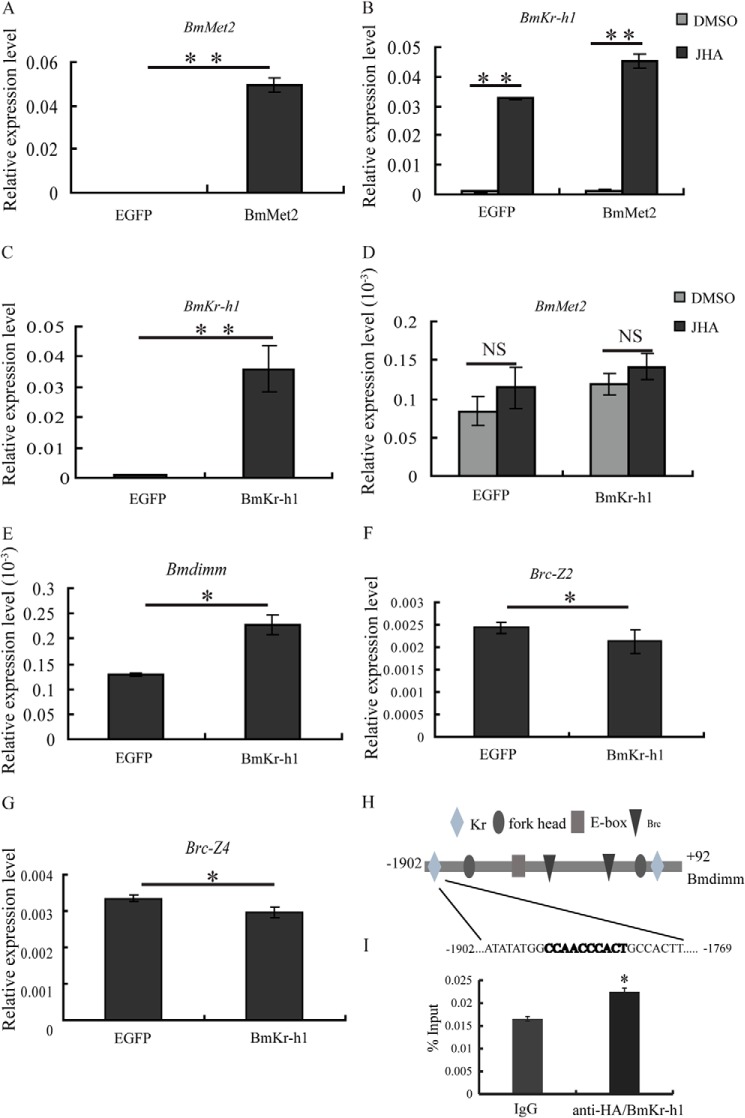
To investigate whether JH signaling pathway exists in cells, the expression of BmKr-h1, BmMet1, BmMet2, and BmSRC was examined in BmE and BmN cells by RT-PCR. The results showed that all genes were expressed in BmE cells (Fig. 7F), implying the presence of a JH signaling pathway. To elucidate the mechanism of regulation, we verified the activity of the JH signal transduction pathway components previously found in silk glands. By qRT-PCR, BmKr-h1 expression was significantly increased after overexpression of BmMet2 in BmE cells and significantly increased by JHA treatment (Fig. 8, A and B). However, BmMet2 expression was not increased by overexpression of BmKr-h1 regardless of JHA treatment (Fig. 8, C and D). These results suggested the existence of a JH-BmMet2-BmKr-h1 cascade in BmE cells. Bmdimm expression was significantly increased after overexpressing BmKr-h1 (Fig. 8E), whereas expression of the ecdysone early response gene Brc (Brc-Z2 and Brc-Z4) was significantly decreased (Fig. 8, F and G). These results indicated that the JH effect on Bmdimm induction was mediated by the JH-Met-Kr-h1 signal pathway.
FIGURE 8.
JH effect on induction of Bmdimm is mediated by the JH-Met-Kr-h1 signaling pathway. A and B, expression of BmKr-h1 after overexpression of BmMet2 assayed by qRT-PCR. C–E, expression of BmMet2 and Bmdimm after overexpression of BmKr-h1 assayed by qRT-PCR. F and G, expression of Brc-Z2 and Brc-Z4 after overexpression of BmKr-h1 assayed by qRT-PCR. BmE cells were transfected with 1 μg of 1180-A4/BmKr-h1 or 1180-A4/BmMet2 and incubated in medium containing 10 μm JHA or DMSO for 12 h. BmRpl3 expression is shown as a control. Results are expressed as means ± S.D. of three independent experiments; *, p < 0.05; **, p < 0.01. H, sequence of the Bmdimm promoter. Potential cis-elements were predicted using the MATINSPECTOR program. I, ChIP assays analyzing BmKr-h1 binding to the Kr response element in the promoter region of Bmdimm. Anti-HA and normal rabbit IgG were used for immunoprecipitation. Enrichment of promoter binding was analyzed by qRT-PCR in triplicate and expressed as a percentage over input. Quantified results are presented as means ± S.D.; *, p < 0.05. NS, no significance.
BmKr-h1 Is an Upstream Regulator of Bmdimm
According to the results of above, BmKr-h1 appeared to be an upstream regulator of Bmdimm. To investigate how BmKr-h1 regulates Bmdimm transcription, a potential regulatory sequence 2000 bp upstream of Bmdimm was cloned and analyzed for the presence of putative regulatory elements using the MatInspector program. Several predicted regulatory elements, including E-box, forkhead, Brc, and Kr, were identified (Fig. 8H). An additional ChIP assay was conducted using an antibody specific against an HA tag after overexpression of HA-BmKr-h1 in BmE cells. To quantify the amount of precipitated DNA, qRT-PCR was performed using primers for the Bmdimm promoter in the −1759 to −1902-bp and −221 to −64-bp regions. The result showed that DNA from the −1759 to −1902-bp region was enriched compared with control IgG (Fig. 8I), but not from the −221- to −64-bp region (data not shown), consistent with the possibility that BmKr-h1 binds to the Kr response element located at −1759 to −1902 bp.
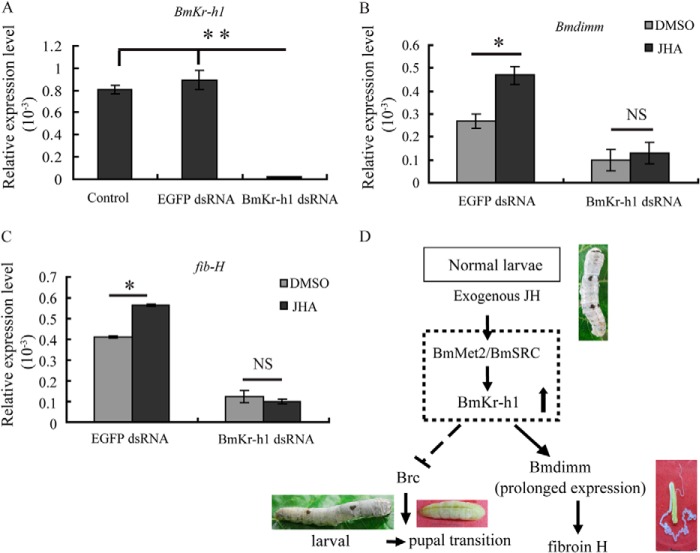
To further investigate whether BmKr-h1 is an upstream regulator of Bmdimm, the effect of BmKr-h1 on Bmdimm expression was then examined by RNAi (Fig. 9A). qRT-PCR results showed that after exposure to JHA, the levels of Bmdimm and fib-H mRNA were increased in BmE cells transfected with GFP double-stranded RNA (dsRNA) as a control. However, the levels of both Bmdimm and fib-H mRNAs were significantly decreased when BmKr-h1 mRNA was knocked down by BmKr-h1 dsRNA, and JHA treatment had no significant effect (Fig. 9, B and C). These results were consistent with the idea that BmKr-h1 is an upstream regulator of Bmdimm.
FIGURE 9.
Knockdown of BmKr-h1 by RNAi in BmE cells and schematic description of hypothesized regulation of fib-H. A, knockdown of BmKr-h1 by RNAi assayed by qRT-PCR. B, expression of Bmdimm after knocking down BmKr-h1 assayed by qRT-PCR. C, expression of fib-H after knocking down BmKr-h1 assayed by qRT-PCR. BmE cells were transfected with 5 μg of BmKr-h1 dsRNA or EGFP dsRNA for 12 h and incubated in medium containing 10 μm JHA or DMSO for 12 h. BmRpl3 expression is shown as a control. Results are expressed as means ± S.D. of three independent experiments; *, p < 0.05; **, p < 0.01. D, schematic representation of the hypothesis for molecular regulation of fib-H. NS, no significance.
DISCUSSION
B. mori silk glands produce large amounts of silk proteins secreted mainly from MSG and PSG cells (53). The genes encoding silk proteins are expressed in all larval instars with maximal expression just prior to the intermolting period of the last larval stage but repressed during molting stages (54), and the proportion of silk protein components encoded by these genes is accurately set (2). Fibroin genes are highly expressed in PSG cells but repressed in MSG cells. In this study, a bHLH transcription factor that shares 74 and 72.5% amino acid sequence identity with dimmed from D. melanogaster and Mist from M. musculus, respectively, was identified and named Bmdimm. A major finding based on several lines of evidence was that this transcription factor mediated JH action on induction of the silk protein gene fib-H. First, the Bmdimm protein was detected only in silk glands and only in MSG and PSG cells, not in ASG cells (Fig. 2B). This indicated that Bmdimm expression might be correlated with silk protein synthesis. Additionally, Bmdimm had a low level of expression in the fourth instar molting stages, with expression increasing gradually to a high level in the feeding stages of the fifth instar (Fig. 2D), a pattern consistent with the expression of fib-H.
The second important finding in this study was that JH regulation of Bmdimm was mediated by the JH-Met-Kr-h1 signaling pathway. JH is necessary for maintaining the larval nature during molting and for repressing metamorphosis (24). Treatment with JH also increases silk protein production (36). This study found that JHA not only extended larval development but also up-regulated Bmdimm expression (Fig. 5A). Several potential JH response elements were predicted from the Bmdimm regulatory regions, including E-box and Kr (Fig. 8H). BmMet2 and BmKr-h1 were expressed earlier than Bmdimm, followed by expression of Brc (Brc-Z2 and Brc-Z4) (Figs. 7 and 8). When BmKr-h1 mRNA was suppressed by dsRNA, Bmdimm expression was significantly decreased and did not respond to JHA; furthermore, expression of fib-H also decreased significantly (Fig. 9). These results indicated that JH regulated Bmdimm expression through the JH-Met-Kr-h1 signaling pathway and that BmKr-h1 acted immediately upstream of Bmdimm. Additional experiments are needed to clarify the precise mechanism of BmKr-h1 action in the regulation of Bmdimm.
The third important discovery from this study is that Bmdimm activated the transcription of fib-H by directly and specifically binding the E-box in its regulatory region. ChIP assays indicated that Bmdimm protein bound to an E-box element with a CAAATG motif (Fig. 3A). EMSA with recombinant Bmdimm and the E-box element probe confirmed this finding (Fig. 3B). Mutation and truncation of the E-box element indicated that Bmdimm binding to the E-box cis-element of the fib-H gene was sequence-specific. The region of −865 to −400 bp upstream of the gene, particularly the motif CAAATG (Fig. 3), was critical for transcription activation of fib-H by Bmdimm binding. The arginine at position 113 of Bmdimm was critical for the protein to bind with the E-box element of the fib-H gene (Fig. 3E). Here, we report that the bHLH transcription factor was directly involved in the regulation of the silk protein gene, and the regulation mechanism is related with the JH signal. Thus, the JH signal pathway, which is the most important and extensive mechanism for insect cells, may be a key in coordinate regulation of the genes encoding silk proteins.
The fourth interesting finding was that Bmdimm interacted with Bmsage. This finding was supported by three experiments. First, far-Western blots demonstrated that Bmdimm bound to Bmsage on membrane (Fig. 4A, panel a1) and Bmsage bound to Bmdimm (Fig. 4A, panel a2). Second, the proteins were co-localized in the nuclei of BmE cells (Fig. 4C). Third, immunoprecipitation showed that Bmdimm protein was present in anti-Bmsage immunoprecipitates (Fig. 4D, panel d1), and Bmsage protein was present in anti-Bmdimm immunoprecipitates (Fig. 4D, panel d2). These results demonstrated that Bmdimm interacted with Bmsage in nuclei. It has been found that an SGF1-forkhead complex acts as a crucial transcription activator that binds to proximal upstream elements of both the fibroin and p25 genes in PSG cells (55). Our previous study demonstrated that Bmsage is involved in regulation of the fib-H gene via interaction with SGF1 (15). Thus, Bmdimm, Bmsage, and SGF1 might form a triple complex to activate the transcription of fib-H, but this needs to be investigated in a future study.
Based on this and previous studies (31), we propose a hypothesis for the regulatory mechanism by which JH induces expression of the fib-H gene (Fig. 9D). JH or JHA binds to a heterodimer of BmMet2-BmSRC, which directly activates the expression of BmKr-h1. BmKr-h1 might suppress larval-pupal metamorphosis by inhibiting the expression of Brc and activating the expression of Bmdimm. Bmdimm, interacting with Bmsage, activates the transcription of fib-H gene in B. mori PSG cells.
Acknowledgments
We thank Dr. Qili Feng (Guangdong Provincial Key Laboratory of Biotechnology for Plant Development, School of Life Sciences, South China Normal University, Guangzhou, China), Dr. Marian Goldsmith (Department of Biological Sciences 291 CBLS-URI, Kingston, RI 02881), and Dr. Shiping Liu (State Key Laboratory of Silkworm Genome Biology, Southwest University, Chongqing) for helpful comments during preparation of this manuscript.
This work was supported by National Basic Research Program of China Grant 2012CB114600, National Hi-Tech Research and Development Program of China Grant 2011AA100306, Program for New Century Excellent Talents NCET-11-0699, and the National Natural Science Foundation of China 31372380.
- ASG
- anterior silk gland
- MSG
- middle silk gland
- PSG
- posterior silk gland
- bHLH
- basic helix-loop-helix transcription factor
- JH
- juvenile hormones
- Met
- methoprene-tolerant
- qRT-PCR
- quantitative real time PCR
- EGFP
- enhanced green fluorescent protein gene
- DIG
- digoxigenin
- JHA
- JH analog.
REFERENCES
- 1. Xia Q., Li S., Feng Q. (2014) Advances in silkworm studies accelerated by the genome sequencing of Bombyx mori. Annu. Rev. Entomol. 59, 513–536 [DOI] [PubMed] [Google Scholar]
- 2. Inoue S., Tanaka K., Arisaka F., Kimura S., Ohtomo K., Mizuno S. (2000) Silk fibroin of Bombyx mori is secreted, assembling a high molecular mass elementary unit consisting of H-chain, L-chain, and p25, with a 6:6:1 molar ratio. J. Biol. Chem. 275, 40517–40528 [DOI] [PubMed] [Google Scholar]
- 3. Maekawa H., Suzuki Y. (1980) Repeated turn-off and turn-on of fibroin gene transcription during silk gland development of Bombyx mori. Dev. Biol. 78, 394–406 [DOI] [PubMed] [Google Scholar]
- 4. Ohta S., Suzuki Y. (1988) Fibroin gene transcription in the embryonic stages of the silkworm, Bombyx mori. Develop. Growth & Differ. 30, 293–299 [DOI] [PubMed] [Google Scholar]
- 5. Mach V. (1995) Silk gland factor-1 involved in the regulation of Bombyx sericin-1 gene contains Fork head motif. J. Biol. Chem. 270, 9340–9346 [DOI] [PubMed] [Google Scholar]
- 6. Weigel D., Jürgens G., Küttner F., Seifert E., Jäckle H. (1989) The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell 57, 645–658 [DOI] [PubMed] [Google Scholar]
- 7. Ohno K., Sawada J., Takiya S., Kimoto M., Matsumoto A., Tsubota T., Uchino K., Hui C.-C., Sezutsu H., Handa H., Suzuki Y. (2013) Silk gland factor-2, involved in Fibroin gene transcription, consists of LIM homeodomain, LIM-interacting, and single-stranded DNA-binding proteins. J. Biol. Chem. 288, 31581–31591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu P.-X., Fukuta M., Takiya S., Matsuno K., Xu X., Suzuki Y. (1994) Promoter of the POU-M1/SGF-3 gene involved in the expression of Bombyx silk genes. J. Biol. Chem. 269, 2733–2742 [PubMed] [Google Scholar]
- 9. Kimoto M., Kitagawa T., Kobayashi I., Nakata T., Kuroiwa A., Takiya S. (2012) Inhibition of the binding of MSG-intermolt-specific complex, MIC, to the sericin-1 gene promoter and sericin-1 gene expression by POU-M1/SGF-3. Dev. Genes Evol. 222, 351–359 [DOI] [PubMed] [Google Scholar]
- 10. Takiya S., Kokubo H., Suzuki Y. (1997) Transcriptional regulatory elements in the upstream and intron of the fibroin gene bind three specific factors POU-M1, BmFkh, and FMBP-1. Biochem. J. 321, 645–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kokubo H., Takiya S., Mach V., Suzuki Y. (1996) Spatial and temporal expression pattern of Bombyx fork head/SGF-1 gene in embryogenesis. Dev. Genes Evol. 206, 80–85 [DOI] [PubMed] [Google Scholar]
- 12. Li M., Wang I. X., Li Y., Bruzel A., Richards A. L., Toung J. M., Cheung V. G. (2011) Widespread RNA and DNA sequence differences in the human transcriptome. Science 333, 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abrams E. W., Mihoulides W. K., Andrew D. J. (2006) Fork head and Sage maintain a uniform and patent salivary gland lumen through regulation of two downstream target genes, PH4&SG1 and PH4&SG2. Development 133, 3517–3527 [DOI] [PubMed] [Google Scholar]
- 14. Fox R. M., Vaishnavi A., Maruyama R., Andrew D. J. (2013) Organ-specific gene expression: the bHLH protein Sage provides tissue specificity to Drosophila FoxA. Development 140, 2160–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao X. M., Liu C., Li Q. Y., Hu W. B., Zhou M. T., Nie H. Y., Zhang Y. X., Peng Z. C., Zhao P., Xia Q. Y. (2014) Basic helix-loop-helix transcription factor bmsage is involved in regulation of fibroin H-chain gene via interaction with SGF1 in Bombyx mori. PLoS One 9, e94091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferré-D'Amaré A. R., Prendergast G. C., Ziff E. B., Burley S. K. (1993) Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363, 38–45 [DOI] [PubMed] [Google Scholar]
- 17. Blackwell T. K., Huang J., Ma A., Kretzner L., Alt F. W., Eisenman R. N., Weintraub H. (1993) Binding of myc proteins to canonical and noncanonical DNA sequences. Mol. Cell. Biol. 13, 5216–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ellenberger T., Fass D., Arnaud M., Harrison S. C. (1994) Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 8, 970–980 [DOI] [PubMed] [Google Scholar]
- 19. Murre C., McCaw P. S., Baltimore D. (1989) A new DNA-binding and dimerization motif in immunoglobulin enhancer binding, daughterless, myod, and myc proteins. Cell 56, 777–783 [DOI] [PubMed] [Google Scholar]
- 20. Ma P. C., Rould M. A., Weintraub H., Pabo C. O. (1994) Crystal structure of myoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell 77, 451–459 [DOI] [PubMed] [Google Scholar]
- 21. Park D. (2008) Mapping peptidergic cells in Drosophila where dimm fits in. PLoS One 3, e1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park D., Shafer O. T., Shepherd S. P., Suh H., Trigg J. S., Taghert P. H. (2008) The Drosophila basic helix-loop-helix protein DIMMED directly activates PHM, a gene encoding a neuropeptide-amidating enzyme. Mol. Cell Biol. 28, 410–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y., Chen K., Yao Q., Wang W., Zhi Z. (2007) The basic helix-loop-helix transcription factor family in Bombyx mori. Dev. Genes Evol. 217, 715–723 [DOI] [PubMed] [Google Scholar]
- 24. Riddiford L. M. (1996) Juvenile hormone: The status of its “status quo” action. Arch. Insect Biochem. Physiol. 32, 271–286 [DOI] [PubMed] [Google Scholar]
- 25. Minakuchi C., Zhou X., Riddiford L. M. (2008) Kruppel homolog 1 (Kr-h1) mediates juvenile hormone action during metamorphosis of Drosophila melanogaster. Mech. Dev. 125, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wilson T. G., Fabian J. (1986) A Drosophila melanogaster mutant resistant to a chemical analog of juvenile hormone. Dev. Biol. 118, 190–201 [DOI] [PubMed] [Google Scholar]
- 27. Miura K., Oda M., Makita S., Chinzei Y. (2005) Characterization of the Drosophila Methoprene-tolerant gene product-Juvenile hormone binding and ligand-dependent gene regulation. FEBS J. 272, 1169–1178 [DOI] [PubMed] [Google Scholar]
- 28. Charles J.-P., Iwema T., Epa V. C., Takaki K., Rynes J., Jindra M. (2011) Ligand-binding properties of a juvenile hormone receptor, Methoprene-tolerant. Proc. Natl. Acad. Sci. U.S.A. 108, 21128–21133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li M., Mead E. A., Zhu J. (2011) Heterodimer of two bHLH-PAS proteins mediates juvenile hormone-induced gene expression. Proc. Natl. Acad. Sci. U.S.A. 108, 638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang H. J., Anderson A. R., Trowell S. C., Luo A. R., Xiang Z. H., Xia Q. Y. (2011) Topological and functional characterization of an insect gustatory receptor. PLoS One 6, e24111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kayukawa T., Minakuchi C., Namiki T., Togawa T., Yoshiyama M., Kamimura M., Mita K., Imanishi S., Kiuchi M., Ishikawa Y., Shinoda T. (2012) Transcriptional regulation of juvenile hormone-mediated induction of Krüppel homolog 1, a repressor of insect metamorphosis. Proc. Natl. Acad. Sci. U.S.A. 109, 11729–11734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kayukawa T., Tateishi K., Shinoda T. (2013) Establishment of a versatile cell line for juvenile hormone signaling analysis in Tribolium castaneum. Sci. Rep. 3, 1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Minakuchi C., Namiki T., Shinoda T. (2009) Kruppel homolog 1, an early juvenile hormone-response gene downstream of Methoprene-tolerant, mediates its anti-metamorphic action in the red flour beetle Tribolium castaneum. Dev. Biol. 325, 341–350 [DOI] [PubMed] [Google Scholar]
- 34. Lozano J., Belles X. (2011) Conserved repressive function of Kruppel homolog 1 on insect metamorphosis in hemimetabolous and holometabolous species. Sci. Rep. 1, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kayukawa T., Murata M., Kobayashi I., Muramatsu D., Okada C., Uchino K., Sezutsu H., Kiuchi M., Tamura T., Hiruma K., Ishikawa Y., Shinoda T. (2014) Hormonal regulation and developmental role of Krüppel homolog 1, a repressor of metamorphosis, in the silkworm Bombyx mori. Dev. Biol. 388, 48–56 [DOI] [PubMed] [Google Scholar]
- 36. Daillie J. (1979) Juvenile-hormone modifies larvae and silk gland development in Bombyx mori. Biochimie 61, 275–281 [DOI] [PubMed] [Google Scholar]
- 37. Kurata S., Koga K., Sakaguchi B. (1978) Nucleolar size in parallel with ribosomal RNA synthesis at diapause termination in the eggs of Bombyx mori. Chromosoma 68, 313–317 [DOI] [PubMed] [Google Scholar]
- 38. Kurata K. (1984) Effect of a juvenile hormon analogue given at various ages of 5th instar larvae on RNA synthesis in the posterior silk gland of the silkworm, Bombyx mori. J. Seric. Sci. Jpn. 53, 421–426 [Google Scholar]
- 39. Pan M. H., Xiao S. Q., Chen M., Hong X. J., Lu C. (2007) Establishment and characterization of two embryonic cell lines of Bombyx mori. In Vitro Cell. Dev. Biol. Anim. 43, 101–104 [DOI] [PubMed] [Google Scholar]
- 40. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCt method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 43. Aslam A. F., Kiya T., Mita K., Iwami M. (2011) Identification of novel bombyxin genes from the genome of the silkmoth Bombyx mori and analysis of their expression. Zool. Sci. 28, 609–616 [DOI] [PubMed] [Google Scholar]
- 44. Wu Y., Li Q., Chen X. Z. (2007) Detecting protein-protein interactions by far Western blotting. Nat. Protoc. 2, 3278–3284 [DOI] [PubMed] [Google Scholar]
- 45. Bobrovnik S. A. (2003) Determination of antibody affinity by ELISA. Theory. J. Biochem. Biophys. Methods 57, 213–236 [DOI] [PubMed] [Google Scholar]
- 46. Liu Q. X., Ueda H., Hirose S. (2000) MBF2 is a tissue- and stage-specific coactivator that is regulated at the step of nuclear transport in the silkworm Bombyx mori. Dev. Biol. 225, 437–446 [DOI] [PubMed] [Google Scholar]
- 47. Lin Y., Meng Y., Wang Y. X., Luo J., Katsuma S., Yang C. W., Banno Y., Kusakabe T., Shimada T., Xia Q. Y. (2013) Vitellogenin receptor mutation leads to the oogenesis mutant phenotype “scanty vitellin” of the silkworm, Bombyx mori. J. Biol. Chem. 288, 13345–13355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kethidi D. R., Perera S. C., Zheng S., Feng Q. L., Krell P., Retnakaran A., Palli S. R. (2004) Identification and characterization of a juvenile hormone (JH) response region in the JH esterase gene from the spruce budworm, Choristoneura fumiferana. J. Biol. Chem. 279, 19634–19642 [DOI] [PubMed] [Google Scholar]
- 49. Chaitanya R. K., Sridevi P., Senthilkumaran B., Dutta Gupta A. (2013) Effect of juvenile hormone analog, methoprene on H-fibroin regulation during the last instar larval development of Corcyra cephalonica. Gen. Comp. Endocrinol. 181, 10–17 [DOI] [PubMed] [Google Scholar]
- 50. Grzelak K., Szczesna E., Sehnal F. (1982) Stimulation of RNA-transcription by juvenile-hormone in degenerating silk glands. Mol. Cell. Endocrinol. 26, 341–351 [DOI] [PubMed] [Google Scholar]
- 51. Tripoulas N. A., Samols D. (1986) Developmental and hormonal regulation of sericin RNA in the silkworm, Bombyx mori. Dev. Biol. 116, 328–336 [Google Scholar]
- 52. Sehnal F., Akai H. (1990) Insect silk glands: their types, development and function, and effects of environmental factors and morphogenetic hormones on them. Int. J. Insect Morphol. Embryol. 19, 79–132 [Google Scholar]
- 53. Suzuki Y., Tsuda M., Takiya S., Hirose S., Suzuki E., Kameda M., Ninaki O. (1986) Tissue-specific transcription enhancement of the fibroin gene characterized by cell-free systems. Proc. Natl. Acad. Sci. U.S.A. 83, 9522–9526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ishizaki H., Suzuki A. (1994) The brain secretory peptides that control moulting and metamorphosis of the silkmoth, Bombyx mori. Int. J. Dev. Biol. 38, 301–310 [PubMed] [Google Scholar]
- 55. Durand B., Drevet J., Couble P. (1992) p25 Gene regulation in Bombyx mori silk gland: two promoter-binding factors have distinct tissue and developmental specificities. Mol. Cell. Biol. 12, 5768–5777 [DOI] [PMC free article] [PubMed] [Google Scholar]