Abstract
Cell transplantation into adult zebrafish has lagged behind mouse due to the lack of immune compromised models. Here, we have created homozygous rag2E450fs mutant zebrafish that have reduced numbers of functional T and B cells but are viable and fecund. Mutant fish engraft zebrafish muscle, blood stem cells, and cancers. rag2E450fs mutant zebrafish are the first immune compromised zebrafish model that permits robust, long-term engraftment of multiple tissues and cancer.
Cell transplantation of human and mouse cells into immune compromised mice has greatly enhanced our understanding of stem cell function, regeneration, and cancer. However, transplantation experiments in mice are expensive, routinely utilize small cohorts of animals, and engraftment is often difficult to visualize directly. By contrast, large-scale cell transplantation of fluorescent blood and cancer cells into syngeneic and irradiated zebrafish has now become routine 1–7. However, these approaches require that donor cells are from the same strain of syngeneic zebrafish or that the recipient immune system is transiently ablated by whole body γ-irradiation two-days prior to transplantation. Irradiated recipients eventually recover their immune system by 20 days post-irradiation and kill engrafted cells 1,2,6, making long-term engraftment studies difficult. To date, immune compromised zebrafish have not been developed as a universal recipient for allograft cell transplantation.
Capitalizing on recently developed gene inactivation methods using genome engineering 8, zinc finger nucleases were engineered to target the Plant Homeodomain (PHD) of the zebrafish recombination activating gene 2 (rag2) at similar residues commonly mutated in the Omenn syndrome 9,10 (Fig. 1a). Mutations in residues of the PHD domain disrupt the RAG2 protein interaction with trimethylated histone H3 to alter chromatin accessibility and to partially impair V(D)J recombination in vivo 11. Omenn syndrome is an autosomal recessive severe combined immunodeficiency (SCID) that results in impaired T and B cell receptor rearrangement leading to reduced numbers of functionally mature lymphocytes in human patients. A mutant zebrafish line was generated in the AB-strain background and contained a frame-shift at amino acid E450 that resulted in premature termination (designated rag2E450fs; Fig. 1b).
Figure 1. rag2E450fs mutant zebrafish lack mature T cells and have a reduced B cell repertoire.
(a) Human RAG2 protein with known SCID mutations denoted. Arrow denotes ZFN target region. (b) Nucleotide (top) and protein sequence (bottom) for the rag2E450fs mutation. Yellow denotes ZFN target sites, blue nucleotide additions, gray nucleotide deletions, and arrow amino acid change at E450. Underlining shows amino acid sequence following frame shift with termination amino acid numbered. (c,d) Thymus sections of 90-day-old wild type (c) and rag2E450fs mutant zebrafish (d). Red dashed lines denote thymus (left). Right panels are amplified views of boxed regions with adipocytes (A) and vacant thymic epithelium (E) shown. (e,f) Whole kidney marrow cytospins of wild type (e) and rag2E450fs zebrafish (f) with lymphocytes (black arrows), erythrocytes (magenta arrows) and granulocytes (cyan arrows) denoted. Scale bars are 200 μm (c,d – left); 50 μm (c,d – right); 20 μm (e,f). (g) Cell counts from cytospins performed on whole kidney marrow. Error bars represent ± 1 s.d., p < 0.05 by Student’s t-test. (h) Gene expression analysis of whole kidney marrow cells. Error bars represent s.e.m, p < 0.05 by Student’s t-test. (i) PCR analysis for tcrb and igm rearrangement of whole kidney marrow cells from wild type (wt) and rag2E450fs zebrafish. negative control (neg).
Heterozygous rag2E450fs mutant fish were incrossed and raised to adulthood. Animals were genotyped at 3 months of age, revealing expected Mendelian ratios (146:wild type, 265:heterozygous, and 129:rag2E450fs mutant). rag2E450fs mutant fish were similar in size to heterozygous and wild type sibling animals (Supplementary Fig. 1), survived fin clip, and remained healthy for >6 months when raised under standard laboratory conditions. Homozygous rag2E450fs mutant zebrafish could reproduce, albeit at reduced fecundity when compared with heterozygous sibling fish (Supplementary Table 1). Histological analysis of 90-day-old rag2E450fs mutant zebrafish revealed a striking reduction in thymic T cells and an altered thymic architecture including reduced numbers of epithelial cells with preponderance of adipocytes (n=5 of 6 mutant animals, Fig. 1c,d). This thymic involution is commonly observed in T cell deficient lines of mice 12 and was not detected in wild type siblings (n=6, p=0.008, Fisher’s exact test). A similar reduction in thymocyte number was also noted in 5-day-old zebrafish (Supplementary Fig. 2). Analysis of whole kidney marrow revealed that adult homozygous rag2E450fs zebrafish contained all blood cell lineages (Fig. 1e,f); however, quantization revealed a striking 75% reduction in lymphocytes of rag2E450fs mutant zebrafish (Fig. 1g and Supplementary Table 2). Transcript expression for mature B and T cell markers was reduced in rag2E450fs mutant marrow, including immunoglobulin m (igm), lymphocyte-specific protein tyrosine kinase (lck), t-cell receptor alpha (tcra) and t-cell receptor beta (tcrb) (Fig. 1h). By contrast, rag1 transcript levels were not reduced in marrow of mutant animals suggesting that early B cell precursors were not altered in rag2E450fs mutant zebrafish (Fig. 1h). Homozygous rag2E450fs mutants did not exhibit expression differences for myeloperoxidase (mpx) and l-plastin (lcp1), confirming that neutrophils, monocyte, macrophages, and other cell lineages were not altered in rag2E450fs mutant zebrafish (Fig. 1h and Supplementary Table 2).
To directly assess the impact of the rag2E450fs mutation on mature T and B cells, whole kidney marrow cells were also assessed for tcrb and igm rearrangements 4,13. Homozygous rag2E450fs mutant fish lacked tcrb rearranged T cells (n=7, Fig. 1i), while the B cell immune repertoire was reduced in 3 of 7 animals tested. Thus, the rag2E450fs mutation results in the production of a hypomorphic protein with reduced receptor recombination, leading to lack mature T cells and a variable reduction of functionally diverse B cells.
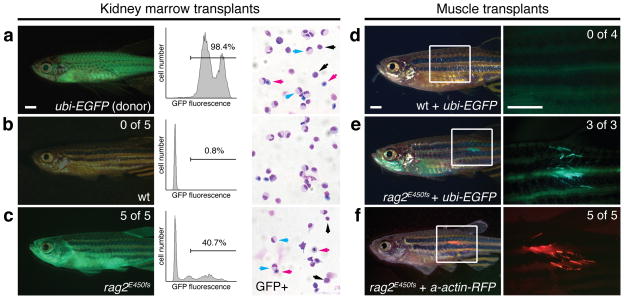
Immune compromised mice, including those deficient in Rag2, have been successfully used for adoptive transfer of mouse and human cells. Yet, immune compromised adult zebrafish have not been developed that permit engraftment of a range of cells and tissues. Indeed, the first example of targeted gene knock out in zebrafish was the rag1-deficient fish 13. rag1 mutant fish lack all mature T and B cells, but have not been widely used for cell transplantation approaches likely due to reduced viability of adult fish and failure to thrive following fin clip and genotyping. To assess if hypomorphic, rag2-deficient zebrafish were amenable to cell transplantation, we first assessed if hematopoietic stem cell engraftment was enhanced in rag2E450fs mutant lines. Specifically, rag2E450fs mutant and wild type sibling adults were sub-lethally irradiated with 10 Gy irradiation to clear the hematopoietic stem cell niche. Animals were transplanted two days after irradiation treatment by intraperitoneal injection with either 5×104 or 3×105 whole kidney marrow cells from unrelated ubiquitin-EGFP donor zebrafish (ubi-EGFP, Fig. 2a)14. As expected, none of the wild type siblings engrafted blood cells (n=10), while all rag2E450fs mutant fish exhibited robust, multi-lineage blood cell engraftment that persisted past 45 days post-transplantation (dpt, n=6) - a full 25 days after immune rejection is normally initiated in wild type, irradiated recipient fish (Fig. 2b,c, p=0.0002, Fisher’s exact test; Supplementary Fig. 3, and Supplementary Table 3) 2,6. Engrafted rag2E450fs homozygous mutant fish also commonly exhibited GFP+ circulating cells by 30 dpt (Supplementary Movie 1). These data show that hypomorphic rag2E450fs can robustly engraft hematopoietic cells from unrelated donors.
Figure 2. rag2E450fs mutant fish engraft hematopoietic and muscle stem cells.
(a) ubi-EGFP transgenic donor fish. (b–c) Wild type (wt, b) or homozygous rag2E450fs recipient fish (c) transplanted with GFP-labeled marrow and imaged at 45 days post-transplantation. Fluorescent image of whole fish (left) with engraftment rates noted. FACS (middle) and cytospin analysis (right) of whole kidney marrow from donor (a) and recipient fish (b–c). FACS sorted GFP+ cells are shown for cytospin analysis in c. Lymphocytes (black arrows), red blood cells (magenta arrows), granulocytes (cyan arrows). (d–e) Fluorescent image of wild type (wt) and rag2E450fs mutant fish engrafted with muscle cells from ubi-EGFP transgenic fish imaged at 30 days post-transplantation. (f) rag2E450fs fish engrafted with muscle cells from alpha-actin-RFP transgenic fish at 30 days post-transplantation. Engraftment rates for d–f are shown on magnified image panels to the right. Scale bars are 2 mm.
To assess the broad utility of the model for engraftment of regenerative tissues, fluorescently labeled muscle cells from adult ubi-EGFP transgenic fish were transplanted by intramuscular injection into non-irradiated recipient animals (5×104 cells/fish, 2μl/animal). Muscle cell viability was >85% following disassociation and single cell preparation (Supplementary Fig. 4). Remarkably, GFP+ muscle fibers were readily detected in all rag2E450fs mutant animals by 30 dpt (n=3 of 3) but not wild type sibling fish (n= 4; p=0.03, Fisher’s exact test; Fig. 2d,e and Supplementary Table 3). Fluorescent fibers persisted in rag2E450fs mutant fish for up to 60 dpt. Histological analysis revealed that GFP+ muscle fibers were viable, contained muscle striations, and were indistinguishable from recipient muscle tissue (Supplementary Fig. 5). We next wanted to verify the specificity of our results using a muscle-specific, fluorescent transgenic line. Muscle cells were isolated from the dorsal musculature of alpha-actin-RFP transgenic fish 15 and injected into the dorsal musculature of recipient fish (2.5×105 cells/fish). Again, robust engraftment of fluorescent muscle was only observed in homozygous rag2E450fs mutant fish by 30 dpt (n=5 of 5, Fig. 2f and Supplementary Table 3), but not heterozygous or wild type siblings (n=5 animals/genotype assessed, p=0.008, Fisher’s exact test). These data show that homozygous rag2E450fs mutant fish can engraft cells even in the absence of preconditioning with low-dose irradiation.
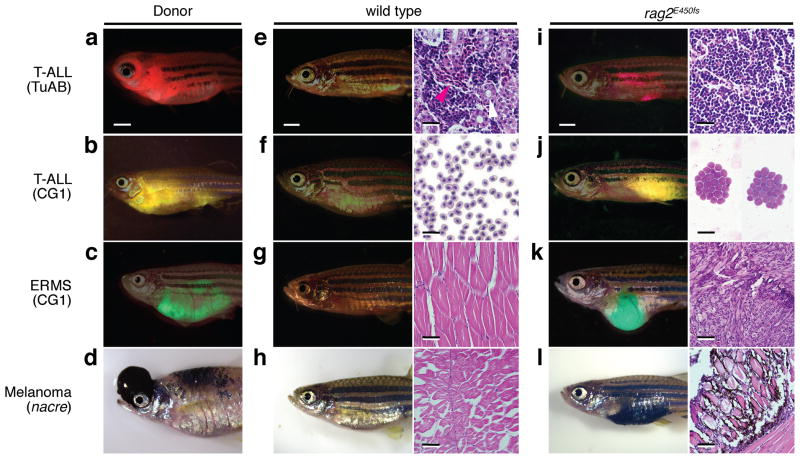
We next wanted to assess the utility of our model to engraft a diversity of transgenic zebrafish cancers including T cell acute lymphoblastic leukemia (TALL), embryonal rhabdomyosarcoma (ERMS), and melanoma. Specifically, primary Myc-induced T-ALL was generated in a mixed Tubingen/AB-strain background 16,17 and transplanted into the peritoneal cavity of recipient animals (Fig. 3a,e,i). Homozygous rag2E450fs mutant fish robustly engrafted T-ALL by 30 dpt even in the absence of prior immune suppression by γ-irradiation (n=2 of 2, 1×105 cells/fish, Fig. 3i). By contrast, heterozygous and wild type siblings failed to engraft T-ALL (n≥7 per genotype, p<0.03, Fisher’s exact test, Fig. 3e). Similar results were observed using three serially-passaged T-ALL from syngeneic CG1-strain zebrafish (1×105 cells/fish). In total, 39 of 39 rag2E450fs mutants engrafted T-ALL arising from CG1-strain fish while 0 of 22 wild type siblings engrafted leukemia (p<0.0001, Fisher’s exact test, Fig. 3b,f,j and Supplementary Table 4), suggesting that engraftment is not restricted to zebrafish strain or major histocompatibility complex (MHC) matching between fish. Homozygous rag2E450fs mutant zebrafish also successfully engrafted fluorescently labeled kRASG12D-induced ERMS generated in both the CG1 and AB-strain background (n=4 independent tumors analyzed, 1×104–1×106 cells/fish). In total, 24 of 27 homozygous rag2E450fs mutant zebrafish engrafted ERMS while 0 of 7 wild type siblings engrafted disease (p<0.0001, Fisher’s exact test, Fig. 3c,g,k and Supplementary Table 4)18. Finally, melanomas that harbor a p53 mutation and over-express both mitfa and BRAFV600E could successfully engraft into rag2E450fs mutant zebrafish but not wild type siblings (n=4 tumors analyzed, 5×105–1×106 cells/fish) 19,20. In total, 25 of 25 homozygous rag2E450fs mutant fish engrafted melanoma while 0 of 16 wild type siblings engrafted disease (p<0.0001, Fisher’s exact test, Fig. 3d,h,l and Supplementary Table 4). No instances of tissue rejection or tumor regression were observed in engrafted rag2E450fs mutant fish (n=104), showing that homozygous rag2E450fs mutant zebrafish can robustly engraft cells from a diversity of genetic backgrounds and even in the absence of preconditioning of recipient animals with γ-irradiation.
Figure 3. Engraftment of zebrafish tumors into rag2E450fs mutant fish.
(a–d) Donor tumors used in cell transplantation studies. dsRED-labeled Myc-induced T-ALL arising in TuAB strain fish (a), zsYellow-labeled T-ALL from CG1-strain fish (b), GFP-labeled embryonal rhabdomyosarcoma (ERMS) from CG1-strain fish (c), and mitfa and BRAFV600E induced melanoma arising in p53-deficient nacre strain fish (d). Merged bright-field and fluorescent images of wild type (e–h) or rag2E450fs mutant fish (i–l) at 30 days post-transplantation. Images of whole kidney marrow sections (e,i), peripheral blood cytospins (f,j), and skeletal muscle (g,k,h,i). Red blood cells denoted by magenta arrows and renal tubules white arrows (e). Scale bars in fish images are 2 mm and histopathology images are 25 μm (e,i), 20 μm (f,j), and 50 μm (g,k,h,l).
Our experiments highlight the use of hypomorphic rag2E450fs mutant fish as the first universal recipient for allograft cell transplantation into adult fish, ushering in a new era of large-scale cell transplantation studies to directly visualize and assess stem cell self-renewal within normal tissues and clonal heterogeneity, therapeutic responses, and growth in cancer.
Online Methods
Animal use and creation of hypomorphic rag2E450fs mutant zebrafish
Zebrafish studies were approved by the Massachusetts General Hospital Subcommittee on Research Animal Care, under protocol #2011N000127.
rag2E450fs mutant zebrafish were created using the previously described Zinc Finger Nuclease (ZFN) pair that targeted the PHD domain of the zebrafish rag2 gene8. Specifically, RNA was prepared for each ZFN arm and microinjected into AB strain zebrafish (500 ng/μl). F0 injected animals were raised to adulthood and single male by female matings performed. Resultant progeny were arrayed into 96-well plates and genomic DNA extracted (n=12 individual embryos per cross). Polymerase Chain Reaction (PCR) was performed using forward primer (5′-ACTGCTCTAGTTGCAATTCCT) and reverse primer (5′-AGCTGGGGTCATCTTCAGT) to produce a 585 base pair (bp) PCR amplicon. The PCR cycle parameters were 1) denaturation: 94°C for 30 sec., 2) annealing: 54°C for 30 sec., and 3) elongation: 68°C for 45 sec., repeated for 35 cycles. PCR samples were purified and sent for Sanger sequencing. From this analysis, one line was identified with a rag2 frame shift mutation that starts at amino acid E450 and results in a premature stop codon mutation (designated rag2E450fs in this manuscript with an official ZFIN.org allele designation of rag2fb101). F1 progeny were subsequently raised to adulthood and heterozygous rag2E450fs fish identified by genotyping (see below).
Genotyping of rag2E450fs mutant zebrafish
The rag2E450fs mutant line is best maintained through heterozygous incrossing (Supplementary Table 1) and produces progeny at the expected Mendelian ratios. The rag2E450fs allele introduces a de novo XcmI site, allowing for restriction enzyme mediated identification of the mutant allele (diagramed with representative results in Supplementary Fig. 6). Specifically, adult 2–4 month-old fish were fin clipped and genomic DNA prepared using the modified HotSHOT method21. Individual genomic DNA was diluted ten-fold and 2 μl used in a standard 25 μl volume PCR using Taq DNA Polymerase (New England Biolabs, Cat# M0273L) with forward primer (5′-ACTGCTCTAGTTGCAATTCCT) and reverse primer (5′-AGCTGGGGTCATCTTCAGT) to produce a 585 base pair (bp) PCR amplicon. The PCR cycle parameters are 1) denaturation: 94°C for 30 sec., 2) annealing: 54°C for 30 sec., and 3) elongation: 68°C for 45 sec., repeated for 35 cycles. For enzymatic digestion, 15 μl of non-purified PCR reaction was combined with 0.3 μl XcmI + 2 μl 10X NEB buffer 2.1 + 2.7 μl water. The 20 μl reaction was incubated at 37°C for ≥4 hrs. Enzymatic digestion was visualized by electorphoresis on a 2% Tris-Acetate-EDTA (TAE) agarose gel containing ethidium bromide. Wild type fish produced a single band at 585 bp. Heterozygous rag2E450fs fish produced three bands at 585, 372 and 212 bp due to digestion of the mutant allele, with the two higher molecular weight bands being most distinctive. Homozygous fish produced two bands at 372 and 212 bp. A representative image of genotyping analysis is shown in Supplementary Fig. 6.
Characterization of the rag2E450fs mutant line
RNA in situ hybridization of larval fish (Supplementary Fig. 2) and histological analysis of thymus of adult fish (Fig. 1c,d) were completed as previously described22. Two-tailed Student’s t-test analysis was performed to assess larval thymus size differences (Supplementary Fig. 1c). For hematopoietic cell quantification (Fig. 1g), cytospins of whole kidney marrow from both rag2E450fs mutant fish and wild type siblings were reviewed by clinical hematopathologists (J.S.B.) and (R.S.L.). At least five 400x magnification fields were analyzed per slide and > 200 cells counted per animal. Sample identification was blinded during counts and genotype only revealed after cell counting was completed. Two-tailed Student’s t-test analysis was performed to assess changes in relative numbers of each blood cell lineage between wild type and rag2E450fs mutant fish. Quantitative real-time PCR (Fig. 1h) and nested PCR for tcra and igm rearrangements (Fig. 1i) were completed using primers shown in Supplementary Tables 5 and 6, essentially as previously described4,23. Two-tailed Student’s t-test analysis was performed to assess expression differences in hematopoietic marker genes using quantitative PCR (Fig. 1h). A threshold of p ≤ 0.05 was considered significant for two-tailed Student’s t-test.
Generation of transgenic zebrafish cancers
T-ALL and ERMS were created by co-injection of either rag2-Myc or rag2-kRASG12D along with fluorescent reporters under the same promoter into one-cell stage animals respectively, which have been previously described16. Tg(mitfa:GFP);mitfa−/−;p53−/−;Tg(mitfa:BRAF(V600E)) melanoma bearing fish were generated as previously described19,20.
Whole kidney marrow transplantation
Donor ubi-EGFP fish were euthanized by tricaine overdose (Tricaine-S, Western Chemical Inc.,). Kidney tissue was dissected, placed into a 1.5 ml Eppendorf tube containing 100 μl of suspension solution (0.9X PBS + 5% FBS), and manually pipetted ≥20 times using a 1 ml pipette tip. The kidney tissue suspension was filtered through a 40 μm strainer (Fisher Scientific, Cat# 352340). Total number of viable cells was calculated by Trypan blue (Life Technologies, Cat# 15250061) staining and hemocytometer counts. Cells were centrifuged at 1,000 g for 10 minutes and then resuspended in desired volumes for cell transplantation (Supplementary Table 3). Recipient fish were preconditioned with 10 Gy γ-irradiation (Cs137 irradiation), 2 days prior to transplantation. Recipient fish were transplanted with the indicated number of viable kidney marrow cells (Supplementary Table 3) injected into the peritoneal cavity using a 26s gauge Hamilton 80366 syringe (Sigma-Aldrich, 20779).
Whole kidney cell engraftment was assessed using whole body epi-fluorescence imaging (Olympus stereo microscope model MVX10, Olympus DP72 microscope digital camera, DP2-BSW software version 2.2), direct visualization of circulating cells within the tail vasculature beginning at 20 days post-transplantation (dpt), and/or FACs. FACs of kidney marrow was completed in the presence of propidium iodide (PI) to exclude dead cells. The GFP+ cell fraction was isolated from the marrow of engrafted rag2E450fs fish and analyzed following cytospin preparation.
Muscle cell transplantation
Dorsal musculature of donor ubi-EGFP or α-actin-RFP fish was excised from tricaine overdosed animals. Specifically, the dorsal musculature of the tail posterior to the anus was harvested for muscle transplantation. The muscle tissue was combined from 10–12 animals and mechanically disassociated by maceration by repeated dicing using a razor blade for 5 minutes on a 10 cm petri dish containing 500 μl of suspension solution (0.9X PBS + 5% FBS). Samples were then supplemented with 5 ml of 0.9X PBS + 5% FBS and a 5 ml serological pipette was used to suspend the homogenized tissue by repeated pipetting ≥30 times. Cells were then filtered through a 40 μm cell strainer and washed with 5 mls of 0.9X PBS + 5% FBS. Total number of viable cells was calculated by Trypan blue staining and hemocytometer counts. Cells were then centrifuged at 1,000 g for 10 minutes and resuspended in desired volumes (Supplementary Table 3) prior to transplantations. No irradiation preconditioning was used prior to transplantation of muscle cells into rag2E450fs mutant fish. Muscle cell transplantations were completed by injecting 2 μl of donor cell preparation into the dorsal musculature on the left side of the recipient fish, using a 26s gauge Hamilton 80366 syringe.
Engraftment was assessed by visualization of fluorescently labeled muscle fibers using epi-fluorescence imaging at 10, 20, 30, 45, and 60 dpt. Engraftment of ubi-EGFP muscle fibers were confirmed by immunohistochemistry on section slides using an anti-EGFP antibody (JL-8, Living Colors, Cat# 632381). Viability of the muscle fibers was confirmed by the absence of cleaved caspase-3 expression (Cell Signaling, Cat# 9664) and TUNEL staining (customized Millipore S7100 kit, Specialized Histopathology & TMI Core at Dana-Farber Cancer Institute).
Cancer cell transplantation
Donor fish with T-ALL, ERMS and melanoma from different transgenic backgrounds were euthanized by tricaine overdose. Animals were imaged using epi-fluorescence microscopy and tumor cells isolated as previously described24. Specifically, tumors were excised from diseased animals and placed in 500 μl of 0.9X PBS + 5% FBS on a 10 cm petri dish. Single-cell suspensions were obtained by maceration of tissue with a razor blade followed by manual pipetting to disassociate cell clumps. Cells were filtered through a 40 μm filter, centrifuged at 1,000g for 10 minutes, and resuspended at the correct volume (Supplementary Table 4) in a total volume of 5 μl per recipient fish for cell transplantation. Cell viability ranged from 58% to 66% assessed by PI staining and FACS analysis of ERMS tumor cells. 5 μl of tumor cells were transplanted into the peritoneal cavity of each recipient fish using a 26s Hamilton 80366 syringe. No irradiation preconditioning was utilized for tumor cell transplantations outlined in this work.
Tumor engraftment was assessed at 10, 20, 30, and 45 dpt by epi-fluorescence microscopy. Recipient fish were sacrificed when moribund or at 45 days post-transplantation for animals that failed to engraft disease (wild type). A subset of transplanted animals were photographed, and either 1) fixed in 4% paraformaldehyde for sectioning or 2) peripheral blood samples were collected for cytospins and histological examination.
Statistics
When possible, cell transplantation experiments were completed using ≥3 animals per arm to facilitate analysis using the Fisher’s exact test. No animals were excluded from our analysis shown in Supplementary Table 3 and 4, with exception to rag2E450fs mutant fish that engrafted ubi-EGFP+ marrow and were scored for circulating GFP+ cells in the tail at ≥20 dpt. We did not include these animals in Supplementary Table 3 because they were sectioned and marrow analysis by FACS was not possible.
Cytospin analysis shown in Fig. 1e–g and Supplementary Table 2 was reviewed by clinical hematopathologists (J.S.B.) and (R.S.L.). Genotype was only revealed after cell counting was completed.
Two-tailed Student’s t-test analysis was performed to assess 1) changes in relative numbers of each blood cell lineage between wild type and rag2E450fs mutant fish (Fig. 1g), 2) expression differences in hematopoietic marker genes using quantitative PCR (Fig. 1h), and 3) thymus size differences (Supplementary Fig. 2c). Fisher’s exact tests were performed to assess 1) differences in adult thymus phenotypes between wild type and rag2E450fs mutant fish (Fig. 1c,d), 2) engraftment rates of whole kidney marrow and muscle transplantations between genotypes (Fig. 2 and Supplementary Table 3) and 3) tumor cell transplantations (Fig. 3 and Supplementary Table 4). A threshold of p ≤ 0.05 was considered significant for all statistical methods.
Supplementary Material
Acknowledgments
This works is supported by Alex Lemonade Stand Foundation (J.S.B., M.S.I., D.M.L.), Leukemia Lymphoma Society (J.S.B.), American Cancer Society (D.M.L.), the MGH Howard Goodman Fellowship (D.M.L.), and US National Institutes of Health grants F32DK098875 (F.E.M.), R24OD016761 and 1R01CA154923 (D.M.L.). Q.T. is funded by the China Scholarship Council. J.N.B. is funded as the Cancer Care Nova Scotia Peggy Davison Clinician. We thank D. Traver, T. North and J. Rawls for their helpful comments and B. Li for advice.
Footnotes
Author Contributions
Q.T., N.S.A., J.S.B., J.C.M., S.A.M., I.M.T., and D.M.L. designed and performed experiments. Q.T., J.C.M., and D.M.L. wrote the manuscript. F.E.M., R.L., I.M.T., M.S.I., and Y.H. contributed reagents and animals. Q.T., N.S.A., J.C.M., J.N.B., R.S.L., and D.M.L. performed data analysis and interpretation.
Competing Financial Interest
The authors declare no competing financial interest.
References
- 1.Traver D, et al. Nature Immunology. 2003;4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- 2.Traver D, et al. Blood. 2004;104:1298–1305. doi: 10.1182/blood-2004-01-0100. [DOI] [PubMed] [Google Scholar]
- 3.White RM, et al. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackburn JS, et al. Leukemia. 2012 [Google Scholar]
- 5.Ignatius MS, et al. Cancer Cell. 2012;21:680–693. doi: 10.1016/j.ccr.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith AC, et al. Blood. 2010 [Google Scholar]
- 7.Moore FE, Langenau DM. Adv Hematol. 2012;2012:478164. doi: 10.1155/2012/478164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sander JD, et al. Nat Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couedel C, et al. J Clin Invest. 2010;120:1337–1344. doi: 10.1172/JCI41305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villa A, et al. Blood. 2001;97:81–88. doi: 10.1182/blood.v97.1.81. [DOI] [PubMed] [Google Scholar]
- 11.Ramon-Maiques S, et al. Proc Natl Acad Sci U S A. 2007;104:18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mombaerts P, et al. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 13.Wienholds E, Schulte-Merker S, Walderich B, Plasterk RH. Science. 2002;297:99–102. doi: 10.1126/science.1071762. [DOI] [PubMed] [Google Scholar]
- 14.Mosimann C, et al. Development. 2011;138:169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G. Dev Biol. 1997;192:289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- 16.Langenau DM, et al. Oncogene. 2008;27:4242–4248. doi: 10.1038/onc.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langenau DM, et al. Science. 2003;299:887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- 18.Langenau DM, et al. Genes Dev. 2007;21:1382–1395. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patton EE, et al. Curr Biol. 2005;15:249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 20.Ceol CJ, et al. Nature. 2011;471:513–517. doi: 10.1038/nature09806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meeker ND, Hutchinson SA, Ho L, Trede NS. Biotechniques. 2007;43:610, 612, 614. doi: 10.2144/000112619. [DOI] [PubMed] [Google Scholar]
- 22.Langenau DM, et al. Proc Natl Acad Sci U S A. 2005;102:6068–6073. doi: 10.1073/pnas.0408708102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blackburn JS, et al. Cancer Cell. 2014;25:366–378. doi: 10.1016/j.ccr.2014.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le X, et al. Proc Natl Acad Sci U S A. 2007;104:9410–9415. doi: 10.1073/pnas.0611302104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.