Abstract
Cholangiocarcinoma (CCA) is an aggressive biliary tract malignancy with limited treatment options and low survival rates. Currently, there are no curative medical therapies for CCA. Recent advances have enhanced our understanding of the genetic basis of this disease, and elucidated therapeutically relevant targets. Therapeutic efforts in development are directed at several key pathways due to genetic aberrations including receptor tyrosine kinase pathways, mutant IDH enzymes, the PI3K-AKT-mTOR pathway, and chromatin remodeling networks. A highly desmoplastic, hypovascular stroma is characteristic of CCAs and recent work has highlighted the importance of targeting this pathway via stromal myofibroblast depletion. Future efforts should concentrate on combination therapies with action against the cancer cell and the surrounding tumor stroma. As the mutational landscape of CCA is being illuminated, molecular profiling of patient tumors will enable identification of specific mutations and the opportunity to offer directed, personalized treatment options.
Keywords: cholangiocarcinoma, molecular pathogenesis, targeted therapy
Cholangiocarcinoma (CCA) is the most common primary biliary malignancy and accounts for 3% of all gastrointestinal malignances.1 Cholangiocarcinomas arise from varying locations within the biliary tree, and are classified according to their anatomic origin into intrahepatic, perihilar, and distal cholangiocarcinoma. The second-order bile ducts are the anatomical boundary between intrahepatic CCA (iCCA) and perihilar CCA (pCCA), whereas the cystic duct is the point of distinction between pCCA and distal CCA (dCCA). Although these subtypes exhibit extensive desmoplasia and markers of cholangiocyte differentiation, each has a unique cancer biology and distinct therapeutic options.1
The desmoplastic response is marked by a prominent tumor microenvironment containing a striking aggregation of cancer-associated fibroblasts (CAF).2 The desmoplastic stroma and the genetic heterogeneity of this malignancy pose therapeutic challenges.3 The desmoplastic stroma may impede access of drugs to the tumor and promote potent survival signals. The genetic heterogeneity of the cancer precludes curative treatment options. Cholangiocarcinoma has an aggressive natural course with medial survival of less than 24 months after diagnosis.4 Potentially curative surgery is an option for all three subtypes, albeit only for patients with early-stage disease. For a select group of patients with pCCA, neoadjuvant chemoradiation followed by liver transplantation is a consideration.1 Currently, there are no curative medical therapies for CCA. Systemic chemotherapy with gemcitabine and cisplatin has become the practice standard for patients with advanced or metastatic disease. However, response to combination chemotherapy in CCA patients is typically limited, and the 5-year survival remains low.5,6 An enhanced understanding of CCA carcinogenesis, tumor-stroma interactions, and key molecular pathways would herald targeted, individualized therapies with improvement in patient survival. We discuss the current knowledge regarding the genetic basis of this disease, essential molecular pathways involved in its carcinogenesis, and review potential targeted therapies that hold promise in CCA therapeutics, emphasizing a personalized medicine approach.
Inflammation and Carcinogenesis
A spectrum of malignancies including CCA arises in the background of chronic inflammation, which suggests that inflammation promotes carcinogenesis by imparting prosurvival signals and inducing genetic aberrations. Inflammatory pathways are not only key components in carcinogenesis, but also promote tumor invasion and migration. Primary sclerosing cholangitis (PSC), characterized by chronic biliary tract inflammation and liver injury, is the most common predis-posing condition for CCA in the Western world.1 The incidence of CCA is particularly high in Southeast Asia, in part due to inflammation secondary to the occurrence of biliary infection by the hepatobiliary flukes Opisthorchis viverrini (O. viverrini) and Clonorchis sinensis. Another common CCA risk factor in Southeast Asia is hepatolithiasis, which is characterized by calculi occurring proximal to the hepatic duct confluence.7 Prolonged biliary inflammation, cholestasis, and bacterial infections induced by these calculi also promote CCA development.7
Inflammatory cells promote oxidative stress, which can promote genetic aberrations. Inducible nitric oxide synthase (iNOS) activation by inflammatory cytokines contributes to nitrosative stress by generation of excess nitric oxide.8 DNA repair enzymes are susceptible to nitric oxide-mediated nitrosylation. Consequently, iNOS activation results in inhibition of DNA repair proteins, and single-stranded, double-stranded, and oxidative DNA lesions.8 Enhanced iNOS expression occurs not only in PSC, but also in CCA indicating that it is involved in tumor formation and progression.8,9
Oxidative stress via generation of oxysterols from biliary cholesterol creates a milieu favorable for tumor development and progression. Oxysterols are cholesterol oxidation products present in human bile, and are increased in the bile of patients with biliary tract inflammation.10,11 Oxysterols are activators of the Hedgehog signaling pathway, a key developmental pathway implicated in CCA cell proliferation, migration, and invasion.1,12,13
Cyclooxygenase-2 is induced by various proinflammatory cytokines, and has been implicated in the carcinogenesis of CCA.14 Oxysterols, bile acids, and iNOS all stimulate over-expression of cyclooxygenase-2.14,15 Secondary bile acids in bile may also contribute to CCA biology. In addition to inducing cyclooxygenase-2, bile acids also activate receptor tyrosine kinases, such as epidermal growth factor receptor (EGFR), which mediate cell proliferation.14
Genetic Studies to Date
Recent advances in genome-wide technologies have provided deeper insight into the genetic basis of this challenging malignancy.16 A recent whole-exome sequencing analysis of eight liver O. viverrini-related tumors and matched normal tissue identified 206 somatic mutations in 187 genes.17 Of these, 15 genes were selected for validation in another 46 O. viverrini-related tumors. Somatic mutations were identified in genes with a known association with malignancy, such as tumor protein 53 (TP53) found to be mutated in 44.4% of cases, Kristen ras sarcoma viral oncogene homolog (KRAS) mutated in 16.7% of cases, and SMAD4 that was mutated in 16.7% of cases. In addition, mutations were found in several newly implicated oncogenes including MLL3 (mutated in 14.8% of cases), ROBO2 (9.3%), RNF43 (9.3%), PEG3 (5.6%), and GNAS (9.3%). The biological functions of these genes broadly include deactivation of histone modifiers, G-protein activation, and loss of genomic stability.17 KRAS mutations were associated with a poor survival in this cohort of patients.17 In another recent whole-exome sequencing analysis, 108 cases of O. viverrini-related tumors and 101 cases of non-O. viverrini-related tumors from Asia and Europe were profiled.18 Recurrent somatic mutations in BAP1 and ARID1A were identified. BAP1 and isocitrate dehydrogenase 1 and 2 (IDH1, IDH2) were noted to be mutated more frequently in nonliver–fluke-related CCAs, and TP53 mutations occurred with increased frequency in liver–fluke-related tumors.18 These findings imply that genetic alterations may vary according to etiological associations. Inactivating mutations of chromatin-remodeling genes such as BAP1, ARID1A, and PBRM1 were also identified in exome sequencing of 32 iCCAs.19 These findings postulate a connection between epigenetic regulation and CCA.
Mutations in genes encoding protein tyrosine phosphatases, particularly PTPN3, have been recently reported in iCCA.20 Whole-exome sequencing of seven pairs of iCCA tumors and the surrounding nontumor tissue identified mutations in nine genes encoding protein tyrosine phosphatases. A prevalence screen of 124 paired samples was then conducted and identified somatic mutations in at least one of these nine protein tyrosine phosphatase genes in 51.6% of iCCAs, including PTPN3 mutations in 41.1% of iCCAs. Higher recurrence rates were noted in patients with tumors containing activating mutations of PTPN3.20
A small number of studies have investigated chromosomal aberrations in CCA. In a recent study, gene-expression profile, high-density single-nucleotide polymorphism array, and mutation analyses were performed in 149 cases of iCCA.21 This analysis identified two main biological classes of CCA: the inflammation class and the proliferation class. Activation of inflammatory pathways, cytokine over-expression, and STAT3 activation were hallmarks of the inflammation class, which comprised 38% of the analyzed iCCAs. Features of the proliferation class, consisting of 62% of the analyzed iCCAs, included activation of oncogenic signaling pathways (such as RAS, mitogen-activated protein kinase, and MET), KRAS and BRAF mutations, DNA amplifications at 11q13.2, and deletions at 14q22.1.21 In a recent integrated genomic analyses of 104 iCCAs and 59 matched controls, unique subclasses of patients were identified on the basis of KRAS mutations, early recurrence, and overall survival time.22 Genes regulating inflammation and proteasome activities were associated with a poor prognosis in these patients.22
Activating mutations of KRAS and loss-of-function mutations of TP53 are a common occurrence in CCA.23 TP53 mutations were reported in 21% of CCAs in a review of studies with an aggregate of 229 CCA patients.24 Direct DNA sequencing analysis of 69 CCAs identified KRAS and BRAF mutations in 22% and 45% of tumors.25 Mutations have also been reported in other genes, albeit less frequently, including EGFR, PIK3CA, NRAS, and APC.1
Pathways and Targeted Therapies
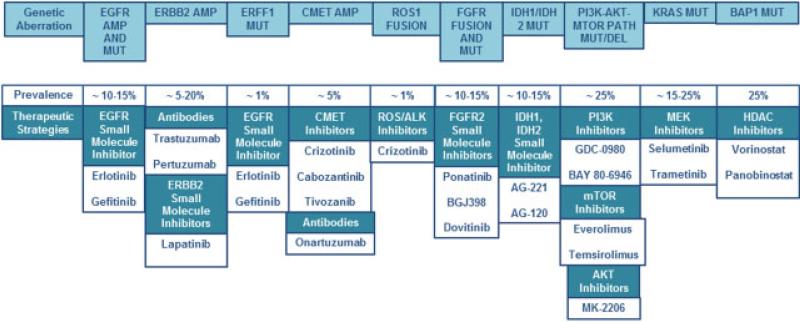
Current standardized regimens available for medical therapy in cholangiocarcinoma have limited effectiveness and considerable side effects. There is a crucial need for development of targeted therapies that interfere with growth and progression of cancer cells while sparing normal cells (Fig. 1).
Fig. 1.
Molecular pathways in cholangiocarcinoma (CCA) and inhibitors. Several molecular pathways are dysregulated in CCA. Inhibitors targeting these pathways are entering human clinical trials. The prevalence of each genetic aberration and potential therapeutic strategies are listed.
Cytokines
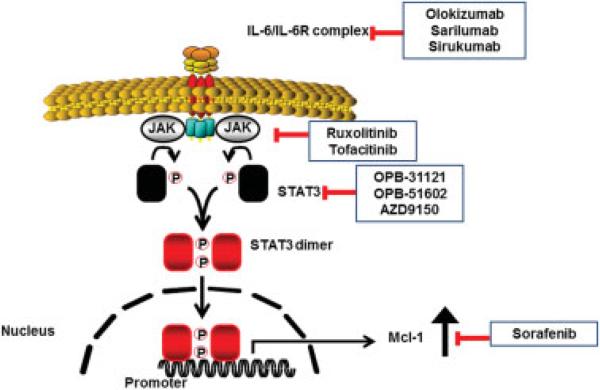
Interleukin- (IL-) 6 is an inflammatory cytokine produced by cholangiocytes in the presence of inflammatory stimuli. It is also secreted by CCA cells and participates in survival and mitogenic signaling in these cells.26,27 The phosphatidylinositol 3-kinase (PI3K)/AKT cell-signaling pathway also generates potent survival signals in several malignancies. AKT is strongly expressed and constitutively active in CCA. IL-6 upregulates the potent antiapoptotic Bcl-2 protein, myeloid cell leukemia sequence 1 (Mcl-1), via an AKT-dependent mechanism.26 Mcl-1 is integral in tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) resistance in CCA.28 Consequently, IL-6 inhibition reduces Mcl-1 expression and enhances TRAIL-mediated apoptosis.26 IL-6 also acts via a signal transducer and activator of transcription- (STAT-) dependent mechanism to enhance Mcl-1 expression.29 A suppressor of cytokine signaling 3 (SOCS) regulates the IL-6/STAT-3 signaling pathway in a feedback manner, and epigenetic silencing of SOCS-3 facilitates sustained IL-6/STAT-3 signaling with resultant enhanced Mcl-1 expression.30 This information suggests several approaches to treating the inflammatory subtype of CCA. For example, neutralizing antibodies to IL-6 are in human clinical trials and may be therapeutic in CCA.31,32 Likewise, inhibitors of the JAK kinases, which activate STAT3 downstream of IL-6 initiated signaling, are also being developed and warrant investigation in preclinical models of CCA (Fig. 2). Finally, analogous to the success of Bcl-2 antagonists in chronic lymphocytic leukemia, Mcl-1 antagonists could prove useful in treating CCA. Such antagonists are in development.33,34 Sorafenib, albeit not a direct Mcl-1 inhibitor, induces apoptosis in cancer cells via downregulation and destabilization of Mcl-1.35
Fig. 2.
IL-6/STAT3 signaling and inhibitors. Binding of IL-6 to its receptor activates Janus kinase (JAK) with subsequent phosphorylation, activation, dimerization, and nuclear translocation of STAT3. STAT3 upregulates Mcl-1 transcription. Inhibitors of IL-6, JAK, STAT3, and Mcl-1 are currently in development.
Notch Signaling Pathway and Inhibitors
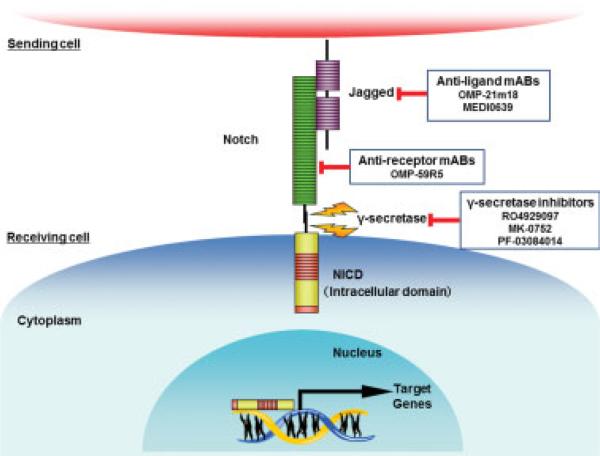
The Notch signaling pathway has a central role in cell-fate determination during embryonic development in several organs including the biliary tree.36 Notch dysregulation is implicated in inflammation and carcinogenesis. Notch expression can be induced by iNOS, and Notch-1 is upregulated in PSC and CCA.37 Two preclinical studies utilizing cell fate-tracing techniques have demonstrated a role for Notch in conversion of mature adult hepatocytes into biliary precursors of iCCA.38,39 Enhanced expression of the Notch intracellular domain (NICD) in hepatic cells is associated with iCCA occurrence.40 Several inhibitors of the Notch pathway are in development. Notch signaling inhibition can occur by various mechanisms including inhibition of Notch receptor activation with γ-secretase inhibitors, inhibition of binding of the ligand with monoclonal antibodies, or inhibition of the transcriptional activity of NICD by blocking peptides (Fig. 3).41
Fig. 3.
Notch signaling pathway and inhibitors. The Notch signaling pathway has been implicated in cholangiocarcinoma (CCA) carcino-genesis. Enhanced expression of NICD occurs in CCA. Several inhibitors of the Notch pathway are currently in development. These include inhibitors of γ-secretase, a key enzyme in the Notch pathway, as well as monoclonal antibodies (mABs) targeting the Notch receptors and ligands.
Receptor Tyrosine Kinase Pathways and Inhibitors
Several growth factor tyrosine kinases are implicated in carcinogenesis and progression of CCAs. These include the ERBB family of receptor tyrosine kinases, fibroblast growth factor receptor (FGFR), and the hepatocyte growth factor (HGF) receptor, c-met.
The ERBB family of receptor tyrosine kinases is comprised of four different receptors, ERBB1 or EGFR, ERBB2 or HER-2/neu, and ERBB3 and 4.42 The EGFR activation leads to downstream activation of mitogen-activated protein kinase (MAPK), a well-known oncogenic signaling pathway. Over-expression of ERBB2, an EGFR family member, has been linked to biliary epithelial tumor formation in mice.43 Moreover, ERBB2 amplification was recently described in a cohort of 100 CCA patients, with a reported prevalence of 3%.44 Erlotinib, an EGFR inhibitor, has had limited success in human cholangiocarcinoma clinical trials.45,46 This may partly be due to an insufficient understanding of EGFR signaling in the molecular pathogenesis of CCA, and failure to select a patient population overexpressing EGFR. Indeed, the reported prevalence of EGFR mutations and amplification is only 15% and 5%, respectively.47 A recent molecular and genomic characterization of 104 surgically resected iCCAs demonstrated significant upregulation of HER2 signaling in tumors with the most malignant phenotype.22 The HER2 upregulation was associated with poor prognosis and frequent coactivation of ERBB3 and EGFR.22 Lapatinib, a dual inhibitor of EGFR and HER2, was significantly more effective in inhibition of CCA cell lines than trastuzumab, which selectively inhibits HER2.22 Loss of function mutation in ERRFI1, an endogenous inhibitor of EGFR, ERBB2, ERBB3, and ERBB4, was detected in a patient with metastatic, treatment refractory iCCA.48 Significant disease regression was observed in this patient after EGFR-targeted therapy with erlotinib.48 Hence, the efficacy of ERBB inhibitors certainly merits further attention as we learn to select and stratify patients for targeted therapies (Fig. 1).
MET tyrosine kinase plays an integral role in carcinogenesis by promoting tumor invasion, protection from apoptosis, and angiogenesis.49 Binding to HGF or scatter factor activates MET. HGF and MET expression is enhanced in CCAs, and is associated with activation of ERBB family members, especially ERBB2.1 An integrative genomic analysis of iCCA identified a proliferation class of iCCA that is characterized by activation of oncogenic signaling pathways such as MET.21 MET amplification has been described in malignancies including gastric, esophageal, ovarian, and nonsmall cell lung cancer, and is associated with a poor clinical outcome.50,51 MET amplification is also linked to resistance to EGFR and ERBB2 inhibitors and may predict sensitivity to MET inhibitors.50,51 A recent study utilizing DNA sequencing of 28 iCCAs detected MET amplification in two cases (7%).51 Crizotinib is a small molecule inhibitor of MET and ROS receptor tyrosine kinases. The U.S. Food and Drug Administration has approved crizotinib for the treatment of metastatic lung cancer in patients with anaplastic lymphoma kinase-positive tumors. Crizotinib, cabozantinib, and tivozanib have therapeutic potential in targeted cancer chemotherapy of CCA tumors with MET amplification (Fig. 1). Tivozanib is currently being evaluated in a phase II trial in adult patients with advanced hepatocellular carcinoma (ClinicalTrials.gov identifier: NCT01807156).
ROS1 Fusion Kinases and Inhibitors
ROS1 fusions proteins have a significant oncogenic role in several malignancies and have been recently identified in CCA.52 Subsequent preclinical studies have validated fused-in-glioblastoma-c-ros-oncogene 1 (FIG-ROS) as a potent oncogene in a mouse model of CCA.53 In this model, FIG-ROS mediated iCCA development was Kras dependent, and led to development of aggressive and metastatic tumors in the presence of Kras.53 In tumors harboring Kras and p53 mutations, FIG-ROS inactivation led to the inhibition of tumor growth. Accordingly, ROS1 inhibitors have potential as targeted therapy in a subset of patients harboring ROS1 fusion proteins. Crizotinib, a ROS1 inhibitor, is approved for use in select patients with non-small cell lung cancer. Recent reports have indicated that a specific ROS1 mutation, ROS1(G2032R), confers resistance to crizotinib. Foretinib, another ROS1 inhibitor, maintains clinical effectiveness in the setting of ROS1 domain mutations including ROS1(G2032R) (Fig. 1).54
FGFR Fusions and Inhibitors
FGFR is a receptor tyrosine kinase involved in a myriad of biological processes including cell transformation, angiogenesis, and tissue repair.55 Fusions of the FGFR gene have been reported in solid cancers. Gene fusions are a class of driver mutations with an essential role in certain cancers, such as the BCR-ABL gene in chronic myeloid leukemia.56 Chronic myeloid leukemia patients with the BCR-ABL fusion gene respond well to imatinib, a small-molecule kinase inhibitor, signifying the value of targeted therapy. Recently, FGFR2-BICC1 gene fusion was described in two cases of CCA by Wu et al.56 A high prevalence (13.6%) of FGFR2 gene fusions was reported in a cohort of 102 CCA cases.55 In addition to detecting several cases with FGFR2-BICC1 fusions, a single case of a novel FGFR gene fusion, FGFR2-AHCYL1, was also identified in this cohort.55 More recently, single cases of FGFR2-MGEA5 and FGFR2-TACC3 gene fusions have been described in iCCA.48
Overexpression of FGFR2-BICC1 is associated with enhanced cell proliferation and altered cell morphology. Based on mechanistic studies, Wu et al proposed that FGFR fusion partners mediate oligomerizations, which initiates activation of the respective FGFR kinase in tumors harboring these mutations.56 Only cells harboring the FGFR gene fusions were sensitive to FGFR inhibitors,55,56 indicating a role for targeted FGFR kinase inhibition in patients with tumors containing these gene fusions (Fig. 1). Indeed, FGFR genetic alterations have been reported to be the most significant predictor for sensitivity to the pan-FGFR inhibitor BGJ398.57 BGJ398 and another small molecule FGFR inhibitor, PD173072, suppressed downstream MAPK signaling and oncogenic activity of FGFR fusion kinases.55 Clinical efficacy of BGJ398 is currently being investigated in a phase I study in adult patients with advanced solid tumors harboring FGFR1/FGFR2 amplification or FGFR3 mutation (ClinicalTrials.gov identifier: NCT01004224). Targeted therapy with ponatinib, another pan-FGFR inhibitor, in an iCCA patient with the FGFR2-MGEA5 fusion resulted in a decrease in tumor necrosis and levels of the tumor marker, CA 19-9.48 Ponatinib also demonstrated preliminary antitumor activity with tumor-size reduction in an iCCA patient harboring the FGFR2-TACC3 fusion.48 Dovitinib, another small molecule FGFR inhibitor, was shown to be an effective and tolerable therapy in a phase II trial in patients with metastatic renal cell cancer.58 Median progression-free survival and overall survival were 3.7 and 11.8 months, respectively.58 Dovitinib is currently in a phase I study in combination with gemcitabine and capecitabine in patients with advanced solid tumors including pancreatic and biliary cancers (ClinicalTrials.gov identifier: NCT01497392).
IDH/IDH2 Mutations and Inhibitors
IDH1 and IDH2 mutations have been reported in 10% to 28% of cholangiocarcinomas, occurring primarily in iCCA.59 In a cohort of 94 CCA cases, the prevalence of IDH1 and IDH2 mutations was 15% and 7%, respectively.60 IDH mutations were associated with clear cell change, poorly differentiated histology, and longer overall survival.60 Similarly, in a separate cohort of 326 iCCA patients, IDH mutations had a prevalence of 10% and were associated with longer overall survival and longer time to recurrence.61 Increased levels of p53 and DNA hypermethylation are observed in the setting of IDH-mutant tumors.61 IDH2 mutations are observed less frequently in iCCA with a prevalence of 27% compared with 73% prevalence of IDH1 mutations.59 A range of IDH1/2 mutations exists and varies according to the IDH amino acid residues mutated. Arginine 132 (R132), IDH1 R132C, is mutated in 44% of iCCA cases whereas IDH1 R132G alteration is seen in 14% iCCAs containing mutated samples.59 The mutated IDH enzymes convert α-ketoglutarate to 2-hydroxyglutarate (2-HG) rather than isocitrate, resulting in an approximate100-fold increase in levels of this oncometabolite in IDH-mutant tumors.59 As 2-HG is present in negligible amounts in normal cells, it has utility as a biomarker in patients with IDH mutations who may be candidates for therapy with IDH inhibitors. This oncometabolite likely promotes carcinogenesis by affecting methylation changes of the genome.59 Small molecule inhibitors of IDH1 and IDH2 are being developed.62,63 AG1–5198, a selective IDH1 inhibitor, blocked the production of 2-HG in a dose-dependent manner and stunted the growth of IDH1-mutant gliomas cells, but not IDH1 wild-type cells.62 The effects of IDH1 inhibition included increased expression of genes associated with gliogenic differentiation and demethylation of histone.62 Similarly, treatment with AGI-6780, a selective IDH2 inhibitor, induced differentiation in IDH2-mutant hematological cell lines.63
Having demonstrated promise in preclinical studies, IDH inhibitors are now being investigated in clinical trials (Fig. 1). Preliminary results from an ongoing phase I trial of AG-221, a first in class inhibitor of mutated IDH2, have indicated promising clinical activity without dose-limiting toxic effects in patients with IDH-mutant driven hematological malignancies (ClinicalTrials.gov identifier: NCT01915498). Clinical benefit has been noted in 7 out of 10 patients in this study thus far. AG-120, a specific IDH1 inhibitor, is the subject of a phase I study in patients with IDH1-mutant associated advanced solid tumors including cholangiocarcinoma (ClinicalTrials.gov identifier: NCT02073994).
PI3K-AKT-mTOR Pathway
Binding of growth factors, such as HGF and FGF, to their respective receptor tyrosine kinases leads to activation of the PI3K cell signaling pathway. Subsequent AKT activation leads to phosphorylation and activation of the mammalian target of rapamycin (mTOR) pathway.47 Deregulation of the PI3K-AKT-mTOR pathway fosters tumor development, cell proliferation and survival, tumor invasion, and angiogenesis. PIK3CA mutations were identified in 5 of 94 resected CCA specimens using MassARRAY technology.47 In a cohort of Chinese CCA patients, PIK3CA mutations were identified in 34.2% of cases.64 Exome sequencing of 32 iCCAs detected somatic mutations in several members of the PI3K pathway including PIK3CA, PIK3C2A, PIK3C2G, and phosphatase and tensin homolog (PTEN).19 Loss of the lipid phosphatase PTEN results in constitutive activation of this pathway. In addition to mutations of PTEN, the oncomiR miR-21 inhibits expression of PTEN in CCA cells causing enhanced AKT/mTOR signaling.65 Various PI3K pathway inhibitors are under investigation in multiple clinical trials of human cancer (Fig. 1).66 GDC-0980, an orally bioavailable potent inhibitor of class I PI3K and mTOR kinase, and BAY 80–6946, a highly selective and potent pan-class PI3K inhibitor, are entering clinical studies.67,68 A phase I trial of everolimus, an mTOR inhibitor, in combination with gemcitabine and cisplatin demonstrated promising clinical activity in patients with treatment refractory solid tumors including cholangiocarcinoma.69
KRAS Mutations
Mutations in KRAS have been frequently described in CCA, along with other mutations in NRAS, BRAF, and downstream MAPK effector pathways.23,25 The proliferation subclass reported by Sia et al was notable for MAPK pathway activation.21 Strategies to therapeutically target tumors with KRAS mutations have focused on targeting downstream effector pathways of KRAS such as Raf/MEK/ERK and PI3K/AKT. Phase II studies of a MEK inhibitor in a mixed population of biliary tract cancers that included CCA have shown some promise (Fig. 1), and studies of sorafenib are underway (SWOG0514). Combination therapy targeting both Raf/MEK/ERK and PI3K/Akt effector pathways similar to that reported for other KRAS dependent tumors is rational, but this approach may be limited by toxicities. Direct inhibitors of KRAS may offer promise if they become available in the future.
BAP1 Mutation and HDAC Inhibitors
Somatic mutations in BAP1, a chromatin-remodeling gene, have previously been reported in renal cell carcinoma, but not in CCA until recently.19 In a cohort of 32 iCCA cases, the prevalence of inactivating mutations in BAP1 was 25%.19 Patients with these mutations had a trend, albeit not statistically significant, toward shorter survival times.19 In a separate cohort of 209 CCAs, BAP1 mutations were detected in 10.5% of non-liver–fluke-related CCAs, compared with 2.8% of liver– fluke-related CCAs.18 Functional studies demonstrated a tumor suppressor role for this gene.18 The discovery of this mutation expands the mutation spectrum of CCA and heralds a role for drugs targeting chromatin remodeling, such as the histone deacetylase (HDAC) inhibitors vorinostat and panobinostat (Fig. 1).19,70
Targeting CAFs in the Tumor Stroma
Cholangiocarcinomas have a characteristic hypovascular, desmoplastic stroma containing α-smooth muscle actin (α-SMA) positive cancer-associated fibroblasts (CAFs).71 Cancer-associated fibroblasts promote tumor progression and migration by production of factors including matricellular proteins, growth factors, chemokines, and matrix metalloproteinases. These factors enhance proliferation, tumor invasion and migration, and induce a profibrogenic response.1 A fibrogenic matrix precedes CCA development and fosters malignant growth.72 It has been postulated that the desmoplasia characteristic of CCA may serve as a niche fostering tumor development rather than as a response to the tumor.2,72 This notion underscores the importance of fibrosis in CCA carcinogenesis, and supports the potential of antifibrotic therapies in CCA chemoprevention. Preclinical studies have demonstrated a reduction in fibrosis and carcinogenesis in CCA with 1D11, a transforming growth factor β (TGF-β) antagonist, as well as curcumin, a nutraceutical agent.73,74 Further preclinical and clinical studies are needed to explore the role of antifibrotic therapies in CCA chemoprevention.
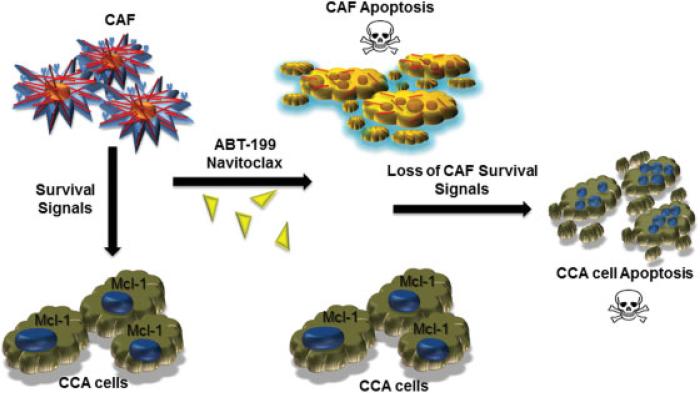
Recent work has highlighted the strong potential of selective targeting of CAF in CCA chemotherapeutic efforts (Fig. 4).75 The BH3 mimetic, Navitoclax, significantly enhanced selective CAF apoptosis, decreased expression of α-SMA, and reduced tumor burden and metastasis while improving survival in an orthotopic model of CCA.75
Fig. 4.
Selective deletion of cancer-associated fibroblasts (CAFs) from cholangiocarcinoma (CCA) tumor microenvironment. Cancer-associated fibroblasts produce factors that promote survival of tumor cells. Cancer-associated fibroblasts are primed or sensitized for apoptosis and hence can be selectively targeted by BH3 mimetics such as navitoclax and ABT-199. Cholangiocarcinoma cells overexpress Mcl-1, which does not bind navitoclax, and hence are not a direct target of navitoclax. However, CAF deletion from the tumor microenvironment leads to secondary cancer cell death due to loss of CAF-mediated prosurvival signals.75
Paracrine sonic Hedgehog signaling, a developmental pathway, promotes the desmoplastic response in CCA.2 Such signaling has a well-established role in pancreatic adenocarcinoma. Reduction of the desmoplastic response via Hedgehog inhibition increased intratumor vascular density, and the intratumoral concentration of gemcitabine, contributing to transient stabilization of the disease.76 Clinical studies using Hedgehog antagonists as combination therapy are underway in pancreatic cancer.2,77 Although results to date have been disappointing in pancreatic cancer, one may still envision a similar role for Hedgehog inhibition in combination chemotherapy in CCA.
Paracrine EGF signaling has also been implicated in tumor-stroma crosstalk. Myofibroblast-derived EGF induced EGFR activation with enhanced migratory and invasive properties in CCA cells.78 An EGF receptor inhibitor, gefitinib, abolished these effects, underscoring the potential of EGFR pathway targeting in CCA. Several studies have demonstrated paracrine platelet-derived growth factor (PDGF) signaling in CCA. Myofibroblast-derived PDGF-BB protects CCA cells from TRAIL-induced apoptosis via a Hedgehog-dependent mechanism.79 Inhibition of the PDGF receptor, PDGFR-β, with imatinib induced CCA cell apoptosis.79 Further preclinical studies demonstrated a reduction in tumor size with imatinib treatment in a rodent model of CCA.80 Despite these promising preclinical studies, erlotinib and imatinib have had been relatively disappointing in limited clinical studies in human CCA.81
Summary
Advances in the molecular and genetic basis of cancer and the ongoing development of tumor-specific therapeutics has heralded the era of personalized or precision medicine. Significant progress has been made in understanding the mutational landscape of cholangiocarcinoma, a notoriously difficult-to-treat malignancy. Continued dissection of the key pathways and genetic drivers of CCA progression will direct efforts at individualized therapy for patients based on the driver mutation for their particular tumor. Limited mutation-directed therapeutic efforts in CCA have been encouraging. Tumor immunotherapy is another frontier in cancer treatment. Inhibition of immune receptors such as programmed cell death 1 has shown promise in certain malignancies.82 Although cholangiocarcinoma is a genetically heterogeneous cancer, its stroma is more genetically uniform. Preclinical studies have demonstrated tumor regression and enhanced survival with selective deletion of CAF from the CCA tumor microenvironment.75 Therefore, the stroma represent an additional potential target in the treatment of CCA. Further work is needed to direct development of combination therapies, which will target not only the cancer cell, but also the cancer stroma in CCA.
Acknowledgments
This work was supported by National Institutes of Health grants DK59427 (G.J.G.), T32DK007198 (S.R.), K12 CA90628 (M.J.B), DK069370 (T.P.), and the Mayo Foundation. S. Rizvi also received support from the American Liver Foundation and International Liver Cancer Association. We thank Ms. Courtney Hoover for excellent secretarial support.
References
- 1.Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 2013;145(6):1215–1229. doi: 10.1053/j.gastro.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sirica AE, Gores GJ. Desmoplastic stroma and cholangiocarcinoma: clinical implications and therapeutic targeting. Hepatology. 2014;59(6):2397–2402. doi: 10.1002/hep.26762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet. 2014;383(9935):2168–2179. doi: 10.1016/S0140-6736(13)61903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farley DR, Weaver AL, Nagorney DM. “Natural history” of unresected cholangiocarcinoma: patient outcome after noncurative intervention. Mayo Clin Proc. 1995;70(5):425–429. doi: 10.4065/70.5.425. [DOI] [PubMed] [Google Scholar]
- 5.Everhart JE, Ruhl CE. Burden of digestive diseases in the United States Part III: liver, biliary tract, and pancreas. Gastroenterology. 2009;136(4):1134–1144. doi: 10.1053/j.gastro.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 6.Valle J, Wasan H, Palmer DH, et al. ABC-02 Trial Investigators Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med. 2010;362(14):1273–1281. doi: 10.1056/NEJMoa0908721. [DOI] [PubMed] [Google Scholar]
- 7.Tyson GL, El-Serag HB. Risk factors for cholangiocarcinoma. Hepatology. 2011;54(1):173–184. doi: 10.1002/hep.24351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaiswal M, LaRusso NF, Burgart LJ, Gores GJ. Inflammatory cytokines induce DNA damage and inhibit DNA repair in cholangio-carcinoma cells by a nitric oxide-dependent mechanism. Cancer Res. 2000;60(1):184–190. [PubMed] [Google Scholar]
- 9.Jaiswal M, LaRusso NF, Shapiro RA, Billiar TR, Gores GJ. Nitric oxide-mediated inhibition of DNA repair potentiates oxidative DNA damage in cholangiocytes. Gastroenterology. 2001;120(1):190–199. doi: 10.1053/gast.2001.20875. [DOI] [PubMed] [Google Scholar]
- 10.Haigh WG, Lee SP. Identification of oxysterols in human bile and pigment gallstones. Gastroenterology. 2001;121(1):118–123. doi: 10.1053/gast.2001.25513. [DOI] [PubMed] [Google Scholar]
- 11.Kuver R. Mechanisms of oxysterol-induced disease: insights from the biliary system. Clin Lipidol. 2012;7(5):537–548. doi: 10.2217/clp.12.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dwyer JR, Sever N, Carlson M, Nelson SF, Beachy PA, Parhami F. Oxysterols are novel activators of the Hedgehog signaling pathway in pluripotent mesenchymal cells. J Biol Chem. 2007;282(12):8959–8968. doi: 10.1074/jbc.M611741200. [DOI] [PubMed] [Google Scholar]
- 13.Nachtergaele S, Mydock LK, Krishnan K, et al. Oxysterols are allosteric activators of the oncoprotein Smoothened. Nat Chem Biol. 2012;8(2):211–220. doi: 10.1038/nchembio.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon JH, Higuchi H, Werneburg NW, Kaufmann SH, Gores GJ. Bile acids induce cyclooxygenase-2 expression via the epidermal growth factor receptor in a human cholangiocarcinoma cell line. Gastroenterology. 2002;122(4):985–993. doi: 10.1053/gast.2002.32410. [DOI] [PubMed] [Google Scholar]
- 15.Yoon JH, Canbay AE, Werneburg NW, Lee SP, Gores GJ. Oxysterols induce cyclooxygenase-2 expression in cholangiocytes: implications for biliary tract carcinogenesis. Hepatology. 2004;39(3):732–738. doi: 10.1002/hep.20125. [DOI] [PubMed] [Google Scholar]
- 16.Andersen JB, Thorgeirsson SS. Genetic profiling of intrahepatic cholangiocarcinoma. Curr Opin Gastroenterol. 2012;28(3):266–272. doi: 10.1097/MOG.0b013e3283523c7e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ong CK, Subimerb C, Pairojkul C, et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet. 2012;44(6):690–693. doi: 10.1038/ng.2273. [DOI] [PubMed] [Google Scholar]
- 18.Chan-On W, Nairismägi ML, Ong CK, et al. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat Genet. 2013;45(12):1474–1478. doi: 10.1038/ng.2806. [DOI] [PubMed] [Google Scholar]
- 19.Jiao Y, Pawlik TM, Anders RA, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45(12):1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Q, Zhao YJ, Wang XY, et al. Activating mutations in PTPN3 promote cholangiocarcinoma cell proliferation and migration and are associated with tumor recurrence in patients. Gastroenterology. 2014;146(5):1397–1407. doi: 10.1053/j.gastro.2014.01.062. [DOI] [PubMed] [Google Scholar]
- 21.Sia D, Hoshida Y, Villanueva A, et al. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology. 2013;144(4):829–840. doi: 10.1053/j.gastro.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersen JB, Spee B, Blechacz BR, et al. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology. 2012;142(4):1021–1031. e15. doi: 10.1053/j.gastro.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sia D, Tovar V, Moeini A, Llovet JM. Intrahepatic cholangiocarcinoma: pathogenesis and rationale for molecular therapies. Oncogene. 2013;32(41):4861–4870. doi: 10.1038/onc.2012.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan SA, Thomas HC, Toledano MB, Cox IJ, Taylor-Robinson SD. p53 Mutations in human cholangiocarcinoma: a review. Liver Int. 2005;25(4):704–716. doi: 10.1111/j.1478-3231.2005.01106.x. [DOI] [PubMed] [Google Scholar]
- 25.Tannapfel A, Sommerer F, Benicke M, et al. Mutations of the BRAF gene in cholangiocarcinoma but not in hepatocellular carcinoma. Gut. 2003;52(5):706–712. doi: 10.1136/gut.52.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi S, Werneburg NW, Bronk SF, Kaufmann SH, Gores GJ. Interleukin-6 contributes to Mcl-1 up-regulation and TRAIL resistance via an Akt-signaling pathway in cholangiocarcinoma cells. Gastroenterology. 2005;128(7):2054–2065. doi: 10.1053/j.gastro.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Park J, Tadlock L, Gores GJ, Patel T. Inhibition of interleukin 6-mediated mitogen-activated protein kinase activation attenuates growth of a cholangiocarcinoma cell line. Hepatology. 1999;30(5):1128–1133. doi: 10.1002/hep.510300522. [DOI] [PubMed] [Google Scholar]
- 28.Taniai M, Grambihler A, Higuchi H, et al. Mcl-1 mediates tumor necrosis factor-related apoptosis-inducing ligand resistance in human cholangiocarcinoma cells. Cancer Res. 2004;64(10):3517–3524. doi: 10.1158/0008-5472.CAN-03-2770. [DOI] [PubMed] [Google Scholar]
- 29.Isomoto H, Kobayashi S, Werneburg NW, et al. Interleukin 6 upregulates myeloid cell leukemia-1 expression through a STAT3 pathway in cholangiocarcinoma cells. Hepatology. 2005;42(6):1329–1338. doi: 10.1002/hep.20966. [DOI] [PubMed] [Google Scholar]
- 30.Isomoto H, Mott JL, Kobayashi S, et al. Sustained IL-6/STAT-3 signaling in cholangiocarcinoma cells due to SOCS-3 epigenetic silencing. Gastroenterology. 2007;132(1):384–396. doi: 10.1053/j.gastro.2006.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Genovese MC, Fleischmann R, Furst D, et al. Efficacy and safety of olokizumab in patients with rheumatoid arthritis with an inadequate response to TNF inhibitor therapy: outcomes of a randomised Phase IIb study. Ann Rheum Dis. 2014;73(9):1607–1615. doi: 10.1136/annrheumdis-2013-204760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka Y, Martin Mola E. IL-6 targeting compared to TNF targeting in rheumatoid arthritis: studies of olokizumab, sarilumab and sirukumab. Ann Rheum Dis. 2014;73(9):1595–1597. doi: 10.1136/annrheumdis-2013-205002. [DOI] [PubMed] [Google Scholar]
- 33.Abulwerdi F, Liao C, Liu M, et al. A novel small-molecule inhibitor of mcl-1 blocks pancreatic cancer growth in vitro and in vivo. Mol Cancer Ther. 2014;13(3):565–575. doi: 10.1158/1535-7163.MCT-12-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abulwerdi FA, Liao C, Mady AS, et al. 3-Substituted-N-(4-hydroxynaphthalen-1-yl)arylsulfonamides as a novel class of selective Mcl-1 inhibitors: structure-based design, synthesis, SAR, and biological evaluation. J Med Chem. 2014;57(10):4111–4133. doi: 10.1021/jm500010b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu C, Bruzek LM, Meng XW, et al. The role of Mcl-1 downregulation in the proapoptotic activity of the multikinase inhibitor BAY 43-9006. Oncogene. 2005;24(46):6861–6869. doi: 10.1038/sj.onc.1208841. [DOI] [PubMed] [Google Scholar]
- 36.Hofmann JJ, Zovein AC, Koh H, Radtke F, Weinmaster G, Iruela-Arispe ML. Jagged1 in the portal vein mesenchyme regulates intrahepatic bile duct development: insights into Alagille syndrome. Development. 2010;137(23):4061–4072. doi: 10.1242/dev.052118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishimura N, Bronk SF, Gores GJ. Inducible nitric oxide synthase up-regulates Notch-1 in mouse cholangiocytes: implications for carcinogenesis. Gastroenterology. 2005;128(5):1354–1368. doi: 10.1053/j.gastro.2005.01.055. [DOI] [PubMed] [Google Scholar]
- 38.Fan B, Malato Y, Calvisi DF, et al. Cholangiocarcinomas can originate from hepatocytes in mice. J Clin Invest. 2012;122(8):2911–2915. doi: 10.1172/JCI63212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sekiya S, Suzuki A. Intrahepatic cholangiocarcinoma can arise from Notch-mediated conversion of hepatocytes. J Clin Invest. 2012;122(11):3914–3918. doi: 10.1172/JCI63065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zender S, Nickeleit I, Wuestefeld T, et al. A critical role for notch signaling in the formation of cholangiocellular carcinomas. Cancer Cell. 2013;23(6):784–795. doi: 10.1016/j.ccr.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 41.Morell CM, Strazzabosco M. Notch signaling and new therapeutic options in liver disease. J Hepatol. 2014;60(4):885–890. doi: 10.1016/j.jhep.2013.11.028. [DOI] [PubMed] [Google Scholar]
- 42.Sirica AE. Role of ErbB family receptor tyrosine kinases in intrahepatic cholangiocarcinoma. World J Gastroenterol. 2008;14(46):7033–7058. doi: 10.3748/wjg.14.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiguchi K, Carbajal S, Chan K, et al. Constitutive expression of ErbB-2 in gallbladder epithelium results in development of adenocarcinoma. Cancer Res. 2001;61(19):6971–6976. [PubMed] [Google Scholar]
- 44.Graham RP, Barr Fritcher EG, Pestova E, et al. Fibroblast growth factor receptor 2 translocations in intrahepatic cholangiocarcinoma. Hum Pathol. 2014;45(8):1630–1638. doi: 10.1016/j.humpath.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 45.Lubner SJ, Mahoney MR, Kolesar JL, et al. Report of a multicenter phase II trial testing a combination of biweekly bevacizumab and daily erlotinib in patients with unresectable biliary cancer: a phase II Consortium study. J Clin Oncol. 2010;28(21):3491–3497. doi: 10.1200/JCO.2010.28.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Philip PA, Mahoney MR, Allmer C, et al. Phase II study of erlotinib in patients with advanced biliary cancer. J Clin Oncol. 2006;24(19):3069–3074. doi: 10.1200/JCO.2005.05.3579. [DOI] [PubMed] [Google Scholar]
- 47.Voss JS, Holtegaard LM, Kerr SE, et al. Molecular profiling of cholangiocarcinoma shows potential for targeted therapy treatment decisions. Hum Pathol. 2013;44(7):1216–1222. doi: 10.1016/j.humpath.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 48.Borad MJ, Champion MD, Egan JB, et al. Integrated genomic characterization reveals novel, therapeutically relevant drug targets in FGFR and EGFR pathways in sporadic intrahepatic cholangiocarcinoma. PLoS Genet. 2014;10(2):e1004135. doi: 10.1371/journal.pgen.1004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat Rev Drug Discov. 2008;7(6):504–516. doi: 10.1038/nrd2530. [DOI] [PubMed] [Google Scholar]
- 50.Appleman LJ. MET signaling pathway: a rational target for cancer therapy. J Clin Oncol. 2011;29(36):4837–4838. doi: 10.1200/JCO.2011.37.7929. [DOI] [PubMed] [Google Scholar]
- 51.Ross JS, Wang K, Gay L, et al. New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next-generation sequencing. Oncologist. 2014;19(3):235–242. doi: 10.1634/theoncologist.2013-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gu TL, Deng X, Huang F, et al. Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PLoS ONE. 2011;6(1):e15640. doi: 10.1371/journal.pone.0015640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saborowski A, Saborowski M, Davare MA, Druker BJ, Klimstra DS, Lowe SW. Mouse model of intrahepatic cholangiocarcinoma validates FIG-ROS as a potent fusion oncogene and therapeutic target. Proc Natl Acad Sci U S A. 2013;110(48):19513–19518. doi: 10.1073/pnas.1311707110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davare MA, Saborowski A, Eide CA, et al. Foretinib is a potent inhibitor of oncogenic ROS1 fusion proteins. Proc Natl Acad Sci U S A. 2013;110(48):19519–19524. doi: 10.1073/pnas.1319583110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arai Y, Totoki Y, Hosoda F, et al. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology. 2014;59(4):1427–1434. doi: 10.1002/hep.26890. [DOI] [PubMed] [Google Scholar]
- 56.Wu YM, Su F, Kalyana-Sundaram S, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3(6):636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guagnano V, Kauffmann A, Wöhrle S, et al. FGFR genetic alterations predict for sensitivity to NVP-BGJ398, a selective pan-FGFR inhibitor. Cancer Discov. 2012;2(12):1118–1133. doi: 10.1158/2159-8290.CD-12-0210. [DOI] [PubMed] [Google Scholar]
- 58.Escudier B, Grünwald V, Ravaud A, et al. Phase II results of Dovitinib (TKI258) in patients with metastatic renal cell cancer. Clin Cancer Res. 2014;20(11):3012–3022. doi: 10.1158/1078-0432.CCR-13-3006. [DOI] [PubMed] [Google Scholar]
- 59.Grassian AR, Pagliarini R, Chiang DY. Mutations of isocitrate dehydrogenase 1 and 2 in intrahepatic cholangiocarcinoma. Curr Opin Gastroenterol. 2014;30(3):295–302. doi: 10.1097/MOG.0000000000000050. [DOI] [PubMed] [Google Scholar]
- 60.Kipp BR, Voss JS, Kerr SE, et al. Isocitrate dehydrogenase 1 and 2 mutations in cholangiocarcinoma. Hum Pathol. 2012;43(10):1552–1558. doi: 10.1016/j.humpath.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 61.Wang P, Dong Q, Zhang C, et al. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene. 2013;32(25):3091–3100. doi: 10.1038/onc.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rohle D, Popovici-Muller J, Palaskas N, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang F, Travins J, DeLaBarre B, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340(6132):622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 64.Xu RF, Sun JP, Zhang SR, et al. KRAS and PIK3CA but not BRAF genes are frequently mutated in Chinese cholangiocarcinoma patients. Biomed Pharmacother. 2011;65(1):22–26. doi: 10.1016/j.biopha.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 65.Meng F, Henson R, Lang M, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangio-carcinoma cell lines. Gastroenterology. 2006;130(7):2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 66.Sheppard K, Kinross KM, Solomon B, Pearson RB, Phillips WA. Targeting PI3 kinase/AKT/mTOR signaling in cancer. Crit Rev Oncog. 2012;17(1):69–95. doi: 10.1615/critrevoncog.v17.i1.60. [DOI] [PubMed] [Google Scholar]
- 67.Liu N, Rowley BR, Bull CO, et al. BAY 80-6946 is a highly selective intravenous PI3K inhibitor with potent p110α and p110δ activities in tumor cell lines and xenograft models. Mol Cancer Ther. 2013;12(11):2319–2330. doi: 10.1158/1535-7163.MCT-12-0993-T. [DOI] [PubMed] [Google Scholar]
- 68.Wallin JJ, Edgar KA, Guan J, et al. GDC-0980 is a novel class I PI3K/mTOR kinase inhibitor with robust activity in cancer models driven by the PI3K pathway. Mol Cancer Ther. 2011;10(12):2426–2436. doi: 10.1158/1535-7163.MCT-11-0446. [DOI] [PubMed] [Google Scholar]
- 69.Costello BA, Borad MJ, Qi Y, et al. Phase I trial of everolimus, gemcitabine and cisplatin in patients with solid tumors. Invest New Drugs. 2014;32(4):710–716. doi: 10.1007/s10637-014-0096-3. [DOI] [PubMed] [Google Scholar]
- 70.Ma X, Ezzeldin HH, Diasio RB. Histone deacetylase inhibitors: current status and overview of recent clinical trials. Drugs. 2009;69(14):1911–1934. doi: 10.2165/11315680-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 71.Sirica AE. The role of cancer-associated myofibroblasts in intrahepatic cholangiocarcinoma. Nat Rev Gastroenterol Hepatol. 2012;9(1):44–54. doi: 10.1038/nrgastro.2011.222. [DOI] [PubMed] [Google Scholar]
- 72.DeClerck YA. Desmoplasia: a response or a niche? Cancer Discov. 2012;2(9):772–774. doi: 10.1158/2159-8290.CD-12-0348. [DOI] [PubMed] [Google Scholar]
- 73.Ling H, Roux E, Hempel D, et al. Transforming growth factor β neutralization ameliorates pre-existing hepatic fibrosis and reduces cholangiocarcinoma in thioacetamide-treated rats. PLoS ONE. 2013;8(1):e54499. doi: 10.1371/journal.pone.0054499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pinlaor S, Prakobwong S, Hiraku Y, Pinlaor P, Laothong U, Yongvanit P. Reduction of periductal fibrosis in liver fluke-infected hamsters after long-term curcumin treatment. Eur J Pharmacol. 2010;638(1-3):134–141. doi: 10.1016/j.ejphar.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 75.Mertens JC, Fingas CD, Christensen JD, et al. Therapeutic effects of deleting cancer-associated fibroblasts in cholangiocarcinoma. Cancer Res. 2013;73(2):897–907. doi: 10.1158/0008-5472.CAN-12-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Olive KP, Jacobetz MA, Davidson CJ, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324(5933):1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garber K. Stromal depletion goes on trial in pancreatic cancer. J Natl Cancer Inst. 2010;102(7):448–450. doi: 10.1093/jnci/djq113. [DOI] [PubMed] [Google Scholar]
- 78.Clapéron A, Mergey M, Aoudjehane L, et al. Hepatic myofibroblasts promote the progression of human cholangiocarcinoma through activation of epidermal growth factor receptor. Hepatology. 2013;58(6):2001–2011. doi: 10.1002/hep.26585. [DOI] [PubMed] [Google Scholar]
- 79.Fingas CD, Bronk SF, Werneburg NW, et al. Myofibroblast-derived PDGF-BB promotes Hedgehog survival signaling in cholangiocarcinoma cells. Hepatology. 2011;54(6):2076–2088. doi: 10.1002/hep.24588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fingas CD, Mertens JC, Razumilava N, Bronk SF, Sirica AE, Gores GJ. Targeting PDGFR-β in cholangiocarcinoma. Liver Int. 2012;32(3):400–409. doi: 10.1111/j.1478-3231.2011.02687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wiedmann MW, Mössner J. Molecular targeted therapy of biliary tract cancer—results of the first clinical studies. Curr Drug Targets. 2010;11(7):834–850. doi: 10.2174/138945010791320818. [DOI] [PubMed] [Google Scholar]
- 82.Ribas A. Tumor immunotherapy directed at PD-1. N Engl J Med. 2012;366(26):2517–2519. doi: 10.1056/NEJMe1205943. [DOI] [PubMed] [Google Scholar]