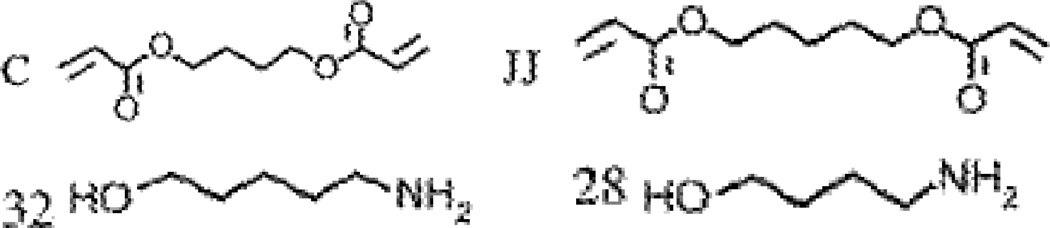Abstract
Gene therapy is an attractive treatment option for diseases of genetic origin, including several cancers and cardiovascular diseases. While viruses are effective vectors for delivering exogenous genes to cells, concerns related to insertional mutagenesis, immunogenicity, lack of tropism, decay and high production costs necessitate the discovery of non-viral methods. Significant efforts have been focused on cationic polymers as non-viral alternatives for gene delivery. Recent studies have employed combinatorial syntheses and parallel screening methods for enhancing the efficacy of gene delivery, biocompatibility of the delivery vehicle, and overcoming cellular level barriers as they relate to polymer-mediated transgene uptake, transport, transcription, and expression. This review summarizes and discusses recent advances in combinatorial syntheses and parallel screening of cationic polymer libraries for the discovery of efficient and safe gene delivery systems.
Keywords: parallel screening, polymer library, transfection, gene delivery, transgene expression, cytotoxicity
INTRODUCTION
Gene therapy is an attractive therapeutic option in order to correct disease at the genetic level by either replacing abnormal genes using exogenous DNA or transiently delivering DNA resulting in the expression of a therapeutically active protein [1–7]. Several strategies using both, viral and non-viral delivery vectors, have been widely employed in order to transfer exogenous genetic material to host cells. Viral vectors include genetically altered retroviruses, lentiviruses, adenoviruses and adeno-associated viruses and have traditionally demonstrated high efficiencies for transgene delivery and expression [8–11]. However, limitations related to immunogenicity, reduced efficacies due to neutralizing antibodies during repeated administration, safety, insertional mutagenesis into the host genome, inflammatory responses, and issues pertaining to scale up of production and purification with viral vectors, necessitate an increasing focus on effective and safe non-viral vectors.
Polymers are synthetic gene delivery vectors which possess the ability to deliver genetic material to target cells [3, 4, 12, 13]. Cationic polymers neutralize the negative charge on plasmid DNA resulting in the formation of nanoscale polymer-DNA complexes (polyplexes). However, a variety of complex biological barriers including those in the extracellular body fluid, extracellular matrix (ECM), and at the cellular level (cellular entry, escape from endo-lysosomal vesicles, cytoplasmic trafficking, DNA unpackaging, and nuclear translocation of the therapeutic DNA, transcription, and translation) limit the transgene expression efficacy associated with polymeric delivery vectors. While viruses have evolved over millions of years for delivering exogenous material to cells, polymeric vectors have only been recently explored for transgene delivery. This limitation associated with polymeric vectors, can be somewhat overcome by designing and synthesizing large sets of these compounds and screening them in parallel for transgene delivery. High-throughput screening techniques have been widely employed in the pharmaceutical industry for facilitating the rapid evaluation of small molecule drugs leading to the identification of novel therapeutic candidates [14–19]. Combinatorial chemistry and parallel screening techniques have recently evolved as attractive options for rapidly generating polymer libraries consisting of candidates that possess a wide range of physicochemical diversities. This review outlines the high-throughput screening and combinatorial approaches for the synthesis of polymers for transgene delivery; small-to-medium sized libraries and derivatives based on poly(ethylenimine), poly-β-amino esters, branched polyaminoesters, polymethacrylate, cyclodextrin, chitosan, dendrimers, polysaccharide-based polymers and polydisulfide amines, and miscellaneous polyamines will be discussed.
POLYETHYLENE IMINE (pEI)
Polyethylene imine or pEI is traditionally considered as the gold standard for polymer-mediated transgene gene delivery, since it was one of the earliest polymers identified to possess high transfection efficacies [20, 21]. This activity is attributed to the ‘proton sponge’ effect, in which, partially protonated polycations absorb protons transported into intracellular endocytic vesicles by the ATPase proton pump. This leads to an accompanying influx of chloride ions to the vesicles resulting in osmotic swelling and rupture of the endocytic vesicles [21]. The ability of pEI to destabilize lysosomal membranes enables pEI-DNA complexes (polyplexes) to escape subsequent degradation in acidic endolysosomal vesicles. Further trafficking through the cytoplasm and nuclear import of the delivered DNA result in high transgene expression in a variety of cell lines [22–26]. Different lengths, molecular weights, and morphologies (branchned and linear) of the polymer are commercially available. It has been demonstrated that low molecular weight (2kDa) pEI is less efficient for transgene expression than the branched high molecular weight (750kDa) pEI. However, polymer cytotoxicity was also found to increase with increasing molecular weights[27]. The dose-dependent toxicity associated with pEI limits the application of the polymer in vivo due to the high concentrations that are typically required for expression of therapeutic transgenes.
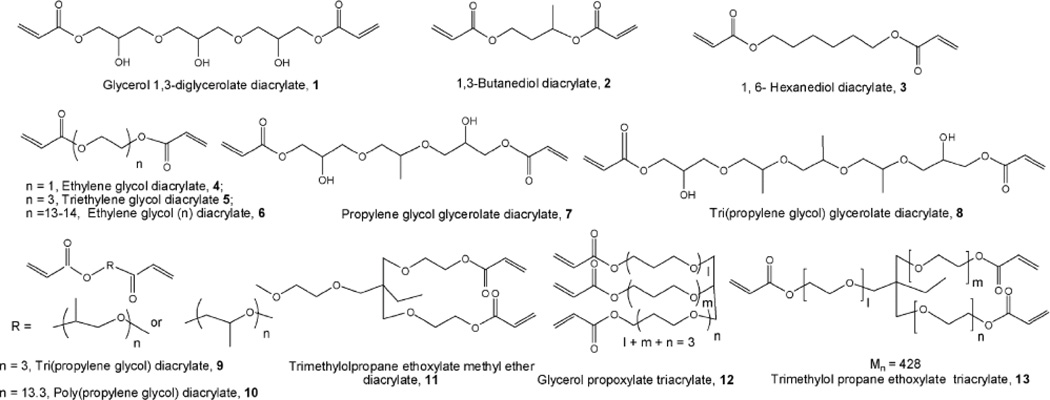
Combinatorial approaches have been employed to optimize the transgene expression efficacy and toxicity of pEI for transgene delivery. Thomas et al. developed a library of 144 pEI polymer derivatives by crosslinking either 423Da linear pEI or a pEI polymer mixture of 423Da linear and 1.8kDa branched pEIs (1:1 w/w ratio) with twenty four bi- and oligoacrylate esters (Figure 1) at three pEI:acrylate molar ratios of 10.5:1, 21:1 and 42:1 [28]. Cross-linked pEI derivatives were employed to transfect COS-7 monkey kidney cells with plasmid DNA (pDNA) containing a transgene expression β-galactosidase at nitrogen:phosphate (N:P) ratios varying from 10 to 20. The highest luciferase expression was found with 423Da pEI cross-linked with tricyclo-5.2.1.0-decane-dimethanol diacrylate at an N:P ratio of 20:1 showing 3670-fold enhancement than the non-crosslinked pEI. The relative transfection efficiency of this polymer was moderately (two-fold) higher than 22kDa linear pEI; molecular weights of the cross-linked polymers were not reported in the study. Eight polymers, derived from crosslinking pEI polymer mixture (1:1 weight ratio of 423Da linear pEI and 1.8kDa branched pEI) and diacrylates, demonstrated up to 850-fold enhancement in transgene expression compared to their parent polymer mixture at nitrogen:phosphate (N:P) ratio of 20:1. Retroorbital administration of the polyplexes, generated from the five most effective polymers and DNA containing luciferase gene, into 6–8 week old Black Swiss male mice resulted in preferential accumulation of the polyplexes in the lungs (1–5×105 RLU/mg) followed by spleen, liver, heart and kidney, indicating the potential use of these polymers in therapeutic gene delivery for lung cancer. Van Vliet et al. investigated functional modalities of branched 25kDa pEI (pEI-25) by functionalizing the primary, secondary or tertiary amines of the polymer by methylation, benzylation and n-dodecylations [29]. A library of 435 polymers was evaluated in a high-throughput microwell format for DNA binding activity, toxicity and transgene expression efficacy; CHO-K1 (chinese hamster ovary) cells were transfected with an enhanced green fluorescent protein (EGFP) reporter gene in order to evaluate transgene expression. Three of the most efficacious pEI derivatives resulted in 19–28% enhanced green fluorescent protein positive (EGFP+) cells at an N:P ratio of 10:1 with negligible toxicities (≥97% cell viability), while jet pEI, Lipofectamine and unmodified pEI-25 expressed ∼30, 10–20 and 0.3% EGFP+ cells, respectively.
Figure 1.
Chemical structures of several bi- and oligoacrylate esters used to crosslink pEI by Thomas et al. [28] Reprinted from Ref. [28] with permission from Elsevier.
POLY-β-AMINO ESTERS (PBAEs)
PBAEs are promising gene delivery vectors because of the presence of hydrolytically degradable ester groups in the building blocks of the polymers, low cytotoxicities and an enormous structural diversity [30, 31]. Anderson et al. synthesized a library of 2350 structurally different PBAEs in a semi-automated fashion by employing 94 different amine monomers and 25 diacrylate monomers [32]. Parallel screening of the polymer library for luciferase expression following transgene delivery to COS-7 cells resulted in forty six polymers that showed luciferase expression levels as high as or better than that demonstrated by pEI-25. In subsequent studies, 486 polymers were re-synthesized at a larger scale than the previous micro-scale synthesis in order to examine the effects of molecular weights of the polymers on their transfection efficiencies [33]. Transfection of COS-7 cells indicated that polymers with molecular weights between 10–61kDa showed the highest transfection levels.
Three polymers from the library, C32, JJ28 and C28, derived from reactions between amino alcohols (5-amino-1-pentanol (32); 4-amino-1-butanol (28)) and diacrylate monomers (1,4-butanediol diacrylate (C); 1,5-pentanediol diacrylate (JJ)) demonstrated the highest transfection efficacies (Figure 2) and were found to be structurally similar while differing from each other by only one carbon in their repeating units. Green et al. studied the effect of several parameters including PBAE polymer type, polymer molecular weights, amount of DNA loading, and size, ζ-potential, and temporal stability of polymer:DNA complexes on the transfection efficacy in human umbilical vein endothelial cells (HUVECs) [34]. It was found that C32, JJ32 and C28 polymers formed complexes with pDNA (encoding the GFP protein) in the size range of ∼190–230nm with ζ-potentials ranging from −6 to −14mV in serum-containing media. GFP expression was observed in 25–47% cells (GFP+ cells) after 48h of transfection [34]. The best performing polymer (C32) was then coated with Arg-Gly-Asp (RGD) peptide through electrostatic polymer-ligand self-assembly in order to target integrin receptors that are overexpressed in many tumor cells [35]. The C32-RGD ligand delivered GFP gene demonstrated approximately 50% GFP+ cells in HUVECs in presence of serum. Following these in vitro studies, the C32 polymer was then used to deliver a suicide gene encoding diphtheria toxin into prostate cancer xenografts in 8-week old nu/nu male mice [36] and 8–16 week old TRAMP mice [37]. Intratumoral injections of the C32-diphtheria toxin polyplexes suppressed tumor growth in 40% of the nu/nu mice [36], whereas delivery to intraprostate tumors induced apoptosis in 80% of the tumor cells at the site of injection in the TRAMP mice [37] .
Figure 2.
Diacrylate esters and amine monomers to synthesize the lead poly-beta-amino esters, C32, JJ28 and C28, as demonstrated by Anderson et al. [33]. Reprinted from Ref. [33] by the permission of Nature Publishing Group.
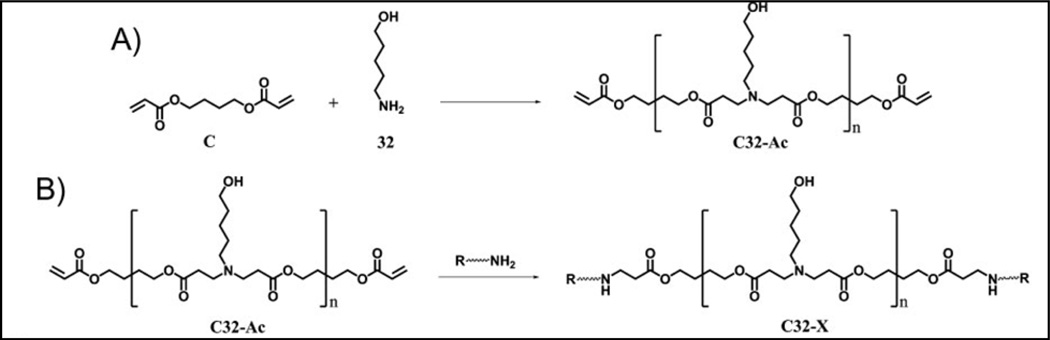
Further studies [38] resulted in the development of a library of end-modified C32 polymers using 36 different amine group containing small molecules as end-capping reagents (Figure 3) [38, 39]. End-group modifications of the C32 polymer with amines enhanced its DNA binding affinities; highest efficacies were seen with moieties that contained a three-carbon spacer between amine end groups (>N-CH2-CH2-CH2-NH2) [38, 39]. Five of these amine modified C32 polymers (named C32–102, −116, −86, −123 and −87) at a polymer:DNA weight ratio of 100:1 exhibited higher luciferase expression levels (1.2×105RLU) than those of both unmodified C32 (9×104RLU) and pEI-25 (5×104RLU). One polymer, C32–108, demonstrated same levels of luciferase expression as unmodified C32 at much lower polymer:DNA ratio (20:1 w/w) than that (100:1) required for the unmodified C32 [38]. Intraperitoneal delivery of polyplexes using nine selected polymers into six-week-old FVB/J male mice indicated that four polymers (C32–103, −116, −117 and −122) resulted in 4 to 12-fold and 15 to 42-fold higher luciferase expression (5×107photons/sec) than that demonstrated by unmodified C32 and pEI-25 polymers, respectively. Structural modifications of the polymers also altered their biodistribution profiles; ten-fold higher accumulation of these polymers was observed in the lungs compared to that seen with unmodified C32. In a different study, a library of 60 polymer derivatives was further generated by modifying the di-amine end-groups of C32 polymer [39]. Polymers with variations in their terminal amines were evaluated with HeLa (cervical cancer cell line), HepG2 (hepatocellular carcinoma cell line) and DC 2.4 (bone marrow derived dendritic cells) cells, in addition to HUVECs, hMSC (human mesenchymal stem cells) and COS-7 cells, in presence of serum. Five lead polymers (named C32–117, −221, −213, −227 and −228) showed 75–90% GFP+ cells in these six different cell types. The end-group modified C32–117 polymer demonstrated 70% GFP+ cells in HUVECs using 100:1 polymer:DNA weight ratio while pEI-25 exhibited ∼1.5%, and adenoviral MOI titers of 100 and 500 expressed approximately 75 and 90% GFP+ cells, respectively [40]. Intraperitoneal injection of polyplexes using the modified polymer C32–117 into MISIIR/TAg female transgenic mice containing ovarian tumors exhibited two-orders of magnitude higher luciferase expression (107 photons/sec) than those (105 photons/sec) in mice injected with polyplexes using the unmodified C32 polymer. Plasmid DNA coding for luciferase protein were used in these studies[40]. The C32–117 polymer also demonstrated moderate levels of GFP expression (approximately 22% GFP+ cells) in undifferentiated human embryonic stem cells (hESC), indicating that transfection efficiency of the terminal group modified C32 polymer was also dependent on the cell line employed [41]
Figure 3.
A reaction schematic of the synthesis of (a) C32 polymer and (b) amine-capping C32 polymer [38, 39]. Reprinted by permission of Nature Publishing Group.
BRANCHED POLY(AMINO ESTER) (PAE)
The lead PBAE polymers described in the previous section are linear PAEs in which tertiary amines remain in the backbones of the polymer structures with hydroxyl ion side chains and primary amine end groups [30, 31, 42]. Branched PAEs containing tertiary amines in the core and primary/ secondary amines at the periphery have also been investigated as gene delivery carriers [43, 44]. The tertiary amines within the branched PAEs provide endosome-buffering activity similar to that with pEI and the primary amines with hydroxyl end-groups increase binding capacities with DNA or any ligand [44]. However, insoluble polymer formation during the synthesis of the branched PAEs and their short half-life of less than thirty minutes in vivo can be of significant concern. Kim et al. employed a library-like approach to identify biodegradable cross-linked PAE vectors at different monomer compositions, temperature gradients and optimum pressure that avoided gel formation of the polymers during the polymerization reactions resulting in excellent solubility in both aqueous condition and organic solvents such as DMF, DMSO etc. [44] . The network-type polymers, nt-PAE were synthesized from two types of monomers containing methyl ester groups in one type and hydroxyl groups in another. The synthesis temperature was increased from 80 to 120°C gradually to avoid any loss of monomers and then maintained at a pressure of 70 cm Hg to obtain average molecular weights of 10–25kDa. Lysine and aminohexanoic acid was used to conjugate primary amines to the hydroxyl groups of the polymers in order to enhance the DNA binding efficacy of the polymers. The amine concentration of the polymer that demonstrated the highest transfection efficacy was four times lower than in pEI-25, indicating lower toxicity of the lead nt-PAE. Concomitantly, the N:P ratios (18:1 and 36:1) at which nt-PAE demonstrated the highest transfection efficiencies were indeed higher than that in case of pEI-25(8:1).
Jere et al. evaluated a mini polymer library of sixteen PAE polymers, composed of PEG and aminosilane. Six different polymer:DNA weight ratios were tested for each of these polymers for transgene expression efficacy in 293T and HeLa cells [45]. The polymers were synthesized using Michael’s addition reaction between amine-containing γ-aminopropyl-triethoxysilane (APES) and polyethylene glycol diacrylate (PEGDA; 258Da) at different mole ratios in which the APES was linked to PEGDA by hydrolysable ester linkages. The lead polymers named sequentially from R117 to R121 contained APES:PEGDA mole ratios from 4:1 to 6:1, exhibited higher luciferase transgene expression (in the range of 1×10505×108 and 1×105–5×106RLU/mg in absence and presence of serum, respectively) but lower cytotoxicities (>70% cell viability) than pEI-25 (1×104–2×106 and 1×104–7×104RLU/mg in absence and presence of serum, respectively with ≤50% cell viability) and lipofectamine standards at polymer:DNA weight ratios of 40, 60 and 90.
POLYMETHACRYLATES
Polymethacrylates are a class of materials that are used in several commercial applications due to their wide range of properties. Polymethacrylate derivatives have been commonly used in biomedical devices utilized for restorative dental composites, bone cement, and contact lenses [46]. One feature that allows the wide range of properties in polymethacrylates is the fact that a single methacrylic repeat unit is capable of having a wide range of functional pendant ester groups incorporated to them. In addition, the ease of synthesis of either homo-, co-, and terpolymers via free radical polymerization and the combining of repeat units with different functionalities, further allows for changes in material properties [46].
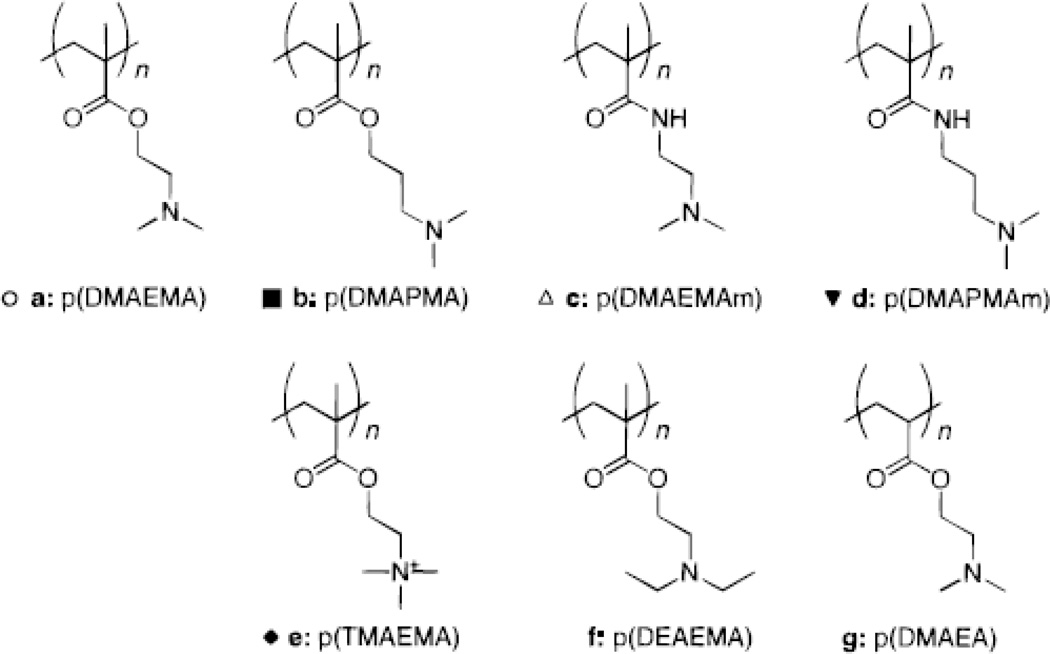
Several studies have explored small sets of polymethacrylate derivatives and their ability to form polyplexes with DNA, for transgene delivery and expression [47]. Poly(2-(dimethylamino)ethyl methacrylate), or p(DMAEMA), has been demonstrated to bind and form polyplexes with DNA, resulting in transgene expression [48]. In a study carried out by van de Wetering et al. a small library of methacrylate/methacrylide polymers with structures similar to that of p(DMAEMA) was synthesized in order to further test transfection efficacy based on structural differences [47]. In this study, either side chains or functional groups of the p(DMAEMA) monomer were replaced with different side chains or functional groups to form six synthetic monomers (Figure 4). Polymers were then generated via radical polymerization and used for transfection in vitro. It was found that even with significant differences in structure, the polymers resulted in the formation of polyplexes with similar particle sizes as well as ζ-potentials compared to those with p(DMAEMA). It was also observed that the p(DMAEMA) analogues were 3–9 fold less cytotoxic than p(DMAEMA) itself. However, it was found that the transfection efficacies of the polymer analogues were 5–400 fold lower than that of p(DMAEMA). Thus, in this study, it was concluded that though the difference in structure may not affect the polyplex formation, it can still greatly affect both cytotoxicity as well as transfection potential.
Figure 4.
General structures of some of cationic methacrylate/methacrylamide polymers investigated by van de Wetering et al. [47]. Reprinted by permission of American Chemical Society.
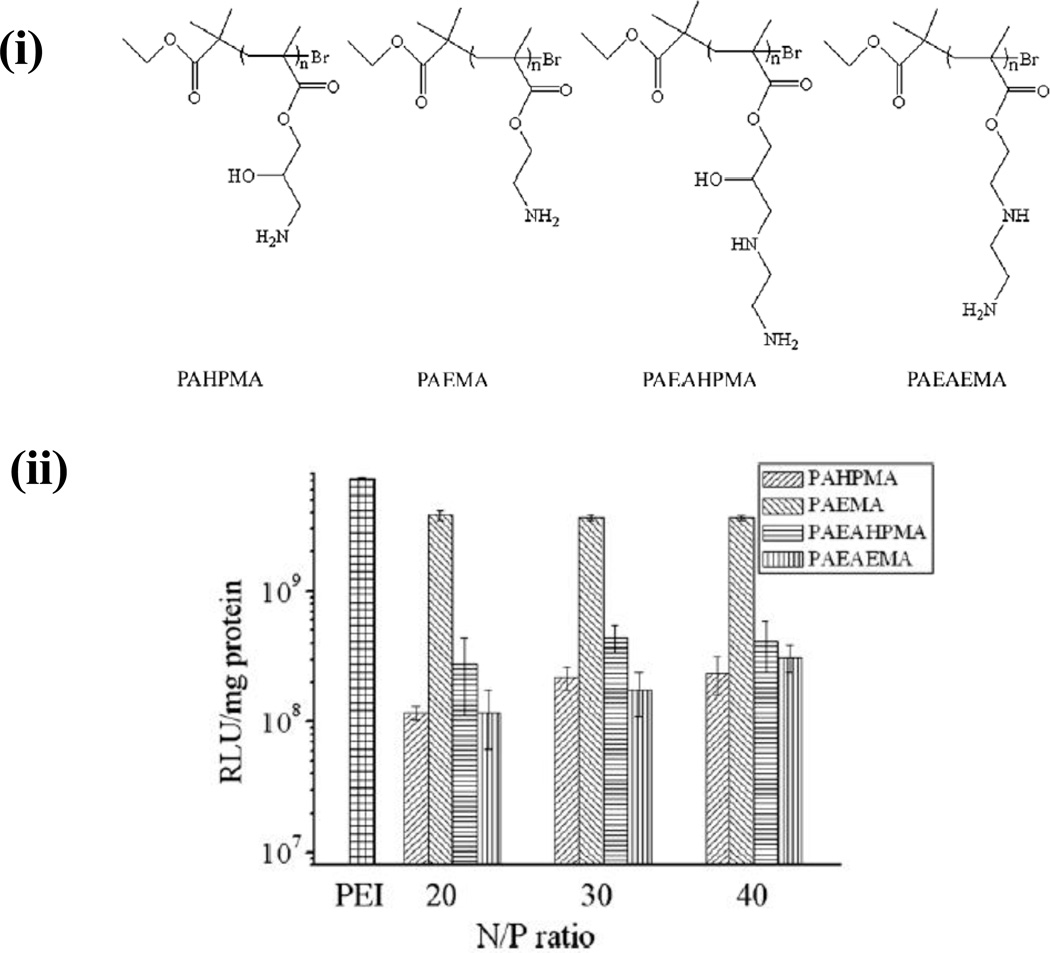
It has been reported that the inclusion of hydroxyl groups in polycations results in a decrease in cytotoxicity as well as an increase in transgene expression (55). Ma et al. synthesized a small library of four polymethacrylate polymers, poly(aminoethyl methacrylate) (PAEMA), poly(3-amino-2-hydroxypropyl methacrylate) (PAHPMA), poly(2-(2-aminoethylamino ethyl methacrylate) (PAEAEMA), and Poly(3(-2-aminoethlamino) 2-hydroxylpropyl methacrylate (PAEAHPMA) (Figure 5 (i)) in order to evaluate the effect of hydroxyl groups on DNA binding, transgene delivery, and cytotoxicities of polymeric vectors [49]. Atom transfer radical polymerization (ATRP) was used to polymerize glycidyl methacrylate (GMA), N-tert-butoxycarbonyl-aminoethyl methacrylate (Boc-AEMA) and, N,N’-di-(tert-butoxycarbonyl)-2-(2-aminoethylamino)ethyl methacrylate (Boc-AEAEMA) in order to obtain polymers with similar degrees of polymerization. The polymers were further characterized with nuclear magnetic resonance (NMR), Fourier transform infrared (FT-IR) spectronomy and gel permeation chromotography (GPC). Heparin displacement assay was employed to demonstrate that hydroxyl groups on the side chains of PAHPMA may facilitate DNA binding; however, data from a gel retardation study indicated that when hydroxylated polymers were used there was no apparent difference in DNA binding compared to polymers without hydroxyl groups. Polymers with hydroxyl groups on the side chains, PAHPMA and PAEAHPMA, exhibited both, lower zeta potentials as well as lower cytotoxicities when compared to similar polymers (PAEMA and PAEAEMA) that did not possess these hydroxyl groups. It was found that PAHPMA polyplexes possessed lower transfection efficacies than PAEMA polyplexes in the absence of serum, whereas PAEAHPMA and PAEAEMA polyplexes possessed similar transfection efficacies (Figure 5 (ii)). However, in the presence of serum, both and PAHPMA and PAEAHPMA retained similar transfection efficacies as that observed in the absence of serum while PAEMA and PAEAEMA showed slightly lower efficacies. Thus, it was found that introduction of hydroxyl groups may result in lower gene transfection efficacies due to the formation of a polyplex with a lower zeta potential and stronger DNA binding via of hydrogen bonding. Due to lower zeta potentials, a shielding effect may occur, in which hydroxyl groups can shield polyplexes from the serum proteins, leading to reduced interactions of the polyplexes with molecules in the medium. Thus, this shielding effect may allow for maintained transfection efficacies due to limited interactions between serum proteins and polyplexes[49].
Figure 5.
(i) Chemical structures of methacrylate containing polymers [49]; (ii) Comparison of transfection efficiency among PAHPMA, PAEMA, PAEAHPMA, PAEAEMA and pEI-25 polymers in 293T cells. The polyplexes were formed at N:P ratio of 20,30 and 40 for the methacrylate based polymers and at an N/P=10 for pEI-25. Transfection was carried out in serum-free DMEM medium [49]. Copyright © 2010, Elsevier; the figure was reprinted with the permission from Elsevier.
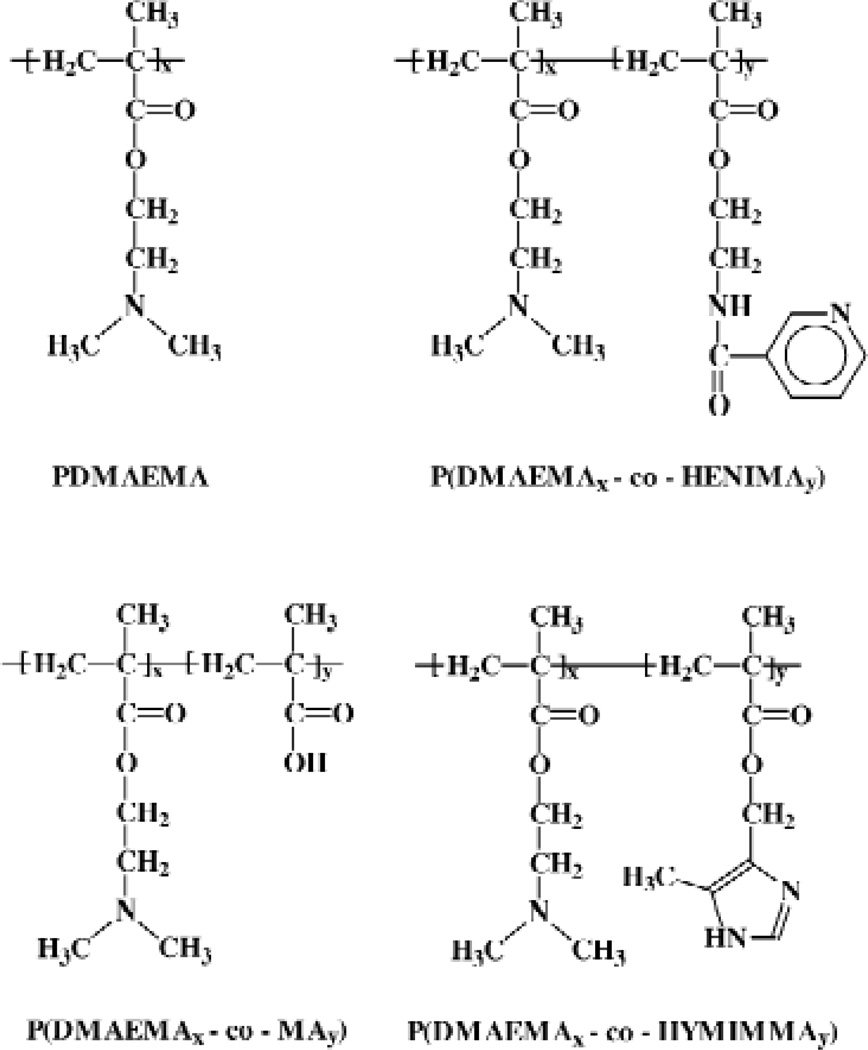
Dubruel et al. evaluated the physicochemical and biological properties of cationic polymethalcrylates possessing functional side groups of varied pKa values to be used as vectors for gene delivery. These side groups consisted of tertiary amines, pyridine and, imidazole groups in addition to methacrylic acid acting as an acid group These groups were chosen in order to mirror the buffering / protonation states of pEI which allow it to act as a “proton sponge” during transgene delivery [50]. A library of polymers was synthesized consisting of PDMAEMA at molecular weights of 93, 110, 166 and 201kDa, P(DMAEMA0.82-co-MA0.18) at 108 kDa, P(DMAEMA0.65-co-MA0.35) at 316 kDa, P(DMAEMA0.9-co-HENIMA0.1) at 140 kDa, P(DMAEMA0.89-co-HENIMA0.11) at 164 kDa, P(DMAEMA0.94-co-HYMIMMA0.06) at 99.5 kDa, P(DMAEMA0.88-co-HYMIMMA0.12) at 72 kDa, and P(DMAEMA0.81-co-HYMIMMA0.19) at 54 kDa. Synthesis was carried out via radical polymerization of a set of co-monomers (Figure 6) [51]. Minor modifications in either the chemical composition or the molecular weight could cause major differences on the DNA condensation abilities of the polymers. Of the polymers tested, only the tertiary amine-containing PDMAEMA (molecular weight of 201kDa) possessed transfection efficacies similar to that of pEI-25 and remained non-toxic to the monkey kidney fibroblast Cos-1 cell line. Polymers with imidazole and pyridine groups or the acid groups were unable to transfect the Cos-1 cells, although they were less cytotoxic than PDMAEMA. Thus, the tertiary amine-containing polymer showed successful transfection when compared to those with the other side groups[50]. A later study demonstrated polymers with imidazole and acid group side chains were taken up by cells via endocytosis, but were slowly released from the acidic vesicles. Thus, not only the buffering properties of the different side chain on the polymethacrylates, but also the kinetics of endosomal release may influence successful transgene delivery and expression[52].
Figure 6.
The chemical structures of polymethacrylates used for gene delivery by Dubruel et al [50]. Reproduced with permission from Elsevier.
CYCLODEXTRIN (CD)-BASED POLYMERS
Cyclodextrins (CD) are natural cyclic oligosaccharides composed of 6, 7 or 8 glucose units (called α-, β-, or γ-CD, respectively) and have been explored for transgene delivery [53]. However, most studies with these polymers have focused on the development of relatively small sets of derivatives, rather than on library / combinatorial approaches. Davis and co-workers[54] developed polymers based on β-cyclodextrin grafted with linear and branched poly(ethyleneimines) for transgene delivery both, in vitro and in vivo. The transfection efficiency of these agents was dependent on the degree of β-cyclodextrin grafting; increased degree of grafting resulting in reduced transfection efficacies. It is possible that the pKa of amines in pEI was influenced by CD grafting resulting in reduced protonation and buffering effect in the endosomal compartment. However, increased CD grafting resulted in reduced cytotoxicities of the resulting polymers.
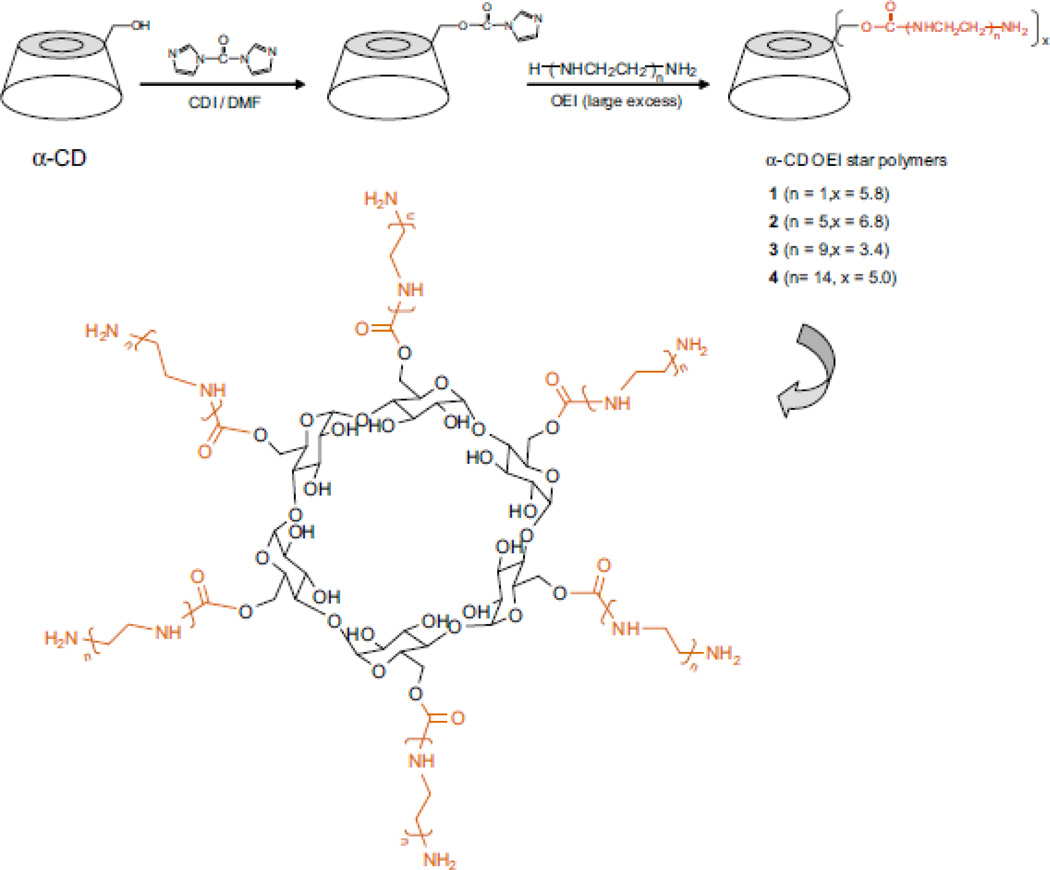
Gonzalez et al.[53] developed three β-cyclodextrin-based linear cationic polymers by condensation of two comonomers (A and B) namely, diamino CD monomer (A) and diimidate comonomer (B). The transfection efficiency of the β-CD-based polymers was comparable to that of pEI-25 and no mortalities in mice were reported after single intravenous (i.v.) and intraperitoneal (i.p.) doses as high as 200 mg/kg. Yang et al. [55] synthesized a small library of four cationic α-CD-oligoethyleneimine (OEI) star polymers by conjugating multiple linear or branched OEI arms onto an α-cyclodextrin core (Figure 7). The molecular structures of the α-CD-OEI star polymers formed complexes with plasmid DNA at N/P ratios from 8–30 where the sizes of the complexes ranged from 100–200 nm in diameter. The polymers showed excellent gene transfection efficiency in HEK293 and COS-7 cells but lower cytotoxicity than that of pEI-25. The LD50 values (50% cell growth inhibition) of the α-CD-OEI polymers were in the range of 88–560µg/ml in HEK293 and COS-7 cells, while the value for pEI-25 was only 12µg/ml in both cell lines. The OEI chain lengths had significant effect on transfection efficacy of the α-CD-OEI polymers by enhancing gene expression with the increase in OEI arm chain length. Increased OEI arm chain length increased the number of primary amines in the star polymers resulting in enhanced binding ability with plasmid DNA and stability of the polyplexes that led to enhanced transfection efficiency. Further studies by Yang et al. [56] focused on the development of four supramolecular cationic polyrotaxanes to enhance transgene delivery. In this study, eight OEI-grafted α-CD rings were threaded over reverse pluronic poly(propylene oxide)-poly(ethylene oxide)-poly(propylene oxide) (PPO-PEO-PPO) amphiphilic triblock copolymer that enabled the α-CD rings to rotate freely along the polymeric chains, enhanced interactions between DNA and polyrotaxanes, and thus led to high transfection efficiencies. These polymers exhibited lower cytotoxicity compared to those of pEI-25 as can be predicted from their LD50 values. The LD50 values of the cationic polyrotaxanes were in between 55 and 155µg/ml, while it was 25µg/ml for pEI-25. Cationic polyrotaxanes with longer OEI chains and at an N/P ratio of 30 exhibited higher gene transfection efficiencies but lower cytotoxicity than pEI-25 in HEK293 cells.
Figure 7.
Synthesis procedures and the structures of α-CD-oligoethyleneimine (OEI) star polymers as described by Yang et al.[55]. The hydroxyl groups of six glucose subunits of α-CD were activated using 1,1’-carbonyldiimidazole (CDI) following the reactions with multiple OEI of different lengths to produce the α-CD-OEI polymers. Adopted from Ref. [55] by permission from Elsevier.
Li et al.[57] developed four cationic β-cyclodextrin-based star-shaped polymers using ATRP reactions. These polymers, containing primary amines, tertiary amines and quaternary ammonium groups, condensed pCMV-Luc plasmid DNA to 80–180 nm sized particles. The polymers with primary and tertiary amino groups exhibited higher cell transfection efficacies in CHSE-214 (chinook salmon embryo) cells than the polymers containing quaternary ammonium groups. Srinivasachari et al. [58] developed five polycationic α-cyclodextrin “click clusters” by linking a per-azido-α-cyclodextrin core moiety to oligoethyleneamine dendrons. These click clusters formed stable complexes with pDNA at N/P ratio 5. Sizes of the polyplexes ranged from 80–130 nm and their zeta potentials ranged from 0 to 25mV. Transfection efficiencies of the polymers depended on the number of oligoethyleneamines in the cluster. Polymers with higher number of oligoethyleneamines showed higher gene transfection efficiencies, presumably due to increased number of amines in the cluster. These polymers were able to effectively deliver Cy5-labeled pDNA and pDNA encoding the luciferase reporter protein into HeLa and H9c2 cells with minimal toxicity (>75% cell viability without any damage in the cell membrane).
Cryan et al. [59] synthesized seven polycationic cyclodextrins with pyridylamino, alkylimidazole, methoxyethylamino groups by the modification of the hydroxyl group attached to the 6th carbon atom of the glucose unit in the cyclodextrins. Among the synthesized polycationic derivatives, heptakispyridylamino functionalized cyclodextrin exhibited 4000-fold higher transfection levels than the uncomplexed (naked) DNA. Pun et al. [60] developed seven β-CD-linear (lpEI; 25kDa) and branched pEI (bpEI; 25kDa) derivatives with varying degree of CD grafting onto pEI polymers that were further modified by PEGylation using admantane-PEG (AD-PEG)moieties in order to increase the stability of polymer:DNA complexes in 150mM Salt concentration. The CD-grafted lpEI (CD-lpEI) and bPEI (CD-bpEI) polyplexes exhibited approximately 25 and 15% EGFP expressing cells in in vitro PC3 cells while the expression levels were ∼15, 15, 10 and 2% using the ungrafted lpEI-25, bpEI, and CD-lpEI with AD-PEG (CD-lpEI+AD-PEG) and (CD-bpEI+AD-PEG), respectively. Taking into account the advantages of reduced cytotoxicity, polyplex stability in physiological salt concentration and comparable transfection efficiency of the (CD-lpEI+AD-PEG) polymers with that of CD-lpEI, polyplexes of (CD-lpEI+AD-PEG) formulated with 120µg pDNA were administered via tail vein injection into female Balb/c mice containing PC3 xenografts. The polyplexes accumulated mostly in the liver and also demonstrated detectable gene expression in the organ.
Reineke et al. [61] synthesized seven amidine-based polycations containing the monomers of hexamethylenediamine, D-trehalose and β-cyclodextrin to study the effects of the carbohydrate size (trehalose vs. β-CD) and its distance from the cationic amidine groups on gene transfection efficiencies and cytotoxicities. Among the polyamidines, the hexamethylenediamine derivative (AP1), which did not have any carbohydrate moiety, demonstrated the lowest transfection efficiency with a concomitant high degree of cell death (IC50=6.6µM) in BHK-21 (Syrian hamster kidney fibroblast) cells. Addition of carbohydrate moieties to the polycation backbone not only improved gene expression levels (106–108 RLU/mg) but decreased the cytotoxicities (IC50=23µM–1.1mM) drastically. Decreasing the length of DNA binding centers (cationic groups) from the carbohydrate moieties resulted in reduced toxicities of the polymers. Polymers containing β-CD showed the highest luciferase expression with minimal toxicities in BHK-21 cells.
CHITOSAN (CS)
Chitosan, a naturally occurring cationic polysaccharide is an attractive candidate for gene delivery because of its several advantages, such as biocompatibility, biodegradability and low toxicity [63, 64]. Mumper et al.[65] introduced chitosan as a non-viral vector for gene delivery. The primary amine groups of chitosan can readily form complexes with negatively charged nucleotides in plasmid DNA by means of electrostatic interactions. Combinatorial-based approaches have not been extensively investigated with chitosan; most studies have focused on a limited set of derivatives. The binding affinities of chitosan derivatives for DNA are dependent on several factors, including molecular weights, degree of deacetylation, N/P ratio, and plasmid concentration [66–69]. Although chitosan possesses several inherent advantages including biocompatibility, biodegradability and low toxicity, properties like poor solubility, low transfection efficiencies, and low cell specificities limit their potential as gene delivery carriers. To improve the solubility and transfection efficiency of chitosan, Zhang et al.[70] developed PEGylated chitosan that exhibited higher transfection efficiency than the CS-DNA complexes in HepG2 (hepatocellular carcinoma) cells both in vitro and in vivo.
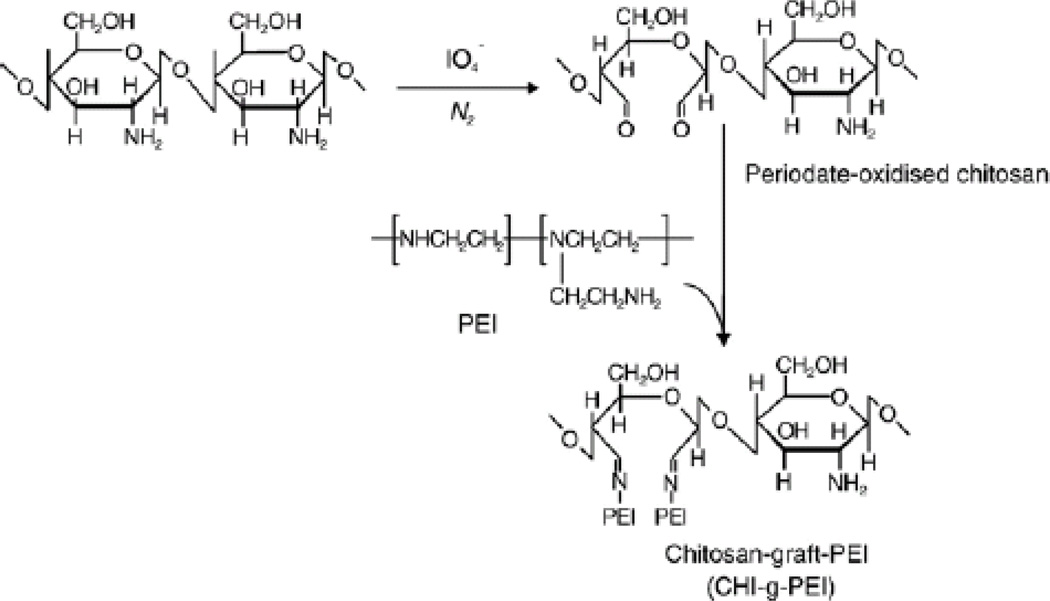
Jiang et al.[62] prepared chitosan grafted-polyethylenimine (CS-g-pEI) copolymers involving an imine reaction between the ketones of periodate-oxidized chitosan and amines of pEI-25 polymers (Figure 8). These copolymers exhibited low cytotoxicity compared to the pEI-25. The CS-g-pEI/DNA complex showed higher transfection efficiency than pEI-25 in HeLa, 293T and HepG2 cell lines. Lu et al.[71] developed a small library of twelve N-maleated chitosan grafted oligoamine (NMC-g-OEI) conjugates by altering the oligoamine side chain and molecular weights. The grafted polymers were synthesized by the Michael addition of N-maleated chitosan with diethylenetriamine (DETA), triethylenetetramine (TETA), tetraethylenepentamine (TEPA) and lpEI. At conjugate/DNA weight ratios from 5–11, the polymers condensed plasmid DNA in a size range of 200–500nm. Transfection studies using the EGFP or luciferase reporter genes showed that NMC(10kDa)-g-pEI (423Da) had higher transfection efficiencies than the unconjugated chitosan and similar efficacy (luciferase expression levels 108-109 RLU/mg) but lower cytotoxicity (∼85% cell viability) than that of pEI-25 (∼70% cell viability) in presence of serum (10%) in 293T and HeLa cells.
Figure 8.
Schematic representation of chitosan grafted pEI copolymer (CS-g-pEI) as proposed by Jiang et al. [62]. The reaction was carried out in two steps in which periodate-oxidized chitosan was prepared in the first step followed by an imine reaction with an amine group of pEI. Adopted from Ref. [62] with permission from Elsevier.
DENDRIMERS
Dendrimers are synthetic, hydrophilic branched polymers with positively charged terminal groups, typically characterized by spherical symmetry [72]. The positively charged groups can bind DNA or antisense oligonucleotides [73, 74] like many other cationic polymers, and are termed as dendriplexes similar to lipoplexes and polyplexes [72]. Dendrimer derivatives have been generated in order to help them overcome cellular obstacles including intracellular internalization through nanoscale hole formation in plasma membranes as induced by cationic dendrimers [75, 76]. Russ et al. synthesized twelve cationic dendrimers containing branched 800 Da oligoethyleneimine (OEI) in the core linked by oligoamines in the surface of the polymers [77]. The dendrimers exhibited lower toxicites and similar levels of reporter gene expression compared to 22kDa linear and 25kDa branched pEI in Neuro2A neuroblastoma cells. The results were consistent in male A/J mice studies following intravenous tail-vein injection; significantly lower accumulation (15000-fold) of the dendriplexes was observed in the lung compared to 22kDa linear pEI. A small set of seven dendrimers was built for gene delivery based on proline branching units by Sanclimens et al. [78]. Second-generation dendrimers were synthesized using two imidazoline-2-carboxylic acid groups and surface-functionalized with PEG. One candidate dendrimer containing arginine residues at its C-terminal surface showed appreciable DNA binding ability, transport of the dendriplexes, accumulation inside cell nucleus and high GFP expression in epithelial HeLa cells.
POLYSACCHARIDE-BASED POLYMERS AND POLYDISULFIDE AMINES
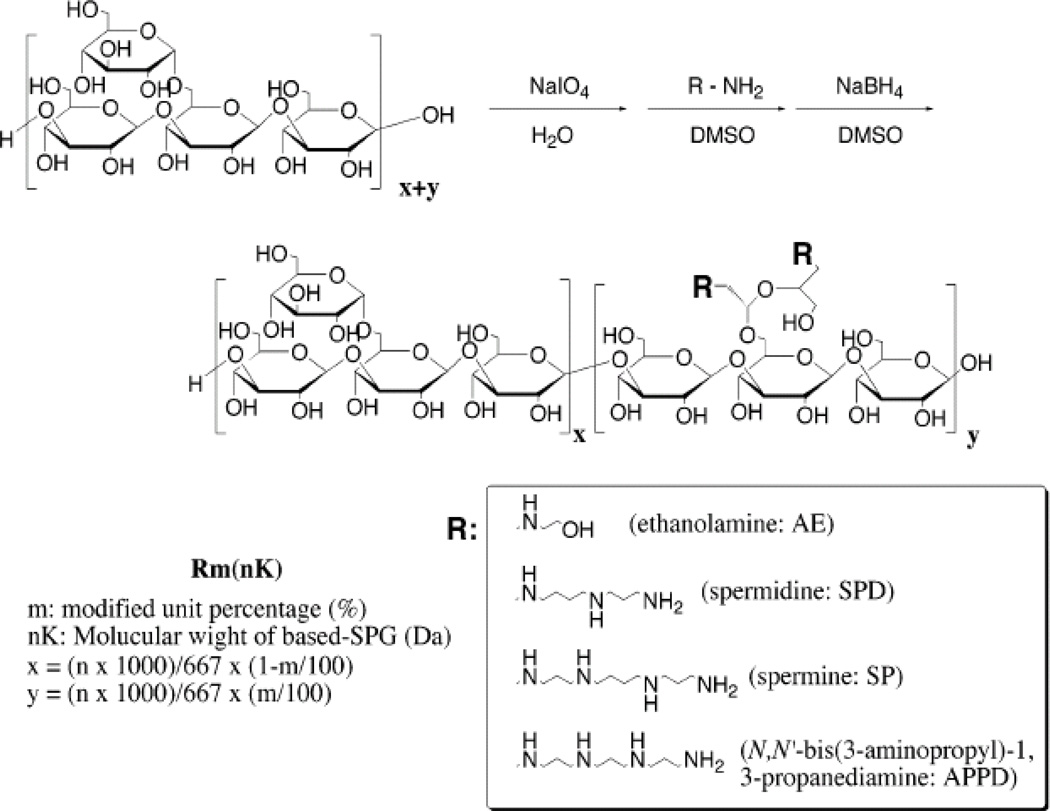
Polysaccharide-based cationic polymers are promising gene delivery polymers due to their high aqueous solubilities and low immunogenicities [79, 80]. Schizophyllan (SPG), a fungus produced extracellular polysaccharide, is a β-(1,3)-glucan binding with glucose side chains via β-(1,6)-glycoside. Schizophyllan has been used as an adjunctive gynecological cancer treatment agent [81, 82] and its oligoamine derivatives have also been investigated for gene delivery by Nagasaki et al. [83]. In 2004, Nagasaki et al. developed a set of eleven schizophyllan-based oligoamine polymers; the oligoamines included ethanolamine (AE), spermidine (SPD), spermine (SP), and N,N’-bis(3-aminopropyl)-1,3-propanediamine (APPD) (Figure 9). The oligoamines were introduced only at the side chain by periodate oxidation and reductive amination. Amine-conjugated schizophyllan derivatives demonstrated a maximal 4.5 fold higher transfection efficiency in vitro than amine-conjugated dextran derivatives (∼ 9×105 RLU/mg), and was suggested to be best suited for long-term expression based on the slow degrading characteristic of the β-(1,3)-glucan. Among all schizophyllan derivatives at the molecular weight of 34kDa, schizophyllan conjugated with APPD oligoamine has the highest in vitro transfection efficacy (∼ 5×105RLU/mg), higher than that of pEI-25 (∼ 1×105RLU/mg). However, APPD-conjugated schizophyllan derivative (34kDa) was more toxic (∼ 70% cell viabilty) than pEI-25 (100% viable cells) under similar conditions. Higher molecular weight schizophyllan derivatives (80kDa) demonstrated higher transfection efficacies (∼ 106RLU/mg protein), but exhibited higher toxicities (60% cell viability) as well. PEGylation of schizophyllan derivatives resulted in a 30% improvement in their cytotoxicity performance (100% cell viability), but resulted in reduced transfection efficacies (1×105RLU/mg protein). Thus, a balance between the degree of PEG conjugation and molecular weight of schizophyllan derivatives is critical for high transfection efficacies, low toxicities and long-term gene expression.
Figure 9.
Synthetic scheme of various oligoamine conjugated cationic schizophyllan (SPG) polysaccharaides as shown by Nagasaki et al. [83]. The figure is reprinted with permission from American Chemical Society.
Liu et al. described the role of the number of amine units in polymers by developing a library of twelve poly(glycoamido)amines [84]. The polymers were synthesized using the poly-condensation reactions of carbohydrates with secondary amines, tested for cytotoxicity and characterized for their abilities to bind, and deliver pDNA to BHK-21 human cervical adenocarcinoma, HeLa human cervix adenocarcinoma, and HepG2 liver carcinoma cells. Three polymers, poly(D-glucaramidopentaethylenetetramine) (D4), poly(galactaramidopentaethylenetetramine) (G4) and poly(D-mannaramidopentaethylenetetramine) (M4), which had four secondary amines between the carbohydrates formed complexes with DNA in the size range of 110–180nm and showed high luciferase reporter transgene expression ranging from 108–109 RLU/mg.
POLYAMINE-BASED AND OTHER LIBRARIES
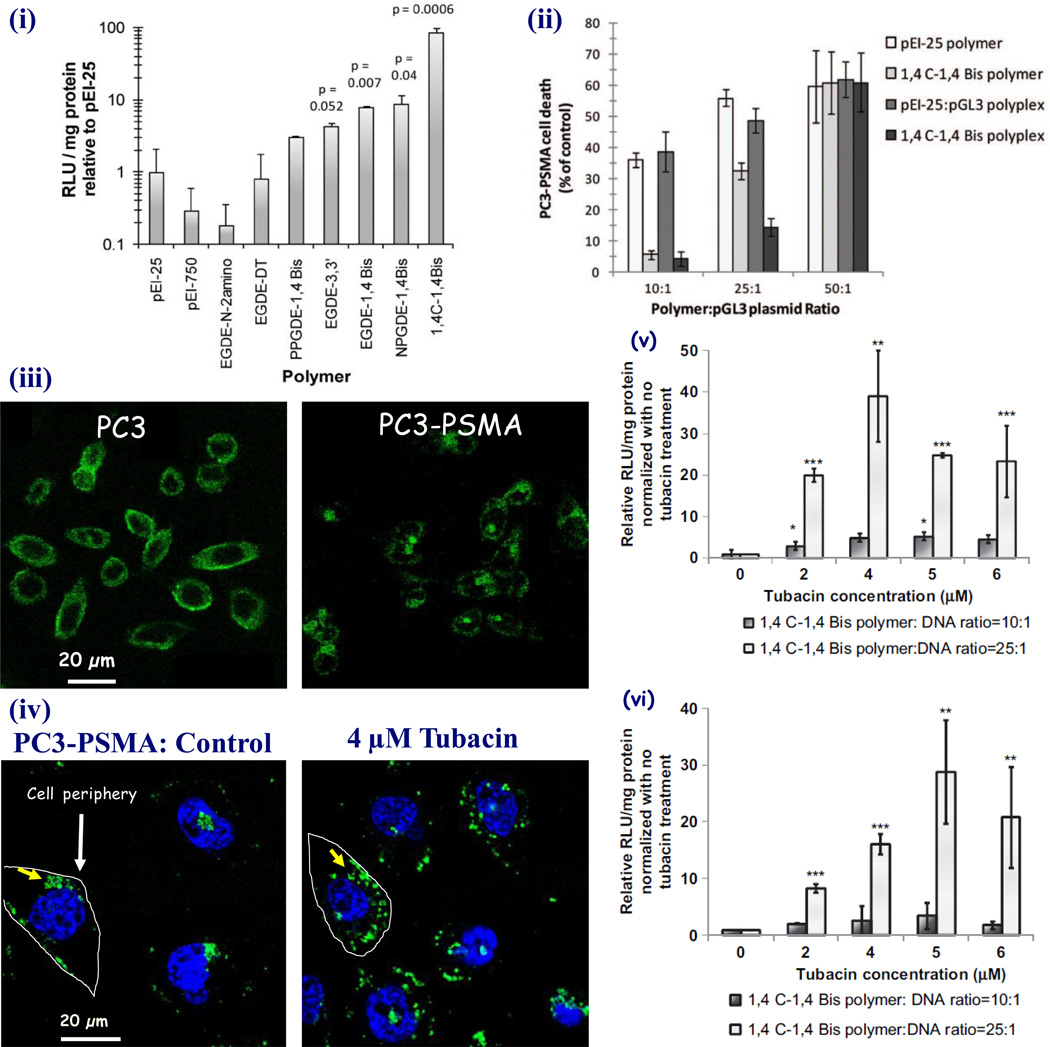
We synthesized a library of 80 cationic polymers by employing a ring-opening reaction of diglycidyl ethers by polyamine monomers [85]. The reaction was carried out at room temperature for 16 h after which, the highly viscous polymers were dissolved in phosphate buffered saline (PBS) solution and adjusted for pH. The reaction kinetics and equilibrium were measured by the percentage decrease of primary amines during the polymerization reaction. Disappearance of epoxide groups and appearance of hydroxyl peaks in fourier transform infrared (FT-IR) spectroscopy were also employed to ascertain polymer formation. The polymers were then rapidly screened for DNA-binding affinities using a fluorescence-based ethidium bromide displacement assay. Transgene expression efficacies of selected polymers were carried out using a luciferase-expression reporter system. One polymer, 1,4C-1,4 Bis (23.5kDa molecular weight) showed the highest luciferase expression in PC3-PSMA cells with up to 80-fold enhancement over pEI-25 (Figure 10(i) and (ii)). Three more polymers, EGDE-3,3’, EGDE-1,4 Bis and NPGDE-1,4 Bis from this library also showed higher transgene expression compared to those observed with pEI-25. The EGDE-3,3’ polymer was further employed for generating stable nanoassemblies with gold nanorods (GNRs) [87].
Figure 10.
(i) Comaprison of transfection efficiency of lead polymers of a diglycidyl ether polyamine library representing their relative luciferase gene expression in PC3-PSMA prostate cancer cells [85]. The polymer, 1,4C-1,4 Bis demonstarted 80-fold higher luciferase expression than that of pEI-25. (ii) Cytotoxicity of 1,4 C-1,4 Bis, pEI-25, and their polyplexes in PC3-PSMA cells at different polymer DNA weight ratios. 1,4C-1,4 Bis polymer had lower toxicity than pEI-25 [85]. Figures are reprinted by permission of Americal Chemical Society. (iii) Subcellular localization of polymer:DNA complexes are important for diffusion of DNA inside nucleus as a necessary step of efficient gene expression [86]. Confocal microscopic images of fluorescein labeled DNA and polymer complexes showed their distribution throughout the cytoplasm in PC3 prostate cancer cells (left) which, in contrast, displayed their accumulation in single spots inside the cytoplasm of a sub-cell line, PC3-PSMA cells (right). The figures are reproduced with permission from Elsevier. (iv) Modulation of the polyplex trafficking behavior using a histone deacetylase 6 inhibitor, tubacin diffused the polyplexes all around the cell nuclei (right) rather transported them to one single spot on top of the nuclei (left) [86]. Following tubcain treatments and simultaneous transfection of both (v) PC3-PSMA and (vi) PC3 cells using luciferase DNA performed 40 and 35-fold higher luciferase gene expression, rescpectively compared to the untreated control cells [86]. Reprinted with permission from Elsevier.
EGDE-3,3’ not only stabilized the complexes in biological media, e.g., PBS buffer, serum-free and serum-containing media, but also demonstrated up to 11-fold higher luciferase expression in PC3-PSMA compared to GNR assemblies generated using pEI-25 [87]. Polymer-mediated transgene expression levels were eight times higher in PC3 prostate cancer cells than that in its sub-clone PC3-PSMA cells for the same polymer employed [86]. The differential expression levels were in part due to differences in sub-cellular distribution of the polyplexes in the two cell types [86]. Polyplexes were distributed throughout the cytoplasm in PC3 cells resulting in higher probability of delivery inside the nucleus for transgene expression than those which were arrested in the single perinuclear recycling compartments (PNRC) in PC3-PSMA cells (Figure 10(iii) and (iv)). Mediators of intracellular trafficking, specifically, tubacin which inhibits the cytoplasmic histone deacetylase 6 (HDAC6), resulted in enhanced transgene expression in both cell types (Figure 10(v) and (vi)). The EGDE-3,3’ polymer also improved adenovirus-mediated gene delivery to bladder cancer cells with low levels or no coxsackie adenovirus receptor (CAR) expression on the cell surface [88]. In addition to enhanced GFP expression, polymer-adenovirus hybrids were able to induce apoptosis in bladder cancer cells, following delivery of TRAIL-expressing adenovirus.
A library of 37 water-soluble cationic polymers was generated by cross-linking low molecular weight polyamines with three different crosslinkers, dithiobis-succinimidyl proprionate (DSP), dimethyl dithiobis-propionamidate (DDBP), and hexanediol diacrylate (HD) [89]. The ester reactive groups in the DSP and DDBP reagents reacted with the polyamines to form amidines which under physiological conditions are completely protonated. In addition, presence of a disulfide bond in DSP and DDBP linkers made the polymers degradable under reducing environments. The HD linker reacted with primary and secondary amines of the polyamine monomer forming esters, which in a secondary reaction, were converted into amides by intermolecular aminolysis. Luciferase expression using the polymer library showed that one polymer resulted in 5–10 fold higher transgene expression compared to that obtained with 22kDa linear pEI, while demonstrating minimal toxicity.
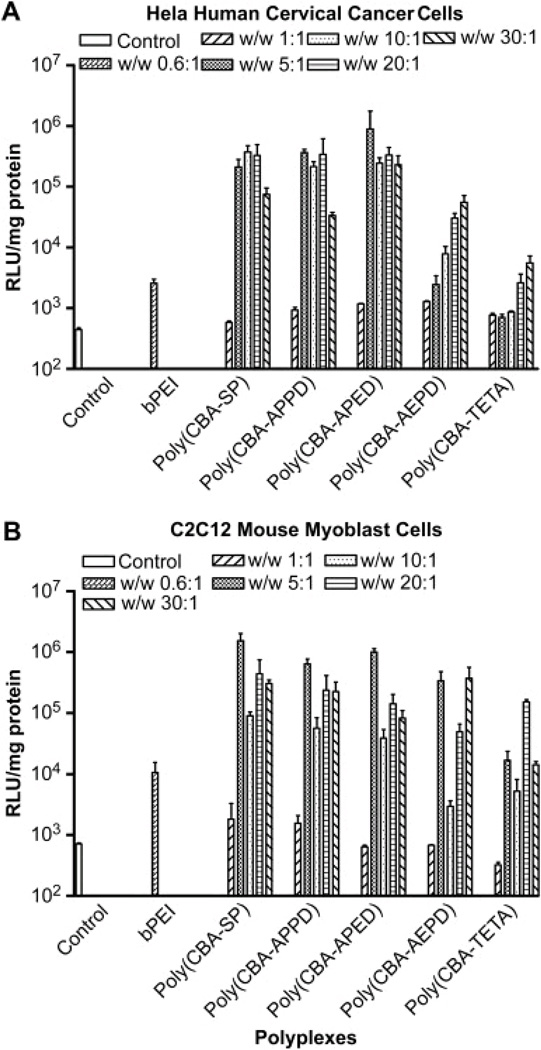
Ou et al. carried out a two-step reaction to synthesize five bioreducible poly(disulfide amine)s differing in the number of oligomethylene (-(CH2)n-, n=2–4) spacers in the main and side chains of the polymers [90]. In the first step of the reaction, five oligoamine monomers (sacrylamidespermine (SP), 3-aminopropyl-1,3-propanediamine (APPD), 3-aminopropyl-ethylenediamine (APED), 2-aminoethyl-1,3-propanediamine (AEPD) and triethylenetetramine (TETA)) were reacted with 2-acetyldimedone (Dde-OH) followed by addition of disulfide containing bisacrylamide in the second step. Introducing disulfide bonds in the polymers was hypothesized to result in the formation of stable complexes with DNA in the oxidative extracellular environment, while ready release of DNA could be facilitated via the cleavage of the disulfide bonds by glutathione and thioredoxin reductases in the cytoplasm in cells [91]. Polymers containing relatively longer oligomethylene side chains showed higher DNA binding activities, buffering capacities, and gene transfection efficacies compared to the polymers with short side chains. All polymers demonstrated greater transfection activities (104–106RLU/mg) than pEI-25 (5×103–104RLU/mg) in C2C12 mouse myoblast and HeLa human cervical cancer cells (Figure 11).
Figure 11.
Transfection efficiencies of polydisulfide amines demonstrating higher activity than 25kDa branched pEI in Hela and C2C12 cancer cells [90]. Reproduced by permission of Elsevier.
Sun et al. reported the synthesis of a small library of amine-functionalized polymers following reaction of poly(2-vinyl-4,4-dimethylazlactone) (PVDMA) with twelve different R-NH2 amines [92]. Most of the twelve polymers employed ranged from 30 to 43kDa in molecular weight and from 1.4–2.0 in polydispersity indices (PDI). The polymers were complexed with DNA at varying polymer:DNA weight ratios of 1:1 to 10:1. Three polymers, P3, P7 and P8, demonstrated maximal luciferase and EGFP expression at polymer:DNA weight ratios of 5:1, 8:1 and 4:1; the transgene expression levels were as high as that with linear pEI and branched pEI. It was suggested that molecular weight of the polymers as well as other physicochemical properties (e.g., size, zeta potential) played an important role in transfection efficiencies of the polymers.
Chen et al. developed a library of 13 polylysine-graft-imidazoleacetic acid conjugates by altering percentage moles of imidazole from 0 to 90 in the side chains of cationic lysine center [93]. The imidazole moieties were employed for their pH buffering / proton-sponge activity in endosomes, while the lysine groups were used for DNA condensation. Transfections using luciferase and EGFP reporter gene revealed that polymers with ∼50% imidazole content had higher buffering capacities compared to the polymers having low imidazole content, high gene expression (500–2500 relative light units and 200–300 GFP expressing cells for luciferase and EGFP gene respectively), and low levels of toxicities.
Wong et al. studied a series of thirty-seven pH sensitive polymers, generated by functionalizing poly(methacryloxysuccimide) with different side groups such as 1°, 2° or 3° amines, imidazole, butyl, hexyl or octyl groups, as gene carriers [94]. Polymers containing 1° amines possessed enough DNA binding efficacies to decrease the fluorescence of ethidium bromide/DNA by 40–60%, and resulted in polyplexes size of 200–300nm; higher transgene transfection efficiencies compared to polymers containing 2° or 3° amines were observed. Transfection efficacies of the polymers were comparable to pEI-25. The polymers possessed minimal cytotoxicity towards NIH/3T3 mouse fibroblast cells; the IC50 values for eight of the ten highest transfecting polymers were > 200µg/ml while that for pEI-25 was 5µg/ml.
MICROARRAY SCREENING
The large number of polymers that can be synthesized for gene delivery necessitates a method of evaluating the polymers that is reproducible and reliable. Traditionally, the synthesis of polymers and their subsequent evaluation have been performed separately but eventually, the process evolved into performing the synthesis and the evaluation in parallel [30]. The sheer size of the polymer libraries that can be synthesized makes it highly desirable to develop screening methods that are high-throughput in nature. In addition, high-throughput screening platforms that integrate polymer synthesis are also of tremendous value. In this section, we will summarize some of the high-throughput technologies that have been used for evaluating gene delivery.
Potential of the DNA microarray based screening is largely based on the work described as “transfected cell microarrays” (TCM) in 2001 [95]. TCMs are created by printing plasmids as spots on a glass slide from an aqueous gel solution. The plasmid spots are then exposed to a transfection reagent and then placed into a cell culture with adherent mammalian cells. In a successful transfection, the cells localized at a particular plasmid spot will exhibit the desired properties of the DNA in that location. Sabatini and co-workers group demonstrated the controlled transfection using plasmids expressing GFP, Cy3, and several drug receptors. The Bradley group described the use of transfection cell microarrays for screening effective transfection reagents in 2004[96]. In these studies, TCMs were first printed with the GFP plasmid and then the plasmid spots on the array were exposed to different transfection reagents. The effectiveness of a transfection was evaluated by the extent to which the cells express GFP. These microarrays were used to measure the gene delivery efficacy of several transfection agents which included: Effectine, Superfect, dendrimers, cationic lipids, and polycationic polymers (G3.0 and G4.0 PAMAM, poly-L-Lys, poly-L-Arg, poly-L-Orn, poly-L-Lys-L-Tyr, polyeneimine, polyeneimine ethoxylated, poly(diallyldimethylammonium chloride), polyarylamide-co-diallyl dimethylammonium chloride and poly(dimethylamine-co-epichlorohydrin-coethylenediamine)). The microarray screen identified five cationic lipids that showed significant transfection activities; traditional transfection methods were also employed to verify these activities. However, certain problems with microarrays as a tool for screening transfection agents were also identified in this study. First, the aqueous gel solution which is used to print the plasmids is more amenable to certain types of transfection agents based on the composition of the solution. For instance, in this study it was determined that the standard Effectine preformed better when the gelatin concentration was increased from 0.1% to 0.4% w/v. Additionally during this study, the authors were unable to show any transfection activity using Superfect, which was used as a positive control in other experiments carried out by the group. Therefore, the performance of the transfection agents used in this screen was not definitive, since in many cases, the gels can interfere with the efficacy of the transfection reagents. Of the five lipids that showed significant transfection activity in this case, all were cholesterol derivatives. Other transfection agents derived from fatty acids showed minimal transfection in the microarray experiment or in well-plate experiments, which indicated a role for the gel composition used in determining transgene expression activity. However, it would be more desirable to develop universal TCMs that could screen transfection agents based on their activity and not be limited by structural interference. Further developments in microarray-based screening are necessary for screening transfection agents in the future.
CONCLUSIONS
Combinatorial library approaches result versatile polymer libraries to identify cationic polymers for transfecting a variety of cell lines in a rapid fashion. Although screening of several cationic polymers and / or their derivatives has been carried out, the diversity of chemical structures among several polymers has made it difficult to correlate between the polymer physicochemical properties, transfection efficiencies, and differential cellular responses following DNA delivery. Further developments in polymer chemistries, elucidation of physicochemical factors influencing uptake, intracellular processing, and transgene expression following delivery of polymer-DNA complexes, cellular responses to delivery and interactions with various cell types in an automated, high-throughput fashion will facilitate the identification of efficient polymeric vectors for gene delivery. It will also enable structure-efficacy correlations for the rational development of effective polymers for transgene delivery and expression. These investigations, in concert with in vivo studies will be critical to advance polymeric vectors towards clinical applications.
REFERENCES
- 1.Boulikas T. Gene therapy of prostate cancer: P53, suicidal genes, and other targets. Anticancer Research. 1997;17(3A):1471–1505. [PubMed] [Google Scholar]
- 2.Fiorucci G, et al. TNF-related apoptosis-inducing ligand (TRAIL) as a pro-apoptotic signal transducer with cancer therapeutic potential. Curr Pharm Des. 2005;11(7):933–944. doi: 10.2174/1381612053381729. [DOI] [PubMed] [Google Scholar]
- 3.Heidel JD, et al. Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(14):5715–5721. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis ME. The First Targeted Delivery of siRNA in Humans via a Self-Assembling, Cyclodextrin Polymer-Based Nanoparticle: From Concept to Clinic. Molecular Pharmaceutics. 2009;6(3):659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 5.Mai JC, et al. A proapoptotic peptide for the treatment of solid tumors. Cancer Res. 2001;61(21):7709–7712. [PubMed] [Google Scholar]
- 6.Edelstein ML, Abedi MR, Wixon J. Gene therapy clinical trials worldwide to 2007 - an update. Journal of Gene Medicine. 2007;9(10):833–842. doi: 10.1002/jgm.1100. [DOI] [PubMed] [Google Scholar]
- 7.Griffith TS, et al. TRAIL Gene Therapy: From Preclinical Development to Clinical Application. Current Gene Therapy. 2009;9(1):9–19. doi: 10.2174/156652309787354612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins SA, et al. Viral vectors in cancer immunotherapy: Which vector for which strategy? Current Gene Therapy. 2008;8(2):66–78. doi: 10.2174/156652308784049345. [DOI] [PubMed] [Google Scholar]
- 9.Coura RD, Nardi NB. A role for adeno-associated viral vectors in gene therapy. Genetics and Molecular Biology. 2008;31(1):1–11. [Google Scholar]
- 10.Tai CK, Kasahara N. Replication-competent retrovirus vectors for cancer gene therapy. Frontiers in Bioscience. 2008;13:3083–3095. doi: 10.2741/2910. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. AAV-mediated TRAIL gene expression driven by hTERT promoter suppressed human hepatocellular carcinoma growth in mice. Life Sciences. 2008;82(23–24):1154–1161. doi: 10.1016/j.lfs.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 12.Davis ME, Brewster ME. Cyclodextrin-based pharmaceutics: Past, present and future. Nature Reviews Drug Discovery. 2004;3(12):1023–1035. doi: 10.1038/nrd1576. [DOI] [PubMed] [Google Scholar]
- 13.Bartlett DW, Davis ME. Impact of tumor-specific targeting and dosing schedule on tumor growth inhibition after intravenous administration of siRNA-containing nanoparticles. Biotechnology and Bioengineering. 2008;99(4):975–985. doi: 10.1002/bit.21668. [DOI] [PubMed] [Google Scholar]
- 14.Eldridge GR, et al. High-throughput method for the production and analysis of large natural product libraries for drug discovery. Analytical Chemistry. 2002;74(16):3963–3971. doi: 10.1021/ac025534s. [DOI] [PubMed] [Google Scholar]
- 15.Harris D. Efficient combinatorial chemistry/high throughput screening paradigm for small molecule drug discovery: Maximizing the discovery of drug candidates and minimizing waste. Abstracts of Papers of the American Chemical Society. 2002;224 008-BTEC. [Google Scholar]
- 16.Seneci P, Miertus S. Combinatorial chemistry and high-throughput screening in drug discovery: Different strategies and formats. Molecular Diversity. 2000;5(2):75–89. doi: 10.1023/a:1013824317218. [DOI] [PubMed] [Google Scholar]
- 17.Congreve MS, Jamieson C. High-throughput analytical techniques for reaction optimization. Drug Discovery Today. 2002;7(2):139–142. doi: 10.1016/s1359-6446(01)02094-3. [DOI] [PubMed] [Google Scholar]
- 18.Shoemaker RH, et al. Application of high-throughput, molecular-targeted screening to anticancer drug discovery. Curr Top Med Chem. 2002;2(3):229–246. doi: 10.2174/1568026023394317. [DOI] [PubMed] [Google Scholar]
- 19.Khandurina J, Guttman A. Microchip-based high-throughput screening analysis of combinatorial libraries. Curr Opin Chem Biol. 2002;6(3):359–366. doi: 10.1016/s1367-5931(02)00323-x. [DOI] [PubMed] [Google Scholar]
- 20.Boussif O, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo polyethyleneimine. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(16):7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonawane ND, Szoka FC, Verkman AS. Chloride accumulation and swelling in endosomes enhances DNA transfer by polyamine-DNA polyplexes. Journal of Biological Chemistry. 2003;278(45):44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 22.Coll JL, et al. In vivo delivery to tumors of DNA complexed with linear polyethylenimine. Human Gene Therapy. 1999;10(10):1659–1666. doi: 10.1089/10430349950017662. [DOI] [PubMed] [Google Scholar]
- 23.Brunner S, et al. Overcoming the nuclear barrier: Cell cycle independent nonviral gene transfer with linear polyethylenimine or electroporation. Molecular Therapy. 2002;5(1):80–86. doi: 10.1006/mthe.2001.0509. [DOI] [PubMed] [Google Scholar]
- 24.Lungwitz U, et al. Polyethylenimine-based non-viral gene delivery systems. European Journal of Pharmaceutics and Biopharmaceutics. 2005;60(2):247–266. doi: 10.1016/j.ejpb.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 25.Jiang G, et al. Target Specific Intracellular Delivery of siRNA/PEI-HA Complex by Receptor Mediated Endocytosis. Molecular Pharmaceutics. 2009;6(3):727–737. doi: 10.1021/mp800176t. [DOI] [PubMed] [Google Scholar]
- 26.Jiang HL, et al. The suppression of lung tumorigenesis by aerosol-delivered folate-chitosan-graft-polyethylenimine/Akt1 shRNA complexes through the Akt signaling pathway. Biomaterials. 2009;30(29):5844–5852. doi: 10.1016/j.biomaterials.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 27.Thomas M, Klibanov AM. Enhancing polyethylenimine’s delivery of plasmid DNA into mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(23):14640–14645. doi: 10.1073/pnas.192581499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas M, et al. Identification of novel superior polycationic vectors for gene delivery by high-throughput synthesis and screening of a combinatorial library. Pharmaceutical Research. 2007;24(8):1564–1571. doi: 10.1007/s11095-007-9279-3. [DOI] [PubMed] [Google Scholar]
- 29.Van Vliet LD, et al. Relating chemical and biological diversity space: A tunable system for efficient gene transfection. Chembiochem. 2008;9(12):1960–1967. doi: 10.1002/cbic.200800003. [DOI] [PubMed] [Google Scholar]
- 30.Lynn DM, et al. Accelerated discovery of synthetic transfection vectors: Parallel synthesis and screening of degradable polymer library. Journal of the American Chemical Society. 2001;123(33):8155–8156. doi: 10.1021/ja016288p. [DOI] [PubMed] [Google Scholar]
- 31.Lynn DM, Langer R. Degradable poly(beta-amino esters): Synthesis, characterization, and self-assembly with plasmid DNA. Journal of the American Chemical Society. 2000;122(44):10761–10768. [Google Scholar]
- 32.Anderson DG, Lynn DM, Langer R. Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angewandte Chemie-International Edition. 2003;42(27):3153–3158. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- 33.Anderson DG, et al. Structure/property studies of polymeric gene delivery using a library of poly(beta-amino esters) Molecular Therapy. 2005;11(3):426–434. doi: 10.1016/j.ymthe.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 34.Green JJ, et al. Biodegradable polymeric vectors for gene delivery to human endothelial cells. Bioconjugate Chemistry. 2006;17(5):1162–1169. doi: 10.1021/bc0600968. [DOI] [PubMed] [Google Scholar]
- 35.Green JJ, et al. Electrostatic ligand coatings of nanoparticles enable ligand-specific gene delivery to human primary cells. Nano Letters. 2007;7(4):874–879. doi: 10.1021/nl062395b. [DOI] [PubMed] [Google Scholar]
- 36.Anderson DG, et al. A polymer library approach to suicide gene therapy for cancer. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(45):16028–16033. doi: 10.1073/pnas.0407218101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng WD, et al. Nanoparticulate delivery of suicide DNA to murine prostate and prostate tumors. Prostate. 2007;67(8):855–862. doi: 10.1002/pros.20576. [DOI] [PubMed] [Google Scholar]
- 38.Zugates GT, et al. Rapid optimization of gene delivery by parallel end-modification of poly(beta-amino ester)s. Molecular Therapy. 2007;15(7):1306–1312. doi: 10.1038/mt.sj.6300132. [DOI] [PubMed] [Google Scholar]
- 39.Sunshine J, et al. Small-Molecule End-Groups of Linear Polymer Determine Cell-Type Gene-Delivery Efficacy. Advanced Materials. 2009;21(48):4947–4951. doi: 10.1002/adma.200901718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Green JJ, et al. Combinatorial modification of degradable polymers enables transfection of human cells comparable to adenovirus. Advanced Materials. 2007;19(19):2836–2842. [Google Scholar]
- 41.Green JJ, et al. Nanoparticles for Gene Transfer to Human Embryonic Stem Cell Colonies. Nano Letters. 2008;8(10):3126–3130. doi: 10.1021/nl8012665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akinc A, et al. Parallel synthesis and biophysical characterization of a degradable polymer library for gene delivery. Journal of the American Chemical Society. 2003;125(18):5316–5323. doi: 10.1021/ja034429c. [DOI] [PubMed] [Google Scholar]
- 43.Wu DC, et al. Evaluation of hyperbranched poly(amino ester)s of amine constitutions similar to polyethylenimine for DNA delivery. Biomacromolecules. 2005;6(6):3166–3173. doi: 10.1021/bm0504983. [DOI] [PubMed] [Google Scholar]
- 44.Kim HJ, et al. Highly effective and slow-biodegradable network-type cationic gene delivery polymer: Small library-like approach synthesis and characterization. Biomaterials. 2006;27(10):2292–2301. doi: 10.1016/j.biomaterials.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 45.Jere D, et al. Poly (amino ester) composed of poly (ethylene glycol) and aminosilane prepared by combinatorial chemistry as a gene carrier. Pharmaceutical Research. 2008;25(4):875–885. doi: 10.1007/s11095-007-9448-4. [DOI] [PubMed] [Google Scholar]
- 46.Holmes PF, Bohrer M, Kohn J. Exploration of polymethacrylate structure-property correlations: Advances towards combinatorial and high-throughput methods for biomaterials discovery. Progress in Polymer Science. 2008;33(8):787–796. doi: 10.1016/j.progpolymsci.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van de Wetering P, et al. Structure-activity relationships of water-soluble cationic methacrylate/methacrylamide polymers for nonviral gene delivery. Bioconjugate Chemistry. 1999;10(4):589–597. doi: 10.1021/bc980148w. [DOI] [PubMed] [Google Scholar]
- 48.Cherng JY, et al. Effect of size and serum proteins on transfection efficiency of poly((2-dimethylamino)ethyl methacrylate)-plasmid nanoparticles. Pharmaceutical Research. 1996;13(7):1038–1042. doi: 10.1023/a:1016054623543. [DOI] [PubMed] [Google Scholar]
- 49.Ma M, et al. Influence of hydroxyl groups on the biological properties of cationic polymethacrylates as gene vectors. Acta Biomaterialia. 2010;6(7):2658–2665. doi: 10.1016/j.actbio.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 50.Dubruel P, et al. Physicochemical and biological evaluation of cationic polymethacrylates as vectors for gene delivery. European Journal of Pharmaceutical Sciences. 2003;18(3–4):211–220. doi: 10.1016/s0928-0987(02)00280-4. [DOI] [PubMed] [Google Scholar]
- 51.Dubruel P, Toncheva V, Schacht EH. pH sensitive vinyl copolymers as vectors for gene therapy. Journal of Bioactive and Compatible Polymers. 2000;15(3):191–213. [Google Scholar]
- 52.Dubruel P, et al. Buffering properties of cationic polymethacrylates are not the only key to successful gene delivery. Biomacromolecules. 2004;5(2):379–388. doi: 10.1021/bm034438d. [DOI] [PubMed] [Google Scholar]
- 53.Gonzalez H, Hwang SJ, Davis ME. New class of polymers for the delivery of macromolecular therapeutics. Bioconjugate Chemistry. 1999;10(6):1068–1074. doi: 10.1021/bc990072j. [DOI] [PubMed] [Google Scholar]
- 54.Davis ME, Bellocq NC. Cyclodextrin-containing polymers for gene delivery. Journal of Inclusion Phenomena and Macrocyclic Chemistry. 2002;44(1–4):17–22. [Google Scholar]
- 55.Yang CA, et al. Cationic star polymers consisting of alpha-cyclodextrin core and oligoethylenimine arms as nonviral gene delivery vectors. Biomaterials. 2007;28(21):3245–3254. doi: 10.1016/j.biomaterials.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 56.Yang CA, et al. A supramolecular gene carrier composed of multiple cationic alpha-cyclodextrins threaded on a PPO-PEO-PPO triblock polymer. Polymer. 2009;50(6):1378–1388. [Google Scholar]
- 57.Li JS, et al. 21-Arm star polymers with different cationic groups based on cyclodextrin core for DNA delivery. Carbohydrate Polymers. 2010;79(2):277–283. [Google Scholar]
- 58.Srinivasachari S, Fichter KM, Reineke TM. Polycationic beta-cyclodextrin “Click Clusters”: Monodisperse and versatile scaffolds for nucleic acid delivery. Journal of the American Chemical Society. 2008;130(14):4618–4627. doi: 10.1021/ja074597v. [DOI] [PubMed] [Google Scholar]
- 59.Cryan SA, et al. Cell transfection with polycationic cyclodextrin vectors. European Journal of Pharmaceutical Sciences. 2004;21(5):625–633. doi: 10.1016/j.ejps.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 60.Pun SH, et al. Cyclodextrin-modified polyethylenimine polymers for gene delivery. Bioconjugate Chemistry. 2004;15(4):831–840. doi: 10.1021/bc049891g. [DOI] [PubMed] [Google Scholar]
- 61.Reineke TM, Davis ME. Structural effects of carbohydrate-containing polycations on gene delivery. 1. Carbohydrate size and its distance from charge centers. Bioconjugate Chemistry. 2003;14(1):247–254. doi: 10.1021/bc025592k. [DOI] [PubMed] [Google Scholar]
- 62.Jiang HL, et al. Chitosan-graft-polyethylenimine as a gene carrier. Journal of Controlled Release. 2007;117(2):273–280. doi: 10.1016/j.jconrel.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 63.Lee KY, et al. Preparation of chitosan self-aggregates as a gene delivery system. Journal of Controlled Release. 1998;51(2–3):213–220. doi: 10.1016/s0168-3659(97)00173-9. [DOI] [PubMed] [Google Scholar]
- 64.Lee MK, et al. The use of chitosan as a condensing agent to enhance emulsion-mediated gene transfer. Biomaterials. 2005;26(14):2147–2156. doi: 10.1016/j.biomaterials.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 65.Mumper RJ, Wang JJ, Claspell JM, Rolland AP. Novel polymeric condensing carriers for gene delivery. Proceedings of the International Symposium on Controlled Release Bioactive Materials. 1995:178–179. [Google Scholar]
- 66.Sato T, Ishii T, Okahata Y. In vitro gene delivery mediated by chitosan. Effect of pH, serum, and molecular mass of chitosan on the transfection efficiency. Biomaterials. 2001;22(15):2075–2080. doi: 10.1016/s0142-9612(00)00385-9. [DOI] [PubMed] [Google Scholar]
- 67.Romoren K, et al. The influence of formulation variables on in vitro transfection efficiency and physicochemical properties of chitosan-based polyplexes. International Journal of Pharmaceutics. 2003;261(1–2):115–127. doi: 10.1016/s0378-5173(03)00301-6. [DOI] [PubMed] [Google Scholar]
- 68.Kiang T, et al. The effect of the degree of chitosan deacetylation on the efficiency of gene transfection. Biomaterials. 2004;25(22):5293–5301. doi: 10.1016/j.biomaterials.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 69.Strand SP, et al. Influence of chitosan structure on the formation and stability of DNA-chitosan polyelectrolyte complexes. Biomacromolecules. 2005;6(6):3357–3366. doi: 10.1021/bm0503726. [DOI] [PubMed] [Google Scholar]
- 70.Zhang YQ, et al. A novel PEGylation of chitosan nanoparticles for gene delivery. Biotechnology and Applied Biochemistry. 2007;46:197–204. doi: 10.1042/BA20060163. [DOI] [PubMed] [Google Scholar]
- 71.Lu B, et al. Chitosan based oligoamine polymers: Synthesis, characterization, and gene delivery. Journal of Controlled Release. 2009;137(1):54–62. doi: 10.1016/j.jconrel.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 72.Shcharbin D, Pedziwiatr E, Bryszewska M. How to study dendriplexes I: Characterization. Journal of Controlled Release. 2009;135(3):186–197. doi: 10.1016/j.jconrel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 73.Bielinska A, et al. Regulation of in vitro gene expression using antisense oligonucleotides or antisense expression plasmids transfected using starburst PAMAM dendrimers. Nucleic Acids Research. 1996;24(11):2176–2182. doi: 10.1093/nar/24.11.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bielinska AU, KukowskaLatallo JF, Baker JR. The interaction of plasmid DNA with polyamidoamine dendrimers: mechanism of complex formation and analysis of alterations induced in nuclease sensitivity and transcriptional activity of the complexed DNA. Biochimica Et Biophysica Acta-Gene Structure and Expression. 1997;1353(2):180–190. doi: 10.1016/s0167-4781(97)00069-9. [DOI] [PubMed] [Google Scholar]
- 75.Majoros IJ, et al. Surface Interaction and Behavior of Poly(amidoamine) Dendrimers: Deformability and Lipid Bilayer Disruption. Journal of Computational and Theoretical Nanoscience. 2009;6(7):1430–1436. doi: 10.1166/jctn.2009.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hong S, et al. The Role of Ganglioside GM(1) in Cellular Internalization Mechanisms of Poly(amidoamine) Dendrimers. Bioconjugate Chemistry. 2009;20(8):1503–1513. doi: 10.1021/bc900029k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Russ V, et al. Novel degradable oligoethylenimine acrylate ester-based pseudodendrimers for in vitro and in vivo gene transfer. Gene Therapy. 2008;15(1):18–29. doi: 10.1038/sj.gt.3303046. [DOI] [PubMed] [Google Scholar]
- 78.Sanclimens G, et al. Synthesis and screening of a small library of proline-based biodendrimers for use as delivery agents. Biopolymers. 2005;80(6):800–814. doi: 10.1002/bip.20301. [DOI] [PubMed] [Google Scholar]
- 79.Yamaoka T, et al. Effect of cation content of polycation-type gene carriers on in vitro gene transfer. Chemistry Letters. 1998;(11):1171–1172. [Google Scholar]
- 80.Azzam T, et al. Polysaccharide-oligoamine based conjugates for gene delivery. Journal of Medicinal Chemistry. 2002;45(9):1817–1824. doi: 10.1021/jm0105528. [DOI] [PubMed] [Google Scholar]
- 81.Kano Y, Kakuta H, Hashimoto J. Effect of sizofiran on regional lymph nodes in patients with head and neck cancer. Biotherapy. 1996;9(4):257–262. doi: 10.1007/BF02620739. [DOI] [PubMed] [Google Scholar]
- 82.Shimizu Y, Hasumi K, Masubuchi K. AUGMENTING EFFECT OF SIZOFIRAN ON THE IMMUNOFUNCTION OF REGIONAL LYMPH-NODES IN CERVICAL-CANCER. Cancer. 1992;69(5):1188–1194. [PubMed] [Google Scholar]
- 83.Nagasaki T, et al. Long-term expression with a cationic polymer derived from a natural polysaccharide: Schizophyllan. Bioconjugate Chemistry. 2004;15(2):249–259. doi: 10.1021/bc034178x. [DOI] [PubMed] [Google Scholar]
- 84.Liu YM, Reineke TM. Hydroxyl stereochemistry and amine number within poly(glycoamidoamine)s affect intracellular DNA delivery. Journal of the American Chemical Society. 2005;127(9):3004–3015. doi: 10.1021/ja0436446. [DOI] [PubMed] [Google Scholar]
- 85.Barua S, et al. Parallel Synthesis and Screening of Polymers for Nonviral Gene Delivery. Molecular Pharmaceutics. 2009;6(1):86–97. doi: 10.1021/mp800151j. [DOI] [PubMed] [Google Scholar]
- 86.Barua S, Rege K. The influence of mediators of intracellular trafficking on transgene expression efficacy of polymer-plasmid DNA complexes. Biomaterials. 2010;31(22):5894–5902. doi: 10.1016/j.biomaterials.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 87.Huang HC, et al. Simultaneous Enhancement of Photothermal Stability and Gene Delivery Efficacy of Gold Nanorods Using Polyelectrolytes (vol 3, pg 2941, 2009) Acs Nano. 2010;4(3):1769–1770. doi: 10.1021/nn900947a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kasman LM, et al. Polymer-Enhanced Adenoviral Transduction of CAR-Negative Bladder Cancer Cells. Molecular Pharmaceutics. 2009;6(5):1612–1619. doi: 10.1021/mp9000958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kloeckner J, Wagner E, Ogris M. Degradable gene carriers based on oligomerized polyamines. European Journal of Pharmaceutical Sciences. 2006;29(5):414–425. doi: 10.1016/j.ejps.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 90.Ou M, et al. A family of bioreducible poly(disulfide amine)s for gene delivery. Biomaterials. 2009;30(29):5804–5814. doi: 10.1016/j.biomaterials.2009.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Neumann H, Smith RA. Cleavage of the disulfide bonds of cystine and oxidized glutathione by phosphorothioate. Archives of Biochemistry and Biophysics. 1967;122(2):354. doi: 10.1016/0003-9861(67)90205-6. &. [DOI] [PubMed] [Google Scholar]
- 92.Sun B, et al. Azlactone-functionalized polymers as reactive templates for parallel polymer synthesis: synthesis and screening of a small library of cationic polymers in the context of DNA delivery. Chemical Communications. 2010;46(12):2016–2018. doi: 10.1039/b921664b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen DJ, et al. Structure-function relationships of gene delivery vectors in a limited polycation library. Journal of Controlled Release. 2005;103(1):273–283. doi: 10.1016/j.jconrel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 94.Wong SY, Sood N, Putnam D. Combinatorial Evaluation of Cations, pH-sensitive and Hydrophobic Moieties for Polymeric Vector Design. Molecular Therapy. 2009;17(3):480–490. doi: 10.1038/mt.2008.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001;411(6833):107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 96.How SE, et al. Polyplexes and lipoplexes for mammalian gene delivery: From traditional to microarray screening. Combinatorial Chemistry & High Throughput Screening. 2004;7(5):423–430. doi: 10.2174/1386207043328616. [DOI] [PubMed] [Google Scholar]