Abstract

Hydrogels provide mechanical support and a hydrated environment that offer good cytocompatibility and controlled release of molecules, and myriad hydrogels thus have been studied for biomedical applications. In the past few decades, research in these areas has shifted increasingly to multicomponent hydrogels that better capture the multifunctional nature of native biological environments and that offer opportunities to selectively tailor materials properties. This review summarizes recent approaches aimed at producing multicomponent hydrogels, with descriptions of contemporary chemical and physical approaches for forming networks, and of the use of both synthetic and biologically derived molecules to impart desired properties. Specific multicomponent materials with enhanced mechanical properties are presented, as well as materials in which multiple biological functions are imparted for applications in tissue engineering, cancer treatment, and gene therapies. The progress in the field suggests significant promise for these approaches in the development of biomedically relevant materials.
1. Introduction
Three-dimensional (3D) hydrogel networks provide mechanical support and hydrophilic properties that are advantageous for myriad applications ranging from those in consumer to biomedical products. The highly porous structure allows for fast diffusion of small molecules,1 and hydrogels thus have been used in separation and purification,2 biosensor,3−5 and tissue regeneration.6−8 Hydrogels provide a hydrated environment for cells, which improves their suitability for tissue engineering applications.8−10 For tissue engineering purposes, hydrogels not only need to provide a physical support for cell growth, but also need to maintain a mechanically active and biochemically appropriate environment that provide cell–matrix interactions to direct cell proliferation and differentiation. Given the variety of properties necessary for optimizing material activity in the biological environment, multicomponent hybrid hydrogels have been of significant research interest.
The formation of a multicomponent hybrid network can be achieved via either chemical or physical means. Many biologically active proteins or peptides can simply be reacted with synthetic polymers via radical polymerization or other conjugation strategies, including click protocols,11−13 yielding multiple opportunities to easily produce multicomponent hydrogels. In particular, highly specific click reactions provide a simple way to produce macromolecules or hydrogel networks with a controllable network structure and patternable design. The nontoxic and mild chemistries enable cell encapsulation and provide opportunities for hydrogel formation in vivo. In addition, the use of physical networks, including those formed from self-assembling peptides and proteins, has expanded the versatility of these physical approaches for producing self-assembling hydrogels.14−16
Both synthetic and natural polymers have been utilized for fabricating scaffolds. For biological application, the materials must be inherently biocompatible, biodegradable, and cell adhesive. Additionally, they must have a porous, mechanically stable, and 3D structure with facile manufacture. Synthetic materials provide a wide range of molecular structures and chemical capability,7,17 while biomimetic materials, and in particular structural proteins such as collagen and elastin, provide mechanical characteristics unique to native tissue.18,19 Hybrid polymeric scaffolds combining natural and synthetic polymers have thus gathered significant and continued interest for their potential to mimic the extracellular matrix (ECM). In addition, to further improve the mechanical robustness of the hydrogel network, composite hybrid hydrogels provide an additional mechanical reinforcement.20−22 Drug delivery can also be enhanced when a second phase, such as drug-loaded nanoparticles and microparticles, is incorporated in the hydrogel matrix.23,24
For most of the biochemically inert polymers, the lack of interaction between cells and hydrogels can limit the utility of the materials for directing cellular behavior, and accordingly, the purposeful design and production of multicomponent hydrogels to fulfill different biological function has grown.6,10,19,25−27 In addition to providing cell adhesion and cell-mediated degradation, incorporation of biofunctional biomolecules, including growth factors28−31 and signaling molecules17,32,33 can also facilitate cell proliferation and differentiation. Controlled delivery of biomolecules to modulate immune response,34−36 with codelivery of therapeutics and DNA, can further expand the functions of hydrogels beyond tissue regeneration to cancer and gene therapies.37−39 The applications of these tunable hydrogels in biomedical engineering are numerous, owing to the ease by which functions can be altered by simple incorporation of the components that are required for particular applications. This review focuses on the recent development and applications of multicomponent hybrid hydrogels.
2. Hydrogel Network Formation
a. Chemical Hydrogels
Stable hydrogel networks are essential to provide structural support, and can be formed by chemical and physical cross-linking; given the wide selection of cross-linking methods available, multiple components can be randomly or selectively incorporated into the hydrogel networks. Chemically cross-linked hydrogel networks, employing covalent bonds, generally provide a stronger and more stable network, although chemical degradation or other strategies are then necessary for elimination of the hydrogels from a biological environment. Covalently cross-linked hydrogels can be formed via various reactions, including free radical polymerization,40−42 click chemistry,12,43−45 and thiol–ene chemistry.46−48 The advantage of radical polymerization is that multiple, vinyl-functionalized components can react and form multicomponent hybrid hydrogels, such as poly(ethylene glycol) dimethacrylate (PEGDMA)/gelatin methacrylate (GelMA)49 and poly(ethylene glycol) diacrylate (PEGDA)/ heparin methacrylate (HepMA)50 in a one-pot reaction. Incorporating bioactive components (e.g., gelatin and heparin) in the matrix imparts desired bioactivity while maintaining necessary mechanical strength. Prepolymerization of the precursor solution before inclusion of cells can reduce free-radical induced cell damage during in situ cell encapsulation,51,52 and there are multiple types of photoinitiators (such as Igracure 295953 and lithium arylphosphinate (LAP)54,55) that maintain high cell viability, and conditions can be employed to make free radical polymerization useful for forming hybrid hydrogels in vivo.56
In addition to free radical polymerization, controlled radical polymerization (CRP), including atom transfer radical polymerization (ATRP)57−59 and reversible addition–fragmentation transfer (RAFT),60−63 have been employed for the formation of hybrid materials and to afford better control over molecular weight, polymer architecture, and controllable incorporation of multiple macromolecules. ATRP polymerizations can be initiated by a chemically functionalized64 or genetically encoded65 initiator(s); the ability to control polymer conjugation with biomolecules is of great interest for producing polymer–peptide and polymer–protein hybrid materials that show stimuli-responsive behavior. In addition, ATRP has permitted the controlled growth of polymers from micropatterned surfaces,66 particles,41 and biomolecules64,65 and has been useful for production of polymer–drug or polymer–protein conjugates. Hydrogels synthesized via CRP show a more homogeneous and ordered network in comparison to networks formed via free radical polymerization, which has been important for providing better control of swelling and deswelling kinetics,67 degradation,60,62 and drug release.59,63 However, given the toxicity of the commonly employed copper- and iron-based catalysts, the materials generally require an extensive purification process prior to use in biomedical applications, including chromatography, precipitation, and dialysis.68 RAFT polymerizations, in contrast, employ chain transfer agents to control the polymerization and thus do not require a special initiator or metal catalyst.40
Click chemistry has been widely used in conjugation due to its fast, highly specific, and efficient reaction, which allows selective modification and incorporation of biologically active molecules (such as cell adhesion and enzymatically degradable peptides) in specific sites, even in the presence of various functional groups and under physiological conditions.11,69 Hydrogels utilizing click chemistry have a well-defined network structure and can show significantly improved mechanical properties.70 The most commonly used click reactions include alkyne–azide, Diels–Alder, and thiol–ene reactions. The popular copper-catalyzed alkyne–azide cycloaddition (CuAAC), which is stable in biological systems, has been widely used in bioconjugation.71−73 To reduce the cytotoxicity of the copper catalyst in biological studies,11 copper-free click chemistries74−77 have been developed that can be readily employed in the presence of cells.74,75 The Diels–Alder cycoladdition reaction, between a conjugated diene and a substituted alkene to form a substituted cyclohexene, is also widely used in hydrogel formation78 and offers the advantage of not requiring an initiator. The reaction is noncytotoxic and maintains cell viability during cell encapsulation,79 but the slow rate of the Diels–Alder chemistry has limited its use for hydrogel systems that require rapid gelation. The development of a fast inverse-electron-demand Diels–Alder reaction, which involves the reaction of a trans-cyclooctene with a tetrazine,80 yields reactions with highly rapid rates and the maintenance of cell viability,45 which has enabled its use for fluorescent labeling of cell surfaces and intracellular labeling of living mammalian cells.81 The Diels–Alder click reaction provides not only a cross-linking chemistry, but also uses imaging agents for live-cell imaging because of its cytocompatibility. Another widely employed class of click reactions, thiol–ene reactions, can proceed via a traditional Michael-type addition or be mediated by radicals and has the advantage of rapid and efficient reaction and the ability to react under ambient conditions. In addition, the availability of a wide variety of thiols, including alkyl thiols, thiophenols, thiol propionate, and thiolglycolates, enables its wide applications in chemical reactions, bioconjugation, surface modification, hydrogel formation, and photopatterning.47 The radical-mediated thiol–ene reaction requires radical initiation, that is, thermal or photolytic, for activation of the thiyl radical that reacts with a broad range of alkenes via a combination of step and chain growth mechanisms.47,71 It shows faster gelation and higher cross-linking density compared to the Michael-type addition,55 and because of the UV initiation, radical thiol–ene can be controlled and triggered spatiotemporally,82 allowing its use in 2D and 3D photopatterning.44,74,76 For example, hydrogels produced with a cross-linker containing available alkyl sulfide functional groups were able to undergo reversible exchange of thiolated biomolecules with photopatterning techniques.83 The unique exchangeable functional groups thus provide dynamic control of hydrogel function.84
b. Physical Hydrogels
Physical hydrogels, in contrast, are formed by secondary interactions, including hydrogen bonding, ionic interactions, and hydrophobic interactions.85 Cooperative physical interactions can be used to form stable hydrogels via crystallization, self-assembly, and thermally induced cross-linking. Although secondary interactions can provide stable hydrogels, the strength of the physical network can be altered by pH, temperature, or organic solvent.86−88 Specific ligand–receptor binding events and self-assembling peptides also can be employed to form physical hydrogels, permitting the elimination of any potential toxic cross-linker or initiator. Although physical gels may suffer from weak mechanical properties and dissociation from the bulk material, physical cross-links formed via multiple methods have been shown to be valuable in the production of multicomponent hydrogels.89−91
One common strategy for the formation of physically cross-linked polymeric gels is through the crystallization of the polymer. Poly(vinyl alcohol) (PVA), in particular, is one of the most widely used polymer hydrogels cross-linked via crystallization induced in a freeze–thawing process.7 The mechanical and swelling properties of these types of hydrogels depend on the crystallinity, which can be well controlled by the processing conditions. Repeated freeze–thawing can improve mechanical properties through the formation of secondary crystallites,92 and the resulting gels are highly elastic and stable at room temperature,92,93 showing consistent compression moduli values after repeated cycles.93 Besides PVA, block copolymers that contains semicrystalline polymer domains can also form crystallite-cross-linked networks. Semicrystalline polymers including poly(caprolactone) (PCL) and poly(lactide) (PLA) have been used to form amphiphilic block copolymers, such as PCL–PEG–PCL and PLA–PEO–PLA.94,95 Heating and cooling cycles induce crystallization of the crystalline block to create a hydrogel network with properties that can be varied by processing to control the crystallinity.
Spontaneous self-assembly, generally driven from cooperative physical interactions,96 has also been widely used in the formation of physical networks. A large range of biomacromolecules, including peptides and proteins can form network structures via formation of coiled-coil, triple helix, and β-sheet structures; canonical examples include collagen-based97−100 and silk-based101−103 hydrogels. Peptide sequences that form self-assembled structures have thus been incorporated into hybrid hydrogels. For example, the peptide sequence (AKAAAKA)2 has been conjugated to Pluronic polymers to form a self-assembled peptide/polymer hybrid hydrogel104,105 that showed a compressive modulus similar to that of native elastin and was capable of supporting cell adhesion. Another approach for synthesizing peptide/polymer hybrids is via polymerization. Functionalized poly-l-glutamate (alkyl-poly-l-EG2Glu) has been produced via ring opening polymerization of the γ-(2-methoxyethoxy)esteryl-l-glutamate N-carboxyanhydride (l-EG2Glu NCA) with alkyl amine; the resulting alkyl polypeptide can spontaneously self-assemble into a hydrogel.106 The alkyl polypeptide with V3A3E3(CO2H) and alkyl end can be self-assembled into aligned hydrogel nanofibers.107 For further details on self-assembling protein and peptide–polymer hybrid hydrogels, the reader is directed to a recent review.15
Thermally responsive polymers, such as poly(N-isopropylacrylamide) (PNIPAAm), have also been employed in self-assembly and the formation of injectable materials for biomedical and drug delivery applications.8 Many peptides and proteins conjugated to PNIPAAm exhibit materials with dual self-assembly and thermally responsive properties.108−111 In one example, hydrogels have been produced via the interactions of coiled-coil domains of PNIPAAm–coiled-coil polypeptide–PNIPAAm triblock polymers. Below the LCST of the PNIPAAm, the hydrogel is only cross-linked by the coiled-coil interactions of the polypeptide (Figure 1) and thus exhibits shear-thinning behavior, which is useful for injection. With an increase of temperature to above 37 °C (e.g., upon injection in vivo), the thermally responsive PNIPAAm segments collapse and aggregate, resulting in a stiff hydrogel with a modulus up to 60 kPa.110 In another example, conjugation of a DNA-binding protein to PNIPAAm was employed to form a biofunctional hydrogel,112 permitting the production of materials that retain the ability to bind specific DNA. The ability to incorporate biomolecules with specific binding properties within the functional hydrogel has also enabled simplified separation and purification of DNA and proteins.
Figure 1.

PNIPAAm–coiled-coil peptide–PNIPAAm thermally responsive self-assembled hydrogel. (a) The hydrogel is cross-linked by the coiled-coil structure formed by the polypeptide and by PNIPAAm after its collapse and aggregation above its LCST. (b) Schematic of PNIPAAm–coiled-coil peptide–PNIPAAm and peptide sequence.110 Reproduced with permission from ref (110). Copyright 2013 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany.
3. Mechanical Improvement
a. Hybrid Networks
The versatility of polymer synthesis and modification enables the production of synthetic polymers in different molecular structures, including star and branched polymers and multiple networks. The widely employed tetra-functionalized PEG has been useful for forming hydrogel networks;25,72,113−117 tetra-PEG hydrogels have become popular owing to their simple, robust, and versatile chemistries.118 The networks have demonstrated improvements in extension and strength compared with conventional hydrogels,119 and more recent reports have shown that there are negligible local defects so that the networks produced from the tetra-PEGs act as a nearly ideal elastic network.120 In another example, a reducible micelle hydrogel has been formed, using a multiarm PEG-containing copolymer, for drug delivery applications. The 8-arm PCL–PEO copolymer was linked by a disulfide core and exhibited a micellar structure;121 the micelles then further cross-linked to form hydrogels. Micelle size could be reduced in the presence of a reducing agent, which cleaved the disulfide core linkage and reduced the sizes of the multiarm polymer by half (to yield a 4-arm architecture). The mechanical strength of the 8-arm hydrogel was nearly 10-fold that of a control hydrogel formed with a cross-linked linear copolymer, and the modulus of the 8-arm micellar hydrogel was decreased 58% when the multiarm polymer was reduced to the 4-arm polymer.
In addition to these variations in polymer architecture, hybrid networks formed with two different polymers have been shown to exhibit excellent mechanical properties. Interpenetrating polymer networks (IPNs), for example, are among the earliest multicomponent, hybrid polymer networks; the concept of IPNs was introduced in the 1960s and remains an active research area.122 Double networks are one unique type of IPN system that contains two types of polymers with an asymmetric network structure123 (Figure 2) and has provided significant improvement in the strength of hydrogels compared to that of single networks.124−127 A double poly(2-acrylamido-2-methylpropanesulfonic acid) (PAMPS)/polyacrylamide (PAAm) network hydrogel, formed via a two-step polymerization, has improved the compressive strength of the hydrogel over 20 times relative to PAMPS and PAAm single network hydrogels while retaining highly elastomeric behavior.127 Other groups have combined biopolymers such as gelatin and bacterial cellulose (BC) to form double network hydrogels with high mechanical strength (up to 5 MPa in compression),126 or PVA/PAAm materials for load-bearing cartilage substitution.128
Figure 2.

PAMPS and PAAm networks of the double network hydrogel under tensile test. The highly cross-linked PAMPS network fractured, while loosely cross-linked PAAm network was still holding the gel stucture during extension.123 Reproduced with permission from ref (123). Copyright 2010 The Royal Society of Chemistry; http://pubs.rsc.org/en/Content/ArticleLanding/2010/SM/b924290b#!divAbstract.
b. Mimics of Natural Proteins
Natural hydrogels, including proteins and polysaccharides, have been used in biological applications and tissue engineering due to their biocompatibility, biodegradability, and biological functions.17 Natural polymers, such as alginate,129 chitosan,130,131 gelatin,99,132,133 and elastin134,135 are able to form physical hydrogels but often have poor mechanical properties.9 However, modification of natural polymers is often more difficult, with fewer chemical options compared to those available with synthetic polymers, and the purification of natural polymers often suffers from batch-to-batch variability. In addition, natural polymers extracted from animals or bacteria raise concerns about immunogenic reactions.118 A recent review includes details regarding polysaccharide-based hydrogels for tissue engineering applications;136 we include here descriptions of protein-based hydrogels based on recombinant polypeptides137 for tissue engineering applications.
i. Elastin
Elastin is one of the most important structural proteins in mammals, providing the elastomeric behavior of most tissues, including tendons and blood vessels.138 The canonical amino acid sequence that gives rise to the mechanical properties of elastin is the flexible VPGXG repeat, where X can be any natural amino acid except proline. Recombinant methods have enabled the development of an enormous variety of biosynthetic elastin-like polypeptides (ELPs).19,134,135,139−144
The inverse transition behavior of elastin, in which ELP forms coacervates above a critical transition temperature, has been widely studied as a function of pH, salt concentration, and temperature.145 The transition temperature can be tuned by variations in the amino acid sequence, where the addition of hydrophobic residues reduces the transition temperature.146 ELP nanoparticles have been produced to encapsulate and release bone morphogenetic proteins (BMP) for potential protein and drug delivery applications.147 With the advantages of ELPs, they have been incorporated into multicomponent materials (both chemically and physically cross-linked) to enhance both the mechanical and biological functions.148 Multiblock elastin polypeptides containing the hydrophobic IPAVG end block for physical cross-linking have shown high extension and tensile strength.148
To further improve the biological properties of ELPs, various cell adhesion peptide and degradation domains have been added to the ELP sequences to improve cell adhesion, spreading, and migration.149 An RGD peptide was incorporated on the surface of a multiblock ELP gel via maleimide–thiol chemistry to promote luminal endothelialization in vascular grafts;134 the surface-specific conjugation enhanced the adhesion and proliferation of both endothelial cells and mesenchymal stem cells. Other groups have taken advantage of the reversible, thermally responsive behavior of ELPs to form low-concentration, injectable hydrogels that can be cross-linked via disulfide bonding of cysteine residues in vivo.150 It has been possible to predict and tune the inverse transition temperature of a wide range of ELPs via sequence design.90,142,143,151−153
In addition to hydrogel matrix materials, ELPs also can form nanoparticles and nanofibers. Silk-elastin multiblock polypeptides can self-assemble into nanoparticles with the silk block in the core.103 Nanoparticles have also been formed from the elastin–mimetic hybrid copolymer PAA-VPGVG;154 in this particular case, the nanoparticles were formed by collective hydrogen binding and hydrophobic interactions, rather than by coacervation of the elastin-like domains, and are of interest in drug delivery applications. ELP electrospun fibers, cross-linked with glutaraldehyde in a vapor-initiated process and then rehydrated in NaCl buffer,155 have provided opportunities for the use of hydrogel fibers to guide cell direction and to mimic the orientations of cells in native tissue. The opportunities for employing ELPs in biomedical fields continue to expand, not only as a result of the mechanical properties that are comparable to those of native elastin, but also due to the responsive behavior of ELPs, which makes them highly versatile for drug delivery applications.
ii. Resilin
Resilin is another structural protein, found in insects, where it is located primarily in active ligament and tendons.156 The excellent resilience and energy storage allows resilin to recover from repetitive high-strain cyclic loading with essentially no hysteresis, even under high frequency conditions, which has an important role in insect flight and jumping157 and in sound production.158 Repetitive constructs of the consensus sequence of resilin from D. melanogaster (GGRPSDSYGAPGGGN) have been produced from the first exon of the Drosophila CG15920 gene via recombinant methods, and the polypeptide showed excellent mechanical properties comparable to those of native resilin.159 The unique resilience of cross-linked resilin-like polypeptide (RLP) and hybrid RLP hydrogels has motivated their use in applications requiring highly elastomeric and biomechanical functions, such as vocal fold therapeutics,160 artificial muscles,161 and cardiovascular applications.162 The RLPs show pH- and temperature-responsive behavior related to that of ELPs, although in addition to the inverse transition temperature, select RLPs can show dual phase transitions with both upper and lower solution critical temperatures.163
To improve the biological functionality of the RLP, our group has produced multiple constructs that incorporate cell adhesion domains (RGD), enzymatic degradation domains (MMP-sensitive), and heparin-binding domains (HBD) to yield a multibiofunctional material (Figure 3).160,164−167 RLP-based hydrogels can be cross-linked by the reaction of amines in the RLP sequence (Lys) with the small-molecule cross-linker tris(hydroxymethyl phosphine) propionic acid (THPP) or tris(hydroxymethyl phosphine) (THP). Hydrogels formed by these methods exhibited excellent mechanical properties characteristic of resilin, while improving cell adhesion and cell-mediated degradation. In studies from other groups, the bone morphogenetic protein-2 (BMP-2) peptide has been incorporated into RLP films derived from A. gambiae; the resulting surfaces promoted osteogenic differentiation of mesenchymal stem cells.168
Figure 3.

Resilin-like polypeptide hydrogels demonstrate useful mechanical properties and biological functions.160 Reproduced with permission from ref (160). Copyright 2013 The Royal Society of Chemistry; http://pubs.rsc.org/en/Content/ArticleLanding/2013/SM/c2sm26812d#!divAbstract.
Other recombinant constructs have combined the properties of multiple structural proteins into a hybrid resilin-elastin-collagen (REC) polypeptide.18 This polypeptide self-assembles into fibrous structures via the interactions of collagen, yield materials with a Young’s modulus between 0.1 and 3 MPa, consistent with those observed for native resilins and elastins. In a related example, the well-characterized GB1 domain was combined with random-coil resilin-like domains to produce multiblock mimics of the passive elastic muscle protein titin.161 The material showed high resilience at low strain and was durable at high strain, consistent with the observed properties of muscle.
We have also explored hybrid RLP materials produced with synthetic polymers as matrices for cardiovascular tissue engineering.162 The RLP was synthesized via biosynthetic methods and contained the RGD integrin-binding domain, MMP degradation domain, and heparin-binding domains of the sequences described above. Four-arm vinyl sulfone-terminated PEG was reacted with the cysteine-containing RLP via Michael-type addition. The resulting hybrid hydrogel maintained the mechanically active and biologically active domains, and supported the spreading of AoAFs during in vivo culture to a significantly greater extent than RLP-only hydrogels. Incorporating RLP and PEG together provides the mechanically durable and resilient hydrogel, with improved cell interactions, that may be useful in the engineering of mechanically active tissues.
c. Composite Matrices for Mechanical Reinforcement
Conventional hydrogels often exhibit weak mechanical strength and poor deformation (e.g., gels from gelatin and agarose),120 and increasing cross-linking density has been a common method for improving mechanical properties both natural and synthetic polymeric hydrogels.7 However, high cross-linking density results in restriction of the chains which yields stiff materials with limited extensibility and reduced water content in the swelled state,85 as well as compromised permeability and slow molecular diffusion.169 Composite hydrogels have thus been investigated as a strategy for improving the mechanical strength of hydrogel-based materials.170 These strategies employ traditional composite approaches in which a filler is either physically entrapped or chemically cross-linked within the hydrogel matrix to produce materials with increased mechanical strength. Mechanically stiff fillers, such as nanoclays, in the composite networks serve as reinforcement and as a multipoint cross-linker to improve the mechanical strength of the composite hydrogel, obviating the requirement for a high network density.171 The reorientation of the filler and polymeric network then serves to maintain the high elasticity of the hydrogel. In one example, nanocomposite hydrogels utilized exfoliated nanoclay to reinforce a PNIPAAm hydrogel; these materials showed both excellent mechanical strength (up to 1000 kPa) and high elasticity (up to 1000% strain-to-break).172−174 Composite hydrogels have since been produced to incorporate a broader scope of inorganic species including silica nanoparticles (SiNPs),175−177 metal nanoparticles,170,178 hydroxyapatite,22,29,179 carbon nanotubes (CNTs),180 and graphene oxide (GO) sheets181 as reinforcement. Although the strength and modulus of these organic–inorganic systems is significantly improved with the addition of the inorganic matrix, leaching of the inorganic species is a concern. In recent decades, the development of organic nanocrystals, organic particles, and electrospun polymer fibers have provided alternatives that avoid the need for the inorganic filler.
i. Nanocrystal-Reinforced Matrices
Polysaccharide nanocrystals, formed primarily by crystal-forming cellulose and chitin, have been utilized to replace inorganic filler in nanoparticle-reinforced hydrogels.21 The rod-like nanocrystals, also referred to as nanowhiskers, can be extracted from natural materials; cellulose nanocrystals are often extracted from cotton or ramie, and chitin nanocrystals are extracted from shrimp or crab.182,183 These nanocrystals have the advantage of being biocompatible and biodegradable, as well as having mechanical strength and moduli that are comparable to those of inorganic fillers (over 100 GPa).182 Different groups have incorporated cellulose nanocrystals (CNC) or chitins as reinforcement fillers for PAAm,184,185 PVA,186 chitosan,187 and carboxymethylcellulose (CMC)/hydroxyethylcellulose (HEC)188 hydrogels. The mechanical properties of the composite hydrogels generally increase with increased nanocrystal content.
CNCs have also been used, in electrospinning of PEO, to reinforce the resulting nanofibers;189 the composite nanofibers showed an increased modulus (38 MPa) compared to that of PEO fiber (15 MPa), and these properties depended on the CNC content. CNC-reinforced, injectable hydrogel comprising a carboxymethyl cellulose and dextran matrix have also been produced;21 chemically cross-linked, CNC-reinforced hydrogels showed a higher modulus compared to physically blended CNC hydrogels. The development of such polysaccharide nanocrystal composites has provided biocompatible and biodegradable fillers, which has enabled the use of nanocrystal composite hydrogels in tissue engineering. However, the sizes of the nanocrystals are limited in scope due to their extraction from naturally occurring materials; thus, the options for engineering properties by altering filler dimensions is also limited.
ii. Particle-Reinforced Matrices
In addition to nanocrystal-containing composite hydrogels, synthetic organic nanoparticles and microparticles also have been incorporated into hydrogels for mechanical reinforcement. For example, the uniform dispersion of monodisperse cationic polystyrene (c-PS) nanoparticles into a PAAm hydrogel improved the compression strength to 40 MPa compared to the original 70 kPa modulus of a PAAm-only hydrogel.190 The improvement in mechanical properties was attributed to the uniform dispersion of monodisperse c-PS that were prefabricated by emulsion polymerization. Another group incorporated the thermoresponsive PNIPAAm microgels into the PAAm matrix and evaluated the mechanical properties below and above the LCST of the PNIPAAm that led to understanding the effect of soft and hard filler on the hydrogel.191 An advantage of the synthetic organic particles in the composite hydrogel is that they can be used not only reinforce the mechanical properties, but can also serve as a vehicle for drug and/or protein delivery. The incorporation of block copolymer micelles (BCMs) in PAAm hydrogels via free radical polymerization resulted in hydrogels that sustain significant elongation (up to 480%),192 and that could also be loaded with hydrophobic drugs (via loading of the hydrophobic core of the BCMs during micelle formation) to permit drug delivery upon mechanical deformation of the hydrogel. Other organic nanoparticles, including hyperbranched polymers,193 polymeric nanoparticles,190,194 micelles,192 and nanogels,178,195,196 have also been used in the production of composite hydrogels for controllable drug delivery. For example, hyperbranched polyester (HPE) hydrogels enabled the entrapment of the hydrophobic drug dexamethasone acetate within the HPE hydrophobic cavities without causing drug aggregation and showed longer sustained release compared to drug encapsulated in a PEG hydrogel.193 The drug-loaded nanoparticle composite hydrogel was able to achieve sustained release and a high drug concentration for local delivery,172 and drug delivery could also be triggered with stimuli such as temperature or mechanical deformation.196
Composite hydrogels are not limited to those formed with nanoparticles; microgel hydrogels have also been shown to improve strength and torsion resistance. Poly(2-acrylamido-2-methylpropanesulfonic sodium) (PNaAMPS) microgel-reinforced PAAm double-network hydrogel films have shown high tensile strength (up to 2.6 MPa with a strain up to approximately 10%; Figure 4).197 Preformed microgels were incorporated into a PAAm hydrogel to form two-phase composite materials. The additional PAAm double network resulted in even greater mechanical enhancement compared to microgel-reinforced single-network hydrogels (e.g., a modulus of nearly 120 kPa compared to the modulus of the reinforced single network of approximately 50 kPa).198
Figure 4.
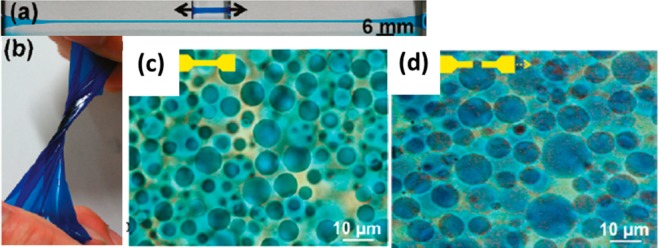
Microgel-reinforced double network PAAm hydrogel that exhibited excellent extension (a) and torsion (b). Microgel before tensile deformation (c) and after deformation (d).197 Reproduced with permission from ref (197). Copyright 2011 American Chemical Society.
Nanoparticles and microparticles can be fabricated via various methods, including emulsion polymerization,60,190,199,200 self-assembly,103,145,147 and phase separation.201−203 In one example, 8-arm PEG has been used to form PEG microspheres via phase-separation in aqueous media.201−203 The PEG microspheres could be cross-linked via the reaction of amines with vinyl sulfone or with acrylate, and the sizes of the microspheres were controllable in different media, with improved cell viability in a microsphere-based scaffold.201 Compared to microspheres formed via emulsion polymerization, these microspheres do not require extensive solvent exchange or washing to remove organic solvent, although the reaction conditions needed to be precisely controlled to prevent bulk gel gelation. Improved control over the reaction kinetics and changes in particle sizes over time will enable better control of the microspheres and properties of the resulting matrices.
iii. Fiber-Reinforced Matrices
The native ECM comprises a complicated and often anisotropic structure, with a combination of fibers and network polymers, such as collagen fibers aligned in tissue.27 Thus, the use of fibrous structures in designed materials has been employed to better mimic native ECM and guide cell direction; electrospinning has been a widely used and simple method to produce controlled nanoscale fibers.204 The applied high-voltage electrostatic force draws a polymer fiber from polymer solutions,205 and the resulting fibers can collected into isotropic or aligned fibrous mats. The activities of cardiomyocytes cultured on random and aligned electrospun biodegradable polyurethane fiber mats were different, with greater multicellular organization on the aligned fiber mats.206 Materials comprising poly(lactic-co-glycolic acid) (PLGA)/gelatin electrospun nanofibrous have also been produced to mimic cardiac tissue;207 after electrospinning, the hydrophilic gelatin could be rehydrated to yield fiber-like hydrogels. Cardiomyocytes cultured on the PLGA/gelatin nanofiber showed enhanced attachment and spreading. Thermoresponsive multiblock poly(PEG/PPG/PCL urethane) hydrogel nanofibers have also been produced for temperature-mediated BSA release from fibers,208 and encapsulated proteins, such as nerve growth factor (NGF)209 and lysozyme,210 maintained their bioactivity after release from PCL-based electrospun fibers.
Nanofibers are also commonly employed fillers used to enhance the mechanical properties of hydrogels. Fibers produced from several biocompatible and biodegradable polymers, including PCL, poly-l-lactide (PLLA), and chitosan, have been studied in different hydrogel systems. Chitosan nanofibers (CNF) incorporated in a PAAm hydrogel improved the mechanical properties of the CNF/PAAm hydrogel compared with those of chitosan/PAAm hydrogels, showing a 2.5-fold higher compressive stress to 50.2 kPa (at 95% strain) than the chitosan/PAAm hydrogels.211 In another example, biodegradable PCL was electrospun with gelatin to form a PCL-gelatin core–shell fiber,20 which was mixed with gelatin and cross-linked to form a composite hydrogel. The fibrous composite hydrogel showed an improvement in modulus to 20.3 kPa from 3.2 kPa (for a gelatin-only hydrogel). In addition, the fibrous structure of the PCL-gelatin alone served to direct cell orientation in a 2D aligned electrospun fiber mat,206 similar to other studies described above. The fibrous composite hydrogel provides a hydrated local environment and 3D support for cells, which is an advantage over traditional fiber mat scaffolds. The construction of aligned fiber hydrogel constructs for cell culture applications remains an active research area owing to its potential in various therapies, including the cardiovascular area.
4. Hybrid Materials with Engineered Biological Functions
Although the strategies described above have provided alternatives for achieving mechanically robust networks, a lack of cell–matrix interaction often leads to the failure of the biomaterials in in vitro and in vivo studies.212−214 Various cell–matrix interactions, including cell adhesion and matrix degradation are required for cell growth and migration,25 and hybrid hydrogels can be employed to capture these properties (Figure 5) in a chemically and mechanically versatile substrate.
Figure 5.

Important materials design considerations for tissue engineering, including cell adhesion peptide, protease sensitive peptide for cell-mediated matrix degradation, and presence of signaling molecules.
a. Cell Adhesion
An inherent limitation of synthetic materials in biological applications is the lack of cell–matrix interactions, which limits cell attachment, remodeling, and migration in a scaffold. Incorporating ECM molecules and cell adhesive peptides (such as those from fibronectin and laminin) in the matrix materials has been widely shown to provide significant enhancement in cellular interactions with various scaffolds.26,27,118,214−216 The integrin-mediated cell adhesion facilitated by these macromolecules provides for cell attachment, spreading, actin organization, and focal adhesion.214 The Arg-Gly-Asp tripeptide (RGD) has been the most commonly employed cell adhesive peptide in hybrid hydrogel systems because of its effective cell adhesion through most integrins.215 Besides the RGD peptide, sequences derived from laminin (LN; such as IKVAV, YIGSR) and fibronectin (FN; such as KQAGDV, REDV) also have been used to induce cell adhesion on hydrogel matrices.118 Table 1 lists additional cell adhesion peptides that have been employed in hydrogel matrices; these sequences, and others, have shown value for stabilizing cells in matrices, as well as facilitating cell migration and maintaining cell functions.217−221
Table 1. Commonly Employed Cell Adhesion Peptides Used in Hydrogels.
| peptides | origin | hydrogel | cells | refs |
|---|---|---|---|---|
| RGD | FN, LN, collagen | PEG, ELP, RLP | endothelial cells, hMSCs, AoAF, islet | (72,134,160,162,222) |
| KQAGDV | FN | PEG | human aortic smooth muscle cells (HASMCs) | (223,224) |
| REDV | FN | PEG | endothelial progenitor cells (EPCs) | (220) |
| PHSRN | FN | PEG, HA | monocyte and valvular interstitial cells (VICs) | (225) |
| IKVAV | LN | SAP, agarose | neural stem cells and PC12 cells | (226) |
| YIGSRG | LN | PEG | endothelial progenitor cells (EPCs) | (220) |
| PDSGR | LN | PEG | murine pancreatic β-cells | (227) |
| LRE | LN | PEG | murine pancreatic β-cells | (227) |
| IKLLI | LN | PEG | murine pancreatic β-cells | (227) |
| GFOGER | collagen-I | PEG | hMSCs | (117) |
| VAPG | elastin | PEG | human aortic smooth muscle cells | (223) |
b. Degradation
Besides cell adhesion, controllable degradation of the matrix material is also important for cell growth and tissue regeneration. The designed scaffold has to degrade at a rate comparable with cell growth and deposition of ECM molecules. Perhaps the most commonly used degradation mechanism for synthetic hydrogels is hydrolytic degradation of ester linkages or polyester segments in polymers.118 Despite the widespread and simple application of these hydrolytic strategies, however, hydrolytic degradation rates are difficult to control in vivo and are not controlled by cell growth.137,228,229 Therefore, cell-mediated degradation strategies have been employed to optimize scaffold degradation with ECM deposition.25,54,218,219,230,231
Matrix metalloproteinase (MMP)-sensitive peptides are a class of enzyme-sensitive peptides derived from native ECM proteins, such as collagen or elastin, that promote cell-mediated matrix degradation;118 Table 2 shows a range of enzyme-sensitive peptides used for these applications. The use of these sequences offers substantive flexibility in controlling matrix degradation, as the substitution of amino acids in a MMP-sensitive peptide modifies degradation kinetics.213 The degradation rates of the materials can extend over a wide range of time scales by simple variations of the amino acids in the sequences, which can provide sufficient control for achieving degradation times that match the needs of a given application. In one example, the morphology of hMSCs encapsulated in MMP-sensitive peptide cross-linked PEG hydrogel depends on the concentration of MMP-sensitive peptide in the hydrogel; variations in the peptide concentration in the hydrogel also permitted the control of hMSC differentiation in different culture media.54
Table 2. Commonly Employed Enzymatically Cleavable Peptides Used in Hydrogels.
| peptides | hydrogel | enzyme | cells | refs |
|---|---|---|---|---|
| GPQG ↓ IAGQ | PEG | MMP-1, collagenase | human foreskin fibroblasts | (213,230) |
| GPQG ↓ IWGQ | PEG, RLP, HA | MMP-1, collagenase | human foreskin fibroblasts, hMSCs | (54,160,213,230,232) |
| GPQG ↓ PAGQ | PEG | collagenase | (230) | |
| L ↓ GPA | PEG | MMP-1 | human dermal fibroblasts (HDFs) and HASMCs | (224) |
| YK ↓ NRD | PEG | plasmin | (233) | |
| VR ↓ N | PEG, HA | plasmin | MSC | (234) |
| CGGY ↓ C | PEG | chymotrypsin | (51) | |
| AAPV ↓ RGGG | PEG | elastase | human neutrophil elastase | (115) |
| AAAAAAAAA | PEG | elastase | human dermal fibroblasts (HDFs) and HASMCs | (224) |
| PEN ↓ FF | PEG | MMP-13 | hMSCs | (235) |
| LVG ↓ LIG | alginate, pluronic | MMP-2 | hMSCs | (236,237) |
In addition to the use of MMP-sensitive peptides for cell-mediated matrix degradation, hydrogels with controlled degradation rates have also been widely employed in drug delivery. The incorporation of a human neutrophil elastase (HNE)-sensitive peptide in a PEG hydrogel via thiol–ene chemistry115,238 was employed to trigger the release of a model protein upon triggered degradation of the HNE-sensitive sequence,238 indicating the potential for cell-mediated degradation in drug delivery applications.239,240 Controllable matrix degradation is also important in 3D cell culture. Relevant examples include the use of a substrate, carboxybetaine methacrylate (CBMA), for reaction with a disulfide containing cross-linker via radical polymerization to form a hydrogel in the presence of cells. During cell culture, this hydrogel rapidly degrades owing to the reaction of the disulfide-containing cross-linker with the cysteine-containing media, permitting recovery of the encapsulated cells.52 Recent exploitation in our laboratories of retro Michael-type addition has also been employed to control hydrogel degradation. In these cases, degradation of select thioether succinimide bonds has been employed to degrade PEG/heparin hydrogels and release heparin at glutathione (GSH) concentrations consistent with intracellular concentrations.241 The degradation mechanism can also be employed for GSH-triggered release of model proteins from PEG-only hydrogels, providing an opportunity for targeted protein delivery over time scales unique from those of disulfide- or hydrolytic-mediated mechanisms.242 A recent review provides a comprehensive description of hydrogel degradation in cellular microenvironments via hydrolytic, enzymatic, thiol-exchange, and photolytic mechanisms.228
c. Immunological Modulation
i. Tissue Regeneration
The recognition of materials by macrophages, which release chemokines to recruit immune cells, and subsequent chronic immune responses often lead to rejection of the implants or scaffolds.35 Recent studies suggest that an active modulated immune response can direct tissue regeneration;243 inflammatory cytokines have an important role in initiation of acute inflammation, cell proliferation, and modulation of tissue healing.35,244 Interleukin-1 (IL-1), granulocytecolony stimulating factor (G-CSF), granulocyte macrophage colony stimulating factor (GM-CSF), CC-chemokine ligand 2 (CCL2), and CCL5 are several of the important factors for tissue healing.25 Hydrogels that deliver GM-CSF topically have been shown to enhance wound healing in patients with second degree burns.245 In addition, chemokines can induce chemotaxis that guides progenitor and stem cell migration and tissue reconstruction. Stromal derived factor 1 (SDF-1), in one such example, was loaded in PEG-heparin hydrogels and showed significant improvement in guiding the migration of early endothelial progenitor cells (eEPCs) compared to gels that did not contain SDF-1;246 the incorporated SDF-1 also reduced scar tissue formation and promoted improved tissue healing.247 Growth factors and tolerance-promoting antigens also have also been shown to enhance tissue regeneration.31,248 For example, regeneration of muscle in a mouse model could be promoted via the use of an RGD-modified alginate hydrogel for codelivery of vascular endothelial growth factor (VEGF), insulin-like growth factor-1 (IGF-1), and myoblasts;249 the VEGF promoted angiogenesis and IGF-1 promoted myogenesis. Hydrogels able to incorporate and controllably release multiple biomolecules, including cells, cytokines, and growth factors, may improve tissue regeneration by the minimization of chronic immune responses and enhancement of tissue growth.
ii. Cancer Therapy
In addition to tissue healing and regeneration, modulated innate immune responses can also be useful for vaccination, treatment of autoimmune disease, or cancer therapies. Studies have shown that PLGA can induce overexpression of TNF-α and IL-6 from dendritic cells (DCs) and enhance immune response; furthermore, PLGA microparticles induced a greater response than PLGA films.250 The enhancement of TNF-α production can also lead to cell death, with possible applications for cancer treatment. An HA/PEG hydrogel was employed to encapsulate a PEGylated tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL).251 The HA/PEG hydrogel showed sustained release of the PEGylated TRAIL in vitro and in vivo, and the PEGylated TRAIL stimulated more apoptosis and greater antitumor efficiency compared to a TRAIL-only hydrogel in an in vivo study on Mia Paca-2 cell-xenografted BALB/c nu/nu mice.251 The tumor volume and tumor weight was significantly less than the blank control at 27 days of treatment, as shown in Figure 6.251 In addition to the release of single classes of molecules, the benefits of codelivery have also been indicated for similar applications. In particular, codelivery of cytokine interleukin-2 (IL-2) and oligodeoxynucleotide immunostimulatory agents, from injectable alginate hydrogels, has been investigated.129 IL-2 recruits T-cells to the local site for immunostimulation by the oligodeoxynucleotides, which stimulate both innate and adaptive immune responses and inhibit metastasis and tumor growth.38
Figure 6.

(a) Mia Paca-2 cell-xenografted BALB/c nu/nu mice treated with blank, TRAIL, or PEG-TRAIL HA hydrogels for 27 days. (b) Removed tumors from each treatment group (n = 4), (c) tumor volume, and (d) tumor weight after 27 days.251 Reproduced with permission from ref (251). Copyright 2014 Elsevier.
The unmethylated, single-stranded cytosine-phosphate-guanine (CpG) oligodeoxynucleotides (ODN) are common activating agents of dendritic cells (DCs),38 which has been the basis for their incorporation into many types of polymeric materials and carriers for stimulation of immune responses, including polymer conjugation252 and matrix incorporation.253 In a particular study, CpG-coated alginate microspheres were encapsulated in an alginate matrix along with IL-2; the microspheres immobilized and modulated the release of the CpG ODN, which enhanced the activation of bone-marrow-derived DCs and further activated tumor-specific, cytotoxic T-cells.253 The hybrid gel was able to modulate the sustained release of the CpG and show enhanced antitumor efficiency compare to CpG-only injections in mice. Such novel hybrid hydrogels offer great promise in modulating the release rates and sequential delivery of antitumor factors. These approaches are additionally powerful when deployed with injectable hydrogels, which allow direct injection at tumor sites for sustained and targeted cancer treatment.
iii. Gene Therapy
Gene therapy aims to treat disease by promotion of essential gene expression by delivered plasmid DNA (pDNA) or by gene silencing by small interfering RNA (siRNA) to target cells.254 Genetic materials such as DNA and RNA, however, are rapidly degraded by DNases and RNases and, thus, require protection; a wide variety of approaches therefore have been developed for producing vectors for gene delivery.255−258 Electrostatic complexation of DNA with polyethylenimine (PEI), prior to release and internalization by the targeted cells, is likely the most commonly employed strategy for complexing and delivering DNA. In a recent example, a PEI–poly(organophosphazene) conjugate was used to bind siRNA and form a thermoresponsive hydrogel owing to the thermosensitive poly(organophosphazene) segment.259 The hydrogel exhibits a gelation temperature of approximately 37 °C and can thus be used in injectable gene therapy, serving as a reservoir for sustained release of PEI–siRNA polyplexes upon degradation of the ester linkage. An acrylated disulfide containing siRNA macromer was employed in particle replication in nonwetting templates (PRINT) technology for formation of nanogels.260 Cleavage of the disulfide linkage in the presence of reducing agents, which are prevalent inside the cell, promoted cleavage and release of siRNA in the intracellular environment. The hydrogel served as additional protection for gene delivery applications.
The delivered vector often activates the innate immune response that leads to activation of antigen-presenting cells (APCs) against the vector and the therapeutic gene.261 Vaccine therapies and gene therapies for cancer and immunodeficiency benefit from the immune reaction that recruits DCs and APCs for immune activation and target transfection, although this response must be controlled in order to be useful. Injectable, composite hydrogels for sequential delivery of chemokines, siRNA, and DNA have thus been developed.262 The siRNA and DNA were loaded into PLGA nanoparticles and the chemokines were encapsulated in a dextran/PEG hydrogel matrix. The chemokine attracted dendritic cells (Figure 7) and promoted an immune response, while the siRNA and DNA induced gene silencing of IL-10 and immune modulation of the DCs by upregulation of phenotypic surface markers.
Figure 7.
(a) Schematic multicomponent hydrogels for immunotherapies. The chemokine signals the migration of the dendritic cells to the hydrogel and siRNA-DNA loaded nanoparticles lead to gene silencing and immune modulation. (b) Primary antigen-presenting cells (APCs) migrating in response to chemokine released from control hydrogels (top), bolus dose (middle), and chemokine loaded hydrogels (bottom) at 0, 4, and 18 h.262 Reproduced with permission from ref (262). Copyright 2009 Elsevier.
Besides allowing the sequestration and release of molecules to attract target cells, peptide-coated nanoparticles or nanogels have also been used for targeted gene delivery and enhancement of cell transfection via receptor-peptide binding.263 PNIPAAm nanogels coated with the YSAYPDSVPMMS (YSA) peptide bound erythro poietin-producing hepatocellular (Eph) A2 receptors, which resulted in localization of the nanogels in cells with high EphA2 expression, a common marker in tumor cells. This specific peptide-receptor binding has the potential to be used more broadly for targeting specific cells and promoting higher gene transfection.
5. Conclusions and Future Perspectives
Multifunctional hydrogels exhibit improved mechanical and biological properties that can be modulated via chemical and physical methods. The existence of well-developed chemistries for bioconjugation and cross-linking, including an expanding range of click reactions, has enabled the controlled incorporation of a variety of multifunctional groups and the design of specialized cross-linked networks containing composite structures and both synthetic and biological materials. Strategies for increasing cross-linking density (to improve modulus), while at the same time maintaining elasticity, have been of enormous interest and promise. The mechanical properties can be enhanced by judicious design of the matrix polymers (and copolymers) and/or the components in the gel; the combination of synthetic and natural polymers offers interesting opportunities to obtain biomechanically active hydrogels. Materials based on elastin and resilin can provide mechanically active function that mimics the biomechanical properties of the native tissue. However, comprehensive studies on the cellular response and in vivo studies of these synthetic and natural hybrid hydrogels remain limited.
The development of composite hydrogels has provided a versatile alternative approach for improving the strength of hydrogels via the use of a stiff second network that reinforces the weak hydrogel network, or via the incorporation of particles in the hydrogel matrix. Hybrid two-phase hydrogels also provide an addition platform for stimuli-induced drug delivery, with the drug stably encapsulated in the second phase until a stimulus is applied. The applications of composite hydrogels as tissue engineering scaffolds has been useful for incorporating drugs into matrices, and modulating the codelivery drugs or molecules at different release rates, while enhancing the mechanical strength. Additional studies that investigate the ratio of the two phases, and the resulting impact on mechanical properties and release kinetics of cargo from the hybrid hydrogel, are needed to inform the design of materials that can control the release of multiple drugs. In addition, while most composite hybrid hydrogels are produced in two steps (particle fabrication and subsequent encapsulation into hydrogel matrix), strategies that would simplify composite gel production into a single step would find significant value, as it would eliminate the need for additional purification of particles prior to their incorporation into hydrogels for biomedical uses. Extensive biological studies are needed to evaluate those materials for such use.
The ability to encapsulate viable cells in 3D formats is a step toward effective cell delivery and tissue regeneration. The incorporation of bioactive peptides has been widely employed to control cell attachment, proliferation and differentiation within synthetic hydrogels, and cell-mediated degradation of these matrices has improved cell growth and spreading. Appropriate design of multicomponent hydrogels has enabled interesting and many untapped opportunities for programming cell behavior to stimulate simultaneous immunotherapeutic treatment and tissue regeneration. While most of the immunomodulating hydrogels studied have been weak physical hydrogels, such as alginate, there is demonstrated and continued need to employ chemically cross-linked and mechanically robust hydrogels for understanding the impact of the matrix on immune response. While it is well-known that the mechanical properties of a matrix modulate cell behavior, the impact of the mechanical properties of a matrix on DCs and their resulting cytokine profile has not yet been studied in detail; further understanding of these processes will inform tissue regeneration, cancer, or gene therapies.
Taken together, the body of work described herein clearly illustrates that the potential of multicomponent hybrid hydrogels for a variety of applications in tissue regeneration and drug delivery. By incorporating and modulating the mechanical functional and bioactive components in the network, the mechanical and biological properties of the hydrogel can be tuned independently without sacrificing one or the other. In the future, hybrid hydrogels are expected to further mimic the microenvironment for cells and tissue reorganization. The mechanically active components should be aimed not only at affecting the bulk mechanical properties, but also should capture the micromechanical properties in native tissue. Multicomponent hydrogels with well-organized domains will offer significant opportunities for these materials.
Acknowledgments
Related work in the authors’ laboratories has been supported by grants from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (P20-RR017716 to K.L.K.), the National Institute on Deafness and Other Communication Disorders (NIDCD, RO1-DC011377A to K.L.K), the National Science Foundation (DMR-0907478 to K.L.K), and the National Heart, Lung, and Blood Institute (RO1-HL108110) and Nemours Foundation.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Patterson J.; Martino M. M.; Hubbell J. A. Mater. Today 2010, 13, 14–22. [Google Scholar]
- La Y.-H.; McCloskey B. D.; Sooriyakumaran R.; Vora A.; Freeman B.; Nassar M.; Hedrick J.; Nelson A.; Allen R. J. Membr. Sci. 2011, 372, 285–291. [Google Scholar]
- Mateescu A.; Wang Y.; Dostalek J.; Jonas U. Membranes 2012, 2, 40–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. J.; Dostalek J.; Knoll W. Biosens. Bioelectron. 2010, 26, 1425–1431. [DOI] [PubMed] [Google Scholar]
- Länge K.; Rapp B. E.; Rapp M. Anal. Bioanal. Chem. 2008, 391, 1509–1519. [DOI] [PubMed] [Google Scholar]
- Jia X.; Kiick K. L. Macromol. Biosci. 2009, 9, 140–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennink W. E. E.; van Nostrum C. F. F. Adv. Drug Delivery Rev. 2012, 64, 223–236. [Google Scholar]
- Lee K. Y.; Mooney D. J. Chem. Rev. 2001, 101, 1869–1880. [DOI] [PubMed] [Google Scholar]
- Nicodemus G. D.; Bryant S. J. Tissue Eng., Part B 2008, 14, 149–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seliktar D. Science 2012, 336, 1124–1128. [DOI] [PubMed] [Google Scholar]
- Moses J. E.; Moorhouse A. D. Chem. Soc. Rev. 2007, 36, 1249–1262. [DOI] [PubMed] [Google Scholar]
- Golas P. L.; Matyjaszewski K. Chem. Soc. Rev. 2010, 39, 1338–1354. [DOI] [PubMed] [Google Scholar]
- Klok H.-A. Macromolecules 2009, 42, 7990–8000. [Google Scholar]
- Kopecek J. Biomaterials 2007, 28, 5185–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopeček J.; Yang J. Angew. Chem., Int. Ed. 2012, 51, 7396–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecek J. J. Polym. Sci., Part A: Polym. Chem. 2009, 47, 5929–5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.; Marchant R. E. Expert Rev. Med. Devices 2011, 8, 607–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracalello A.; Santopietro V.; Vassalli M.; Marletta G.; Del Gaudio R.; Bochicchio B.; Pepe A. Biomacromolecules 2011, 12, 2957–2965. [DOI] [PubMed] [Google Scholar]
- DiMarco R. L.; Heilshorn S. C. Adv. Mater. 2012, 24, 3923–3940. [DOI] [PubMed] [Google Scholar]
- Kai D.; Prabhakaran M. P.; Stahl B.; Eblenkamp M.; Wintermantel E.; Ramakrishna S. Nanotechnology 2012, 23, 095705. [DOI] [PubMed] [Google Scholar]
- Yang X.; Bakaic E.; Hoare T.; Cranston E. D. Biomacromolecules 2013, 14, 4447–4455. [DOI] [PubMed] [Google Scholar]
- Gaharwar A. K.; Dammu S. A.; Canter J. M.; Wu C.-J.; Schmidt G. Biomacromolecules 2011, 12, 1641–1650. [DOI] [PubMed] [Google Scholar]
- Sabnis A.; Wadajkar A. S.; Aswath P.; Nguyen K. T. Nanomedicine 2009, 5, 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyondi D.; Webster T. J.; Banerjee R. Int. J. Nanomed. 2013, 8, 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice J. J.; Martino M. M.; De Laporte L.; Tortelli F.; Briquez P. S.; Hubbell J. A. Adv. Healthc. Mater. 2013, 2, 57–71. [DOI] [PubMed] [Google Scholar]
- Aizawa Y.; Owen S. C.; Shoichet M. S. Prog. Polym. Sci. 2012, 37, 645–658. [Google Scholar]
- Place E. S.; Evans N. D.; Stevens M. M. Nat. Mater. 2009, 8, 457–470. [DOI] [PubMed] [Google Scholar]
- Kim S. H.; Kiick K. L. Macromol. Rapid Commun. 2010, 31, 1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudalla G. A.; Murphy W. L. Adv. Funct. Mater. 2011, 21, 1754–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall J. D.; Lin C.-C.; Anseth K. S. Biomacromolecules 2011, 12, 1051–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tayalia P.; Mooney D. J. Adv. Mater. 2009, 21, 3269–3285. [DOI] [PubMed] [Google Scholar]
- Callahan L. a S.; Ganios A. M.; McBurney D. L.; Dilisio M. F.; Weiner S. D.; Horton W. E.; Becker M. L. Biomacromolecules 2012, 13, 1625–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H. J.; Park T. G. Adv. Drug Delivery Rev. 2007, 59, 249–262. [DOI] [PubMed] [Google Scholar]
- Lin C.-C.; Metters A. T.; Anseth K. S. Biomaterials 2009, 30, 4907–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehler R.; Graham J.; Shea L. Biotechniques 2011, 51, 239–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz S.; Rammelt S.; Scharnweber D.; Simon J. C. Biomaterials 2011, 32, 6692–6709. [DOI] [PubMed] [Google Scholar]
- Schroeder A.; Heller D. A.; Winslow M. M.; Dahlman J. E.; Pratt G. W.; Langer R.; Jacks T.; Anderson D. G. Nat. Rev. Cancer 2012, 12, 39–50. [DOI] [PubMed] [Google Scholar]
- Nishikawa M.; Mizuno Y.; Mohri K.; Matsuoka N.; Rattanakiat S.; Takahashi Y.; Funabashi H.; Luo D.; Takakura Y. Biomaterials 2011, 32, 488–494. [DOI] [PubMed] [Google Scholar]
- Moon J. J.; Huang B.; Irvine D. J. Adv. Mater. 2012, 24, 3724–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas J.; Mantovani G.; Haddleton D. M. Macromol. Rapid Commun. 2007, 28, 1083–1111. [Google Scholar]
- Pokorski J.; Breitenkamp K. J. Am. Chem. Soc. 2011, 133, 9242–9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev G. V.; Mogilevich M. M.. Three-Dimensional Free-Radical Polymerization Cross-Linked and Hyper-Branched Polymers; Springer: Berlin, Heidelberg, 2009; pp 33–81. [Google Scholar]
- Azagarsamy M. A.; Anseth K. S. ACS Macro Lett. 2013, 2, 5–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeForest C.; Anseth K. Nat. Chem. 2011, 3, 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmo C. M.; Owen S. C.; Shoichet M. S. Biomacromolecules 2011, 12, 824–830. [DOI] [PubMed] [Google Scholar]
- Hoyle C. E.; Lowe A. B.; Bowman C. N. Chem. Soc. Rev. 2010, 39, 1355–1387. [DOI] [PubMed] [Google Scholar]
- Hoyle C. E.; Bowman C. N. Angew. Chem., Int. Ed. 2010, 49, 1540–1573. [DOI] [PubMed] [Google Scholar]
- Lowe A.; Hoyle C.; Bowman C. J. Mater. Chem. 2010, 4745–4750. [Google Scholar]
- Hutson C. B.; Ph D.; Nichol J. W.; Aubin H.; Bae H.; Khademhosseini A. Tissue Eng., Part A 2011, 17, 1713–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durst C. A.; Cuchiara M. P.; Mansfield E. G.; West J. L.; Grande-Allen K. J. Acta Biomater. 2011, 7, 2467–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.-C.; Raza A.; Shih H. Biomaterials 2011, 32, 9685–9695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien H.-W.; Tsai W.-B.; Jiang S. Biomaterials 2012, 33, 5706–5712. [DOI] [PubMed] [Google Scholar]
- Williams C. G.; Malik A. N.; Kim T. K.; Manson P. N.; Elisseeff J. H. Biomaterials 2005, 26, 1211–1218. [DOI] [PubMed] [Google Scholar]
- Anderson S. B.; Lin C.-C.; Kuntzler D. V.; Anseth K. S. Biomaterials 2011, 32, 3564–3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih H.; Lin C.-C. Biomacromolecules 2012, 13, 2003–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elisseeff J.; Anseth K. Proc. Natl. Acad. Sci. U.S.A. 1999, 96, 3104–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegwart D. J.; Oh J. K.; Matyjaszewski K. Prog. Polym. Sci. 2012, 37, 18–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.; Matyjaszewski K. Polym. Chem. 2012, 3, 1813–1819. [Google Scholar]
- Forbes D. C.; Creixell M.; Frizzell H.; Peppas N. A. Eur. J. Pharm. Biopharm. 2013, 84, 472–478. [DOI] [PubMed] [Google Scholar]
- Oliveira M. A. M.; Boyer C.; Nele M.; Pinto J. C.; Zetterlund P. B.; Davis T. P. Macromolecules 2011, 44, 7167–7175. [Google Scholar]
- Boyer C.; Bulmus V.; Davis T.; Ladmiral V. Chem. Rev. 2009, 109, 5402–5436. [DOI] [PubMed] [Google Scholar]
- Ercole F.; Thissen H.; Tsang K.; Evans R. A.; Forsythe J. S. Macromolecules 2012, 45, 8387–8400. [Google Scholar]
- Liu J.; Cui L.; Kong N.; Barrow C. J.; Yang W. Eur. Polym. J. 2014, 50, 9–17. [Google Scholar]
- Gao W.; Liu W. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 15231–15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeler J. C.; Woodman B. F.; Averick S.; Miyake-Stoner S. J.; Stokes A. L.; Hess K. R.; Matyjaszewski K.; Mehl R. A. J. Am. Chem. Soc. 2010, 132, 13575–13577. [DOI] [PubMed] [Google Scholar]
- Chirra H. D.; Biswal D.; Hilt J. Z. Polym. Adv. Technol. 2011, 22, 773–780. [Google Scholar]
- Yoon J. A.; Kowalewski T.; Matyjaszewski K. Macromolecules 2011, 44, 2261–2268. [Google Scholar]
- Chang C.-W.; Bays E.; Tao L.; Alconcel S. N. S.; Maynard H. D. Chem. Commun. 2009, 3580–3582. [DOI] [PubMed] [Google Scholar]
- Hu X.; Li D.; Zhou F.; Gao C. Acta Biomater. 2011, 7, 1618–1626. [DOI] [PubMed] [Google Scholar]
- Malkoch M.; Vestberg R.; Gupta N.; Mespouille L.; Dubois P.; Mason A. F.; Hedrick J. L.; Liao Q.; Frank C. W.; Kingsbury K.; Hawker C. J. Chem. Commun. 2006, 2774–2776. [DOI] [PubMed] [Google Scholar]
- Nimmo C. M.; Shoichet M. S. Bioconjugate Chem. 2011, 22, 2199–2209. [DOI] [PubMed] [Google Scholar]
- Liu S. Q.; Ee P. L. R.; Ke C. Y.; Hedrick J. L.; Yang Y. Y. Biomaterials 2009, 30, 1453–1461. [DOI] [PubMed] [Google Scholar]
- Van Dijk M.; van Nostrum C. F.; Hennink W. E.; Rijkers D. T. S.; Liskamp R. M. J. Biomacromolecules 2010, 11, 1608–1614. [DOI] [PubMed] [Google Scholar]
- DeForest C. A.; Polizzotti B. D.; Anseth K. S. Nat. Mater. 2009, 8, 659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin J. M.; Prescher J. A.; Laughlin S. T.; Agard N. J.; Chang P. V.; Miller I. A.; Lo A.; Codelli J. A.; Bertozzi C. R. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 16793–16797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeForest C. A.; Anseth K. S. Angew. Chem., Int. Ed. 2012, 124, 1852–1855. [Google Scholar]
- Sletten E. M.; Bertozzi C. R. Angew. Chem., Int. Ed. 2009, 48, 6974–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A. Macromol. Chem. Phys. 2010, 211, 1417–1425. [Google Scholar]
- Zheng J.; Callahan L. A. S.; Hao J.; Guo K.; Wesdemiotis C.; Weiss R. A.; Becker M. L. ACS Macro Lett. 2012, 1, 1071–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman M. L.; Royzen M.; Fox J. M. J. Am. Chem. Soc. 2008, 130, 13518–13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. S.; Tangpeerachaikul A.; Selvaraj R.; Taylor M. T.; Fox J. M.; Ting A. Y. J. Am. Chem. Soc. 2012, 134, 792–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelhuis N. Ten; Diehl C.; Schlaad H. Macromolecules 2008, 9946–9947. [Google Scholar]
- Gandavarapu N. R.; Azagarsamy M. a; Anseth K. S. Adv. Mater. 2014, 26, 2521–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall J. D.; Anseth K. S. Biomacromolecules 2012, 13, 2410–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman A. S. Adv. Drug Delivery Rev. 2012, 64, 18–23. [Google Scholar]
- Dong Y.; Saeed A. O.; Hassan W.; Keigher C.; Zheng Y.; Tai H.; Pandit A.; Wang W. Macromol. Rapid Commun. 2011, 120–126. [DOI] [PubMed] [Google Scholar]
- Stahl P. J.; Romano N. H.; Wirtz D.; Yu S. M. Biomacromolecules 2010, 11, 2336–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbern J. C.; Minami E.; Stayton P. S.; Murry C. E. Biomaterials 2011, 32, 2407–2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altunbas A.; Lee S. J.; Rajasekaran S. A.; Schneider J. P.; Pochan D. J. Biomaterials 2011, 32, 5906–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulyasasmita W.; Lee J. S.; Heilshorn S. C. Biomacromolecules 2011, 12, 3406–3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolambkar Y. M.; Dupont K. M.; Boerckel J. D.; Huebsch N.; Mooney D. J.; Hutmacher D. W.; Guldberg R. E. Biomaterials 2011, 32, 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan C.; Peppas N. Adv. Polym. Sci. 2000, 153, 28–65. [Google Scholar]
- Holloway J. L.; Lowman A. M.; Palmese G. R. Soft Matter 2013, 9, 826–833. [Google Scholar]
- Sanabria-delong N.; Crosby A. J.; Tew G. N. Biomacromolecules 2008, 2784–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S.; Zhang L.; Ma D. J. Phys. Chem. B 2006, 110, 12225–12229. [DOI] [PubMed] [Google Scholar]
- Wu Z. L.; Gong J. P. NPG Asia Mater. 2011, 3, 57–64. [Google Scholar]
- O’Leary L.; Fallas J.; Bakota E. Nat. Chem. 2011, 3, 821–828. [DOI] [PubMed] [Google Scholar]
- Krishna O. D.; Jha A. K.; Jia X.; Kiick K. L. Biomaterials 2011, 32, 6412–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camci-Unal G.; Cuttica D.; Annabi N.; Demarchi D.; Khademhosseini A. Biomacromolecules 2013, 14, 1085–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T.; Kiick K. L. Eur. Polym. J. 2013, 49, 2998–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X.; Park S.-H.; Gil E. S.; Xia X.-X.; Weiss A. S.; Kaplan D. L. Biomaterials 2011, 32, 8979–8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W.; He J.; Nichol J. W.; Wang L.; Hutson C. B.; Wang B.; Du Y.; Fan H.; Khademhosseini A. Acta Biomater. 2011, 7, 2384–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X.-X.; Xu Q.; Hu X.; Qin G.; Kaplan D. L. Biomacromolecules 2011, 12, 3844–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieshaber S. E.; Farran A. J. E.; Bai S.; Kiick K. L.; Jia X. Biomacromolecules 2012, 13, 1774–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieshaber S. E.; Nie T.; Yan C.; Zhong S.; Teller S. S.; Clifton R. J.; Pochan D. J.; Kiick K. L.; Jia X. Macromol. Chem. Phys. 2011, 212, 229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.; Wu D.; Fu W.; Li Z. Biomacromolecules 2013, 14, 2494–2498. [DOI] [PubMed] [Google Scholar]
- Zhang S.; Greenfield M. a; Mata A.; Palmer L. C.; Bitton R.; Mantei J. R.; Aparicio C.; de la Cruz M. O.; Stupp S. I. Nat. Mater. 2010, 9, 594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassman M. J.; Olsen B. D. Soft Matter 2013, 9, 6814–6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam C. N.; Olsen B. D. Soft Matter 2013, 9, 2393–2402. [Google Scholar]
- Glassman M. J.; Chan J.; Olsen B. D. Adv. Funct. Mater. 2013, 23, 1182–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H.; Olsen B. D.; Khademhosseini A. Biomaterials 2012, 33, 3143–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piluso S.; Cassell H. C.; Gibbons J. L.; Waller T. E.; Plant N. J.; Miller A. F.; Cavalli G. Soft Matter 2013, 9, 6752–6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz K. M.; Anseth K. S. Soft Matter 2013, 9, 1570–1579. [Google Scholar]
- Liu S. Q.; Tian Q.; Wang L.; Hedrick J. L.; Hui J. H. P.; Yang Y. Y.; Ee P. L. R. Macromol. Rapid Commun. 2010, 31, 1148–1154. [DOI] [PubMed] [Google Scholar]
- Aimetti A. A.; Tibbitt M. W.; Anseth K. S. Biomacromolecules 2009, 10, 1484–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutolf M. P.; Raeber G. P.; Zisch A. H.; Tirelli N.; Hubbell J. A. Adv. Mater. 2003, 15, 888–892. [Google Scholar]
- Liu S. Q.; Tian Q.; Hedrick J. L.; Po Hui J. H.; Ee P. L. R.; Yang Y. Y. Biomaterials 2010, 31, 7298–7307. [DOI] [PubMed] [Google Scholar]
- Zhu J. Biomaterials 2010, 31, 4639–4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibayama M. Polym. J. 2010, 43, 18–34. [Google Scholar]
- Shibayama M. Soft Matter 2012, 8, 8030–8038. [Google Scholar]
- Sun L.; Liu W.; Dong C.-M. Chem. Commun. 2011, 47, 11282–11284. [DOI] [PubMed] [Google Scholar]
- Sperling L.; Mishra V. Polym. Adv. Technol. 1996, 7, 197–208. [Google Scholar]
- Gong J. P. Soft Matter 2010, 6, 2583–2590. [Google Scholar]
- Wu Z. L.; Kurokawa T.; Liang S.; Gong J. P. Macromolecules 2010, 43, 8202–8208. [Google Scholar]
- Tanaka Y.; Gong J. P.; Osada Y. Prog. Polym. Sci. 2005, 30, 1–9. [Google Scholar]
- Nakayama A.; Kakugo A.; Gong J. P.; Osada Y.; Takai M.; Erata T.; Kawano S. Adv. Funct. Mater. 2004, 14, 1124–1128. [Google Scholar]
- Gong J. P. P.; Katsuyama Y.; Kurokawa T.; Osada Y. Adv. Mater. 2003, 15, 1155–1158. [Google Scholar]
- Bodugoz-Senturk H.; Macias C. E.; Kung J. H.; Muratoglu O. K. Biomaterials 2009, 30, 589–596. [DOI] [PubMed] [Google Scholar]
- Hori Y.; Winans A. M.; Irvine D. J. Acta Biomater. 2009, 5, 969–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai N.; Gunn J.; Zhang M. Adv. Drug Delivery Rev. 2010, 62, 83–99. [DOI] [PubMed] [Google Scholar]
- Tan H.; Chu C. R.; Payne K. A.; Marra K. G. Biomaterials 2009, 30, 2499–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichol J. W.; Koshy S. T.; Bae H.; Hwang C. M.; Yamanlar S.; Khademhosseini A. Biomaterials 2010, 31, 5536–5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.; Xu K.; Zheng X.; Giacomin A. J.; Mix A. W.; Kao W. J. Biomaterials 2012, 33, 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi S.; Krishnamurthy V. R.; Caves J. M.; Haller C. A.; Chaikof E. L. Acta Biomater. 2012, 8, 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi S.; Caves J. M.; Martinez A. W.; Haller C. A.; Chaikof E. L. J. Biomed. Mater. Res., Part A 2013, 101, 1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan F.; Ahmad S. R. Macromol. Biosci. 2013, 13, 395–421. [DOI] [PubMed] [Google Scholar]
- Straley K. S.; Heilshorn S. C. Soft Matter 2009, 5, 114–124. [Google Scholar]
- Krishna O. D.; Kiick K. L. Biopolymers 2010, 94, 32–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong E.; Tirrell D. A. Adv. Mater. 2010, 22, 5271–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu I.-L.; Patterson M. A.; Carpenter Desai H. E.; Mehl R. A.; Giorgi G.; Conticello V. P. ChemBioChem 2013, 14, 968–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srokowski E. M.; Woodhouse K. A. J. Mater. Sci.: Mater. Med. 2013, 24, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel J. R.; Radford D. C.; Chilkoti A. Biomacromolecules 2013, 14, 2866–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen T.; Hassouneh W.; Trabbic-Carlson K.; Chilkoti A. Biomacromolecules 2013, 14, 1514–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe K. J.; Antaris A. L.; Heilshorn S. C. Acta Biomater. 2013, 9, 5590–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacEwan S. R.; Chilkoti A. Biopolymers 2010, 94, 60–77. [DOI] [PubMed] [Google Scholar]
- Trabbic-Carlson K.; Meyer D. E.; Liu L.; Piervincenzi R.; Nath N.; LaBean T.; Chilkoti A. Protein Eng. Des. Sel. 2004, 17, 57–66. [DOI] [PubMed] [Google Scholar]
- Bessa P. C.; Machado R.; Nürnberger S.; Dopler D.; Banerjee A.; Cunha A. M.; Rodríguez-Cabello J. C.; Redl H.; van Griensven M.; Reis R. L.; Casal M. J. Controlled Release 2010, 142, 312–318. [DOI] [PubMed] [Google Scholar]
- Sallach R. E.; Cui W.; Wen J.; Martinez A.; Conticello V. P.; Chaikof E. L. Biomaterials 2009, 30, 409–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. C.; Tirrell D. A. Biomacromolecules 2008, 9, 2984–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai D.; Xu D.; Liu W.; Garcia Quiroz F.; Callahan D. J.; Zalutsky M. R.; Craig S. L.; Chilkoti A. Biomaterials 2012, 33, 5451–5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoorchian A.; Holland N. B. Biomacromolecules 2011, 12, 4022–4029. [DOI] [PubMed] [Google Scholar]
- Le D. H. T.; Hanamura R.; Pham D.-H.; Kato M.; Tirrell D. A.; Okubo T.; Sugawara-Narutaki A. Biomacromolecules 2013, 14, 1028–1034. [DOI] [PubMed] [Google Scholar]
- Chu H.-S.; Park J.-E.; Kim D.-M.; Kim B.-G.; Won J.-I. Protein Expr. Purif. 2010, 74, 298–303. [DOI] [PubMed] [Google Scholar]
- Grieshaber S. E.; Paik B. A.; Bai S.; Kiick K. L.; Jia X. Soft Matter 2013, 9, 1589–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez P. L.; Sweet J. a; Fink H.; Chennazhi K. P.; Nair S. V.; Enejder A.; Heilshorn S. C. Adv. Healthc. Mater. 2013, 2, 114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis-Fogh T. J. Exp. Biol. 1960, 37, 889–907. [Google Scholar]
- Burrows M. Nature 2003, 424, 509. [DOI] [PubMed] [Google Scholar]
- Bennet-Clark H. J. Exp. Biol. 2007, 210, 3879–3881. [DOI] [PubMed] [Google Scholar]
- Elvin C. M.; Carr A. G.; Huson M. G.; Maxwell J. M.; Pearson R. D.; Vuocolo T.; Liyou N. E.; Wong D. C. C.; Merritt D. J.; Dixon N. E. Nature 2005, 437, 999–1002. [DOI] [PubMed] [Google Scholar]
- Li L.; Tong Z.; Jia X.; Kiick K. K. L. Soft Matter 2013, 9, 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S.; Dudek D. M.; Cao Y.; Balamurali M. M.; Gosline J.; Li H. Nature 2010, 465, 69–73. [DOI] [PubMed] [Google Scholar]
- McGann C. L.; Levenson E. A.; Kiick K. L. Macromol. Chem. Phys. 2013, 214, 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta N. K.; Truong M. Y.; Mayavan S.; Choudhury N. R.; Elvin C. M.; Kim M.; Knott R.; Nairn K. M.; Hill A. J. Angew. Chem., Int. Ed. 2011, 50, 4428–4431. [DOI] [PubMed] [Google Scholar]
- Charati M. B.; Ifkovits J. L.; Burdick J. A.; Linhardt J. G.; Kiick K. L. Soft Matter 2009, 5, 3412–3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Teller S.; Clifton R. J.; Jia X.; Kiick K. L. Biomacromolecules 2011, 12, 2302–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Kiick K. L. Front. Chem. 2014, 2, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Charati M. B.; Kiick K. L. Polym. Chem. 2010, 1, 1160–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.; Renner J. N.; Liu J. C. Biomater. Sci. 2014, 2, 1110–1119. [DOI] [PubMed] [Google Scholar]
- Myung D.; Waters D. Polym. Adv. Technol. 2008, 19, 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabiri K.; Omidian H.; Zohuriaan-Mehr M.; Doroudiani S. Polym. Compos. 2011, 32, 277–289. [Google Scholar]
- Haraguchi K. Curr. Opin. Solid State Mater. Sci. 2007, 11, 47–54. [Google Scholar]
- Haraguchi K.; Takehisa T.; Fan S. Macromolecules 2002, 35, 10162–10171. [Google Scholar]
- Haraguchi K.; Li H.; Matsuda K.; Takehisa T. Macromolecules 2005, 38, 3482–3490. [Google Scholar]
- Haraguchi K.; Li H.-J. Macromolecules 2006, 39, 1898–1905. [Google Scholar]
- Wu L.; Zeng L.; Chen H.; Zhang C. Polym. Bull. 2011, 68, 309–316. [Google Scholar]
- Lin W.-C.; Fan W.; Marcellan A.; Hourdet D.; Creton C. Macromolecules 2010, 43, 2554–2563. [Google Scholar]
- Li J.; Suo J.; Jia L. Polym. Eng. Sci. 2010, 4, 689–696. [Google Scholar]
- Schexnailder P.; Schmidt G. Colloid Polym. Sci. 2008, 287, 1–11. [Google Scholar]
- Fu S.; Ni P.; Wang B.; Chu B.; Zheng L.; Luo F.; Luo J.; Qian Z. Biomaterials 2012, 33, 4801–4809. [DOI] [PubMed] [Google Scholar]
- Luo Y. L.; Zhang C. H.; Chen Y. S.; Yang W. Mater. Res. Innov. 2009, 13, 18–27. [Google Scholar]
- Zhang N.; Li R.; Zhang L.; Chen H.; Wang W.; Liu Y.; Wu T.; Wang X.; Wang W.; Li Y.; Zhao Y.; Gao J. Soft Matter 2011, 7, 7231–7239. [Google Scholar]
- Lin N.; Huang J.; Dufresne A. Nanoscale 2012, 4, 3274–3294. [DOI] [PubMed] [Google Scholar]
- Zhou C.; Wu Q. Nanocryst.: Synth., Charact. Appl. 2012, 103–120. [Google Scholar]
- Zhou C.; Wu Q.; Yue Y.; Zhang Q. J. Colloid Interface Sci. 2011, 353, 116–123. [DOI] [PubMed] [Google Scholar]
- Zhou C.; Wu Q.; Zhang Q. Colloid Polym. Sci. 2010, 289, 247–255. [Google Scholar]
- Abitbol T.; Johnstone T.; Quinn T. M.; Gray D. G. Soft Matter 2011, 7, 2373–2379. [Google Scholar]
- Araki J.; Yamanaka Y.; Ohkawa K. Polym. J. 2012, 44, 713–717. [Google Scholar]
- Dai Q.; Kadla J. F. J. Appl. Polym. Sci. 2009, 114, 1664–1669. [Google Scholar]
- Zhou C.; Chu R.; Wu R.; Wu Q. Biomacromolecules 2011, 12, 2617–2625. [DOI] [PubMed] [Google Scholar]
- Wu Y.; Zhou Z.; Fan Q.; Chen L.; Zhu M. J. Mater. Chem. 2009, 19, 7340–7346. [Google Scholar]
- Meid J.; Dierkes F.; Cui J.; Messing R.; Crosby A. J.; Schmidt A.; Richtering W. Soft Matter 2012, 8, 4254–4263. [Google Scholar]
- Xiao L.; Liu C.; Zhu J.; Pochan D. Soft Matter 2010, 6, 5293–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Patel A.; Gaharwar A. K.; Mihaila S. M.; Iviglia G.; Mukundan S.; Bae H.; Yang H.; Khademhosseini A. Biomacromolecules 2013, 14, 1299–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Xia M.; Fan Q.; Zhu M. Chem. Commun. 2010, 46, 7790–7792. [DOI] [PubMed] [Google Scholar]
- Molinos M.; Carvalho V.; Silva D. M.; Gama F. M. Biomacromolecules 2012, 13, 517–527. [DOI] [PubMed] [Google Scholar]
- Moghadam M. N.; Kolesov V.; Vogel A.; Klok H.-A.; Pioletti D. P. Biomaterials 2014, 35, 450–455. [DOI] [PubMed] [Google Scholar]
- Hu J.; Hiwatashi K.; Kurokawa T.; Liang S. M.; Wu Z. L.; Gong J. P. Macromolecules 2011, 44, 7775–7781. [Google Scholar]
- Hu J.; Kurokawa T.; Hiwatashi K.; Nakajima T.; Wu Z. L.; Liang S. M.; Gong J. P. Macromolecules 2012, 45, 5218–5228. [Google Scholar]
- Headen D. M.; Aubry G.; Lu H.; García A. J. Adv. Mater. 2014, 26, 3003–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei R.; George J. T.; Park J.; Grunlan M. A. Soft Matter 2012, 8, 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott E. A.; Nichols M. D.; Kuntz-Willits R.; Elbert D. L. Acta Biomater. 2010, 6, 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbert D. L. Acta Biomater. 2011, 7, 31–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. W.; Segar C. E.; Nguyen P. K.; MacEwan M. R.; Efimov I. R.; Elbert D. L. Acta Biomater. 2012, 8, 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. M.; Oyen M. L. JOM 2013, 65, 505–516. [Google Scholar]
- Bhardwaj N.; Kundu S. C. Biotechnol. Adv. 2010, 28, 325–347. [DOI] [PubMed] [Google Scholar]
- Rockwood D. N.; Akins R. E.; Parrag I. C.; Woodhouse K. A.; Rabolt J. F. Biomaterials 2008, 29, 4783–4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakaran M. P.; Kai D.; Ghasemi-Mobarakeh L.; Ramakrishna S. Biomed. Mater. 2011, 6, 055001. [DOI] [PubMed] [Google Scholar]
- Loh X. J.; Peh P.; Liao S.; Sng C.; Li J. J. Controlled Release 2010, 143, 175–182. [DOI] [PubMed] [Google Scholar]
- Chew S. Y.; Wen J.; Yim E. K. F.; Leong K. W. Biomacromolecules 2005, 6, 2017–2024. [DOI] [PubMed] [Google Scholar]
- Jiang H.; Hu Y.; Li Y.; Zhao P.; Zhu K.; Chen W. J. Controlled Release 2005, 108, 237–243. [DOI] [PubMed] [Google Scholar]
- Zhou C.; Wu Q. Colloids Surf., B 2011, 84, 155–162. [DOI] [PubMed] [Google Scholar]
- Morais J. M.; Papadimitrakopoulos F.; Burgess D. J. AAPS J. 2010, 12, 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutolf M. P.; Lauer-Fields J. L.; Schmoekel H. G.; Metters a. T.; Weber F. E.; Fields G. B.; Hubbell J. a. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 5413–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBaron R. G.; Athanasiou K. A. Tissue Eng. 2000, 6, 85–103. [DOI] [PubMed] [Google Scholar]
- Hersel U.; Dahmen C.; Kessler H. Biomaterials 2003, 24, 4385–4415. [DOI] [PubMed] [Google Scholar]
- Rahmany M. B.; Van Dyke M. Acta Biomater. 2013, 9, 5431–5437. [DOI] [PubMed] [Google Scholar]
- Mosiewicz K. A.; Johnsson K.; Lutolf M. P. J. Am. Chem. Soc. 2010, 132, 5972–5974. [DOI] [PubMed] [Google Scholar]
- Kraehenbuehl T. P.; Ferreira L. S.; Zammaretti P.; Hubbell J. A.; Langer R. Biomaterials 2009, 30, 4318–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bott K.; Upton Z.; Schrobback K.; Ehrbar M.; Hubbell J. A.; Lutolf M. P.; Rizzi S. C. Biomaterials 2010, 31, 8454–8464. [DOI] [PubMed] [Google Scholar]
- Seeto W. J.; Tian Y.; Lipke E. A. Acta Biomater. 2013, 9, 8279–8289. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Ni J.; Chen L.; Yu L.; Xu J.; Ding J. J. Biomed. Mater. Res., Part B: Appl. Biomater. 2012, 100, 1599–1609. [DOI] [PubMed] [Google Scholar]
- Weber L. M.; Lopez C. G.; Anseth K. S. J. Biomed. Mater. Res., Part A 2009, 90, 720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann B.; West J. J. Biomed. Mater. Res. 2002, 60, 86–93. [DOI] [PubMed] [Google Scholar]
- Mann B. K.; Gobin A. S.; Tsai A. T.; Schmedlen R. H.; West J. L. Biomaterials 2001, 22, 3045–3051. [DOI] [PubMed] [Google Scholar]
- Schmidt D.; Kao W. J. Biomed. Mater. Res., Part A 2007, 83A, 617–625. [DOI] [PubMed] [Google Scholar]
- Cheng T.-Y.; Chen M.-H.; Chang W.-H.; Huang M.-Y.; Wang T.-W. Biomaterials 2013, 34, 2005–2016. [DOI] [PubMed] [Google Scholar]
- Weber L. M.; Hayda K. N.; Haskins K.; Anseth K. S. Biomaterials 2007, 28, 3004–3011. [DOI] [PubMed] [Google Scholar]
- Kharkar P. M.; Kiick K. L.; Kloxin A. M. Chem. Soc. Rev. 2013, 42, 7335–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick J. A.; Murphy W. L. Nat. Commun. 2012, 3, 1269. [DOI] [PubMed] [Google Scholar]
- Miller J. S.; Shen C. J.; Legant W. R.; Baranski J. D.; Blakely B. L.; Chen C. S. Biomaterials 2010, 31, 3736–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau Y.; Luo Y.; Cheung A. C. Y.; Nagai Y.; Zhang S.; Kobler J. B.; Zeitels S. M.; Langer R. Biomaterials 2008, 29, 1713–1719. [DOI] [PubMed] [Google Scholar]
- Lei Y.; Gojgini S.; Lam J.; Segura T. Biomaterials 2011, 32, 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo Y. S.; Rizzi S. C.; Ehrbar M.; Weber F. E.; Hubbell J. A.; Lutolf M. P. J. Biomed. Mater. Res., Part A 2010, 93, 870–877. [DOI] [PubMed] [Google Scholar]
- West J. L.; Hubbell J. A. Macromolecules 1999, 32, 241–244. [Google Scholar]
- Salinas C. N.; Anseth K. S. Biomaterials 2008, 29, 2370–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca K. B.; Bidarra S. J.; Oliveira M. J.; Granja P. L.; Barrias C. C. Acta Biomater. 2011, 7, 1674–1682. [DOI] [PubMed] [Google Scholar]
- Garripelli V. K.; Kim J.-K.; Son S.; Kim W. J.; Repka M. A.; Jo S. Acta Biomater. 2011, 7, 1984–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aimetti A. A.; Machen A. J.; Anseth K. S. Biomaterials 2009, 30, 6048–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekaran A.; García J. R.; Clark A. Y.; Kavanaugh T. E.; Lin A. S.; Guldberg R. E.; García A. J. Biomaterials 2014, 35, 5453–5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway J. L.; Ma H.; Rai R.; Burdick J. A. J. Controlled Release 2014, 188, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin A. D.; Kiick K. L. Polym. Chem. 2013, 4, 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharkar P. M.; Kloxin A. M.; Kiick K. L. J. Mater. Chem. B 2014, 2, 5511–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan G.; Mooney D. J. Trends Biotechnol. 2008, 26, 382–392. [DOI] [PubMed] [Google Scholar]
- Anderson J. Annu. Rev. Mater. Res. 2001, 31, 81–110. [Google Scholar]
- Zhang L.; Chen J.; Han C. Wound Repair Regen. 2009, 17, 685–689. [DOI] [PubMed] [Google Scholar]
- Prokoph S.; Chavakis E.; Levental K. R.; Zieris A.; Freudenberg U.; Dimmeler S.; Werner C. Biomaterials 2012, 33, 4792–4800. [DOI] [PubMed] [Google Scholar]
- Rabbany S.; Pastore J.; Yamamoto M.; Miller T.; Rafii S.; Aras R.; Penn M. Cell Transplant. 2010, 19, 399–408. [DOI] [PubMed] [Google Scholar]
- Lee K.; Silva E. A.; Mooney D. J. J. R. Soc., Interface 2011, 8, 153–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borselli C.; Cezar C. A.; Shvartsman D.; Vandenburgh H. H.; Mooney D. J. Biomaterials 2011, 32, 8905–8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M.; Mata J.; Babensee J. J. Biomed. Mater. Res., Part A 2007, 80, 7–12. [DOI] [PubMed] [Google Scholar]
- Byeon H. J.; Choi S. H.; Choi J. S.; Kim I.; Shin B. S.; Lee E. S.; Park E.-S.; Lee K. C.; Youn Y. S. Acta Biomater. 2014, 10, 142–150. [DOI] [PubMed] [Google Scholar]
- Levenson E. A.; Kiick K. L. Acta Biomater. 2014, 10, 1134–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahrsdörfer B.; Weiner G. Update Cancer Ther 2008, 3, 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz C.; Wagner E. J. Controlled Release 2012, 161, 554–565. [DOI] [PubMed] [Google Scholar]
- Green M. D.; Foster A. A.; Greco C. T.; Roy R.; Lehr R. M.; Epps T. H. III; Sullivan M. O. Polym. Chem. 2014, 10.1039/C4PY00638K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y.; Rahim M.; Ng Q.; Segura T. J. Controlled Release 2011, 153, 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd M. E.; Shin S.; Shea L. D. J. Controlled Release 2012, 157, 80–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard J. A.; Wesson P. J.; Wang C. E.; Stevans A. C.; Holland S. J.; Shikanov A.; Grzybowski B. A.; Shea L. D. Biomaterials 2011, 32, 5092–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.; Park M.; Song S. Nano Lett. 2012, 6, 5757–5766. [DOI] [PubMed] [Google Scholar]
- Dunn S. S.; Tian S.; Blake S.; Wang J.; Galloway A. L.; Murphy A.; Pohlhaus P. D.; Rolland J. P.; Napier M. E. J. Am. Chem. Soc. 2012, 134, 7423–7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessis N.; GarciaCozar F.; Boissier M.-C. Gene Ther. 2004, 11, S10–17. [DOI] [PubMed] [Google Scholar]
- Singh A.; Suri S.; Roy K. Biomaterials 2009, 30, 5187–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn W. H.; Dickerson E. B.; Smith M. H.; Mcdonald J. F.; Lyon L. A. Bioconjugate Chem. 2009, 20, 960–968. [DOI] [PMC free article] [PubMed] [Google Scholar]



