This first systematic and comprehensive screen of human phosphatases for a regulatory role in the survival motor neuron (SMN) complex identifies the catalytically inactive, non–receptor-type tyrosine phosphatase PTPN23/HD-PTP as a novel SMN complex regulator. PTPN23 maintains a highly phosphorylated state of SMN, which is important for its function in snRNP assembly.
Abstract
The survival motor neuron (SMN) complex fulfils essential functions in the assembly of snRNPs, which are key components in the splicing of pre-mRNAs. Little is known about the regulation of SMN complex activity by posttranslational modification despite its complicated phosphorylation pattern. Several phosphatases had been implicated in the regulation of SMN, including the nuclear phosphatases PPM1G and PP1γ. Here we systematically screened all human phosphatase gene products for a regulatory role in the SMN complex. We used the accumulation of SMN in Cajal bodies of intact proliferating cells, which actively assemble snRNPs, as a readout for unperturbed SMN complex function. Knockdown of 29 protein phosphatases interfered with SMN accumulation in Cajal bodies, suggesting impaired SMN complex function, among those the catalytically inactive, non–receptor-type tyrosine phosphatase PTPN23/HD-PTP. Knockdown of PTPN23 also led to changes in the phosphorylation pattern of SMN without affecting the assembly of the SMN complex. We further show interaction between SMN and PTPN23 and document that PTPN23, like SMN, shuttles between nucleus and cytoplasm. Our data provide the first comprehensive screen for SMN complex regulators and establish a novel regulatory function of PTPN23 in maintaining a highly phosphorylated state of SMN, which is important for its proper function in snRNP assembly.
INTRODUCTION
Noncoding sequences in nascent eukaryotic mRNA transcripts (introns) need to be removed by splicing to generate the open reading frame for translation. This reaction is catalyzed by the spliceosome, a macromolecular machine containing uridine (U)-rich small nuclear ribonucleoprotein particles (snRNPs) as major components. snRNPs consist of a (U)-rich small nuclear RNA (snRNA) entity and a large set of proteins that are either specific to one snRNP or common to all of them. The biogenesis—that is, the formation of U snRNPs from snRNA and protein subunits—occurs predominantly in the cytoplasm after snRNA export from the nucleus (for reviews see Wachtel and Manley, 2009; Will and Luhrmann, 2011). Purified snRNP proteins and snRNAs assemble spontaneously to snRNPs, whereas the same reaction in cell extracts or intact cells depends on the activity of the survival motor neuron (SMN) complex (Meister et al., 2001). The SMN complex enables binding of the seven common proteins (also termed Sm proteins) onto a single-stranded Sm site of the snRNA, which results in the formation of the toroidal Sm core domain (Kambach et al., 1999; Meister et al., 2001; Stark et al., 2001; Pellizzoni et al., 2002; Chari et al., 2008). The human SMN complex consist of nine core subunits, including the eponymous SMN protein, Gemins 2–8, and the unr interacting protein (unrip, also termed serine/threonine kinase adaptor protein [STRAP]; for reviews see Pellizzoni, 2007; Fischer et al., 2011). Partial loss of SMN activity causes spinal muscular atrophy, a neurological disorder characterized by the loss of α-motor neurons and muscle degeneration (for reviews see Burghes and Beattie, 2009; Farrar et al., 2009; Coady and Lorson, 2011). SMN-mediated steps of snRNP assembly precede reimport of the snRNPs into the nucleus. On nuclear entry, the snRNP particles concentrate in Cajal (coiled) bodies (CBs; Lafarga et al., 2009), where they further mature, receive posttranslational modifications, and eventually become integrated into the splicing system (Sleeman and Lamond, 1999; Stanek et al., 2008).
The SMN complex has been shown to associate with snRNPs for nuclear import (Narayanan et al., 2004) and direct their subsequent targeting into CBs (reviewed in Stanek and Neugebauer, 2006; Shaw et al., 2008). As intranuclear subcompartments, CBs accumulate a variety of factors to increase the efficiency of nuclear steps of snRNP biogenesis (Sleeman and Lamond, 1999; Darzacq et al., 2002; Kiss et al., 2002; Schaffert et al., 2004). SMN complex generally concentrates in CBs in highly proliferating cells, which correlates with high snRNP biogenesis activity. Inhibition of efficient snRNP biogenesis—for instance, upon reducing the levels of the snRNA nuclear export adaptor PHAX (Lemm et al., 2006) or global inhibition of nuclear protein export using leptomycin B (Carvalho et al., 1999)—quickly abolishes SMN accumulation in CBs in these cells. Consistently, the deregulated activity of nuclear phosphatases such as PPM1G (Petri et al., 2007), PP1γ (Renvoise et al., 2012), and PPP4 (Carnegie et al., 2003) was shown to affect accumulation of SMN in CBs. These data suggest that reversible phosphorylation of SMN plays a role in the regulation of the SMN complex in snRNP assembly. It was shown previously that the degree of phosphorylation of the SMN protein in the nucleus is clearly lower than in the cytoplasm (Grimmler et al., 2005). Assuming that the SMN complex (or parts thereof) shuttles between nucleus and cytoplasm, this would imply that SMN becomes dephosphorylated by phosphatase(s) in the nucleus (Grimmler et al., 2005).
Recently we uncovered an enormous complexity of phosphorylation of SMN complex proteins. Systematic phosphorylation analysis identified not only 12 serine/threonine phosphorylation sites in SMN itself, but also an additional 17 in other SMN complex proteins with high confidence (Husedzinovic et al., 2014). Moreover, using nonphosphorylatable variants of SMN, we showed that these sites, as well as three recently identified tyrosine phosphorylations in the TUDOR domain of SMN (Rikova et al., 2007), regulate the accumulation of SMN in nuclear CBs. Together, these data strongly suggest a complex regulatory network that influences the functions of the SMN complex in the cytoplasm and in the nucleus. Understanding this network not only requires systematic survey of phosphorylated sites in SMN complex proteins, but also calls for a comprehensive survey of kinases and phosphatases that modify and regulate the SMN complex.
Here we describe a microscopy-based, high-throughput assay that allowed the screening of all human phosphatase gene products for their potential role in the regulation of the SMN complex. We used accumulation of SMN in nuclear CBs in proliferating human HeLa cells as an optical readout that can be automatically scored in a large number of samples. In our primary screen, knockdown of 29 human protein phosphatases led to changes in the accumulation of SMN in nuclear CBs. We carried out further validation of 21 of these and confirmed 13 potential SMN complex regulators, including the previously described nuclear SMN complex regulators PPM1G and PP1γ. Surprisingly, our screen identified the catalytically inactive phosphatase His domain–containing tyrosine phosphatase (HD-PTP)/tyrosine-protein phosphatase non–receptor type 23 (PTPN23) as a novel interactor and regulator of the SMN complex. Knockdown of the shuttling protein PTPN23 caused loss of SMN accumulation in CBs and reduced SMN phosphorylation. This phenotype supports a model in which PTPN23 acts to protect a highly phosphorylated and hence assembly-active cytosolic state of SMN. Our approach not only represents the first systematic screen of phosphatases regulating the SMN complex, but also points to PTPN23 as a novel, unusual regulator of SMN complex localization.
RESULTS
Systematic identification of human phosphatases as potential regulators of SMN complex function
The number of phosphosites identified in all SMN complex components suggests complicated regulatory cues to adjust SMN complex functions to cellular signals (Husedzinovic et al., 2014). However, identification of kinases and phosphatases has been sporadic. To identify regulators of SMN localization and function systematically, we decided to test phosphatases, which may exert regulatory functions on SMN, in a genome-wide approach. The analysis of subcellular SMN localization by indirect immunofluorescence (Husedzinovic et al., 2014) and automated microscopy served as a readout. After knockdown of individual phosphatases by solid-phase reverse small interfering RNA (siRNA) transfection in 96-well format, we fixed cells and stained them with 4′,6-diamidino-2-phenylindole (DAPI) to detect the DNA and with an antibody that recognizes SMN. As a technical positive control for successful transfection, we knocked down the inner centromere protein (INCENP), whose loss quickly leads to obviously multinucleated cells that can easily be recognized by DAPI staining (Neumann et al., 2006). Imaging of the DAPI and the SMN signal was performed by automatized microscopy on an Olympus ScanR screening microscope using a 10× objective. In every well, four images were obtained for each respective fluorescence channel, resulting in a data set of 19,968 images (Supplemental Figure S1). After 72 h, close to 100% of all cells displayed the phenotype characteristic for INCENP knockdown in all 96-well plates analyzed, indicating efficient siRNA uptake and knockdown (Figure 1A, top). In contrast, nontargeting siRNAs did not interfere with cell growth, cell proliferation, or the expected diffuse localization of SMN in the cytoplasm and its accumulations in nuclear CBs (Figure 1A, bottom). From pilot experiments, we realized that loss of SMN accumulation in CBs was by far the most prominent alteration in the subcellular distribution of SMN, exemplified best by the knockdown of the nuclear phosphatase PPM1G (Figure 1A, bottom; Petri et al., 2007).
FIGURE 1:
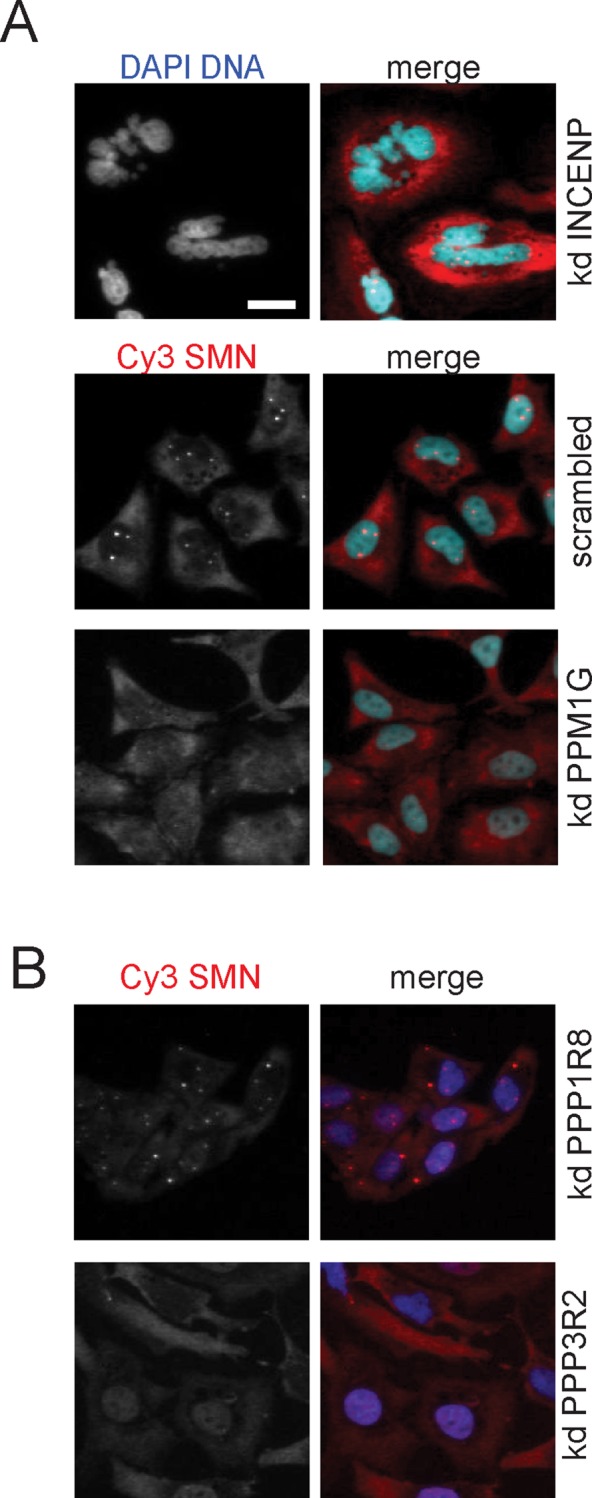
Subcellular distribution of SMN after genome-wide knockdown of human phosphatases. At 72 h after cell seeding and inverse siRNA transfection in 96-well plates, HeLa cells were fixed and immunostained using an α-SMN antibody; nuclei were stained using DAPI. An Olympus ScanR High-content Screening Station was used to analyze the subcellular distribution of the SMN protein. Images were collected using a 10× objective; for better visibility, only a small part of the image is shown here, whereas the entire image is displayed in Supplemental Figure S1. Scale bar, 20 μm. (A) Knockdown (kd) of inner centromeric protein (INCENP) was used as a positive control for transfection and knockdown efficiency, and scrambled siRNA (nontargeting any known gene) was used as negative control. Loss of nuclear SMN accumulations after PPM1G knockdown exemplifies the identification of protein phosphatases the knockdown of which leads to the loss of SMN accumulations in CBs. (B) Additional examples of disturbed SMN localization. Top, cytoplasmic accumulations of SMN after PPP1R8 knockdown. Bottom, loss of nuclear SMN accumulations accompanied by an increase of nucleoplasmic signal after PPP3R2 knockdown; blue, DNA; red, hSMN.
To systematically identify phosphatase knockdown affecting accumulation of SMN in CBs, we developed an automatic image algorithm to quantify the number of cells with at least one or without nuclear SMN accumulation in CBs. After automatic image evaluation, we scored 24 protein phosphatases (see Materials and Methods) that showed significant (twofold higher than the median of controls in an individual 96-well plate; see Materials and Methods) loss of SMN accumulation in CBs. Further visual inspection of automatically generated images identified an additional 4 phosphatases that upon knockdown led to the loss of SMN accumulation in CBs (Supplemental Table S1). Visual inspection also revealed prominent alterations of SMN subcellular localization after knockdown of PPP1R8 (cytoplasmic accumulation; Figure 1B, top) and loss of SMN accumulation from CBs accompanied by increase of nucleoplasmic SMN signal as seen upon depletion of PPP3R2 (Figure 1B, bottom).
For further validation of initially scored phosphatases and to detect changes in the subcellular localization of SMN with high resolution, we reevaluated the knockdown of 21 phosphatases by forward transfection. Table 1 summarizes these results and displays the changes in the subcellular localization of SMN, as well as the number of siRNAs that induced the respective phenotypes. The percentage of cells without any detectable SMN accumulation in CBs increased by at least 40, up to 85% compared with negative controls. Loss of SMN accumulation in the nucleus observed by knockdown of two phosphatases, PPP3R2 (calcineurin regulatory subunit B) and NAP1L1 (nucleosome assembly protein 1 like 1), was reproducible with two of three different, or three of four, siRNAs, respectively (Table 1). In addition to PPP3R2, the loss of SMN accumulation in CBs could be observed upon knockdown of PPP3CB (Table 1), the catalytic subunit of calcineurin, suggesting a functional relation of these two proteins in the regulation of the SMN complex. Our analysis also identified PP1γ/PPP1CC (protein phosphatase 1, catalytic subunit, gamma), as well as several of its regulators (PPP1R8, PPP1R12B, PPP1R12C, PPP1R14D; Table 1), which are known to be involved in cell cycle regulation, glycogen metabolism, mRNA processing, RNA splicing, and regulation of transcription (Cohen, 2002). Only the knockdown of PPP1R8 (nuclear inhibitor of protein phosphatase 1) showed cytoplasmic accumulation (Table 1 and Figure 1B).
TABLE 1:
Changes in the subcellular distribution of SMN upon knockdown of human phosphatases.
 |
Gene products were further evaluated as potential SMN regulators by three siRNAs in forward transfection. Specific phenotypes in columns 3 (blue) and 4 (green) vs. global cellular phenotypes in columns 5 (orange) and 6 (red). Numbers indicate siRNAs that showed the respective cellular phenotypes.
Validation of PTPN23
Besides the aforementioned phosphatases, knockdown of the non–receptor-type protein tyrosine phosphatase 23 (HD-PTP/PTPN23) led to loss of SMN accumulation in CBs. This was of particular interest since PTPN23 was shown to be catalytically inactive (Gingras et al., 2009b). We rationalized that PTPN23 could instead act as a phosphotyrosine-binding module, which might regulate another kinase involved in the SMN complex phosphoregulation, or might prevent dephosphorylation of the SMN complex itself.
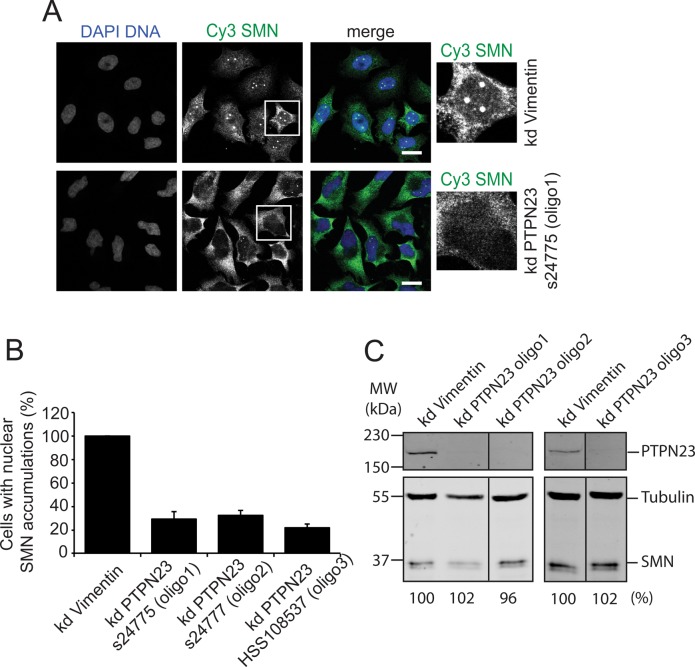
To validate and further characterize a potential role of PTPN23 in the regulation of SMN biology, we used three sets of siRNA oligonucleotides specific for PTPN23 and confirmed the loss of SMN accumulation in CBs in a quantitative manner by indirect immunofluorescence (Figure 2). To evaluate the RNA interference (RNAi)–mediated knockdown of PTPN23, we performed a Western blot with PTPN23-specific antibodies (see Materials and Methods; Figure 2C). All siRNAs significantly reduced PTPN23 protein levels (Figure 2C) but left SMN protein amounts unaffected. By counting cells showing SMN accumulation in CBs after imaging with high resolution (63× objective), we were able to determine the decrease in the relative number of cells with SMN accumulation in CBs to 30–35% compared with controls (Figure 2B).
FIGURE 2:
siRNA-based knockdown of PTPN23. Validation of the loss of nuclear SMN accumulation upon knockdown of PTPN23 (kd PTPN23) by forward transfection using three different siRNA oligonucleotides: Ambion silencer select s24775 (oligo1), s24777 (oligo2), and Stealth select HSS108537 (oligo3); siRNAs against vimentin were used as negative controls. Cells were transfected and incubated for 72 h. (A) Immunostaining of HeLa cells after siRNA transfection (oligo1) using α-SMN and α-coilin antibodies. Blue, DNA; green, SMN; red, coilin; scale bar, 20 μm. (B) Quantification of the loss of nuclear SMN accumulations observed in A. The relative numbers of cells bearing SMN accumulations (one or more) in Cajal bodies were calculated as mean values from three independent experiments (N = 200) and are represented in columns; error bars, SEM. (C) Western blot analysis of SMN and PTPN23 protein amounts after PTPN23 knockdown; HeLa total lysates were separated on a high-TEMED SDS gel (see Materials and Methods), and the SMN protein amount was analyzed by quantitative immunoblot using specific antibodies against SMN; numbers indicate relative SMN protein amounts; tubulin was used as a loading control. PTPN23 knockdown was analyzed using a 10% SDS–PAGE gel, flowed by Western blot using an α-PTPN23 antibody.
Next we investigated the kinetics of the reduction of PTPN23 after siRNA knockdown and correlated it with the number of cells losing SMN accumulation in CBs. We took samples for Western blot analysis and immunofluorescence every 24 h after transfection for a period of 3 d. PTPN23 and SMN protein levels were quantified and normalized using α-tubulin signals (Figure 3, A and B). The quantification indicated that the kinetics of PTPN23 depletion was similar with all siRNA oligonucleotides used (Figure 3B, top). Again, SMN protein levels remained stable (Figure 3B, bottom). All three knockdown conditions consistently reduced the relative number of cells showing SMN accumulation linearly to ∼30% compared with controls 3 d after transfection (Figure 3C).
FIGURE 3:
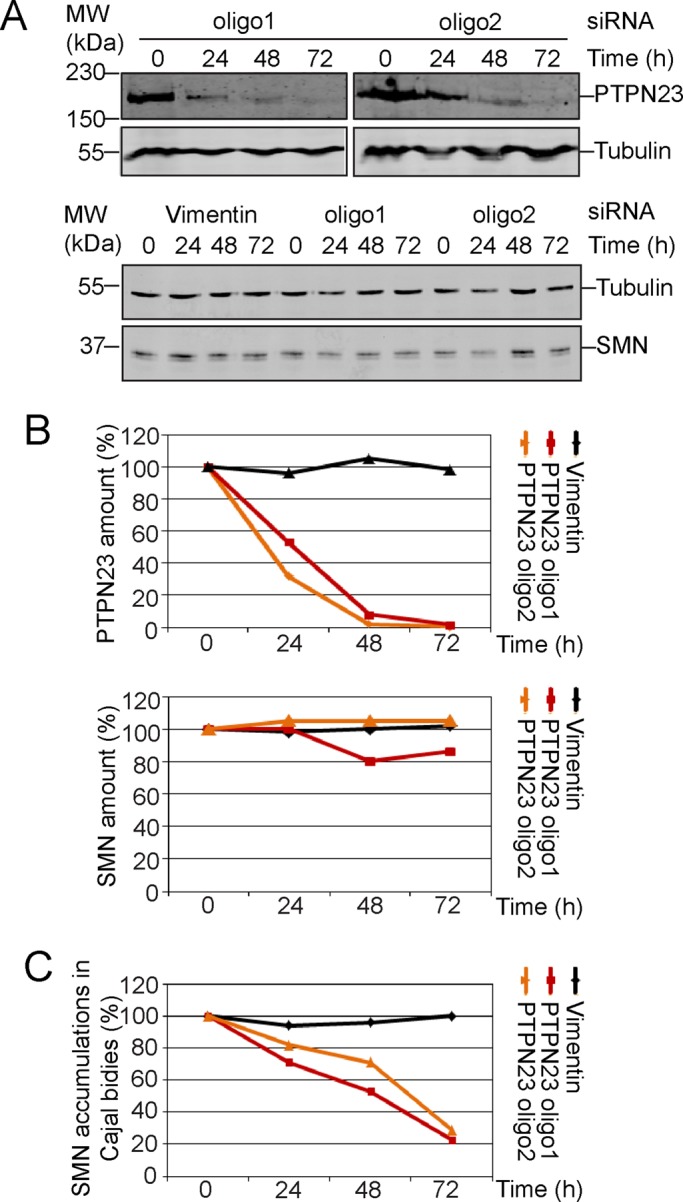
Kinetics of PTPN23 depletion upon siRNA. Kinetics of PTPN23 protein depletion using different siRNAs as explained for Figure 2; siRNAs against vimentin were used as negative controls. Cells were transfected, and samples were taken every 24 h for a period of 3 d. (A) Western blot analysis of SMN and PTPN23 protein amounts after siRNA-mediated PTPN23 knockdown; the PTPN23 and SMN protein amounts were analyzed by quantitative immunoblot using specific antibodies against SMN and PTPN23; numbers indicate time points after siRNA transfection. Tubulin was used as a loading control. (B) Graphs represent amounts of PTPN23 or SMN over a 3-d period evaluated as determined by quantitative Western blot. (C) Increase in number of cells losing SMN accumulations in CBs.
Interaction of PTPN23 and SMN
At this point, our data clearly suggested a functional interaction between SMN and PTPN23. To test a possible physical interaction between the two components, we used affinity-purified PTPN23 antibodies to purify PTPN23 under native conditions (Figure 4A). The prominent band at 180 kDa (Figure 4A) was analyzed by mass spectrometry to test the identity of the PTPN23 protein (unpublished data). Strikingly, the SMN protein, the SMN interaction partners Gemin2 and 3, and the SMN complex component unrip were specifically coprecipitated with PTPN23 in immunoprecipitates using α- PTPN23 antibodies (Figure 4B).
FIGURE 4:
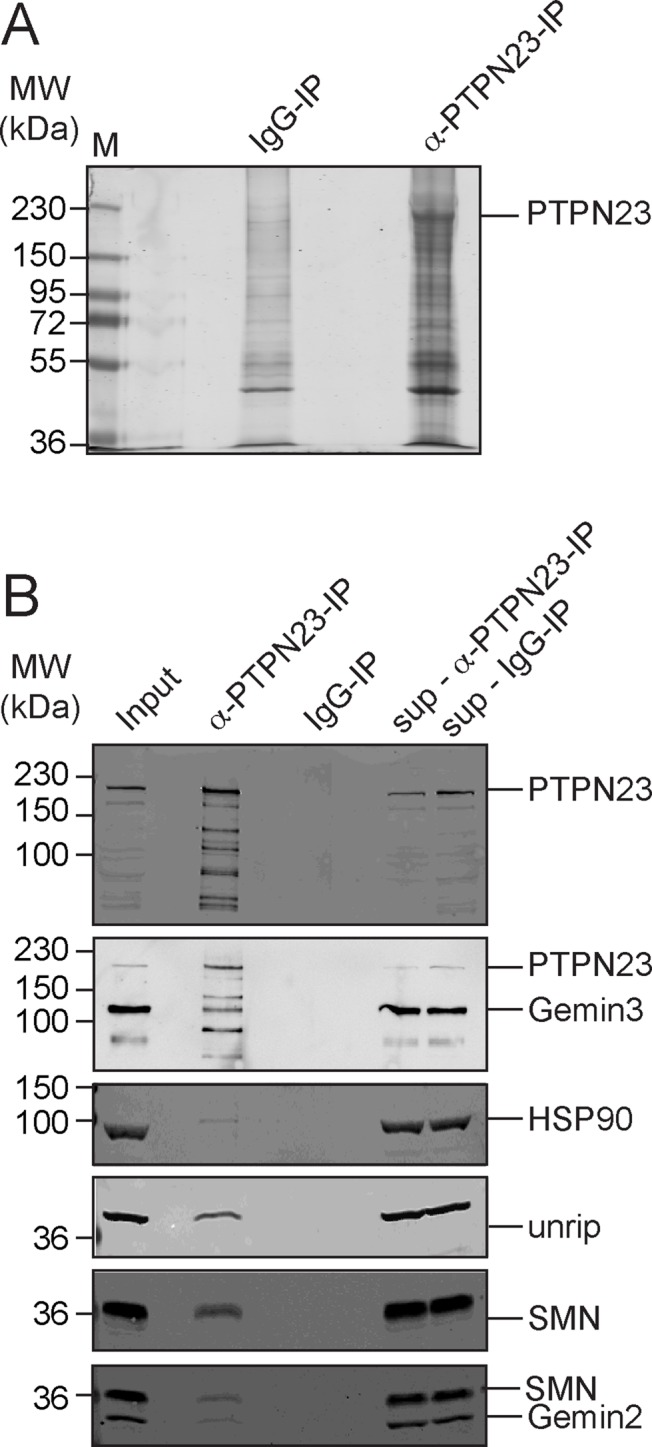
Interaction of SMN and PTPN23. Immunoprecipitation was performed in HeLa total cell lysates using an α-PTPN23 antibody (A, B) covalently coupled to protein A or G–Sepharose; an unspecific rabbit IgG coupled to protein A–Sepharose was used as a control. (A) SDS–PAGE and colloidal Coomassie stain of α-PTPN23 and α-IgG immunoprecipitation; the most prominent band was identified as PTPN23 by mass spectrometry. (B) Immunoblot analysis of immunoprecipitations using α-PTPN23 antibodies or an unspecific IgG as a controls. SMN, unrip, PTPN23, and tubulin were detected. sup, supernatant. The asterisk indicates an unspecific band.
Localization of exogenous and endogenous PTPN23
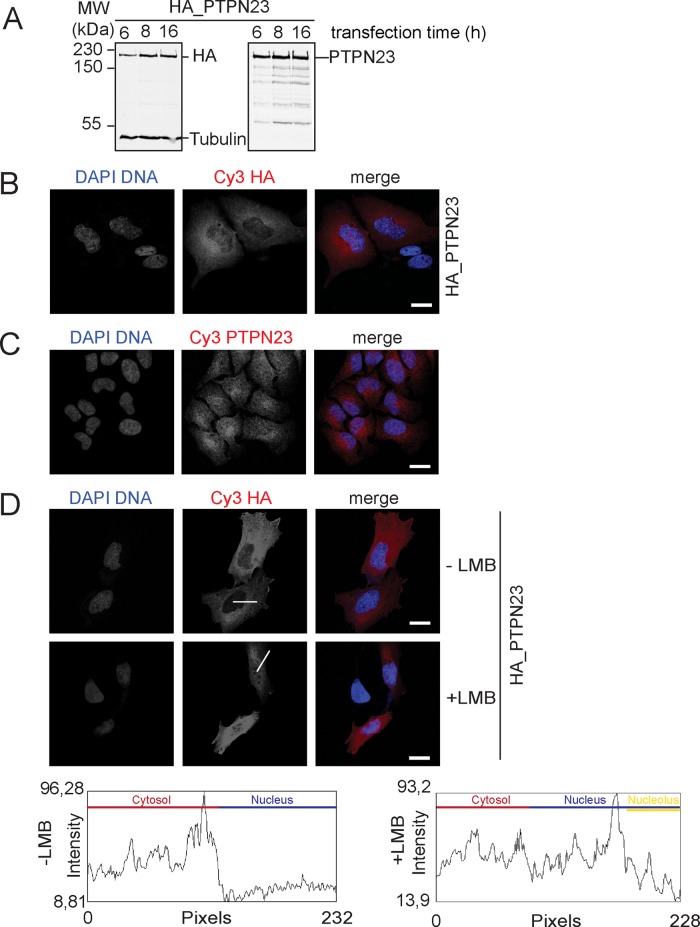
The loss of the accumulation of SMN in nuclear CBs raised the question of the localization of PTPN23. To analyze this, we fused the PTPN23 cDNA to a triple-hemagglutinin (HA) tag, transfected the HA-tagged PTPN23, and detected the expressed protein after 6, 8, and 16 h. In Western blot, a specific antibody against the HA tag recognized a band at ∼180 kDa, which matched to the band recognized by the α-PTPN23 antibody and the calculated 179-kDa molecular weight of PTPN23 (Figure 5A).
FIGURE 5:
Subcellular localization of exogenous and endogenous PTPN23. (A) HA-tagged PTPN23 was expressed in HeLa cells for 6, 8, and 16 h and detected by Western blot using specific antibodies against the HA tag (left) or PTPN23 (right). (B) Localization analysis of HA-PTPN23 by indirect immunofluorescence in cells expressing the protein for 8 h (blue, DAPI; red, HA; scale bar, 20 μm). (C) Localization analysis of endogenous PTPN23 by indirect immunofluorescence using an α-PTPN23 antibody; blue, DAPI; red, PTPN23; scale bar, 20 μm. (D) Localization analysis of HA-PTPN23 (8-h expression) before and after addition of the nuclear protein export inhibitor LMB. LMB was applied at 45 min; indirect immunofluorescence was performed using an α-HA antibody; blue, DAPI; red, HA; scale bar, 20 μm. White lines indicate fluorescence intensity measurements; plots show the respective intensity analysis after ImageJ-based quantification. Measured sections in the cytoplasm, within the nucleoplasm, and in the nucleolus are indicated within the plots in different colors.
Detecting HA-tagged PTPN23 by indirect immunofluorescence, we observed that the exogenous PTPN23 mostly localized to the cytoplasm, in the perinuclear region, suggesting localization of the protein in the endoplasmic reticulum, Golgi apparatus, or endosomes, as well as possibly the nucleus (Figure 5B). Moreover, indirect immunofluorescence using an affinity- purified PTPN23 yielded a signal in cytoplasm and nucleus (Figure 5C). To further validate our observation, we tested whether PTPN23 shuttles between nucleus and cytoplasm. The HA-tagged variant was expressed for 8 h in human cells, which were subsequently treated with leptomycin B (LMB) in order to block CRM1-mediated nuclear protein export. As a control, HA-PTPN23 expressing cells were left untreated. The subcellular distribution of the PTPN23 was analyzed by indirect immunofluorescence (Figure 5D, top) and quantified using ImageJ. Plots of the quantification (Figure 5D, bottom) showed higher signal intensity in the nucleus upon LMB treatment compared with untreated cells. Moreover, the signal in the nucleolus was reduced, suggesting loss of the protein from nucleoli upon LMB treatment. These data indicated that PTPN23 localizes in the cytoplasm as well as in the organelles around the nucleus but also shuttles between nucleus and cytoplasm.
Analysis of assembly and phosphorylation state of the SMN complex
At this point, our data suggested a direct regulatory function of PTPN23 for the SMN complex. PTPN23, for instance, may play a critical role in the assembly of the complex. In PTPN23-knockdown cells, however, SMN coprecipitated at least as efficiently as in controls with Gemin 3, Gemin 2, and unrip (Figure 6A). This indicated efficient SMN complex assembly after PTPN23 knockdown. We then assayed a direct effect of PTPN23 phosphatase on the phosphorylation pattern of SMN protein and performed a two-dimensional (2D) gel analysis of SMN. Cells were knocked down of PTPN23, corresponding cell lysates prepared for 2D gel electrophoresis, and SMN analyzed by Western blot. Several acidic species of SMN were lost upon knockdown of PTPN23 (Figure 6, red arrows in knockdown panel), indicating reduced phosphorylation of SMN after loss of PTPN23. This showed that PTPN23 does not critically influence assembly of the SMN complex but regulates its phosphorylation state.
FIGURE 6:
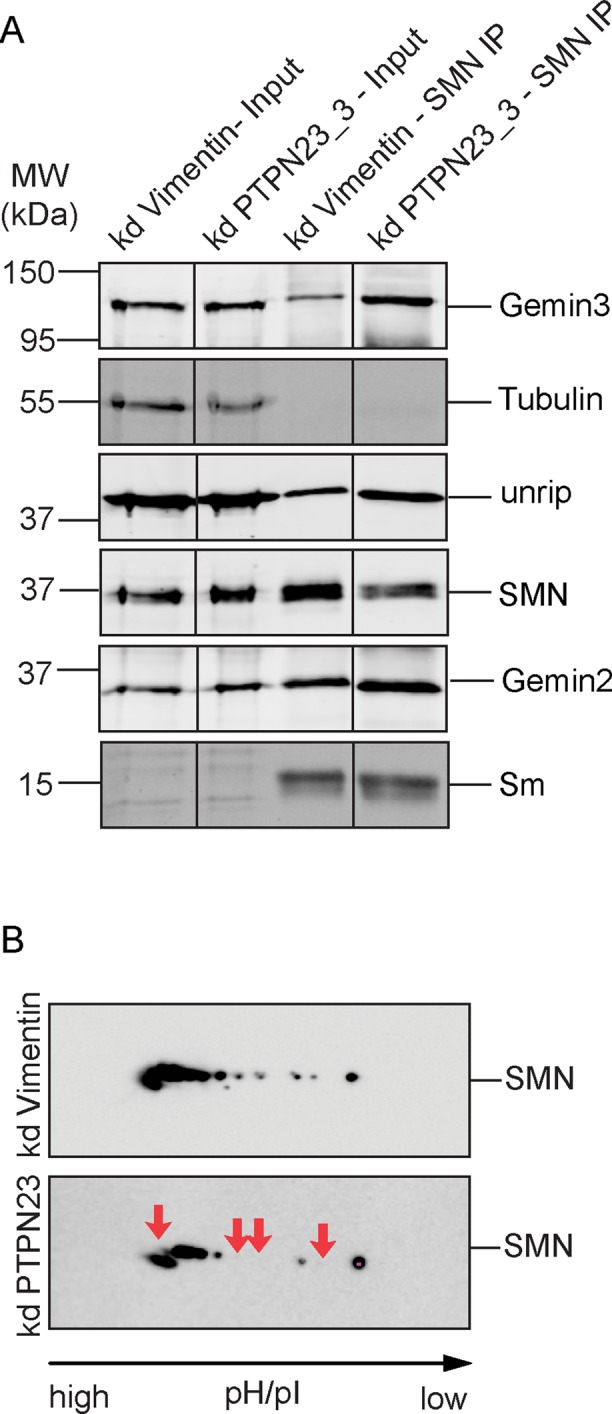
Assembly and phosphorylation state of SMN after PTPN23 knockdown. (A) Association of SMN with interaction partners in the complex in control cell lysates or lysates after PTPN23 knockdown. SMN was immunoprecipitated from total lysates using specific SMN-antibodies, and immunoprecipitates were purified and probed for the presence of SMN, Gemin3, Gemin 4, and unrip as indicated. (B) Two-dimensional gel electrophoresis and immunoblot analysis of SMN after PTPN23 knockdown (kd). Cells were transfected with siRNAs against the indicated phosphatases; vimentin was used as a negative control. At 72 h after transfection, cells were resuspended in isoelectric focusing buffer and lysed; proteins corresponding to 10-mg cell pellet were used for isoelectric focusing, subsequent SDS–PAGE, and Western blot analysis. The phosphorylation state of the SMN protein was analyzed using an antibody specific to SMN in controls or after PTPN23 knockdown. Red arrows in bottom (PTPN23 knockdown) indicate disappearing phosphorylated SMN species.
DISCUSSION
Although mechanistic aspects of SMN complex function in cytoplasmic Sm core assembly catalysis are understood in some detail, the regulation of its function and its localization remains largely unclear. SMN shows a complex pattern of posttranslational modifications. Apart from the large number (12) of phosphorylation sites in SMN itself, almost all other complex components were found to be phosphorylated (Husedzinovic et al., 2014). This indicated a key role of phosphorylation in the modulation of SMN complex formation, activity, and localization.
To identify regulators of SMN that act by reversible phosphorylation, we exploited siRNA-based high-throughput analysis. We performed the first systematic analysis of a potential function of 279 human phosphatases and phosphatase-related gene products in the regulation of SMN localization and function. A workflow of indirect immunofluorescence, automated imaging, and image analysis enabled us to generate and analyze a large data set of SMN subcellular localization. Applying computational analysis of obtained images further allowed quantification of the penetrance of loss-of-function phenotypes, which were defined as either the reduction or the increase in number of cells showing SMN accumulation in CBs. Here, to reduce the influence of the variability on hit selection, we used median and the median absolute deviation (MAD) for hit selection. The robustness of median and MAD against outliers allows identification of weak hits, which would be otherwise reported as false negatives (Stone et al., 2007; Chung et al., 2008). Our screen confirmed the nuclear phosphatase PPM1G, as well as the recently identified SMN regulator PP1γ/PPP1CC (Renvoise et al., 2012). Of interest, in our hands, knockdown of PP1γ resulted in impaired accumulation of SMN in nuclear CB. In contrast, down-regulation of its regulatory subunit, NIPP1/PPP1R8, led to the generation of cytoplasmic SMN accumulations and an increase in the number of CB-like SMN accumulations in the nucleus. These observations suggest a function of PPP1CC as a positive regulator of SMN complex activity, which is antagonized by NIPP1/PPP1R8.
We also identified and validated the PTPN23 as a novel SMN complex regulator. PTPN23 was initially described as a tumor suppressor candidate gene involved in cell migration and endosomal trafficking (Gingras et al., 2009a) and, more recently, was shown to regulate sorting of the epidermal growth factor receptor to multivesicular bodies (Ali et al., 2013). Moreover, PTPN23 was implicated in the regulation of cell migration and cell invasion in cancer cells (Mariotti et al., 2009a, b; Lin et al., 2011). However, little is known about molecular details of PTPN23 function. PTPN23 is a member of the Bro1 domain–containing protein family but also contains several other structural motifs, such as a histidine-rich domain (HIS), the protein tyrosine domain (PTP), and a proteolytic degradation targeting sequence (PEST motif; Toyooka et al., 2000). The multidomain structure of PTPN23 suggests activities in numerous cellular functions. Of interest, PTPN23 was shown to be a catalytically inactive phosphatase, which could stably bind to tyrosine-phosphorylated target proteins, preventing them from dephosphorylation (Barr et al., 2009; Gingras et al., 2009a, b). Our data show that the knockdown of PTPN23 leads to a significant reduction in the number of cells showing SMN accumulation in CBs. The analysis of both endogenous and exogenous PTPN23 revealed that this protein mostly localizes to the cytoplasm but also shuttles between nucleus and cytoplasm. According to this observation, PTPN23 could be involved in the regulation of the SMN complex in both compartments and/or in the regulation of SMN exchange between nucleus and cytoplasm. Of note, we clearly observed a shift of the SMN protein toward hypophosphorylated variants, suggesting that PTPN23 interacts with SMN and thereby protects its tyrosine-phosphorylated state. We showed recently that tyrosine phosphorylation modulates not only SMN complex localization but most likely also its functions. An SMN variant lacking all three tyrosine phosphorylation sites in the TUDOR domain of SMN fails to accumulate in nuclear CBs (Husedzinovic et al., 2014). It is therefore tempting to speculate that PTPN23 interacts with SMN to maintain the tyrosine phosphorylation in the TUDOR domain. The latter may regulate the activity of SMN in snRNP assembly or its trafficking between the nucleus and the cytoplasm. Careful analysis of single-site mutations in the tyrosines of the TUDOR domains after PTPN23 knockdown will be required to make precise the functions of these modifications.
Taken together, our data systematically identify phosphatases that critically determine SMN complex localization and, most likely, the functions of the complex in intact cells. Our experiments not only pinpoint PTPN23 as a novel, unusual regulator of SMN, but they also provide an entry point for further mechanistic studies to unravel the regulation of the SMN in molecular detail.
MATERIALS AND METHODS
cDNA construction
A PTPN23 cDNA clone was obtained from German Resource Center (RZPD, Berlin, Germany; now Source Biosciences, Nottingham, UK). PTPN23 bearing a triple-HA tag was constructed using pcDNA3.1 (Life Technologies, Darmstadt, Germany).
PTPN23 antibody generation
Three different PTPN23 peptides (amino acids 710–743, 1220–1243, and 1609–1623 of the human sequence) were chosen and synthesized for antibody production (Peptide Specialties Laboratories, Heidelberg, Germany). The peptide antibodies were purified from serum of immunized rabbits using peptide-coupled columns according to standard procedures.
Phosphatome-wide siRNA screen
The phosphatome-wide siRNA screen was based on reverse transfection using the Ambion Silencer Select phosphatase library (Ambion Silencer Select Human Phosphatase siRNA Library, 4397919-AMO01Y9M; Life Technologies). The library consisted of three different siRNAs targeting one gene of interest, that is, 894 siRNAs against 298 human phosphatases and phosphatase-related gene products. All siRNAs of the libraries were validated in silico and on the bench to individually target the multiple transcripts of a particular gene (Neumann et al., 2010). Transfections were performed in 96-well plate format (Neumann et al., 2006, 2010; Erfle et al., 2008).
For the primary phosphatome-wide siRNA screen, cells were seeded in 100 μl DMEM medium with antibiotics using a Multidrop Combi cell dispenser (Thermo Scientific, Waltham, MA). After 72 h of incubation, cells were fixed using 3% paraformaldehyde for 10 min. Subsequently, cells were washed three times with 1× phosphate-buffered saline (PBS) and blocked and permeabilized with 1× PBS/Triton X-100 for 1 h. SMN staining was performed using the α-SMN (7B10) primary antibody (1.5 μg/ml) and goat α-mouse Cy3–labeled secondary antibody (2 μg/ml) with incubations of 1 h each. DNA was stained with DAPI at a concentration of 0.25 μg/ml. For the analysis of the subcellular SMN distribution, four images per well were collected on an Olympus ScanR screening microscope (Olympus, Hamburg, Germany) using a 10× objective (UPSLAPO 10×/0.4 numerical aperture [NA]; Olympus).
Automatic image evaluation
To analyze the cellular SMN distribution automatically, we developed an algorithm for the analysis of large-scale microscopy data. In the first step, nuclei and SMN accumulations are segmented. The morphological characteristics of the objects are extracted via several image-processing steps using Matlab (MathWorks, Natick, MA) and Dipimage (DIPlib 1.6 Scientific Image Processing Library; Quantitative Imaging Group, Delft University of Technology, Netherlands; www.diplib.org). From the raw images (tif file, 12 bit), a smoothing function corresponding to the radii of the objects of interest (nuclei: DAPI channel/spots: Cy3 channel) was used to obtain background-corrected images. The global threshold was calculated using Otsu's method, which returned normalized intensity values lying in the range [0, 1]. After that, binary images were generated, and only objects not touching the image boarder (edge objects) were considered. After labeling of the binary image, objects were classified and thus identified as nuclei/spots according to the following criteria: 1) object size, 2) shape (eccentricity, perimeter, Feret diameter), 3) intensity, and 4) relation between eccentricity and object size.
Objects featuring parameters lying outside of these classifications, such as objects that are too large or small, display atypical shape, or have too low or high intensities, were excluded from further analysis. Moreover, we applied a very sensitive edge detection function (Sobel filter using two 3 × 3 kernels) before the generation of binary images. This differentiation operator was introduced in order to detect very weak gradients of the image intensity function (spots with intensity levels lying marginally above the background level).
The border pixels of the identified nuclei were calculated, resulting in corresponding polygons, and the overlay with the identified spots returned polygons containing spots or polygons without spots. A spot was defined as lying within a nucleus if its calculated center of mass was lying inside the polygon. Segmented images were processed further statistically to identify the possible regulators of SMN localization and function. The image processing provided parameters such as 1) number of nuclei without spots (defined as number of nuclei with 0 or 1 spot), 2) number of nuclei with spots (defined as number of nuclei with >1 spot), and 3) number of nuclei with x spots, where x = 2, 3,…, 20.
Statistical data analysis
These parameters were used to evaluate normalized hit scorings with respect to reference samples. Hit scoring focused on the relative change (gain/loss) of these parameters in terms of 1) percentage of nuclei with spots and 2) percentage of nuclei without spots. To extract hits despite experimental variations, we normalized all data points to the median of loss of SMN accumulations in each plate individually. We calculated the MAD and defined the hit threshold for each plate respectively as 2 × MAD. The relative change of the parameters was calculated as a mean of two replicates for each of the 96-well plates. Based on this analysis, the number of selected hits was 61. To verify the selected hits, the microscopy images were visually inspected. Twenty-three of 61 hits were discarded as false positives due to low signal-to-noise ratio. Therefore, the automatic analysis scored 38 primary hits, which led to the loss of SMN accumulations from CBs, including PPM1G itself. Images with <10 nuclei were excluded from statistical analysis. In addition, images with high cell density and out-of-focus images were also excluded because of the possible inaccurate segmentation.
Cell culture: RNA interference and protein overexpression
HeLa CCl2 cells were cultivated in DMEM medium (Life Technologies) supplemented with 10% (vol/vol) fetal calf serum (FCS), 1% (vol/vol) l-glutamine, 1% penicillin, and 1% streptomycin at 37°C and 5% CO2. For immunofluorescence, cells were grown on glass cover slips. All transfection experiments were carried out in DMEM without antibiotics. siRNA-based knockdown of PTPN23 in HeLa CCl2 cells was performed using Ambion Silencer Select oligos (oligo 1: 5′-AGUUUGUCCUGAAGAAUUAtt-3′, 5′-UAAUUCUUCAGGACAAACUtc-3′; oligo 2: 5′-GACCAAAAGUGGAACUCCAtt-3′, 5′-UGGAGUUCCACUUUUGGUCca-3′) at a final concentration of 20 nM and Stealth siRNA (oligo 3: 5′- GAGGUCCUGGACCAGUUCAUGGAUU-3′, 5′-AAUCCAUGAACUGGUCCAGGACCUC 3′) at a final concentration of 50 nM). As a negative control, siRNAs against vimentin were used. For the siRNA transfection, Lipofectamine RNAiMAX (Life Technologies) transfection reagent was used as described in the manufacturer's protocol. For transient plasmid transfection, Lipofectamine 2000 (Life Technologies) was used. The transfection was performed according to the manufacturer's protocol.
Cell fixation, immunofluorescence, imaging, and image processing
For immunofluorescence, cells were grown on cover slips, washed with 1× PBS, and fixed in 3% paraformaldehyde for 10 min at room temperature. Afterward, cells were washed with 1× PBS and blocked using 1× PBS containing 0.1% Triton X-100 and 10% FCS (PBSTS). Fixed cells were incubated with primary antibodies for 1 h, washed, and then incubated with secondary antibodies for 1 h. Primary and secondary antibodies were diluted in 1× PBSTS (primary: α-SMN 7B10, mouse, from 1.5 µg/ml 1:700, α-coilin H300 rabbit [sc-32860; Santa Cruz Biotechnology, Heidelberg, Germany] from 1 μg/ml 1:200, α-coilin F-7 mouse [sc-55594; Santa Cruz Biotechnology] from 1 μg/ml 1:500, α-PTPN23, rabbit, from 1 μg/ml 1:100). DNA was stained using DAPI at a concentration of 0.25 μg/ml. Cells were washed and mounted on object slides using Fluoromount-G mounting medium (Southernbiotech, Birmingham, AL). Analysis was done on Leica SP2 confocal microscope using a 63×/1.2 NA water immersion objective (Leica Microsystems, Mannheim, Germany). Images were processed using the Leica confocal software and Photoshop CS4.
SDS–PAGE and Western blot
Proteins were separated by SDS–PAGE according to Laemmli (1970). For protein separation, cells were lysed by the addition of protein loading buffer (Laemmli, 1970). The samples were boiled for 10 min at 95°C. To analyze the running behavior of proteins based on their difference in posttranslational modifications such as phosphorylation, high-tetramethylethylenediamine [TEMED] gels were used (running gel: 13 ml of 30% acrylamide (37.5:1), 7.5 ml of 1.5 M Tris, pH 8.8, 9.25 ml of double-distilled H2O, 250 μl of 10% SDS, 100 μl of 10% ammonium persulfate, and 100 μl of TEMED; stacking gel: 1.5 ml of 30% acrylamide (37.5:1), 1.7 ml of 1 M Tris, pH 6.8, 5.8 ml of double-distilled H2O, 75 μl of 10% SDS, 45 μl of 10% ammonium persulfate and 22.5 μl of TEMED). After SDS–PAGE, proteins were transferred to the nitrocellulose membranes. Primary antibodies used for the protein immunodetection were the same as for immunofluorescence (dilutions: α-SMN 7B10 mouse, 1:4000, from 1.5 μg/ml, α-PTPN23 from 1 μg/ml, 1:1000, α-tubulin mouse [DM1A, T9026; Sigma-Aldrich, Taufkirchen, Germany], 1:5.000). Secondary antibodies used were IRDye 800 α-mouse immunoglobulin G (IgG) and IRDye 700 α-rabbit IgG (Life Technologies), both at 1:10,000 dilution. Fluorescence signals were detected using an infrared scanner (Odyssey; LICOR Biosciences, Lincoln, NE).
2D gel electrophoresis
For 2D gel electrophoresis, cells were washed and harvested with ice-cold 1× PBS.
After centrifugation at 5000 rpm at 4°C for 5 min, pellets were resuspended in isoelectric focusing buffer at room temperature and additionally lysed by sonification. Insoluble particles were removed by centrifugation at 60,000 rpm (TLA 100.3 Rotor; Beckman Coulter, Krefeld, Germany) at room temperature for 30 min. Proteins of ∼10-mg cell pellet were separated by isoelectric focusing in the first dimension and by SDS–PAGE in the second dimension. Afterward, Western blot (wet blot transfer) was performed. The SMN protein was detected using a mouse monoclonal anti-SMN (7B10; Meister et al., 2000) antibody and a horseradish peroxidase–coupled anti-mouse antibody. The signals were detected using ECL Plus Western Blotting Substrate (Pierce Biotechnology, Thermo Scientific, Rockford, IL) and analyzed by ImageQuant LAS 4000 (GE Healthcare, Munich, Germany).
HeLa cell lysate preparation for immunoprecipitation experiments
We resuspended 5 × 109 cells (HeLa whole-cell pellet; Cilbiotech, Mons, Belgium) in 3× volumes of 1× PBS with 10 mM Na3VO4, 5 mM NaF, and protease inhibitors (complete EDTA free; Roche, Mannheim, Germany) and lyzed them by sonification. Insoluble particles were removed by centrifugation at 1500 × g for 10 min. For the lipid extraction, N-heptane was added at a dilution of 1:15. Lysates were gently mixed and centrifuged at 25,000 rpm (SW41 Ti rotor; Beckman Coulter). The middle phase was collected using a syringe and a needle. The protein concentration was ∼1.5 mg/ml.
α-SMN immunoprecipitation
We added 3 ml of cell lysate with a protein concentration of 1.5 mg/ml to 50 μl of α-SMN antibody (7B10; Meister et al., 2000) covalently coupled to protein G–Sepharose beads (GE Healthcare) and incubated by rotation at 4°C for 2 h. The beads were extensively washed 10 times for 20 min with 1× PBS and 5 mM NaF. The immunoprecipitated proteins were eluted with 100 mM glycine, pH 2.2, and precipitated by chloroform–methanol precipitation as described (Wessel and Flugge, 1984).
α-PTPN23 immunoprecipitation
We loaded 3 mg of total protein from HeLa lysate onto 50 μl of PTPN23 peptide antibody (amino acids 710–743) covalently coupled to protein A–Sepharose beads (Life Technologies) and incubated for 2 h by rotation at 4°C. The beads were extensively washed 10 times for 10 min with ice-cold 1× PBS supplied with protease inhibitors and 5 mM NaF. Bound proteins were eluted using 2 M MgCl2. Subsequently, proteins were eluted using 100 mM glycine, pH 2.2. The proteins from the MgCl2 fraction were precipitated using five volumes of ice-cold isopropanol, and the proteins from the glycine fraction were precipitated using five volumes of acetone, both at −20°C.
Supplementary Material
Acknowledgments
This work was supported by grants of the Deutsche Forschungsgemeinschaft to O.J.G. (OG1737/6-1), U.F. (FI 573/6-1), and the ViroQuant-CellNetworks RNAi screening facility through the CellNetworks-Cluster of Excellence (EXC81).
Abbreviations used:
- CB
Cajal (coiled) body
- HD-PTP
His domain–containing tyrosine phosphatase
- PTPN23
tyrosine-protein phosphatase non–receptor type 23
- SMN
survival motor neuron
- snRNA
(uridine-rich) small nuclear RNA
- snRNP
(uridine-rich) small nuclear ribonucleoprotein particle.
Footnotes
Present addresses: *Nationales Centrum für Tumorerkrankungen, D-69120 Heidelberg; Germany;
†Max-Planck-Institute for Biophysical Chemistry, D-37077 Göttingen, Germany.
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-06-1151) on November 12, 2014.
REFERENCES
- Ali N, Zhang L, Taylor S, Mironov A, Urbe S, Woodman P. Recruitment of UBPY and ESCRT exchange drive HD-PTP-dependent sorting of EGFR to the MVB. Curr Biol. 2013;23:453–461. doi: 10.1016/j.cub.2013.02.033. [DOI] [PubMed] [Google Scholar]
- Barr AJ, Ugochukwu E, Lee WH, King ON, Filippakopoulos P, Alfano I, Savitsky P, Burgess-Brown NA, Muller S, Knapp S. Large-scale structural analysis of the classical human protein tyrosine phosphatome. Cell. 2009;136:352–363. doi: 10.1016/j.cell.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghes AH, Beattie CE. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick. Nat Rev Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnegie GK, Sleeman JE, Morrice N, Hastie CJ, Peggie MW, Philp A, Lamond AI, Cohen PT. Protein phosphatase 4 interacts with the Survival of Motor Neurons complex and enhances the temporal localisation of snRNPs. J Cell Sci. 2003;116:1905–1913. doi: 10.1242/jcs.00409. [DOI] [PubMed] [Google Scholar]
- Carvalho T, Almeida F, Calapez A, Lafarga M, Berciano MT, Carmo-Fonseca M. The spinal muscular atrophy disease gene product, SMN: A link between snRNP biogenesis and the Cajal (coiled) body. J Cell Biol. 1999;147:715–728. doi: 10.1083/jcb.147.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari A, Golas MM, Klingenhager M, Neuenkirchen N, Sander B, Englbrecht C, Sickmann A, Stark H, Fischer U. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell. 2008;135:497–509. doi: 10.1016/j.cell.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Chung N, Zhang XD, Kreamer A, Locco L, Kuan PF, Bartz S, Linsley PS, Ferrer M, Strulovici B. Median absolute deviation to improve hit selection for genome-scale RNAi screens. J Biomol Screen. 2008;13:149–158. doi: 10.1177/1087057107312035. [DOI] [PubMed] [Google Scholar]
- Coady TH, Lorson CL. SMN in spinal muscular atrophy and snRNP biogenesis. Wiley Interdiscip Rev. RNA. 2011;2:546–564. doi: 10.1002/wrna.76. [DOI] [PubMed] [Google Scholar]
- Cohen PT. Protein phosphatase 1—targeted in many directions. J Cell Sci. 2002;115:241–256. doi: 10.1242/jcs.115.2.241. [DOI] [PubMed] [Google Scholar]
- Darzacq X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2’-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erfle H, Neumann B, Rogers P, Bulkescher J, Ellenberg J, Pepperkok R. Work flow for multiplexing siRNA assays by solid-phase reverse transfection in multiwell plates. J Biomol Screening. 2008;13:575–580. doi: 10.1177/1087057108320133. [DOI] [PubMed] [Google Scholar]
- Farrar MA, Johnston HM, Grattan-Smith P, Turner A, Kiernan MC. Spinal muscular atrophy: molecular mechanisms. Curr Mol Med. 2009;9:851–862. doi: 10.2174/156652409789105516. [DOI] [PubMed] [Google Scholar]
- Fischer U, Englbrecht C, Chari A. Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscip Rev. RNA. 2011;2:718–731. doi: 10.1002/wrna.87. [DOI] [PubMed] [Google Scholar]
- Gingras MC, Kharitidi D, Chenard V, Uetani N, Bouchard M, Tremblay ML, Pause A. Expression analysis and essential role of the putative tyrosine phosphatase His-domain-containing protein tyrosine phosphatase (HD-PTP) Int J Dev Biol. 2009a;53:1069–1074. doi: 10.1387/ijdb.082820mg. [DOI] [PubMed] [Google Scholar]
- Gingras MC, Zhang YL, Kharitidi D, Barr AJ, Knapp S, Tremblay ML, Pause A. HD-PTP is a catalytically inactive tyrosine phosphatase due to a conserved divergence in its phosphatase domain. PLoS One. 2009b;4:e5105. doi: 10.1371/journal.pone.0005105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmler M, Bauer L, Nousiainen M, Korner R, Meister G, Fischer U. Phosphorylation regulates the activity of the SMN complex during assembly of spliceosomal U snRNPs. EMBO Rep. 2005;6:70–76. doi: 10.1038/sj.embor.7400301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husedzinovic A, Oppermann FS, Draeger-Meurer S, Chari A, Fischer U, Daub H, Gruss OJ. Phosphoregulation of the human SMN Complex. Eur J Cell Biol. 2014;93:106–117. doi: 10.1016/j.ejcb.2014.01.006. [DOI] [PubMed] [Google Scholar]
- Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, Luhrmann R, Li J, Nagai K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- Kiss AM, Jady BE, Darzacq X, Verheggen C, Bertrand E, Kiss T. A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res. 2002;30:4643–4649. doi: 10.1093/nar/gkf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lafarga M, Casafont I, Bengoechea R, Tapia O, Berciano MT. Cajal's contribution to the knowledge of the neuronal cell nucleus. Chromosoma. 2009;118:437–443. doi: 10.1007/s00412-009-0212-x. [DOI] [PubMed] [Google Scholar]
- Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonne R, Luhrmann R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell. 2006;17:3221–3231. doi: 10.1091/mbc.E06-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Aranda V, Muthuswamy SK, Tonks NK. Identification of PTPN23 as a novel regulator of cell invasion in mammary epithelial cells from a loss-of-function screen of the “PTP-ome.”. Genes Dev. 2011;25:1412–1425. doi: 10.1101/gad.2018911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti M, Castiglioni S, Garcia-Manteiga JM, Beguinot L, Maier JA. HD-PTP inhibits endothelial migration through its interaction with Src. Int J Biochem Cell Biol. 2009a;41:687–693. doi: 10.1016/j.biocel.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Mariotti M, Castiglioni S, Maier JA. Inhibition of T24 human bladder carcinoma cell migration by RNA interference suppressing the expression of HD-PTP. Cancer Lett. 2009b;273:155–163. doi: 10.1016/j.canlet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- Meister G, Buhler D, Laggerbauer B, Zobawa M, Lottspeich F, Fischer U. Characterization of a nuclear 20S complex containing the survival of motor neurons (SMN) protein and a specific subset of spliceosomal Sm proteins. Hum Mol Genet. 2000;9:1977–1986. doi: 10.1093/hmg/9.13.1977. [DOI] [PubMed] [Google Scholar]
- Meister G, Buhler D, Pillai R, Lottspeich F, Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- Narayanan U, Achsel T, Luhrmann R, Matera AG. Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein. Mol Cell. 2004;16:223–234. doi: 10.1016/j.molcel.2004.09.024. [DOI] [PubMed] [Google Scholar]
- Neumann B, Held M, Liebel U, Erfle H, Rogers P, Pepperkok R, Ellenberg J. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat Methods. 2006;3:385–390. doi: 10.1038/nmeth876. [DOI] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep. 2007;8:340–345. doi: 10.1038/sj.embor.7400941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- Petri S, Grimmler M, Over S, Fischer U, Gruss OJ. Dephosphorylation of survival motor neurons (SMN) by PPM1G/PP2Cgamma governs Cajal body localization and stability of the SMN complex. J Cell Biol. 2007;179:451–465. doi: 10.1083/jcb.200704163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renvoise B, Querol G, Verrier ER, Burlet P, Lefebvre S. A role for protein phosphatase PP1gamma in SMN complex formation and subnuclear localization to Cajal bodies. J Cell Sci. 2012;125:2862–2874. doi: 10.1242/jcs.096255. [DOI] [PubMed] [Google Scholar]
- Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- Schaffert N, Hossbach M, Heintzmann R, Achsel T, Luhrmann R. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 2004;23:3000–3009. doi: 10.1038/sj.emboj.7600296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw DJ, Eggleton P, Young PJ. Joining the dots: production, processing and targeting of U snRNP to nuclear bodies. Biochim Biophys Acta. 2008;1783:2137–2144. doi: 10.1016/j.bbamcr.2008.07.025. [DOI] [PubMed] [Google Scholar]
- Sleeman JE, Lamond AI. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr Biol. 1999;9:1065–1074. doi: 10.1016/s0960-9822(99)80475-8. [DOI] [PubMed] [Google Scholar]
- Stanek D, Neugebauer KM. The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma. 2006;115:343–354. doi: 10.1007/s00412-006-0056-6. [DOI] [PubMed] [Google Scholar]
- Stanek D, Pridalova-Hnilicova J, Novotny I, Huranova M, Blazikova M, Wen X, Sapra AK, Neugebauer KM. Spliceosomal small nuclear ribonucleoprotein particles repeatedly cycle through Cajal bodies. Mol Biol Cell. 2008;19:2534–2543. doi: 10.1091/mbc.E07-12-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H, Dube P, Luhrmann R, Kastner B. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature. 2001;409:539–542. doi: 10.1038/35054102. [DOI] [PubMed] [Google Scholar]
- Stone DJ, Marine S, Majercak J, Ray WJ, Espeseth A, Simon A, Ferrer M. High-throughput screening by RNA interference: control of two distinct types of variance. Cell Cycle. 2007;6:898–901. doi: 10.4161/cc.6.8.4184. [DOI] [PubMed] [Google Scholar]
- Toyooka S, Ouchida M, Jitsumori Y, Tsukuda K, Sakai A, Nakamura A, Shimizu N, Shimizu K. HD-PTP: A novel protein tyrosine phosphatase gene on human chromosome 3p21.3. Biochem Biophys Res Commun. 2000;278:671–678. doi: 10.1006/bbrc.2000.3870. [DOI] [PubMed] [Google Scholar]
- Wachtel C, Manley JL. Splicing of mRNA precursors: the role of RNAs and proteins in catalysis. Mol Biosyst. 2009;5:311–316. doi: 10.1039/b820828j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a003707. a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.