ER mannosidase I (ERManI) was found recently in the Golgi. This result is found to arise artificially from membrane disturbance in immunofluorescence methods. ERManI is located in novel vesicles to which substrates traffic and that converge at the ER-derived quality control compartment under ER stress.
Abstract
Endoplasmic reticulum α1,2 mannosidase I (ERManI), a central component of ER quality control and ER-associated degradation (ERAD), acts as a timer enzyme, modifying N-linked sugar chains of glycoproteins with time. This process halts glycoprotein folding attempts when necessary and targets terminally misfolded glycoproteins to ERAD. Despite the importance of ERManI in maintenance of glycoprotein quality control, fundamental questions regarding this enzyme remain controversial. One such question is the subcellular localization of ERManI, which has been suggested to localize to the ER membrane, the ER-derived quality control compartment (ERQC), and, surprisingly, recently to the Golgi apparatus. To try to clarify this controversy, we applied a series of approaches that indicate that ERManI is located, at the steady state, in quality control vesicles (QCVs) to which ERAD substrates are transported and in which they interact with the enzyme. Both endogenous and exogenously expressed ERManI migrate at an ER-like density on iodixanol gradients, suggesting that the QCVs are derived from the ER. The QCVs are highly mobile, displaying dynamics that are dependent on microtubules and COP-II but not on COP-I vesicle machinery. Under ER stress conditions, the QCVs converge in a juxtanuclear region, at the ERQC, as previously reported. Our results also suggest that ERManI is turned over by an active autophagic process. Of importance, we found that membrane disturbance, as is common in immunofluorescence methods, leads to an artificial appearance of ERManI in a Golgi pattern.
INTRODUCTION
Glycoprotein quality monitoring is a highly controlled process in which the folding states of glycoproteins are constantly evaluated. Folding states are communicated to the quality control machinery by the structure of the N-linked oligosaccharides these proteins bear (Spiro, 2000; Aebi et al., 2010; Benyair et al., 2011). Nascent glycoproteins receive an oligosaccharide consisting of three glucose, nine mannose, and two GlcNAc residues (Glc3Man9GlcNAc2) in a single transfer reaction. As glycoproteins progress in their folding attempts, they undergo sequential cleavage of their oligosaccharides, starting with the two terminal glucose residues, to produce Glc1Man9GlcNAc2. Glycoproteins bearing this oligosaccharide can interact with the lectin-chaperone calnexin (CNX) or its soluble homologue calreticulin (CRT), an association that allows such glycoproteins time to achieve proper folding (Hebert et al., 1997; Parodi, 2000). The terminal glucose residue is then trimmed, producing a nonglucosylated oligosaccharide, Man9GlcNAc2. Glycoproteins with this oligosaccharide dissociate from CNX and are scrutinized by the folding sensor UDPGlc:glycoprotein glycosyl transferase (UGGT). Glycoproteins that are found to be incompletely folded or misfolded will be reglucosylated by UGGT, allowing them to renew their association with CNX or CRT and commence further folding attempts. The cyclical action of glucose trimming, examination, and reglucosylation is known as the CNX cycle and is an important part of glycoprotein quality control (Caramelo and Parodi, 2008). A crucial step in the CNX cycle is its termination, by which a glycoprotein is deemed either properly folded or terminally misfolded. As glycoproteins go through the CNX cycle, their mannose residues are sequentially and extensively trimmed from Man9GlcNAc2 to Man5-6GlcNAc2 (Herscovics et al., 2002; Frenkel et al., 2003; Hosokawa et al., 2003), until a critical mannose residue, the glucose acceptor, is removed, thus precluding the glycoprotein from association with CNX and targeting it to endoplasmic reticulum–associated degradation (ERAD; Hammond and Helenius, 1994; Burda and Aebi, 1998; Frenkel et al., 2003; Singh et al., 2012). Because ER α1,2 mannosidase I (ERManI) was found compartmentalized in the ER-derived quality control compartment (ERQC), it was suggested that its high concentration in this compartment could be sufficient for this extensive trimming (Avezov et al., 2008). This requirement for ERManI and mannose trimming is essential for glycoprotein ERAD, except under ER stress, which causes up-regulation and concentration of ER degradation–enhancing α-mannosidase-like protein 1 in the ERQC and abrogation of the mannose-trimming requirement (Ron et al., 2011). Initially characterized in Saccharomyces cerevisiae, ERManI (Mns1p in yeast) is a type 2 transmembrane protein that trims α1,2-linked mannose residues, with preference for the B-branch terminal mannose of N-linked oligosaccharides (Camirand et al., 1991; Grondin and Herscovics, 1992; Tremblay and Herscovics, 1999). ERManI was first observed as a transmembrane yeast enzyme that localizes to the ER (Esmon et al., 1984; Burke et al., 1996). This localization depends on yeast Rer1p (Massaad et al., 1999), as ERManI has no known ER retention signals (H/KDEL, dilysine or diarginine). When studied in mammalian cells, ERManI was also described as an ER membrane protein, as in yeast (Gonzalez et al., 1999; Tremblay and Herscovics 1999). However, further studies showed that ERManI is concentrated in the ERQC (Avezov et al., 2008), to which ERAD substrates cycle. It was claimed that ERManI resides in the Golgi apparatus (Pan et al., 2011), and a COPI-dependent cycling mechanism was postulated by which ERAD substrates reach the Golgi and recycle to the ER for degradation (Pan et al., 2013). However, most ERAD substrates do not reach the Golgi (Shenkman et al., 1997; Hebert et al., 2010; Izawa et al., 2012), raising a requirement for a different mechanism of quality control. Here we show that ERManI resides in quality control vesicles (QCVs) with ER-like density, to which ERAD substrates are delivered. Under stress conditions, these QCVs converge to form the previously described ERQC. Our results further suggest that ERManI is finally delivered for degradation by an autophagic pathway. Moreover, we conclude that the sighting of ERManI in the Golgi apparatus in fixed cells is artifactual and is caused by disturbance of intracellular membranes associated with common fixation methods.
RESULTS
ERManI localizes to juxtanuclear accumulations and dynamic vesicles with ER-like density
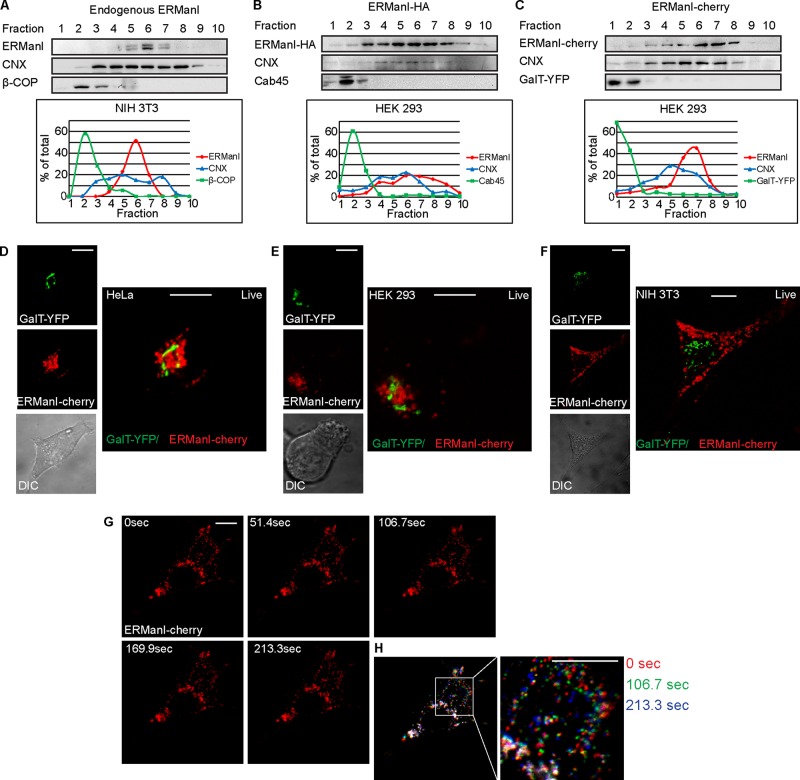
Studies conducted in several labs, including our own, have described different subcellular localizations of ERManI under various conditions (Esmon et al., 1984; Burke et al., 1996; Avezov et al., 2008; Pan et al., 2011; Smirle et al., 2013). To clarify the controversy surrounding the subcellular localization of ERManI, we used equilibrium centrifugation density gradients in which murine NIH 3T3 cells expressing endogenous ERManI levels were homogenized and run through a continuous iodixanol gradient. Western blot analysis using anti-ERManI antibodies showed a distinct separation between ERManI, which migrated in the middle to heavy fractions of the gradient, and a protein localized mainly in the Golgi, β-COP, which migrated in characteristic low-density fractions (Figure 1A and Supplemental Figure S1). ERManI-containing fractions overlapped with those including the ER marker CNX but in a much more restricted pattern. To expand our observation, we then analyzed another cell type, human HEK 293 cells, with exogenous expression of transiently transfected hemagglutinin (HA)-tagged ERManI. The results were similar to those of endogenous ERManI, with ERManI-HA peaking in fractions 6 and 7, whereas a different endogenous Golgi marker, Cab45, migrated similarly to β-COP, peaking at fraction 2 (Figure 1B). In addition, a fusion protein that we constructed, of ERManI linked to mCherry (ERManI-cherry), migrated similarly, peaking in fractions 6 and 7, compared with the migration of an exogenously expressed Golgi marker, GalT–yellow fluorescent protein (YFP), which appeared in the light fractions (Figure 1C). Therefore endogenous or exogenously expressed ERManI behaved similarly in different cell types, localizing to a subset of ER-density fractions and clearly separated from Golgi fractions. The finding that ERManI was absent from the light fractions contradicts the recent suggestion that ERManI is a Golgi-resident enzyme. Furthermore, the migration of ERManI in fractions that coincide with a subset of heavier fractions containing CNX is consistent with our previous results, showing ERManI to reside in an ER-derived compartment, the ERQC (Avezov et al., 2008). Several proteins that reside in the ERQC or accumulate in this compartment upon ER stress were shown to migrate at these densities (Leitman et al., 2014). To visualize directly ERManI localization in cells, we conducted live-cell confocal microscopy experiments in which HeLa, HEK 293, and NIH 3T3 cells were transfected with ERManI-cherry and the Golgi marker GalT-YFP. The results obtained in live HeLa (Figure 1D) and HEK 293 (Figure 1E) cells show ERManI-cherry mostly concentrated in juxtanuclear accumulations localized near, but distinct from, the Golgi marker GalT-YFP, which is consistent with ERQC localization (Avezov et al., 2008). A fraction of ERManI-cherry was also seen in a punctate pattern, likely disperse vesicles, and close examination of the juxtanuclear accumulations suggested that they also consist mainly of vesicles. In NIH 3T3 cells, ERManI-cherry was observed mostly in a disperse vesicular pattern and less so in juxtanuclear accumulations, which did not colocalize with the Golgi marker GalT-YFP (Figure 1F). Time-lapse live microscopy showed ERManI-containing vesicles in two characteristic states. Whereas some vesicles converged around the nucleus and were relatively immobile, others were very mobile, moving along the cell periphery (Supplemental Movies S1–S3). Vesicles in NIH 3T3 cells (Figure 1G and Supplemental Movie S3) were less mobile than those in the other cell types. To better visualize ERManI vesicle dynamics in NIH 3T3, we selected three time frames, which were overlaid and pseudocolored. All three colors can be observed where the ERManI containing vesicles are mobile, whereas immobile vesicles appear white (Figure 1H). In most of the cell periphery, all three colors can be discerned (Figure 1H, inset), pointing toward high levels of vesicle mobility, whereas areas that appear white are visible around the nucleus, suggesting that, on the acquired time scale, vesicles in this region are immobile or at least not as mobile as peripheral ones.
FIGURE 1:
ERManI is in subcellular compartments with ER-like density, which include dynamic peripheral vesicles and less mobile juxtanuclear accumulations. (A–C) Iodixanol equilibrium centrifugation gradients show comigration of a subset of fractions containing CNX with endogenous ERManI in NIH 3T3 cells (A) and with exogenously expressed ERManI with a small HA tag (B) or fused to a larger fluorescent protein, mCherry (C), in HEK 293 cells. ERManI is in denser fractions than those of the Golgi, as visualized by both endogenous (β-COP and Cab45) and exogenous Golgi markers (GalT-YFP). Antibodies used were mouse anti-ERManI, anti–β-COP, and anti-GFP and rabbit anti-CNX and anti-Cab45. A representative experiment out of three repeat experiments of each is shown, and its quantification is shown in the graphs at the bottom. (D–F) Live-cell imaging of HeLa (D), HEK 293 (E), and NIH 3T3 cells (F) expressing ERManI-cherry and GalT-YFP shows that ERManI localization is distinct from the Golgi. Bars, 10 μm. (G) Time-lapse images of an NIH 3T3 cell expressing ERManI-cherry. (H) Three time points from G were overlaid and pseudocolored as indicated to illustrate the high levels of mobility of ERManI-containing vesicles.
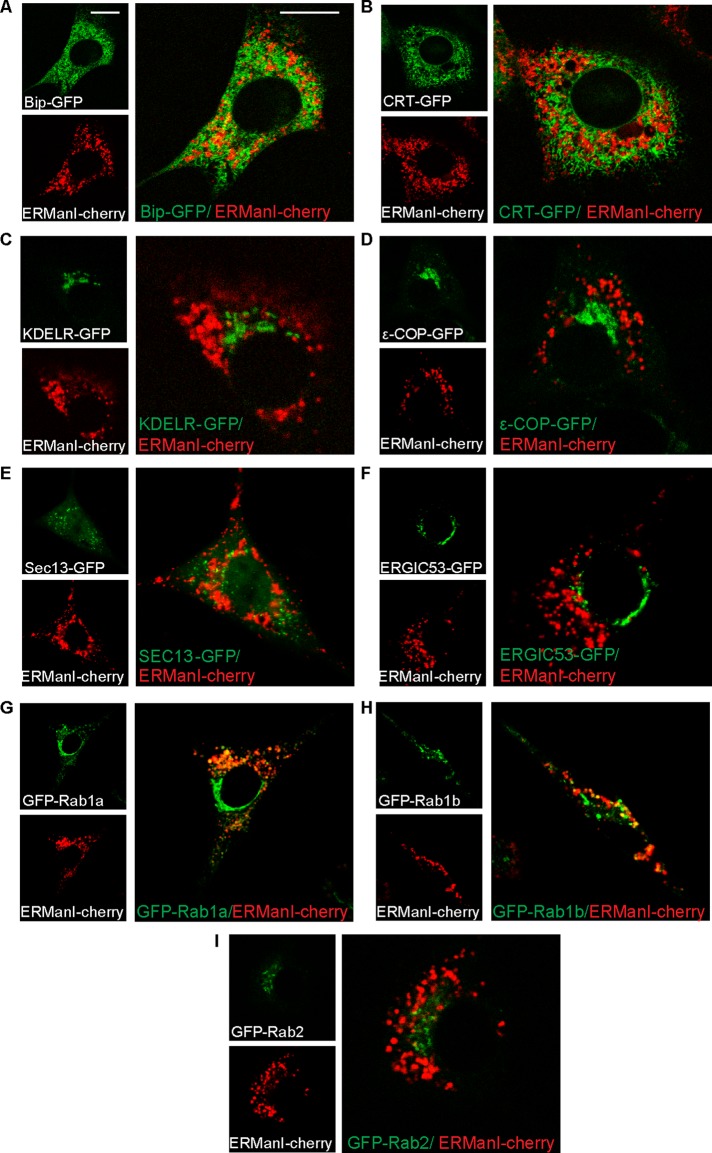
ERManI-containing vesicles do not colocalize with markers of other organelles, yet some are Rab1 positive
To better characterize the ERManI-containing vesicles, which we will call “quality control vesicles” (QCVs), we concentrated our efforts on NIH 3T3 cells, as in these cells the vesicular pattern of ERManI was more disperse. Many subcellular organelles are vesicular or partly vesicular in nature, and thus it became important to examine the distinction of QCVs from such organelles. To this end, we compared the localization of ERManI-cherry–containing QCVs to various organelle and vesicle markers. We observed little or no colocalization between QCVs and markers of the ER—BiP–green fluorescent protein (GFP) and CRT-GFP (Figure 2, A and B). No colocalization was observed between the QCVs and markers of the Golgi apparatus—KDEL receptor (Figure 2C) and ε-COP (Figure 2D), which is also present in COP-I vesicles. There was no colocalization with ER exit sites (Sec13-GFP; Figure 2E) or with the ER-to-Golgi intermediate compartment (ERGIC53-GFP) either (Figure 2F). However, when imaging ERManI-cherry along with Ras-related proteins in brain (Rabs), which are associated with vesicular trafficking, there was a certain degree of colocalization. Rab proteins function not only in the docking of vesicles to their targets, but also in vesicle budding and association of vesicles with the cytoskeleton (Nuoffer et al., 1994; Echard et al., 1998; Nielsen et al., 1999; Allan et al., 2000). Although Rab1a and Rab1b have been associated with COP-II vesicle formation at the ER, they also arrive at the Golgi apparatus, where they mitigate vesicle tethering. Rab2, on the other hand, has been implicated in COP-I vesicle formation and is found both in the ER and Golgi (Zerial and McBride, 2001). When coexpressing ERManI-cherry and GFP-Rab1a, two subpopulations of Rab1a were apparent, one that appears in a Golgi-like pattern and another that was disperse in vesicles. The latter subpopulation partially colocalized with ERManI-containing vesicles (Figure 2G). The same was true when coexpressing ERManI-cherry with GFP-Rab1b, although the colocalization was not as extensive as with Rab1a (Figure 2H). GFP-Rab2 was mainly in a vesicular pattern, which was distinct from ERManI-containing QCVs (Figure 2I). These results suggest a relationship between Rab1 and QCVs, although the exact nature of this relationship is unclear.
FIGURE 2:
QCVs containing ERManI are distinct from other organelles. (A–F) Live-cell imaging of NIH 3T3 cells expressing ERManI-cherry and various organelle markers shows distinction of ERManI-containing vesicles from the ER (A, BiP-GFP; B, CRT-GFP), the Golgi (C, KDELR-GFP), COP-I vesicles (D, εCOP-GFP), and ER exit sites and early COP-II vesicles (E, Sec13-GFP). QCVs are also distinct from the ERGIC (F, ERGIC53-GFP). Bars, 10 μm. (G–I) QCVs show partial colocalization with GFP-Rab1a (G) and GFP-Rab1b (H) but not with GFP-Rab2 (I).
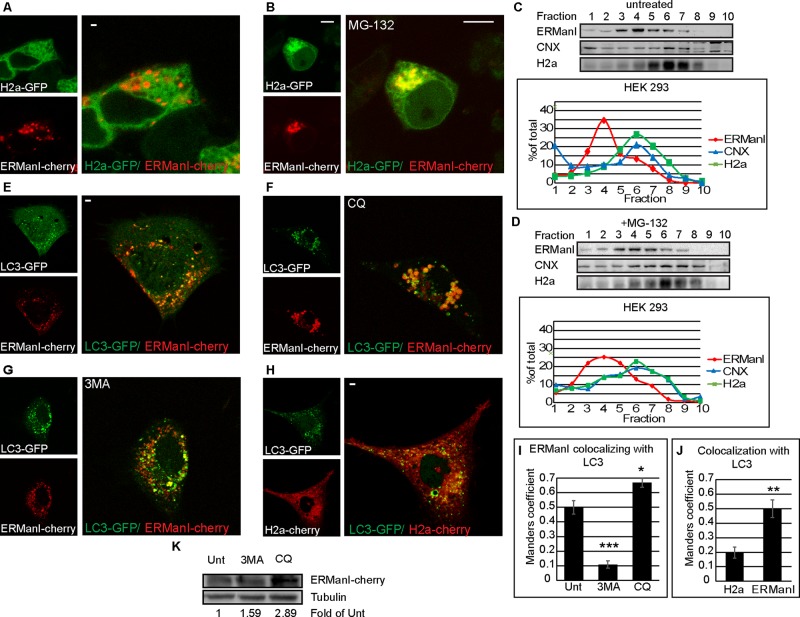
ERManI accumulates with an ERAD substrate near the nucleus under proteasomal inhibition (ERQC), whereas a subpopulation colocalizes with microtubule-associated protein1 light chain 3
When examining the ERAD substrate uncleaved precursor of asialoglycoprotein receptor H2a linked to GFP (H2a-GFP; Leitman et al., 2013) in live-cell microscopy, very little colocalization could be observed with ERManI-cherry–containing QCVs (Figure 3A), and this is likely due to the fact that ERManI accelerates the proteasomal degradation of H2a (Avezov et al., 2008). However, upon proteasomal inhibition, it is clear that a large portion (but not all) of the QCVs converged near the nucleus, colocalizing with the substrate H2a-GFP (Figure 3B and Supplemental Figure S2) in the ERQC, as we saw previously (Avezov et al., 2008). In untreated cells, only a subpopulation of ERManI comigrated with the substrate in iodixanol gradients (Figure 3C). The comigration increased to some extent upon proteasomal inhibition, with a larger portion of ERManI in heavier fractions (Figure 3D).
FIGURE 3:
ERManI concentrates at the ERQC under proteasomal inhibition, and some ERManI-containing vesicles colocalize with LC3-GFP. (A, B) Live-cell imaging of HEK 293 cells expressing ERManI-cherry and the ERAD substrate H2a-GFP. When left untreated (A), no colocalization is apparent, but under proteasomal inhibition by 20 μM MG-132 for 3 h (B), ERManI and the ERAD substrate colocalize at a juxtanuclear region characterized before as the ERQC. Bars, 10 μm. (C, D) Iodixanol equilibrium centrifugation gradients show overlap of fractions containing endogenous ERManI and transiently expressed H2a in untreated HEK 293 cells (C), with some increased overlap after MG-132 treatment (40 μM for 3 h; D). ERManI and H2a are found in fractions containing the ER marker CNX; quantification is shown in the graphs. (E–G) Live-cell imaging of NIH 3T3 cells expressing ERManI-cherry and LC3-GFP. There is a degree of colocalization in untreated cells (E). Lysosomal inhibition by CQ at 100 μM for 24 h increases colocalization (F), whereas inhibition of autophagy by 3MA at 5 mM for 24 h decreases it (G). (H) NIH 3T3 cells expressing LC3-GFP and H2a-cherry show little to no colocalization between the ERAD substrate and the autophagosomes. (I) Quantitation of ERManI colocalization with LC3, inferred from the Manders coefficient. ***p (vs. untreated) = 0.0002; *p = 0.045. (J) Comparisons of the quantitation of H2a or ERManI colocalization with LC3, inferred from the Manders coefficient. **p = 0.0021. (K) HEK 293 cells transiently expressing ERManI-Cherry were incubated for 6 h with 3MA (5 mM) or CQ (100 μM) or left untreated. Lysates were immunoblotted with primary rabbit anti-dsRed or mouse anti-tubulin and secondary antibodies conjugated to HRP.
ERManI is a short-lived protein degraded at the lysosomes (Wu et al., 2007; Avezov et al., 2008), although the pathway by which ERManI arrives at the lysosomes is unknown. We compared ERManI-cherry localization to that of microtubule-associated protein1 light chain 3 (LC3), an autophagosome marker (Tanida et al., 2008). Under normal conditions, some colocalization between LC3 and ERManI vesicles was apparent under live-cell microscopy (Figure 3E). On treatment with the lysosomal inhibitor chloroquine (CQ), the intensity of ERManI-cherry fluorescence increased significantly as it accumulated in structures larger than those observed in untreated cells (Figure 3F). Colocalization with LC3 also increased noticeably (Figure 3I), suggesting that ERManI may arrive via autophagosomes to the lysosomes for degradation. The autophagy inhibitor 3-methyladenine had the reverse effect, reducing LC3-ERManI colocalization quite drastically but without affecting fluorescence intensity (Figure 3, G and I). H2a-cherry showed little to no colocalization with LC3-GFP, suggesting that in untreated cells, the fraction of vesicles of ERManI that colocalizes with LC3-GFP is distinct from the compartments containing the ERAD substrate (Figure 3, H and J). ERManI levels were increased by cell treatment with 3-methyladenine, and a much larger increase was obtained by treatment with chloroquine, as judged by immunoblot (Figure 3K). Altogether the results are consistent with an autophagic pathway for degradation of ERManI.
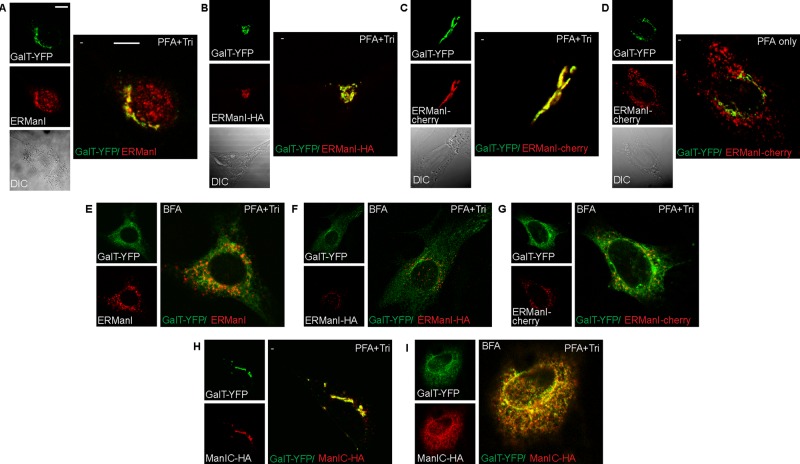
The ERManI localization pattern is artifactually altered by cell fixation and permeabilization
Previous research reporting Golgi-localization of ERManI was carried out under conditions of fixation using 4% paraformaldehyde (PFA) and permeabilization with 0.5% Triton X-100 (Pan et al., 2011). Those results are in stark contrast with the results of our live-cell imaging experiments and of the iodixanol gradients with endogenous or exogenous ERManI expression (Figures 1–3). This led us to examine ERManI under the conditions used by the Sifers group and of other widely used fixation and permeabilization methods. NIH 3T3 cells were transfected with GalT-YFP and left with endogenous levels of ERManI (Figure 4A) or cotransfected with either ERManI-HA (Figure 4B) or ERManI- cherry (Figure 4C). These cells were then fixed and permeabilized either with PFA and 0.5% Triton (PFA+Tri) or with methanol-acetone (Met:Ac) or left unpermeabilized after PFA fixation (PFA only). Surprisingly, in PFA+Tri–treated (Figure 4, A–C) or Met:Ac-treated cells (Supplemental Figure S3, A–C) there was a high degree of colocalization between the various forms of ERManI and GalT-YFP. This was also true in HeLa and HEK 293 cells (Supplemental Figure S4, A–E) and also using a different anti-ERManI antibody with an endogenous Golgi marker (Supplemental Figure S4C). The colocalization was not 100%, leaving visible amounts of nonoverlapping ERManI and GalT-YFP, especially in HEK 293 cells (Supplemental Figure S4, D and E). Nevertheless, in all cells that were fixed with 4% PFA but left unpermeabilized, ERManI-cherry was strikingly found in vesicles, as in the live microscopy experiments, distinct from the Golgi marker GalT-YFP (Figure 4D and Supplemental Figures S3G and S4, F and G). This suggests that the convergence of ERManI with the Golgi, as seen in PFA+Tri–treated cells, is not a biological process but rather a physical artifact that arises from membrane perturbation. In the absence of detergents or organic solvents (live-cell imaging, iodixanol gradients, or PFA treatment without detergent), ERManI does not localize to the Golgi. To investigate further the apparent ERManI colocalization with the Golgi apparatus when cells are fixed and permeabilized, we analyzed cells treated with brefeldin A (BFA). Under such treatment, both resident and itinerant Golgi membrane proteins are known to redistribute to the ER membrane (Doms et al., 1989). NIH 3T3 cells were transfected with GalT-YFP and either ERManI-HA (Figure 4F and Supplemental Figure S3E) or ERManI-cherry (Figure 4G and Supplemental Figure S3F) or left with endogenous levels of ERManI (Figure 4E and Supplemental Figure S3D) and were then fixed and permeabilized with either PFA+Tri or Met:Ac. After BFA treatment, ERManI remained in a vesicular pattern (Figure 4, E–G, and Supplemental Figure S3, D–F) even under fixation/permeabilization conditions that we had seen to cause artifactual ERManI colocalization with the Golgi marker GalT-YFP. As a control, we analyzed the localization of the Golgi-resident type II transmembrane enzyme Man1C. The high degree of homology between Man1C and ERManI makes this enzyme an excellent control. In untreated cells, Man1C-HA colocalized with the Golgi marker GalT-YFP (Figure 4H). This colocalization persisted in BFA-treated cells, showing both GalT-YFP and Man1C-HA in an ER pattern (Figure 4I). Schnell et al. (2012) recently reviewed various immunolabeling artifacts that arise from differences in cell fixation methods and the effect of these differences on antibody-epitope recognition. They stressed the importance of validating results gathered by immunofluorescence with live-cell microscopy. The localization of ERManI in live cells is consistent with that obtained by density gradient centrifugation. In contrast, ERManI localization in fixed and permeabilized cells and the results obtained from BFA treatment led us to believe that the permeabilization methods, which include lipid extraction, shift the localization of the QCVs, yielding untrustworthy results.
FIGURE 4:
ERManI localization is altered in fixed and permeabilized cells. (A–C) NIH 3T3 cells transfected with GalT-YFP only (A) or together with ERManI-HA (B) or ERManI-cherry (C) were fixed with 3% PFA and permeabilized with 0.5% Triton. They were then stained with mouse anti-HA (B) or mouse anti-ERManI (A; for endogenous ERManI), followed by goat anti-mouse Dylight 594 or left unstained (C). There is colocalization between all versions of ERManI and the Golgi marker. Bars, 10 μm. (D) When fixed with 3% PFA but left unpermeabilized, cells show ERManI-cherry in vesicles, distinct from GalT-YFP. (E–G) Cells treated with 5 μg/ml BFA for 1 h show an expected redistribution of the Golgi marker GalT-YFP to an ER pattern, whereas endogenous ERManI (E), ERManI-HA (F), and ERManI-cherry (G) are all in a vesicular pattern. Staining as in A–C. (H, I) Cells expressing GalT-YFP and the Golgi mannosidase Man1C-HA were either left untreated (H) or treated with BFA (I). They were then fixed as in A. Both proteins colocalize before (Golgi pattern) and after BFA treatment (ER pattern).
The mobility of ERManI-containing vesicles is dependent on microtubules and COP-II machinery
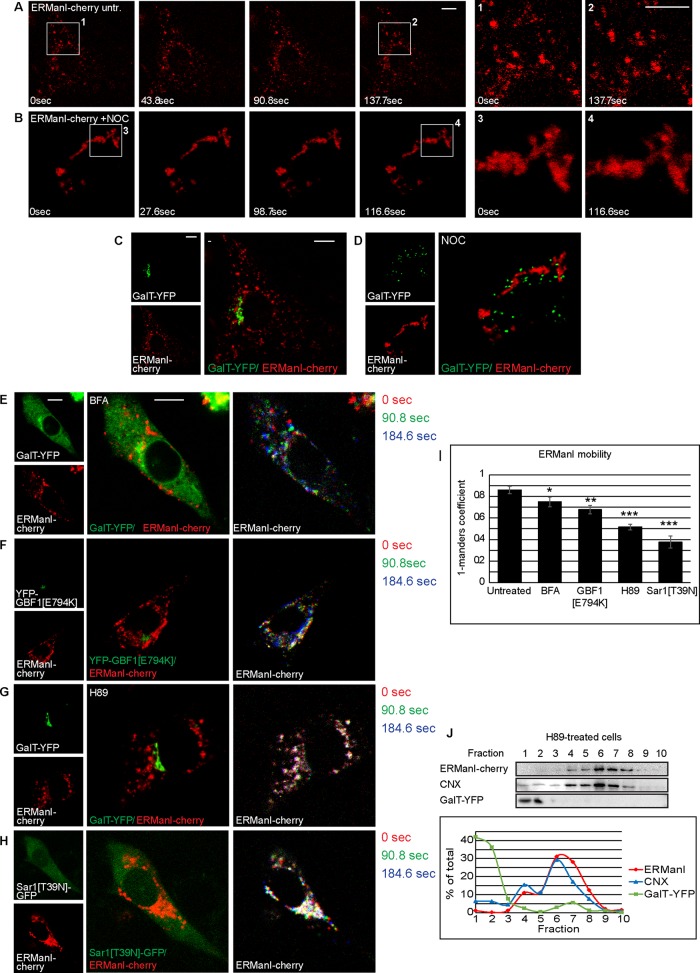
As we mentioned, in live, untreated cells, most QCVs display high levels of mobility, whereas others are less mobile, concentrating next to the nucleus. This becomes evident by time-lapse live-cell microscopy (Figure 5A and Supplemental Movie S4) and by comparing the first and last images of this time lapse (Figure 5A, 1 and 2). To analyze whether QCVs are transported by an active mechanism, we used known inhibitors of vesicle transport. In cells treated with the microtubule polymerization inhibitor nocodazole (Noc), not only was vesicle mobility arrested, but ERManI-cherry was observed in aggregated, immobile patches (Figure 5B, 3 and 4, and Supplemental Movie S5), not colocalizing with the Golgi marker GalT-YFP, which relocalized to characteristic Golgi ministacks under this treatment (Rogalski and Singer, 1984; Thyberg and Moskalewski, 1985; Figure 5, C and D). These results indicate that the observed mobility of QCVs is in fact an active, microtubule-dependent process. The requirement of microtubules for QCV mobility raised the possibility that these vesicles were in fact cargo vesicles, carrying ERManI to some final destination, rather than being distinct, functional vesicles. To investigate this point, we incubated cells with BFA, a known inhibitor of COP-I vesicular transport. Under BFA treatment, no change in vesicle dynamics or dispersion was observed, suggesting that the COP-I machinery does not play a role in the movement of these vesicles (Figure 5E, Supplemental Figure S5A, and Supplemental Movie S6). Quantitation of vesicle mobility was done by comparing two time points from the time-lapse experiment using the Manders coefficients (M2 in M1), showing only a small reduction in mobility (Figure 5I). A more specific examination of the involvement of COP-I machinery was afforded by the use of a dominant-negative mutant form of Golgi BFA-resistant guanine nucleotide exchange factor 1 (GBF1[E794K]) fused to enhanced YFP. GBF1 acts as the guanine exchange factor (GEF) for Arf1 (Claude et al., 1999), and overexpression of its dominant- negative form causes arrest in COP-I vesicle formation (Garcia-Mata et al., 2003). Expression of YFP-GBF1[E794K] had very little effect on the vesicular pattern or the mobility of ERManI-cherry (Figure 5, F and I, Supplemental Figure S5B, and Supplemental Movie S7), consistent with the results obtained using BFA, suggesting that COP-I machinery does not play any significant role in ERManI vesicle mobility. In contrast, in cells treated with the PKA inhibitor H89, an inhibitor of COPII vesicular transport (Aridor and Balch, 2000), the mobility of QCVs was significantly affected, despite maintaining a disperse pattern (Figure 5, G and I, Supplemental Figure S5C, and Supplemental Movie S8), suggesting a possible role for COP-II machinery in QCV mobility. This effect was not identical to that of nocodazole treatment, in which the immobile vesicles converged into patches around the nucleus (Figure 5D). Despite the change in vesicle dynamics under H89 treatment, no shift in vesicle density was observed, as ERManI was still found in fractions with an ER-like density in an iodixanol gradient assay (Figure 5J). Because H89 is a general protein kinase A inhibitor, a different approach was needed in order to directly implicate COP-II machinery in QCV mobility. To this end, we constructed a plasmid coding for a dominant-negative mutant form of the COP-II coat GTPase Sar1 in an IRES-GFP vector (Sar1[T39N]-IRES-GFP). This allowed us to confirm Sar1[T39N] expression in live-cell imaging without fusing a fluorescent protein to Sar1[T39N]. Cells expressing this dominant-negative form of Sar1 showed a large, immobile fraction of QCVs next to the nucleus, whereas vesicles in the periphery retained their mobility (Figure 5, H and I, Supplemental Figure S5D, and Supplemental Movie S9), suggesting that two distinct populations of QCVs exist, only one of them requiring COP-II for their mobility. The complete lack of mobility under nocodazole treatment suggests that both populations require microtubules for vesicle movement.
FIGURE 5:
QCV movement is dependent on microtubules and COPII. (A, B) Time-lapse images of ERManI-cherry in untreated (A) and Noc-treated (6 μg/ml for 2 h) (B) cells; regions of interest were enlarged at two chosen time points to show QCV movement in untreated cells (A, 1 and 2) in contrast to movement arrest and a patchy appearance in a Noc-treated cell (B, 3 and 4). Bars, 10 μm. (C, D) Golgi localization in the cells in A as indicated by GalT-YFP (C) and Golgi disruption by Noc (D). ERManI-cherry and GalT-YFP do not colocalize either before or after treatment. (E–H) Live-cell imaging of NIH 3T3 cells expressing ERManI-cherry and GalT-YFP. Three time points were chosen, pseudocolored, and overlaid to illustrate vesicle movement. Interference with COPI vesicle trafficking does not affect QCV vesicle movement, as seen by BFA treatment (5 μg/ml for 1 h; E) or coexpression with dominant-negative GBF1 (F). Interference with COPII vesicle trafficking blocks QCV movement, as observed with H89 treatment (50 μM for 1 h; G) and with coexpression of dominant-negative Sar1 (H). (I) Quantitation of ERManI-cherry mobility under the various treatments using the Manders coefficient, comparing ERManI after 90 s to initial ERManI. We show the 1 − Manders coefficient, with lower values indicating less mobility. p(BFA) = 0.041, p(GBF1[E794K]) = 0.005, p((H89)) = 0.00037, and p(Sar1[T39N]) = 0.00012. (J) Iodixanol equilibrium centrifugation gradient of HEK 293 cells treated with H89 shows that despite arrest in vesicle movement, ERManI remains in fractions of ER-like density.
ERManI interacts with an ERAD substrate in vesicles that converge to form the ERQC
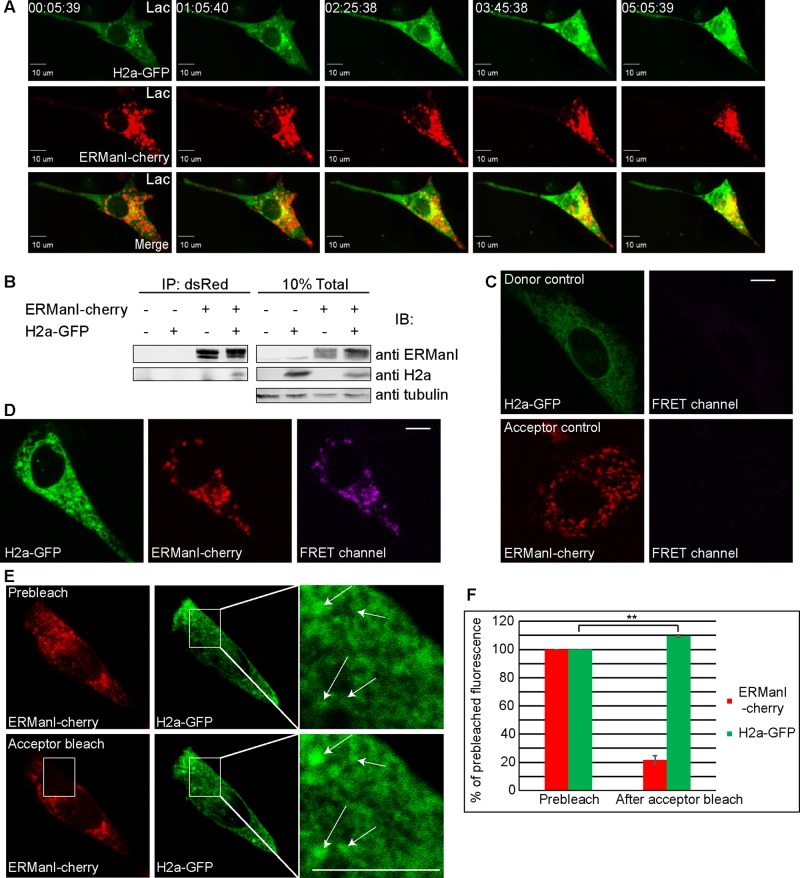
We saw that ERManI appears mainly in a vesicular pattern in untreated cells, in contrast to its accumulation together with its substrates in the juxtanuclear ERQC when proteasomes are inhibited (Figure 3, A and B). Therefore we analyzed the changes that take place with time upon proteasomal inhibition. During a 5-h time lapse under proteasomal inhibition, QCVs converged from the cell periphery to the juxtanuclear region. Concomitantly, H2a-GFP accumulated in the juxtanuclear region at the ERQC, as seen before, where it colocalized with ERManI (Figure 6A and Supplemental Movie S10). Before the concentration at the ERQC, we could observe transient colocalization of ERManI and the substrate in punctate regions.
FIGURE 6:
QCVs are the site of ERManI interaction with its substrate and upon substrate accumulation converge at the juxtanuclear ERQC. (A) Live-cell imaging of an NIH 3T3 cell expressing ERManI-cherry and H2a-GFP showing both proteins accumulating and colocalizing next to the nucleus over time of treatment with a proteasomal inhibitor (17 μM Lac). Time is indicated (hours:minutes:seconds). (B) Association of H2a and ERManI fused to fluorescent proteins, as seen by immunoprecipitation of H2a-GFP with ERManI-cherry with anti-dsRed, followed by immunoblotting with the indicated antibodies, in HEK 293 cells (left). Right, a fraction of the total lysate. (C, D) The interaction between ERManI-cherry and H2a-GFP takes place in vesicles as shown by direct FRET in live NIH 3T3 cells. Donor and acceptor controls are H2a-GFP or ERManI-cherry expressed individually; note little to no emission in control FRET channels (C). When the fluorescent fusion proteins were coexpressed, direct FRET was observed (D; FRET channel) and coincided with most but not all ERManI-cherry–containing vesicles. Bars, 10 μm. (E, F) Acceptor bleach FRET of 3% PFA–fixed NIH 3T3 cells. Acceptor bleach was done using a diode-pumped solid-state laser at 561-nm wavelength and induced an increase in donor fluorescence. Regions of interest were enlarged; arrows indicate donor fluorescence increase in a vesicular pattern (E). Quantification of three acceptor bleach FRET experiments showed an average increase of 9% in donor fluorescence with p = 0.0097 (F).
We wondered whether we could determine the localization of the interaction between ERManI-cherry and H2a-GFP and whether it takes place before their convergence at the ERQC. First we had to determine that fusion to the fluorescent proteins does not hinder interaction of the enzyme with its substrate. H2a-GFP coimmunoprecipitated with ERManI-cherry (Figure 6B), indicating that ERManI and its substrate interact even when both are fused to fluorophores. It is interesting to note the lower levels of total H2a-GFP in cells that also express ERManI-cherry, due to the acceleration of ERAD in cells overexpressing ERManI. To localize this interaction in cells, we implemented direct and indirect Förster resonance energy transfer (FRET) assays. NIH 3T3 cells expressing ERManI-cherry, H2a-GFP, or both were imaged live. Donor (H2a-GFP) and acceptor (ERManI-cherry) controls showed little to no spectral bleedthrough from these fluorophores to the FRET channel (Figure 6C). In cells transfected with both H2a-GFP and ERManI-cherry, FRET is visible in a pattern resembling that of ERManI-cherry QCVs (Figure 6D). These results suggest that ERManI-H2a interaction occurs in these vesicles and not in the ER. A second approach to localize the ERManI-H2a interaction is indirect FRET, in which the increase in donor fluorescence is measured before and after acceptor bleach in fixed cells. In this experiment, H2a-GFP fluorescence levels were measured before and after photobleaching of ERManI-cherry (Figure 6E). The bleached area (white rectangle) was enlarged and white arrows overlaid to show localization of FRET before and after acceptor bleach. Because the indirect FRET was done on fixed (but not permeabilized) cells, this approach avoids possible ambiguity due to vesicle movement during the course of the experiment. Quantitation of fluorescence levels indicated that under conditions of acceptor bleach (22% of prebleach remaining), donor fluorescence increased by an average of 9%, typical for FRET, indicating interaction between ERManI-cherry and H2a-GFP (Figure 6F). Under examination of postbleach H2a-GFP, it is evident that the increase in fluorescence occurs in a vesicular pattern (Figure 6E, arrows in inset), strengthening the results obtained in live cells by direct FRET.
Taken together, the results suggest that ERManI resides in COPII-dependent vesicles, where it interacts with its substrates, concentrating in the ERQC upon accumulation of the misfolded glycoproteins.
DISCUSSION
ERManI was proposed to function as an ERAD timer in the quality control of glycoproteins (Helenius and Aebi, 2004). The relatively high concentration of ERManI in an ER-derived compartment, the ERQC (Avezov et al., 2008), suggested that by itself this enzyme could be responsible for the extensive trimming required to target a misfolded glycoprotein to ERAD (Frenkel et al., 2003; Avezov et al., 2008). Most glycoproteins are retained in the ER and not delivered to the Golgi until properly folded. If they cannot attain correct folding, their N-glycans are progressively trimmed until the Man5-6GlcNAc2 structures are formed, which cannot be reglucosylated and therefore cannot reenter the CNX folding cycle (Frenkel et al., 2003). The Man5-6GlcNAc2 structures are recognized by the OS-9 and XTP3-B lectins that trap the misfolded glycoproteins in the ERQC and deliver them to ERAD (Groisman et al., 2011; Leitman et al., 2014). The importance of the compartmentalization and exclusion from the Golgi is apparent when considering the Man5-6GlcNAc2 N-glycan structures, which are formed in the ER/ERQC by extensive mannose trimming (Frenkel et al., 2003; Avezov et al., 2008). The same oligosaccharides are formed in the Golgi apparatus, by Golgi mannosidases, as part of a glycoprotein's normal maturation pathway (Lal et al., 1994; Moremen et al., 1994). Premature delivery of an incompletely folded glycoprotein to the Golgi and trimming of its N-glycans to Man5-6GlcNAc2 at this location would release it from the grip of the ER-Golgi lectins ERGIC53, VIPL, and VIP36 and allow its untimely trafficking through the secretory pathway (Kamiya and Kato, 2008; Lederkremer, 2009).
Here we deepened our understanding of the pre-Golgi events leading to the compartmentalization and interaction of newly made glycoproteins with ERManI. ERManI is localized at the steady state mainly in dynamic vesicles with ER-like density. These distinct vesicles are divided into at least two subpopulations, one of which engages with newly made glycoproteins, suggesting its role in quality control and targeting to ERAD, whereas the other subpopulation is involved in the turnover of ERManI by autophagy or autophagy-like mechanisms, as suggested for other short-lived ERAD components (Bernasconi and Molinari, 2011; Merulla et al., 2013). We named the first subpopulation quality control vesicles. A proteomics study of rat liver (Gilchrist et al., 2006), reevaluated later (Smirle et al., 2013), identified most of ERManI in a vesicular fraction containing COPI vesicles, which might have included other, then-uncharacterized, vesicles as well (QCVs). We determined that it is in the QCVs that ERManI interacts transiently with its glycoprotein substrates. These brief interactions of compartmentalized ERManI with the substrates would lead to a slow progressive trimming of their N-glycans, giving time for them to fold properly and escape to the Golgi. Recently it was reported that ERManI is able to trim glucosylated N-glycans (Aikawa et al., 2014). After this trimming, the glycans become less sensitive to glucosidase II (Stigliano et al., 2011), and they would then remain in the CNX cycle. Therefore initial exposure to ERManI would allow for more time in the folding process.
ERManI-containing vesicles colocalize partially with Rab1, suggesting the presence of transport machinery in these vesicles. Such machinery may serve in the cycling of ERAD substrates between the ER and QCVs. Alternatively, Rab1 may be involved in the delivery of ERManI to autophagosomes, the formation of which was reported to be Rab1 dependent (Zoppino et al., 2010). Of interest, we also show that the mobility of QCVs is dependent on microtubules and COPII machinery. Whereas the requirement for functional microtubules for QCV mobility is understandable, the COPII machinery dependence requires further examination. These vesicles could be a small minority of COPII vesicles, even though Sec13-GFP does not seem to colocalize with the QCVs (Figure 2). Alternatively, it is possible that QCVs are not COPII coated but that, under inhibition of COPII vesicle formation, necessary proteins such as motor protein adaptors do not reach the QCVs, causing their arrest. This issue and higher-resolution electron microscopy imaging of the vesicles should be addressed in future studies.
The stark differences in ERManI localization and pattern when analyzed in density gradients or viewed in live cells as opposed to fixed, permeabilized cells are remarkable. The artifactual localization of ERManI in fixed, permeabilized cells seems to reflect an unusual physical artifact. This can be concluded from the fact that PFA fixation does not cause ERManI shift in localization, but subsequent permeabilization with Triton does. At the point in which Triton is introduced, the cells have already been fixed and do not possess active transport pathways by which ERManI localization could be altered. The shift in localization suggests a lack of strong ERManI tethering in the natural localization, the QCVs, and a high affinity of ERManI for Golgi-localized proteins in the absence of membrane lipids.
Under proteasomal inhibition, the QCVs converge from the cell periphery along with ERAD substrate glycoproteins to merge at the juxtanuclear ERQC. This concentration might possibly enhance the mannose trimming of the ERAD substrates when they accumulate, namely under ER stress, enhancing delivery to ERAD. It is noteworthy that in some cell types, such as HEK 293 and HeLa, ERManI is mainly concentrated at the juxtanuclear ERQC also at the steady state in untreated cells. This and other lines of evidence suggest that the ERQC exists under steady-state, unstressed conditions. OS-9 resides constitutively in this compartment (Ron et al., 2011; Leitman et al., 2014). We recently showed that the mechanism for recruitment of ERAD substrates and of other quality control and ERAD components at the ERQC involves activation of the PERK branch of the UPR and phosphorylation of its target eIF2α, which leads to the transcriptional up-regulation of Herp, a protein that is also relocalized to the ERQC (Leitman et al., 2014). Herp then initiates assembly of an ERAD complex by association with HRD1. Overexpression of a phosphomimetic mutant of eIF2α (S51D; Kondratyev et al., 2007), inhibition of eIF2α-P dephosphorylation, or overexpression of Herp (Leitman et al., 2014) is sufficient to allow observation of protein accumulation at the ERQC. This suggests that even in unstressed conditions, transient accumulation of unfolded protein molecules probably also causes temporary activation of the PERK pathway and induction of Herp, leading to dynamic protein recruitment to the ERQC. We can envision a model in which ERAD substrate glycoproteins would be targeted to the ERQC for ERAD after trimming by ERManI in QCVs. Under high protein load, ERManI would be directly delivered to the ERQC, accelerating glycoprotein trimming and delivery to ERAD.
MATERIALS AND METHODS
Materials
Lactacystin (Lac) was from Calbiochem (La Jolla, CA). Protein A Sepharose was from Repligen (Waltham, MA), MG-132. BFA, H89, nocodazole (Noc), leupeptin, 3-methyladenine (3MA), chloroquine, and other common reagents were from Sigma-Aldrich (St. Louis, MO). HindIII, XhoI, and XmaI restriction enzymes were from NEB (Ipswich, MA).
Plasmids and constructs
ERManI cDNA was subcloned into mCherry N1, using the primers CGCGCGTAATACGATCACTATAG and CATGCACCCGGGCTGATCCTCCACCTCCAGACATGGCAGGGGTCCAGATAGGC with HindIII and XmaI restriction sites and with a SGGGGS flexible linker. GalT-YFP (first 60 amino acids of β1,3-galactosyltransferase linked to YFP in pEYFP) was used in Avezov et al. (2008). LC3-GFP was used before (Kondratyev et al., 2007). Man1C-HA in PMH (Roche Diagnostics, Basel, Switzerland) was a kind gift of A. Herscovics (McGill University, Montreal, Canada). H2a-GFP was previously described (Leitman et al., 2013), KDELR-GFP, ε-COP-GFP, and Sec13-GFP were kind gifts of Tom Kirchhausen (Harvard Children's Hospital, Boston, MA), ERGIC53-GFP was a kind gift of H. Ben-Tekaya (University of Basel, Basel, Switzerland), GFP-Rab1a, GFP-Rab1b, and GFP-Rab2 were kind gifts of Eran Bachrach (Tel Aviv University, Tel Aviv, Israel), and YFP-GBF1[E749K] and Sar1[T39N] were kind gifts of J. Bonifacino (National Institutes of Health, Bethesda, MD). Sar1[T39N] cDNA was subcloned into IRES-GFP (Clontech) using the primers CTCGAGATGTCTTTCATCTTT and CTCGAGTCAGTCAATATACT with XhoI restriction sites.
Antibodies
Mouse anti-HA tag, anti–β-COP, and anti-tubulin were from Sigma-Aldrich, and anti-GFP was from Santa Cruz Biotechnology (Dallas, TX). Rabbit anti–GM-130 was from Abcam (Cambridge, England). Mouse anti-human ERManI was a kind gift from Richard Sifers (Baylor College, Houston, TX) (used only for human cells) or was from Santa Cruz Biotechnology (used for mouse cells). Rabbit anti-calnexin was from Sigma-Aldrich, and anti-dsRED was from MBL. Goat anti-mouse immunoglobulin G (IgG) linked to horseradish peroxidase (HRP) and goat anti-rabbit IgG-HRP were from Jackson Labs (West Grove, PA). Goat anti-mouse Dylight 594 and goat anti-rabbit Dylight 488 were from Thermo Scientific (Barrington, IL). Rabbit polyclonal anti-H2 N-terminus was previously described (Tolchinsky et al., 1996), Goat anti-mouse IgG linked to agarose beads was from Sigma-Aldrich. Rabbit anti-Cab45 was described before (Scherer et al., 1996).
Cell culture, media, and transfections
NIH 3T3, HEK 293, and HeLa cells were grown in DMEM supplemented with 10% bovine calf serum at 37°C under 5% CO2. Transfections were carried out using a Neon MP-100 microporator system (Life Technologies, Carlsbad, CA) for NIH 3T3 (1400 V, 20 ms, one pulse), HEK 293 (1150 V, 20 ms, two pulses), and HeLa (1005 V, 35 ms, two pulses) cells. Transfections for 293 cells for gradient centrifugation were carried out according to the calcium phosphate method.
Gradient fractionation
Cells were scraped and rinsed. For endogenous ERManI detection, ∼2.5-fold cells were used compared with samples with exogenous expression. After Dounce cell homogenization in an iso-osmotic buffer (10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.4, and 250 mM sucrose), debris and nuclei were pelleted at 1000 × g at 4°C for 10 min, and the supernatants were loaded on top of an iodixanol gradient (10–34%). Iodixanol solutions were prepared in 60 mM HEPES, pH 7.4, and 250 mM sucrose iso-osmotic buffer. The gradients were ultracentrifuged at 24,000 rpm (98,500 × g, Beckman SW41 rotor) for 16 h at 4°C. Eleven fractions were collected from gradient top to bottom and subjected to immunoblotting. Density was calculated from the refractive index of the fractions.
Treatments, coimmunoprecipitation, and immunoblotting
Cells were treated with 5 μg/ml BFA or 50 μM H89 for 1 h or 6 μg/ml Noc, 17 μM Lac, or 40 μM MG-132 for the stated times. Treatment with 100 μM chloroquine was for 24 h and with 5 mM 3MA for 24 h. SDS–PAGE and detection by enhanced chemiluminescence were done as previously described (Groisman et al., 2011). For coimmunoprecipitation, cell lysis was done in 1% NP-40, 50 mM Tris (Indianapolis, IN)/HCl (pH 8), 150 mM NaCl, and protease inhibitor cocktail (Roche) for 30 min on ice. After centrifugation at 25,000 × g in 4°C for 30 min, lysates were immunoprecipitated overnight with rabbit anti dsRED using protein A–Sepharose beads. After three washes with lysis buffer diluted 1:5, bound proteins were eluted with sample buffer at 100°C for 5 min and immunoblotted.
Fluorescence microscopy
For fixation purposes, cells were transfected and grown for 24 h on glass coverslips. Cells were then fixed with 3% paraformaldehyde and either permeabilized with 0.5% Triton or left unpermeabilized. Another fixation method used is methanol:acetone, as described in Kamhi-Nesher et al. (2001). For live-cell microscopy, cells were transfected and grown for 24 h on glass-bottom 35-mm plates (Greiner Bio One, Vilvoorde, Belgium) and treated as stated. Images and time-lapse videos were captured using a Zeiss 510 Meta laser scanning microscope. Quantitations were carried out using ImageJ software and JACoP plug-in (Bolte and Cordelieres, 2006). For acceptor bleach FRET analysis, ERManI-cherry was bleached using a diode-pumped solid-state 561-nm laser at full output for 50 iterations. Long-duration live-cell imaging was done using a spinning disk confocal microscope (CSU-22 Confocal Head, Yokogawa; Axiovert 200M; Carl Zeiss MicroImaging, Jena, Germany) under control of SlideBook (Intelligent Imaging Innovations, Göttingen, Germany) with 63× or 100× oil immersion objectives (Plan Apochromat, numerical aperture 1.4) and an Evolve electron-multiplying charge-coupled device camera (Photometrics, Tucson, AZ). Samples were illuminated with 473- or 561-nm 40-mW solid-state lasers. Typical exposure times were 0.1–0.5 s.
Supplementary Material
Acknowledgments
We are grateful to J. Bonifacino, T. Kirchhausen, H. Ben-Tekaya, E. Bachrach, R. Sifers, and A. Herscovics for reagents and A. Hizi for help with density determination. This work was supported by grants from the Israel Science Foundation (1070/10) and German–Israeli Project Cooperation (Deutsch-Israelische Projektkooperation K 5-1).
Abbreviations used:
- 3MA
3-methyladenine
- BFA
brefeldin A
- CNX
calnexin
- CRT
calreticulin
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum–associated degradation
- ERManI
ER α1,2 mannosidase I
- ERQC
ER-derived quality control compartment
- FRET
Förster resonance energy transfer
- Lac
lactacystin
- LC3
microtubule-associated protein1 light chain 3
- Noc
nocodazole
- PFA
paraformaldehyde
- QCV
quality control vesicle
- Rab
Ras-related protein in brain
- UGGT
UDPGlc:glycoprotein glycosyl transferase.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-06-1152) on November 19, 2014.
REFERENCES
- Aebi M, Bernasconi R, Clerc S, Molinari M. N-glycan structures: recognition and processing in the ER. Trends Biochem Sci. 2010;35:74–82. doi: 10.1016/j.tibs.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Aikawa J, Takeda Y, Matsuo I, Ito Y. Trimming of glucosylated N-glycans by human ER alpha1,2-mannosidase I. J Biochem. 2014;155:375–384. doi: 10.1093/jb/mvu008. [DOI] [PubMed] [Google Scholar]
- Allan BB, Moyer BD, Balch WE. Rab1 recruitment of p115 into a cis-SNARE complex: programming budding COPII vesicles for fusion. Science. 2000;289:444–448. doi: 10.1126/science.289.5478.444. [DOI] [PubMed] [Google Scholar]
- Aridor M, Balch WE. Kinase signaling initiates coat complex II (COPII) recruitment and export from the mammalian endoplasmic reticulum. J Biol Chem. 2000;275:35673–35676. doi: 10.1074/jbc.C000449200. [DOI] [PubMed] [Google Scholar]
- Avezov E, Frenkel Z, Ehrlich M, Herscovics A, Lederkremer GZ. Endoplasmic reticulum (ER) mannosidase I is compartmentalized and required for N-glycan trimming to Man5-6GlcNAc2 in glycoprotein ER-associated degradation. Mol Biol Cell. 2008;19:216–225. doi: 10.1091/mbc.E07-05-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyair R, Ron E, Lederkremer GZ. Protein quality control, retention, and degradation at the endoplasmic reticulum. Int Rev Cell Mol Biol. 2011;292:197–280. doi: 10.1016/B978-0-12-386033-0.00005-0. [DOI] [PubMed] [Google Scholar]
- Bernasconi R, Molinari M. ERAD and ERAD tuning: disposal of cargo and of ERAD regulators from the mammalian ER. Curr Opin Cell Biol. 2011;23:176–183. doi: 10.1016/j.ceb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Burda P, Aebi M. The ALG10 locus of Saccharomyces cerevisiae encodes the alpha-1,2 glucosyltransferase of the endoplasmic reticulum: the terminal glucose of the lipid-linked oligosaccharide is required for efficient N-linked glycosylation. Glycobiology. 1998;8:455–462. doi: 10.1093/glycob/8.5.455. [DOI] [PubMed] [Google Scholar]
- Burke J, Lipari F, Igdoura S, Herscovics A. The Saccharomyces cerevisiae processing alpha 1,2-mannosidase is localized in the endoplasmic reticulum, independently of known retrieval motifs. Eur J Cell Biol. 1996;70:298–305. [PubMed] [Google Scholar]
- Camirand A, Heysen A, Grondin B, Herscovics A. Glycoprotein biosynthesis in Saccharomyces cerevisiae. Isolation and characterization of the gene encoding a specific processing alpha-mannosidase. J Biol Chem. 1991;266:15120–15127. [PubMed] [Google Scholar]
- Caramelo JJ, Parodi AJ. Getting in and out from calnexin/calreticulin cycles. J Biol Chem. 2008;283:10221–10225. doi: 10.1074/jbc.R700048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claude A, Zhao BP, Kuziemsky CE, Dahan S, Berger SJ, Yan JP, Armold AD, Sullivan EM, Melancon P. GBF1: A novel Golgi-associated BFA-resistant guanine nucleotide exchange factor that displays specificity for ADP-ribosylation factor 5. J Cell Biol. 1999;146:71–84. [PMC free article] [PubMed] [Google Scholar]
- Doms RW, Russ G, Yewdell JW. Brefeldin A redistributes resident and itinerant Golgi proteins to the endoplasmic reticulum. J Cell Biol. 1989;109:61–72. doi: 10.1083/jcb.109.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echard A, Jollivet F, Martinez O, Lacapere JJ, Rousselet A, Janoueix-Lerosey I, Goud B. Interaction of a Golgi-associated kinesin-like protein with Rab6. Science. 1998;279:580–585. doi: 10.1126/science.279.5350.580. [DOI] [PubMed] [Google Scholar]
- Esmon B, Esmon PC, Schekman R. Early steps in processing of yeast glycoproteins. J Biol Chem. 1984;259:10322–10327. [PubMed] [Google Scholar]
- Frenkel Z, Gregory W, Kornfeld S, Lederkremer GZ. Endoplasmic reticulum-associated degradation of mammalian glycoproteins involves sugar chain trimming to Man6-5GlcNAc2. J Biol Chem. 2003;278:34119–34124. doi: 10.1074/jbc.M305929200. [DOI] [PubMed] [Google Scholar]
- Garcia-Mata R, Szul T, Alvarez C, Sztul E. ADP-ribosylation factor/COPI-dependent events at the endoplasmic reticulum-Golgi interface are regulated by the guanine nucleotide exchange factor GBF1. Mol Biol Cell. 2003;14:2250–2261. doi: 10.1091/mbc.E02-11-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist A, Au CE, Hiding J, Bell AW, Fernandez-Rodriguez J, Lesimple S, Nagaya H, Roy L, Gosline SJ, Hallett M. Quantitative proteomics analysis of the secretory pathway. Cell. 2006;127:1265–1281. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- Gonzalez DS, Karaveg K, Vandersall-Nairn AS, Lal A, Moremen KW. Identification, expression, characterization of a cDNA encoding human endoplasmic reticulum mannosidase I, the enzyme that catalyzes the first mannose trimming step in mammalian Asn-linked oligosaccharide biosynthesis. J Biol Chem. 1999;274:21375–21386. doi: 10.1074/jbc.274.30.21375. [DOI] [PubMed] [Google Scholar]
- Groisman B, Shenkman M, Ron E, Lederkremer GZ. Mannose trimming is required for delivery of a glycoprotein from EDEM1 to XTP3-B and to late endoplasmic reticulum-associated degradation steps. J Biol Chem. 2011;286:1292–1300. doi: 10.1074/jbc.M110.154849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin B, Herscovics A. Topology of ER processing alpha-mannosidase of Saccharomyces cerevisiae. Glycobiology. 1992;2:369–372. doi: 10.1093/glycob/2.4.369. [DOI] [PubMed] [Google Scholar]
- Hammond C, Helenius A. Folding of VSV G protein: sequential interaction with BiP and calnexin. Science. 1994;266:456–458. doi: 10.1126/science.7939687. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Bernasconi R, Molinari M. ERAD substrates: which way out. Semin Cell Dev Biol. 2010;21:526–532. doi: 10.1016/j.semcdb.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Hebert DN, Zhang JX, Chen W, Foellmer B, Helenius A. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J Cell Biol. 1997;139:613–623. doi: 10.1083/jcb.139.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Herscovics A, Romero PA, Tremblay LO. The specificity of the yeast and human class I ER alpha 1,2-mannosidases involved in ER quality control is not as strict previously reported. Glycobiology. 2002;12:14G–15G. [PubMed] [Google Scholar]
- Hosokawa N, Tremblay LO, You Z, Herscovics A, Wada I, Nagata K. Enhancement of endoplasmic reticulum (ER) degradation of misfolded Null Hong Kong alpha1-antitrypsin by human ER mannosidase I. J Biol Chem. 2003;278:26287–26294. doi: 10.1074/jbc.M303395200. [DOI] [PubMed] [Google Scholar]
- Izawa T, Nagai H, Endo T, Nishikawa S. Yos9p and Hrd1p mediate ER retention of misfolded proteins for ER-associated degradation. Mol Biol Cell. 2012;23:1283–1293. doi: 10.1091/mbc.E11-08-0722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamhi-Nesher S, Shenkman M, Tolchinsky S, Fromm SV, Ehrlich R, Lederkremer GZ. A novel quality control compartment derived from the endoplasmic reticulum. Mol Biol Cell. 2001;12:1711–1723. doi: 10.1091/mbc.12.6.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya Y, Kato K. Structural basis for trafficking and quality control of glycoproteins by intracellular lectins [in Japanese]. Tanpakushitsu Kakusan Koso. 2008;53(12 Suppl):1662–1669. [PubMed] [Google Scholar]
- Kondratyev M, Avezov E, Shenkman M, Groisman B, Lederkremer GZ. PERK-dependent compartmentalization of ERAD and unfolded protein response machineries during ER stress. Exp Cell Res. 2007;313:3395–3407. doi: 10.1016/j.yexcr.2007.07.006. [DOI] [PubMed] [Google Scholar]
- Lal A, Schutzbach JS, Forsee WT, Neame PJ, Moremen KW. Isolation and expression of murine and rabbit cDNAs encoding an alpha 1,2-mannosidase involved in the processing of asparagine-linked oligosaccharides. J Biol Chem. 1994;269:9872–9881. [PubMed] [Google Scholar]
- Lederkremer GZ. Glycoprotein folding, quality control and ER-associated degradation. Curr Opin Struct Biol. 2009;19:515–523. doi: 10.1016/j.sbi.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Leitman J, Shenkman M, Gofman Y, Shtern NO, Ben-Tal N, Hendershot LM, Lederkremer GZ. Herp coordinates compartmentalization and recruitment of HRD1 and misfolded proteins for ERAD. Mol Biol Cell. 2014;25:1050–1060. doi: 10.1091/mbc.E13-06-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitman J, Ulrich Hartl F, Lederkremer GZ. Soluble forms of polyQ-expanded huntingtin rather than large aggregates cause endoplasmic reticulum stress. Nat Commun. 2013;4:2753. doi: 10.1038/ncomms3753. [DOI] [PubMed] [Google Scholar]
- Massaad MJ, Franzusoff A, Herscovics A. The processing alpha1,2-mannosidase of Saccharomyces cerevisiae depends on Rer1p for its localization in the endoplasmic reticulum. Eur J Cell Biol. 1999;78:435–440. doi: 10.1016/s0171-9335(99)80070-3. [DOI] [PubMed] [Google Scholar]
- Merulla J, Fasana E, Solda T, Molinari M. Specificity and regulation of the endoplasmic reticulum-associated degradation machinery. Traffic. 2013;14:767–777. doi: 10.1111/tra.12068. [DOI] [PubMed] [Google Scholar]
- Moremen KW, Trimble RB, Herscovics A. Glycosidases of the asparagine-linked oligosaccharide processing pathway. Glycobiology. 1994;4:113–125. doi: 10.1093/glycob/4.2.113. [DOI] [PubMed] [Google Scholar]
- Nielsen E, Severin F, Backer JM, Hyman AA, Zerial M. Rab5 regulates motility of early endosomes on microtubules. Nat Cell Biol. 1999;1:376–382. doi: 10.1038/14075. [DOI] [PubMed] [Google Scholar]
- Nuoffer C, Davidson HW, Matteson J, Meinkoth J, Balch WE. A GDP-bound of rab1 inhibits protein export from the endoplasmic reticulum and transport between Golgi compartments. J Cell Biol. 1994;125:225–237. doi: 10.1083/jcb.125.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S, Cheng X, Sifers RN. Golgi-situated endoplasmic reticulum alpha-1, 2-mannosidase contributes to the retrieval of ERAD substrates through a direct interaction with gamma-COP. Mol Biol Cell. 2013;24:1111–1121. doi: 10.1091/mbc.E12-12-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S, Wang S, Utama B, Huang L, Blok N, Estes MK, Moremen KW, Sifers RN. Golgi localization of ERManI defines spatial separation of the mammalian glycoprotein quality control system. Mol Biol Cell. 2011;22:2810–2822. doi: 10.1091/mbc.E11-02-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parodi AJ. Protein glucosylation and its role in protein folding. Annu Rev Biochem. 2000;69:69–93. doi: 10.1146/annurev.biochem.69.1.69. [DOI] [PubMed] [Google Scholar]
- Rogalski AA, Singer SJ. Associations of elements of the Golgi apparatus with microtubules. J Cell Biol. 1984;99:1092–1100. doi: 10.1083/jcb.99.3.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron E, Shenkman M, Groisman B, Izenshtein Y, Leitman J, Lederkremer GZ. Bypass of glycan-dependent glycoprotein delivery to ERAD by up-regulated EDEM1. Mol Biol Cell. 2011;22:3945–3954. doi: 10.1091/mbc.E10-12-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer PE, Lederkremer GZ, Williams S, Fogliano M, Baldini G, Lodish HF. Cab45, a novel (Ca2+)-binding protein localized to the Golgi lumen. J Cell Biol. 1996;133:257–268. doi: 10.1083/jcb.133.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell U, Dijk F, Sjollema KA, Giepmans BN. Immunolabeling artifacts and the need for live-cell imaging. Nat Methods. 2012;9:152–158. doi: 10.1038/nmeth.1855. [DOI] [PubMed] [Google Scholar]
- Shenkman M, Ayalon M, Lederkremer GZ. Endoplasmic reticulum quality control of asialoglycoprotein receptor H2a involves a determinant for retention and not retrieval. Proc Natl Acad Sci USA. 1997;94:11363–11368. doi: 10.1073/pnas.94.21.11363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Chaudhry P, Parent S, Asselin E. Ubiquitin-proteasomal degradation of COX-2 in TGF-beta stimulated human endometrial cells is mediated through endoplasmic reticulum mannosidase I. Endocrinology. 2012;153:426–437. doi: 10.1210/en.2011-1438. [DOI] [PubMed] [Google Scholar]
- Smirle J, Au CE, Jain M, Dejgaard K, Nilsson T, Bergeron J. Cell biology of the endoplasmic reticulum and the Golgi apparatus through proteomics. Cold Spring Harb Perspect Biol. 2013;5:a015073. doi: 10.1101/cshperspect.a015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro RG. Glucose residues as key determinants in the biosynthesis and quality control of glycoproteins with N-linked oligosaccharides. J Biol Chem. 2000;275:35657–35660. doi: 10.1074/jbc.R000022200. [DOI] [PubMed] [Google Scholar]
- Stigliano ID, Alculumbre SG, Labriola CA, Parodi AJ, D’Alessio C. Glucosidase II and N-glycan mannose content regulate the half-lives of monoglucosylated species in vivo. Mol Biol Cell. 2011;22:1810–1823. doi: 10.1091/mbc.E11-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Ueno T, Kominami E. LC3 and Autophagy. Methods Mol Biol. 2008;445:77–88. doi: 10.1007/978-1-59745-157-4_4. [DOI] [PubMed] [Google Scholar]
- Thyberg J, Moskalewski S. Microtubules and the organization of the Golgi complex. Exp Cell Res. 1985;159:1–16. doi: 10.1016/s0014-4827(85)80032-x. [DOI] [PubMed] [Google Scholar]
- Tolchinsky S, Yuk MH, Ayalon M, Lodish HF, Lederkremer GZ. Membrane-bound versus secreted forms of human asialoglycoprotein receptor subunits. Role of a juxtamembrane pentapeptide. J Biol Chem. 1996;271:14496–14503. doi: 10.1074/jbc.271.24.14496. [DOI] [PubMed] [Google Scholar]
- Tremblay LO, Herscovics A. Cloning and expression of a specific human alpha 1,2-mannosidase that trims Man9GlcNAc2 to Man8GlcNAc2 isomer B during N-glycan biosynthesis. Glycobiology. 1999;9:1073–1078. doi: 10.1093/glycob/9.10.1073. [DOI] [PubMed] [Google Scholar]
- Wu Y, Termine DJ, Swulius MT, Moremen KW, Sifers RN. Human endoplasmic reticulum mannosidase I is subject to regulated proteolysis. J Biol Chem. 2007;282:4841–4849. doi: 10.1074/jbc.M607156200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- Zoppino FC, Militello RD, Slavin I, Alvarez C, Colombo MI. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic. 2010;11:1246–1261. doi: 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.