Although the protein components at the kinetochore have been largely identified, there are limited data on how these proteins assemble into a higher-order structure. Biochemical analysis of the nematode kinetochore protein KNL-1 reveals that it oligomerizes in a structurally specific way, such that it could contribute to kinetochore organization.
Abstract
The kinetochore is a large, macromolecular assembly that is essential for connecting chromosomes to microtubules during mitosis. Despite the recent identification of multiple kinetochore components, the nature and organization of the higher-order kinetochore structure remain unknown. The outer kinetochore KNL-1/Mis12 complex/Ndc80 complex (KMN) network plays a key role in generating and sensing microtubule attachments. Here we demonstrate that Caenorhabditis elegans KNL-1 exists as an oligomer, and we identify a specific domain in KNL-1 responsible for this activity. An N-terminal KNL-1 domain from both C. elegans and the related nematode Caenorhabditis remanei oligomerizes into a decameric assembly that appears roughly circular when visualized by electron microscopy. On the basis of sequence and mutational analysis, we identify a small hydrophobic region as responsible for this oligomerization activity. However, mutants that precisely disrupt KNL-1 oligomerization did not alter KNL-1 localization or result in the loss of embryonic viability based on gene replacements in C. elegans. In C. elegans, KNL-1 oligomerization may coordinate with other kinetochore activities to ensure the proper organization, function, and sensory capabilities of the kinetochore–microtubule attachment.
INTRODUCTION
The kinetochore is a macromolecular protein assembly that forms the primary connection between chromosomes and spindle microtubules (Cheeseman and Desai, 2008). The major group of proteins responsible for the ability of the kinetochore to capture a microtubule is the conserved KNL-1/Mis12 complex/Ndc80 complex (KMN) network (Cheeseman et al., 2004, 2006). The Ndc80 complex acts as the critical microtubule-binding element within the KMN network (Cheeseman et al., 2006; DeLuca et al., 2006; Wei et al., 2007; Ciferri et al., 2008), with the Mis12 complex acting to connect the KMN network to the inner kinetochore (Gascoigne et al., 2011; Przewloka et al., 2011; Screpanti et al., 2011). Finally, KNL-1 is a large protein that is required to assemble the KMN network (Cheeseman et al., 2006). KNL-1 possesses a weak microtubule-binding activity (Cheeseman et al., 2006; Welburn et al., 2010; Espeut et al., 2012) and provides a scaffold for multiple signaling proteins at kinetochores, including PP1 (Liu et al., 2010), Bub1, and Bub3 (Kiyomitsu et al., 2007, 2011; Krenn et al., 2012; Caldas et al., 2013; Vleugel et al., 2013; Zhang et al., 2014).
Although the protein components at the kinetochore have been largely identified, there is limited data on how these proteins assemble into a productive higher-order conformation to facilitate microtubule interactions and kinetochore integrity. Because prior studies demonstrated that at least ∼8–20 copies of the KMN network proteins are bound to each microtubule at kinetochores (Joglekar et al., 2006, 2008; Lawrimore et al., 2011), the organization of these multiple complexes is a critical task. One possibility is that the microtubule itself imparts a higher-order organization to the kinetochore elements that bind the microtubule lattice. This could occur through the intrinsic symmetry of the microtubule or simply due to spatial constraints in binding sites. Alternatively, a subset of kinetochore proteins may act to organize kinetochore proteins into the higher-order structure. For example, at centrioles, the oligomerization of the central hub element Sas6 provides the organization and ninefold symmetry to the centriole barrel (Kitagawa et al., 2011; van Breugel et al., 2011). In this way, a single component of a complex could organize the remaining components to bring them into close proximity. However, it not known whether any kinetochore components self-associate in a defined way that would provide such an organization to the kinetochore. Our prior work reconstituting the C. elegans KMN network found that KNL-1 behaved as a much larger species than would be expected based on its molecular weight (Cheeseman et al., 2006). We interpreted this as a potential oligomerization for KNL-1, but the basis for and nature of this behavior were unclear.
Here we investigate this apparent KNL-1 oligomerization activity. Our work demonstrates that nematode KNL-1 oligomerizes to a defined state at physiologically relevant concentrations. The oligomeric region forms a roughly circular structure when visualized by electron microscopy. Biochemical experiments and sequence analysis identified a small region that is conserved in nematodes as containing the oligomerization activity. However, interfering with KNL-1 oligomerization by deletion of this region or specific point mutants did not result in dramatic defects in C. elegans replacement experiments. We propose that nematode KNL-1 oligomerization may act in concert with other, unidentified organizational elements within the kinetochore to generate a higher-order kinetochore structure to organize the microtubule-binding interface or signaling networks at kinetochores.
RESULTS
The nematode KNL-1 N-terminus oligomerizes
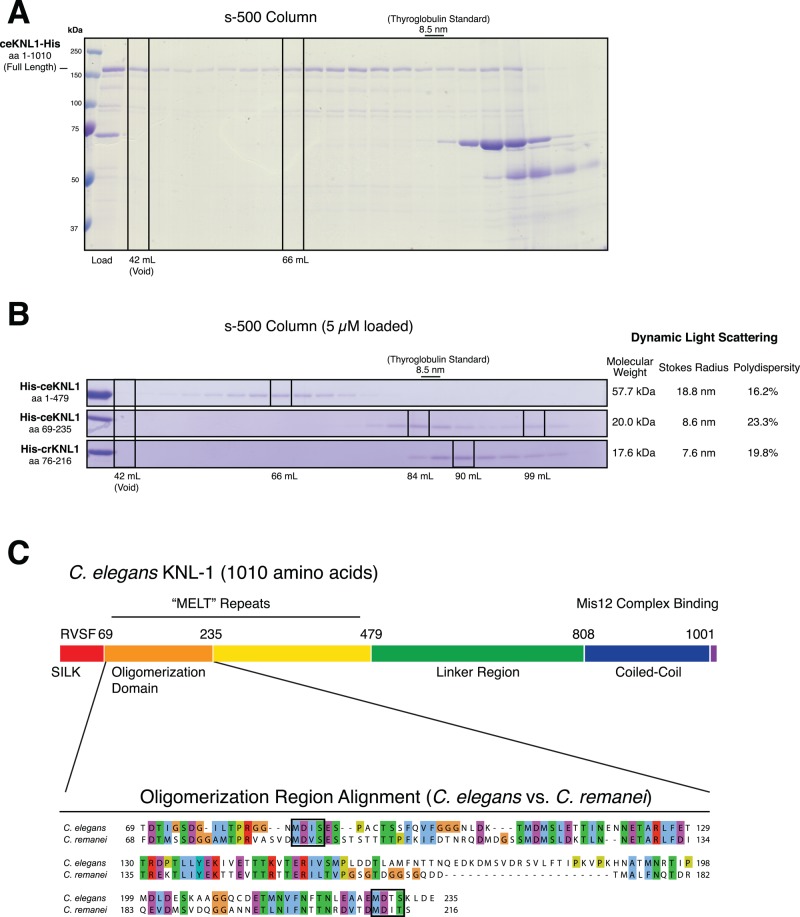
We found previously that recombinant, full-length C. elegans KNL-1 behaved as a much larger species than expected based on its predicted molecular weight in size exclusion chromatography (SEC; see Figure 1A) and sucrose gradients (Cheeseman et al., 2006). We reasoned that this behavior could be due to a combination of possibilities: 1) KNL-1 aggregates nonspecifically, 2) KNL-1 is highly elongated, or 3) KNL-1 oligomerizes in a structurally specific manner. To investigate the basis for this behavior, we began by creating truncations for C. elegans KNL-1 (Supplemental Figure 1, A and B). On the basis of the migration of these truncations by SEC, we found that the N-terminal half of KNL-1 was sufficient to display this large apparent behavior (Figure 1B). The N-terminus of KNL-1 acted as a single large species as revealed by both defined peaks in SEC and low polydispersity as assessed by dynamic light scattering (DLS) (Figure 1B). We further refined the region responsible for this activity to a small, ∼150–amino acid domain in the N-terminus of KNL-1, which we will refer to as the “oligomerization domain.” This 150–amino acid construct was well behaved biochemically but acted as a much larger assembly (8.6-nm Stokes radius) than expected based on its predicted molecular weight (20 kDa). For comparison, the globular thyroglobulin size standard has a similar Stokes radius of 8.5 nm but a molecular mass of 670 kDa.
FIGURE 1:
Identification of an N-terminal oligomerization domain in nematode KNL-1. (A) Coomassie-stained SDS–PAGE gel showing size exclusion chromatography analysis for full-length ceKNL-1 purified from bacteria. The load volume is shown as is, but fractions were trichloroacetic acid precipitated and resuspended to concentrate the samples before gel loading. (B) Coomassie-stained SDS–PAGE gels showing fractions from size exclusion chromatography analysis of C. elegans and C. remanei KNL-1 protein fragments. We loaded 5 μM concentrations of the indicated proteins on an s-500 column, with the relative elution volumes indicated. Right, predicted molecular weights, Stokes radii, and percentage polydispersity measured by dynamic light scattering. (C) Top, schematic of the C. elegans KNL-1 protein, with the previously defined motifs and regions indicated. Bottom, sequence alignment of the C. elegans and C. remanei oligomerization domains with conserved residues indicated and “MELT” repeats indicated with boxes.
To test whether this apparent KNL-1 oligomerization activity was conserved in diverse nematode species, we analyzed the behavior of the C. remanei KNL-1 protein, which has diverged significantly from C. elegans KNL-1 (31% amino acid identity along the entire length) but displays clear homology, including in the N-terminal oligomerization domain (Figure 1C). After purification of a recombinant C. remanei KNL-1 fragment with homology to the C. elegans oligomerization domain, we found that the C. remanei protein was also oligomeric based on SEC and DLS (Figure 1B), with the 17.6-kDa domain of C. remanei KNL-1 behaving like a 7.6-nm species. Thus both C. elegans and C. remanei KNL-1 display apparent oligomerization behavior in this conserved N-terminal region.
The KNL-1 oligomerization domain forms a defined higher-order oligomer
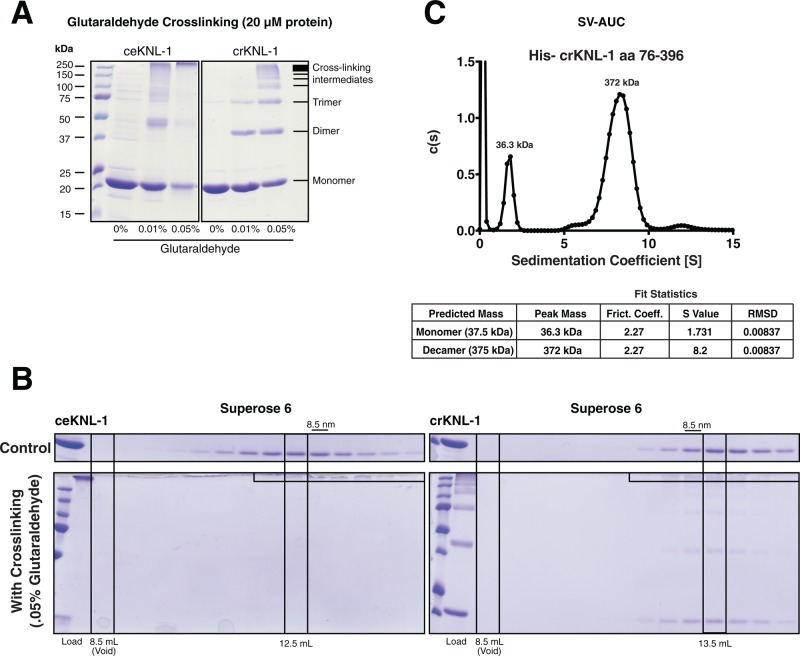
Because both the C. elegans and C. remanei KNL-1 oligomerization domains behaved similarly like large defined species, we sought to determine whether this large size was due to specific higher-order oligomerization or whether the protein has a highly elongated shape or is aggregation prone. To test this, we first analyzed the effect of the cross-linker glutaraldehyde on the KNL-1 oligomerization domains. At appropriate protein concentrations and time scales, glutaraldehyde will generate covalent linkages (usually between lysine residues) but only between proteins that are present in close proximity (<7.5 Å; Wine et al., 2007). We found that both the C. elegans and C. remanei KNL-1 oligomerization domains could be readily cross-linked with glutaraldehyde (Figure 2A). At high glutaraldehyde concentrations, the proteins were almost completely cross-linked into a single large species that likely corresponds to the fully cross-linked oligomer. However, at lower glutaraldehyde concentrations, we observed incompletely cross-linked species. On the basis of the migration of these cross-linked forms in SDS–PAGE gels, we were able to detect the presence of a ladder of incompletely cross-linked species with clear bands detected for dimers and trimers of KNL-1. Owing to the apparent large size of the cross-linked domains observed by SDS–PAGE, we sought to ensure that the glutaraldehyde was not artificially generating a large oligomer through spurious interactions. To test this, we cross-linked each domain using glutaraldehyde and compared the behavior of control and cross-linked proteins by SEC (Figure 2B). Of importance, the cross-linked KNL-1 proteins migrated similarly to the non–cross-linked proteins by SEC, and we did not observe any large cross-linked aggregates in the void volume of the column. These cross-linking experiments demonstrate that KNL-1 subunits are in close proximity and self-associate into a higher-order complex.
FIGURE 2:
The KNL-1 N-terminal domain oligomerizes into a decameric assembly. (A) Coomassie-stained SDS–PAGE gels showing the C. elegans and C. remanei KNL-1 oligomerization domains (at a concentration of 20 μM) treated with the indicated concentrations of the cross-linking agent glutaraldehyde. The shift in migration SDS–PAGE gel reflects the formation of multimeric cross-linked assemblies, as indicated on the right. (B) Coomassie-stained SDS–PAGE gels showing fractions from the size exclusion chromatography analysis of the C. elegans and C. remanei oligomerization domains. The native oligomerization domains (top) and the domains cross-linked using 0.05% glutaraldehyde (bottom) display similar migration, indicating that this treatment does not result in protein aggregation. We note that cross-linking appears to make the oligomers slightly smaller, potentially from stabilizing disordered regions of protein. The fully cross-linked C. elegans protein migrates just below the stacking gel. (C) Trace from the SV-AUC analysis of C. remanei KNL-1 amino acids 76–396. Fitting of the migration behavior (bottom) is consistent with the presence of a monomeric and decameric form.
We next sought to determine the stoichiometry of the KNL-1 oligomer using sedimentation velocity analytical ultracentrifugation (SV-AUC). For this analysis, we observed the best fit and behavior for a larger N-terminal fragment of C. remanei KNL-1. The SV-AUC analysis indicated that C. remanei KNL-1 formed a decamer, as well as having a monomeric form (Figure 2C). Although this protein behaved primarily as a single defined species, we observed some apparent disassociation of the larger assembly during the sedimentation run based on the spread of the oligomeric peak and the fitted frictional coefficient of ∼2. We also analyzed the C. elegans KNL-1 oligomerization domain by AUC, but we were unable to obtain a consistent fit for this protein due to a larger spread of the primary peak (unpublished data), likely due to its disassociation during the assay. On the basis of these SV-AUC data, together with the SEC and DLS analysis, we conclude that nematode KNL-1 N-terminus forms a defined high-order oligomer composed of ∼10 subunits.
The KNL-1 oligomerization domain forms a circular structure when visualized by electron microscopy
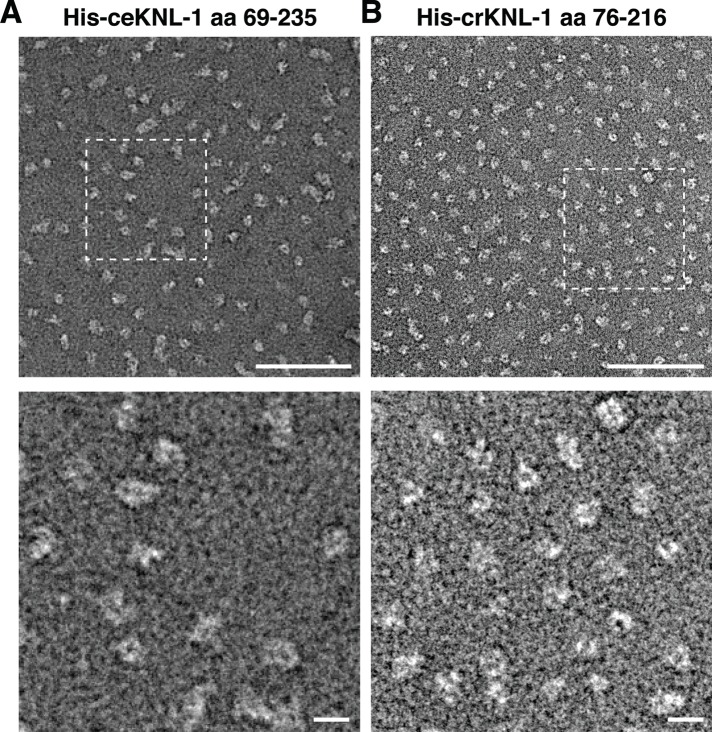
To visualize directly KNL-1 oligomerization, we analyzed the C. elegans and C. remanei oligomerization domains by negative-stain transmission electron microscopy. We found that KNL-1 was present as particles of roughly similar size and shape. Although there was some variability in individual particles, the C. elegans KNL-1 oligomerization domain formed a low-resolution circular or ring-like structure with a diameter of ∼15 nm (Figure 3). Similarly, the C. remanei oligomerization domain was present as a circular structure with a diameter of ∼11 nm (Figure 3). Thus the KNL-1 N-terminus oligomerizes into a particle with a roughly cylindrical shape.
FIGURE 3:
Visualization of the KNL-1 oligomerization domain by transmission electron microscopy. (A) Top, a field of ceKNL-1 oligomerization domain particles detected using transmission electron microscopy with negative staining. Scale bar, 100 nm. Bottom, a zoomed-in view from the boxed region above. Scale bar, 20 nm. (B) Top, a field of crKNL-1 oligomerization domain particles. Scale bar, 100 nm. Bottom, zoomed-in view from the boxed region above. Scale bar, 20 nm.
KNL-1 oligomerization occurs through a small hydrophobic region
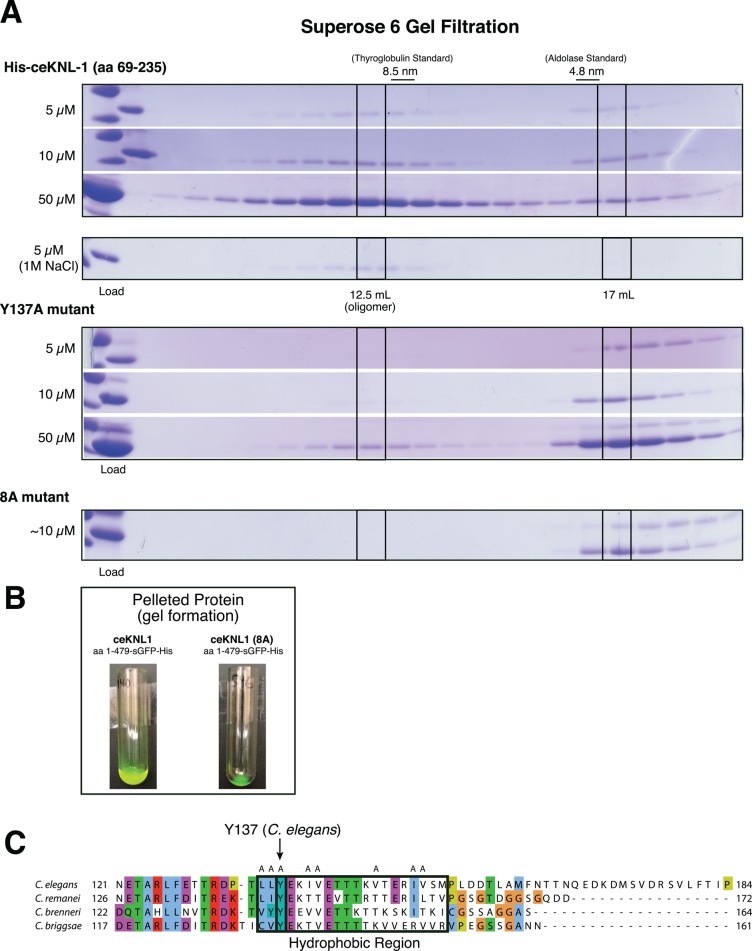
We next sought to determine the structural basis and specific residues required for the oligomerization of the KNL-1 N-terminal domain. We reasoned that KNL-1 self-association could occur through hydrogen bonding, charge–charge interactions, or hydrophobic interactions. To test this, we analyzed behavior of the C. elegans oligomerization domain by SEC under high-salt conditions (1 M NaCl). Such conditions will negate charge–charge interactions but strengthen hydrophobic interactions. We found that KNL-1 self-association was enhanced in 1 M NaCl (Figure 4A), suggesting that it is dependent on hydrophobic interactions. Through sequence analysis, we identified a small region within KNL-1 that contains multiple hydrophobic residues and is conserved among Caenorhabditis species (Figure 4C). Mutating the combination of the hydrophobic residues in this region to alanine (KNL-1 8A) abolished the oligomerization activity based on altered migration in size exclusion chromatography (Figure 4A). Mutation of a single conserved tyrosine residue (Y137A) within this hydrophobic region strongly reduced KNL-1 oligomerization without an obvious effect on protein expression or behavior (Figure 4A).
FIGURE 4:
KNL-1 oligomerization requires a conserved, hydrophobic patch. (A) Coomassie-stained SDS–PAGE gels showing fractions from size exclusion chromatography analysis of the C. elegans KNL-1 oligomerization domain tested at increasing concentrations or in the presence of 1 M NaCl as indicated. Top, migration behavior of the wild-type oligomerization domain. Middle, migration behavior of the Y137A mutant, which severely compromises the oligomerization activity. Bottom, migration behavior of the 8A mutant construct (concentration is approximate due to the presence of a contaminating protein). (B) Pellets from the wild-type and 8A mutant sfGFP-tagged proteins. The wild-type protein produces a substantial amount of a protein gel substance after nickel bead elution. This gel can be pelleted at low speed (22,000 × g; not shown) and high speed (100,000 × g; shown here). (C) Alignment of Caenorhabditis KNL-1 proteins showing the conservation of the residues in the oligomerization domain, highlighting the presence of a hydrophobic patch and the presence of the conserved tryptophan residue. Mutations included in the 8A mutant are indicated by A's above the residues.
Furthermore, we found that when our larger N-terminal ceKNL-1 construct (amino acids 1–479) was tagged with superfolder green fluorescent protein (sfGFP; Pedelacq et al., 2006) at its C-terminus, we obtained dramatically higher protein expression compared with the untagged version (Supplemental Figure 1C). At these high protein concentrations, we found that the KNL-1 protein formed a gel-like material after bead elution that pelleted efficiently in a centrifuge tube (Figure 4B) and expanded the apparent bead volume during its purification (Supplemental Figure 1C). The formation of this gel-like material, as well as the observed increase in bead volume, was disrupted by the KNL-1 8A mutation (Figure 4B and Supplemental Figure 1C). Therefore nematode KNL-1 oligomerizes using specific residues in a small conserved hydrophobic protein region.
KNL-1 oligomerization mutants do not dramatically disrupt chromosome segregation
Based on the foregoing biochemical analyses, nematode KNL-1 proteins undergo oligomerization. To test the contributions of this oligomerization domain to kinetochore function, we analyzed the effect of these mutants in vivo. For these experiments, we generated transgenic C. elegans strains using single-copy mos insertions expressing RNA interference (RNAi)–resistant wild-type KNL-1-mCherry (Espeut et al., 2012) or mutants designed to disrupt the KNL-1 oligomerization. This includes mutations in the hydrophobic residues that are required for KNL-1 oligomerization (KNL-1 8A) or a deletion of the defined oligomerization domain (∆102–236). Each of these KNL-1 mutants localized to the holocentric C. elegans kinetochores during mitosis similar to wild-type KNL-1 (Figure 5A). To test the effects of these mutants, we depleted endogenous KNL-1 by RNAi in the transgenic strains. In the absence of transgene expression, KNL-1 depletion resulted in penetrant embryonic lethality (Figure 5B) and eliminated kinetochore–microtubule interactions based on the rapid and premature separation of spindle poles (Figure 5C; Desai et al., 2003; Cheeseman et al., 2004). Expression of wild-type KNL-1-mCherry was able to fully rescue embryonic lethality (Figure 5B) and mitotic spindle elongation behavior (Figure 5C). Despite the significant defects in oligomerization observed in our biochemical assays, expression of the KNL-1 8A hydrophobic mutant did not result in obvious defects in embryonic lethality (Figure 5B) or spindle pole elongation (Figure 5C). Deletion of the entire oligomerization domain in KNL-1 (∆102–236) did not result in embryonic lethality (Figure 5B) but did display a small but reproducible delay in spindle pole elongation (Figure 5C). We note that the ∆102-236 deletion likely also reduces BUB-1 recruitment (Moyle et al., 2014), in addition to perturbing KNL-1 oligomerization. Finally, to test whether oligomerization activity is required for the function of KNL-1 as a signaling scaffold, we generated KNL-1 mutants that disrupt both oligomerization (8A mutant) and diminish BUB-1 recruitment through mutation of the MELT sequence repeats (Moyle et al., 2014). However, the 8A+MELT double mutant displayed normal embryonic viability (Figure 5B). Overall these data suggest that the KNL-1 oligomerization domain is not essential. However, we speculate that this activity may synergize with other, unidentified features of the nematode kinetochore to promote proper chromosome segregation.
FIGURE 5:
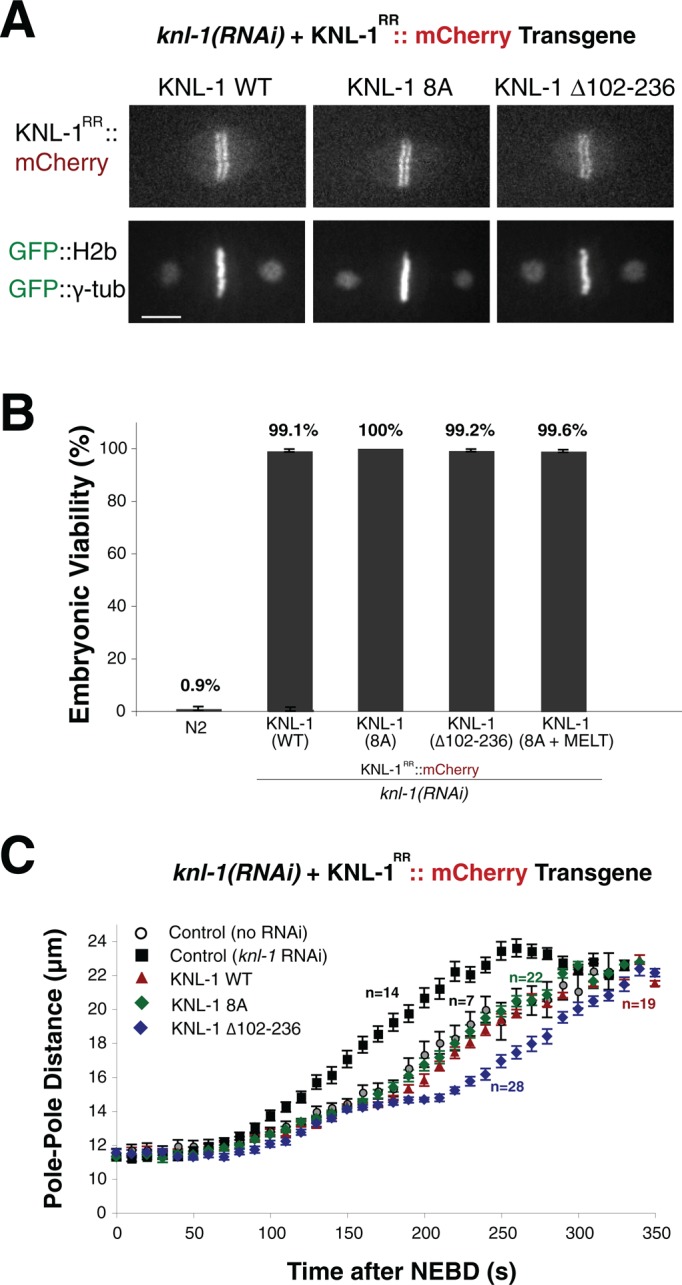
KNL-1 oligomerization is not essential for C. elegans viability. (A) Fluorescence images showing the localization of the mCherry-KNL-1 proteins (wild type or the indicated mutants) expressed in the first-cell-division C. elegans embryo. The bar-like localization reflects localization to the holocentric C. elegans kinetochores. Scale bar, 5 μm. (B) Graph indicating the embryonic viability after KNL-1 RNAi for N2 worms (7 worms and 1512 embryos) and worms stably expressing wild-type KNL-1 (7 worms and 733 embryos), KNL-1 8A (10 worms and 1041 embryos), KNL-1 ∆102-236 (6 worms and 676 embryos), and KNL-1 8A + MELT (13 worms and 1269 embryos). The graph shows the percentage viability ± SE. (C) Graph showing spindle pole separation over time during the first embryonic cell division for control embryos (control, no RNAi; n = 7), KNL-1 RNAi embryos (control, KNL-1 RNAi; n = 14), or KNL-1 RNAi embryos expressing KNL-1 wild type (n = 19), KNL-1 8A (n = 22), or KNL-1 ∆102–236 (n = 28). Error bars represent SE. The curves are aligned with respect to nuclear envelope breakdown (NEBD).
DISCUSSION
Our prior work suggested a potential self-association for KNL-1 (Cheeseman et al., 2006). Here we demonstrated that an N-terminal domain of nematode KNL-1 oligomerizes as a defined decameric assembly. Although this oligomerization activity is not essential for viability in C. elegans, it may function coordinately with additional factors to organize elements of the kinetochore. Although the oligomerization region we identified is conserved in nematode species, we did not detect obvious conservation of this domain in other organisms. We note that recent work on the human KNL1 protein has suggested the potential for its self-association through its N-terminal region based on immunoprecipitation from cells (Petrovic et al., 2014). This self-interaction may indicate the binding of hKNL1 to itself or may be mediated by one of its binding partners, such as Bub1. We note that the oligomerization that we have defined is early within the “MELT” repeat region of nematode KNL-1 (Figure 1C), similar to the position of the Bub1-interacting “KI” motifs in human KNL1 (Kiyomitsu et al., 2011). In both cases, this suggests the formation of a higher-order complex of KNL1 and its spindle assembly checkpoint-binding partners at its N-terminus. Different organisms may also have distinct requirements for these features of kinetochore organization and function. For example, we note that in contrast with the human kinetochore, in C. elegans, the Ska1 complex and the N-terminal tail of Ndc80 are dispensable for kinetochore function (Schmidt et al., 2012; Cheerambathur et al., 2013). It is also possible that other kinetochore proteins may self-associate, such as has been proposed for CENP-Q (Amaro et al., 2010), to contribute to kinetochore organization.
We propose that there are three principal functions for self-association of kinetochore components. First, interactions between kinetochore components may be critical for the structural integrity of kinetochores. Second, self-association of kinetochore components may be important to organize the microtubule interface. Finally, such a self-association may help to cluster signaling molecules at kinetochores. We hypothesize that the observed oligomerization for the C. elegans KNL-1 may play a role in organizing the N-terminus of the protein. The N-terminus of KNL-1 in all organisms is predicted to be largely disordered (Caldas and DeLuca, 2014; our unpublished analysis). Recent work has demonstrated that KNL-1 binds to Bub1 using its “MELT” repeats in this region (Krenn et al., 2012; Caldas et al., 2013; Vleugel et al., 2013; Zhang et al., 2014). Given that multiple repeats are present throughout the N-terminus of KNL-1, this may allow a single molecule of KNL-1 to recruit multiple Bub1 proteins (Vleugel et al., 2013). Self-association of KNL-1 would act to further locally concentrate Bub1, potentially amplifying this signal for its roles in the spindle assembly checkpoint and recruiting Aurora B to centromeres. Generating a focus of signaling activity may be especially important in a holocentric kinetochore since a diffuse kinetochore poses different signaling requirements compared with a localized kinetochore. It is likely that other kinetochore components possess properties that promote kinetochore structure and organization in parallel to KNL-1.
MATERIALS AND METHODS
Plasmids
Hexahistidine (6xHis) Escherichia coli expression constructs for the KNL-1 oligomerization domains (His-ceKNL-1 amino acids 69–235, His-crKNL-1 amino acids 76–216, and His-crKNL-1 amino acids 76–396) were amplified from C. elegans cDNA or synthesized by Genewiz and cloned into pRSETa to add an N-terminal His tag (MRGSHHHHHHGMAS-). The 6xHis-ceKNL-1 1–479 expression construct was generated using a modified pET3aTr vector to add a PreScission cleavable, N-terminal His tag (MRGSHHHHHHGMASMTGGQQMGRDLYDDDDKLEVLFQGPH-). sfGFP constructs were cloned with a custom C-terminal sfGFP-His tag. Mutations for ceKNL-1 constructs were introduced using PCR.
Protein production and purification
Proteins were produced using 3–12 l of BL21 (DE3) E. coli. Generally, bacteria were grown to OD 0.6–1 at 30°C in lysogeny broth (LB) medium containing antibiotic and 0.4% glucose. The temperature was reduced to 18°C, and protein production was induced with 100 mM isopropyl-β-d-thiogalactoside. The bacteria were harvested 6 h postinduction (20 h for GFP constructs) with lysis buffer (50 mM sodium phosphate, pH 8, 300 mM NaCl, 40 mM imidazole) and frozen at −80°C. The bacterial pellet was then thawed and lysed using 1 mg/ml lysozyme and sonication. Then 10 mM β-mercaptoethanol (BME) was added. The lysate was pelleted at 40,000 × g for 30 min. The supernatant was bound to nickel-nitriloacetic acid resin (Qiagen, Venlo, Netherlands) for 1 h at 4°C. The resin was washed with wash buffer (50 mM sodium phosphate, pH 8, 500 mM NaCl, 40 mM imidazole, 10 mM BME, 0.1% Tween-20). Bound protein was then eluted with elution buffer (50 mM sodium phosphate, pH 7, 500 mM NaCl, 250 mM imidazole, 10 mM BME). Elutions were loaded onto Superose 6 or Superdex 200 columns for gel filtration into Schwartz buffer (20 mM sodium phosphate, pH 7, 150 mM NaCl, 1 mM dithiothreitol [DTT]). Peak fractions were checked using SDS–PAGE gels stained with Coomassie. The peak fractions were then pooled and spin concentrated (Vivaspin; GE Healthcare, Little Chalfont, UK). Protein concentrations were determined using the Bio-Rad Assay kit. Protein was used fresh (within a few days on ice and never freeze/thawed) for all experiments.
Gel filtration
Proteins were loaded at indicated concentrations onto either a Sephacryl S-500 HR 16/60 column or a Superose 6 10/30 GL column equilibrated in Schwartz buffer. Size standards run with matching loading volumes are marked as indicated in the figures. Runs were analyzed using representative fractions spanning the column runs with SDS–PAGE gels stained with Coomassie. Note that due to the low absorption coefficient at 280 nm for the KNL-1 protein fragments, we used large Coomassie-stained gels instead of ultraviolet traces for visualization.
Dynamic light scattering
Measurements were taken using a Protein Solutions (now Wyatt Technology, Santa Barbara, CA) Dynapro instrument and Dynamics V6 software. The measurements were taken using 10 reads each with a 10-s averaging time.
Glutaraldehyde cross-linking
Proteins were cross-linked at the indicated concentrations of protein. Glutaraldehyde (70% stock solution, EM grade; Sigma-Aldrich, St. Louis, MO) was diluted in double-distilled H2O (ddH2O) to 0.2 or 1% (1% was also used for Superose 6 runs) and mixed with protein at 1:20 to the indicated final concentrations. Mock cross-linking was performed using the equivalent volume of ddH2O. The proteins were cross-linked for 10 min at room temperature and then quenched with 1:10 volume of 1 M Tris, pH 8. For Figure 2A, the protein was loaded onto a 12% SDS–PAGE gel for visualization with Coomassie. For Figure 2B, after the quenching, the proteins were pelleted at 18,000 × g and then loaded onto the Superose 6 gel filtration column in Schwartz buffer, and runs were visualized using 12% SDS–PAGE gels stained with Coomassie.
Analytical ultracentrifugation
The sedimentation-velocity experiment for the His-crKNL-1 76-396 construct was conducted using purified protein at ∼20 μM in Schwartz buffer using a Beckman (Pasadena, CA) Optima XL-I analytical ultracentrifuge in interference mode (MIT Biophysical Instrumentation Facility, Cambridge, MA). Data were collected at 20°C at 25,000 rpm. The data were fitted using SEDFIT to a model for continuous sedimentation coefficient distribution, assuming a single frictional coefficient. The molecular weights were estimated using the best-fit frictional coefficients.
Election microscopy
To prevent disassembly of the oligomers under the conditions used for election microscopy, His-ceKNL-1 69-235 and His-crKNL-1 76-216 were cross-linked at ∼5 and ∼10 μM, respectively, using a final concentration of 0.1% glutaraldehyde. Following quenching, protein was dialyzed into EM Buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1 mM DTT) for 5 h at 4ºC. Samples were then kept on ice until grid preparation. For grid preparation, 4-μl samples were applied to freshly glow-discharged continuous carbon grids and stained with 0.75% uranyl formate. Images were collected on an FEI (Hillsborough, OR) Tecnai F-20 electron microscope with a Gatan US4000 charge-coupled device detector using a nominal magnification of 62,000× (83,701× at detector) and a defocus of −3 μm.
Sequence analysis
Sequences were aligned using ClustalX and Jalview software.
Worm strains
The worm strains used in this study are listed in Supplemental Table S1. The KNL-1 mutations were engineered into a vector expressing KNL-1::mCherry (Espeut et al., 2012). Plasmids were injected into strain EG4322 to obtain stable single-copy integrants (Frokjaer-Jensen et al., 2008). Integration of transgenes was confirmed by PCR. For live imaging, transgenes were crossed into a strain expressing GFP::H2b/GFP::γ-tubulin, and the transgene as well as both markers were homozygosed before analysis.
RNA-mediated interference
Double-stranded RNAs used in this study are listed in Supplemental Table S2. All RNAi was performed by microinjection. L4 worms were injected with double-stranded RNAs (dsRNAs) and incubated for 38–43 h at 20°C before imaging of the embryos. For lethality assays, L4 worms were injected with dsRNA and singled onto plates at 24 h postinjection; adult worms were removed from the plates at 48 h postinjection, and hatched larvae and unhatched embryos were counted at 72 h postinjection.
Time-lapse microscopy
For imaging of chromosomes and pole tracking analysis, images were acquired on a deconvolution microscope (DeltaVision; Applied Precision/GE Healthcare) equipped with a charge-coupled device camera (CoolSnap; Roper Scientific, Sarasota, FL) with 5 × 2–μm z-stacks, 2 × 2 binning, and a 60×/1.3 numerical aperture (NA) U-planApo objective (Olympus, Tokyo, Japan) at 10-s intervals and 100-ms exposure at 18°C. Spindle pole separation was quantified as described (Desai et al., 2003).
For KNL-1 localization, embryos expressing GFP::H2b/GFP::γ-tubulin/KNL-1::mCherry were filmed every 20 s with 5 × 2–μm z-stacks on an Andor Revolution XD Confocal System (Andor Technology, Belfast, UK) and a confocal scanner unit (CSU-10; Yokogawa) mounted on an inverted microscope (TE2000-E; Nikon, Tokyo, Japan) equipped with 100×/1.4 NA Plan Apochromat lens and outfitted with an electron multiplying, back-thinned charge-coupled device camera (binning 1 × 1; iXon; Andor Technology) at 20°C. Exposure was 100 ms for GFP and 300 ms for mCherry.
Supplementary Material
Acknowledgments
We gratefully acknowledge Debby Pheasant and the Biophysical Instrumentation Facility for the Study of Complex Macromolecular Systems (NSF-007031). We thank Nikolaus Grigorieff, Mathijs Vleugel, Bob Sauer, Thomas Schwartz, Ellen Kloss, and members of the Cheeseman laboratory for their support, input, and helpful discussions. This work was supported by a Scholar Award to I.M.C. from the Leukemia and Lymphoma Society, grants from the National Institutes of Health/National Institute of General Medical Sciences to I.M.C. (GM088313) and A.D. (GM074215), and a Research Scholar Grant to I.M.C. (121776) from the American Cancer Society. M.R. was supported by funding from the Howard Hughes Medical Institute to Nikolaus Grigorieff.
Abbreviations used:
- BME
β-mercaptoethanol
- ddH2O
double-distilled water
- DLS
dynamic light scattering
- dsRNAs
double-stranded RNAs
- EM
electron microscopy
- KMN
KNL-1/Mis12 complex/Ndc80 complex
- SEC
size exclusion chromatography
- sfGFP
superfolder green fluorescent protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-06-1125) on November 19, 2014.
REFERENCES
- Amaro AC, Samora CP, Holtackers R, Wang E, Kingston IJ, Alonso M, Lampson M, McAinsh AD, Meraldi P. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat Cell Biol. 2010;12:319–329. doi: 10.1038/ncb2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas GV, DeLuca JG. KNL1: bringing order to the kinetochore. Chromosoma. 2014;123:169–181. doi: 10.1007/s00412-013-0446-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas GV, DeLuca KF, DeLuca JG. KNL1 facilitates phosphorylation of outer kinetochore proteins by promoting Aurora B kinase activity. J Cell Biol. 2013;203:957–969. doi: 10.1083/jcb.201306054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur DK, Gassmann R, Cook B, Oegema K, Desai A. Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Science. 2013;342:1239–1242. doi: 10.1126/science.1246232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, III, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Shevchenko A, Hyman A, Oegema K. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeut J, Cheerambathur DK, Krenning L, Oegema K, Desai A. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J Cell Biol. 2012;196:469–482. doi: 10.1083/jcb.201111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck D, Finley K, Liu X, Wan Y, Berman J, He X, Salmon ED, Bloom KS. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, Olieric V, Bortfeld M, Erat MC, Fluckiger I, Gonczy P, Steinmetz MO. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011;144:364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Murakami H, Yanagida M. Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1. Mol Cell Biol. 2011;31:998–1011. doi: 10.1128/MCB.00815-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Obuse C, Yanagida M. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev Cell. 2007;13:663–676. doi: 10.1016/j.devcel.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Krenn V, Wehenkel A, Li X, Santaguida S, Musacchio A. Structural analysis reveals features of the spindle checkpoint kinase Bub1-kinetochore subunit Knl1 interaction. J Cell Biol. 2012;196:451–467. doi: 10.1083/jcb.201110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore J, Bloom KS, Salmon ED. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J Cell Biol. 2011;195:573–582. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Vleugel M, Backer CB, Hori T, Fukagawa T, Cheeseman IM, Lampson MA. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol. 2010;188:809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle MW, Kim T, Hattersley N, Espeut J, Cheerambathur DK, Oegema K, Desai A. A Bub1-Mad1 interaction targets the Mad1-Mad2 complex to unattached kinetochores to initiate the spindle checkpoint. J Cell Biol. 2014;204:647–657. doi: 10.1083/jcb.201311015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- Petrovic A, Mosalaganti S, Keller J, Mattiuzzo M, Overlack K, Krenn V, De Antoni A, Wohlgemuth S, Cecatiello V, Pasqualato S, et al. Modular assembly of RWD domains on the Mis12 complex underlies outer kinetochore organization. Mol Cell. 2014;53:591–605. doi: 10.1016/j.molcel.2014.01.019. [DOI] [PubMed] [Google Scholar]
- Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, Glover DM. CENP-C is a structural platform for kinetochore assembly. Curr Biol. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Schmidt JC, Arthanari H, Boeszoermenyi A, Dashkevich NM, Wilson-Kubalek EM, Monnier N, Markus M, Oberer M, Milligan RA, Bathe M, et al. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev Cell. 2012;23:968–980. doi: 10.1016/j.devcel.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screpanti E, De Antoni A, Alushin GM, Petrovic A, Melis T, Nogales E, Musacchio A. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breugel M, Hirono M, Andreeva A, Yanagisawa HA, Yamaguchi S, Nakazawa Y, Morgner N, Petrovich M, Ebong IO, Robinson CV, et al. Structures of SAS-6 suggest its organization in centrioles. Science. 2011;331:1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- Vleugel M, Tromer E, Omerzu M, Groenewold V, Nijenhuis W, Snel B, Kops GJ. Arrayed BUB recruitment modules in the kinetochore scaffold KNL1 promote accurate chromosome segregation. J Cell Biol. 2013;203:943–955. doi: 10.1083/jcb.201307016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- Welburn JP, Vleugel M, Liu D, Yates JR, 3rd, Lampson MA, Fukagawa T, Cheeseman IM. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wine Y, Cohen-Hadar N, Freeman A, Frolow F. Elucidation of the mechanism and end products of glutaraldehyde crosslinking reaction by X-ray structure analysis. Biotechnol Bioeng. 2007;98:711–718. doi: 10.1002/bit.21459. [DOI] [PubMed] [Google Scholar]
- Zhang G, Lischetti T, Nilsson J. A minimal number of MELT repeats supports all the functions of KNL1 in chromosome segregation. J Cell Sci. 2014;127:871–884. doi: 10.1242/jcs.139725. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.