Unlike the six other Rho GEFs in fission yeast, Gef3 colocalizes with and depends on septins to localize during late cytokinesis. Gef3 binds to GDP-Rho4 and accelerates its nucleotide exchange in vitro. Moreover, Gef3 modulates Rho4 activity in vivo and works in the same genetic pathways as Rho4 to regulate septation in fission yeast cytokinesis.
Abstract
Rho GTPases, activated by Rho guanine nucleotide exchange factors (GEFs), are conserved molecular switches for signal transductions that regulate diverse cellular processes, including cell polarization and cytokinesis. The fission yeast Schizosaccharomyces pombe has six Rho GTPases (Cdc42 and Rho1–Rho5) and seven Rho GEFs (Scd1, Rgf1–Rgf3, and Gef1–Gef3). The GEFs for Rho2–Rho5 have not been unequivocally assigned. In particular, Gef3, the smallest Rho GEF, was barely studied. Here we show that Gef3 colocalizes with septins at the cell equator. Gef3 physically interacts with septins and anillin Mid2 and depends on them to localize. Gef3 coprecipitates with GDP-bound Rho4 in vitro and accelerates nucleotide exchange of Rho4, suggesting that Gef3 is a GEF for Rho4. Consistently, Gef3 and Rho4 are in the same genetic pathways to regulate septum formation and/or cell separation. In gef3∆ cells, the localizations of two potential Rho4 effectors—glucanases Eng1 and Agn1—are abnormal, and active Rho4 level is reduced, indicating that Gef3 is involved in Rho4 activation in vivo. Moreover, overexpression of active Rho4 or Eng1 rescues the septation defects of mutants containing gef3∆. Together our data support that Gef3 interacts with the septin complex and activates Rho4 GTPase as a Rho GEF for septation in fission yeast.
INTRODUCTION
Cytokinesis is the last step of the cell cycle. It results in a mother cell splitting into two daughter cells. The basic mechanism for cytokinesis is conserved from yeast to humans. Cytokinesis requires the coordination of several key events: cleavage-site selection, assembly of the actomyosin contractile ring, constriction and disassembly of the contractile ring, plasma membrane deposition, septum formation or extracellular matrix remodeling, and daughter cell separation/midbody abscission (Balasubramanian et al., 2004; Barr and Gruneberg, 2007; Pollard and Wu, 2010; Green et al., 2012). Most of the events are controlled by Rho GTPases and their regulators (Hall, 1998, 2012; Garcia et al., 2006b; Jordan and Canman, 2012). Rho GTPases are highly conserved molecular switches that control signal transduction pathways. Rho guanine nucleotide exchange factors (GEFs) catalyze the exchange from GDP to GTP and thus activate Rho GTPases (Jaffe and Hall, 2005). Rho GTPase–activating proteins (GAPs) turn off Rho GTPases by catalyzing their intrinsic hydrolase activity (Jaffe and Hall, 2005).
The fission yeast Schizosaccharomyces pombe is an excellent model system for studying cytokinesis (Gould and Simanis, 1997; Roberts-Galbraith and Gould, 2008; Lee et al., 2012). The anillin-related protein Mid1 plays a crucial role in division-site selection together with the Polo kinase Plo1 and DYRK kinase Pom1 (Sohrmann et al., 1996; Bähler and Pringle, 1998; Bähler et al., 1998a; Paoletti and Chang, 2000; Celton-Morizur et al., 2006; Almonacid et al., 2011; Ye et al., 2012). Mid1 is essential for the assembly of cytokinesis nodes, the precursors of the contractile ring (Wu et al., 2006; Hachet and Simanis, 2008; Coffman et al., 2009; Almonacid et al., 2011; Laporte et al., 2011; Padmanabhan et al., 2011; Lee and Wu, 2012). The cytokinesis nodes condense into the contractile ring through a search, capture, pull, and release mechanism (Vavylonis et al., 2008; Laporte et al., 2010, 2012; Ojkic et al., 2011). The ring matures during anaphase B by recruiting more proteins under the regulation of the septation initiation network pathway (Wu et al., 2003; Hachet and Simanis, 2008; Huang et al., 2008). Then the ring constricts, and the division septum is formed (Wu et al., 2003; Proctor et al., 2012). The septum contains a primary septum flanked by two secondary septa (Johnson et al., 1973). During cell separation, the primary septum and the surrounding cell wall on cell sides are degraded by hydrolytic enzymes, including β‑glucanase Eng1 and α-glucanase Agn1, and the secondary septa remodel to become the cell wall at the new ends of the daughter cells (Martin-Cuadrado et al., 2003, 2008; Dekker et al., 2004; Sipiczki, 2007).
Both budding yeast and fission yeast have six homologous Rho GTPases, named Rho1– Rho5 and Cdc42 (Garcia et al., 2006b; Park and Bi, 2007; Perez and Rincon, 2010). Cdc42 is essential for cell-polarity establishment and maintenance, actin organization, and septation (septum formation and/or cell separation) during cytokinesis (Adams et al., 1990; Johnson and Pringle, 1990; Caviston et al., 2003; Rincon et al., 2007; Howell et al., 2012; Atkins et al., 2013; Onishi et al., 2013; Slaughter et al., 2013). Rho1 is essential for cell-wall formation and integrity, actomyosin-ring assembly, and septation (Arellano et al., 1997; Nakano et al., 1997; Tolliday et al., 2002; Yoshida et al., 2006; Levin, 2011). Rho5 is suggested to be a functional homologue of Rho1 in S. pombe (Nakano et al., 2005; Rincon et al., 2006), whereas in Saccharomyces cerevisiae it is involved in mediating stress response (Schmitz et al., 2002; Singh et al., 2008). Rho2 is nonessential and also involved in cell-wall synthesis by regulating α-glucan synthase (Madaule et al., 1987; Calonge et al., 2000). In budding yeast, Rho3 is crucial for polarized cell growth by regulating exocytosis and formin activation, and its roles are often shared with Rho4 (Matsui and Toh, 1992; Imai et al., 1996; Adamo et al., 1999; Doignon et al., 1999; Robinson et al., 1999; Wu et al., 2010a). Similarly, in fission yeast, Rho3 functions in polarized cell growth and modulates the exocyst complex for cell separation (Nakano et al., 2002; Wang et al., 2003). It has been suggested that S. pombe Rho4 participates in the delivery of certain secretory vesicles and regulates the localization of glucanases Eng1 and Agn1 during cell separation (Nakano et al., 2003; Santos et al., 2003, 2005). However, the relationship between Rho3 and Rho4 is unknown in fission yeast. The exocyst is an octameric protein complex (containing Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70, and Exo84) that tethers secretory vesicles to the plasma membrane during polarized cell growth and cytokinesis (TerBush et al., 1996; Guo et al., 1999; Hsu et al., 2004; He et al., 2007a, b; Mukherjee et al., 2014). The exocyst complex is involved in transporting and secreting the hydrolytic enzymes Eng1 and Agn1 for daughter cell separation in fission yeast cytokinesis (Wang et al., 2002; Martin-Cuadrado et al., 2005).
Seven Rho GEFs—Scd1, Gef1–Gef3, and Rgf1–Rgf3—have been identified in S. pombe (Iwaki et al., 2003; Garcia et al., 2006b; Wu et al., 2010b). Among these, Scd1 and Gef1 are GEFs for Cdc42 (Coll et al., 2003; Hirota et al., 2003), and Rgf1–Rgf3 function as GEFs for Rho1 and possibly Rho5 (Tajadura et al., 2004; Morrell-Falvey et al., 2005; Mutoh et al., 2005; Garcia et al., 2006a, 2009). Recently we and others found that Gef2 localizes to cortical nodes and the contractile ring and plays roles in division-site positioning and septation (Moseley et al., 2009; Ye et al., 2012; Guzman-Vendrell et al., 2013; Jourdain et al., 2013; Zhu et al., 2013; Akamatsu et al., 2014). Gef2 binds to Rho4, Rho1, and Rho5 in vitro and regulates Rho4 localization in vivo (Zhu et al., 2013). However, the functions and substrate GTPase(s) of Gef3 were still unknown.
Besides Rho GTPases and their GEFs, septins are also essential for cytokinesis in many cell types (Longtine et al., 1996; Gladfelter et al., 2001; Caudron and Barral, 2009; Oh and Bi, 2011). Septins are GTP-binding proteins that usually associate with the plasma membrane and function as scaffolds and diffusion barriers (Hall et al., 2008; Oh and Bi, 2011). In budding yeast, septins localize to the mother-bud neck as filaments and are required for the localizations of ∼100 proteins (Byers and Goetsch, 1976; Gladfelter et al., 2001; McMurray and Thorner, 2009; Oh and Bi, 2011). S. pombe has seven septins, and four of them (Spn1–Spn4) are expressed in vegetative cells and localize to the division site during cytokinesis (Longtine et al., 1996; Berlin et al., 2003; Tasto et al., 2003; Wu et al., 2003, 2010b; An et al., 2004; Petit et al., 2005; Onishi et al., 2010). Spn1 and Spn4 are the core members for septin-complex assembly (An et al., 2004; McMurray and Thorner, 2008). Mid2, an anillin homologue related to S. cerevisiae Bud4 and Candida albicans Int1, colocalizes with septins and stabilizes septin rings during cytokinesis in fission yeast (Berlin et al., 2003; Tasto et al., 2003). Deletion mutants of septins or mid2 are viable but display a delay in the separation of daughter cells (Berlin et al., 2003; Tasto et al., 2003). One function of the septin ring is to regulate the localization of glucanases Eng1 and Agn1, delivered by exocytosis during cell separation (Martin-Cuadrado et al., 2005). However, unlike budding yeast, very few fission yeast proteins have been reported to localize to the division site in a septin-dependent manner, and thus our knowledge on septin functions in fission yeast cytokinesis is quite limited. Of note, a Rho GEF (SA-RhoGEF) was identified to be a binding partner of a mammalian septin Sept9b and act as a link between septins and Rho signaling (Nagata and Inagaki, 2005). Hence the relationship between septins and Rho GEFs is of great interest.
Here we show that Gef3 localizes to the division site in a septin-dependent manner in fission yeast. Gef3 and the Rho GTPase Rho4 bind to each other and are in the same genetic pathways to regulate septation. Gef3 possesses GEF activity toward Rho4 in vitro and activates Rho4 in vivo. Rho4 overexpression can rescue cell-separation defects in mutants containing gef3∆. Thus we conclude that the Gef3 is a GEF for Rho4 and is involved in the final stages of cytokinesis.
RESULTS
Gef3 colocalizes with septins at the cell-division site
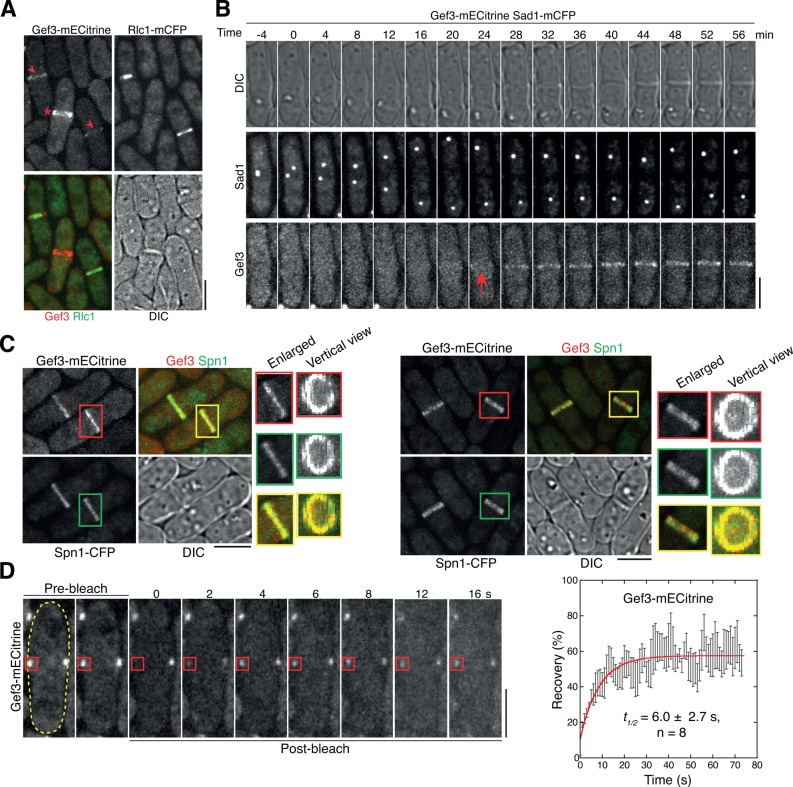
The putative Rho GEF Gef3 was the smallest and least-studied Rho GEFs in S. pombe (Iwaki et al., 2003; Garcia et al., 2006b; Perez and Rincon, 2010). To explore the functions of Gef3, we first tagged it with the yellow fluorescent protein (YFP) variant monomeric enhanced Citrine (mECitrine; Griesbeck et al., 2001; Ye et al., 2012) at its C-terminus and examined its localization (Figure 1). Gef3-mECitrine concentrated at the cell-division site with varied intensity during cytokinesis (Figure 1A). Using monomeric cyan fluorescent protein (mCFP)–tagged myosin regulatory light chain Rlc1 and SUN-domain protein Sad1 as the contractile-ring and spindle-pole body (SPB) marker (Wu et al., 2003), respectively, we found that Gef3 appeared at the cell equator as a nonconstricting ring at the end of anaphase B (Figure 1, A and B, and Supplemental Figure S1, A and B). Gef3 intensity at the division site seemed to increase during septum formation (Figure 1, A and B, and Supplemental Figure S1, A and B). However, its diameter remained the same until cell separation, when it spread to new cell ends (Supplemental Figure S1B, 57 min).
FIGURE 1:
Gef3 colocalizes with septins at the cell-division site. (A) Gef3-mECitrine localizes to the cell-division site but does not constrict with Rlc1-mCFP–labeled contractile ring. DIC, differential interference contrast. Arrowheads mark cells with constricting contractile rings. The asterisk marks a cell with a complete septum after contractile-ring disassembly. (B) Time course (in minutes) of Gef3 localization at the division site using the SPB protein Sad1 as a cell-cycle marker. Time 0 marks SPB separation. The arrow marks the appearance of Gef3 at the division site. (C) Colocalization of Gef3 and the septin Spn1 in septin single (left) and double rings (right). Enlarged and vertical views of the ring structures within the boxed regions are shown to the right. (D) FRAP analysis of Gef3 in cells with complete septa. Left, representative cell before (prebleach) and after (postbleach) photobleaching. The boxed region was bleached at time 0. The cell boundary is marked with broken lines. Right, mean recovery of eight cells. Mean ± SEM is plotted, and the curve fit is in red. Bars, 5 μm.
The timing and pattern of Gef3 localization resemble those of septins (Berlin et al., 2003; Tasto et al., 2003; Wu et al., 2003). Thus we examined whether Gef3 colocalizes with Spn1, a core component of the septin complex (An et al., 2004). Indeed, Gef3 colocalized with Spn1 as either single (Figure 1C, left) or double rings (Figure 1C, right) from its appearance to just before cell separation (Supplemental Figure S1B). To determine Gef3 dynamics, we performed fluorescence recovery after photobleaching (FRAP) assays in cells with complete septa to avoid interference from protein recruiting to the division site. Different from septins, which form stable rings at the division site (Berlin et al., 2003), Gef3 was quite dynamic, with a recovery half-time of 6.0 ± 2.7 s after photobleaching (Figure 1D). Taken together, the results show that Gef3 colocalizes with septins at the division site during septum formation and cell separation, but Gef3 is more dynamic than septins.
Gef3 localization depends on septins
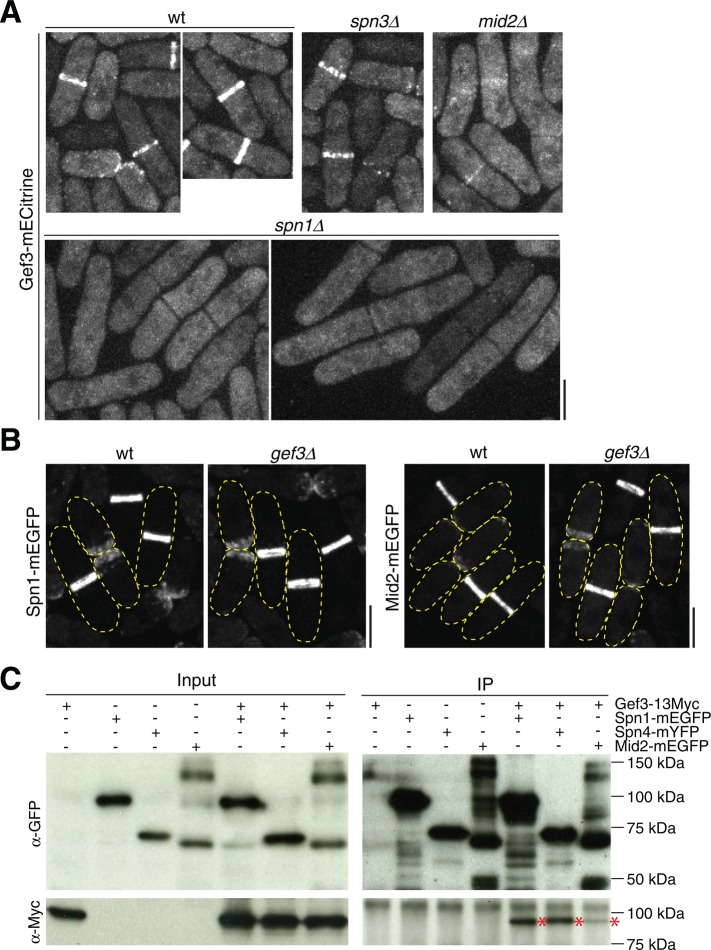
Septins are scaffolding proteins and essential for localization of ∼100 proteins to the bud neck in budding yeast (Versele and Thorner, 2005; Caudron and Barral, 2009; Oh and Bi, 2011; Spiliotis and Gladfelter, 2012; Wloka and Bi, 2012). It is still a mystery why septins are not essential and very few proteins depend on septins to localize in fission yeast (Berlin et al., 2003; Tasto et al., 2003; An et al., 2004; Martin-Cuadrado et al., 2005; Wu et al., 2010b). Given the colocalization of Gef3 and septins, we tested their interdependence for localization to the division site. Of interest, Gef3 localization at the division site was abolished in spn1∆ cells (Figure 2A), in which no other septins can localize (An et al., 2004). Gef3 intensity was also reduced in spn3∆ cells (Figure 2A), a deletion in which the septin structure is slightly compromised (An et al., 2004). The anillin-like protein Mid2 regulates septin dynamics and localization in S. pombe (Berlin et al., 2003; Tasto et al., 2003; Wu et al., 2010b). In mid2∆ cells, the septin level at the division site is reduced, and septins spread to the septum disk instead of the double rings (Berlin et al., 2003; Tasto et al., 2003). Consistently, Gef3 intensity at the division site was dramatically reduced and spread to the septum as septins in mid2∆ cells (Figure 2A and unpublished data). In contrast, the localizations of Spn1 and Mid2 were not affected in gef3∆ cells (Figure 2B). Thus septins are essential for Gef3 localization.
FIGURE 2:
Gef3 depends on septins to localize and physically interacts with the septin complex. (A) Gef3 localization in wt, spn1∆, spn3∆, and mid2∆ cells. (B) Spn1 (left) and Mid2 (right) localization in wt and gef3∆ cells. The cell boundary of most cells is marked with broken lines. (C) Gef3-13Myc co-IP with Spn1, Spn4, and Mid2. Asterisks mark the Gef3-13Myc bands after pull down. Bars, 5 μm.
The localization dependence suggests that Gef3 and septins may form protein complexes. Indeed, Gef3-13Myc was pulled down by the septins Spn1–monomeric enhanced green fluorescent protein (mEGFP) and Spn4-mYFP and Mid2-mEGFP in coimmunoprecipitation (co-IP) assays (Figure 2C). Together these data suggest that Gef3 is recruited to the division site through physical interactions with septins and/or anillin Mid2, although the nature of the interactions needs further study.
Gef3 regulates septation in later stages of cytokinesis
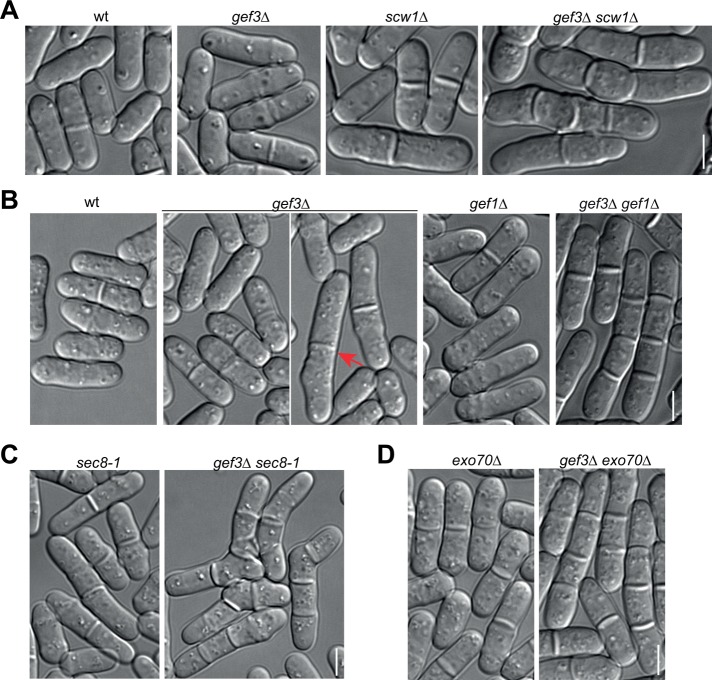
Septins are involved in septation by regulating the localizations of β-glucanase Eng1 and α-glucanase Agn1 (Martin-Cuadrado et al., 2005). Gef3 localization and its dependence on septins suggest that it has a function in later stages of cytokinesis. Indeed, gef3∆ cells had mild septation defects at 36°C (Figure 3, A and B; for quantification, see later discussion of Figure 6, B and D): higher percentage of septating cells, some long, septating cells (Figure 3B, arrow), and occasionally a few multiseptated cells (unpublished data). The phenotype indicates that Gef3 functions redundantly in late cytokinesis. To identify the proteins that play an overlapping role with Gef3, we tested genetic interactions between gef3∆ and other cytokinetic mutations (Table 1). Of interest, gef3∆ had strong synthetic genetic interactions with scw1∆, gef1∆, and exocyst mutants (Figure 3 and Table 1), as described later.
FIGURE 3:
Synthetic genetic interactions of gef3∆ with other cytokinesis mutations. (A–D) DIC images showing synthetic interactions between gef3∆ and scw1∆ (A), gef1∆ (B), and exocyst mutants sec8-1 (C) and exo70∆ (D) at at 25°C (A, C) or after shifting to 36°C for 6 h (B, D). The arrow marks a long gef3∆ cell likely with delay in cell separation. Bars, 5 μm.
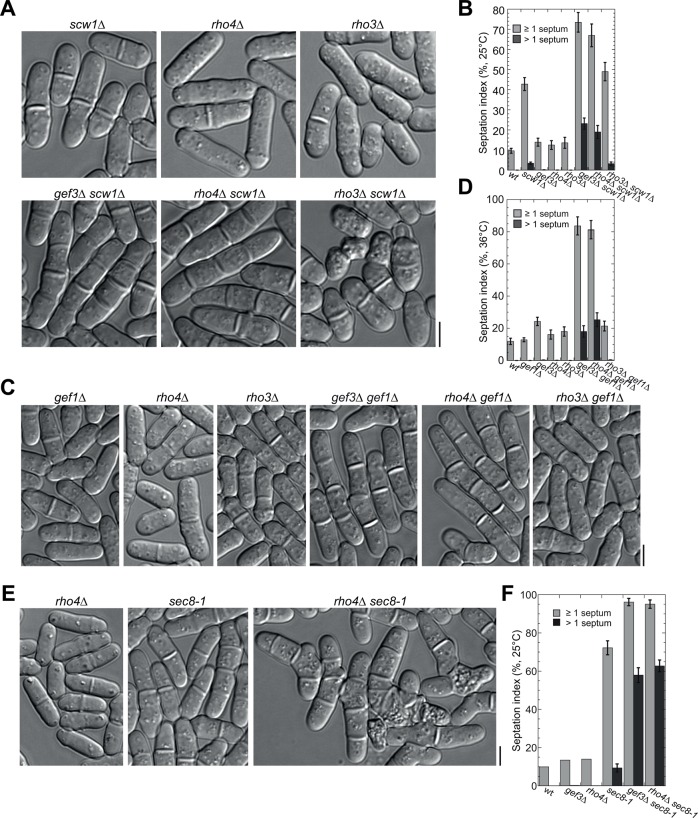
FIGURE 6:
Gef3 and Rho4 are in the same genetic pathways. (A–F) rho4∆ shows similar genetic interactions as gef3∆. DIC images (A, C) and septation indices (B, D) showing that gef3∆ and rho4∆, but not rho3∆, have similar synthetic genetic interactions with scw1∆ at 25°C (A, B) or with gef1∆ at 36°C (C, D; grown at 36°C for 6 h before imaging). (E, F) DIC images (E) and quantification (F) of synthetic interactions of exocyst mutant sec8-1 with gef3∆ and rho4∆ at 25°C. Mean ± 1 SD from three independent experiments is plotted for sec8-1 and double mutants, and only one set of representative data is shown for wt, gef3∆, and rho4∆, as they are repetitive to B. For each strain, n > 600 cells in each of the three experiments in B, D, and F. Bars, 5 μm.
TABLE 1:
Genetic interactions of double mutants described in this study.
| Mutation 1 | Mutation 2 | Genetic interaction at 25ºCa |
|---|---|---|
| gef3Δ | scw1Δ | +++ |
| gef3Δ | gef1Δ | +++b |
| gef3Δ | exo70Δ | +++b |
| gef3Δ | sec8-1 | +++ |
| gef3Δ | for3Δ | +++c |
| gef3Δ | cdc42-1625(A158V) | ++c |
| gef3Δ | myo52Δ | ++ |
| gef3Δ | art1Δ | * |
| gef3Δ | spn1Δ | +b |
| gef3Δ | spn4Δ | +b |
| gef3Δ | plo1-ts18 | – |
| gef3Δ | bgs1-191 | – |
| gef3Δ | gef2Δ | +b |
| gef3Δ | rgf1Δ | – |
| gef3Δ | rgf2Δ | – |
| gef3Δ | rgf3-s44 | * |
| gef3Δ | scd1Δ | – |
| gef3Δ | rho1-596 | – |
| gef3Δ | rho2Δ | – |
| gef3Δ | rho3Δ | +b |
| gef3Δ | rho4Δ | +b |
| gef3Δ | rho5Δ | – |
| gef3Δ | agn1Δ | ++b |
| gef3(1-270) | gef1Δ | +++b |
| gef3(1-69) | gef1Δ | +++b |
| gef3(70-525) | gef1Δ | +++b |
| gef3(ΔDH) | gef1Δ | +++b |
| gef3(ΔDH) | scw1Δ | +++ |
| gef3(ΔDH) | exo70Δ | +++b |
| gef3(ΔDH) | sec8-1 | +++ |
| rho4Δ | scw1Δ | +++ |
| rho4Δ | gef1Δ | +++b |
| rho4Δ | exo70Δ | +++b |
| rho4Δ | sec8-1 | +++ |
| rho4Δ | for3Δ | +++c |
| rho4Δ | myo52Δ | ++ |
| rho4Δ | art1Δ | * |
| rho4Δ | plo1-ts18 | – |
| rho4Δ | bgs1-191 | – |
| rho4Δ | rgf1Δ | – |
| rho4Δ | rgf2Δ | – |
| rho4Δ | rgf3-s44 | * |
| rho4Δ | scd1Δ | – |
| rho4Δ | rho1-596 | – |
| rho4Δ | rho2Δ | – |
| rho4Δ | rho5Δ | – |
| rho4Δ | agn1Δ | ++b |
| rho3Δ | scw1Δ | +++d |
| rho3Δ | gef1Δ | – |
aCells were grown exponentially in YE5S liquid medium before checking the morphology under DIC. The severity of defects compared with the parental mutants (in terms of abnormal septation) was classified as follows: +++, very strong synthetic interaction and very severe septation defects; ++, strong synthetic interaction and prominent septation defects; +, mild synthetic interaction and moderate septation defects; –, no additive septation defects; *, no additive septation defects and mild suppression of cell lysis in mutation 2.
bThe synthetic interaction is (even) more obvious at 36ºC.
cThe synthetic interaction is reflected by loss of polarity instead of septation defects.
dThe synthetic interaction is reflected by increased cell lysis instead of septation defects.
Scw1 is an RNA-binding protein, and scw1∆ has strong synthetic genetic interaction with spn1∆ (Wu et al., 2010b). gef3∆ scw1∆ double mutant displayed very strong additive defects in septation (see Table 1 for classification of genetic interactions), with more septating and multiseptated cells than single mutants (Figure 3A; see Figure 6B for quantification). Similarly, gef3∆ also had very strong synthetic interaction with gef1∆, a deletion mutant of the Cdc42 GEF gef1 (Coll et al., 2003; Hirota et al., 2003). Most gef3∆ gef1∆ cells contained one or more septa (Figure 3B; see Figure 6D for quantification). Intriguingly, gef3∆ had strong synthetic interactions with formin for3∆ mutant and cdc42-1625, a temperature-sensitive (ts) mutant of cdc42, in cell polarization (Table 1 and Supplemental Figure S2A). In addition, gef3∆ had strong synthetic interaction in cell separation and cell polarization with myo52∆ (Supplemental Figure S2B), a myosin V deletion defective in exocytic vesicle transport to the sites of polarized growth (Win et al., 2001; Bendezú and Martin, 2011). In contrast, gef3∆ had no or mild genetic interactions with mutations in arrestin art1 (which has strong synthetic interaction with spn1∆; Wu et al., 2010b), in septins spn1 and spn4, in Polo kinase plo1 (which has strong synthetic interaction with gef2∆; Ye et al., 2012), in β-glucan synthase bgs1, in other Rho GEFs gef2, rgf1-3, and scd1, and in Rho GTPases rho1 to rho5 (Table 1). Together these data suggest that Gef3 may regulate late cytokinesis together with Scw1 and Gef1.
The exocyst complex is involved in daughter cell separation by targeting the hydrolytic enzymes Eng1 and Agn1 to the septum (Martin-Cuadrado et al., 2005). We then tested whether gef3∆ had genetic interactions with exocyst mutations. The ts mutant sec8-1 was defective in cell separation even at the permissive temperature, but multiseptated sec8-1 cells are rare (Wang et al., 2002, 2003; Martin-Cuadrado et al., 2005). However, most gef3∆ sec8-1 cells contained multiple septa (Figure 3C). In addition, gef3∆ had very strong synthetic interaction with another exocyst mutant, exo70∆ (Figure 3D). Because the GTPase Rho3 modulates exocyst activity during cell separation and rho3∆ is synthetic lethal with exocyst mutants in fission yeast (Wang et al., 2003), the genetic interactions between gef3∆ and exocyst mutants suggested that Rho3 could be a potential substrate for Gef3. However, unlike Rho3, which partially depends on the exocyst complex for localization (Wang et al., 2003), no obvious defect in Gef3 localization was observed in exocyst mutants at 25 (unpublished data) or 36°C (Supplemental Figure S2C). Collectively these genetic interactions indicate that Gef3 is involved in later stages of cytokinesis to regulate septum formation and/or cell separation.
Gef3 preferentially interacts with Rho4 GTPase in vitro
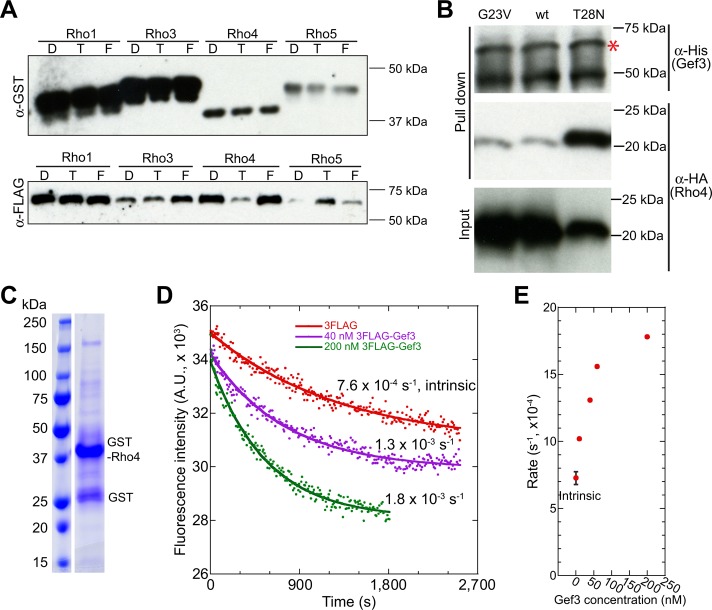
To identify the potential Rho GTPase(s) that Gef3 regulates, we tested the interactions between purified full-length (FL) Gef3 and all six Rho GTPases in vitro. The hexahistidine (6His)-tagged Gef3 was purified from Escherichia coli (Supplemental Figure S3A, asterisk) and then was pulled down by purified recombinant glutathione S-transferase (GST)–tagged Rho proteins under the nucleotide-depletion condition (Supplemental Figure S3B). Gef3 interacted with Rho1, Rho4, and Rho5 but not with Rho3 or Cdc42 (Supplemental Figure S3, B and C). Apparently, the co-IP between Rho4 and Gef3 was the most efficient. Compared to the GST control, GST-Rho4 pulled down ∼35 times more 6His-Gef3 (Supplemental Figure S3C).
Because Rho GEF functions are often regulated by posttranslational modifications such as phosphorylation (Cherfils and Zeghouf, 2013), we also purified 3FLAG-Gef3 from S. pombe (Figure 4A, asterisk) and repeated the Rho GTPase pull-down assay (Figure 4, B and C). Consistently, Gef3 still preferentially bound to Rho4 (Figure 4C), although it was also pulled down by other Rho GTPases less efficiently. Together these data suggest that Gef3 has the strongest interaction with Rho4 GTPase.
FIGURE 4:
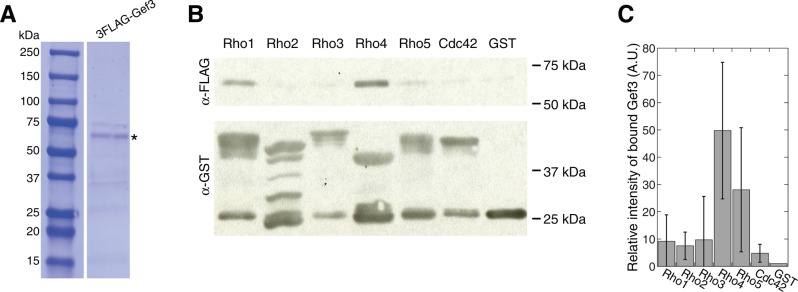
Gef3 binds to the Rho GTPase Rho4 in vitro. (A) Coomassie blue staining of SDS–PAGE showing purified 3FLAG-Gef3 from S. pombe. The asterisk marks the 3FLAG-Gef3 band with the expected size. (B, C) Gef3 binds to Rho4 in pull-down assays. Purified GST-Rho GTPases and GST control were bound to beads, depleted for nucleotides, and then incubated with purified 3FLAG-Gef3. The amount of pulled-down Gef3 was detected by Western blotting (B; representative of four independent assays) and quantified (C; mean ± 1 SD). The intensities of 3FLAG-Gef3 bands were normalized by the intensities of pulled-down Rho GTPases. The intensity of 3FLAG-Gef3 band in GST control was set as 1.
Gef3 is a Rho4 GEF in vitro
To determine whether Gef3 might act as a Rho GEF, we first tested whether Gef3 prefers GDP-bound Rho GTPase, a prerequisite for being a GEF. GST-Rho1, 3, 4, and 5 preloaded with GTPγS (a nonhydrolyzable analogue of GTP) or GDP or nucleotide depleted were incubated with yeast extract from cells expressing 3FLAG-Gef3. For Rho1 and Rho3, GTP or GDP had no obvious effect on the amount of Gef3 that was pulled down (Figure 5A). Gef3 coprecipitated with GTPγS-Rho5 more efficiently than GDP‑ or nucleotide-free Rho5 (Figure 5A). In contrast, Gef3 coprecipitated with GDP-bound and nucleotide-free Rho4 much more efficiently than GTPγS-Rho4 (Figure 5A), which suggests that Gef3 associates preferentially with inactive Rho4. Consistently, Gef3 pulled down more GDP-locked constitutive inactive Rho4-T28N compared with wt Rho4 or GTP-locked constitutive active Rho4-G23V (Figure 5B). Together the results indicate that Gef3 may function as a GEF for Rho4, but may have other GEF-independent functions with other Rho GTPases.
FIGURE 5:
Gef3 is a Rho GEF of Rho4 in vitro. (A) Gef3 coprecipitates more efficiently with GDP-Rho4 than GTPγS-Rho4. Purified GST-Rho1, 3, 4, and 5 were bound to beads, preloaded with either GDP (D) or GTPγS (T), or depleted for nucleotides (F) and incubated with yeast extract from cells expressing 3FLAG-Gef3. Experiments were performed three times for Rho4 and twice for Rho1, Rho3, and Rho5, and a representative Western blotting is shown. (B) Gef3 pulls down Rho4-T28N more efficiently than wt and Rho4-G23V. Purified 6His-Gef3 was bound to beads and incubated with yeast extracts from cells expressing 3HA-tagged Rho4-G23V, Rho4 (wt), or Rho4-T28N (Santos et al., 2003). Asterisk marks the expected 6His-Gef3 band. (C) Coomassie blue staining of SDS–PAGE showing purified GST-Rho4 from E. coli. (D, E) Gef3 accelerates nucleotide exchange of mant-GDP bound GST-Rho4 in GEF activity assays. Fluorescence intensity was monitored to reflect the disassociation of mant-GDP from Rho4. (D) Nucleotide exchanges in the presence of 3FLAG peptide as a control (intrinsic) or two concentrations of 3FLAG-Gef3 shown with the calculated rates after curve fit (see Materials and Methods). (E) Fluorescence decrease rate vs. Gef3 concentration. The intrinsic rate is the mean ± 1 SD from three independent assays.
Next we tested directly whether Gef3 stimulates the release of GDP from Rho4 in vitro. Purified GST-Rho4 (Figure 5C) was preloaded with N-methylanthraniloyl-GDP (mant-GDP), a fluorescent GDP analogue, before addition of Gef3. Both 6His-Gef3 purified from E. coli and 3FLAG-Gef3 purified from S. pombe accelerated mant-GDP release from Rho4 in a Gef3 concentration–dependent manner (Figure 5, D and E, and Supplemental Figure S3D), indicating that Gef3 has GEF activity toward Rho4 in vitro.
Gef3 and Rho4 are in the same genetic pathways to regulate cytokinesis
To elucidate the relationship between Gef3 and Rho4 in vivo, we test whether Gef3 and Rho4 function in the same genetic pathway (Figure 6 and Table 1). We expected gef3∆ and rho4∆ to have similar genetic interactions with other mutations if Gef3 activates Rho4 in vivo. Indeed, rho4∆ had very strong synthetic interactions with scw1∆, gef1∆, and exocyst mutants. The double mutants displayed severe defects in septum formation and/or cell separation (Figure 6, Table 1, and Supplemental Figure S2, D and E). The septation indices of rho4∆ scw1∆, rho4∆ gef1∆, rho4∆ sec8-1, and rho4∆ exo70∆ double mutants were almost identical to those of gef3∆ scw1∆, gef3∆ gef1∆, gef3∆ sec8-1, and gef3∆ exo70∆, respectively (Figure 6, B, D, and F, and Supplemental Figure S2E). The synthetic interaction of gef3∆ and rho4∆ with myo52∆, for3∆, and exocyst mutants suggests that Gef3 and Rho4 may play roles in actin cable– and exocyst- dependent delivery of vesicles for cell separation and polarity establishment (Figures 3, C and D, and 6, E and F, Table 1, and Supplemental Figure S2, B, D, and E). On the other hand, like gef3∆, rho4∆ had no or mild genetic interactions with mutations in arrestin art1, polo kinase plo1, β-glucan synthase bgs1, other Rho GEFs rgf1-3, scd1, and Rho GTPases rho1, rho2, and rho5 (Table 1).
Because rho3∆ is synthetic lethal with exocyst mutant sec8-1 and plays roles in late cytokinesis (Nakano et al., 2002; Wang et al., 2003; Kita et al., 2011), we also tested the genetic interactions between rho3∆ and scw1∆ or gef1∆ (Figure 6, A–D, and Table 1). rho3∆ did not show similar genetic interactions with these two mutations as gef3∆ and rho4∆. rho3∆ scw1∆ cells were shorter and rounder and had more cell lysis than single mutants (Figure 6A), whereas gef3∆ scw1∆ and rho4∆ scw1∆ had higher septation indices and more multiseptated cells (Figure 6, A and B). In addition, rho3∆ had no synthetic genetic interaction with gef1∆ (Figure 6, C and D). Taken together, the results indicate that Rho4 but not Rho3 is in the same genetic pathways as Gef3 in regulating septum formation and/or cell separation.
Recruitment and maintenance of Rho4 at the division site are affected in gef3∆ and spn1∆ cells
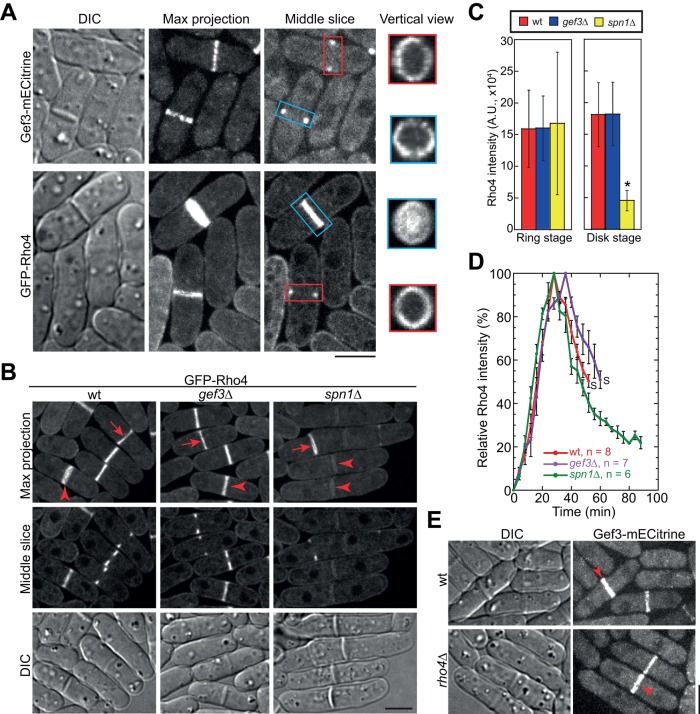
Besides activating Rho GTPases, GEFs are often involved in recruiting Rho GTPases to the functioning site in vivo (Garcia et al., 2006b; Yoshida et al., 2009; Zhu et al., 2013), so we tested whether Gef3 affects Rho4 localization to the division site. Because Rho3 modulates exocytosis and regulates cell separation (Wang et al., 2003; Kita et al., 2011), we tested its localization in gef3∆, although our in vitro binding and genetic data do not support that Rho3 is Gef3 substrate. Rho4 localizes to the cell-division site during septum formation and cell separation (Figure 7A; Nakano et al., 2003; Santos et al., 2003). Rho3 localizes to the division site, Golgi, endosomes, and plasma membrane (Supplemental Figure S4A; Nakano et al., 2002; Wang et al., 2003; Kita et al., 2011). Like Gef3, both Rho4 and Rho3 localized to ring structures when they first appeared at the division site (Figure 7A and Supplemental Figure S4A, red boxes). As the septum formed, Rho3 and Rho4 formed disk-like structures (Figure 7A and Supplemental Figure S4A, blue boxes), which is different from Gef3, as it stayed as the hollow rings even in cells with a complete septum (Figure 7A, blue box). The difference in localization between Rho4 and Gef3 implies that Rho4 activation by Gef3 may be spatially restricted.
FIGURE 7:
Recruitment and maintenance of Rho4 at the division site are affected in gef3∆ and spn1∆ cells. (A) Localization of Gef3 and Rho4 in maximum-intensity projection, the middle slice of z-stacks, or vertical view of color-boxed regions. Red boxes, cells at the start of septum formation; blue boxes, cells with complete septa. (B, C) Localization (B) and intensity quantification (C) of Rho4 at the division site in wt, gef3∆, and spn1∆ cells. (B) The maximum projection and middle slice of z-stacks are shown. Arrows, cells at the start of septum formation (ring stage); arrowheads, cells with complete septa (disk stage). (C) Fluorescence intensities (mean ± 1 SD) at the ring and disk stage. *p < 0.001 compared with wt from t test. For each strain, n > 40 cells at each stage. (D) Time-lapse analysis of GFP-Rho4 intensity at the division site during cytokinesis. The peak intensity of Rho4 for each measured cell is set as 100%. Time 0 marks the time point just before Rho4 appears at the division site (see Supplemental Figure S4B for example). Mean ± SEM for each time point. S, cell separation. Note that most spn1∆ cells did not separate at the last time point shown in the graph (88 min). (E) Gef3 localization is not affected by rho4∆. Arrowheads mark cells with complete septa. Bars, 5 μm.
Although we detected no obvious changes in Rho4 localization and intensity in either the ring (arrows) or the disk (arrowheads) in unsynchronized gef3∆ cells (Figure 7, B and C), spn1 deletion did partially affect Rho4 localization. Whereas Rho4 localization and intensity at the ring were not obviously affected in spn1∆ cells (Figure 7, B and C, arrow), Rho4 intensity at the division site in cells with a complete septum (the disk stage) was significantly reduced (Figure 7, B and C, arrowheads). To determine whether this effect is caused by the delayed cell separation in spn1∆ cells, we followed the localization and intensity of Rho4 with time-lapse microscopy (Figure 7D and Supplemental Figure S4B). In wt cells, Rho4 intensity reached the peak ∼24 min after its appearance at the division site (Figure 7D, red line). Then the intensity dropped to ∼50% of the peak value when the daughter cells separated (Figure 7D, red line). In spn1∆ cells, Rho4 recruitment was similar to wt cells, whereas Rho4 intensity decreased faster and further (Figure 7D, green line). Of interest, in gef3∆ cells, Rho4 levels at the division site peaked at ∼32 min, but the intensity dropped at the similar rate as in wt cells (Figure 7D, purple line). On the other hand, the localization and intensity of Rho3 in neither the ring (arrows) nor the disk (arrowheads) were affected by gef3∆ or spn1∆ (Supplemental Figure S4, C and D). Moreover, Gef3 localization and intensity at the division site were not affected by rho4∆ or rho3∆ (Figure 7E and Supplemental Figure S4E). Together these data suggest that septins regulate the localization of both Rho4 and Gef3 during cytokinesis and that Gef3 only plays a minor role in Rho4 recruitment.
Gef3 modulates the localization of glucanases Eng1 and Agn1 at the division site
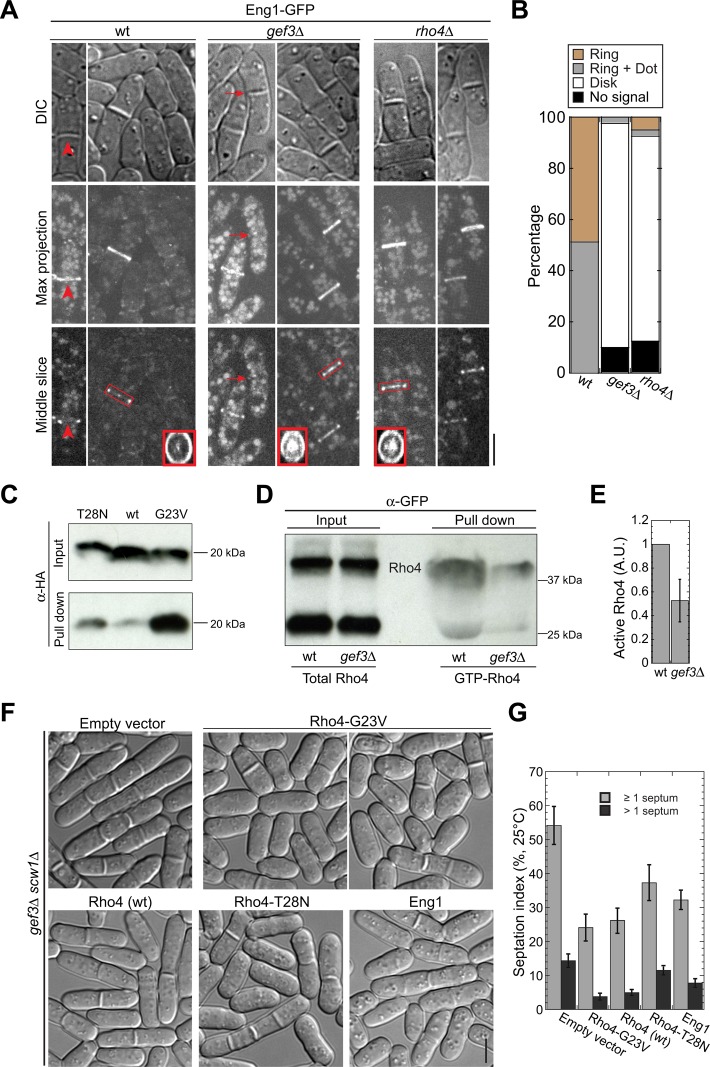
Next we tested whether Gef3 affects the localizations of potential Rho4 downstream effectors, which may reflect the localization of active Rho4. Because Rho4 regulates the secretion of β-glucanase Eng1 and α-glucanase Agn1 during cell separation (Santos et al., 2005), we checked the localizations of Eng1 and Agn1 in gef3∆. In wt cells with complete septa, Eng1 localized to nonconstricting rings in ∼50% of cells (Figure 8A, arrowhead), whereas the rest of the cells also had a weak Eng1 dot signal in the center of the septa besides the ring signal (ring plus dot; Figure 8, A and B; wt, red box). However, Eng1 ring signal was lost at the division site in 10% of gef3∆ cells with complete septa (Figure 8, A and B, arrows). In the majority of gef3∆ cells (90%), Eng1 formed a nonuniform disk at the septum, with brighter signals at the ring and the center (Figure 8, A and B, red box). Consistent with a previous report (Santos et al., 2003, 2005), rho4∆ cells showed the same mislocalization pattern for Eng1 (Figure 8, A and B), which is also similar to reported Eng1 localization in exocyst mutants (Martin-Cuadrado et al., 2005). In addition, as in rho4∆ cells (Santos et al., 2005), Agn1 localization at the division site was undetectable or significantly reduced in gef3∆ cells (Supplemental Figure S5A). Moreover, both gef3∆ and rho4∆ were synthetic sick with agn1∆ in terms of cell separation (Supplemental Figure S5B), which was expected, given that agn1∆ eng1∆ displayed additive defects in cell separation (Dekker et al., 2004). Thus Gef3 modulates the normal localizations of glucanases Eng1 and Agn1, likely by regulating Rho4 activity.
FIGURE 8:
Gef3 regulates the localization of glucanases and Rho4 activity in vivo. (A, B) Eng1 localization is affected in gef3∆ cells. Micrographs (A) and quantification (B) showing that Eng1 localization at division site is affected in gef3∆ and rho4∆ cells. The arrowhead indicates a wt cell with Eng1 in the nonconstricting ring. Vertical views of the regions marked with red boxes are shown at the bottom corners. Arrows mark a gef3∆ cell without an intact Eng1 ring at the division site. (B) Only cells with complete septa were counted. For each strain, n > 40 cells. (C) RBD pulls down constitutively active Rho4-G23V more efficiently than wt and constitutively inactive Rho4-T28N. Yeast extracts from cells expressing 3HA tagged wt or mutant Rho4 were incubated with 200 μg of GST-RBD. The amount of Rho4 was detected by Western blotting. (D, E) Western blotting (D) and quantification (E) showing that gef3∆ cells have less active Rho4 than wt cells. (E) Mean ± 1 SD from three independent experiments. The level in wt is normalized to 1. (F, G) Overexpression of Rho4 or Eng1 rescues the septation defects in gef3∆ scw1∆ cells. DIC images (F) and septation index (G) of gef3∆ scw1∆ cells with plasmids grown under inducing conditions in liquid EMM5S – leucine for 24 h. (G) Mean ± 1 SD from three independent experiments, and n > 600 for each strain in each experiment. Bars, 5 μm.
Rho GEF Gef3 activates Rho4 GTPase in vivo
The identical effects of gef3∆ and rho4∆ on Eng1 and Agn1 localization suggest that Gef3 activates Rho4 for cell separation in vivo. To test this idea, we performed pull-down experiments using cell extracts to compare the amount of active Rho4 using Rho-binding domain (RBD) of human Rhotekin protein. Because RBD preferentially binds to GTP-RhoA in mammalian cells (Reid et al., 1996; Ren et al., 1999), we first tested whether GST-RBD prefers active Rho4 in S. pombe. GST-RBD pulled down active GTP-locked Rho4-G23V more efficiently than wt Rho4 and inactive Rho4-T28N (Figure 8C). Next we compared active Rho4 in wt and gef3∆ cells. gef3∆ cells had about half the amount of GTP-Rho4 as wt cells (Figure 8, D and E), suggesting that Gef3 activates Rho4 in vivo.
Our genetic data provide further support that Gef3 activates Rho4 in vivo. Overexpression of constitutively active Rho4-G23V reduced the septation index of gef3∆ scw1∆ cells from ∼54% (with empty vector) to ∼24% after 24 h of induction. The percentage of multiseptated cells was also reduced from ∼14 to ∼4% (Figure 8, F and G). Overexpression of wt and the constitutively inactive Rho4-T28N also partially rescued gef3∆ scw1∆ mutant (Figure 8, F and G), probably due to the basal or residual Rho GTPase activity. Moreover, overexpression of Eng1 (Figure 8, F and G) and Agn1 (unpublished data) also partially rescued the septation defects in gef3∆ scw1∆. In contrast, overexpression of wt Rho4, Rho4-G23V, and Rho4-T28N did not rescue scw1∆ cells (Supplemental Figure S5, C and D), indicating that the rescue of gef3∆ scw1∆ is specific to the Gef3-Rho4 pathway. Thus high levels of active Rho4 and its potential effectors Eng1 and Agn1 can compensate the loss of Gef3. Taken together, these data suggest that Gef3 activates Rho4 GTPase as a GEF in vivo.
N-terminus of Gef3 is important for its localization, and DBL homology domain and C-terminus of Gef3 are critical for its function
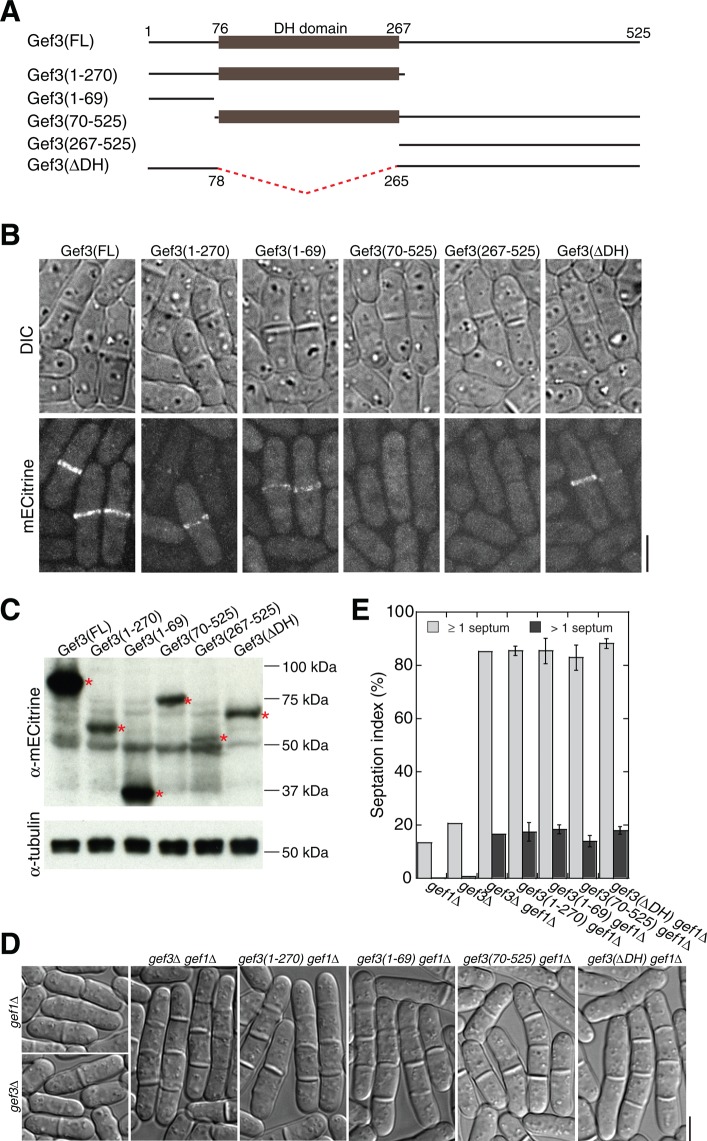
To understand how Gef3 localizes and functions, we performed domain analyses. Most GEFs of Rho GTPases contain both DBL homology (DH) and pleckstrin homology (PH) domains (Cherfils and Zeghouf, 2013), whereas Gef3 only contains the DH domain (amino acids [aa] 76–267, Figure 9A; Garcia et al., 2006b). Of interest, both Gef3 C-terminal truncations, Gef3(1–270) and Gef3(1–69), together with the truncation without the DH domain (Gef3∆DH), still localized to the rings at the division site, although the signals were weaker than that of FL Gef3 (Figure 9, A and B). In contrast, neither of the N-terminal truncations Gef3(70–525) and Gef3(267–525) localized to the division site (Figure 9, A and B), although all the truncations were expressed and detected by Western blotting (Figure 9C).
FIGURE 9:
Domain analyses for Gef3. (A) Schematics of Gef3 domains and truncations constructed. (B, C) Localizations (B) and protein levels (C) of FL and truncated Gef3. Asterisks on Western blotting mark the bands with the expected sizes for FL and truncated Gef3. Tubulin was blotted as a loading control. (D, E) DIC images (D) and septation index (E) showing synthetic interactions between gef3 truncations and gef1∆. Cells were grown at 36°C for 6 h before imaging. (E) Mean ± 1 SD from three independent experiments, and n > 600 cells for each strain in each experiment. Bars, 5 μm.
Consistent with the idea that Gef3 regulates cytokinesis as a Rho4 GEF, Gef3∆DH was not sufficient for Gef3 function, although it still localized to the division site. gef3∆DH had synthetic interactions with gef1∆, scw1∆, and exocyst mutant exo70∆ and sec8-1 mutants (Figure 9, D and E, and Table 1), and the phenotypes of the double mutants resembled those of gef1∆ gef3∆ (Figure 9, D and E), scw1∆ gef3∆, and sec8-1 gef3∆. Of interest, the two C-terminal truncation mutants gef3(1-270) and gef3(1-69) also had synthetic interactions with gef1∆, scw1∆, and sec8-1 mutants (Figure 9, D and E, and Table 1). Thus aa 271–525 and the DH domain are both important for Gef3 function. Given that Gef3 lacks the conventional PH domain, it is possible that the C-terminus may become necessary to facilitate Gef3's function as a GEF. Together these data indicate that the N-terminal aa 1–69 are required for Gef3 localization, and the C-terminus including the DH domain is required for Gef3 function during septation.
DISCUSSION
In this study, we find that Gef3, unlike the other six predicted (or proven) Rho GEFs, depends on septins for localization to the division site. Gef3 plays roles in the regulation of septation during fission yeast cytokinesis. With combined genetic and biochemical experiments, we identify GTPase Rho4 as a substrate for the Rho GEF Gef3.
Gef3 is a Rho GEF for the GTPase Rho4
The putative Rho GEF Gef3 was the least characterized among the predicted Rho GEFs in S. pombe. In this study, several lines of evidence support that Gef3 has GEF activity toward the GTPase Rho4: 1) purified Gef3 coprecipitates most efficiently with purified Rho4 in vitro when nucleotides are depleted (Figure 4 and Supplemental Figure S3); 2) Gef3 prefers to bind GDP‑bound Rho4 (Figure 5, A and B); 3) Gef3 accelerates nucleotide exchange of Rho4 in vitro (Figure 5, D and E); 4) Gef3 and Rho4 are in the same genetic pathways (Figure 6 and Table 1); 5) gef3∆ and rho4∆ have similar effects on the localization of Eng1 (Figure 8, A and B), a potential effector of active Rho4; 6) gef3∆ cells have less active Rho4 than wt cells (Figure 8, C–E); 7) overexpression of constitutively active and wt Rho4 rescues septation defects in gef3∆ scw1∆ but not scw1∆ cells (Figure 8, F and G, and Supplemental Figure S5, C and D); and 8) the DH domain of Gef3 is important for its function (Figure 9). Together our data indicate that Gef3 is a GEF of Rho4 GTPase.
We previously found that putative Rho GEF Gef2 regulates Rho4 localization in vivo and binds to Rho4 in vitro, suggesting that Gef2 could be another GEF of Rho4 (Zhu et al., 2013). Here we find that gef2∆ is slightly synthetic sick with gef3∆ (Table 1) in septation. This suggests that both Gef2 and Gef3 are involved in regulating Rho4 during septation. However, they may use different mechanisms, given that gef2∆ reduces Rho4 localization at the division site (Zhu et al., 2013) but gef3∆ only mildly delays Rho4 recruitment (Figure 7D). Moreover, Gef2 localizes to the contractile ring and Gef3 to the septin ring, and thus their regulations of Rho4 may be spatially and temporally separated.
Cell separation in S. pombe is achieved by the concerted actions of β-glucanase Eng1 and α-glucanase Agn1 to digest the primary septum and the surrounding cell wall on cell sides (Martin-Cuadrado et al., 2003; Dekker et al., 2004; Sipiczki, 2007). The proper localizations of Eng1 and Agn1 require the functions of the exocyst complex, septins, and active Rho4 (Martin-Cuadrado et al., 2005; Santos et al., 2005). Rho4 also regulates septum formation, given that rho4∆ cells have thicker secondary septa with abnormal structure by electron microscopy (Nakano et al., 2003) and rho4∆ has very strong synthetic interaction with gef1∆ (Figure 6, C and D), a deletion mutant of the Cdc42 GEF involved in septum formation (Hirota et al., 2003). Further studies are needed to distinguish whether the defects in gef3∆ and gef3∆-containing double mutants are caused by cell separation, septum formation, or both.
Gef3 may have Rho4-independent functions
The mild synthetic septation defects in gef3∆ rho4∆ double mutant (Table 1) and partial colocalization of Gef3 and Rho4 (Figure 7A) suggest that Gef3 may have Rho4-independent functions, which may or may not depend on its GEF activity. Besides Rho4, Gef3 also interacts with Rho1 and Rho5 in in vitro binding assays (Figure 4 and Supplemental Figure S3). Both Rho1 and Rho5 are activated by several Rho GEFs (Tajadura et al., 2004; Mutoh et al., 2005; Nakano et al., 2005; Garcia et al., 2006a, 2009; Rincon et al., 2006) and have multiple downstream effectors (Arellano et al., 1996, 1999; Nakano et al., 2005; Rincon et al., 2006; Prosser et al., 2011). However, the fact that the GDP-bound Rho1 and Rho5 are not preferred in the binding assay (Figure 5A) does not support that Gef3 is a GEF for Rho1 and Rho5. Consistently, the lack of synthetic genetic interactions between gef3∆ and mutations in rho1 and rho5, in their known Rho GEFs (rgf1, rgf2, and rgf3), and in their effectors or regulators (bgs1 and art1) further indicates that Gef3 is not a major GEF for Rho1 or Rho5 (Table 1).
During the final preparation of our manuscript, Muñoz et al. (2014) reported that Gef3 physically interacts with active, GTP-bound Rho3 when they were overexpressed, but the level of active Rho3 in vivo was not affected by Gef3 levels, indicating that Gef3 acts as a scaffold but not a GEF to stabilize GTP-Rho3. Our in vitro binding assays agree with their finding that Gef3 is not a GEF of Rho3. However, our genetic data do not support strong functional connection between Gef3 and Rho3 in vivo. Further studies on Gef3–Rho3 physical interaction would be of interest.
Gef3 has the least efficient physical interaction with nucleotide-free Cdc42 (Figure 4 and Supplemental Figure S3); thus it is unlikely that Gef3 is a GEF for Cdc42. Gef1 and Scd1 are known GEFs for Cdc42 (Coll et al., 2003; Hirota et al., 2003). The very strong synthetic interaction between gef3∆ and gef1∆ in septation indicates that Gef3 and Gef1/Cdc42 may play an overlapping function in septum formation. The polarity defects in gef3∆ cdc42-1625 and gef3∆ for3∆ (Table 1 and Supplemental Figure S2A) suggest that Gef3 also has a function in cell polarity. Although Gef3 only localizes to the division site and transiently to the new cell ends, its role in cell polarization is consistent with growth-polarity defects reported in mutants with defects in final stages of cytokinesis (Bohnert and Gould, 2012). Alternatively, Gef3 and Rho4 may indirectly regulate cell polarity by affecting exocyst-dependent delivery of vesicles to polarized growth sites, especially given that gef3∆ and rho4∆ are both synthetic sick with myo52∆, a mutant defective in long-distance vesicle transport along actin cables (Win et al., 2001; Bendezú and Martin, 2011).
Conserved septins-anillin-Rho GEF module at cell-division site
Septins are usually associated with the plasma membrane and function as a scaffold or diffusion barrier to regulate cellular processes, including cytokinesis and cell polarization (Kinoshita, 2003; Bertin et al., 2008; Oh and Bi, 2011). In budding yeast, septins are essential and critical for localization of many proteins to the mother-bud neck/division site (Oh and Bi, 2011). In fission yeast, very few proteins have been identified to be septin dependent at the division site, which agrees with the mild phenotype of septin deletion in this yeast (Longtine et al., 1996; Berlin et al., 2003; Tasto et al., 2003; An et al., 2004). Consistent with a recent report (Muñoz et al., 2014), we found that Gef3 localization is completely abolished in spn1∆ and severely compromised without anillin Mid2 (Figure 2A). The spn1∆ mutant also affects Rho4 maintenance at the septum (Figure 7, B–D). Moreover, our co-IP data are consistent with the idea that Gef3, septins, and anillin Mid2 form a protein complex (Figure 2C).
It is well known that septins-Bud3-Bud4 (anillin-like) and septins–Cdc24 (Cdc42 GEF)–Cdc42 modules are essential for division-site selection and cell polarization (Chant et al., 1995; Gladfelter et al., 2002; Iwase et al., 2006; Bi and Park, 2012; Kang et al., 2013; Sadian et al., 2013). Recently Kang et al. (2014) reported that Bud3 is a GEF of Cdc42 during polarity establishment. Although the functions of budding yeast Bud3 and fission yeast Gef3 are apparently different and Gef3 is only one-third as long as Bud3, both of them contain only a DH domain instead of typical DH-PH domain as in other Rho GEFs. Considering the high divergence between budding yeast and fission yeast, the septins-anillin-Rho GEF complex might present one of the most conserved modules for septin functions. Consistently, Bud3 homologues in other fungi have been suggested to function as a Rho4 GEF for contractile-ring and septum formation (Justa-Schuch et al., 2010; Si et al., 2010). In animal cells, interactions between septins, anillin, and the Rho GEFs Ect2 and Pebble during cytokinesis have also been reported (Oegema et al., 2000; Hickson and O’Farrell, 2008; Silverman-Gavrila et al., 2008; Frenette et al., 2012). Moreover, a mammalian septin Sept9b was found to bind to a Rho GEF (SA-RhoGEF) and regulate its localization and GEF activity (Nagata and Inagaki, 2005). Thus the septins-anillin-Rho signaling is a highly conserved module in cell polarization and cytokinesis.
In conclusion, we revealed the role of Gef3 in septation during cytokinesis and found that Gef3 physically interacts with the septins–anillin complex and its localization depends on septins. We found that Rho4 GTPase is the substrate of Gef3. It will be interesting to test whether Gef3 also regulates other Rho GTPases, although not necessarily acting as a GEF. Finally, we propose that septins-anillin-Rho GEFs is a highly conserved module functioning at the cell-division site.
MATERIALS AND METHODS
Strains and genetic, molecular, and cellular methods
Strains used in this study are listed in Supplemental Table S1. PCR-based gene targeting and standard yeast genetics were used to construct strains (Moreno et al., 1991; Bähler et al., 1998b). All tagged and truncated strains for gef3 and rho3 are regulated under their endogenous promoters and integrated into native chromosomal loci, except the overexpression strain JW6030 for purifying 3FLAG-Gef3 from S. pombe, which is integrated at gef3 native locus under the control of inducible 3nmt1 promoter (Maundrell, 1990). GFP-Rho4 strain contains Prho4-GFP-rho4 integrated at leu1 locus with the endogenous rho4 deleted (a gift from Beatriz Santos, Universidad de Salamanca, Spain; Santos et al., 2003).
The N-terminal tagging and truncations of gef3 and rho3 were made by first cloning the promoter regions of gef3 (−439 to +6) and rho3 (−193 to +6) into the pFA6a-kanMX6-P3nmt1-mECitrine plasmid at BglII and PacI sites to replace the 3nmt1 promoter. The resulting plasmids JQW755 and JQW756 were sequenced and used as templates to amplify kanMX6-Pgef3-mECitrine or kanMX6-Prho3-mECitrine fragments flanked with homologous sequences from the designated positions at gef3 or rho3 locus, which were then transformed into wt strain as described (Bähler et al., 1998b). We used PCR to confirm the positive transformants. The functionalities of Gef3 tagged at N- or C-terminus were tested by crossing the strains to gef1∆. The growth and morphology of the double mutants were similar to those of gef1∆ single mutant at different temperatures, indicating that the tagged proteins are functional.
The Gef3(∆DH) truncation was made as follows: Pgef3-gef3-mECitrine-kanMX6 was cloned into Topo vector (JQW769). Then the sequence encoding aa 79–264 of Gef3 was removed by PCR as described (Lee and Wu, 2012) to obtain the plasmid JQW770. This resulting plasmid was sequenced and then used as the template to amplify Pgef3-gef3(∆DH)-mECitrine-kanMX6 fragment. The purified PCR product was transformed into gef3∆ strain (JW5492) to obtain the gef3(∆DH) truncation mutant with the mECitrine tag at its C-terminus.
Cells were grown in exponential phase in yeast extract medium with five supplements (YE5S) or Edinburgh minimal medium plus five supplements (EMM5S) at 25°C for 36–48 h before microscopy or other experiments as previously described (Wu et al., 2006; Coffman et al., 2013), except where noted. Plasmids for overexpression of Eng1 (Santos et al., 2005) and wt, constitutively active, and constitutively inactive Rho4 (gifts from Beatriz Santos; Santos et al., 2003) were under the control of 3nmt1 promoter. The plasmids were transformed into yeast as described (Bähler et al., 1998b) and EMM5S – leucine was used to induce the expression of 3nmt1 promoter.
Microscopy and data analysis
Microscopy was performed as previously described (Coffman et al., 2009, 2013; Wang et al., 2014). Briefly, cells in exponential phase in liquid cultures were collected by centrifugation at 5000 rpm for 30 s. For fluorescence microscopy, cells were washed twice with EMM5S to reduce autofluorescence. Live-cell microscopy was performed using a thin layer of liquid EMM5S with 20% gelatin (Sigma-Aldrich, St. Louis, MO) and 5 μM n-propyl-gallate and observed at 23–25°C except where noted.
Three microscopy systems were used to acquire images with Nikon 100×/1.4 numerical aperture Plan-Apo objective lenses (Nikon, Melville, NY). For fluorescence images and time-lapse movies, a spinning disk confocal system (UltraVIEW Vox CSUX1 system; PerkinElmer, Waltham, MA) with 440-, 488-, 515-, and 561-nm solid-state lasers and a back-thinned electron-multiplying charge-coupled device (CCD) camera (C9100-13; Hamamatsu, Bridgewater, NJ) on a Nikon Ti-E microscope without binning was used. For most differential interference contrast (DIC) images used for the quantifications of septation defects, a Nikon Eclipse Ti inverted microscope equipped with a Nikon cooled digital camera DS-Ql1 was used. Owing to the high cytoplasmic signals (and/or autofluorescence), we imaged cells expressing Eng1-GFP or Agn1-GFP using the UltraVIEW ERS system, which has lower sensitivity but better spatial resolution. The spinning disk confocal system (PerkinElmer) is equipped with 488-nm argon ion laser and a cooled CCD camera (ORCA-AG; Hamamatsu) on a Nikon Eclipse TE2000-U microscope.
Image analyses were performed using Volocity (PerkinElmer) and ImageJ (National Institutes of Health, Bethesda, MD). Images in figures are maximum-intensity projections of z-sections spaced at 0.3–0.5 μm except where noted. For Figure 8A, the z-spacing is 0.1 μm. To quantify the fluorescence intensity of Rho3 and Rho4 at the division site, we summed the intensity from 13 z-sections spaced at 0.4 μm (Coffman et al., 2011). Then a rectangular region of interest (ROI) that is big enough to include >90% of ring or septum signal was used to measure the intensity. The intensity of the cytoplasmic signal in a concentric rectangular ROI (approximately twice as big as the ROI that elongated along the cell long axis) was used for background subtraction (Wu and Pollard, 2005; Wu et al., 2008; Coffman and Wu, 2012). The p values in this study were calculated using two-tailed Student's t tests.
FRAP analysis
FRAP assays were performed using the photokinesis unit on UltraVIEW Vox confocal system, similar to the assays described previously (Coffman et al., 2011; Zhu et al., 2013; Wang et al., 2014). Briefly, the middle focal plane of cells with complete septa was chosen to bleach the fluorescence signal at the division site. Selected ROIs were bleached to <50% of the original fluorescence intensity after four prebleach images were collected. One hundred images with 1-s delay were collected after photobleaching. The images were then corrected for background and photobleaching during image acquisition. The prebleach intensity of the ROI was normalized to 100% and the intensity just after bleaching to 0%. The end time of the bleach was set as time 0. Intensities of every three consecutive postbleaching time points were averaged to reduce noise. Then the intensity data were plotted and fitted using the exponential equation y = m1 + m2 exp(−m3x), where m3 is the off-rate (KaleidaGraph; Synergy Software, PA). The half-time of recovery was calculated using the equation t1/2 = (ln2)/m3.
IP and Western blotting
IP assays and Western blottings were carried out as previously described (Laporte et al., 2011; Lee and Wu, 2012; Wang et al., 2014) with the following modifications: 1) 200 mg (Figure 2C) and 40 mg (Figure 9C) of lyophilized cells for each strain were used for IPs and Western blottings, respectively, due to the low abundance of Gef3; 2) The monoclonal anti-Myc antibody (1:1000 dilution; 9E10; Santa Cruz Biotechnology, Santa Cruz, CA) was used to detect Gef3-13Myc. Rho GTPases were detected by monoclonal anti-GST antibody (3G10/1B3, 1:10,000 dilution; NB600-446; Novus Biologicals, Littleton, CO). A monoclonal antibody against FLAG (1:2000 dilution; F1804; Sigma-Aldrich) was used to detect 3FLAG-Gef3, and 6His-Gef3 was detected by anti-His antibody (1:10,000 dilution; 631212; Clontech, Mountain View, CA). Secondary anti-mouse antibody was used at 1:10,000 dilution for all the primary antibodies mentioned. In addition, triple-hemagglutinin (HA)–tagged wt and mutant Rho4 were detected by anti-HA antibody (1:2000 dilution; 3F10; Roche, Mannheim, Germany), and a secondary anti-rat antibody (A5795; Sigma-Aldrich) was used at 1:4000 dilution.
Protein purification
Purification of 6His-tagged protein from E. coli was carried out as previously described (Zhu et al., 2013) with the following modifications: BL21(DE3)pLysS cells (69451; Novagen, EMD Chemicals, Darmstadt, Germany) carrying 6His-Gef3 expression plasmid (JQW734) were grown at 37ºC for 6 h after adding 1 mM isopropyl-β-d-thiogalactoside (IPTG; Saha and Pollard, 2012).
Purification of 3FLAG-Gef3 from S. pombe was performed similarly to previously described (Wang et al., 2014). Briefly, 3FLAG-Gef3 was overexpressed in spn1∆ background, since Gef3 was released to cytoplasm (Figure 2A). The 3nmt1-3FLAG-Gef3 spn1∆ cells (JW6030) were induced in EMM5S at 30ºC for 48 h and collected for lyophilization. Approximately 3 g of lyophilized cells were broken by grinding, and then the cell powder was mixed with 40 ml of cold HK extraction buffer (25 mM Tris, pH 7.5, 1% NP40, 300 mM NaCl, 5 mM EDTA, 15 mM ethylene glycol tetraacetic acid [EGTA], 60 mM β-glycerophosphate, 500 μM Na3VO4, 10 mM NaF, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], and protease inhibitors [Roche]). The cell extract was cleared by two rounds of centrifugation at 4°C (21,000 rpm for 10 min; 38,000 rpm for 30 min). Then cell extract (∼35 ml) was incubated with 600 μl of beads covalently coated with anti-FLAG antibody (F2426; Sigma-Aldrich) at 4°C for 1.5 h. The beads were collected by centrifugation at 4000 rpm for 2 min and washed once with equal volume (35 ml) of HK extraction buffer, four times with an equal volume (35 ml) of washing buffer (25 mM Tris, pH 7.5, 300 mM NaCl, 5 mM EDTA, 500 μM Na3VO4, 10 mM NaF, 1 mM PMSF, and 1 mM DTT), and twice with 1 ml of washing buffer. To elute 3FLAG-Gef3, we incubated beads with 1 ml of 3FLAG peptide (F4799; Sigma-Aldrich) at a final concentration of 200 μg/ml in the washing buffer at 4°C for 30 min.
GST and GST-tagged Rho1–Rho5 and Cdc42 were expressed and purified from BL21(DE3)pLysS cells (induced with 0.5 mM IPTG at 15°C for 6 h) using glutathione–Sepharose beads (17-5132-01; GE Healthcare, Chalfont St Giles, United Kingdom) as described previously (Zhu et al., 2013). Bead-bound GST or GST-Rhos were treated to deplete nucleotides or load GDP/GTPγS for the following pull-down assays as described next. The GST-Rho4 used in the GEF assays was eluted by 100 mM reduced glutathione.
In vitro pull-down assays for Gef3 and Rho GTPases
For pull-down assays of nucleotide-depleted Rho GTPases, the purified 6His-Gef3 or 3FLAG-Gef3 was dialyzed into the final binding buffer (25 mM 3-(N-morpholino)propanesulfonic acid [MOPS], pH 7.2, 60 mM β-glycerophosphate, 15 mM p-nitrophenyl phosphate, 1 mM DTT, 1% Triton X-100, 1 mM PMSF, and protease inhibitor tablets [Roche]) as previously described (Iwaki et al., 2003; Zhu et al., 2013). The beads with GST control and GST-Rho proteins were incubated with buffer containing 50 mM Tris, pH 7.5, 1 mM DTT, and 5 mM EDTA to deplete nucleotides at 30°C for 10 min. Then 500 μl of ∼0.25 μM 3FLAG-Gef3 or 6His-Gef3 in binding buffer was added to 30 μl of beads with each nucleotide-depleted Rho protein and incubated at 4°C for 1 h. After incubation, glutathione beads were washed with 1 ml of binding buffer three times, and the bound proteins were detected by Western blotting.
The pull-down assays for GDP- or GTPγS-preloaded Rho1, Rho3, Rho4, and Rho5 were performed as described (Kozminski et al., 2003; Singh et al., 2008) with minor modification. Briefly, 30 μl of GST or GST-Rho bound beads were first incubated in Buffer I (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM DTT, 10 mM EDTA, 10% glycerol, 0.1% Triton X-100, and protease inhibitors [Roche]) for 1 h at room temperature to deplete nucleotide. Then beads were washed with Buffer I containing 5 mM MgCl2 and then incubated with Buffer I containing 5 mM MgCl2 plus 0.5 mM GTPγS or 0.5 mM GDP (Roche) for 30 min at room temperature to load the desired nucleotide. To stabilize the nucleotide-bound Rho GTPases, the beads were resuspended in Buffer I without EDTA but containing 10 mM MgCl2 plus 0.5 mM GTPγS or 0.5 mM GDP and incubated 20 min at room temperature. To pull down Gef3, the beads were incubated with yeast extracts prepared from 50 mg of lyophilized cells expressing 3FLAG-Gef3 (JW6030) at 4°C for 1.5 h. After incubation, beads were washed with 1 ml of Buffer I without EDTA but containing 10 mM MgCl2 three times, and the bound proteins were detected by Western blotting.
To test the preference of Gef3 in binding with Rho4 (Figure 5B), we induced cells transformed with 3HA-Rho4 plasmids (wt, G23V, and T28N) in EMM5S – leucine for 20 h before collection for cell extracts. We incubated 30 μl of Talon metal affinity resin (635501; Clontech) carrying 6His-Gef3 with yeast extracts from 30 mg of lyophilized cells at 4°C for 1.5 h.
GEF activity assay
In vitro nucleotide exchange assay was performed as described (Kang et al., 2014) with modifications. Briefly, purified GST-Rho4 was incubated with 50-fold molar excess of mant-GDP (Invitrogen, Carlsbad, CA) in a nucleotide-loading buffer (20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 5 mM EDTA, and 2 mM DTT) for 25 min at 25°C. After termination of nucleotide loading with 20 mM MgCl2, excess mant-GDP was removed by running through NAP™-5 gravity column (17-0853-01; GE Healthcare), and mant-GDP–loaded GST-Rho4 was switched into the GEF assay buffer (20 mM Tris-HCl, pH 7.5, 50 mM NaCl, 10 mM MgCl2, 1% glycerol, and 1 mM DTT). A GEF assay was performed by mixing 40 μl of ∼2 μM mant-GDP-loaded Rho4 with 5 μl of GEF assay buffer containing 3FLAG-Gef3 (with different concentrations) or 3FLAG in the presence of 400 μM GDP. Fluorescence intensity was monitored at ∼20°C every 10 s using the Infinite M1000Pro plate reader (Tecan Group, Männedorf, Switzerland), with excitation at 360 nm and emission at 440 nm. Data were fitted with a monophasic exponential curves using y = m1 − m2 exp(−m3x), where m3 is the fluorescence decrease rate plotted in Figure 5, D and E (KaleidaGraph; Synergy Software, PA).
Active Rho4 pull-down assay
We carried out active Rho pull-down assays using commercial GST-RBD kit (16116; Thermo Scientific, Waltham, MA). Briefly, 50 mg of lyophilized cells were lysed with 600 μl of the lysis buffer (25 mM Tris-HCl, pH 7.2, 150 mM NaCl, 5 mM MgCl2, 1% NP40, 5% glycerol, and protease inhibitors). The cell extracts were cleared by two rounds of centrifugation at 4°C (5000 rpm for 5 min; 15,000 rpm for 30 min). Protein concentration was measured using the Bradford method. Then 200 μg of GST-RBD protein coupled to glutathione–Sepharose beads was incubated with 3 mg of proteins from the cell extracts at 4°C for 1.5 h to immunoprecipitate active Rho GTPases. The beads were then washed three times with lysis buffer, and the bound proteins were detected by Western blotting. To test whether GST-RBD prefers active Rho4 (Figure 8C), we incubated bead-bound 200 μg of GST-RBD at 4°C for 1.5 h with yeast extracts from 30 mg of lyophilized cells expressing 3HA-Rho4 plasmids (wt, G23V, and T28N). Cells transformed with the Rho4 plasmids were induced in EMM5S – leucine for 20 h before collection.
Supplementary Material
Acknowledgments
We thank Mohan Balasubramanian, Sophie Martin, Masaaki Miyamoto, Pilar Pérez, and Beatriz Santos for strains and plasmids; the James Hopper, Anita Hopper, Paul Herman, and Stephen Osmani laboratories for sharing equipment and reagents; and Pil Jung Kang, Hay-Oak Park, and members of the Wu laboratory for helpful suggestions and discussion. We appreciate I-Ju Lee for critical reading of the manuscript. This work was supported by the Ohio State University start-up fund to D.S.K., a Pelotonia Graduate Fellowship to Y.-H. Z., and National Institutes of Health Grant R01GM086546 to J.-Q.W.
Abbreviations used:
- aa
amino acid
- DH
DBL homology
- DIC
differential interference contrast
- EMM5S
Edinburgh minimal medium plus five supplements
- FL
full length
- FRAP
fluorescence recovery after photobleaching
- GEF
guanine nucleotide exchange factor
- IP
immunoprecipitation
- mant-GDP
N-methylanthraniloyl-GDP
- mCFP
monomeric cyan fluorescent protein
- mECitrine
monomeric enhanced Citrine
- mEGFP
monomeric enhanced green fluorescent protein
- PH
pleckstrin homology
- RBD
Rho-binding domain
- ROI
region of interest
- SPB
spindle pole body
- ts
temperature sensitive
- wt
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-07-1196) on November 19, 2014.
*Present address: Department of Plant Pathology, The Ohio State University, Columbus, OH 43210.
REFERENCES
- Adamo JE, Rossi G, Brennwald P. The Rho GTPase Rho3 has a direct role in exocytosis that is distinct from its role in actin polarity. Mol Biol Cell. 1999;10:4121–4133. doi: 10.1091/mbc.10.12.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams AE, Johnson DI, Longnecker RM, Sloat BF, Pringle JR. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu M, Berro J, Pu KM, Tebbs IR, Pollard TD. Cytokinetic nodes in fission yeast arise from two distinct types of nodes that merge during interphase. J Cell Biol. 2014;204:977–988. doi: 10.1083/jcb.201307174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almonacid M, Celton-Morizur S, Jakubowski JL, Dingli F, Loew D, Mayeux A, Chen J-S, Gould KL, Clifford DM, Paoletti A. Temporal control of contractile ring assembly by Plo1 regulation of myosin II recruitment by Mid1/anillin. Curr Biol. 2011;21:473–479. doi: 10.1016/j.cub.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Morrell JL, Jennings JL, Link AJ, Gould KL. Requirements of fission yeast septins for complex formation, localization, and function. Mol Biol Cell. 2004;15:5551–5564. doi: 10.1091/mbc.E04-07-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arellano M, Duran A, Perez P. Rho 1 GTPase activates the (1–3)β-D-glucan synthase and is involved in Schizosaccharomyces pombe morphogenesis. EMBO J. 1996;15:4584–4591. [PMC free article] [PubMed] [Google Scholar]
- Arellano M, Duran A, Perez P. Localisation of the Schizosaccharomyces pombe rho1p GTPase and its involvement in the organisation of the actin cytoskeleton. J Cell Sci. 1997;110:2547–2555. doi: 10.1242/jcs.110.20.2547. [DOI] [PubMed] [Google Scholar]
- Arellano M, Valdivieso MH, Calonge TM, Coll PM, Duran A, Perez P. Schizosaccharomyces pombe protein kinase C homologues, pck1p and pck2p, are targets of rho1p and rho2p and differentially regulate cell integrity. J Cell Sci. 1999;112:3569–3578. doi: 10.1242/jcs.112.20.3569. [DOI] [PubMed] [Google Scholar]
- Atkins BD, Yoshida S, Saito K, Wu CF, Lew DJ, Pellman D. Inhibition of Cdc42 during mitotic exit is required for cytokinesis. J Cell Biol. 2013;202:231–240. doi: 10.1083/jcb.201301090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Pringle JR. Pom1p, a fission yeast protein kinase that provides positional information for both polarized growth and cytokinesis. Genes Dev. 1998;12:1356–1370. doi: 10.1101/gad.12.9.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Steever AB, Wheatley S, Wang Y-L, Pringle JR, Gould KL, McCollum D. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J Cell Biol. 1998a;143:1603–1616. doi: 10.1083/jcb.143.6.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Wu J-Q, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998b;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, Bi E, Glotzer M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr Biol. 2004;14:R806-R818. doi: 10.1016/j.cub.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Bendezú FO, Martin SG. Actin cables and the exocyst form two independent morphogenesis pathways in the fission yeast. Mol Biol Cell. 2011;22:44–53. doi: 10.1091/mbc.E10-08-0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin A, Paoletti A, Chang F. Mid2p stabilizes septin rings during cytokinesis in fission yeast. J Cell Biol. 2003;160:1083–1092. doi: 10.1083/jcb.200212016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, McMurray MA, Grob P, Park SS, Garcia G, III, Patanwala I, Ng HL, Alber T, Thorner J, Nogales E. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc Natl Acad Sci USA. 2008;105:8274–8279. doi: 10.1073/pnas.0803330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Park H-O. Cell polarization and cytokinesis in budding yeast. Genetics. 2012;191:347–387. doi: 10.1534/genetics.111.132886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert KA, Gould KL. Cytokinesis-based constraints on polarized cell growth in fission yeast. PLoS Genet. 2012;8:e1003004. doi: 10.1371/journal.pgen.1003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. A highly ordered ring of membrane-associated filaments in budding yeast. J Cell Biol. 1976;69:717–721. doi: 10.1083/jcb.69.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calonge TM, Nakano K, Arellano M, Arai R, Katayama S, Toda T, Mabuchi I, Perez P. Schizosaccharomyces pombe rho2p GTPase regulates cell wall α-glucan biosynthesis through the protein kinase pck2p. Mol Biol Cell. 2000;11:4393–4401. doi: 10.1091/mbc.11.12.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron F, Barral Y. Septins and the lateral compartmentalization of eukaryotic membranes. Dev Cell. 2009;16:493–506. doi: 10.1016/j.devcel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Caviston JP, Longtine M, Pringle JR, Bi E. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol Biol Cell. 2003;14:4051–4066. doi: 10.1091/mbc.E03-04-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celton-Morizur S, Racine V, Sibarita JB, Paoletti A. Pom1 kinase links division plane position to cell polarity by regulating Mid1p cortical distribution. J Cell Sci. 2006;119:4710–4718. doi: 10.1242/jcs.03261. [DOI] [PubMed] [Google Scholar]
- Chant J, Mischke M, Mitchell E, Herskowitz I, Pringle JR. Role of Bud3p in producing the axial budding pattern of yeast. J Cell Biol. 1995;129:767–778. doi: 10.1083/jcb.129.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- Coffman VC, Nile AH, Lee I-J, Liu H, Wu J-Q. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol Biol Cell. 2009;20:5195–5210. doi: 10.1091/mbc.E09-05-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman VC, Sees JA, Kovar DR, Wu J-Q. The formins Cdc12 and For3 cooperate during contractile ring assembly in cytokinesis. J Cell Biol. 2013;203:101–114. doi: 10.1083/jcb.201305022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman VC, Wu J-Q. Counting protein molecules using quantitative fluorescence microscopy. Trends Biochem Sci. 2012;37:499–506. doi: 10.1016/j.tibs.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman VC, Wu P, Parthun MR, Wu J-Q. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J Cell Biol. 2011;195:563–572. doi: 10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll PM, Trillo Y, Ametzazurra A, Perez P. Gef1p, a new guanine nucleotide exchange factor for Cdc42p, regulates polarity in Schizosaccharomyces pombe. Mol Biol Cell. 2003;14:313–323. doi: 10.1091/mbc.E02-07-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker N, Speijer D, Grun CH, van den Berg M, de Haan A, Hochstenbach F. Role of the α-glucanase Agn1p in fission-yeast cell separation. Mol Biol Cell. 2004;15:3903–3914. doi: 10.1091/mbc.E04-04-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doignon F, Weinachter C, Roumanie O, Crouzet M. The yeast Rgd1p is a GTPase activating protein of the Rho3 and Rho4 proteins. FEBS Lett. 1999;459:458–462. doi: 10.1016/s0014-5793(99)01293-4. [DOI] [PubMed] [Google Scholar]
- Frenette P, Haines E, Loloyan M, Kinal M, Pakarian P, Piekny A. An anillin-Ect2 complex stabilizes central spindle microtubules at the cortex during cytokinesis. PLoS One. 2012;7:e34888. doi: 10.1371/journal.pone.0034888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P, Garcia I, Marcos F, de Garibay GR, Sanchez Y. Fission yeast rgf2p is a rho1p guanine nucleotide exchange factor required for spore wall maturation and for the maintenance of cell integrity in the absence of rgf1p. Genetics. 2009;181:1321–1334. doi: 10.1534/genetics.108.094839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P, Tajadura V, Garcia I, Sanchez Y. Rgf1p is a specific Rho1-GEF that coordinates cell polarization with cell wall biogenesis in fission yeast. Mol Biol Cell. 2006a;17:1620–1631. doi: 10.1091/mbc.E05-10-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P, Tajadura V, Garcia I, Sanchez Y. Role of Rho GTPases and Rho-GEFs in the regulation of cell shape and integrity in fission yeast. Yeast. 2006b;23:1031–1043. doi: 10.1002/yea.1409. [DOI] [PubMed] [Google Scholar]
- Gladfelter AS, Bose I, Zyla TR, Bardes ES, Lew DJ. Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J Cell Biol. 2002;156:315–326. doi: 10.1083/jcb.200109062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter AS, Pringle JR, Lew DJ. The septin cortex at the yeast mother-bud neck. Curr Opin Microbiol. 2001;4:681–689. doi: 10.1016/s1369-5274(01)00269-7. [DOI] [PubMed] [Google Scholar]
- Gould KL, Simanis V. The control of septum formation in fission yeast. Genes Dev. 1997;11:2939–2951. doi: 10.1101/gad.11.22.2939. [DOI] [PubMed] [Google Scholar]
- Green RA, Paluch E, Oegema K. Cytokinesis in animal cells. Annu Rev Cell Dev Biol. 2012;28:29–58. doi: 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J Biol Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- Guo W, Roth D, Walch-Solimena C, Novick P. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 1999;18:1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman-Vendrell M, Baldissard S, Almonacid M, Mayeux A, Paoletti A, Moseley JB. Blt1 and Mid1 provide overlapping membrane anchors to position the division plane in fission yeast. Mol Cell Biol. 2013;33:418–428. doi: 10.1128/MCB.01286-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O, Simanis V. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 2008;22:3205–3216. doi: 10.1101/gad.1697208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Hall A. Rho family GTPases. Biochem Soc Trans. 2012;40:1378–1382. doi: 10.1042/BST20120103. [DOI] [PubMed] [Google Scholar]
- Hall PA, Russell SE, Pringle JR. The Septins. New York: John Wiley & Sons; 2008. [Google Scholar]
- He B, Xi F, Zhang J, TerBush D, Zhang X, Guo W. Exo70p mediates the secretion of specific exocytic vesicles at early stages of the cell cycle for polarized cell growth. J Cell Biol. 2007a;176:771–777. doi: 10.1083/jcb.200606134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Xi F, Zhang X, Zhang J, Guo W. Exo70 interacts with phospholipids and mediates the targeting of the exocyst to the plasma membrane. EMBO J. 2007b;26:4053–4065. doi: 10.1038/sj.emboj.7601834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson GR, O’Farrell PH. Rho-dependent control of anillin behavior during cytokinesis. J Cell Biol. 2008;180:285–294. doi: 10.1083/jcb.200709005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Tanaka K, Ohta K, Yamamoto M. Gef1p and Scd1p, the Two GDP-GTP exchange factors for Cdc42p, form a ring structure that shrinks during cytokinesis in Schizosaccharomyces pombe. Mol Biol Cell. 2003;14:3617–3627. doi: 10.1091/mbc.E02-10-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell AS, Jin M, Wu CF, Zyla TR, Elston TC, Lew DJ. Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell. 2012;149:322–333. doi: 10.1016/j.cell.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SC, TerBush D, Abraham M, Guo W. The exocyst complex in polarized exocytosis. Int Rev Cytol. 2004;233:243–265. doi: 10.1016/S0074-7696(04)33006-8. [DOI] [PubMed] [Google Scholar]
- Huang Y, Yan H, Balasubramanian MK. Assembly of normal actomyosin rings in the absence of Mid1p and cortical nodes in fission yeast. J Cell Biol. 2008;183:979–988. doi: 10.1083/jcb.200806151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai J, Toh-e A, Matsui Y. Genetic analysis of the Saccharomyces cerevisiae RHO3 gene, encoding a rho-type small GTPase, provides evidence for a role in bud formation. Genetics. 1996;142:359–369. doi: 10.1093/genetics/142.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaki N, Karatsu K, Miyamoto M. Role of guanine nucleotide exchange factors for Rho family GTPases in the regulation of cell morphology and actin cytoskeleton in fission yeast. Biochem Biophys Res Commun. 2003;312:414–420. doi: 10.1016/j.bbrc.2003.10.140. [DOI] [PubMed] [Google Scholar]
- Iwase M, Luo J, Nagaraj S, Longtine M, Kim HB, Haarer BK, Caruso C, Tong Z, Pringle JR, Bi E. Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol Biol Cell. 2006;17:1110–1125. doi: 10.1091/mbc.E05-08-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Johnson BF, Yoo BY, Calleja GB. Cell division in yeasts: movement of organelles associated with cell plate growth of Schizosaccharomyces pombe. J Bacteriol. 1973;115:358–366. doi: 10.1128/jb.115.1.358-366.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DI, Pringle JR. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan SN, Canman JC. Rho GTPases in animal cell cytokinesis: an occupation by the one percent. Cytoskeleton (Hoboken) 2012;69:919–930. doi: 10.1002/cm.21071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdain I, Brzezinska EA, Toda T. Fission yeast Nod1 is a component of cortical nodes involved in cell size control and division site placement. PLoS One. 2013;8:e54142. doi: 10.1371/journal.pone.0054142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justa-Schuch D, Heilig Y, Richthammer C, Seiler S. Septum formation is regulated by the RHO4-specific exchange factors BUD3 and RGF3 and by the landmark protein BUD4 in Neurospora crassa. Mol Microbiol. 2010;76:220–235. doi: 10.1111/j.1365-2958.2010.07093.x. [DOI] [PubMed] [Google Scholar]
- Kang PJ, Hood-DeGrenier JK, Park H-O. Coupling of septins to the axial landmark by Bud4 in budding yeast. J Cell Sci. 2013;126:1218–1226. doi: 10.1242/jcs.118521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang PJ, Lee ME, Park H-O. Bud3 activates Cdc42 to establish a proper growth site in budding yeast. J Cell Biol. 2014;206:19–28. doi: 10.1083/jcb.201402040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M. Assembly of mammalian septins. J Biochem (Tokyo) 2003;134:491–496. doi: 10.1093/jb/mvg182. [DOI] [PubMed] [Google Scholar]
- Kita A, Li C, Yu Y, Umeda N, Doi A, Yasuda M, Ishiwata S, Taga A, Horiuchi Y, Sugiura R. Role of the small GTPase Rho3 in Golgi/endosome trafficking through functional interaction with adaptin in fission yeast. PLoS One. 2011;6:e28000. doi: 10.1371/journal.pone.0016842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Beven L, Angerman E, Tong AH, Boone C, Park H-O. Interaction between a Ras and a Rho GTPase couples selection of a growth site to the development of cell polarity in yeast. Mol Biol Cell. 2003;14:4958–4970. doi: 10.1091/mbc.E03-06-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Coffman VC, Lee I-J, Wu J-Q. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J Cell Biol. 2011;192:1005–1021. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Ojkic N, Vavylonis D, Wu J-Q. α-Actinin and fimbrin cooperate with myosin II to organize actomyosin bundles during contractile-ring assembly. Mol Biol Cell. 2012;23:3094–3110. doi: 10.1091/mbc.E12-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Zhao R, Wu J-Q. Mechanisms of contractile-ring assembly in fission yeast and beyond. Semin Cell Dev Biol. 2010;21:892–898. doi: 10.1016/j.semcdb.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I-J, Coffman VC, Wu J-Q. Contractile-ring assembly in fission yeast cytokinesis: Recent advances and new perspectives. Cytoskeleton (Hoboken) 2012;69:751–763. doi: 10.1002/cm.21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I-J, Wu J-Q. Characterization of Mid1 domains for targeting and scaffolding in fission yeast cytokinesis. J Cell Sci. 2012;125:2973–2985. doi: 10.1242/jcs.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics. 2011;189:1145–1175. doi: 10.1534/genetics.111.128264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, DeMarini DJ, Valencik ML, Al-Awar OS, Fares H, De Virgilio C, Pringle JR. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- Madaule P, Axel R, Myers AM. Characterization of two members of the rho gene family from the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1987;84:779–783. doi: 10.1073/pnas.84.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Cuadrado AB, Duenas E, Sipiczki M, Vazquez de Aldana CR, del Rey F. The endo-β-1,3-glucanase eng1p is required for dissolution of the primary septum during cell separation in Schizosaccharomyces pombe. J Cell Sci. 2003;116:1689–1698. doi: 10.1242/jcs.00377. [DOI] [PubMed] [Google Scholar]
- Martin-Cuadrado AB, Encinar del Dedo J, de Medina-Redondo M, Fontaine T, del Rey F, Latge JP, Vazquez de Aldana CR. The Schizosaccharomyces pombe endo-1,3-β-glucanase Eng1 contains a novel carbohydrate binding module required for septum localization. Mol Microbiol. 2008;69:188–200. doi: 10.1111/j.1365-2958.2008.06275.x. [DOI] [PubMed] [Google Scholar]
- Martin-Cuadrado AB, Morrell JL, Konomi M, An H, Petit C, Osumi M, Balasubramanian M, Gould KL, Del Rey F, de Aldana CR. Role of septins and the exocyst complex in the function of hydrolytic enzymes responsible for fission yeast cell separation. Mol Biol Cell. 2005;16:4867–4881. doi: 10.1091/mbc.E04-12-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y, Toh EA. Yeast RHO3 and RHO4 ras superfamily genes are necessary for bud growth, and their defect is suppressed by a high dose of bud formation genes CDC42 and BEM1. Mol Cell Biol. 1992;12:5690–5699. doi: 10.1128/mcb.12.12.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- McMurray MA, Thorner J. Septin stability and recycling during dynamic structural transitions in cell division and development. Curr Biol. 2008;18:1203–1208. doi: 10.1016/j.cub.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray MA, Thorner J. Septins: molecular partitioning and the generation of cellular asymmetry. Cell Div. 2009;4:18. doi: 10.1186/1747-1028-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Morrell-Falvey JL, Ren L, Feoktistova A, Haese GD, Gould KL. Cell wall remodeling at the fission yeast cell division site requires the Rho-GEF Rgf3p. J Cell Sci. 2005;118:5563–5573. doi: 10.1242/jcs.02664. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Mayeux A, Paoletti A, Nurse P. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 2009;459:857–860. doi: 10.1038/nature08074. [DOI] [PubMed] [Google Scholar]
- Mukherjee D, Sen A, Aguilar RC. RhoGTPase-binding proteins, the exocyst complex and polarized vesicle trafficking. Small GTPases. 2014;5:e28453. doi: 10.4161/sgtp.28453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh T, Nakano K, Mabuchi I. Rho1-GEFs Rgf1 and Rgf2 are involved in formation of cell wall and septum, while Rgf3 is involved in cytokinesis in fission yeast. Genes Cells. 2005;10:1189–1202. doi: 10.1111/j.1365-2443.2005.00908.x. [DOI] [PubMed] [Google Scholar]
- Muñoz S, Manjón E, Sánchez Y. The putative exchange factor Gef3p interacts with Rho3p GTPase and the septin ring during cytokinesis in fission yeast. J Biol Chem. 2014;289:21995–22007. doi: 10.1074/jbc.M114.548792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K, Inagaki M. Cytoskeletal modification of Rho guanine nucleotide exchange factor activity: identification of a Rho guanine nucleotide exchange factor as a binding partner for Sept9b, a mammalian septin. Oncogene. 2005;24:65–76. doi: 10.1038/sj.onc.1208101. [DOI] [PubMed] [Google Scholar]
- Nakano K, Arai R, Mabuchi I. The small GTP-binding protein Rho1 is a multifunctional protein that regulates actin localization, cell polarity, and septum formation in the fission yeast Schizosaccharomyces pombe. Genes Cells. 1997;2:679–694. doi: 10.1046/j.1365-2443.1997.1540352.x. [DOI] [PubMed] [Google Scholar]
- Nakano K, Arai R, Mabuchi I. Small GTPase Rho5 is a functional homologue of Rho1, which controls cell shape and septation in fission yeast. FEBS Lett. 2005;579:5181–5186. doi: 10.1016/j.febslet.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Nakano K, Imai J, Arai R, Toh EA, Matsui Y, Mabuchi I. The small GTPase Rho3 and the diaphanous/formin For3 function in polarized cell growth in fission yeast. J Cell Sci. 2002;115:4629–4639. doi: 10.1242/jcs.00150. [DOI] [PubMed] [Google Scholar]
- Nakano K, Mutoh T, Arai R, Mabuchi I. The small GTPase Rho4 is involved in controlling cell morphology and septation in fission yeast. Genes Cells. 2003;8:357–370. doi: 10.1046/j.1365-2443.2003.00639.x. [DOI] [PubMed] [Google Scholar]
- Oegema K, Savoian MS, Mitchison TJ, Field CM. Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis. J Cell Biol. 2000;150:539–552. doi: 10.1083/jcb.150.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y, Bi E. Septin structure and function in yeast and beyond. Trends Cell Biol. 2011;21:141–148. doi: 10.1016/j.tcb.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojkic N, Wu J-Q, Vavylonis D. Model of myosin node aggregation into a contractile ring: the effect of local alignment. J Phys Condens Matter. 2011;23:374103. doi: 10.1088/0953-8984/23/37/374103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Koga T, Hirata A, Nakamura T, Asakawa H, Shimoda C, Bähler J, Wu J-Q, Takegawa K, Tachikawa H, et al. Role of septins in the orientation of forespore membrane extension during sporulation in fission yeast. Mol Cell Biol. 2010;30:2057–2074. doi: 10.1128/MCB.01529-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Ko N, Nishihama R, Pringle JR. Distinct roles of Rho1, Cdc42, and Cyk3 in septum formation and abscission during yeast cytokinesis. J Cell Biol. 2013;202:311–329. doi: 10.1083/jcb.201302001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan A, Bakka K, Sevugan M, Naqvi NI, D'souza V, Tang X, Mishra M, Balasubramanian MK. IQGAP-related Rng2p organizes cortical nodes and ensures position of cell division in fission yeast. Curr Biol. 2011;21:467–472. doi: 10.1016/j.cub.2011.01.059. [DOI] [PubMed] [Google Scholar]
- Paoletti A, Chang F. Analysis of mid1p, a protein required for placement of the cell division site, reveals a link between the nucleus and the cell surface in fission yeast. Mol Biol Cell. 2000;11:2757–2773. doi: 10.1091/mbc.11.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H-O, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez P, Rincon SA. Rho GTPases: regulation of cell polarity and growth in yeasts. Biochem J. 2010;426:243–253. doi: 10.1042/BJ20091823. [DOI] [PubMed] [Google Scholar]
- Petit CS, Mehta S, Roberts RH, Gould KL. Ace2p contributes to fission yeast septin ring assembly by regulating mid2+ expression. J Cell Sci. 2005;118:5731–5742. doi: 10.1242/jcs.02687. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Wu J-Q. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor SA, Minc N, Boudaoud A, Chang F. Contributions of turgor pressure, the contractile ring, and septum assembly to forces in cytokinesis in fission yeast. Curr Biol. 2012;22:1601–1608. doi: 10.1016/j.cub.2012.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser DC, Drivas TG, Maldonado-Baez L, Wendland B. Existence of a novel clathrin-independent endocytic pathway in yeast that depends on Rho1 and formin. J Cell Biol. 2011;195:657–671. doi: 10.1083/jcb.201104045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid T, Furuyashiki T, Ishizaki T, Watanabe G, Watanabe N, Fujisawa K, Morii N, Madaule P, Narumiya S. Rhotekin, a new putative target for Rho bearing homology to a serine/threonine kinase, PKN, and rhophilin in the rho-binding domain. J Biol Chem. 1996;271:13556–13560. doi: 10.1074/jbc.271.23.13556. [DOI] [PubMed] [Google Scholar]
- Ren XD, Kiosses WB, Schwartz MA. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 1999;18:578–585. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon S, Coll PM, Perez P. Spatial regulation of Cdc42 during cytokinesis. Cell Cycle. 2007;6:1687–1691. doi: 10.4161/cc.6.14.4481. [DOI] [PubMed] [Google Scholar]
- Rincon SA, Santos B, Perez P. Fission yeast Rho5p GTPase is a functional paralogue of Rho1p that plays a role in survival of spores and stationary-phase cells. Eukaryot Cell. 2006;5:435–446. doi: 10.1128/EC.5.3.435-446.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Gould KL. Stepping into the ring: the SIN takes on contractile ring assembly. Genes Dev. 2008;22:3082–3088. doi: 10.1101/gad.1748908. [DOI] [PubMed] [Google Scholar]
- Robinson NG, Guo L, Imai J, Toh EA, Matsui Y, Tamanoi F. Rho3 of Saccharomyces cerevisiae, which regulates the actin cytoskeleton and exocytosis, is a GTPase which interacts with Myo2 and Exo70. Mol Cell Biol. 1999;19:3580–3587. doi: 10.1128/mcb.19.5.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadian Y, Gatsogiannis C, Patasi C, Hofnagel O, Goody RS, Farkasovsky M, Raunser S. The role of Cdc42 and Gic1 in the regulation of septin filament formation and dissociation. Elife. 2013;2:e01085. doi: 10.7554/eLife.01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Pollard TD. Characterization of structural and functional domains of the anillin-related protein Mid1p that contribute to cytokinesis in fission yeast. Mol Biol Cell. 2012;23:3993–4007. doi: 10.1091/mbc.E12-07-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B, Gutierrez J, Calonge TM, Perez P. Novel Rho GTPase involved in cytokinesis and cell wall integrity in the fission yeast Schizosaccharomyces pombe. Eukaryot Cell. 2003;2:521–533. doi: 10.1128/EC.2.3.521-533.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B, Martin-Cuadrado AB, Vazquez de Aldana CR, del Rey F, Perez P. Rho4 GTPase is involved in secretion of glucanases during fission yeast cytokinesis. Eukaryot Cell. 2005;4:1639–1645. doi: 10.1128/EC.4.10.1639-1645.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz HP, Huppert S, Lorberg A, Heinisch JJ. Rho5p downregulates the yeast cell integrity pathway. J Cell Sci. 2002;115:3139–3148. doi: 10.1242/jcs.115.15.3139. [DOI] [PubMed] [Google Scholar]
- Si H, Justa-Schuch D, Seiler S, Harris SD. Regulation of septum formation by the Bud3-Rho4 GTPase module in Aspergillus nidulans. Genetics. 2010;185:165–176. doi: 10.1534/genetics.110.114165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman-Gavrila RV, Hales KG, Wilde A. Anillin-mediated targeting of peanut to pseudocleavage furrows is regulated by the GTPase Ran. Mol Biol Cell. 2008;19:3735–3744. doi: 10.1091/mbc.E08-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Kang PJ, Park H-O. The Rho5 GTPase is necessary for oxidant-induced cell death in budding yeast. Proc Natl Acad Sci USA. 2008;105:1522–1527. doi: 10.1073/pnas.0707359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M. Splitting of the fission yeast septum. FEMS Yeast Res. 2007;7:761–770. doi: 10.1111/j.1567-1364.2007.00266.x. [DOI] [PubMed] [Google Scholar]
- Slaughter BD, Unruh JR, Das A, Smith SE, Rubinstein B, Li R. Non-uniform membrane diffusion enables steady-state cell polarization via vesicular trafficking. Nat Commun. 2013;4:1380. doi: 10.1038/ncomms2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrmann M, Fankhauser C, Brodbeck C, Simanis V. The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 1996;10:2707–2719. doi: 10.1101/gad.10.21.2707. [DOI] [PubMed] [Google Scholar]
- Spiliotis ET, Gladfelter AS. Spatial guidance of cell asymmetry: septin GTPases show the way. Traffic. 2012;13:195–203. doi: 10.1111/j.1600-0854.2011.01268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajadura V, Garcia B, Garcia I, Garcia P, Sanchez Y. Schizosaccharomyces pombe Rgf3p is a specific Rho1 GEF that regulates cell wall β-glucan biosynthesis through the GTPase Rho1p. J Cell Sci. 2004;117:6163–6174. doi: 10.1242/jcs.01530. [DOI] [PubMed] [Google Scholar]
- Tasto JJ, Morrell JL, Gould KL. An anillin homologue, Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J Cell Biol. 2003;160:1093–1103. doi: 10.1083/jcb.200211126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush DR, Maurice T, Roth D, Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Tolliday N, VerPlank L, Li R. Rho1 directs formin-mediated actin ring assembly during budding yeast cytokinesis. Curr Biol. 2002;12:1864–1870. doi: 10.1016/s0960-9822(02)01238-1. [DOI] [PubMed] [Google Scholar]
- Vavylonis D, Wu J-Q, Hao S, O'Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- Versele M, Thorner J. Some assembly required: yeast septins provide the instruction manual. Trends Cell Biol. 2005;15:414–424. doi: 10.1016/j.tcb.2005.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Tang X, Balasubramanian MK. Rho3p regulates cell separation by modulating exocyst function in Schizosaccharomyces pombe. Genetics. 2003;164:1323–1331. doi: 10.1093/genetics/164.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Tang X, Liu J, Trautmann S, Balasundaram D, McCollum D, Balasubramanian MK. The multiprotein exocyst complex is essential for cell separation in Schizosaccharomyces pombe. Mol Biol Cell. 2002;13:515–529. doi: 10.1091/mbc.01-11-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Lo Presti L, Zhu Y-H, Kang M, Wu Z, Martin SG, Wu J-Q. The novel proteins Rng8 and Rng9 regulate the myosin-V Myo51 during fission yeast cytokinesis. J Cell Biol. 2014;205:357–375. doi: 10.1083/jcb.201308146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win TZ, Gachet Y, Mulvihill DP, May KM, Hyams JS. Two type V myosins with non-overlapping functions in the fission yeast Schizosaccharomyces pombe: Myo52 is concerned with growth polarity and cytokinesis, Myo51 is a component of the cytokinetic actin ring. J Cell Sci. 2001;114:69–79. doi: 10.1242/jcs.114.1.69. [DOI] [PubMed] [Google Scholar]
- Wloka C, Bi E. Mechanisms of cytokinesis in budding yeast. Cytoskeleton (Hoboken) 2012;69:710–726. doi: 10.1002/cm.21046. [DOI] [PubMed] [Google Scholar]
- Wu H, Turner C, Gardner J, Temple B, Brennwald P. The Exo70 subunit of the exocyst is an effector for both Cdc42 and Rho3 function in polarized exocytosis. Mol Biol Cell. 2010a;21:430–442. doi: 10.1091/mbc.E09-06-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-Q, Kuhn JR, Kovar DR, Pollard TD. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- Wu J-Q, McCormick C, Pollard TD. Counting proteins in living cells by quantitative fluorescence microscopy with internal standards. Methods Cell Biol. 2008;89:253–273. doi: 10.1016/S0091-679X(08)00609-2. [DOI] [PubMed] [Google Scholar]
- Wu J-Q, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Wu J-Q, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-Q, Ye Y, Wang N, Pollard TD, Pringle JR. Cooperation between the septins and the actomyosin ring and role of a cell-integrity pathway during cell division in fission yeast. Genetics. 2010b;186:897–915. doi: 10.1534/genetics.110.119842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Lee I-J, Runge KW, Wu J-Q. Roles of putative Rho-GEF Gef2 in division-site positioning and contractile-ring function in fission yeast cytokinesis. Mol Biol Cell. 2012;23:1181–1195. doi: 10.1091/mbc.E11-09-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Bartolini S, Pellman D. Mechanisms for concentrating Rho1 during cytokinesis. Genes Dev. 2009;23:810–823. doi: 10.1101/gad.1785209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Kono K, Lowery DM, Bartolini S, Yaffe MB, Ohya Y, Pellman D. Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science. 2006;313:108–111. doi: 10.1126/science.1126747. [DOI] [PubMed] [Google Scholar]
- Zhu Y-H, Ye Y, Wu Z, Wu J-Q. Cooperation between Rho-GEF Gef2 and its binding partner Nod1 in the regulation of fission yeast cytokinesis. Mol Biol Cell. 2013;24:3187–3204. doi: 10.1091/mbc.E13-06-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.