The fission yeast F-BAR proteins Cdc15 and Imp2 and their combined SH3-domain partners appear to act as “molecular glue” to stabilize the interaction between the plasma membrane and a complex network of proteins at the division site that mediates cell division.
Abstract
Schizosaccharomyces pombe cdc15 homology (PCH) family members participate in numerous biological processes, including cytokinesis, typically by bridging the plasma membrane via their F-BAR domains to the actin cytoskeleton. Two SH3 domain–containing PCH family members, Cdc15 and Imp2, play critical roles in S. pombe cytokinesis. Although both proteins localize to the contractile ring, with Cdc15 preceding Imp2, only cdc15 is an essential gene. Despite these distinct roles, the SH3 domains of Cdc15 and Imp2 cooperate in the essential process of recruiting other proteins to stabilize the contractile ring. To better understand the connectivity of this SH3 domain–based protein network at the CR and its function, we used a biochemical approach coupled to proteomics to identify additional proteins (Rgf3, Art1, Spa2, and Pos1) that are integrated into this network. Cell biological and genetic analyses of these SH3 partners implicate them in a range of activities that ensure the fidelity of cell division, including promoting cell wall metabolism and influencing cell morphogenesis.
INTRODUCTION
Cytokinesis requires an actomyosin-based contractile apparatus linked to the plasma membrane in many eukaryotic cells (reviewed in Guertin et al., 2002; Glotzer, 2005). Studies in multiple organisms have shown that the composition of the division apparatus changes dramatically during its lifetime; relay teams of proteins build, constrict, and disassemble the contractile ring (CR). However, much remains to be learned about which specific protein–protein interactions and protein–membrane interactions are vital to the cytokinetic process and how this complex network of interactions is regulated spatially and temporally.
Proteins required for division apparatus assembly have been identified and extensively characterized in Schizosaccharomyces pombe (Wolfe and Gould, 2005). One of the first proteins to be detected at the incipient CR (Wu et al., 2003) is the founding member of the S. pombe Cdc15 homology (PCH) family of membrane-binding F-BAR proteins, Cdc15 (reviewed in Roberts-Galbraith and Gould, 2010). Because Cdc15 is also one of the most abundant proteins at the division site (Wu and Pollard, 2005), it is in a position to interact with multiple CR components involved in different functions at a substoichiometric ratio. In line with this possibility, known cdc15 functions include recruiting early secretory compartments to the division site (Vjestica et al., 2008), playing a role in endocytosis (Arasada and Pollard, 2011), and stabilizing the CR during anaphase (Wachtler et al., 2006; Roberts-Galbraith et al., 2009; Arasada and Pollard, 2014). However, the molecular mechanisms by which Cdc15 performs these essential functions are not known.
Although Cdc15 is essential for cytokinesis (Nurse et al., 1976; Fankhauser et al., 1995), its C-terminal SH3 domain is dispensable, unless the SH3 domain of the related F-BAR protein, Imp2 (Demeter and Sazer, 1998), is also deleted (Roberts-Galbraith et al., 2009). In the absence of both SH3 domains, CRs form in metaphase but then unravel during anaphase. Thus these two SH3 domains are functionally redundant, a conclusion supported by the finding that the Imp2 SH3 domain substituted for the Cdc15 SH3 domain in a domain-swapping experiment (Roberts-Galbraith et al., 2009). Because both Cdc15 and Imp2 homodimerize and at least Cdc15 oligomerizes (Roberts-Galbraith et al., 2009, 2010), their SH3 domains can stabilize the CR, presumably by tethering proline-rich proteins to the division site, which in turn mediate other interactions to create an extensive protein meshwork. Reciprocally, Cdc15 lacking its SH3 domain is more mobile than wild-type Cdc15 at the CR, indicating that SH3-domain partners stabilize it there (Roberts-Galbraith et al., 2009).
Previously we identified two proteins that interact with the Cdc15 and Imp2 SH3 domains, Fic1 and Pxl1 (Roberts-Galbraith et al., 2009). Fic1 is a C2-domain protein with an extensive proline-rich C-terminus, whereas Pxl1 contains proline-rich sequences in its N-terminus and LIM domains in its C-terminus. Neither Fic1 nor Pxl1 is essential, but the deletion mutants display mild to moderate cytokinetic defects and genetic interactions with other cytokinetic mutants, and the double fic1∆ pxl1∆ mutant is inviable (Ge and Balasubramanian, 2008; Pinar et al., 2008; Roberts-Galbraith et al., 2009). Independent of its interaction with Cdc15 and Imp2, Fic1 also interacts with the SH3 domain of Cyk3 (Bohnert and Gould, 2012), a multidomain protein likely to interact with several ring components (Pollard et al., 2012). Thus Cdc15 and Imp2 and their combined SH3 domain interactors appear to act as “molecular glue” to stabilize a complex interaction network at the division site and mediate CR assembly and function and ultimately cell division.
Of the ∼150 S. pombe proteins that have been reported to localize to the cell division site (Matsuyama et al., 2006), ∼60 contain proline-rich sequences corresponding to potential SH3-binding motifs. Therefore we predicted that additional proteins involved in CR function would be capable of binding the Cdc15SH3 and Imp2SH3 domains. Here we describe experiments that yielded two new Cdc15SH3 and Imp2SH3 binding partners at the CR—the RhoGEF Rgf3 and the previously uncharacterized Spa2 (SPAC3G9.05). We examined the relevance of these new Cdc15SH3- and Imp2SH3-domain interactors to the formation of the network and investigated their functions in cell division.
RESULTS
Binding specificity of the Cdc15 and Imp2 SH3 domains
The C-terminal SH3 domains (Figure 1A) of Cdc15 and Imp2 share 66% sequence identity (Figure 1B), and we found previously that they appeared to be functionally interchangeable (Roberts-Galbraith et al., 2009). However, to determine whether the few amino acid differences between the two SH3 domains have the potential to modulate their binding specificities, we took an unbiased approach to study their specificities in vitro. Glutathione S-transferase (GST) fusions of the Cdc15SH3 and Imp2SH3 domains were produced in Escherichia coli and used to scan phage-displayed peptide libraries, as described for the entire complement of Saccharomyces cerevisiae SH3 domains (Tonikian et al., 2009). In this assay, both domains selected class I ligands (specifically K/RXLPXΦP; reviewed in Mayer, 2001; Figure 1, C and D), in accordance with their functional overlap (Roberts-Galbraith et al., 2009). Thus we used just one of the two SH3 domains to identify additional CR binding partners.
FIGURE 1:
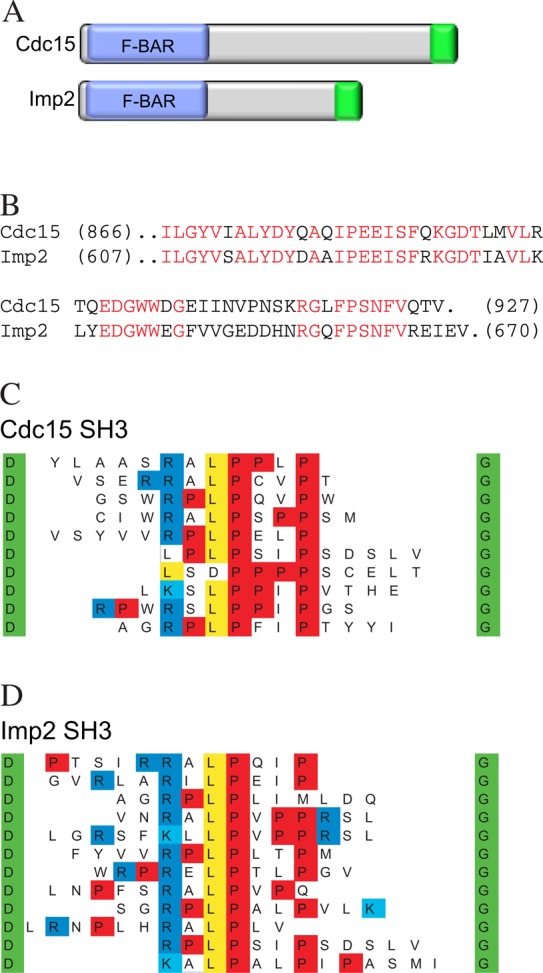
Binding specificity of Cdc15 and Imp2 SH3 domains. (A) Schematic diagram of Cdc15 and Imp2, with F-BAR domains in blue and SH3 domains in green. (B) Sequence alignment of the Cdc15SH3 and Imp2SH3 domains. (C, D) Unique sequences selected by the GST-Cdc15SH3 (C) and GST-Imp2 SH3 (D) domains are displayed in alignment form, with prolines in red, basic residues in blue, and leucines in yellow. The anchor amino acids at the ends of selected sequences are in green. Multiple copies of most sequences were obtained. For each domain, >20 sequences were obtained.
Identification of Cdc15SH3-interacting proteins
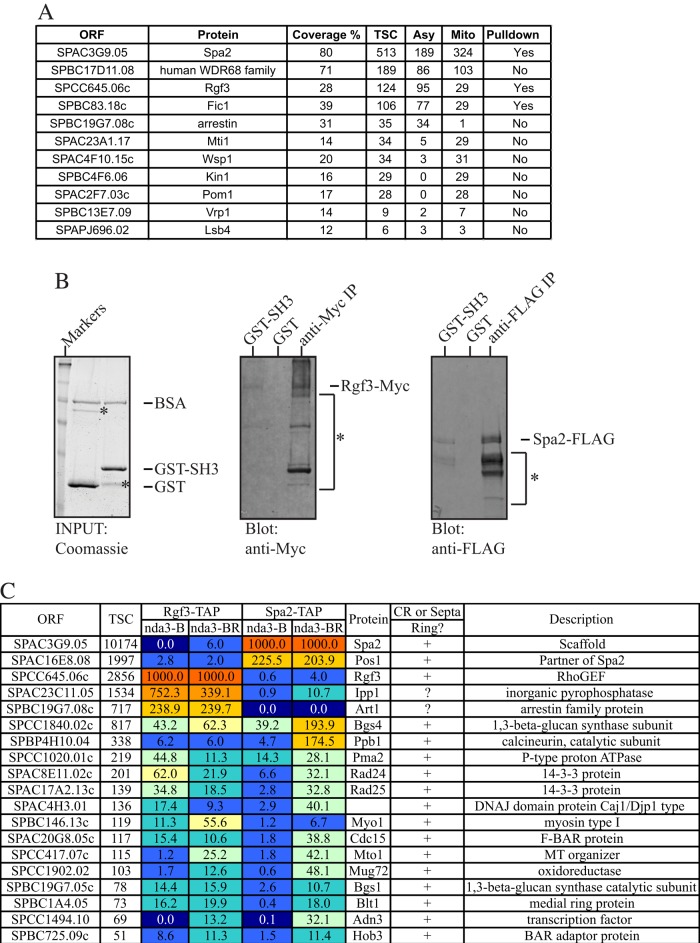
An MBP-Cdc15SH3 fusion expressed in E. coli and purified on amylose beads was used as bait. Protein lysates from S. pombe cells growing asynchronously or arrested in prometaphase with the cold-sensitive nda3-KM311 tubulin mutant were incubated with the amylose beads. The beads were then extensively washed before elution of interactors with buffers containing increasing salt concentrations and finally maltose. Selected salt-eluted fractions from the 500 mM NaCl elution that we had determined contained Fic1-FLAG by immunoblotting (unpublished data) were trichloroacetic acid (TCA) precipitated, digested with trypsin, and identified by two-dimensional (2D) liquid chromatography–tandem mass spectrometry (LC-MS/MS; Supplemental Table S1). Potential direct interactors of the Cdc15SH3 domain (i.e., those that contain a class I SH3 binding motif) are listed in Figure 2A. To validate these interactions, we tagged genes encoding the putative binding partners at their endogenous loci to produce C-terminal fusions with green fluorescent protein (GFP), FLAG3, or Myc13 and then tested them for their ability to bind GST-Cdc15SH3, but not GST, from cells arrested in mitosis (Figure 2B and unpublished data). The results of these experiments are given in Figure 2A. In addition to Fic1, which served as a positive control, Cdc15SH3 associated with Rgf3 and Spa2 (Figure 2B). We were unable to confirm an association with the other proteins listed in Figure 2A using this approach (unpublished data). Rgf3 is a guanine nucleotide exchange factor for Rho1 and affects CR formation and cell separation by regulating cell wall metabolism (Tajadura et al., 2004; Morrell-Falvey et al., 2005; Mutoh et al., 2005; Wu et al., 2010). Spa2 has not been previously characterized in S. pombe; it is the apparent orthologue of S. cerevisiae Spa2, which is required for pheromone-induced cell polarization and normal bud site selection (Snyder, 1989; Gehrung and Snyder, 1990).
FIGURE 2:
Identification of Cdc15SH3 binding partners. (A) Summary of mass spectrometry results. Percentage sequence coverage and total spectral counts (TSCs) are shown. TSC is a combination of spectral counts from experiments using lysate from mitotic (Mito) and asynchronously (Asy) growing cells. The ability of GST-Cdc15SH3 to pull out the indicated proteins is listed in the Pulldown column. (B) Recombinant GST or GST-Cdc15SH3 on glutathione beads was added to protein lysates from rgf3-Myc13 or spa2-FLAG strains and washed, and bound proteins were detected by immunoblotting with appropriate antibodies. Anti-Myc and anti-FLAG immunoprecipitates from the relevant strains served as positive controls. *: nonspecific bands and/or degradation products. (C) Heatmap of proteins identified in Rgf3-TAP and Spa2-TAP in prometaphase (Block, B) and after a 30-min release into anaphase (Block and Release, BR) by mass spectrometry. Only proteins that localize to the cell division site and were identified with >50 TSCs are listed. See Supplemental Table S2 for a complete list of all proteins identified. The values shown for each protein represent the relative abundance (normalized to bait) for each purification.
As a further test of Cdc15 and Imp2 interaction, we performed reciprocal purifications of Rgf3-TAP and Spa2-TAP from cells arrested in prometaphase or released from this arrest for 30 min to allow septum formation and identified associated proteins by 2D LC-MS/MS. As predicted, both proteins copurified Cdc15 and Imp2, among other possible CR partners (Figure 2C and Supplemental Table S2).
Rgf3 and Spa2 colocalize with SH3 proteins Cdc15 and Imp2
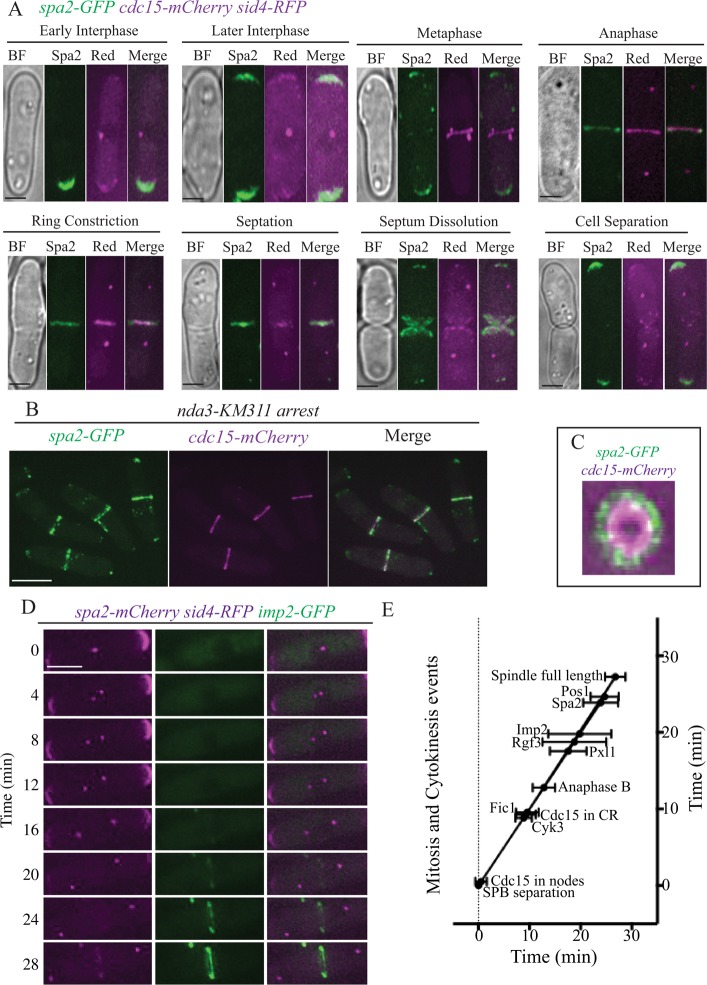
Cdc15 localizes to puncta at cell tips during interphase and then forms medial cortical nodes, which condense into CRs during mitosis and cytokinesis (Fankhauser et al., 1995; Carnahan and Gould, 2003). Imp2 also localizes to the cell division site but arrives ∼20 min later than Cdc15 (Demeter and Sazer, 1998; Figure 3E). We reasoned that Spa2 and Rgf3 would at least partially colocalize with Cdc15 and/or Imp2 at these locations over the course of the cell cycle. As shown previously, Rgf3 localized exclusively to the CR (Tajadura et al., 2004; Morrell-Falvey et al., 2005; Mutoh et al., 2005), where it colocalized with Cdc15 (Supplemental Figure S1A) and Imp2 (Supplemental Figure S1B). Considering SPB separation as time 0, live-cell imaging showed that Cdc15 arrived at the cell middle at 0.5 ± 1.1 min and formed rings at 9.3 ± 1.9 min (n = 5) and Imp2 localized to the CR at 19.8 ± 6.2 min (n = 11; Figure 3, D and E, and Supplemental Figure S2B), in excellent agreement with previous reports (Wu et al., 2003; Roberts-Galbraith et al., 2010; Arasada and Pollard, 2011). Rgf3 was detected in rings at 18.8 ± 6.2 min (n = 8), indicating that it closely mimics the timing of Imp2-GFP recruitment to the CR during anaphase B and the CR recruitment of another Cdc15 and Imp2 SH3-interacting component, Pxl1 (Roberts-Galbraith et al., 2009; Figure 3E and Supplemental Figure S1C).
FIGURE 3:
Spa2 colocalizes with Cdc15 and Imp2 in cytokinetic rings. (A) Representative live- cell images of the spa2-GFP cdc15-mCherry sid4-RFP at different cell cycle stages. Scale bar, 2 μm. (B) Arrested spa2-GFP cdc15-mCherry nda3-KM311 cells were fixed in ice-cold ethanol and then imaged. Scale bar, 5 μm. (C) Representative spa2-GFP cdc15-mCherry CR in initial stages of constriction rotated 90° and enlarged. (D) Images at 4-min intervals from a representative movie for spa2-mCherry3 imp2-GFP sid4-RFP. Scale bars, 5 μm. (E) Time line showing the detection of various proteins at the division site. Time 0 was defined as SPB separation, and the mean time of detection of each protein ± SD is plotted. The onset of anaphase B was determined in n = 16 cells to be 12.7 ± 2.0 min, and maximum SPB separation (end of anaphase B) was at 26.7 ± 1.9 min, determined with n = 14 cells. Fic1 joined the CR at 9.6 ± 2.2, Cyk3 at 8.8 ± 1.6, Pos1 at 24.7 ± 2.7, and Pxl1 at 17.6 ± 3.6 min. n = 5 for Cdc15; 12 for Cyk3; 9 for Fic1; 9 for Pxl1; 8 for Rgf3; 11 for Imp2; 8 for Spa2; and 6 for Pos1.
On the other hand, the localization of S. pombe Spa2 has not been previously described, and therefore we tagged the 3′ end of the open reading frame (ORF) with sequences encoding GFP or mCherry and imaged it with the spindle pole body (SPB) protein Sid4 as a marker of cell cycle stage (Chang and Gould, 2000). Spa2-GFP was detected in bright arcs at one or both cell tips during interphase; it localized first to the “old” cell ends (the cell ends opposite of the CR) after cytokinesis, and then, after new end take off (NETO), it localized to both cell tips (Figure 3A). At active sites of polarized growth (cell tips), Spa2-GFP overlapped with Cdc15-mCherry. During mitosis, Spa2 relocalized to the cell division site, where it colocalized with Cdc15-mCherry (Figure 3A), and was especially apparent in prometaphase-arrested cells that organize CRs during the cell cycle block (Figure 3B). Time-lapse imaging showed that Spa2 arrived at CRs at 23.9 ± 3.4 min (n = 8), later than Imp2 but before the end of anaphase B (Figure 3, D and E). Only some Spa2 appeared to constrict with the CR, accumulating at a constriction point, but most appeared to form a ring around the Cdc15 ring and lined the septum as it formed (Figure 3, A and C, and Supplemental Figure S2A).
Functional analysis of SH3-domain interaction with Rgf3 and Spa2
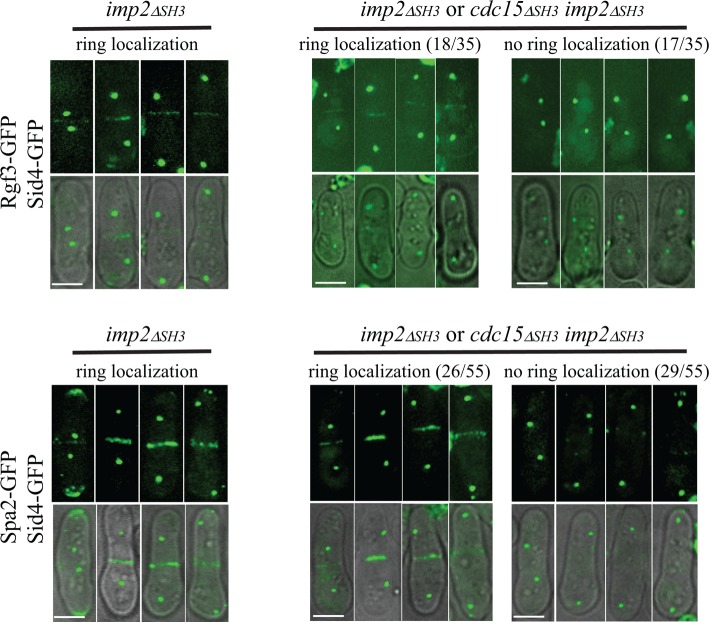
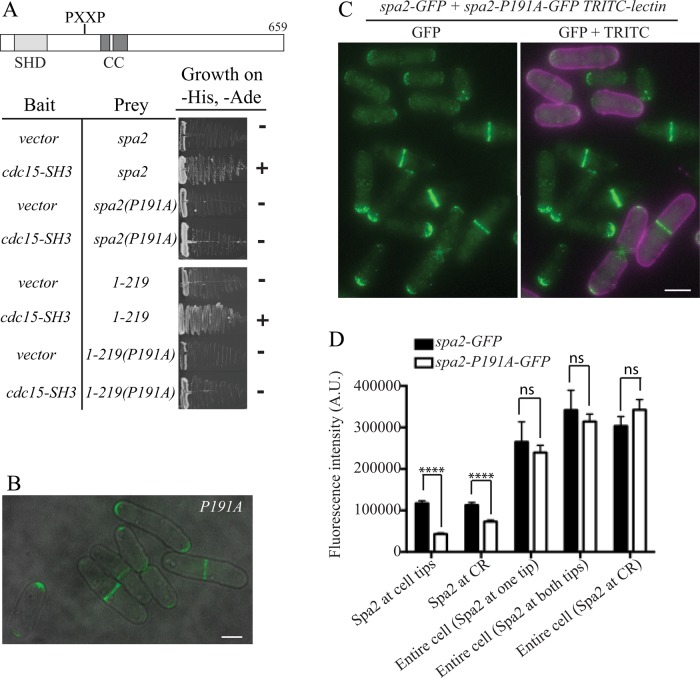
The SH3 domains of Cdc15 and Imp2 are collectively essential and functionally interchangeable (Roberts-Galbraith et al., 2009). Thus, to determine whether the SH3 domains of Cdc15 and Imp2 are necessary for recruiting Rgf3 and/or Spa2 to CRs, we used a spore germination assay as we did previously to assess the role of these domains in recruitment of Fic1 and Pxl1 to the CR (Roberts-Galbraith et al., 2009). In brief, sid4-GFP cdc15∆SH3 cells were crossed with sid4-GFP imp2∆SH3 cells. These strains also produced either Rgf3-GFP or Spa2-GFP. Spores from the resultant diploids were collected and germinated in medium containing hygromycin to select for the imp2∆SH3 allele. The relevant genotypes of germinating spores would therefore be either imp2∆SH3 or imp2∆SH3 cdc15∆SH3 in approximately equal amounts. As a control, spores containing only the imp2∆SH3 allele were germinated in parallel. In imp2∆SH3 cells with two SPBs, 97% contained either Rgf3 (n = 31) or Spa2 (n = 40) in CRs, as expected (Figure 4, left panels). In the mixture of single- and double-mutant cells, only about half of the cells with two SPBs showed Rgf3 (18 of 35 cells) or Spa2 (26 of 55 cells) at CRs (Figure 4, right panels). We conclude that the SH3 domains of Cdc15 and Imp2 are together required directly or indirectly for Spa2 and Rgf3 recruitment to the division site.
FIGURE 4:
SH3 domains of Cdc15 and Imp2 cooperatively recruit CR components. Spores with relevant genotype imp2∆SH3-FLAG-hygR or imp2∆SH3-FLAG-hygR cdc15∆SH3-FLAG-kanR containing the indicated tagged proteins were germinated in the presence of hygromycin overnight and imaged live. Scale bars, 5 μm.
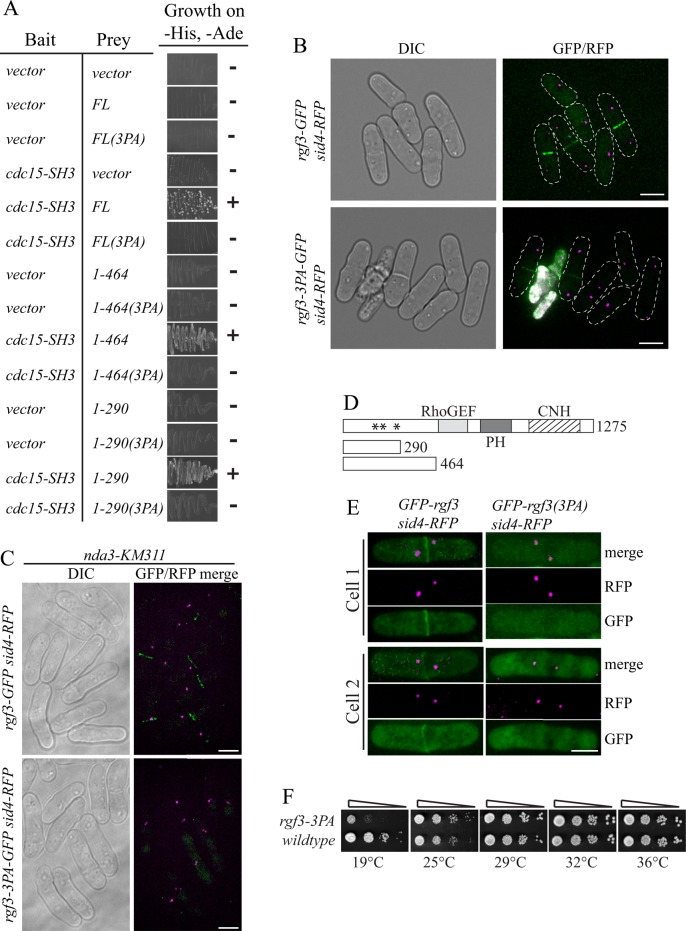
Because the SH3 domains of Cdc15 and Imp2 recruit several proteins, the absence of Rgf3 and Spa2 at CRs in the previous experiments may be due to failure to recruit other proteins that also interact with Rgf3 and Spa2. To test the importance of a direct association between these proteins and the Cdc15 and Imp2 SH3 domains, we first defined the motifs within Rgf3 and Spa2 that bind Cdc15SH3, using our knowledge of the consensus binding sites for Cdc15SH3 and Imp2SH3 (Figure 1C). Rgf3 residues 1–290 contain three close matches to the consensus binding motif (Supplemental Figure S3), and all Rgf3 fragments that included these sites interacted with Cdc15SH3 in a yeast two-hybrid assay (Figure 5, A and D). When the first or second proline in all three motifs was mutated to alanine (3PA), the two-hybrid interactions were abolished (Figure 5A). To test whether interaction with Cdc15 and Imp2 SH3 domains was important for Rgf3 recruitment to the CR, we replaced rgf3+ with rgf3-3PA-GFP. Although Rgf3-3PA-GFP localized to the site of cell division, the signal appeared only after cells had begun ring constriction and septation (Figure 5B). To confirm that cells in the early stages of cytokinesis lacked Rgf3-3PA-GFP rings, we blocked cells in early mitosis using the nda3-KM311 mutant. All (100%) early mitotic cells contained Rgf3-GFP rings, but Rgf3-3PA rings were never detected at this stage (Figure 5C). Accordingly, we observed that exogenously produced N-terminal Rgf3 fragments containing residues 1–290 localized to CRs in early mitotic cells (as judged by the short distance between SPBs), but their ability to do so was eliminated by the 3PA mutations (Figure 5, D and E, Supplemental Figure S4A). We conclude from these results that the Cdc15 and Imp2 SH3 domains recruit Rgf3 to preconstriction CRs. We noted that a significant fraction of rgf3-3PA cells lysed at the end of cell division, a phenotype reminiscent of rgf3 loss (Morrell-Falvey et al., 2005; Figure 5B), and rgf3-3PA cells were cold sensitive for growth (Figure 5F). Thus preconstriction recruitment of Rgf3 via the Cdc15 and Imp2 SH3 domains is ultimately important for the fidelity of cell division.
FIGURE 5:
Characterization of SH3 domain–Rgf3 interaction. (A–C) All three predicted Cdc15SH3 binding sites in Rgf3 at amino acids 154–160, 169–175, and 264–270 were mutated at their first proline position to alanines to make the 3PA mutant in full-length (FL) rgf3 as well as in fragments of rgf3 encoding residues 1–464 or 1–290. These were tested for interaction in the yeast two-hybrid assay (A) and for their localization as GFP fusion proteins (B–E). Scale bars, 5 μm (B), 3 μm (C). Asterisks in D indicate locations of PXXP sites. (F) The indicated strains were spotted in 10-fold serial dilutions on YE plates and incubated at the indicated temperatures.
Spa2 contains only one match to the Imp2/Cdc15SH3 consensus binding motif at residues 188–194 (Supplemental Figure S5). Both full-length spa2 and a fragment encoding residues 1–219 interacted with sequences encoding Cdc15SH3 in the yeast two-hybrid assay (Figure 6A). Mutation of P191 to alanine eliminated the two-hybrid interactions (Figure 6A). We replaced spa2+ with spa2-P191A and tagged spa2-P191A with sequences encoding GFP in order to study mutant protein localization. Spa2-P191A localized to the correct places but seemed dimmer (Figure 6B). To quantitate this and directly compare wild-type and mutant Spa2 localization in the same field of view, we labeled Spa2-P191A-GFP cells with a fluorescent lectin, mixed them with Spa2-GFP cells (or vice versa), and imaged them together (Figure 6C). By comparing total fluorescence intensities of the two strains, we found that significantly less Spa2-P191A-GFP than wild type localized to CRs and tips, with the effect on tip localization more pronounced (Figure 6, C and D). However, there was no decrease in the overall level of the mutant protein relative to wild type (Figure 6D). Thus the interaction with the Cdc15 and Imp2 SH3 domains is a strong determinant of Spa2 intracellular localization.
FIGURE 6:
Characterization of SH3 domain–Spa2 interaction. (A) The predicted Cdc15SH3 binding site in Spa2 was mutated at its first proline position to alanine to make the P191A mutant in full-length (FL) spa2 and an N-terminal fragment of spa2 encoding residues 1–219. These were tested for interaction with Cdc15SH3 in the yeast two-hybrid assay. (B) spa2-191A-GFP cells imaged live. Scale bar, 3 μm. (C, D) spa2-P191A-GFP cells were labeled with tetramethylrhodamine isothiocyanate (TRITC)–lectin and mixed with spa2-GFP cells (or vice versa) and imaged live. Representative image in B, and quantitation of fluorescence intensity of Spa2-GFP at cell tips or CRs and whole cells with Spa2 at one tip, two tips, or CRs in C. The graph represents results of three biological replicates and n ≥ 200 for cell tip and CR localization and n ≥ 30 for each entire cell condition. Half of the measurements were from experiments with spa2-GFP cells labeled with lectin and half from experiments with spa2-P191A-GFP cells labeled with lectin. ****p < 0.0001. Error bars represent SEM. Scale bar, 5 μm.
Rgf3 forms a complex with Art1
To better understand the function(s) of the SH3 domain–centered network, we further investigated the roles of Rgf3 and Spa2. A prominent hit in the MS analyses of Rgf3 complexes was Art1 (Figure 2C), a nonessential, arrestin-like protein that localizes to the division site (Wu et al., 2010). A GFP-Art1 fusion produced in Rgf3-FLAG cells coimmunoprecipitated Rgf3 (Figure 7A), confirming our MS results. We found that art1∆ and any C-terminally tagged art1 allele exacerbated the cell lysis phenotype of a temperature-sensitive rgf3 mutation (Morrell-Falvey et al., 2005), rgf3lad1, and reduced its viability (Figure 7, B and C, and unpublished data). rgf3lad1 cells lyse at division under restrictive conditions due to the failure of the mutant protein to localize to the CR and promote secondary septum formation in a Rho1-dependent manner (Morrell-Falvey et al., 2005). Of interest, art1∆ also showed negative genetic interactions with Rgf3-GFP (Figure 7D). This indicates that rgf3-GFP is likely a hypomorphic allele and loss of art1 further decreases its function. Like rgf3lad1 cells (Morrell-Falvey et al., 2005), art1∆ is sensitive to compounds that affect cell wall integrity, indicating it has cell wall defects (Figure 7E).
FIGURE 7:
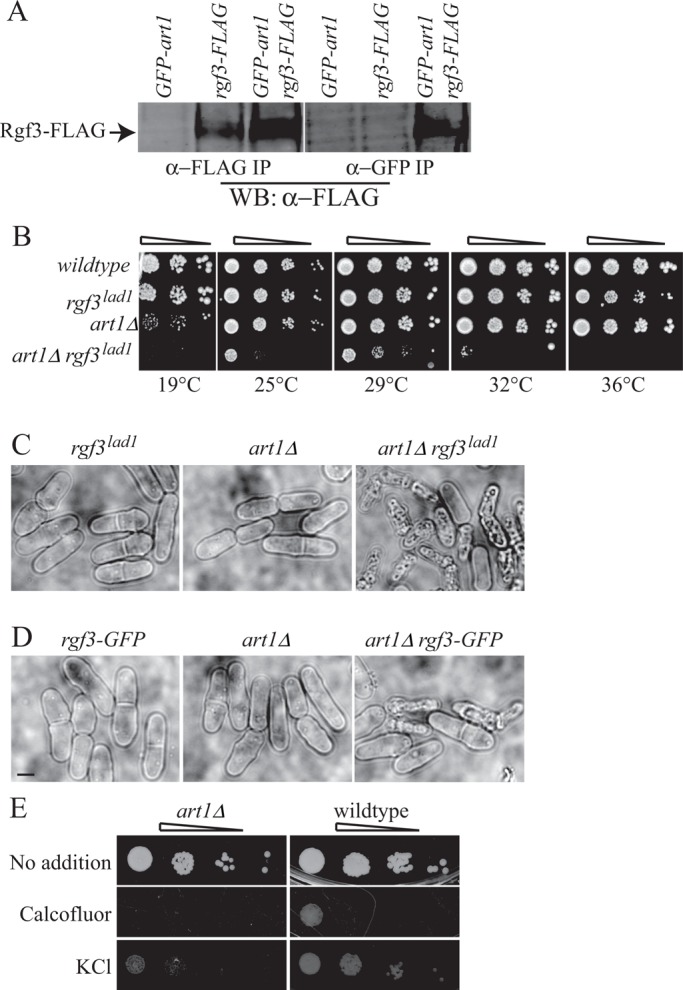
Art1 interaction with Rgf3. (A) GFP-Art1 was expressed or not in wild-type or Rgf3-FLAG cells. An anti-FLAG immunoblot of anti-FLAG (left) or anti-GFP (right) immunoprecipitations from the indicated strains. (B) The indicated strains were spotted in 10-fold serial dilutions on YE plates and incubated at the indicated temperatures. (C, D) Live-cell images of the indicated strains grown in YE at 25°C. Scale bars, 3 μm. (E) The indicated strains were spotted in 10-fold serial dilutions on YE plates containing 0.5 mg/ml calcofluor or 1 M KCl and incubated at 29°C.
Characterization of spa2+ function
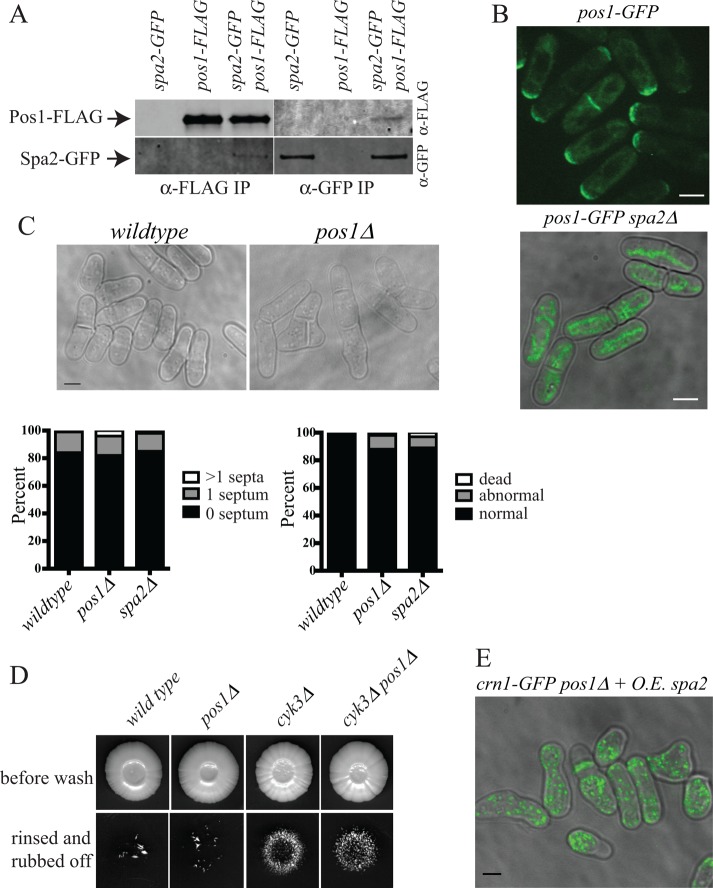
In S. cerevisiae and Candida albicans, Spa2 is involved in polarizing cells for budding and mating (Snyder, 1989; Gehrung and Snyder, 1990; Zheng et al., 2003). We found that S. pombe spa2∆ cells are viable, form normal mating projections when exposed to cells of opposite mating type, and fused and sporulated like wild-type cells (unpublished data). At 25–36°C, spa2∆ cells were morphologically normal, established a normal bipolar growth pattern after cell division, and exhibited normal patterns of F-actin distribution (Supplemental Figure S4, B–D). This is in contrast to fic1∆ cells, which delay polarization of growth at new ends after cytokinesis (Bohnert and Gould, 2012). However, at 19°C, some spa2∆ cells lost polarity and exhibited septation defects (Figure 8A). This result prompted us to explore whether Spa2 might be involved in morphogenesis under atypical growth conditions. S. pombe can undergo a dimorphic switch from single-celled growth to a pseudohyphal invasive form in which polarized growth is restrained to a single cell end (Dodgson et al., 2010; Pohlmann and Fleig, 2010). We tested whether Spa2 was required for this dimorphic switch in growth pattern. spa2∆ cells were incapable of invasive growth and suppressed invasion of two strains proficient in pseudohyphal growth, fic1∆ and cyk3∆ (Bohnert and Gould, 2012; Figure 8B). Further, spa2 overexpression depolarized the actin cytoskeleton and altered cell growth patterns (Figure 8C). Taken together, these results indicate that Spa2 can influence the cell morphogenesis machinery.
FIGURE 8:
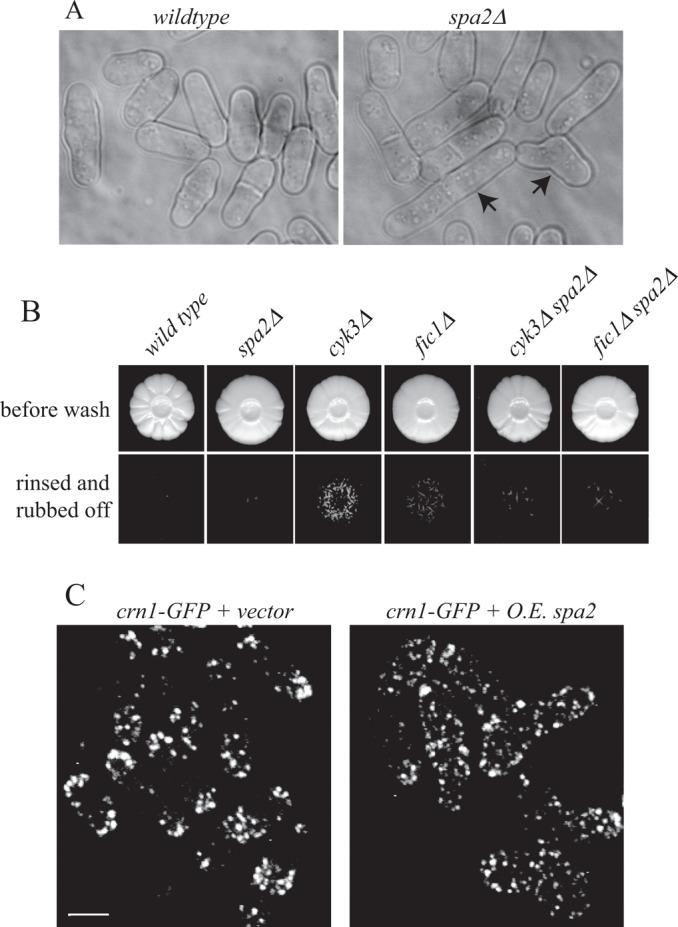
spa2∆ is defective in polarized growth under special circumstances. (A) Representative bright-field live-cell images of the indicated strains grown at 19°C. Arrows point to cells displaying polarity or septation defects. Scale bar, 3 μm. (B) Invasive growth assays for strains of the indicated genotypes on 2% agar. Cells were spotted on rich medium and incubated for 20 d at 29°C (top). Colonies were then rinsed under a stream of water and rubbed off (bottom). (C) crn1-GFP cells containing a vector control (left) or overexpressing spa2+ (right) were imaged live. Scale bar, 5 μm.
In S. cerevisiae, Spa2 is considered to be part of a defined complex termed the polarisome (Fujiwara et al., 1998; Sheu et al., 1998). Polarisome components, as defined in S. cerevisiae, were not identified at appreciable levels in Spa2-TAPs (Figure 2C and Supplemental Table S2). Instead, the uncharacterized sequence orphan encoded by ORF SPAC16E8.08 was reproducibly detected at high levels (Figure 2C). This protein was named Pos1 for partner of Spa2. To examine Pos1-Spa2 association, we tagged pos1 with sequences encoding GFP, TAP, or FLAG3 at the 3′ end of its protein-coding region. Pos1-FLAG and Pos1-TAP copurified Spa2-GFP and Spa2 and vice versa (Figure 9A and Supplemental Tables S2 and S3). Pos1-GFP localized to the cortex at cell tips and arrived at the division site coincidently with Spa2 (Figures 9B, left, and 3E). Its localization was Spa2 dependent because in spa2∆ cells, Pos1-GFP was observed solely on mitochondria (Figure 9B, bottom, and Supplemental Figure S6A). pos1+ is not an essential gene, and pos1∆ cells, in which Spa2-GFP localizes normally, have wild-type morphology and growth patterns at 25–36°C (Supplemental Figure S6B and unpublished data). However, pos1∆ cells have growth defects at 19°C, similar to spa2∆ cells (Figure 9C). To discern whether inhibition of cyk3∆ pseudohyphal growth by spa2 deletion (Figure 8B) was due to Pos1 loss at sites of polarized growth, we asked whether pos1∆ affected cyk3∆ pseudohyphal growth. Of interest, we found that it did not (Figure 9D), nor did it affect the phenotype of spa2+ overexpression (Figure 9E). Thus Spa2 affects polarized growth independently of its interaction with Pos1.
FIGURE 9:
Pos1 interaction with Spa2. (A) An anti-FLAG immunoblot (top) and an anti-GFP immunoblot (bottom) of anti-FLAG (left) or anti-GFP (right) immunoprecipitations from the indicated strains. (B) Representative live-cell images of the indicated strains grown at 19°C. Scale bars, 3 μm. Bottom is merged with differential interference contrast image. (C) Representative bright-field live-cell images of the indicated strains. Scale bar, 3 μm. The indicated phenotypes of these strains grown at 19°C are scored on 300 cells and shown in the graphs below the images. (D) Invasive growth assays for strains of the indicated genotypes on 2% agar. Cells were spotted on rich medium and incubated for 20 d at 29°C (top). Colonies were then rinsed under a stream of water and rubbed off (bottom). (E) crn1-GFP pos1∆ cells overexpressing spa2+ were imaged live at 25°C. Scale bar, 5 μm.
Building a functional interaction network emanating from F-BAR proteins
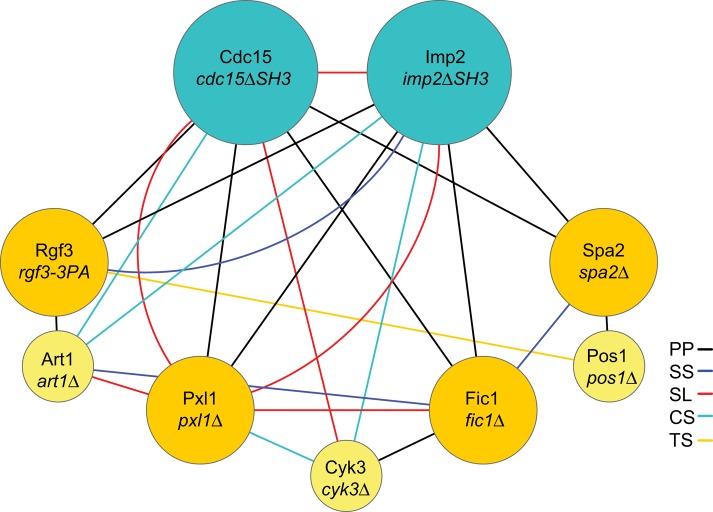
As shown in the foregoing and previously (Roberts-Galbraith et al., 2009), the SH3 domains of Cdc15 and Imp2, through their homodimerization and oligomerization, promote the formation of a network of proteins (minimally Fic1, Pxl1, Spa2, Pos1, Rgf3, and Art1), most of which are nonessential, linked through modular domains and multivalent interactions. To begin determining their functional relationships, we performed pairwise genetic crosses between cdc15∆SH3, imp2∆SH3, rgf3-3PA, spa2∆, fic1∆, pxl1∆, cyk3∆, pos1∆, and art1∆ and tested the growth of the resultant double mutants at a variety of temperatures. We included cyk3∆ in this analysis because Cyk3 binds directly to Fic1 (Bohnert and Gould, 2012). We included pos1∆ and art1∆ because Pos1 associates with Spa2 and Art1 with Rgf3 (Figures 2C, 6A, and 8A and Supplemental Tables S2 and S3). Several new interactions among these mutations that provided insight were identified (Table 1, Figure 10, and Supplemental Figure S7A). First, imp2∆SH3 was synthetically lethal with pxl1∆ and displayed negative genetic interactions with cyk3∆ and rgf3-3PA (Table 1, Figure 10, and Supplemental Figure S7A). These results provide additional evidence that the Imp2 SH3 domain independently functions to organize the division site. Second, among the SH3-domain binding partners, pxl1∆ showed negative interactions with almost all mutants except spa2∆ and pos1∆ (Table 1, Figure 10, and Supplemental Figure S7A), suggesting that Pxl1 plays a particularly distinctive role in cell division. Third, fic1∆ and cyk3∆ showed different genetic interaction profiles (Table 1 and Figure 10). This was unexpected because in S. cerevisiae, the homologues Inn1p and Cyk3p, respectively, appear to work together to activate the Chs2 chitin synthase for septum formation (Nishihama et al., 2009; Devrekanli et al., 2012). Our genetic results indicate that Fic1 and Cyk3 must have at least one distinct molecular mechanism in S. pombe cell division, despite arriving simultaneously to the CR (Figure 3E). Fourth, a significant negative interaction was detected between fic1∆ and spa2∆ at 19°C (Table 1 and Supplemental Figure S7B), indicating that Fic1 and Spa2 perform overlapping functions in cell division despite their very different kinetics of recruitment to the division site (Figure 3E). Because fic1∆ spa2∆ cells were significantly compromised in their ability to separate at the end of division (Supplemental Figure S7B), it is likely that both contribute in some manner to cell wall metabolism or membrane trafficking. Physical and genetic interactions of SH3-domain proteins and their binding partners were integrated into a network diagram to provide visualization of the relevant interactions within this protein network (Figure 10).
TABLE 1:
Pairwise genetic interactions in SH3-domain network.
| Parent 1 | Parent 2 | Growth at 29°C | Parent 1 | Parent 2 | Growth at 29˚C |
|---|---|---|---|---|---|
| cdc15ΔSH3(aa869) | imp2ΔSH3 | SL | spa2Δ | cdc15ΔSH3(aa869) | + |
| cdc15ΔSH3(aa869) | spa2Δ | + | spa2Δ | imp2ΔSH3 | + |
| cdc15ΔSH3(aa869) | pos1Δ | + | spa2Δ | pos1Δ | + |
| cdc15ΔSH3(aa869) | rgf3-3PA | + | spa2Δ | rgf3-3PA | + |
| cdc15ΔSH3(aa869) | fic1Δ | + | spa2Δ | fic1Δ | SS |
| cdc15ΔSH3(aa869) | cyk3Δ | SL | spa2Δ | cyk3Δ | + |
| cdc15ΔSH3(aa869) | pxl1Δ | SL | spa2Δ | pxl1Δ | + |
| cdc15ΔSH3(aa869) | art1Δ | CS | spa2Δ | art1Δ | + |
| imp2ΔSH3 | cdc15ΔSH3(aa869) | SL | pos1Δ | cdc15ΔSH3(aa869) | + |
| imp2ΔSH3 | spa2Δ | + | pos1Δ | imp2ΔSH3 | + |
| imp2ΔSH3 | pos1Δ | + | pos1Δ | spa2Δ | + |
| imp2ΔSH3 | rgf3-3PA | SS | pos1Δ | rgf3-3PA | TS |
| imp2ΔSH3 | fic1Δ | + | pos1Δ | fic1Δ | + |
| imp2ΔSH3 | cyk3Δ | CS | pos1Δ | cyk3Δ | + |
| imp2ΔSH3 | pxl1Δ | SL | pos1Δ | pxl1Δ | + |
| imp2ΔSH3 | art1Δ | CS | pos1Δ | art1Δ | + |
| rgf3-3PA | cdc15ΔSH3(aa869) | + | art1Δ | spa2Δ | + |
| rgf3-3PA | imp2ΔSH3 | SS | art1Δ | pos1Δ | + |
| rgf3-3PA | spa2Δ | + | art1Δ | fic1Δ | SS |
| rgf3-3PA | pos1Δ | TS | art1Δ | cyk3Δ | + |
| rgf3-3PA | fic1Δ | + | art1Δ | cdc15ΔSH3 | CS |
| rgf3-3PA | cyk3Δ | + | art1Δ | imp2ΔSH3 | CS |
| rgf3-3PA | pxl1Δ | + | art1Δ | rgf3-3PA | ND |
| fic1Δ | cdc15ΔSH3(aa869) | + | cyk3Δ | cdc15ΔSH3(aa869) | SL |
| fic1Δ | imp2ΔSH3 | + | cyk3Δ | imp2ΔSH3 | CS |
| fic1Δ | spa2Δ | SS | cyk3Δ | spa2Δ | + |
| fic1Δ | pos1Δ | + | cyk3Δ | pos1Δ | + |
| fic1Δ | rgf3-3PA | + | cyk3Δ | rgf3-3PA | + |
| fic1Δ | cyk3Δ | + | cyk3Δ | fic1Δ | + |
| fic1Δ | pxl1Δ | SL | cyk3Δ | pxl1Δ | CS |
| fic1Δ | art1Δ | SS | cyk3Δ | art1Δ | + |
SL, synthetic lethality; SS, synthetic sick; CS and TS, cold (19°C) and heat (36°C) sensitive, respectively; +, no detected genetic interaction; ND, not done.
FIGURE 10:
Protein and genetic connectivity of SH3-domain network. Proteins are denoted in nodes (circles), with the specific alleles used to assay genetic interactions indicated in italics directly below. Each edge (connection between nodes) is color coded as follows: black is a protein–protein interaction (PP), red is synthetic lethal (SL), blue is synthetic sick (SS), teal denotes cold sensitive (CS), and orange denotes temperature-sensitive (TS) interactions.
DISCUSSION
More than 100 proteins localize to the cell division site in S. pombe, and most contain multiple protein-interaction domains, including SH3 domains. At least a dozen SH3 domain–containing proteins are present at the cell division site, and the SH3 ligand (PxxP) motif is significantly enriched in cytokinetic proteins compared with the complete proteome (74 vs. 57%, respectively; Wood et al., 2012). On the basis of our results, we infer that disruption of specific interactions between the SH3 domains of Cdc15 and Imp2 and their partners (e.g., Rgf3 and Spa2) alters the timing and/or completeness of protein recruitment. Our results underline the complexity and redundancy of cytokinetic protein recruitment, revealing the web of interactions that mediates the robustness of cytokinesis.
Using peptide libraries, we determined that Cdc15SH3 and Imp2SH3 preferentially select class I polyproline ligands in vitro (Figure 1, C and D). It is satisfying that this consensus preference matches the single binding site within Fic1 described previously, LPPVP; Bohnert and Gould, 2012), and also the motifs in Rgf3 and Spa2 that bind Cdc15SH3. It is also similar to the single polyproline motif (LPPLP) that confers binding between S. cerevisiae Hof1SH3 and Inn1p (Nishihama et al., 2009), the homologue of S. pombe Fic1. Thus the SH3 domains of Cdc15, Imp2, and Hof1 likely bind similar sets of proteins in vivo.
The previously uncharacterized protein identified here as an SH3 interactor, Spa2, localizes to arc-like structures at cell tips during interphase and the cell division site during cytokinesis and septation. This pattern is reminiscent of its homologues in other yeast and fungi, including S. cerevisiae (Snyder, 1989), Aspergillus nidulans (Virag and Harris, 2006), Ashbya gossypii (Knechtle et al., 2003), Ustilago maydis (Knechtle et al., 2003), Neurospora crassa (Araujo-Palomares et al., 2009), and C. albicans (Zheng et al., 2003). Spa2's localization pattern is largely due to its interactions with the SH3 domain of Cdc15 (at tips and CRs) and Imp2 (at CRs). However, these molecular interactions are not solely responsible for recruiting Spa2 to its sites of function, as Spa2-191A still localizes to the CR. We speculate that Spa2 is also recruited indirectly via other proteins connected to the Cdc15 and Imp2 SH3 network. Further study, particularly of Spa2's conserved N-terminal SHD, may provide a more complete picture of Spa2's mechanism of intracellular targeting.
Although S. pombe Spa2 localizes to sites of morphogenesis, its loss does not generate detectable defects in polarized growth, polarization of new cell ends formed after cytokinesis, or the formation of mating projections under typical laboratory growth conditions; however, Spa2 is required for cells to adopt a pseudohyphal growth pattern. Although these results contrast with the established role of S. cerevisiae Spa2 in regulation of morphogenesis rate and pattern in S. cerevisiae (Snyder, 1989; Gehrung and Snyder, 1990; Chenevert et al., 1994), they are in accord with the role of Spa2 in hyphal and/or invasive growth in C. albicans (Zheng et al., 2003), A. gossypii (Knechtle et al., 2003), U. maydis (Carbo and Perez-Martin, 2008), N. crassa (Araujo-Palomares et al., 2009), and A. nidulans (Virag and Harris, 2006). The functional difference with S. cerevisiae Spa2 may reflect the existence of redundant mechanisms controlling growth polarity under laboratory growth conditions and/or a rewiring of components into different signaling modules in S. pombe. In S. cerevisiae, Spa2 together with Pea2, Bud6, and the formin Bni1 have been defined as the polarisome (Fujiwara et al., 1998; Sheu et al., 1998), although S. cerevisiae Spa2 also associates with components of two mitogen-activated protein kinase pathways (Sheu et al., 1998; van Drogen and Peter, 2002) and other actin-binding proteins (Fujiwara et al., 1998; Shih et al., 2005). We did not detect physical or genetic interactions between Spa2 and the S. pombe homologues of Bni1, Bud6, or MAP kinases (Supplemental Table S2 and unpublished data). Despite these differences, it seems plausible that S. pombe Spa2 also performs a scaffolding function at sites of morphogenesis since we found that it is required for localizing the newly described Pos1. Although we do not know precisely what role either protein plays, our genetic analyses and others (Dixon et al., 2008; Ryan et al., 2012) implicate Spa2 in cell wall metabolism, a likely function, given that the protein is found only in cell-walled organisms.
Our data showing that the Cdc15 and Imp2 SH3 domains are required for timely and complete Rgf3 recruitment implicate these F-BAR proteins directly in the activation of cell wall formation, which is tightly coupled to cell division. Rgf3 is a specific guanine nucleotide exchange factor for Rho1 (Tajadura et al., 2004), and its loss leads to cell lysis at the time of division due to defects in secondary septum formation (Tajadura et al., 2004; Morrell-Falvey et al., 2005; Mutoh et al., 2005). Rho1 is required to activate glucan synthase activities (Arellano et al., 1996), including Bgs1 (required for primary septum formation; Liu et al., 1999; Cortes et al., 2007) and Bgs4 (required for primary septum completion and secondary septum formation; Munoz et al., 2013). In this regard, it is interesting that Pxl1, another Cdc15 and Imp2 SH3- interacting protein (Roberts-Galbraith et al., 2009), has also been implicated in modulating Rho1 activity during cytokinesis (Pinar et al., 2008). A previous study implicated the middle domain of Cdc15 as important in recruitment of Bgs1 to the cell division site, ascribing this as a key role for Cdc15 in cell division (Arasada and Pollard, 2014). However, such a role would not explain the CR defects observed in cells lacking just the Cdc15 C-terminal SH3 domain (Roberts-Galbraith et al., 2009; Arasada and Pollard, 2014), which are similar to those lacking Bgs4 (Munoz et al., 2013). Our data suggest that an important role of the F-BAR proteins Cdc15 and Imp2 is to form a network of proteins via their SH3 domains that coordinate the changes in cell wall metabolism necessary for the division process. These roles could include reorganization of the actin cytoskeleton or trafficking of appropriate cell wall synthesis enzymes in concert with CR constriction.
Other SH3 domain–containing proteins at the cell division site, including Cyk3 and at least 10 others, may be “branches” off the Cdc15SH3-Imp2SH3 recruitment network (Figure 10). This would explain why Spa2-191A and Rgf3-3PA still localize to the cell division site, albeit poorly, through indirect interactions. The range of genetic interactions detected among components of the SH3 domain network also supports this idea (Figure 10). Thus we have advanced our understanding of the complex web of protein interactions that mediate CR maturation and ultimately cell division. However, we note that we did not identify a known Cdc15SH3 interactor, Pxl1 (Roberts-Galbraith et al., 2009), using this approach. Thus there are likely more interactions mediated by the SH3 domains of the F-BAR scaffolding proteins in the CR yet to be discovered.
MATERIALS AND METHODS
Peptide phage display
Sequences encoding the SH3 domains of Cdc15 (amino acids 867–927) and Imp2 (amino acids 608–670) were cloned into the bacterial expression vector pGEX2T. GST fusion proteins were produced in E. coli and purified on GST-Bind beads (EMD Chemicals, San Diego, CA) according to the manufacturer's instructions.
Strains and media
S. pombe strains (Supplemental Table S4) were grown in yeast extract medium or minimal medium with appropriate supplements (Moreno et al., 1991). Transformations were performed by the lithium acetate method (Keeney and Boeke, 1994; Gietz et al., 1995). Epitope-tagged strains were constructed as described previously (Wach et al., 1994; Bahler et al., 1998), so that open reading frames were tagged at the 3′ end of endogenous loci with a GFP-KanR, mCherry-KanR, or TAP-KanR cassette. Appropriate tagging was confirmed by PCR, live-cell imaging, and/or immunoblotting. Strain construction and tetrad analysis were accomplished through standard methods. Overexpression of the art1, rgf3, or spa2 genes or fragments was accomplished using the nmt1 (strong) or nmt81 (weak) promoters in the pREP series of vectors (Basi et al., 1993). Transformants were grown in the presence of thiamine to dampen expression and then washed into thiamine-free medium to promote gene expression. To assay pseudohyphal invasion into 2% agar, 5 μl containing a total of 105 cells was spotted on 2% YE agar and incubated at 29°C for 20 d. Colonies were subsequently placed under a steady stream of water, and surface growth was wiped off using a paper towel.
Purification of Cdc15SH3 binding partners
Cell pellets were prepared from asynchronously growing untagged or fic1-FLAG cells or from arrested nda3-KM311 or nda3-KM311 fic1-FLAG cells (∼1.3 × 1011 cells/pellet). Lysates were prepared in NP-40 buffer and incubated with MBP or MBP-Cdc15SH3 on amylose beads (New England Biolabs, Ipswich, MA) as per the manufacturer's instructions. Beads with bound proteins were separated from the remaining lysate on Poly-Prep Chromatography columns (Bio-Rad, Hercules, CA). Beads were washed with 10 ml of ice-cold NP-40 lysis buffer (150 mM NaCl) and then with 1 ml of NP-40 LB with 200 mM NaCl. Bound protein was eluted with a series of 15 100-μl aliquots of NP-40 lysis buffer with 500 mM NaCl, followed by elution buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA, 10 mM maltose). A portion of each elution was analyzed by SDS–PAGE and immunoblotting (anti-FLAG) or silver staining. Fractions 7–9 from the 500 mM NaCl elutions were combined, TCA precipitated, and analyzed by MS as described in the following.
TAP and MS analysis
Proteins were purified by TAP as described (Tasto et al., 2001) and subjected to mass spectrometric analysis on a Thermo LTQ as previously detailed (McDonald et al., 2002; Roberts-Galbraith et al., 2009). Thermo RAW files were converted to MZXML or DTA files using Scansifter (version 2.1.25; Ma et al., 2011). Spectra with <20 peaks were excluded from our analysis. The remaining spectra were searched using Myrimatch (Tabb et al., 2007) or the SEQUEST algorithm (TurboSequest, version 27, revision 12, University of Washington, Seattle, WA) against the S. pombe protein database (created in May 2011 from pombase.org). Common contaminants were added, and all sequences were reversed to estimate the false discovery rate, yielding 10,352 total entries. Variable modifications (C+57; M+16; S,T,Y+80; for all spectra), strict trypsin cleavage, <10 missed cleavages, fragment mass tolerance 0.00 Da (due to rounding in SEQUEST, this results in 0.5-Da tolerance), and parent mass tolerance 2.5 Da were allowed. Peptide identifications were assembled and filtered in IDPicker (Ma et al., 2009) or Scaffold (version 4.3.2; Proteome Software, Portland, OR), using the following criteria: minimum of 99.9% protein identification probability; minimum of two unique peptides; minimum of 95% peptide identification probability; minimum peptide length of five amino acids; and minimum of one tryptic terminus.
Microscopy
Fixed- and live-cell images of S. pombe cells were acquired using one of the following: 1) a spinning disk confocal microscope (Ultraview LCI; PerkinElmer, Waltham, MA), which is equipped with a 100×/numerical aperture (NA) 1.40 PlanApo oil immersion objective, a 488-nm argon ion laser (GFP), a 594-nm helium neon laser (red fluorescent protein, mCherry), a charge-coupled device camera (Orca-ER; Hamamatsu Phototonics, Hamamatsu City, Japan), and MetaMorph 7.1 software (MDS Analytical Technologies; Molecular Devices, Sunnyvale, CA); or 2) a personal DeltaVision microscope system (GE Healthcare, Issaquah, WA), which includes an Olympus IX71 microscope, 60×/NA 1.42 PlanApo and 100×/NA 1.40 UPlanSApo objectives, fixed- and live-cell filter wheels, a Photometrics CoolSnap HQ2 camera, and softWoRx imaging software.
Time-lapse imaging was performed using an ONIX microfluidics perfusion system (CellASIC, Hayward, CA). Cells were loaded into Y04C plates for 5 s at 8 psi, and yeast extract (YE) liquid medium flowed into the chamber at 5 psi throughout imaging. For time-lapse and static images, Z-series optical sections were taken at 0.5-mm spacing. Time-lapse images were obtained at an interval between 2 and 4 min.
To directly compare wild-type and mutant populations within the same field of view, we incubated one population with fluorescently conjugated lectin (Sigma-Aldrich, St. Louis, MO), which labels cell walls. Specifically, 1 μl of a 5 mg/ml stock of fluorescein isothiocyanate–lectin in water was added to 1 ml of cells for a final concentration of 5 μg/ml. Cells were then incubated for 10 min at room temperature, washed three times, and resuspended in media. The lectin-labeled cell population and unlabeled cell population were mixed 1:1 immediately before imaging. The reciprocal labeling of populations was also done to account for any signal bleedthrough.
Intensity measurements were made with ImageJ software (National Institutes of Health, Bethesda, MD). For all intensity measurements of Spa2-GFP, the background was subtracted by creating a region of interest (ROI) in the same image where there were no cells (Waters, 2009). The area of the background was divided by the raw intensity of the background to give average intensity of background per pixel. This number was multiplied by the area of the ROI measuring Spa2-GFP localization and then subtracted from the raw intensity measurement of that ROI measuring Spa2-GFP localization to give the final intensity measurement (Waters, 2009). Measurements of Spa2-GFP localization were done in three biological replicates, and images were acquired on the DeltaVision microscope using Z-series optical sections of 0.2-mm spacing. Images for quantification were not deconvolved.
Immunoprecipitations and immunoblotting
Whole-cell lysates were prepared in NP-40 buffer in native conditions as previously described (Gould et al., 1991). Proteins were immunoprecipitated from protein lysates using anti-GFP (Roche, Nutley, NJ), anti-FLAG (Sigma-Aldrich), or anti-Myc (9E10), followed by Protein G Sepharose beads (GE Healthcare).
For immunoblotting, proteins were resolved by 3–8% Tris-acetate PAGE or 4–12% NuPAGE, transferred by electroblotting to a polyvinylidene difluoride membrane (Immobilon P; Millipore, Bedford, MA), and incubated with the set of primary antibodies indicated at 1 μg/ml. Primary antibodies were detected with secondary antibodies coupled to Alexa Fluor 680 (Life Technologies, Grand Island, NY) or IRDye800 (LI-COR Biosciences, Lincoln, NE) and visualized using an Odyssey Infrared Imaging System (LI-COR Biosciences).
Supplementary Material
Acknowledgments
We thank Raffi Tonikian for his assistance with the peptide array experiments and Fred Chang for the crn1-GFP strain. This work was supported by National Institutes of Health Grant GM101035 to K.L.G.
Abbreviations used:
- CR
contractile ring
- MS
mass spectrometry
- NETO
new end take off
- TRITC
tetramethylrhodamine isothiocyanate
- YE
yeast extract.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-10-1451) on November 26, 2014.
*Present address: Department of Cell and Developmental Biology, University of Illinois at Urbana–Champaign, Urbana, IL 61801.
REFERENCES
- Arasada R, Pollard TD. Distinct roles for F-BAR proteins Cdc15p and Bzz1p in actin polymerization at sites of endocytosis in fission yeast. Curr Biol. 2011;21:1450–1459. doi: 10.1016/j.cub.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasada R, Pollard TD. Contractile ring stability in S. pombe depends on F-BAR protein Cdc15p and Bgs1p transport from the Golgi complex. Cell Rep. 2014;8:1533–1544. doi: 10.1016/j.celrep.2014.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo-Palomares CL, Riquelme M, Castro-Longoria E. The polarisome component SPA-2 localizes at the apex of Neurospora crassa and partially colocalizes with the Spitzenkorper. Fungal Genet Biol. 2009;46:551–563. doi: 10.1016/j.fgb.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Arellano M, Duran A, Perez P. Rho 1 GTPase activates the (1–3)beta-D-glucan synthase and is involved in Schizosaccharomyces pombe morphogenesis. EMBO J. 1996;15:4584–4591. [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- Bohnert KA, Gould KL. Cytokinesis-based constraints on polarized cell growth in fission yeast. PLoS Genet. 2012;8:e1003004. doi: 10.1371/journal.pgen.1003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbo N, Perez-Martin J. Spa2 is required for morphogenesis but it is dispensable for pathogenicity in the phytopathogenic fungus Ustilago maydis. Fungal Genet Biol. 2008;45:1315–1327. doi: 10.1016/j.fgb.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Carnahan RH, Gould KL. The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe. J Cell Biol. 2003;162:851–862. doi: 10.1083/jcb.200305012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Gould KL. Sid4p is required to localize components of the septation initiation pathway to the spindle pole body in fission yeast. Proc Natl Acad Sci USA. 2000;97:5249–5254. doi: 10.1073/pnas.97.10.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenevert J, Valtz N, Herskowitz I. Identification of genes required for normal pheromone-induced cell polarization in Saccharomyces cerevisiae. Genetics. 1994;136:1287–1296. doi: 10.1093/genetics/136.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes JC, Konomi M, Martins IM, Munoz J, Moreno MB, Osumi M, Duran A, Ribas JC. The (1,3)beta-D-glucan synthase subunit Bgs1p is responsible for the fission yeast primary septum formation. Mol Microbiol. 2007;65:201–217. doi: 10.1111/j.1365-2958.2007.05784.x. [DOI] [PubMed] [Google Scholar]
- Demeter J, Sazer S. imp2, a new component of the actin ring in the fission yeast Schizosaccharomyces pombe. J Cell Biol. 1998;143:415–427. doi: 10.1083/jcb.143.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devrekanli A, Foltman M, Roncero C, Sanchez-Diaz A, Labib K. Inn1 and Cyk3 regulate chitin synthase during cytokinesis in budding yeasts. J Cell Sci. 2012;125:5453–5466. doi: 10.1242/jcs.109157. [DOI] [PubMed] [Google Scholar]
- Dixon SJ, Fedyshyn Y, Koh JL, Prasad TS, Chahwan C, Chua G, Toufighi K, Baryshnikova A, Hayles J, Hoe KL, et al. Significant conservation of synthetic lethal genetic interaction networks between distantly related eukaryotes. Proc Natl Acad Sci USA. 2008;105:16653–16658. doi: 10.1073/pnas.0806261105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodgson J, Brown W, Rosa CA, Armstrong J. Reorganization of the growth pattern of Schizosaccharomyces pombe in invasive filament formation. Eukaryotic Cell. 2010;9:1788–1797. doi: 10.1128/EC.00084-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Reymond A, Cerutti L, Utzig S, Hofmann K, Simanis V. The S. pombe cdc15 gene is a key element in the reorganization of F- actin at mitosis. Cell. 1995;82:435–444. doi: 10.1016/0092-8674(95)90432-8. [correction published in Cell (1997). 89, 1185] [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Tanaka K, Mino A, Kikyo M, Takahashi K, Shimizu K, Takai Y. Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1221–1233. doi: 10.1091/mbc.9.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W, Balasubramanian MK. Pxl1p, a paxillin-related protein, stabilizes the actomyosin ring during cytokinesis in fission yeast. Mol Biol Cell. 2008;19:1680–1692. doi: 10.1091/mbc.E07-07-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrung S, Snyder M. The SPA2 gene of Saccharomyces cerevisiae is important for pheromone-induced morphogenesis and efficient mating. J Cell Biol. 1990;111:1451–1464. doi: 10.1083/jcb.111.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS- DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- Gould KL, Moreno S, Owen DJ, Sazer S, Nurse P. Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 1991;10:3297–3309. doi: 10.1002/j.1460-2075.1991.tb04894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Trautmann S, McCollum D. Cytokinesis in eukaryotes. Microbiol Mol Biol Rev. 2002;66:155–178. doi: 10.1128/MMBR.66.2.155-178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney JB, Boeke JD. Efficient targeted integration at leu1–32 and ura4–294 in Schizosaccharomyces pombe. Genetics. 1994;136:849–856. doi: 10.1093/genetics/136.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knechtle P, Dietrich F, Philippsen P. Maximal polar growth potential depends on the polarisome component AgSpa2 in the filamentous fungus Ashbya gossypii. Mol Biol Cell. 2003;14:4140–4154. doi: 10.1091/mbc.E03-03-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang H, McCollum D, Balasubramanian MK. Drc1p/Cps1p, a 1,3-beta-glucan synthase subunit, is essential for division septum assembly in Schizosaccharomyces pombe. Genetics. 1999;153:1193–1203. doi: 10.1093/genetics/153.3.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma ZQ, Dasari S, Chambers MC, Litton MD, Sobecki SM, Zimmerman LJ, Halvey PJ, Schilling B, Drake PM, Gibson BW, Tabb DL. IDPicker 2.0: Improved protein assembly with high discrimination peptide identification filtering. J Proteome Res. 2009;8:3872–3881. doi: 10.1021/pr900360j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma ZQ, Tabb DL, Burden J, Chambers MC, Cox MB, Cantrell MJ, Ham AJ, Litton MD, Oreto MR, Schultz WC, et al. Supporting tool suite for production proteomics. Bioinformatics. 2011;27:3214–3215. doi: 10.1093/bioinformatics/btr544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- Mayer BJ. SH3 domains: complexity in moderation. J Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- McDonald WH, Ohi R, Miyamoto DT, Mitchison TJ, Yates JR., III Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int J Mass Spectrom. 2002;219:245–251. [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Morrell-Falvey JL, Ren L, Feoktistova A, Haese GD, Gould KL. Cell wall remodeling at the fission yeast cell division site requires the Rho-GEF Rgf3p. J Cell Sci. 2005;118:5563–5573. doi: 10.1242/jcs.02664. [DOI] [PubMed] [Google Scholar]
- Munoz J, Cortes JC, Sipiczki M, Ramos M, Clemente-Ramos JA, Moreno MB, Martins IM, Perez P, Ribas JC. Extracellular cell wall beta(1,3)glucan is required to couple septation to actomyosin ring contraction. J Cell Biol. 2013;203:265–282. doi: 10.1083/jcb.201304132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh T, Nakano K, Mabuchi I. Rho1-GEFs Rgf1 and Rgf2 are involved in formation of cell wall and septum, while Rgf3 is involved in cytokinesis in fission yeast. Genes Cells. 2005;10:1189–1202. doi: 10.1111/j.1365-2443.2005.00908.x. [DOI] [PubMed] [Google Scholar]
- Nishihama R, Schreiter JH, Onishi M, Vallen EA, Hanna J, Moravcevic K, Lippincott MF, Han H, Lemmon MA, Pringle JR, Bi E. Role of Inn1 and its interactions with Hof1 and Cyk3 in promoting cleavage furrow and septum formation in S. cerevisiae. J Cell Biol. 2009;185:995–1012. doi: 10.1083/jcb.200903125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Pinar M, Coll PM, Rincon SA, Perez P. Schizosaccharomyces pombe Pxl1 is a paxillin homologue that modulates Rho1 activity and participates in cytokinesis. Mol Biol Cell. 2008;19:1727–1738. doi: 10.1091/mbc.E07-07-0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmann J, Fleig U. Asp1, a conserved 1/3 inositol polyphosphate kinase, regulates the dimorphic switch in Schizosaccharomyces pombe. Mol Cell Biol. 2010;30:4535–4547. doi: 10.1128/MCB.00472-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard LW, Onishi M, Pringle JR, Lord M. Fission yeast Cyk3p is a transglutaminase-like protein that participates in cytokinesis and cell morphogenesis. Mol Biol Cell. 2012;23:2433–2444. doi: 10.1091/mbc.E11-07-0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Chen JS, Wang J, Gould KL. The SH3 domains of two PCH family members cooperate in assembly of the Schizosaccharomyces pombe contractile ring. J Cell Biol. 2009;184:113–127. doi: 10.1083/jcb.200806044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Gould KL. Setting the F-BAR: functions and regulation of the F-BAR protein family. Cell Cycle. 2010;9:4091–4097. doi: 10.4161/cc.9.20.13587. [DOI] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Ohi MD, Ballif BA, Chen JS, McLeod I, McDonald WH, Gygi SP, Yates JR, 3rd, Gould KL. Dephosphorylation of F-BAR protein Cdc15 modulates its conformation and stimulates its scaffolding activity at the cell division site. Mol Cell. 2010;39:86–99. doi: 10.1016/j.molcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CJ, Roguev A, Patrick K, Xu J, Jahari H, Tong Z, Beltrao P, Shales M, Qu H, Collins SR, et al. Hierarchical modularity and the evolution of genetic interactomes across species. Mol Cell. 2012;46:691–704. doi: 10.1016/j.molcel.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Santos B, Fortin N, Costigan C, Snyder M. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol Cell Biol. 1998;18:4053–4069. doi: 10.1128/mcb.18.7.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih JL, Reck-Peterson SL, Newitt R, Mooseker MS, Aebersold R, Herskowitz I. Cell polarity protein Spa2P associates with proteins involved in actin function in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:4595–4608. doi: 10.1091/mbc.E05-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. The SPA2 protein of yeast localizes to sites of cell growth. J Cell Biol. 1989;108:1419–1429. doi: 10.1083/jcb.108.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, Fernando CG, Chambers MC. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res. 2007;6:654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajadura V, Garcia B, Garcia I, Garcia P, Sanchez Y. Schizosaccharomyces pombe Rgf3p is a specific Rho1 GEF that regulates cell wall beta-glucan biosynthesis through the GTPase Rho1p. J Cell Sci. 2004;117:6163–6174. doi: 10.1242/jcs.01530. [DOI] [PubMed] [Google Scholar]
- Tasto JJ, Carnahan RH, Hayes McDonald W, Gould KL. Vectors and gene targeting modules for tandem affinity purification in Schizosaccharomyces pombe. Yeast. 2001;18:657–662. doi: 10.1002/yea.713. [DOI] [PubMed] [Google Scholar]
- Tonikian R, Xin X, Toret CP, Gfeller D, Landgraf C, Panni S, Paoluzi S, Castagnoli L, Currell B, Seshagiri S, et al. Bayesian modeling of the yeast SH3 domain interactome predicts spatiotemporal dynamics of endocytosis proteins. PLoS Biol. 2009;7:e1000218. doi: 10.1371/journal.pbio.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Drogen F, Peter M. Spa2p functions as a scaffold-like protein to recruit the Mpk1p MAP kinase module to sites of polarized growth. Curr Biol. 2002;12:1698–1703. doi: 10.1016/s0960-9822(02)01186-7. [DOI] [PubMed] [Google Scholar]
- Virag A, Harris SD. Functional characterization of Aspergillus nidulans homologues of Saccharomyces cerevisiae Spa2 and Bud6. Eukaryotic Cell. 2006;5:881–895. doi: 10.1128/EC.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vjestica A, Tang XZ, Oliferenko S. The actomyosin ring recruits early secretory compartments to the division site in fission yeast. Mol Biol Cell. 2008;19:1125–1138. doi: 10.1091/mbc.E07-07-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Poehlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wachtler V, Huang Y, Karagiannis J, Balasubramanian MK. Cell cycle-dependent roles for the FCH-domain protein Cdc15p in formation of the actomyosin ring in Schizosaccharomyces pombe. Mol Biol Cell. 2006;17:3254–3266. doi: 10.1091/mbc.E05-11-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC. Accuracy and precision in quantitative fluorescence microscopy. J Cell Biol. 2009;185:1135–1148. doi: 10.1083/jcb.200903097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe BA, Gould KL. Split decisions: coordinating cytokinesis in yeast. Trends Cell Biol. 2005;15:10–18. doi: 10.1016/j.tcb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, Aslett M, Lock A, Bahler J, Kersey PJ, Oliver SG. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 2012;40:D695–699. doi: 10.1093/nar/gkr853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Kuhn JR, Kovar DR, Pollard TD. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Ye Y, Wang N, Pollard TD, Pringle JR. Cooperation between the septins and the actomyosin ring and role of a cell-integrity pathway during cell division in fission yeast. Genetics. 2010;186:897–915. doi: 10.1534/genetics.110.119842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XD, Wang YM, Wang Y. CaSPA2 is important for polarity establishment and maintenance in Candida albicans. Mol Microbiol. 2003;49:1391–1405. doi: 10.1046/j.1365-2958.2003.03646.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.