Abstract
Clinical classification of sequence variants identified in hereditary disease genes directly affects clinical management of patients and their relatives. The International Society for Gastrointestinal Hereditary Tumours (InSiGHT) undertook a collaborative effort to develop, test and apply a standardized classification scheme to constitutional variants in the Lynch Syndrome genes MLH1, MSH2, MSH6 and PMS2. Unpublished data submission was encouraged to assist variant classification, and recognized by microattribution. The scheme was refined by multidisciplinary expert committee review of clinical and functional data available for variants, applied to 2,360 sequence alterations, and disseminated online. Assessment using validated criteria altered classifications for 66% of 12,006 database entries. Clinical recommendations based on transparent evaluation are now possible for 1,370 variants not obviously protein-truncating from nomenclature. This large-scale endeavor will facilitate consistent management of suspected Lynch Syndrome families, and demonstrates the value of multidisciplinary collaboration for curation and classification of variants in public locus-specific databases.
Identification of a high-risk disease-causing constitutional mutation in a cancer patient guides the clinical management of the whole family, with implications for counselling, cancer treatment options, pre-symptomatic surveillance, and consideration of risk-reducing surgery and/or medication regimes1. Carriers of mutations in the mismatch repair (MMR) genes MLH1, MSH2, MSH6 and PMS2 causing Lynch Syndrome (LS)1 have a substantially increased risk of colorectal and endometrial cancer, along with increased risk of ovarian, gastric, small bowel, urothelial, brain, hepatobiliary, pancreatic, bladder, kidney, prostate, and breast cancers1-8. However, intensive management reduces mortality9.
Sequence variants of uncertain functional and clinical significance are common in genetic test reports. Although several lines of evidence can be evaluated to assess their significance, usually none of them can be used on its own to obtain clinically useful variant interpretation, and for many variants comprehensive data are lacking. Laboratories are generally conservative in designating pathogenic variants, assigning variants as “uncertain significance” unless overwhelming evidence of pathogenicity exists. Several schemes for classifying variants in genes associated with Mendelian conditions have been proposed for use in the clinical setting. Since clinically useful actions are currently only considered for high-penetrant mutations, all of these systems are aimed at differentiating high-penetrant from low-penetrant/neutral variants and do not consider intermediate risk variants. They differ in the range and format of data used for classification, and the number of variant classes10-12. The International Agency for Research on Cancer (IARC) classification system, endorsed by the Human Variome Project (HVP), facilitates standardized categorization by defining classes that can be linked to validated quantitative measures of causality/pathogenicity from statistical models13-16, or from validated interpretation of qualitative data17. Importantly, only the 5-class IARC system has been linked to clinical recommendations for all classes: clinical testing and full high-risk surveillance guidelines for Class 5 “pathogenic” and Class 4 “likely pathogenic”; advice to treat as “no mutation detected for this disorder” for Class 1 “not pathogenic” and Class 2 “likely not pathogenic”; and acquisition of additional data to provide more robust classifications for Class 2, Class 4 and Class 3 “uncertain”.
Locus-specific databases (LSDBs) are an important source of information for clinicians and researchers to assess data as well as opinion on the clinical relevance of disease gene sequence variants, and have a fundamental role in variant classification due to the added value of aggregated data. Consistent and normalized data curation is critical to the value of databases for categorizing the relationship between genetic variation and disease – especially for clinical application. It has previously been recommended by the IARC Working Group that a panel covering a range of expertise in variant classification provide consensus opinion on variant pathogenicity prior to publicly accessible display of such information18. Another important component of the classifications provided by LSDBs is transparency regarding the criteria and supporting information used for classification, so that LSDB users can consider the information for their own application in the research and clinical setting18.
The International Society for Gastrointestinal Hereditary Tumours (InSiGHT) has merged multiple gene mutation/variant repositories to create the InSiGHT Colon Cancer Gene Variant Database for MMR and other colon cancer susceptibility genes19-23, hosted by Leiden Open Variation Database (LOVD). Following recommendations for LSDB curation18, InSiGHT formed an international panel of researchers and clinicians to review MMR gene variants submitted to the database. To encourage submission of unpublished clinical and research data to further facilitate variant classification, the microattribution approach24 was implemented using Open Researcher and Contributor Identifications (ORCID). Here we present the results of the InSiGHT Variant Interpretation Committee (VIC) effort to develop, test and apply a five-tiered scheme to classify 2,360 unique constitutional MMR gene variants.
Curation of MMR gene variants submitted to the InSiGHT Colon Cancer Gene Variant Databases
Through December 2012, after 3,458 alterations to standardize nomenclature, there were 12,635 submissions of 2,730 unique MMR gene variants lodged in the InSiGHT database. Furthermore, 370 (13.6%) unique variants were not identified in constitutional (germline) DNA (see Supplementary Fig. 1 and Supplementary Table 1 for details), and were excluded from further analyses since: (i) no evidence exists that these occur as constitutional variants, and (ii) no clinical information was available to assess their potential role in hereditary disease. The 2,360 constitutional variants included: 932 MLH1 (39%), 842 MSH2 (36%), 449 MSH6 (19%), and 137 PMS2 (6%). Most variants were nonsense/frameshift predicted to cause protein truncation (800, 34%), followed by “not obviously truncating” non-synonymous variants as the next largest group, including missense substitutions, small in-frame insertions/deletions (indels) and read-through alterations of the translation termination codon (746, 32%).
Variants had originally been assigned a classification by submitters according to the following classes: pathogenic; probably pathogenic; no known pathogenicity; probably no pathogenicity; effect unknown. No information was recorded to document the rationale for classification, or standards to classify variants. Considering 1,382 constitutional variants with multiple entries in the InSiGHT database, there were discordances in classification between submitters for 869 variants. Some of these discordances arose because of classification based on single data points or references, such as single functional assay results22, or inferences from individual publications originally lodged in the Mismatch Repair Genes Variant Database23 (see example in Supplementary Table 2).
Development of a five-tiered system for consistent classification of MMR gene variants
The InSiGHT VIC (see Methods) was established in 2007 to address discrepancy in classification of MMR gene variants lodged on the InSiGHT database. Since March 2011 the VIC has made a concerted effort to develop standardized criteria for variant classification, employing a modified “Delphi consensus process”25 to evaluate current scientific evidence and reach consensus. In line with the HVP26, the IARC classification system10 for variant categorization (see Table 1) was adopted by InSiGHT for MMR variant classification. Briefly, multiple lines of evidence are required for classification, and evidence for each variant must include data associating the variant with both clinical and functional consequences (see Methods).
Table 1. InSiGHT variant classification scheme with accompanying recommendations for family management, adapted from the IARC 5-tiered classification system*.
| InSiGHT MMR variant class definition for Lynch syndrome** |
Predictive testing of at-risk relatives |
Surveillance for positive at-risk relatives | Research testing of relatives |
|---|---|---|---|
| 5: Pathogenic | Yes | Full high-risk guidelines | Not indicated |
| 4: Likely pathogenic | Yes*** | Full high-risk guidelines | Yes |
| 3: Uncertain | No*** | Based on family history & other risk factors |
Yes |
| 2: Likely not pathogenic | No*** | Based on family history & other risk factors. Treat as “no mutation detected” in this gene for this disorder |
Yes |
| 1: Not pathogenic | No*** | Based on family history & other risk factors. Treat as “no mutation detected” in this gene for this disorder |
Not indicated |
Adapted from Plon et al10. Full high-risk surveillance guidelines for cancers in the Lynch spectrum are outlined in Vasen et al1. Research testing entails cascade testing for the variant in affected and unaffected family members to facilitate segregation analysis, and is indicated for variants in classes 2-4 to refine classification. Consent from subjects through a protocol approved by a human subjects committee should be obtained.
Class definition is described in detail in the Supplementary Note and Supplementary Table 4, and is based on quantitative evidence defined by multifactorial likelihood posterior probability (with cut points >0.99 for Class 5; 0.95-0.99 for Class 4; 0.05-0.949 for Class 3; 0.001-0.049 for Class 2; <0.001 for Class 1) or combined qualitative evidence determined by consensus opinion as defined by the InSiGHT Variant Interpretation Committee. “Pathogenic” is defined as “clinically relevant in a genetic counseling setting such that germline variant status will be used to inform patient and family management.”
Recommend continued testing of proband for any additional available testing modalities available e.g. rearrangements.
The scheme was first tested on a subset of 117 MMR gene variants, and the criteria evolved and were refined by consensus to accommodate new data and inconsistencies over multiple classification teleconferences and face-to-face meetings. Final criteria were then applied retrospectively and to all remaining unique variants listed in the database (see Supplementary Table 3). Figure 1 shows an overview of the InSiGHT classification criteria (see Supplementary Note, Supplementary Table 4 for detailed criteria and justifications). At the close of each VIC teleconference/meeting, consensus classifications were noted. Where necessary, action items to improve or clarify classifications included:
Calls for missing clinical and functional information for specific variants to committee members and the general InSiGHT membership.
Requests for more detailed data or data clarifications from the authors of original publications.
Re-assessment of classification after additional data were obtained.
At the end of the process, the InSiGHT database was updated with the final consensus classifications and the supporting data, to ensure transparency.
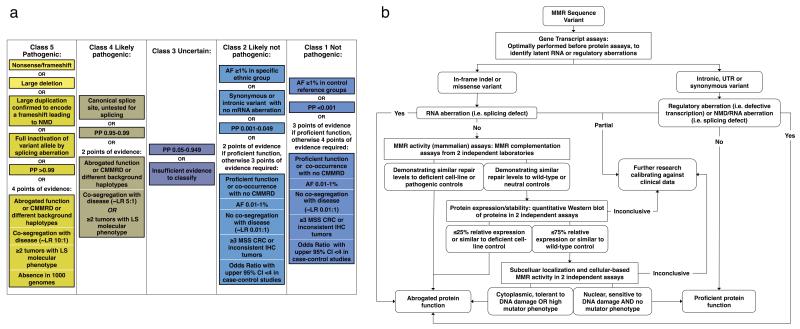
Figure 1.
Overview of 5-tiered InSiGHT classification guidelines.
(a) Simplified guidelines describing levels and types of evidence required to reach different classes. See the supplementary information for the full guidelines (Supplementary Note) and detailed rationale behind each criterion (Supplementary Table 3). The Lynch Syndrome molecular phenotype described in Classes 5 and 4 includes microsatellite instability and/or loss of expression of relevant protein(s) as determined by immunohistochemistry. In this study, variants resulting in a premature termination codon or large genomic deletions of functionally important domains, generally considered pathogenic on the basis of DNA sequence alone, are referred to as Class 5a “assumed pathogenic” variants. All other variants reaching class 5 are termed Class 5b. (b) Flowchart used to assist in interpretation of available functional assay data. Assays reviewed for classification are shown in Supplementary Table 4, and the values used to define abrogated or normal function are shown in Supplementary Table 5. The cut-offs <25% and >75% set for protein expression, as used in previous publications47,48, are very conservative given reported abrogated function associated with MLH1 expression defects of ~50% or lower49. For variants that had normal/inconclusive/intermediate MMR activity in 2 independent assays, but deficient protein function in 2 independent assays, abrogated function was assigned. AF – allele frequency; PP – posterior probability of pathogenicity derived by multifactorial likelihood analysis; CMMRD – constitutional mismatch repair deficiency (MIM 276300); LR – likelihood ratio; LS – Lynch Syndrome; MSS – microsatellite stable; CRC – colorectal cancer; IHC – immunohistochemistry; NMD – nonsense mediated decay.
The major issues faced by the committee in the review process were determining data redundancy across multiple sources (resolved through discussion with authors), paucity of information, incomplete/inaccurate data, and difficulties in interpretation of functional assays. To facilitate functional assay interpretation, supporting information and flowcharts were developed (Fig. 1b, Supplementary Tables 5-6), and multiple meetings dedicated to review variants with apparently discordant functional assay results were coordinated (Supplementary Table 3).
Validation of the InSiGHT qualitative classification criteria
Nonsense or frameshift alterations, or large genomic deletions interrupting functionally important domains are generally considered pathogenic on the basis of DNA sequence alone; here these are referred to as “assumed pathogenic” variants (termed Class 5a in figures). There were 990 “assumed pathogenic” variants in the database, 640 of which were private mutations. In order to demonstrate the robustness of the qualitative classification criteria, 170 “assumed pathogenic” variants (68 MLH1, 75 MSH2, 13 MSH6, 14 PMS2) were reviewed as a validation set against the Class 5 (pathogenic) qualitative criteria required for the variants termed Class 5b in figures (see Methods, Supplementary Table 7). Class 5b designation required: evidence of abrogated protein function; at least two tumors with microsatellite instability (MSI) or appropriate loss of MMR protein expression; and a segregation likelihood ratio >10:1 (incorporating gene-specific cumulative risks27) or variant co-segregation with disease reported in at least two Amsterdam criteria positive families. Class 5b was attained by all 60 validation set variants that had sufficient clinical data to assess these required criteria. The other 110 validation set variants could not be assigned to Class 5b largely because family co-segregation and tumor data were scarce or unobtainable - presumably because these variants are accepted as disease-causing and routinely used for clinical presymptomatic testing in families (see Implementing Microattribution). Of these, 72 were assigned to Class 4 due to lack of only one point of evidence, and 38 variants fell in Class 3 due to insufficient data. However, only 2/13 MSH6 and 2/14 PMS2 variants fulfilled Class 5, reflecting the lower penetrances and later ages of onset of PMS228 and MSH629 deleterious variants. Together these results indicate that the criteria for classification using qualitative data were sufficiently stringent to ensure conservative classification.
Application of InSiGHT classification guidelines to 2,360 individual constitutional MMR variants
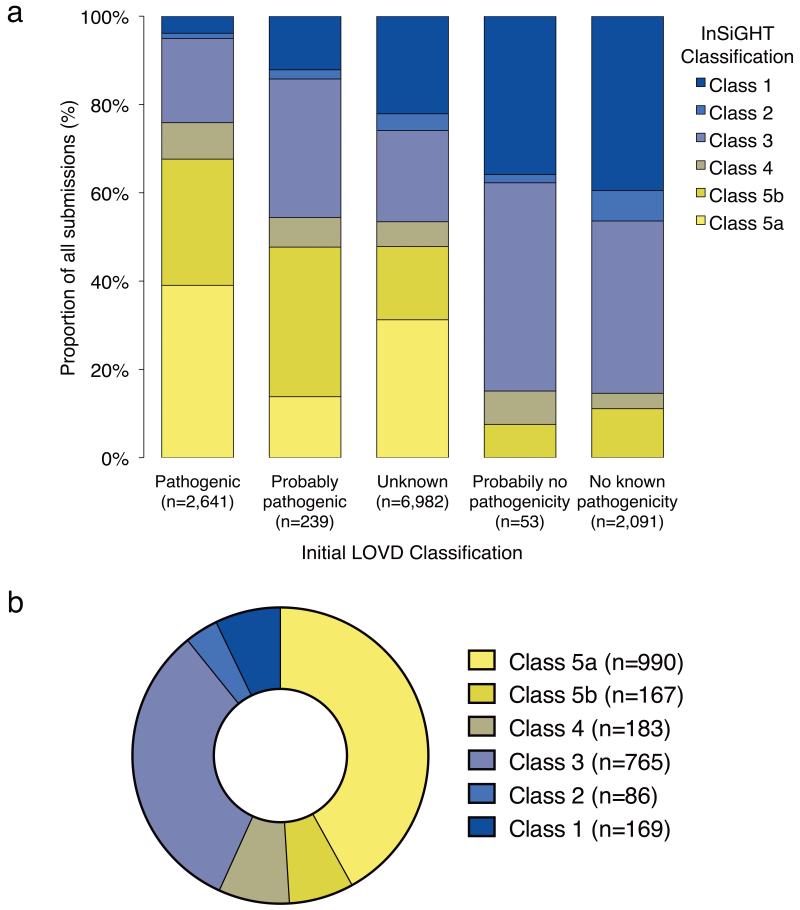
Of the 12,006 eligible variant entries in the InSIGHT database, submitter versus final classification differed for 7,935 (66%), including changes from “no known pathogenicity” to Class 5 (pathogenic) and vice versa (Fig. 2a). The overall breakdown of final classifications is shown in Figure 2b. In addition to the 990 “assumed pathogenic” truncating/large deletion variants (Class 5a), consistent medical management is now also possible for the remaining 1,370 “not obviously truncating” variants; these include 167 (12%) Class 5 (pathogenic) variants (Class 5b), 183 (14%) Class 4 (likely pathogenic) variants, 86 (6%) Class 2 (likely not pathogenic) variants and 169 (12%) Class 1 (not pathogenic) variants.
Figure 2. Outcome of standardized 5-tiered InSiGHT classification of constitutional MMR gene variants.
a) The plot represents the proportion of the 5-tiered classifications for all documented constitutional variants in the database, against the original LOVD database classifications assigned by submitters for each entry. Class 5a is a subset of Class 5 containing the assumed pathogenic nonsense mutations, small frameshift indels, and large deletions. Class 5b includes not-obviously truncating variants considered to be pathogenic on the basis of combined evidence (See Supplementary Note). Results show that standardized classification led to altered classifications for a considerable proportion of variant entries, including downgrading for variants submitted as pathogenic (24%), and upgrading of variants with unknown pathogenicity to likely pathogenic (5.6%) or pathogenic (48%). In addition, clinically important misclassifications were identified for unique variants initially submitted as not pathogenic (54 unique variants reclassified as Class 5b, and 25 reclassified as Class 4) and unique variants submitted as pathogenic (28 unique variants reclassified as Class 1, 16 reclassified as Class 2, and 218 reclassified as Class 3). b) Pie chart showing distribution of final InSiGHT VIC classifications.
As shown in Figure 3 and Supplementary Figure 2, non-synonymous variants (see Fig. 3 footnote) made up the majority of Class 3 variants (524/765; 68%) and newly assigned Class 5b variants (91/167; 54%; see Supplementary Table 8 for detailed information supporting classifications). Substitutions of canonical dinucleotide splice sites fell predominantly in Class 4, due to lack of functional RNA analyses; however if experimentally tested these would likely move to Class 5b. Intronic variants outside conserved splice sites were the most prevalent variant type in Class 1.
Figure 3. Classifications of all documented unique variants by variant type.
The plot represents the proportion of the different variant types within the 5 classes. Class 5a is a subset of Class 5 containing the assumed pathogenic mutations (nonsense mutations, small frameshift indels, and large deletions). All other variants reaching class 5 are termed Class 5b (see Supplementary Note). The different variant types are: PTC – variants that introduce premature terminating codons, i.e. nonsense mutations and small frameshift indels; LGDel – large genomic deletions or disrupting inversions; LGDup – large genomic duplications; SS – variants in the canonical splice site dinucleotides; NS – not obviously truncating non-synonymous variants outside the Kozak consensus sequence i.e. missense, small in-frame insertion/deletions, and read-through translation termination codon alterations; S – synonymous variants; I – intronic variants outside the canonical splice site dinucleotides; ATG/UTR – variants in the initiation codon, and the 5′ or 3′ untranslated regions. See Supplementary Figure 2 for further details of variant types, by MMR gene.
Final categorization (see methods) of “not obviously truncating” variants as Class 1, 2, 4 or 5 was achieved by applying qualitative criteria for 391 variants, using quantitative multifactorial likelihood analysis methodology for 192 variants (based on bioinformatic prior probability plus evidence from segregation and/or tumour data, see Thompson et al16), and either quantitative or qualitative criteria for 26 variants. Where classifications derived using quantitative versus qualitative criteria differed, this reflected the amount of data available rather than deficiencies in the classification criteria, with no variants considered Class 1/2 using one approach and Class 4/5 using the other. Six synonymous variants reached Class 5b due to their effects on splicing. For substitutions occurring in initiation codons (often assumed to be pathogenic30-32), only 1/9 had sufficient evidence to determine pathogenicity.
Implementing Microattribution
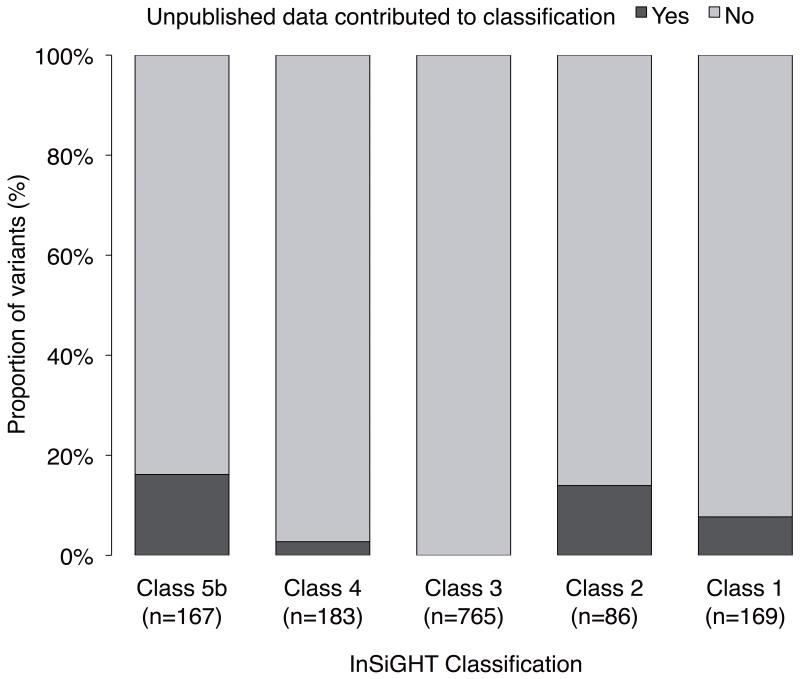
Microattribution is a means to incentivize placement of unpublished data in the public domain, by assigning scholarly contribution to authors similar to citations conventions afforded to journal articles33. Retrospective and prospective microattribution was implemented to acknowledge and encourage the submission of unpublished data to the InSiGHT database, including submission of additional detailed clinical information from authors of published reports. Microattribution was assigned for initial variant submission, segregation and family history data, pathology (MSI, immunohistochemistry) information, in vitro functional assays (mainly RNA splicing), and variant frequency in normal individuals. As of July 2013, a total of 6,015 microattributions were conferred, including 3,763 for variant submission, 2,111 for family and tumor pathology data, 97 for in vitro assays, and 25 for frequency data. Notably, 19% of the clinical and functional microattributions contributed additional information critical to classification of the “assumed pathogenic” validation subset (Class 5a). These data also highlighted that clinical testing for “assumed pathogenic” variants is mostly undertaken in the presymptomatic setting (see above). The contribution of microattribution to final classification of “not obviously truncating” variants is shown in Figure 4. Importantly, classification was altered for 57/169 (34%) variants for which novel unpublished data were obtained. Moreover, implementation of microattribution stimulated submission of 128 novel MMR variants, yet to be classified.
Figure 4. Contribution of microattribution to classification of “not obviously truncating” variants.
Dark shading (YES) indicates the proportion of variants for each class, where the additional data obtained through microattribution contributed to their final classification.
Preliminary Analysis of Class 3 Variants of Uncertain Significance
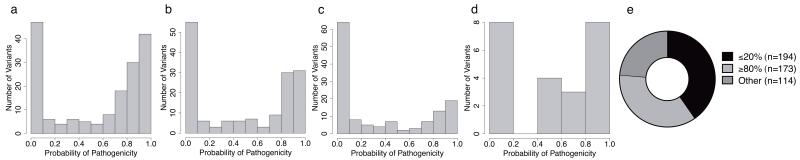
Missense variants in MMR genes are abundant among Class 3 (uncertain) variants and present a considerable clinical problem. Quantitative multifactorial likelihood analysis is an effective approach to missense variant classification, since validated bioinformatic predictions34 based on amino acid conservation and physicochemical properties can be used as a surrogate for in vitro variant effect on protein function. In silico analyses previously shown to have high accuracy (area under receiving operator characteristic [ROC] curve 0.93)34 were used to estimate the prior probability of pathogenicity for all 481 Class 3 (uncertain) missense variants (see Fig. 5), to prioritize requests for data to facilitate future multifactorial analysis. The distribution of prior probabilities for MLH1 and MSH2 Class 3 variants is clearly bimodal, suggesting that ~50% of MLH1/MSH2 missense variants may be classified as pathogenic after further investigation. In total, 401 missense variants had extreme prior probabilities of 20% or ≥80%, 270 of which were <10% or >90%, indicating that Class 1 or Class 5 could be easily reached by incorporating segregation or tumor information. It is also possible that some Class 3 variants with low prior probability of pathogenicity based on predicted missense alteration will cause splicing aberrations, as already observed for 42/746 of not obviously truncating non-synonymous variants. Incorporation of validated bioinformatic splicing prediction tools into the MMR gene multifactorial model, as is under development for BRCA1/235, will assist prioritization of such likely spliceogenic variants.
Figure 5. Probabilities of pathogenicity for 481 Class 3 “uncertain” missense variants, derived by in silico analysis.
Distribution of probabilities of pathogenicity as estimated from a calibrated algorithm based on customized MAPP and PolyPhen2 scores34, for (a) MLH1, n=186; (b) MSH2, n=169; (c) MSH6, n=145; (d) PMS2, n=24; (e) all four MMR genes, showing stratification of variants with prior probabilities ≤20% or ≥80% to prioritize variants for further investigation and classification.
Investigating potential effects of Class 3 regulatory variants (see methods) showed that all 15 5′UTR variants fell within multiple transcription factor binding sites, but no evidence for miRNA binding interruption for six 3′UTR variants was found (data not shown). Multifactorial analyses and transcription assays would help elucidate if these variants affect gene function.
Discussion
The InSiGHT VIC has successfully undertaken a collaborative effort to establish standardized variant interpretation guidelines using a modified Delphi process, encourage data submission, and provide objective assessment of MMR gene variants involved in Lynch Syndrome. The criteria developed provide a basis for standardised clinical classification of variants to inform patient and family management by genetic counselling10. This initiative has achieved the systematic evaluation of 2,360 constitutional variants, which will benefit thousands of families internationally. Importantly, 605 variants not resulting in premature termination codons, including 217 non-synonymous substitutions, have now been assigned to Class 5 (pathogenic) and Class 4 (likely pathogenic), or Class 1 (not pathogenic) and Class 2 (likely not pathogenic). These can now also be used as standards for the calibration of functional assays36,37.
The clinical significance of 32% of the variants investigated remains uncertain. A large proportion (71%) of these were “private” variants occurring in only one family, which are problematic to classify due to the paucity of available clinical information. Clinicians play a fundamental role in promoting collection of segregation and other information relevant for classification. We anticipate that development of this interpretation scheme, plus the implementation of microattribution, will create incentive to assist in accumulating clinical data. The value of microattribution for data accrual has previously been demonstrated for hemoglobinopathies24, and the InSiGHT initiative now demonstrates the clinical utility of data collection. The promotion of standardised data formats will assist transition into fully quantitative unbiased classification, eventually incorporating other components of the qualitative guidelines. In addition, the difficulties experienced in interpreting apparently discordant functional assays emphasize the importance of assay validation and standardization38,39. Such experience will be directly applicable to functional analysis of deep intronic and regulatory variants, which are increasingly detected with the advancement of DNA sequencing technologies.
To accommodate the lower penetrance and reported lesser degrees of tumor MSI associated with MSH6 and PMS2 mutations28,29,40-44, gene-specific criteria should also be considered for future iterations of the classification guidelines e.g., stipulating inclusion of segregation odds for MSH6 and PMS2 variants for classification, and use of modified panels to detect MSI status.
Another challenging issue to contemplate will be incorporating intermediate risk alleles45 into classification schemes, including the clinical recommendations that might be linked to such variants. The identification of a subset of MMR gene alleles with apparently discordant clinical and functional features that renders them “resistant” to classification will provide the basis for future studies to define the most appropriate methodology and criteria to identify such variants. Further studies will also be required to assess if variants with abrogated DNA damage response but normal mismatch repair46 are associated with the same clinical features as classical pathogenic mutations in MMR genes.
The InSiGHT database is a well-recognized resource for the clinical and research community, receiving over 20,000 hits/month. The development and adoption of standard templates allows transparency in the review process. Database users can view relevant information and sources in relation to guideline interpretation when considering the classification provided by the committee. The guidelines must evolve to accommodate additional kinds of evidence, but we anticipate no clinical issues as long as the variant classifications are dated and linked to a dated set of guidelines with the supporting information used to derive the classification. The final classifications have also been submitted to NCBI’s ClinVar for higher exposure, but expert classifications and underlying data rest with InSiGHT.
This is the first large-scale comprehensive classification effort demonstrating the value of expert panel evaluation to curation of an LSDB, and providing summary information used to assign variant pathogenicity. It also shows how classification may be assisted by promoting standardized data submission from stake-holders in the clinical and research setting, in order to access unpublished clinical and functional information to facilitate variant classification. Therefore, the InSiGHT initiative provides an important model of LSDB-centric multidisciplinary collaboration for transparent DNA variant interpretation.
Online Methods
InSiGHT Variant Interpretation Committee expertise
The InSiGHT Variant Interpretation Committee (VIC) (current chair author MG), includes 40 multidisciplinary experts from 5 continents (See Supplementary Table 9 for disciplines covered by VIC members). The Committee is responsible to its Governance Committee, which in turn is responsible to the InSiGHT Council. InSiGHT has recently joined the Human Variome Project and is a founding member of its Gene and Disease Specific Council. The InSiGHT Council specifically considered the need and responsibility associated with classification assignment on its database, and took all reasonable steps to both invite the highest possible expertise to contribute to the classification process and to ensure its processes and legal standing are robust.
InSiGHT database curation
Mutalyzer50 was used to standardize the nomenclature of all variants present on the database as of December 2012. Variants with multiple submissions that were originally assigned a classification of pathogenic/probably pathogenic, and no known pathogenicity/probably no pathogenicity, were included in the group of “discordant” variants used to test the classification criteria. All unique variants identified in the database were assigned to one of the following sources: constitutional, somatic, artificial, and unknown.
Development of 5-tier InSiGHT classification criteria
The InSiGHT classification criteria were developed using the Delphi method25. A 5-tiered classification system originally developed for consistent classification of MMR gene variants identified amongst Colon Cancer Family Registry participants16,34 was selected as a baseline for the InSiGHT classification criteria. This system included the option of classification based on posterior probabilities arising from multifactorial likelihood analysis15,16,51,52, and also multiple combinations of qualitative data not yet calibrated for inclusion in quantitative analyses but which are often reported in the literature or available from clinical sources. These baseline classification criteria were critically reviewed by InSiGHT members attending the InSiGHT San Antonio meeting in April 2011, and by VIC members via email. In response to comment, the rules were amended for clarity, to apply more stringent interpretation of functional assay data, and to consider additional points of evidence. These InSiGHT rules were used for variant classification over a series of 11 meetings (10 teleconferences and one face-to-face meeting), with further changes incorporated after each meeting to include additional points of evidence identified to be relevant during the review process as the committee encountered different combinations of useful data from published and unpublished sources. For example, after discussion, co-occurrence of a variant with a pathogenic mutation in the same gene with clinical information regarding constitutional MMR deficiency phenotype53 was included as an in vivo test of MMR function, and the 1000 genomes data54 was accepted as a test for population frequency. Consistency of the accumulative evidence required for a given class was reviewed by presentation of the rules at a face-to-face meeting of committee members. Supporting documentation was developed to assist the interpretation of splicing and functional assay results, by author BAT in consultation with a subset of committee members with specific expertise in this field (see Fig. 1b, Supplementary Tables 4-5). Where necessary, rule alterations were applied retrospectively to variants evaluated in previous meetings. The finalised rules (shown in simplified format in Fig. 1, detailed in the Supplementary Note) were then used to assess all remaining variants lodged in the InSiGHT database.
Classification of MMR gene variants by literature review and data collation
Variants occurring in 1000 genomes54 with allele frequency greater than 1% were automatically classified as Class 1. Committee members were invited to participate in at least one classification meeting. A core group participated in each meeting, with attendance invited from Committee membership to make up the balance. Before each meeting, participants were assigned, through randomisation, a subset of variants to be assessed. Each attendee was provided with literature pertaining to the list of variants to be discussed, and where relevant, additional unpublished clinical or research information submitted by committee members to InSiGHT curator JPP. Meeting attendees were requested to thoroughly review and summarize all information pertaining to the subset of variants in a spreadsheet template, and provide a class assignment based on their interpretation of the information accessed. All reviewer summaries, submitted clinical information, and causality analysis results were compiled into a single file to allow comparison of data and class assignments for each variant, and circulated to the teleconference participants. During committee meetings, variants were discussed one at a time, assessing the following: class assigned by each reviewer; rationale for classification according to the classification guidelines; difficulties in interpreting specific data sources; assessment of possible redundancy of information due to multiple publications including all or part of the same information pertaining to a variant; differences in interpretation of the guidelines as provided and adjustments required to improve their clarity; consensus view on variant class considering the preceding discussion; action required to obtain additional information for refining classification of variants that remained in Class 2, 3 or 4 at the close of discussion. Where classifications differed using qualitative versus quantitative criteria, this was due to differences in availability of specific data types for the two approaches, and the most extreme classification was assigned for relevant variants. Author BAT applied rules-based classification for variants that were truncating/large deletion from nomenclature, canonical splice site with no splicing data, or frequency >1% in a control reference group. Author BAT then collated all information for all unique “not obviously truncating” variants (including those reviewed in teleconferences previously), and determined which variants had sufficient information to allow classification outside of class 3. Summary information for these variants were circulated for independent class assignment by at least three reviewers from the VIC, and classification finalized at teleconferences or by email.
Validation of Qualitative Criteria
A subset of truncating variants and large genomic deletions were selected to validate the qualitative classification criteria. The variants were selected on the basis of availability of data from the first point of evidence in the qualitative Class 5 criterion, i.e. in vitro functional assay results (e.g. protein truncation test or genomic/mRNA confirmation of large deletions); Constitutional MMR Deficiency Syndrome phenotype; or different haplotypes across multiple families. Published and unpublished data for these variants were then used to validate the other points of evidence required for Class 5 “pathogenic”.
Preliminary Analysis of Class 3 “uncertain” Variants
In silico probabilities of pathogenicity were estimated for all Class 3 missense variants, as described elsewhere34. Preliminary bioinformatic analysis of Class 3 regulatory variants was undertaken using the ENCODE data55 on UCSC genome browser.
Implementation of the microattribution process
The variant interpretation process utilizes both published and unpublished data. For published literature the pubmed ID was used to reference the original work. Some unpublished data was recorded in the InSiGHT database at study initiation, and InSiGHT members were also requested by email to contribute information important for variant classification using a standardized submission template. Data submitters were requested to provide a permanent unique publicly searchable ID, preferably from the ORCID system to facilitate adoption of the microattribution approach. Microattribution was assigned for the different types of information corresponding to the points of evidence required for classification, namely submitters were allocated one credit of microattribution for each type of information received:
Variant (Mandatory)
Family History/Pedigree
MSI
Immunohistochemistry
In-vitro functional
RNA splicing assays
Population frequency
All unpublished data received by the VIC was recorded in microattribution tables for each element type, where each microattribution table lists a unique researcher ID along with submitted information. Microattribution counts for submitters are publically available on the InSiGHT website. Additionally, the data will be made available in nanopublication format.
Statistical Analysis
Multifactorial likelihood analysis was done for variants with appropriate tumor and segregation data available, using methods previously reported16,34,51, described briefly as follows. Bayes factor analysis was conducted by author BAT to assess MLH1, MSH2, MSH6, and PMS2 variant causality from segregation data16,51, for both published and unpublished pedigrees with sufficient relevant information on cancer and variant carrier status. Penetrance estimates from Senter et al28 were used in the Bayes segregation analysis27 of PMS2 variants. Where family relationship status was unknown, a conservative segregation likelihood ratio (LR) was derived i.e. setting affected carriers as first-degree relatives, which is less informative than segregation between second-degree relatives. Colorectal tumor MSI and somatic BRAF mutation status were used to assign LRs according to tumor phenotype, derived as previously reported from the ratio of these characteristics in known mutation carrier cases vs non-mutation carrier cases16. For each variant, the individual LRs (co-segregation, tumor) were multiplied to calculate the odds for causality. Then, a posterior probability was calculated from combining the prior probability (in silico for missense variants34 or based on sequence location for all other variants13) and the odds for causality using Bayes rule: posterior = (prior × odds × (1/(1-prior)))/(prior × odds × (1/(1-prior))+1).
STATA 11 was used to calculate sample size of truncating variant validation set: H0: p=0.01 with the following assumptions =0.05 (one-sided) and power=0.95.
All other analyses were completed using the statistical package R and GraphPad Prism 6. For meta-analysis of population frequency data, the proportions were combined using an inverse variance random effects model, to account for heterogeneity between studies.
Supplementary Material
Acknowledgements
We are extremely grateful to the Hicks Foundation (Australia) for inaugural support of InSiGHT database curator JPP. Funding for VIC teleconferences was provided by Cancer Council of Victoria. BAT is supported by a Cancer Council of Queensland PhD scholarship and Queensland Institute of Medical Research PhD Top-Up award. ABS is a National Health and Medical Research Council Senior Research Fellow. The work done by ABS and BAT was additionally supported by Cancer Australia (1010859). MG is supported by a grant from the Tuscan Tumor Institute (ITT). InSiGHT database curator JPP is currently supported by The Royal Melbourne Hospital Foundation. SVT, MSG, ABS, LJR, and RS are supported by grant 1R01CA164944 from the National Cancer Institute/National Institutes of Health (NCI/NIH). GC and MP were supported by Ministerio de Ciencia e Innovación (SAF 12-33636) and Fundación Científica de la AECC. AF is supported by the French National Cancer Institute and INCa French MMR Committee. SMF is supported by grants from the Association of International Cancer Research (12-1087) and Medical Research Council UK (MR/K018647/1). NHS Wales National Institute for Health and Social Care (NIHSCR) funding to IMF, via Cardiff & Vale University Health Board. DEG is supported by funding from a Mayo SPORE grant P50CA11620106 (PI Jim Ingle). CDH is funded by NIH grant CA115783. EH-K and MM are supported by German Cancer Aid (Deutsche Krebshilfe) and Wilhelm-Sander-Foundation. MK-C is funded by Cancer Institute NSW. SYL is supported by the Hong Kong Cancer Fund. AM is supported by the French National Cancer Institute and the Direction Générale de l’Offre des Soins (INCa/DGOS). The Sigrid Juselius Foundation funds MN. Funding for PP is provided by the European Research Council (FP7-ERC-232635). LJR is funded by Nordea-fonden. BR-P is supported by German Cancer Aid. MOW was supported by the Canadian Cancer Society Research Institute (grant #18223). We thank all the submitters of data to the InSiGHT database (retrospective and prospective), the Colon Cancer Family Registry and the German HNPCC Consortium for their contribution of unpublished data, acknowledged formally through microattribution. We would also like to acknowledge Louise Marquart for providing statistical advice and Tracy O’Mara for providing advice and assistance with the statistical package R.
InSiGHT Collaborators
Adela Castillejo47, Adrienne Sexton48, A.K.W. Chan28, Alessandra Viel49, Amie Blanco50, Amy French51, Andreas Laner22, Anja Wagner52, Ans van den Ouweland52, Arjen Mensenkamp53, Artemio Payá54, Beate Betz37, Bert Redeker55, Betsy Smith56, Carin Espenschied57, Carole Cummings58, Christoph Engel59, Claudia Fornes60, Cristian Valenzuela61, Cristina Alenda54, Daniel Buchanan62, Daniela Barana63, Darina Konstantinova64, Dianne Cairns65, Elizabeth Glaser66, Felipe Silva67, Fiona Lalloo68, Francesca Crucianelli44, Frans Hogervorst69, Graham Casey70, Ian Tomlinson71, Ignacio Blanco10, Isabel López Villar72, Javier Garcia-Planells73, Jeanette Bigler74, Jinru Shia75, Joaquin Martinez-Lopez76, Johan J.P. Gille77, John Hopper78, John Potter79, José Luis Soto47, Jukka Kantelinen31, Kate Ellis80, Kirsty Mann48, Liliana Varesco81, Liying Zhang82, Loic Le Marchand83, Makia J. Marafie84, Margareta Nordling85, Maria Grazia Tibiletti86, Mariano Ariel Kahan87, Marjolijn Ligtenberg53, Mark Clendenning62, Mark Jenkins78, Marsha Speevak88, Martin Digweed89, Matthias Kloor90, Megan Hitchins91, Megan Myers50, Melyssa Aronson92, Mev Dominguez Valentin93, Michael Kutsche94, Michael Parsons1, Michael Walsh62, Minttu Kansikas31, Mohd Nizam Zahary95, Monica Pedroni96, Nao Heider97, Nicola Poplawski98, Nils Rahner99, Noralane M. Lindor100, Paola Sala101, Peng Nan102, Peter Propping103, Polly Newcomb79, Rajiv Sarin104, Robert Haile70, Robert Hofstra52, Robyn Ward91, Rossella Tricarico44, Ruben Bacares75, Sean Young105, Sergio Chialina60, Serguei Kovalenko106, Shanaka R. Gunawardena51, Sira Moreno107, S.L. Ho28, S.T. Yuen28, Stephen N. Thibodeau51, Steve Gallinger108, Terrilea Burnett83, Therese Teitsch109, T.L. Chan28, Tom Smyrk51, Treena Cranston110, Vasiliki Psofaki111, Verena Steinke-Lange99, Victor-Manuel Barbera112
47Department of Molecular Genetics, Elche University General Hospital, Elche, Spain. 48Familial Cancer Centre, Royal Melbourne Hospital, Australia. 49Oncological Referral Center, IRCCS, Aviano, Italy. 50Hereditary GI Cancer Prevention Program, University of California, San Francisco, USA. 51Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, Minnesota, USA. 52Department of Clinical Genetics, Erasmus Medical Center, Rotterdam, The Netherlands. 53Department of Human Genetics, Radboud University Medical Center, Nijmegen, The Netherlands. 54Department of Pathology, Hospital Universitario Alicante, Spain. 55Department of Clinical Genetics, Academic Medical Center, Amsterdam, The Netherlands. 56Benefis Sletten Cancer Institute, Great Falls, MT, USA. 57Division of Clinical Cancer Genetics, City of Hope, Duarte, CA, USA. 58The Family Cancer Clinic, St Mark’s Hospital, Harrow, UK. 59Institute for Medical Informatics, Statistics and Epidemiology, University of Leipzig, Germany. 60Molecular genetics, STEM Lab, Rosario, Argentina. 61School of Medicine, New York University, USA. 62Department of Population Health, QIMR Berghofer Medical Research Institute, Brisbane, Australia. 63U.O.C. Oncologia ULSS5 Ovest Vicentino, Ospedale di Montecchio Maggiore (VI), Italy. 64Molecular Medicine Center, Medical University of Sofia, Bulgaria. 65Liverpool Women’s Hospital, Liverpool, UK. 66International Society for Gastrointestinal Hereditary Tumours. 67Laboratory of Genomics and Molecular Biology, A C Camargo Cancer Center, Brazil. 68Manchester Centre for Genomic Medicine, Central Manchester University Hospitals NHS Foundation Trust, UK. 69Family Cancer Clinic and Department of Pathology, The Netherlands Cancer Institute, The Netherlands. 70Department of Preventive Medicine, University of Southern California, Los Angeles, CA, USA. 71Molecular and Population Genetics Laboratory, London Research Institute, Cancer Research UK, UK. 72Hospital 12 de Octubre, Universidad Complutense, Madrid, Spain. 73Institute of Genomic Medicine, University of Valencia, Spain. 74Medical Sciences, Amgen Inc., Seattle, WA, USA. 75Department of Pathology, Memorial Sloan-Kettering Cancer Center, USA. 76Molecular Biology Laboratory, Hospital Universitario 12 de Octubre, Madrid, Spain. 77Clinical Genetics, VU University Medical Centre, The Netherlands. 78Centre for Molecular Environmental, Genetic and Analytic (MEGA) Epidemiology, University of Melbourne, Victoria, Australia. 79Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA. 80Hunter Family Cancer Service, Waratah, Australia. 81Center for Hereditary Tumours, National Institute for Cancer Research, Genoa, Italy. 82Diagnostic Molecular Genetics Laboratory, Memorial Sloan-Kettering Cancer Center, USA. 83University of Hawaii Cancer Center, Honolulu, Hawaii, USA. 84Cancer Genetics Unit, Kuwait Medical Genetics Centre, Kuwait. 85Department of Molecular and Clinical Genetics, Institute of Biomedicine, Sahlgrenska Academy, University of Gothenburg, Sweden. 86Unit of Pathology, Varese Hospital, Varese, Italy. 87Molecular Oncology ICBME, Hospital Italiano de Buenos Aires, Argentina. 88Division of Genetics, Trillium Health Partners, Credit Valley Hospital, Mississauga, Ontario, Canada. 89Charité Berlin, Institute of Human Genetics, Germany. 90Department of Applied Tumor Biology, Institute of Pathology, University of Heidelberg, Germany. 91Lowy Cancer Research Centre, Prince of Wales Clinical School, Faculty of Medicine University of New South Wales, Australia. 92Familial GI Cancers Unit, Mount Sinai Hospital, Toronto, Canada. 93Department of Oncology, Clinical Science, Lund University, Sweden. 94Laboratory for Molecular Medicine Genetic, Praenatalzentrum laboratories, Hamburg, Germany. 95Human Genome Centre, School of Medical Sciences, Universiti Sains Malaysia, Malaysia. 96Department of Medicine and Medical Specialties, Modena University Hospital, Italy. 97RIKEN, Japan. 98SA Pathology, Women’s and Children’s Hospital, Australia. 99Medical Faculty, Institute of Human Genetics, University of Duesseldorf, Germany. 100Department of Health Science Research, Mayo Clinic, Scottsdale, Arizona, USA. 101Hereditary Cancers of the Digestive Tract Unit, Predictive and Preventive Medicine, National Tumor Institute IRCCS Foundation, Milan, Italy. 102School of Life Sciences, Fudan University, China. 103Institute of Human Genetics, University of Bonn, Germany. 104SARIN LAB, ACTREC, Tata Memorial Centre, Mumbai, India. 105Cancer Genetics Laboratory, British Columbia Cancer Agency, Vancouver, Canada. 106Genetic Technologies Limited, Australia. 107Genetics Service, Hospital Virgen del Camino, Spain. 108Zane Cohen Centre for Digestive Diseases, Toronto, Canada. 109Dartmouth Medical School, Dartmouth College, Lebanon, New Hampshire, USA. 110Oxford Medical Genetics Laboratories, Oxford University Hospitals NHS Trust, The Churchill Hospital, Oxford, UK. 111Biochemical Laboratory, University Hospital of Ioannina, Greece. 112Research Laboratory, University Hospital of Elche, Elche, Spain.
Footnotes
URLs
Clinical Molecular Genetics Society (CMGS) classification system, http://cmgsweb.shared.hosting.zen.co.uk/BPGs/Best_Practice_Guidelines.htm; Human Variome Project (HVP), www.humanvariomeproject.org; Leiden Open Variation Database (LOVD), www.lovd.nl; ORCID, orcid.org; InSiGHT, www.insight-group.org/classifications; NCBI ClinVar, http://www.ncbi.nlm.nih.gov/clinvar/; Mutalyzer, mutalyzer.nl; HCI LOVD for MMR gene missense substitution prior probabilities of pathogenicity, http://hci-lovd.hci.utah.edu; UCSC Genome Browser, http://genome.ucsc.edu; nanopublication, http://www.nanopub.org; R-project, http://www.r-project.org/.
Note: Supplementary information is available on the Nature Genetics website.
Accession codes
All data can be accessed by the InSiGHT website. Variants have been submitted to the Leiden Open Variome Database (LOVD) and ClinVar, and are searchable by the gene names MLH1, MSH2, MSH6 and PMS2. The RefSeq and RefSeqGene accessions (respectively) for the MMR genes are: MLH1 – NM_000249.3, NG_007109.2; MSH2 – NM_000251.2, NG_00710.2; MSH6 – NM_000179.2, NG_00711.1; PMS2 – NM_000535.5, NG_008466.1.
Author contributions
ABS and BAT drafted the manuscript. BAT conducted InSiGHT database nomenclature standardization and data-cleaning, systematic literature and data review, statistical analyses, final data analyses and assisted in presentation of data in web-based format. BAT, ABS, SVT, MSG, DEG and MG formulated the baseline guidelines for consideration by VIC members. BAT, ABS developed the functional flowchart, and with LJR, CDH, GC, MP, AM, BR-P, EH-F, MSG, MM, TF, MN formed the functional subcommittee contributing to the supporting documents for functional assay interpretation. DEG provided statistical input. JPP provided data management, organised teleconferences, collated information post-teleconference, co-ordinated microattribution, and was responsible for presentation of data in web-based format. JTDD provided support for the LOVD database and created the LOVD nanopublications. FM is the responsible InSiGHT Councilor who initiated the concept of the VIC in 2007, and has been responsible for advocating for funding, and organizing the face-to-face meeting in Paris. MG co-ordinated the VIC and chaired teleconferences and face-to-face meetings. All authors provided critique on the classification criteria, and/or participated in review of variants at teleconferences, face-to-face meetings or by email. All authors provided critical review of the manuscript.
Competing financial interests
The authors declare no competing financial interests
References
- 1.Vasen HF, et al. Revised guidelines for the clinical management of Lynch syndrome (HNPCC): recommendations by a group of European experts. Gut. 2013 doi: 10.1136/gutjnl-2012-304356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Umar A, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–8. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Oers JM, et al. PMS2 endonuclease activity has distinct biological functions and is essential for genome maintenance. Proc Natl Acad Sci U S A. 2010;107:13384–9. doi: 10.1073/pnas.1008589107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Win AK, et al. Risks of primary extracolonic cancers following colorectal cancer in lynch syndrome. J Natl Cancer Inst. 2012;104:1363–72. doi: 10.1093/jnci/djs351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buerki N, et al. Evidence for breast cancer as an integral part of lynch syndrome. Genes Chromosomes Cancer. 2012;51:83–91. doi: 10.1002/gcc.20935. [DOI] [PubMed] [Google Scholar]
- 6.Scott RJ, et al. Hereditary nonpolyposis colorectal cancer in 95 families: differences and similarities between mutation-positive and mutation-negative kindreds. Am J Hum Genet. 2001;68:118–127. doi: 10.1086/316942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grindedal EM, et al. Germ-line mutations in mismatch repair genes associated with prostate cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:2460–7. doi: 10.1158/1055-9965.EPI-09-0058. [DOI] [PubMed] [Google Scholar]
- 8.Win AK, et al. Colorectal and other cancer risks for carriers and noncarriers from families with a DNA mismatch repair gene mutation: a prospective cohort study. J Clin Oncol. 2012;30:958–64. doi: 10.1200/JCO.2011.39.5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarvinen HJ, et al. Ten years after mutation testing for Lynch syndrome: cancer incidence and outcome in mutation-positive and mutation-negative family members. J Clin Oncol. 2009;27:4793–7. doi: 10.1200/JCO.2009.23.7784. [DOI] [PubMed] [Google Scholar]
- 10.Plon SE, et al. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008;29:1282–91. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tavtigian SV, Greenblatt MS, Goldgar DE, Boffetta P. Assessing pathogenicity: overview of results from the IARC Unclassified Genetic Variants Working Group. Hum Mutat. 2008;29:1261–4. doi: 10.1002/humu.20903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards CS, et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 13.Easton DF, et al. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet. 2007;81:873–83. doi: 10.1086/521032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldgar DE, et al. Genetic evidence and integration of various data sources for classifying uncertain variants into a single model. Hum Mutat. 2008;29:1265–72. doi: 10.1002/humu.20897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldgar DE, et al. Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am J Hum Genet. 2004;75:535–44. doi: 10.1086/424388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson BA, et al. A multifactorial likelihood model for MMR gene variant classification incorporating probabilities based on sequence bioinformatics and tumor characteristics: a report from the Colon Cancer Family Registry. Hum Mutat. 2013;34:200–9. doi: 10.1002/humu.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spurdle AB, Couch FJ, Hogervorst FB, Radice P, Sinilnikova OM. Prediction and assessment of splicing alterations: implications for clinical testing. Hum Mutat. 2008;29:1304–13. doi: 10.1002/humu.20901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenblatt MS, et al. Locus-specific databases and recommendations to strengthen their contribution to the classification of variants in cancer susceptibility genes. Hum Mutat. 2008;29:1273–81. doi: 10.1002/humu.20889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plazzer JP, et al. The InSiGHT database: utilizing 100 years of insights into Lynch Syndrome. Fam Cancer. 2013 doi: 10.1007/s10689-013-9616-0. [DOI] [PubMed] [Google Scholar]
- 20.Peltomaki P, Vasen H. Mutations associated with HNPCC predisposition -- Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269–76. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peltomaki P, Vasen HF, The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. Gastroenterology. 1997;113:1146–58. doi: 10.1053/gast.1997.v113.pm9322509. [DOI] [PubMed] [Google Scholar]
- 22.Ou J, et al. Functional analysis helps to clarify the clinical importance of unclassified variants in DNA mismatch repair genes. Hum Mutat. 2007;28:1047–54. doi: 10.1002/humu.20580. [DOI] [PubMed] [Google Scholar]
- 23.Woods MO, et al. A new variant database for mismatch repair genes associated with Lynch syndrome. Hum Mutat. 2007;28:669–73. doi: 10.1002/humu.20502. [DOI] [PubMed] [Google Scholar]
- 24.Giardine B, et al. Systematic documentation and analysis of human genetic variation in hemoglobinopathies using the microattribution approach. Nat Genet. 2011;43:295–301. doi: 10.1038/ng.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fox BI, et al. Developing an expert panel process to refine health outcome definitions in observational data. J Biomed Inform. 2013 doi: 10.1016/j.jbi.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 26.Kohonen-Corish MR, et al. Deciphering the colon cancer genes--report of the InSiGHT-Human Variome Project Workshop, UNESCO, Paris 2010. Hum Mutat. 2011;32:491–4. doi: 10.1002/humu.21450. [DOI] [PubMed] [Google Scholar]
- 27.Thompson D, Easton DF, Goldgar DE. A full-likelihood method for the evaluation of causality of sequence variants from family data. Am J Hum Genet. 2003;73:652–5. doi: 10.1086/378100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Senter L, et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology. 2008;135:419–28. doi: 10.1053/j.gastro.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baglietto L, et al. Risks of Lynch syndrome cancers for MSH6 mutation carriers. J Natl Cancer Inst. 2010;102:193–201. doi: 10.1093/jnci/djp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonadona V, et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. Jama. 2011;305:2304–10. doi: 10.1001/jama.2011.743. [DOI] [PubMed] [Google Scholar]
- 31.Mangold E, et al. Spectrum and frequencies of mutations in MSH2 and MLH1 identified in 1,721 German families suspected of hereditary nonpolyposis colorectal cancer. Int J Cancer. 2005;116:692–702. doi: 10.1002/ijc.20863. [DOI] [PubMed] [Google Scholar]
- 32.Barnetson RA, et al. Identification and survival of carriers of mutations in DNA mismatch-repair genes in colon cancer. N Engl J Med. 2006;354:2751–63. doi: 10.1056/NEJMoa053493. [DOI] [PubMed] [Google Scholar]
- 33.Patrinos GP, et al. Microattribution and nanopublication as means to incentivize the placement of human genome variation data into the public domain. Hum Mutat. 2012;33:1503–12. doi: 10.1002/humu.22144. [DOI] [PubMed] [Google Scholar]
- 34.Thompson BA, et al. Calibration of multiple in silico tools for predicting pathogenicity of mismatch repair gene missense substitutions. Hum Mutat. 2013;34:255–65. doi: 10.1002/humu.22214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vallee MP, et al. Classification of missense substitutions in the BRCA genes: a database dedicated to Ex-UVs. Hum Mutat. 2012;33:22–8. doi: 10.1002/humu.21629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drost M, et al. A rapid and cell-free assay to test the activity of lynch syndrome-associated MSH2 and MSH6 missense variants. Hum Mutat. 2011 doi: 10.1002/humu.22000. [DOI] [PubMed] [Google Scholar]
- 37.Heinen CD, Juel Rasmussen L. Determining the functional significance of mismatch repair gene missense variants using biochemical and cellular assays. Hered Cancer Clin Pract. 2012;10:9. doi: 10.1186/1897-4287-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Couch FJ, et al. Assessment of functional effects of unclassified genetic variants. Hum Mutat. 2008;29:1314–26. doi: 10.1002/humu.20899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rasmussen LJ, et al. Pathological assessment of mismatch repair gene variants in lynch syndrome: past, present and future. Hum Mutat. 2012 doi: 10.1002/humu.22168. [DOI] [PubMed] [Google Scholar]
- 40.Leenen CH, et al. Pitfalls in molecular analysis for mismatch repair deficiency in a family with biallelic pms2 germline mutations. Clin Genet. 2011;80:558–65. doi: 10.1111/j.1399-0004.2010.01608.x. [DOI] [PubMed] [Google Scholar]
- 41.Mead LJ, et al. Microsatellite instability markers for identifying early-onset colorectal cancers caused by germ-line mutations in DNA mismatch repair genes. Clin Cancer Res. 2007;13:2865–9. doi: 10.1158/1078-0432.CCR-06-2174. [DOI] [PubMed] [Google Scholar]
- 42.Plaschke J, et al. Lower incidence of colorectal cancer and later age of disease onset in 27 families with pathogenic MSH6 germline mutations compared with families with MLH1 or MSH2 mutations: the German Hereditary Nonpolyposis Colorectal Cancer Consortium. J Clin Oncol. 2004;22:4486–94. doi: 10.1200/JCO.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 43.Wu Y, et al. Association of hereditary nonpolyposis colorectal cancer-related tumors displaying low microsatellite instability with MSH6 germline mutations. Am J Hum Genet. 1999;65:1291–8. doi: 10.1086/302612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.You JF, et al. Tumours with loss of MSH6 expression are MSI-H when screened with a pentaplex of five mononucleotide repeats. Br J Cancer. 2010;103:1840–5. doi: 10.1038/sj.bjc.6605988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spurdle AB, et al. BRCA1 R1699Q variant displaying ambiguous functional abrogation confers intermediate breast and ovarian cancer risk. J Med Genet. 2012;49:525–32. doi: 10.1136/jmedgenet-2012-101037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie J, et al. An MLH1 Mutation Links BACH1/FANCJ to Colon Cancer, Signaling, and Insight toward Directed Therapy. Cancer Prev Res (Phila) 2010;3:1409–1416. doi: 10.1158/1940-6207.CAPR-10-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kosinski J, Hinrichsen I, Bujnicki JM, Friedhoff P, Plotz G. Identification of Lynch syndrome mutations in the MLH1-PMS2 interface that disturb dimerization and mismatch repair. Hum Mutat. 2010;31:975–82. doi: 10.1002/humu.21301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi M, et al. Functional analysis of human MLH1 variants using yeast and in vitro mismatch repair assays. Cancer Res. 2007;67:4595–604. doi: 10.1158/0008-5472.CAN-06-3509. [DOI] [PubMed] [Google Scholar]
- 49.Hinrichsen I, et al. Expression defect size among unclassified MLH1 variants determines pathogenicity in Lynch syndrome diagnosis. Clin Cancer Res. 2013;19:2432–41. doi: 10.1158/1078-0432.CCR-12-3299. [DOI] [PubMed] [Google Scholar]
- 50.Wildeman M, van Ophuizen E, den Dunnen JT, Taschner PE. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum Mutat. 2008;29:6–13. doi: 10.1002/humu.20654. [DOI] [PubMed] [Google Scholar]
- 51.Arnold S, et al. Classifying MLH1 and MSH2 variants using bioinformatic prediction, splicing assays, segregation, and tumor characteristics. Hum Mutat. 2009;30:757–70. doi: 10.1002/humu.20936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spurdle AB. Clinical relevance of rare germline sequence variants in cancer genes: evolution and application of classification models. Curr Opin Genet Dev. 2010;20:315–23. doi: 10.1016/j.gde.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 53.Wimmer K, Etzler J. Constitutional mismatch repair-deficiency syndrome: have we so far seen only the tip of an iceberg? Hum Genet. 2008;124:105–22. doi: 10.1007/s00439-008-0542-4. [DOI] [PubMed] [Google Scholar]
- 54.A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.