ABSTRACT
DNA double-strand breaks (DSBs) are a type of lethal DNA damage. The repair of DSBs requires tight coordination between the factors modulating chromatin structure and the DNA repair machinery. BRG1, the ATPase subunit of the chromatin remodelling complex Switch/Sucrose non-fermentable (SWI/SNF), is often linked to tumorigenesis and genome instability, and its role in DSB repair remains largely unclear. In the present study, we show that BRG1 is recruited to DSB sites and enhances DSB repair. Using DR-GFP and EJ5-GFP reporter systems, we demonstrate that BRG1 facilitates homologous recombination repair rather than nonhomologous end-joining (NHEJ) repair. Moreover, the BRG1–RAD52 complex mediates the replacement of RPA with RAD51 on single-stranded DNA (ssDNA) to initiate DNA strand invasion. Loss of BRG1 results in a failure of RAD51 loading onto ssDNA, abnormal homologous recombination repair and enhanced DSB-induced lethality. Our present study provides a mechanistic insight into how BRG1, which is known to be involved in chromatin remodelling, plays a substantial role in the homologous recombination repair pathway in mammalian cells.
KEY WORDS: DNA double-strand break, BRG1, Homologous recombination, RAD52, RAD51
INTRODUCTION
DNA double-strand breaks (DSBs), one of the most severe forms of DNA damage, are generated by exogenous factors (e.g. exposure to ionising radiation) or arise from endogenous by-products (e.g. DNA replication fork collapse) (Peterson and Almouzni, 2013). DSBs can lead to mutagenesis, genomic instability and even cell death if left unrepaired or misrepaired (Rich et al., 2000). In cells that encounter DSBs, there is a sophisticated DNA damage response (DDR). In brief, DSBs are detected by the MRE11–RAD50–NBS1 (MRN complex) and Ku70–Ku80 (also known as XRCC6–XRCC5) complexes, which in turn recruit the apical phosphoinositide 3-kinase-like kinases (PIKKs) (Falck et al., 2005). Then, Ser139 at the C-terminus of histone variant H2AX is phosphorylated by PIKK, and the phosphorylated variant is referred to as γH2AX (Rogakou et al., 1998). γH2AX can be propagated along chromatin near DSBs, forming megabase-sized foci that accumulate downstream repair factors (van Attikum and Gasser, 2009). There are two major processes for the DNA DSB repair in mammalian cells – homologous recombination and nonhomologous end-joining (NHEJ) (Khanna and Jackson, 2001; van Gent et al., 2001). Homologous recombination repairs DSBs precisely using the intact copy of a homologous sequence from a sister chromatid during the late S and G2 phases of the cell cycle. In the G1 phase, NHEJ repairs the majority of DSBs without relying on extensive sequence homology (Johnson and Jasin, 2000; Lim et al., 2000).
In eukaryotes, DSBs occur in the chromatin context, and the repair machinery must have access to naked DNA. Thus, the process of DSB repair is more complicated than is portrayed by the current working models. However, little is known regarding how the proteins that are involved in nucleosome remodelling and that are required for the homologous recombination or NHEJ pathways coordinate to accomplish DSB repair. The SWI/SNF complex is an evolutionarily conserved ATP-dependent chromatin remodelling complex, the involvement of which in gene transcription and DNA damage response is well established (Smith and Peterson, 2004; Osley et al., 2007). However, the function of SWI/SNF in DSB repair is still elusive. It has been reported that SWI/SNF promotes the phosphorylation of H2AX Ser139 and is vital for the cell response to DSBs in human cells (Park et al., 2006; Ray et al., 2009; Lee et al., 2010). Yeast mutants lacking SWI/SNF activity are sensitive to DNA damaging agents that cause DSBs, and the synapsis between the invading MATa ssDNA and HMLα DNA is blocked in these mutants (Cruz et al., 2012; Chai et al., 2005). BRG1, the core ATPase subunit of the SWI/SNF complex, is recruited to DSB sites, where it directly interacts with γH2AX-containing nucleosomes. This interaction involves the binding of the bromodomain of BRG1 to acetylated histone 3 (H3), which in turn induces additional H3 acetylation through recruitment of the acetyltransferase GCN5 (also known as KAT2A) (Lee et al., 2010). Additionally, BRG1 is further phosphorylated by ATM in response to DNA damage (Kwon et al., 2014), and global deletion of BRG1 in mice leads to embryonic lethality (Bultman et al., 2000). During V(D)J recombination, an endogenous DSB repair process, human (h)SWI/SNF stimulates V(D)J cleavage by RAG proteins on reconstituted mononucleosomes and nucleosomal arrays in vitro. In vivo, the Brg1 catalytic subunit of hSWI/SNF is broadly associated with immunoglobulin loci that are poised for rearrangement (Patenge et al., 2004; Morshead et al., 2003).
Although several reports have contributed to our understanding of the essential role of BRG1 in DSB repair, the precise molecular mechanism through which BRG1 is implicated in DSB repair in mammalian cells is still obscure. In this study, we demonstrate that BRG1 is recruited to DNA damage sites. Furthermore, we identify that BRG1 promotes homologous recombination repair through interacting with RAD52 and then regulates the formation of RAD51-binding nucleofilaments. This novel function of BRG1 is consistent with its role in maintaining genome stability and preventing carcinogenesis.
RESULTS
BRG1 is essential for DSB repair
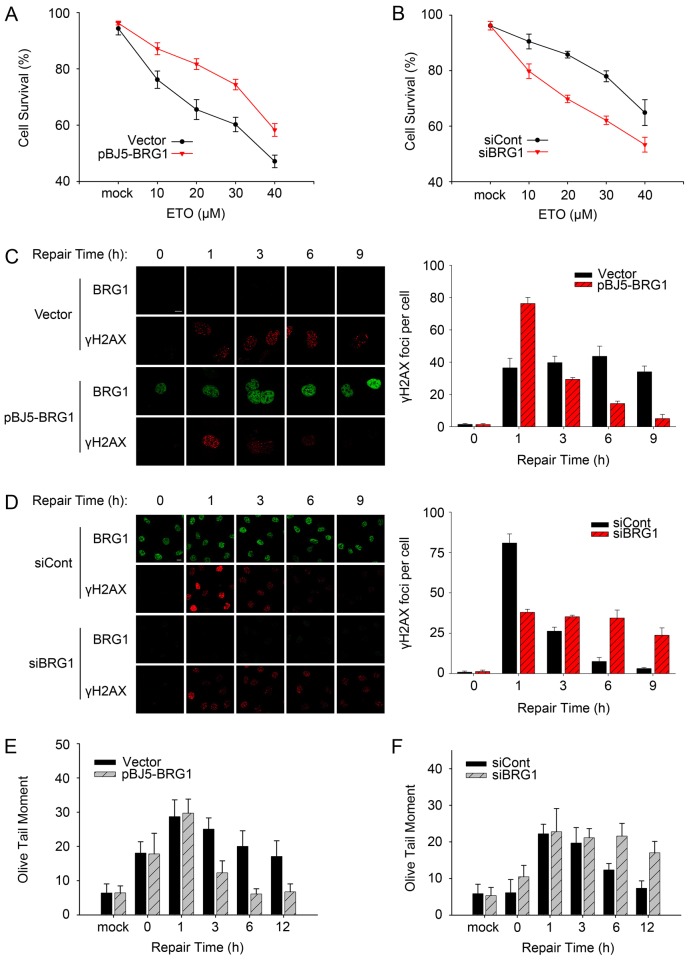
To establish the significance of BRG1 in DSB repair, we first examined the influence of BRG1 expression on cell viability following treatment with etoposide (ETO), which is well accepted as a DSB-inducing agent (Sung et al., 2006). SW13 human adrenal adenocarcinoma cells, which are deficient in BRM and BRG1 (Wong et al., 2000), were transfected with pBJ5-BRG1 or an empty vector; U2OS cells were transfected with BRG1-specific small interfering (si)RNA or control siRNA. Then, the cells were mock-treated or treated with increased concentrations of ETO, and cell viability was detected using the cell colony formation assay. Fig. 1A shows that SW13 cells expressing BRG1 were more resistant to drug treatment than the cells transfected with empty vector. Similarly, U2OS cells transfected with BRG1 siRNA displayed increased sensitivity to ETO treatment compared with control cells (Fig. 1B). Additionally, we treated cells with bleomycin, which has also been reported to induce DSBs (Xu et al., 2010). The results showed that BRG1 depletion resulted in lower cell viability after bleomycin treatment (supplementary material Fig. S1A,B). These data indicate that BRG1 is essential for cell survival after DNA damage.
Fig. 1.
BRG1 is a vital factor in the DDR. (A) SW13 cells transfected with pBJ5-BRG1 or empty vector and (B) U2OS cells transfected with BRG1 siRNA (siBRG1) or control siRNA (siCont) were mock treated or exposed to the indicated doses of ETO for 20 min and then supplied with fresh medium. After 14 days, cells were stained with Crystal Violet. Colonies containing >50 cells were counted. Each value represents the mean±s.d. (three independent experiments). (C) SW13 cells transfected with pBJ5-BRG1 or empty vector were treated with 10 µM ETO and allowed to repair DSBs for different time intervals. Cells were fixed and subjected to confocal immunofluorescence analysis. Representative confocal microscopy images are shown on the left. Scale bar: 10 µm. Quantitative analysis of γH2AX foci (>10 pixels) from three independent experiments is shown on the right, and for each experiment, >50 cells were analysed per time-point. (D) H2AX phosphorylation level at the indicated time-points in U2OS cells transfected with the indicated siRNAs was detected and analysed as in C. Scale bar: 10 µm. (E,F) The repair kinetics of ETO-induced DSBs were detected using a comet assay in SW13 and U2OS cells in the presence or absence of BRG1 after ETO treatment. The olive tail moment was determined as the end point of DSBs; 100 individual comets were counted per time-point for each experiment. Data in C−F show the mean±s.d.
Next, we explored whether the low viability of BRG1-depleted cells after DNA damage was coupled with impaired DSB repair signalling. We examined the changes in the formation of γH2AX foci following ETO treatment. Compared with ETO-treated control cells (transfected with empty vector), SW13 cells with expression of BRG1 showed more γH2AX foci (1 h), which decreased to background level at 9 h (Fig. 1C). In U2OS cells, BRG1 knockdown resulted in reduced formation of γH2AX foci at the early phase (<1 h) and the persistence of accumulated unrepaired foci at the later time-points (3–9 h; Fig. 1D). To detect the occurrence of DSBs and exclude the possibility that the lower γH2AX signals observed in BRG1-deficient cells was due to reduced DSB induction, we performed neutral comet assays. Fig. 1E and supplementary material Fig. S1C show that ETO treatment induced comparable DSB levels in SW13 cells with or without BRG1 expression. Notably, DSB repair could be achieved within 12 h in SW13 cells expressing BRG1, whereas the control cells accumulated more DSBs. In U2OS cells, we also found that BRG1 knockdown led to more persistent DSBs at the late repair phase after ETO treatment (Fig. 1F; supplementary material Fig. S1D). The combined data suggest that BRG1 loss leads to deficient damage signalling and impeded DSB repair. The transfection and interference efficiency were determined by immunoblotting (supplementary material Fig. S1C,D). Moreover, we obtained similar results when SW13 and U2OS cells were treated with bleomycin (supplementary material Fig. S1E,F). Taken together, these data clearly show that BRG1 is a key effector in the DNA damage response as well as in DSB repair.
BRG1 is recruited to DSB-associated chromatin
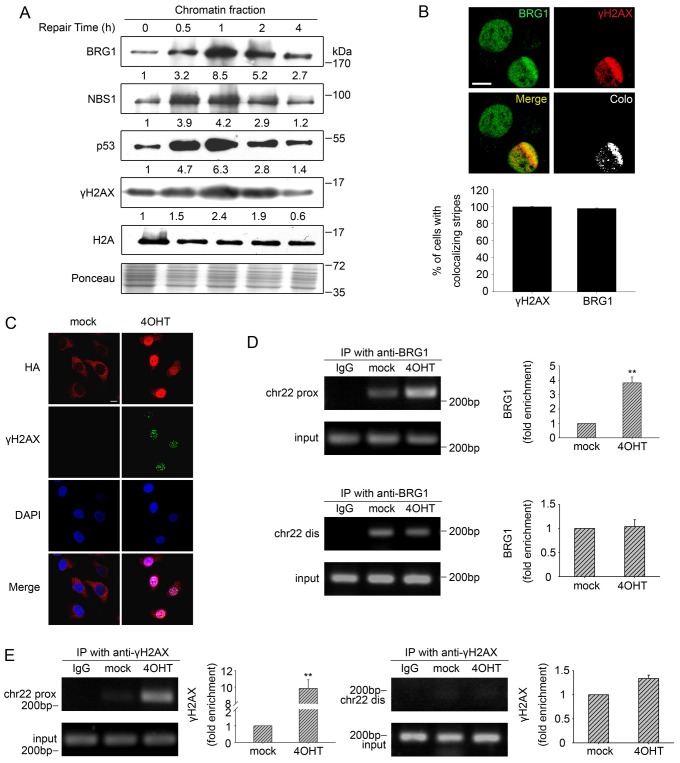
To monitor the dynamics of BRG1 recruitment to DSB sites, we purified the chromatin-enriched fractions from ETO-treated U2OS cells. As shown in Fig. 2A and supplementary material Fig. S2, during DSB repair, BRG1 accumulated at chromatin, and this accumulation culminated at 1 h and then decreased gradually. Likewise, two important DSB response factors, NBS1 (also known as nibrin) and p53, as well as the DSB marker γH2AX behaved in a similar manner to BRG1. To verify that the chromatin-associated BRG1 localised to DNA damage sites, we examined the distribution of BRG1 after microlaser irradiation by using confocal microscopy. We observed that BRG1 was enriched at the laser-induced DSB sites that were marked by γH2AX (Fig. 2B).
Fig. 2.
BRG1 is enriched on the chromatin at DSBs. (A) U2OS cells were incubated with 10 µM ETO and then allowed to repair DSBs. The chromatin fractions were extracted from cell pellets collected at the indicated time-points and were subjected to immunoblot analysis. Ponceau S staining showed the equal loading of total proteins, and H2A was used to show equal quantities of chromatin fractions from different samples. (B) U2OS cells were subjected to multiphoton laser irradiation. After 1 h, cells were immunostained for endogenous BRG1 and γH2AX. Colocalisation (Colo) of BRG1 and γH2AX in merged images was analysed by using the confocal microscopy software to generate white dots. The percentage of cells with colocalisation of γH2AX with BRG1 is indicated as the mean±s.d. (lower panel). Scale bar: 10 µm. (C) AsiSI–ER-U2OS cells were subjected to 4OHT treatment (3 h) or were mock treated. Then, the cells were fixed and incubated with antibodies against HA–AsiSI and γH2AX. Scale bar: 10 µm. (D) ChIP analysis was performed using AsiSI–ER-U2OS cells with or without 4OHT treatment using an antibody against BRG1 or isotype IgG control antibody. Precipitated DNA was assessed by PCR amplification using primers located in close proximity to (prox) and distal from (dis) the AsiSI site as described in Materials and Methods. IP, immunoprecipitation; chr22, chromosome 22. (E) A ChIP assay was performed using antibody against γH2AX. Quantification of immunoprecipitated material is shown. The values represent the mean±s.d. (three independent experiments); **P<0.01.
To further confirm the above results, we next performed a chromatin immunoprecipitation (ChIP) assay. To induce DSBs in cells, we utilised U2OS cells stably expressing the HA-tagged AsiSI–ER restriction enzyme, an 8-bp cutter that, on average, generates fragments longer than 1 Mb in the human genome (Iacovoni et al., 2010). The fused AsiSI contains a modified oestrogen receptor hormone-binding domain (ER), which only binds to 4OHT, and 4OHT treatment induces nuclear localisation of AsiSI–ER and generates DSBs (Littlewood et al., 1995; Agger et al., 2005). We first tested the efficiency of DSB induction after 4OHT exposure in AsiSI–ER-U2OS cells by γH2AX staining. Fig. 2C shows that 4OHT treatment resulted in notable nuclear localisation of the AsiSI enzyme and formation of γH2AX foci, indicating that abundant DNA DSBs were induced, and the DNA damage response was activated. Then, a ChIP assay was performed using a BRG1-specific antibody. Fig. 2D shows that BRG1 accumulation in proximity to AsiSI-dependent DSBs on chromosome 22 was significantly increased after 4OHT treatment, whereas there was less chromatin enrichment of BRG1 distal to the DSB sites. In addition, γH2AX showed enrichment at damage sites similar to BRG1 (Fig. 2E). Therefore, our combined data suggest that BRG1 indeed accumulates at DSB-associated chromatin.
Loss of BRG1 impairs homologous recombination repair
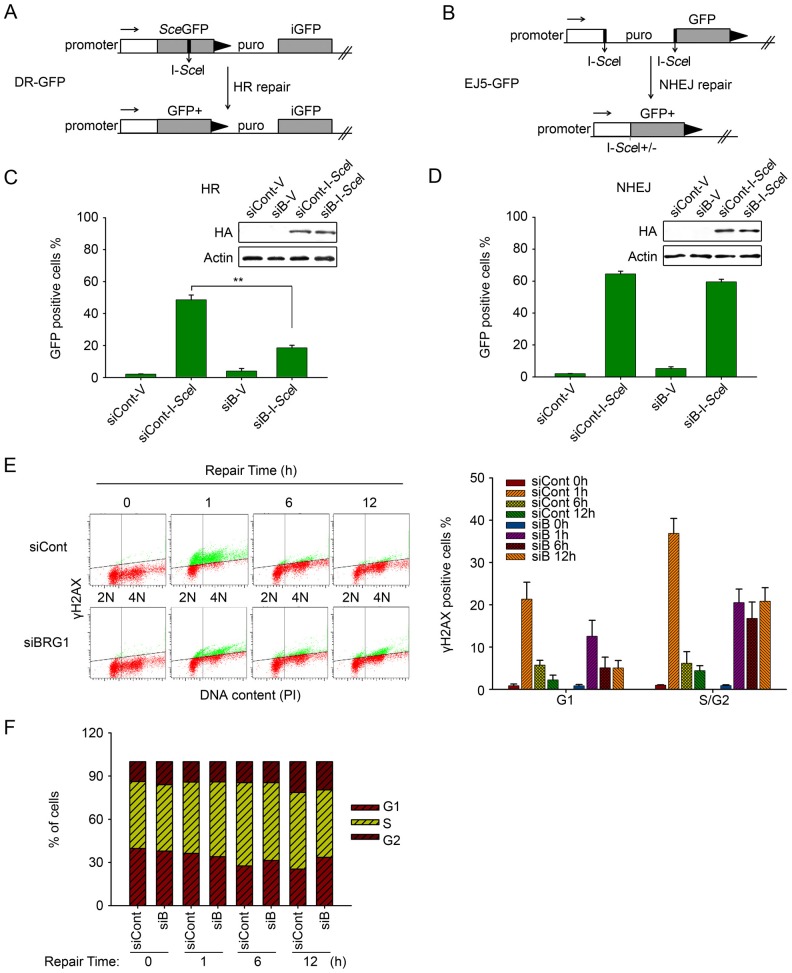
DSB repair involves two distinct pathways; thus, we examined the impact of BRG1 on the homologous recombination and NHEJ repair pathways by utilising two well-characterised GFP-based reporter systems – DR-GFP and EJ5-GFP. The DR-GFP reporter contains an upstream GFP gene with insertion of an I-SceI recognition site (SceGFP) and a downstream internal GFP repeat (iGFP). Expression of the I-SceI endonuclease leads to DSB induction in the upstream GFP. Then, homologous recombination repair using the downstream GFP repeat as the template restores the functional GFP expression and the GFP-positive cells can be quantified using flow cytometry (Fig. 3A) (Weinstock et al., 2006; Helfricht et al., 2013). The EJ5-GFP reporter contains a non-functional GFP gene carrying a puromycin gene that separates the GFP coding sequence from its promoter. The puromycin gene is flanked by two I-SceI recognition sites. Transfection of plasmid encoding I-SceI induces two DSBs flanking the puromycin gene. Once the puromycin gene is excised by NHEJ repair, the promoter is ligated to the GFP coding gene, leading to GFP expression (Fig. 3B) (Bennardo et al., 2008; Helfricht et al., 2013).
Fig. 3.
BRG1 depletion impairs homologous recombination repair. (A) Schematic of the DR-GFP reporter used to monitor homologous recombination (HR). (B) Schematic of the EJ5-GFP reporter used to monitor NHEJ in U2OS cells (see text for details). puro, puromycin gene. (C,D) DR-GFP-U2OS cells and EJ5-GFP-U2OS cells with or without BRG1 knockdown were transfected with pCBASceI or empty vector plasmids. After 72 h, the cells were analysed for GFP-positive cells by flow cytometry to demonstrate the repair efficiency of homologous recombination and NHEJ. The values represent the mean±s.d. (three independent experiments). The inner immunoblot shows the expression of the HA–I-SceI enzyme. siCont, control siRNA; siB, BRG1 siRNA. (E) U2OS cells transfected with BRG1 siRNA or control siRNA were treated with 10 µM ETO and allowed to repair for the indicated time intervals. Flow cytometric analysis of DNA content and γH2AX level was obtained by using propidium iodide (PI) and γH2AX staining; 1×104 events were analysed from a single experiment. The data shown are from a single representative experiment out of three replicates. Quantification of γH2AX-positive cells at G1 or S/G2 phase is shown on the right as the mean±s.d. (F) Analysis of cell cycle distribution after ETO treatment as in E is shown.
DR-GFP-U2OS and EJ5-GFP-U2OS cells with or without prior BRG1 depletion were transfected with pCBASceI or empty vector plasmids to introduce DSBs. After 72 h, the cells were analysed by flow cytometry to detect GFP-positive cells. Interestingly, we found that BRG1 knockdown significantly decreased the fraction of GFP-positive DR-GFP cells (Fig. 3C) but not the EJ5-GFP cells (Fig. 3D), suggesting that BRG1 primarily participates in the homologous recombination repair pathway. Moreover, flow cytometric analysis of the cell cycle and γH2AX levels after DSB induction showed that the persistent γH2AX signal at the late repair stage resulting from BRG1 depletion mainly occurred in the S/G2 phase (Fig. 3E), indicating that the effects of BRG1 on DNA repair are more pronounced in S/G2, in agreement with a role in homologous recombination repair. At the same time, similar cell cycle distributions were observed between the BRG1-knockdown cells and control cells during DSB repair, thereby indicating that BRG1 directly affects homologous recombination frequency without affecting the cell cycle (Fig. 3F). These results therefore suggest that BRG1 mainly participates in the homologous recombination repair pathway and plays a key role in the homologous recombination repair of DSBs.
BRG1 depletion leads to ssDNA retention and impaired RAD51 loading
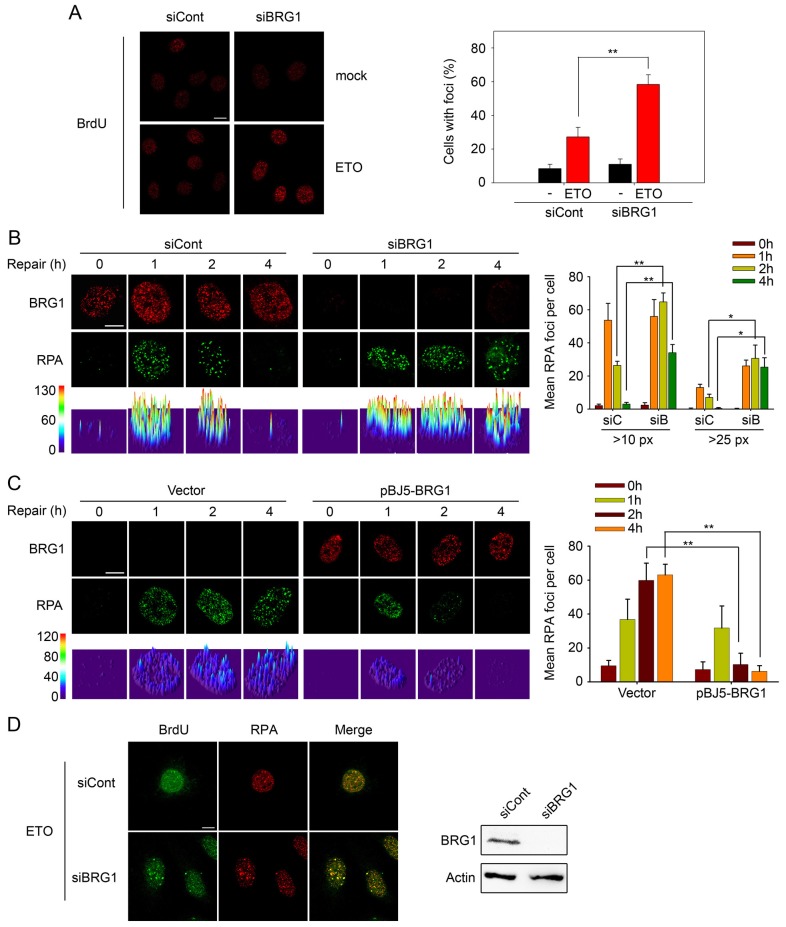
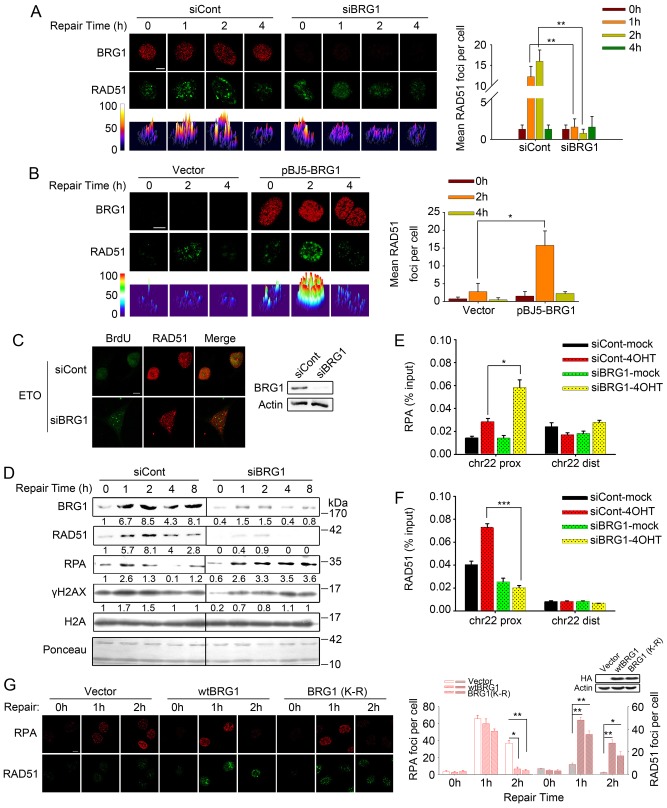
To perform homologous recombination repair, upon DSB occurrence, the broken DNA ends are resected to form ssDNAs, which are further coated with RPA. Then, RPA facilitates the recruitment of RAD51 to initiate homologous DNA recombination and complete the homologous recombination repair process (Lisby et al., 2004). To explore the specific role of BRG1 in the homologous recombination process, we performed immunofluorescence analysis to determine the effect of BRG1 depletion on the kinetics of ssDNAs. We found that BRG1 knockdown led to more BrdU foci staining after ETO treatment (2 h), suggesting more ssDNA retention during DSB repair (Fig. 4A). Next, we observed that BRG1 depletion also resulted in persistent and large RPA foci during the late phase of repair (2–4 h), which coincided with the accumulation of ssDNAs. However, the early recruitment of RPA was comparable to that of controls, which implied that ssDNA resection, occurring at the early phase of homologous recombination, was not influenced by BRG1 knockdown (Fig. 4B). A similar observation was noted in SW13 cells with or without BRG1 expression (Fig. 4C). Furthermore, double immunofluorescent staining with anti-BrdU and anti-RPA antibodies showed that the majority of cells that were positive for RPA also displayed ssDNA foci, and BRG1 knockdown increased the percentage of double-stained cells (Fig. 4D). Previous studies have reported that retained RPA strongly inhibits strand invasion in homologous recombination repair by limiting the access of RAD51 to ssDNAs (Symington, 2002; Wang and Haber, 2004). We next examined the RAD51 foci formation with or without BRG1 knockdown. Notably, we found that BRG1 knockdown resulted in a significant decrease in the formation of RAD51 foci (Fig. 5A). Similarly, SW13 cells transfected with empty vector showed markedly impaired RAD51 foci formation compared with that of SW13 cells transfected with pBJ5-BRG1 (Fig. 5B). Furthermore, we observed that in cells treated with ETO, BRG1 ablation led to a significant decrease in the number of RAD51 foci and, accordingly, increased staining of BrdU foci, indicating that the loading of RAD51 to ssDNAs is blocked by BRG1 depletion (Fig. 5C). The combined data suggest that BRG1 might direct the onsite replacement of RPA with RAD51 on ssDNAs.
Fig. 4.
Loss of BRG1 results in ssDNA accumulation and retention of RPA foci. (A) U2OS cells transfected with BRG1 siRNA (siBRG1) or control siRNA (siCont) were pre-labelled with BrdU and then treated with 10 µM ETO for 20 min. After 2 h, the cells were fixed under non-denaturing conditions, and BrdU in ssDNA was detected with an anti-BrdU antibody. Representative images are presented. Quantitative analysis of BrdU-positive cells is shown on the right. Cells with more than five foci were considered positive, with an average of 100 cells counted per experiment (three independent experiments). (B) U2OS cells transfected with BRG1 siRNA (siBRG1, siB) or control siRNA (siCont, siC) were exposed to 10 µM ETO for 20 min. At the indicated time-points, cells were pre-extracted with buffer D to release non-chromatin binding proteins, and detected by immunostaining with antibodies recognising BRG1 (red) and RPA (green), respectively. The lower row shows the three-dimensional plot of the intensity of RPA shown in the upper panels, as determined by using Image J software. Quantification of RPA foci [>10 or >25 pixels (px)] is shown on the right. (C) SW13 cells transfected with pBJ5-BRG1 or empty vector were treated and analysed as in B. Foci with >10 pixels were counted. All quantitative data show the mean±s.d.; *P<0.05; **P<0.01. (D) U2OS cells were treated as in A. After 2 h, cells were fixed and detected by immunostaining using anti-BrdU and anti-RPA antibodies. BRG1 expression was analysed by immunoblotting. Scale bars: 10 µm.
Fig. 5.
BRG1 is required for RAD51 recruitment to DNA damage sites. (A,B) U2OS cells transfected with BRG1 siRNA (siBRG1) or control siRNA (siCont) (A) and SW13 cells transfected with pBJ5-BRG1 or empty vector (B) were treated with ETO for 20 min. At the indicated repair time-points, cells were pre-extracted with buffer D to release non-chromatin binding proteins and samples were analysed by immunostaining with antibodies recognising BRG1 (red) and RAD51 (green). The lower row shows the three-dimensional plot of the intensity of RAD51 shown in the upper panels, as determined by using Image J software. Quantification of RAD51 foci is shown on the right. Foci with >10 pixels were counted, with an average of 100 cells counted per experiment. (C) U2OS cells transfected with BRG1 siRNA or control siRNA were incubated in ETO for 20 min. After 2 h, the cells were fixed and detected by using anti-BrdU and anti-RAD51 antibodies (left panel). The expression of BRG1 was analysed by immunoblotting (right panel). (D) At 48 h post transfection with BRG1 siRNA or control siRNA, U2OS cells were treated with 10 µM ETO and cultured for the indicated time intervals before lysis. Chromatin fractions were analysed by immunoblotting using the indicated antibodies. (E,F) AsiSI–ER-U2OS cells were transfected with the BRG1 siRNA or control siRNA. After 48 h, cells were treated with 4OHT or were mock treated. A ChIP assay was performed using antibodies against RPA and RAD51. Immunoprecipitated and input DNA were analysed by real-time qPCR using the indicated primers. Three independent experiments were performed. chr22, chromosome 22; prox, proximal; dist, distal. (G) SW13 cells transfected with HA-tagged pBJ5-wtBRG1, ATPase mutant BRG1 (K798R) or empty vector were exposed to 10 µM ETO for 20 min. The cells were allowed to repair DSBs for the indicated time intervals, then pre-extracted with buffer D and stained with RPA and RAD51 antibodies. Images were captured by using confocal microscopy (60×). Scale bars: 10 µm. The expression of HA-tagged BRG1 was analysed by immunoblotting. All quantitative data show the mean±s.d.; *P<0.05; **P<0.01; ***P<0.001.
To support the above findings, we isolated chromatin fractions at the indicated repair time intervals after DSB induction. Immunoblot analysis showed that BRG1 depletion resulted in lower RAD51 recruitment to chromatin and persistent accumulation of RPA on chromatin (Fig. 5D; supplementary material Fig. S3). To determine whether the loss of RAD51 recruitment and persistent RPA retention were restricted to DSB sites, we performed a ChIP assay on chromosome 22 using RAD51- and RPA-specific antibodies in the AsiSI DSB induction system. The result showed that BRG1 deficiency led to sustained accumulation of RPA on chromatin proximal to DSBs (Fig. 5E), whereas the recruitment of RAD51 to chromatin around DSBs decreased significantly in BRG1-knockdown cells (Fig. 5F). However, the assembly of RAD51 and RPA was only detected on the chromatin domains flanking the DSBs (Fig. 5E,F). Next, we questioned whether the replacement of RPA by RAD51 is related to the ATPase activity of BRG1. We transfected the wild-type BRG1, ATPase mutant BRG1 (K798R) and empty vector plasmids into SW13 cells separately. In response to DNA damage, cells expressing wild-type and mutant BRG1 both showed normal accumulation of RAD51 and the replacement of RPA (Fig. 5G), implying that, independent of its ATPase activity, BRG1 regulates RAD51 assembly after DSB induction through another mechanism. Collectively, these data strongly suggest that loss of BRG1 leads to deficient RAD51 binding to DSB sites and a corresponding increase in both RPA binding and ssDNA retention.
BRG1 modulates RAD51 assembly by interacting with the mediator RAD52
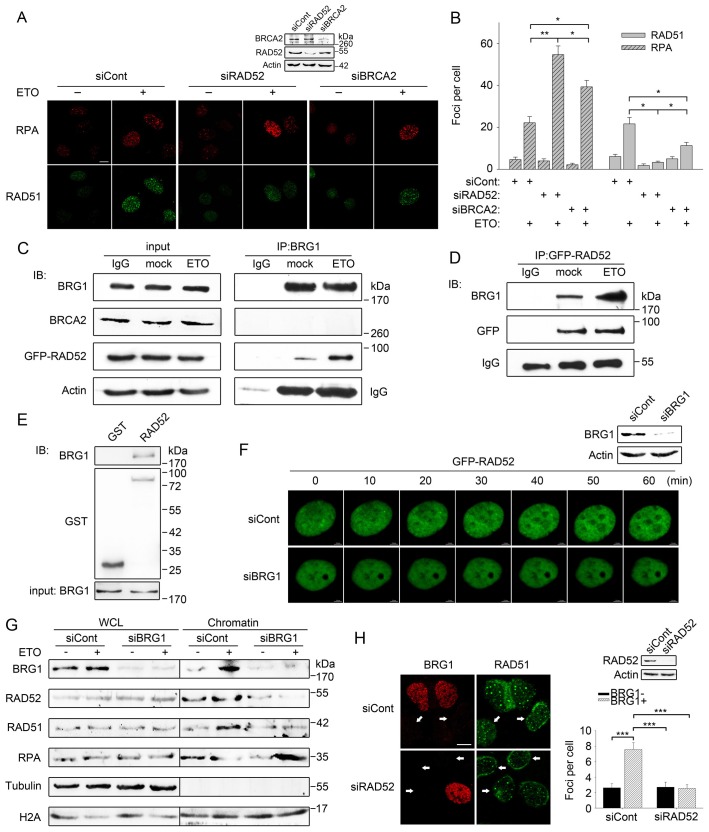
The above data show that BRG1 regulates the replacement of RPA with RAD51, but we failed to detect the existence of RPA and RAD51 in the BRG1 immunocomplex (supplementary material Fig. S4A), implying that there might be other mediators manipulated by BRG1. Previous studies have reported that BRCA2 mediates the replacement of RPA with RAD51 and promotes strand annealing in mammalian cells, whereas Rad52 plays the same role in yeast (Liu et al., 2010; Liu and Heyer, 2011; New et al., 1998; Sugiyama and Kowalczykowski, 2002). However, several recent reports have demonstrated that human RAD52 also functions as a mediator in homologous recombination repair (Wray et al., 2008; Liu and Heyer, 2011). Therefore, we first tested whether RAD52 or BRCA2 is the mediator for the replacement of RPA with RAD51 in the present study. As shown in Fig. 6A,B, either BRCA2 knockdown or RAD52 knockdown led to a persistent RPA signal and impaired recruitment of RAD51 after ETO treatment (with RAD52 depletion resulting in more severe effects), suggesting that both are crucial mediators. A co-immunoprecipitation assay showed that BRCA2 did not exist in the BRG1-associated immunoprecipitate, whereas GFP–RAD52 and BRG1 could be co-immunoprecipitated (Fig. 6C,D), indicating that BRG1 manipulates RAD52 to accomplish the replacement of RPA with RAD51. Moreover, a GST pull-down assay demonstrated that BRG1 interacted with RAD52 in vitro (Fig. 6E).
Fig. 6.
BRG1 interacts with RAD52 and regulates its accumulation at DSB sites during homologous recombination repair. (A) U2OS cells transfected with BRCA2 siRNA (siBRCA2), RAD52 siRNA (siRAD52) or control siRNA (siCont) were exposed to 10 µM ETO for 20 min. After 2 h, cells were fixed and detected by immunostaining with antibodies recognising RPA (red) and RAD51 (green). Scale bar: 10 µm. The expression of BRCA2 and RAD52 was examined by immunoblotting. (B) The number of RPA and RAD51 foci in A was analysed with Image J software. Foci with more than ten pixels were counted, with an average of 100 cells counted per experiment. (C) U2OS cells transfected with GFP–RAD52 were exposed to ETO or were mock treated, and the cell pellets were lysed 1 h later. Cell lysates were incubated with a BRG1-specific antibody. The immunoprecipitated (IP) proteins were separated by SDS-PAGE and probed for BRCA2 and GFP. IB, immunoblot. (D) U2OS cells treated as in C were lysed and incubated with GFP-specific antibody. The immunoprecipitated proteins were separated by SDS-PAGE and probed for BRG1. (E) Untreated U2OS cells were lysed, and the lysates were incubated with GST or GST–RAD52. Bound proteins were separated by SDS-PAGE and immunoblotted with an anti-BRG1 antibody. The equivalent amount of BRG1 added to the binding reactions is shown in the input panel. (F) U2OS cells were pre-treated with BRG1 siRNA (siBRG1) or control siRNA for 48 h and then transfected with GFP–RAD52. Cells treated with ETO or mock treated were analysed by using time-lapse microscopy in a Zigmond chamber, with images taken at 60-s intervals over a 60-min timecourse (see supplementary material Movies 1, 2). Scale bar: 3.5 µm. Immunoblot analysis of BRG1 expression is also shown. (G) U2OS cells transfected with BRG1 siRNA or control siRNA were treated with 10 µM ETO or were mock treated. After repair for 2 h, chromatin fractions and whole cell lysate (WCL) were analysed by immunoblotting with the indicated antibodies. (H) SW13 cells pre-treated with control siRNA or a RAD52 siRNA pool were transfected with the pBJ5-BRG1 plasmid. After 24 h, the cells were treated with ETO for 20 min. Then, the cells were fixed 2 h later and detected by immunostaining with antibodies recognising BRG1 (red) and RAD51 (green). The white arrows indicate SW13 cells without BRG1 expression. Scale bar: 10 µm. RAD52 expression was detected by immunoblotting. Quantification of RAD51 foci (>10 pixels) is shown on the right. Quantitative data in B,H show the mean±s.d.; *P<0.05; **P<0.01; ***P<0.001.
We next assessed the distribution of GFP–RAD52 during DSB repair through time-lapse microscopy with and without knockdown of BRG1 expression. Fig. 6F illustrates that in control cells, the GFP–RAD52 foci increased in a time-dependent manner, whereas knockdown of BRG1 resulted in diminished formation of RAD52 foci after DNA damage (supplementary material Movie 1 for control cell and Movie 2 for BRG1-siRNA-treated cell). Furthermore, both chromatin extraction and laser-track immunofluorescence analysis demonstrated that BRG1 depletion decreased the recruitment of RAD52 and RAD51 to damaged chromatin (Fig. 6G; supplementary material Fig. S4C,D). Importantly, BRG1 expression led to increased formation of RAD51 foci in SW13 cells after ETO treatment, which was abrogated by the silencing of RAD52 (Fig. 6H). By contrast, RAD52 depletion had no significant effect on RAD51 foci formation in SW13 cells lacking BRG1 expression. The result suggested that BRG1 is an upstream regulator of RAD52. These data collectively indicate that BRG1 modulates the dynamics of RAD52 in response to DSBs, which is crucial for the replacement of RPA and for the association of RAD51 with DNA.
DISCUSSION
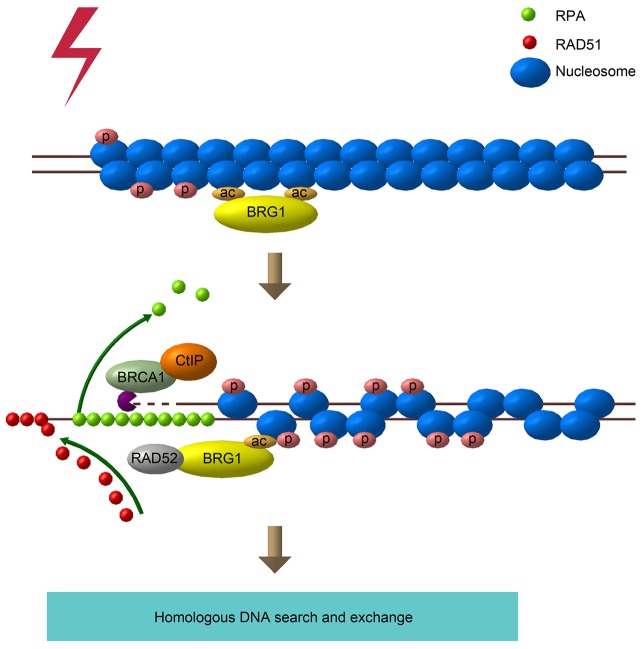
Although it is well accepted that BRG1 plays important roles in DNA damage repair (Martens and Winston, 2003), the precise role of BRG1 in DSB repair has not been fully addressed. Here, we describe a new function of BRG1 in the homologous recombination repair pathway of DSBs. As in the model shown in Fig. 7, BRG1 is recruited to DSB sites at an early stage of the DDR. Then, BRG1 primarily participates in homologous recombination repair by facilitating the replacement of RPA with RAD51 at DSB sites. Specifically, BRG1 interacts with the mediator RAD52 and regulates its recruitment to DSBs, which is crucial for the loading of RAD51 to ssDNAs and the homologous DNA invasion step. Taken together, these results indicate that BRG1 plays a crucial role in the efficient execution of homologous recombination repair by regulating RAD51 assembly.
Fig. 7.
A model for the role of BRG1 in regulating DSB repair. When DSBs occur, BRG1 is recruited to the DNA damage sites. Subsequently, BRG1 interacts with RAD52 and promotes its recruitment to DSB sites, which facilitates RAD51 assembly on RPA-bound ssDNAs. Finally, the RAD51-bound nucleofilament invades into homologous DNA to promote homologous recombination repair. P, phosphorylation; ac, acetylation.
During DSB repair, the condensed chromatin structure prevents the access of repair factors to the broken DNA. Swi/Snf in yeast </emph>has been defined as an important chromatin remodeller and transcriptional regulator in DSB repair (Cruz et al., 2012). Recent studies using mammalian cells have shown that BRG1 (the ATPase subunit of SWI/SNF) can be recruited to DSBs by interacting with γH2AX-containing nucleosomes (Lee et al., 2010). In this study, we show that BRG1 can be recruited to DSB sites and contribute to the DSB repair process (Fig. 2). BRG1-depleted cells are more sensitive to DNA-damaging drugs (Fig. 1; supplementary material Fig. S1). In addition, the expression levels of most DSB repair proteins are not changed in BRG1-knockdown cells in the present study (supplementary material Fig. S3). Thus, we can conclude that BRG1 plays a crucial role in DSB repair rather than the regulation of gene expression.
Homologous recombination and NHEJ are the two main repair pathways of DSB damage (Khanna and Jackson, 2001; van Gent et al., 2001). There is evidence for a role for Swi2/Snf2, the yeast homologue of BRG1, in the mating type locus DSB repair through the homologous recombination pathway (Chai et al., 2005). However, it is still unclear in which pathway BRG1 participates during mammalian DSB repair. The present study utilising two GFP reporter systems (DR-GFP-U2OS and EJ5-GFP-U2OS) showed that BRG1 primarily facilitated mammalian homologous recombination repair rather than NHEJ repair. Moreover, BRG1 ablation led to the accumulation of unrepaired DSBs in the S/G2 phase of the cell cycle (Fig. 3). The results indicate that BRG1 is a crucial factor in homologous recombination repair of DSBs.
Our findings led us to elucidate the mechanism through which BRG1 regulates the homologous recombination process. Previous studies have shown that the DNA ends at DSBs need to be resected by the CtIP–MRN–BRCA1 complex (CtIP is also known as RBBP8) to form ssDNAs, which is crucial to initiate homologous recombination repair (Yun and Hiom, 2009), and that the chromatin remodelling protein SMARCAD1 (Fun30 in yeast) promotes the resection of DNA DSB ends (Chen et al., 2012; Costelloe et al., 2012; Eapen et al., 2012). Then, the resection of DSB ends allows the binding of RPA to the single-stranded DNA (Nimonkar et al., 2011), and the strand exchange protein RAD51 replaces RPA to form the presynaptic filament (New et al., 1998; Lisby et al., 2004), a process mediated by BRCA2 or RAD52 (Shinohara and Ogawa, 1998; Wray et al., 2008; Liu and Heyer, 2011) and the yeast Rad55p–Rad54p heterodimer (Symington, 2002; Wolner et al., 2003). Our results show for the first time that BRG1 depletion leads to increased ssDNA retention and deviant distribution and kinetics of RPA in response to DNA damage (Fig. 4). Intriguingly, we further found a corresponding significant decrease in RAD51 recruitment to DSB sites, along with an increase in the accumulation of ssDNA in BRG1-depleted cells (Fig. 5). The absence of BrdU foci in control cells after ETO treatment might result from the structure of RAD51-coated ssDNA or the large proteins involved in strand invasion. These data indicate that BRG1 depletion results in impaired replacement of RPA with RAD51 and a delayed homologous DNA strand exchange.
Because RAD51 is required for the initiation of homologous recombination repair (San Filippo et al., 2008), we further explored how BRG1 directed the replacement of RPA with RAD51. In this study, we did not find a direct relationship between BRG1 and RAD51 or RPA (supplementary material Fig. S4A), implying that there might be other factors involved. BRCA2 is known as the main mediator of RPA replacement by RAD51 in human cells (Liu et al., 2010) and, in yeast, Rad52 functions in the same way as BRCA2 (Shinohara and Ogawa, 1998). Recent studies have further reported that mammalian RAD52 shares biochemical activities with yeast Rad52 and compensates for BRCA2 when BRCA2 is deficient (Feng et al., 2011; Lok et al., 2013). RAD52-dependent homologous recombination occurs at a later stage to restart a subset of blocked or collapsed replication forks (Wray et al., 2008). In the present study, we found that BRCA2 and RAD52 knockdown both led to a decrease in the number of RAD51 foci and increased RPA foci persistence, indicating that BRCA2 and RAD52 appear to act in parallel pathways. Moreover, we documented an interaction between BRG1 and RAD52 rather than BRCA2, and we found that the recruitment of RAD52 to DSB sites decreased dramatically upon the loss of BRG1, which accordingly led to impaired assembly of RAD51 on ssDNAs (Fig. 6; supplementary material Fig. S4). Simultaneously, we also detected that BRCA2 kinetics in response to DNA damage were not significantly affected by BRG1 knockdown (supplementary material Fig. S4B). These results suggest that BRG1 might modulate the replacement of RPA with RAD51 through interacting with RAD52. However, the possibility that there might be a more complicated relationship between BRCA2 and RAD52 cannot be excluded, because RAD52 depletion resulted in a more severe effect on the replacement of RPA with RAD51 than did BRCA2 depletion, and RAD52 depletion blocked the recruitment of RAD51 to laser-induced DSB sites. Further studies will be necessary to define the exact molecular mechanisms underlying the roles of BRG1, BRCA2 and RAD52. Given that the SWI/SNF complex contains more than ten subunits besides BRG1 and some of them are related to DNA damage repair (Rai et al., 2006; Peng et al., 2009), whether the interaction between BRG1 and RAD52 is direct and which domains are responsible will be the focus of future investigations.
In conclusion, we have provided evidence that BRG1 plays an important role in DSB repair. Multiple congenital human disorders and tumours are associated with genetic defects in DNA repair and damage response pathways (O'Driscoll, 2012), and BRG1 is also found to be mutated in many types of human cancer cell lines (Roberts and Orkin, 2004). Our present study brought to light an important role of BRG1 in promoting DNA damage repair and maintaining genomic integrity. Data from our study also imply that BRG1 could be a promising target in cancer prevention and therapy.
MATERIALS AND METHODS
Antibodies, plasmids and reagents
The following antibodies were used in this study: mouse monoclonal anti-SMARCA4 (clone 20C3.2), anti-γH2AX (05-636) and rabbit polyclonal anti-H2A (07-146) (Millipore Corporation, CA); mouse monoclonal anti-HA (H9658), anti-GFP (G1546), anti-actin (A5441) and anti-BrdU (B2531) (Sigma-Aldrich, St Louis, MO); rabbit polyclonal anti-γH2AX (#9718S) (Cell Signaling Technology, Boston, MA); rabbit polyclonal anti-BRG1 (ab70558), anti-NBS1 (ab32074), anti-RAD51 (ab63801) and mouse monoclonal anti-RPA (ab2175) (Abcam, San Francisco, CA); rabbit polyclonal anti-H2AX (10856-1-AP) (Proteintech, Chicago, IL); rabbit polyclonal anti-RAD51 (sc-8349), anti-RAD52 (sc-365341), anti-BRCA2 (sc-28235) and mouse monoclonal anti-p53 (sc-126) (Santa Cruz Biotechnology, Dallas, TX). Puromycin, etoposide (ETO), bleomycin, micrococcal nuclease (MNase), Hoechst 33342 and 4-hydroxy tamoxifen (4OHT) were purchased from Sigma-Aldrich. The pBABE HA-AsiSI-ER plasmid was provided generously by Dr Gaelle Legube (Laboratoire de Biologie Cellulaire et Moleculaire du Controle de la Proliferation, France). The pBJ5-vector, pBJ5-HA-wtBRG1 and pBJ5-HA-ATPase-mutant BRG1 (K798R) were constructed by Dr Weidong Wang (National Institute on Aging, Baltimore, MD) and Dr Gerald Crabtree (Stanford University, CA) and provided generously by Dr Anthony N. Imbalzano (University of Massachusetts Medical School, MA). The pCBASceI and pCAGGS vector plasmids were kindly provided by Dr Xingzhi Xu (Capital Normal University, China). The YFP-N1-RAD52 plasmid was kindly provided by Dr Claudia Lukas (Institute of Cancer Biology and Centre for Genotoxic Stress Research, Danish Cancer Society, Denmark), and the GFP–RAD52-expressing plasmid was constructed by inserting the RAD52 coding sequence into the pEGFP-N1 vector in our laboratory. The sequences encoding RAD52 were amplified by PCR using the YFP-RAD52 as template, and the fragments were subcloned into the BamHI and EcoRI restriction sites of pGEX 4T-1 vector.
Cell culture and DSB induction
Cells were grown in a 5% CO2 humidified incubator at 37°C. U2OS, DR-GFP-U2OS, EJ5-GFP-U2OS and AsiSI-ER-U2OS cells were grown in DMEM supplemented with 10% FBS. SW13 cells were cultured in IMDM supplemented with 10% FBS. For DSB induction, cells were incubated with the indicated concentrations of ETO (DNA topoisomerase inhibitor) or bleomycin (radio-mimetic drug) for 20 min and released for repair. In addition, AsiSI-ER-U2OS cells were treated with 300 nM 4OHT for 3 h to induce DSBs.
For multiphoton laser micro-irradiation, U2OS cells were grown on coverslips and sensitised with 2 µg/ml Hoechst 33342 for 5 min as described previously (Kruhlak et al., 2006; Helfricht et al., 2013). Then, the coverslips were placed in a Chamlide TC-A live-cell imaging chamber that was mounted on the stage of a confocal microscope (FluoView FV1000, Olympus) integrated with a pulsed nitrogen laser (OLYMPUS, U-RFL-T). The pulsed nitrogen laser (50 Hz, 405 nm) was directly coupled to the epifluorescence path of the microscope and focused through a 40× 2 HCX PLAN APO 1.25–0.75 oil-immersion objective. The laser output power was set to 85% to generate strictly localised subnuclear DNA damage.
Transfections and siRNA-mediated interference
Plasmid transfections were performed using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. For stable transfection, the pBABE HA-AsiSI-ER plasmid was transfected into U2OS cells using the standard Lipofectamine 2000 protocol, and AsiSI–ER-U2OS cells were selected using 1 mg/ml puromycin. Isolated individual clones were further validated by immunoblotting.
siRNA duplex oligoribonucleotides were synthesised by GenePharma (Shanghai, China). Transfections of siRNA duplexes were performed using the Lipofectamine 2000 according to the manufacturer's instructions. Experiments were performed 48 h after transfection. The sense sequences were as follows: control siRNA (nonsense oligo), 5′-UUCUCCGAACGUGUCACGUTT-3′; BRG1 siRNA, 5′-UGGAGAAGCAGCAGAAGAUU-3′; RAD52 siRNA pool, 5′-CAGAAGGUGUGCUACAUUG-3′, 5′-GGUCAUCGGGUAAUUAAUC-3′, 5′-GGCCCAGAAUACAUAAGUA-3′ and 5′-GGAAGAGCCAGGACAUGAA-3′; and BRCA2 siRNA, 5′-GGACUUAUUUACCAAGCAU-3 and 5′-GCAGGACUCUUAGGUCCAA-3′.
Immunoblot analysis
Cells were harvested, washed with PBS and lysed in RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 1% SDS, 1 mM EDTA, 1 mM Na3VO4, 2 mM NaF, 1 mM β-glycerophosphate, 2.5 mM sodium pyrophosphate) containing 1 mM PMSF and protease inhibitors. Whole-cell lysates were mixed with SDS sample buffer, separated by 5–15% SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked overnight with TBS containing 0.1% Tween 20 (TBST) and 5% fat-free milk powder, and probed with primary antibodies. After washing with TBST, the membranes were incubated with secondary antibodies and detected by using ECL Plus immunoblot reagents.
Chromatin isolation
A total of 4×106 cells were washed with PBS. Then, 10% of the cell pellets was lysed in RIPA buffer to produce a whole-cell lysate control; the remaining portion was resuspended in 200 µl of solution A (10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 10 mM NaF, 1 mM Na2VO3 and protease inhibitors). Triton X-100 was added to a final concentration of 0.1%, and the cells were incubated for 10 min on ice. Cytoplasmic proteins were separated from nuclei by low-speed centrifugation (4 min at 1300 g, 4°C). The isolated nuclei were washed once with solution A and then lysed in 200 µl of solution B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT and protease inhibitors) for 30 min. Insoluble chromatin was collected by centrifugation (4 min at 1700 g, 4°C), washed once in solution B and centrifuged again at high speed (10,000 g) for 1 min. The final chromatin pellet was resuspended in 200 µl of Laemmli buffer and sonicated for 15 s. Chromatin was digested by resuspending in solution A containing 1 mM CaCl2 and 50 units of MNase and incubating at 37°C for 1 min, after which the nuclease reaction was halted by the addition of 1 mM EGTA (Rai et al., 2006). The lysates were analysed by immunoblotting.
Microscopic imaging
Cells grown on glass coverslips were fixed with 10% (w/v) formaldehyde in PBS for 10 min, and then permeabilised with 0.5% (v/v) Triton X-100 for 5 min. After permeabilisation, cells were washed and blocked in 10% BSA for 30 min. The cells were incubated with the primary antibody, washed and stained with a secondary antibody. For RPA and RAD51 staining, prior to fixation by ice-cold methanol on ice for 10 min, cells were pre-extracted in buffer D (10 mM PIPES pH 7.0, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 0.5% Triton X-100) to exclude the soluble non-chromatin binding proteins. For BrdU staining, cells were incubated in BrdU (10 mM) for 24 h followed by ETO (10 µM) treatment for 20 min. Cells were then washed and fixed as described above. Cells were then observed by using a laser-scanning confocal microscope equipped with a 60× oil-immersion objective lens. Fluorescent images were collected using Olympus FV10-ASW 1.7 software.
Chromatin immunoprecipitation assay
The ChIP assay was performed as described previously (Tyteca et al., 2006; Iacovoni et al., 2010), with slight modifications. Briefly, formaldehyde was added to the medium at a final concentration of 1% for 10 min and the reaction was halted by adding 0.125 M glycine. Then, the cells were washed and harvested by scraping. Pelleted cells were incubated in lysis buffer (50 mM Tris-HCl pH 8.1, 10 mM EDTA, 1% SDS) and sonicated. Samples were diluted in dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl pH 8.1, 167 mM NaCl) and precleared with blocked Protein A/G beads (Sigma-Aldrich). Precleared samples were incubated with the indicated antibodies or isotype IgG overnight at 4°C. Immune complexes were then recovered by incubating the samples with blocked beads and were eluted twice by incubation in elution buffer (1% SDS, 100 mM NaHCO3) for 15 min. Crosslinking was reversed by adding 5 M NaCl and RNase A to the samples and incubating overnight at 62°C. After proteinase K treatment, immunoprecipitated and input DNA were purified with phenol/chloroform, precipitated and analysed in duplicate by real-time qPCR performed on a StepOnePlus Real-Time PCR System (Applied Biosystems) using the SYBR Green qPCR SuperMix (Invitrogen), according to the manufacturer's instructions. We used primers located in close proximity to (3.7 kb) and distal (2 Mb) from the AsiSI site on chromosome 22 at position 19180307 (Iacovoni et al., 2010). All samples were calibrated to amplification from input DNA. Three biological replicates were performed. The primer sequences were as follows: chr22:19180307_dist_FW, 5′-CCCATCTCAACCTCCACACT-3′; chr22:19180307_dist_REV, 5′-CTTGTCCAGATTCGCTGTGA-3′; chr22:19180307_prox_FW, 5′-CCTTCTTTCCCAGTGGTTCA-3′; and chr22:19180307_prox_REV, 5′-GTGGTCTGACCCAGAGTGGT-3′.
Colony formation assay
U2OS and SW13 cells were transfected with siRNAs or pBJ5-BRG1 respectively, incubated for 48 h and then trypsinised, counted and plated in six-well dishes (1000 cells/dish). Cells were grown for 5 h before treatment with the indicated concentrations of ETO or bleomycin for 20 min. Following a 14-day recovery period, the cells were fixed in 50% (v/v) methanol/0.01% (w/v) Crystal Violet solution for 5 min. Colonies were defined as groups of 50 or more cells (Franken et al., 2006). Survival points divided by 1000 were assayed in triplicate.
Neutral cell comet assay
The neutral comet assay was performed using the Comet Assay kit from Trevigen (Gaithersburg, MD) following the manufacturer's instructions, and images were captured under a fluorescent microscope (ECLIPSE 80i, Nikon, Japan). Comets were analysed using CometScore software (TriTek, Sumerduck, VA).
Homologous recombination and NHEJ repair analysis
Homologous recombination and NHEJ were measured in DR-GFP-U2OS and EJ5-GFP-U2OS cells as described previously (Zhang et al., 2005). The cells with or without BRG1 depletion were transfected with pCBASceI, an empty vector or construct pEGFP, containing the full-length GFP cDNA, as a calibration control for GFP-positive cells. After 72 hours, cells were processed for flow cytometric analysis. For each analysis, 1×104 cells were collected, and each experiment was repeated three times.
Flow cytometry
Cell cycle progression and γH2AX levels were monitored in U2OS cells after ETO treatment, incubation for the indicated time intervals and fixation in 70% ethanol overnight. Cells were permeabilised (0.25% Triton X-100 on ice for 15 min), washed, incubated overnight in PBS containing 0.1% BSA and γH2AX antibodies, and then incubated with the secondary antibody for 30 min at room temperature. Cells were incubated in 50 mg/ml propidium iodide and 100 mg/ml RNase A for 30 min, and 1×104 cells per sample were analysed on a BD FACSarray flow cytometer.
Live-cell imaging
Cells grown in 35-mm dishes with 14-mm glass bottoms were transiently transfected with a plasmid encoding GFP–RAD52 for 24 h, treated with 10 µM ETO for 20 min and then placed on a 37°C heated stage within a 5% CO2 atmosphere chamber. The sequential images were captured by using an UltraVIEW Vox (PerkinElmer Inc., Waltham, MA) spinning-disk confocal microscope with a Ti-E microscope (Nikon, Japan) in fluorescence channel at 60 s intervals over a 60-min timecourse using a 40× objective.
Immunoprecipitation and GST pull-down assay
Cells were lysed on ice using RIPA lysis buffer for 30 min. The lysates were subjected to centrifugation at 13,000 g for 30 min. A total of 10% of the supernatant was reserved as input, and the remaining portion was precleared with 20 µl of Protein G beads at 4°C for 3 h and incubated with the indicated antibodies at 4°C overnight prior to incubation with 30 µl of Protein G beads at 4°C for 3 h. The beads were washed three times with lysis buffer. Bound proteins were eluted by boiling the beads in SDS sample buffer for 5 min. Eluted proteins were resolved by 5–15% gradient SDS-PAGE and transferred to nitrocellulose membrane. Immunoblotting was performed with the appropriate antibodies. GST pull-down assay was performed as described previously (Tong et al., 2013).
Statistical analysis
All statistical analyses were performed using one-tailed Student's t-tests in SPSS software between pairs of conditions. Error bars on the figures correspond to standard deviations. Quantifications are based on at least three independent experiments. In all figures, significant differences between specified pair of conditions, as judged by t-test, are indicated using asterisks (*P<0.05; **P<0.01; ***P<0.001).
Supplementary Material
Acknowledgments
The authors thank Dr Gaelle Legube for kindly supplying the pBABE-HA-AsiSI-ER plasmid; Dr Weidong Wang and Dr Gerald Crabtree for the pBJ5-vector, pBJ5-HA-wtBRG1 and pBJ5-HA-mutant BRG1 (K798R) plasmids; Dr Claudia Lukas for the YFP-N1-RAD52 plasmid; Dr Xingzhi Xu for the pCBASceI and pCAGGS vector plasmids and the EJ5-GFP-U2OS cell line; and Dr Yungui Yang (Beijing Institute of Genomics, Chinese Academy of Sciences) for the DR-GFP-U2OS cell line.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
W.Q. and X.Z. designed the study, analysed the data and wrote the manuscript. W.Q., R.W. and H.C. performed most of the experiments. X.W. and T.X. performed some of the experiments. X.B., I.B. and L.H. contributed to critical reading of the manuscript, scientific advice and guidance during the study.
Funding
This work was supported by the grants from National Nature Science Foundation of China [grant numbers 31371313 to L.H., 31371293 to X.B. and 81372288 to X.Z.]; the Program for Introducing Talents to Universities [grant number B07017 to X.Z.]; and the National Institute of Environmental Health Sciences [grant number R01 ES018948 to I.B.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.159103/-/DC1
References
- Agger K., Santoni-Rugiu E., Holmberg C., Karlström O., Helin K. (2005). Conditional E2F1 activation in transgenic mice causes testicular atrophy and dysplasia mimicking human CIS. Oncogene 24, 780–789 10.1038/sj.onc.1208248 [DOI] [PubMed] [Google Scholar]
- Bennardo N., Cheng A., Huang N., Stark J. M. (2008). Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 4, e1000110 10.1371/journal.pgen.1000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman S., Gebuhr T., Yee D., La Mantia C., Nicholson J., Gilliam A., Randazzo F., Metzger D., Chambon P., Crabtree G. <et al. (2000). A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6, 1287–1295 10.1016/S1097-2765(00)00127-1 [DOI] [PubMed] [Google Scholar]
- Chai B., Huang J., Cairns B. R., Laurent B. C. (2005). Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 19, 1656–1661 10.1101/gad.1273105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Cui D., Papusha A., Zhang X., Chu C. D., Tang J., Chen K., Pan X., Ira G. (2012). The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 489, 576–580 10.1038/nature11355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costelloe T., Louge R., Tomimatsu N., Mukherjee B., Martini E., Khadaroo B., Dubois K., Wiegant W. W., Thierry A., Burma S. <et al. (2012). The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature 489, 581–584 10.1038/nature11353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz L. A., Guecheva T. N., Bonato D., Henriques J. A. (2012). Relationships between chromatin remodeling and DNA damage repair induced by 8-methoxypsoralen and UVA in yeast Saccharomyces cerevisiae. Genet. Mol. Biol. 35 Suppl. 4, 1052–1059 10.1590/S1415-47572012000600021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eapen V. V., Sugawara N., Tsabar M., Wu W. H., Haber J. E. (2012). The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation. Mol. Cell. Biol. 32, 4727–4740 10.1128/MCB.00566-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J., Coates J., Jackson S. P. (2005). Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 434, 605–611 10.1038/nature03442 [DOI] [PubMed] [Google Scholar]
- Feng Z., Scott S. P., Bussen W., Sharma G. G., Guo G., Pandita T. K., Powell S. N. (2011). Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc. Natl. Acad. Sci. USA 108, 686–691 10.1073/pnas.1010959107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken N. A., Rodermond H. M., Stap J., Haveman J., van Bree C. (2006). Clonogenic assay of cells in vitro. Nat. Protoc. 1, 2315–2319 10.1038/nprot.2006.339 [DOI] [PubMed] [Google Scholar]
- Helfricht A., Wiegant W. W., Thijssen P. E., Vertegaal A. C., Luijsterburg M. S., van Attikum H. (2013). Remodeling and spacing factor 1 (RSF1) deposits centromere proteins at DNA double-strand breaks to promote non-homologous end-joining. Cell Cycle 12, 3070–3082 10.4161/cc.26033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovoni J. S., Caron P., Lassadi I., Nicolas E., Massip L., Trouche D., Legube G. (2010). High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 29, 1446–1457 10.1038/emboj.2010.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. D., Jasin M. (2000). Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19, 3398–3407 10.1093/emboj/19.13.3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna K. K., Jackson S. P. (2001). DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27, 247–254 10.1038/85798 [DOI] [PubMed] [Google Scholar]
- Kruhlak M. J., Celeste A., Dellaire G., Fernandez-Capetillo O., Müller W. G., McNally J. G., Bazett-Jones D. P., Nussenzweig A. (2006). Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 172, 823–834 10.1083/jcb.200510015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S. J., Park J. H., Park E. J., Lee S. A., Lee H. S., Kang S. W., Kwon J. (2014). ATM-mediated phosphorylation of the chromatin remodeling enzyme BRG1 modulates DNA double-strand break repair. Oncogene 10.1038/onc.2013.556 [DOI] [PubMed] [Google Scholar]
- Lee H. S., Park J. H., Kim S. J., Kwon S. J., Kwon J. (2010). A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 29, 1434–1445 10.1038/emboj.2010.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D. S., Vogel H., Willerford D. M., Sands A. T., Platt K. A., Hasty P. (2000). Analysis of ku80-mutant mice and cells with deficient levels of p53. Mol. Cell. Biol. 20, 3772–3780 10.1128/MCB.20.11.3772-3780.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Barlow J. H., Burgess R. C., Rothstein R. (2004). Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118, 699–713 10.1016/j.cell.2004.08.015 [DOI] [PubMed] [Google Scholar]
- Littlewood T. D., Hancock D. C., Danielian P. S., Parker M. G., Evan G. I. (1995). A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 23, 1686–1690 10.1093/nar/23.10.1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Heyer W. D. (2011). Who's who in human recombination: BRCA2 and RAD52. Proc. Natl. Acad. Sci. USA 108, 441–442 10.1073/pnas.1016614108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Doty T., Gibson B., Heyer W. D. (2010). Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1260–1262 10.1038/nsmb.1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok B. H., Carley A. C., Tchang B., Powell S. N. (2013). RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene 32, 3552–3558 10.1038/onc.2012.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens J. A., Winston F. (2003). Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13, 136–142 10.1016/S0959-437X(03)00022-4 [DOI] [PubMed] [Google Scholar]
- Morshead K. B., Ciccone D. N., Taverna S. D., Allis C. D., Oettinger M. A. (2003). Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc. Natl. Acad. Sci. USA 100, 11577–11582 10.1073/pnas.1932643100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- New J. H., Sugiyama T., Zaitseva E., Kowalczykowski S. C. (1998). Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391, 407–410 10.1038/34950 [DOI] [PubMed] [Google Scholar]
- Nimonkar A. V., Genschel J., Kinoshita E., Polaczek P., Campbell J. L., Wyman C., Modrich P., Kowalczykowski S. C. (2011). BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25, 350–362 10.1101/gad.2003811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Driscoll M. (2012). Diseases associated with defective responses to DNA damage. Cold Spring Harb. Perspect. Biol. 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley M. A., Tsukuda T., Nickoloff J. A. (2007). ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat. Res. 618, 65–80 10.1016/j.mrfmmm.2006.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. H., Park E. J., Lee H. S., Kim S. J., Hur S. K., Imbalzano A. N., Kwon J. (2006). Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J. 25, 3986–3997 10.1038/sj.emboj.7601291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenge N., Elkin S. K., Oettinger M. A. (2004). ATP-dependent remodeling by SWI/SNF and ISWI proteins stimulates V(D)J cleavage of 5 S arrays. J. Biol. Chem. 279, 35360–35367 10.1074/jbc.M405790200 [DOI] [PubMed] [Google Scholar]
- Peng G., Yim E. K., Dai H., Jackson A. P., Burgt I., Pan M. R., Hu R., Li K., Lin S. Y. (2009). BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nat. Cell Biol. 11, 865–872 10.1038/ncb1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C. L., Almouzni G. (2013). Nucleosome dynamics as modular systems that integrate DNA damage and repair. Cold Spring Harb. Perspect. Biol. 5, a012658 10.1101/cshperspect.a012658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R., Dai H., Multani A. S., Li K., Chin K., Gray J., Lahad J. P., Liang J., Mills G. B., Meric-Bernstam F. <et al. (2006). BRIT1 regulates early DNA damage response, chromosomal integrity, and cancer. Cancer Cell 10, 145–157 10.1016/j.ccr.2006.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., Mir S. N., Wani G., Zhao Q., Battu A., Zhu Q., Wang Q. E., Wani A. A. (2009). Human SNF5/INI1, a component of the human SWI/SNF chromatin remodeling complex, promotes nucleotide excision repair by influencing ATM recruitment and downstream H2AX phosphorylation. Mol. Cell. Biol. 29, 6206–6219 10.1128/MCB.00503-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich T., Allen R. L., Wyllie A. H. (2000). Defying death after DNA damage. Nature 407, 777–783 10.1038/35037717 [DOI] [PubMed] [Google Scholar]
- Roberts C. W., Orkin S. H. (2004). The SWI/SNF complex—chromatin and cancer. Nat. Rev. Cancer 4, 133–142 10.1038/nrc1273 [DOI] [PubMed] [Google Scholar]
- Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. (1998). DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- San Filippo J., Sung P., Klein H. (2008). Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 10.1146/annurev.biochem.77.061306.125255 [DOI] [PubMed] [Google Scholar]
- Shinohara A., Ogawa T. (1998). Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391, 404–407 10.1038/34943 [DOI] [PubMed] [Google Scholar]
- Smith C. L., Peterson C. L. (2004). ATP-dependent chromatin remodeling. Curr. Top. Dev. Biol. 65, 115–148 10.1016/S0070-2153(04)65004-6 [DOI] [PubMed] [Google Scholar]
- Sugiyama T., Kowalczykowski S. C. (2002). Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 277, 31663–31672 10.1074/jbc.M203494200 [DOI] [PubMed] [Google Scholar]
- Sung P. A., Libura J., Richardson C. (2006). Etoposide and illegitimate DNA double-strand break repair in the generation of MLL translocations: new insights and new questions. DNA Repair (Amst.) 5, 1109–1118 10.1016/j.dnarep.2006.05.018 [DOI] [PubMed] [Google Scholar]
- Symington L. S. (2002). Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66, 630–670 10.1128/MMBR.66.4.630-670.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H., Zhao B., Shi H., Ba X., Wang X., Jiang Y., Zeng X. (2013). c-Abl tyrosine kinase plays a critical role in β2 integrin-dependent neutrophil migration by regulating Vav1 activity. J. Leukoc. Biol. 93, 611–622 10.1189/jlb.1012487 [DOI] [PubMed] [Google Scholar]
- Tyteca S., Vandromme M., Legube G., Chevillard-Briet M., Trouche D. (2006). Tip60 and p400 are both required for UV-induced apoptosis but play antagonistic roles in cell cycle progression. EMBO J. 25, 1680–1689 10.1038/sj.emboj.7601066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H., Gasser S. M. (2009). Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 19, 207–217 10.1016/j.tcb.2009.03.001 [DOI] [PubMed] [Google Scholar]
- van Gent D. C., Hoeijmakers J. H., Kanaar R. (2001). Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2, 196–206 10.1038/35056049 [DOI] [PubMed] [Google Scholar]
- Wang X., Haber J. E. (2004). Role of Saccharomyces single-stranded DNA-binding protein RPA in the strand invasion step of double-strand break repair. PLoS Biol. 2, E21 10.1371/journal.pbio.0020021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock D. M., Nakanishi K., Helgadottir H. R., Jasin M. (2006). Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 409, 524–540 10.1016/S0076-6879(05)09031-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolner B., van Komen S., Sung P., Peterson C. L. (2003). Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell 12, 221–232 10.1016/S1097-2765(03)00242-9 [DOI] [PubMed] [Google Scholar]
- Wong A. K., Shanahan F., Chen Y., Lian L., Ha P., Hendricks K., Ghaffari S., Iliev D., Penn B., Woodland A. M. <et al. (2000). BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 60, 6171–6177. [PubMed] [Google Scholar]
- Wray J., Liu J., Nickoloff J. A., Shen Z. (2008). Distinct RAD51 associations with RAD52 and BCCIP in response to DNA damage and replication stress. Cancer Res. 68, 2699–2707 10.1158/0008-5472.CAN-07-6505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Sun Y., Jiang X., Ayrapetov M. K., Moskwa P., Yang S., Weinstock D. M., Price B. D. (2010). The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J. Cell Biol. 191, 31–43 10.1083/jcb.201001160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun M. H., Hiom K. (2009). CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 459, 460–463 10.1038/nature07955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ma Z., Treszezamsky A., Powell S. N. (2005). MDC1 interacts with Rad51 and facilitates homologous recombination. Nat. Struct. Mol. Biol. 12, 902–909 10.1038/nsmb991 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.