ABSTRACT
Most human cancers depend on the telomerase to maintain telomeres; however, about 10% of cancers are telomerase negative and utilize the alternative lengthening of telomeres (ALT) mechanism. Mutations in the DAXX gene have been found frequently in both telomerase-positive and ALT cells, and how DAXX mutations contribute to cancers remains unclear. We report here that endogenous DAXX can localize to Cajal bodies, associate with the telomerase and regulate telomerase targeting to telomeres. Furthermore, disease mutations that are located in different regions of DAXX differentially impact on its ability to interact with its binding partners and its targeting to Cajal bodies and telomeres. In addition, DAXX knockdown by RNA interference led to reduced telomerase targeting to telomeres and telomere shortening. These findings collectively support a DAXX-centric pathway for telomere maintenance, where DAXX interaction with the telomerase regulates telomerase assembly in Cajal bodies and telomerase targeting to telomeres.
KEY WORDS: DAXX, Cancer, Telomerase, Telomere
INTRODUCTION
To prevent the progressive loss of genetic information from linear chromosomes, the telomerase specifically adds telomeric repeat sequences to the 3′ end of chromosomes (Harley et al., 1990; Blackburn, 1992). In humans, the core telomerase complex consists of the catalytic subunit TERT and the RNA template TERC (Collins, 2006). In addition, several proteins including DKC1 and DKC1-associated proteins such as NHP2, GAR1, NOP10, Nopp140, NAF1, Pontin and Reptin have been shown to associate with the core complex to form the telomerase complex, thereby ensuring the activity and function of the telomerase (Dragon et al., 2000; Pogacić et al., 2000; Hoareau-Aveilla et al., 2006; Fu and Collins, 2007; Venteicher et al., 2008). TERC maturation and the assembly of mature TERC with TERT into active telomerase complex take place in Cajal bodies, the coilin-rich nuclear compartment (Gall, 2000; Jády et al., 2004; Zhu et al., 2004; Cioce and Lamond, 2005; Cristofari et al., 2007). Active telomerase complexes are subsequently targeted to telomeres by a poorly understood mechanism.
Although TERC expression appears to be abundant and ubiquitous in human cells, the expression of TERT is tightly regulated (Kanaya et al., 1998; Takakura et al., 1998). TERT is predominantly expressed in highly proliferative tissues and stem cells, but its levels appear to be low or undetectable in somatic cells (Greenberg et al., 1998). Many human cancers exploit the telomerase by upregulating TERT expression and telomerase activity, to avoid telomere shortening and promote cell growth and proliferation (Kim et al., 1994; Greider, 1996; Shay and Bacchetti, 1997; Hanahan and Weinberg, 2000; Cong et al., 2002). Interestingly, telomeres can also be extended by a homologous recombination-based pathway termed alternative lengthening of telomeres (ALT) in ∼10–15% of cancers (Dunham et al., 2000).
Genomic studies using cancer cells from differentiated pancreatic neuroendocrine tumors and pediatric glioblastoma have revealed a strong correlation between the cancers having the ALT phenotype and the presence of mutations in three proteins – DAXX, ATRX and histone H3.3 – indicating a possible role for these proteins in repressing the ALT pathway (Dunham et al., 2000; Heaphy et al., 2011; Jiao et al., 2011; Lovejoy et al., 2012; Schwartzentruber et al., 2012). Through literature search and database analysis, we compiled a list of naturally occurring DAXX mutant alleles in human cancers (Heaphy et al., 2011; Jiao et al., 2011; Lovejoy et al., 2012; Schwartzentruber et al., 2012), and noted that DAXX mutations have been identified in both ALT and telomerase-positive cancers (Stransky et al., 2011; Ding et al., 2012; Imielinski et al., 2012; Dulak et al., 2013; Assié et al., 2014). Such observations suggest that DAXX might not only function in ALT regulation but also in telomerase regulation. However, the molecular connections between DAXX mutations and telomere dysfunction have yet to be explored.
Here, we show that endogenous DAXX can localize to Cajal bodies, associate with the telomerase complex, and facilitate telomerase assembly and targeting to telomeres. Furthermore, these activities of DAXX are differentially disrupted by disease mutations located in different regions of the DAXX protein. Knockdown of DAXX by RNA interference (RNAi) led to reduced telomerase targeting to telomeres as well as telomere shortening. Our study has revealed a new function of DAXX in telomerase-positive cells, and suggests that DAXX dysfunction might forestall telomerase-dependent telomere maintenance.
RESULTS
DAXX is a new telomerase-interacting protein
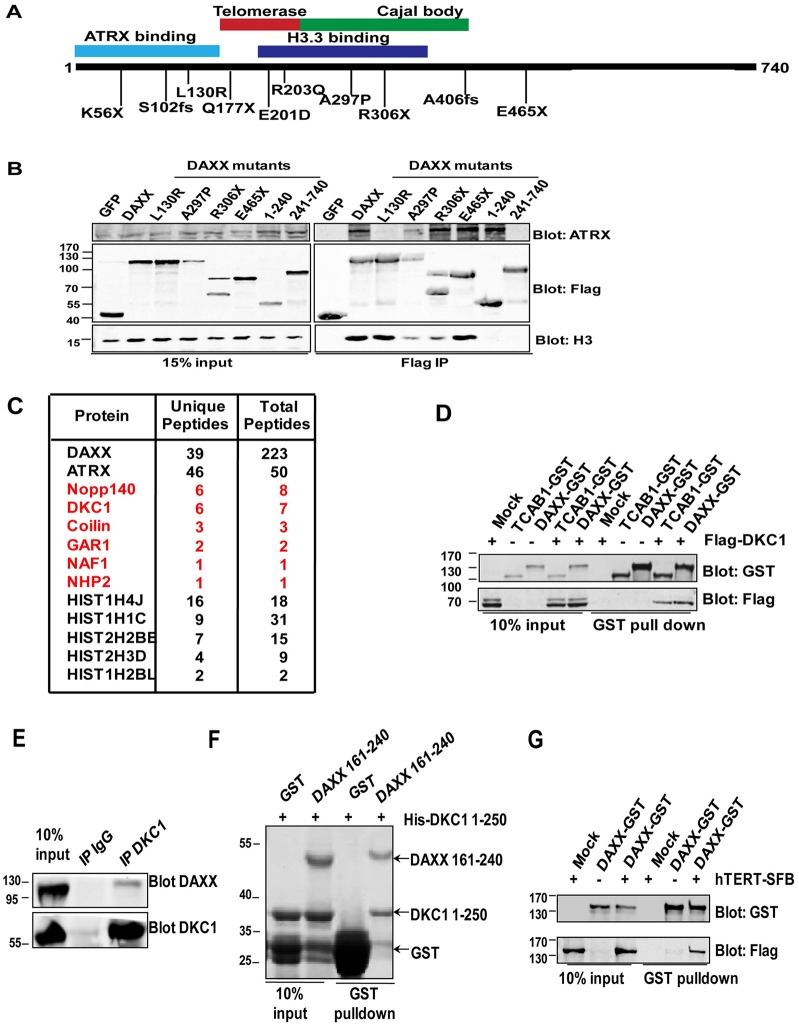
DAXX interaction with ATRX and histones has been well documented (Xue et al., 2003; Drané et al., 2010; Lewis et al., 2010). Many DAXX mutations are located within ATRX and histone H3.3-interacting domains (Tang et al., 2004; Elsässer et al., 2012; Liu et al., 2012), which might result in reduced binding of DAXX to ATRX and histone H3.3. We checked some mutants and found that L130R in the ATRX-binding domain indeed caused the loss of the interaction with ATRX, whereas mutants carrying A297P and R306X mutations in the H3.3-binding domain showed a decreased interaction with H3.3. (Fig. 1A,B; supplementary material Fig. S1A; supplementary material Table S1). Although ATRX and H3.3 mutations have been found primarily in telomerase-negative ALT cancers (Heaphy et al., 2011; Lovejoy et al., 2012; Schwartzentruber et al., 2012), DAXX mutations are seen in both telomerase-positive and -negative cancers. These observations suggest that defective association between such DAXX mutants and ATRX and H3.3 alone cannot account for the pathogenesis of these telomerase-positive cancers, and that there are as yet unexplored pathways that might be crucial for DAXX function.
Fig. 1.
DAXX interacts with the telomerase. (A) Schematic representation of the domain organization of DAXX. Functional domains defined previously and in this study are highlighted with colored boxes. DAXX mutations frequently found in human cancers are also indicated. More detailed information on DAXX disease mutations is listed in supplementary material Table S1. (B) 293T cells transiently expressing SFB-tagged wild-type DAXX, DAXX disease mutants and DAXX truncation mutants were harvested for immunoprecipitation (IP) using anti-Flag antibodies. The immunoprecipitates were then blotted with the indicated antibodies. SFB-tagged GFP was used as negative control. (C) Ectopically expressed SFB-tagged wild-type DAXX proteins were sequentially immunoprecipitated using streptavidin and S-tag beads, and the immunoprecipitates were then sent for mass spectrometry sequencing. The number of unique and total peptides for each protein is listed. Proteins implicated in telomerase biogenesis and regulation are highlighted in red. (D) 293T cells transiently co-expressing GST-tagged DAXX and Flag-tagged DKC1 were used for GST pulldown experiments and probed with the indicated antibodies. GST-tagged TCAB1 was used as a positive control for DKC1 binding. (E) Co-immunoprecipitation of endogenous DKC1 and DAXX was performed using anti-DKC1 antibodies in 293T cells. The precipitates were resolved by SDS-PAGE and western blotted with anti-DAXX or anti-DKC1 antibodies. (F) In vitro GST pulldown assays were performed using bacterially expressed GST-tagged DAXX (residues 161–240) and His-tagged DKC1 (residues 1–250). The precipitates were resolved by SDS-PAGE and stained with Coomassie Blue. GST alone served as a negative control. (G) 293T cells transiently co-expressing GST-tagged DAXX and SFB-tagged hTERT were used for GST pulldown experiments and probed with the indicated antibodies.
We reasoned that elucidating the composition of the DAXX protein complex should help uncover additional signaling pathways mediated by DAXX, and performed large-scale tandem affinity purifications of the DAXX complex followed by mass spectrometry sequencing using 293T cells stably expressing DAXX proteins tagged with SFB (S-tag, Flag and streptavidin-binding peptide). As expected, ATRX and core histones were identified in the DAXX complex (Fig. 1C). Interestingly, we also found multiple subunits of the telomerase complex (e.g. DKC1, GAR1, NHP2, NAF1 and Nopp140), as well as proteins important for telomerase maturation (e.g. coilin).
To further study the association between DAXX and telomerase components, we carried out bi-molecular fluorescence complementation (BiFC/PCA) assays in live cells. BiFC reveals protein–protein interactions when binding between two proteins that are respectively tagged with split halves of a fluorescence protein brings the fragments together for co-folding and fluorescence complementation (Lee et al., 2011) (supplementary material Fig. S1B). Here, DKC1 and DAXX were respectively tagged with the N- and C-terminal fragment of YFP (DKC1–YFPn and DAXX–YFPc) and co-expressed in HTC75 cells for fluorescence detection by flow cytometry. YFPc-tagged PDK1 and TCAB1 were used, respectively, as negative and positive controls for DKC1 (Venteicher et al., 2009). As expected, cells co-expressing DKC1 and TCAB1 displayed fluorescence complementation (∼17% cells), and ∼35% of DAXX and DKC1 co-expressing cells were also YFP positive (supplementary material Fig. S1C,D).These data further support our large-scale immunoprecipitation and mass spectrometry results, and indicate that DAXX might be an integral component of the telomerase complex.
Next, we generated 293T cells that co-expressed GST-tagged DAXX and Flag-tagged DKC1. Consistent with our immunoprecipitation and mass spectrometry data, GST–DAXX was able to co-precipitate with Flag–DKC1 at a level comparable to TCAB1 (Fig. 1D). Importantly, endogenous DKC1 could also precipitate with endogenous DAXX (Fig. 1E). The binding between DKC1 and DAXX might be direct as bacterially expressed GST-tagged DAXX (residues 161–240) was able to pull down His-tagged DKC1 (amino acids 1–250) in in vitro binding assays (Fig. 1F). Notably, we also found that DAXX could interact with the telomerase catalytic subunit TERT when we co-expressed GST–DAXX with SFB-tagged human TERT (hTERT) in 293T cells (Fig. 1G). Taken together, these findings indicate that DAXX can indeed associate with core members of the telomerase holoenzyme.
The N-terminal region of DAXX is required for DAXX association with telomerase
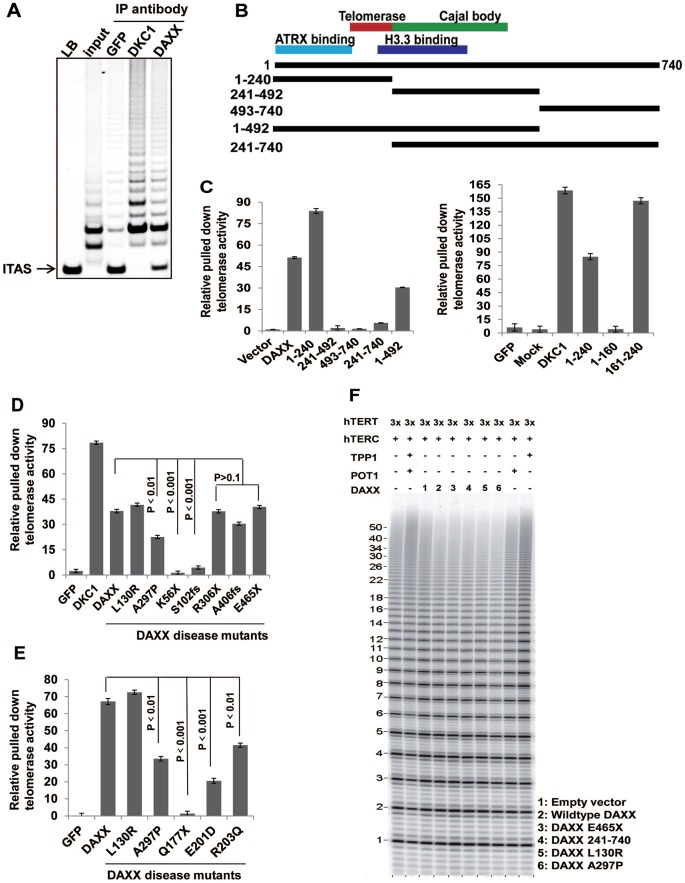
We then tested whether DAXX could associate with active telomerase by immunoprecipitating endogenous DAXX proteins for TRAP assays. As shown in Fig. 2A, endogenous DAXX, similar to DKC1, could precipitate telomerase activities in vivo. Next, we carried out quantitative PCR-based TRAP (Q-TRAP) assays (Herbert et al., 2006) using 293T cells expressing SFB-tagged full-length or truncation mutants of DAXX to pinpoint the region(s) of DAXX responsible for telomerase association (Fig. 2B). Total telomerase activities appeared to be similar among these cell lines (supplementary material Fig. S2). As shown in Fig. 2C, the DAXX N-terminal region (amino acids 1–240) was highly efficient at co-precipitating telomerase activities, and removing the first 240 amino acids abrogated the ability of DAXX to bring down telomerase activities. In fact, the region encompassing amino acids 161–240 was sufficient to pull down substantial amount of telomerase activity. Taken together, these observations suggest that DAXX interacts with the telomerase, with the sequence from 161–240 being the most crucial.
Fig. 2.
DAXX disease mutants are defective in telomerase association. (A) 293T cells were incubated with the indicated antibodies and the immunoprecipitates (IP) were used in nonradioactive TRAP assays to detect co-precipitated telomerase activities. The anti-GFP antibody served as a negative control, and the anti-DKC1 antibody was used as a positive control. LB, lysis buffer; ITAS, 36 bp internal TRAP assay standards. (B) Schematic representation of various DAXX truncation mutants used in the experiments. Expression of these mutants was confirmed by western blotting (supplementary material Figs S2 and S3). (C) SFB-tagged full-length and truncation mutants of DAXX were immunoprecipitated (IP) by anti-Flag antibodies and the associated telomerase activity was analyzed by quantitative-PCR-based TRAP assays (Q-TRAP). The relative amount of precipitated telomerase activities was normalized to the amount of DAXX proteins in the immunoprecipitates (supplementary material Fig. S2). Results are mean±s.e.m. (n = 3). (D,E) SFB-tagged DAXX disease mutants were immunoprecipitated (IP) by anti-Flag antibodies and the associated telomerase activity was analyzed by Q-TRAP. The relative amount of precipitated telomerase activities was normalized to the amount of DAXX proteins in the immunoprecipitates (supplementary material Fig. S3). Results are mean±s.e.m. (n = 3). P-values were determined by Student's t-test. (F) Primer extension assays were performed using 293T cells expressing wild-type or mutant DAXX, as indicated, with ectopically expressed hTERT (3×) and human TERC (hTERC). TPP1 and POT1 were included as controls.
Based on the results thus far, we postulated that some of the mutations might affect the ability of DAXX to interact with the telomerase, and we tested this hypothesis in 293T cells expressing SFB-tagged wild-type and mutant DAXX proteins, with DKC1 serving as a positive control. DAXX mutants were expressed at comparable levels, and total cellular telomerase activities in various DAXX mutant cells were similar to control cells (supplementary material Figs S2 and S3). All the mutants tested did not alter endogenous DAXX expression (supplementary material Fig. S4). Interestingly, several mutations in the N-terminal region such as K56X, S102fs, Q177X, E201D and R203Q (where fs denotes a frameshift and X a stop codon), led to lost or reduced abilities of DAXX to bring down telomerase activities (Fig. 2D,E).
Altered association between telomerase and its regulatory components might result from changes in telomerase processivity. In this scenario, DAXX would interact with the telomerase and act as a telomerase processing factor in a manner similar to TPP1 (Wang et al., 2007; Xin et al., 2007; Nandakumar et al., 2012). We therefore co-expressed human TERC and hTERT in the presence or absence of various DAXX proteins in 293T cells and performed direct primer extension assays (Fig. 2F). The expression of various DAXX proteins appeared to have little effect on telomerase processivity, suggesting that DAXX is not a processing factor.
Endogenous DAXX localizes to Cajal bodies in a cell-cycle-dependent manner
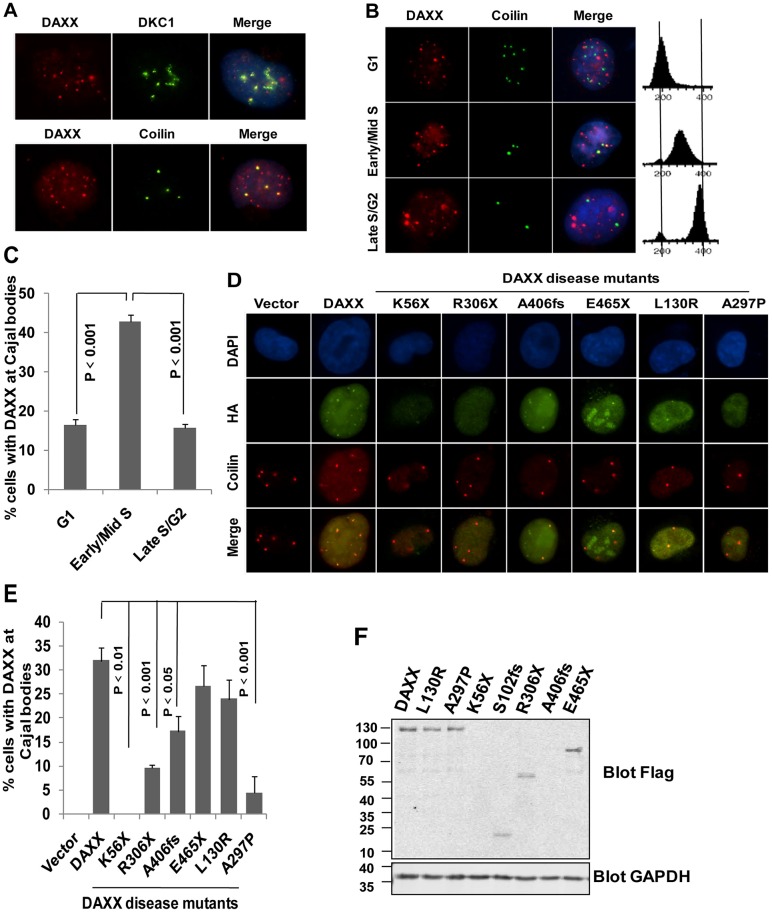
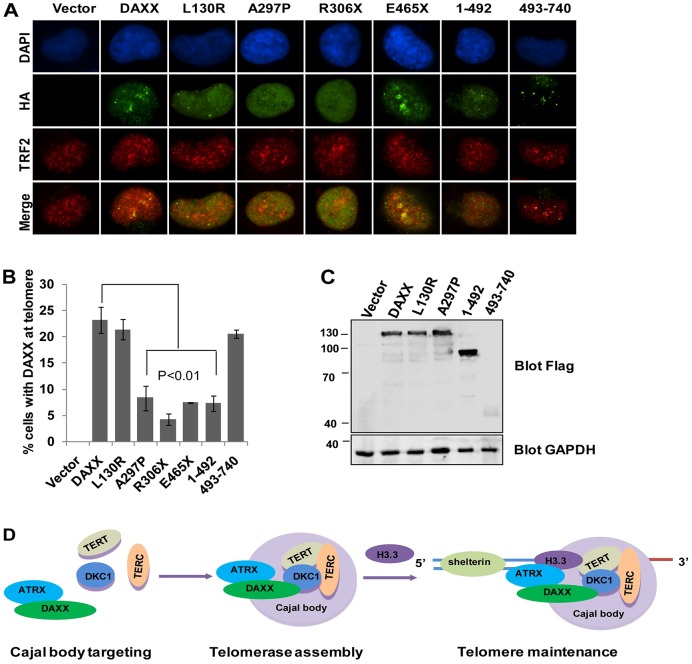
The finding that DAXX interacts with DKC1 and TERT suggests that DAXX might also localize to Cajal bodies, where telomerase assembly occurs. In telomerase-positive HTC75 cells, we found that endogenous DAXX colocalized with DKC1 (Fig. 3A), consistent with its association with endogenous DKC1. In addition, a portion of endogenous DAXX could co-stained with coilin, a Cajal body marker, in ∼25% of the cells (Fig. 3A). When we further examined DAXX targeting in synchronized cells, we observed that co-staining of DAXX and coilin peaked during early and mid S phase (Fig. 3B,C), which coincided with the time window for telomerase assembly. These spatial and temporal characteristics of DAXX suggest a role for DAXX in telomerase biogenesis or assembly in Cajal bodies. In support of this idea, several DAXX disease mutants (e.g. K56X, R306X and A297P) exhibited reduced localization to Cajal bodies (Fig. 3D–F).
Fig. 3.
Endogenous DAXX localizes to Cajal bodies in a cell cycle-dependent manner. (A) Immunostaining analysis of HTC75 cells using antibodies against endogenous DAXX (red) and DKC1 (green) or the Cajal body marker coilin (green). DAPI was used to stain the nuclei. (B) HTC75 cells were synchronized by a double thymidine block. The colocalization of endogenous DAXX (red) with coilin (green) was then assessed by immunostaining at different time points following cell cycle release. Cell cycle profiles are shown on the right. G1, early and mid S phase, and late S and G2 phase cells were enriched, respectively, at 0, ∼2–4 and ∼6–8 hours following release. (C) Quantification of data from B. Results are mean±s.e.m. (n = 3). (D) Representative images of indirect immunofluorescence analysis of HTC75 cells expressing HA–FLAG-tagged wild-type and mutant DAXX proteins, using the indicated antibodies. (E) Quantification of immunostaining data from D. Results are mean±s.e.m. (n = 3). (F) Expression levels of HA–FLAG-tagged wild-type and mutant DAXX proteins in HTC75 cells were analyzed by western blots using the indicated antibodies. GAPDH were used as loading control. P-values were determined by Student's t-test.
The N-terminal deletion mutant of DAXX (missing the first 240 amino acids) was still able to localize to Cajal bodies (data not shown), implicating the region between 241 and 406 in Cajal body targeting of DAXX. Notably, the L130R mutant that failed to associate with ATRX still localized to Cajal bodies, suggesting that DAXX Cajal body targeting is independent of ATRX. The N-terminal half of DAXX contains several regions that mediate its interaction with other proteins and target it to subcellular compartments (Fig. 1A). The H3.3 domain overlaps with the Cajal-body-targeting domain in primary sequences but nonetheless functions distinctly. These findings suggest that DAXX-dependent regulation of telomeres is likely multi-pronged, requiring its interaction with the telomerase as well as ATRX and H3.3.
DAXX is required for telomerase targeting to telomeres
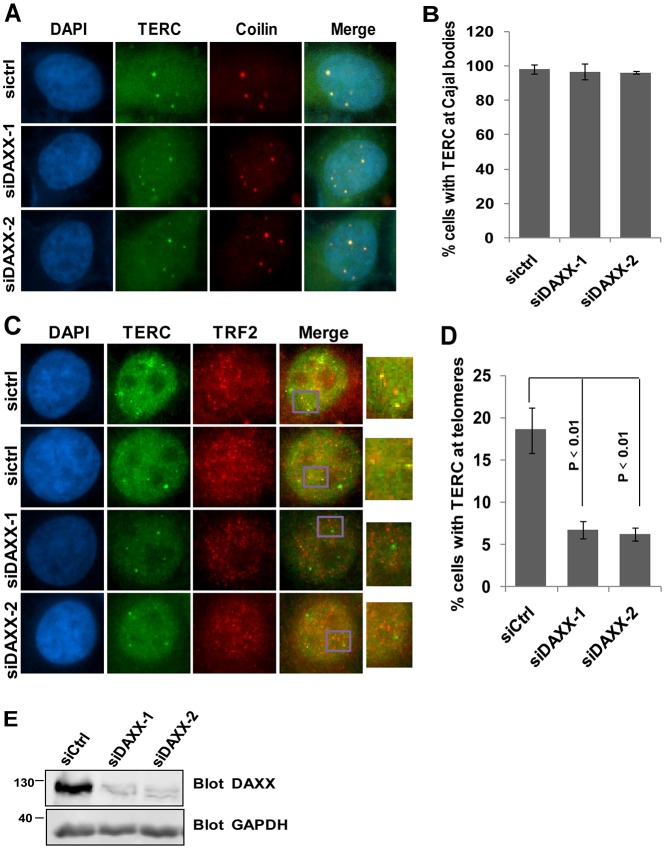
DAXX–telomerase association and DAXX targeting to Cajal bodies raises the possibility that DAXX is required for targeting of telomerase to Cajal bodies and/or telomeres. To test this hypothesis, we probed how DAXX knockdown would impact on endogenous telomerase RNA (TERC) targeting in HTC75 cells (Fig. 4). Two different siRNA oligonucleotides against DAXX (denoted siDAXX-1 and siDAXX2) were examined, both of which could efficiently knockdown DAXX (∼90%) (Fig. 4E). Although the ability of TERC to localize to Cajal bodies was unaffected by changes in DAXX expression (Fig. 4A,B), we observed a significant decrease in co-staining of TERC with TRF2 in cells knocked down for DAXX (Fig. 4C,D), indicating reduced telomere localization of TERC in DAXX-depleted cells and implicating DAXX in the regulation of targeting telomerase to telomeres.
Fig. 4.
DAXX is required for telomerase targeting to telomeres but not Cajal bodies. (A) HTC75 cells were transfected with control siRNA oligonucleotides (siCtrl) or siRNA oligonucleotides targeting DAXX, and analyzed (after 48 h) by RNA FISH combined with indirect immunofluorescence. Green, telomerase RNA TERC that was detected by FISH. Red, coilin that was detected by immunofluorescence. (B) Quantification of data from A. Results are mean±s.e.m. (n = 3). (C) Cells from A were similarly examined by RNA FISH combined with immunofluorescence for co-staining of TERC (green) and TRF2 (red). A magnified view of the boxed area in the Merge image is shown on the right. (D) Quantification of data from C. Results are mean±s.e.m. (n = 3). (E) Endogenous DAXX protein levels in cells from A were analyzed by western blotting, with GAPDH as a loading control. P-values were determined by Student's t-test.
DAXX positively regulates telomere length in telomerase positive cells
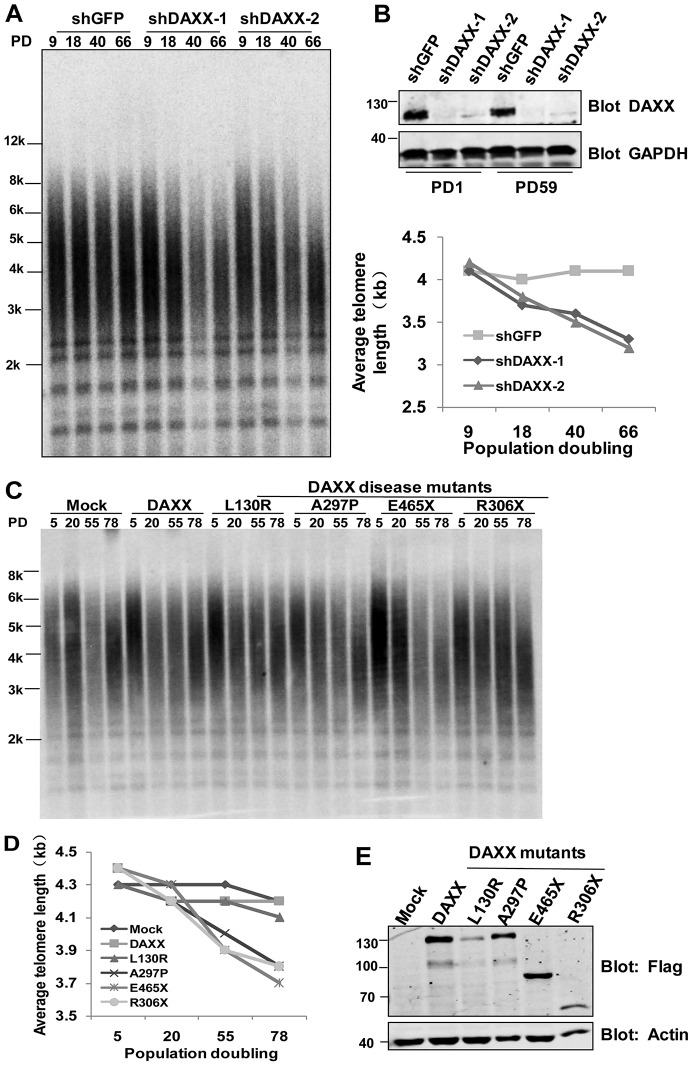
If DAXX participated in telomerase targeting, DAXX inhibition would be expected to negatively impact telomerase-mediated telomere length control, leading to shorter telomeres over time. To test this scenario, we analyzed the average telomere length in HTC75 cells stably expressing two DAXX short hairpin (sh)RNAs (denoted shDAXX-1 and shDAXX-2) (Fig. 5A,B). Both shRNAs achieved >90% knockdown efficiency (Fig. 5B). As we predicted, the two DAXX-knockdown cell lines displayed progressive shortening of telomeres (Fig. 5A), supporting a role of DAXX in telomere length maintenance through its regulation of the telomerase. Shortened telomeres were also apparent in HTC75 cells stably expressing several DAXX disease mutants, including A297P, R306X and E465X (Fig. 5C–E). We postulated that even though some DAXX mutants retained the ability to target to Cajal bodies and associate with the telomerase (e.g. E465X), their ability to target the telomerase to telomeres for telomere length regulation would still be compromised if the mutants themselves were defective in localizing to telomeres. To test this hypothesis, we examined the telomere localization of various DAXX mutants in HTC75 cells (Fig. 6A–C). Indeed, the mutants A297P, R306X and E465X were severely affected in their ability to localize to telomeres (Fig. 6A–C). Most DAXX mutations associated with human cancers result in defective C-terminal expression due to frameshift mutations or advanced stop codons. (Fig. 1A). Interestingly, we found that co-staining with TRF2 remained relatively intact for the C-terminal region of DAXX (DAXX 493–740) but was drastically reduced for the N-terminal region (DAXX 1–492), indicating that the DAXX C-terminus is required for its localization to telomeres. Taken together with our data on the ability of DAXX to interact with the telomerase in Cajal bodies, these observations point to a role for DAXX in facilitating telomerase assembly in Cajal bodies and the subsequent targeting of the assembled telomerase to telomeres. Our results underscore the functional significance of DAXX–telomerase interaction, the disruption of which might be manifested in patients with DAXX mutations.
Fig. 5.
DAXX positively regulates telomere length in telomerase-positive cells. (A) HTC75 cells stably expressing control (shGFP) and shRNA sequences against DAXX (shDAXX-1 and shDAXX-2) were passaged over time (PD, population doubling) and examined for average telomere length by a telomere restriction fragment assay. (B) Top, endogenous DAXX levels in cells from A (PD1 and PD59) were analyzed by western blotting, with actin as a loading control. Bottom, telomere length was quantified using TeloRun. (C) HTC75 cells stably expressing SFB-tagged wild-type DAXX or disease mutants were analyzed by using a telomere restriction fragment assay. (D) Telomere length data from C were quantified using TeloRun. (E) Expression levels of different DAXX proteins were analyzed by western blotting. Actin was used as a loading control.
Fig. 6.
DAXX C-terminal is required for telomere targeting. (A) Representative images of indirect immunofluorescence analysis of HTC75 cells expressing HA–FLAG-tagged wild-type and mutant DAXX proteins, using the indicated antibodies. (B) Quantification of immunostaining data from F. Results are mean±s.e.m. (n = 3). P-values were determined by Student's t-test. (C) Expression levels of HA–FLAG-tagged wild-type and mutant DAXX proteins in HTC75 cells were analyzed by western blotting using the indicated antibodies. GAPDH were used as loading control. (D) Our proposed model for DAXX function in telomerase regulation and telomere maintenance. Shelterin is the six-protein complex made up of TRF1, TRF2, TIN2, RAP1, TPP1 and POT1.
DISCUSSION
Based on our systematic analysis of DAXX disease mutations, we propose a model in which DAXX facilitates the assembly of telomerase holoenzyme in Cajal bodies and the targeting of telomerase to telomeres (Fig. 6D). In this model, DAXX interacts with telomerase subunits and helps with their assembly in Cajal bodies; subsequently, DAXX assists with shuttling the fully assembled telomerase holoenzyme to telomeres. During this process, ATRX is also recruited by DAXX to facilitate H3.3 deposition at telomeres for altering or maintaining telomere chromatin structure. Several lines of evidence support this model. Our large-scale immunoprecipitation and mass spectrometry studies identified several members of the telomerase complex (including DKC1) as associating with DAXX. The interaction between DAXX and DKC1 was confirmed by co-immunoprecipitation and in vitro pulldown assays. DAXX was also able to bring down both the TERT protein and endogenous telomerase activities. Moreover, immunostaining analysis indicated that there was a cell-cycle-dependent localization of endogenous DAXX to Cajal bodies, where telomerase maturation and assembly occur. DAXX-depletion in telomerase-positive cells led to reduced targeting of telomerase to telomeres, which was accompanied by telomere shortening. These findings indicate that DAXX might act as a scaffold that facilitates the assembly and targeting of the active telomerase complex and positively regulates telomere length during S phase.
Our domain-mapping studies revealed that distinct domains mediated DAXX interaction with the telomerase, ATRX and H3.3 (Fig. 1A), and that disease mutations could selectively affect DAXX interaction with its binding partners. Although these binding regions often overlap in primary sequences, structural analysis should help differentiate the crucial residues or peptides responsible for interactions between DAXX and each of its partners.
The fact that mutations that affect DAXX interaction with ATRX, H3.3 or the telomerase can independently lead to DAXX dysfunction argues for an indispensable role of each pathway in DAXX-dependent telomere maintenance. Although the essential function of the ATRX–H3.3–DAXX complex in telomere regulation is beginning to be appreciated, DAXX-mediated telomerase regulation has never before been explored. Our studies offer new insight into the underlying mechanisms for DAXX mutations in cancer cells. The DAXX disease mutants that we tested have defective targeting to Cajal bodies and telomeres. Such DAXX dysfunction might lead to compromised telomerase targeting and subsequent telomere shortening. According to the COSMIC database, many DAXX mutations have also been found in telomerase-positive cancers. It is interesting to postulate that these DAXX mutations might condition telomerase-positive DAXX mutant cells to use the telomerase-independent pathways for survival. In fact, the transition from the telomerase-positive state to telomerase-independent telomere maintenance might prove particularly significant in cancer therapy, because resistance to telomerase-targeting therapies might drive cancer cells to adopt ALT (Hu et al., 2012). The precise steps and signals that lead to complete adoption of ALT by cancer cells warrant further investigation.
MATERIALS AND METHODS
Vectors and cell lines
cDNAs encoding human TERT (hTERT), dyskerin/DKC1, as well as wild-type, truncation (1–160, 161–240, 1–240, 241–492, 493–740, 1–492 and 241–740) and disease mutants (S33fs, K56X, S102fs, L130R, Q177X, E201D, R203Q, R306X, A297P, A406fs, E465X, S588fs, K658fs, G660fs and A680fs) of DAXX were cloned into pBabe or pCL-based retroviral vectors, for C-terminal epitope tagging with either SFB (S-, Flag- and streptavidin-binding tag) or Flag, pDEST-27 (Invitrogen), for GST tagging, or pMSCV-Flag-HA retroviral vectors, for N-terminal tagging with Flag or HA. The DAXX truncation mutant 161–240 and DKC1 truncation mutant 1–250 were also respectively cloned into pDEST-15 (GST tag, Invitrogen) and pDEST-17 (His tag, Invitrogen) for prokaryotic expression and in vitro binding assays.
HTC75 and 293T cells were used for transient or stable expression of various proteins. Synchronization was performed by using double thymidine block and the DNA content of the cells was analyzed by flow cytometry. shRNA sequences were cloned into pcl-mU6 retroviral vectors. The targeting sequences for various shRNAs and siRNA oligonucleotides are: shGFP, 5′-CACAAGCTGGAGTACAACT-3′; shDAXX-1, 5′-AAGGAGTTGGATCTCTCAGAA-3′; shDAXX-2, 5′-GGTAAAGATGGAGACAAGA-3′; and siDAXX-1, 5′-GCCAGGGCCUGAUACCUUCdTdT-3′; siDAXX-2, 5′-GGUAAAGAUGGAGACAAGAdTdT-3′.
Large-scale tandem affinity purification and mass spectrometry analysis
Affinity purification and mass spectrometry analysis was carried out essentially as previously described (Feng et al., 2009). Briefly, 293T cells stably expressing SFB-tagged DAXX were lysed in 1× NETN buffer (40 mM Tris-HCl pH 8.0, 1 mM EDTA, 100 mM NaCl, 0.5% NP-40, 1 mM DTT, with phosphatase and proteinase inhibitors) for 30 min and centrifuged at 20,000 g for 20 min at 4°C. The supernatant was subsequently incubated with 300 µl streptavidin–Sepharose beads (Amersham Biosciences) for 2 h at 4°C, followed by incubation with 2 mg/ml biotin (Sigma) for 2 h at 4°C. The eluate was then incubated with 40 µl S-protein–agarose beads (Novagen) overnight at 4°C. The immunoprecipitates were then resolved by SDS-PAGE, visualized by Coomassie Blue staining, and analyzed by mass spectrometry (Taplin Biological Mass Spectrometry facility, Harvard University, MA).
Bi-molecular fluorescence complementation assay
Pair-wise examination of protein–protein interactions by BiFC was carried out as previously described (Lee et al., 2011). HTC75 cells stably co-expressing two proteins respectively tagged with YFP fragments (YFPn and YFPc) were generated for fluorescence complementation assessment in live cells by flow cytometry.
Antibodies used for co-immunoprecipitation, western blot, and indirect immunofluorescence
Co-immunoprecipitation and western blotting were carried out as previously described (Liu et al., 2004). The antibodies used were: rabbit polyclonal anti-Flag (F7425, Sigma), mouse monoclonal anti-GST (M20007, Abmart), mouse monoclonal anti-actin (P30002, Abmart), mouse monoclonal anti-GAPDH (M20006M, Abmart), rabbit polyclonal anti-tubulin (A302-631A, Bethyl), rabbit polyclonal anti-DKC1 (sc-48794, Santa Cruz Biotechnology), mouse polyclonal anti-DKC1 (Santa Cruz Biotechnology, sc-373956), rabbit polyclonal anti-H3 (9701S, Cell Signaling Technology), rabbit polyclonal anti-ATRX (sc-15408, Santa Cruz Biotechnology), and rabbit polyclonal anti-DAXX (sc-7152, Santa Cruz Biotechnology) antibodies.
Indirect immunofluorescence was carried out as previously described (Liu et al., 2004). The antibodies used were: mouse polyclonal anti-DKC1 (Santa Cruz Biotechnology, sc-373956; 1∶200), mouse monoclonal anti-coilin (Abcam, ab11822; 1∶2000), rabbit polyclonal anti-Flag (Sigma, F7425; 1∶1000), rabbit polyclonal anti-DAXX (Santa Cruz Biotechnology, sc-7152; 1∶200), mouse monoclonal anti-HA (Sigma, H3663; 1∶500), rabbit polyclonal anti-Flag (Sigma, F7425; 1∶500), rabbit polyclonal anti-coilin (Santa Cruz Biotechnology, sc-32860; 1∶200), mouse monoclonal anti-TRF2 (Millipore, 05-521; 1∶200), rabbit monoclonal anti-TRF2 (Abcam, ab108997; 1∶500), Alexa-Fluor-488 goat-anti-mouse (Life Technologies; 1∶1000), and VRDye-549-conjuagted goat-anti-rabbit (Multi Sciences; 1∶2000) antibodies.
IP-TRAP, Q-TRAP and telomerase primer extension assays
Immunoprecipitation followed by TRAP assays (IP-TRAP) was performed as described (Chai et al., 2002). Briefly, cell lysates were incubated with the antibody (3 µg) for 4 hours at 4°C with constant rotation, followed by incubation with protein A/Gagarose beads (Thermo Scientific) at 4°C for another 2 hours. The agarose bead were then resuspended with 40 µl lysis buffer, with 2 µl used for non-radioactive TRAP assays to detect telomerase activity as previously described (Xin et al., 2007). The products were resolved on polyacrylamide gels (8%) and visualized with Gel Red (Biotium). Relative telomerase activity was calculated using the ImageQuant software (GE Healthcare).
Real-time quantitative PCR-based TRAP (Q-TRAP) assays were carried out as described (Herbert et al., 2006). Briefly, cells (3–5×106) were lysed on ice for 30 min in 5×pellet volume of high-salt buffer [20 mM HEPES (pH 7.9), 0.42 mM KCl, 25% glycerol, 0.2% NonidetP-40, 0.1 mM EDTA, 1 mM DTT and protease inhibitors], and then diluted with 5×volume of low-salt buffer [20 mM HEPES (pH 7.9), 100 mM KCl, 25% glycerol, 0.1 mM EDTA, 1 mM DTT and protease inhibitors], before centrifugation at 14,000 rpm for 10 min at 4°C. The supernatant was incubated with anti-Flag M2-agarose beads (Sigma) and the immunoprecipitated proteins were eluted in elution buffer (150–180 µl) [25 mM Tris-HCl (pH 7.4), 136 mM NaCl, 2.6 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10% glycerol, 1 mM DTT and protease and RNase inhibitors] containing 3×Flag peptides (200 µg/ml) (Sigma). The eluate was diluted 2–5-fold before being for PCR. Each 25 µl Q-TRAP reaction contained 2 µl of the eluted proteins, 100 ng each of TS primer (5′-AATCCGTCGAGCAGAGTT-3′) and ACX primer (5′-GCGCGGCTTACCCTTACCCTTACCCTAACC-3′), and 1 mM EGTA, in SYBR Green PCR Master Mix (Applied Biosystems). The reaction mixtures were incubated at 30°C for 30 min and then PCR-amplified (40 cycles of 95°C for 15 s and 60°C for 60 s) using an ABI StepOne Plus real-time PCR system (Applied Biosystems).
Telomerase activity primer extension assays were performed as described previously (Wang et al., 2007; Xin et al., 2007; Nandakumar et al., 2012). Briefly, 293T cells transiently expressing 3×-FLAG-hTERT and hTERC were immunopurified using anti-FLAG beads. A synthetic telomeric DNA primer was then extended in the presence of α-32P-dGTP and unlabeled dATP and TTP. Telomerase activity and processivity were normalized to the amount of hTERT as determined by western blotting.
In vitro GST pulldown assay
Recombinant GST, GST-DAXX 161–240 and His-DKC1 1–250 were extracted from BL21(DE3) bacteria under denaturing conditions, refolded through dialysis, and purified using Glutathione 4B Sepharose or Ni Sepharose High Performance beads (both from GE Healthcare). For pull-down experiments, the purified recombinant proteins were mixed together and incubated at 4°C overnight, before incubation with 20 µl of Glutathione 4B Sepharose beads (GE Healthcare) for 2 hours at 4°C and analysis by SDS-PAGE and Coomassie Blue staining.
Telomere restriction fragment assay
HTC75 cells stably expressing constructs were generated and passaged for telomere restriction fragment analysis as previously described (Xin et al., 2007). Average telomere length was calculated using Telorun (Ouellette et al., 2000).
Human TERC FISH combined with indirect immunofluorescence
Probes complementary to different regions of telomerase RNA were used as previously reported (Tomlinson et al., 2006). Probes were synthesized and conjugated with FITC (TAKARA). Fluorescence in situ hybridization (FISH) was performed with several modifications from previously described protocols (Tomlinson et al., 2006). Briefly, following the last wash with 1× PBS, cells were fixed again in 4% formaldehyde at room temperature for 10 min, rehydrated in 2× SSC and 50% formamide for 5 min at room temperature, followed by pre-hybridization at 37°C for ≥1 h in 1× hybridization solution (2×SSC, 50% deionized formamide, 10% dextran sulfate, 2 mM vanadylribonucleotide complex, 0.002 mg/ml nuclease-free BSA, and 1 mg/ml E. coli tRNA). The cells were subsequently incubated in 1× hybridization solution containing the three human TERC probes (each 25 ng) at 37°C overnight, washed with 2× SSC with 50% formamide at 37°C twice (30 min each wash) and with 1× PBS at room temperature five times (5 min each wash), before mounting the coverslips onto slides.
Supplementary Material
Acknowledgments
We thank our colleagues in Zhou Songyang's laboratory for their insightful discussions and technical assistance.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
M.T., W.M. and Z.S. designed the experiments; M.T., Y.L., Y.Zhang, Y.C., W.H., D.W. and A.J.Z. performed the experiments and provided technical support; Y.Zhao, T.R.C., W.M. and Z.S. advised on the design of the experiments; M.T., D.L. and Z.S. were responsible for the preparation of the manuscript.
Funding
This work was supported by the National Basic Research Program (973 Program) [grant numbers 2010CB945400 and 2012CB911201]; the National Natural Science Foundation of China (NSFC) [grant numbers 91019020, 81330055, 31171397, 31271533, 31371508 and 91213302]; National Institute of General Medical Sciences (NIGMS) [grant numbers GM081627, NCI CA133249 and GM095599]; and the Welch Foundation [grant numbers Q-1673, CPRIT RP130135, CDMRP BC123368, DOD 3351021323]. T.R.C. is an investigator of the Howard Hughes Medical Institute. We would also like to acknowledge the support of the C-BASS core and the Dan L. Duncan Cancer Center [grant number P30CA125123]. Deposited in PMC for release after 6 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.159467/-/DC1
References
- Assié G., Letouzé E., Fassnacht M., Jouinot A., Luscap W., Barreau O., Omeiri H., Rodriguez S., Perlemoine K., René-Corail F. <et al. (2014). Integrated genomic characterization of adrenocortical carcinoma. Nat. Genet. 46, 607–612 10.1038/ng.2953 [DOI] [PubMed] [Google Scholar]
- Blackburn E. H. (1992). Telomerases. Annu. Rev. Biochem. 61, 113–129 10.1146/annurev.bi.61.070192.000553 [DOI] [PubMed] [Google Scholar]
- Chai W., Ford L. P., Lenertz L., Wright W. E., Shay J. W. (2002). Human Ku70/80 associates physically with telomerase through interaction with hTERT. J. Biol. Chem. 277, 47242–47247 10.1074/jbc.M208542200 [DOI] [PubMed] [Google Scholar]
- Cioce M., Lamond A. I. (2005). Cajal bodies: a long history of discovery. Annu. Rev. Cell Dev. Biol. 21, 105–131 10.1146/annurev.cellbio.20.010403.103738 [DOI] [PubMed] [Google Scholar]
- Collins K. (2006). The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell Biol. 7, 484–494 10.1038/nrm1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y. S., Wright W. E., Shay J. W. (2002). Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 66, 407–425 10.1128/MMBR.66.3.407-425.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G., Adolf E., Reichenbach P., Sikora K., Terns R. M., Terns M. P., Lingner J. (2007). Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol. Cell 27, 882–889 10.1016/j.molcel.2007.07.020 [DOI] [PubMed] [Google Scholar]
- Ding L., Ley T. J., Larson D. E., Miller C. A., Koboldt D. C., Welch J. S., Ritchey J. K., Young M. A., Lamprecht T., McLellan M. D. <et al. (2012). Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510 10.1038/nature10738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon F., Pogacić V., Filipowicz W. (2000). In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 20, 3037–3048 10.1128/MCB.20.9.3037-3048.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drané P., Ouararhni K., Depaux A., Shuaib M., Hamiche A. (2010). The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24, 1253–1265 10.1101/gad.566910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulak A. M., Stojanov P., Peng S., Lawrence M. S., Fox C., Stewart C., Bandla S., Imamura Y., Schumacher S. E., Shefler E. <et al. (2013). Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat. Genet. 45, 478–486 10.1038/ng.2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham M. A., Neumann A. A., Fasching C. L., Reddel R. R. (2000). Telomere maintenance by recombination in human cells. Nat. Genet. 26, 447–450 10.1038/82586 [DOI] [PubMed] [Google Scholar]
- Elsässer S. J., Huang H., Lewis P. W., Chin J. W., Allis C. D., Patel D. J. (2012). DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature 491, 560–565 10.1038/nature11608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Huang J., Chen J. (2009). MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 23, 719–728 10.1101/gad.1770609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D., Collins K. (2007). Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol. Cell 28, 773–785 10.1016/j.molcel.2007.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J. G. (2000). Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol. 16, 273–300 10.1146/annurev.cellbio.16.1.273 [DOI] [PubMed] [Google Scholar]
- Greenberg R. A., Allsopp R. C., Chin L., Morin G. B., DePinho R. A. (1998). Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene 16, 1723–1730 10.1038/sj.onc.1201933 [DOI] [PubMed] [Google Scholar]
- Greider C. W. (1996). Telomere length regulation. Annu. Rev. Biochem. 65, 337–365 10.1146/annurev.bi.65.070196.002005 [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A. (2000). The hallmarks of cancer. Cell 100, 57–70 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- Harley C. B., Futcher A. B., Greider C. W. (1990). Telomeres shorten during ageing of human fibroblasts. Nature 345, 458–460 10.1038/345458a0 [DOI] [PubMed] [Google Scholar]
- Heaphy C. M., de Wilde R. F., Jiao Y., Klein A. P., Edil B. H., Shi C., Bettegowda C., Rodriguez F. J., Eberhart C. G., Hebbar S. <et al. (2011). Altered telomeres in tumors with ATRX and DAXX mutations. Science 333, 425 10.1126/science.1207313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert B. S., Hochreiter A. E., Wright W. E., Shay J. W. (2006). Nonradioactive detection of telomerase activity using the telomeric repeat amplification protocol. Nat. Protoc. 1, 1583–1590 10.1038/nprot.2006.239 [DOI] [PubMed] [Google Scholar]
- Hoareau-Aveilla C., Bonoli M., Caizergues-Ferrer M., Henry Y. (2006). hNaf1 is required for accumulation of human box H/ACA snoRNPs, scaRNPs, and telomerase. RNA 12, 832–840 10.1261/rna.2344106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Hwang S. S., Liesa M., Gan B., Sahin E., Jaskelioff M., Ding Z., Ying H., Boutin A. T., Zhang H. <et al. (2012). Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell 148, 651–663 10.1016/j.cell.2011.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M., Berger A. H., Hammerman P. S., Hernandez B., Pugh T. J., Hodis E., Cho J., Suh J., Capelletti M., Sivachenko A. <et al. (2012). Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107–1120 10.1016/j.cell.2012.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády B. E., Bertrand E., Kiss T. (2004). Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell Biol. 164, 647–652 10.1083/jcb.200310138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Shi C., Edil B. H., de Wilde R. F., Klimstra D. S., Maitra A., Schulick R. D., Tang L. H., Wolfgang C. L., Choti M. A. <et al. (2011). DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331, 1199–1203 10.1126/science.1200609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya T., Kyo S., Takakura M., Ito H., Namiki M., Inoue M. (1998). hTERT is a critical determinant of telomerase activity in renal-cell carcinoma. Int. J. Cancer 78, 539–543 [DOI] [PubMed] [Google Scholar]
- Kim N. W., Piatyszek M. A., Prowse K. R., Harley C. B., West M. D., Ho P. L., Coviello G. M., Wright W. E., Weinrich S. L., Shay J. W. (1994). Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015 10.1126/science.7605428 [DOI] [PubMed] [Google Scholar]
- Lee O. H., Kim H., He Q., Baek H. J., Yang D., Chen L. Y., Liang J., Chae H. K., Safari A., Liu D. <et al. (2011). Genome-wide YFP fluorescence complementation screen identifies new regulators for telomere signaling in human cells. Mol. Cell. Proteomics 10, M110 001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P. W., Elsaesser S. J., Noh K. M., Stadler S. C., Allis C. D. (2010). Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. USA 107, 14075–14080 10.1073/pnas.1008850107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Safari A., O'Connor M. S., Chan D. W., Laegeler A., Qin J., Songyang Z. (2004). PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 6, 673–680 10.1038/ncb1142 [DOI] [PubMed] [Google Scholar]
- Liu C. P., Xiong C., Wang M., Yu Z., Yang N., Chen P., Zhang Z., Li G., Xu R. M. (2012). Structure of the variant histone H3.3-H4 heterodimer in complex with its chaperone DAXX. Nat. Struct. Mol. Biol. 19, 1287–1292 10.1038/nsmb.2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy C. A., Li W., Reisenweber S., Thongthip S., Bruno J., de Lange T., De S., Petrini J. H., Sung P. A., Jasin M. ALT Starr Cancer Consortium(2012). Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 8, e1002772 10.1371/journal.pgen.1002772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J., Bell C. F., Weidenfeld I., Zaug A. J., Leinwand L. A., Cech T. R. (2012). The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492, 285–289 10.1038/nature11648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette M. M., Liao M., Herbert B. S., Johnson M., Holt S. E., Liss H. S., Shay J. W., Wright W. E. (2000). Subsenescent telomere lengths in fibroblasts immortalized by limiting amounts of telomerase. J. Biol. Chem. 275, 10072–10076 10.1074/jbc.275.14.10072 [DOI] [PubMed] [Google Scholar]
- Pogacić V., Dragon F., Filipowicz W. (2000). Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 20, 9028–9040 10.1128/MCB.20.23.9028-9040.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J., Korshunov A., Liu X. Y., Jones D. T., Pfaff E., Jacob K., Sturm D., Fontebasso A. M., Quang D. A., Tönjes M. <et al. (2012). Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 10.1038/nature10833 [DOI] [PubMed] [Google Scholar]
- Shay J. W., Bacchetti S. (1997). A survey of telomerase activity in human cancer. Eur. J. Cancer 33, 787–791 10.1016/S0959-8049(97)00062-2 [DOI] [PubMed] [Google Scholar]
- Stransky N., Egloff A. M., Tward A. D., Kostic A. D., Cibulskis K., Sivachenko A., Kryukov G. V., Lawrence M. S., Sougnez C., McKenna A. <et al. (2011). The mutational landscape of head and neck squamous cell carcinoma. Science 333, 1157–1160 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takakura M., Kyo S., Kanaya T., Tanaka M., Inoue M. (1998). Expression of human telomerase subunits and correlation with telomerase activity in cervical cancer. Cancer Res. 58, 1558–1561. [PubMed] [Google Scholar]
- Tang J., Wu S., Liu H., Stratt R., Barak O. G., Shiekhattar R., Picketts D. J., Yang X. (2004). A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J. Biol. Chem. 279, 20369–20377 10.1074/jbc.M401321200 [DOI] [PubMed] [Google Scholar]
- Tomlinson R. L., Ziegler T. D., Supakorndej T., Terns R. M., Terns M. P. (2006). Cell cycle-regulated trafficking of human telomerase to telomeres. Mol. Biol. Cell 17, 955–965 10.1091/mbc.E05-09-0903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher A. S., Meng Z., Mason P. J., Veenstra T. D., Artandi S. E. (2008). Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 132, 945–957 10.1016/j.cell.2008.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher A. S., Abreu E. B., Meng Z., McCann K. E., Terns R. M., Veenstra T. D., Terns M. P., Artandi S. E. (2009). A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323, 644–648 10.1126/science.1165357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Podell E. R., Zaug A. J., Yang Y., Baciu P., Cech T. R., Lei M. (2007). The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445, 506–510 10.1038/nature05454 [DOI] [PubMed] [Google Scholar]
- Xin H., Liu D., Wan M., Safari A., Kim H., Sun W., O'Connor M. S., Songyang Z. (2007). TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature 445, 559–562 10.1038/nature05469 [DOI] [PubMed] [Google Scholar]
- Xue Y., Gibbons R., Yan Z., Yang D., McDowell T. L., Sechi S., Qin J., Zhou S., Higgs D., Wang W. (2003). The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl. Acad. Sci. USA 100, 10635–10640 10.1073/pnas.1937626100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Tomlinson R. L., Lukowiak A. A., Terns R. M., Terns M. P. (2004). Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol. Biol. Cell 15, 81–90 10.1091/mbc.E03-07-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.