ABSTRACT
The pentameric WASH complex is best known for its role in regulating receptor trafficking from retromer-rich endosomal subdomains. FAM21 functions to stabilize the WASH complex through its N-terminal head domain and localizes it to endosomes by directly binding the retromer through its extended C-terminal tail. Herein, we used affinity purification combined with mass spectrometry to identify additional FAM21-interacting proteins. Surprisingly, multiple components of the nuclear factor κB (NF-κB) pathway were identified, including the p50 and p65 (RelA) NF-κB subunits. We show that FAM21 interacts with these components and regulates NF-κB-dependent gene transcription at the level of p65 chromatin binding. We further demonstrate that FAM21 contains a functional monopartite nuclear localization signal sequence (NLS) as well as a CRM1/exportin1-dependent nuclear export signal (NES), both of which work jointly with the N-terminal head domain and C-terminal retromer recruitment domain to regulate FAM21 cytosolic and nuclear subcellular localization. Finally, our findings indicate that FAM21 depletion sensitizes pancreatic cancer cells to gemcitabine and 5-fluorouracil. Thus, FAM21 not only functions as an integral component of the cytoplasmic WASH complex, but also modulates NF-κB gene transcription in the nucleus.
KEY WORDS: FAM21, WASH complex, Retromer, NF-κB, Pancreatic cancer
INTRODUCTION
The Wiskott–Aldrich syndrome protein and Scar homologue (WASH) is a recently discovered member of the Wiskott–Aldrich syndrome protein (WASP) superfamily of nucleation-promoting factors (NPFs) (Linardopoulou et al., 2007). The WASP family of NPFs stimulates filamentous (F)-actin polymerization through activation of the ubiquitous actin-related protein 2/3 (Arp2/3) complex (Linardopoulou et al., 2007; Liu et al., 2009; Rottner et al., 2010; Takenawa and Suetsugu, 2007). The WASH complex is a major Arp2/3 NPF on the surface of endosomes, where it participates in endosomal protein trafficking by direct interaction with the retromer complex subunit VPS35 (Bear, 2009; Derivery et al., 2009; Duleh and Welch, 2010; Gomez and Billadeau, 2009; Harbour et al., 2012; Helfer et al., 2013; Jia et al., 2012; Seaman et al., 2013). The WASH complex has been shown to regulate endosome-to-Golgi retrieval of the cation-independent mannose 6-phosphate receptor (Gomez and Billadeau, 2009) as well as the endosome-to-cell surface recycling of certain cargo proteins, such as the transferrin receptor (Derivery et al., 2009), α5β1 integrin (Zech et al., 2011), the β2-adrenergic receptor (Temkin et al., 2011), T cell receptor, glucose transporter GLUT1 (Piotrowski et al., 2013) and major histocompatibility complex (MHC) II molecules (Graham et al., 2014).
In addition to its well-established activity on endosomes, recent reports have further implicated the WASH complex in the regulation of lysosomal function and autophagosome formation. Indeed, the WASH complex interacts with the biogenesis of lysosome-related organelles complex-1 (BLOC-1) and its cargos, suggesting that it might also function in the formation of specialized lysosome-related organelles (Monfregola et al., 2010; Ryder et al., 2013). In addition, the WASH complex is involved in lysosome maturation through recycling of the vacuolar (V)-ATPase (Carnell et al., 2011; Park et al., 2013), exocytosis through its interaction with the exocyst complex (Monteiro et al., 2013) and autophagosome formation in conjunction with the retromer protein VPS35 (Zavodszky et al., 2014), as well as autophagic and phagocytic digestion (King et al., 2014).
The WASH complex exists as a highly conserved pentameric protein complex consisting of WASH1, FAM21, strumpellin/KIAA0196, SWIP/KIAA1033 and CCDC53 (Derivery and Gautreau, 2010; Derivery et al., 2009; Gomez and Billadeau, 2009; Jia et al., 2010), and the binary interaction has been delineated by biochemical and yeast two-hybrid analyses (Harbour et al., 2012; Jia et al., 2010). The largest member of the WASH complex is FAM21, which contains a globular head domain of ∼220 amino acids that is necessary for WASH complex assembly and stability that is mediated by binding to WASH and SWIP (Harbour et al., 2012; Jia et al., 2010). Following the head domain, FAM21 contains a long unstructured tail of ∼1100 amino acids, which contains a series of repeats known as LFa motifs (L-F-acidic3–10-L-F), which are involved in direct binding of VPS35 (Jia et al., 2012). The precise functions of the WASH complex proteins strumpellin, SWIP and CCDC53 are currently unknown. Importantly, mutations in strumpellin result in an autosomal dominant form of hereditary spastic paraplegia (HSP) (de Bot et al., 2013; Vardarajan et al., 2012), and mutations in SWIP have been associated with autosomal recessive intellectual disability and late-onset Alzheimer's disease (Ropers et al., 2011; Vardarajan et al., 2012). The association of these phenotypes with the role of the WASH complex in the regulation of receptor trafficking or organelle biogenesis remains to be further characterized.
Accumulating evidence suggests that FAM21 is the most crucial and versatile component of the WASH complex. FAM21–VPS35 interaction is necessary for recruitment of WASH complex to endosomal membranes (Gomez and Billadeau, 2009; Harbour et al., 2010; Harbour et al., 2012; Helfer et al., 2013). Notably, a rare autosomal-dominant VPS35 mutation (D620N) has recently been identified as a cause of Parkinson's disease owing to destabilized retromer–WASH complex association and impaired autophagy (McGough et al., 2014; Zavodszky et al., 2014). In addition, biochemical characterization has indicated that the FAM21 tail is capable of binding to the capping protein CAPZ and inhibiting its actin-capping activity (Hernandez-Valladares et al., 2010). In this regard, the minimal region within the FAM21 tail responsible for binding to the capping protein CAPZ has been identified (Jia et al., 2010). Besides VPS35 and CAPZ, the FAM21 tail also interacts with RME-8 (Freeman et al., 2014), and FKBP15, CCDC22 and CCDC93 (Harbour et al., 2012). Therefore, taking advantage of the finely mapped binding regions in FAM21, we generated a FAM21 deletion mutant incapable of interacting with known binding partners (e.g. other WASH-complex members and CAPZ) to facilitate identification of new interacting protein(s). This strategy led to the identification of several nuclear factor κB (NF-κB) components as new FAM21-interacting proteins. Our results reveal a new role for FAM21 in the regulation of NF-κB-dependent gene transcription in the nucleus, and reveal the mechanism regulating FAM21 nuclear shuttling, thereby expanding on the previously known cytoplasmic function of FAM21 in WASH-complex-dependent vesicular trafficking.
RESULTS
FAM21 interacts with NF-κB p65 and p50
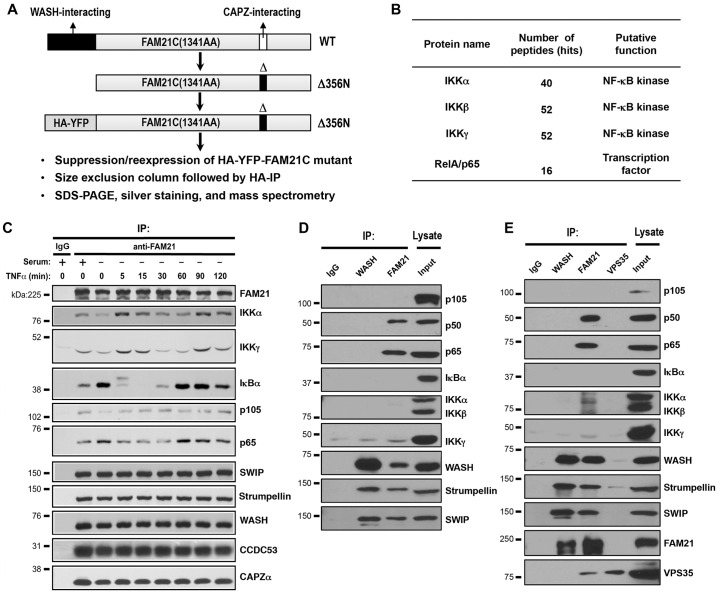
To identify FAM21-interacting proteins, a biochemical screen was performed based on the suppression and re-expression vector system, which allows depletion of endogenous FAM21 along with simultaneous expression of an HA–YFP-tagged FAM21 truncation mutant (Gomez and Billadeau, 2009). Briefly, the compound deletion mutant (deficient in WASH and CAPZ binding) was transiently expressed in HeLa cells and purified by size-exclusion chromatography followed by anti-HA immunoprecipitation (Fig. 1A). Protein bands were excised following SDS-PAGE, digested with trypsin, and identified by nano-liquid-chromatography–tandem mass spectrometry. This strategy allowed for the enrichment of FAM21 tail-interacting proteins. Interestingly, several NF-κB-related proteins were identified, including inhibitor of NF-κB (IκB) kinase (IKK)α, IKKβ, NEMO (also known as IKKγ) and the p65 (RelA) NF-κB subunit (Fig. 1B). Besides NF-κB signaling components, several other top hits include triple functional domain protein (TRIO), Ras GTPase-activating-like protein (IQGAP1), and TBC1 domain family member 4 (TBCD4).
Fig. 1.
FAM21 interacts with multiple NF-κB components. (A) Schematic view of strategies used to identify FAM21-interacting proteins in HeLa cells. Cells were transfected with the depicted FAM21 mutant construct and cell lysate was prepared at 72 h post-transfection. (B) Summary of NF-κB components associated with FAM21 identified through mass spectrometry. (C) Co-immunoprecipitation (IP) of endogenous NF-κB proteins and FAM21 in HeLa cells following TNFα treatment for the indicated period of time. (D,E) Co-immunoprecipitation of endogenous NF-κB with FAM21 in BXPC3 (D) and Panc1 cells (E).
The canonical NF-κB is predominantly a heterodimer of the p65 and p50 subunits (encoded by RELA and NFKB1, respectively), which are bound and sequestered by IκB proteins in the cytosol. Various intracellular and extracellular stimuli such as TNFα activate the IKK complex, which phosphorylates IκBα leading to its ubiquitylation and proteasomal degradation. The activated p65–p50 dimer translocates into the nucleus where it binds and activates transcription of many NF-κB target genes, including those involved in cell survival, proliferation, the inflammatory response and anti-apoptotic factors (Wajant and Scheurich, 2011). As expected, TNFα treatment resulted in a substantial IκBα phosphorylation by 5 min and loss of total IκBα protein by 15 min (supplementary material Fig. S1). By 60 min, IκBα protein levels were largely restored as a result of NF-κB-dependent transcription and synthesis. Interestingly, FAM21 co-immunoprecipitated IKKα, IKKγ, p105, p65 and IκBα from TNFα-stimulated HeLa cells (Fig. 1C). FAM21 also co-immunoprecipitated p65 and p50 from the pancreatic cancer cell lines Panc1 and BXPC3 (Fig. 1D,E). However, we could not reproducibly detect FAM21 interaction with IKKα, IKKβ, IKKγ, p105 or IκBα in either pancreatic cancer cell, suggesting that these interactions are likely not as stable or universally present. Interestingly, we did not detect WASH or VPS35 binding to p50, p65 or other NF-κB-related proteins (Fig. 1D,E), suggesting that FAM21 might be forming new complexes independently of its association with WASH or VPS35.
Depletion of FAM21, but not WASH, decreases NF-κB transcriptional activity
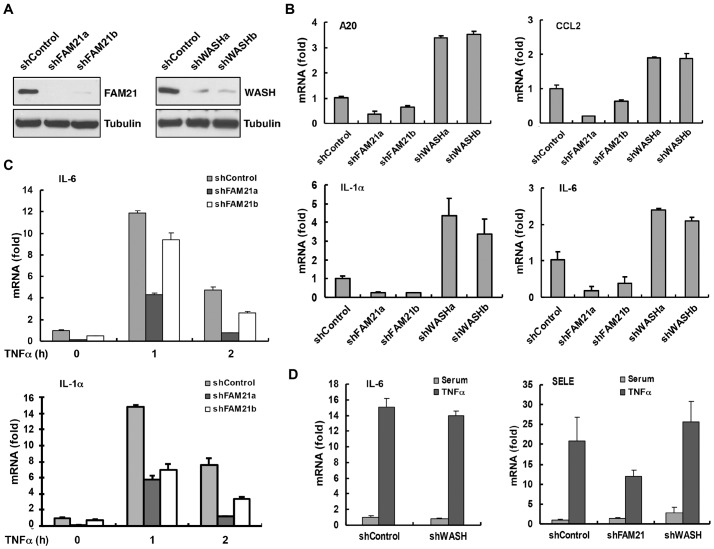
To investigate whether FAM21 or WASH regulates NF-κB transcriptional activities, we generated stable FAM21 or WASH knockdown cells lines using multiple lentiviral short hairpin RNA (shRNA) constructs. As expected, expression of FAM21 and WASH were markedly diminished in Panc1 cells as revealed by immunoblotting (Fig. 2A). Quantitative real-time PCR analysis of cDNA obtained from cells expressing control and FAM21 shRNA (shControl and shFAM21, respectively) revealed that the basal mRNA levels of several NF-κB target genes {A20, CCL2, IL-1a [encoding interleukin (IL)-1α] and IL6} were reduced up to 5-fold (Fig. 2B). Interestingly, WASH-suppressed cells displayed a 2–4-fold increase in NF-κB target gene expression (Fig. 2B). Consistent with this, depletion of FAM21 in HeLa cells diminished TNFα-dependent activation of NF-κB target genes, including those encoding IL-1α, IL-6 and SELE (Fig. 2C,D), whereas no significant change was observed in WASH-suppressed HeLa cells (Fig. 2D). These data indicate that in contrast to WASH, FAM21 regulates NF-κB transcriptional activity.
Fig. 2.
FAM21 depletion reduces NF-κB transcriptional activity. (A) Lysate prepared from cells stably transduced with specific lentiviral shRNAs or scrambled control was subjected to immunoblotting to demonstrate specific and efficient knockdown of endogenous FAM21 and WASH in Panc1 cells. (B) Quantitative RT-PCR was performed to analyze basal mRNA expression of selected NF-κB targets in Panc1 cells depleted of FAM21 or WASH expression. Note that FAM21 depletion significantly decreased basal mRNA levels of indicated genes. In contrast, loss of WASH increased the mRNA levels of these same genes. (C) HeLa scrambled control and two FAM21 knockdown stable cells were treated with TNFα (15 ng/ml) for the indicated periods of time and analyzed for NF-κB targets expression by qRT-PCR. (D) The levels of NF-κB target gene mRNA expression was determined by qRT-PCR in FAM21- or WASH-depleted HeLa cells following TNFα treatment for 1 h.
FAM21 does not affect the phosphorylation status of IκBα or nuclear translocation of NF-κB
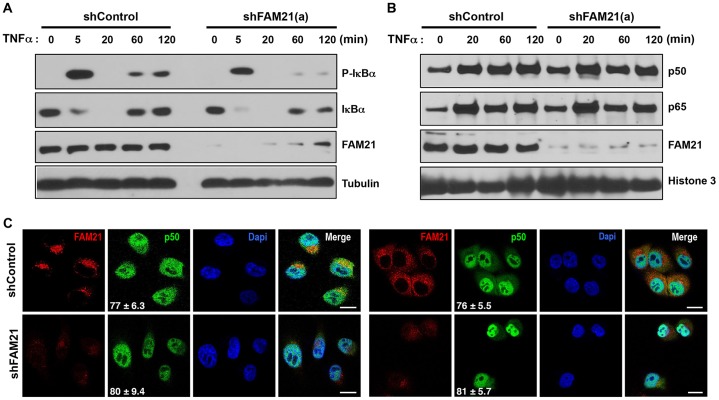
Given that FAM21 interacts with NF-κB proteins and modulates their transcriptional activity, an important question was how and where FAM21 might exert this effect. Therefore, we first investigated whether FAM21 participated in NF-κB activation upstream of IKK activity by examining TNFα-mediated IκBα phosphorylation. As shown in Fig. 3A, TNFα-induced IκBα phosphorylation was slightly decreased in FAM21-depleted HeLa cells when compared to shControl-expressing cells. However, the basal and TNFα-induced levels of IκBα were reduced in shFAM21 HeLa cells, suggesting that although TNFα-induced IκBα phosphorylation was likely intact in the absence of FAM21, there was a defect in the transcriptional regulation of IκBα, a well-known and rapidly induced target of NF-κB (Sun et al., 1993). Importantly, FAM21 depletion did not significantly change the levels of p105, p50 or p65 in the cytoplasm (supplementary material Fig. S2). We next investigated whether FAM21 affected the translocation of NF-κB from the cytosol to the nucleus, a defining step in NF-κB-dependent transcription. Significantly, FAM21 depletion did not impact on TNFα-stimulated nuclear translocation of p65 or p50 at the indicated time points (Fig. 3B). To further validate these observations, we examined the expression and nuclear localization of p50 and p65 following TNFα treatment using fluorescence microscopy. In resting cells, p65 was predominantly localized in the cytosol as expected, and there was no significant change in FAM21-depleted Panc1 cells compared to cells expressing control shRNA (supplementary material Fig. S3A). Consistent with the fractionation data, p50 and p65 both translocate to the nucleus upon TNFα stimulation in FAM21-depleted cells (Fig. 3C). Taken together, these results suggest that FAM21 affects NF-κB transcriptional activation at a point distal to p65 and p50 nuclear entry.
Fig. 3.
FAM21 does not significantly affect phosphorylation status of IκBα or nuclear translocation of NF-κB. (A) HeLa cells stably expressing lentiviral shRNA against FAM21 or control shRNA were treated with TNFα for the indicated periods of time. Cell lysates were collected and examined for the phosphorylated (P-IκBα) and total IκBα levels by immunoblotting. (B) Immunoblotting of nuclear protein extracts from HeLa cells treated with TNFα for the indicated periods of time. Note the efficient suppression of FAM21 by FAM21-specific shRNA and but similar levels of nuclear localized of p65 in shControl and shFAM21 cell at various time periods following TNFα treatment. Histone 3 serves as a nuclear marker and loading control. (C) Panc1 cells stably expressing the indicated shRNA were treated with TNFα (100 ng/ml) for 30 min and subcellular distribution of endogenous p50 and p65 were determined by immunofluorescence. Mean percentages of 100 cells with predominantly nuclear localized NF-κB p65 are shown in the lower left corner. Results are mean±s.d. Scale bars: 20 µm.
FAM21 is present in the nucleus and contains both a functional NLS and NES
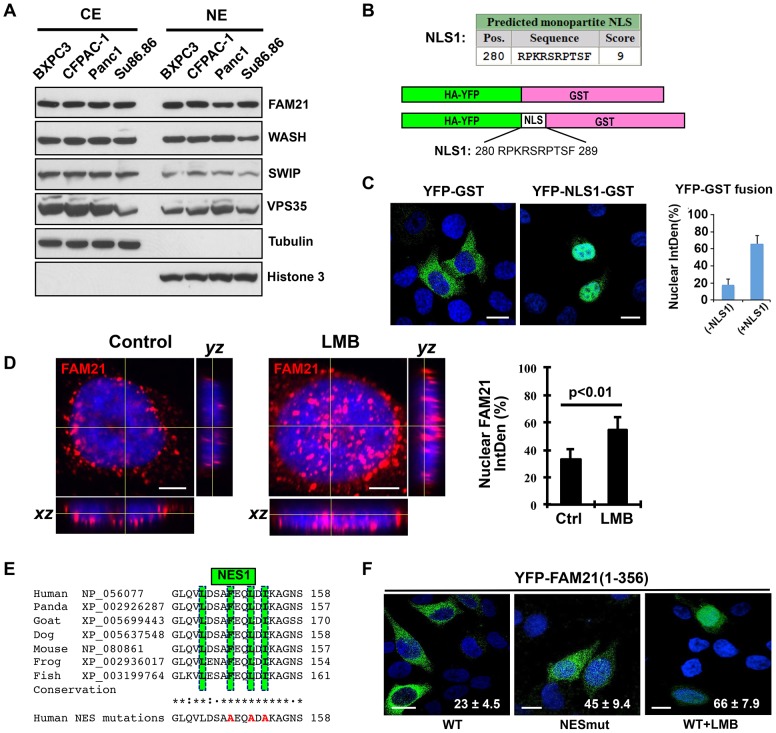
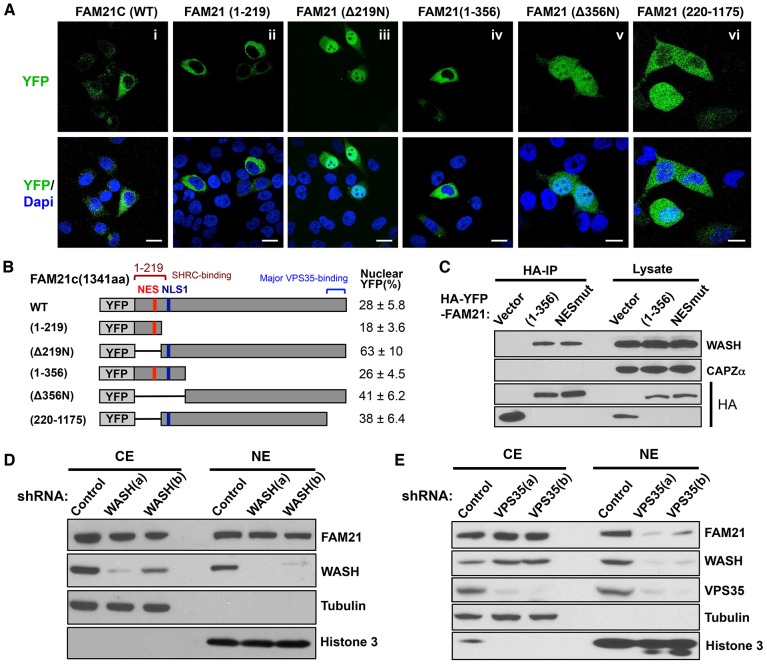
Although there is no direct experimental evidence that FAM21 is present or has a function in the nucleus, mass-spectrometry-based proteomics analysis has previously identified phosphopeptides derived from FAM21 in both the cytoplasm and nucleus (http://www.phosida.com/; Hernandez-Valladares et al., 2010). These findings, together with our above data suggest that FAM21 might modulate NF-κB activity in the nucleus. To investigate this possibility, we first examined whether FAM21 exhibited nuclear localization by preparing subcellular fractions from several pancreatic cancer lines (BXPC3, CFPAC-1, Panc1 and Su86.86). As shown in Fig. 4A, FAM21 was present in both the nuclear and cytosolic fractions, as were the FAM21-interacting proteins WASH, SWIP and VPS35. We next performed confocal imaging analysis of endogenous FAM21 in HeLa as well as in Panc1 cells. These results prompted us to inspect the FAM21 sequence for a potential nuclear localization signal (NLS) that could contribute to its nuclear translocation. Using multiple NLS prediction algorithms, a total of seven putative NLS sequences were found within the FAM21 tail. Among them, a strong monopartite NLS with a near perfect score was identified between amino acids 280 and 289 (designated as NLS1, Fig. 4B). Six additional bipartite NLS sequences were also found in the middle fragment and C-terminus of FAM21, albeit with lower scores (supplementary material Fig. S3B). To experimentally determine the function of NLS1 without the interference of passive diffusion of small proteins (<50 kDa) (Görlich and Kutay, 1999), we engineered an YFP–GST fusion protein expression vector and inserted the monopartite NLS1 (R280PKRSRPTSF289) between YFP and GST (Fig. 4B). The control YFP–GST fusion protein was not imported into the nucleus and showed a predominant cytoplasmic fluorescent pattern, whereas YFP–NLS1–GST was mainly localized to the cell nucleus, suggesting NLS1 is a bona fide NLS (Fig. 4C).
Fig. 4.
FAM21 is present in the nucleus and contains both a functional NLS and NES. (A) The cytosolic (CE) and nuclear extracts (NE) from four types of pancreatic cancer cell lines were analyzed by immunoblotting with the indicated antibodies. Tubulin and Histone 3 serve as cytosolic and nuclear markers, respectively. Note the abundant presence of FAM21 and indicated additional WASH complex members in the nucleus. (B) Putative NLS1 in FAM21 tail domain as predicted by ‘cNLS Mapper’. Note the near perfect score for FAM21 monopartite NLS1. The predicted FAM21 NLS1 motif R280PKRSRPTSF289 was inserted between YFP and GST, and HA–YFP fusion GST serves as a control. (C) The control YFP–GST or YFP–NLS1–GST fusion proteins were expressed in the HeLa cells and used for confocal microscopy. The quantitative result of YFP signal calculated as the percentage present in the nucleus relative to that in the whole cell is shown to the right. Scale bars: 20 µm. (D) HeLa cells were control- or LMB-treated and stained for immunofluorescence as indicated. Confocal z-stack images and orthogonal views were acquired (slice 10 of 21) for each treated cell. The graph depicts the quantitative FAM21 integrated density (IntDen) for 100 cells, calculated as the percentage present in the nucleus relative to that in the whole cell. Scale bars: 5 µm. Results are mean±s.d. (E) NetNES1.1 and ValidNESs programs predicted a putative NES domain within the N-terminal globular head domain of FAM21. (F) YFP–FAM21(1–356) (WT) with or without LMB or an NES mutant (E) were transfected into HeLa cells and nuclear FAM21 is shown (as a percentage of total signal; mean±s.d.). Scale bars: 20 µm.
We next investigated whether endogenous FAM21 would accumulate in the nucleus following leptomycin B (LMB) treatment, an inhibitor of CRM1/Exportin1-dependent nuclear export. HeLa cells were control-treated or treated with LMB for 20 h and then stained for FAM21 or DNA and imaged. Confocal z-stack images were generated for each treatment and slice 10 of 21 for each sample is shown (Fig. 4D). As can be seen, FAM21 accumulates in the nucleus of control cells (Fig. 4D, left image) and appears to accumulate more in the LMB-treated sample (Fig. 4D, right image). Orthogonal views for each z-stack projection are also shown, which clearly demonstrate the accumulation of FAM21 inside the control nucleus and to a greater extent within the LMB-treated nucleus. Quantification of the nuclear FAM21 integrated density (Intden) was obtained from 100 nuclei in each treatment. These data show that roughly 33% of FAM21 is in the nucleus in control cells, whereas 55% of FAM21 is accumulated in the nucleus in the cells treated with LMB (Fig. 4D). Taken together, these data suggest that FAM21 can accumulate in the nucleus and it likely occurs in a CRM1-dependent manner.
In addition to the multiple NLS(s), a putative nuclear export signal (NES) was also identified using NetNES 1.1 (http://www.cbs.dtu.dk/services/NetNES/) and ValidNESs (http://validness.ym.edu.tw/) prediction tools (designated as NES1, Fig. 4E). The FAM21 NES1 sequence (144LDSAFEQLDI153) fits the CRM1-dependent NES consensus {L-x(2,3)-[LIVFM]-x(2,3)-L-x-[LI]} (Kosugi et al., 2008) with all the key hydrophobic residues (shown in bold) fully conserved across several species (Fig. 4E). To evaluate whether this sequence, indeed, has nuclear exporting function, we generated an NES1 mutant (Fig. 4E) in the context of YFP-fused FAM21(1–356). This portion of FAM21 contains the N-terminal WASH regulatory complex (SHRC)-binding domain and NLS1 (Jia et al., 2012; Jia et al., 2010). As expected, the fluorescent signal for YFP–FAM21(1–356) is mainly localized to the cytosol, whereas the NES1 mutant nearly doubled the fluorescence intensity in the cell nucleus (Fig. 4F). Moreover, treatment with LMB led to increased levels of nuclear fluorescence (Fig. 4F). Taken together, these results demonstrate that FAM21 is present in the nucleus and contains a functional NES in addition to the NLS.
Assembly of the WASH complex and retromer both influence FAM21 nuclear localization
To delineate whether FAM21 localization is regulated by its interactions with the WASH complex or retromer, we generated a series of YFP–FAM21 truncation mutants and examined their subcellular localization by fluorescence imaging (Fig. 5A,B). Full-length FAM21 as well as its N-terminal fragments containing the SHRC-binding region (amino acids 1–219 or 1–356) were primarily cytosolic suggesting that NES1 predominates over NLS1 activity (Fig. 5Ai,ii,iv). In support of this notion, FAM21 (Δ219N) lacking the NES was mainly localized in the nucleus (Fig. 5Aiii). Additionally a FAM21 Δ356N deletion mutant devoid of SHRC-binding and NLS1 remained also largely localized to the nucleus (Fig. 5Av), suggesting that the bipartite NLS sequences might also contribute to nuclear translocation of FAM21. Of note, a variant of FAM21 with a mutation of NES1, which is located within the minimal SHRC-binding region, did not bind CAPZ as expected. (Jia et al., 2010), but still retained WASH binding (Fig. 5C). This suggests that FAM21 interaction with WASH and CRM1 are not mutually exclusive. To further investigate the potential effect of SHRC assembly on endogenous FAM21 nuclear localization, we used lentiviral shRNA suppression vectors to determine the effect of WASH knockdown on FAM21 expression and nuclear localization. We found that suppression of WASH has no significant effect on the level of nuclear FAM21 (Fig. 5D). Taken together, these data suggest that the NLS1 contributes to FAM21 nuclear translocation and is sufficient to drive nuclear import, but is not essential for nuclear translocation of endogenous FAM21, whereas the NES1 and interaction with SHRC facilitates its cytoplasmic localization.
Fig. 5.
Both the WASH complex and retromer assembly influence FAM21 nuclear localization. (A) HeLa cells were transfected with the indicated HA–YFP–FAM21 mutants for YFP fluorescence imaging. Scale bars: 20 µm. (B) Diagram depicting the FAM21 deletion mutants. The percentage of nuclear fluorescence relative to that in the whole cell for each construct is shown on the right (mean±s.d.). (C) HeLa cells were transfected with the indicated HA–YFP–FAM21 vectors for immunoprecipitation. Co-immunoprecipitated (IP) endogenous WASH and CAPZα were determined by immunoblot analysis with the indicated antibodies. (D,E) The cytosolic (CE) and nuclear extracts (NE) of Panc1 cells stably expressing the indicated shRNA were analyzed by immunoblot analyses. Note the efficient suppression of WASH (D) or VPS35 (E) each by two unique shRNA target sequences (denoted a and b). Note that VPS35, but not WASH suppression, highly reduced the levels of nuclear FAM21.
The retromer complex recruits the WASH complex to endosomes through direct binding of VPS35 with acidic LFa repeats within the FAM21 tail (Jia et al., 2012). We, as well as others, have previously shown that retromer protein VPS35 regulates FAM21 endosomal accumulation (Harbour et al., 2012; Jia et al., 2012). To determine whether retromer endosomal recruitment of FAM21 might contribute to its nuclear translocation, we generated a FAM21(220–1175) truncation mutant lacking both the C-terminal LFa repeats, which are crucial for VPS35 interaction (Jia et al., 2012), as well as the WASH/SWIP-binding region. Interestingly, this mutant also localized in the nucleus and cytoplasm, but its nuclear intensity was substantially reduced compared to that of FAM21(Δ219N) suggesting that nuclear translocation of FAM21 is at least partially dependent on VPS35 interaction (Fig. 5Avi,B). Consistent with these data, depletion of VPS35 in Panc1 cells resulted in a substantial decrease in nuclear localized FAM21 as well as WASH, whereas their cytosolic levels were unchanged (Fig. 5E). These results suggest that retromer interaction is required for FAM21 as well as WASH nuclear accumulation.
FAM21 interacts with p65 and regulates its target gene transcription
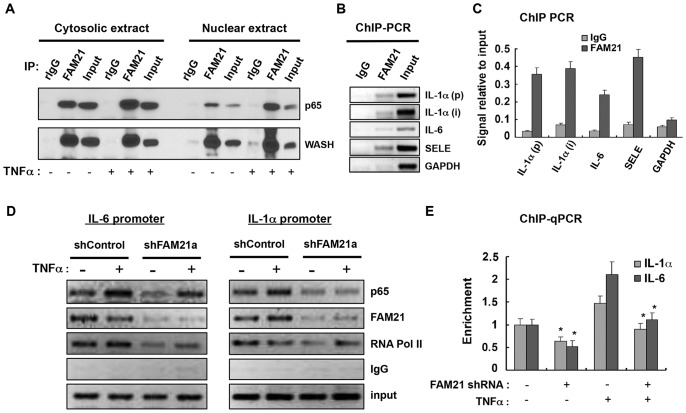
To address whether FAM21 associates with nuclear NF-κB we immunoprecipitated FAM21 from cytosolic and nuclear extracts. As shown in Fig. 6A, FAM21 interacted with p65 as well as WASH in both the cytoplasm and nucleus. Moreover, the FAM21 interaction with p65 was enhanced upon TNFα treatment. This FAM21–p65 nuclear interaction prompted us to test the possibility that FAM21 regulates p65 loading onto chromatin. Using ChIP, we found that FAM21 was recruited to NF-κB-responsive DNA regions of IL-1a, IL6 and SELE in HeLa cells (Fig. 6B,C). Interestingly, FAM21-depletion decreased the recruitment of p65 to these NF-κB-responsive chromatin regions in the presence or absence of TNFα stimulation (Fig. 6D,E). These results demonstrated that FAM21 could interact with p65 in the nucleus and affects transcriptional activity in part by impacting on p65 chromatin recruitment.
Fig. 6.
Nuclear FAM21 associates with p65 and affects its target gene transcription. (A) Immunoprecipitations (IP) were performed with cytosolic and nuclear extracts, respectively, on HeLa cells in the absence or presence of TNFα treatment for 1 h to determine the endogenous interactions of FAM21 with p65 and WASH. (B,C) ChIP was performed with anti-FAM21 serum and chromatin of nonstimulated HeLa cells to demonstrate FAM21 binding to the indicated target gene chromatin. (D,E) Control or FAM21 shRNA knockdown Panc1 cells were subjected to ChIP followed by semi-quantitative PCR (D) as well as qRT-PCR (E) analyses to determine p65 binding to IL-6 or IL-1α promoters following 1 h of TNFα treatment. The values were normalized to p65 binding over input in control cells without TNFα stimulation and expressed as mean±s.d. derived from three independent experiments (E). Note the reduced p65 binding to the target gene promoters in FAM21 suppressed cells. *P<0.05.
FAM21 depletion sensitizes pancreatic cancer cells to gemcitabine and 5-FU- induced apoptosis
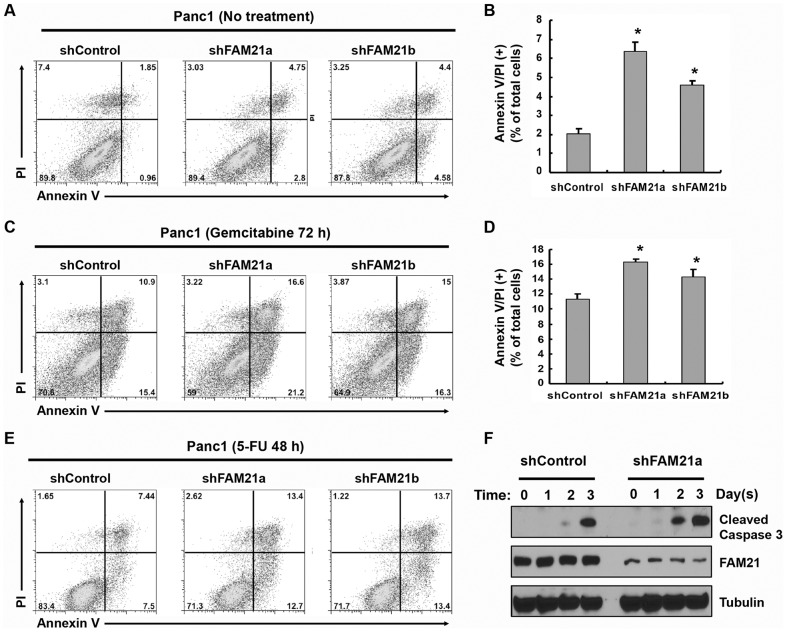
Considering that NF-κB is a key apoptotic regulator and contributes to cell survival in pancreatic and many other types of cancer cells, we examined the potential effect of FAM21 on cell survival in response to chemotherapeutic drugs known to induce apoptosis in pancreatic cancer cells. For gemcitabine, we observed an IC50 ranging between 1 to 5 µM in Panc1 cells (data not shown). Interestingly, Panc1 cells with stable suppression of FAM21 revealed a significantly increased basal level of apoptosis compared to control shRNA (shFAM21a, 6.4%±0.47; shFAM21b, 4.6%±0.20; shControl, 2.0%±0.26, mean±s.d.) (Fig. 7A,B). Likewise, FAM21 deletion also enhanced gemcitabine-induced apoptosis after 48 h (data not shown) and 72 h of treatment (Fig. 7C,D). Similar enhancement was observed in FAM21-supressed cells following 5-FU treatment (Fig. 7E; supplementary material Fig. S4A). Likewise, drug sensitization occurred in BXPC3 cells (supplementary material Fig. S4B–D). We also noted increased accumulation of cleaved caspase 3 following gemcitabine treatment, consistent with the notion that FAM21 depletion increased apoptosis (Fig. 7F). Taken together, these data suggest that FAM21 contributes to chemoresistance in pancreatic cancer cells, which is likely mediated in part by FAM21 regulation of NF-κB target genes.
Fig. 7.
FAM21 depletion sensitizes pancreatic cancer cells to anticancer drug-induced apoptosis. (A,B) Panc1 cells stably expressing control or FAM21 shRNA were simultaneous stained with Annexin V and propidium iodide (PI) and examined by flow cytometry and the results were analyzed with FlowJo software. (B) Results from three independent experiments were quantified and shown as mean±s.d. in the right panel. *P<0.05 compared to shControl. (C,D) Panc1 cells stably expressing control or FAM21 shRNA were treated with 5 µM gemcitabine for 72 h and stained with Annexin V and propidium iodide followed by flow cytometry analysis. (E) Panc1 cells stably expressing the indicated shRNA were treated with 5-FU (5 µg/ml) for 48 h and examined by flow cytometry. (F) Panc1 cells stably expressing the indicated shRNA were treated with gemcitabine (5 µM) for indicated times. Whole-cell extracts were collected and subjected to immunoblotting against cleaved caspase 3 with β-tubulin as loading control.
DISCUSSION
In this work, we have identified interactions between FAM21 and multiple components of the NF-κB signaling pathway. We further show that nuclear localized FAM21 is associated with p65 and contributes to NF-κB target gene regulation in pancreatic cancer cells. This is, to our knowledge, the first direct demonstration that FAM21 is present and functions in the cell nucleus. Our findings have therefore extended the role of FAM21 from its WASH and retromer-based regulation of endocytic cargo trafficking in the cytosol to transcriptional regulation mediated by interaction with NF-κB in the nucleus. Given that constitutive NF-κB activation is a signature occurrence in many human cancers, including pancreatic cancer, our findings suggest that FAM21 likely participates in the regulation of cancer cell survival and drug responses.
FAM21, as the largest WASH subunit, has a unique role in that it is not only essential in assembling and stabilizing the WASH complex, but also serves to bridge the WASH complex to the retromer complex through direct interaction with VPS35 (Harbour et al., 2012; Seaman et al., 2013). To date, multiple proteins have been shown to interact with FAM21 including CCDC22, CCDC93 and FKBP15, to name a few (Freeman et al., 2014; Harbour et al., 2012). Significantly, our results indicate that FAM21 can also interact with p65 and other components of the NF-κB pathway. Interestingly, CCDC22 is associated with X-linked intellectual disability (XLID) and its disease-causing mutation has previously been shown to impair NF-κB activation owing to decreased IκBα ubiquitylation and degradation. Surprisingly, even in HeLa cells treated with TNFα, we did not observe significant defects in the initial wave of IκBα phosphorylation or degradation, or p65–p50 nuclear translocation in the absence of FAM21. Nonetheless, the level of IκBα, one of the earliest responsive NF-κB target genes, decreased at later time points following stimulation, suggesting impaired NF-κB activity downstream of p65–p50 nuclear translocation. NF-κB is constitutively activated in pancreatic cancer, and its activation can lead to resistance to apoptosis through the expression of several potent anti-apoptotic genes (Carbone and Melisi, 2012; Wang et al., 1999). Our study demonstrated an interaction of FAM21 with the NF-κB p65 and p50 subunits and revealed that FAM21 depletion diminished constitutively activated NF-κB target gene expression in pancreatic cancer cells, resulting in increased spontaneous and drug-induced apoptosis. Further delineation of the mechanism by which FAM21 promotes p65 transcriptional activity could uncover new modes of regulation and new drug targets.
Consistent with a nuclear effect of FAM21 on NF-κB, we show that FAM21 exists in the nucleus of cancer cells and can be co-immunoprecipitated with p65. Furthermore, our data show that FAM21 can interact with the promoter regions of key NF-κB target genes and that loss of FAM21 results in decreased p65 chromatin loading. It is noteworthy that other actin NPFs such as JMY, WASP, neuronal WASP (N-WASP) and WAVE1 have previously been observed in the nucleus and involved in gene transcription (Coutts et al., 2009; Miyamoto et al., 2013; Sadhukhan et al., 2014; Taylor et al., 2010; Wu et al., 2006; Zuchero et al., 2009). Nuclear N-WASP can regulate RNA-polymerase-II-dependent transcription through a direct interaction with the PSF-NonO (polypyrimidine-tract-binding-protein-associated splicing factor-non-Pou-domain octamer-binding protein, also known as p54nrb) complex, and this process is associated with polymerization of actin (Wu et al., 2006). By contrast, nuclear WASP modulates the transcription of the T helper 1 (TH1) regulator gene TBX21 by association with H3K4 trimethyltransferase and H3K9/H3K36 tridemethylase, which is independent of Arp2/3-dependent actin polymerization (Sadhukhan et al., 2014; Taylor et al., 2010). A recent report has demonstrated that nuclear WAVE1 is required for transcriptional reprogramming in oocytes and embryonic development (Miyamoto et al., 2013). Thus, the WASP family proteins are emerging as crucial regulators of gene expression in the nucleus.
Vertebrate WASH has been predicted to contain an NLS and an NES (Linardopoulou et al., 2007). In fact, the nuclear localization of mouse WASH has recently been observed in hematopoietic stem cells (HSCs) (Xia et al., 2014). Importantly, nuclear WASH was shown to play an essential role in cell differentiation through transcriptional regulation of c-Myc and its target genes. Although the mechanism by which WASH accumulates in the nucleus was not examined in this study, our work suggests that FAM21 is involved. In the present study, FAM21 depletion dramatically decreased basal and TNFα-induced NF-κB-dependent transcriptional activity, which we did not observe upon WASH knockdown. It remains possible that nuclear FAM21 modulation of NF-κB activity is either independent of WASH and Arp2/3 activation or involves a different mode of regulation. In fact, whereas FAM21 depletion led to a nearly complete loss of WASH, WASH depletion did not significantly affect FAM21 protein levels, suggesting the existence of a separate pool of FAM21 that might function independently of WASH in the regulation of NF-κB (Gomez and Billadeau, 2009; Jia et al., 2010).
In an effort to understand the molecular mechanism(s) regulating FAM21 cytosolic-to-nuclear translocation, we have identified and experimentally characterized FAM21 nuclear localization signal 1 (NLS1) and nuclear export signal 1 (NES1). We could demonstrate that the FAM21 sequence (R280PKRSRPTSF289) represents a bona fide NLS capable of efficiently driving nuclear translocation of a heterologous YFP–GST fusion protein. Similarly, NES1 (L144DSAFEQLDI153) is also functional for nuclear export based on mutagenesis analysis and LMB treatment. One of our striking findings is that a FAM21 truncation mutant devoid of the FAM21 globular head (Δ219N) dramatically increased protein nuclear localization. This FAM21 head domain (amino acids 1–219) represents the minimal region responsible for WASH binding and also contains NES1 (Jia et al., 2012; Jia et al., 2010). We suggest that FAM21 assembly into the WASH complex can override NLS1 and lead to endosomal retention. This is further facilitated by its NES, which likely remains exposed in the WASH-complex, as mutations of key NES residues do not interfere with WASH binding. Interestingly, the FAM21 Δ356N deletion mutant with further deletion of NLS1 retained nuclear localization, albeit at a reduced level (41%±6.2, mean±s.d.) compared to FAM21 Δ219N (63%±10, Fig. 5B) suggesting that NLS1, as well as the remaining bipartite NLS sequences may all contribute to nuclear translocation of FAM21.
As FAM21 is largely known to reside in the cytoplasm on the surface of endosomes, NLSs are likely subjected to specific mechanisms of regulation in order to allow their exposure and subsequent nuclear translocation of FAM21. In fact, previous studies have shown that the LFa motifs within the FAM21 tail directly interact with VPS35 (Gomez and Billadeau, 2009; Harbour et al., 2010; Jia et al., 2012) promoting endosomal accumulation and the regulation of endosome to plasma membrane as well as retrograde endosome to Golgi trafficking. Importantly, our results suggest that recruitment by the retromer is required for normal FAM21 nuclear accumulation (Fig. 5Avi,E). The exact mechanism by which the retromer facilitates this nuclear shuttling remains to be determined, but endosome-to-Golgi-to-ER retrograde trafficking has been found to regulate the nuclear accumulation of EGFR (Du et al., 2014; Wang et al., 2010). In addition, whether other signal inputs at the endosome lead to exposure of the NLS1 or bipartite NLSs, and, ultimately, nuclear trafficking of FAM21 complexes remains to be determined. Overall, our results suggest that FAM21 association with WASH and retromer complexes jointly governs the subcellular localization of endogenous FAM21. Whether and how the other identified NLS motifs within the FAM21 tail also contribute to regulating its subcellular localization will require further experiments.
In summary, the finding that FAM21 is a nuclear protein and participates in regulation of NF-κB signaling extends our understanding of FAM21 from its previous WASH- and retromer-complex-dependent role in endosomal cargo trafficking. Unlike many other nuclear regulators of NF-κB, such as histone acetylases and deacetylases, FAM21 itself has no intrinsic enzymatic activity, but appears to function as a scaffolding protein to facilitate p65 chromatin binding. Given the steady presence of FAM21 in the cell nucleus, we speculate that nuclear FAM21 has additional interacting partners and might regulate targets other than NF-κB. Future work will be required to further elucidate the detailed mechanism underlying FAM21 regulation of NF-κB and its additional potential nuclear activities. Finally, because WASH is also found in the nucleus and is associated with FAM21 there, it will be important to clarify whether FAM21 indeed functions independently of WASH (or the WASH complex) or adopts a unique mode of action within the nuclear environment. Equally important will be examining the role of nuclear FAM21 and WASH in the various identified neurological pathologies and cancer where some of their effects might reside in the nucleus and not on endosomes. Therefore, nuclear FAM21 offers a new avenue of research into this highly versatile protein and its contribution to disease.
MATERIALS AND METHODS
Regents, plasmids and generation of stable cell lines
Reagents were purchased from Sigma-Aldrich (St Louis, MO) unless otherwise specified. Recombinant human TNFα was purchased from R&D Systems (Minneapolis, MN). Anti-HA affinity matrix was obtained from Roche (Indianapolis, IN). Anti-FAM21, anti-WASH, anti-VPS35 and anti-SWIP antibodies were described previously (Gomez et al., 2012). Anti-histone H3 antibody was purchased from Abcam (Cambridge, MA). Antibodies from Santa Cruz Biotechnology (Santa Cruz, CA) include rabbit polyclonal anti-p65, -p50, IκBα, IKK α/β antibodies, and mouse monoclonal anti-p50 and -p65 antibody. Fluorescein isothiocyanate (FITC)-conjugated annexin V and IKKγ antibodies were from BD Biosciences (San Diego, CA). Phosphorylated IκBα (phosphorylated at Ser32 and Ser36) was purchased from Cell Signaling Technology (Beverly, MA). FAM21 truncations used in shFAM21 and HA–YFP–FAM21 rescue vector system were described previously (Gomez and Billadeau, 2009) and included the following: 1–219; Δ219N (amino acids 220–1341); 1–356; Δ356N (amino acids 357–1341); and 219–1275. For knockdown of endogenously expressed FAM21, WASH and VPS35, we used a lentivirus-mediated short hairpin RNA (shRNA) expression system (Sigma). The plasmids were constructed in pLKO.1 vector, and the shRNA targeting sequences (listed in the 5′ to 3′ direction) were CCCCACAGCAAACCTTCTAAA (shFAM21a), GCTGTGAACTATGGCTTACAA (shFAM21b), GCGCCACTGTGTTCTTCTCTA (WASHa), CCAGAGAACTACTTCTATGTG (WASHb), AACAGAGCAGATTAACAAACA (VPS35a), and AGGGGTCTGTTTCTTCGAAAT (VPS35b). Lentiviral packaging, transduction and selection of stable cells were performed as previously described following institutional biosafety regulations (Zhang et al., 2011). All plasmids were sequence verified in the Mayo Clinic Gene Analysis Shared Resource.
Cell culture, transfection and stimulation
HeLa, Panc1, CFPAC1 and HEK293T cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. BXPC3 and SU86.86 cells were cultured in RPMI 1640 medium with 10% fetal bovine serum as described previously (Herreros-Villanueva et al., 2013). HeLa cells were transfected using electroporation as described previously (Gomez and Billadeau, 2009). For imaging analysis, cells were transfected with a reduced level of Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA). Transfected HeLa and Panc1 cells were treated with TNFα (15 ng/ml and 100 ng/ml, respectively) after overnight serum starvation.
Immunoprecipitation and protein extraction
For immunoprecipitation, cells were lysed in lysis buffer (20 mM HEPES pH 7.2, 50 mM potassium acetate, 1 mM EDTA, 200 mM D-sorbitol, 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 10 µg/ml leupeptin, 10 µg/ml aprotinin and 1 mM Na3VO4), and 500–1000 µg protein for immunoprecipitation and 50–100 µg protein for lysates were prepared and analyzed by immunoblotting. Nuclear and cytosolic fractionated protein extracts were prepared as previously described (Dignam et al., 1983).
Identification of FAM21-binding proteins from HeLa cells
A FAM21 truncated mutant in which amino acids 1–356 (the WASH-complex-binding domain) and 1029–1047 (minimal CAPZ-binding motif) were removed was constructed in pCMS3-H1P-HA.YFP vector-based suppression and re-expression system as described previously (Gomez and Billadeau, 2009). HeLa cells were transfected by electroporation. At 72 h post transfection, the cells were collected in 1.5 ml of 1× PBS and homogenized using Dounce pestle B. Homogenates were clarified by high-speed ultracentrifugation and were subjected to size-exclusion chromatography as previously described (Gomez and Billadeau, 2009). Fractions containing the HA–FAM21 truncation mutant were incubated at 4°C for 1 h with anti-HA beads. The beads were washed four times with 1 ml of lysis buffer after incubation, and the obtained material was loaded onto SDS-PAGE gels followed by silver staining. Protein bands were then excised, digested with trypsin, and identified by nano-liquid-chromatography–tandem mass spectrometry methods at the Mayo Clinic Proteomics Core Shared Resource.
Immunofluorescence and confocal microscopy
HeLa and Panc1 cells were grown on coverslips, fixed in 4% paraformaldehyde and prepared for immunofluorescence as described previously (Zhang et al., 2014). In some cases, cells were pretreated with TNFα for the indicated periods of time. Images were obtained using an LSM-710 laser scanning confocal microscope (Carl Zeiss, Jena, Germany). To quantify the percentage of fluorescence in the nucleus compared to the whole cell, we started by drawing a region of interest (ROI) around the nucleus and measured the integrated intensity of ROI. Next, we drew an ROI around the entire cell and measured total integrated density within the cell. Using the integrated intensities obtained from the whole cell and nuclear ROIs, we obtained the percentage of nuclear FAM21 or NF-κB fluorescence. Leptomycin B (LMB) was used to block CRM1-mediated nuclear export. HeLa cells were treated with 40 nM LMB for 20 h, whereas YFP–FAM21(1–356)-transfected HeLa cells were treated with 20 nM LMB for 2 h. Quantitative fluorescence expression in cells and the nucleus was analyzed with Fiji software and 20–100 transfected or treated cells were scored from multiple microscopic fields from three independent experiments.
RNA extraction and qRT-PCR
RNA was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA), and reverse transcription was processed with the Superscript III RT-PCR Kit (Invitrogen). Quantitative real-time PCR (qRT-PCR) was performed with the SYBR Green PCR Master Mix (Invitrogen) using the ABI Prism 7900TM Sequence Detection System (Applied Biosystems, Grand Island, NY). Experiments were performed in triplicate using three independent cDNAs, and the results were calculated following the 2−ΔΔCt method (Schmittgen and Livak, 2008). The qRT-PCR results were all normalized to GAPDH and experiments were performed in triplicate using three independent cDNAs and expressed as mean±s.d. Primer sequences are presented in supplementary material Table S1.
Cell proliferation and apoptosis analyses
Cell growth was measured by MTS assay and cell apoptosis analysis was performed as described previously (Zhang et al., 2011). Briefly, ∼105 Panc1 and BXPC3 stable cells were seeded in 6-well plates and incubated overnight and treated with gemcitabine (5 µM for Panc1 and 0.25 µM for BXPC3) or 5-fluorouracil (5-FU, 5 µg/ml for Panc1 and 0.25 µg/ml for BXPC3) for 48 or 72 h. The cells were harvested by trypsinization, washed twice with cold PBS, followed by staining with FITC–Annexin-V and propidium iodide. Stained cells were then analyzed on a Becton Dickinson FACSCanto II flow cytometer (Franklin Lakes, NJ) followed by calculation using FlowJo 8.8.7 software (Tree Star, Ashland, OR). The extent of caspase 3 activation was determined by immunoblotting as an additional indicator of apoptosis.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) was performed using the EZ ChIP kit (Millipore, Billerica, MA) following the manufacturer's instructions as described previously (Zhang et al., 2011). Pre-cleared chromatin was immunoprecipitated with rabbit anti-FAM21 and mouse anti-p65 antibody using normal mouse or rabbit IgG as control. ChIP DNA was used for semi-quantitative PCR or qRT-PCR analyses. The specific primer sequences used for ChIP qRT-PCR are provided in supplementary material Table S2.
Statistical analysis
Data are reported as mean±s.d., and statistical analysis was performed using unpaired two-tailed Student's t-tests. Results were considered statistically significant when the P-value was less than 0.05 (P<0.05).
Supplementary Material
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Z.-H.D. and T.S.G. designed, performed experiments and analyzed the results. D.G.O. and C.A.P.-K. helped to perform flow cytometry and confocal imaging experiments. J.-S.Z. and D.D.B. conceived, designed the experiments, and analyzed the results. Z.-H.D., J.-S.Z. and D.D.B. wrote the manuscript.
Funding
This work was supported in part by the National Institutes of Health [grant number R01AI065474 to D.D.B.]; the Mayo Clinic Pancreatic Cancer Specialized Program of Research Excellence (SPORE) [grant number P50CA102701 to D.D.B.]; an Allergic Diseases Training grant [grant number T32AI07047 to D.G.O.]; a Tumor Microenvironment Training grant [grant number T32CA148073 to. C.A.P-K.]; the Education Department of Helongjiang Province, China [grant number 12541907 to Z.H.D.]; the National Natural Science Foundation of China [grant number 81472601 to J.-S.Z.]; and an American Cancer Society Institutional New Investigator Award (to J.-S.Z.). The Gene Analysis and Proteomics Core Shared Resources were supported in part by the Mayo Clinic Comprehensive Cancer Center support grant [grant number P30CA15083]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.161513/-/DC1
References
- Bear J. E. (2009). Sorting out endosomes in the WASH. Dev. Cell 17, 583–584 10.1016/j.devcel.2009.10.019 [DOI] [PubMed] [Google Scholar]
- Carbone C., Melisi D. (2012). NF-κB as a target for pancreatic cancer therapy. Expert Opin. Ther. Targets 16 Suppl. 2, S1–S10 10.1517/14728222.2011.645806 [DOI] [PubMed] [Google Scholar]
- Carnell M., Zech T., Calaminus S. D., Ura S., Hagedorn M., Johnston S. A., May R. C., Soldati T., Machesky L. M., Insall R. H. (2011). Actin polymerization driven by WASH causes V-ATPase retrieval and vesicle neutralization before exocytosis. J. Cell Biol. 193, 831–839 10.1083/jcb.201009119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts A. S., Weston L., La Thangue N. B. (2009). A transcription co-factor integrates cell adhesion and motility with the p53 response. Proc. Natl. Acad. Sci. USA 106, 19872–19877 10.1073/pnas.0906785106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bot S. T., Vermeer S., Buijsman W., Heister A., Voorendt M., Verrips A., Scheffer H., Kremer H. P., van de Warrenburg B. P., Kamsteeg E. J. (2013). Pure adult-onset spastic paraplegia caused by a novel mutation in the KIAA0196 (SPG8) gene. J. Neurol. 260, 1765–1769 10.1007/s00415-013-6870-x [DOI] [PubMed] [Google Scholar]
- Derivery E., Gautreau A. (2010). Evolutionary conservation of the WASH complex, an actin polymerization machine involved in endosomal fission. Commun. Integr. Biol. 3, 227–230 10.4161/cib.3.3.11185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E., Sousa C., Gautier J. J., Lombard B., Loew D., Gautreau A. (2009). The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev. Cell 17, 712–723 10.1016/j.devcel.2009.09.010 [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. (1983). Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 10.1093/nar/11.5.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Shen J., Hsu J. L., Han Z., Hsu M. C., Yang C. C., Kuo H. P., Wang Y. N., Yamaguchi H., Miller S. A. <et al. (2014). Syntaxin 6-mediated Golgi translocation plays an important role in nuclear functions of EGFR through microtubule-dependent trafficking. Oncogene 33, 756–770 10.1038/onc.2013.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duleh S. N., Welch M. D. (2010). WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton (Hoboken) 67, 193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman C. L., Hesketh G., Seaman M. N. (2014). RME-8 coordinates the activity of the WASH complex with the function of the retromer SNX dimer to control endosomal tubulation. J. Cell Sci. 127, 2053–2070 10.1242/jcs.144659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez T. S., Billadeau D. D. (2009). A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell 17, 699–711 10.1016/j.devcel.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez T. S., Gorman J. A., de Narvajas A. A., Koenig A. O., Billadeau D. D. (2012). Trafficking defects in WASH-knockout fibroblasts originate from collapsed endosomal and lysosomal networks. Mol. Biol. Cell 23, 3215–3228 10.1091/mbc.E12-02-0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Kutay U. (1999). Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 607–660 10.1146/annurev.cellbio.15.1.607 [DOI] [PubMed] [Google Scholar]
- Graham D. B., Osborne D. G., Piotrowski J. T., Gomez T. S., Gmyrek G. B., Akilesh H. M., Dani A., Billadeau D. D., Swat W. (2014). Dendritic cells utilize the evolutionarily conserved WASH and retromer complexes to promote MHCII recycling and helper T cell priming. PLoS ONE 9, e98606 10.1371/journal.pone.0098606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour M. E., Breusegem S. Y., Antrobus R., Freeman C., Reid E., Seaman M. N. (2010). The cargo-selective retromer complex is a recruiting hub for protein complexes that regulate endosomal tubule dynamics. J. Cell Sci. 123, 3703–3717 10.1242/jcs.071472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour M. E., Breusegem S. Y., Seaman M. N. (2012). Recruitment of the endosomal WASH complex is mediated by the extended ‘tail’ of Fam21 binding to the retromer protein Vps35. Biochem. J. 442, 209–220 10.1042/BJ20111761 [DOI] [PubMed] [Google Scholar]
- Helfer E., Harbour M. E., Henriot V., Lakisic G., Sousa-Blin C., Volceanov L., Seaman M. N., Gautreau A. (2013). Endosomal recruitment of the WASH complex: active sequences and mutations impairing interaction with the retromer. Biol. Cell 105, 191–207 10.1111/boc.201200038 [DOI] [PubMed] [Google Scholar]
- Hernandez-Valladares M., Kim T., Kannan B., Tung A., Aguda A. H., Larsson M., Cooper J. A., Robinson R. C. (2010). Structural characterization of a capping protein interaction motif defines a family of actin filament regulators. Nat. Struct. Mol. Biol. 17, 497–503 10.1038/nsmb.1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herreros-Villanueva M., Zhang J. S., Koenig A., Abel E. V., Smyrk T. C., Bamlet W. R., de Narvajas A. A., Gomez T. S., Simeone D. M., Bujanda L. <et al. (2013). SOX2 promotes dedifferentiation and imparts stem cell-like features to pancreatic cancer cells. Oncogenesis 2, e61 10.1038/oncsis.2013.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D., Gomez T. S., Metlagel Z., Umetani J., Otwinowski Z., Rosen M. K., Billadeau D. D. (2010). WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc. Natl. Acad. Sci. USA 107, 10442–10447 10.1073/pnas.0913293107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D., Gomez T. S., Billadeau D. D., Rosen M. K. (2012). Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Mol. Biol. Cell 23, 2352–2361 10.1091/mbc.E11-12-1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. S., Gueho A., Hagedorn M., Gopaldass N., Leuba F., Soldati T., Insall R. H. (2013). WASH is required for lysosomal recycling and efficient autophagic and phagocytic digestion. Mol. Biol. Cell 24, 2714–2726 10.1091/mbc.E13-02-0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Hasebe M., Tomita M., Yanagawa H. (2008). Nuclear export signal consensus sequences defined using a localization-based yeast selection system. Traffic 9, 2053–2062 10.1111/j.1600-0854.2008.00825.x [DOI] [PubMed] [Google Scholar]
- Linardopoulou E. V., Parghi S. S., Friedman C., Osborn G. E., Parkhurst S. M., Trask B. J. (2007). Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet. 3, e237 10.1371/journal.pgen.0030237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Abreu-Blanco M. T., Barry K. C., Linardopoulou E. V., Osborn G. E., Parkhurst S. M. (2009). Wash functions downstream of Rho and links linear and branched actin nucleation factors. Development 136, 2849–2860 10.1242/dev.035246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough I. J., Steinberg F., Jia D., Barbuti P. A., McMillan K. J., Heesom K. J., Whone A. L., Caldwell M. A., Billadeau D. D., Rosen M. K. <et al. (2014). Retromer binding to FAM21 and the WASH complex is perturbed by the Parkinson disease-linked VPS35(D620N) mutation. Curr. Biol. 24, 1670–1676 10.1016/j.cub.2014.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K., Teperek M., Yusa K., Allen G. E., Bradshaw C. R., Gurdon J. B. (2013). Nuclear Wave1 is required for reprogramming transcription in oocytes and for normal development. Science 341, 1002–1005 10.1126/science.1240376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monfregola J., Napolitano G., D'Urso M., Lappalainen P., Ursini M. V. (2010). Functional characterization of Wiskott-Aldrich syndrome protein and scar homolog (WASH), a bi-modular nucleation-promoting factor able to interact with biogenesis of lysosome-related organelle subunit 2 (BLOS2) and gamma-tubulin. J. Biol. Chem. 285, 16951–16957 10.1074/jbc.M109.078501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro P., Rossé C., Castro-Castro A., Irondelle M., Lagoutte E., Paul-Gilloteaux P., Desnos C., Formstecher E., Darchen F., Perrais D. <et al. (2013). Endosomal WASH and exocyst complexes control exocytosis of MT1-MMP at invadopodia. J. Cell Biol. 203, 1063–1079 10.1083/jcb.201306162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park L., Thomason P. A., Zech T., King J. S., Veltman D. M., Carnell M., Ura S., Machesky L. M., Insall R. H. (2013). Cyclical action of the WASH complex: FAM21 and capping protein drive WASH recycling, not initial recruitment. Dev. Cell 24, 169–181 10.1016/j.devcel.2012.12.014 [DOI] [PubMed] [Google Scholar]
- Piotrowski J. T., Gomez T. S., Schoon R. A., Mangalam A. K., Billadeau D. D. (2013). WASH knockout T cells demonstrate defective receptor trafficking, proliferation, and effector function. Mol. Cell. Biol. 33, 958–973 10.1128/MCB.01288-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropers F., Derivery E., Hu H., Garshasbi M., Karbasiyan M., Herold M., Nürnberg G., Ullmann R., Gautreau A., Sperling K. <et al. (2011). Identification of a novel candidate gene for non-syndromic autosomal recessive intellectual disability: the WASH complex member SWIP. Hum. Mol. Genet. 20, 2585–2590 10.1093/hmg/ddr158 [DOI] [PubMed] [Google Scholar]
- Rottner K., Hänisch J., Campellone K. G. (2010). WASH, WHAMM and JMY: regulation of Arp2/3 complex and beyond. Trends Cell Biol. 20, 650–661 10.1016/j.tcb.2010.08.014 [DOI] [PubMed] [Google Scholar]
- Ryder P. V., Vistein R., Gokhale A., Seaman M. N., Puthenveedu M. A., Faundez V. (2013). The WASH complex, an endosomal Arp2/3 activator, interacts with the Hermansky-Pudlak syndrome complex BLOC-1 and its cargo phosphatidylinositol-4-kinase type IIα. Mol. Biol. Cell 24, 2269–2284 10.1091/mbc.E13-02-0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhukhan S., Sarkar K., Taylor M., Candotti F., Vyas Y. M. (2014). Nuclear role of WASp in gene transcription is uncoupled from its ARP2/3-dependent cytoplasmic role in actin polymerization. J. Immunol. 193, 150–160 10.4049/jimmunol.1302923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Seaman M. N., Gautreau A., Billadeau D. D. (2013). Retromer-mediated endosomal protein sorting: all WASHed up! Trends Cell Biol. 23, 522–528 10.1016/j.tcb.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S. C., Ganchi P. A., Ballard D. W., Greene W. C. (1993). NF-kappa B controls expression of inhibitor I kappa B alpha: evidence for an inducible autoregulatory pathway. Science 259, 1912–1915 10.1126/science.8096091 [DOI] [PubMed] [Google Scholar]
- Takenawa T., Suetsugu S. (2007). The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8, 37–48 10.1038/nrm2069 [DOI] [PubMed] [Google Scholar]
- Taylor M. D., Sadhukhan S., Kottangada P., Ramgopal A., Sarkar K., D'Silva S., Selvakumar A., Candotti F., Vyas Y. M. (2010). Nuclear role of WASp in the pathogenesis of dysregulated TH1 immunity in human Wiskott-Aldrich syndrome. Sci. Transl. Med. 2, 37ra44 10.1126/scitranslmed.3000813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temkin P., Lauffer B., Jäger S., Cimermancic P., Krogan N. J., von Zastrow M. (2011). SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat. Cell Biol. 13, 717–723 10.1038/ncb2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardarajan B. N., Bruesegem S. Y., Harbour M. E., Inzelberg R., Friedland R., St George-Hyslop P., Seaman M. N., Farrer L. A. (2012). Identification of Alzheimer disease-associated variants in genes that regulate retromer function. Neurobiol. Aging 33, 2231 e15–2231.e30 10.1016/j.neurobiolaging.2012.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajant H., Scheurich P. (2011). TNFR1-induced activation of the classical NF-κB pathway. FEBS J. 278, 862–876 10.1111/j.1742-4658.2011.08015.x [DOI] [PubMed] [Google Scholar]
- Wang W., Abbruzzese J. L., Evans D. B., Larry L., Cleary K. R., Chiao P. J. (1999). The nuclear factor-kappa B RelA transcription factor is constitutively activated in human pancreatic adenocarcinoma cells. Clin. Cancer Res. 5, 119–127. [PubMed] [Google Scholar]
- Wang Y. N., Wang H., Yamaguchi H., Lee H. J., Lee H. H., Hung M. C. (2010). COPI-mediated retrograde trafficking from the Golgi to the ER regulates EGFR nuclear transport. Biochem. Biophys. Res. Commun. 399, 498–504 10.1016/j.bbrc.2010.07.096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Yoo Y., Okuhama N. N., Tucker P. W., Liu G., Guan J. L. (2006). Regulation of RNA-polymerase-II-dependent transcription by N-WASP and its nuclear-binding partners. Nat. Cell Biol. 8, 756–763 10.1038/ncb1433 [DOI] [PubMed] [Google Scholar]
- Xia P., Wang S., Huang G., Zhu P., Li M., Ye B., Du Y., Fan Z. (2014). WASH is required for the differentiation commitment of hematopoietic stem cells in a c-Myc-dependent manner. J. Exp. Med. 211, 2119–2134 10.1084/jem.20140169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodszky E., Seaman M. N., Moreau K., Jimenez-Sanchez M., Breusegem S. Y., Harbour M. E., Rubinsztein D. C. (2014). Mutation in VPS35 associated with Parkinson's disease impairs WASH complex association and inhibits autophagy. Nat. Commun. 5, 3828 10.1038/ncomms4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech T., Calaminus S. D., Caswell P., Spence H. J., Carnell M., Insall R. H., Norman J., Machesky L. M. (2011). The Arp2/3 activator WASH regulates α5β1-integrin-mediated invasive migration. J. Cell Sci. 124, 3753–3759 10.1242/jcs.080986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. S., Koenig A., Young C., Billadeau D. D. (2011). GRB2 couples RhoU to epidermal growth factor receptor signaling and cell migration. Mol. Biol. Cell 22, 2119–2130 10.1091/mbc.E10-12-0969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. S., Herreros-Villanueva M., Koenig A., Deng Z., de Narvajas A. A., Gomez T. S., Meng X., Bujanda L., Ellenrieder V., Li X. K. <et al. (2014). Differential activity of GSK-3 isoforms regulates NF-κB and TRAIL- or TNFα induced apoptosis in pancreatic cancer cells. Cell Death Dis. 5, e1142 10.1038/cddis.2014.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchero J. B., Coutts A. S., Quinlan M. E., Thangue N. B., Mullins R. D. (2009). p53-cofactor JMY is a multifunctional actin nucleation factor. Nat. Cell Biol. 11, 451–459 10.1038/ncb1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.