Abstract
Bacterial second messenger cyclic di-AMP (c-di-AMP) is implicated in signaling DNA damage and cell wall stress through interactions with several protein receptors and a widespread ydaO-type riboswitch. We report the crystal structures of c-di-AMP riboswitches from Thermoanaerobacter pseudethanolicus and Thermovirga lienii determined at ~3.0-Å resolution. In both species, the RNA adopts an unforeseen ‘square’ shaped pseudosymmetrical architecture that features two three-way junctions, a turn and a pseudoknot, positioned in the square corners. Uncharacteristically for riboswitches, the structure is stapled by two ligand molecules that span the interior of the structure and employ similar noncanonical interactions for RNA recognition. Mutations in either ligand-binding site negatively affect c-di-AMP binding, suggesting that the riboswitch-triggered genetic response requires contribution of both ligands. Our data provide what are to our knowledge the first insights into specific sensing of c-di-AMP and a molecular mechanism underlying the common c-di-AMP–dependent control of essential cellular processes in bacteria.
Nucleotide-based second messengers trigger a broad range of physiological responses in all forms of life1,2. Bacteria have developed their own signaling systems based on cyclic dinucleotides c-di-GMP, cyclic 3′,5′-GMP-AMP (3′3′-cGAMP) and c-di-AMP. C-di-AMP was discovered during a structural study3 and later detected in human pathogenic bacteria4–7 and in the cytosol of host cells after invasion by Listeria monocytogenes8. C-di-AMP is synthesized in response to unidentified stimuli from two molecules of ATP (Supplementary Results, Supplementary Fig. 1a) and is sensed by specific receptors9 that affect various cellular processes.
In many bacteria, c-di-AMP signaling involves an RNA motif identified in the 5′ untranslated region of the ydaO gene as a putative riboswitch10. The riboswitch was first suggested to respond to ATP11 and most recently to c-di-AMP12. The ydaO motif is one of the most common riboswitches, with over 3,000 representatives in various bacterial species. The motif is found in the vicinity of genes involved in cell wall metabolism, synthesis and transport of osmoprotectants, sporulation and other important biological processes10,13 and therefore is anticipated to either control different processes or at least be involved in the control of different responses associated with these processes. Some of these cellular activities involve c-di-AMP signaling (reviewed in ref. 2), suggesting a major role of the riboswitch in c-di-AMP–dependent gene expression control.
Metabolite-sensing domains of riboswitches form intricate three-dimensional structures to ensure highly specific binding of cognate ligands. c-di-AMP riboswitches have been suggested to bind a single c-di-AMP molecule with subnanomolar-to-nanomolar affinity and discriminate against natural compounds with similarities to c-di-AMP moieties by at least three orders of magnitude12. The consensus secondary structure of the c-di-AMP–sensing domain of the ydaO-type riboswitches comprises two three-way junctions connected by two helices and a large conserved interior loop12. A long-range pseudoknot formed by pairing of the middle and 3′ terminal regions closes the domain. In-line probing experiments12 revealed ligand-induced changes across both highly and less conserved regions, implying ligand-dependent formation of a riboswitch structure. The combination of extensive ligand-induced modulations, multiple conserved nucleotides, poor prediction of base-pairing interactions in the center of the riboswitch and the lack of similarities to other riboswitches has made identification of ligand-binding determinants by conventional genetic and biochemical techniques nearly impossible.
To obtain insights on the specific c-di-AMP sensing and molecular mechanism underlying riboswitch-dependent gene expression control, we determined the crystal structures of the c-di-AMP–bound sensing domain of the ydaO riboswitch from two species. The structures reveal a unique RNA architecture and unforeseen binding of two ligand molecules to a single riboswitch domain. Furthermore, in vitro binding experiments suggest that both ligand molecules are required for the formation of the riboswitch structure and for riboswitch-mediated gene expression control.
RESULTS
The c-di-AMP riboswitch structure
We determined the 3.05- Å and 3.00-Å crystal structures of the c-di-AMP–bound sensing domain of typical ydaO riboswitches from thermophilic bacteria T. pseudethanolicus and T. lienii (Supplementary Table 1 and Supplementary Fig. 1b, c). Although the crystal structures generally conform to the secondary structure consensus, the riboswitches form a unique RNA architecture with structural elements not envisioned from bioinformatics predictions (Fig. 1a and Supplementary Fig. 2a).
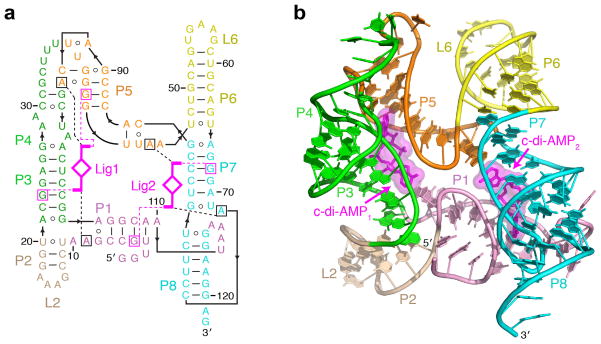
Figure 1. Overall structure and schematics of the T. pseudethanolicus c-di-AMP riboswitch.
(a) Secondary structure schematic of the riboswitch fold observed in the crystal structure of the complex. Key base-specific hydrogen bonding and stacking interactions between ligand (magenta) and RNA nucleotides (squared) are shown as dashed magenta and black lines, respectively. (b) Overall c-di-AMP riboswitch structure in a ribbon representation showing front view. Ligands are in stick and surface representations.
The c-di-AMP riboswitches adopt an architecture best described as a pseudosymmetrical square (Fig. 1a, b and Supplementary Fig. 2b, c) composed of four structural elements in the corners of the square and helical stems P1, P3, P5 and P7 connecting the corners. Pseudosymmetrical three-way junctions 3WJ1 and 3WJ2 respectively form the opposite first and third corners. The second corner constitutes a double turn between helices P4 and P5 built by base-pairing of nucleotides in the large asymmetrical loop12. A long-range pseudoknot, P7-P8, linked to P1 via a linker, J1-8, is positioned in the fourth corner. The predicted pairing, P4 (ref. 12), was not observed in the structures, and the P4 label was assigned to the new pairing within the internal loop. An additional hairpin, P4a, emerges from the P4-P5 turn in other species, such as Bacillus subtilis, and converts the turn into a three-way junction (Supplementary Figs. 1d and 2a). The T. pseudethanolicus and T. lienii riboswitch structures are in excellent agreement and exhibit only minor species-specific differences (Supplementary Fig. 3a, b).
Despite the report of a 1:1 binding stoichiometry between the riboswitch and c-di-AMP12, we observe two c-di-AMP molecules positioned along P3-P4 and P7-P8 stems in the interior of both riboswitch structures. The c-di-AMP molecules bind RNA in an extended conformation that allows both ligands to span the distance between the corners of the square, fastening two halves of the structure.
Pseudosymmetry in the riboswitch
The two halves of the c-di-AMP riboswitch display remarkable similarity if superimposed on the bound ligands (Supplementary Fig. 2d). The junctions and adjacent helices adopt almost identical conformations, and some similarity extends to parts of the turn and the pseudoknot, with the exception of regions connecting P4-P5 and P8-P1.
The two ligands are sandwiched between P4-P5 and P3 (site 1) and P1 and P7-P8 (site 2), respectively (Fig. 2a, b). The c-di-AMP1 molecule connects 3WJ1 and the P4-P5 turn, whereas the c-di-AMP2 molecule bridges 3WJ2 and the J1-8 loop. The four adenine bases of the c-di-AMP molecules are capped by adenine bases extending from the three-way junctions (A9 and A45) and either the turn (A93) or the pseudoknot loop (A72). Thus, one base of each ligand is engaged in binding to highly similar junctions, whereas the second base interacts with distinct RNA elements that nevertheless share some similarity. Notably, adenosyl moieties of each ligand molecule form virtually identical interactions with RNA (see below), and therefore c-di-AMP–riboswitch interactions exhibit double pseudosymmetry: the first between the two halves of each ligand and the second between the two ligand molecules.
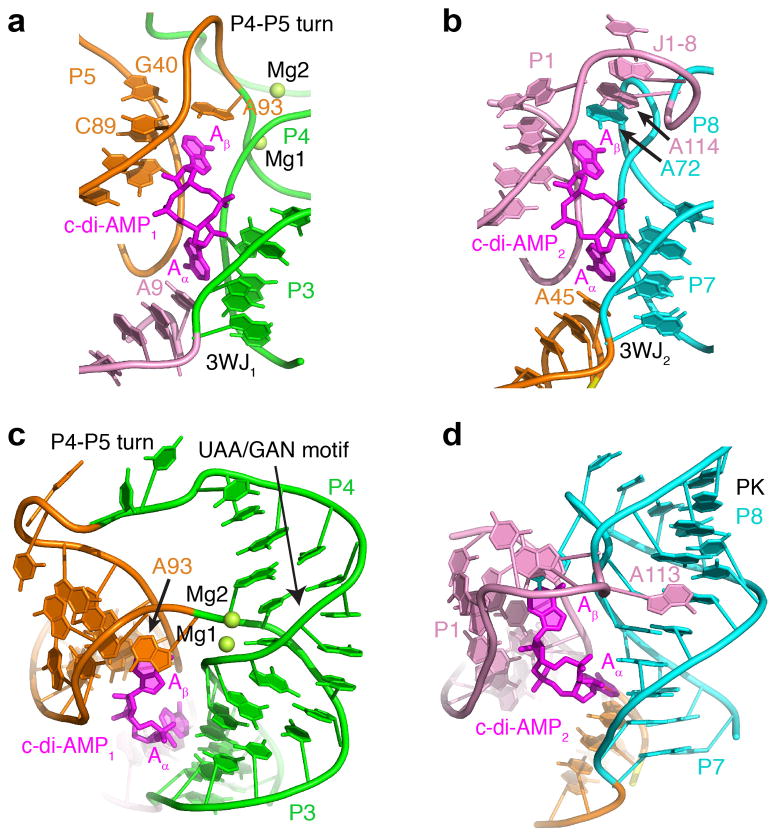
Figure 2. Structural elements of the T. pseudethanolicus c-di-AMP riboswitch.
(a, b) Structures of the ligand binding sites 1 (a) and 2 (b) shown with nucleotides that either interact with ligands directly or depend on ligand binding. (c) Structure of the double turn bound to c-di-AMP. Mg2+ cations are shown as green spheres. (d) Structure of the pseudoknot and J1-8 region bound to c-di-AMP.
Riboswitch structure is composed of recurrent motifs
The three-way junctions of the ydaO riboswitch are formed by the two stacked helices P2-P3 in 3WJ1 and P6-P7 in 3WJ2 attached to a third helix P1 or P5, respectively, at a roughly right angle (Fig. 2a, b and Supplementary Fig. 4). Helices P1 and P5 are connected to the pseudocontinuous helices by two stacked adenines packed against the minor groove of P3 and P7 via A-minor interactions14 and hydrogen bonding between riboses (Supplementary Fig. 4a, d). Orientation of the helices and stacking interactions suggest that the riboswitch junctions belong to family C junctions15. However, in contrast to the consensus family C motif characterized by a longer loop reinforced by a noncanonical base pair (Supplementary Fig. 4c, f), the riboswitch junctions contain shorter junctional loops composed of only two adenines. Such abbreviated junctions may be unstable and require external reinforcement to reduce movements of the nonstacked helices P1 and P5, as observed in a similar junction locked up by interactions with a protein (Supplementary Fig. 4b, e)16.
The double turn of the c-di-AMP riboswitch is composed of the recurrent UAA/GAN motif (Supplementary Fig. 5)17,18, which bends stem 4, and an additional sharp turn between P4 and P5 (Fig. 2c), reminiscent of kink-turn19 and Ω-turn20 motifs. The well-defined half of the P4-P5 turn is observed in other RNAs and most likely represents a previously uncharacterized recurrent turn motif (Supplementary Fig. 6). The less well-defined pyrimidine-rich half of the P4-P5 turn does not participate in base pairing and apparently does not form a stable conformation (Fig. 2c). The double turn is reinforced by a pair of Mg2+ cations, confirmed by the anomalous signal of Mn2+ cations (Supplementary Fig. 7). The Mg2+ cations bind RNA strongly and specifically and are not displaced by high concentrations of ammonium cations during crystallization. These Mg2+ cations were found in both riboswitch structures and are likely to be important for maintaining the overall RNA conformation as the riboswitch does not bind c-di-AMP in the absence of magnesium (Supplementary Fig. 9f).
The riboswitch pseudoknot contains two coaxially stacked helices, P7 and P8 (Fig. 2d), which correspond to two stems of a standard H-type pseudoknot. The helices are connected by a typical single-adenine loop, A72 (Fig. 2b), that crosses the major groove in a standard pseudoknot. However, A72 is not placed inside the groove of the stem, and instead it loops out for interactions with the Aβ base of c-di-AMP2 and stacking interactions with nucleotides of J1-8. With respect to the standard pseudoknot structure, the J1-8 region can be viewed as part of the longer loop that crosses the minor grove of the pseudoknot. Residues from this region, A113 and A114, interact with the minor groove of P7 (Fig. 2d) and stack on the base of A72 (Fig. 2b), respectively, contributing to stabilization of the pseudoknot structure.
Intermolecular stacking interactions stabilize riboswitch
The two c-di-AMP molecules are positioned between helical stems and form multiple interactions with their minor grooves. When viewed from the top, the adenine bases of ligands are turned away from each other (Fig. 2c, d) to maximize interactions with adjacent helices. Each base of the ligands stacks on the adenine base of the RNA and makes a number of tertiary interactions critical for the conformation of the riboswitch elements. The Aα bases of ligands are positioned within the clefts between junctional stems, where they stack on the adenines from the dinucleotide junctional loops (Fig. 2a, b). These stacking interactions hold A9 and A45 in place for the formation of the A-minor C22-G103•A9 and C83-G67•A45 triples, which involve interactions between sugar edges of adenines and the minor groove functionalities of the C-G base pairs (Supplementary Fig. 8a, b). The A-minor triples rigidify junctional conformations and most likely prevent movement of P1 and P5. The stacking interactions extend into helices P1 and P5 and position the helices at the correct angle in respect to the pseudocontinuous junctional helices.
The Aβ bases of the ligands stack on the adenine bases from different RNA elements. The Aβ base of c-di-AMP1 stacks on A93 from the P4-P5 turn (Fig. 2c) and stabilizes the A-amino–kissing21 G40-C89•A93 triple, characterized by interactions of the Watson-Crick edge of A93 with the minor groove of a C-G base pair (Fig. 3a). This triple is formed by residues from the two halves of the P4-P5 turn and is most likely critical for stabilizing the sharply kinked conformation of the turn. The Aβ base of c-di-AMP2 stacks on A72 from the pseudoknot loop, which in turn stacks on residues of the J1-8 linker (Fig. 2b). Notably, A72 participates in the formation of a triple base platform but, in contrast to the other three triple alignments, does not interact with the corresponding base pair, U4-A109. Therefore, stacking of the Aβ base with A72 is most likely important for clamping the two loops of the pseudoknot but is less critical for stabilizing pseudoknot conformation. In line with this observation, in the T. lienii riboswitch, the J1-8 connector is shorter so that U114, not A72, stacks on Aβ of c-di-AMP2 (Supplementary Figs. 3b and 8f). Low nucleotide conservation further emphasizes variability in the J1-8 region (Supplementary Fig. 2a). Despite some differences, stacking of ligand bases with RNA serves the same goal: formation of adjacent triple base platforms that help stabilize RNA elements. Notably, stacking interactions between ligand and RNA bases are not perfect. The planes of ligand and RNA bases are not parallel and approach each other at a ~10–30° angle (Fig. 2b, c). Moreover, the planes of almost all of the ligand bases are rotated about 90° with respect to the RNA bases so that the N6 groups of the RNA bases are located over the six-membered ring of the ligand bases (Fig. 3a, b and Supplementary Fig. 8a, b). Thus, stacking interactions between bound ligands and RNA are probably not sufficiently strong to maintain the overall RNA conformation without contribution of additional interactions.
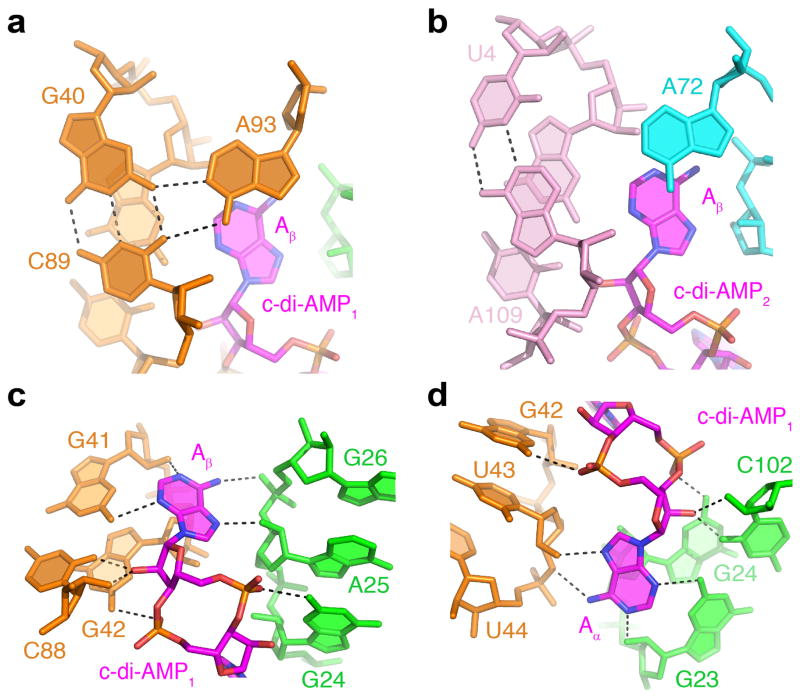
Figure 3. Recognition of c-di-AMP molecules by the T. pseudethanolicus riboswitch.
Putative hydrogen bonds are depicted by black dashed lines. (a, b) Triple alignments stabilized by stacking with the Aβ bases of c-di-AMP1 (a) and c-di-AMP2 (b). (c, d) Interactions of RNA with Aβ (c) and Aα (d) adenosyl moieties of c-di-AMP1.
Intermolecular hydrogen bonding reinforce the structure
Although the described interactions between base planes of ligands and RNA most likely contribute to stability of RNA elements, the majority of binding affinity and specificity is provided by hydrogen bonding of the ligands to the riboswitch helices. All four adenosine moieties of the ligands are packed against minor grooves of the adjacent stems and form four similar sets of hydrogen bond interactions. These contacts involve type I A-minor interactions14 of the ligand base and ribose with G-C base pairs and hydrogen bonding between the Hoogsteen edge of the ligand bases and the sugar-phosphate backbone of RNA (Fig. 3c, d and Supplementary Fig. 8c, d). Nonbridging oxygen atoms in phosphate moieties of ligands, except the Aβ adenosyl moiety of c-di-AMP2, form hydrogen bonds with N2 of guanosines from the respective helices. Most nucleotides that use bases for ligand recognition are highly conserved (Supplementary Fig. 8e). Tight packing of ligands between RNA helices and multiple base-specific interactions ensure specific recognition of c-di-AMP and discrimination against related molecules in cells12.
Specificity and stoichiometry of ligand binding
We measured binding affinity of the ydaO riboswitch to nucleotides and cyclic dinucleotides by isothermal titration calorimetry (ITC). The T. pseudethanolicus riboswitch titration did not yield sufficient heat for accurate determination of KD. The T. lienii riboswitch showed a stronger heat signal at 20 mM Mg2+ and approximately 2:1 stoichiometry (N = 1.74 ± 0.02, mean ± s.d.) of ligand-to-RNA binding with a macroscopic KD = 300 ± 166 nM (Supplementary Fig. 9a) when data were fitted to the binding model with equivalent ligand-binding sites. To determine binding stoichiometry in mesophilic bacteria and compare ITC data with published in-line probing results11,12, we conducted measurements with the B. subtilis ydaO riboswitch. We determined 2:1 stoichiometry (N = 1.93 ± 0.15 and N = 1.97 ± 0.12 at 20 mM and 5 mM Mg2+, respectively) of c-di-AMP binding to the wild-type B. subtilis riboswitch and KD = 41.7 ± 16.7 nM in 20 mM Mg2+ and KD = 7.4 ± 2.2 nM in 5 mM Mg2+ (Fig. 4a and Supplementary Fig. 9b). These binding affinities are lower than the previously measured KD ≤ 700 pM by in-line probing12 at 10 mM Mg2+ but are in range of KD values for ydaO-type riboswitches from other bacteria12. We could not detect binding of ATP and AMP to the B. subtilis riboswitch, suggesting that, despite observed weak binding by other techniques11, ATP is not the primary riboswitch ligand (Supplementary Fig. 9g, h). We also did not observe binding to cyclic dinucleotides 3′3′-cGAMP and c-di-GMP (Supplementary Fig. 9i, j).
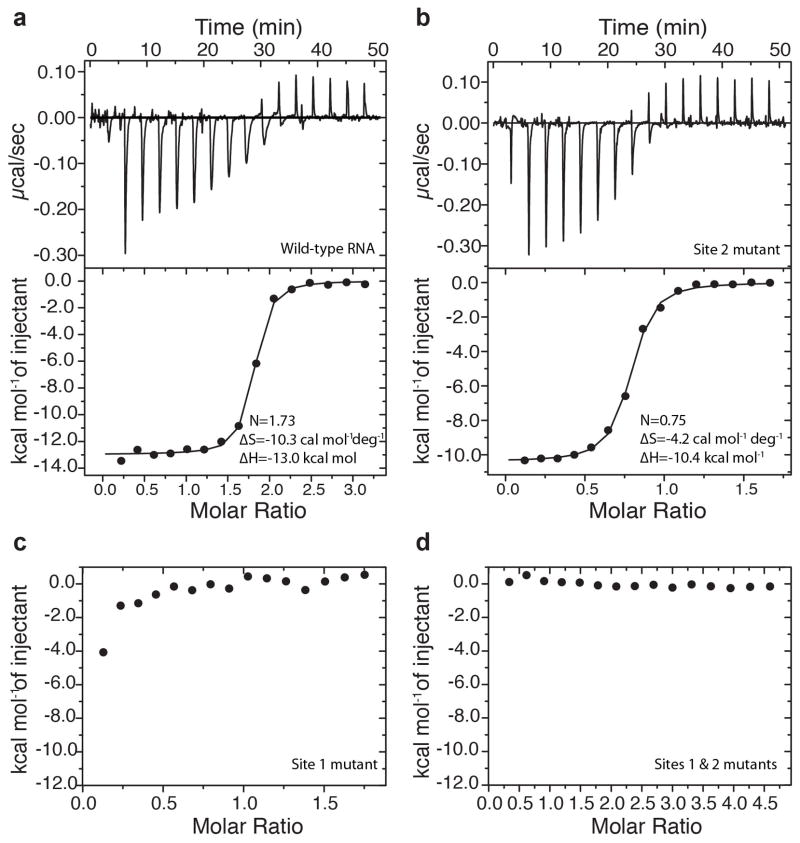
Figure 4. Binding of c-di-AMP molecules to the riboswitch.
(a–d) Representative ITC titrations with integrated fitted heat plots of c-di-AMP binding to the B. subtilis c-di-AMP riboswitch12 at 20 mM Mg2+ determined for wild-type RNA (a), site 2 mutant (b), site 1 mutant (c) and RNA with mutations in both sites (d). KD values are 56.2 nM and 195 nM for the wild type and the site 2 mutant, respectively.
Contribution of c-di-AMP to structure formation
To understand the contribution of each ligand molecule to riboswitch structure formation and genetic control, we prepared three mutants in the context of the B. subtilis ydaO riboswitch. This riboswitch contains all of the important elements observed in the structures of the riboswitches from thermophiles and provides a more robust heat signal than the thermophilic counterparts used in crystallization. In two mutants, we disrupted c-di-AMP binding to either site 1 or 2 by converting G-C base pairs in the two A-minor motifs to C-G base pairs (Supplementary Fig. 1d). These mutations do not disrupt helices and should not affect the structure. The third mutant combined mutations in both sites.
Elimination of site 2 with the mutations G5C, C108G, G68C and C82G (T. pseudethanolicus riboswitch numbering) decreases ligand binding approximately fivefold (KD = 195 ± 22 nM), showing approximately 1:1 stoichiometry (N = 0.73 ± 0.01) at 20 mM Mg2+ (Fig. 4b). At the physiological 5 mM Mg2+ concentration, binding is more affected because, despite a moderate, approximately fivefold reduction in affinity (KD = 37.0 ± 20.8 nM), only ~20% of RNA (N = 0.21 ± 0.02) is capable of ligand binding (Supplementary Fig. 9c). Such a low fraction of ligand-bound RNA most likely reflects difficulties in adopting ligand-competent conformations for the majority of the RNA population. These data suggest that disruption of ligand binding in site 2 negatively affects ligand binding to the riboswitch but does not abolish binding to site 1. Disruption of site 1 by the mutations G23C, C102G, G41C and C88G greatly decreased c-di-AMP binding such that only the first point of the titration showed heat signal during ITC experiments (Fig. 4c and Supplementary Fig. 9d). Simultaneous mutations in both binding sites abolished ligand binding entirely (Fig. 4d and Supplementary Fig. 9e). Thus, c-di-AMP binding to the riboswitch requires unequal contribution of the evolutionarily conserved site 1 and less conserved site 2 (Supplementary Fig. 3c).
DISCUSSION
Riboswitches typically control expression of genes associated with metabolism and transport of cognate ligands22. Riboswitches that sense the second messengers c-di-GMP23,24 and c-di-AMP12 modulate expression of a broad range of genes and are of special interest to biologists. Two structures determined in the course of our study revealed that c-di-AMP riboswitches adopt architectures not only dissimilar to those of c-di-GMP riboswitches but also markedly different from those of other riboswitches and RNAs. Although two types of c-di-GMP riboswitches adopt junctional folds that accommodate a single compact c-di-GMP molecule in the junctional core25–27, the c-di-AMP riboswitch forms an unprecedented pseudosymmetrical architecture involving four RNA elements connected into a ‘squared’ scaffold. Moreover, two ligand molecules symmetrically interact with two modules of a single RNA domain, a feature not yet observed in RNA structures. Unlike proteins, RNA molecules are rarely composed of similar modules or display internal symmetry, with the pseudosymmetrical FMN riboswitch being the most notable exception28. In contrast to the single-liganded FMN riboswitch, the ydaO riboswitch presents a unique example of double pseudosymmetry in the RNA structure and in the ligand-binding pockets for four adenosyl moieties of two ligands. Binding of more than one ligand to a single RNA domain is not characteristic for riboswitches and was observed only in magnesium29 and tetrahydrofolate30 riboswitches. Tandem glycine31–33 riboswitches also bind two ligands but use two similar adjoined RNA domains for interactions with each ligand.
Although a number of riboswitches contain recurrent motifs34, the ydaO riboswitch is almost entirely composed of such motifs, which are either well known or awaiting detailed characterization. These motifs seem to be intrinsically unstable and therefore require binding of ligand molecules for stabilization. For instance, the three-way junctions with abbreviated loops from other RNAs are reinforced externally by binding to proteins15,16. In the ydaO riboswitch, similar junctions are most likely stabilized by insertions of c-di-AMP bases. Stacking interactions of c-di-AMP with A93 should also strengthen the P4-P5 turn, which contains a flexible pyrimidine-rich region. Lastly, the ligand base contributes to locking two loops of the P7-P8 pseudoknot, thus improving stability of the pseudoknot. This feature can be paralleled with pseudoknots from the fluoride35, preQ1-I21,36,37 and preQ1-II38 riboswitches, whose conformations are reinforced by interactions of long pseudoknot loops with ligands and helical stems. Currently, the lack of a ligand-unbound riboswitch structure and data on stability of structural elements preclude our understanding of the exact contribution of c-di-AMP binding to formation and stabilization of the riboswitch structure. However, projection of the in-line probing data for B. subtilis riboswitch on the three-dimensional structure of the T. pseudethanolicus riboswitch reveals that RNA sites with decreased cleavage upon c-di-AMP binding are present in all structural motifs of the riboswitch (Supplementary Fig. 3d), supporting our suggestion that c-di-AMP facilitates formation or at least stabilizes all of the key elements of the riboswitch structure.
Ligand molecules in an extended conformation glue together the helical stems by similar binding to the minor grooves of the adjacent helices. Such binding sharply contrasts with two known structures of c-di-GMP riboswitches, in which the ligand adopts a compact conformation stabilized by intercalation of an adenine between guanine bases of the ligand and nonequivalent hydrogen bonds between these bases and junctional residues25–27. A ‘sandwich’ between two helical stems and an extended ligand has not been observed in other riboswitch structures and can be best compared to the S-adenosyl methionine (SAM)-I riboswitch39, where SAM in a compact conformation is positioned between two helices, and the thiamin pyrophosphate (TPP) riboswitch40–42, where the ligand in an extended conformation binds two helices in perpendicular orientation.
As in other riboswitches, practically all of the heteroatoms of the c-di-AMP bases participate in specific hydrogen bonding. Somewhat unexpectedly, but in line with some other riboswitches, these interactions do not involve Watson-Crick pairing of ligand and RNA bases43. Instead, c-di-AMP molecules make use of A-minor interactions for anchoring adenosyl moieties to the helical stems. The A-minor interactions typically participate in packing single-stranded regions with helices14,44, and the c-di-AMP riboswitch structure expands the repertoire of these interactions to ligand-RNA binding.
Our ITC binding assay confirms the 2:1 stoichiometry of the c-di-AMP–riboswitch complex observed in the crystal structures. Such binding stoichiometry could have been envisioned from the steep binding curves in in-line probing experiments, which were nevertheless treated as 1:1 binding12. The binding affinities determined by ITC are lower but still comparable with reported KD values12. The KD values, however are much smaller than experimentally determined intracellular c-di-AMP concentrations of 1.7 μM and 2.1 μM in cytoplasmic extracts of B. subtilis and Staphylococcus aureus, respectively5,7. These concentrations may increase up to 15-fold above wild-type levels, suggesting that at least some c-di-AMP riboswitches do not equilibrate with their cognate ligands and are most likely driven kinetically by concentrations of c-di-AMP higher than their KD value45.
Pseudosymmetry of c-di-AMP binding has been observed in the structure of a c-di-AMP–synthesizing protein, diadenylate cyclase DisA3. The protein functions as an octamer, and, unlike the riboswitch, two protein molecules bind a single c-di-AMP located at the interface of the protein dimer (Supplementary Fig. 10a). As DisA evolved for binding ATP substrates and releasing c-di-AMP product, c-di-AMP recognition by the protein and riboswitch differs markedly. The protein forms base-specific hydrogen bonds to two groups on the Watson-Crick edge of adenine, whereas the riboswitch recognizes all edges of c-di-AMP (Supplementary Fig. 10b, c). In addition, the conformation to which c-di-AMP binds the riboswitch has a more extended conformation than the conformation with which it binds the protein (Supplementary Fig. 10d). At the moment, the ydaO riboswitch structure is the only available structure of a specific c-di-AMP receptor, and more relevant comparison with protein receptors of c-di-AMP cannot be done.
Our structural and biochemical data demonstrate that the c-di-AMP riboswitch structure is tuned for exquisite binding to c-di-AMP molecules. The riboswitch effectively discriminates against guanine-containing c-di-GMP and 3′3′-cGAMP because guanine bases would clash with G-C base pairs and distort A-minor triples28. In contrast, deoxyriboses of c-di-dAMP can be easily accommodated but cannot form several hydrogen bonds with RNA, resulting in lower binding affinity12. Smaller ligands and linear derivatives of c-di-AMP, such as AMP, ATP and pApA, can make many observed interactions but cannot rigidify the structure of the ligand-binding module. Thus, as in some other riboswitches28,40–42,46,47, several different principles account for discrimination against related molecules.
The mutational study suggests that formation of ligand binding site 1 is a prerequisite for formation of site 2. C-di-AMP binding to RNA is reduced substantially if site 1 is incapable of ligand binding, in agreement with detrimental effects on ligand binding of mutations in the UAA/GAN motif and P4-P5 turn12. These data suggest that the ligand-dependent stabilization of the ligand-binding module 1, composed of 3WJ1 and the P4-P5 turn, is essential for the formation of the pseudoknot-containing module 2. Inactivation of site 2 or even disruption of the pseudoknot12 decreases but does not abolish ligand binding to site 1. Thus, our data suggest a model in which c-di-AMP initially binds site 1 and organizes 3WJ1 and the double turn. Then, the second c-di-AMP molecule binds site 2 and stabilizes 3WJ2 and the long-range pseudoknot. This sequence of binding events is indirectly supported by higher conservation of nucleotides around site 1, especially in the P4-P5 turn, than in site 2. Formation of a stable pseudoknot critically affects genetic control as the pseudoknot may sequester a Shine-Dalgarno sequence and prevent translation initiation, as we suggest for the T. pseudethanolicus riboswitch (Supplementary Fig. 1b), or sequester an antiterminator region in transcriptionally controlled riboswitches. The precise mechanism of transcription termination by c-di-AMP riboswitches in B. subtilis, however, remains to be investigated. Our model suggests the contribution of two ligand molecules to the mechanism of riboswitch-mediated gene control and hints at cooperativity between two c-di-AMP–binding sites that will be addressed in future studies.
The crystal structures of the widespread c-di-AMP riboswitches revealed several unusual and unforeseen features used by RNA for structure formation and small ligand sensing. These new principles of ligand recognition encourage further exploration of c-di-AMP riboswitches and their role in c-di-AMP signaling in cells. As c-di-AMP riboswitches seem to be involved in triggering critical processes in pathogenic bacteria, such as transition of Mycobacterium tuberculosis from stasis to growth12, our data may provide additional targets for pharmacological explorations.
ONLINE METHODS
RNA and complex preparation
The 122-nucleotide sensing domain of the T. pseudethanolicus ATCC 33223 c-di-AMP riboswitch and the 124-nucleotide domain from the T. lienii DSM 17291 c-di-AMP riboswitch followed by a hepatitis delta virus (HDV) ribozyme were prepared by PCR amplification from the DNA template composed of chemically synthesized oligonucleotides. The PCR fragments were digested by HindIII and cloned into the pUT7 vector prepared with HindIII and StuI48. The 138-nucleotide B. subtilis riboswitch fragment comprising nucleotides 8–141 (ref. 5) with two additional guanines at the 5′ end and UC at the 3′ end were cloned into the pUT7 vector upstream of the hammerhead ribozyme. The RNA constructs do not carry any internal mutations in addition to changes in the termini (Supplementary Fig. 1b–d). The plasmids were amplified in the Escherichia coli strain DH5α and purified using PureLink HiPure Plasmid Gigaprep Kit (Life Technologies). The plasmids were linearized by HindIII at 37 °C overnight and purified by phenol-chloroform extraction. The plasmids were then precipitated by ethanol and dissolved in water. The plasmid DNAs were used as templates for preparative in vitro T7 RNA polymerase transcription. Transcription reactions were conducted in a 50-ml mixture of 0.1 M Tris-HCl buffer, pH 7.9, 30 mM DTT, 20 mM MgCl2, 30 mM DTT, 2 mM spermidine, 4 mM of each ribonucleotide triphosphates, 75 μg/ml DNA template and 0.06 mg/ml RNA polymerase. Transcription reactions were typically incubated for 4 h at 37 °C, and, after adjusting MgCl2 to 30–50 mM, incubation continued for 45 min to complete the ribozyme cleavage. Transcription mixtures were frozen, thawed and centrifuged at 15,000g for 30 min to remove precipitate. RNA was then purified by denaturing PAGE followed by electroelution and ethanol precipitation. To remove remaining contaminants, RNA was further purified by anion-exchange chromatography on a DEAE column (GE Healthcare)49 and precipitated by ethanol.
C-di-AMP was dissolved in water at a concentration of 40 mM and stored in aliquots at −20 °C. Riboswitch complexes were prepared by mixing 0.5 mM RNA and 1.0 mM c-di-AMP in a buffer containing 20 mM Tris-HCl, pH 7.0, 50 mM KCl and 5 mM MgCl2. The complex was heated at 90 °C for 1 min, chilled on ice and incubated at 37 °C for 15 min before crystallization.
Crystallization and structure determination
Riboswitch crystals were grown by hanging-drop vapor diffusion after mixing 1 μl complex with 1 μl reservoir solution at 20 °C. The reservoir solution for the T. pseudethanolicus contained 1.5 M (NH4)2SO4, 0.1M HEPES, pH 7.3, and 0.2 M Li2SO4. The solution for the T. lienii riboswitch contained 0.05 M sodium cacodylate, pH 6.0, 0.5 mM spermine, 20 mM magnesium acetate, 100 mM NaCl and 25% 2-methyl-2,4-pentanediol (v/v). For heavy atom soaking, crystals of the T. pseudethanolicus riboswitch were gradually transferred into stabilizing solution containing 0.1 M HEPES, pH 7.3, 20% PEG 3350, 0.2 M (NH4)2SO4 and 0.2 M Li2SO4 supplemented with 2 mM [Ir(NH3)6]Cl3 and soaked for 2 h. For Mn2+ soaking, [Ir(NH3)6]Cl3 was replaced by MnCl2. For data collection, crystals were passed through a cryoprotectant solution composed of stabilizing solution and 20% (v/v) glycerol and flash-frozen in liquid nitrogen. X-ray diffraction data for the T. pseudethanolicus riboswitch were collected at 100 K (Supplementary Table 1) at X25 beamline at the Brookhaven National Laboratory at wavelength 1.105 Å, and for the T. lienii riboswitch, data were collected at the 24-ID-E beamline at the Argonne National Laboratory at wavelength 0.9792 Å. The data were reduced using HKL2000 (HKL Research). The structure was phased by single anomalous dispersion (SAD) using PHENIX50 AutoSol routine (Supplementary Fig. 11). The RNA was partially traced using PHENIX Autobuild routine. Tracing was completed manually in Coot51. Electron density maps for residues 35–36 and 54–56 were partial but allowed structure modeling. The structure of the T. lienii riboswitch was determined by molecular replacement using PHENIX and the T. pseudethanolicus riboswitch-based search model with omitted loops. Residues 1–2 and 38–42 did not have good density maps and were not added to the final model. The structures were refined with PHENIX. C-di-AMP and metal cations were added at the later stages of refinement. Figures were prepared with PyMol (http://pymol.org/).
Preparation of mutant RNAs
Mutagenesis was conducted by inverse PCR using oligonucleotides with desired mutations and original DNA plasmids as templates. The PCR mixtures were treated by DpnI to remove DNA template, ligated and transformed into the E. coli DH5α. Mutant plasmid DNAs were verified by sequencing. Clones with correct plasmid DNAs were grown in a large volume, and plasmid DNAs were extracted by the Gigaprep Kit. Mutant RNAs were prepared by in vitro transcription and purified by the same procedure as wild-type RNAs except that ion-exchange chromatography was omitted for some samples. We verified that omission of this purification step did not affect binding affinity.
ITC measurements
Experiments were performed 2–5 times with independently prepared riboswitch samples using the Microcal calorimeter ITC200 at 25 °C. Prior to titration, 0.01- to 0.06-mM RNA samples were dialyzed overnight at 4 °C against experimental buffer containing 50 mM HEPES-KOH, pH 6.8, 100 mM KCl and 5 mM or 20 mM MgCl2. The RNA samples were heated at 94 °C for 1 min, chilled on ice, incubated at 37 °C for 15 min, centrifuged at 12,000g and at 25 °C for 5 min and incubated at 25 °C for 5 min. In the conditions without MgCl2, dialysis was performed in the buffer supplemented with 0.5 mM EDTA. For measurements, the ligands, dissolved in the dialysis buffer at concentrations ~2.5- to 40-fold higher than the RNA concentration, were typically titrated into the RNA solution in the sample cell (V = 207 μl) by 16–20 serial injections of ~2.4 μl each, with 180-s intervals between injections and a reference power of 8 μcal s−1. Thermograms were corrected by subtracting ligand-induced heat changes resulting from dilution of the ligand. The data were integrated and analyzed by Origin 7.0 software (Microcal, Inc.) assuming a simple binding model with equivalent binding sites (‘one set of sites’ model). The determined macroscopic KD values report general binding of ligands with RNA and do not report microscopic KD values for each binding site, which cannot be reliably determined from our data. Measured values from 2–5 independent experiments were averaged in Origin and are reported as average ± s.d.
Supplementary Material
Acknowledgments
We thank the personnel of beamlines X25 at the Brookhaven National Laboratory and 24-ID at the Argonne National Laboratory funded by the US Department of Energy. We thank O. Ouerfelli for the synthesis of iridium hexamine and L. Jaeger for discussions. A.S. was supported by funds from the New York University School of Medicine.
Footnotes
Author contributions
A.G. crystallized the riboswitches, determined their structures and conducted ITC experiments. A.G. and A.S. refined the structures and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Supplementary information is available in the online version of the paper.
Accession codes: Protein Data Bank: Coordinates for the c-di-AMP riboswitch structures from T. pseudethanolicus and T. lienii have been deposited under accession codes 4QK8 (T. pseudethanolicus, native), 4QKA (T. pseudethanolicus, iridium hexamine soak) and 4QK9 (T. lienii, native).
References
- 1.Pesavento C, Hengge R. Bacterial nucleotide-based second messengers. Curr Opin Microbiol. 2009;12:170–176. doi: 10.1016/j.mib.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Corrigan RM, Grundling A. Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol. 2013;11:513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- 3.Witte G, Hartung S, Buttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30:167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 4.Kamegaya T, Kuroda K, Hayakawa Y. Identification of a Streptococcus pyogenes SF370 gene involved in production of c-di-AMP. Nagoya J Med Sci. 2011;73:49–57. [PMC free article] [PubMed] [Google Scholar]
- 5.Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 2011;12:594–601. doi: 10.1038/embor.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker JR, et al. STING-dependent recognition of cyclic di-AMP mediates type I interferon responses during Chlamydia trachomatis infection. MBio. 2013;4:e00018–13. doi: 10.1128/mBio.00018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Grundling A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corrigan RM, et al. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci USA. 2013;110:9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrick JE, et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson PY, Fedor MJ. The ydaO motif is an ATP-sensing riboswitch in Bacillus subtilis. Nat Chem Biol. 2012;8:963–965. doi: 10.1038/nchembio.1095. [DOI] [PubMed] [Google Scholar]
- 12.Nelson JW, et al. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol. 2013;9:834–839. doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Block KF, Hammond MC, Breaker RR. Evidence for widespread gene control function by the ydaO riboswitch candidate. J Bacteriol. 2010;192:3983–3989. doi: 10.1128/JB.00450-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hainzl T, Huang S, Sauer-Eriksson AE. Structure of the SRP19 RNA complex and implications for signal recognition particle assembly. Nature. 2002;417:767–771. doi: 10.1038/nature00768. [DOI] [PubMed] [Google Scholar]
- 17.Lee JC, Gutell RR, Russell R. The UAA/GAN internal loop motif: a new RNA structural element that forms a cross-strand AAA stack and long-range tertiary interactions. J Mol Biol. 2006;360:978–988. doi: 10.1016/j.jmb.2006.05.066. [DOI] [PubMed] [Google Scholar]
- 18.Jaeger L, Verzemnieks EJ, Geary C. The UA_handle: a versatile submotif in stable RNA architectures. Nucleic Acids Res. 2009;37:215–230. doi: 10.1093/nar/gkn911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wadley LM, Pyle AM. The identification of novel RNA structural motifs using COMPADRES: an automated approach to structural discovery. Nucleic Acids Res. 2004;32:6650–6659. doi: 10.1093/nar/gkh1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang M, Peterson R, Feigon J. Structural insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Mol Cell. 2009;33:784–790. doi: 10.1016/j.molcel.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 22.Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sudarsan N, et al. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science. 2010;329:845–848. doi: 10.1126/science.1190713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kulshina N, Baird NJ, Ferre-D’Amare AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat Struct Mol Biol. 2009;16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith KD, et al. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith KD, Shanahan CA, Moore EL, Simon AC, Strobel SA. Structural basis of differential ligand recognition by two classes of bis-(3′-5′)-cyclic dimeric guanosine monophosphate-binding riboswitches. Proc Natl Acad Sci USA. 2011;108:7757–7762. doi: 10.1073/pnas.1018857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature. 2009;458:233–237. doi: 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dann CE, III, et al. Structure and mechanism of a metal-sensing regulatory RNA. Cell. 2007;130:878–892. doi: 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 30.Trausch JJ, Ceres P, Reyes FE, Batey RT. The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer. Structure. 2011;19:1413–1423. doi: 10.1016/j.str.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandal M, et al. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 32.Huang L, Serganov A, Patel DJ. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol Cell. 2010;40:774–786. doi: 10.1016/j.molcel.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butler EB, Xiong Y, Wang J, Strobel SA. Structural basis of cooperative ligand binding by the glycine riboswitch. Chem Biol. 2011;18:293–298. doi: 10.1016/j.chembiol.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serganov A, Patel DJ. Metabolite recognition principles and molecular mechanisms underlying riboswitch function. Annu Rev Biophys. 2012;41:343–370. doi: 10.1146/annurev-biophys-101211-113224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren A, Rajashankar KR, Patel DJ. Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch. Nature. 2012;486:85–89. doi: 10.1038/nature11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein DJ, Edwards TE, Ferre-D’Amare AR. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat Struct Mol Biol. 2009;16:343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roth A, et al. A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat Struct Mol Biol. 2007;14:308–317. doi: 10.1038/nsmb1224. [DOI] [PubMed] [Google Scholar]
- 38.Liberman JA, Salim M, Krucinska J, Wedekind JE. Structure of a class II preQ1 riboswitch reveals ligand recognition by a new fold. Nat Chem Biol. 2013;9:353–355. doi: 10.1038/nchembio.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 40.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 42.Edwards TE, Ferre-D’Amare AR. Crystal structures of the Thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 43.Peselis A, Serganov A. Themes and variations in riboswitch structure and function. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbagrm.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doherty EA, Batey RT, Masquida B, Doudna JA. A universal mode of helix packing in RNA. Nat Struct Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- 45.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 46.Serganov A, Huang L, Patel DJ. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature. 2008;455:1263–1267. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garst AD, Heroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem. 2008;283:22347–22351. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Serganov A, et al. Ribosomal protein S15 from Thermus thermophilus—cloning, sequencing, overexpression of the gene and RNA-binding properties of the protein. Eur J Biochem. 1997;246:291–300. doi: 10.1111/j.1432-1033.1997.00291.x. [DOI] [PubMed] [Google Scholar]
- 49.Pikovskaya O, Serganov AA, Polonskaia A, Serganov A, Patel DJ. Preparation and crystallization of riboswitch-ligand complexes. Methods Mol Biol. 2009;540:115–128. doi: 10.1007/978-1-59745-558-9_9. [DOI] [PubMed] [Google Scholar]
- 50.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 51.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.