Abstract
Purpose
Since many transferred, good morphology embryos fail to implant, technologies to identify embryos with high developmental potential would be beneficial. The Eeva™ (Early Embryo Viability Assessment) Test, a prognostic test based on automated detection and analysis of time-lapse imaging information, has been shown to benefit embryo selection specificity for a panel of three highly experienced embryologists (Conaghan et al., 2013). Here we examined if adjunctive use of Eeva Test results following morphological assessment would allow embryologists with diverse clinical backgrounds to consistently improve the selection of embryos with high developmental potential.
Methods
Prospective, double-blinded multi-center study with 54 patients undergoing blastocyst transfer cycles consented to have embryos imaged using the Eeva System, which automatically measures key cell division timings and categorizes embryos into groups based on developmental potential. Five embryologists of diverse clinical practices, laboratory training, and geographical areas predicted blastocyst formation using day 3 morphology alone and day 3 morphology followed by Eeva Test results. Odds ratio (OR) and diagnostic performance measures were calculated by comparing prediction results to true blastocyst outcomes.
Results
When Eeva Test results were used adjunctively to traditional morphology to help predict blastocyst formation among embryos graded good or fair on day 3, the OR was 2.57 (95 % CI=1.88–3.51). The OR using morphology alone was 1.68 (95 % CI=1.29–2.19). Adjunct use of the Eeva Test reduced the variability in prediction performance across all five embryologists: the variability was reduced from a range of 1.06 (OR=1.14 to 2.20) to a range of 0.45 (OR=2.33 to 2.78).
Conclusions
The Eeva Test, an automated, time-lapse enabled prognostic test, used adjunctively with morphology, is informative in helping embryologists with various levels of experience select embryos with high developmental potential.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-014-0366-1) contains supplementary material, which is available to authorized users.
Keywords: In-vitro fertilization, Time-lapse imaging, Cell division timings, Embryo selection, Elective single embryo transfer, Prognostic test
Introduction
A major hurdle facing assisted reproductive technology (ART) professionals is the adoption of elective single embryo transfer (eSET), which has been stymied by a lack of tools to confidently select the embryo with highest potential for implantation [1–3]. Today, embryologists select embryos using morphological assessment at distinct time-points. However, morphological assessment is prone to inter- and intra-observer variation and has been shown to have limited predictive ability [4, 5]. Although culture to the blastocyst stage is increasingly practiced to narrow down which embryo(s) should be transferred [2, 6, 7], it prolongs the embryo’s exposure to in vitro conditions, which is unsuitable for patients who have few embryos to culture and has been linked to a potentially increased risk of epigenetic disorders and preterm delivery [8–12]. Overall, despite efforts to transfer only good morphology embryos on day 3 or day 5, more than 50 % of embryos selected by morphology alone fail to implant [13].
In order to drive confidence for eSET, approaches that can add quantitative and reliable information regarding embryo developmental potential must be demonstrated [14]. One such approach is to subject the embryo to an invasive, diagnostic test, such as pre-implantation genetic screening (PGS), to increase the chances of selecting a genetically normal and therefore implantable embryo [15–17]. Another option is to non-invasively extract prognostic information in order to improve the odds of selecting an embryo with high developmental potential [17–19]. In the latter case, time-lapse imaging has shown significant promise by enabling assessment of embryo dynamics and critical milestone achievements over multiple embryo development stages in vitro [18]. The potential of time-lapse imaging was demonstrated by Wong et al., who first established that early cell divisions could be used for embryo assessment and prediction of embryo development [19], and Meseguer et al., who later correlated early cell division timings with embryo implantation data [20]. The critical time-lapse biomarkers that were first shown to be predictive of successful embryo development were P2 (duration of the 2-cell stage [19], also called cc2 and t3-t2 [20]) and P3 (duration of the 3-cell stage [19], also called S2 and t4-t3 [20]). Subsequent studies confirmed the importance of these early non-invasive biomarkers and added other parameters (e.g., ICSI to 5-cell, ICSI to 8-cell) that, together with P2 and/or P3, could differentiate viable embryos defined by blastocyst quality, implantation after day 3 transfer, and chromosomal abnormality (aneuploidy) at the cleavage-stage [21–23, 18, 24]. For those time-lapse markers that have been consistently predictive across platforms and clinics, hierarchical selection models have been proposed [21, 14, 25, 26]. However, the majority of these embryo selection models should be considered with caution as they have been generated with data from a single clinic, have not been validated on independent data for their original endpoint, and have not been demonstrated to improve selection over traditional morphology.
While the prevailing literature has anticipated the potential of using time-lapse information to aid embryo selection, only one report has tested this hypothesis directly. Conaghan et al. described: (1) development of a novel, automated, time-lapse-based predictive test (The Eeva™ Test), (2) validation of the automated and predictive test on a large, independent set of data, and (3) embryo assessment results after three experienced embryologists used the test adjunctively to morphology, compared to morphology alone [14]. The Conaghan study focused on evaluating the specificity and positive predictive value (PPV) of blastocyst predictions made by three experienced embryologists who participated in the development of the Eeva Test. This was an important and pioneering first step in the validation of the Eeva Test; however, confirmation that the test is reproducible in the hands of a broader range of users with varied backgrounds and experience levels is also needed.
Here, we present results from a new panel of five embryologists representing a diverse range of practices, laboratory training, and geographical areas, who used the Eeva Test adjunctively to morphology to make embryo assessments. The purpose of the study was to evaluate if adjunct use of the Eeva Test was consistently informative in predicting blastocyst formation, for these five embryologists. Therefore, odds ratios, indicating whether adjunctive assessment was informative (better than random prediction), were calculated for each embryologist and compared. Diagnostic performance measures (specificity, sensitivity, PPV, negative predictive value (NPV)) were also evaluated. Because the Eeva Test is intended for use as an adjunct to traditional morphology, odds ratios and diagnostic performance measures were evaluated for all embryos, and for a subset of morphologically good/fair embryos that are candidates for embryo selection.
Materials and methods
Study design
This was a prospective, double-blinded, multi-center study, designed to evaluate the odds ratios for methodologies used for embryo assessment: day 3 morphology alone and day 3 morphology followed by Eeva Test results. Data used for the study were collected from June 2011–April 2012 as reported in Conaghan et al. [ClinicalTrials.gov NCT01369446] [14]. Briefly, the data included N = 758 embryos from 54 patients who were undergoing blastocyst transfer cycles and consented to have their embryos imaged using the Eeva System. Women who were undergoing fresh in vitro fertilization (IVF) treatment using their own eggs or donor eggs, were ≥18 years of age and had a total antral follicle count of at least 8 as imaged and measured by ultrasound prior to stimulation were enrolled. Other inclusion criteria were fertilization using only fresh or frozen ejaculated sperm (no surgically removed sperm) and embryos cultured to day 5. Patients were excluded if they were gestational carriers, planned preimplantation genetic diagnosis or preimplantation genetic screening, had reinseminated eggs, had a history of cancer, or were participating concurrently in another clinical study.
Embryo culture and Eeva imaging
Embryo image sequences (videos) were collected using the Eeva System as described previously by Conaghan et al. [14]. Specifically, on the day of retrieval (Day 0), oocytes were fertilized using conventional insemination or intracytoplasmic sperm injection (ICSI). Immediately following the fertilization assessment, successfully fertilized oocytes (2PNs) were transferred to a multi-well Eeva Dish for group-culture and monitoring in a standard incubator. For the collection of these videos, embryos were cultured per the standard laboratory protocols of three separate IVF clinics. The embryo data used for the technology’s development included diversity in the type of stimulation protocol, culture media and incubation environment used (5 % and 20 % Oxygen tension) in order to create a generalizable prediction model that could be applied across multiple centers. Routine day 3 embryo grading was performed by laboratory embryologists who were blinded to the Eeva information.
Computer-automated assessment of embryo videos was performed using the Eeva Test, which automatically measures cell division timings P2 (time between first and second mitosis) and P3 (time between second and third mitosis), and provides a High (Eeva High) or Low (Eeva Low) probability of blastocyst formation depending on the P2 value (High range: 9.33–11.47 h) and the P3 value (High range: 0.00–1.73 h).
Embryologist panel and embryo assessment
The Eeva System is indicated to provide adjunctive information on events occurring during the first 2 days of embryo development that may predict further development to the blastocyst stage on day 5 of embryo culture. This adjunctive information aids in the selection of embryo(s) for transfer on day 3 when, following morphological assessment on day 3, there are multiple embryos deemed suitable for transfer or freezing. To evaluate the impact of this intended use, a panel of five embryologists who were not involved in the embryo culture and Eeva imaging phase described above was assembled. Instead, these embryologists were naïve Eeva Test evaluators, representing a separate and diverse range of clinical practices, laboratory training, and geographical areas within the United States. Embryologists 1, 2 and 5 were Senior Embryologists with more than 10 years of clinical embryology experience each, and Embryologists 3 and 4 were Junior Embryologists with less than 3 years of clinical embryology experience. The clinics where the embryologists worked ranged in practice volume from <300 cycles per year to >1,000 cycles per year, based on 2011 SART reporting data (www.sartcorsonline.com) [27].
For this study, each of the five embryologists initially assessed embryos by predicting blastocyst formation using day 3 morphology alone, then by using day 3 morphology followed by Eeva Test results. Specifically, for day 3 morphology alone assessment, each embryologist reviewed the day 3 morphology data for each patient’s cohort of embryos, including cell number, degree of fragmentation (0 %, <10 %, 10–25 %, >25 %), degree of symmetry (perfect, moderately asymmetrical, severely asymmetrical), and oocyte age. Each embryologist then assigned: (1) an overall grade based on day 3 morphology data (A: Good, B: Fair+, C: Fair-, D: Poor), and (2) a prediction of the outcome as “blastocyst” or “arrested”. For the adjunct (day 3 morphology followed by Eeva Test) assessment, the embryologists were provided the same day 3 morphology data, as well as additional Eeva Test results (Eeva P2 and P3 values, Eeva High or Low scores); they then assigned a prediction of the outcome as “blastocyst” or “arrested” to those embryos with a morphological grade of good or fair.
All data were presented to embryologists as full cohorts of embryos for each patient. Embryologists made their assessment in isolation from one another. They were blinded to the clinical site where each subject was enrolled, the fate of the embryos, the outcomes of the IVF procedures, and the results from the other embryologists on the panel.
Performance evaluation methods
To quantitatively evaluate the performance of the adjunct assessment, the odds ratio (OR) for predicting blastocyst formation was calculated by comparing the embryologist’s prediction of blast (probability of an event happening) to whether a blastocyst formed. In this study, blastocyst formation was defined by blastocysts that were transferred or frozen on day 5 (quality blastocyst formation). The overall OR for blastocyst prediction was estimated using a logistic mixed effects model with both patient and embryologist inputted as random effects in the model [28]. The individual OR for blastocyst prediction was calculated for all five embryologists. Confidence intervals (CI) of the overall OR were calculated by incorporating the variance within individual embryologist estimates, as well as the variance between all embryologists.
Diagnostic performance measures of specificity, sensitivity, positive predictive value (PPV) and negative predictive value (NPV) and quality blastocyst formations rates were also calculated for each embryologist. Averages were calculated using all five embryologists, and confidence intervals were estimated by bootstrapping the observed results to account for correlation among embryos within cohorts.
Statistical analyses
Data and statistical analyses were performed using SAS software version 9.2. To obtain the overall OR and calculate the confidence intervals, the SAS procedure GLIMMIX was used to perform a logistic mixed effects regression, and the “empirical” option was invoked to obtain a GEE type analysis and make inferences that are not sensitive to the choice of the covariance models. A logistic mixed effects model was selected to account for the embryologist and patient as potentially confounding factors in this embryo-level analysis. The OR and associated confidence intervals significantly greater than 1 indicated whether a methodology for blastocyst prediction was informative and not due to chance alone. A Chi-square proportions test was used to compare blastocyst formation rates for Eeva High vs. Eeva Low groups, and a p-value <0.05 was considered to be statistically significant.
Results
For this embryo assessment study, data for N = 758 embryos were prospectively collected from 54 patients who had received IVF treatment, blastocyst culture and Eeva System imaging at three U.S. clinical sites [14]. The clinical characteristics of the 54 patients are provided in Table 1.
Table 1.
Clinical characteristics of the 54 study patients
| Demographics (n = 54 patients) | Mean ± SD |
|---|---|
| Egg age (years) | 32.2 ± 5.1 |
| Body mass index (kg/m2) | 24.1 ± 4.7 |
| Antral follicle count | 22.0 ± 9.2 |
| Number of follicles | 21.5 ± 7.8 |
| Number of oocytes | 21.5 ± 7.8 |
| Number of 2PN | 14.3 ± 5.0 |
Eeva Test scores correlate with blastocyst formation rates
Grading assignments of A, B, C and D were made so that embryologists could simulate the intended use of the Eeva Test for all embryos (those they graded as A, B, C or D), followed by a subset of the good/fair morphology embryos (those they graded as A, B or C). The prevalence of embryos graded as A, B, C, and D by all five individual embryologists is provided in Supplementary Figure 1.
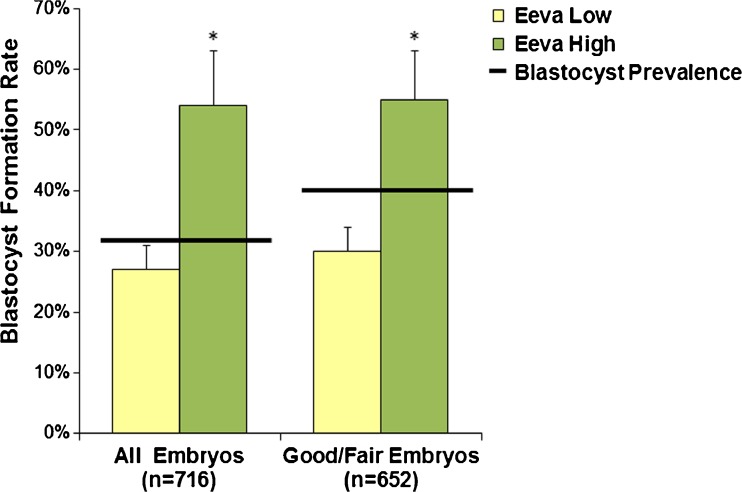
Separately, the Eeva Test automatically generated an Eeva High or Eeva Low score for each embryo, indicating a high or low probability of blastocyst formation. From the assessed embryos, 22 % (167/758) were Eeva High, 72 % (548/758) were Eeva Low, and 6 % (42/758) did not receive a result. For all embryos graded as A, B, C or D with Eeva scores (n = 716), the blastocyst formation rate (defined as quality blastocysts that were transferred or frozen on day 5) was 33 % overall, 27 % for Eeva Lows and 54 % for Eeva Highs. For the good/fair embryos graded as A, B, or C by at least one panelist with Eeva scores (n = 652), the average blastocyst formation rate was 36 % overall, 30 % for Eeva Lows, and 55 % for Eeva Highs (Fig. 1). For both populations of embryos, blastocyst formation rates were significantly higher in the Eeva High group than in the Eeva Low group (p < 0.0001).
Fig. 1.
Eeva High and Eeva Low scores correlate to a high or low probability of blastocyst formation for all embryos (n = 716) and for those denoted as morphologically good/fair (n = 652). For both populations, blastocyst formation rates were significantly higher in the Eeva High group than in the Eeva Low group. *p < 0.0001 (error bars represent upper 95 % confidence intervals)
Adjunct use of the Eeva Test following day 3 morphology is informative
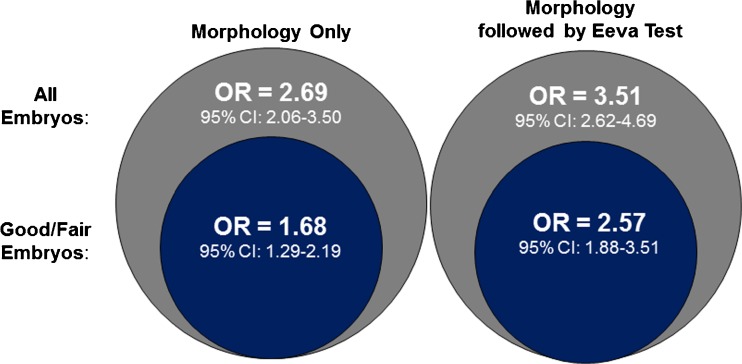
Among all embryos, the overall odds ratio across all embryologists using day 3 morphology only to predict blastocyst formation was 2.69 (95 % CI=2.06–3.50). When day 3 morphology followed by Eeva Test results were used to predict blastocyst formation among all embryos, the overall odds ratio across all embryologists was 3.51 (95 % CI=2.62–4.69) (Fig. 2). Among good/fair morphology embryos, the overall odds ratio across all embryologists using day 3 morphology only to predict blastocyst formation was 1.68 (95 % CI=1.29–2.19). When day 3 morphology followed by Eeva Test results were used to predict blastocyst formation among the good/fair morphology embryos, the overall odds ratio across all embryologists was 2.57 (95 % CI=1.88–3.51) (Figs. 2 and 3a).
Fig. 2.
Odds ratio for predicting blastocyst formation using Morphology Only (left) and Morphology followed by Eeva Test (right). Odds ratios and 95 % confidence intervals were calculated for all embryos (represented in gray) and for the subset of embryos graded as good/fair (represented in blue)
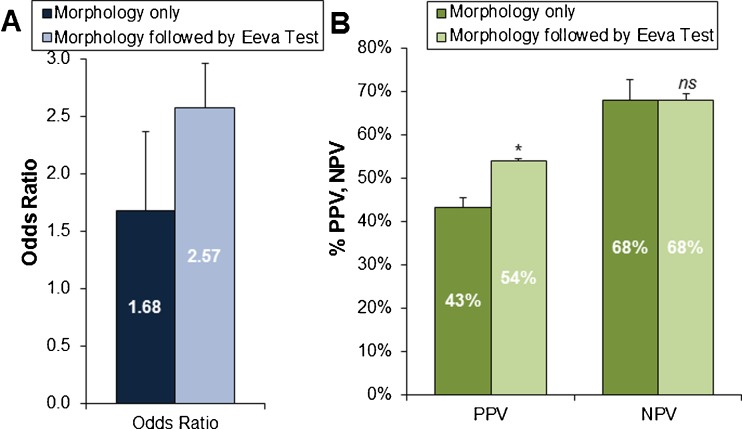
Fig. 3.
a Overall odds ratio, b mean positive predictive value (PPV) and mean negative predictive value (NPV) across all embryologists predicting blastocyst formation using Morphology Only and Morphology followed by Eeva Test, among good/fair embryos. *p = 0.02, ns not significant (error bars represent upper 95 % confidence intervals)
Adjunct use of the Eeva Test following day 3 morphology is informative for good/fair embryos
The good/fair morphology embryos were analyzed further, since these are the embryos for which embryologists need further prognostic information to aid embryo selection. In this group of good/fair morphology embryos, the overall odds ratio for embryologists using day 3 morphology alone was significantly >1 or random prediction (1.68 vs 1.0, p < 0.0001). The overall odds ratio for embryologists using day 3 morphology followed by Eeva Test was higher than morphology alone and significantly >1 or random prediction (2.57 vs. 1.0, p < 0.0001) (Fig. 3a).
Evaluating other diagnostic measures, embryologists using the Eeva Test as an adjunct to traditional morphology to predict blastocyst formation significantly improved their average specificity (76 % vs. 39 % for day 3 morphology alone, p < 0.0001). Sensitivity declined as expected (45 % vs. 72 % for day 3 morphology alone, p < 0.0001) because the Eeva Test was designed to achieve high specificity to help distinguish among the good/fair morphology embryos which have lower potential of development (false positives). Thus, positive predictive value (PPV) was significantly improved (54 % vs. 43 % for day 3 morphology alone, p = .02), while negative predictive value (NPV) was maintained (68 % vs. 68 %) (Fig. 3b).
Consistency among embryologists
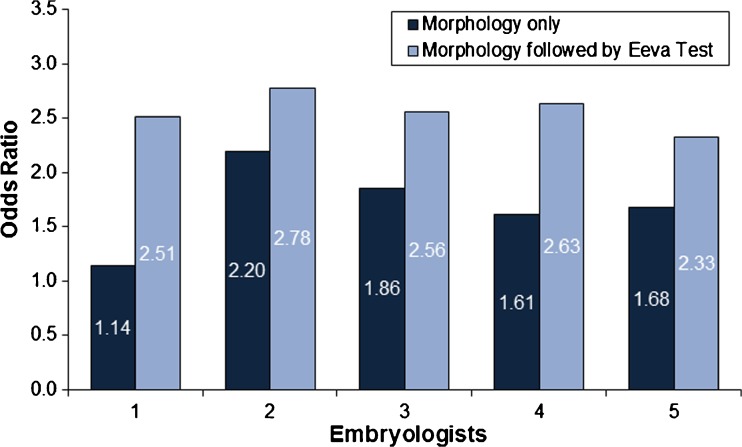
To determine how consistently informative the day 3 morphology followed by adjunct use of Eeva Test was among individual embryologists, assessments among the good/fair morphology embryos were further evaluated using the odds ratio. Adjunct use of day 3 morphology followed by Eeva Test resulted in odds ratios of 2.51, 2.78, 2.56, 2.63 and 2.33 for embryologists 1 through 5, respectively. In contrast, using day 3 morphology resulted in odds ratios of 1.14, 2.20, 1.86, 1.61 and 1.68 for embryologists 1 through 5, respectively (Fig. 4). Thus, when the Eeva Test was used adjunctively to morphology, the odds ratio was improved for each embryologist, and the variability in blastocyst prediction across all five embryologists was reduced from a range of 1.06 (OR=1.14 to 2.20) to a range of 0.45 (OR=2.33 to 2.78).
Fig. 4.
Consistent improvement in odds ratios for individual embryologists who predicted blastocyst formation using Morphology Only and Morphology followed by Eeva Test, among good/fair embryos
Discussion
Although further research is needed, elective single embryo transfer may be more widely adopted with new prognostic information that can assist embryologists in their selection of the embryo most likely to develop. The objective of this study was to determine whether adjunctive use of the prognostic Eeva Test with traditional morphology was consistently informative for embryologists seeking to select embryos with higher developmental potential. Our results demonstrate that five embryologists who used the Eeva Test adjunctively with day 3 morphology each benefited from prognostic information that helped them select the embryos with higher developmental potential.
The degree of benefit to embryologists was quantified by calculating the odds ratio and other diagnostic measures for methodologies applied to two sets of embryos: (1) all embryos, and (2) a sub-group of good/fair morphology embryos that are candidates for embryo selection. The odds ratio is an important quantitative indicator of performance that determines whether a prognostic test is informative [29]. In our case, we used odds ratio to quantify the relative odds of successful blastocyst formation for an embryo predicted to develop (i.e. an embryo that might be selected on day 3), compared to an embryo not predicted to develop (i.e. an embryo that might be de-selected). For example, at baseline (using traditional morphology for all embryos), the odds of an embryo forming a blastocyst was 2.69 times higher for those embryos predicted to form a blastocyst than among those embryos predicted to arrest. This suggests that traditional morphology is highly informative in assessing embryos overall. However, a more clinically relevant challenge arises when traditional morphology must select among the morphologically good/fair embryos; in this sub-group, the odds ratio of an embryo forming a blastocyst predicted by traditional morphology was 1.68, slightly better than random prediction (p < 0.0001). In contrast, adding results from the Eeva Test to aid in the selection among good/fair morphology embryos increased the odds ratio of an embryo forming a blastocyst to 2.57, which was a 53 % increase over traditional morphology and significantly better than random prediction (p < 0.0001). The Eeva Test therefore adds new prognostic information that is particularly informative for identifying which embryo(s) should be selected among good/fair morphology embryos.
Along with odds ratio, we found that the average PPV was significantly improved while the average NPV was maintained for embryologists using morphology followed by the Eeva Test. The predictive and prognostic information provided by the Eeva Test is based on a combination of unique features, including: (1) scientifically grounded cell division timings that have been shown to correlate to blastocyst prediction, gene expression analysis and clinical outcomes [14, 19]; (2) real-time, automated extraction of these timing parameters from time-lapse videos [14]; and (3) an ultimate prognostic score of High or Low developmental potential [14]. In this study, we confirmed that Eeva High embryos have a significantly higher likelihood of forming a blastocyst than Eeva Low embryos. The difference was significant even for a subset of good/fair morphology embryos, suggesting that the Eeva Test result can help to further distinguish among similar-looking embryos that are evaluated first by morphological criteria. Since successful development to the blastocyst stage is a critical milestone that is associated with higher implantation potential relative to cleavage-stage embryos [2, 30], our results suggest that the Eeva Test may improve overall success rates when used to select embryos with traditional morphology.
Adjunct use of the Eeva Test following day 3 morphology was consistently informative for each of five embryologists representing a separate and diverse range of clinical practices, laboratory training (<3 to >10 years), and geographical areas within the United States. Notably, the embryologist with the greatest improvement in odds ratio was one of the senior embryologists with more than 10 years of training in morphology grading. All embryologists in our study experienced an increase in odds ratio when the Eeva Test information was added to their traditional morphology assessment and blastocyst predictions. The odds ratio among the embryologists was also less variable for adjunct predictions compared to traditional morphology alone, suggesting that the Eeva Test may help reduce inter- and intra-observer variability commonly associated with morphological assessment [4, 14, 31].
The introduction of any new prognostic test into the IVF laboratory requires rigorous validation and demonstration of reproducibility [32, 33]. Validation of any time-lapse finding or time-lapse enabled selection methodology should include biological validation, clinical validation, performance characterization and comparison to standard of care [34]. Reproducibility of the technology should be demonstrated in clinical settings that are independent from the clinics that collected and initially developed the proposed test. From a technical standpoint, the predictive test must therefore be developed using diverse data, and then tested on a large, unbiased sampling of clinical data. One of the unique features of the Eeva Test is that it was built using multi-clinic data with substantial heterogeneity in embryo culture media, environmental conditions and insemination technique [14]. Other predictive models that have been proposed have been created using small samplings of data from a single clinic [21, 25, 26, 35]. Successful demonstration of validation and generalizability can form the basis for approval from a regulatory body and give confidence to clinicians and patients that the technology is safe, effective and beneficial [33]. Our study uniquely addresses both requirements of validation and generalizability by prospectively testing the impact of the Eeva Test in the hands of five embryologists from five distinct IVF clinics. It also considered a practical intended use model by focusing on the good/fair morphology embryos that are pre-selected with the expertise of the embryologist.
Conclusions
Adjunctive use of the Eeva™ (Early Embryo Viability Assessment) Test, a prognostic test based on automated detection and analysis of time-lapse imaging information, is highly informative and allows embryologists from diverse clinical backgrounds to consistently improve the selection of embryos with high developmental potential. Irrespective of the clinic practice, experience level, and training, embryologists were able to consistently improve their ability to select embryos with higher developmental potential, particularly among good/fair morphology embryos. Therefore, the Eeva Test can assist clinical embryologists in making informed decisions when selecting embryos for transfer or freezing. As a next step of a rigorous path to demonstrating utility of this test, prospective evaluation focused on implantation and pregnancy outcomes after using the Eeva Test as an adjunct to morphology grading is underway.
Electronic supplementary material
Morphological grade assignments (A, B, C, and D) made by five embryologists using day 3 morphology data for n=758 embryos. (GIF 52 kb)
Acknowledgments
We acknowledge Auxogyn, Inc. for sponsoring this study, the clinical, scientific and algorithm groups at Auxogyn for insightful discussion, and the physicians, embryologists and patients who participated in the development and validation of the Eeva Test.
Footnotes
Capsule The Eeva Test is highly informative and, when used adjunctively to morphology, allows embryologists from diverse clinical backgrounds to consistently improve embryo selection.
References
- 1.Van Montfoort AP, Dumoulin JC, Kester AD, Evers JL. Early cleavage is a valuable addition to existing embryo selection parameters: a study using single embryo transfers. Hum Reprod. 2004;19(9):2103–8. doi: 10.1093/humrep/deh385. [DOI] [PubMed] [Google Scholar]
- 2.Diamond MP, Willman S, Chenette P, Cedars MI. The clinical need for a method of identification of embryos destined to become a blastocyst in assisted reproductive technology cycles. J Assist Reprod Genet. 2012;29(5):391–6. doi: 10.1007/s10815-012-9732-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Practice Committee of Society for Assisted Reproductive T. Practice Committee of American Society for Reproductive M Elective single-embryo transfer. Fertil Steril. 2012;97(4):835–42. doi: 10.1016/j.fertnstert.2011.11.050. [DOI] [PubMed] [Google Scholar]
- 4.Paternot G, Devroe J, Debrock S, D’Hooghe TM, Spiessens C. Intra- and inter-observer analysis in the morphological assessment of early-stage embryos. Reprod Biol Endocrinol: RB&E. 2009;7:105. doi: 10.1186/1477-7827-7-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paternot G, Debrock S, D’Hooghe T, Spiessens C. Computer-assisted embryo selection: a benefit in the evaluation of embryo quality? Reprod Biomed Online. 2011;23(3):347–54. doi: 10.1016/j.rbmo.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Cruz M, Gadea B, Garrido N, Pedersen KS, Martinez M, Perez-Cano I, et al. Embryo quality, blastocyst and ongoing pregnancy rates in oocyte donation patients whose embryos were monitored by time-lapse imaging. J Assist Reprod Genet. 2011;28(7):569–73. doi: 10.1007/s10815-011-9549-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gleicher N, Barad DH. A review of, and commentary on, the ongoing second clinical introduction of preimplantation genetic screening (PGS) to routine IVF practice. J Assist Reprod Genet. 2012;29(11):1159–66. doi: 10.1007/s10815-012-9871-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niemitz EL, Feinberg AP. Epigenetics and assisted reproductive technology: a call for investigation. Am J Hum Genet. 2004;74(4):599–609. doi: 10.1086/382897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horsthemke B, Ludwig M. Assisted reproduction: the epigenetic perspective. Hum Reprod Update. 2005;11(5):473–82. doi: 10.1093/humupd/dmi022. [DOI] [PubMed] [Google Scholar]
- 10.Manipalviratn S, DeCherney A, Segars J. Imprinting disorders and assisted reproductive technology. Fertil Steril. 2009;91(2):305–15. doi: 10.1016/j.fertnstert.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sazonova A, Kallen K, Thurin-Kjellberg A, Wennerholm UB, Bergh C. Neonatal and maternal outcomes comparing women undergoing two in vitro fertilization (IVF) singleton pregnancies and women undergoing one IVF twin pregnancy. Fertil Steril. 2013;99(3):731–7. doi: 10.1016/j.fertnstert.2012.11.023. [DOI] [PubMed] [Google Scholar]
- 12.Practice Committees of the American Society for Reproductive M, the Society for Assisted Reproductive T. Blastocyst culture and transfer in clinical-assisted reproduction: a committee opinion. Fertility and sterility. 2013;99(3):667–72. doi:10.1016/j.fertnstert.2013.01.087. [DOI] [PubMed]
- 13.Clua E, Tur R, Coroleu B, Boada M, Rodriguez I, Barri PN, et al. Elective single-embryo transfer in oocyte donation programmes: should it be the rule? Reprod Biomed Online. 2012;25(6):642–8. doi: 10.1016/j.rbmo.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 14.Conaghan J, Chen AA, Willman SP, Ivani K, Chenette PE, Boostanfar R, et al. Improving embryo selection using a computer-automated time-lapse image analysis test plus day 3 morphology: results from a prospective multicenter trial. Fertil Steril. 2013;100(2):412–9 e5. doi: 10.1016/j.fertnstert.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 15.Yang Z, Liu J, Collins GS, Salem SA, Liu X, Lyle SS, et al. Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: results from a randomized pilot study. Mol Cytogenet. 2012;5(1):24. doi: 10.1186/1755-8166-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forman EJ, Upham KM, Cheng M, Zhao T, Hong KH, Treff NR, et al. Comprehensive chromosome screening alters traditional morphology-based embryo selection: a prospective study of 100 consecutive cycles of planned fresh euploid blastocyst transfer. Fertil Steril. 2013;100(3):718–24. doi: 10.1016/j.fertnstert.2013.04.043. [DOI] [PubMed] [Google Scholar]
- 17.Chen AA, Shen S. Predicting Embryo Developmental Potential and Viability Using Automated Time-Lapse Analysis (Eeva Test). 2013:377–89. doi:10.1007/978-1-4614-8376-2_22.
- 18.Chen AA, Tan L, Suraj V, Reijo Pera R, Shen S. Biomarkers identified with time-lapse imaging: discovery, validation, and practical application. Fertil Steril. 2013;99(4):1035–43. doi: 10.1016/j.fertnstert.2013.01.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong CC, Loewke KE, Bossert NL, Behr B, De Jonge CJ, Baer TM, et al. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat Biotechnol. 2010;28(10):1115–21. doi: 10.1038/nbt.1686. [DOI] [PubMed] [Google Scholar]
- 20.Meseguer M, Rubio I, Cruz M, Basile N, Marcos J, Requena A. Embryo incubation and selection in a time-lapse monitoring system improves pregnancy outcome compared with a standard incubator: a retrospective cohort study. Fertil Steril. 2012;98(6):1481–9 e10. doi: 10.1016/j.fertnstert.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 21.Meseguer M, Herrero J, Tejera A, Hilligsoe KM, Ramsing NB, Remohi J. The use of morphokinetics as a predictor of embryo implantation. Hum Reprod. 2011;26(10):2658–71. doi: 10.1093/humrep/der256. [DOI] [PubMed] [Google Scholar]
- 22.Dal Canto M, Coticchio G, Mignini Renzini M, De Ponti E, Novara PV, Brambillasca F, et al. Cleavage kinetics analysis of human embryos predicts development to blastocyst and implantation. Reprod Biomed Online. 2012;25(5):474–80. doi: 10.1016/j.rbmo.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 23.Rubio I, Kuhlmann R, Agerholm I, Kirk J, Herrero J, Escriba MJ, et al. Limited implantation success of direct-cleaved human zygotes: a time-lapse study. Fertil Steril. 2012;98(6):1458–63. doi: 10.1016/j.fertnstert.2012.07.1135. [DOI] [PubMed] [Google Scholar]
- 24.Chavez SL, Loewke KE, Han J, Moussavi F, Colls P, Munne S, et al. Dynamic blastomere behaviour reflects human embryo ploidy by the four-cell stage. Nat Commun. 2012;3:1251. doi: 10.1038/ncomms2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Hickman CF. Modelling a risk classification of aneuploidy in human embryos using non-invasive morphokinetics. Reprod Biomed Online. 2013;26(5):477–85. doi: 10.1016/j.rbmo.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Basile N, Nogales Mdel C, Bronet F, Florensa M, Riqueiros M, Rodrigo L, et al. Increasing the probability of selecting chromosomally normal embryos by time-lapse morphokinetics analysis. Fertil Steril. 2014;101(3):699–704. doi: 10.1016/j.fertnstert.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 27.Racowsky C, Stern JE, Gibbons WE, Behr B, Pomeroy KO, Biggers JD. National collection of embryo morphology data into society for assisted reproductive technology clinic outcomes reporting system: associations among day 3 cell number, fragmentation and blastomere asymmetry, and live birth rate. Fertil Steril. 2011;95(6):1985–9. doi: 10.1016/j.fertnstert.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Rubin DB. Multiple Imputation for Nonresponse in Surveys. 1987.
- 29.Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PM. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol. 2003;56(11):1129–35. doi: 10.1016/S0895-4356(03)00177-X. [DOI] [PubMed] [Google Scholar]
- 30.Stacey D, Bennett CL, Barry MJ, Col NF, Eden KB, Holmes-Rovner M, et al. Decision aids for people facing health treatment or screening decisions. Cochrane Database Syst Rev. 2011;10:CD001431. doi: 10.1002/14651858.CD001431.pub3. [DOI] [PubMed] [Google Scholar]
- 31.Baxter Bendus AE, Mayer JF, Shipley SK, Catherino WH. Interobserver and intraobserver variation in day 3 embryo grading. Fertil Steril. 2006;86(6):1608–15. doi: 10.1016/j.fertnstert.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 32.Harper JC, Harton G. The use of arrays in preimplantation genetic diagnosis and screening. Fertil Steril. 2010;94(4):1173–7. doi: 10.1016/j.fertnstert.2010.04.064. [DOI] [PubMed] [Google Scholar]
- 33.Palmer SS, Barnhart KT. Biomarkers in reproductive medicine: the promise, and can it be fulfilled? Fertil Steril. 2013;99(4):954–62. doi: 10.1016/j.fertnstert.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harper J, Cristina Magli M, Lundin K, Barratt CLR, Brison D. When and how should new technology be introduced into the IVF laboratory? Hum Reprod. 2012;27(2):303–13. doi: 10.1093/humrep/der414. [DOI] [PubMed] [Google Scholar]
- 35.Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Thornton S. Retrospective analysis of outcomes after IVF using an aneuploidy risk model derived from time-lapse imaging without PGS. Reprod Biomed Online. 2013;27(2):140–6. doi: 10.1016/j.rbmo.2013.04.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Morphological grade assignments (A, B, C, and D) made by five embryologists using day 3 morphology data for n=758 embryos. (GIF 52 kb)