Abstract
Understanding how brain activation mediates behaviors is a central goal of systems neuroscience. Here we apply an automated method for mapping brain activation in the mouse in order to probe how sex-specific social behaviors are represented in the male brain. Our method uses the immediate early gene c-fos, a marker of neuronal activation, visualized by serial two-photon tomography: the c-fos-GFP-positive neurons are computationally detected, their distribution is registered to a reference brain and a brain atlas, and their numbers are analyzed by statistical tests. Our results reveal distinct and shared female and male interaction-evoked patterns of male brain activation representing sex discrimination and social recognition. We also identify brain regions whose degree of activity correlates to specific features of social behaviors and estimate the total numbers and the densities of activated neurons per brain areas. Our study opens the door to automated screening of behavior-evoked brain activation in the mouse.
Introduction
Central to the understanding of brain functions is insight into the distribution of neuronal activity which drives behavior. Local measurements of brain activity in behaving mice can be made with electrodes and fluorescent calcium indicators (Buzsaki, 2004; Grewe and Helmchen, 2009), but such approaches provide information regarding only a very small fraction of the approximately 70 millions neurons that comprise the mouse brain. The detection of elevated levels of the immediate-early genes (IEGs) linked to recent neuronal activity (Clayton, 2000; Guzowski et al., 2005) is a more spatially comprehensive technique. While it lacks the time resolution of electrophysiological recordings or calcium imaging, it does have the potential of providing a complete view of recent whole-brain activity. Once determined, the whole brain IEG-based map can be used to generate structure-function hypotheses to be probed by high-resolution recordings, as well as optogenetic and chemogenetic methods (Fenno et al., 2011; Lee et al., 2014).
Here we use a pipeline of computational methods that permits automated unbiased mapping of c-fos induction in mouse brains at single-cell resolution, in a similar way as recently described for mapping the induction of the IEG Arc (Vousden et al., 2014). Specifically, we use serial two-photon (STP) tomography (Ragan et al., 2012) to image the expression of c-fos-GFP, a transgenic c-fos-green fluorescent protein reporter (Reijmers et al., 2007), across the entire mouse brain. The activated c-fos-GFP-positive (c-fos-GFP+) cells are computationally detected, their location is mapped at stereotaxic coordinates within a reference brain, and their numbers and densities per anatomical brain areas are determined within the Allen Mouse Brain Atlas. Finally, region of interest (ROI)-based and voxel-based statistical tests are applied to identify brain areas with behaviorally evoked c-fos-GFP activation.
To demonstrate the application of the computational pipeline to the mapping of behavior-evoked brain activation, we focus on mouse social behavior and generate activation maps representing sex-specific social behaviors in the male brain. Rodent social behavior is an area of intense research and c-fos mapping, lesion studies and other functional approaches have been used to identify brain regions that are activated and contribute to male and female sexual behaviors as well as male-male aggressive behaviors (Anderson, 2012; Bialy and Kaczmarek, 1996; Brennan and Zufall, 2006; Coolen et al., 1996; Pfaus and Heeb, 1997; Veening et al., 2005; Yang and Shah, 2014). Much less is known, on the other hand, about the brain areas activated during the initial period of sex discrimination and social recognition before the manifestation of the correct behavioral response.
Here we explore the question of sex discrimination and social recognition by limiting the male-female and male-male interactions to a brief 90 sec period, during which the behavioral repertoire comprises only social exploratory activity, such as anogenital sniffing, close following, and nose-to-nose sniffing, without mating or aggression. A side-by-side comparison of the female and male interaction-evoked whole-brain activation revealed 1) a broad activation of areas downstream of both the main and accessory olfactory bulb (MOB and AOB) in the male-female interaction and a bias toward structures downstream of the MOB in the male-male interaction; 2) activation of structures related to behavioral motivation during the male-female but not male-male interaction, and 3) sex-specific as well as shared hypothalamic activation. Taking advantage of the cellular resolution of the whole-brain data, we then identified brain regions whose level of activation was correlated to specific features of the social behaviors, including regions linked to anogenital sniffing that lie downstream of the pheromone-activated AOB, and regions linked to close following that belong to the striato-pallido-thalamo-cortical circuitry. Finally, we calculated the total numbers and the densities of c-fos-GFP+ cells per activated brain regions of the female- and male-specific brain datasets, providing a quantitative estimate of whole-brain activation evoked by social behaviors.
RESULTS
Whole-brain detection of c-fos-GFP+ cells in STP tomography datasets
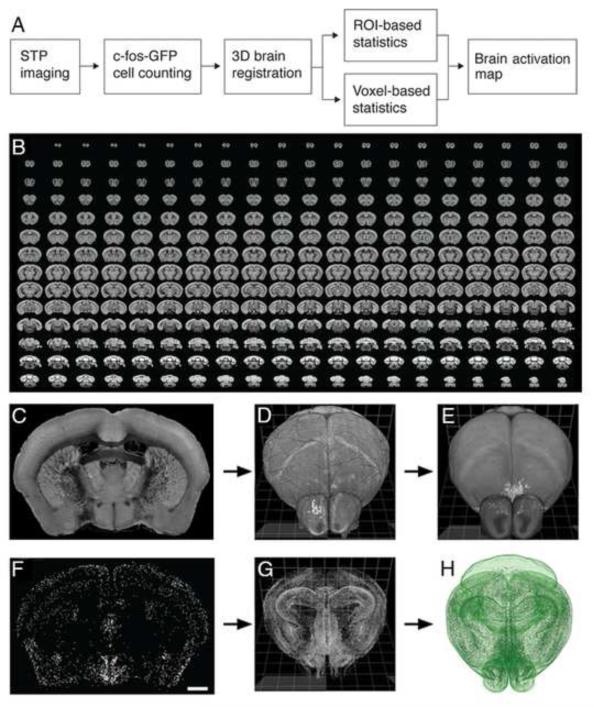
We have established an automated and quantitative whole-brain method for mapping behaviorally-evoked c-fos induction in transgenic reporter mice expressing c-fos-GFP from a recombinant c-fos promoter (Reijmers et al., 2007) (Methods). This necessitated the development and optimization of: 1) computational detection of c-fos-GFP+ cells in the mouse brain imaged by STP tomography (Ragan et al., 2012), 2) 3D registration of the STP datasets to a reference mouse brain, and 3) statistical analyses of the whole-brain distribution of the c-fos-GFP+ cells (Figure 1A).
Figure 1. STP tomography and computational detection of c-fos-GFP positive cells.
(A) Imaging and data processing pipeline for mapping whole-brain activation in c-fos-GFP mice. (B) A sample 280-serial section dataset of a c-fos-GFP mouse brain imaged by STP tomography. (C-H) Registration of CN-detected c-fos-GFP+ cells in the RSTP brain. (C) A coronal section shows the autofluorescence signal, which is used for registering the 3D reconstructed sample brain (D) onto the RSTP brain (E). (F) 2,177 c-fos-GFP+ cells were detected in the same coronal section; scale bar = 1 mm. (G) 360,183 c-fos-GFP+ cells were detected in the whole brain, reconstructed in 3D and (H) registered onto the RSTP brain using the image registration parameters established in the (D-E) step.
The mouse brains were imaged by STP tomography as datasets of 280 serial coronal sections, with x-y resolution 1.0 μm and z-spacing 50 μm, which required an imaging time of ~21 hours per brain (Figure 1B) (Ragan et al., 2012). To achieve a reliable computational detection of the c-fos-GFP+ cells throughout the whole brain, we used convolutional networks (CNs) that can learn to recognize image features in complex datasets (Jain et al., 2007; Turaga et al., 2010) (Figure S1; Methods). Since nearby c-fos-GFP+ cells were sometimes merged in the CN output, a post-processing step was devised that could separate such “touching” cells (Figure S1). The CN performance was then quantified on a new set of marked up FOVs from a second c-fos-GFP brain using the F-score measure, which represents the harmonic mean of the precision and recall (i.e. the false positive and false negative error rate), where F score 1 is the best and 0 the worst. The CN performance reached F-score 0.88 (precision 0.86, recall 0.90), which was comparable to human inter-user variability represented by F-score 0.90 (precision 0.90, recall 0.90) (Figure S1; Methods). We conclude that the trained CN provides an automated and highly accurate method for detection of c-fos-GFP+ cells in whole mouse brains imaged by STP tomography.
Anatomical registration of the whole-brain c-fos-GFP data
Results from the CN-based cell counting produce a number of c-fos-GFP+ cells per the individual 280-section datasets, with each cell having an xyz location. To be able to compare patterns of c-fos activation between experimental groups in one common brain volume, we created a Reference STP (RSTP) brain co-registered to the digital Allen Brain Atlas (ABA) for 8-week old C57BL/6 mouse (Sunkin et al., 2013) (Figure S2A; Methods; Movie S1). The image registrations were done by a 3D affine transformation, followed by a 3D B-spline transformation with Mattes Mutual information as the similarity measure (Mattes et al., 2003). The 3D registration accuracy was calculated to 65.0 ± 39.9 □m for (mean ± SD) (Figure S2B-C; Methods), which is also the accuracy for the registration of all STP experimental datasets to the RSTP brain for data analysis. The alignment of the RSTP and ABA Nissl brains was further improved by 2D affine and B-spline transformations using STP tomography-imaged CAG-Keima brain, which has a Nissl-like fluorescent labeling from the broadly expressing CAG (Cytomegalovirus-IE/chicken □-actin) promoter (Figure S3A; Methods). Finally, the alignment of many ABA anatomical labels was validated, and in some cases manually corrected, based on a comparison to brain structures delineated by tissue autofluorescence or FP expression in parvalbumin-, glutamic acid decarboxylase (GAD)-, and somatostatin-specific transgenic reporters (Taniguchi et al., 2011) (Figure S3B-D).
Calculation of the sample size for c-fos-GFP-based mapping of mouse brain activation
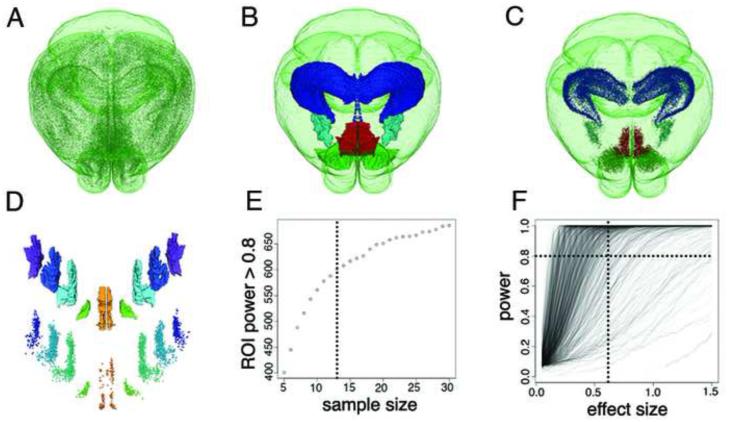
The RSTP brain allows us to calculate the number of c-fos-GFP+ cells per anatomical ABA regions in the 280-section datasets. To estimate the required sample size for statistical comparisons, we used power analysis on data from a baseline group of mice (Methods). The brains of 7 c-fos-GFP mice (no experimental manipulation) were imaged by STP tomography, warped to the RSTP brain and the c-fos-GFP+ cells were counted per each anatomical ROI. To determine the optimal sample size, Monte Carlo methods were applied to this data to simulate ROI counts for two groups at various effect sizes. As shown in Figure 2, the number (N) of sufficiently powered ROIs (α < 0.05, power > 0.80) increased at an approximately constant rate until N = 10, where it started to plateau. We chose N = 12-13 as sample size per group, which assures high statistical power for most ROIs.
Figure 2. ROI-based segmentation and sample size calculation.
(A-D) ROI-based segmentation of the whole-brain c-fos-GFP+ cell count. (A) Whole-brain view of 360,183 c-fos-GFP+ cells (same brain as in Figure 1H). (B) Examples of ABA ROI segmentation and (C) the corresponding c-fos-GFP+ cell counts: hippocampus—dark blue; 33,508 cells; medial amygdalar nucleus—light blue; 3,035 cells; nucleus accumbens—green; 13,627 cells; and infralimbic cortical area—red; 4,665 cells. (D) Further segmentation of the infralimbic region by cortical layers: top shows the layer ROIs, from layer 1 (orange) to layer 6 (purple); bottom shows the c-fos-GFP+ cell counts: ILA1 = 223, ILA2 = 243, ILA2/3 = 1,572, ILA5 = 1,731, ILA6 = 896 c-fos-GFP+ cells. The spacing between the layers was enlarged for better visualization. (E-F) Estimation of the sample size based on power analysis of c-fos-GFP+ cell counts. (E) The simulation of the relationship between the number of sufficiently powered ROIs and the sample size shows a steep increase till about N = 10, which then begins to plateau. For the current study we chose a sample size of N = 13 (dashed line). (F) The plot of the relationship between the statistical power of each ROI and the effect size for N = 13 group. Of the total 763 ROIs analyzed, 601 (78.8%) showed sufficient statistical power at the effect size 0.6 and 699 (91.6%) at the effect size 1.0.
The selection of the social behavioral protocols and characterization of c-fos-GFP induction
Interactions between a male and a female mouse, and a male and a male mouse include initial common social behaviors, such as anogenital sniffing and close following, and consequent sex-specific behaviors, such mounting and fighting. In the current study we wished to focus on the comparison of brain activation evoked during the initial social exploration-based phase of the male-female and male-male interactions, during which the male is expected to recognize the social stimulus and to discriminate the sex of the interacting partner.
The social comparison was based on two experimental groups. In the male-female interaction group an ovariectomized (OVX) conspecific female was introduced for 90 sec in the home-cage of a naïve c-fos-GFP+ male, while in the male-male interaction group a conspecific male was used as the 90 sec stimulus (Movie S2). As described before in studies of social recognition (Ferguson et al., 2001; Ferguson et al., 2000), the brief interaction period included exploratory behavioral activities of anogenital sniffing, close following, and nose-to-nose sniffing, but no sexual behavior or aggression (Figure S4). The OVX female, which was recognized by the male as a social stimulus comparable to an intact female (Figure S4), was chosen to limit experimental variability due to the estrous cycle (Ferguson et al., 2000; Winslow, 2003).
For control, we included four groups. Baseline group included mice that were not handled or otherwise manipulated. Handling group included mice that were transferred to the experimental area for 90 sec. Object group included mice that received a novel object for 90 sec. And finally olfactory group included mice that were exposed for 90 sec to a novel object enriched with banana-like odor (ISO; isoamyl acetate; note that ISO is a monomolecular odor and as such it is likely to induce simpler activation patterns compared to complex volatile odors.).
In order to characterize the time course of c-fos-GFP induction, we used the 90 sec ISO stimulation and tested c-fos-GFP increase in the main olfactory bulb at 0.5, 1.5, 3 and 5 hours post-stimulus. This protocol revealed a peak induction at 3 hours after the stimulation, which returned to the baseline level at 5 hours (Figure S5A-C). The time of 3 hours post-stimulus was selected for analysis of all behavioral experiments.
In order to compare the c-fos-GFP signal to native c-fos signal, we analyzed female interaction-driven induction in 8 selected brain regions by anti-c-fos immunohistochemistry in wild type C57BL/6 mice and by STP tomography in c-fos-GFP mice (note that the c-fos signal was analyzed at 1 hour post-stimulus because of the short half-life of the native c-fos protein). Overall, the c-fos-GFP signal represented 59 ± 6% (mean ± SEM) of anti-c-fos immuno-signal, indicating that the direct c-fos-GFP fluorescence detects approximately 60% of all c-fos induced cells (Figure S5D-I). Importantly, the female interaction-driven increase was also highly comparable between the wild type and c-fos-GFP mice (Figure S5D-I).
ROI- and voxel-based statistical analyses
The distribution of the c-fos-GFP+ cells among the different behavioral groups was compared using ROI- and voxel-based statistical tests corrected for multiple comparisons by false discovery rate (FDR) (Methods). The 694 ROIs analyzed represent the segmentation of the RSTP Brain volume by the ABA anatomical regions and the c-fos-GFP cell counts are compared ROI-to-ROI between the experimental groups (Methods). The RSTP Brain voxelization (done by □m diameter) generates discrete digitization unbiased of anatomical regions and the c-fos-GFP cell counts are compared voxel-to-voxel (Methods). The voxel-based statistics can reveal “hotspot” areas of activation and sub-regional differences within the anatomical ROIs (Figure S6).
In the first ROI analysis, the comparison of the male-female, male-male, olfactory, and handling groups to the baseline group revealed broad patterns of brain activation, with ~69%, 76%, 79%, and 35% of ROIs activated by the respective manipulations (Table S1). Since all ROIs activated in the handling group were also activated in the other three groups, the handling-induced brain activation represents non-specific shared stimuli, such as moving the cage to the experimental area. In order to determine the stimulus-specific brain activations, we next compared the male-female, male-male, and olfactory groups to the handling group by both ROI and voxel-based analysis.
Female and male interaction-evoked brain activation
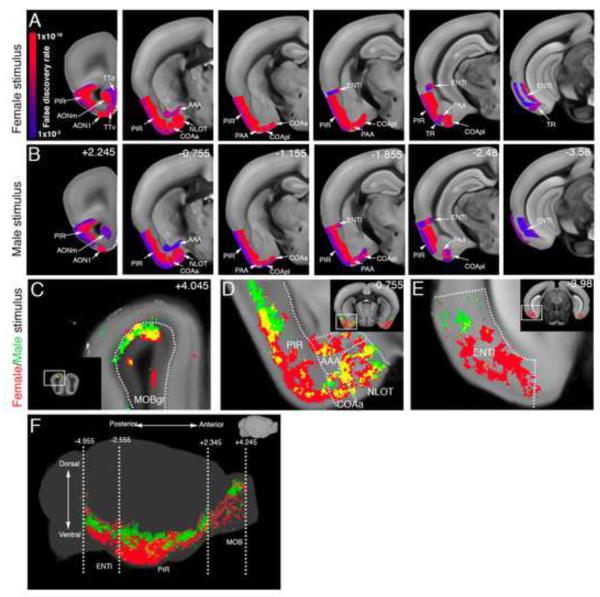
It has been proposed that the detection of volatile pheromones by the main olfactory epithelium (MOE) and MOB is necessary for sex discrimination (Baum and Kelliher, 2009) (see Discussion). However, the mechanism of such detection at the level of downstream brain structures is not known. A comparison of the female and male interaction-evoked activation by ROI statistics revealed largely overlapping c-fos-GFP induction among MOB-connected brain regions, including the anterior olfactory nucleus (AON), piriform cortex (PIR), nucleus of the lateral olfactory tract (NLOT), anterior amygdala area (AAA), piriform-amygdala area (PAA), anterior and posterior lateral cortical amygdala (COAa, COApl), and entorhinal cortex lateral (ENTl), in addition to a female specific activation of taenia tecta (TT) and postpiriform transition area (TR) (Figure 3; Table S2; note that the heat map data in Figures 3-5 show statistical significance, while the magnitude of c-fos upregulation is provided in Figure S7). A further analysis by voxel-based statistics revealed a mainly dorsal MOB activation by both stimuli and a clear dorsal-ventral separation between the two stimuli in the PIR and ENT (Figure 3C-F; It should be noted that overlapping voxel activation between the male and female datasets, seen as yellow areas in 3C-F, represents activation of the same area but not necessarily the same neurons). These data suggest that spatial organization of the dorsal MOB outputs leads to activation of distinct neuronal populations in the PIR and ENT, which may contribute to sex discrimination in the male brain.
Figure 3. Social behavior-activated areas: the MOB and its direct downstream circuitry.
(A-B) ROI analysis: The male-female (A) and male-male groups (B) are compared to the handling group and significantly activated ROIs downstream of the MOB are displayed. Most of the regions were activated by both stimuli. Heatmap in (A) represents FDR corrected statistical significance. Numbers in (B) represent bregma A/P coordinates. See Table S2 for ROI full names. (C-F) Voxel based analysis revealed activation pattern selective for the female stimulus (red), the male stimulus (green), and shared by both stimuli (yellow). (C) Both male and female stimuli induced dorsal activation in the MOB. (D-F) Dorso-ventral separation was detected between the male- and female-evoked activation in the PIR (D, F) and ENT (E, F). See also Movie S3 for full dataset.
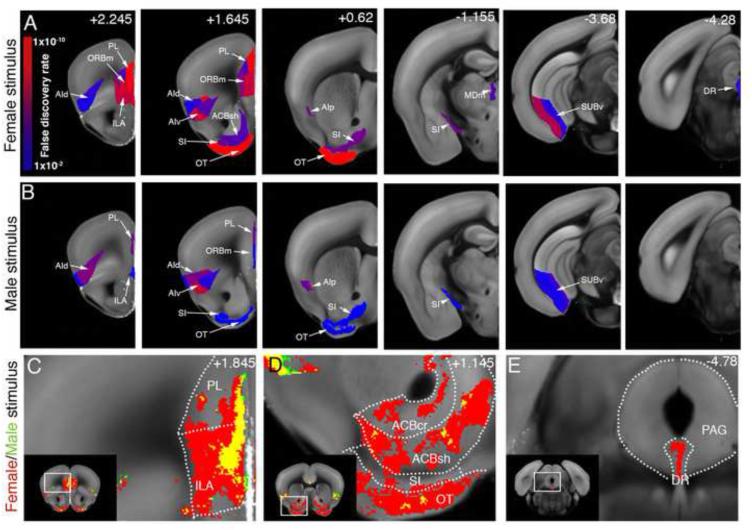
Figure 5. Social behavior-activated areas: Motivational circuitry.
(A-B) ROI analysis: The male-female (A) and male-male groups (B) are compared to the handling group and significantly activated ROIs previously implicated in behavioral motivation are displayed. The female stimulus activated frontal cortical areas (PL, ORBm, ILA, AI), ventral striatum (OT, ACB, SI), midline thalamus (MDm), ventral hippocampus (SUBv), and seretonergic DR while the male stimulus activated AI, only superficial layer of PL, ORBm, and ILA, and weakly SI, OT, and SUBv. Heatmap in (A) represents FDR corrected statistical significance. Numbers in (B) represent bregma A/P coordinates. See Table S2 for ROI full names. (C-E) Voxel analysis showed that (C) the entire ventral part of PL and dorsal half of ILA was activated by the female stimulation, while only the upper layers of the same regions were activated by male stimulation. (D) Ventral striatum (ACB, SI, OT) showed patch shaped strong activation pattern by female stimulus, but not by male stimulus. (E) Voxel analysis pinpointed the maximal activation in the DR by the female stimulus at A/P coordinate −4.78.
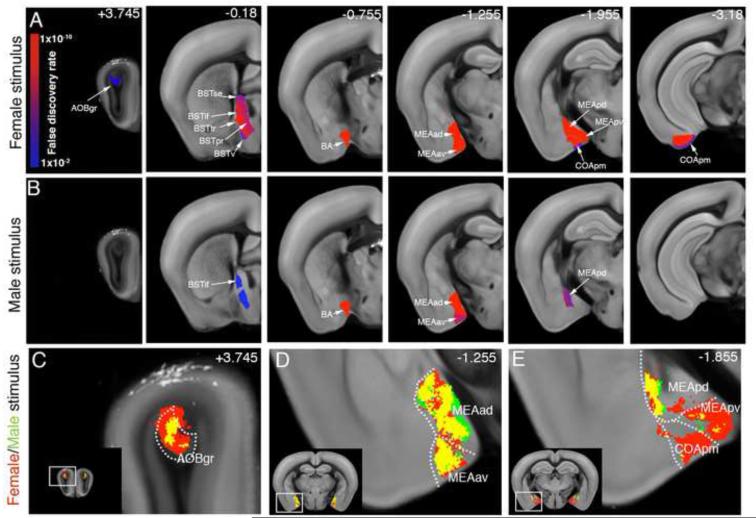
The sensing of non-volatile pheromones by the vomeronasal organ (VNO) and AOB has been proposed to play a critical role in mate recognition and behavioral motivation (Baum and Kelliher, 2009). Our analysis of brain regions downstream of the AOB revealed a strong bias toward the female interaction-evoked brain activation, including the AOB granular cell layer (AOBgr), the posterior medial cortical amygdala (COApm), the entire medial amygdala (MEA), bed nucleus of the accessory olfactory tract (BA) and bed nuclei of the stria terminalis (BST) (Figure 4; Table S2; Movie S3). In contrast, male-male interaction induced activation in fewer AOB-linked areas, including the BA and MEA anterior dorsal (ad), anterior ventral (av), and posterior dorsal (pd) (Figure 4; Table S2). Voxel analysis revealed focal activation in the AOB in the male-male interaction (Figure 4C) and a largely overlapping activation in the MEAad, av, and pd in the male-female and male-male datasets (Figure 4D-E; Table S2; Movie S3).
Figure 4. Social behavior-activated areas: the AOB and its direct downstream circuitry.
(A-B) ROI analysis: The male-female (A) and male-male groups (B) are compared to the handling group and significantly activated ROIs downstream of the AOB are displayed. The female stimulus activated all AOB downstream regions, while the male stimulus induced only a partial activation of these areas. Heatmap in (A) represents FDR corrected statistical significance. Numbers in (B) represent bregma A/P coordinates. See Table S2 for ROI full names. (C-E) Voxel based analysis revealed a largely overlapping activation pattern (yellow) in the co-activated AOB (C), MEAad and MEAav (D), and MEApd (E), and selective female-evoked activation in the MEApv and COApm (E). See also Movie S3 for full dataset.
The male-female interaction also showed strongly evoked activation of brain areas linked to behavioral motivation, including the olfactory tubercle (OT) and nucleus accumbens shell (ACBsh) of the ventral striatum, prelimbic, infralimbic, and orbital medial (PL, ILA, and ORBm) prefrontal cortical areas, agranular insular cortex (AI), substantia innominata (SI; also known as ventral pallidum), medial dorsal thalamus (MDm), hippocampal ventral subiculum (SUBv), and the serotonergic dorsal raphe (DR) (Figure 5A; Table S2). In contrast, the male-male interaction had a comparable induction only in the AI, much weaker activation in the prefrontal cortices, SI, OT, and SUBv, and no significant activation in the MDm and DR (Figure 5B; Table S2). Voxel based analysis revealed that activation in the medial prefrontal cortices in the male-male interaction was limited to superficial cortical layers (Figure 5C; Movie S3). We also observed a focal activation in the DR at a specific A/P bregma location in the male-female dataset (Figure 5E; Movie S3).
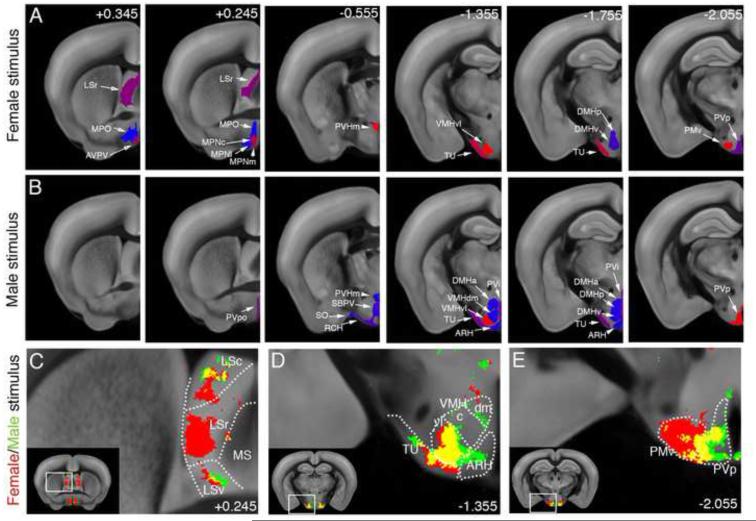
The activation of the septal and hypothalamic nuclei is known to mediate both sexual and defensive/aggressive behaviors (Anderson, 2012; Swanson, 2000). We therefore asked whether the brief interaction used in our experiments was sufficient to activate these regions even though it lacked overt mating and fighting. The ROI analysis revealed that the female stimulus induced activation of the rostral lateral septum (LSr) and neuroendocrine nuclei, including the medial preoptic nucleus (MPN), medial preoptic area (MPO), ventral premammillary nucleus (PMv), ventrolateral part of the ventromedial nucleus (VMHvl), paraventricular hypothalamic nucleus (PVH), dorsomedial hypothalamus (DMH), anteroventral periventricular nucleus (AVPV), posterior periventricular hypothalamic nucleus (PVp), and tuberal nucleus (TU) (Figure 6A; Table S2). The male stimulus activated the VMHvl, DMH, PVH, PVp, and TU from the structures of the male-female dataset, in addition to a male specific activation of the dorsomedial part of the ventromedial nucleus (VMHdm), the anterior, preoptic, and intermediate periventricular nuclei (PVa, PVpo, PVi), retrochiasmatic area (RCH), subparaventricular zone (SBPV), supraoptic nucleus (SO), and arcuate nucleus (ARH) (Figure 6B; Figure S7; Table S2). Voxel based analysis revealed very distinct and focal LSr activation at A/P coordinates between +0.345 and −0.145 (Figure 6C; Movie S3). The activation in the VMHvl, which was previously shown to play a role in both sexual and aggressive behaviors (Lin et al., 2011), was highly overlapping between the male-female and male-male datasets (Figure 6D; Movie S3). In addition, only a medial part of the PMv was activated in the male-male dataset, suggesting a functional subdivision within this structure (Figure 6E; Movie S3).
Figure 6. Social behavior-activated areas: Septal and Hypothalamic activation.
(A-B) ROI analysis: The male-female (A) and male-male groups (B) are compared to the handling group and significantly activated ROIs of the septum and hypothalamus are displayed. The female stimulus activated the rostral lateral septum (LSr), AVPV, medial preoptic area (MPO, MPN), PVH, TU, VMHvl, posterior and ventral DMH, PMv, and PVp. The male stimulus also activated the PVH, VMHvl, DMH (anterior part), TU, and PVp, in addition to a selective activation of the periventricular hypothalamic nuclei (PVpo, PVi), SBPV, RCH, SO, ARH, and VMHdm. Heatmap in (A) represents FDR corrected statistical significance. Numbers in (B) represent bregma A/P coordinates. See Table S2 for ROI full names. (C-D) Voxel analysis; (C) A distinct voxel activation was observed in the LSr only by female stimulation. (D) VMHvl showed largely overlapping activation by both stimuli, while VMHdm and ARH showed activation only by the male stimulus. (E) PMv is highly activated by the female stimulus, while the medial part of PMv was also activated by the male stimulus. See also Movie S3 for full dataset.
Finally, among additional brain areas the claustrum (CLA), basomedial amygdala (BMA), and intercalated amygdala (IA) were activated by both the female and male interactions, capsular central amygdala (CEAc), basolateral amygdala (BLA), and the thalamic parataenial nucleus (PT) were activated only in response to the female stimulus, and the temporal associational, perirhinal, and ectorhinal (TEa, PERI, and ECT) cortical areas were activated only in response to the male stimulus (Table S2). The activation of the hippocampal CA2 region linked to social memory (Hitti and Siegelbaum, 2014) was also detected in both the male-female and male-male datasets (Table S2).
Social behavior-specific brain activation
In addition to the male versus female comparison described above, we also asked which of the activated brain regions are specific to social behavior, i.e. are shared between the male-female and male-male datasets and are not activated in response to a non-social stimulus represented by a novel object enriched with a volatile odor (banana-like isoamyl acetate; ISO).
First, we compared the ISO dataset to the handling control. This analysis revealed the expected activation of the PIR and other areas downstream of the MOB, which was similar to the social behavior-evoked activation (Table S2). The activation throughout the rest of the brain, however, was highly divergent from the pattern evoked by the social stimuli, as it included many cortical areas, the entire hippocampus, and the hypothalamic subfornical organ (SFO) regulating autonomic functions (Smith and Ferguson, 2010), suprachiasmatic nucleus (SCH) regulating sleep, waking and locomotor activity (Saper et al., 2005), and the arcuate nucleus (ARH) linked to feeding (Sternson, 2013) (Table S2).
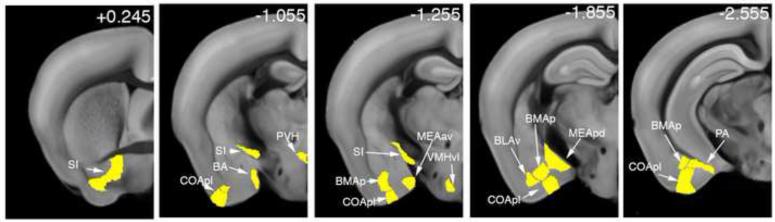
Second, we compared the shared male-female and male-male brain activation to the ISO dataset. This analysis revealed the subset of areas specific to social behavior, which included the amygdalar regions BA, COApl, MEAav, MEApd, BMAp, BLAv, and PA, the hypothalamic VMHvl and PVH, and the SI (Figure 7; Table S3).
Figure 7. Social behavior-specific brain areas.
Brain areas activated by both female and male stimulus, but not by ISO stimulation, are displayed as ROIs. Unique social behavior-activated areas included the amygdalar BA, COApl, MEAav, MEApd, BLAv, BMAp, PA, hypothalamic PVH, VMHvl, and ventral pallidum (SI).
Correlation of c-fos activation to time spent in social behaviors
The time spent in a specific behavioral activity may be expected to correlate to the number of c-fos-GFP+ cells in brain regions driving this activity. We next tested whether this correlation may be used to functionally link the activated brain areas in the male-female and male-male datasets to specific features of the social behavior.
The correlation to the time spent in anogenital sniffing identified mainly areas connected to volatile and non-volatile olfactory signaling, such as the COAa, COApl, COApm, MEA, and BST, and hypothalamic neuroendocrine areas including the MPN, PMv, and VMHvl (Table 1). Correlation to the time spent in close following identified some of the same areas, such as the MEA and BST, but also areas linked to behavioral motivation, including the ACB, OT, SI, ILA, PL, ORBm, MDm, and DR (Table 1). Finally, the correlation to the time spent in nose-to-nose sniffing did not identify any positive association, suggesting that this behavioral feature is not quantitatively linked to any brain regions in our datasets. These data suggest that distinct aspects of the social behavior engage distinct sets of brain areas and that whole-brain cellular c-fos-GFP analysis is able to reveal this structure-function relationship.
Table 1.
c-fos-GFP count to the social behavioral correlation
| ROIs | Anogentical sniffing |
close following | |
|---|---|---|---|
| ILA | + | Isocortex | |
| PL | + | ++ | |
| ORBm | + | ++ | |
| TT | ++ | ||
| DP | ++ | Olfactory Area |
|
| AOBgr | ++ | ||
| COAa | + | ||
| COApl | +++ | + | |
| COApm | +++ | + | |
| PAA | +++ | + | |
| TR | + | ||
| ENTmv | ++ | Hippocampal Formation |
|
| CA3 | + | ||
| EP | + | + | Cortical Subplate |
| BLAa | ++ | + | |
| BLAp | + | ||
| BLAv | ++ | ||
| BMAa | ++ | ||
| BMAp | +++ | ||
| PA | +++ | ||
| ACBsh | + | Cerebral Nuclei |
|
| OT | + | ++ | |
| AAA | + | ||
| LSr | + | ||
| CEAc | + | + | |
| IA | ++ | ||
| MEAad | +++ | ||
| MEAav | +++ | ||
| MEApd | +++ | + | |
| MEApv | +++ | + | |
| SI | + | ||
| BSTmg | + | ++ | |
| BSTv | + | ++ | |
| BSTp | ++ | ||
| MDm | + | Thalamus | |
| PT | + | ||
| AVPV | ++ | + | Hypothalamus |
| MPN | ++ | + | |
| PMv | +++ | + | |
| VMHvl | + | ||
| TU | ++ | ||
| DR | + | Midbrain |
Calculation of the density of c-fos-GFP+ cells per ROIs
While the above analyses identified the activated brain areas, the cellular resolution of our data allowed us to also estimate the total numbers and the densities of c-fos-GFP+ cells per anatomical ROIs. Since the z planes in the 280-section datasets are spaced 50 μm apart, we transformed the serial 2D data into 3D whole-brain estimates using a stereological method (Williams and Rakic, 1988) applied to a high-resolution 5,600-section dataset with z spacing of 2.5 μm (Methods). The obtained 2D-to-3D conversion factor of 2.5 was then used to multiply the 2D ROI counts in order to estimate the total numbers of c-fos-GFP+ cells, and the total counts were divided by the ROI volumes in order to estimate the densities of c-fos-GFP+ cells per activated ROIs (Figure S7).
The average cell density in the structures significantly activated in the female and male datasets were, respectively, 4993 ± 400 and 4519 ± 283 per cubic mm (mean ± SEM), whereas the average density in these structures in the handling control was 3127 ± 201 per cubic mm (Figure S7). Therefore the social interactions evoked on average ~1,500 to 2,000 c-fos-GFP+ cells per cubic mm compared to the handling control, suggesting a sparse activation of a few percent of neurons per brain areas (see Discussion).
DISCUSSION
While the general organization of the brain structures regulating sexual and aggressive behavior is beginning to be understood (Anderson, 2012; Sokolowski and Corbin, 2012), much remains unknown about how information is processed from the sensory periphery (the olfactory system in rodents) to give rise to sex-specific behavioral responses. Here utilizing a pipeline of computational methods, including ROI-based whole-brain mapping of c-fos activation, voxel-based mapping of sub-regional differences in c-fos activation, and correlation analysis linking ROI activation to behavior, we compared brief female interaction-evoked activation in the brain of a male mouse to the activation evoked by brief interaction with a male. Some more salient findings from our analyses are discussed below following the method discussion, while the complete ROI- and voxel-based results are provided as a resource in Table S1-S3 and Movie S3.
The method pipeline for c-fos-GFP-based mouse brain screening
The entire method pipeline is automated, highly standardized and operates at a reasonably high-throughput: the imaging time per one brain is ~21 hours, while the imaged processing and computational analyses take ~24 hours that occurs in parallel with the STP imaging (Ragan et al., 2012; Vousden et al., 2014).
The first key part of the computational pipeline is the detection of c-fos-GFP+ cells in the STP datasets. We chose to use CNs, because these algorithms rely on the learning procedure to account for signal to noise ratio (SNR) variability and improved performance is achieved by simply increasing the training dataset (Jain et al., 2007; Turaga et al., 2010). The trained CN performance (F-score 0.88) was in fact close to human expert performance (F-score 0.9), demonstrating the power of this approach for analysis of fluorescent labeling in STP tomography-imaged mouse brains. We have also tested two other cell detection methods—cell counting in the Volocity Image analysis software (Perkin Elmer) and cell counting based on watershed algorithm (Kopec et al., 2011)—but these were considerable less reliable (F score < 0.5) compared to the CN-based detection.
The second critical step of the method is the registration of the datasets to the RSTP Brain and the Allen Mouse Brain Atlas. Fixation-induced tissue autofluorescence provides rich image content for the registration by the warping algorithm Elastix (Mattes et al., 2003). As a result, we were able to achieve a high level of precision (~60 μm jitter) for the registration of the experimental datasets to the RSTP brain (Figure S2). The alignment of the ABA Nissl-stained sections to the RSTP brain was further helped by the use of the transgenic CAG-Keima brain with a cellular FP labeling that matched in most brain regions the cellular Nissl signal, and by several interneuron-specific reporter mice (Taniguchi et al., 2011) that helped to validate and improve the matching of the labels to specific brain nuclei (Figure S3). Consequently, the precision ABA labels became closely aligned to the RSTP brain, as judged based on brain landmarks, such as the corpus callosum, hippocampal pyramidal layers, and many structural borders visible in the autofluorescence signal (Figure S3).
The last part of the method pipeline includes statistical analyses of the brainwide c-fos-GFP+ cell counts. Since it was first established in rat models of seizure, the inducibility of c-fos has been utilized to map neuronal activation in many behavioral and pharmacological experiments, demonstrating that c-fos can be used as an activity reporter in most if not all areas of the brain (Dragunow and Robertson, 1987; Morgan et al., 1987). The ROI- and voxel-based statistical analyses established here transform the traditional laborious immunostaining or in situ hybridization based c-fos mapping into an automated whole-brain assay.
In addition to the current application, these methods can also be used to detect and quantify other fluorescent protein-expressing transgenic mouse brains by simply training new CN on a different ground-truth data. This makes our pipeline easily adaptable to many other applications in quantitative whole-brain mapping, such as the generation of whole-brain cell counts in cell type-specific GFP reporter mice (Taniguchi et al., 2011).
Female- and male-evoked maps of whole-brain activation in the male brain
By focusing on the initial period of social exploratory behaviors between a naïve male and a novel conspecific female or male mouse, we set out to determine the brain activation patterns that underlie social recognition and sex discrimination in the male brain. Our results revealed that while the brief interactions led to an activation of the expected sex-specific response at the hypothalamic level (indicating that the behaviors were sufficient for correct sex discrimination), the upstream patterns of brain activation strongly diverged between the two stimuli.
At the level of the AOB and MOB signaling, the female stimulus evoked activation of all downstream connected brain structures, while the male stimulus showed activation of all MOB-linked structures but only a subset of the AOB-linked structures. The strong MOB-driven brain activation in both behaviors agrees with the role of volatile signaling in sex discrimination proposed by studies using chemical lesion of the MOE (Keller et al., 2006) or genetic disruption of cellular signaling in the MOE (Mandiyan et al., 2005). The finding that the male and female stimuli activate different parts of the PIR and ENT areas suggests that topologically distinct MOB cortical outputs may discriminate the sex-specific stimuli. This dorso-ventral separation is an example of a novel spatial organization in the piriform cortex, which until now has been considered to lack gross sensory input-based topology (Ghosh et al., 2011; Sosulski et al., 2011).
The role of the VNO and AOB-driven activation in social behaviors appears to be less clear than that of the MOE/MOB signaling. Lesioning of the VNO failed to affect sex discrimination in male mice (Pankevich et al., 2004), even though it did impair vocalization after nasal contact with female urine (Bean, 1982), while genetic disruption of VNO signaling caused male-male mounting instead of aggressive behavior without affecting male-female behavior (Stowers et al., 2002). Our data point to a more prominent role of the AOB-connected brain structures in the male-female interaction, as MEA, BST, BA and COApm were all activated in the male-female dataset, but only MEA and BA were activated in the male-male dataset. The selective BST activation in the male-female dataset included the posterior division nuclei (principal, interfascicular, and transverse) proposed to function in reproductive behaviors (Dong and Swanson, 2004), and the magnocellular nucleus of the anterior division proposed to control neuroendocrine functions and pelvic functions, including penile erection (Dong and Swanson, 2006).
The female, but not the male, stimulus also evoked activation of brain areas of the striato-pallido-thalamo-cortical circuit known to positively regulate behavioral motivation (Ikemoto, 2007; Sesack and Grace, 2010), including the ventral striatum (OT, ACB), ventral pallidum (SI), thalamus (MDm), and prefrontal cortex (ILA, PL, ORB). While we did not detect activation of the dopaminergic neurons of the ventral tegmental area (VTA), which are known to reinforce ACB functions within this circuit during sexual behavior (Ikemoto, 2007; Sesack and Grace, 2010), we did detect activation of the serotonergic DR, which was recently shown to be necessary for ACB functions in social reward (Dolen et al., 2013); The switch between the DR and VTA modulation of ventral striatum may contribute to a transition between exploratory and consummatory male-female behavior.
The analysis of the hypothalamic brain areas revealed activation of structures regulating sexual and aggressive behaviors (Anderson, 2012; Swanson, 2000): the MPN and PMv regulating male reproductive behavior (Simerly, 2002; Yang and Shah, 2014) were selectively activated in the male-female dataset, the VMHvl regulating both male sexual behavior and aggression (Anderson, 2012; Lin et al., 2011; Yang et al., 2013) was activated in response to both female and male stimuli, and the VMHdm regulating male defensive behaviors (Lin et al., 2011; Sokolowski and Corbin, 2012) was activated only in the male-male dataset. Since the brief social interactions did not comprise mating or aggression, these data show that the activation of the hypothalamic nuclei can precede the manifestation of these behaviors as part of the male-female and male-male social exploration-based behaviors.
The quantification of the whole-brain activation maps at cellular level
The cellular resolution of our data also allowed us to search for correlations between behavioral activity and brain activation, and to estimate the density of activated cells per brain area.
The correlation between behavior and c-fos activation can be expected to identify the most behaviorally relevant brain regions in which the number of c-fos activated cells reflects the behavioral performance in individual animals. In agreement with this hypothesis, regions correlated to the time spent in anogenital sniffing included mainly amygdalar and hypothalamic areas of the vomeronasal sensory-motor system transforming the chemosensory information into sexual or aggressive behavior (Swanson, 2000), while the brain areas correlated to the time spent in following included the structures linked to behavioral motivation and described above as part of the striato-pallido-thalamo-cortical circuit.
The correlation analysis can be used to add functional significance to activated regions which were not previously known to be involved in social behaviors. For example, the activation of the amygdalar IA and CEAc nuclei was correlated to the anogenital sniffing time, while the PT thalamus activation was correlated to following. Since both IA and CEAc can inhibit the medial central amygdala (CEAm), which is the output fear pathway (Pitkanen et al., 1997), these data suggest that the IA and CEAc are activated by chemosensory cues and may act to modulate fear behaviors during social exploration. The PT, a part of the dorsal group of thalamic nuclei, projects to the ACB (Kelley and Stinus, 1984) and may play a role in motivational modulation of the male-female social behavior.
Finally, the quantification of the numbers of c-fos-GFP+ cells per brain area can provide information about the approximate percentage of neurons behaviorally recruited in the identified brain areas. For example, we observed on average two-fold increase (~3,600 c-fos-GFP+ cells per cubic mm) in the prefrontal cortical areas in the male-female datasets, compared to the handling control (Figure S7). Since neuronal density in the mouse cortex is estimated at ~80,000 to 100,000 per cubic mm (Herculano-Houzel et al., 2006; Keller and Carlson, 1999; Meyer et al., 2010), these data suggest less than 5% of neurons is recruited in response to the female social stimulus. Further, as most brain areas showed similar c-fos-GFP+ densities, the behavioral recruitment of a few percent of neurons is likely a general feature of c-fos activation. This may represent c-fos induction occurring only in the most strongly activated cells and such sparse c-fos induction may be relevant for the proposed sparse coding of sensory inputs (Olshausen and Field, 2004).
Caveats of the current study
There are several caveats associated with our study. First, while the behavior is limited to 90 sec, our assay cannot determine whether the observed c-fos-GFP induction occurred entirely during this brief time period or whether some downstream activation occurred during a longer time interval. Second, the behavioral paradigm includes both the introduction and removal of the stimulus animal from the home cage of the c-fos-GFP male and some of the observed activation pattern thus may reflect stress induced by these manipulations. Third, the use of the OVX females in our study restricts the interpretation of the male-female activation data to social exploration that lacks the effects of estrous hormones. Thus male-female interaction with, for example, estradiol-induced OVX mice may be expected to induce brain activation partially distinct from the one described in the current study. Fourth, since the c-fos-GFP reporter labels ~60% of all c-fos+ cells detected by immunostaining, it is possible that some areas with native c-fos activation were missed in our assay. Finally, fifth, c-fos is a member of a family of IEGs regulated by neuronal activity, and the detection of other IEGs, such as Arc, homer-1A or zif-268, can be expected to reveal partially overlapping activation maps compared to the c-fos-GFP map identified in our paper. Because neuronal activation in some brain areas may induce other IEGs but fail to induce c-fos, the c-fos-GFP-based network of brain areas described here should not be interpreted as a complete brain activation map evoked by social behavior.
Conclusions
Our method of c-fos-GFP-based screening generates cellular-resolution maps of behaviorally evoked whole-brain activation in the mouse. The patterns of female and male interaction-evoked brain activation revealed clear separation between the two stimuli, including at the level of brain structures downstream of both volatile and non-volatile chemosensory signaling. These activation patterns were also markedly different from the activation pattern evoked during non-social olfactory-enhanced exploratory behavior. These findings demonstrate that our method can be used for screening behavior-evoked whole-brain activation, and we envision that future experiments will yield brain map-like descriptions for other innate behaviors, such as aggression and defensive behaviors, or cognitive behaviors, such as attention and decision making. Further, the same method can be applied to genetic mouse models of neurodevelopmental disorders with the aim of identifying circuit deficits underlying changes in social, cognitive and other higher order brain functions.
METHODS
Animals
Animal procedures were approved by the Cold Spring Harbor Laboratory Animal Care and Use Committee. The c-fos-GFP mice, Tg(Fos-tTA,Fos-EGFP) line, were obtained from the Jackson Laboratory. In our study we used the direct c-fos-GFP signal, whereas several other studies used the tTA protein to drive other reporter molecules (Garner et al., 2012; Liu et al., 2012; Matsuo et al., 2008; Reijmers et al., 2007).
Behavioral tests and cfos-GFP induction time course
Heterozygous c-fos-GFP male mice (8 – 11 weeks old mice) were individually housed for one week before the test. The behavioral stimuli were: transfer of the animal to the experimental arena (handling control) or plus introduction of an OVX conspecific female (male-female group), conspecific male (male-male group), 50 ml falcon tube (object group), and 50 ml falcon tube with a side-opening in which was cotton ball with isoamyl acetate (1:100 in mineral oil, 40 μl per experiment, freshly made each day). The stimulus was placed in the home cage for 90 seconds and then removed. The behavior was video-recorded and was manually scored. After the behavioral stimulus was removed, the mice remained in the home cage for additional 3 hrs and then killed by transcardial perfusion. For the time course of c-fos-GFP induction, isoamylacetate was introduced into the mouse home cage for a brief period of 90 sec. The mice were killed at selected time points of 0.5, 1.5, 3 and 5 hours post stimulation.
Brain preparation, STP tomography imaging and data processing
The brains were prepared as described in our previous study (Ragan et al., 2012). Briefly, The brains were embedded in oxidized 4% agarose, crosslinked, and imaged as 280 serial sections. The raw image tiles were corrected for illumination and stitched in 2D in matlab and aligned in 3D in Fiji (Ragan et al., 2012). The CNs for detection of c-fos-GFP+ cells was trained based on ground truth data marked up by an expert biologist. The CN performance was scored based on the F-score; F score = the harmonic mean of the precision and recall. Stereological procedure was used to calculate how CNs 2D based counting can be converted into 3D counting to calculate the densities of c-fos-GFP+ cells per activated ROIs. 3D registration methods using Elastix were the same as described (Ragan et al., 2012), but with modified parameters. See Supplementary Methods for more details.
c-fos immunohistochemistry and comparison to c-fos-GFP+ cell counting
Wild type C57BL/6 mice (8 to 10 week old) underwent the same behaviors as the c-fos-GFP mice of the male-to-female and handling groups. The mice were killed and perfused 1 hour later and the brains were fixed overnight in 4% PFA, then cut as 50 micron coronal sections. For immunohistochemistry, sections were exposed to rabbit anti-c-fos antibody (1:10000, Santa Cruz SC052) and labeled by DAB solution. FIJI (imageJ) and Volocity (Perkin-Elmer) were used for cell counting.
Statistics
We ran statistical comparisons between different behavioral groups based on either ROIs or evenly spaced voxels. Voxels were overlapping 3D spheres with 100 μm diameter each and spaced 20 μm apart from each other. The cell count of each voxel was calculated as the number of nuclei found within 100 μm from the center of the voxel in all 3D. To account for multiple comparisons across all voxel/ROI locations, we thresholded the p-values and reported false discovery rates. For correlation between c-fos-GFP cell counts and social behavior, Pearson correlation R values were calculated between c-fos-GFP cell counts and time spent in social behaviors. See Supplementary Methods for more details.
Supplementary Material
ACKNOWLEDGMENTS
We thank G. Fitzgerald, R. Palaniswamy, and J. Zambratto for expert technical assistance, J. Kuhl for comments and graphics work, W. Denk and K. Baldwin for comments on the manuscript, and members of the Osten lab for helpful discussions. PO is supported by the National Institute of Mental Health grant 1R01MH096946-01, Simons Foundation for Autism Research grants 204719 and 253447, and CSHL; YK is supported by NARSAD Young Investigator fellowship; MH is supported by the Allen Institute for Brain Science; HSS is supported by Howard Hughes Medical Institute and Gatsby Charitable Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anderson DJ. Optogenetics, sex, and violence in the brain: implications for psychiatry. Biological psychiatry. 2012;71:1081–1089. doi: 10.1016/j.biopsych.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum MJ, Kelliher KR. Complementary roles of the main and accessory olfactory systems in mammalian mate recognition. Annu Rev Physiol. 2009;71:141–160. doi: 10.1146/annurev.physiol.010908.163137. [DOI] [PubMed] [Google Scholar]
- Bean NJ. Olfactory and vomeronasal mediation of ultrasonic vocalizations in male mice. Physiology & behavior. 1982;28:31–37. doi: 10.1016/0031-9384(82)90097-x. [DOI] [PubMed] [Google Scholar]
- Bialy M, Kaczmarek L. c-Fos expression as a tool to search for the neurobiological base of the sexual behaviour of males. Acta neurobiologiae experimentalis. 1996;56:567–577. doi: 10.55782/ane-1996-1162. [DOI] [PubMed] [Google Scholar]
- Brennan PA, Zufall F. Pheromonal communication in vertebrates. Nature. 2006;444:308–315. doi: 10.1038/nature05404. [DOI] [PubMed] [Google Scholar]
- Buzsaki G. Large-scale recording of neuronal ensembles. Nature neuroscience. 2004;7:446–451. doi: 10.1038/nn1233. [DOI] [PubMed] [Google Scholar]
- Clayton DF. The genomic action potential. Neurobiology of learning and memory. 2000;74:185–216. doi: 10.1006/nlme.2000.3967. [DOI] [PubMed] [Google Scholar]
- Coolen LM, Peters HJ, Veening JG. Fos immunoreactivity in the rat brain following consummatory elements of sexual behavior: a sex comparison. Brain research. 1996;738:67–82. doi: 10.1016/0006-8993(96)00763-9. [DOI] [PubMed] [Google Scholar]
- Dolen G, Darvishzadeh A, Huang KW, Malenka RC. Social reward requires coordinated activity of nucleus accumbens oxytocin and serotonin. Nature. 2013;501:179–184. doi: 10.1038/nature12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong HW, Swanson LW. Projections from bed nuclei of the stria terminalis, posterior division: implications for cerebral hemisphere regulation of defensive and reproductive behaviors. The Journal of comparative neurology. 2004;471:396–433. doi: 10.1002/cne.20002. [DOI] [PubMed] [Google Scholar]
- Dong HW, Swanson LW. Projections from bed nuclei of the stria terminalis, magnocellular nucleus: implications for cerebral hemisphere regulation of micturition, defecation, and penile erection. The Journal of comparative neurology. 2006;494:108–141. doi: 10.1002/cne.20789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragunow M, Robertson HA. Kindling stimulation induces c-fos protein(s) in granule cells of the rat dentate gyrus. Nature. 1987;329:441–442. doi: 10.1038/329441a0. [DOI] [PubMed] [Google Scholar]
- Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annual review of neuroscience. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson JN, Aldag JM, Insel TR, Young LJ. Oxytocin in the medial amygdala is essential for social recognition in the mouse. J Neurosci. 2001;21:8278–8285. doi: 10.1523/JNEUROSCI.21-20-08278.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson JN, Young LJ, Hearn EF, Matzuk MM, Insel TR, Winslow JT. Social amnesia in mice lacking the oxytocin gene. Nature genetics. 2000;25:284–288. doi: 10.1038/77040. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Larson SD, Hefzi H, Marnoy Z, Cutforth T, Dokka K, Baldwin KK. Sensory maps in the olfactory cortex defined by long-range viral tracing of single neurons. Nature. 2011;472:217–220. doi: 10.1038/nature09945. [DOI] [PubMed] [Google Scholar]
- Grewe BF, Helmchen F. Optical probing of neuronal ensemble activity. Current opinion in neurobiology. 2009;19:520–529. doi: 10.1016/j.conb.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Timlin JA, Roysam B, McNaughton BL, Worley PF, Barnes CA. Mapping behaviorally relevant neural circuits with immediate-early gene expression. Current opinion in neurobiology. 2005;15:599–606. doi: 10.1016/j.conb.2005.08.018. [DOI] [PubMed] [Google Scholar]
- Herculano-Houzel S, Mota B, Lent R. Cellular scaling rules for rodent brains. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12138–12143. doi: 10.1073/pnas.0604911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitti FL, Siegelbaum SA. The hippocampal CA2 region is essential for social memory. Nature. 2014;508:88–92. doi: 10.1038/nature13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemoto S. Dopamine reward circuitry: two projection systems from the ventral midbrain to the nucleus accumbens-olfactory tubercle complex. Brain research reviews. 2007;56:27–78. doi: 10.1016/j.brainresrev.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain V, Murray J, Roth F, Turaga S, Zhigulin V, Briggman K, Helmstaedter M, Denk W, Seung HS. Supervised Learning of Image Restoration with Convolutional Networks. Paper presented at: Computer Vision, 2007 ICCV 2007 IEEE 11th International Conference on (IEEE).2007. [Google Scholar]
- Keller A, Carlson GC. Neonatal whisker clipping alters intracortical, but not thalamocortical projections, in rat barrel cortex. The Journal of comparative neurology. 1999;412:83–94. doi: 10.1002/(sici)1096-9861(19990913)412:1<83::aid-cne6>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Keller M, Douhard Q, Baum MJ, Bakker J. Sexual experience does not compensate for the disruptive effects of zinc sulfate--lesioning of the main olfactory epithelium on sexual behavior in male mice. Chem Senses. 2006;31:753–762. doi: 10.1093/chemse/bjl018. [DOI] [PubMed] [Google Scholar]
- Kelley AE, Stinus L. The distribution of the projection from the parataenial nucleus of the thalamus to the nucleus accumbens in the rat: an autoradiographic study. Experimental brain research Experimentelle Hirnforschung. 1984;54:499–512. doi: 10.1007/BF00235475. [DOI] [PubMed] [Google Scholar]
- Kopec CD, Bowers AC, Pai S, Brody CD. Semi-automated atlas-based analysis of brain histological sections. Journal of neuroscience methods. 2011;196:12–19. doi: 10.1016/j.jneumeth.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HM, Giguere PM, Roth BL. DREADDs: novel tools for drug discovery and development. Drug discovery today. 2014;19:469–473. doi: 10.1016/j.drudis.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Boyle MP, Dollar P, Lee H, Lein ES, Perona P, Anderson DJ. Functional identification of an aggression locus in the mouse hypothalamus. Nature. 2011;470:221–226. doi: 10.1038/nature09736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandiyan VS, Coats JK, Shah NM. Deficits in sexual and aggressive behaviors in Cnga2 mutant mice. Nature neuroscience. 2005;8:1660–1662. doi: 10.1038/nn1589. [DOI] [PubMed] [Google Scholar]
- Mattes D, Haynor DR, Vesselle H, Lewellen TK, Eubank W. PET-CT image registration in the chest using free-form deformations. IEEE Transactions in Medical Imaging. 2003;22:120–128. doi: 10.1109/TMI.2003.809072. [DOI] [PubMed] [Google Scholar]
- Meyer HS, Wimmer VC, Oberlaender M, de Kock CP, Sakmann B, Helmstaedter M. Number and laminar distribution of neurons in a thalamocortical projection column of rat vibrissal cortex. Cereb Cortex. 2010;20:2277–2286. doi: 10.1093/cercor/bhq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JI, Cohen DR, Hempstead JL, Curran T. Mapping patterns of c-fos expression in the central nervous system after seizure. Science. 1987;237:192–197. doi: 10.1126/science.3037702. [DOI] [PubMed] [Google Scholar]
- Olshausen BA, Field DJ. Sparse coding of sensory inputs. Current opinion in neurobiology. 2004;14:481–487. doi: 10.1016/j.conb.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Pankevich DE, Baum MJ, Cherry JA. Olfactory sex discrimination persists, whereas the preference for urinary odorants from estrous females disappears in male mice after vomeronasal organ removal. J Neurosci. 2004;24:9451–9457. doi: 10.1523/JNEUROSCI.2376-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaus JG, Heeb MM. Implications of immediate-early gene induction in the brain following sexual stimulation of female and male rodents. Brain research bulletin. 1997;44:397–407. doi: 10.1016/s0361-9230(97)00219-0. [DOI] [PubMed] [Google Scholar]
- Pitkanen A, Savander V, LeDoux JE. Organization of intra-amygdaloid circuitries in the rat: an emerging framework for understanding functions of the amygdala. Trends in neurosciences. 1997;20:517–523. doi: 10.1016/s0166-2236(97)01125-9. [DOI] [PubMed] [Google Scholar]
- Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J, Arganda-Carreras I, Kim Y, Seung HS, Osten P. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nature methods. 2012;9:255–258. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijmers LG, Perkins BL, Matsuo N, Mayford M. Localization of a stable neural correlate of associative memory. Science. 2007;317:1230–1233. doi: 10.1126/science.1143839. [DOI] [PubMed] [Google Scholar]
- Saper CB, Lu J, Chou TC, Gooley J. The hypothalamic integrator for circadian rhythms. Trends in neurosciences. 2005;28:152–157. doi: 10.1016/j.tins.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Sesack SR, Grace AA. Cortico-Basal Ganglia reward network: microcircuitry. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2010;35:27–47. doi: 10.1038/npp.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simerly RB. Wired for reproduction: organization and development of sexually dimorphic circuits in the mammalian forebrain. Annual review of neuroscience. 2002;25:507–536. doi: 10.1146/annurev.neuro.25.112701.142745. [DOI] [PubMed] [Google Scholar]
- Smith PM, Ferguson AV. Circulating signals as critical regulators of autonomic state--central roles for the subfornical organ. American journal of physiology. 2010;299:R405–415. doi: 10.1152/ajpregu.00103.2010. [DOI] [PubMed] [Google Scholar]
- Sokolowski K, Corbin JG. Wired for behaviors: from development to function of innate limbic system circuitry. Front Mol Neurosci. 2012;5:55. doi: 10.3389/fnmol.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosulski DL, Bloom ML, Cutforth T, Axel R, Datta SR. Distinct representations of olfactory information in different cortical centres. Nature. 2011;472:213–216. doi: 10.1038/nature09868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternson SM. Hypothalamic survival circuits: blueprints for purposive behaviors. Neuron. 2013;77:810–824. doi: 10.1016/j.neuron.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers L, Holy TE, Meister M, Dulac C, Koentges G. Loss of sex discrimination and male-male aggression in mice deficient for TRP2. Science. 2002;295:1493–1500. doi: 10.1126/science.1069259. [DOI] [PubMed] [Google Scholar]
- Sunkin SM, Ng L, Lau C, Dolbeare T, Gilbert TL, Thompson CL, Hawrylycz M, Dang C. Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic acids research. 2013;41:D996–D1008. doi: 10.1093/nar/gks1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson LW. Cerebral hemisphere regulation of motivated behavior. Brain research. 2000;886:113–164. doi: 10.1016/s0006-8993(00)02905-x. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsani D, Fu Y, Lu J, Lin Y, et al. A Resource of Cre Driver Lines for Genetic Targeting of GABAergic Neurons in Cerebral Cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turaga SC, Murray JF, Jain V, Roth F, Helmstaedter M, Briggman K, Denk W, Seung HS. Convolutional networks can learn to generate affinity graphs for image segmentation. Neural computation. 2010;22:511–538. doi: 10.1162/neco.2009.10-08-881. [DOI] [PubMed] [Google Scholar]
- Veening JG, Coolen LM, de Jong TR, Joosten HW, de Boer SF, Koolhaas JM, Olivier B. Do similar neural systems subserve aggressive and sexual behaviour in male rats? Insights from c-Fos and pharmacological studies. European journal of pharmacology. 2005;526:226–239. doi: 10.1016/j.ejphar.2005.09.041. [DOI] [PubMed] [Google Scholar]
- Vousden DA, Epp J, Okuno H, Nieman BJ, van Eede M, Dazai J, Ragan T, Bito H, Frankland PW, Lerch JP, et al. Whole-brain mapping of behaviourally induced neural activation in mice. Brain structure & function. 2014 doi: 10.1007/s00429-014-0774-0. [DOI] [PubMed] [Google Scholar]
- Williams RW, Rakic P. Three-dimensional counting: an accurate and direct method to estimate numbers of cells in sectioned material. The Journal of comparative neurology. 1988;278:344–352. doi: 10.1002/cne.902780305. [DOI] [PubMed] [Google Scholar]
- Winslow JT. Mouse social recognition and preference. Current protocols in neuroscience. 2003 doi: 10.1002/0471142301.ns0816s22. editorial board, Jacqueline N Crawley [et al Chapter 8, Unit 8 16. [DOI] [PubMed] [Google Scholar]
- Yang CF, Chiang MC, Gray DC, Prabhakaran M, Alvarado M, Juntti SA, Unger EK, Wells JA, Shah NM. Sexually dimorphic neurons in the ventromedial hypothalamus govern mating in both sexes and aggression in males. Cell. 2013;153:896–909. doi: 10.1016/j.cell.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CF, Shah NM. Representing Sex in the Brain, One Module at a Time. Neuron. 2014;82:261–278. doi: 10.1016/j.neuron.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.