Abstract
Background
Identifying sources of variation in the nicotine and nitrosamine metabolic inactivation pathways is important to understanding the relationship between smoking and cancer risk. Numerous UGT1A and UGT2B enzymes are implicated in nicotine and nitrosamine metabolism in vitro; however, little is known about their roles in vivo.
Methods
Within UGT1A1, UGT1A4, UGT1A9, UGT2B7, UGT2B10, and UGT2B17, 47 variants were genotyped, including UGT2B10*2 and UGT2B17*2. The association between variation in these UGTs and glucuronidation activity within European and African American current smokers (n=128), quantified as urinary ratios of the glucuronide over unconjugated compound for nicotine, cotinine, trans-3’-hydroxycotinine, and NNAL, was investigated in regression models assuming a dominant effect of variant alleles.
Results
Correcting for multiple testing, three UGT2B10 variants were associated with cotinine glucuronidation, rs2331559 and rs11726322 in European Americans and rs835309 in African Americans (P≤.0002). Additional variants predominantly in UGT2B10 were nominally associated with nicotine (P=.008-.04) and cotinine (P=<.001-.02) glucuronidation in both ethnicities in addition to UGT2B10*2 in European Americans (P=.01, P<.001). UGT2B17*2 (P=.03) in European Americans and UGT2B7 variants (P=.02-.04) in African Americans were nominally associated with 3HC glucuronidation. UGT1A (P=.007-.01), UGT2B10 (P=.02) and UGT2B7 (P=.02-03) variants in African Americans were nominally associated with NNAL glucuronidation.
Conclusions
Findings from this initial in vivo study support a role for multiple UGTs in the glucuronidation of tobacco-related compounds in vivo, in particular UGT2B10 and cotinine glucuronidation.
Impact
Findings also provide insight into ethnic differences in glucuronidation activity, which could be contributing to ethnic disparities in the risk for smoking-related cancers.
Keywords: UGT1A, UGT2B, nicotine, NNK, cigarette smoking
Introduction
Cigarette smoking is the leading risk factor for lung cancer (1, 2), and is also associated with numerous other cancers including those of the respiratory tract, digestive tract, bladder, pancreas and kidney (3, 4). Genetic factors are associated with differences in the susceptibility to tobacco-related cancers including genes involved in the metabolism of nicotine and tobacco smoke carcinogens (5-7). For instance, CYP2A6 gene variants are associated with reduced lung cancer risk within smokers of diverse ethnicities (8-13), as is a gene variant in CYP2A13 (14) – key enzymes in the nicotine inactivation pathway and in the activation pathway of tobacco specific nitrosamines (TSNAs) such as 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), respectively (15, 16).
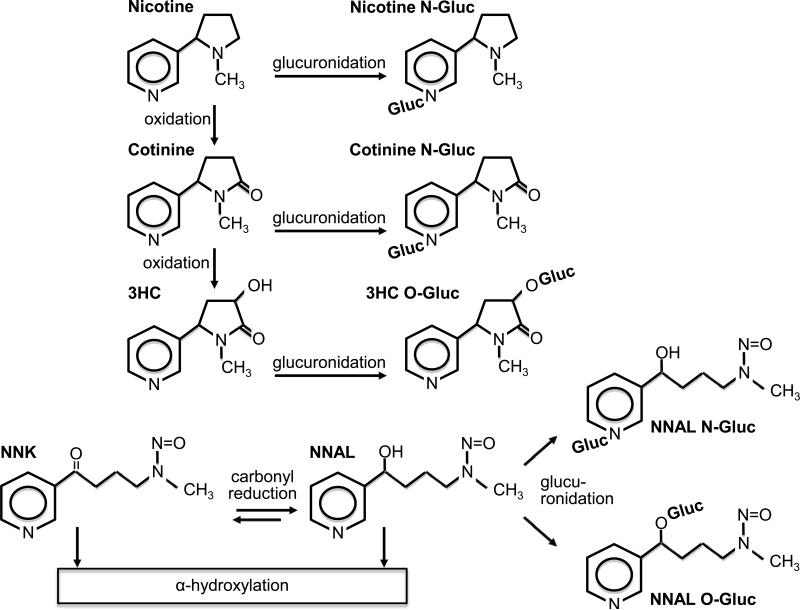
UDP-glucuronosyltransferases (UGTs) represent another group of enzymes with the potential to influence the relationship between smoking and cancer risk via their contribution to the metabolism of both nicotine and nitrosamines, including NNK (Figure 1). In addition to the major metabolic pathway of nicotine to cotinine and further to trans-3’-hydroxycotinine (3HC) chiefly mediated by CYP2A6, nicotine, cotinine and 3HC are also substrates of UGTs (17). Glucuronide conjugates account for 25-30% of recovered nicotine metabolites in urine (18-20). NNK is extensively metabolized by carbonyl reduction to 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), which, like NNK, is also carcinogenic (21, 22). NNAL is then metabolically detoxified by glucuronidation, and the non-carcinogenic glucuronide conjugates account for approximately 60% of NNAL detected in urine (23, 24).
Figure 1.
Simplified schematic of nicotine and NNK metabolism to glucuronide conjugates.
UGT gene variants may have utility in cancer risk prediction – deficient activity of some UGT enzymes enhances susceptibility to chemical carcinogenesis in animals (25, 26), and genetic variants in UGT1A7, UGT1A10 and UGT2B17 are associated with the risk for tobacco-related cancers in humans (27-30). Additionally, it is important to understand the impact of variable glucuronidation on biomarkers of nitrosamine exposure and on the ratio of NNAL-glucuronide to NNAL, a biomarker of nitrosamine detoxification (31) and a potential marker of cancer risk (32). Variable glucuronidation could also influence smoking through effects on nicotine clearance and could impact the interpretation of biomarkers of nicotine exposure and metabolism (33-35).
Many polymorphisms have been identified in the genes encoding the UGT1A and UGT2B enzymes (36); however, there are limited data on the impact of UGT variants on nicotine and nitrosamine metabolism in vivo, and in current cigarette smokers (37, 38). In vivo investigations have focused on two alleles, UGT2B10*2 and UGT2B17*2. UGT2B10*2 is associated with impaired nicotine and cotinine glucuronidation, while UGT2B17*2 is associated with impaired 3HC and NNAL glucuronidation (20, 27, 34, 35, 39).
In vitro studies of human liver microsomes and expressed UGTs implicate additional UGT1A and UGT2B enzymes in the glucuronidation of nicotine, its metabolites and NNAL. In human liver microsomes: UGT2B10*2 is associated with reduced glucuronidation of nicotine and cotinine, but also 3HC and NNAL (33, 40, 41), UGT2B17*2 is associated with reduced glucuronidation of 3HC and NNAL (33, 42), and UGT1A4*2 and UGT2B7*2 are associated with reduced glucuronidation of NNAL (42, 43). Inhibition studies in human liver microsomes implicate UGT1A1 in nicotine glucuronidation (44), UGT1A4 in nicotine, cotinine and 3HC glucuronidation (44-46), and UGT1A9 in nicotine and cotinine glucuronidation (44, 46). Recombinant UGTs from baculovirus-infected insect cells or over-expressed UGTs in human cell lines also implicate UGT1A4 in the glucuronidation of nicotine, cotinine and NNAL (47, 48) and provide evidence for the involvement of UGT1A9 in the glucuronidation of 3HC and NNAL (45, 49) and for UGT2B7 in the glucuronidation of nicotine and 3HC (45, 47).
Given the data suggesting the involvement of multiple UGT1A and UGT2B enzymes in vitro, and the limited knowledge of the contribution of UGT gene variants to nicotine and nitrosamine glucuronidation in vivo, we investigated the association between variation in UGT1A1, UGT1A4, UGT1A9, UGT2B7, UGT2B10, and UGT2B17, and nicotine, cotinine, 3HC and NNAL glucuronidation in European and African American current smokers. Since few UGT gene variants have been characterized with respect to these substrates, and the functional impact of UGT gene variants can be substrate specific (50), we genotyped 43 tag single nucleotide polymorphisms (SNPs) within the candidate genes. We also genotyped two UGT1A9 insertion/deletion variants and, importantly, UGT2B10*2 and UGT2B17*2, the only alleles previously associated with impaired in vivo glucuronidation in smokers (20, 35, 39). To gauge the suitability of the data set for genetic association analyses of urinary metabolite ratios, we genotyped participants for altered activity CYP2A6 variants to confirm the well-established association between CYP2A6 and the ratio of 3HC over cotinine, a biomarker for CYP2A6 activity and nicotine clearance (Supplementary Results) (51). In addition to identifying which UGT gene variants influence the glucuronidation of tobacco-related compounds among European Americans, a population in which sources of variation in glucuronidation are better characterized (in vitro studies almost exclusively utilize livers from individuals of European descent), the present study evaluated whether the same UGT variants were associated with glucuronidation among African Americans to provide pilot data focused on understanding the higher risk of smoking-related lung cancer observed among African Americans compared to European Americans (52, 53).
Materials and Methods
Study description
Participant recruitment and characteristics are detailed elsewhere (54). Briefly, 128 current smokers required to be 18–65 years old, healthy and to have smoked an average of 10 cigarettes per day or more for the past year or longer were recruited for a cross-sectional biomarker study. Subjects had to be self-identified non-Hispanic white (referred to as European American) or African American, with 4 grandparents of the same ethnicity. Smoking measures were collected and the time from smoking the last cigarette prior to urine sampling was recorded. One European American and one African American participant were excluded from the main genotype-phenotype analysis due to the absence of a urine sample and insufficient UGT genotyping results, respectively. Participant characteristics for the remaining 126 individuals are provided (Table 1). The study was approved by Institutional Review Boards at the University of California San Francisco, the University of Chicago and the University of Toronto.
Table 1.
Characteristics of participants included in analyses, n = 126a
| Characteristic | European American n = 66 | African American n = 60 | P valueb |
|---|---|---|---|
| Male, no. (%) | 38 (58) | 35 (58) | .93 |
| Age in years, median (IQR) | 34 (25-45) | 41 (36-49) | <.01 |
| Body mass index, median (IQR) | 24 (22-28) | 27 (24-33) | <.01 |
| Menthol cigarette use, no. (%) | 18 (27) | 41 (68) | <.01 |
| Cigarettes/dayc, median (IQR) | 20 (14-20) | 15 (10-20) | .04 |
| Nicotine equiv. (nmol/mg-creat)d, median (IQR) | 63 (54-75) | 44 (37-53) | <.01 |
| Min from last cig, median (IQR) | 80 (50-107) | 90 (60-140) | .08 |
| Activity ratios (geometric mean, 95% CI) | |||
| NIC-Gluc/ free NIC | 0.33 (0.25-0.44) | 0.17 (0.13-0.23) | <.01 |
| COT-Gluc/ free COT | 1.03 (0.87-1.23) | 0.40 (0.28-0.57) | <.01 |
| 3HC-Gluc/ free 3HC | 0.19 (0.17-0.22) | 0.26 (0.23-0.30) | <.01 |
| NNAL-Gluc/ free NNAL | 2.06 (1.87-2.27) | 1.85 (1.61-2.12) | .33 |
128 participants in the original study, 67 European American and 61 African American: One sample dropped due to absence of urinary biomarkers and another due to poor genotyping call rate
P values: Non-parametric Wilcoxon ranksum test or chi square
Average of self-reported cigarettes/day in the three days before biomarker collection
Nicotine equivalents: Urinary sum of nicotine, cotinine, trans-3-hydroxycotinine and their glucuronide conjugates normalized to creatinine
IQR: interquartile range
UGT Genotyping
Tag SNPs were selected within the candidate genes (+/− 10 kb) by determining the minimal set of common SNPs (minor allele frequency ≥ 5%) that captured the pairwise linkage disequilibrium (r2 > 0.8) for all common SNPs within the HapMap CEU population. In HapMap2 release 24, the set of SNPs capture (at an r2 ≥ 0.8) on average over 80% of existing common SNPs within the CEU population, and in the YRI population they capture on average 37% of the existing common SNPs (Supplementary Table 1).
UGT2B10*2 (rs61750900) was genotyped using a PCR-restriction fragment length polymorphism in which the amplified product was subjected to digestion with HinfI (40). The UGT2B17*2 deletion allele was assessed using the TaqMan gene expression assay, HS00854486_sH, with copy number reference assay TaqMan RNase P for the internal control, determined by TaqMan real-time quantitative PCR (55-57). Genotyping for 41 tag SNPs in UGT1A4, UGT1A9, UGT2B7 and UGT2B10 was done using the KASPar SNP genotyping system by LGC Genomics (formerly KBiosciences, LGC Limited, Middlesex, UK). Genotyping for the UGT1A9 indels, rs45625337 and rs10538910, was performed using the in-house GeneScan (PCR-Sizing) assays run on ABI 3700 as described previously (58). Genotyping for UGT1A4_135498 C>A (rs6755571) and UGT1A4_135876 T>C (rs12468274) was performed using a SNaPshot (Applied Biosystems, Life Technologies, Carlsbad, CA) single base extension 2-plex assay as described (59). The extension products were run on an ABI 3130xl (Applied Biosystems), and the data analyzed by the GeneMapper software (Applied Biosystems).
Analytical Chemistry
Urine concentrations of nicotine and its metabolites cotinine, 3HC, and their respective glucuronide metabolites were measured by LC-MS/MS from spot urine with glucuronide conjugates calculated from the difference in total concentration before and after alkaline hydrolysis (nicotine, cotinine) or hydrolysis by ß-glucuronidase (3HC, NNAL) as previously described (18, 51, 54, 60). Urine creatinine was measured in the San Francisco General Hospital clinical laboratory using a standard colorimetric assay.
Metabolite Phenotypes
To investigate UGT genotype-phenotype relationships, the urinary ratios of nicotine-glucuronide over nicotine, cotinine-glucuronide over cotinine, 3HC-glucuronide over 3HC, and NNAL-glucuronide over NNAL were used as glucuronidation activity phenotypes (27, 35, 39). The urinary ratio of total 3HC (free and glucuronide conjugated) over free cotinine was used as a phenotype of CYP2A6 oxidative metabolism (51). Nicotine equivalents, a measure of the total intake of nicotine, were calculated as the molar sum of nicotine, cotinine, 3HC, and their glucuronide metabolites in urine corrected for creatinine concentration (61).
Statistical Analyses
Comparison of demographic, smoking, and metabolite phenotypes in African versus European Americans was performed by Wilcoxon's rank-sum test (continuous variables) or χ2 (categorical variables). Correlations between metabolite phenotypes were performed on non-transformed values by Spearman's. Geometric mean values are presented for non-normally distributed values unless indicated otherwise. Linkage disequilibrium between the UGT1A and UGT2B variants was assessed using Haploview (62), while other statistical analyses were performed using Stata13 (StataCorp, College Station, TX). Hardy-Weinberg equilibrium tests were performed using the Stata program hwsnp (Author: Mario A. Cleves, University of Arkansas for Medical Sciences). Regression association analyses between UGT variants and glucuronidation phenotypes were performed using the Stata program qtlsnp assuming a dominant effect of the variant allele (Author: Mario A. Cleves, University of Arkansas for Medical Sciences). All regression models were performed separately within each ethnicity and were adjusted for age, gender and menthol smoking. Due to individuals missing genotypes and/or biomarker data, the number of observations within each model ranged from N=60-66 among European Americans, and N=51-60 among African Americans. In total 36 UGT variants were tested against four phenotypes in European Americans and 43 UGT variants were tested against four phenotypes in African Americans, hence the significance thresholds were set at P ≤.0003 and .0002, respectively, as determined by a Bonferroni correction for multiple testing. Nominally significant associations are also presented (P ≤.05).
Results
Glucuronidation phenotypes
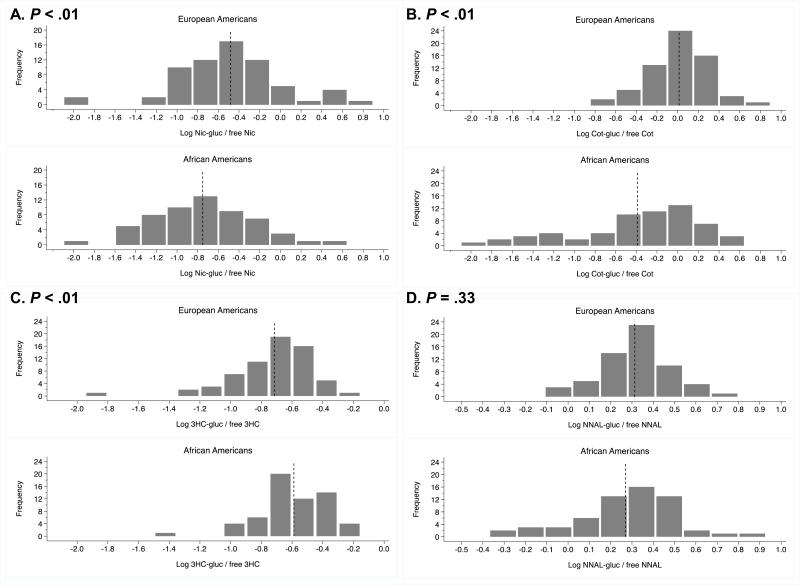
The distribution of each of the four glucuronide phenotype ratios by ethnicity was assessed (Figure 2A-D). African Americans had lower nicotine and cotinine glucuronide ratios compared to European Americans, as reported previously (39, 63). 3HC glucuronide ratios were higher among African Americans compared to European Americans, while no ethnic differences in NNAL glucuronide ratios were observed. Within-subject nicotine and cotinine glucuronide ratios were correlated in each ethnic group, but neither was correlated with the 3HC glucuronide ratio (Table 2), consistent with previous studies using 24 hour urine (18, 39). The NNAL glucuronide ratio was correlated with the cotinine glucuronide ratio and with the 3HC glucuronide ratio in each ethnicity (Table 2).
Figure 2.
Glucuronidation phenotypes among European Americans and African Americans expressed as the logarithm transformed ratio of glucuronide conjugate over free nicotine in A, cotinine in B, trans-3-hydroxycotinine in C, and NNAL in D. Dotted lines indicate mean values of logarithm transformed phenotypes (refer to Table 1 for non-transformed means). P values, A-D, from nonparametric Wilcoxon ranksum test comparing the phenotype distribution by ethnicity.
Table 2.
Spearman correlations between glucuronidation activity measures (non-transformed)
| EA | rho, P value | NIC-Gluc ratio | COT-Gluc ratio | 3HC-Gluc ratio | NNAL-Gluc ratio | AA |
| NIC-Gluc ratio | -- | 0.49, <.001 | −0.20, .13 | 0.19, .15 | ||
| COT-Gluc ratio | 0.34, .005 | -- | −0.05, .71 | 0.56, <.001 | ||
| 3HC-Gluc ratio | 0.10, .41 | 0.13, .31 | -- | 0.32, .01 | ||
| NNAL-Gluc ratio | 0.04, .76 | 0.34, .007 | 0.38, .002 | -- |
EA: European American; AA: African American
Demographic characteristics and urinary sampling parameters were evaluated as potential covariates of the glucuronide ratios. Demographic characteristics differed by ethnicity – African Americans were older (P<.01), had a higher BMI (P<.01), reported fewer cigarettes smoked per day (P=.04) and a greater prevalence of menthol cigarette use (P<.01) (Table 1). Correlations between glucuronide ratios and time from last cigarette, creatinine concentration and nicotine equivalents were also assessed, since metabolites were quantified from spot urine. No consistent patterns emerged between glucuronide ratios and age, BMI, creatinine, nicotine equivalents or time from last cigarette (Supplementary Table 2). No differences in glucuronide ratios by gender were noted among either ethnicity, but 3HC and NNAL glucuronide ratios were lower among African American menthol versus non-menthol smokers (Supplementary Table 3). Menthol is glucuronidated by enzymes such as UGT2B7 and UGT2B17 (64, 65), which are also capable of glucuronidating 3HC and NNAL (33, 42); thus, menthol could act as a competitive inhibitor, and menthol was included as a covariate in genotype-phenotype analyses. Age and gender were also included as covariates due to a priori evidence that these variables may influence the activity of specific UGTs (37, 66-68).
UGT genotyping results
The UGT2B10*2 allele, rs61750900T, had a minor allele frequency of 9% and 4% among European and African Americans, respectively, and the UGT2B17*2 allele had a minor allele frequency of 30% and 21% among European and African Americans, respectively, consistent with published frequencies (27, 35, 39). Among European Americans, rs3771342 and rs835310, and among African Americans, rs12468274 and rs12468543, were not consistent with Hardy-Weinberg Equilibrium and were excluded. Among European Americans, 3 variants in the UGT1A locus and 6 variants in the UGT2B locus displayed high linkage disequilibrium (R2 > 0.9) (Supplementary Figure 1A and 2A, respectively). Among African Americans, no variants in the UGT1A locus and only 2 variants in the UGT2B locus displayed high linkage disequilibrium (Supplementary Figure 1B and 2B, respectively). Variants in high linkage disequilibrium were also excluded leaving 36 and 43 UGT variants in subsequent genotype-phenotype analyses in European and African Americans, respectively (Supplementary Table 4).
Glucuronidation associations
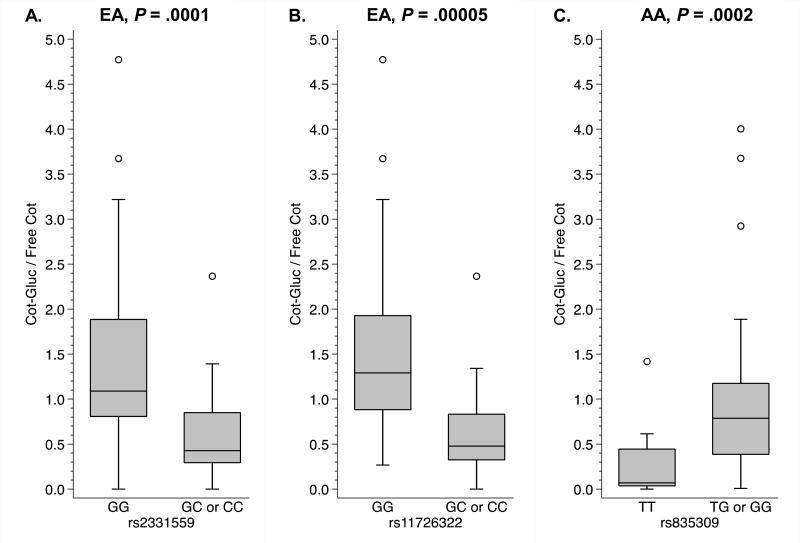
Three UGT2B10 gene variants were statistically significantly associated with cotinine glucuronidation (Figure 3) following correction for multiple testing (outlined in statistical methods). Among European Americans, individuals heterozygous or homozygous for the minor alleles of rs2331559 and rs11726322 had significantly lower cotinine glucuronide ratios compared to those homozygous for the major alleles (Figure 3A-B). Among African Americans, individuals heterozygous or homozygous for the minor allele of rs835309 had significantly higher cotinine glucuronide ratios compared to those homozygous for the major allele (Figure 3C). Nominally significant associations are also reported in the following sections, as this was the first investigation of multiple candidate genes chosen based on in vitro data with the goal of providing a relative sense of the potential importance of these candidate genes in the glucuronidation of nicotine and nitrosamines in smokers.
Figure 3.
UGT2B10 variants significantly associated with cotinine glucuronidation following correction for multiple testing. Individuals heterozygous or homozygous for the minor alleles of rs2331559 in A and rs11726322 in B had lower cotinine glucuronide ratios, whereas individuals heterozygous or homozygous for the minor allele of rs835309 in C had higher cotinine glucuronide ratios. Genotype-phenotype relationships illustrated with box plots displaying the median value and interquartile range (25th to the 75th percentile) and whiskers displaying the upper and lower values within 1.5 times the interquartile range, and open circles displaying outlying individuals. P values from multivariate logistic regression modeling assuming a dominant effect of the minor allele. EA: European American; AA: African American.
Nicotine glucuronidation
Two UGT2B10 variants were nominally associated with impaired nicotine glucuronidation activity in European Americans, while in African Americans, two UGT2B10 variants were nominally associated with enhanced and two with impaired activity (Table 3A). A variant in the UGT1A4 locus and two in the UGT1A1 locus were also nominally associated with nicotine glucuronidation in African Americans.
Table 3A.
UGT variants nominally associated with the nicotine glucuronide ratio
| UGT Variant | European American Smokers | African American Smokers | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Alleles | MAF | Pdom | Coef (95% CI) | Alleles | MAF | Pdom | Coef (95% CI) | |||
| UGT1A4 locus | ||||||||||
| rs13401281 | T : G | .409 | T : G | ↑ | .475 | .012 | 0.35 (0.08, 0.62) | |||
| UGT1A1 locus | ||||||||||
| rs6742078 | G : T | .408 | G : T | ↑ | .379 | .049 | 0.26 (.001, 0.52) | |||
| rs3771342a | C : A | .112 | C : A | ↓ | .117 | .024 | −0.38 (−0.72, −0.05) | |||
| UGT2B10 locus | ||||||||||
| rs61750900 (*2) | G : T | ↓ | .090 | .013 | −0.42 (−0.76, −0.09) | G : T | .043 | |||
| rs2331559 | G : C | ↓ | .119 | .015 | 0.38 (0.07, 0.68) | C : G | ↑ | .362 | .008 | 0.34 (0.09, 0.60) |
| rs835309b | G : T | .106 | T : G | ↑ | .491 | .026 | 0.34 (0.04, 0.64) | |||
| rs11726322 | G : C | .129 | G : C | ↓ | .254 | .010 | −0.34 (−0.60, −0.09) | |||
| rs7673996 | C : T | .031 | C : T | ↓ | .225 | .042 | −0.27 (−0.53, −0.01) | |||
Variant excluded from analyses in European Americans
P value not reported in European Americans as variant in high linkage disequilibrium with rs2331559.
Alleles indicated as major : minor with impaired allele in bolded italics and an arrow indicating direction of effect for the minor allele. Pdom and coefficient with 95% confidence intervals from multivariate logistic regression modeling assuming a dominant effect of the minor allele and reported when P < .05. Variants ordered by location along chromosome 2 (UGT1A) and chromosome 4 (UGT2B). MAF: Minor allele frequency
Cotinine glucuronidation
In addition to the two UGT2B10 variants significantly associated with impaired cotinine glucuronidation (Figure 3A-B), UGT2B10*2 was nominally associated with impaired cotinine glucuronidation activity, while a single variant in UGT2B7 was nominally associated with enhanced glucuronidation in European Americans (Table 3B). Among African Americans, in addition to the UGT2B10 variant significantly associated with enhanced cotinine glucuronidation (Figure 3C), one other UGT2B10 variant was nominally associated with enhanced and two with impaired activity (Table 3B). Two variants in the UGT1A4 locus and single variants in the common UGT1A exons and 3’ flanking region of UGT1A were also associated with cotinine glucuronidation in African Americans (Table 3B).
Table 3B.
UGT variants nominally/significantly associated with the cotinine glucuronide ratio
| UGT SNP/Variant | European American Smokers | African American Smokers | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Alleles | MAF | Pdom | Coef (95% CI) | Alleles | MAF | Pdom | Coef (95% CI) | |||
| UGT1A4 locus | ||||||||||
| rs3732220 | C : T | .092 | C : T | ↑ | .108 | .018 | 0.50 (0.09, 0.91) | |||
| rs871514 | G : A | .478 | G : A | ↓ | .359 | .027 | −0.36 (−0.68, −0.04) | |||
| UGT1A exon 2-5 | ||||||||||
| rs1018124 | T : C | .056 | T : C | ↑ | .108 | .013a | 0.48 (0.10, 0.86) | |||
| UGT1A 3’ flanking | ||||||||||
| rs10209214 | T : C | .313 | T : C | ↓ | .142 | .023a | −0.39 (−0.72, −0.05) | |||
| UGT2B10 locus | ||||||||||
| rs61750900 (*2) | G : T | ↓ | .090 | .0004 | −0.37 (−0.56, −0.17) | G : T | .043 | |||
| rs294765 | G : A | .015 | G : A | ↓ | .217 | .017 | −0.39 (−0.71, −0.07) | |||
| rs2331559 | G : C | ↓ | .119 | .0001 | −0.36 (−0.53, −0.19) | C : G | ↑ | .362 | .006 | 0.43 (0.13, 0.74) |
| rs835309b | G : T | .106 | T : G | ↑ | .491 | .0002 | 0.66 (0.33, 0.99) | |||
| rs835310c | C : G | .023 | C : G | ↓ | .408 | .005 | −0.46 (−0.78, −0.14) | |||
| rs11726322 | G : C | ↓ | .129 | .00005 | −0.35 (−0.51, −0.19) | G : C | .254 | |||
| UGT2B7 locus | ||||||||||
| rs4356975 | C : T | ↑ | .381 | .042 | 0.17 (0.01, 0.33) | C : T | .397 | |||
Absence of individuals homozygous for the minor allele
P value not reported in European Americans as variant in high LD with rs2331559
Variant excluded from analyses in European Americans.
Alleles indicated as major : minor with impaired allele in bolded italics and an arrow indicating direction of effect for the minor allele. Pdom and coefficient with 95% confidence intervals from multivariate logistic regression modeling assuming a dominant effect of the minor allele and reported when P < .05. Variants ordered by location along chromosome 2 (UGT1A) and chromosome 4 (UGT2B). MAF: Minor allele frequency
3HC glucuronidation
Among European Americans, the UGT2B17 copy number variant was nominally associated with impaired 3HC glucuronidation activity (Table 3C). Among African Americans, two variants in the UGT2B7 locus were nominally associated with impaired 3HC glucuronidation activity (Table 3C).
Table 3C.
UGT variants nominally associated with the 3HC glucuronide ratio
| UGT SNP/Variant | European American Smokers | African American Smokers | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Alleles | MAF | Pdom | Coef (95% CI) | Alleles | MAF | Pdom | Coef (95% CI) | |||
| UGT2B17 locus | ||||||||||
| copy number | 1 : 0 | ↓ | .303 | .028 | −0.15 (−0.29, −0.02) | 1 : 0 | .215 | |||
| UGT2B7 locus | ||||||||||
| rs57216626 | C : G | .448 | C : G | ↓ | .250 | .016 | −0.14 (−0.25, −0.03) | |||
| rs4535394 | C : T | .440 | C : T | ↓ | .339 | .026 | −0.13 (−0.25, −0.02) | |||
UGT2B17 0 allele: deletion allele. Alleles indicated as major : minor with impaired allele in bolded italics and an arrow indicating direction of effect for the minor allele. Pdom and coefficient with 95% confidence intervals from multivariate logistic regression modeling assuming a dominant effect of the minor allele and reported when P < .05. Variants ordered by location along chromosome 2 (UGT1A) and chromosome 4 (UGT2B). MAF: Minor allele frequency
NNAL glucuronidation
No UGT variants reached nominal significance with NNAL glucuronidation activity among European Americans (Table 3D). Among African Americans, two UGT2B7 variants were nominally associated with NNAL glucuronidation, one with enhanced and one with impaired activity (Table 3D). Single variants in UGT1A1, the 3’ flanking region of UGT1A, and UGT2B10 were also associated with NNAL glucuronidation in African Americans (Table 3D).
Table 3D.
UGT variants nominally associated with the NNAL glucuronide ratio
| UGT SNP/Variant | European American Smokers | African American Smokers | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Alleles | MAF | Pdom | Coef (95% CI) | Alleles | MAF | Pdom | Coef (95% CI) | ||
| UGT1A1 locus | |||||||||
| rs3771342a | C : A | .112 | C : A | ↑ | .117 | .012 | 0.19 (0.04, 0.34) | ||
| UGT1A 3’ flanking | |||||||||
| rs10203853 | A : T | .431 | A : T | ↓ | .292 | .007 | −0.16 (−0.38, −0.05) | ||
| UGT2B10 locus | |||||||||
| rs835310a | C : G | .023 | C : G | ↓ | .408 | .019 | −0.15 (−0.28, −0.03) | ||
| UGT2B7 locus | |||||||||
| rs12506592 | A : G | .162 | A : G | ↓ | .275 | .030 | −0.14 (−0.26, −0.01) | ||
| rs4356975 | C : T | .381 | C : T | ↑ | .397 | .017 | 0.15 (0.03, 0.28) | ||
Variant excluded from analyses in European Americans.
Alleles indicated as major : minor with impaired allele in bolded italics and an arrow indicating direction of effect for the minor allele. Pdom and coefficient with 95% confidence intervals from multivariate logistic regression modeling assuming a dominant effect of the minor allele and reported when P < .05. Variants ordered by location along chromosome 2 (UGT1A) and chromosome 4 (UGT2B). MAF: Minor allele frequency
Discussion
This is the first study to investigate variation in multiple candidate UGT1A and UGT2B genes and glucuronidation activity within the nicotine and nitrosamine metabolic pathways among European and African American current smokers. Prior to this investigation, only UGT2B10*2 and UGT2B17*2 had been shown to influence the glucuronidation of tobacco-related compounds in vivo (20, 27, 34, 35, 39); whereas additional UGT1A and UGT2B enzymes were implicated in these pathways in vitro (33, 40-42, 44-49). Since few UGT variants have been functionally characterized with respect to our substrates of interest, we focused this initial investigation on tag SNPs to provide a more comprehensive examination of the contribution of UGT genetic variation to variation in the glucuronidation of tobacco-related compounds in European Americans. We also examined whether these tag SNPs chosen in European Americans were associated with glucuronidation among African American smokers.
Our findings confirm an important contribution of genetic variation in UGT2B10 to nicotine and cotinine glucuronidation with multiple UGT2B10 variants nominally associated with nicotine and with cotinine glucuronidation in both ethnicities. Three variants in UBT2B10 remained statistically significantly associated with cotinine glucuronidation following correction for multiple testing. Of note, the two variants significantly associated with cotinine glucuronidation in European Americans, rs2331559 and rs11726322, had similar effect sizes to UGT2B10*2 (Table 3B). UGT2B10*2 likely did not reach the Bonferroni corrected threshold of P≤.0003 due to the lower prevalence of the *2 allele (~9%) compared to rs2331559 (~12%) and rs11726322 (13%). The minor (less frequent) alleles of the UGT2B10 variants surviving correction for multiple testing in European Americans were associated with impaired cotinine glucuronidation; whereas, among African Americans the minor allele of the UGT2B10 variant surviving correction for multiple testing was associated with enhanced activity. Hence, in contrast to European Americans, the major (more frequent) allele in African would have reduced activity (versus enhanced activity) potentially contributing to the lower levels of nicotine and cotinine glucuronide conjugates observed among African Americans (Figure 2A and 2B) (39, 63).
In addition to UGT2B10, we also observed nominal associations between variants in UGT1A1, UGT1A4 and UGT2B7 and nicotine and/or cotinine glucuronidation. Consistent with inhibition studies of UGT1A1 in human liver microsomes, which demonstrate an impact on nicotine but not cotinine glucuronidation (44), UGT1A1 was only associated with the nicotine glucuronide ratio. UGT1A4 is implicated in the glucuronidation of nicotine and cotinine in vitro and is considered to be the second most active UGT in the N-glucuronidation of these compounds after UGT2B10 (44, 46, 47). We observed associations between UGT1A4 variants and both the nicotine and cotinine glucuronide ratios potentially reflecting the minor contribution of this enzyme. UGT2B7 is capable of nicotine glucuronidation in vitro (47); however, the nominal association that we report is between UGT2B7 and cotinine glucuronidation and may represent a chance finding.
As the nicotine and cotinine glucuronide ratios were correlated, we anticipated that the same UGT variants might be associated with both glucuronidation phenotypes. While we observed overlap in variants associated with both pathways, particularly in UGT2B10, more UGT variants were associated with the cotinine glucuronide ratio than with the nicotine glucuronide ratio and the only variants surviving correction for multiple testing were associated with cotinine glucuronidation. A greater genetic contribution to inter-individual variation in cotinine versus nicotine glucuronidation has been proposed based on twin studies (69). Alternatively, this difference may be the result of a more stable glucuronidation phenotype for cotinine.
Comparatively few UGT variants were associated with the 3HC glucuronide ratio. We replicated the in vivo association of UGT2B17*2 with lower 3HC glucuronidation (20, 35) among European Americans. In line with the O-glucuronidation of 3HC by UGT2B7 observed in vitro (45), two UGT2B7 variants were associated with the 3HC glucuronide ratio among African Americans. We observed a high degree of LD in the UGT2B7 locus among European Americans (Supplementary Figure 2A); thus, the absence of association with UGT2B7 among European Americans could reflect a lower prevalence and/or diversity of altered activity UGT2B7 variants or inadequate tagging of functional variants.
Our findings do not support a dominant contribution of genetic variation in any single UGT to the overall level of NNAL glucuronidation, which may reflect the formation of both N- and O-glucuronide conjugates of NNAL in vivo (24, 43). Hence, multiple UGT enzymes, and their variants, may each make a relatively small contribution to the overall NNAL glucuronide ratio. Consistent with the correlations that we observed between the NNAL and cotinine glucuronide ratios, and the NNAL and 3HC glucuronide ratios (Table 2), the UGT2B10 rs835310 G allele showed impaired glucuronidation of both NNAL and cotinine and the UGT2B7 rs12506592 G allele showed impaired glucuronidation of both NNAL and 3HC (Tables 3D and 3B). We did not replicate the in vivo or in vitro association of UGT2B17*2 with impaired glucuronidation of NNAL (27, 42). However, Gallagher et al. reported an association only among women (27), and our study size was insufficient to test for genotype-gender interactions, and Lazarus et al. measured O-glucuronide formation specifically (42), whereas our analytical method did not distinguish between N and O-glucuronide conjugates. The N- and O-glucuronides are formed in near equal amounts in vivo (24), so a small genetic effect on either pathway may not be detectable in the current study. Consistent with a smaller effect of any one UGT on the overall level of NNAL glucuronidation, the effect sizes that we observed between UGT variants and NNAL glucuronidation (Table 3D) were approximately half the magnitude of the observed associations with cotinine glucuronidation (Table 3B). The overall low levels of NNAL in the urine of smokers (free or glucuronidated) compared to cotinine may also have hindered genotype-phenotype associations.
The ratio of NNAL-glucuronide to NNAL is a biomarker of nitrosamine detoxification (31) and a potential marker of cancer risk (32). There is conflicting evidence regarding ethnic differences in this ratio, specifically whether it is lower among African Americans (70, 71). We did not observe a significant difference in NNAL glucuronide ratios by ethnicity, while replicating differences in other ratios (Figure 2). However, 10% of African Americans had ratios below the lowest ratio observed among European Americans potentially putting these individuals at greater risk for cancer, as Chung et al. found that lower NNAL glucuronide ratios were associated with an increased risk of cancer (32). Variation in the UGT2B10 and UGT2B7 loci may be of particular interest in terms of cancer risk disparities, since these variants were only nominally associated with NNAL glucuronidation activity among African Americans.
In addition to altered carcinogen detoxification, variation in UGTs could influence lung cancer indirectly through altered smoking. Berg et al. reported significantly lower nicotine equivalents (a measure of nicotine intake) in smokers with the UGT2B10 *1/*2 genotype in a study of European Americans (34) and in a mixed analysis of European and African American smokers (39), and speculated that slower nicotine glucuronidation may lead to reduced nicotine consumption. However, in a larger study of African American smokers, Zhu et al. did not find a significant association between UGT2B10*2 and lower nicotine equivalents (35). Consistent with glucuronidation as a minor pathway for nicotine inactivation (17), and with Zhu et al., neither UGT2B10*2 genotype (data not shown), nor importantly the actual nicotine glucuronidation ratio (Supplementary Table 2), were associated with nicotine equivalents in either European or African Americans.
Many nominally significant associations between UGT variants and glucuronidation phenotypes were observed despite both the relatively small sample size for a genetic association study and the quantification of metabolites from spot urine with variable time from last cigarette. Biomarker assessment from spot urine is unlikely to have biased findings, since we observed similar correlations between glucuronide ratios as reported for 24 hour urine (18, 39). Furthermore, urinary cotinine, NNAL and nicotine equivalents, which are all biomarkers of nicotine consumption, were correlated as expected (data not shown) and neither nicotine equivalents nor the glucuronide ratios were systemically associated with time from last cigarette (Supplementary Table 2). Only one nominally significant variant, UGT1A1 rs3771342, displayed an inconsistent direction of effect with the nicotine and NNAL glucuronidation phenotypes potentially reflecting an indirect genotype-phenotype association or simply a chance observation. The smaller sample size precluded interactions analyses. In particular, the interaction between menthol and UGT2B7 and UGT2B17 gene variants would be worthwhile exploring in a larger dataset given the conflicting evidence concerning menthol smoking and lung cancer risk (reviewed in (72)). More associations were observed among African Americans than among European Americans, as is seen in other genomic regions displaying lower linkage disequilibrium in African populations (e.g. chromosome 15q25 and lung cancer risk (73)). Of note, the gene variants investigated were initially chosen to provide relatively good coverage of European smokers and provide relatively low coverage of common variation in Africans (Supplementary Table 1) suggesting that even more variation may be identified among this ethnic group. Alternatively, a single untested variant may underlie multiple associations observed in a gene region through linkage disequilibrium.
Overall, study findings confirmed a role for multiple UGTs in the glucuronidation of tobacco-related compounds in vivo and contributed to the understanding of sources of variation in the nicotine and nitrosamine metabolic inactivation pathways. Concurrently examining genetic sources of variation in European and African American smokers also provided insight into ethnic differences in glucuronidation activity, which could be contributing to ethnic disparities in the risk for smoking-related cancers.
Supplementary Material
Acknowledgments
Financial support: This work was supported by a University Endowed Chair in Addictions for the Department of Psychiatry (RF Tyndale); the National Institutes of Health (U01DA020830 to RF Tyndale and NL Benowitz); the Canadian Institutes of Health Research (TMH109787 to RF Tyndale, Doctoral Award to CA Wassenaar); the Centre for Addiction and Mental Health and the CAMH foundation; the Canada Foundation for Innovation (#20289 and #16014); the Ontario Ministry of Research and Innovation; and US Public Health Service grants (DA02277 and DA12393 to NL Benowitz) and from the National Institute on Drug Abuse, National Institutes of Health. The clinical studies were carried out in part at the Clinical Research Center at San Francisco General Hospital Medical Center (NIH/NCRR UCSF-CTSI UL1 RR024131). The effort of the University of Chicago authors was supported by the National Institutes of Health (GM61393 to S Das, P Chen, MJ Ratain).
Footnotes
Conflict of Interest statements: Rachel F. Tyndale has served as a consultant to pharmaceutical companies, primarily on smoking cessation. Neal L. Benowitz has been a paid consultant to Pfizer, GlaxoSmithKline and McNeil, and has served as a paid expert witness in litigation against tobacco companies.
Contributor Information
Catherine A. Wassenaar, Department of Pharmacology and Toxicology, University of Toronto, Toronto, Ontario, Canada (catherine.wassenaar@utoronto.ca)
David V. Conti, Department of Preventive Medicine, University of Southern California, Los Angeles, CA (dconti@med.usc.edu)
Soma Das, Department of Human Genetics, University of Chicago, Chicago, IL (sdas@bsd.uchicago.edu).
Peixian Chen, Department of Human Genetics, University of Chicago, Chicago, IL (pchen@bsd.uchicago.edu).
Edwin H. Cook, Department of Psychiatry, University of Illinois at Chicago, Chicago, IL (edcook@uic.edu)
Mark J. Ratain, Department of Medicine, Committee on Clinical Pharmacology and Pharmacogenomics, Cancer Research Center, University of Chicago, Chicago, Illinois, USA (mratain@medicine.bsd.uchicago.edu)
Neal L. Benowitz, Division of Clinical Pharmacology and Experimental Therapeutics, Departments of Medicine and Bioengineering & Therapeutic Science, University of California San Francisco, San Francisco, California, USA (nbenowitz@medsfgh.ucsf.edu)
Rachel F. Tyndale, Campbell Family Mental Health Research Institute, Centre for Addiction and Mental Health, Departments of Psychiatry, Pharmacology and Toxicology, University of Toronto, Toronto, Ontario, Canada (r.tyndale@utoronto.ca)
References
- 1.Alberg AJ, Ford JG, Samet JM. Epidemiology of lung cancer: ACCP evidence-based clinical practice guidelines. Chest. (2nd edition) 2007;132:29S–55S. doi: 10.1378/chest.07-1347. [DOI] [PubMed] [Google Scholar]
- 2.Peto R, Lopez AD, Boreham J, Thun M, Heath C., Jr. Mortality from tobacco in developed countries: indirect estimation from national vital statistics. Lancet. 1992;339:1268–78. doi: 10.1016/0140-6736(92)91600-d. [DOI] [PubMed] [Google Scholar]
- 3.U.S. Department of Health & Human Services . The Health Consequences of Smoking: A Report of the Surgeon General. U.S. Department of Health and Human Services; Atlanta: 2004. [Google Scholar]
- 4.World Health Organization IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Tobacco Smoke and Involuntary Smoking. 2004:1179–87. [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez-Antona C, Gomez A, Karlgren M, Sim SC, Ingelman-Sundberg M. Molecular genetics and epigenetics of the cytochrome P450 gene family and its relevance for cancer risk and treatment. Hum Genet. 2010;127:1–17. doi: 10.1007/s00439-009-0748-0. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz AG, Prysak GM, Bock CH, Cote ML. The molecular epidemiology of lung cancer. Carcinogenesis. 2007;28:507–18. doi: 10.1093/carcin/bgl253. [DOI] [PubMed] [Google Scholar]
- 7.Taioli E. Gene-environment interaction in tobacco-related cancers. Carcinogenesis. 2008;29:1467–74. doi: 10.1093/carcin/bgn062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wassenaar CA, Dong Q, Wei Q, Amos CI, Spitz MR, Tyndale RF. Relationship between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 variation and smoking behaviors and lung cancer risk. J Natl Cancer Inst. 2011;103:1342–6. doi: 10.1093/jnci/djr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rotunno M, Yu K, Lubin JH, Consonni D, Pesatori AC, Goldstein AM, et al. Phase I metabolic genes and risk of lung cancer: multiple polymorphisms and mRNA expression. PLoS One. 2009;4:e5652. doi: 10.1371/journal.pone.0005652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gemignani F, Landi S, Szeszenia-Dabrowska N, Zaridze D, Lissowska J, Rudnai P, et al. Development of lung cancer before the age of 50: the role of xenobiotic metabolizing genes. Carcinogenesis. 2007;28:1287–93. doi: 10.1093/carcin/bgm021. [DOI] [PubMed] [Google Scholar]
- 11.Fujieda M, Yamazaki H, Saito T, Kiyotani K, Gyamfi MA, Sakurai M, et al. Evaluation of CYP2A6 genetic polymorphisms as determinants of smoking behavior and tobacco-related lung cancer risk in male Japanese smokers. Carcinogenesis. 2004;25:2451–8. doi: 10.1093/carcin/bgh258. [DOI] [PubMed] [Google Scholar]
- 12.Liu ZB, Shu J, Wang LP, Jin C, Lou ZX. Cytochrome P450 2A6 deletion polymorphism and risk of lung cancer: a meta-analysis. Mol Biol Rep. 2013;40:5255–9. doi: 10.1007/s11033-013-2625-0. [DOI] [PubMed] [Google Scholar]
- 13.Wassenaar CA, Ye Y, Cai Q, Aldrich M, Knight J, Spitz MR, et al. CYP2A6 Variation and Lung Cancer in African American Smokers – Findings from Two Independent Populations. Cancer Research - under review. 2014 doi: 10.1093/carcin/bgu235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Tan W, Hao B, Miao X, Zhou G, He F, et al. Substantial reduction in risk of lung adenocarcinoma associated with genetic polymorphism in CYP2A13, the most active cytochrome P450 for the metabolic activation of tobacco-specific carcinogen NNK. Cancer Res. 2003;63:8057–61. [PubMed] [Google Scholar]
- 15.Mwenifumbo JC, Tyndale RF. Genetic variability in CYP2A6 and the pharmacokinetics of nicotine. Pharmacogenomics. 2007;8:1385–402. doi: 10.2217/14622416.8.10.1385. [DOI] [PubMed] [Google Scholar]
- 16.Jalas JR, Hecht SS, Murphy SE. Cytochrome P450 enzymes as catalysts of metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, a tobacco specific carcinogen. Chem Res Toxicol. 2005;18:95–110. doi: 10.1021/tx049847p. [DOI] [PubMed] [Google Scholar]
- 17.Hukkanen J, Jacob P, 3rd, Benowitz NL. Metabolism and disposition kinetics of nicotine. Pharmacol Rev. 2005;57:79–115. doi: 10.1124/pr.57.1.3. [DOI] [PubMed] [Google Scholar]
- 18.Benowitz NL, Jacob P, 3rd, Fong I, Gupta S. Nicotine metabolic profile in man: comparison of cigarette smoking and transdermal nicotine. J Pharmacol Exp Ther. 1994;268:296–303. [PubMed] [Google Scholar]
- 19.Byrd GD, Chang KM, Greene JM, deBethizy JD. Evidence for urinary excretion of glucuronide conjugates of nicotine, cotinine, and trans-3'-hydroxycotinine in smokers. Drug Metab Dispos. 1992;20:192–7. [PubMed] [Google Scholar]
- 20.Chen G, Giambrone NE, Jr., Dluzen DF, Muscat JE, Berg A, Gallagher CJ, et al. Glucuronidation genotypes and nicotine metabolic phenotypes: importance of functional UGT2B10 and UGT2B17 polymorphisms. Cancer Res. 2010;70:7543–52. doi: 10.1158/0008-5472.CAN-09-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hecht SS. Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem Res Toxicol. 1998;11:559–603. doi: 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- 22.Castonguay A, Lin D, Stoner GD, Radok P, Furuya K, Hecht SS, et al. Comparative carcinogenicity in A/J mice and metabolism by cultured mouse peripheral lung of N′-nitrosonornicotine, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, and their analogues. Cancer Res. 1983;43:1223–9. [PubMed] [Google Scholar]
- 23.Upadhyaya P, Kenney PM, Hochalter JB, Wang M, Hecht SS. Tumorigenicity and metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol enantiomers and metabolites in the A/J mouse. Carcinogenesis. 1999;20:1577–82. doi: 10.1093/carcin/20.8.1577. [DOI] [PubMed] [Google Scholar]
- 24.Carmella SG, Le Ka KA, Upadhyaya P, Hecht SS. Analysis of N- and O-glucuronides of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) in human urine. Chem Res Toxicol. 2002;15:545–50. doi: 10.1021/tx015584c. [DOI] [PubMed] [Google Scholar]
- 25.Kim PM, Wells PG. Genoprotection by UDP-glucuronosyltransferases in peroxidase-dependent, reactive oxygen species-mediated micronucleus initiation by the carcinogens 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and benzo[a]pyrene. Cancer Res. 1996;56:1526–32. [PubMed] [Google Scholar]
- 26.Vienneau DS, DeBoni U, Wells PG. Potential genoprotective role for UDP-glucuronosyltransferases in chemical carcinogenesis: initiation of micronuclei by benzo(a)pyrene and benzo(e)pyrene in UDP-glucuronosyltransferase-deficient cultured rat skin fibroblasts. Cancer Res. 1995;55:1045–51. [PubMed] [Google Scholar]
- 27.Gallagher CJ, Muscat JE, Hicks AN, Zheng Y, Dyer AM, Chase GA, et al. The UDP-glucuronosyltransferase 2B17 gene deletion polymorphism: sex-specific association with urinary 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol glucuronidation phenotype and risk for lung cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:823–8. doi: 10.1158/1055-9965.EPI-06-0823. [DOI] [PubMed] [Google Scholar]
- 28.Araki J, Kobayashi Y, Iwasa M, Urawa N, Gabazza EC, Taguchi O, et al. Polymorphism of UDP-glucuronosyltransferase 1A7 gene: a possible new risk factor for lung cancer. Eur J Cancer. 2005;41:2360–5. doi: 10.1016/j.ejca.2005.04.043. [DOI] [PubMed] [Google Scholar]
- 29.Zheng Z, Park JY, Guillemette C, Schantz SP, Lazarus P. Tobacco carcinogen-detoxifying enzyme UGT1A7 and its association with orolaryngeal cancer risk. J Natl Cancer Inst. 2001;93:1411–8. doi: 10.1093/jnci/93.18.1411. [DOI] [PubMed] [Google Scholar]
- 30.Elahi A, Bendaly J, Zheng Z, Muscat JE, Richie JP, Jr., Schantz SP, et al. Detection of UGT1A10 polymorphisms and their association with orolaryngeal carcinoma risk. Cancer. 2003;98:872–80. doi: 10.1002/cncr.11587. [DOI] [PubMed] [Google Scholar]
- 31.Carmella SG, Akerkar SA, Richie JP, Jr., Hecht SS. Intraindividual and interindividual differences in metabolites of the tobacco-specific lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) in smokers' urine. Cancer Epidemiol Biomarkers Prev. 1995;4:635–42. [PubMed] [Google Scholar]
- 32.Chung CJ, Lee HL, Yang HY, Lin P, Pu YS, Shiue HS, et al. Low ratio of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol-glucuronides (NNAL-Gluc)/free NNAL increases urothelial carcinoma risk. Sci Total Environ. 2011;409:1638–42. doi: 10.1016/j.scitotenv.2011.01.046. [DOI] [PubMed] [Google Scholar]
- 33.Chen G, Giambrone NE, Lazarus P. Glucuronidation of trans-3′-hydroxycotinine by UGT2B17 and UGT2B10. Pharmacogenet Genomics. 2012;22:183–90. doi: 10.1097/FPC.0b013e32834ff3a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berg JZ, von Weymarn LB, Thompson EA, Wickham KM, Weisensel NA, Hatsukami DK, et al. UGT2B10 genotype influences nicotine glucuronidation, oxidation, and consumption. Cancer Epidemiol Biomarkers Prev. 2010;19:1423–31. doi: 10.1158/1055-9965.EPI-09-0959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu AZ, Zhou Q, Cox LS, Ahluwalia JS, Benowitz NL, Tyndale RF. Variation in trans-3′-hydroxycotinine glucuronidation does not alter the nicotine metabolite ratio or nicotine intake. PLoS One. 2013;8:e70938. doi: 10.1371/journal.pone.0070938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.UGT Allele Nomenclature Committee [16-May-2014];UGT UDP-Glucuronosyltransferase Alleles Nomenclature page. Available from: http://www.pharmacogenomics.pha.ulaval.ca/cms/ugt_alleles/
- 37.Court MH. Interindividual variability in hepatic drug glucuronidation: studies into the role of age, sex, enzyme inducers, and genetic polymorphism using the human liver bank as a model system. Drug Metab Rev. 2010;42:209–24. doi: 10.3109/03602530903209288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guillemette C. Pharmacogenomics of human UDP-glucuronosyltransferase enzymes. Pharmacogenomics J. 2003;3:136–58. doi: 10.1038/sj.tpj.6500171. [DOI] [PubMed] [Google Scholar]
- 39.Berg JZ, Mason J, Boettcher AJ, Hatsukami DK, Murphy SE. Nicotine metabolism in African Americans and European Americans: variation in glucuronidation by ethnicity and UGT2B10 haplotype. J Pharmacol Exp Ther. 2010;332:202–9. doi: 10.1124/jpet.109.159855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen G, Dellinger RW, Gallagher CJ, Sun D, Lazarus P. Identification of a prevalent functional missense polymorphism in the UGT2B10 gene and its association with UGT2B10 inactivation against tobacco-specific nitrosamines. Pharmacogenet Genomics. 2008;18:181–91. doi: 10.1097/FPC.0b013e3282f4dbdd. [DOI] [PubMed] [Google Scholar]
- 41.Chen G, Blevins-Primeau AS, Dellinger RW, Muscat JE, Lazarus P. Glucuronidation of nicotine and cotinine by UGT2B10: loss of function by the UGT2B10 Codon 67 (Asp>Tyr) polymorphism. Cancer Res. 2007;67:9024–9. doi: 10.1158/0008-5472.CAN-07-2245. [DOI] [PubMed] [Google Scholar]
- 42.Lazarus P, Zheng Y, Aaron Runkle E, Muscat JE, Wiener D. Genotype-phenotype correlation between the polymorphic UGT2B17 gene deletion and NNAL glucuronidation activities in human liver microsomes. Pharmacogenet Genomics. 2005;15:769–78. doi: 10.1097/01.fpc.0000175596.52443.ef. [DOI] [PubMed] [Google Scholar]
- 43.Wiener D, Fang JL, Dossett N, Lazarus P. Correlation between UDP-glucuronosyltransferase genotypes and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone glucuronidation phenotype in human liver microsomes. Cancer Res. 2004;64:1190–6. doi: 10.1158/0008-5472.can-03-3219. [DOI] [PubMed] [Google Scholar]
- 44.Nakajima M, Tanaka E, Kwon JT, Yokoi T. Characterization of nicotine and cotinine N-glucuronidations in human liver microsomes. Drug Metab Dispos. 2002;30:1484–90. doi: 10.1124/dmd.30.12.1484. [DOI] [PubMed] [Google Scholar]
- 45.Yamanaka H, Nakajima M, Katoh M, Kanoh A, Tamura O, Ishibashi H, et al. Trans-3′-hydroxycotinine O- and N-glucuronidations in human liver microsomes. Drug Metab Dispos. 2005;33:23–30. doi: 10.1124/dmd.104.001701. [DOI] [PubMed] [Google Scholar]
- 46.Kuehl GE, Murphy SE. N-glucuronidation of nicotine and cotinine by human liver microsomes and heterologously expressed UDP-glucuronosyltransferases. Drug Metab Dispos. 2003;31:1361–8. doi: 10.1124/dmd.31.11.1361. [DOI] [PubMed] [Google Scholar]
- 47.Kaivosaari S, Toivonen P, Hesse LM, Koskinen M, Court MH, Finel M. Nicotine glucuronidation and the human UDP-glucuronosyltransferase UGT2B10. Mol Pharmacol. 2007;72:761–8. doi: 10.1124/mol.107.037093. [DOI] [PubMed] [Google Scholar]
- 48.Wiener D, Doerge DR, Fang JL, Upadhyaya P, Lazarus P. Characterization of N-glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) in human liver: importance of UDP-glucuronosyltransferase 1A4. Drug Metab Dispos. 2004;32:72–9. doi: 10.1124/dmd.32.1.72. [DOI] [PubMed] [Google Scholar]
- 49.Ren Q, Murphy SE, Zheng Z, Lazarus P. O-Glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) by human UDP-glucuronosyltransferases 2B7 and 1A9. Drug Metab Dispos. 2000;28:1352–60. [PubMed] [Google Scholar]
- 50.Ehmer U, Vogel A, Schutte JK, Krone B, Manns MP, Strassburg CP. Variation of hepatic glucuronidation: Novel functional polymorphisms of the UDP-glucuronosyltransferase UGT1A4. Hepatology. 2004;39:970–7. doi: 10.1002/hep.20131. [DOI] [PubMed] [Google Scholar]
- 51.Dempsey D, Tutka P, Jacob P, 3rd, Allen F, Schoedel K, Tyndale RF, et al. Nicotine metabolite ratio as an index of cytochrome P450 2A6 metabolic activity. Clin Pharmacol Ther. 2004;76:64–72. doi: 10.1016/j.clpt.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 52.Haiman CA, Stram DO, Wilkens LR, Pike MC, Kolonel LN, Henderson BE, et al. Ethnic and racial differences in the smoking-related risk of lung cancer. N Engl J Med. 2006;354:333–42. doi: 10.1056/NEJMoa033250. [DOI] [PubMed] [Google Scholar]
- 53.Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF, et al. SEER Cancer Statistics Review, 1975-2011. National Cancer Institute; Bethesda, MD: 2014. [Google Scholar]
- 54.Benowitz NL, Dains KM, Dempsey D, Wilson M, Jacob P. Racial Differences in the Relationship Between Number of Cigarettes Smoked and Nicotine and Carcinogen Exposure. Nicotine & Tobacco Research. 2011;13:772–83. doi: 10.1093/ntr/ntr072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 56.Wilson W, 3rd, Pardo-Manuel de Villena F, Lyn-Cook BD, Chatterjee PK, Bell TA, Detwiler DA, et al. Characterization of a common deletion polymorphism of the UGT2B17 gene linked to UGT2B15. Genomics. 2004;84:707–14. doi: 10.1016/j.ygeno.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 57.Xue Y, Sun D, Daly A, Yang F, Zhou X, Zhao M, et al. Adaptive evolution of UGT2B17 copy-number variation. Am J Hum Genet. 2008;83:337–46. doi: 10.1016/j.ajhg.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Innocenti F, Liu W, Chen P, Desai AA, Das S, Ratain MJ. Haplotypes of variants in the UDP-glucuronosyltransferase1A9 and 1A1 genes. Pharmacogenet Genomics. 2005;15:295–301. doi: 10.1097/01213011-200505000-00004. [DOI] [PubMed] [Google Scholar]
- 59.Innocenti F, Ramirez J, Obel J, Xiong J, Mirkov S, Chiu YL, et al. Preclinical discovery of candidate genes to guide pharmacogenetics during phase I development: the example of the novel anticancer agent ABT-751. Pharmacogenet Genomics. 2013;23:374–81. doi: 10.1097/FPC.0b013e3283623e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jacob P, 3rd, Havel C, Lee DH, Yu L, Eisner MD, Benowitz NL. Subpicogram per milliliter determination of the tobacco-specific carcinogen metabolite 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol in human urine using liquid chromatography-tandem mass spectrometry. Anal Chem. 2008;80:8115–21. doi: 10.1021/ac8009005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng S, Kapur S, Sarkar M, Muhammad R, Mendes P, Newland K, et al. Respiratory retention of nicotine and urinary excretion of nicotine and its five major metabolites in adult male smokers. Toxicol Lett. 2007;173:101–6. doi: 10.1016/j.toxlet.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 62.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 63.Benowitz NL, Perez-Stable EJ, Fong I, Modin G, Herrera B, Jacob P., 3rd. Ethnic differences in N-glucuronidation of nicotine and cotinine. J Pharmacol Exp Ther. 1999;291:1196–203. [PubMed] [Google Scholar]
- 64.Coffman BL, King CD, Rios GR, Tephly TR. The glucuronidation of opioids, other xenobiotics, and androgens by human UGT2B7Y(268) and UGT2B7H(268). Drug Metab Dispos. 1998;26:73–7. [PubMed] [Google Scholar]
- 65.Turgeon D, Carrier JS, Chouinard S, Belanger A. Glucuronidation activity of the UGT2B17 enzyme toward xenobiotics. Drug Metab Dispos. 2003;31:670–6. doi: 10.1124/dmd.31.5.670. [DOI] [PubMed] [Google Scholar]
- 66.Court MH, Hao Q, Krishnaswamy S, Bekaii-Saab T, Al-Rohaimi A, von Moltke LL, et al. UDP-glucuronosyltransferase (UGT) 2B15 pharmacogenetics: UGT2B15 D85Y genotype and gender are major determinants of oxazepam glucuronidation by human liver. J Pharmacol Exp Ther. 2004;310:656–65. doi: 10.1124/jpet.104.067660. [DOI] [PubMed] [Google Scholar]
- 67.Greenblatt DJ, Divoll M, Harmatz JS, Shader RI. Oxazepam kinetics: effects of age and sex. J Pharmacol Exp Ther. 1980;215:86–91. [PubMed] [Google Scholar]
- 68.Strasser SI, Smid SA, Mashford ML, Desmond PV. Sex hormones differentially regulate isoforms of UDP-glucuronosyltransferase. Pharm Res. 1997;14:1115–21. doi: 10.1023/a:1012130118186. [DOI] [PubMed] [Google Scholar]
- 69.Lessov-Schlaggar CN, Benowitz NL, Jacob P, Swan GE. Genetic influences on individual differences in nicotine glucuronidation. Twin Res Hum Genet. 2009;12:507–13. doi: 10.1375/twin.12.5.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Muscat JE, Djordjevic MV, Colosimo S, Stellman SD, Richie JP., Jr Racial differences in exposure and glucuronidation of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK). Cancer. 2005;103:1420–6. doi: 10.1002/cncr.20953. [DOI] [PubMed] [Google Scholar]
- 71.Richie JP, Jr., Carmella SG, Muscat JE, Scott DG, Akerkar SA, Hecht SS. Differences in the urinary metabolites of the tobacco-specific lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in black and white smokers. Cancer Epidemiol Biomarkers Prev. 1997;6:783–90. [PubMed] [Google Scholar]
- 72.Kabat GC, Shivappa N, Hebert JR. Mentholated cigarettes and smoking-related cancers revisited: an ecologic examination. Regul Toxicol Pharmacol. 2012;63:132–9. doi: 10.1016/j.yrtph.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Amos CI, Gorlov IP, Dong Q, Wu X, Zhang H, Lu EY, et al. Nicotinic acetylcholine receptor region on chromosome 15q25 and lung cancer risk among African Americans: a case-control study. J Natl Cancer Inst. 2010;102:1199–205. doi: 10.1093/jnci/djq232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.