Abstract
The NME1 gene represents the prototypical metastasis suppressor, whose expression inhibits cell motility and metastasis without impact on primary tumor growth in a number of different human cancers. This report outlines our recent efforts to define the molecular mechanisms through which NME1 both suppresses cell motility and promotes genomic integrity in the setting of human melanoma. Forced NME1 expression in a variety of melanoma-derived cell lines was shown to induce dynamic changes in cell morphology and reorganization of the actin cytoskeleton, with formation of a network of thick stress fibers and assembly of fibronectin fibrils at large focal adhesions. Moreover, NME1 expression results in adhesion reprogramming through an impact on integrin repertoire and focal adhesion dynamics. Having previously demonstrated that NME1 expression promotes repair of DNA damage induced by ultraviolet radiation (UVR) in both yeast and mammalian cells, probably via the nucleotide excision repair pathway, we have more recently demonstrated that NME1 is rapidly recruited to double-strand breaks. This preliminary result represents the first evidence of direct interactions between NME1 and DNA in the context of DNA repair, and has set the stage for current efforts to probe its functional interactions with double-strand break repair pathways. Discussed herein are molecular models to explain the interactions of NME1 with such diverse cellular functions as cell motility and DNA repair, potentially through its nucleoside diphosphate kinase and 3′-5′ exonuclease activities.
Keywords: NME1, melanoma, metastasis, integrin, fibronectin, DNA repair
Introduction
Cutaneous malignant melanoma (CMM) is the most lethal form of skin cancer, and its incidence is increasing rapidly in the U.S. and around the world (http://seer.cancer.gov/statfacts/html/melan.html). A particularly disturbing increase has been noted for young women exposed to UV radiation in tanning beds (Purdue et al., 2008). Despite recent improvements in targeted therapies for management of metastatic melanoma (e.g. BRAF inhibitors and anti-CTL4), recurrence usually occurs. A better understanding of the metastatic process is acutely needed to prevent or inhibit melanoma in its advanced forms.
Metastasis suppressor genes have provided exceptional opportunities for understanding molecular events that specifically drive the metastatic cascade. The NME1 gene (aka nucleoside diphosphate kinase/NDPK, NM23-H1) represents the first prototype of a metastasis suppressor, and was initially identified in the mouse melanoma cell line K-1735 as an underexpressed transcript in a metastatic variant of the line (Leone et al., 1991). Confirmation of metastasis suppressor activity has been obtained in cell lines and tumors of melanoma, breast carcinoma and other cancers (Hartsough and Steeg, 2000). In fact, expression of NME1 RNA has been cited as one of the most reliable prognostic indices for positive outcome in CMM (Winnepenninckx et al., 2006). The protein exhibits three enzymatic activities that could mediate its suppressor function. First described was its nucleoside diphosphate kinase (NDPK) activity, proposed to maintain balance in nucleotide pools by catalyzing transfer of -phosphate between NDPs and NTPs (Agarwal et al., 1978). A substrate channeling mechanism has been proposed in which NTPs are directly transferred from the NDPK active site to effector molecules (Crawford et al., 2006). NME1 also exhibits a histidine kinase activity that may mediate its anti-motility function (Wagner et al., 1997). Our laboratory described its 3′-5′ exonuclease activity (Ma et al., 2004) and its essentiality for metastasis suppressor function (Zhang et al., 2010). Consistent with the known roles of these enzymes in DNA replication and repair (Shevelev and Hübscher, 2002), we demonstrated antimutator activity of NME1 in S. cerevisiae (Yang et al., 2009) and UVR-treated melanoma cells, via enhancement of the NER pathway (Jarrett et al., 2011). We recently validated metastasis suppressor activity of NME1 and the closely-related NME2 gene in melanoma in vivo, using a transgenic mouse model of UV-induced melanoma (Jarrett et al., 2013).
NME1 suppresses the motility and invasive characteristics of a number of cancer-derived cell lines (for a review, see (Salerno et al., 2003), and our laboratory has observed this activity in multiple human melanoma lines (Zhang et al., 2011; Jarrett et al., 2011). A prevailing view is that NME1 exerts an inhibitory influence on a number of intracellular substrates implicated in cell motility, probably involving direct protein-protein contacts (Marino et al., 2011). In addition, considerable evidence indicates that NME1 exerts broad-ranging effects on the transcriptome, although the underlying mechanisms have yet to be fully elucidated. Studies have demonstrated binding of NME1 and NME2 to single-stranded motifs in the promoter regions of the CMYC (Postel et al., 1993) and PDGFA (Ma et al., 2004; Ma et al., 2005) genes, and these interactions and others have been validated by chromatin immunoprecipitation (Cervoni et al., 2006; Cervoni et al., 2003; Egistelli et al., 2009), suggesting NME proteins may modulate transcription of genes that effect its suppressor function. In this regard, NME1 has been shown to regulate global gene expression profiles in the breast carcinoma cell line, MDA-MB-435, with expression of the lysophosphatidic acid receptor EDG2 identified as a motility-driving target of NME1-mediated suppression (Horak et al., 2007). Finally, our demonstration that NME expression promotes DNA repair pathways such as NER suggests the possibility that part of its metastasis suppressor activity is mediated through inhibiting the acquisition of metastasis-driving mutations. Clearly, the cellular functions of NME proteins are myriad and its potential impacts on the metastatic process are probably complex as well. The purpose of this report is to outline recent progress made in our laboratory to define the molecular mechanisms underlying the impact of NME1 on metastasis in melanoma. In particular, we focus on its ability to 1) regulate cytoskeletal and focal adhesion dynamics and 2) promote genomic stability through direct interactions with at least two DNA repair pathways, nucleotide excision repair (NER) and double-strand break repair (DSBR).
Motility suppressing effects of NME1 in melanoma cells are associated with induction of thick actin stress fibers and altered focal adhesion dynamics
Suppression of melanoma cell motility by NME1 is associated with profound changes in cell morphology and reorganization of the actin cytoskeleton
An early observation we made upon forced expression of NME1 in aggressive melanoma cell lines (e.g. 1205Lu, M14 and WM793) was a marked alteration in cell morphology. This change was characterized by a switch from a bipolar, spindle shape with multiple filopodia in the motile parent lines to a more spread out and polygonal appearance in the NME1 transfectants. As cell morphology and migration are well-recognized to be regulated through mechanical functions of the actin cytoskeleton (Lazaro-Dieguez et al., 2007), we conducted phalloidin staining for filamentous actin (F-actin) in these melanoma cell lines. The NME1-induced morphological transition was shown to be associated with generation of a network of thick F-actin bundles (stress fibers), contrasting with more finely punctate or diffuse actin staining in the parent cell line counterparts (Fig. 1). This is consistent with the observation that melanoma cells of low metastatic potential exhibit prominent stress fibers terminating in large focal adhesions, and are associated with enhanced adhesion and reduced motility (Volk et al., 1984). We have recently observed that fibronectin fibrils are assembled in NME1-expressing melanoma cells at large focal adhesions visualized with fluorescent paxillin. Moreover, our preliminary results suggest NME1 modestly induces expression of fibronectin mRNA and protein, which may contribute to formation of large focal adhesions. Nevertheless, localized secretion, proteolytic processing and matrix assembly also have profound effects on the function of fibronectin, and we are currently actively investigating the effect of NME1 on these processes as well. Further complexity of the role of fibronectin in metastasis is indicated by a report that fibronectin production by fibroblasts at future organ sites of metastasis promotes recruitment of bone marrow-derived cells, which serve to prime the pre-metastatic niche for tumor cell colonization (Barkan et al., 2010). Our results reinforce the notion that cell migration requires an optimal level of substrate adhesion, determined by the type and concentrations of adhesion ligands (e.g. fibronectin) in the extracellular matrix. NME1 appears to suppress motility by inducing localized deposition of fibronectin to cell-substrate adhesions at levels high enough to inhibit their turnover. Studies are underway to validate this hypothesis by using forced expression and shRNA approaches to assess the impact of endogenous fibronectin expression and matrix assembly on motility and metastatic potential of melanoma cells.
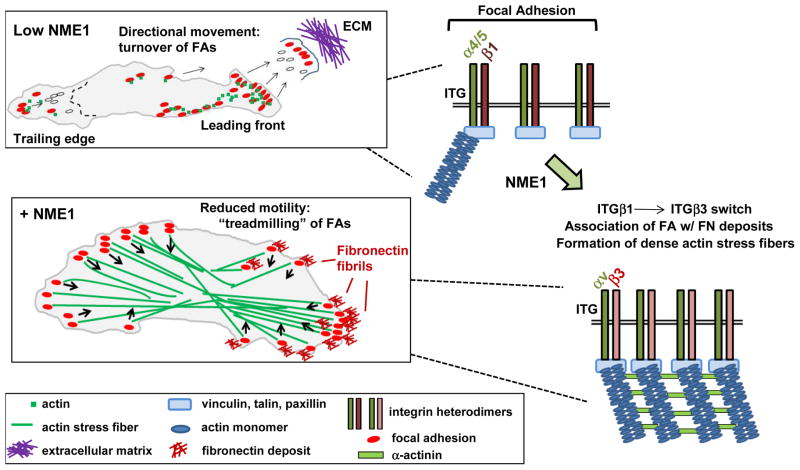
Fig. 1.
Model describing potential impact of NME1 expression on focal adhesion (FA) dynamics in melanoma cells. In the low NME1 condition shown at top, the motile melanoma cell is shown with its leading front at right and trailing edge at left. Forward membrane movement is accompanied by formation of new FAs and disassembly of old ones at both the leading and trailing edges. The dotted line shown in the cell represents the boundary of a future trailing edge. In the NME1-replete condition (“+NME1”), cell motility is reduced and associated with formation of robust stress fibers that terminate at FAs associated with arrays of short, extracellular FN fibrils. Importantly, FAs flow interiorly (arrows) from all points on the cell perimeter, are disassembled, and replaced at the perimeter with new FAs. The latter results in “treadmilling in place” with minimal net cell movement. At the right of each cell are shown the theoretical impact of NME1 expression on integrin β repertoire at FAs, and assembly of stress fibers.
NME1 expression results in adhesion reprogramming through alterations in integrin repertoire and focal adhesion dynamics
To assess the effect of NME1 on the dynamics of focal adhesion (FA) formation and turnover, we have recently employed time-lapse video TIRF (total internal reflection fluorescence) microscopy. Visualization of FAs in metastatic melanoma cell lines with a paxillin-DsRed fusion protein has revealed rapid FA turnover at both the leading and trailing edges of cells, with new FAs appearing continuously within newly-formed pseudopodia at the leading edge (Fig. 1). In contrast, forced NME1 expression was associated with larger and more stable FAs that were somewhat enriched at opposing poles but also seen on lateral edges. The FAs migrated from the edges towards the cell interior, and were continuously replaced by new FAs at all edges. The net result was a “treadmilling” of the FAs, which resulted in little or no net cell movement. Interestingly, preliminary studies indicate the effects of NME1 are not accompanied by cell-wide activation of such well-known small G protein modulators of cell motility as RhoA, Rac1 and Cdc42, nor did NME1 expression modulate the activation of these proteins in response to the chemokine SDF-1/CXCL12. This contrast with reported NME-mediated modulation of small G protein activation (for a review, see (Marino et al., 2011)) is somewhat unexpected and indicates alternate mechanisms may be operative in melanoma. We are also addressing whether the impact of NME1 on small G protein activation may be directed to specific subcellular locations depending upon sites of integrin-ligand interactions. Localized small G protein activation, particularly in relation to the leading and trailing edges of cells has been described (O’Connor et al., 2012), and might have been undetectable in our immunoblot analysis of whole cell extracts.
One potential clue to underlying mechanisms has been provided by a microarray analysis we conducted in WM793 melanoma cells, which identified NME1-mediated downregulation of the integrin 1 (ITGB1) subunit of the α5β1 fibronectin receptor. Increased ITGB1 expression has been associated with increased metastatic potential (Danen et al., 1994; Shibue and Weinberg, 2009; Huck et al., 2010; Grzesiak et al., 2011) and shortened patient survival (Nikkola et al., 2004; Yao et al., 2007; Oshita et al., 2004) in melanoma and other human cancers, and is receiving attention as a potential therapeutic target in metastatic disease (Barkan and Chambers, 2011). Efforts are ongoing in our laboratory to address the underlying mechanism through which NME1 regulates expression of ITGB1 and perhaps other members of the integrin family of proteins. In addition to its regulation of ITGB1 gene expression, NME1 may exert an impact on the spatiotemporal dynamics of integrin adhesion complexes at the cell surface. Of potential relevance is the equilibrium between two key fibronectin receptors, the α5β1 and αvβ3 integrins, which can have profound influence on adhesion strength, FA stability, mechanosensing, matrix assembly and, ultimately, motile and invasive behavior of tumor cells. This equilibrium has been shown to be maintained to a large extent through opposing profiles of dynamin-mediated endocytosis (Morgan et al., 2013).
Focal adhesions with rapid turnover rates, common in actively motile cells, have been shown to be enriched for the α5β1 complex. Conversely, stable FAs with slower endocytosis rates preferentially display the αvβ3 integrin complex. The conversion between slow and fast recycling beta integrin complexes is facilitated by activation of the small G protein Arf6 that in turn promotes dynamin-mediated endocytosis of FAs. The NDPK activity of NME1 has been proposed to provide GTP directly to Arf6 and dynamin to promote endocytosis (D’SouzaSchorey and Chavrier, 2006; Veluthakal et al., 2013), possibly through the direct process of “substrate channeling”, as described (Crawford et al., 2006). More recently, a “GTP fueling” hypothesis for NME1-mediated endosomal recycling has been proposed as a metastasis-suppressing mechanism in breast carcinoma via maintenance of E-cadherin expression at the cell surface. We are currently addressing the relevance of these NME1-driven mechanisms to possible switching of integrin subunits at the surface of melanoma cells (Fig. 1). It will also be of interest to determine whether the effects of NME1 in melanoma cells on integrin expression are cooperative with, or exclusive of, its reported interactions with intracellular modulators of cell motility and invasion (e.g. Tiam1, Dbl-1, Lbc, etc.)(Marino et al., 2011).
NME1 and DNA repair
Our entrance into the field of NME1 research was initiated by the identification of the protein as a DNA binding factor, revealed through the screening of a lambda phage cDNA expression library with a 5′-radiolabeled 33-mer oligodeoxynucleotide. Further study revealed NME1 exhibits a Mg+2-dependent 3′-5′ exonuclease activity on single-stranded DNA substrates (Ma et al., 2004). 3′-5′ exonucleases provide DNA proofreading and trimming functions during DNA repair and replication (Shevelev and Hübscher, 2002). These are functions which are known to be required in the double strand break repair (DSBR) and translesion synthesis (TLS) pathways of DNA repair/replication, the latter in particular by virtue of its use of low fidelity DNA polymerases (e.g. DNA polymerase) as part of its molecular mechanism. Our first evidence in support of a DNA repair function for NME1 was obtained in the yeast, Saccharomyces cerevisiae, in which ablation of the NME1 homolog YNK1 was shown to result in delayed and error-prone repair of DNA lesions induced by ultraviolet radiation (UVR) and the DNA topoisomerase II inhibitor, etoposide (Yang et al., 2009). Subsequently, we demonstrated that human melanoma cell lines with coordinately low expression of NME1 and NME2 (e.g. WM793) repaired UV-induced 6-4 photoproducts and other DNA polymerase-blocking lesions at a slow rate, and that the kinetics of repair were accelerated significantly upon forced expression of NME1 (Jarrett et al., 2011). These results were corroborated in embryo fibroblasts derived from transgenic mouse strains (MEFs) rendered null in the NME1 and/or NME2 loci, which exhibited much slower kinetics of repair of UV-induced DNA lesions than MEFs from wild-type mice. NME1 mutants deficient in NDPK activity (K12Q, H118F) exhibited impaired abilities to promote repair of UV-induced 6-4 photoproducts, while little impact was observed with a 3′-5′ exonuclease-inactivating mutation (E5A). This result was not surprising in light of the dominant role played by the nucleotide excision repair (NER) pathway in 6-4 photoproduct repair. The pathway involves a DNA polymerase fill-in step after excision of a DNA segment bearing the lesion, to which the NDPK and substrate channeling activity might contribute, but which has no known requirement for a 3′-5′ exonuclease activity (Fig. 2). Interestingly, the 3′-5′ exonuclease-dead mutant E5A was impaired in the ability to promote repair of DNA polymerase-blocking lesions induced by UV, as well as spontaneous and UV-induced mutations in the HPRT locus, strongly suggesting alternative repair mechanisms requiring the 3′-5′ exonuclease activity of NME1 may be operative in those settings.
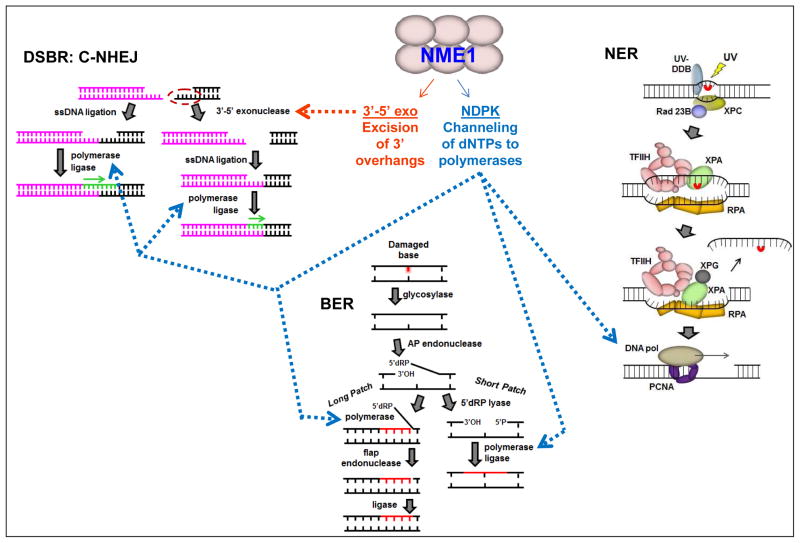
Fig. 2.
Potential contributions of enzymatic activities of NME1 to DNA repair pathways. Three examples of many known DNA repair pathways are represented: left, the classical non-homologous endjoining (C-NHEJ) pathway of double-strand break repair (DSBR); below center, the long and short patch pathways of base excision repair (BER); right, nucleotide excision repair (NER). Blue arrows are directed to those processes for which the NDPK activity of NME1 may contribute nucleotides to DNA polymerases through a substrate channeling mechanism, while the brown arrow represents one step in the C-NHEJ pathway at which the 3′-5′ exonuclease activity could participate.
More recently, to more directly address the relative contributions of the NDPK and 3′-5′ exonuclease activities of NME1 to DNA repair we have begun to focus on the DSBR pathway, which requires both DNA polymerase and 3′-5′ exonuclease activities (Fig. 2). One approach we are currently using is the retroviral delivery system for expression of the homing endonuclease I-PpoI as pioneered in the laboratory of Michael Kastan (Berkovich et al., 2007), which introduces DSBs at a relatively small number of defined loci. The approach is appealing because it can be used in conjunction with chromatin immunoprecipitation to assess the recruitment of NME1 to a specific DSB repair site. Preliminary results indicate that NME1 is indeed recruited rapidly (within 30 min) to I-PpoI-catalyzed DSBs, suggesting it may be part of the initial wave of proteins (e.g. ATM) recruited as part of the DSB-sensing machinery. Consistent with this notion, we have also observed co-immunoprecipitation of NME1 and ATM following treatment with the DSB inducer bleomycin. Current efforts are being directed to fully exploiting the I-PpoI system in a variety of cancer and nontransformed cell lines to measure the kinetics and topology of NME1 recruitment to DSBs. In addition, DSBR kinetics can be determined by quantitative PCR using primers spanning known I-PpoI cleavage sites. Experiments are also underway to dissect the relative contributions of NME proteins to the two primary forms of DSBR, homologous recombination (HR) and non-homologous end-joining (NHEJ). Cell line panels such as those we have constructed in the WM793 melanoma line to express wild-type and enzymatically-deficient forms of NME1 will be used to dissect the specific contributions of the NDPK and 3′-5′ exonuclease activities to these DSBR pathways.
Addressing the potential impact of the genome stabilizing activity of NME1 on cancer progression will probably prove more challenging than evaluating its effect on the motile and invasive properties of cancer cells. Nevertheless, we have already demonstrated that ablation of its 3′-5′ exonuclease activity is correlated with loss of metastasis suppressor function (Zhang et al., 2011), at least strongly suggesting the prospect that its DNA repair function is required. We are currently applying the powerful techniques of whole genome sequencing to our NME knockout mouse model of metastatic melanoma in an effort to identify metastasis-driving mutations that may arise as a consequence of NME loss. Further analysis of the time course of their appearance and their potential interactions with known melanoma drivers would be highly informative. In fact, the possibility exists that mutations secondary to NME1 deficiency could provide evidence that NME1 loss may be initiated quite early in a subpopulation of cells within the primary tumor, perhaps playing a role in genesis or progression of a cancer stem cell compartment.
Conclusions
One implication of our studies is that NME proteins interact physically and functionally with very disparate processes in melanoma cells. On one hand, expression of NME1 has dramatic effects on morphology and motility phenotypes, at least some of which is directed to promoting strong focal adhesions via targeted secretion of fibronectin and suppressing presentation of the motility-driving integrin ITGB1 at the cell surface. The mechanisms underlying these events remain obscure at present, although we have uncovered some evidence that NME1 may regulate expression of genes that play key roles. On the other hand, NME1 clearly promotes genomic stability through interactions with at least two DNA repair pathways, NER and DSBR, and we have obtained preliminary evidence to indicate it participates directly in those processes. Moreover, NME1 clearly regulates gene expression in a host of different cellular settings, although it is not yet clear whether this is a direct effect on gene transcription and/or RNA export/stability, or is secondary to signaling events initiated in the cytoplasm. An overall model summarizing the potential impacts of NME1 on cancer progression is presented in Fig. 3. Thus, at least two major questions come to mind: how does NME1 participate in such disparate processes, and how is such a singular and profound activity as metastasis suppression executed through so many regulatory targets? While the answers are not yet within our grasp, the substrate channeling hypothesis for NDPK activity would appear to provide some commonality. For example, the NDPK activity could “fuel” both the ARF6/dynamin machinery with GTP for recycling of motility-regulating proteins at the cell surface, as well as with dNTPs for DNA polymerase function in the nucleus during DNA repair. It seems likely that much of the specificity of the overall actions of NME proteins reside in the molecular partners with which they interact. Although NME1 has been labeled a “sticky” protein (Marino et al., 2011), careful and skilled application of biochemical and molecular biological approaches should provide insights needed for the eventual decoding of its biological functions and roles in cancer progression.
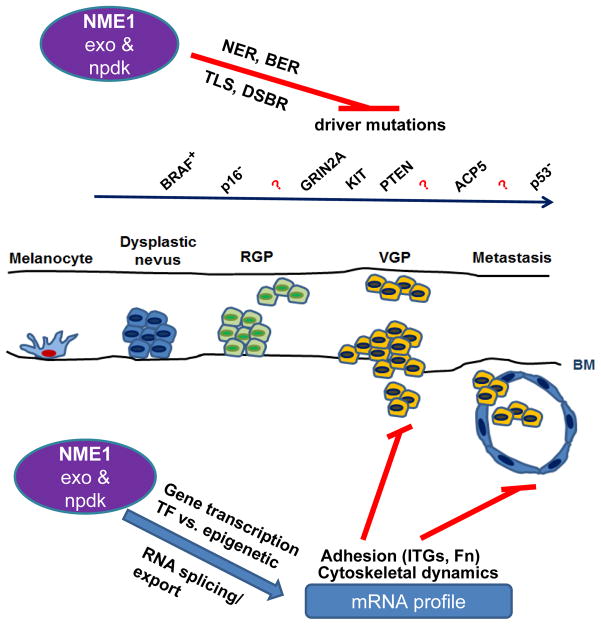
Fig. 3.
Hypothetical model of how NME1 expression and enzymatic activities acutely impacts the adhesive and cytoskeletal properties of melanoma cells, while chronically suppressing progression-driving mutations through its DNA repair function. RGP, radial growth phase; VGP, vertical growth phase.
Acknowledgments
This work was supported by the United States National Institutes of Health, National Cancer Institute grants CA83237 and CA159871 (D.M. Kaetzel), and training grant T32CA15474 (M.K. Leonard).
Contributor Information
David M. Kaetzel, Email: DKaetzel@som.umaryland.edu, Department of Biochemistry and Molecular Biology, and Greenebaum Cancer Center, School of Medicine, University of Maryland-Baltimore, Baltimore MD 21201, USA
Mary K. Leonard, Department of Biochemistry and Molecular Biology, and Greenebaum Cancer Center, School of Medicine, University of Maryland-Baltimore, Baltimore MD 21201, USA
Gemma S. Cook, Department of Biochemistry and Molecular Biology, and Greenebaum Cancer Center, School of Medicine, University of Maryland-Baltimore, Baltimore MD 21201, USA
Marian Novak, Department of Molecular and Biomedical Pharmacology, College of Medicine, University of Kentucky, Lexington, KY 40536, USA.
Stuart G. Jarrett, Department of Molecular and Biomedical Pharmacology, College of Medicine, University of Kentucky, Lexington, KY 40536, USA
Xiuwei Yang, Department of Molecular and Biomedical Pharmacology, College of Medicine, University of Kentucky, Lexington, KY 40536, USA.
Alexey M. Belkin, Department of Biochemistry and Molecular Biology, and Greenebaum Cancer Center, School of Medicine, University of Maryland-Baltimore, Baltimore MD 21201, USA
Reference List
- Agarwal RP, Robinson B, Parks RE. Nucleoside diphosphokinase from erythrocytes. Methods Enzymol. 1978;51:376–386. doi: 10.1016/s0076-6879(78)51051-3. [DOI] [PubMed] [Google Scholar]
- Barkan D, Chambers AF. beta1-integrin: a potential therapeutic target in the battle against cancer recurrence. Clin Cancer Res. 2011;17:7219–7223. doi: 10.1158/1078-0432.CCR-11-0642. [DOI] [PubMed] [Google Scholar]
- Barkan D, Green JE, Chambers AF. Extracellular matrix: a gatekeeper in the transition from dormancy to metastatic growth. Eur J Cancer. 2010;46:1181–1188. doi: 10.1016/j.ejca.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- Cervoni L, Egistelli L, Eufemi M, d’Abusco AS, Altieri F, Lascu I, Turano C, Giartosio A. DNA sequences acting as binding sites for NM23/NDPK proteins in melanoma M14 cells. J Cell Biochem. 2006;98:421–428. doi: 10.1002/jcb.20808. [DOI] [PubMed] [Google Scholar]
- Cervoni L, Pietrangeli P, Chichiarelli S, Altieri F, Egistelli L, Turano C, Lascu I, Giartosio A. In vivo cross-linking of nm23/nucleoside diphosphate kinase to the PDGF-A gene promoter. Mol Biol Rep. 2003;30:33–40. doi: 10.1023/a:1022261009207. [DOI] [PubMed] [Google Scholar]
- Crawford RM, Treharne KJ, Arnaud-Dabernat S, Daniel JY, Foretz M, Viollet B, Mehta A. Understanding the molecular basis of the interaction between NDPK-A and AMPK alpha 1. Mol Cell Biol. 2006;26:5921–5931. doi: 10.1128/MCB.00315-06. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- Danen EH, Ten Berge PJ, van Muijen GN, Van ‘t Hof-Grootenboer AE, Brocker EB, Ruiter DJ. Emergence of alpha 5 beta 1 fibronectin- and alpha v beta 3 vitronectin-receptor expression in melanocytic tumour progression. Histopathology. 1994;24:249–256. doi: 10.1111/j.1365-2559.1994.tb00517.x. [DOI] [PubMed] [Google Scholar]
- Egistelli L, Chichiarelli S, Gaucci E, Eufemi M, Schinina ME, Giorgi A, Lascu I, Turano C, Giartosio A, Cervoni L. IFI16 and NM23 bind to a common DNA fragment both in the P53 and the cMYC gene promoters. J Cell Biochem. 2009;106:666–672. doi: 10.1002/jcb.22053. [DOI] [PubMed] [Google Scholar]
- Grzesiak JJ, Tran Cao HS, Burton DW, Kaushal S, Vargas F, Clopton P, Snyder CS, Deftos LJ, Hoffman RM, Bouvet M. Knockdown of the beta(1) integrin subunit reduces primary tumor growth and inhibits pancreatic cancer metastasis. Int J Cancer. 2011;129:2905–2915. doi: 10.1002/ijc.25942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsough MT, Steeg PS. Nm23/nucleoside diphosphate kinase in human cancers. J Bioenerg Biomembr. 2000;32:301–308. doi: 10.1023/a:1005597231776. [DOI] [PubMed] [Google Scholar]
- Horak CE, Lee JH, Elkahloun AG, Boissan M, Dumont S, Maga TK, Arnaud-Dabernat S, Palmieri D, Stetler-Stevenson WG, Lacombe ML, Meltzer PS, Steeg PS. Nm23-H1 suppresses tumor cell motility by down-regulating the lysophosphatidic acid receptor EDG2. Cancer Res. 2007;67:7238–7246. doi: 10.1158/0008-5472.CAN-07-0962. [DOI] [PubMed] [Google Scholar]
- Huck L, Pontier SM, Zuo DM, Muller WJ. beta1-integrin is dispensable for the induction of ErbB2 mammary tumors but plays a critical role in the metastatic phase of tumor progression. Proc Natl Acad Sci U S A. 2010;107:15559–15564. doi: 10.1073/pnas.1003034107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett SG, Novak M, Dabernat S, Daniel JY, Mellon I, Zhang Q, Harris N, Ciesielski MJ, Fenstermaker RA, Kovacic D, Slominski A, Kaetzel DM. Metastasis suppressor NM23-H1 promotes repair of UV-induced DNA damage and suppresses UV-induced melanomagenesis. Cancer Res. 2011 doi: 10.1158/0008-5472.CAN-11-1795. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett SG, Novak M, Harris N, Merlino G, Slominski A, Kaetzel DM. NM23 deficiency promotes metastasis in a UV radiation-induced mouse model of human melanoma. Clin Exp Metastasis. 2013;30:25–36. doi: 10.1007/s10585-012-9495-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro-Dieguez F, Colonna C, Cortegano M, Calvo M, Martinez SE, Egea G. Variable actin dynamics requirement for the exit of different cargo from the trans-Golgi network. FEBS Lett. 2007;581:3875–3881. doi: 10.1016/j.febslet.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Leone A, Flatow U, King CR, Sandeen MA, Margulies IM, Liotta LA, Steeg PS. Reduced tumor incidence, metastatic potential, and cytokine responsiveness of nm23-transfected melanoma cells. Cell JID - 0413066. 1991;65:25–35. doi: 10.1016/0092-8674(91)90404-m. [DOI] [PubMed] [Google Scholar]
- Ma D, McCorkle JR, Kaetzel DM. The metastasis suppressor NM23-H1 possesses 3′-5′ exonuclease activity. J Biol Chem. 2004;279:18073–18084. doi: 10.1074/jbc.M400185200. [DOI] [PubMed] [Google Scholar]
- Ma D, Nutt CL, Shanehsaz P, Peng X, Louis DN, Kaetzel DM. Autocrine PDGF-dependent gene expression in glioblastoma cells is mediated largely by activation of the transcription factor SRE-BP, and is associated with altered genotype and patient survival in human brain tumors. Cancer Res. 2005;65:5523–5534. doi: 10.1158/0008-5472.CAN-04-2582. [DOI] [PubMed] [Google Scholar]
- Marino N, Marshall JC, Steeg PS. Protein-protein interactions: a mechanism regulating the anti-metastatic properties of Nm23-H1. Naunyn Schmiedebergs Arch Pharmacol. 2011;384:351–362. doi: 10.1007/s00210-011-0646-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MR, Hamidi H, Bass MD, Warwood S, Ballestrem C, Humphries MJ. Syndecan-4 phosphorylation is a control point for integrin recycling. Dev Cell. 2013;24:472–485. doi: 10.1016/j.devcel.2013.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikkola J, Vihinen P, Vlaykova T, Hahka-Kemppinen M, Heino J, Pyrhonen S. Integrin chains beta1 and alphav as prognostic factors in human metastatic melanoma. Melanoma Res. 2004;14:29–37. doi: 10.1097/00008390-200402000-00005. [DOI] [PubMed] [Google Scholar]
- O’Connor KL, Chen M, Towers LN. Integrin alpha6beta4 cooperates with LPA signaling to stimulate Rac through AKAP-Lbc-mediated RhoA activation. Am J Physiol Cell Physiol. 2012;302:C605–C614. doi: 10.1152/ajpcell.00095.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshita F, Kameda Y, Hamanaka N, Saito H, Yamada K, Noda K, Mitsuda A. High expression of integrin beta1 and p53 is a greater poor prognostic factor than clinical stage in small-cell lung cancer. Am J Clin Oncol. 2004;27:215–219. doi: 10.1097/01.coc.0000054894.64867.80. [DOI] [PubMed] [Google Scholar]
- Postel EH, Berberich SJ, Flint SJ, Ferrone CA. Human c-myc transcription factor PuF identified as nm23-H2 nucleoside diphosphate kinase, a candidate suppressor of tumor metastasis. Science. 1993;261:478–480. doi: 10.1126/science.8392752. [DOI] [PubMed] [Google Scholar]
- Purdue MP, Freeman LE, Anderson WF, Tucker MA. Recent trends in incidence of cutaneous melanoma among US Caucasian young adults. J Invest Dermatol. 2008;128:2905–2908. doi: 10.1038/jid.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salerno M, Ouatas T, Palmieri D, Steeg PS. Inhibition of signal transduction by the nm23 metastasis suppressor: possible mechanisms. Clin Exp Metastasis. 2003;20:3–10. doi: 10.1023/a:1022578000022. [DOI] [PubMed] [Google Scholar]
- Shevelev IV, Hübscher U. The 3′-5′ exonucleases. Nat Rev Mol Cell Biol. 2002;3:1–12. doi: 10.1038/nrm804. [DOI] [PubMed] [Google Scholar]
- Shibue T, Weinberg RA. Integrin beta1-focal adhesion kinase signaling directs the proliferation of metastatic cancer cells disseminated in the lungs. Proc Natl Acad Sci U S A. 2009;106:10290–10295. doi: 10.1073/pnas.0904227106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veluthakal R, Kaetzel D, Kowluru A. Nm23-H1 regulates glucose-stimulated insulin secretion in pancreatic beta-cells via Arf6-Rac1 signaling axis. Cell Physiol Biochem. 2013;32:533–541. doi: 10.1159/000354457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk T, Geiger B, Raz A. Motility and adhesive properties of high- and low-metastatic murine neoplastic cells. Cancer Res. 1984;44:811–824. [PubMed] [Google Scholar]
- Wagner PD, Steeg PS, Vu ND. Two-component kinase-like activity of nm23 correlates with its motility-suppressing activity. Proc Natl Acad Sci U S A. 1997;94:9000–9005. doi: 10.1073/pnas.94.17.9000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnepenninckx V, Lazar V, Michiels S, Dessen P, Stas M, Alonso SR, Avril MF, Ortiz Romero PL, Robert T, Balacescu O, Eggermont AM, Lenoir G, Sarasin A, Tursz T, van den Oord JJ, Spatz A. Gene expression profiling of primary cutaneous melanoma and clinical outcome. J Natl Cancer Inst. 2006;98:472–482. doi: 10.1093/jnci/djj103. [DOI] [PubMed] [Google Scholar]
- Yang M, Jarrett SG, Craven R, Kaetzel DM. YNK1, the yeast homolog of human metastasis suppressor NM23, is required for repair of UV radiation- and etoposide-induced DNA damage. Mutat Res. 2009;660:74–78. doi: 10.1016/j.mrfmmm.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao ES, Zhang H, Chen YY, Lee B, Chew K, Moore D, Park C. Increased beta1 integrin is associated with decreased survival in invasive breast cancer. Cancer Res. 2007;67:659–664. doi: 10.1158/0008-5472.CAN-06-2768. [DOI] [PubMed] [Google Scholar]
- Zhang Q, McCorkle JR, Novak M, Yang M, Kaetzel DM. Metastasis suppressor function of NM23-H1 requires its 3′;-5′ exonuclease activity. Int J Cancer. 2010 doi: 10.1002/ijc.25307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, McCorkle JR, Novak M, Yang M, Kaetzel DM. Metastasis suppressor function of NM23-H1 requires its 3′;-5′ exonuclease activity. Int J Cancer. 2011;128:40–50. doi: 10.1002/ijc.25307. [DOI] [PMC free article] [PubMed] [Google Scholar]