Abstract
Background
Breast cancer (BC) is considered to be one of the most important causes of death worldwide, and it affects the Iranian female population a decade earlier than female in other parts of the world. Human telomerase reverse transcriptase (hTERT) is a main subunit of the telomerase complex. MNS16A is located downstream of the hTERT gene and is recognized as the regulator of hTERT promoter activity. The aim of the present study was to access the possible impact of hTERT variants on BC risk in an Iranian population in southeast Iran.
Methods
A total of 491 subjects including 266 BC patients and 225 healthy women participated in the study. Polymerase chain reaction (PCR) was used to genotype the MNS16A variable number of tandem repeats and 177 bp ins/del polymorphisms in the hTERT gene. PCR-RFLP and ARMS-PCR were used to genotype hTERT rs2736098 and 2735940, respectively. The association between genotypes and BC was assessed by computing the odds ratio (OR) and 95% confidence intervals (95% CI) from logistic regression analyses. A p-value of <0.05 was considered statistically significant.
Results
The MNS16A genotype frequency distribution in BC patients was: LL, 43.2%; LS, 51.1%; and SS, 5.7%, and in controls: LL, 29.5%; LS, 68.3%; and SS, 2.2%. The LS genotype decreased the risk of BC compared with LL (OR = 0.51, 95% CI = 0.35-0.75, p < 0.001). The hTERT 177 bp ins/del polymorphism was not polymorphic in our population. All subjects had the ins/ins genotype. Our findings indicate that the MNS16A genotype and hTERT rs2736098 variant were associated with BC risk in the study. We also showed that the rs2736098 A/G polymorphism increased the risk of BC (OR = 1.80, 95% CI = 1.12-2.88, p = 0.017, AG vs AA; OR = 1.80, 95% CI = 1.06-3.06, p = 0.033, GG vs AA; OR = 1.87, 95% CI = 1.19-2.94, p = 0.006, AG + GG vs AA). No significant association was found between the rs2735940 C/T variant and BC.
Conclusion
Our findings indicate that the MNS16A genotype and the hTERT rs2736098 variant influence the risk of BC in an Iranian population in southeast Iran.
Keywords: hTERT, Breast cancer, Polymorphism, Genotyping, MNS16A
Background
Many women are diagnosed with breast cancer (BC) each day worldwide. Globally, BC may be considered the most common cancer among females and it is also the leading cause of cancer-related deaths in many countries [1]. BC is recognized as an important health care problem worldwide, affecting approximately 1 million women annually [1–3]. BC is also reported to be one of the most frequent malignancies among Iranian women, and it comprises 21.4% of female cancers in this population [4]. Interestingly, it has been reported that BC affects Iranian women about a decade earlier than women in Western countries [5], which highlights the importance of research on BC in the Iranian population. Several different factors are involved in BC pathogenesis, but its exact etiology is complicated and is not clearly identified. Our previous investigations provided solid evidence that genetic factors play important roles in the pathogenesis and progression of this malignancy in the population in southeast Iran [6–11].
Telomeres are repeat sequences of TTAGGG at the end of linear chromosomes and are responsible for protecting against loss of genetic information during the process of cellular division [12, 13]. Repeated cell cycles cause telomere shortening, drive the cells into the senescence condition, and finally trigger programmed cell death I, which potentially protects the cells against genomic instability and carcinogenesis [14]. Therefore, telomeres could be considered to be a key factors in cellular genomic maintenance and a potential candidate for carcinogenesis [12]. Human telomerase reverse transcriptase (hTERT) is a catalytic subunit of the telomerase [15], which together with the telomerase RNA component (TERC), is the main subunit of the telomerase complex [16]. Human TERT (hTERT) is located in 5p15.33 [17] and consists of 16 exons. Telomerase is a ribonucleoprotein that maintains integrity in the telomere regions, which subsequently shorten each replication cycle [18, 19]. Without telomeres, genomes would increasingly lose their information and would be truncated after cell division because enzymes that replicate DNA cannot continue duplication all the way to the chromosome ends [20]. It has been proposed that genetic instability is a driving force for transformation of normal cells into malignant cells during carcinogenesis [21].
MNS16A is known as a polymorphic tandem repeat minisatellite that is located downstream of the hTERT gene and it was first shown to be involved in promoter activity in lung cancer cell lines [22]. The variants that contain short tandem repeats have more effective promoter activity than those with long repeats, highlighting the importance of the number of tandem repeats in the risk of lung cancer. Many other groups have investigated the role of MNS16A in the etiology of different malignancies including cerebral [23], lung [24], breast [25], and colorectal cancer [26], but their results were inconsistent.
Because hTERT is the key molecular complex that maintains telomere stability, genetic variants in hTERT might impact on the risk of BC. However, considering the important role of MNS16A in hTERT gene promoter activity, we evaluated the MNS16A genotype and the impact of hTERT polymorphisms on BC susceptibility in a sample of the Iranian population.
Methods
Patients
This case-control study enrolled 266 pathologically confirmed BC patients who were referred to the Ali Ebneh Abitaleb hospital (Iran) and 225 age- and population-matched healthy women who participated in a screening project for metabolic syndrome; they were unrelated to the patients and had no history of any type of cancer. The clinicopathologic characteristics of the patients are summarized in Table 1. Ethical approvals for recruitment were obtained from the local Ethics Committee of Zahedan University of Medical Sciences, and informed consent was obtained from all patients and healthy individuals. Blood samples from patients and healthy controls were collected in EDTA-containing tubes and DNA was extracted using the salting out method, as described previously [27]. The quality of the isolated DNA was verified using electrophoresis on 1% agarose gel, quantitated spectrophotometrically and stored at -20°C until further use.
Table 1.
Clinical and pathological characteristics of breast cancer patients
| Characteristics | Patients n (%) |
|---|---|
| Age (years) | |
| ≤50 | 148 (55.6) |
| >50 | 111 (41.7) |
| Unknown | 7 (2.6) |
| Pathological type | |
| Ductal | 175 (65.8) |
| Others | 91 (34.2) |
| Tumor Size (cm) | |
| ≤ 2 | 88 (33.1) |
| >2 | 163 (61.3) |
| Unknown | 15 (5.6) |
| Histological grade | |
| I | 46 (17.3) |
| II | 138 (51.9) |
| III | 45 (16.9) |
| IV | 1 (0.4) |
| Unknown | 36 (13.5) |
| Stage | |
| I | 44 (16.5) |
| II | 99 (37.2) |
| III | 70 (26.3) |
| IV | 39 (14.7) |
| Unknown | 14 (5.3) |
| Estrogen receptor | |
| Positive | 154 (57.9) |
| Negative | 83 (31.2) |
| Unknown | 29 (10.9) |
| Progesterone Receptor | |
| Positive | 147 (55.3) |
| Negative | 88 (33.1) |
| Unknown | 31 (11.6) |
| HER2 status | |
| Positive | 128 (48.1) |
| Negative | 124 (46.6) |
| Unknown | 14 (5.3) |
Polymerase chain reaction
Polymerase chain reaction (PCR) was used to genotype the MNS16A variable number of tandem repeat polymorphisms with the primer set, as previously reported [22]. The forward and reverse primer sequences were 5′-AGGATTCTGATCTCTGAAGGGTG-3′ and 5′-TCTGCCTGAGGAAGGACGTATG-3′, respectively. PCR was performed using 2X Prime Taq Premix (Genet Bio, Korea). The amplification procedure consisted of an initial denaturing step for 5 min at 95°C followed by 30 cycles for 30 s at 95°C, 20 s at 67.5°C, and 17 s at 72°C, as well as a final extension step for 10 min at 72°C. The PCR products were visualized on 3% agarose gel containing 0.5 μg/ml of ethidium bromide (Figure 1a) and genotypes were assigned as previously reported [22]: the 243 bp and 272 bp bands were classified as the short (S*) allele, and the 333 bp and 302 bp bands were classified as the long (L*) allele, thus defining the MNS16A genotypes as L*/L*, L*/S* and S*/S*.
Figure 1.
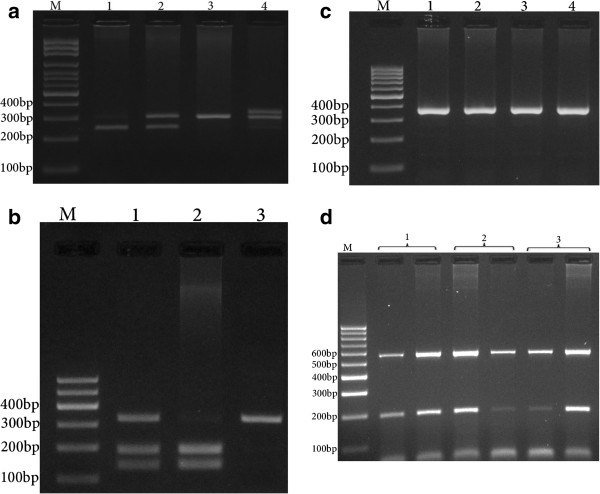
Photograph of DNA electophoresis for detection of hTERT polymorphisms. a: MNS16A. Lane 1, SS; lane 2, L/S; lanes 3 and 4, LL. b: rs1718119. Lane 1, GA; Lane 2, GG; lane 3, AA. c: hTERT 177 bp ins/del polymorphism. Lanes 1, 2, 3 and 4, ins/ins. d: rs2735940. Lane 1, TC; lane 2 TT; lane 3, CC. M = DNA marker.
hTERT 2736098 genotyping was accomplished using RFLP. The forward and reverse primers were 5′AGGACGCGTGGACCGAGTGA-3′ and 5′- GGAACCCAGAAAGATGGTCTC-3′, respectively. In each 0.20 ml reaction, 1 μl of genomic DNA (~100 ng/ml), 1 μl of each primer and 10 μl of 2X Prime Taq Premix (Genet Bio, Korea) and 7 μl ddH2O were added. The PCR conditions were set as follows: 95°C for 5 min, 30 cycles of 95°C for 30 s, 67°C for 30 s, and 72°C for 28 s and a final extension step of 72°C for 10 min. The PCR product (10 μl) was digested using Bsp120I restriction enzyme. The G allele was digested and produced 137 bp and 187 bp fragments while the A allele was undigested and produced a 324 bp fragment (Figure 1b).
hTERT 177 bp insertion/deletion genotyping was performed using PCR with forward (5′-GACCATCCTGGACTGATGGC-3′) and reverse (5′-AGGGGTGAACAATGGCGAAT-3′) primers, which can produce 366 bp and 189 bp insertion and deletion alleles, respectively. The PCR cycling conditions were 95°C for 5 min, 30 cycles of 95°C for 30 s, 66°C for 30 s, and 72°C for 26 s and a final extension step of 72°C for 10 min. The PCR products were verified on 2% agarose gels containing 0.5 μg/ml ethidium bromide, and observed under UV light (Figure 1c).
The hTERT rs2735940 polymorphism was genotyped using the amplification refractory mutation system polymerase chain reaction (ARMS-PCR) method. The primer sequences were as follows: generic primer, 5′-CGGCAAACACTGAAATGCTA-3′; T allele, 5′-GGGATTTCTAGAAGAGCGACAT-3′; and C allele, 5′-GGGATTTCTAGAAGAGCGACAC-3′. The product size for the allele was 197 bp. Beta-2 microglobulin (B2MF: 5′-TGTAAACACTTGGTGCCTGATATAGCTTGA-3′, B2MR: 5′-CATCAGTATCTCAGCAGGTGCCACTAATCT-3′), which produces 574 bp fragments, was used as an internal control.
In each 0.20 ml reaction solution, 1.3 μl of genomic DNA (~100 ng/ml), 0.5 μl of each primer and 10 μl of 2X Prime Taq Premix (Genet Bio, Korea) and 6.7 μl ddH2O were added. The PCR cycling condition was an initial denaturation at 95°C for 5 min followed by 30 cycles of 30 s at 95°C, annealing temperature for 23 s at 60°C, and 30 s at 72°C, with a final extension of 72°C for 10 min. The PCR products were verified on 2% agarose gels containing 0.5 μg/ml ethidium bromide, and observed under UV light (Figure 1d). Product sizes were 252 bp for either of the Ins or Del alleles, and 574 bp for the internal control. The presence of a 252 bp allele-specific band, in conjunction with a 574 bp control band, was considered to be positive evidence for each particular allele. The absence of an allele-specific band and the presence of a control band were considered to be evidence for the absence of an allele (Figure 1d).
Statistical analysis
Statistical analysis was performed using the statistical software package SPSS 18. The association between genotypes and BC were assessed by computing the odds ratio (OR) and 95% confidence intervals (95% CI) from logistic regression analyses. Haplotype analysis was performed using SNPStats software [28, 29]. The Hardy–Weinberg equilibrium was tested for polymorphisms. A p-value of <0.05 was considered statistically significant.
Results and discussion
Results
The study groups included 266 BC patients with a mean age of 48.9 ± 11.1 years and 225 healthy women with a mean age of 50.0 ± 12.9 years. The patient group demographic information is summarized in Table 1. No significant difference in age was found between the groups (p = 0.306). The frequency distribution of the MNS16A genotypes in BC patients was: LL, 43.2%; LS, 51.1%; and SS, 5.7%, and the distribution in controls was: LL, 29.5%; LS, 68.3%; and SS, 2.2% (Table 2). Our finding showed that the L/S and L/S + S/S decreased the risk of BC (OR = 0.51, 95% CI = 0.35-0.75, p < 0.001 and OR = 0.55, 95% CI = 0.38-0.81, p = 0.002, respectively) compared with the L/L genotype.
Table 2.
The genotypes and allele distribution of hTERT variants in breast cancer patients and the control group
| Variants | Patients n (%) | Controls n (%) | OR (95% CI) | p |
|---|---|---|---|---|
| MNS16A Genotype | ||||
| L/L | 115 (43.2) | 66 (29.5) | 1.00 | - |
| L/S | 136(51.1) | 153(68.3) | 0.51 (0.35-0.75) | <0.001 |
| S/S | 15 (5.7) | 5 (2.2) | 1.72 (0.61-4.95) | 0.460 |
| L/S + S/S | 151 (56.8) | 158 (65.5) | 0.55 (0.38-0.81) | 0.002 |
| Allele | ||||
| L | 366 (68.8) | 285 (63.6) | 1.00 | - |
| S | 166 (31.2) | 163 (36.7) | 0.79 (0.61-1.03) | 0.090 |
| rs2736098 | ||||
| AA | 40 (15.8) | 58 (26.1) | 1.00 | - |
| AG | 140 (55.3) | 113 (50.9) | 1.80 (1.12-2.88) | 0.017 |
| GG | 72 (28.5) | 51 (23.0) | 1.80 (1.06-3.06) | 0.033 |
| AG + GG | 212 (83.8) | 164 (73.9) | 1.87 (1.19-2.94) | 0.006 |
| Allele | ||||
| A | 220 (43.7) | 229 (51.6) | 1.00 | - |
| G | 284 (56.3) | 215 (48.4) | 1.38 (1.06-1.78) | 0.016 |
| rs2735940 | ||||
| CC | 45 (17.0) | 39 (17.3) | 1.00 | - |
| CT | 124 (47.0) | 138 (61.3) | 0.78 (0.48-1.28) | 0.380 |
| TT | 95 (36.0) | 48 (21.3) | 1.72 (0.99-2.98 | 0.066 |
| CT + TT | 219 (83.0) | 186 (82.6) | 1.02 (0.64-1.64) | 0.991 |
| Allele | ||||
| C | 214 (40.5) | 216 (48.0) | 1.00 | - |
| T | 314 (59.5) | 234 (52.0) | 1.35 (1.05-1.75) | 0.020 |
The hTERT rs2736098 A/G variant was associated with BC risk (Table 2). Our results indicated that AG as well as GG and AG + GG increased the risk of BC (OR = 1.80, 95% CI = 1.12-2.88, p = 0.017; OR = 1.80, 95%CI = 1.06-3.06, p = 0.033 and OR = 1.87, 95% CI = 1.19-2.94, p = 0.006, respectively) compared with the AA genotype. The rs2736098 G allele increased the risk of BC compared with the A allele (OR = 1.38, 95% CI = 1.06-1.78, p = 0.016). Our results also demonstrated that the hTERT rs2735940 polymorphism was not associated with BC risk/protection, while the rs2735940 T allele increased the risk of BC compared with the C allele (OR = 1.35, 95% CI = 1.05-1.75, p = 0.020). The 177 bp ins/del polymorphism was not polymorphic in our population so that all patients and controls had the insertion allele for the hTERT 177 bp ins/del polymorphism.
Haplotype analysis is shown in Table 3. Haplotypes LCA and STA decreased the risk of BC compared with LTG (MNS16A T/rs2735940 T/rs2736098 G). No significant association was observed among the hTERT polymorphisms and clinicopathologic parameters, including tumor stage, tumor grade, estrogen and progesterone receptors (ER, PgR), tumor size, and human growth factor receptor 2 (HER2) (Table 4).
Table 3.
MNS16A, rs2735940 and rs2736098 haplotype frequencies of hTERT polymorphisms in breast cancer patients and the control group
| MNS16A | rs2735940 | rs2736098 | Patient | Control | OR (95% CI) | p |
|---|---|---|---|---|---|---|
| L | T | G | 0.2316 | 0.1423 | 1.00 | - |
| L | C | G | 0.1638 | 0.2163 | 0.56 (0.22-1.42 | 0.220 |
| L | T | A | 0.1862 | 0.1259 | 0.92 (0.46-1.86) | 0.810 |
| L | C | A | 0.1064 | 0.1514 | 0.50 (0.25-0.99) | 0.048 |
| S | T | A | 0.1644 | 0.1047 | 0.35 (0.16-0.72) | 0.005 |
| S | T | G | 0.0975 | 0.0875 | 0.78 (0.23-2.61) | 0.680 |
| S | C | A | 0.0630 | 0.0739 | 0.41 (0.13-1.26) | 0.120 |
| S | C | G | 0.0720 | 0.0385 | 1.00 (0.18-5.68) | 0.970 |
Table 4.
Association between hTERT polymorphisms and clinicopathological characteristics
| Variables | MNS16A | p | rs2735940 | p | rs2736098 | p | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LL | LS | SS | CC | CT | TT | AA | AG | GG | ||||
| Age (years) | 0.123 | 0.181 | 0.893 | |||||||||
| ≤50 | 58 | 83 | 7 | 21 | 70 | 57 | 21 | 82 | 39 | |||
| >50 | 56 | 48 | 7 | 24 | 52 | 33 | 17 | 57 | 30 | |||
| Pathological type | 0.812 | 0.704 | ||||||||||
| Ductal | 78 | 87 | 10 | 30 | 85 | 60 | 30 | 91 | 45 | 0.351 | ||
| Others | 37 | 49 | 5 | 15 | 39 | 35 | 10 | 49 | 28 | |||
| Tumor size (cm) | 0.429 | 0.427 | 0.575 | |||||||||
| ≤2 | 35 | 47 | 6 | 12 | 44 | 32 | 14 | 48 | 19 | |||
| >2 | 77 | 79 | 7 | 33 | 76 | 54 | 22 | 89 | 46 | |||
| TNM Stage | 0.850 | 0.237 | 0.595 | |||||||||
| I | 19 | 22 | 3 | 7 | 25 | 12 | 8 | 27 | 8 | |||
| II | 47 | 48 | 5 | 19 | 41 | 40 | 12 | 51 | 32 | |||
| III | 32 | 35 | 3 | 14 | 38 | 17 | 12 | 37 | 17 | |||
| IV | 13 | 24 | 2 | 5 | 17 | 17 | 4 | 21 | 11 | |||
| Grade | 0.437 | 0.072 | 0.286 | |||||||||
| I | 23 | 21 | 2 | 10 | 19 | 17 | 10 | 20 | 14 | |||
| II | 64 | 67 | 7 | 18 | 62 | 58 | 17 | 81 | 34 | |||
| III | 14 | 30 | 1 | 11 | 26 | 8 | 6 | 22 | 15 | |||
| IV | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |||
| ER status | 0.858 | 0.090 | 0.260 | |||||||||
| Positive | 68 | 78 | 8 | 26 | 81 | 47 | 23 | 79 | 46 | |||
| Negative | 37 | 43 | 3 | 15 | 32 | 36 | 14 | 47 | 16 | |||
| PgR status | 0.627 | 0.620 | 0.137 | |||||||||
| Positive | 62 | 79 | 6 | 28 | 71 | 48 | 18 | 80 | 42 | |||
| Negative | 41 | 42 | 5 | 13 | 42 | 33 | 19 | 44 | 20 | |||
| HER2 status | 0.155 | 0.404 | 0.186 | |||||||||
| Positive | 50 | 69 | 9 | 27 | 59 | 42 | 15 | 75 | 32 | |||
| Negative | 61 | 59 | 4 | 18 | 60 | 45 | 22 | 59 | 36 | |||
Discussion
Telomeres are involved in maintaining genomic stability [30]. In the current study, we investigated the impact of hTERT variants on BC risk in a sample of the Iranian population in southeast Iran. Our data demonstrated that MNS16A LS and L/S + S/S decreased the risk of BC. AG as well as GG and AG + GG increased the risk of BC for the rs2736098 A/G polymorphism. The rs2736098 G allele was associated with an increased risk of BC. Although the hTERT rs2735940 C/T polymorphism was not associated with BC risk/protection, the rs2735940 T allele was significantly associated with BC risk. The 177 bp ins/del polymorphism was not polymorphic in our population (all individuals were the ins/ins genotype).
Earlier studies showed that hTERT mRNA expression is regulated by MNS16A in lung cancer [31], while studies in BC patients showed that MNS16A and BC risk association are strongly related to the geographic area of the study and the selection of the patient population [25, 32]. Glioblastoma multiforme studies have also confirmed that the MNS16A association with the risk of cancer incidence is highly dependent on the population’s ethnicity [23, 33, 34]. Overall, it can be concluded that there is much controversy regarding the association of MNS16A with different cancers, which highlights the importance of cancer origin and ethnicity in the results.
Studies on the hTERT rs2736098 variant also showed a significant controversy in association of the variant and BC risk [35–37]. Haiman et al. [37] observed a positive association between the 5p15 locus and the increased risk of BC while Savage et al. [35] suggested a protective effect of three correlated SNPs in this region, including rs2736098, among Polish women with a positive family history. For familial cancers, association was also observed, although not statistically significant, after Bonferroni adjustment. In the present study, we found that the rs2736098 polymorphism increased the risk of BC in our population, and it can be concluded that an association between BC risk and hTERT rs2736098 variant is generally related to ethnicity of the study population and the geographical location of the sample.
A functional variant located in the promoter of the hTERT gene, -1327C > T (rs2735940), is associated with telomere length [38]. There are few reports about the correlation between the hTERT rs2735940 variant and BC. Recently, Pellatt et al. found no association between the hTERT rs2735940 polymorphism and BC risk. They found that this variant was associated with estrogen receptor negative/progesterone receptor positive (ER-/PR-) tumors (OR = 0.73, 95% CI = 0.59-0.91) [39].
Conclusion
Our findings indicate that the MNS16A genotype and the hTERT rs2736098 variant influence the risk of BC in an Iranian population in southeast Iran. A limitation of this study is the relatively small sample size. Further research on hTERT polymorphisms is required to validate our findings in other ethnic groups in the Iranian population and in Middle Eastern countries. Because BC is a prevalent disease among the female population worldwide, identifying potential markers that can identify the possibility of this cancer is of significant importance in identifying BC in these patients.
Acknowledgement
This research was funded as a dissertation grant (SA’s MSc thesis) from the Deputy for Research, Zahedan University of Medical Sciences. SG was supported by a start-up grant from University of Manitoba, Canada.
Abbreviations
- BC
Breast cancer
- PCR
Polymerase chain reaction
- hTERT
Human telomerase reverse transcriptase
- ER
Estrogen receptor
- PgR
Progesterone receptor
- HER2
Human epidermal growth factor 2 receptor.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MH designed the study concepts, analyzed the data and prepared the manuscript. SA and ME conducted the study and drafted the manuscript. SMH and MT were involved in sample and data collection, and final approval of the manuscript. SG was involved in data analysis, drafting the manuscript and final approval of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Mohammad Hashemi, Email: mhd.hashemi@gmail.com.
Shadi Amininia, Email: shadi_am65@yahoo.com.
Mahboubeh Ebrahimi, Email: ebrahimi.m22@gmail.com.
Seyed Mehdi Hashemi, Email: hashemimahdi@zaums.ac.ir.
Mohsen Taheri, Email: amirt112@yahoo.com.
Saeid Ghavami, Email: saeid.ghavami@gmail.com.
References
- 1.Youlden DR, Cramb SM, Dunn NA, Muller JM, Pyke CM, Baade PD. The descriptive epidemiology of female breast cancer: an international comparison of screening, incidence, survival and mortality. Cancer Epidemiol. 2012;36(3):237–248. doi: 10.1016/j.canep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Wu JT, Kral JG. The NF-kappaB/IkappaB signaling system: a molecular target in breast cancer therapy. J Surg Res. 2005;123(1):158–169. doi: 10.1016/j.jss.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Turashvili G, Bouchal J, Burkadze G, Kolar Z. Differentiation of tumours of ductal and lobular origin: II. Genomics of invasive ductal and lobular breast carcinomas. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2005;149(1):63–68. doi: 10.5507/bp.2005.006. [DOI] [PubMed] [Google Scholar]
- 4.Babu GR, Samari G, Cohen SP, Mahapatra T, Wahbe RM, Mermash S, Galal OM. Breast cancer screening among females in Iran and recommendations for improved practice: a review. Asian Pac J Cancer Prev. 2011;12(7):1647–1655. [PubMed] [Google Scholar]
- 5.Kolahdoozan S, Sadjadi A, Radmard AR, Khademi H. Five common cancers in Iran. Arch Iran Med. 2010;13(2):143–146. [PubMed] [Google Scholar]
- 6.Hashemi M, Fazaeli A, Ghavami S, Eskandari-Nasab E, Arbabi F, Mashhadi MA, Taheri M, Chaabane W, Jain MV, Los MJ. Functional polymorphisms of FAS and FASL gene and risk of breast cancer - pilot study of 134 cases. PLoS One. 2013;8(1):e53075. doi: 10.1371/journal.pone.0053075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashemi M, Eskandari-Nasab E, Fazaeli A, Taheri M, Rezaei H, Mashhadi M, Arbabi F, Kaykhaei MA, Jahantigh M, Bahari G. Association between polymorphisms of glutathione S-transferase genes (GSTM1, GSTP1 and GSTT1) and breast cancer risk in a sample Iranian population. Biomark Med. 2012;6(6):797–803. doi: 10.2217/bmm.12.61. [DOI] [PubMed] [Google Scholar]
- 8.Hashemi M, Eskandari-Nasab E, Fazaeli A, Rezaei H, Mashhadi MA, Arbabi F, Taheri M. Bi-directional PCR allele-specific amplification (bi-PASA) for detection of caspase-8–652 6 N ins/del promoter polymorphism (rs3834129) in breast cancer. Gene. 2012;505(1):176–179. doi: 10.1016/j.gene.2012.05.043. [DOI] [PubMed] [Google Scholar]
- 9.Amininia S, Hashemi M, Ebrahimi M, Mashhadi MA, Hashemi SM, Taheri M, Ghavami S. Association between CCNE1 polymorphisms and the risk of breast cancer in a sample of southeast Iranian population. Med Oncol. 2014;31(10):189. doi: 10.1007/s12032-014-0189-z. [DOI] [PubMed] [Google Scholar]
- 10.Eskandari-Nasab E, Hashemi M, Ebrahimi M, Amininia S, Bahari G, Mashhadi MA, Taheri M. Evaluation of CCL5–403 G > A and CCR5 Delta32 gene polymorphisms in patients with breast cancer. Cancer Biomark. 2014;14(5):343–351. doi: 10.3233/CBM-140411. [DOI] [PubMed] [Google Scholar]
- 11.Hashemi M, Omrani M, Eskanadri-Nasab E, Hasani SS, Mashhadi M, Taheri M. A 40-bp Insertion/Deletion Polymorphism of Murine Double Minute2 (MDM2) Increased the Risk of Breast Cancer in Zahedan, Southeast Iran. Iran Biomed J. 2014;18(4):245–249. doi: 10.6091/ibj.13332.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basu N, Skinner HG, Litzelman K, Vanderboom R, Baichoo E, Boardman LA. Telomeres and telomere dynamics: relevance to cancers of the GI tract. Expert Rev Gastroenterol Hepatol. 2013;7(8):733–748. doi: 10.1586/17474124.2013.848790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mason PJ, Perdigones N. Telomere biology and translational research. Transl Res. 2013;162(6):333–342. doi: 10.1016/j.trsl.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vodenicharov MD, Wellinger RJ. The cell division cycle puts up with unprotected telomeres: cell cycle regulated telomere uncapping as a means to achieve telomere homeostasis. Cell Cycle. 2007;6(10):1161–1167. doi: 10.4161/cc.6.10.4224. [DOI] [PubMed] [Google Scholar]
- 15.Qian Y, Yang L, Cao S. Telomeres and telomerase in T cells of tumor immunity. Cell Immunol. 2014;289(1–2):63–69. doi: 10.1016/j.cellimm.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Gomez DE, Armando RG, Farina HG, Menna PL, Cerrudo CS, Ghiringhelli PD, Alonso DF. Telomere structure and telomerase in health and disease (review) Int J Oncol. 2012;41(5):1561–1569. doi: 10.3892/ijo.2012.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leem SH, Londono-Vallejo JA, Kim JH, Bui H, Tubacher E, Solomon G, Park JE, Horikawa I, Kouprina N, Barrett JC, Larionov V. The human telomerase gene: complete genomic sequence and analysis of tandem repeat polymorphisms in intronic regions. Oncogene. 2002;21(5):769–777. doi: 10.1038/sj.onc.1205122. [DOI] [PubMed] [Google Scholar]
- 18.Bacchetti S, Counter C. Telomeres and telomerase in human cancer (review) Int J Oncol. 1995;7(3):423–432. [PubMed] [Google Scholar]
- 19.Meyerson M. Role of telomerase in normal and cancer cells. J Clin Oncol. 2000;18(13):2626–2634. doi: 10.1200/JCO.2000.18.13.2626. [DOI] [PubMed] [Google Scholar]
- 20.O'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11(3):171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Soria JC, Chang YS, Lee HY, Wei Q, Mao L. Association of a functional tandem repeats in the downstream of human telomerase gene and lung cancer. Oncogene. 2003;22(46):7123–7129. doi: 10.1038/sj.onc.1206852. [DOI] [PubMed] [Google Scholar]
- 23.Andersson U, Osterman P, Sjostrom S, Johansen C, Henriksson R, Brannstrom T, Broholm H, Christensen HC, Ahlbom A, Auvinen A, Feychting M, Lönn S, Kiuru A, Swerdlow A, Schoemaker M, Roos G, Malmer B. MNS16A minisatellite genotypes in relation to risk of glioma and meningioma and to glioblastoma outcome. Int J Cancer. 2009;125(4):968–972. doi: 10.1002/ijc.24363. [DOI] [PubMed] [Google Scholar]
- 24.Jin G, Yoo SS, Cho S, Jeon HS, Lee WK, Kang HG, Choi YY, Choi JE, Cha SI, Lee EB, Kim CH, Jung TH, Kim YT, Park JY. Dual roles of a variable number of tandem repeat polymorphism in the TERT gene in lung cancer. Cancer Sci. 2011;102(1):144–149. doi: 10.1111/j.1349-7006.2010.01782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Hu Z, Liang J, Wang Z, Tang J, Wang S, Wang X, Qin J, Wang X, Shen H. A tandem repeat of human telomerase reverse transcriptase (hTERT) and risk of breast cancer development and metastasis in Chinese women. Carcinogenesis. 2008;29(6):1197–1201. doi: 10.1093/carcin/bgn099. [DOI] [PubMed] [Google Scholar]
- 26.Hofer P, Baierl A, Feik E, Fuhrlinger G, Leeb G, Mach K, Holzmann K, Micksche M, Gsur A. MNS16A tandem repeats minisatellite of human telomerase gene: a risk factor for colorectal cancer. Carcinogenesis. 2011;32(6):866–871. doi: 10.1093/carcin/bgr053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hashemi M, Eskandari-Nasab E, Fazaeli A, Bahari A, Hashemzehi NA, Shafieipour S, Taheri M, Moazeni-Roodi A, Zakeri Z, Bakhshipour A, Ghavami S. Association of genetic polymorphisms of glutathione-S-transferase genes (GSTT1, GSTM1, and GSTP1) and susceptibility to nonalcoholic fatty liver disease in Zahedan, Southeast Iran. DNA Cell Biol. 2012;31(5):672–677. doi: 10.1089/dna.2011.1343. [DOI] [PubMed] [Google Scholar]
- 28.Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 29.Hashemi M, Eskandari-Nasab E, Moazeni-Roodi A, Naderi M, Sharifi-Mood B, Taheri M. Association of CTSZ rs34069356 and MC3R rs6127698 gene polymorphisms with pulmonary tuberculosis. Int J Tuberc Lung Dis. 2013;17(9):1224–1228. doi: 10.5588/ijtld.12.0762. [DOI] [PubMed] [Google Scholar]
- 30.Hoch NC, Lai X, Heierhorst J. Genomic stability disorders: from budding yeast to humans. Front Biosci. 2013;5:396–411. doi: 10.2741/S379. [DOI] [PubMed] [Google Scholar]
- 31.Xia X, Rui R, Quan S, Zhong R, Zou L, Lou J, Lu X, Ke J, Zhang T, Zhang Y, Liu L, Yan J, Miao X. MNS16A tandem repeats minisatellite of human telomerase gene and cancer risk: a meta-analysis. PLoS One. 2013;8(8):e73367. doi: 10.1371/journal.pone.0073367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zagouri F, Sergentanis TN, Gazouli M, Tsigginou A, Dimitrakakis C, Eleutherakis-Papaiakovou E, Papaspyrou I, Chrysikos D, Theodoropoulos G, Zografos GC, Antsaklis A, Dimopoulos AM, Papadimitriou CA. HTERT MNS16A polymorphism in breast cancer: a case-control study. Mol Biol Rep. 2012;39(12):10859–10863. doi: 10.1007/s11033-012-1982-4. [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Wei Q, Wang LE, Aldape KD, Cao Y, Okcu MF, Hess KR, El-Zein R, Gilbert MR, Woo SY, Prabhu SS, Fuller GN, Bondy ML. Survival prediction in patients with glioblastoma multiforme by human telomerase genetic variation. J Clin Oncol. 2006;24(10):1627–1632. doi: 10.1200/JCO.2005.04.0402. [DOI] [PubMed] [Google Scholar]
- 34.Carpentier C, Lejeune J, Gros F, Everhard S, Marie Y, Kaloshi G, Laigle-Donadey F, Hoang-Xuan K, Delattre JY, Sanson M. Association of telomerase gene hTERT polymorphism and malignant gliomas. J Neurooncol. 2007;84(3):249–253. doi: 10.1007/s11060-007-9378-3. [DOI] [PubMed] [Google Scholar]
- 35.Savage SA, Chanock SJ, Lissowska J, Brinton LA, Richesson D, Peplonska B, Bardin-Mikolajczak A, Zatonski W, Szeszenia-Dabrowska N, Garcia-Closas M. Genetic variation in five genes important in telomere biology and risk for breast cancer. Br J Cancer. 2007;97(6):832–836. doi: 10.1038/sj.bjc.6603934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mocellin S, Verdi D, Pooley KA, Landi MT, Egan KM, Baird DM, Prescott J, De Vivo I, Nitti D. Telomerase reverse transcriptase locus polymorphisms and cancer risk: a field synopsis and meta-analysis. J Natl Cancer Inst. 2012;104(11):840–854. doi: 10.1093/jnci/djs222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haiman CA, Chen GK, Vachon CM, Canzian F, Dunning A, Millikan RC, Wang X, Ademuyiwa F, Ahmed S, Ambrosone CB, Baglietto L, Balleine R, Bandera EV, Beckmann MW, Berg CD, Bernstein L, Blomqvist C, Blot WJ, Brauch H, Buring JE, Carey LA, Carpenter JE, Chang-Claude J, Chanock SJ, Chasman DI, Clarke CL, Cox A, Cross SS, Deming SL, Diasio RB, et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat Genet. 2011;43(12):1210–1214. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burke LS, Hyland PL, Pfeiffer RM, Prescott J, Wheeler W, Mirabello L, Savage SA, Burdette L, Yeager M, Chanock S, De Vivo I, Tucker MA, Goldstein AM, Yang XR. Telomere length and the risk of cutaneous malignant melanoma in melanoma-prone families with and without CDKN2A mutations. PLoS One. 2013;8(8):e71121. doi: 10.1371/journal.pone.0071121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pellatt AJ, Wolff RK, Torres-Mejia G, John EM, Herrick JS, Lundgreen A, Baumgartner KB, Giuliano AR, Hines LM, Fejerman L, Cawthon R, Slattery ML. Telomere length, telomere-related genes, and breast cancer risk: the breast cancer health disparities study. Genes Chromosomes Cancer. 2013;52(7):595–609. doi: 10.1002/gcc.22056. [DOI] [PMC free article] [PubMed] [Google Scholar]


