Abstract
Background:
β-lactams resistant Streptococcus pneumoniae are an emerging problem throughout the world. Several resistance mechanisms have been reported, including expression of drug-destroying enzymes such as β-lactamases, altered drug targets such as conformational changes in PBPs, decreased bacterial permeability, and increased drug efflux.
Objectives:
The present study aimed to determine the relationship between the results of polymerase chain reaction identification of the Pbp1a, Pbp2b and Pbp2x genes (penicillin-binding proteins) and susceptibilities of β-lactam antibiotics against S. pneumoniae.
Materials and Methods:
Fifty five isolates of S. pneumoniae were obtained from clinical samples with antimicrobial tests. The susceptibilities of isolates to benzylpenicillin, imipenem, oxacillin, ceftazidime were determined. The resistance genotype was determined by the polymerase chain reaction with primers designed for the PBP genes.
Results:
The number of S. pneumoniae isolates resistant to benzylpenicillin, imipenem, oxacillin and ceftazidime were 94.5%, 100%, 100%, and 21.8%, respectively. Analysis of mutation in the genes for pbp showed that 85% of isolates had mutations in pbp2x, pbp2b and pbp1a. Susceptibility to benzylpenicillin was decreased once the number of mutated pbp genes in S. pneumonia increased. According to the results of this study, S. pneumoniae isolates showed reduced susceptibility due to accumulation of resistance genes.
Conclusions:
We suggest that studies should be performed to evaluate changes in Minimum Inhibitory Concentration (MIC) values as well as genetic mutations in order to determine prevalence of S. pneumoniae resistance against antimicrobial agents.
Keywords: S. pneumoniae, PCR, lytA, Pbp1a, Pbp2b, Pbp2x
1. Background
Streptococcus pneumoniae is a common etiological agent of serious invasive infections and the most common cause of pneumonia, which is a major cause of mortality throughout the world (1, 2). On the verge of the antibiotic era, clinical isolates of S. pneumoniae showed a high sensitivity to antibiotics such as benzylpenicillin, which had an exceptionally low (5-10 ng/mL) Minimum Inhibitory Concentration (MIC) against these strains. Penicillin has been commonly recommended as the antibiotic for pneumococcal infections. The emergence of drug-resistant S. pneumoniae posed a new difficult challenge for treatment of these infections (3). The initial incidence of clinically important penicillin-resistant isolates has since been reported worldwide.
The spread of penicillin and multidrug resistance among S. pneumonia has become an important universal issue and a challenge for the present treatment strategies (4). β-lactams are the most widely used antibiotics; however, extensive resistance has evolved among most common pathogens such as S. pneumoniae (5, 6). Antimicrobial susceptibility records on common pathogens including Escherichia coli, Klebsiella spp., Haemophilus influenzae, Staphylococcus aureus, S. pneumoniae, and S. pyogenes confirmed that antimicrobial resistance is still a limited issue (7). Penicillin and macrolide resistance in S. pneumoniae are considered as a serious concern globally, particularly, in Asian countries (8). The β-lactam antibiotics are broadly utilized to treat community and healthcare-associated infections, and the appearance and spread of antimicrobial resistance to this family of drugs is a significant threat to human health (9).
Several resistance mechanisms have been reported, including expression of drug-destroying enzymes such as β-lactamases (10, 11), altered drug targets such as conformational changes in PBPs (10), decreased bacterial permeability (10), and increased drug efflux (12). Understanding the principles for resistance mechanisms is essential and important for surveillance and control of infections. Furthermore, appropriate antimicrobial use is also of importance, for instance penicillins and cephalosporins (members of B-lactam antibiotics) inhibit PBPs, which are vital for bacterial cell wall biogenesis. Pathogenic bacteria have developed efficient antibiotic resistance mechanisms which in gram-positive bacteria, includes mutations in PBP genes that facilitate avoidance of B-lactam inhibition (13).
Resistance of S. pneumoniae to β -lactams is due to reduced affinity of PBPs, particularly PBP1A, PBP2B and PBP2X. S. pneumoniae contains the mosaic pbp sequence, which has high homology with pbp in oral streptococci (14). Resistance of S. pneumoniae to Β-lactams results from homologous recombination of pbp with the pbp genes of β-lactam-resistant oral streptococci (15-19). Thus, molecular epidemiological analysis, antimicrobial susceptibilities and resistance genes analysis are crucial in prevention and therapy of infectious diseases such as drug resistance in S. pneumoniae. Therefore, clinical laboratories should consider screening selected isolates to determine their susceptibility to macrolides, β-lactams, vancomycin or clindamycin.
2. Objectives
This study aimed to investigate the true prevalence of β-lactam resistance among clinically important isolates of S. pneumoniae and to examine susceptibility of S. pneumoniae to various antimicrobial agents including benzylpenicillin, imipenem, oxacillin and ceftazidim. In addition, the distribution of PBP genes of pbp1a, pbp2b, and pbp2x was investigated. Furthermore, the changes in MIC and dispensation of the antimicrobial-resistance genes were compared.
3. Materials and Methods
3.1. Bacterial Strains
Isolates of S. pneumoniae (n = 55) were collected from outpatients seen at the Medical Center of Hamadan. Clinical samples were cerebro-spinal fluid (CSF) (6), sputum (20), otorrhea (1), pharynx (8) and others (2).
3.2. Identification of Bacterial Isolates
All isolates were identified according to the method summarized by the Centers for Disease Control (21). Gram-positive cocci α- hemolytic colonies on blood agar plates supplemented with 5 mg/ mL gentamycin were collected with a wire loop and subcultured on blood agar plates. An optochin disk with a diameter of 6 mm (5 mg ethylhydrocupreine) was placed aseptically on the streak of inoculum. The plate was incubated at 37˚C in a 5% CO2 atmosphere for 18-24 hours. Isolates with an inhibition zone of 14 mm in diameter, which were susceptible to optochin, were selected. Strains with inhibition zones ranging from 9 mm to 13 mm were tested for bile solubility using 2% sodium deoxycholate (bile salt) (Oxoid, Basingstoke, Hamsphire, England). Finally isolates were confirmed by polymerase chain reaction (PCR) for autolysin (lytA) gene (22-24). Isolates were stored in 15% glycerol at -80˚ C until used for PCR and E-Test (25, 26).
3.3. Antimicrobial Test
The drug susceptibility of confirmed samples with microbial tests was determined using antibiotic E-test strips (Himedia Laboratories pvt. Ltd, India). Antibiotics employed in this study were: benzylpenicillin, imipenem, oxacillin, and ceftazidim. Inocula were prepared by direct suspension of colonies grown overnight on sheep blood agar in Muller-Hinton broth to achieve turbidity equivalent to a 0.5 Mc-Farland opacity standard. The 100-mm-diameter agar plates were inoculated by confluent swabbing of the surface with the adjusted inoculum suspensions. After application of the E-test strips, pneumococcal test plates were incubated in ambient air at 35˚C for 20 to 22 hours (25-28). E-test MICs were interpreted by noting the point of intersection of the growth ellipse margin with the MIC scale on the E-test strip when viewed from the upper agar surface with the plate lids removed (Figure 1).
Figure 1. Antibiogram Procedure by E-test.
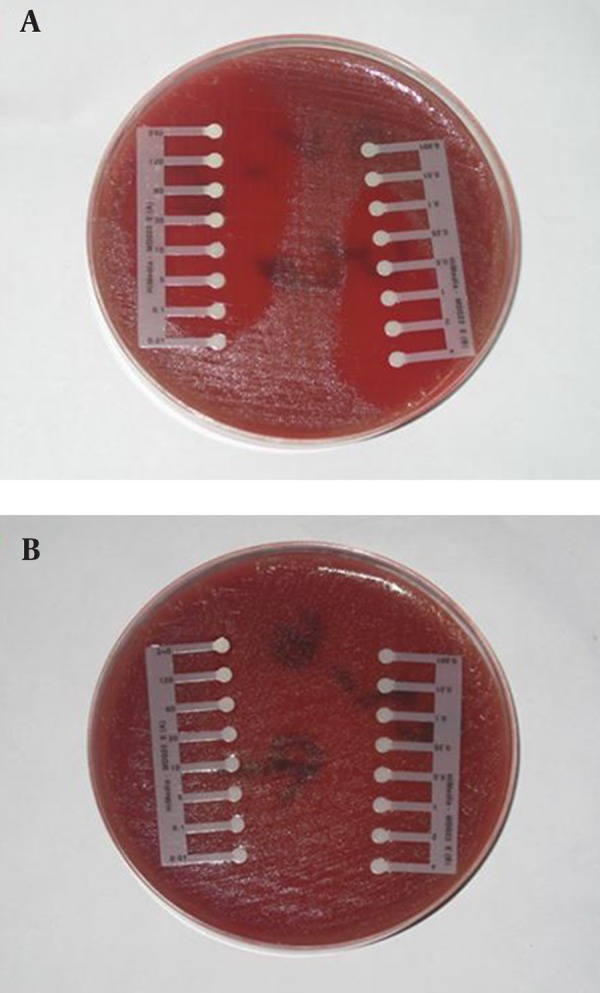
a, sensitive; b, resistant.
3.4. Polymerase Chain Reaction Analysis
Isolates were grown in blood agar medium containing 5% sheep blood agar (Merck, Darmstadt, Germany) with 5% CO2 at 37˚C for 48 hours. A single colony of S. pneumoniae was removed and suspended in lysis solution as reported previously and cells were lysed for 10 minutes at 60˚C followed by 5 minutes at 94˚C (22, 28). Furthermore, 1 µL of supernatant was used for the PCR reaction. Each reaction tube contained 20 µL solutions that included 10 x buffer (2 µL), MgCl2 25 Mm (0.4 µL), dNTPs 10 Mm (0.8 µL), distilled water (13.7 µL), Taq 5 U/µL (0.2 µL), DNA (1 µL), primer 1 (10 pm) (1 µL) and primer 2 (10 pm) (1 µL). Amplification of pbp1a, pbp2b, and pbp2x were examined by PCR using the primers reported by Ubukata and co-workers (22, 29). To raise the percentage of PRSP strains and to compensate for the variety of gene mutations, primers were designed to amplify the genes of susceptible strains but not of resistant strains.
Primer pairs used included: for lytA, 5'-TGAAGCGGATTATCATGGC-3' and 3'-GCTAAACTCCCT GTATCAAGCG-5' (273bp); for pbp1A, 5'-AAACAA GGTCGGACTCAACC-3' and 5-'AGGTGCTACAAATTGAGAGG-3' (430 bp); for Pbp2X, 5'-CCAGGTTCC ACTATGAAAGTG-3' and 5'-CATCCGTCAAACC GAAACGG-3' (292bp); for Pbp2B, 5'-CAATCTAGAGTC TGCTATGGA- 3' and 5-'GGTCAATTCCTGTCGC AGTA-3' (77 bp). PCR conditions were as follows: initial denaturation step at 94˚C for 3 minutes followed by 30 cycles at 94˚C for 20 seconds , 57˚C for 20 seconds and 72˚C for 15 seconds, and a final extension step at 72˚C for 7 minutes (21). The amplified DNA fragments were separated on a 3% agarose gel. S. pneumoniae PTCC 1240 was used as the control strain.
4. Results
Fifty five out of 400 samples were positive for S. pneumoniae with microbial test and PCR (Figure 2).
Figure 2. Polymerase Chain Reaction for Identification of lytA Gene (273 bp) in Isolates.
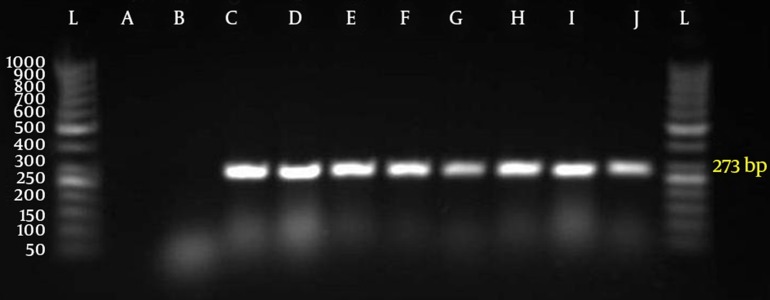
Gel electrophoresis with autolysin gene (L = marker 50 bp, agarose LE 3%, A = negative control with distilled water, B = negative control with E. coli, C = positive control with S. pneumoniae (PTCC NO.1240), D-J = lytA gene (273bp))
All 55 strains were classified into following six genotypes, according to PCR for pbp1a, pbp2x, pbp2b genes:
PSSP with three normal genes (n = 0%);
PISP with abnormal pbp2x (n = 100%);
PISP with abnormal pbp2b (n = 100%);
PISP with abnormal pbp1a (n = 85.5%);
PISP (pbp2x+2b) with abnormal pbp2x and pbp2b (n = 100%);
PISP (pbp1a+2x) with abnormal pbp1a and pbp2x (n = 100%);
PISP (pbp1a+2b) with abnormal pbp1a and pbp2b (n = 100%);
PRSP with three abnormal PBP genes (n = 85.5%). E-test results revealed that 94.5% of isolates were resistant to penicillin, 100% to oxacillin, 100% to imipenem, and 21.8% to ceftazidime.
The most frequent genotype of penicillin resistance genes were related to pbp2x and pbp2b. Furthermore, 85.5 % of the samples with PRSP genotype had all three mutant genes and none of the samples were sensitive to penicillin. All samples had at least two genes related to penicillin resistance. Samples with one or two penicillin resistance genes had intermediate resistance to penicillin. Figure 3 indicates the relationship between distributions of antimicrobial minimum inhibitory concentrations for S. pneumoniae in mutations of PBP genes.
Figure 3. Antimicrobial Minimum Inhibitory Concentrations for Streptococcus pneumoniae in PBP Genes Mutants.
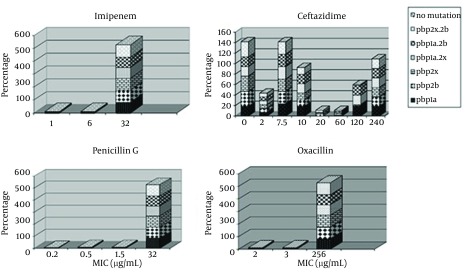
The highest percentage of antibiotic resistance was towards oxacillin and imipenem (100%). In addition, 94.5 % of samples were resistant to penicillin, imipenem and oxacillin, while 21% of samples were resistant to all tested antibiotic E-test strips. The most frequent penicillin resistance genes were pbp2x and pbp2b while 85.5 % of the samples had the PRSP genotype (had all three mutated genes). Furthermore, no penicillin sensitive strain was detected and 100% of the samples had at least two penicillin resistance genes and those with one or two penicillin resistance genes had intermediate penicillin resistance (PISP).
5. Discussion
Pneumococci have remained as important human pathogens despite the introduction of penicillin and the new generation of antibiotics. Our results showed that the incidence of penicillin-resistant strains among Iranian clinical isolates is alarmingly high. The rate of resistance to penicillin in our isolates was higher than the resistance rates reported from other countries, underlining the necessity for more attention to be paid to antibiotic therapy for pneumococcal infections. A detailed study from Brazil indicated that the prevalence of resistant strains increased during the six years of the study period according to analysis of MICs (30).
The present study showed a 94.5% prevalence rate for penicillin resistant S. pneumoniae. This is higher than the 67.8% reported from Tanzania (31) and 64% from Kuwait (32) and very higher than the 9.5% reported from Zahedan, Iran (33). PCR is a rapid and simple technique with high sensitivity and specificity for detection of microorganisms (34-37). Primarily, a microbial test and PCR were conducted to determine S. pneumoniae. PCR analysis confirmed the microbial test results. Since the first cases of invasive pneumococcal infections caused by PRSP were reported in 1977, penicillin-nonsusceptible strains have become a global concern (38). Several reports have determined a high prevalence of pneumococcal resistance to penicillin as well as other antibiotics, such as cephalosporins and macrolides.
A recent survey in the United States showed that 18.4% of isolates were resistant to penicillin (20). Furthermore, the overall proportion of the isolates which were resistant to three or more classes of drugs was reported to be increased (39). Some studies have documented the emergence of decreased susceptibility of S. pneumoniae to fluoroquinolones; in addition, a failure in therapy of cases with pneumococcal pneumonia treated with oral levofloxacin has been reported (40, 41). A number of factors have been reported to play a role in carriage and transmission of penicillin resistant. S. pneumonia; however, the most important factor is probably recent antibiotic use. Other risk factors for resistant pneumococcal carriage include young age, attendance to day care centers, and human immunodeficiency virus (HIV) infections in some populations (42, 43).
Autolysins are enzymes that degrade different bonds in the peptidoglycan which ultimately result in the lysis and death of the cell. The S. pneumoniae contains a powerful autolytic enzyme that has been described as an N-acetylmuramoyl-L-alanine amidase (44, 45). The autolysin LytA is responsible for release of lipoteichoic and teichoic acids, which are host inflammatory response mediators. Neuraminidase (45) is a choline-binding protein which is found in the cytoplasm of S. pneumoniae and released when the cells undergo autolysis. Cell wall autolysin may have a function in pathogenesis of S. pneumoniae through lysing a proportion of the invading pneumococci, which causes the release of potentially lethal toxins. Earlier studies have revealed that autolysin releases extremely inflammatory cell wall breakdown products, which eventually contribute to pathogenesis (44, 45). In addition, autolysin-deficient S. pneumoniae were shown to have a degree of attenuated virulence in one of the previous reports (44). Therefore, it can be presumed that autolysin contributes to early pathogenesis of the pneumococcal disease.
In the present study, all of our isolates were positive for lytA, irrespective of the kind of disease they were causing. All the isolates from both invasive and ocular infections were lytA positive suggesting that irrespective of site of isolation and kind of infection, autolysin is a necessity for the S. pneumoniae isolates. In the current work, examination of MICs for β-lactams, including penicillin, imipenem, oxacillin, ceftazidim, showed a trend in S. pneumoniae resistance to β-lactams. The existence of mutant PBP genes affected the MIC of β-lactam resistance since the occurrence of these genes in resistant was higher compared to the sensitive isolates (Figure 3).
The lower affinity of PBPs, 1A, 2B and 2X are involved in β-lactams-resistance in S. pneumoniae. Furthermore, we used PCR with primers specific to susceptible alleles for the detection of mutations in PBP genes, which demonstrated that 85% of isolates had mutations in the PBP genes. The comparison of these results with results of E-test demonstrated that there are other PBP genes, which are involved in low frequency of resistance. Previous studies have shown strong associations between the use of antimicrobial agents in the community and emergence of antimicrobial resistance in a number of organisms (46, 47). This might be due to selection of the resistant strains which have mutations in the PBP genes that result from β-lactam utilization. Another possibility is the implantation of transformation between S. pneumoniae and related streptococcal species. The increase in β-lactam resistant S. pneumoniae might be associated with the increase in highly resistant strains and rapid transfer of cloned resistance which in turn could be due to introduction and administration of new β-lactam and macrolide antibiotics as well as high population and travel to affected areas.
A number of strategies including simultaneous multiple drug therapy, which have been reported to be effective in reducing the development of resistance in pathogens such as HIV and mycobacterium tuberculosis are not practical for S. pneumoniae due to within-host development of resistance during treatment (48). In the United States and Europe, decreased resistance by judicious antibiotic use has been reported (48). However, this strategy is not practical in for developing country such as Iran where antibiotics are sold without prescription over the counter. The present results suggest that it is important to appraise the changes in MIC values as well as genetic mutations in order to evaluate the prevalence of resistance to antimicrobial agents in S. pneumoniae. Furthermore, penicillin is not suitable for treatment of pneumococcal infections; instead a range of macrolides can be administrated to successfully treat strains with intermediate resistance to penicillin in serious infections. However, the necessity of using macrolides should be considered due to the increased macrolides resistance in this family.
For strains with high resistance to penicillin, vancomycin and fluoroquinolones including sparfloxacin are selected as antimicrobial agents. In addition, selected antibiotics should be administrated to treat pneumococcal bacterial infection based on the pattern of pneumococcal drug resistance in the area. Thus, in isolates assumed to be S. pneumoniae, examination of three PBP and lytA together gives value to predict the susceptibility within two hours which finally leads to a selective treatment for infectious diseases caused by S. pneumonia (37). Consequently, the increase in antibiotic resistance among S. pneumoniae strains in Iran seems to be alerting and the treatment of infections should be further analyzed and investigated and thus presently antibiotics used for practical treatment of certain S. pneumoniae infections may need to be further reviewed.
Acknowledgments
The authors wish to thank the Vice-chancellor of Research of Hamadan University of Medical Sciences for their financial support. We also thank Mr. Haydarbarghi for his assistance in culture and isolation of Streptococcus pneumoniae.
Footnotes
Authors’ Contributions:Study concept and design: Marzieh Gharibi. Analysis and interpretation of data: Marzieh Gharibi. Drafting of the manuscript: Marzieh Gharibi and Giti Kalantarian. Critical revision of the manuscript for important intellectual content: Mohammad Yousef Alikhani, Mohammad Najafi Mosleh and Massoud Saidijam.
Funding/Support:This study was supported by grant 8610046531 provided by the Hamadan University of Medical Sciences.
References
- 1.van der Poll T, Opal SM. Pathogenesis, treatment, and prevention of pneumococcal pneumonia. Lancet. 2009;374(9700):1543–56. doi: 10.1016/S0140-6736(09)61114-4. [DOI] [PubMed] [Google Scholar]
- 2.Chiavolini D, Pozzi G, Ricci S. Animal models of Streptococcus pneumoniae disease. Clin Microbiol Rev. 2008;21(4):666–85. doi: 10.1128/CMR.00012-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moreillon P, Wenger A, Caldelari I. [Pneumococcal antibiotic resistance]. Rev Med Suisse Romande. 2000;120(8):651–9. [PubMed] [Google Scholar]
- 4.Friedland IR, McCracken GH, Jr. Management of infections caused by antibiotic-resistant Streptococcus pneumoniae. N Engl J Med. 1994;331(6):377–82. doi: 10.1056/NEJM199408113310607. [DOI] [PubMed] [Google Scholar]
- 5.Appelbaum PC. Resistance among Streptococcus pneumoniae: Implications for drug selection. Clin Infect Dis. 2002;34(12):1613–20. doi: 10.1086/340400. [DOI] [PubMed] [Google Scholar]
- 6.Perez-Llarena FJ, Bou G. Beta-lactamase inhibitors: the story so far. Curr Med Chem. 2009;16(28):3740–65. doi: 10.2174/092986709789104957. [DOI] [PubMed] [Google Scholar]
- 7.Consumption of antimicrobial agents and occurrence of antimicrobial resistance in norway. Norway: Nowegian Zoonosis Centre, TromsQ/Oslo; 2002. [Google Scholar]
- 8.Song JH, Chang HH, Suh JY, Ko KS, Jung SI, Oh WS, et al. Macrolide resistance and genotypic characterization of Streptococcus pneumoniae in Asian countries: a study of the Asian Network for Surveillance of Resistant Pathogens (ANSORP). J Antimicrob Chemother. 2004;53(3):457–63. doi: 10.1093/jac/dkh118. [DOI] [PubMed] [Google Scholar]
- 9.Woodford N, Livermore DM. Infections caused by Gram-positive bacteria: a review of the global challenge. J Infect. 2009;59 Suppl 1:S4–16. doi: 10.1016/S0163-4453(09)60003-7. [DOI] [PubMed] [Google Scholar]
- 10.Livermore DM. Mechanisms of resistance to cephalosporin antibiotics. Drugs. 1987;34 Suppl 2:64–88. doi: 10.2165/00003495-198700342-00007. [DOI] [PubMed] [Google Scholar]
- 11.Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657–86. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006;19(2):382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol Rev. 2006;30(5):673–91. doi: 10.1111/j.1574-6976.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 14.Hakenbeck R. Transformation in Streptococcus pneumoniae: mosaic genes and the regulation of competence. Res Microbiol. 2000;151(6):453–6. doi: 10.1016/s0923-2508(00)00170-4. [DOI] [PubMed] [Google Scholar]
- 15.Hakenbeck R. Mosaic genes and their role in penicillin-resistant Streptococcus pneumoniae. Electrophoresis. 1998;19(4):597–601. doi: 10.1002/elps.1150190423. [DOI] [PubMed] [Google Scholar]
- 16.Baquero F, Blazquez J, Loza E, Canton R. [Molecular basis of resistance to beta-lactams in infections by Streptococcus pneumoniae]. Med Clin (Barc). 1998;110 Suppl 1:8–11. [PubMed] [Google Scholar]
- 17.Coffey TJ, Dowson CG, Daniels M, Spratt BG. Genetics and molecular biology of beta-lactam-resistant pneumococci. Microb Drug Resist. 1995;1(1):29–34. doi: 10.1089/mdr.1995.1.29. [DOI] [PubMed] [Google Scholar]
- 18.Dowson CG, Coffey TJ, Spratt BG. Origin and molecular epidemiology of penicillin-binding-protein-mediated resistance to beta-lactam antibiotics. Trends Microbiol. 1994;2(10):361–6. doi: 10.1016/0966-842x(94)90612-2. [DOI] [PubMed] [Google Scholar]
- 19.Dowson CG, Hutchison A, Woodford N, Johnson AP, George RC, Spratt BG. Penicillin-resistant viridans streptococci have obtained altered penicillin-binding protein genes from penicillin-resistant strains of Streptococcus pneumoniae. Proc Natl Acad Sci U S A. 1990;87(15):5858–62. doi: 10.1073/pnas.87.15.5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karlowsky JA, Thornsberry C, Jones ME, Evangelista AT, Critchley IA, Sahm DF, et al. Factors associated with relative rates of antimicrobial resistance among Streptococcus pneumoniae in the United States: results from the TRUST Surveillance Program (1998-2002). Clin Infect Dis. 2003;36(8):963–70. doi: 10.1086/374052. [DOI] [PubMed] [Google Scholar]
- 21.Centers for Disease Control and Prevention. Manual for the laboratory identification and antimicrobial susceptibility testing of bacterial pathogens of public health importance in the developing world. 2003
- 22.Ubukata K, Muraki T, Igarashi A, Asahi Y, Konno M. Identification of penicillin and other beta-lactam resistance inStreptococcus pneumoniae by polymerase chain reaction. J Infect Chemother. 1997;3(4):190–7. doi: 10.1007/BF02490033. [DOI] [PubMed] [Google Scholar]
- 23.Ubukata K, Asahi Y, Yamane A, Konno M. Combinational detection of autolysin and penicillin-binding protein 2B genes of Streptococcus pneumoniae by PCR. J Clin Microbiol. 1996;34(3):592–6. doi: 10.1128/jcm.34.3.592-596.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia P, Garcia JL, Garcia E, Lopez R. Nucleotide sequence and expression of the pneumococcal autolysin gene from its own promoter in Escherichia coli. Gene. 1986;43(3):265–72. doi: 10.1016/0378-1119(86)90215-5. [DOI] [PubMed] [Google Scholar]
- 25.Jorgensen JH, Howell AW, Maher LA. Quantitative antimicrobial susceptibility testing of Haemophilus influenzae and Streptococcus pneumoniae by using the E-test. J Clin Microbiol. 1991;29(1):109–14. doi: 10.1128/jcm.29.1.109-114.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker CN, Stocker SA, Culver DH, Thornsberry C. Comparison of the E Test to agar dilution, broth microdilution, and agar diffusion susceptibility testing techniques by using a special challenge set of bacteria. J Clin Microbiol. 1991;29(3):533–8. doi: 10.1128/jcm.29.3.533-538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jorgensen JH, Maher LA, Howell AW. Use of Haemophilus test medium for broth microdilution antimicrobial susceptibility testing of Streptococcus pneumoniae. J Clin Microbiol. 1990;28(3):430–4. doi: 10.1128/jcm.28.3.430-434.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Committee for Clinical Laboratory Standards. Performance Standards for Antimicrobial Disk Susceptibility Test: Approved Standards. Villanova: NCCLS; 2001. [Google Scholar]
- 29.Ubukata K, Iwata S, Sunakawa K. In vitro activities of new ketolide, telithromycin, and eight other macrolide antibiotics against Streptococcus pneumoniae having mefA and ermB genes that mediate macrolide resistance. J Infect Chemother. 2003;9(3):221–6. doi: 10.1007/s10156-003-0258-2. [DOI] [PubMed] [Google Scholar]
- 30.Barroso DE, Godoy D, Castineiras TM, Tulenko MM, Rebelo MC, Harrison LH. beta-Lactam resistance, serotype distribution, and genotypes of meningitis-causing Streptococcus pneumoniae, Rio de Janeiro, Brazil. Pediatr Infect Dis J. 2012;31(1):30–6. doi: 10.1097/INF.0b013e31822f8a92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moyo SJ, Steinbakk M, Aboud S, Mkopi N, Kasubi M, Blomberg B, et al. Penicillin resistance and serotype distribution of Streptococcus pneumoniae in nasopharyngeal carrier children under 5 years of age in Dar es Salaam, Tanzania. J Med Microbiol. 2012;61(Pt 7):952–9. doi: 10.1099/jmm.0.042598-0. [DOI] [PubMed] [Google Scholar]
- 32.Mokaddas EM, Rotimi VO, Albert MJ. Implications of Streptococcus pneumoniae penicillin resistance and serotype distribution in Kuwait for disease treatment and prevention. Clin Vaccine Immunol. 2008;15(2):203–7. doi: 10.1128/CVI.00277-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bokaeian M, Khazaei HA, Javadimehr M. Nasopharyngeal Carriage, Antibiotic Resistance and Serotype Distribution of Streptococcus Pneumoniae among Healthy Adolescents in Zahedan. Iran Red Crescent Med J. 2011;13(5):328–33. [PMC free article] [PubMed] [Google Scholar]
- 34.Amini Bavil Olyaee S, Sabahi F, Karimi M. PCR optimization: Improving of human cytomegalovirus (HCMV) PCR to achieve a highly sensitive detection method. Iran J Biotech. 2003;1(1):59–64. [Google Scholar]
- 35.Dehnavi E, Ahani Azari M, Hasani S, Nassiry MR, Mohajer M, Khan Ahmadi AR. Genetic variability of calpastatin and calpain genes in Iranian Zel sheep using PCR-RFLP and PCR-SSCP methods. Iran J Biotech. 2012;10(2):136–9. doi: 10.1155/2012/472307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fahami S, Kordbacheh P, Moazeni M, Mahmoodi M, Mirhendi H. Species Identification and Strain Typing of Candida Isolates by PCR-RFLP and RAPD-PCR Analysis for Determining the Probable Sources of Nosocomial Infections. Iran Red Crescent Med J. 2014;12(5):539–47. [Google Scholar]
- 37.Soleimani M, Zolfaghari MR, Morovvati A. Development and Comparison of Conventional PCR and SYBR Green Real Time PCR for Detection of Aggregatibacter actinomycetemcomitans and Tannerella forsythensis. Jundishapur J Microbiol. 2013;6(8) doi: 10.5812/jjm.6757. [DOI] [Google Scholar]
- 38.Ubukata K, Chiba N, Hasegawa K, Kobayashi R, Iwata S, Sunakawa K. Antibiotic susceptibility in relation to penicillin-binding protein genes and serotype distribution of Streptococcus pneumoniae strains responsible for meningitis in Japan, 1999 to 2002. Antimicrob Agents Chemother. 2004;48(5):1488–94. doi: 10.1128/AAC.48.5.1488-1494.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitney CG, Farley MM, Hadler J, Harrison LH, Lexau C, Reingold A, et al. Increasing prevalence of multidrug-resistant Streptococcus pneumoniae in the United States. N Engl J Med. 2000;343(26):1917–24. doi: 10.1056/NEJM200012283432603. [DOI] [PubMed] [Google Scholar]
- 40.Chen DK, McGeer A, de Azavedo JC, Low DE. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. Canadian Bacterial Surveillance Network. N Engl J Med. 1999;341(4):233–9. doi: 10.1056/NEJM199907223410403. [DOI] [PubMed] [Google Scholar]
- 41.Davidson R, Cavalcanti R, Brunton JL, Bast DJ, de Azavedo JCS, Kibsey P, et al. Resistance to levofloxacin and failure of treatment of pneumococcal pneumonia. N Engl J Med. 2002;346(10):747–50. doi: 10.1056/NEJMoa012122. [DOI] [PubMed] [Google Scholar]
- 42.Henderson FW, Gilligan PH, Wait K, Goff DA. Nasopharyngeal carriage of antibiotic-resistant pneumococci by children in group day care. J Infect Dis. 1988;157(2):256–63. doi: 10.1093/infdis/157.2.256. [DOI] [PubMed] [Google Scholar]
- 43.Jones N, Huebner R, Khoosal M, Crewe-Brown H, Klugman K. The impact of HIV on Streptococcus pneumoniae bacteraemia in a South African population. AIDS. 1998;12(16):2177–84. doi: 10.1097/00002030-199816000-00013. [DOI] [PubMed] [Google Scholar]
- 44.Ng EW, Costa JR, Samiy N, Ruoff KL, Connolly E, Cousins FV, et al. Contribution of pneumolysin and autolysin to the pathogenesis of experimental pneumococcal endophthalmitis. Retina. 2002;22(5):622–32. doi: 10.1097/00006982-200210000-00014. [DOI] [PubMed] [Google Scholar]
- 45.Paton JC, Berry AM, Lock RA. Molecular analysis of putative pneumococcal virulence proteins. Microb Drug Resist. 1997;3(1):1–10. doi: 10.1089/mdr.1997.3.1. [DOI] [PubMed] [Google Scholar]
- 46.Nava JM, Bella F, Garau J, Lite J, Morera MA, Marti C, et al. Predictive factors for invasive disease due to penicillin-resistant Streptococcus pneumoniae: a population-based study. Clin Infect Dis. 1994;19(5):884–90. doi: 10.1093/clinids/19.5.884. [DOI] [PubMed] [Google Scholar]
- 47.Quach C, Weiss K, Moore D, Rubin E, McGeer A, Low DE. Clinical aspects and cost of invasive Streptococcus pneumoniae infections in children: resistant vs. susceptible strains. Int J Antimicrob Agents. 2002;20(2):113–8. doi: 10.1016/s0924-8579(02)00127-9. [DOI] [PubMed] [Google Scholar]
- 48.Schrag SJ, Beall B, Dowell SF. Limiting the spread of resistant pneumococci: biological and epidemiologic evidence for the effectiveness of alternative interventions. Clin Microbiol Rev. 2000;13(4):588–601. doi: 10.1128/cmr.13.4.588-601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


