Abstract
Fenofibrate (FF) is a common lipid-lowering drug and a potent agonist of the peroxisome proliferator-activated receptor alpha (PPARα). FF and several other agonists of PPARα have interesting anticancer properties, and our recent studies demonstrate that FF is very effective against tumor cells of neuroectodermal origin. In spite of these promising anticancer effects, the molecular mechanism(s) of FF-induced tumor cell toxicity remains to be elucidated. Here we report a novel PPARα-independent mechanism explaining FF's cytotoxicity in vitro and in an intracranial mouse model of glioblastoma. The mechanism involves accumulation of FF in the mitochondrial fraction, followed by immediate impairment of mitochondrial respiration at the level of complex I of the electron transport chain. This mitochondrial action sensitizes tested glioblastoma cells to the PPARα-dependent metabolic switch from glycolysis to fatty acid β-oxidation. As a consequence, prolonged exposure to FF depletes intracellular ATP, activates the AMP-activated protein kinase–mammalian target of rapamycin–autophagy pathway, and results in extensive tumor cell death. Interestingly, autophagy activators attenuate and autophagy inhibitors enhance FF-induced glioblastoma cytotoxicity. Our results explain the molecular basis of FF-induced glioblastoma cytotoxicity and reveal a new supplemental therapeutic approach in which intracranial infusion of FF could selectively trigger metabolic catastrophe in glioblastoma cells.
INTRODUCTION
Fenofibrate (FF) is a common lipid-lowering drug and a potent agonist of peroxisome proliferator-activated receptor alpha (PPARα). Multiple reports indicate a beneficial role for lipid-lowering drugs, including fibrates and statins, as anticancer agents (1–7). For example, a 10-year, all-cause mortality study involving 7,722 patients treated with different fibrates revealed that the use of these drugs is associated with a significantly lower total mortality rate and a reduced probability of death from cancer (8). In cell culture and animal studies, various members of the fibrate family, which are all agonists of PPARα, demonstrate interesting anticancer effects, which are not fully understood. FF inhibited tumor growth by reducing both inflammation and angiogenesis in host tissue (5). Clofibrate attenuated ovarian cancer cell proliferation (9, 10), and gemfibrozil (GEM) inhibited the invasiveness of glioblastoma cells (11). In our previous work, FF synergized with staurosporine to reduce melanoma lung metastases (3, 12), significantly reduced glioblastoma invasiveness (13), and triggered apoptotic death in medulloblastoma (14) and human glioblastoma cell lines by inducing the FOXO3A-Bim apoptotic pathway (15). All of these studies encouraged the use of FF as a supplemental anticancer drug, a concept supported by recent clinical trials in which chronic administration of FF along with chemotherapeutic agents used at relatively low doses minimizes the toxicity and acute side effects of chemotherapy while maintaining efficacy for patients with recurrent brain malignancies and leukemias (16, 17). In spite of these very promising results, the mechanism(s) of the exceptional anticancer effects of FF relative to other metabolic compounds, including other PPAR agonists or metformin (Met), remains largely unknown.
The primary and conventional function of FF is the activation of PPARα transcriptional activity. In this process, FF must first be converted to fenofibric acid (FA) by blood and tissue esterases. FA then binds and activates PPARα, which triggers the expression of numerous metabolic enzymes involved in fatty acid β-oxidation (18–20). In addition, activated PPARα decreases glucose uptake by repressing the insulin-dependent glucose transporter GLUT4 (19, 21) and elevated oxidation of the fatty acids and ketone bodies further suppresses the expression of glycolytic enzymes (22, 23). This metabolic switch could initiate a gradual decline in energy metabolism in tumor cells (24–26), which is consistent with the fundamental observation by Otto Warburg that tumor cells are distinctly dependent on glycolysis (27, 28) for both energy production and biosynthesis of intermediate metabolites (29). However, in comparison with the anticancer effects of other potent agonists of PPARα, those of FF are much more pronounced, implying that FF may also act in a PPARα-independent manner. In this regard, FF was shown to alter the expression of growth differentiation factor 15 (20); affect cell membrane fluidity in a manner similar to that of cholesterol (30); and interfere with the respiratory function of isolated liver and heart mitochondria (31, 32). Here we report the novel observation that FF, but not its PPARα-active metabolite FA, accumulates in the mitochondrial fraction of human glioblastoma cells. As a consequence, these neoplastic cells respond with a sudden and severe inhibition of mitochondrial respiration and an immediate but transient increase in glycolysis. We further demonstrate that complex I of the electron transport chain (ETC) is the preferred target of mitochondrial FF. The subsequent decline in intracellular ATP preceded the activation of AMP-activated protein kinase (AMPK) and inhibition of mammalian target of rapamycin (mTOR) activity. As a consequence, this energy-sensitive pathway activated autophagy, which, according to our data, could explain a delay in the onset of FF-induced apoptosis. Intriguingly, augmentation of autophagy by rapamycin counteracted FF-induced cytotoxicity, while autophagy inhibitors potentiated its antiglioblastoma efficacy. In summary, our findings may encourage future clinical trials that include FF as a supplemental metabolic drug that directly and effectively triggers energetic catastrophe in glioblastoma cells with significantly reduced toxicity in normal astrocytes.
MATERIALS AND METHODS
Cell culture.
We used human glioblastoma cell lines LN-229 (ATCC CRL-2611) and U-87MG (ATCC HTB-14), which, according to our previous studies, are highly sensitive to FF (13, 15). Both cell lines were maintained as semiconfluent monolayer cultures in Dulbecco's modified Eagle's medium supplemented with 50 U/ml penicillin, 50 ng/ml streptomycin, and 10% fetal bovine serum (FBS) at 37°C in a 5% CO2 atmosphere. The cells were treated with different agonists of PPARα, including FF (Sigma-Aldrich, St. Louis, MO), FA (Sigma-Aldrich, St. Louis, MO), GEM (Sigma-Aldrich, St. Louis, MO), and WY146,43 (WY; Cayman Chemical, Ann Arbor, MI) at concentrations ranging from 10 to 50 μM; with Met (Sigma-Aldrich, St. Louis, MO) at concentrations ranging from 50 to 500 μM; with GW6471 at 1 μM; and with GW7647 at concentrations ranging from 1 nM to 10 μM. Control cultures were treated with the corresponding volumes of dimethyl sulfoxide (DMSO) (vehicle). Normal human astrocytes (NHA) were cultured according to the manufacturer's recommendations (Lonza/Clonetics, Walkersville, MD). All metabolic and cell culture assays involving NHA were performed between passages 2 and 6. Primary glioblastoma multiforme (GBM) astrosphere cultures were derived directly from GBM patients and were obtained according to the IRB protocol (IRB 6774, LSU Health Sciences Center [HSC], New Orleans, LA). Fresh tumor tissue explants were cut with a sharp scalpel, and the resulting small tissue fragments were incubated in Hanks' balanced salt solution containing Liberase (96 μg/ml) and DNase (100 μg/ml) (Roche, Indianapolis, IN) at 37°C for 30 min. The enzymatic digestion of the tumor was stopped by using 1 volume of medium containing 10% FBS, and the cells obtained were centrifuged at 1,600 rpm at 4°C for 5 min. The cell pellet was suspended in NeuroCult NS-A Basal Medium (Human) supplemented with l-glutamine (2 mM), N2, B27, bovine serum albumin (75 μg/ml), recombinant human epidermal growth factor (10 ng/ml), basic fibroblast growth factor (10 ng/ml), and heparin (2 μg/ml). The cells obtained were then plated on culture dishes covered with poly-2-hydroxyethyl methacrylate (20 mg/ml; Sigma-Aldrich). This procedure was used previously in our lab to force cell growth in suspension (33) and the formation of neurospheres (34).
Detection of FF and FA by HPLC.
All high-performance liquid chromatography (HPLC) data were obtained with the Agilent 1100 apparatus equipped with a line degasser, a binary pump (high-pressure mixer), an autosampler, a column thermostat, and a diode array detector (DAD) (Agilent Technologies, Santa Clara, CA). The analytical column (3 μm, 4.6 by 150 mm, octyl silane C8; YMC America, Inc.), solvent A (50 mM acetic acid in water), and solvent B (acetonitrile) with isocratic flow were used to detect and quantify the FF and FA contents of subcellular fractions. The flow rate was set to 1 ml/min, the column temperature was 20°C, and the sample volume was 5 μl. The DAD wavelength was set to 285 nm. Sample preparation by detergent-free subcellular fractionation was based on hypotonic sucrose buffer and ultracentrifugation. An aliquot (150 μl) of each cytosolic or mitochondrial fraction (isolated from 1 × 106 cells) was mixed with 150 μl of acetonitrile, centrifuged, passed through 0.22-μm filters, and placed in HPLC sample vials. FF and FA standards (both from Sigma-Aldrich) were used to verify their exact retention times (RTs) and unique UV-Vis absorbance. Mitochondria were isolated with a mitochondrial isolation kit according to the manufacturer's recommendations (AMRESCO Inc. Solon, OH).
Western blotting.
Total protein extracts were isolated from LN-229 cells and NHA. Sample preparation and immunoblotting were performed according to the standard procedures described previously (13). The primary antibodies utilized were an anti-AMPKα(pThr-172) rabbit polyclonal, an anti-p70S6K(pThr-389) rabbit monoclonal, and an anti-LC3β rabbit polyclonal antibody (all from Cell Signaling Technology, Danvers, MA), and an anti-Grb-2 mouse monoclonal antibody (BD Bioscience, San Jose, CA) was used as a loading marker. To evaluate the purity of cytosolic and mitochondrial fractions, corresponding protein extracts were prepared and the resulting blots were analyzed with an anti-α-tubulin mouse monoclonal antibody (clone B7; Santa Cruz Biotechnology, Dallas, TX)-cytosolic fraction marker and with an anti-Cox IV rabbit monoclonal antibody (clone 3E11; Cell Signaling)-mitochondrial fraction marker.
Evaluation of metabolic parameters.
Metabolic responses of LN-229 and U-87MG cells, primary human glioblastoma cells, and NHA were evaluated with an Extracellular Flux Analyzer (XF24; Seahorse Biosciences, North Billerica, MA). Prior to each assay, the cells were plated in growth-supporting medium in 24-well plates (4 × 104 cells/well). At the time of measurement, the growth media were replaced with serum-free XF medium (Seahorse) and cartridges equipped with oxygen-sensitive and pH-sensitive probes (Seahorse) were placed above the cells. The oxygen consumption rate (OCR; indicative of mitochondrial respiration) and extracellular acidification rate (ECAR; indicative of glycolysis) were evaluated after injecting the following metabolic toxins: oligomycin (inhibitor of ATP synthase; complex V of the ETC), carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP; uncoupling factor), rotenone (inhibitor of mitochondrial complex I of the ETC), 2-deoxyglucose (2-DG; inhibitor of hexokinase). The metabolic effects of the selected lipid-lowering drugs were determined by comparing OCR and ECAR values with those of the control (cells preincubated with DMSO [vehicle] for 24 h). In some experiments, immediate responses to the selected lipid-lowering drugs were also tested.
Clonogenic growth.
Exponentially growing cultures of multiple tumor cell lines (see Fig. S2 in the supplemental material), including LN-229 and U-87MG, were washed with fresh serum-free medium, trypsinized, and plated at 1 × 103 cells/35-mm dish. Clonogenic growth was evaluated 2 weeks after continuous cell growth in medium containing 10% FBS with or without FF, FA, GEM, WY, GW7647, or Met. All compounds were used at 50 μM, with the exception of GW7647, which was used at 1 and 10 μM. The resulting clones were fixed and stained with 0.25% crystal violet in methanol as described previously (14).
PPAR luciferase assay.
PPAR transcriptional activity was determined by utilizing the J3TkpGL3 reporter plasmid, which contains a luciferase gene driven by the PPAR-responsive element (PPRE), which consists of three copies of the J site from the apo-AII gene promoter (35). The activation of PPAR elements was evaluated in LN-229 cells following stimulation with FF (50 μM), FA (50 μM), GEM (50 μM), WY (50 μM), and GW7647 (1 μM) with a firefly-Renilla dual-luciferase reporter system (Promega, Madison, WI) and a Synergy 2 microplate reader (BioTek, Winooski, VT).
Intracellular ATP levels.
Intracellular ATP levels were measured with the ApoSENSOR ATP assay kit according to the manufacturer's recommendations (BioVision, Milpitas, CA). Cells were treated either with the vehicle (DMSO) or 50 μM FF or FA and harvested at the time points indicated. Aliquots of 1 × 104 cells were resuspended in 100 μl of nucleotide-releasing buffer containing 1 μl of ATP-monitoring enzyme. Luminescence was measured by a Synergy 2 microplate reader and calculated with Gen5 software (BioTek).
Mitochondrial potential and cell death assay.
We used a flow cytometry-based MitoPotential kit according to the manufacturer's protocol (Guava EasyCyte). Loss of mitochondrial inner transmembrane potential (ΔΨm) was measured with the cationic dye JC-1, which gives either green or orange fluorescence, depending upon mitochondrial membrane depolarization. Following cell exposure to FF (50 μM; Sigma-Aldrich), chloroquine (2.5 μM; Sigma-Aldrich), bafilomycin A1 (1 nM; Sigma-Aldrich), rapamycin (100 nM; Sigma-Aldrich), or the indicated combinations of the compounds, cells were harvested by trypsinization, loaded with JC-1 reagent for 30 min, and immediately analyzed by Guava EasyCyte with MitoPotential software (Millipore, Billerica, MA). Cell treatment with valinomycin (Sigma-Aldrich), which fully depolarizes mitochondria, was utilized as a positive control (36). In some cases, JC-1-loaded cells were also labeled with 7-aminoactinomycin D (7-AAD), which allows the detection and quantification of dead cells in the same experiment (Millipore).
Intracranial tumor growth and treatment.
All of the procedures described that involved experimental animals were performed in accordance with IACUC protocol 3146 at LSU HSC, New Orleans, LA. The human glioblastoma cell line U-87MG (ATCC HTB-14) stably expressing a luciferase reporter (U-87MG–luc) was used in this experiment. Female nude-Foxn1nu athymic mice, 6 to 8 weeks of age (Harlan Laboratories, Inc., Indianapolis, IN), were anesthetized with 4% isoflurane and secured in a stereotaxic head frame (Harvard Apparatus, Holliston, MA). Tumor cells (1 × 105 in 2 μl of artificial cerebrospinal fluid) were injected into the left striatum (0.26 mm anterior to the bregma, 0.15 mm lateral to the sagittal suture, and 0.3 mm down from the surface) through a burr hole in the skull with a 10-μl Hamilton syringe. Following tumor cell implantation, mice were injected intracranially (at the same coordinates) with 5 μl of DMSO (control) or 5 μl of 1 mM FF in DMSO. Biophotonic images of the skull were captured with a Xenogen IVIS 200 imaging system (PerkinElmer, Palo Alto, CA) 2 weeks after initial cell implantation. Prior to imaging, each mouse received an intraperitoneal injection of 100 μl of d-luciferin (30 mg/ml solution; PerkinElmer, Waltham, MA) and was anesthetized by isoflurane inhalation. The resulting images were quantified with Living Image 4.1 software (Xenogen), and the data are expressed as average radiance (photons/s/cm2/sr) according to the manufacturer's recommendations.
Statistical analysis.
The data were analyzed with a homoscedastic Student t test. Differences between control and experimental groups were considered significant at P values of <0.05.
RESULTS
Detection of FF in mitochondrial fraction.
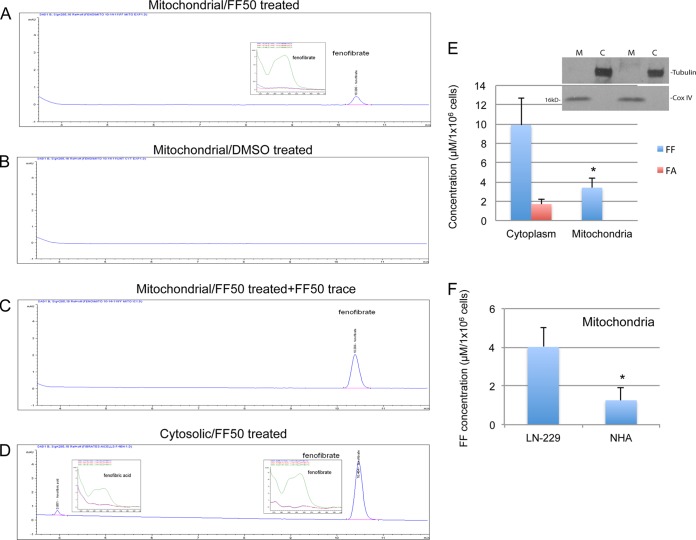
Several reports indicate that FF may act in a PPARα-independent manner (5, 12, 14, 18, 20, 30, 37). To analyze these off-target effects, we subjected LN-229 human glioblastoma cells and NHA to subcellular fractionation following treatment with 50 μM FF. The results in Fig. 1 demonstrate the detection of FF and its PPAR-active metabolite FA by HPLC. A single peak from the purified mitochondrial fraction of FF-treated cells was detected at the same RT as the FF standard (Fig. 1A). This peak was not detected in the control DMSO-treated sample (Fig. 1B). To further verify that this peak represents FF, the fraction was spiked with the FF standard, and this procedure again generated a single peak reflecting the sum of mitochondrial FF and the spike (Fig. 1C). Two distinct peaks were detected in the cytosolic fraction (Fig. 1D). The first peak was at an RT of 3.9 min, corresponding to FA, and the second was at an RT of 10.4 min, corresponding to unprocessed FF. In addition, spectral analyses of peaks (insets in Fig. 1A and D) matched the corresponding spectral analyses of the FF and FA standards. Figure 1E demonstrates the quantification of the peaks detected and reflects the amounts of FF and FA calculated per 1 × 106 cells exposed to 50 μM FF for 24 h. The purity of subcellular fractions was monitored by Western blotting with anti-α-tubulin (marker of the cytosolic fraction) and anti-Cox IV (marker of the mitochondrial fraction) antibodies. Note also a significant difference between the concentrations of unprocessed FF accumulating in the mitochondrial fractions obtained from NHA and LN-229 cells (Fig. 1F).
FIG 1.
The mitochondrial membrane fraction contains unprocessed FF. HPLC was used to detect FF and its PPARα-active metabolite FA in the mitochondrial membrane fraction (A to C) and the cytosolic fraction (D) isolated from the FF-treated human glioblastoma cell line LN-229. Under the HPLC conditions described in Materials and Methods, FF was eluted at 10.4 min and FA was eluted at 3.9 min. (A) Mitochondrial membranes isolated from LN-229 cells treated with 50 μM FF (FF50) for 24 h. (B) Mitochondrial membranes isolated from control DMSO-treated cells. (C) Mitochondrial membranes isolated from FF-treated cells additionally treated (spiked) with the FF standard prior to HPLC analysis. The insets in panels A and D represent the unique UV-Vis absorbance spectra of the peaks obtained corresponding to FF and FA, respectively (the three different traces in the insets demonstrate three different values for the diode array detector [DAD] to obtain FF and FA spectral analyses). (E) Quantitative analysis of FF and FA in mitochondrial membrane and cytosolic fractions isolated from LN-229 cells exposed to 50 μM FF for 24 h. Concentrations of FF and FA were calculated from the corresponding calibration curves and are presented in μM FF or amounts of FA per 1 × 106 cells. The data are average values and standard deviations from three separate measurements performed in triplicate (n = 9). (Inset) Western blot analysis demonstrating the purity of cytosolic (C) and mitochondrial membrane (M) fractions in which α-tubulin and Cox IV were use as cytosolic and mitochondrial markers, respectively. (F) Quantification of unprocessed FF in mitochondrial fractions isolated from LN-229 cells and NHA. The data are average values and standard deviations from two measurements performed in triplicate (n = 6). An asterisk indicates a statistically significant difference between LN-229 and NHA.
FF triggers severe and immediate inhibition of oxygen consumption.
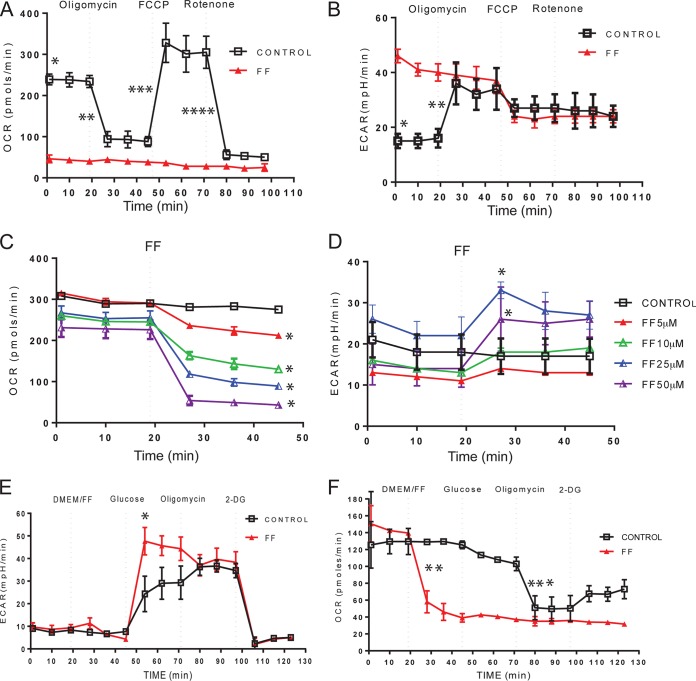
Detection of FF in the mitochondrial fraction prompted further investigations of the putative effect(s) of FF on glioblastoma mitochondrial function and energy metabolism. We used an Extracellular Flux Analyzer (XF24; Seahorse Biosciences) to measure the real-time OCR (indicative of mitochondrial respiration) and ECAR (indicative of glycolytic activity) in the same sample. These metabolic parameters were initially measured in monolayer cultures of LN-229 cells in which sequential injections of the metabolic toxins oligomycin (inhibitor of complex V [ATP synthase]), FCCP (uncoupling factor), and rotenone (inhibitor of mitochondrial complex I [NADH dehydrogenase]) were tested. In control samples (Fig. 2A, squares), oligomycin injection repressed oxygen consumption because of the inhibition of ATP synthase (complex V), thus preventing discharge of the proton gradient normally formed by the ETC. In response to FCCP, the accumulated proton gradient was discharged, triggering a sharp increase in oxygen consumption without ATP production. Finally, injection of rotenone blocked FCCP-induced oxygen consumption by repressing the formation of the proton gradient at the level of ETC complex I. The corresponding ECAR values of the control (Fig. 2B, squares) increased >2-fold following oligomycin injection, which represents an increase in glycolysis in response to an immediate and severe impairment of respiration. These metabolic responses of the control cells were dramatically altered in LN-229 cells cultured in the presence of 50 μM FF for 24 h. The continuous presence of FF resulted in a severe impairment of OCR and a failure to respond to any of the metabolic toxins tested as described above (Fig. 2A, red triangles). In contrast, the initial ECAR values for FF-pretreated cells were almost 3-fold higher than those of controls (Fig. 2B, triangles). Importantly, the viability of the cells tested was not affected at the 24-h time point (see also Fig. 4A). Further analyses demonstrated that FF-induced suppression of OCR was immediate (less than 10 min), was dose dependent (Fig. 2C), and was accompanied by a corresponding increase in glycolysis (Fig. 2D).
FIG 2.
FF inhibits mitochondrial respiration. Effects of FF on OCR (indicative of respiration) and ECAR (indicative of glycolysis) in human glioblastoma cell line LN-229 determined with an extracellular flux analyzer (XF24; Seahorse Bioscience). (A, B) Cells were plated at 4 × 104/well and cultured in the presence of 10% FBS with or without 50 μM FF for 24 h. Metabolic responses to FF or the vehicle (DMSO) were evaluated after sequential injection of the following metabolic toxins: oligomycin at 0.3 μM, FCCP at 0.5 μM, and rotenone at 0.3 μM. In panel A, a single asterisk indicates a statistically significant difference (homoscedastic Student t test, P ≤ 0.05) between control and FF-treated samples (initial OCR values), two asterisks indicate a significant percent decrease in OCR after oligomycin injection, three asterisks indicate a significant percent increase in OCR after FCCP injection, and four asterisks indicate a significant percent decrease in OCR after rotenone injection. (B) An asterisk indicates a statistically significant difference between control and FF-treated samples (initial ECAR values), and two asterisks indicate a significant percent increase in ECAR after oligomycin injection. (C, D) Immediate OCR and ECAR responses to 0, 5, 10, 25, and 50 μM FF injected at the indicated time points. (E, F) Evaluation of the glycolytic capacity of LN-229 cells. In all of the panels, the data are average values ± the standard deviations from five measurements and each experiment was repeated at least three times (n = 15). In panels C and D, an asterisk indicates a statistically significant difference between control and FF-treated samples calculated for the first time point following FF injection. In panels E and F, an asterisk indicates a statistically significant increase in ECAR between control and FF-treated samples following the addition of glucose, two asterisks indicate a statistically significant decrease in OCR following FF treatment, and three asterisks indicate a statistically significant decrease in OCR between control and FF-treated samples following the injection of oligomycin.
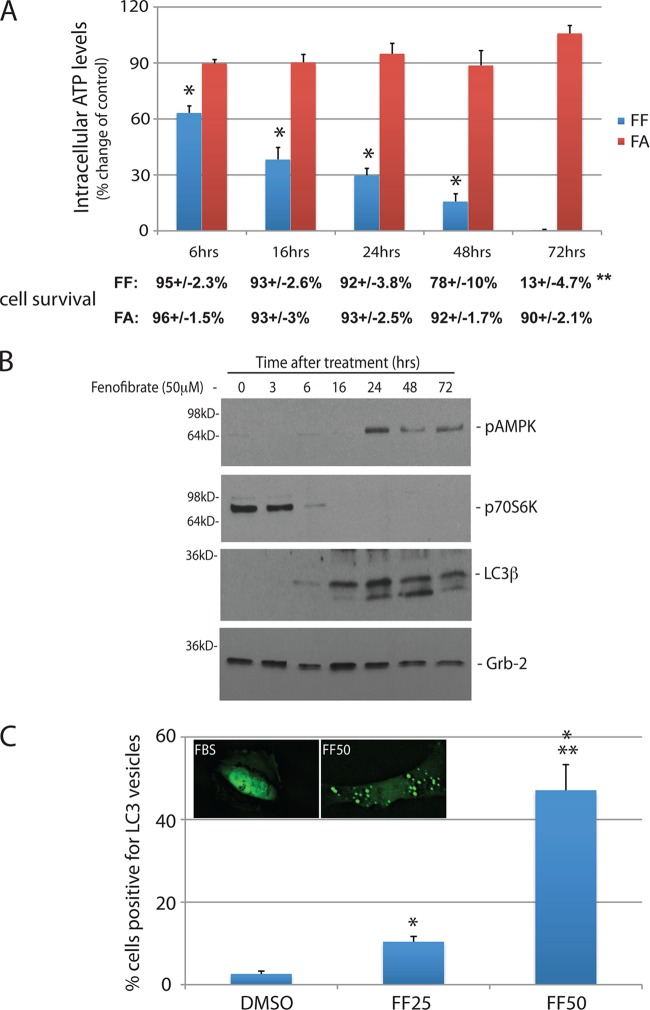
FIG 4.
Prolonged FF treatment induces an energy deficit and autophagy. (A) Intracellular ATP levels evaluated in LN-229 cells stimulated with either 50 μM FF or 50 μM FA for 6, 16, 24, 48, or 72 h. The values below the bars are the corresponding survival rates following FF or FA treatment. Cell viability was measured with a flow cytometry-based 7-AAD assay (Millipore). All data are average values ± the standard deviations calculated from two experiments performed in triplicate (n = 6). An asterisk indicates a statistically significant difference between FF- and FA-treated samples (P ≤ 0.05). Data on cell survival are average values calculated from three independent measurements (n = 3) ± the standard deviations. Two asterisks indicate a statistically significant difference between cell survival at 24 and that at 72 h following FF treatment (P = 0.00011). The difference between the 24- and 48-h values was not significant (P = 0.09). (B) Immunoblot assay-based detection of phosphorylated forms of AMPK (sensor of intracellular energy deficit), p70S6K (substrate for mTOR), and the autophagy marker LC3β. Grb-2 was used as a loading control in FF-treated LN-229 cells at the time points indicated. (C) Detection and quantification of FF-induced autophagy with LN-229 cells expressing the enhanced GFP (EGFP)-LC3β vector (Addgene 11546; Karla Kirkegaard). The cells were treated with 25 and 50 μM FF or left untreated for 24 h (DMSO). The data are average values of cells with LC3-positive intracellular vesicles ± the standard deviations. An asterisk indicates a value statistically significantly different from that obtained with FBS, and two asterisks indicate a value statistically significantly different from that obtained with FF25.
The observed increase in glycolysis following FF-induced inhibition of respiration prompted further analysis of this process. Figure 2E (ECAR) and F (OCR) demonstrate the results of a typical experimental design to evaluate glycolytic capacity. ECAR values are stabilized at low levels in both control and FF-treated glucose-starved LN-229 cells. Subsequent injection of 10 mM glucose into the FF-treated sample triggered an immediate increase in ECAR, indicating a strong glycolytic response to glucose addition in FF-treated samples (triangles). In control cells (squares), ECAR responses to glucose injection were noticeably delayed. The corresponding OCR values (Fig. 2F) dropped down following FF injection and remained low following glucose, oligomycin, and 2-DG injections (as expected). These results establish the baseline glycolytic capacity of LN-229 cells and indicate that cells treated with FF immediately switch their energy metabolism toward glucose utilization.
FF blocks respiratory complex I.
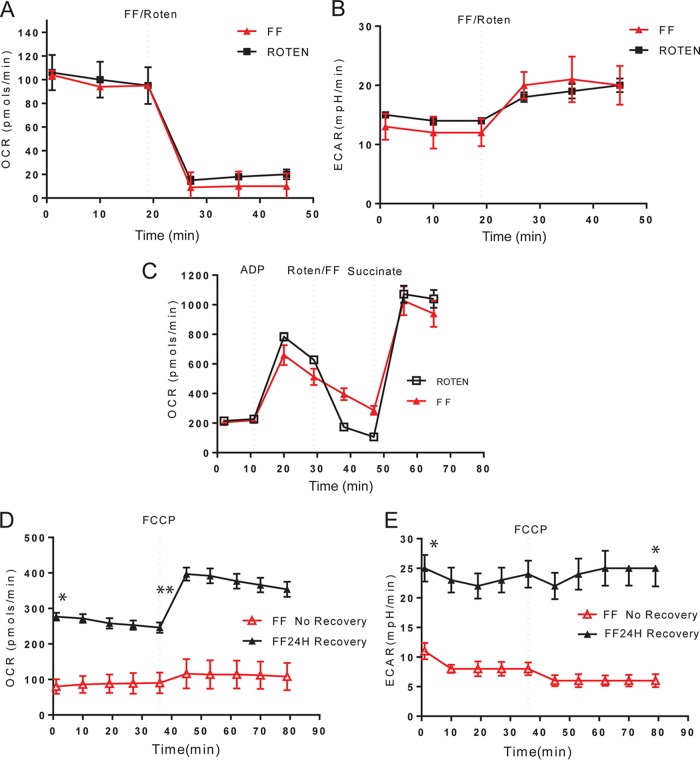
Since FF-treated cells are characterized by a very low respiration rate and do not respond to FCCP with an immediate increase in oxygen consumption (Fig. 2A), we hypothesized that a putative mitochondrial target for FF could be located at the beginning of the ETC. To test this possibility, we compared the OCR and ECAR kinetics of LN-229 cells treated with either FF or the well-known complex I inhibitor rotenone. The results in Fig. 3 demonstrate almost identical OCR (Fig. 3A) and ECAR (Fig. 3B) traces for FF and rotenone, which may suggest that these two chemicals have the same mitochondrial target. To further investigate this possibility, we have attempted to rescue OCR in FF-treated cells and rotenone-treated cells by circumventing complex I (NADH dehydrogenase) via the direct utilization of complex II with its direct substrate, succinate. Since succinate does not penetrate intact cells, we used isolated mitochondria supplemented with exogenous ADP, which is required to sustain oxygen consumption in this experimental setting. As shown in Fig. 3C, injection of succinate restored oxygen consumption in both FF- and rotenone-treated mitochondria, further indicating that FF preferentially targets complex I without disrupting complex II. While these data indicate that FF and rotenone act on the same mitochondrial target, FF is considered to have low systemic toxicity and rotenone is a pesticide and a potent metabolic toxin. This discrepancy prompted us to test if we could reverse the FF-induced inhibition of mitochondrial respiration. In this paradigm, LN-229 cells were pretreated with 50 μM FF for 24 h and then they were continuously exposed to FF or the FF-containing medium was replaced with fresh medium and the cells were allowed to recover for 24 h. The results in Fig. 3D demonstrate that the initial OCR values after recovery (FF24H recovery) are similar to the initial OCR values in control cultures, ranging between 200 and 300 pmol/min (Fig. 2A). Additionally, after recovery, cells responded to FCCP with an immediate increase in OCR, indicating that the proton gradient across the inner mitochondrial membrane was restored after FF removal. In contrast, cells continuously exposed to FF (FF no recovery) had a very low initial OCR and did not respond to FCCP. These results indicate that the FF-induced inhibition of mitochondrial respiration is fully reversible and that the continuous presence of unprocessed FF may be necessary to sustain its antirespiratory effect. We also expected to see a significant increase in ECAR values in the presence of FF, as already demonstrated in Fig. 2D following an immediate response to FF or in Fig. 2B after 24 h of FF incubation. However, this effect was completely lost in LN-229 cells exposed to FF for 48 h (Fig. 3E, red triangles), in which we observed a >2-fold drop in ECAR values at the 48-h time point. This could indicate that the cells exposed to FF for 48 h already activated a PPARα-dependent transcriptional switch from glycolysis to fatty acid β-oxidation (22, 23).
FIG 3.
FF-mediated inhibition of ETC complex I. Comparison of immediate OCR (A) and ECAR (B) responses to 0.3 μM rotenone (Roten) and 50 μM FF. The experimental conditions are described in the legend to Fig. 2. (C) Direct activation of ETC complex II by 5 mM succinate rescues mitochondrial respiration blocked by FF. Isolated mitochondria (60) were measured in the presence of exogenous 4 mM ADP. The data are average values from five measurements per experiment ± the standard deviations, and each experiment was repeated at least three times (n = 15). (D, E) FF-induced inhibition of mitochondrial respiration is reversible. OCR and ECAR values were measured either in cell cultures continuously exposed to FF for 48 h (FF No Recovery) or following removal of the drug, where the initial 24-h incubation with FF was followed by 24 h of cell recovery in the absence of the drug (FF24H Recovery). The data are average values from five measurements ± the standard deviations, and each experiment was repeated at least three times (n = 15). In panel D, an asterisk indicates a statistically significant difference between FF-treated samples with or without recovery (initial OCR values) and two asterisks indicate a statistically significant percent OCR increase after FCCP injection. In panel E, the asterisks indicate the initial and final ECAR values.
FF activates energy deficit-dependent signaling responses.
Although we have previously reported that FF decreases intracellular ATP, those measurements were performed only after 48 h and we did not consider immediate mitochondrial effects of this lipid-lowering drug during that initial work (13). The results in Fig. 4A demonstrate that the intracellular ATP levels start to drop as early as 6 h after FF treatment and continue to decline, reaching very low levels at 72 h. During this time, cell viability was not affected up to 24 h; it declined from 92% ± 4% to 78% ± 10% at 48 h (not significant) and dropped to 13% ± 5% at 72 h (P = 0.00011). In contrast, FA had no discernible effect on ATP levels and did not impact cell survival (Fig. 4A). We also found that the observed depletion of intracellular ATP is accompanied by the phosphorylation of AMPK, inhibition of mTOR-dependent phosphorylation of p70S6K (mTOR substrate), and the expression of the autophagy marker LC3β, which all happened between 16 and 24 h of FF treatment (Fig. 4B). LN-229 cells stably expressing an LC3-green fluorescent protein (GFP) fusion (inset in Fig. 4C) were used to monitor autophagy. The results in Fig. 4C demonstrate that, in comparison to the control (DMSO), 5-fold and almost 50-fold increases in the formation of autophagosomes were observed in LN-229/LC3-GFP cells exposed to 25 and 50 μM FF, respectively. These data indicate that the FF-induced energy deficit triggers an early onset of autophagy, which precedes apoptotic death by about 48 h in LN-229 cells.
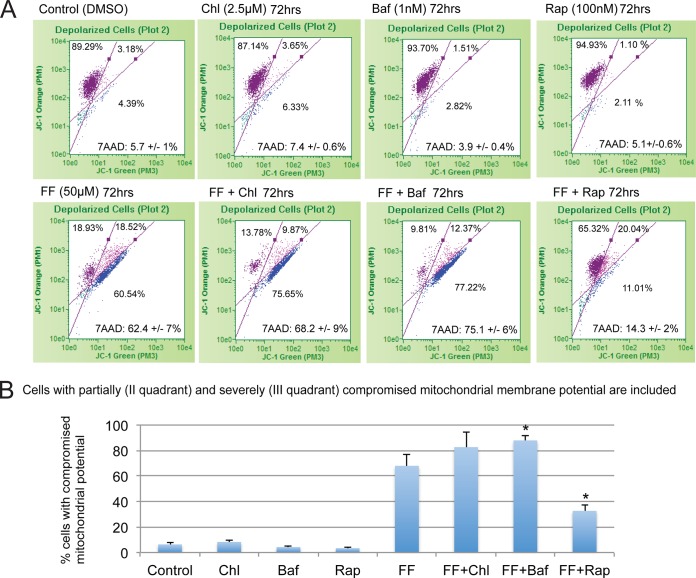
Since FF-induced autophagy occurs before apoptosis, we sought to determine whether autophagy modulators could affect the sensitivity of these cancer cells to this lipid-lowering drug. The results in Fig. 5A are flow cytometry-based measurements of mitochondrial membrane potential (JC-1 assay) and cell viability (7-AAD assay) when FF-treated LN-229 cells were exposed to noncytotoxic doses of the autophagy enhancer rapamycin or the autophagy inhibitors chloroquine and bafilomycin A1 (Fig. 5A, first row). This approach allowed us to establish a direct correlation between cell death, cell survival, and the severity of mitochondrial injury in the same sample. In all of the parts of Fig. 5A, the cells with intact mitochondria are located in quadrant I, the cells with partially compromised mitochondria are in quadrant II, and the cells with depolarized mitochondria are in quadrant III. As shown in the second row of Fig. 5A, 50 μM FF triggered both severe mitochondrial dysfunction and cell death after 72 h of continuous treatment. Surprisingly, FF-induced cytotoxicity was significantly reduced in the presence of rapamycin. In contrast, chloroquine and bafilomycin A1 slightly enhanced FF-induced cytotoxicity. Quantification of the effects of FF on mitochondrial potential is presented by the histogram depicted in Fig. 5B, in which cell populations with partially (quadrant II) and severely (quadrant III) compromised mitochondria were combined. In summary, these results indicate that autophagy plays a significant role in glioblastoma responses to FF, and the observed early onset of autophagy could be responsible for the delayed glioblastoma cell death observed in the presence of FF.
FIG 5.
Autophagy delays FF-induced glioblastoma cytotoxicity. (A) Flow cytometry-based measurements of mitochondrial membrane potential and cell death in FF-treated LN-229 cells. The treatment was accompanied either by rapamycin (Rap; 100 nM) to induce autophagy or by the autophagy inhibitors chloroquine (Chl; 2.5 μM) and bafilomycin A1 (Baf; 1 nM). Cells with intact mitochondrial potential are located in quadrant I, those with partially compromised mitochondrial potential are in quadrant II, and those with depolarized mitochondria are in quadrant III. The cells were also labeled with 7-AAD, which allows the detection and quantification of cell death in the same sample. Average values for 7-AAD-positive cells are indicated at the bottom of each III quadrant. (B) Quantification of the percentages of cells with compromised mitochondrial potential depicted in panel A. The data are average values ± the standard deviations from two separate experiments performed in triplicate (n = 6). An asterisk indicates a value significantly different from that of cells treated with FF only (P ≤ 0.05).
Anticancer efficacy of FF.
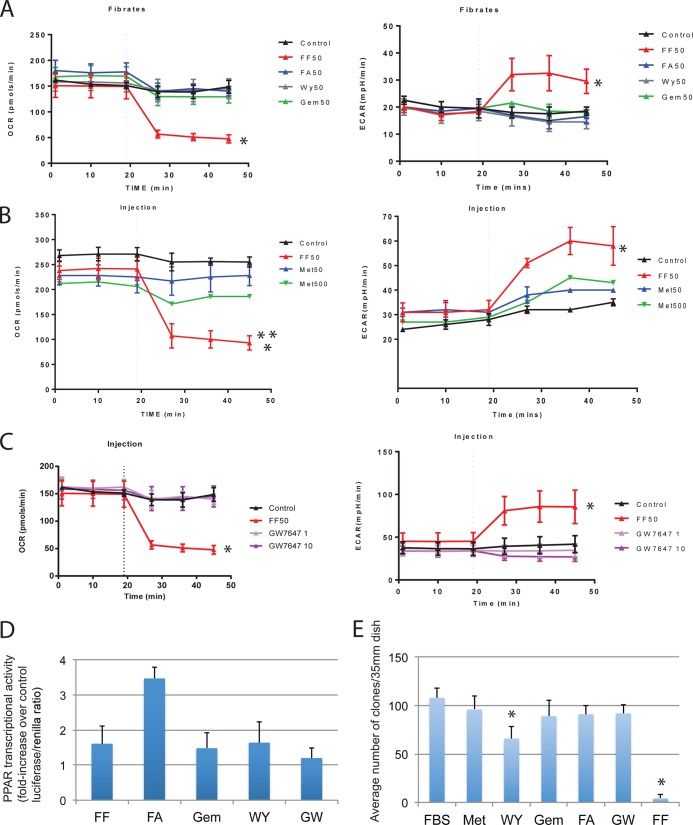
Since our results in Fig. 4A demonstrate that FA is ineffective in targeting glioblastoma energy metabolism and in compromising cell survival, we asked if other PPARα agonists, including GEM, WY, and GW7647, could imitate the mitochondrial action of FF. We also tested Met for its reported effects against diabetes and possibly cancer (38). Further supporting this notion, Met is currently being tested against breast cancer in clinical trials (39). The results in Fig. 6A to C show that of all of the lipid-lowering drugs tested, only FF was capable of interfering with oxygen consumption (OCR). In addition, 50 μM FF was a significantly more potent OCR repressor than 500 μM Met (** in Fig. 6B). We have also verified that PPARα agonists used in this experiment were transcriptionally active (Fig. 6D), further indicating that the observed mitochondrial effects are unique to FF and most likely independent of PPARα. All of the agonists tested were used at a concentration of 50 μM, with the exception of GW7647, which was used at 1 and 10 μM (the 50% effective concentration [EC50] of GW7647 is 6 nM, and that of FF/FA is 12 μM).
FIG 6.
Effects of PPARα agonists on mitochondrial respiration and clonal growth. (A) OCR and ECAR were registered by using the experimental procedure described in the legend to Fig. 2. LN-229 cells were preincubated with 50 μM FF, FA, GEM, or WY. (B) Comparison of OCR and ECAR in LN-229 cells treated with either 50 μM FF or Met (antidiabetic drug with known anticancer potential) at either 50 μM (Met50) or 500 μM (Met500). In panels A and B, the data represent average OCR and ECAR values from three separate experiments performed in triplicate (n = 9) ± the standard deviations. (A) An asterisk indicates a statistically significant difference between control and FF-treated samples. (B) Two asterisks indicate a statistically significant difference between 50 μM FF and 500 μM Met. (C) OCR and ECAR values for LN-229 cells preincubated with either 50 μM FF (EC50 = 12 μM) or the highly specific PPARα agonist GW7647 (EC50 = 6 nM) at 1 or 10 μM. An asterisk indicates a statistically significant difference between the control and FF-treated samples. Please note that GW7647 does not affect OCR at 10 μM. (D) PPAR transcriptional activity evaluated in LN-229 cells expressing the J3TkpGL3 reporter plasmid, which contains the luciferase gene driven by PPRE (13). The cells were stimulated with 50 μM FF, FA, GEM, or WY and 1 μM GW7647 (GW). The samples were analyzed with a firefly-Renilla dual-luciferase reporter system. Data are presented as average percent increases over the control (DMSO) value ± the standard deviations calculated from two experiments performed in triplicate (n = 6). (E) Clonogenic growth of LN-229 cells tested in the presence of Met, WY, GW7647 (GW), FA, or FF. Following 72 h of continuous cell growth, the cells were exposed to the PPARα agonists indicated and Met, all at 50 μM (because of unspecific cytotoxicity, GW7647 was used at 1 μM), and allowed to grow for an additional 2 weeks. The data are average numbers of clones ± the standard deviations calculated from three separate experiments performed in duplicate (n = 6). An asterisk indicates a value statistically significantly different from that of the control (DMSO).
Since all of the PPARα agonists tested and Met failed to reproduce the acute mitochondrial effects of FF, we compared their antiglioblastoma effects by using a clonogenic assay. The results in Fig. 6E show clonogenic growth of LN-229 cells in 10% FBS supplemented with WY (50 μM), FA (50 μM), Met (50 μM), GEM (50 μM), GW7647 (1 μM), and FF (50 μM). As expected, FF almost completely blocked the clonogenic growth of LN-229 cells (96% inhibition). In contrast, Met, FA, GEM, and GW7647 were only marginally effective and WY demonstrated a significant (39%) reduction in clonal growth. In this experiment, all of the PPARα agonists and Met were applied 72 h after the initial plating of the cells (1 × 103 cells/35-mm dish) and the culture media containing fresh drugs were replaced every 72 h. For an additional comparison of the dose-response effects of FF and GW7647 on LN-229 clonogenic growth, see Fig. S3 in the supplemental material.
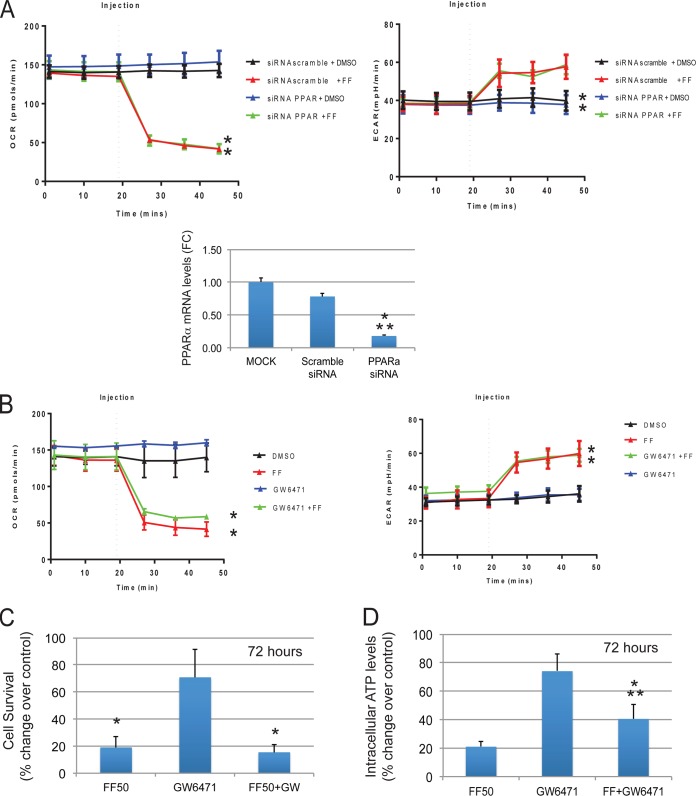
To verify further that the observed mitochondrial effects of FF are PPARα independent, we analyzed both PPARα cellular knockdown by using a PPARα small interfering RNA (siRNA) previously validated in our lab (13) and a highly specific PPARα antagonist, GW6471 (40). Importantly, LN-229 cells pretreated with either PPARα siRNA (Fig. 7A) or GW6471 (Fig. 7B) were still fully responsive to FF and demonstrated an immediate inhibition of mitochondrial respiration (OCR). The histogram in Fig. 7A (bottom) demonstrates a >5-fold decrease in PPARα mRNA levels in LN-229 cells pretreated with PPARα-specific siRNA for 48 h compared to mock-treated and scrambled-siRNA-treated controls. We have also tested the effects of the GW6471 antagonist on the FF-induced decline in intracellular ATP and cell survival. In comparison to controls, LN-229 cells treated with FF in the presence or absence of GW6471 (72 h of treatment) both demonstrated extensive tumor cell death (Fig. 7C), which indicates that the impairment of PPARα did not rescue LN-229 cells from FF treatment. Interestingly, the same treatment with GW6471 partially attenuated the FF-induced decline in intracellular ATP (Fig. 7D), further implicating PPARα in the long-term responses to this lipid-lowering drug.
FIG 7.
Effects of PPARα cellular knockout on FF-induced mitochondrial responses. (A) OCR and ECAR values following 48 h of cell preincubation with 100 nM PPARα-specific siRNAs (mixture of four at 25 nM each; Thermo Scientific) or 100 nM scrambled siRNA (Santa Cruz). The cells were injected with either the vehicle (DMSO) or a single dose of 50 μM FF. The data are average OCR and ECAR values ± the standard deviations calculated from two separate experiments performed in triplicate (n = 6). An asterisk indicates a statistically significant difference between DMSO- and FF-treated samples (P ≤ 0.05). Histogram: real-time PCR detection of PPARα mRNA expressed as the fold difference (FC) between mock-transfected cells and cells transfected with scrambled or PPARα-specific siRNA. The PCR values (cycle threshold, CT) of PPARα mRNA were normalized to the glyceraldehyde 3-phosphate dehydrogenase reference gene, and this resulted in a ΔCT value. Relative quantifications of PPARα mRNA from PPARα siRNA-treated and scrambled-siRNA-treated samples were then calculated as FC with the formula 2−ΔΔCT (61). An asterisk indicates a value statistically significantly different from that of mock-treated samples, and two asterisks indicate values statistically significantly different from those of scrambled-siRNA-treated samples. (B) OCR and ECAR values following 48 h of cell preincubation with 1 μM GW6471. Cells were injected with either the vehicle (DMSO) or a single dose of 50 μM FF. The data are average OCR and ECAR values ± the standard deviations calculated from two separate experiments performed in triplicate (n = 6). An asterisk indicates a statistically significant difference between DMSO-treated and FF-treated samples. Panels C and D demonstrate intracellular ATP levels and cell survival, respectively. In both panels, LN-229 cells were exposed to a GW6471 antagonist (1 μM) for 72 h in the presence or absence of 50 μM FF. The data are average values from two experiments performed in triplicate (n = 6) ± the standard deviations. In panel C, an asterisk indicates a value statistically significantly different from that of samples treated exclusively with GW6471. In panel D, an asterisk indicates a value statistically significantly different from that of samples treated exclusively with GW6471 and two asterisks indicate a value statistically significantly different from that obtained with 50 μM FF.
We also tested the long-term anticlonogenic effects of FF in B16 melanoma cells stably expressing PPARα shRNA (see Fig S1 in the supplemental material). In these studies, the partial cellular knockdown of neither PPARα nor the PPARα antagonist GW6471 affected FF-induced inhibition of clonogenic growth in this model.
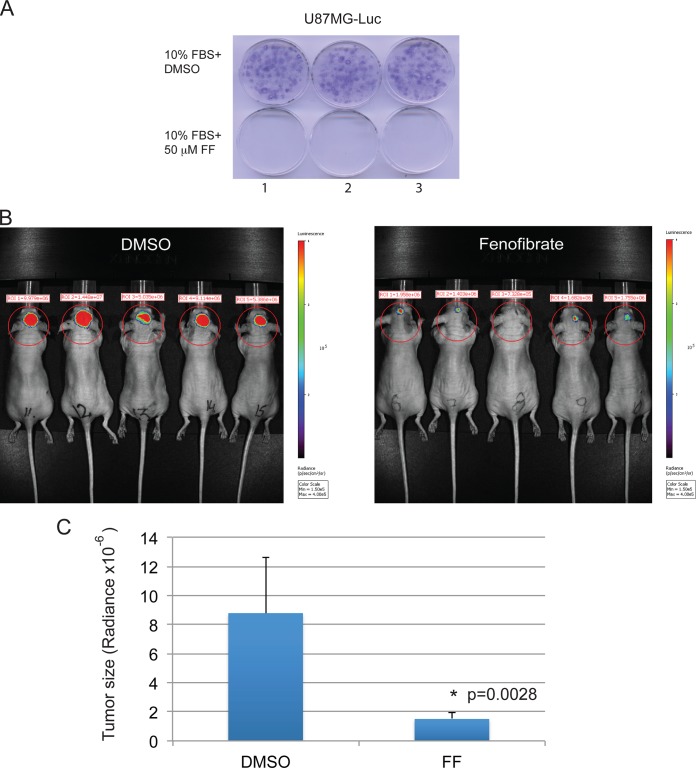
Considering the remarkable antiglioblastoma effects of FF in vitro, we also tested if FF could attenuate glioblastoma growth in vivo by using an orthotopic xenograft mouse model (immunodeficient mice with brain tumors that developed as a result of intracranial implantation of highly invasive U-87MG–luc). The results in Fig. 8A confirmed the high sensitivity of U-87MG–luc cells to FF treatment in vitro. Next, using mice with U-87MG–luc intracranial tumors, we tested the oral administration of FF (50 mg/kg/day). Because we were unable to detect FF or FA in brain tumor tissue (not shown) following oral administration, we tested the antiglioblastoma efficacy of FF by using intracranial delivery. Biophotonic images of the skull obtained with a Xenogen IVIS 200 imaging system (Fig. 8B) clearly demonstrate inhibition of tumor growth in FF-treated mice (5 μl of 1 mM FF in DMSO) in comparison to the controls (5 μl of DMSO). After selecting equivalent regions of the skull (circles in Fig. 8B), luminescence values where recorded and correlated with tumor size. The calculated average size of the tumors in FF-treated mice was 1.51 × 10−6 ± 0.47 × 10−6 radiance, and in the controls it was 8.79 × 10−6 ± 3.86 × 10−6 radiance, which represents an almost 6-fold reduction of tumor growth. The data were collected from two experiments with five mice per group (n = 10). Since we did not observed any side effects associated with the intracranial injection of 1 mM FF during the experiment, we suspected that the potential neurotoxic effects of intracranial FF are minor, at least at this dose.
FIG 8.
Effect of intracranially delivered FF on glioblastoma tumor growth. (A) Effect of FF on the clonogenic growth of U-87MG human glioblastoma cells stably expressing the luciferase reporter (U-87MG–luc), which were used in intracranial glioblastoma model. (B) Measurement of intracranial tumor growth after intracranial injection of FF. U-87MG–luc cells (1 × 105) were implanted into the brains of immunodeficient mice (Foxn1nu; Harlan Laboratories). Tumor-bearing mice were subsequently treated with 5 μl of DMSO (control) or 5 μl of 1 mM FF in DMSO by injection at the same place where the tumor cells were implanted. Two weeks later, bioluminescence imaging was done with the Xenogen IVIS 200 system. Tumor size is expressed as radiance (photons/s/cm2/sr) and was quantified with the Living Image 4.1 software according to the manufacturer's recommendations (Xenogen). Data in panel C represent the average radiance from 10 mice ± the standard deviation calculated from two separate experiments. An asterisk indicates a value statistically significantly different from that of DMSO-treated control mice.
Glioblastoma cells and normal astrocytes differ in sensitivity to FF.
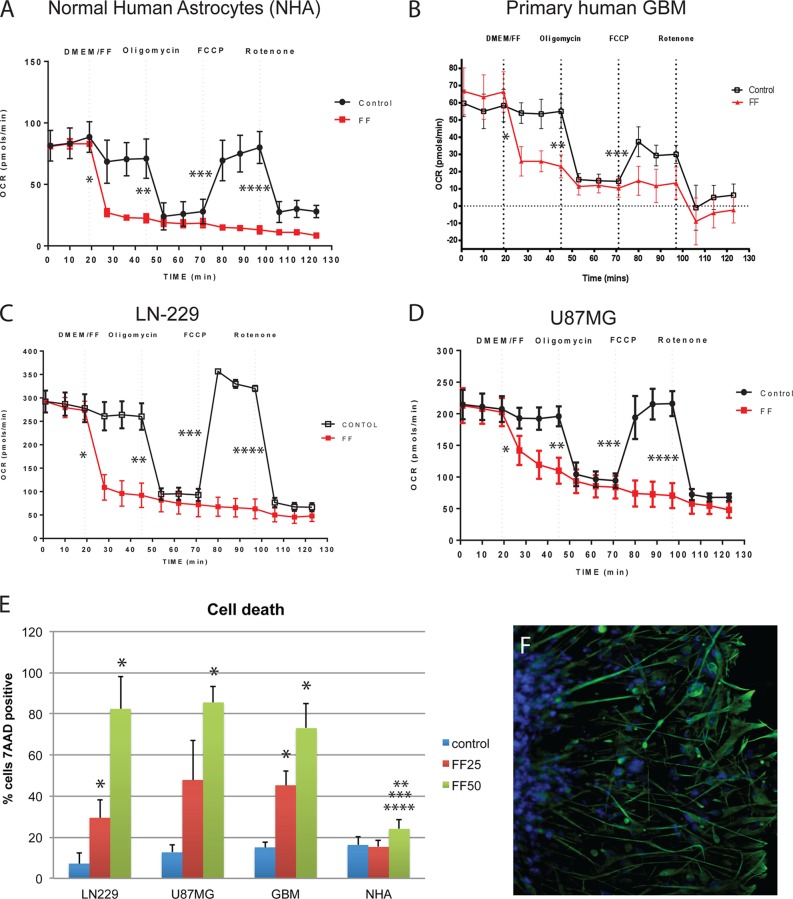
Since the adverse effects of FF on mitochondria mimic those of the common metabolic toxin rotenone, it is reasonable to ask why FF has low systemic toxicity. To address this intriguing question, we have tested FF-induced cytotoxicity in NHA (Lonza/Clonetics), two human glioblastoma cell lines, LN-229 and U-87MG, and primary cultures of human GBM cells obtained after tumor resection from patients diagnosed with GBM. The results in Fig. 9A to D demonstrate that all of the cells tested, including NHA, responded to FF with a rapid decline in mitochondrial respiration (OCR). In spite of these similar mitochondrial responses, NHA survived FF treatment much better than the glioblastoma cells tested. The results in Fig. 9E demonstrate that 22% of NHA, 81% of LN-229, 82% of U-87MG, and 73% of primary human GBM cells died following exposure to 50 μM FF for 72 h. Although we still do not fully understand why NHA are less sensitive to FF, we have observed certain cell type-specific differences in the baseline levels of respiration and glycolysis, which could contribute to their sensitivity to FF. For instance, the initial OCR values of NHA are between 3- and 4-fold lower than those of U-87MG and LN-229, respectively (Fig. 9A to D). Interestingly, the OCR values of primary human GBM cells were more similar to those of NHA than to those of the glioblastoma cell lines, but these primary tumor cells were still highly sensitive to the treatment (Fig. 9E). In addition, results in Fig. 9F demonstrate that the primary human GBM cells used in this experiment are positive for the marker glial fibrillary acidic protein (GFAP), which is abundantly expressed in normal astrocytes and glial tumors but is frequently lost in human glioblastoma cell lines, including U-87MG (41–43). Although these primary GBM cells were more similar to NHA with respect to GFAP expression (Fig. 9F) and the baseline OCR and ECAR values (Fig. 9A to D), they were still more sensitive to FF than NHA were (Fig. 9E).
FIG 9.
Metabolic and survival responses of glial cells to FF. (A to D) Effects of FF on OCR in monolayer cultures of NHA, primary human GBM cells, and two human glioblastoma cell lines, LN-229 and U-87MG. OCR values were registered in serum-free XF assay medium following the sequential injection of FF (50 μM), oligomycin (0.3 μM), FCCP (0.5 μM), and rotenone (0.3 μM). Control cells were injected with DMSO in XF medium. The results are average OCR values from three experiments performed in triplicate (n = 9) ± the standard deviations. An asterisk indicates a statistically significant difference between control and FF-treated samples (percent decrease in OCR after FF injection), two asterisks indicate a statistically significant difference between percent decreases in OCR after oligomycin injection, three asterisks indicate a statistically significant difference between percent increases in OCR after FCCP injection, and four asterisks indicate a statistically significant difference between percent decreases in OCR after rotenone injection. (E) Survival of LN-229, U-87MG, and primary GBM cells and NHA evaluated with the flow cytometry-based 7-AAD assay. Cells were cultured in the corresponding growth-promoting media in the presence or absence of 25 or 50 μM FF for 72 h. The data are average values from three experiments performed in triplicate (n = 9). An asterisk indicates a statistically significant difference from the corresponding control, two asterisks indicate a statistically significant difference between NHA and LN-229, three asterisks indicate a statistically significant difference between NHA and U-87MG, and four asterisks indicate a statistically significant difference between NHA and GBM. (F) GFAP/DAPI-labeled primary gliosphere that was allowed to adhere and spread following 5 weeks of continuous growth in suspension. Note that all tumor cells are positive for the glial marker GFAP.
DISCUSSION
In this study, we attempted to explain the molecular mechanisms responsible for the metabolic catastrophe triggered in glioblastoma cells by a common lipid-lowering drug, FF, and to evaluate its anticancer efficacy by using a xenograft model of intracranial glioblastoma in mice. Glial tumors account for nearly 50% of all adult primary intracranial neoplasms, among which GBM is the most aggressive and remains practically incurable (44, 45). GBM originates from glial cells in the brain and spinal cord and is characterized by rapid cell growth, resistance to radiotherapy and chemotherapy, and relentless invasion of the central nervous system (44). Current therapeutic approaches to GBM include invasive surgery and aggressive radiotherapy, followed by chemotherapy (46, 47) and/or immunotherapy (48). Collectively, these approaches prolong survival by only 12 to 24 months and incur significant morbidity (13, 45, 48).
We have previously reported that FF exerts strong antiproliferative, antimetastatic, and proapoptotic effects in various tumors of neuroectodermal origin, including, melanoma, medulloblastoma, and glioblastoma (3, 12–15). This could be highly desirable for new therapeutic strategies against malignant glial tumors, since FF is a commonly used and FDA-approved lipid-lowering drug that was originally used to normalize the plasma lipid and lipoprotein profiles of patients with hypercholesterolemia (18, 49). Although these beneficial metabolic effects of FF could be partially explained by the activation of PPARα-dependent transcription (18), the anticancer dynamics of this drug are much more complex and may involve both PPARα-dependent and PPARα-independent mechanisms. Parallel to the long-term metabolic effects, we have observed several anticancer responses to FF that are difficult to explain by using a PPARα-dependent mechanism of action (12–15). Other examples of a possible off-target action of FF include the effects of cell membrane fluidity in endothelial cells (5, 20) and the inhibition of oxygen consumption by isolated cardiac and liver mitochondria (31, 32).
In this study, we detected and quantified the accumulation of unprocessed FF in the mitochondrial fraction of glioblastoma cells (Fig. 1A) and an immediate blockade of mitochondrial respiration (Fig. 2C), which was followed by a continuous decline in intracellular ATP (Fig. 4A). Interestingly, we did not observe any changes in either respiration or ATP levels when FF was replaced with its PPARα-active metabolite, FA. Since we did not detect any traces of FA in the mitochondrial fraction (Fig. 1A), this could indicate that the observed effects require unprocessed FF and its direct interaction with mitochondria. Although the interaction of unprocessed FF with mitochondrial membranes is necessary for its exceptional anticancer effects, it may not be sufficient, since PPARα siRNA partially attenuated some of the glioblastoma responses to FF (13, 15). In addition, our results in Fig. 3E demonstrate that glioblastoma cells exposed to FF for 48 h are no longer capable of compensating for impaired respiration by increasing glycolysis and that the PPARα antagonist GW6471 partially attenuated the FF-induced decline in intracellular ATP (Fig. 7D). One logical explanation could be that FA-mediated activation of PPARα, which triggers fatty acid β-oxidation and inhibits glycolysis (18), could already be initiated at the 48-h time point. Therefore, it is conceivable that FF can trigger two independent pathways. (i) Unprocessed FF blocks mitochondrial respiration, and (ii) FA attenuates glycolysis, which leaves the affected tumor cells with only a few alternatives for survival. Interestingly, we also demonstrated that the events that precede FF-induced cytotoxicity include phosphorylation of AMPK, inhibition of mTOR-dependent phosphorylation of p70S6K, and derepression of autophagy (Fig. 4B and C). These multiple cell signaling changes/adaptations occurred between 6 and 24 h of treatment and were followed by massive tumor cell death, which in our experiments always occurred at least 72 h posttreatment. Since autophagy has been shown to have contradictory effects on tumor cell growth and survival (50–53), we investigated how the observed early onset of autophagy affects FF-induced cytotoxicity. Surprisingly, additional activation of autophagy by rapamycin clearly attenuated FF cytotoxicity, while noncytotoxic doses of autophagy inhibitors enhanced FF-induced glioblastoma cell death. These findings prompted us to speculate that in response to an FF-induced energy deficit, glioblastoma cells can activate autophagy as an alternative source of energy, which in turn could be responsible for the observed delay in FF cytotoxicity.
We also attempted to answer why FF is preferentially toxic to tumor cells and, in contrast to rotenone, has low systemic toxicity. One possibility is that normal cells metabolize FF to FA more effectively than malignant cells do, and we have shown that FA does not inhibit mitochondrial respiration (Fig. 6A). Deesterification of FF, which generates the active PPARα agonist FA, can be carried out by cellular esterases such as carboxylesterases CES1 and CES2 or arylesterases such as paraoxonase 1 (54, 55). In contrast to arylesterases, carboxylesterases are not present in human blood but are highly expressed in tissues, including those of the intestine, liver, kidney, and lung, and are responsible for drug metabolism and detoxification (56, 57). Interestingly, CES1 and CES2 expression is frequently downregulated in hepatic and colon carcinomas and their levels are significantly lower than in normal tissues (56, 58, 59). Therefore, cancer cells might be less capable of processing FF to FA than normal cells are. In such a case, normal cells would rather be exposed to FA-activating PPARα and with minimal harm to their mitochondria. Conversely, cancer cells would be exposed to both unprocessed FF and FA, which, working together, could compromise both mitochondrial respiration and glycolysis, ultimately leading to a metabolic catastrophe and tumor cell death. Indeed, the results in Fig. 1F demonstrate a significantly lower quantity of unprocessed FF in the mitochondrial fraction of NHA than in that of LN-229.
In summary, the results of this study demonstrate a new interplay between metabolic and signaling pathways triggered in glioblastoma cells by a common lipid-lowering drug, FF. Importantly, these antitumor effects are delivered by a drug that has low systemic toxicity and could be introduced as a regimen used to support conventional glioblastoma therapies, many of which are highly toxic.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of Stanley S. Scott Cancer Center for helpful discussions and Jay Dunn from Seahorse Bioscience for technical assistance and guidance in the design XF24-based metabolic assays.
This work has been supported by NIH grants R01-CA095518 (K.R.) and P20-GM103501 (A.O. and K.R.) and LSU HSC startup funds to K.R. M.G. is supported by the Foundation for Polish Science-POMOST Program cofinanced by the European Union within the European Regional Development Fund. F.P. is supported by RO1-MH079751, and C.P. is supported by R01-CA142362.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00562-14.
REFERENCES
- 1.Cemeus C, Zhao TT, Barrett GM, Lorimer IA, Dimitroulakos J. 2008. Lovastatin enhances gefitinib activity in glioblastoma cells irrespective of EGFRvIII and PTEN status. J Neurooncol 90:9–17. doi: 10.1007/s11060-008-9627-0. [DOI] [PubMed] [Google Scholar]
- 2.Egerod FL, Nielsen HS, Iversen L, Thorup I, Storgaard T, Oleksiewicz MB. 2005. Biomarkers for early effects of carcinogenic dual-acting PPAR agonists in rat urinary bladder urothelium in vivo. Biomarkers 10:295–309. doi: 10.1080/13547500500218682. [DOI] [PubMed] [Google Scholar]
- 3.Grabacka M, Placha W, Plonka PM, Pajak S, Urbanska K, Laidler P, Slominski A. 2004. Inhibition of melanoma metastases by fenofibrate. Arch Dermatol Res 296:54–58. doi: 10.1007/s00403-004-0479-y. [DOI] [PubMed] [Google Scholar]
- 4.Grabacka M, Reiss K. 2008. Anticancer properties of PPARalpha—effects on cellular metabolism and inflammation. PPAR Res 2008:930705. doi: 10.1155/2008/930705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panigrahy D, Kaipainen A, Huang S, Butterfield CE, Barnes CM, Fannon M, Laforme AM, Chaponis DM, Folkman J, Kieran MW. 2008. PPARalpha agonist fenofibrate suppresses tumor growth through direct and indirect angiogenesis inhibition. Proc Natl Acad Sci U S A 105:985–990. doi: 10.1073/pnas.0711281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saidi SA, Holland CM, Charnock-Jones DS, Smith SK. 2006. In vitro and in vivo effects of the PPAR-alpha agonists fenofibrate and retinoic acid in endometrial cancer. Mol Cancer 5:13. doi: 10.1186/1476-4598-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yanae M, Tsubaki M, Satou T, Itoh T, Imano M, Yamazoe Y, Nishida S. 2011. Statin-induced apoptosis via the suppression of ERK1/2 and Akt activation by inhibition of the geranylgeranyl-pyrophosphate biosynthesis in glioblastoma. J Exp Clin Cancer Res 30:74. doi: 10.1186/1756-9966-30-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardette V, Bongard V, Dallongeville J, Arveiler D, Bingham A, Ruidavets JB, Amouyel P, Haas B, Ducimetiere P, Ferrieres J. 2009. Ten-year all-cause mortality in presumably healthy subjects on lipid-lowering drugs (from the Prospective Epidemiological Study of Myocardial Infarction [PRIME] prospective cohort). Am J Cardiol 103:381–386. doi: 10.1016/j.amjcard.2008.09.092. [DOI] [PubMed] [Google Scholar]
- 9.Shigeto T, Yokoyama Y, Xin B, Mizunuma H. 2007. Peroxisome proliferator-activated receptor alpha and gamma ligands inhibit the growth of human ovarian cancer. Oncol Rep 18:833–840. doi: 10.3892/or.18.4.833. [DOI] [PubMed] [Google Scholar]
- 10.Yokoyama Y, Xin B, Shigeto T, Mizunuma H. 2011. Combination of ciglitazone, a peroxisome proliferator-activated receptor gamma ligand, and cisplatin enhances the inhibition of growth of human ovarian cancers. J Cancer Res Clin Oncol 137:1219–1228. doi: 10.1007/s00432-011-0993-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strakova N, Ehrmann J, Bartos J, Malikova J, Dolezel J, Kolar Z. 2005. Peroxisome proliferator-activated receptors (PPAR) agonists affect cell viability, apoptosis and expression of cell cycle related proteins in cell lines of glial brain tumors. Neoplasma 52:126–136. [PubMed] [Google Scholar]
- 12.Grabacka M, Plonka PM, Urbanska K, Reiss K. 2006. Peroxisome proliferator-activated receptor alpha activation decreases metastatic potential of melanoma cells in vitro via down-regulation of Akt. Clin Cancer Res 12:3028–3036. doi: 10.1158/1078-0432.CCR-05-2556. [DOI] [PubMed] [Google Scholar]
- 13.Drukala J, Urbanska K, Wilk A, Grabacka M, Wybieralska E, Del Valle L, Madeja Z, Reiss K. 2010. ROS accumulation and IGF-IR inhibition contribute to fenofibrate/PPARalpha-mediated inhibition of glioma cell motility in vitro. Mol Cancer 9:159. doi: 10.1186/1476-4598-9-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urbanska K, Pannizzo P, Grabacka M, Croul S, Del Valle L, Khalili K, Reiss K. 2008. Activation of PPARalpha inhibits IGF-I-mediated growth and survival responses in medulloblastoma cell lines. Int J Cancer 123:1015–1024. doi: 10.1002/ijc.23588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilk A, Urbanska K, Grabacka M, Mullinax J, Marcinkiewicz C, Impastato D, Estrada JJ, Reiss K. 2012. Fenofibrate-induced nuclear translocation of FoxO3A triggers Bim-mediated apoptosis in glioblastoma cells in vitro. Cell Cycle 11:2660–2671. doi: 10.4161/cc.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sterba J, Valik D, Mudry P, Kepak T, Pavelka Z, Bajciova V, Zitterbart K, Kadlecova V, Mazanek P. 2006. Combined biodifferentiating and antiangiogenic oral metronomic therapy is feasible and effective in relapsed solid tumors in children: single-center pilot study. Onkologie 29:308–313. doi: 10.1159/000093474. [DOI] [PubMed] [Google Scholar]
- 17.Zapletalova D, Andre N, Deak L, Kyr M, Bajciova V, Mudry P, Dubska L, Demlova R, Pavelka Z, Zitterbart K, Skotakova J, Husek K, Martincekova A, Mazanek P, Kepak T, Doubek M, Kutnikova L, Valik D, Sterba J. 2012. Metronomic chemotherapy with the COMBAT regimen in advanced pediatric malignancies: a multicenter experience. Oncology 82:249–260. doi: 10.1159/000336483. [DOI] [PubMed] [Google Scholar]
- 18.Grabacka M, Pierzchalska M, Reiss K. 2013. Peroxisome proliferator activated receptor α ligands as anticancer drugs targeting mitochondrial metabolism. Curr Pharm Biotechnol 14:342–356. doi: 10.2174/1389201011314030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmed W, Ziouzenkova O, Brown J, Devchand P, Francis S, Kadakia M, Kanda T, Orasanu G, Sharlach M, Zandbergen F, Plutzky J. 2007. PPARs and their metabolic modulation: new mechanisms for transcriptional regulation? J Intern Med 262:184–198. doi: 10.1111/j.1365-2796.2007.01825.x. [DOI] [PubMed] [Google Scholar]
- 20.Araki H, Tamada Y, Imoto S, Dunmore B, Sanders D, Humphrey S, Nagasaki M, Doi A, Nakanishi Y, Yasuda K, Tomiyasu Y, Tashiro K, Print C, Charnock-Jones DS, Kuhara S, Miyano S. 2009. Analysis of PPARalpha-dependent and PPARalpha-independent transcript regulation following fenofibrate treatment of human endothelial cells. Angiogenesis 12:221–229. doi: 10.1007/s10456-009-9142-8. [DOI] [PubMed] [Google Scholar]
- 21.Finck BN, Kelly DP. 2002. Peroxisome proliferator-activated receptor alpha (PPARalpha) signaling in the gene regulatory control of energy metabolism in the normal and diseased heart. J Mol Cell Cardiol 34:1249–1257. doi: 10.1006/jmcc.2002.2061. [DOI] [PubMed] [Google Scholar]
- 22.Randle PJ. 1998. Regulatory interactions between lipids and carbohydrates: the glucose fatty acid cycle after 35 years. Diabetes Metab Rev 14:263–283. doi:. [DOI] [PubMed] [Google Scholar]
- 23.Wolfe RR. 1998. Metabolic interactions between glucose and fatty acids in humans. Am J Clin Nutr 67:519S–526S. [DOI] [PubMed] [Google Scholar]
- 24.Beckner ME, Gobbel GT, Abounader R, Burovic F, Agostino NR, Laterra J, Pollack IF. 2005. Glycolytic glioma cells with active glycogen synthase are sensitive to PTEN and inhibitors of PI3K and gluconeogenesis. Lab Invest 85:1457–1470. doi: 10.1038/labinvest.3700355. [DOI] [PubMed] [Google Scholar]
- 25.Jelluma N, Yang X, Stokoe D, Evan GI, Dansen TB, Haas-Kogan DA. 2006. Glucose withdrawal induces oxidative stress followed by apoptosis in glioblastoma cells but not in normal human astrocytes. Mol Cancer Res 4:319–330. doi: 10.1158/1541-7786.MCR-05-0061. [DOI] [PubMed] [Google Scholar]
- 26.Marie SK, Shinjo SM. 2011. Metabolism and brain cancer. Clinics (Sao Paulo) 66(Suppl 1):S33–S43. doi: 10.1590/S1807-59322011001300005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warburg O. 1956. On the origin of cancer cells. Science 123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 28.Warburg O. 1956. On respiratory impairment in cancer cells. Science 124:269–270. [PubMed] [Google Scholar]
- 29.Dell' Antone P. 2012. Energy metabolism in cancer cells: how to explain the Warburg and Crabtree effects? Med Hypotheses 79:388–392. doi: 10.1016/j.mehy.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 30.Gamerdinger M, Clement AB, Behl C. 2007. Cholesterol-like effects of selective COX inhibitors and fibrates on cellular membranes and amyloid-beta production. Mol Pharmacol 72:141–151. doi: 10.1124/mol.107.034009. [DOI] [PubMed] [Google Scholar]
- 31.Nadanaciva S, Dykens JA, Bernal A, Capaldi RA, Will Y. 2007. Mitochondrial impairment by PPAR agonists and statins identified via immunocaptured OXPHOS complex activities and respiration. Toxicol Appl Pharmacol 223:277–287. doi: 10.1016/j.taap.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Zungu M, Felix R, Essop MF. 2006. Wy-14,643 and fenofibrate inhibit mitochondrial respiration in isolated rat cardiac mitochondria. Mitochondrion 6:315–322. doi: 10.1016/j.mito.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Valentinis B, Reiss K, Baserga R. 1998. Insulin-like growth factor-I-mediated survival from anoikis: role of cell aggregation and focal adhesion kinase. J Cell Physiol 176:648–657. [DOI] [PubMed] [Google Scholar]
- 34.Gualco E, Urbanska K, Perez-Liz G, Sweet T, Peruzzi F, Reiss K, Del Valle L. 2010. IGF-IR-dependent expression of survivin is required for T-antigen-mediated protection from apoptosis and proliferation of neural progenitors. Cell Death Differ 17:439–451. doi: 10.1038/cdd.2009.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vu-Dac N, Schoonjans K, Kosykh V, Dallongeville J, Fruchart JC, Staels B, Auwerx J. 1995. Fibrates increase human apolipoprotein A-II expression through activation of the peroxisome proliferator-activated receptor. J Clin Invest 96:741–750. doi: 10.1172/JCI118118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Troiano L, Ferraresi R, Lugli E, Nemes E, Roat E, Nasi M, Pinti M, Cossarizza A. 2007. Multiparametric analysis of cells with different mitochondrial membrane potential during apoptosis by polychromatic flow cytometry. Nat Protoc 2:2719–2727. doi: 10.1038/nprot.2007.405. [DOI] [PubMed] [Google Scholar]
- 37.Wybieralska E, Szpak K, Gorecki A, Bonarek P, Miekus K, Drukala J, Majka M, Reiss K, Madeja Z, Czyz J. 2011. Fenofibrate attenuates contact-stimulated cell motility and gap junctional coupling in DU-145 human prostate cancer cell populations. Oncol Rep 26:447–453. doi: 10.3892/or.2011.1321. [DOI] [PubMed] [Google Scholar]
- 38.Whitaker-Menezes D, Martinez-Outschoorn UE, Flomenberg N, Birbe RC, Witkiewicz AK, Howell A, Pavlides S, Tsirigos A, Ertel A, Pestell RG, Broda P, Minetti C, Lisanti MP, Sotgia F. 2011. Hyperactivation of oxidative mitochondrial metabolism in epithelial cancer cells in situ: visualizing the therapeutic effects of metformin in tumor tissue. Cell Cycle 10:4047–4064. doi: 10.4161/cc.10.23.18151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin-Castillo B, Vazquez-Martin A, Oliveras-Ferraros C, Menendez JA. 2010. Metformin and cancer: doses, mechanisms and the dandelion and hormetic phenomena. Cell Cycle 9:1057–1064. doi: 10.4161/cc.9.6.10994. [DOI] [PubMed] [Google Scholar]
- 40.El-Sisi A, Hegazy S, El-Khateeb E. 2013. Effects of three different fibrates on intrahepatic cholestasis experimentally induced in rats. PPAR Res 2013:781348. doi: 10.1155/2013/781348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawamata H, Tachibana M, Fujimori T, Imai Y. 2006. Differentiation-inducing therapy for solid tumors. Curr Pharm Des 12:379–385. doi: 10.2174/138161206775201947. [DOI] [PubMed] [Google Scholar]
- 42.Kruczynski A, Pasteels J, Rombaut K, Salmon I, Camby I, Limouzy A, Delsol G, Brotchi J, Kiss R. 1995. The characterization of nuclear-DNA content, the proliferative activity and the immunohistochemical expression of gfap, vim, leu-7, s-100, p53 and cathepsin-d in human glioblastoma multiformes (hgbms) versus human gbm cell-lines grafted into the brains of nude-mice. Int J Oncol 6:473–481. [DOI] [PubMed] [Google Scholar]
- 43.Qu J, Rizak JD, Fan Y, Guo X, Li J, Huma T, Ma Y. 2014. Establishment and partial characterization of a human tumor cell line, GBM-HSF, from a glioblastoma multiforme. Hum Cell 27:129–136. doi: 10.1007/s13577-013-0086-3. [DOI] [PubMed] [Google Scholar]
- 44.Nakada M, Nakada S, Demuth T, Tran NL, Hoelzinger DB, Berens ME. 2007. Molecular targets of glioma invasion. Cell Mol Life Sci 64:458–478. doi: 10.1007/s00018-007-6342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terzis AJ, Niclou SP, Rajcevic U, Danzeisen C, Bjerkvig R. 2006. Cell therapies for glioblastoma. Expert Opin Biol Ther 6:739–749. doi: 10.1517/14712598.6.8.739. [DOI] [PubMed] [Google Scholar]
- 46.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. 2005. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 47.Stupp R, Weber DC. 2005. The role of radio- and chemotherapy in glioblastoma. Onkologie 28:315–317. doi: 10.1159/000085575. [DOI] [PubMed] [Google Scholar]
- 48.Wen PY, Yung WK, Lamborn KR, Dahia PL, Wang Y, Peng B, Abrey LE, Raizer J, Cloughesy TF, Fink K, Gilbert M, Chang S, Junck L, Schiff D, Lieberman F, Fine HA, Mehta M, Robins HI, DeAngelis LM, Groves MD, Puduvalli VK, Levin V, Conrad C, Maher EA, Aldape K, Hayes M, Letvak L, Egorin MJ, Capdeville R, Kaplan R, Murgo AJ, Stiles C, Prados MD. 2006. Phase I/II study of imatinib mesylate for recurrent malignant gliomas: North American Brain Tumor Consortium Study 99-08. Clin Cancer Res 12:4899–4907. doi: 10.1158/1078-0432.CCR-06-0773. [DOI] [PubMed] [Google Scholar]
- 49.Saurav A, Kaushik M, Mohiuddin SM. 2012. Fenofibric acid for hyperlipidemia. Expert Opin Pharmacother 13:717–722. doi: 10.1517/14656566.2012.658774. [DOI] [PubMed] [Google Scholar]
- 50.Carew JS, Nawrocki ST, Cleveland JL. 2007. Modulating autophagy for therapeutic benefit. Autophagy 3:464–467. doi: 10.4161/auto.4311. [DOI] [PubMed] [Google Scholar]
- 51.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, White E. 2006. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kondo Y, Kanzawa T, Sawaya R, Kondo S. 2005. The role of autophagy in cancer development and response to therapy. Nat Rev 5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 53.Shimizu S, Takehara T, Hikita H, Kodama T, Tsunematsu H, Miyagi T, Hosui A, Ishida H, Tatsumi T, Kanto T, Hiramatsu N, Fujita N, Yoshimori T, Hayashi N. 2011. Inhibition of autophagy potentiates the antitumor effect of the multikinase inhibitor sorafenib in hepatocellular carcinoma. Int J Cancer 131:548–557. doi: 10.1002/ijc.26374. [DOI] [PubMed] [Google Scholar]
- 54.Bahar FG, Ohura K, Ogihara T, Imai T. 2012. Species difference of esterase expression and hydrolase activity in plasma. J Pharm Sci 101:3979–3988. doi: 10.1002/jps.23258. [DOI] [PubMed] [Google Scholar]
- 55.Najib J. 2002. Fenofibrate in the treatment of dyslipidemia: a review of the data as they relate to the new suprabioavailable tablet formulation. Clin Ther 24:2022–2050. doi: 10.1016/S0149-2918(02)80095-9. [DOI] [PubMed] [Google Scholar]
- 56.Guichard S, Terret C, Hennebelle I, Lochon I, Chevreau P, Fretigny E, Selves J, Chatelut E, Bugat R, Canal P. 1999. CPT-11 converting carboxylesterase and topoisomerase activities in tumour and normal colon and liver tissues. Br J Cancer 80:364–370. doi: 10.1038/sj.bjc.6690364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Redinbo MR, Potter PM. 2005. Mammalian carboxylesterases: from drug targets to protein therapeutics. Drug Discov Today 10:313–325. doi: 10.1016/S1359-6446(05)03383-0. [DOI] [PubMed] [Google Scholar]
- 58.Na K, Lee EY, Lee HJ, Kim KY, Lee H, Jeong SK, Jeong AS, Cho SY, Kim SA, Song SY, Kim KS, Cho SW, Kim H, Paik YK. 2009. Human plasma carboxylesterase 1, a novel serologic biomarker candidate for hepatocellular carcinoma. Proteomics 9:3989–3999. doi: 10.1002/pmic.200900105. [DOI] [PubMed] [Google Scholar]
- 59.Tang X, Wu H, Wu Z, Wang G, Wang Z, Zhu D. 2008. Carboxylesterase 2 is downregulated in colorectal cancer following progression of the disease. Cancer Invest 26:178–181. doi: 10.1080/07357900701560786. [DOI] [PubMed] [Google Scholar]
- 60.Frezza C, Cipolat S, Scorrano L. 2007. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc 2:287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 61.Pacifici M, Delbue S, Ferrante P, Jeansonne D, Kadri F, Nelson S, Velasco-Gonzalez C, Zabaleta J, Peruzzi F. 2013. Cerebrospinal fluid miRNA profile in HIV-encephalitis. J Cell Physiol 228:1070–1075. doi: 10.1002/jcp.24254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.